Abstract
MicroRNAs (miRNAs) have been found to play important regulatory roles in certain neurodegenerative diseases. The aim of the present study was to investigate the effect of miRNA-153 (miR-153) on the neural differentiation of HT-22 cells. Overexpression of miR-153 induced the differentiation of HT-22 cells, increasing the number of protrusions and branches, reducing the S phase distribution of the cell cycle, and attenuating the cell proliferation rate as determined using the Cell Counting Kit-8 assay. Furthermore, miR-153 increased the expression of neuron-specific γ-enolase (NSE), neuronal nuclei (NeuN), and N-ethylmaleimide-sensitive fusion attachment protein 23 (SNAP23) and SNAP25 at the transcriptional and protein level by PCR and western blot analysis. Moreover, miR-153 caused obvious upregulation of peroxiredoxin 5 (PRX5), which has been found to protect neural cells from death and apoptosis. miR-153 promoted neural differentiation and protected neural cells by upregulating the neuron markers γ-enolase, neuronal nuclei, and the functional proteins SNAP23, SNAP25 and PRX5. Therefore, miR-153 may be a potential target for the treatment of certain neurodegenerative diseases.
Keywords: miR-153, HT-22 cell line, neuron-specific enolase, neuronal nuclei, N-ethylmaleimide-sensitive fusion attachment proteins 23 and 25, peroxiredoxin 5
Introduction
Recent studies indicate that microRNAs (miRNAs) play an important role in neural growth and development (1,2). Dicer deficiency may result in the occurrence of neurodegenerative diseases, and inhibition of miRNA synthesis leads to loss of dopaminergic neurons (3). Loss of miRNA activity in Dicer1-deficient mice was found to cause spinal motor neuron disease, which was attributed to dysregulation of miR-9 (4). miRNA profiling revealed differential miRNA expression between healthy subjects and patients with neurodegenerative diseases. Pre-miR-133b, −218-2, −15b, −101-1, −107, −335 and −345 were found to be downregulated in the midbrain of patients with Parkinson's disease (5). miRNA-145 was found to be markedly increased during neural stem cell differentiation, and inhibition of miRNA-145 downregulated the expression of neuronal markers (6). In another study, miRNA-124 was found to increase neuron formation in the subventricular zone of adult mouse brain (7). In addition, the role of miR-9 was found to be crucial for neural progenitor proliferation and differentiation in the developing telencephalon (8).
Mouse miRNA-153 (miR-153) is located on chromosome 12. Certain disease-related proteins were found to be targets of miR-153. α-synuclein accumulation was shown to play a central role in the pathogenesis of Parkinson's disease. Doxakis et al reported that miRNA-7 and miR-153 can downregulate α-synuclein at the mRNA and protein levels (9). It was also reported that overexpression of miR-153 caused SNAP25 downregulation and resulted in near complete paralysis in zebrafish embryos (10). Another study on miR-153 demonstrated its inhibitory effect on gliogenesis by targeting Nfia/b in mouse neural stem/progenitor cells (11).
During neural differentiation, miR-153 may regulate certain neurogenesis-related genes, including N-ethylmaleimide-sensitive fusion attachment proteins (SNAPs). SNAPs and their receptors (SNAREs) constitute the core machinery for membrane fusion and are essential for intracellular vesicular trafficking. SNAP23 and SNAP25 are involved in neural regeneration and differentiation (12). Peroxiredoxin (PRX)5 was found to protect neural cells from amyloid-beta oligomer (AβO) damage (13). However, the function of miR-153 in neural development and differentiation and its relevance to the mechanism of neurodegenerative diseases has not been fully elucidated.
The aim of the present study was to investigate the role of miR-153 in neural development. A series of tests were performed in vitro to evaluate cell morphology, cell growth, cell cycle distribution, neural development-related genes and protein expression in the HT-22 cell line, in order to determine whether miR-153 induces neural differentiation and elucidate its effect on the expression of neuron-specific γ-enolase (NSE), neuronal nuclei (NeuN), PRX5, SNAP23 and SNAP25.
Materials and methods
Construction of the miR-153 plasmid and lentivirus packaging
Mouse precursor miR-153 (pre-miR153, miRbase accession no. MI0000175) was synthesized and further constructed into the pLVX-ZsGreen-miRNA-Puro vector, and packaged into lentivirus by Wuhan Viraltherapy Technologies Co., Ltd. The final titers ranged between 107 and 108 transducing units (TU)/ml.
Cell culture, infection and monoclone screen
The mouse hippocampal HT-22 cell line was purchased from Shanghai Xiaoying Biotech Company. HT-22 cells were maintained at 37°C and 5% CO2 in DMEM/high glucose (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS and 100 U/ml penicillin/streptomycin (Gibco; Thermo Fisher Scientific, Inc.). HT-22 cells were infected separately with miR-153 viral particles and control viral particles at 5х106 TU/106 cells. The green fluorescent protein (GFP) expression in the vector was used to determine transfection efficiency. After 48 h of infection, HT-22 cells were passaged and several monoclones were selected and further cultured into stably infected HT-22 cells. The stably miR-153-infected and control HT-22 cells were used in all further analyses.
miRNA extraction, quantitative polymerase chain reaction (qPCR) and regular PCR analysis (14)
The kits for miRNA isolation (DP501), miRNA First-Strand cDNA Synthesis (KR211) and miRNA qPCR Detection (FP411) were purchased from Tiangen Biotech Co., Ltd. For miRNA isolation, the main procedures were as follows. The cells were lysed in lysis buffer and maintained at room temperature for 5 min; 200 µl chloroform was added, vortexed for 15 sec and kept at room temperature for 5 min. The mixture was then centrifuged at 4°C at 10,000 × g for 15 min, and the upper layer of the aqueous phase was transferred to a new tube. Next, a proper volume of ethanol was added, and the solution was vortexed and transferred to a spin column. The solution was centrifuged and the eluate was saved; a proper volume of ethanol was added, vortexed and transferred to a spin column, followed by centrifugation at 10,000 g for 30 sec at room temperature. After rinsing twice, the miRNA in the spin column was dissolved in RNase-free ddH2O. For miRNA First-Strand cDNA Synthesis, up to 2 µg of miRNA was mixed with reaction buffer and enzymes to a final volume of 20 µl for each test; the mixtures were treated at 42°C for 60 min and then at 95°C for 5 min to denature the enzymes. For miRNA qPCR, the StepOnePlus PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.) was applied. A total of 20 µl of reaction system was prepared, containing 10 µl 2Х miRNA Premix (with SYBR & ROX), forward primers (synthesized by Shanghai Sangon Biotech Company) and reverse primers (provided in the kit, 10 µm for each) and 2 µl miRNA First-Strand cDNA. qPCR reactions were carried out in triplicate, and the data were normalized against the levels of U6 RNA. The sequences of mature mmu-miR-153-5p (accession no. MIMAT0016992) and mmu-miR-153-3p (accession no. MIMAT0000163) were used as forward primers in the qPCR analysis. Regular PCR was performed to detect mouse pre-miR-153 expression. The primers of regular PCR were as follows: Forward, CGGTGTCATTTTTGTGACGT and reverse, CAATGATCACTTTTGTGACT. The expression of pre-miR-153 was analyzed after 38 cycles of regular PCR.
