Abstract
Recovery of the blood supply is the most effective treatment against ischemic heart disease; however, it is also a major cause of myocardial ischemia/reperfusion injury in clinical therapy. Curcumin has been reported to possess beneficial effects against hypoxia/reoxygenation (H/R)-induced cardiomyocyte injury by regulating cell proliferation, apoptosis and antioxidant enzyme activity. The aim of the present study was to investigate the molecular mechanisms underlying the effects of curcumin on H/R-injured cardiomyocytes. H9C2 cardiomyocytes were pretreated with curcumin, and then cultured under H/R conditions. The viability of H9C2 cells was measured using a Cell Counting kit-8 assay, and the levels of intracellular lactate dehydrogenase (LDH), malondialdehyde (MDA) and superoxide dismutase (SOD) were measured to assess cell injury. Levels of reactive oxygen species (ROS) and apoptosis were evaluated by flow cytometry. The expression levels of Notch intracellular domain (NICD) and numerous downstream genes were analyzed via reverse transcription-quantitative polymerase chain reaction and western blotting. The results revealed that curcumin protected H9C2 cells against H/R-induced injury, reversing the H/R-induced increases in LDH and MDA levels, and decreases in SOD levels. ROS levels in H/R-induced cells were also significantly downregulated by curcumin treatment (P<0.01), and the apoptotic rate was significantly decreased from 15.13% in the H/R group to 7.7% in the H/R + curcumin group (P<0.01). The expression levels of NICD, hairy and enhancer of split (Hes)-1, Hes-5 and hairy/enhancer-of-split related with YRPW motif protein 1 (Hey-1) were significantly decreased in H/R-treated cells following curcumin treatment. Treatment with Jagged1 attenuated the effects of curcumin on cell viability, ROS levels and apoptosis; the Notch pathway was also reactivated. The present study indicated that there was a role for the Notch pathway in the protective effects of curcumin against H/R-induced cardiomyocyte injury, suggesting that downregulation of the Notch pathway may alleviate H/R-induced injury in H9C2 cells.
Keywords: myocardial ischemia/reperfusion injury, Notch intracellular domain, Jagged1, ischemic heart disease
Introduction
Ischemic heart disease (IHD) is one of the leading causes of mortality worldwide (1). At present, recovering cardiac blood supply is the most effective treatment against IHD; however, it is also a major cause of myocardial ischemia/reperfusion injury (MIRI) in clinical therapy (2). MIRI can result in further myocardial damage, subsequently worsening the therapeutic outcomes of reperfusion therapy. Currently, methods for treating MIRI are insufficiently effective (3,4). Cardiomyocyte apoptosis is an important characteristic of MIRI that is observed during early phases of reperfusion. Therefore, further investigation of the mechanisms underlying reperfusion-induced cardiomyocyte injury is required to develop treatments for IHD.
Jagged1 belongs to the Delta/Serrate/LAG-2 (DSL) family of ligands for Notch receptors, which regulate cell proliferation and differentiation in mammals (5). Following ligand-mediated activation of the Notch pathway, the Notch intracellular domain (NICD) is released and binds with the transcription factor CBF-1/suppressor of hairless/lag to form a transcription complex, and then induces downstream gene transcription; for example, hairy and enhancer of split (Hes)-1, Hes-5 and hairy/enhancer-of-split related with YRPW motif protein 1 (Hey-1) (6,7). The Notch pathway is an evolutionarily conserved pathway that is widely involved in cell proliferation, differentiation and apoptosis (8). Jagged1, a DSL ligand for the mammalian Notch receptor, activates gene expression and the suppression of differentiation by binding to Notch and inducing the proteolytic release of NICD (9). Activation of the Notch pathway induces downstream proteins involved in the cell cycle and apoptosis, including NICD, Hes-1, Hes-5 and Hey-1 (5,9). Previous studies have reported that the Notch signaling pathway is involved in hypoxia/reoxygenation (H/R)-induced cardiomyocyte injury (10–12). Additionally, a previous study indicated that inhibition of microRNA-449a protects H9C2 cells against H/R-induced damage by silencing the Notch1 signaling pathway (12).
Curcumin, or 1,7-bis(4-hydroxy-3-methoxyphenyl)-1,6-heptadiene-3,5-dione, is a natural phenolic substance present in the rhizome of Curcumae longae (13,14). Curcumin has received increasing scientific attention due to its range of reported biological effects, including anti-inflammatory, antioxidant, anticarcinogenic and cardioprotective effects (15,16). Previous studies have reported that by regulating cell proliferation, apoptosis and antioxidant enzymes, curcumin induces positive effects on ischemia/reperfusion (I/R) injury in various organs (17,18). Additionally, a number of studies have demonstrated that curcumin attenuates I/R injury by regulating various signaling pathways. In 2017, Liu et al (19) demonstrated that curcumin inhibits nitric oxide (NO) signaling to protect kidney tubules against renal I/R injury. Similarly, curcumin also exhibits positive effects on hepatic I/R injury by suppressing the Toll-like receptor (TLR)4 pathway (20). Furthermore, Kim et al (21) suggested that curcumin modulates the TLR2/NF-κB signaling pathway to mitigate cardiomyocyte I/R-induced injury. Additional studies have reported that curcumin acts as a G-quadruplex-specific ligand to regulate telomerase activity, thereby regulating apoptosis (22–24). However, the protective mechanisms underlying the protective effects of curcumin against I/R injury are yet to be fully determined.
Focusing on the regulation of apoptosis, the present study aimed to determine the underlying mechanisms of curcumin on H/R-induced cardiomyocyte injury. Additionally, the role of the Notch signaling pathway in the actions of curcumin on cardiomyocyte injury were investigated.
