Abstract
Tree shrews are most closely related to the primates and so possess a number of advantages in experimental studies; they have been used as an animal model in bacterial and virus infection, cancer, endocrine system disease, and certain nervous system diseases. Their olfactory ensheathing cells (OECs) are able to release several cytokines to promote neuronal survival, regeneration and remyelination. The present study used western blot analysis to identify antibody specificity in protein extracts from whole tree shrew brains to identify the specificity of p75 nerve growth factor receptor (NGFR) derived from rabbits (75 kDa). OECs were cultured and isolated, then stained and identified using the antibodies for p75NGFR. To investigate the capacity of OECs to express cytokines and growth factors, microarray technology was used, and the analysis revealed that OECs were able to express 9,821 genes. Of these genes, 44 genes were from the neurotrophic factor family, which may indicate their potential in transplantation in vivo. The present study considered the function of OECs as revealed by other studies, and may contribute to future research.
Keywords: tree shrew, olfactory ensheathing cells, microarray analysis
Introduction
Tree shrews (Tupaia belangeri, Wagner, 1841) are small animals originally regarded as either primates or insectivores and later classified as a separate order, Scandentia (1,2). They are most closely related to the primates, with the advantages of being readily available, easy to tame and possessing a strong reproductive capacity (3). Due to these advantages they have been successfully employed as an animal model for bacterial (4) or viral infection (5,6), tumors (7), endocrine system disease (3,8), and certain nervous system diseases, including myopia (9–11), psychosocial stress (12,13), cerebral ischemia (14) and aging (15).
Olfactory ensheathing cells (OECs) are the glial cells that derive from the olfactory placode and envelop olfactory axons in the course of migration from the olfactory epithelium to the bulb (16). A number of studies have identified that OECs can not only release various cytokines (17,18) that can promote neuronal survival (19), regeneration (20,21) and remyelination (22,23), in vitro and in vivo, but can also migrate into lesion regions (24) and facilitate functional repair (25,26) following injuries. In the present study, OECs of tree shrews were isolated and purified successfully for the first time, to the best of the authors' knowledge. Cytokine or the receptor mRNA expression was also investigated using a microarray. The results of the present study may contribute to the use of OECs in transplantation and the evaluation of their curative effect in fundamental research and future clinical trials.
Materials and methods
Animals
Tree shrews (2 newborn for cell culture and 1 adult male, weight 140±10 g at the start of the experiment) and 1 adult male Sprague-Dawley rats (for western blot analysis, weight 140±10 g at the start of the experiment) were provided by Kunming Medical University Animal Center (Kunming, China) and employed in this study. The animals were raised at 20±5°C, 40–60% humidity and a 12-h light/dark cycle with food and water ad libitum. All animal work was conducted according to the guidelines for the care and use of experimental animals established by the Ministry of Science and Technology of the People's Republic of China (approval no. 2006-398), and was approved by the ethics committee of Institute of Neuroscience, Kunming Medical University (Kunming, China). All animals were bred in separated cages.
Primary cell cultures
Primary cultures of OECs were set up from newborn tree shrews and the animals were sacrificed. Olfactory bulbs were aseptically removed. The meningeal layer was stripped off with fine forceps and the tissue was enzymatically (trypsin 0.25%; 1:30 tissues to trypsin ratio) and mechanically dissociated, then incubated in 5% CO2 at 37°C for 15 min. The cells were recovered by centrifugation (100 × g for 5 min at 37°C) in culture medium [Dulbecco's modified Eagle's medium/F12 (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific, Inc.)] and seeded at 1×105 cells/ml into culture plates and incubated in 5% CO2 at 37°C. Cells were cultured for 10 h and then the supernatant was removed to another culture plate.
Antibodies
Antibodies against p75 neurotrophin receptor (p75NGFR) and BrdU were used to study the purity and proliferation of OECs. For each antibody, a minimum of two whole series of different animals were studied. Western blot analysis with protein extracts from whole tree shrew and rat brains ensured specificity of the antibodies used. The tissues were homogenized in RIPA buffer (Beyotime Institute of Biotechnology, Haimen, China) that contained a mixture of proteinase inhibitors and the supernatant was collected by centrifuging at 4°C at 22,500 × g for 15 min. The protein concentration was determined by the DC protein assay from Bio-Rad Laboratories, Inc. (Hercules, CA, USA), and protein samples (100 µg) of each cell lysate in SDS loading buffer (Biosharp) were electrophoresed on 10% SDS-PAGE gel. Electrophoresis was performed and the proteins were transferred onto polyvinylidene difluoride membranes (Pall Life Sciences, Port Washington, NY, USA) using electroblotting apparatus (Bio-Rad Laboratories, Inc.). The membranes were blocked at 37°C for 1 h in TBS containing 0.1% Tween-20 and 5% dried milk, and were then incubated at 4°C overnight with NGFRp75 antibody (1:1,000; cat. no. AB1554; EMD Millipore, Billerica, MA, USA) They were incubated with the IRDye 800-conjugated affinity purified goat anti-rabbit immunoglobulin (Ig)G (1:5,000; cat. no. 611-1302; Rockland Immunochemicals, Inc., Pottstown, PA, USA) for 1 h at room temperature and visualization using an Odyssey laser scanning system (LI-COR Biosciences, Lincoln, NE, USA). Blots were reprobed with the monoclonal mouse anti-β actin antibody (1:5,000; cat. no. A2228; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), followed by reaction with IRDye 800-conjugated affinity purified goat anti-mouse IgG (1:5,000; cat. no. 610-1302; Rockland Immunochemicals, Inc.) at 37°C for 2 h to confirm equal protein loading.