Analysis of cell viability
Cell viability was assessed with the CCK-8 assay. The CCK-8 kit was purchased from Signalway Antibody Co. (Lot: 8620). HT-22 cells stably expressing rLV-miR and rLV-miR-153 were cultured in 96-well plates (1х104 cells per well). After incubation for 24 and 48 h, the cells were then incubated with CCK-8 (10 µl CCK-8 added to each well) for 1 h at 37°C. Absorbance was measured at 450 nm by using SpetraMax M3 (Molecular Devices LLC).
Cell cycle analysis
The Cell Cycle Analysis Kit was purchased from Beyotime Institute of Biotechnology. Cells were cultured in 6-well plates and were then collected, washed with cold PBS, fixed in iced 70% ethanol at 4°C for 24 h, washed again with cold PBS and stained in a propidium iodide (PI)/RNase A mixture at 37°C for 30 min in the dark. Finally, the cells were analyzed using a fluorescence-activated cell sorting (FACS) Caliber system (FACSVerse, BD Biosciences).
RNA extraction, reverse transcription (RT)-PCR and qPCR
Total RNA was purified using TRIzol reagent (Takara). RT was performed using 5X Primescript RT Master Mix (Takara) at 37°C for 15 min and at 85°C for 5 sec to denature the enzymes. cDNA samples were analyzed by regular PCR (KT211, Tiangen Biotech Co., Ltd.) or qPCR (SuperReal SYBR Green PreMix Plus, FP205, Tiangen Biotech Co., Ltd.) with appropriate primers. The PCR products were analyzed by electrophoresis in agarose gels. For qPCR, 10 ng RNA was used for each test, and the reactions were performed in triplicate on a Step One Plus thermocycler (Applied Biosystems; Thermo Fisher Scientific, Inc.). The data were normalized against the levels of β-actin mRNA. The primers were as follows: CDK12: Forward, AACAGCTAATGGAAGGACTGG and reverse, CAGAGTTATAGAGCCGAGCAAG; PRX5: Forward, CCCTCAGTGGAGGTATTTGAAG and reverse, CAGAGTTGAGAGAGGATGTTGG; NSE: Forward, GAACTGGATGGGACTGAGAATAA and reverse, CTTCAATGGAGACCACAGGATAG; β-actin: Forward, GAGGTATCCTGACCCTGAAGTA and reverse: GCTCGAAGTCTAGAGCAACATAG.
Western blotting
Whole cell protein extracts were harvested with RIPA lysis buffer (P0013B; Beyotime Institute of Biotechnology) consisting of 50 mM Tris (pH 7.4), 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, sodium orthovanadate, sodium fluoride, EDTA and leupeptin. For electrophoresis, 20 µg of each protein extract was loaded onto denatured 12% polyacrylamide gel and transferred to a PVDF membrane (EMD Millipore). The membrane was incubated with 5% skimmed milk for 1 h at room temperature followed by overnight incubation at 4°C with PRX5 (17724-1-AP), NSE (55235-1-AP), SNAP25 (14093-1-AP) and β-actin (60008-1-lg) primary antibodies (all from ProteinTech Group, Inc.). The abovementioned primary antibodies were incubated at a dilution ratio of 1:1,000. For SNAP23 (ProteinTech Group, Inc., 10825-1-AP), the dilution ratio was 1:500. For NeuN detection, the primary antibody was purchased from Cell Signaling Technology, Inc. (no. 12943) and the dilution ratio was 1:1,000. On the following day, the membranes were incubated at room temperature for 1 h with horseradish peroxidase (HRP)-conjugated anti-mouse secondary antibody (ProteinTech Group, Inc., SA00001-1), or HRP-conjugated anti-rabbit secondary antibody (ProteinTech Group, Inc., SA00001-2). HRP signals were detected by enhancing chemical luminol reagent (ProteinTech Group, Inc.) on Amersham Imager 600 (GE Healthcare).
Immunofluorescence
Cells were seeded into 6-well plates, fixed in methanol for 10 min at room temperature, and then permeabilized in 0.5% TritonX-100 for 10 min at room temperature. The samples were then washed in PBS before blocking overnight at 4°C with primary antibodies against NSE, SNAP23 and SNAP25, which were the same with the primary antibodies used in western blotting. The cells were then incubated with Cy3-conjugated goat anti-rabbit antibody (ProteinTech Group, Inc., SA00009-2) for 2 h at room temperature. Prior to visualization, the cells were incubated with DAPI (Invitrogen; Thermo Fisher Scientific, Inc.) for 5 min, washed with PBS and then visualized under an EVOS microscope (Thermo Fisher Scientific, Inc.).
Statistical analysis
Statistical significance was determined by t-test with GraphPad Prism 7 (GraphPad Software, Inc.). Cell cycle data analysis was performed by FlowJo.7 software (FlowJo LLC). For cell cycle and immunofluorescence analysis, statistical significance was determined by multiple t-tests. For morphology and immunostaining analysis, cells were quantified in a blinded manner using ImageJ 8.0 software (National Institutes of Health, Bethesda, MD, USA). A minimum of 5 randomly selected fields were quantified for the assays. For western blot analysis, the quantification was performed by ImageJ 8.0 software. In all cases, P<0.05 was considered to indicate a statistically significant difference.