Materials and methods
Cell culture
H9C2 cells (ATCC® CRL-1446™; American Type Culture Collection) were cultured in 6-well plates (2×104 cells/well) with Dulbecco's modified Eagle's medium (DMEM; cat. no. D5030; Sigma-Aldrich; Merck KGaA) containing 10% fetal bovine serum (FBS; cat. no. 10099141; Thermo Fisher Scientific, Inc.); cells were maintained at 37°C in a humidified incubator containing 5% CO2.
Establishment of the H/R model
According to a previous study (25), H9C2 cells cultured in phosphate-buffered saline (PBS) alone were exposed to low oxygen (95% N2 + 5% CO2/O2) for 4 h in a humidified hypoxia chamber (Stemcell Technologies, Inc.), followed by reoxygenation (0–12 h) in DMEM supplemented with 0.5% FBS under normal culture conditions. Cells were harvested to measure cell viability at 4, 8 and 12 h. Control cells were maintained under normoxic conditions.
Cell viability assay
The viability of H9C2 cardiomyocytes was evaluated using a Cell Counting kit-8 (CCK-8) assay (Dojindo Molecular Technologies, Inc.) according to the manufacturer's protocol. Briefly, after cells were treated in the aforementioned way, cells were seeded into 96-well plates (3×105 cells/well) and incubated at 37°C with 5% CO2 for 24 h. Subsequently, CCK-8 reagent was added to each well, and cardiomyocytes were cultured at room temperature for 4 h. Absorbance at 450 nm was detected using a microplate reader (Cany Precision Instruments Co., Ltd.).
Determination of cell injury
H9C2 cells were digested with trypsin and collected by centrifugation after washing with PBS. Following centrifugation at 8,000 × g for 10 min at 4°C, the supernatant was collected for testing. According to the manufacturer's protocols, intracellular lactate dehydrogenase (LDH), malondialdehyde (MDA) and superoxide dismutase (SOD) activity levels were detected using LDH (cat. no. BC0680), MDA (cat. no. BC0020) and SOD (cat. no. BC0170) assay kits (all Beijing Solarbio Science & Technology Co., Ltd.), respectively.
Effects of curcumin pretreatment on cardiomyocytes subjected to H/R
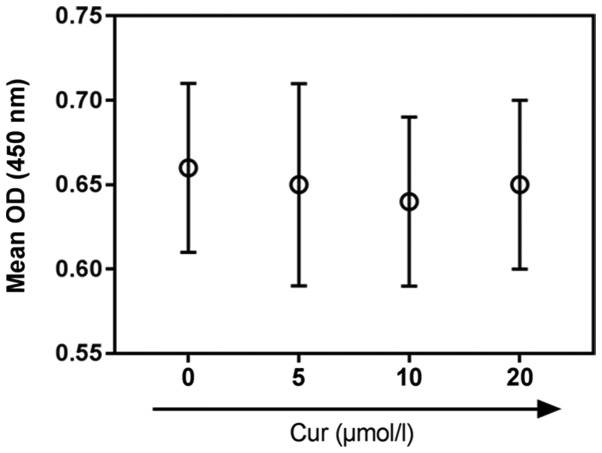
Curcumin (purity >98%; cat. no. 08511; Sigma-Aldrich; Merck KGaA) was dissolved in dimethyl sulfoxide. H9C2 cardiomyocytes were pretreated at 37°C for 2 h in a humidified incubator containing 5% CO2 with different concentrations of curcumin (0, 5, 10 and 20 µM) for 24 h, as previously described (26,27), to determine a non-toxic concentration. The viability of pretreated H9C2 cells was examined using a CCK-8 assay; 10 µM was then selected for use in subsequent experiments.
To determine the effects of curcumin on H/R-induced cardiomyocyte injury, cells were assigned to the following four groups: i) H9C2 cells cultured under normoxic conditions (Control); ii) H9C2 cells cultured with curcumin (10 µM) at 37°C for 2 h under normoxic conditions (Cur); iii) H9C2 cells subjected to 4 h of hypoxia followed by 8 h of reoxygenation (H/R); and iv) H9C2 cells pretreated with 10 µM curcumin for 2 h, then subjected to 4 h of hypoxia followed by 8 h of reoxygenation (H/R + Cur).
Cardiomyocyte viability was assessed following the various treatments using CCK-8 assays, and indicators of cardiomyocyte injury (LDH, MDA and SOD activity) were measured using the aforementioned assay kits.
Assessment of reactive oxygen species (ROS) and apoptosis
The levels of intracellular ROS were determined using a Reactive Oxygen Species Assay kit (cat. no. S0033; Beyotime Institute of Biotechnology). Trypsinized cells were first collected by centrifugation in the aforementioned manner, and then washed with PBS. Cells were cultured with 10 mM dichlorodihydrofluorescein diacetate at 37°C for 30 min. Subsequently, cells were harvested and analyzed using a BD FACScalibur flow cytometer (BD Biosciences) and the data was analyzed using Summit Software V4.3 (Dako; Agilent Technologies, Inc.).
Apoptosis was evaluated using an Annexin V-FITC/propidium iodide (PI) staining kit (Thermo Fisher Scientific, Inc.). Briefly, following incubation for 48 h, the cells were trypsinized and collected by centrifugation in the aforementioned manner. Subsequently, the cells were resuspended in binding buffer (10 mM HEPES, pH 7.4, 140 mM NaCl, 2.5 mM CaCl2) and cultured with Annexin V (1:20) for 3 min at room temperature. The cells were then stained with PI (20 µg/ml) in the dark for 15 min at room temperature. Cell apoptosis was determined immediately using flow cytometry and the data were analyzed using BD CellQuest™ Pro Software version 1.2 (BD Biosciences).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from H9C2 cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and purified with RNase-free DNase (Takara Bio, Inc.). RNA aliquots (1 µg) from each sample were reversed transcribed into cDNA using a PrimeScript™ RT reagent kit (Takara Bio, Inc.). The RT reaction was conducted as follows: 65°C for 5 min, 30°C for 6 min and 50°C for 60 min. The cDNA was then used for qPCR. Relative mRNA expression levels were analyzed using an ABI 7500 Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.) with a SYBR-Green kit (Takara Bio, Inc.). All primer sequences are presented in Table I. qPCR amplification was conducted as follows: Denaturation at 94°C for 2 min, followed by 35 cycles of denaturation at 94°C for 30 sec, annealing at 63°C for 30 sec and 72°C for 1 min, and a final extension step at 72°C for 7 min. All experiments were conducted in triplicate, and relative expression levels were determined using the 2−∆∆Cq method, using GAPDH for normalization (28).