Immunofluorescence
Immunofluorescence was performed on OECs. The cells were fixed with 4% formalin solution at room temperature for 5 min then washed with PBS and permeabilized with 0.1% Triton X-100. The cells were then blocked with 1% bovine serum albumin (10082-139; Gibco; Thermo Fisher Scientific, Inc.) at 37°C for 30 min and incubated for 18 h at 4°C with anti-p75NGFR (1:500; AB1554; EMD Millipore) and anti-BrdU (1:200; mAb #5292; Cell Signaling Technology, Inc., Danvers, MA, USA) antibodies. Following washing with PBS, the cells were incubated at 37°C for 2 h with respective anti-Mouse IgG H&L (Alexa Fluor 647; 1:500; ab150115; Abcam, Cambridge, MA, USA). Following incubation, the slides were washed with PBS, mounted and examined under a fluorescence microscope (Leica TCS SP2; Leica Microsystems GmbH, Wetzlar, Germany). Fluoroshield mounting medium with DAPI (Sigma-Aldrich; Merck KGaA) was used for DAPI staining and the staining was performed according to the manufacturer's protocol.
Microarray analysis
Total RNA from OECs was quantified using a NanoDrop ND-1000 (Thermo Fisher Scientific, Inc.) and RNA integrity was assessed using standard denaturing agarose gel electrophoresis (27) (performed by KangChen Biotech Co., Ltd., Shanghai, China). Total RNA from each sample was used for labeling and array hybridization as follows: i) Reverse transcription using the Superscript ds-cDNA Synthesis kit (Invitrogen; Thermo Fisher Scientific, Inc.); ii) ds-cDNA labeling using the NimbleGen One-Color DNA Labeling kit (Roche NimbleGen, Inc., Madison, WI, USA); iii) array hybridization using the NimbleGen Hybridization System, followed by washing with the NimbleGen wash buffer kit (Roche NimbleGen, Inc.); and iv) array scanning using the Axon GenePix 4000B microarray scanner (Molecular Devices, LLC, Sunnyvale, CA, USA). Scanned images (TIFF format) were then imported into NimbleScan software (version 2.5; Roche NimbleGen, Inc.) for grid alignment and expression data analysis. Expression data were normalized through quantile normalization and the Robust Multichip Average (RMA) algorithm included in the NimbleScan software. The Probe level (*_norm_RMA.pair) files and Gene level (*_RMA.calls) files were generated following normalization. All gene level files were imported into Agilent GeneSpring GX software (version 11.5; Agilent Technologies, Inc., Santa Clara, CA, USA) for further analysis. Finally, hierarchical clustering was performed using Gene Ontology (http://www.geneontology.org) to show distinguishable gene expression profiling among samples.
Results
Specificity of the antibody
Western blotting with protein extracts from whole tree shrew and rat brains ensured specificity of the antibodies used in the present study. Western blotting demonstrated that the antibodies produced bands that were located at the same level in the gel for rat and tree shrew brain protein extracts. Notably, the western blots also demonstrated that these bands had the proper molecular weight for the p75NGFR antibody examined (Fig. 1A).
Figure 1.
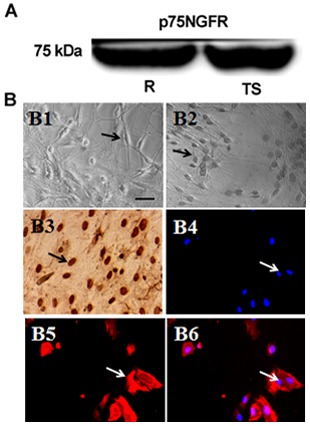
(A) Specificity of the antibody. The bands of p75NGFR with protein extracts from whole rat and tree shrew brain. The molecular marker was a restrained maker and did not react with ECL. (B) OEC biological characteristics in vitro. (B1) Morphology of OECs cultured for 24 h (magnification, ×200). (B2) Morphology of OECs cultured for 6 days (magnification, ×200). (B3) Immunocytochemical staining of BrdU (magnification, ×200). (B4) p75NGFR positive cells (magnification, ×400). (B5) DAPI positive cells (magnification, ×400). (B6) Merged view of the p75NGFR and DAPI double positive cells (magnification, ×400). OECs, olfactory ensheathing cells; NGFR, nerve growth factor receptor; R, rat; TS, tree shrew.