Results
Generation of a lentivirus expressing miR-153
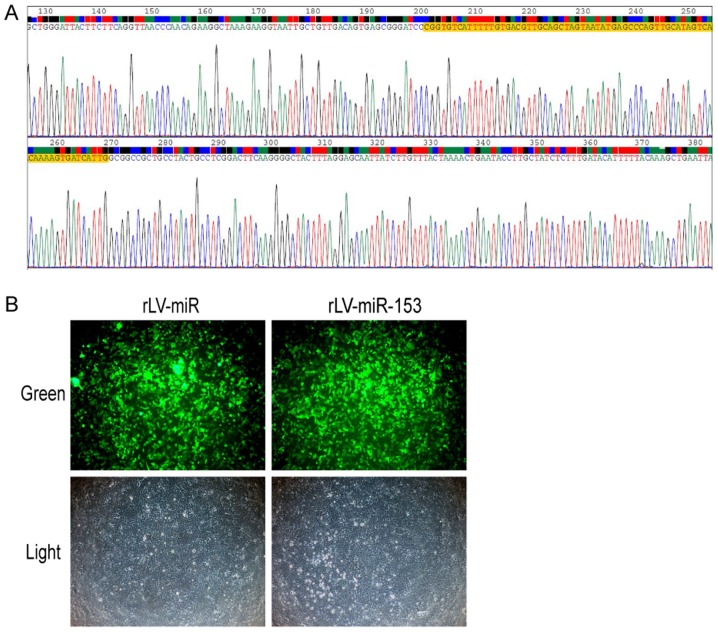
In order to observe the effects of miR-153, we first generated a lentivirus expressing miR-153. The sequence of miR-153 was obtained from www.mirbase.org. The sequence of mouse stem-loop precursor miR-153 (pre-miR153, MI0000175) was selected. The results revealed that the sequence of pre-miR153 inserted in the lentivirus plasmid was correct (Fig. 1A). Moreover, a lentivirus expressing miR-153 (rLV-miR-153) was produced using 293 cells (Fig. 1B). These observations suggested that the lentivirus expressing rLV-miR-153 was successfully constructed.
Figure 1.
Construction of the pLV-miR-153 plasmid and rLV-miR-153 virus. (A) Sequencing result of the pLV-miR-153 plasmid. The part highlighted in yellow indicates the correct mmu pre-miR-153 sequence (69 bp). (B) Infection of 293 cells by the rLV-miR-153 lentivirus. The images are of live cells. Top row, observation under green fluorescent light; bottom row, observation under white light. Magnification, ×100.
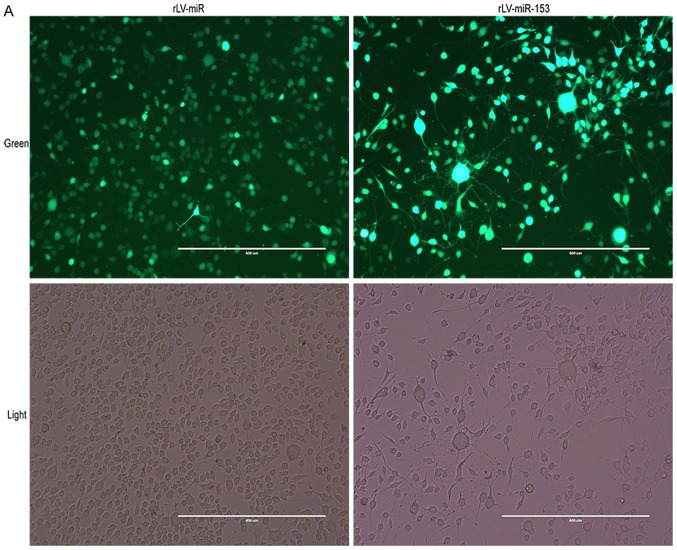
Establishment of miR-153-overexpressing HT-22 stable cell lines
To assess the effect of miR-153 on neurons, stable HT-22 cell lines with miR-153 overexpression were established. First, HT-22 cells were infected by rLV-miR-153 or rLV-miR, and then the positive monoclonal cells expressing GFP were selected for culture (Fig. 2A). Next, total RNA was extracted from the above mentioned two stable cell lines, RT-PCR was performed, and miRNA was extracted for qPCR analysis. As shown in Fig. 2B, pre-miR-153 was significantly increased in the rLV-miR153 group compared with the rLV-miR group. In particular, the qPCR results demonstrated that both mature miR-153-3 prime and mature miR-153-5 prime (Fig. 2C) were significantly increased compared with the rLV-miR group (miR-153-3 prime: 0.0316±0.0019 vs. 0.0015±0.0007, respectively, P=0.0007<0.01; and miR-153-5 prime: 0.0038±0.0007 vs. 0.0004±0.00036, respectively, P=0.0043). These results suggested that miR-153-overexpressing HT-22 stable cell lines were established.
Figure 2.
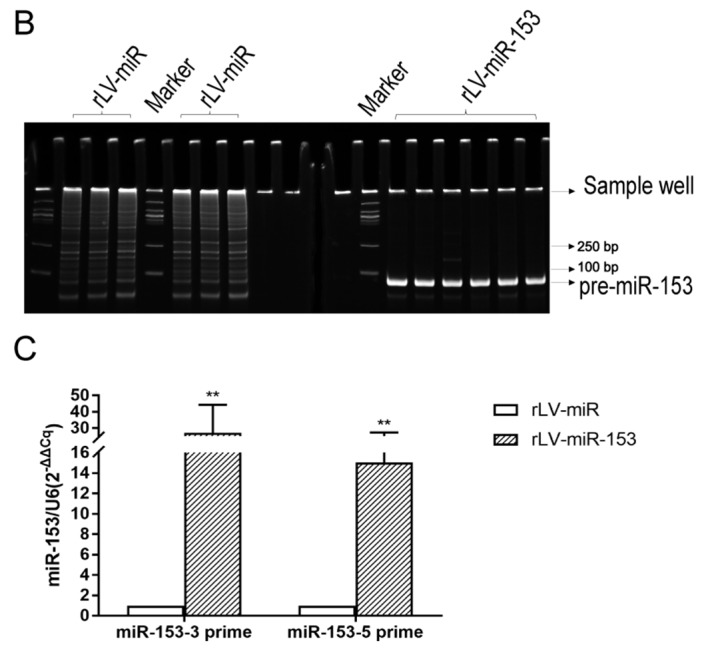
Identification of the overexpression of mmu miR-153 in HT-22 cells. (A) Selected monoclonal HT-22 cells after infection with the rLV-miR-153 virus and rLV-miR virus. The images are of living cells. Top row, observation under green fluorescent light; bottom row, observation under white light. Scale bar 400 µm. (B) Polymerase chain reaction (PCR) analysis of pre-miR-153 expression in monoclonal HT-22 cells infected by rLV-miR and rLV-miR-153. The products were subjected to PAGE. The arrow indicates mmu pre-miR-153 band (69 bp). (C) qPCR analysis of mature mmu miR-153 expression in selected monoclonal HT-22 cells. The relative expression of mature mmu miR-153-3 prime (MIMAT0000163) and mmu miR-153-5 prime (MIMAT0016992) is shown. Relative expression was calculated using the relative cycle quantification method (2−ΔΔCq) and normalized to U6. Significance was determined by comparing rLV-miR-153 to rLV-miR control with two-tailed paired t-test. Error bars represent standard deviation. **P<0.01.