Table I.
Primers for reverse transcription-quantitative polymerase chain reaction.
| Gene name | Primer sequence |
|---|---|
| NICD | F: 5′-ATGACTGCCCAGGAAACAAC-3′ |
| R: 5′-GTCCAGCCATTGACACACAC-3′ | |
| Hes-1 | F: 5′-CAACACGACACCGGACAAAC-3′ |
| R: 5′-CGGAGGTGCTTCACTGTCAT-3′ | |
| Hes-5 | F: 5′-ACATGGCCTTGGCTGTCTGA-3′ |
| R: 5′-TGCACCCACCCATACAAAGG-3′ | |
| Hey-1 | F: 5′-GCCGACGAGACCGAATCAAT-3′ |
| R: 5′-GGAGACCAGGCGAACACGA-3 | |
| GAPDH | F: 5′-GACATGCCGCCTGGAGAAAC-3′ |
| R: 5′-AGCCCAGGATGCCCTTTAGT-3 |
Hes, hairy and enhancer of split; Hey-1, hairy/enhancer-of-split related with YRPW motif protein 1; NICD, Notch intracellular domain; F, forward; R, reverse.
Western blot analysis
Total protein was extracted from cells using RIPA Lysis Buffer (Beyotime Institute of Biotechnology) and centrifuged at 12,000 × g for 20 min at 4°C. Protein quantification was performed using a bicinchoninic acid protein assay kit (Beyotime Institute of Biotechnology). Subsequently, total protein (20 µg) was subjected to 10% SDS-PAGE and transferred to polyvinylidene difluoride membranes (EMD Millipore). The membranes were blocked at 37°C for 1.5 h with 5% non-fat milk in TBS-Tween-20 buffer (100 mM NaCl, 10 mM Tris-HCl, pH 7.4, 0.1% Tween-20) prior to incubation with primary antibodies at 4°C overnight. Primary antibodies against NICD (1:100; cat. no. 4147) and GAPDH (1:1,000; cat. no. 2118) were purchased from Cell Signaling Technology, Inc. Membranes were then incubated with horseradish peroxidase-conjugated goat anti-rabbit secondary antibodies (cat. no. 7074; 1:2,000; Cell Signaling Technology, Inc.) at room temperature for 1 h. Protein bands were visualized using Pierce™ ECL substrate (cat. no. 32106; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol, and the results were normalized to GAPDH expression. Quantity One software version 4.6.2 (Bio-Rad Laboratories) was used to quantify western blots.
Effects of Jagged1 co-treatment with curcumin on H/R-induced H9C2 cell injury
The recombinant mouse Jagged1 protein was obtained from Abcam (cat. no. ab109346). H9C2 cells were co-treated with Jagged1 (50 ng/l) and curcumin (10 µM) in a humidified incubator with 5% CO2 at 37°C. To investigate the effects of curcumin on H/R-induced cardiomyocyte injury, cells were assigned to the following six groups: i) H9C2 cells cultured under normoxic conditions (Control); ii) H9C2 cells subjected to 4 h of hypoxia followed by 8 h of reoxygenation (H/R); iii) H9C2 cells cultured with Jagged1 for 1 h under normoxic conditions (Jagged1); iv) H9C2 cells pretreated with Jagged1 for 1 h and then subjected to 4 h of hypoxia followed by 8 h of reoxygenation (H/R + Jagged1); v) H9C2 cells pretreated with curcumin for 2 h and then subjected to 4 h of hypoxia, followed by 8 h of reoxygenation (H/R + Cur); and vi) H9C2 cells co-treated with Jagged1 and curcumin for 3 h and then subjected to 4 h of hypoxia, followed by 8 h of reoxygenation (H/R + Cur + Jagged1).
The viability of H9C2 cells from each group was evaluated using the CCK-8 assay as aforementioned. Flow cytometry was used to detect the ROS levels and apoptosis of cardiomyocytes. Transcriptional and post-transcriptional levels of NICD were determined via RT-qPCR and western blot analysis, respectively. Altered mRNA expression of downstream genes in the Notch signaling pathway was also investigated.
Statistical analysis
Statistical analysis was performed using GraphPad Prism version 6.0 software (GraphPad Software, Inc.). All data are presented as the mean ± standard deviation. Differences were analyzed using one-way analysis of variance followed by Tukey's multiple comparison post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
H/R reduces H9C2 cardiomyocyte viability and contributes to cell injury
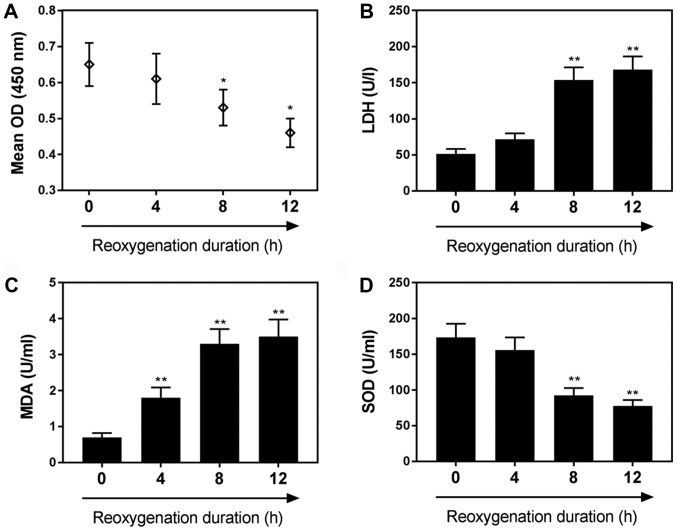
As shown in Fig. 1A, H9C2 cell viability was decreased as the duration of reoxygenation increased. The viability of cardiomyocytes subjected to 4 h of hypoxia followed by 8 h of reoxygenation was markedly reduced compared with the control group (P<0.05). The levels of LDH, an indicator of cardiomyocyte injury, were significantly increased following 8 h of reoxygenation compared with the control (Fig. 1B). MDA content was significantly increased following 4 h of reoxygenation, (Fig. 1C), whereas SOD levels were significantly decreased following 8 h of reoxygenation, compared with in the control group (P<0.01). Specific values for these assays are presented in Table II.