OEC biological characteristics in vitro
To investigate the method of culturing OECs from tree shrews, the bulbus olfactorius of newborn tree shrews was used. Cultured for 24 h, certain cells began to demonstrate adherence and assume a round or star-like shape (Fig. 1B1). Then, 6 days following adherent culture, three classic morphologies of OECs were observed: Bipolar or fusiform and oblate (Fig. 1B2). Among these shapes, fusiform and oblate were more common, and an evident three-dimensional appearance. After 14 days, the number of cells did not significantly increase, although the cells were still growing rapidly (data not shown). Cells were stained against BrdU (Fig. 1B3) to confirm their capacity for proliferation, which demonstrated that all stages of OECs nuclei were positively stained, demonstrating that they possessed proliferative ability. Immunocytochemical staining against p75NGFR (Fig. 1B4) demonstrated that the fusiform cells or their nuclei were positively stained, demonstrating that they were OECs. All cell nuclei were positively stained with DAPI (Fig. 1B5) and the merged image is demonstrated in Fig. 1B6.
Gene chip assay
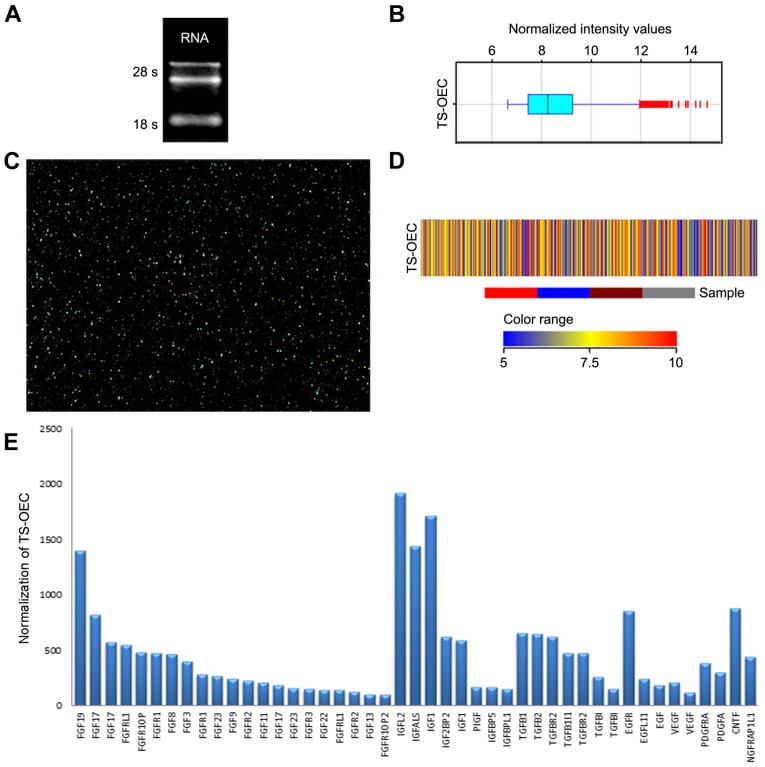
In order to investigate RNA integrity and genomic DNA contamination, the denaturing agarose gel electrophoresis test was used. The result demonstrated that optical density (OD) A260/A280 ratio was 2.00 and the OD A260/A230 ratio was 2.35. These results confirmed that the RNA was pure [for spectrophotometer, the O.D. A260/A280 ratio should be close to 2.0 for pure RNA (ratios between 1.8 and 2.1 are acceptable) The OD A260/A230 ratio should be >1.8]. The 28S and 18S ribosomal RNA bands were fairly sharp, intense bands and there was a diffuse smear of ethidium bromide-stained material migrating between the 18S and 28S ribosomal bands, possibly comprising mRNA and other heterogeneous RNA species (Fig. 2A). Prior to analysis a boxplot was performed (Fig. 2B). Then, gene chip assay suggested that OECs expressed 9,821 genes (Fig. 2C) and 45 important genes are listed in Table I and Fig. 2D and E.
Figure 2.
Gene chip assay. (A) Bands of RNA agarose gel electrophoresis. (B) Boxplot of the samples. (C) Map of gene chip assay. (D) Heat map of OECs expressed genes. (E) Bar chart of 45 important genes. OECs, olfactory ensheathing cells; NGFR, nerve growth factor receptor.
Table I.