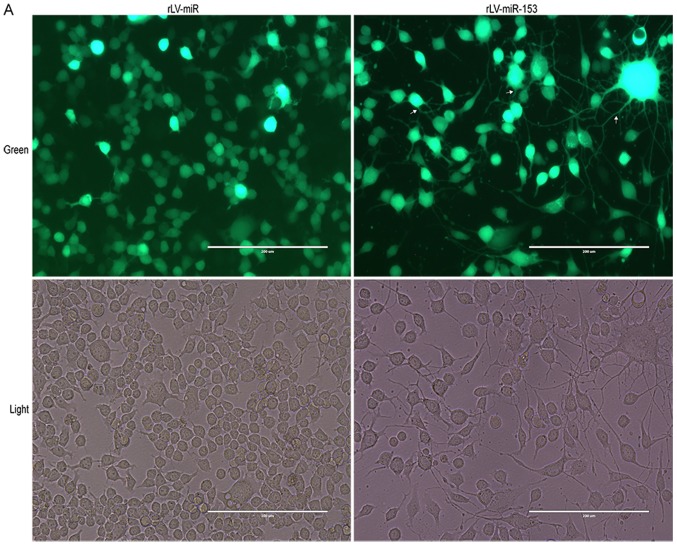
miR-153 induces neural differentiation of HT-22 cells
To validate whether miR-153 affects the differentiation of HT-22 cells, in vitro assays were performed to assess cell differentiation and growth in HT-22 cells infected with rLV-miR and rLV-miR-153. The results demonstrated that HT-22 cell differentiation, with protrusions and branches, was markedly enhanced in the rLV-miR-153 group compared with the rLV-miR group (Fig. 3A). As shown in Fig. 3B, the percentage of differentiated cells was increased by ~2-fold in the rLV-miR-153 group compared with the rLV-miR group (62.80±6.55 vs. 29.26±8.17, respectively, P=0.0079). The results confirmed that miR-153 promotes cell differentiation.
Figure 3.
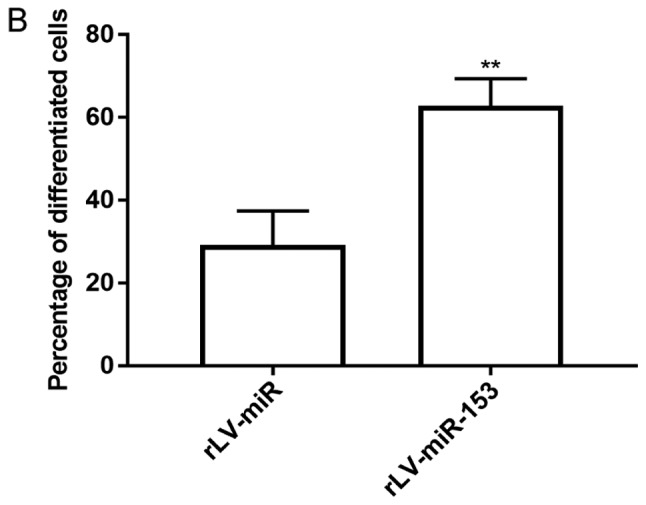
Morphology of HT-22 cells after rLV-miR-153 overexpression. (A) Green, green fluorescent protein (GFP) in infected HT-22 cells. Light, observation under white light. Cells with protrusions were identified as differentiated cells (arrows). Scale bar 200 µm. The images are of live cells. (B) Significance was determined by comparing rLV-miR-153 overexpression to control cells with two-tailed unpaired t-test; n=5. Error bars represent standard deviation. **P<0.01. As indicated by the arrows in A, cells with protrusions and branches were identified as positive and counted. Five visual fields per cell group were examined and the positive percentage of cells among GFP-positive cells was calculated. The difference was found to be statistically significant.
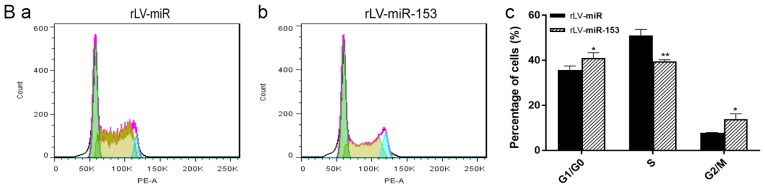
miR-153 attenuates the growth of HT-22 cells due to increased differentiation
Given that miR-153 induces differentiation of HT-22 cells, it was investigated whether the effect of miR-153 on cell growth is dependent upon the increased differentiation. To support our observations regarding the differentiation of HT-22 cells, a cell proliferation assay was performed. First, cell proliferation was measured with the CCK-8 assay in the two stable HT-22 cell lines, namely the rLV-miR and rLV-miR-153 groups. As shown in Fig. 4A, although the number of HT-22 cells increased rapidly in the rLV-miR-153 group compared with the rLV-miR group on the first day (1.5308±0.0151 vs. 1.0652±0.0138, P=0.0000245), the proliferation ability of the HT-22 cells was attenuated on the second day in the rLV-miR-153 group compared with the rLV-miR group (1.7496±0.0466 vs. 1.8604±0.1149, P=0.0533).
Figure 4.
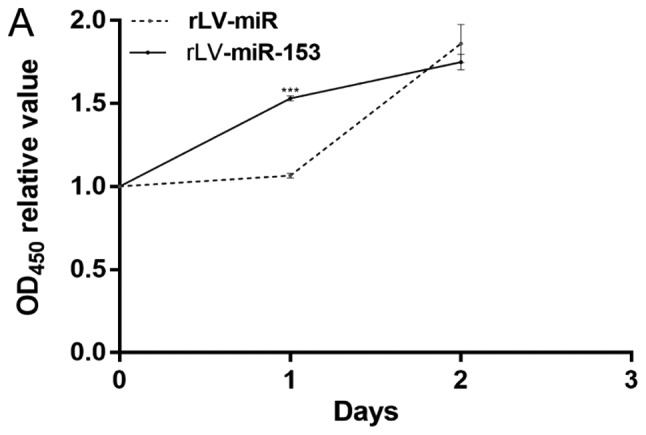
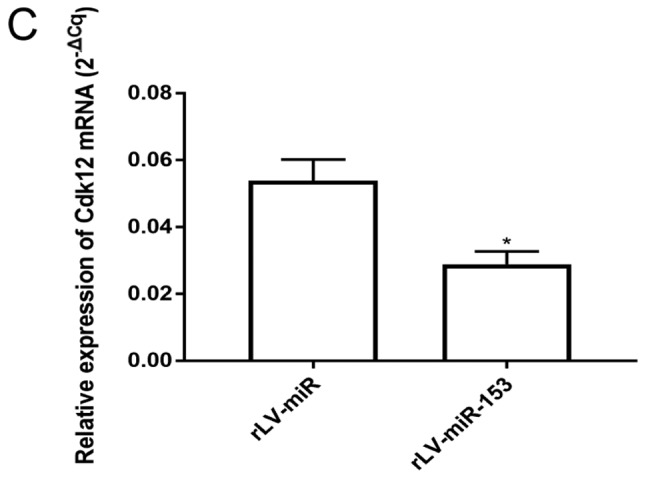
miR-153 regulates cell growth. (A) Overexpression of miR-153 caused changes in the proliferation rate of HT-22 cells, ***P<0.001. (B) Cell cycle distribution of HT-22 cells with rLV-miR (a) and rLV-miR-153 overexpression (b) was determined by flow cytometry. Significance was determined by comparing G1/G0, S and G2 phase cells with miR-153 overexpression to control cells with multiple unpaired t-tests (c). *P<0.05, **P<0.01. (C) Overexpression of miR-153 caused reduction in CDK12 mRNA in HT-22 cells. Significance was determined by comparing miR-153 overexpression to control cells with two-tailed paired t-test. Error bars represent standard deviation. *P<0.05. CDK, cyclin-dependent kinase.