Figure 1.
H/R reduces H9C2 cardiomyocyte viability and contributes to cell injury. (A) Effects of H/R on H9C2 cell viability following 0, 4, 8 and 12 h of reoxygenation, as determined by a Cell Counting Kit-8 assay. Effects of H/R on the following markers of cardiomyocyte injury: (B) LDH, (C) MDA and (D) SOD, following 0, 4, 8 and 12 h of reoxygenation. Data are presented as the mean ± standard deviation (n=3). *P<0.05, **P<0.01 vs. 0 h. H/R, hypoxia/reoxygenation; LDH, lactate dehydrogenase; MDA, malondialdehyde; OD, optical density; SOD, superoxide dismutase.
Table II.
Effects of reoxygenation duration on MDA, LDH and SOD levels.
| Reoxygenation duration | 0 h | 4 h | 8 h | 12 h |
|---|---|---|---|---|
| LDH (U/l) | 50.1±4.7 | 73.8±5.6 | 150.5±48.5a | 164.6±41.6a |
| MDA (U/ml) | 0.8±0.1 | 1.8±0.3a | 3.1±0.5a | 3.4±0.5a |
| SOD (U/ml) | 174.3±39.6 | 149.5±38.4 | 82.1±19.6a | 64.5±13.2a |
P<0.01 vs. 0 h. LDH, lactate dehydrogenase; MDA, malondialdehyde; SOD, superoxide dismutase.
Curcumin attenuates H/R-induced H9C2 cell apoptosis
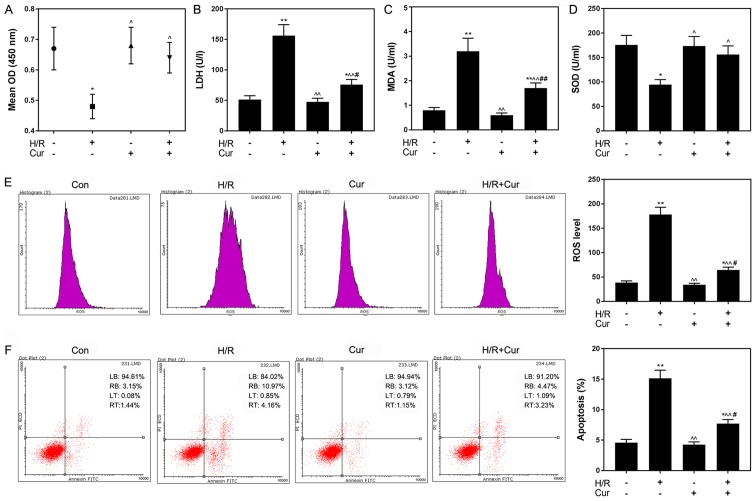
Curcumin treatment did not affect H9C2 cell viability under normoxic conditions (Fig. 2). Therefore, 10 µM curcumin was selected for subsequent experiments. As presented in Fig. 3A, curcumin significantly increased the viability of H/R-treated cardiomyocytes compared with H/R treatment alone, whereas it induced no significant effects on the viability of cells cultured under normoxic conditions. Cardiomyocyte injury was induced by H/R treatment, as indicated by the significantly upregulated release of LDH and MDA, and the significantly decreased release of SOD, compared with in the control group. Following curcumin pretreatment, these effects were significantly attenuated compared with in the H/R group (Fig. 3B-D). Additionally, ROS levels were significantly increased in the H/R group compared with in the control group (P<0.01; Fig. 3E). Curcumin significantly reduced the H/R-induced increase in ROS levels (P<0.05). As determined using flow cytometry, the percentage of apoptotic H/R-treated H9C2 cells was 15.13%, which was significantly increased compared with the control group (4.59%; P<0.01); conversely, curcumin pretreatment significantly decreased the rate of apoptosis in H/R-treated cells to 7.7% (P<0.05; Fig. 3F).
Figure 2.
Curcumin does not affect cardiomyocyte viability. Effects of various concentrations of curcumin (0, 5, 10 and 20 µM) on H9C2 cell viability, as determined using a Cell Counting kit-8 assay. Data are presented as the mean ± standard deviation (n=3). Cur, curcumin; OD, optical density.
Figure 3.
Curcumin attenuates H/R-induced injury and apoptosis in H9C2 cells. (A) Curcumin reversed the inhibitory effects of H/R on H9C2 cell viability. Curcumin pretreatment attenuated the effects of H/R on the levels of (B) LDH, (C) MDA and (D) SOD. Effects of curcumin on H/R-induced increases in (E) ROS levels and (F) apoptosis. Data are presented as the mean ± standard deviation (n=3). *P<0.05, **P<0.01 vs. Con; ^P<0.05, ^^P<0.01 vs. H/R; #P<0.05, ##P<0.01 vs. Cur. Con, control (normoxic culture conditions); Cur, curcumin; H/R, hypoxia/reoxygenation; LDH, lactate dehydrogenase; MDA, malondialdehyde; OD, optical density; PI, propidium iodide; ROS, reactive oxygen species; SOD, superoxide dismutase.