Genes with significant expression difference.
| Sequence identification | Normalized tree shrew OECs | Gene name | Synonyms |
|---|---|---|---|
| NM_001002915 | 1914.4823 | IGFL2 | VPRI645 |
| M37484 | 1709.5454 | IGF1 | IGFI |
| NM_004970 | 1441.7056 | IGFALS | ALS |
| NM_005117 | 1399.2097 | FGF19 | n/a |
| NM_201284 | 849.4621 | EGFR | ERBB, ERBB1, mENA |
| NM_000614 | 879.9232 | CNTF | HCNTF |
| BC105131 | 818.3038 | FGF17 | FGF-13 |
| BC000125 | 658.60596 | TGFB1 | CED, DPD1, TGFB |
| M19154 | 643.18677 | TGFB2 | MGC116892, TGF-β2 |
| BC040499 | 623.899 | TGFBR2 | AAT3, HNPCC6, MFS2, RIIC, TGFR-2, TGFβ-RII |
| NM_001007225 | 620.82745 | IGF2BP2 | IMP-2, IMP2, VICKZ2, p62 |
| M29644 | 592.314 | IGF1 | IGFI |
| AY358869 | 576.01324 | FGF17 | FGF-13 |
| NM_001004356 | 551.01306 | FGFRL1 | FGFR5, FHFR |
| BC037785 | 484.15298 | FGFR1OP | FOP |
| AK024388 | 476.04077 | FGFR1 | BFGFR, C-FGR, CD331, CEK, FLG, FLT2, H2, H3, H4, H5, KAL2, N-SAM |
| NM_015927 | 474.31787 | TGFB1I1 | ARA55, HIC-5, HIC5, TSC-5 |
| NM_001024847 | 474.17993 | TGFBR2 | AAT3, HNPCC6, MFS2, RIIC, TGFR-2, TGFβ-RII |
| NM_006119 | 463.30542 | FGF8 | AIGF, HBGF-8 |
| NM_001012978 | 436.89304 | NGFRAP1L1 | BEX5, MGC104434, MGC126446 |
| NM_005247 | 398.42584 | FGF3 | HBGF-3, INT2 |
| NM_006206 | 383.68063 | PDGFRA | CD140A, MGC74795, PDGFR2 |
| NM_002607 | 299.87314 | PDGFA | PDGF-A, PDGF1 |
| M63888 | 280.45587 | FGFR1 | BFGFR, C-FGR, CD331, CEK, FLG, FLT2, H2, H3, H4, H5, KAL2, N-SAM |
| NM_020638 | 267.7028 | FGF23 | ADHR, HPDR2, HYPF, PHPTC |
| BC004972 | 260.64957 | TGFBI | BIGH3, CDB1, CDG2, CDGG1, CSD, CSD1, CSD2, CSD3, LCD1 |
| NM_198283 | 246.34807 | EGFL11 | KIAA0663, dJ1018A4.2 |
| BC069692 | 240.3946 | FGF9 | GAF, HBFG-9, MGC119914, MGC119915 |
| NM_023028 | 225.70663 | FGFR2 | BEK, BFR-1, CD332, CEK3, CFD1, ECT1, JWS, K-SAM, KGFR, TK14, TK25 |
| BC032502 | 210.7425 | FGF11 | FHF3, FLJ16061, MGC102953, MGC45269, |
| S85192 | 207.95761 | VEGF | MGC70609, VEGFA, VPF |
| BC093731 | 185.4517 | EGF | URG |
| NM_003867 | 183.04973 | FGF17 | FGF-13 |
| NM_002643 | 168.185 | PIGF | MGC32646, MGC33136 |
| BC011453 | 165.53284 | IGFBP5 | IBP5 |
| BC098252 | 157.41748 | FGF23 | ADHR|HPDR2|HYPF|PHPTC| |
| BC026352 | 151.22711 | TGFBI | BIGH3, CDB1, CDG2, CDGG1, CSD, CSD1, CSD2, CSD3, LCD1 |
| NM_000142 | 151.22061 | FGFR3 | ACH, CD333, CEK2, HSFGFR3EX, JTK4 |
| NM_001007563 | 149.79308 | IGFBPL1 | IGFBP-RP4, bA113O24.1 |
| NM_020637 | 140.37721 | FGF22 | n/a |
| AK172829 | 139.57779 | FGFRL1 | FGFR5, FHFR |
| NM_022976 | 125.33181 | FGFR2 | BEK, BFR-1, CD332, CEK3, CFD1, ECT1, JWS, K-SAM, KGFR, TK14, TK25 |
| M27281 | 120.924126 | VEGF | MGC70609, VEGFA, VPF |
| NM_004114 | 102.02009 | FGF13 | FGF2, FHF2 |
| NM_015633 | 100.07872 | FGFR1OP2 | DKFZp564O1863, HSPC123-like |
OECs, olfactory ensheathing cells; n/a, not applicable.