Subsequently, to identify whether miR-153 inhibits cell growth by altering the cell cycle (G1, S and G2/M phase), FACS was performed. As shown in Fig. 4B-a to -c, the percentage of cells in the S phase was significantly decreased in the rLV-miR-153 group compared with the rLV-miR group (39.58±0.64 vs. 51.01±2.72, respectively, P=0.0063). The percentage of cells in the G0/G1 phase was significantly increased in the rLV-miR-153 group compared with the rLV-miR group (41.03±2.34 vs. 35.70±1.76, respectively, P=0.034). The percentage of cells in the G2/M phase was significantly increased in the rLV-miR-153 group compared with the rLV-miR group (13.94±2.32 vs. 7.79±0.31, respectively, P=0.02).
Furthermore, qPCR analysis of several cell cycle-related genes revealed that the expression of CDK12 was significantly decreased in the rLV-miR-153 group compared with the rLV-miR group (Fig. 4C, 0.0288±0.0039 vs. 0.0538±0.0064, respectively, P=0.0294), and miR-153 may affect the cell cycle by downregulating CDK12. Taken together, these observations indicated that miR-153 attenuated the growth of HT-22 cells by altering the cell cycle, which was associated with the process of cell differentiation.
miR-153 regulates neuron-specific factors in HT-22 cells
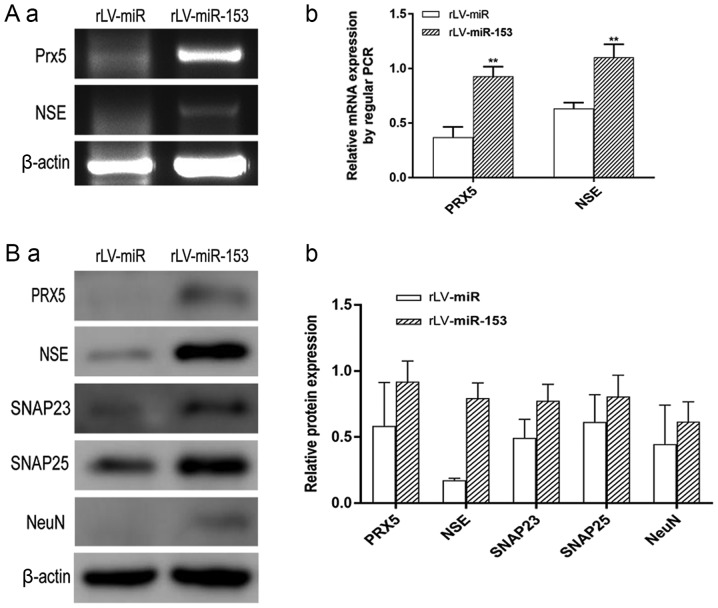
To elucidate the mechanism through which miR-153 controls the differentiation of HT-22 cells, several neuron differentiation-related markers were investigated. The RT-PCR results demonstrated that the transcriptional level of PRX5 was significantly increased in the rLV-miR-153 group compared with the rLV-miR group (0.9280±0.0905 vs. 0.3713±0.0940, respectively, P=0.0018; Fig. 5A-a, upper panel, and 5A-b). In addition, the transcriptional level of NSE was significantly increased in the rLV-miR-153 group compared with the rLV-miR group (1.1030±0.1198 vs. 0.6333±0.0545, respectively, P=0.0035; Fig. 5A-a, middle panel, and 5A-b). Moreover, the results of the western blot analysis demonstrated that the protein expression level of PRX5, NSE, SNAP23, SNAP25 and NeuN were significantly increased in the rLV-miR-153 group compared with these levels in the rLV-miR group (Fig. 5B-a and -b).
Figure 5.
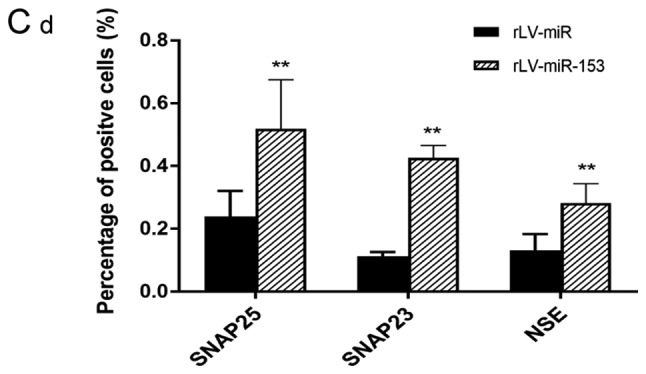
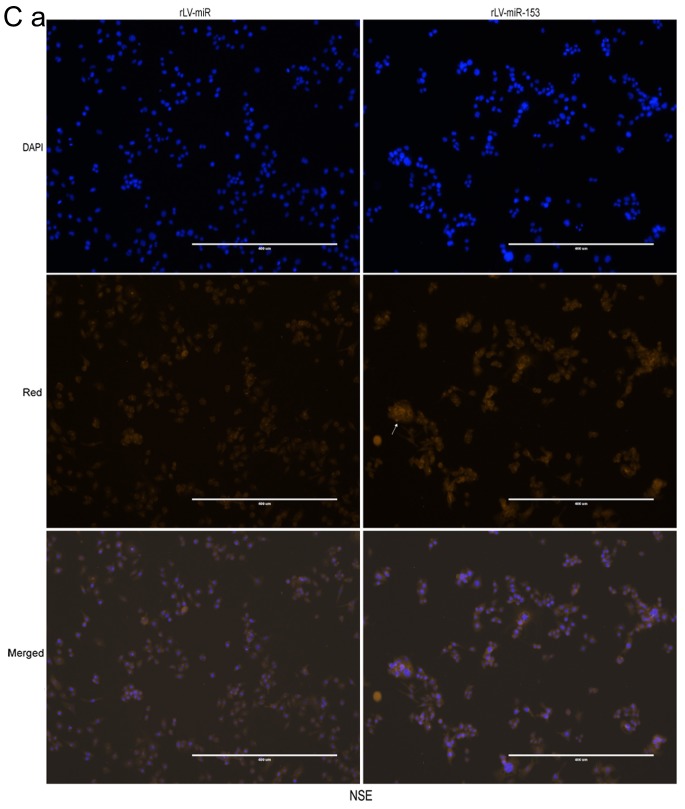
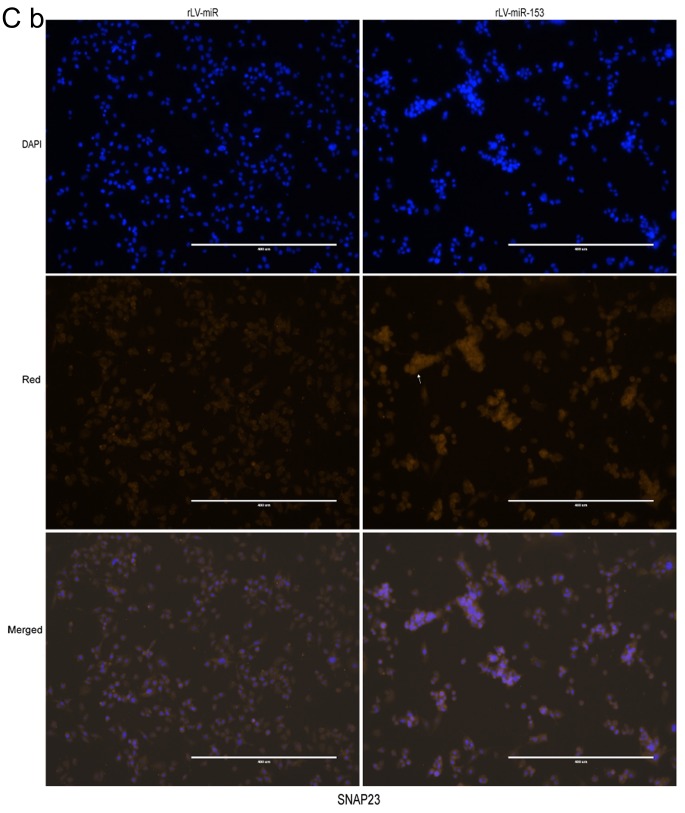
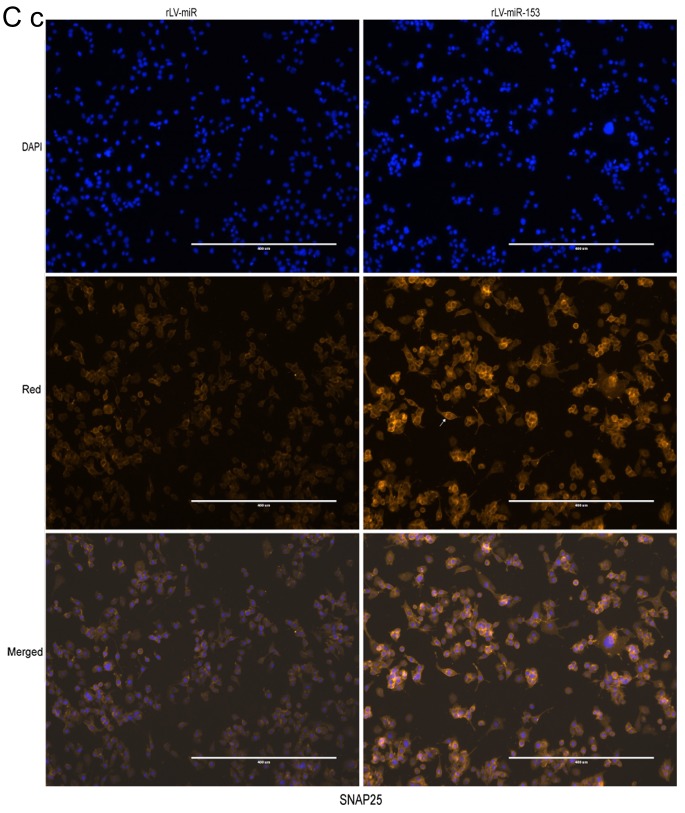