Curcumin pretreatment inhibits the Notch signaling pathway
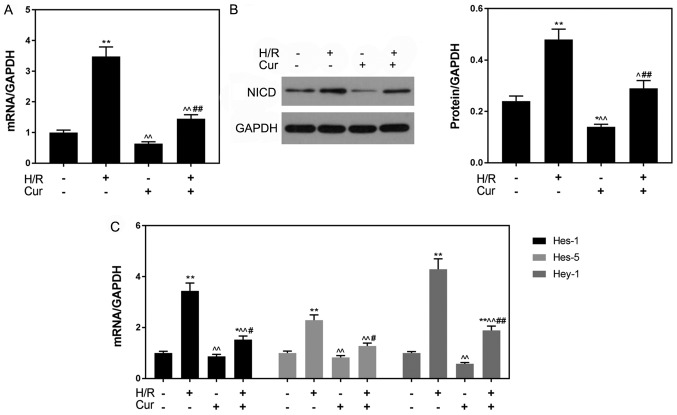
The levels of NICD, Hes-1, Hes-5 and Hey-1 were measured in H9C2 cells following H/R-induced injury to investigate the effects of curcumin on H/R-induced cardiomyocytes. As presented in Fig. 4A and B, the transcriptional and post-transcriptional levels of NICD were significantly increased following H/R treatment compared with in the control group (P<0.01). Conversely, the expression levels of NICD in the Cur + H/R group were significantly decreased compared with in the H/R group. Additionally, as presented in Fig. 4C, the mRNA expression levels of Hes-1, Hes-5 and Hey-1 following H/R were significantly upregulated compared with in the control group, whereas curcumin pretreatment significantly attenuated this H/R-induced upregulation of genes downstream of the Notch pathway.
Figure 4.
Curcumin pretreatment inhibits the Notch signaling pathway. (A) mRNA and (B) protein expression levels of NICD as determined via RT-qPCR and western blotting, respectively. (C) Expression of downstream genes (Hes-1, Hes-5 and Hey-1) of the Notch pathway as determined via RT-qPCR analysis. GAPDH was used as an internal control. Data are presented as the mean ± standard deviation (n=3). *P<0.05, **P<0.01 vs. Con; ^P<0.05, ^^P<0.01 vs. H/R; #P<0.05, ##P<0.01 vs. Cur. Con, control (normoxic culture conditions); Cur, curcumin; H/R, hypoxia/reoxygenation; Hes, hairy and enhancer of split; Hey-1, hairy/enhancer-of-split related with YRPW motif protein 1; NICD, Notch intracellular domain; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Jagged1 suppresses the protective effects of curcumin against H/R-induced H9C2 cell death
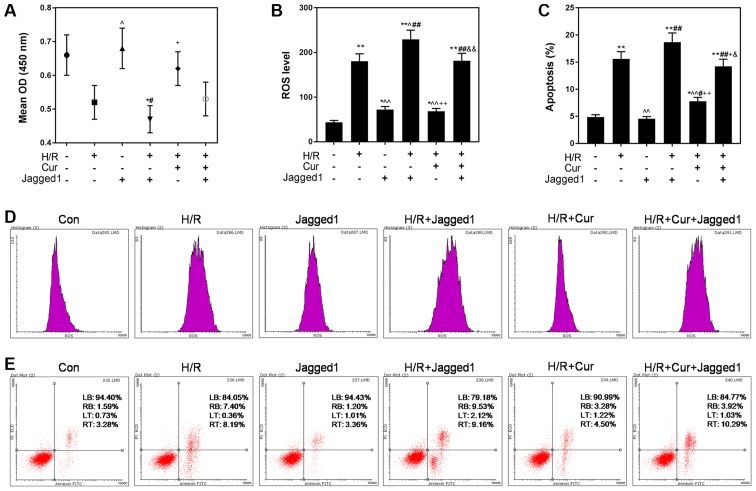
Notch signaling was activated using Jagged1 (50 ng/l), and Jagged1 affected the protective effects of curcumin on H/R-induced H9C2 cardiomyocytes. As presented in Fig. 5A, activation of the Notch signaling pathway by Jagged1 partially reversed the protective effects of curcumin on the viability of H/R-damaged cells. Furthermore, it was observed that ROS levels were also significantly increased in the H/R + Cur + Jagged1 group compared with in the H/R + Cur group (P<0.01). Additionally, the apoptotic rate of cells was significantly increased from 7.78% in the H/R + Cur group to 14.21% in the H/R + Cur + Jagged1 group (Fig. 5E).
Figure 5.
Jagged1 attenuates the protective effects of curcumin against H/R-induced cardiomyocyte cell death. (A) Jagged1 reversed the inhibitory effects of curcumin against H/R-induced reductions in H9C2 cell viability. (B and D) Jagged1 treatment attenuated the downregulation of ROS levels induced by curcumin. (C and E) Jagged1 alleviated the anti-apoptotic effects of curcumin. Data are presented as the mean ± standard deviation (n=3). *P<0.05, **P<0.01 vs. Con; ^P<0.05, ^^P<0.01 vs. H/R; #P<0.05, ##P<0.01 vs. Jagged1; +P<0.05, ++P<0.01 vs. H/R + Jagged1; &P<0.05, &&P<0.01 vs. H/R + Cur. Con, control (normoxic culture conditions); Cur, curcumin; H/R, hypoxia/reoxygenation; OD, optical density; PI, propidium iodide; ROS, reactive oxygen species.