Discussion
Specificity of the antibodies
Tree shrews possess a number of advantages as animal models, thus, studies on them are popular. However, there are no specific reagents for tree shrews, particularly antibodies. The present study used western blot analysis on protein extracts from whole tree shrew and rat brains to ensure specificity of the antibodies used. Protein extracts from rat brain were used as a positive control, as the specificity of the antibodies has already been verified in this species. The results demonstrated that the antibodies produced bands that were located at the same level. Notably, the molecular weight for p75NGFR was ~75 kDa (28). These data suggested that the antibodies used were specific and the results reliable.
OECs biological characteristics in vitro
The present study was the first to successfully isolate and purify tree shrew OECs, to the best of the authors' knowledge. As in a previous study, the cultured cells exhibited the three classical morphologies of OECs: Bipolar or fusiform, multi-ecphyma shape and oblate (26). Previous studies have demonstrated that p75NGFR was the specific marker of OEC (29,30) and that the antibody to BrdU is useful in detecting S-phase cells, as BrdU-positive cells are proliferating (31,32). The present study demonstrated that the majority of cells were p75NGFR positive and BrdU positive. These data further proved that the majority of the cultured cells were OECs and the majority of them proliferated well.
Gene chip assay
The microarray technique allows the monitoring of the expression of tens of thousands of genes simultaneously in one hybridization experiment. DNA segments are closely arranged on a slide. cDNA was reverse transcribed from mRNA derived from tree shrew OECs, labelled with a Cyanine3 (Cy3) fluorophore and hybridized on the microarray slide. Through this technique, the detection of differentially expressed genes is greatly facilitated. In the present study, prior to the microarray analysis, RNA quantification and quality assurance was performed to ensure reliability of the results (33–35). Prior to the statistical analysis, a boxplot test was performed. The boxplot is a traditional method for visualizing the distribution of a dataset (36) and is useful for comparing the distributions of several datasets (37). In the present study, a boxplot was used to observe the distributions of expression values for the samples in the experiment following normalization. The data demonstrated that tree shrew OECs can express 9,821 genes, containing 44 cytokines, including the fibroblast growth factors family (38), the insulin-like growth factor family (39–42), the transforming growth factor family (38), epidermal growth factor (43,44), ciliary neurotrophic factor (40), vascular endothelial growth factor (45) and platelet derived growth factor receptor α (46,47), which were hypothesized as possessing the capacity to promote neuronal survival, regeneration, remyelination or function improvement.
In conclusion, tree shrew OECs were isolated and purified successfully for the first time, to the best of the authors' knowledge, and the cells were able to secrete a number of cytokines that may contribute to nervous system diseases. Although there was no specific kit, tests were performed to ensure that the data were reliable. These results may lead to future fundamental research or clinical trials.
Acknowledgements
Not applicable.
Funding
The authors of the present study would like to thank the Chinese Ministry of the National Science-Technology Support Plan Project for financial support (grant no. 2014BAI01B01).
Availability of data and materials
All the materials included in the manuscript, including all relevant raw data, will be made freely available to any researchers who wish to use them for non-commercial purposes, while preserving any necessary confidentiality and anonymity.
Authors' contributions
T-HW, NL and H-YW designed the study. T-YW performed cell cultures, X-JD, W-JH, and T-YW performed immunofluorescence, NL and JW performed microarray analysis, western blotting and wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All animal work was conducted according to the guidelines for the care and use of experimental animals established by the Ministry of Science and Technology of the People's Republic of China (approval no. 2006-398), and was approved by the ethics committee of Institute of Neuroscience, Kunming Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Diogo R. The head and neck muscles of the Philippine colugo (Dermoptera: Cynocephalus volans), with a comparison to tree-shrews, primates, and other mammals. J Morphol. 2009;270:14–51. doi: 10.1002/jmor.10666. [DOI] [PubMed] [Google Scholar]