Neuron-specific markers expressed in rLV-miR and rLV-miR-153 cells. (A) Relative PRX5 and NSE expression as detected by regular PCR (a). Significance was determined by comparing the miR-153 overexpression group to the control with unpaired t-test; n=3 (b). (B) The protein expression of PRX5, NSE, SNAP23, SNAP25 and NeuN was analyzed by western blotting (a). Band density was determined in the miR-153 overexpression group and control, the error bars represent standard deviation (b). (C) The protein expression of (a) NSE was analyzed by immunofluorescence staining. Scale bar 400 µm. (C) The protein expression of (b) SNAP23 and (c) SNAP25 was analyzed by immunofluorescence staining. NSE (a), SNAP23 (b) and SNAP25 (c) were found to be significantly increased in rLV-miR-153-overexpressing cells. Arrows indicate positively stained cells. Significance was determined by comparing NSE, SNAP23 and SNAP25 protein expression in the rLV-miR-153-overexpressing group to control with multiple unpaired t-tests; n=5. Error bars represent standard deviation. **P<0.01. Cells exhibiting a strong orange color were identified as positive cells expressing NSE, SNAP23 and SNAP25 (arrows). (C-d) Five visual fields of each cell group were examined and the positive percentage of cells was calculated. NSE, neuron-specific enolase; PRX5, peroxiredoxin 5; SNAP, N-ethylmaleimide-sensitive fusion attachment protein; NeuN, neuronal nuclei.
Furthermore, immunofluorescence staining was also performed to detect the protein expression of NSE, SNAP23 and SNAP25. The number of cells expressing NSE was markedly higher in the rLV-miR-153 group compared with that in the rLV-miR group (Fig. 5C-a) and the positive rate of NSE expression was significantly increased in the rLV-miR-153 group compared with the rLV-miR group (0.2816±0.0623 vs. 0.1311±0.0526, respectively, P=0.0075; Fig. 5C-d). Similarly, the positive rate of SNAP23 expression was significantly increased in the rLV-miR-153 group compared with the rLV-miR group (0.4254±0.0404 vs. 0.1123±0.0142, respectively, P=8х10−6; Fig. 5C-b and -d). In addition, the positive rate of SNAP25 expression was also significantly increased in the rLV-miR-153 group compared with the rLV-miR group (0.5181±0.1571 vs. 0.2392±0.0815, respectively, P=3х10−5; Fig. 5C-c and -d). Collectively, these results suggest that miR-153 promoted the expression of neuron-specific factors, including PRX5, NSE, SNAP23, SNAP25 and NeuN.