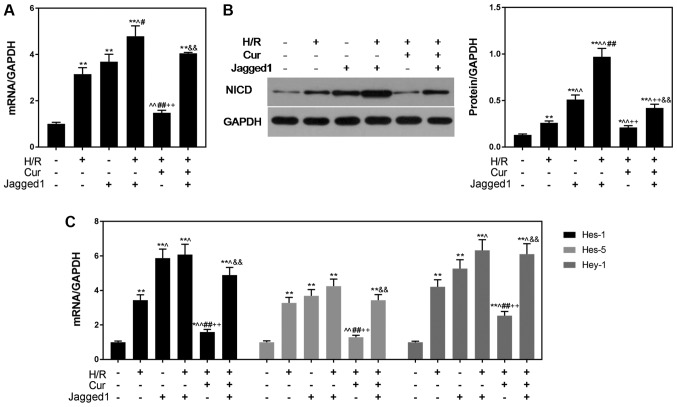
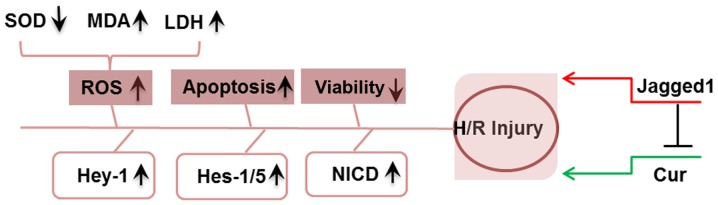
The increased expression of NICD and downstream genes of the Notch pathway in the Jagged1 group indicated that Jagged1 alone activated Notch signaling (Fig. 6). In addition, NICD, Hes-1, Hes-5 and Hey-1 levels were significantly upregulated in the H/R + Cur + Jagged1 group compared with in the H/R + Cur group (Fig. 6), suggesting that curcumin protected cardiomyocytes from H/R-induced apoptosis by modulating the Notch signaling pathway. A summary of the proposed mechanisms underlying the protective effects of curcumin on H/R injury reported during the present study is presented in Fig. 7.
Figure 6.
Jagged1 promotes the expression of Notch pathway-associated genes. Jagged1 upregulated the (A) mRNA and (B) protein expression levels of NICD. (C) Expression of downstream genes (Hes-1, Hes-5 and Hey-1) in the Notch pathway was upregulated by Jagged1. GAPDH was used as an internal control. Data are presented as the mean ± standard deviation (n=3). *P<0.05, **P<0.01 vs. Con; ^P<0.05, ^^P<0.01 vs. H/R; #P<0.05, ##P<0.01 vs. Jagged1; ++P<0.01 vs. H/R + Jagged1; &&P<0.01 vs. H/R + Cur. Con, control (normoxic culture conditions); Cur, curcumin; H/R, hypoxia/reoxygenation; Hes, hairy and enhancer of split; Hey-1, hairy/enhancer-of-split related with YRPW motif protein 1; NICD, Notch intracellular domain.
Figure 7.
Schematic of the proposed mechanisms underlying the effects of curcumin and Jagged1 reported in the present study. H/R treatment induced cardiomyocyte injury via the elevation of oxidative stress (including ROS production, reduced SOD activity, enhanced MDA content and increased LDH activity), increased apoptosis and decreased cell viability. Curcumin attenuated H/R injury, whereas Jagged1 exacerbated H/R-induced injury and reversed the effects of curcumin. Cur, curcumin; H/R, hypoxia/reoxygenation; Hes, hairy and enhancer of split; Hey-1, hairy/enhancer-of-split related with YRPW motif protein 1; LDH, lactate dehydrogenase; MDA, malondialdehyde; NICD, Notch intracellular domain; ROS, reactive oxygen species; SOD, superoxide dismutase.
Discussion
Reperfusion treatment substantially alleviates myocardial ischemia; however, it can lead to irreversible cardiomyocyte apoptosis and even cardiac remodeling, and as a result, MIRI is a leading cause of IHD (29). The prevention of cardiomyocyte apoptosis is hypothesized to be one of the key approaches to preventing MIRI (30). A number of studies have reported that curcumin effectively reduces H/R-induced H9C2 cell injury by modulating various pathways (31–33); however, the mechanisms underlying the protective effects of curcumin on cardiomyocytes remain unclear, and the contribution of the Notch pathway to cardiomyocyte injury induced by H/R is yet to be fully elucidated. In the present study, an important role for the Notch signaling pathway in the protective effects of curcumin against H/R-induced H9C2 cell death was observed. Curcumin significantly decreased cardiomyocyte apoptosis following H/R, potentially by downregulating the Notch pathway.
Firstly, the viability, and the levels of LDH, MDA and SOD were evaluated to determine the effects of curcumin on a cardiomyocyte model of H/R. As biomarkers of lipid peroxidation and oxidative stress, LDH, MDA and SOD levels are closely associated with the degree of cell injury (34). In the present study, LDH and MDA contents progressively increased as the duration of reoxygenation was increased, suggesting that the injury of H9C2 cells, and oxidative stress and lipid peroxidation, were aggravated as the period of reoxygenation was extended. Previous studies have suggested that curcumin protects cardiomyocytes against MIRI by inhibiting the leakage of LDH, enhancing SOD activity and reducing MDA production (33,35). These results were consistent with the present study, in which curcumin modulated the levels of LDH, MDA and SOD to improve H/R-induced oxidative stress and increase antioxidative activity in H9C2 cells. In addition, MIRI promotes the production of ROS, elevated levels of which are sufficiently toxic to damage all cellular components (36); however, the present findings revealed that curcumin treatment significantly decreased H/R-induced ROS production, reducing the toxic effects of ROS on cardiomyocytes. It has been reported that MIRI induces cardiomyocyte apoptosis via a number of apoptotic pathways, including the Janus kinase 2/STAT3 and TLR4/NF-κB pathways (10,20,37). In the present study, curcumin pretreatment reduced the H/R-induced apoptosis of H9C2 cells. Collectively, the present findings demonstrated the positive effects of curcumin on H9C2 cardiomyocyte apoptosis induced by H/R.