- 2.Xu L, Chen SY, Nie WH, Jiang XL, Yao YG. Evaluating the phylogenetic position of Chinese tree shrew (Tupaia belangeri chinensis) based on complete mitochondrial genome: Implication for using tree shrew as an alternative experimental animal to primates in biomedical research. J Genet Genomics. 2012;39:131–137. doi: 10.1016/j.jgg.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Collins PM, Tsang WN, Urbanski HF. Endocrine correlates of reproductive development in the male tree-shrew (Tupaia belangeri) and the effects of infantile exposure to exogenous androgens. Gen Comp Endocrinol. 2007;154:22–30. doi: 10.1016/j.ygcen.2007.06.024. [DOI] [PubMed] [Google Scholar]
- 4.Li SA, Lee WH, Zhang Y. Two bacterial infection models in tree shrew for evaluating the efficacy of antimicrobial agents. Dongwuxue Yanjiu. 2012;33:1–6. doi: 10.3724/SP.J.1141.2012.01001. [DOI] [PubMed] [Google Scholar]
- 5.Yang ZF, Zhao J, Zhu YT, Wang YT, Liu R, Zhao SS, Li RF, Yang CG, Li JQ, Zhong NS. The tree shrew provides a useful alternative model for the study of influenza H1N1 virus. Virol J. 2013;10:111. doi: 10.1186/1743-422X-10-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu G, Gao Z, He W, Ma Y, Feng X, Cai T, Lu F, Liu L, Li W. microRNA expression in hepatitis B virus infected primary treeshrew hepatocytes and the independence of intracellular miR-122 level for de novo HBV infection in culture. Virology. 2014;448:247–254. doi: 10.1016/j.virol.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 7.Xu Y, Köck J, Lu Y, Yang D, Lu M, Zhao X. Suppression of hepatitis B virus replication in Tupaia hepatocytes by tumor necrosis factor alpha of Tupaia belangeri. Comp Immunol Microbiol Infect Dis. 2011;34:361–368. doi: 10.1016/j.cimid.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 8.van der Hart MG, de Biurrun G, Czeh B, Rupniak NM, den Boer JA, Fuchs E. Chronic psychosocial stress in tree shrews: Effect of the substance P (NK1 receptor) antagonist L-760735 and clomipramine on endocrine and behavioral parameters. Psychopharmacology (Berl) 2005;181:207–216. doi: 10.1007/s00213-005-2260-0. [DOI] [PubMed] [Google Scholar]
- 9.Arumugam B, McBrien NA. Muscarinic antagonist control of myopia: Evidence for M4 and M1 receptor-based pathways in the inhibition of experimentally-induced axial myopia in the tree shrew. Invest Ophthalmol Vis Sci. 2012;53:5827–5837. doi: 10.1167/iovs.12-9943. [DOI] [PubMed] [Google Scholar]
- 10.Guo L, Frost MR, He L, Siegwart JT, Jr, Norton TT. Gene expression signatures in tree shrew sclera in response to three myopiagenic conditions. Invest Ophthalmol Vis Sci. 2013;54:6806–6819. doi: 10.1167/iovs.13-12551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu X, McBrien NA, Smith EL, III, Troilo D, Wallman J. Eyes in various species can shorten to compensate for myopic defocus. Invest Ophthalmol Vis Sci. 2013;54:2634–2644. doi: 10.1167/iovs.12-10514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Czeh B, Simon M, Schmelting B, Hiemke C, Fuchs E. Astroglial plasticity in the hippocampus is affected by chronic psychosocial stress and concomitant fluoxetine treatment. Neuropsychopharmacology. 2006;31:1616–1626. doi: 10.1038/sj.npp.1300982. [DOI] [PubMed] [Google Scholar]
- 13.Zambello E, Fuchs E, Abumaria N, Rygula R, Domenici E, Caberlotto L. Chronic psychosocial stress alters NPY system: Different effects in rat and tree shrew. Prog Neuropsychopharmacol Biol Psychiatry. 2010;34:122–130. doi: 10.1016/j.pnpbp.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 14.Li S, Meng Q, Zhang L. Experimental therapy of a platelet-activating factor antagonist (ginkgolide B) on photochemically induced thrombotic cerebral ischaemia in tree shrews. Clin Exp Pharmacol Physiol. 1999;26:824–825. doi: 10.1046/j.1440-1681.1999.03146.x. [DOI] [PubMed] [Google Scholar]
- 15.Keuker JI, de Biurrun G, Luiten PG, Fuchs E. Preservation of hippocampal neuron numbers and hippocampal subfield volumes in behaviorally characterized aged tree shrews. J Comp Neurol. 2004;468:509–517. doi: 10.1002/cne.10996. [DOI] [PubMed] [Google Scholar]
- 16.Yang H, He BR, Hao DJ. Biological roles of olfactory ensheathing cells in facilitating neural regeneration: A systematic review. Mol Neurobiol. 2014;51:168–179. doi: 10.1007/s12035-014-8664-2. [DOI] [PubMed] [Google Scholar]