Discussion
A number of studies have investigated the emerging role of miRNAs in nervous system growth, development and pathogenesis of neurodegenerative diseases. Typical neurodegenerative diseases, such as Alzheimer's disease, Parkinson's disease, amyotrophic lateral sclerosis and Huntington's disease, are the most extensively investigated and indicate a potential role of miRNAs in neuronal development and function (15–17). miRNA profile comparison and individual miRNA studies have demonstrated that the brain has a specific miRNA expression profile, and each miRNA may perform a specific function to maintain brain integrity (18–20).
The results of the present study indicated that miR-153 promotes the neuronal differentiation of HT-22 cells, which is reflected by the morphological changes, such as the increasing number of protrusions and branches, and the upregulation of NSE, NeuN, PRX5, SNAP23 and SNAP25 expression. Mouse hippocampal HT-22 cells are a sub-line cloned from HT4 cells that has been grown without establishing synaptic connections (21); thus, there were no obvious protrusions and branches in HT-22 cells. The change in morphology was likely due to the effects of miR-153 overexpression on cellular growth and differentiation. Similarly, another study reported that miR-153 inhibited astrogliogenesis of neural stem/progenitor cells, while the neuronal differentiation of tertiary neurospheres was increased significantly by miR-153 (11). These findings strongly support the association of miR-153 with neuronal differentiation.
Moreover, we observed that the effect of miR-153 on cell proliferation was associated with cell cycle changes and cell differentiation. Our data indicated that the rapid growth on day 1 in the rLV-miR-153 group may have been due to the increasing percentage of cells in the G2/M phase and the decreasing percentage of cells in the S phase, which resulted in the slow growth of cells in the rLV-miR-153 group on day 2. Of note, we found that miR-153 decreased the expression of CDK12. CDK12 can promote mammalian cell proliferation, including the murine brain, liver and H1299 cancer cells, by regulating pre-replicative complex assembly (22). Mouse CDK12 is a multifunctional protein that was reported to maintain genomic stability and the pluripotency of murine embryonic stem cells (23). Chen et al (24), reported that neural progenitor cells of mice with CDK12 mutation accumulated at the G2/M phase with neuron misalignment, which demonstrated that CDK12 is crucial for cell proliferation. Notably, the decreased number of S phase cells and increased number of G2/M phase cells were associated with decreased CDK12 mRNA in the miR-153 overexpression group. This may suggest that miR-153 promoted neuron differentiation and synchronously delayed cell proliferation. Therefore, it may be hypothesized that miR-153 has different functions at different stages. In addition, different cell types may respond differently to miR-153 modulation. Further studies are required to fully elucidate the effect of miR-153 on neural cell cycle and related genes.
In particular, the findings of the present study indicated that miR-153 regulated the expression of neuron-specific genes. For example, miR-153 increased the expression of NSE at the transcriptional and protein level in HT-22 cells. γ-enolase is one isozyme of three glycolytic enolases, which is neuron-specific and used as a marker for all types of neurons. The appearance of γ-enolase is a late event during neural differentiation, making it a useful index of neural maturation. The remaining two enolases are α-enolase, which is ubiquitous, and β-enolase, which is muscle-specific (25). HT-22 cells are not mature neural cells; therefore, the upregulation of NSE after miR-153 overexpression indicates that further neural differentiation and maturation occurred in HT-22 cells. The upregulation of enolase may be a secondary effect of miR-153. As this study is a preliminary research, the association between miR-153 and enolase will be further explored in the following study. In addition, NeuN is a neuronal nuclear antigen that is commonly used as a biomarker for neurons, particularly mature neurons. In the present study, the upregulation of the NeuN protein, as demonstrated by western blot analysis, further confirmed the neural differentiation of HT-22 cells induced by miR-153 overexpression. Importantly, miR-153 also enhanced the expression of PRX5 in HT-22 cells. PRX5 was previously reported to protect neural cells from AβO damage (13); PRX5 may also play an important role during miR-153-induced neural differentiation.
Intriguingly, bioinformatics prediction demonstrated that miR-153 targets the 3′ untranslated region (UTP) of SNAP25, Chunyao et al demonstrated that increased miR-153 levels caused decreased SNAP25 expression resulting in movement defects in zebrafish embryos, and SNAP25 was identified as the target of miR-153 (10). However, a unique aspect of our study was that both SNAP23 and SNAP25 were increased in miR-153-overexpressing HT-22 cells. SNAP23 and SNAP25 have been found to modulate exocytosis and neuronal development (12). Indeed, other miRNAs also exhibit such inconsistencies. For example, Nr2e1 mRNA has a miR-9 responsive element in its 3′ UTR. Nr2e1 protein expression was found to be reduced in mouse embryos with miR-9 mutation, and miR-9 did not suppress luciferase expression from a reporter conjugated to the 3′ UTR of Nr2e1 in P19 cells. Moreover, miR-9-2 upregulated the expression of endogenous Nr2e1 in P19 cells (8). This discrepancy between an miRNA and the expression of its target genes may have some possible explanations. On one hand, this discrepancy may be due to differences in cell type, differentiation stage, animal species or RNA-binding protein repertoires. Some RNA-binding proteins that facilitate binding of miR-153 to its target may be missing or insufficient in HT-22 cells. On the other hand, miR-153 may increase SNAP25 expression indirectly by suppressing another gene that inhibits SNAP25 expression. Further studies are required to elucidate the precise mechanism underlying the regulation of SNAP25 and SNAP23 by miR-153. In addition, SNAP25 misregulation was found to play a key role in some human diseases including attention deficit-hyperactivity disorder, schizophrenia, bipolar I disorder, Huntington's disease and Alzheimer's disease (26–29). Therefore, the upregulation of SNAP25 by miR-153 in the present study may be a potential therapy target for some neurodegenerative diseases.
Collectively, these results on miRNA expression in neural cells indicate that miR-153 may function differently in different regions of the central nervous system, or during different developmental stages. The HT-22 cell line was used to analyze the effects of miR-153 in the present study, and the results may differ in embryonic or animal models in response to miR-153 overexpression.
Although our study demonstrated that miR-153 is implicated in neuronal differentiation and normal function by regulating multiple proteins, a series of issues regarding miR-153 controlling neurogenesis should be further explored. First, the exact role of miR-153 in neural development must be confirmed in vivo. Second, the detailed molecular mechanism underlying the function of miR-153 in neurogenesis requires further investigation. Third, the clinical applicability of miR-153 must be investigated, particularly in neurodegenerative diseases.
Acknowledgements
Not applicable.