Furthermore, to investigate the potential role of the Notch pathway in MIRI, the expression levels of NICD and certain downstream genes were determined. Previous studies have demonstrated that the Notch signaling pathway serves a positive role in inhibiting MIRI, protecting cardiomyocytes against H/R damage (38–40). Additionally, certain medicines exert inhibitory effects on MIRI via the Notch signaling pathway; for example, relaxin protects myocardial cells against H/R injury by upregulating Notch1 signaling (41). Conversely, opposing findings were reported in the present study, as it was observed that the expression levels of NICD, Hes-1, Hes-5 and Hey-1 were significantly increased in response to H/R treatment. Following curcumin treatment, the expression levels of Notch pathway genes were downregulated, as was H/R-induced cardiomyocyte apoptosis. Therefore, it was hypothesized that Notch pathway activation promoted H/R-induced cardiomyocyte injury. To test this hypothesis, Jagged1 protein, a ligand that interacts with four receptors in the Notch pathway, was used to reactivate the Notch1 receptor and NICD release following curcumin treatment. Notably, it was observed that ROS levels and the apoptotic rate were significantly increased following Jagged1 treatment, and that the mRNA expression levels of Hes-1, Hes-5 and Hey-1 were also significantly unregulated. These results were consistent with the hypothesis that the Notch pathway not only contributed to H/R-induced H9C2 cardiomyocyte apoptosis, but also exacerbated H/R-induced injury. It has been reported that Jagged1 is involved in cell injury (42–44). Therefore, the role of Jagged1 in H/R-induced H9C2 cardiomyocyte injury, and the involvement of Notch signaling in the effects of curcumin were investigated. The results revealed that curcumin possessed the ability to suppress H/R-induced H9C2 cell apoptosis, and that Jagged1 attenuated the inhibitory effects of curcumin by activating the Notch pathway. Therefore, curcumin and Jagged1 induced opposing effects on apoptotic processes in H9C2 cells. Previous studies have also indicated that the ligand Jagged2 exhibits anti-apoptotic effects (45,46). In addition, it was previously reported that inhibition of the Delta1/Notch1 pathway affects apoptosis (47). Pelullo et al (48) revealed that Jagged1 potentially contributes to the occurrence of acute lymphoblastic leukemia via Notch3. The exact mechanisms underlying the interaction/competition between the two molecules were not investigated in the present study. In addition, various signaling pathways, including the NO (49), TLR4 (50) and TLR/NF-κB signaling pathways (51) have also been reported to be involved in the regulation of IR injury; however, the involvement of these pathways in the effects of curcumin or Jagged1 were not investigated in the present study.
In conclusion, the present study revealed that H/R injury activated the Notch signaling pathway, and induced H9C2 cardiomyocyte injury and apoptosis; however, in vivo experiments are required to validate these findings. Additionally, curcumin not only inhibited the association between H/R injury and Notch signaling by inhibiting NICD expression, but also reduced the H/R-induced apoptosis of H9C2 cells. Furthermore, Jagged1 recombinant protein treatment further suggested that the Notch pathway contributed to H/R damage in cardiomyocytes. These findings provide improved understanding of the mechanisms via which curcumin affects H/R-induced cardiomyocyte apoptosis.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
PZ and MY made substantial contributions to the conception and design of the study. HH, ML, SL, ZC, CS, ZK, AL and QW performed the acquisition and analysis of data, and interpreted the data. PZ and MY drafted the manuscript. All authors approved of the final version of the manuscript to be published. All authors agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Soriano JB, Rojas-Rueda D, Alonso J, Anto JM, Cardona PJ, Fernandez E, Garcia-Basteiro AL, Benavides FG, Glenn SD, Krish V, et al. The burden of disease in Spain: Results from the global burden of disease 2016. Med Clin (Barc) 2018;151:171–190. doi: 10.1016/j.medcli.2018.05.011. (In English, Spanish) [DOI] [PubMed] [Google Scholar]
- 2.Bell MT, Puskas F, Agoston VA, Cleveland JC, Jr, Freeman KA, Gamboni F, Herson PS, Meng X, Smith PD, Weyant MJ, et al. Toll-like receptor 4-dependent microglial activation mediates spinal cord ischemia-reperfusion injury. Circulation. 2013;128(Suppl 1):S152–S156. doi: 10.1161/CIRCULATIONAHA.112.000024. [DOI] [PubMed] [Google Scholar]
- 3.Hausenloy DJ, Yellon DM. Myocardial ischemia-reperfusion injury: A neglected therapeutic target. J Clin Invest. 2013;123:92–100. doi: 10.1172/JCI62874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang D, Chen TY, Liu FJ. Che-1 attenuates hypoxia/reoxygenation-induced cardiomyocyte apoptosis by upregulation of Nrf2 signaling. Eur Rev Med Pharmacol Sci. 2018;22:1084–1093. doi: 10.26355/eurrev_201802_14395. [DOI] [PubMed] [Google Scholar]