- 17.Lang BC, Zhang Z, Lv LY, Liu J, Wang TY, Yang LH, Liao DQ, Zhang WS, Wang TH. OECs transplantation results in neuropathic pain associated with BDNF regulating ERK activity in rats following cord hemisection. BMC Neurosci. 2013;14:80. doi: 10.1186/1471-2202-14-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pastrana E, Moreno-Flores MT, Avila J, Wandosell F, Minichiello L, Diaz-Nido J. BDNF production by olfactory ensheathing cells contributes to axonal regeneration of cultured adult CNS neurons. Neurochem Int. 2007;50:491–498. doi: 10.1016/j.neuint.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 19.He BR, Xie ST, Wu MM, Hao DJ, Yang H. Phagocytic removal of neuronal debris by olfactory ensheathing cells enhances neuronal survival and neurite outgrowth via p38MAPK Activity. Mol Neurobiol. 2014;49:1501–1512. doi: 10.1007/s12035-013-8588-2. [DOI] [PubMed] [Google Scholar]
- 20.Huo SJ, Li Y, Raisman G, Yin ZQ. Transplanted olfactory ensheathing cells reduce the gliotic injury response of Muller cells in a rat model of retinitis pigmentosa. Brain Res. 2011;1382:238–244. doi: 10.1016/j.brainres.2010.12.079. [DOI] [PubMed] [Google Scholar]
- 21.Sandvig I, Hoang L, Sardella TC, Barnett SC, Brekken C, Tvedt K, Berry M, Haraldseth O, Sandvig A, Thuen M. Labelling of olfactory ensheathing cells with micron-sized particles of iron oxide and detection by MRI. Contrast Media Mol Imaging. 2012;7:403–410. doi: 10.1002/cmmi.1465. [DOI] [PubMed] [Google Scholar]
- 22.Sasaki M, Lankford KL, Radtke C, Honmou O, Kocsis JD. Remyelination after olfactory ensheathing cell transplantation into diverse demyelinating environments. Exp Neurol. 2011;229:88–98. doi: 10.1016/j.expneurol.2011.01.010. [DOI] [PubMed] [Google Scholar]
- 23.Radtke C, Kocsis JD. Peripheral nerve injuries and transplantation of olfactory ensheathing cells for axonal regeneration and remyelination: Fact or fiction? Int J Mol Sci. 2012;13:12911–12924. doi: 10.3390/ijms131012911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andrews MR, Stelzner DJ. Evaluation of olfactory ensheathing and schwann cells after implantation into a dorsal injury of adult rat spinal cord. J Neurotrauma. 2007;24:1773–1792. doi: 10.1089/neu.2007.0353. [DOI] [PubMed] [Google Scholar]
- 25.Sun T, Ye C, Zhang Z, Wu J, Huang H. Cotransplantation of olfactory ensheathing cells and Schwann cells combined with treadmill training promotes functional recovery in rats with contused spinal cords. Cell Transplant. 2013;22(Suppl 1):S27–S38. doi: 10.3727/096368913X672118. [DOI] [PubMed] [Google Scholar]
- 26.Wang YC, Xia QJ, Ba YC, Wang TY, Lin N, Zou Y, Shang FF, Zhou XF, Wang TH, Fu XM, Qi JG. Transplantation of olfactory ensheathing cells promotes the recovery of neurological functions in rats with traumatic brain injury associated with downregulation of Bad. Cytotherapy. 2014;16:1000–1010. doi: 10.1016/j.jcyt.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 27.Aranda PS, LaJoie DM, Jorcyk CL. Bleach gel: A simple agarose gel for analyzing RNA quality. Electrophoresis. 2012;33:366–369. doi: 10.1002/elps.201100335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vanella L, Barbagallo I, Acquaviva R, Di Giacomo C, Cardile V, Abraham NG, Sorrenti V. Ellagic acid: Cytodifferentiating and antiproliferative effects in human prostatic cancer cell lines. Curr Pharm Des. 2013;19:2728–2736. doi: 10.2174/1381612811319150008. [DOI] [PubMed] [Google Scholar]
- 29.Woodhouse A, Vincent AJ, Kozel MA, Chung RS, Waite PM, Vickers JC, West AK, Chuah MI. Spinal cord tissue affects ensheathing cell proliferation and apoptosis. Neuroreport. 2005;16:737–740. doi: 10.1097/00001756-200505120-00016. [DOI] [PubMed] [Google Scholar]
- 30.Raucci F, Tiong JD, Wray S. P75 nerve growth factor receptors modulate development of GnRH neurons and olfactory ensheating cells. Front Neurosci. 2013;7:262. doi: 10.3389/fnins.2013.00262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dowling MR, Kan A, Heinzel S, Zhou JH, Marchingo JM, Wellard CJ, Markham JF, Hodgkin PD. Stretched cell cycle model for proliferating lymphocytes. Proc Natl Acad Sci USA. 2014;111:6377–6382. doi: 10.1073/pnas.1322420111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gallaher ZR, Johnston ST, Czaja K. Neural proliferation in the dorsal root ganglia of the adult rat following capsaicin-induced neuronal death. J Comp Neurol. 2014;522:3295–3307. doi: 10.1002/cne.23598. [DOI] [PubMed] [Google Scholar]