Funding
The present study received funding from ‘Outstanding Leaders Training Program of Pudong Health Bureau of Shanghai’ (grant no. PWRI2016-02).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
YZ and JL designed the study. SJ and JX prepared the reagents and analyzed data. CW and QM cultured the cells and drafted the manuscript. CX and JL performed the molecular biology experiments. CX, YG, QW, WX, YQ and YH analyzed and interpreted the data, and revised the manuscript for important intellectual content. CX and JL wrote the manuscript. YZ and JL reviewed and revised the document. All authors reviewed and approved the final version of the manuscript. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Choi PS, Zakhary L, Choi WY, Caron S, Alvarez-Saavedra E, Miska EA, McManus M, Harfe B, Giraldez AJ, Horvitz RH, et al. Members of the miRNA-200 family regulate olfactory neurogenesis. Neuron. 2008;57:41–55. doi: 10.1016/j.neuron.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, Hannon G, Abeliovich A. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haramati S, Chapnik E, Sztainberg Y, Eilam R, Zwang R, Gershoni N, McGlinn E, Heiser PW, Wills AM, Wirguin I, et al. miRNA malfunction causes spinal motor neuron disease. Proc Natl Acad Sci USA. 2010;107:13111–13116. doi: 10.1073/pnas.1006151107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harraz MM, Dawson TM, Dawson VL. MicroRNAs in Parkinson's disease. J Chem Neuroanat. 2011;42:127–130. doi: 10.1016/j.jchemneu.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morgado AL, Rodrigues CM, Sola S. MicroRNA-145 regulates neural stem cell differentiation through the Sox2-Lin28/let-7 signaling pathway. Stem Cells. 2016;34:1386–1395. doi: 10.1002/stem.2309. [DOI] [PubMed] [Google Scholar]
- 7.Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shibata M, Nakao H, Kiyonari H, Abe T, Aizawa S. MicroRNA-9 regulates neurogenesis in mouse telencephalon by targeting multiple transcription factors. J Neurosci. 2011;31:3407–3422. doi: 10.1523/JNEUROSCI.5085-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doxakis E. Post-transcriptional regulation of alpha-synuclein expression by mir-7 and mir-153. J Biol Chem. 2010;285:12726–12734. doi: 10.1074/jbc.M109.086827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei C, Thatcher EJ, Olena AF, Cha DJ, Perdigoto AL, Marshall AF, Carter BD, Broadie K, Patton JG. miR-153 regulates SNAP-25, synaptic transmission, and neuronal development. PLoS One. 2013;8:e57080. doi: 10.1371/journal.pone.0057080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsuyama J, Bunt J, Richards LJ, Iwanari H, Mochizuki Y, Hamakubo T, Shimazaki T, Okano H. MicroRNA-153 regulates the acquisition of gliogenic competence by neural stem cells. Stem Cell Reports. 2015;5:365–377. doi: 10.1016/j.stemcr.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zylbersztejn K, Galli T. Vesicular traffic in cell navigation. FEBS J. 2011;278:4497–4505. doi: 10.1111/j.1742-4658.2011.08168.x. [DOI] [PubMed] [Google Scholar]
- 13.Kim B, Park J, Chang KT, Lee DS. Peroxiredoxin 5 prevents amyloid-beta oligomer-induced neuronal cell death by inhibiting ERK-Drp1-mediated mitochondrial fragmentation. Free Radic Biol Med. 2016;90:184–194. doi: 10.1016/j.freeradbiomed.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.O'Brien RJ, Wong PC. Amyloid precursor protein processing and Alzheimer's disease. Annu Rev Neurosci. 2011;34:185–204. doi: 10.1146/annurev-neuro-061010-113613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qiu L, Tan EK, Zeng L. microRNAs and neurodegenerative diseases. Adv Exp Med Biol. 2015;888:85–105. doi: 10.1007/978-3-319-22671-2_6. [DOI] [PubMed] [Google Scholar]
- 17.Leggio L, Vivarelli S, L'Episcopo F, Tirolo C, Caniglia S, Testa N, Marchetti B, Iraci N. microRNAs in Parkinson's disease: From pathogenesis to novel diagnostic and therapeutic approaches. Int J Mol Sci. 2017;18:E2698. doi: 10.3390/ijms18122698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bak M, Silahtaroglu A, Moller M, Christensen M, Rath MF, Skryabin B, Tommerup N, Kauppinen S. MicroRNA expression in the adult mouse central nervous system. RNA. 2008;14:432–444. doi: 10.1261/rna.783108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu J, Li L, Suo WZ. HT22 hippocampal neuronal cell line possesses functional cholinergic properties. Life Sci. 2009;84:267–271. doi: 10.1016/j.lfs.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 22.Lei T, Zhang P, Zhang X, Xiao X, Zhang J, Qiu T, Dai Q, Zhang Y, Min L, Li Q, et al. Cyclin K regulates prereplicative complex assembly to promote mammalian cell proliferation. Nat Commun. 2018;9:1876. doi: 10.1038/s41467-018-04258-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Juan HC, Lin Y, Chen HR, Fann MJ. Cdk12 is essential for embryonic development and the maintenance of genomic stability. Cell Death Differ. 2016;23:1038–1048. doi: 10.1038/cdd.2015.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen HR, Juan HC, Wong YH, Tsai JW, Fann MJ. Cdk12 regulates neurogenesis and late-arising neuronal migration in the developing cerebral cortex. Cereb Cortex. 2017;27:2289–2302. doi: 10.1093/cercor/bhw081. [DOI] [PubMed] [Google Scholar]
- 25.Isgro MA, Bottoni P, Scatena R. Neuron-specific enolase as a biomarker: Biochemical and clinical aspects. Adv Exp Med Biol. 2015;867:125–143. doi: 10.1007/978-94-017-7215-0_9. [DOI] [PubMed] [Google Scholar]
- 26.Corradini I, Verderio C, Sala M, Wilson MC, Matteoli M. SNAP-25 in neuropsychiatric disorders. Ann N Y Acad Sci. 2009;1152:93–99. doi: 10.1111/j.1749-6632.2008.03995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scarr E, Gray L, Keriakous D, Robinson PJ, Dean B. Increased levels of SNAP-25 and synaptophysin in the dorsolateral prefrontal cortex in bipolar I disorder. Bipolar Disord. 2006;8:133–143. doi: 10.1111/j.1399-5618.2006.00300.x. [DOI] [PubMed] [Google Scholar]
- 28.Smith R, Klein P, Koc-Schmitz Y, Waldvogel HJ, Faull RL, Brundin P, Plomann M, Li JY. Loss of SNAP-25 and rabphilin 3a in sensory-motor cortex in Huntington's disease. J Neurochem. 2007;103:115–123. doi: 10.1111/j.1471-4159.2007.04703.x. [DOI] [PubMed] [Google Scholar]
- 29.Bereczki E, Francis PT, Howlett D, Pereira JB, Hoglund K, Bogstedt A, Cedazo-Minguez A, Baek JH, Hortobagyi T, Attems J, et al. Synaptic proteins predict cognitive decline in Alzheimer's disease and Lewy body dementia. Alzheimers Dement. 2016;12:1149–1158. doi: 10.1016/j.jalz.2016.04.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.