- 33.Lubelchek RJ, Max B, Sandusky CJ, Hota B, Barker DE. Reliability at the lower limits of HIV-1 RNA quantification in clinical samples: A comparison of RT-PCR versus bDNA assays. PLoS One. 2009;4:e6008. doi: 10.1371/journal.pone.0006008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Senechal B, James VL. Ten years of external quality assessment of human immunodeficiency virus type 1 RNA quantification. J Clin Microbiol. 2012;50:3614–3619. doi: 10.1128/JCM.01221-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodriguez RA, Polston PM, Wu MJ, Wu J, Sobsey MD. An improved infectivity assay combining cell culture with real-time PCR for rapid quantification of human adenoviruses 41 and semi-quantification of human adenovirus in sewage. Water Res. 2013;47:3183–3191. doi: 10.1016/j.watres.2013.03.022. [DOI] [PubMed] [Google Scholar]
- 36.Hong Y, Davis B, Marron JS, Kwitt R, Singh N, Kimbell JS, Pitkin E, Superfine R, Davis SD, Zdanski CJ, Niethammer M. Statistical atlas construction via weighted functional boxplots. Med Image Anal. 2014;18:684–698. doi: 10.1016/j.media.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gleiss A, Sanchez-Cabo F, Perco P, Tong D, Heinze G. Adaptive trimmed t-statistics for identifying predominantly high expression in a microarray experiment. Stat Med. 2011;30:52–61. doi: 10.1002/sim.4093. [DOI] [PubMed] [Google Scholar]
- 38.Diemel LT, Jackson SJ, Cuzner ML. Role for TGF-beta1, FGF-2 and PDGF-AA in a myelination of CNS aggregate cultures enriched with macrophages. J Neurosci Res. 2003;74:858–867. doi: 10.1002/jnr.10837. [DOI] [PubMed] [Google Scholar]
- 39.Cannella B, Pitt D, Capello E, Raine CS. Insulin-like growth factor-1 fails to enhance central nervous system myelin repair during autoimmune demyelination. Am J Pathol. 2000;157:933–943. doi: 10.1016/S0002-9440(10)64606-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Copelman CA, Cuzner ML, Groome N, Diemel LT. Temporal analysis of growth factor mRNA expression in myelinating rat brain aggregate cultures: Increments in CNTF, FGF-2, IGF-I, and PDGF-AA mRNA are induced by antibody-mediated demyelination. Glia. 2000;30:342–351. doi: 10.1002/(SICI)1098-1136(200006)30:4<342::AID-GLIA30>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 41.Saito F, Kanagawa M, Ikeda M, Hagiwara H, Masaki T, Ohkuma H, Shimizu T, Sonoo M, Toda T, Matsumura K. Overexpression of LARGE suppresses muscle regeneration via down-regulation of insulin-like growth factor 1 and aggravates muscular dystrophy in mice. Hum Mol Genet. 2014;23:4543–4558. doi: 10.1093/hmg/ddu168. [DOI] [PubMed] [Google Scholar]
- 42.ShengM H, Lau KH, Baylink DJ. Role of osteocyte-derived insulin-like growth factor I in developmental growth, modeling, remodeling, and regeneration of the bone. J Bone Metab. 2014;21:41–54. doi: 10.11005/jbm.2014.21.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gezginci-Oktayoglu S, Karatug A, Bolkent S. The relation among NGF, EGF and insulin is important for triggering pancreatic beta cell apoptosis. Diabetes Metab Res Rev. 2012;28:654–662. doi: 10.1002/dmrr.2339. [DOI] [PubMed] [Google Scholar]
- 44.Munemasa Y, Chang CS, Kwong JM, Kyung H, Kitaoka Y, Caprioli J, Piri N. The neuronal EGF-related gene Nell2 interacts with Macf1 and supports survival of retinal ganglion cells after optic nerve injury. PLoS One. 2012;7:e34810. doi: 10.1371/journal.pone.0034810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nowacka M, Obuchowicz E. BDNF and VEGF in the pathogenesis of stress-induced affective diseases: An insight from experimental studies. Pharmacol Rep. 2013;65:535–546. doi: 10.1016/S1734-1140(13)71031-4. [DOI] [PubMed] [Google Scholar]
- 46.Awuah PK, Nejak-Bowen KN, Monga SP. Role and regulation of PDGFRalpha signaling in liver development and regeneration. Am J Pathol. 2013;182:1648–1658. doi: 10.1016/j.ajpath.2013.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McGowan SE, McCoy DM. Platelet-derived growth factor-A and sonic hedgehog signaling direct lung fibroblast precursors during alveolar septal formation. Am J Physiol Lung Cell Mol Physiol. 2013;305:L229–L239. doi: 10.1152/ajplung.00011.2013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the materials included in the manuscript, including all relevant raw data, will be made freely available to any researchers who wish to use them for non-commercial purposes, while preserving any necessary confidentiality and anonymity.