- 5.Bash J, Zong WX, Banga S, Rivera A, Ballard DW, Ron Y, Gelinas C. Rel/NF-kappaB can trigger the Notch signaling pathway by inducing the expression of Jagged1, a ligand for Notch receptors. EMBO J. 1999;18:2803–2811. doi: 10.1093/emboj/18.10.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: Cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 7.Geisler F, Strazzabosco M. Emerging roles of Notch signaling in liver disease. Hepatology. 2015;61:382–392. doi: 10.1002/hep.27268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo J, Wang P, Wang R, Wang J, Liu M, Xiong S, Li Y, Cheng B. The Notch pathway promotes the cancer stem cell characteristics of CD90(+) cells in hepatocellular carcinoma. Oncotarget. 2016;7:9525–9537. doi: 10.18632/oncotarget.6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zavadil J, Cermak L, Soto-Nieves N, Bottinger EP. Integration of TGF-beta/Smad and Jagged1/Notch signalling in epithelial-to-mesenchymal transition. EMBO J. 2004;23:1155–1165. doi: 10.1038/sj.emboj.7600069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai W, Yang X, Han S, Guo H, Zhao Z, Wang H, Guan H, Jia Y, Gao J, Yang T, et al. Notch1 pathway protects against burn-induced myocardial injury by repressing reactive oxygen species production through JAK2/STAT3 signaling. Oxid Med Cell Longev. 2016;2016:5638943. doi: 10.1155/2016/5638943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kopan R, Ilagan MX. The canonical Notch signaling pathway: Unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng J, Wu Q, Lv R, Huang L, Xu B, Wang X, Chen A, He F. MicroRNA-449a inhibition protects H9C2 cells against hypoxia/reoxygenation-induced injury by targeting the Notch-1 signaling pathway. Cell Physiol Biochem. 2018;46:2587–2600. doi: 10.1159/000489686. [DOI] [PubMed] [Google Scholar]
- 13.Esatbeyoglu T, Huebbe P, Ernst IM, Chin D, Wagner AE, Rimbach G. Curcumin--from molecule to biological function. Angew Chem Int Ed Engl. 2012;51:5308–5332. doi: 10.1002/anie.201107724. [DOI] [PubMed] [Google Scholar]
- 14.Yeh CH, Chen TP, Wu YC, Lin YM, Jing Lin P. Inhibition of NFkappaB activation with curcumin attenuates plasma inflammatory cytokines surge and cardiomyocytic apoptosis following cardiac ischemia/reperfusion. J Surg Res. 2005;125:109–116. doi: 10.1016/j.jss.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 15.Park BH, Lim JE, Jeon HG, Seo SI, Lee HM, Choi HY, Jeon SS, Jeong BC. Curcumin potentiates antitumor activity of cisplatin in bladder cancer cell lines via ROS-mediated activation of ERK1/2. Oncotarget. 2016;7:63870–63886. doi: 10.18632/oncotarget.11563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kunnumakkara AB, Bordoloi D, Padmavathi G, Monisha J, Roy NK, Prasad S, Aggarwal BB. Curcumin, the golden nutraceutical: Multitargeting for multiple chronic diseases. Br J Pharmacol. 2017;174:1325–1348. doi: 10.1111/bph.13621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen SQ, Zhang Y, Xiang JJ, Xiong CL. Protective effect of curcumin against liver warm ischemia/reperfusion injury in rat model is associated with regulation of heat shock protein and antioxidant enzymes. World J Gastroenterol. 2007;13:1953–1961. doi: 10.3748/wjg.v13.i13.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yucel AF, Kanter M, Pergel A, Erboga M, Guzel A. The role of curcumin on intestinal oxidative stress, cell proliferation and apoptosis after ischemia/reperfusion injury in rats. J Mol Histol. 2011;42:579–587. doi: 10.1007/s10735-011-9364-0. [DOI] [PubMed] [Google Scholar]
- 19.Liu F, Ni W, Zhang J, Wang G, Li F, Ren W. Administration of curcumin protects kidney tubules against renal ischemia-reperfusion injury (RIRI) by modulating nitric oxide (NO) signaling pathway. Cell Physiol Biochem. 2017;44:401–411. doi: 10.1159/000484920. [DOI] [PubMed] [Google Scholar]
- 20.Wang L, Li N, Lin D, Zang Y. Curcumin protects against hepatic ischemia/reperfusion induced injury through inhibiting TLR4/NF-κB pathway. Oncotarget. 2017;8:65414–65420. doi: 10.18632/oncotarget.18676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim YS, Kwon JS, Cho YK, Jeong MH, Cho JG, Park JC, Kang JC, Ahn Y. Curcumin reduces the cardiac ischemia-reperfusion injury: involvement of the toll-like receptor 2 in cardiomyocytes. J Nutr Biochem. 2012;23:1514–1523. doi: 10.1016/j.jnutbio.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 22.Jha N, Mishra S, Mamidi A, Mishra A, Jha S, Surolia A. Targeting Human Telomeric G- Quadruplexes DNA with curcumin and its synthesized analogues under molecular crowding condition. RSC Advances. 2016;6:7474–7487. doi: 10.1039/C5RA17390F. [DOI] [Google Scholar]
- 23.Lin S, Gao W, Tian Z, Yang C, Lu L, Mergny JL, Leung CH, Ma DL. Luminescence switch-on detection of protein tyrosine kinase-7 using a G-quadruplex-selective probe. Chem Sci. 2015;6:4284–4290. doi: 10.1039/C5SC01320H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang M, Mao Z, Kang TS, Wong CY, Mergny JL, Leung CH, Ma DL. Conjugating a groove-binding motif to an Ir(iii) complex for the enhancement of G-quadruplex probe behavior. Chem Sci. 2016;7:2516–2523. doi: 10.1039/C6SC00001K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park M, Youn B, Zheng XL, Wu D, Xu A, Sweeney G. Globular Adiponectin, acting via adipoR1/APPL1, protects H9c2 cells from hypoxia/reoxygenation-induced apoptosis. Plos One. 2011;6:e19143. doi: 10.1371/journal.pone.0019143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Magalhães LG, Machado CB, Morais ER, Moreira EB, Soares CS, Da SS, Da SFA, Rodrigues V. In vitro schistosomicidal activity of curcumin against Schistosoma mansoni adult worms. Parasitology Research. 2009;104:1197–1201. doi: 10.1007/s00436-008-1311-y. [DOI] [PubMed] [Google Scholar]
- 27.Gu Q, Cai Y, Huang C, Shi Q, Yang H. Curcumin increases rat mesenchymal stem cell osteoblast differentiation but inhibits adipocyte differentiation. Pharmacognosy Magazine. 2012;8:202–208. doi: 10.4103/0973-1296.99285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Yamamoto T, Sadoshima J. Protection of the heart against ischemia/reperfusion by silent information regulator 1. Trends Cardiovasc Med. 2011;21:27–32. doi: 10.1016/j.tcm.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu H, Jing X, Dong A, Bai B, Wang H. Overexpression of TIMP3 protects against cardiac ischemia/reperfusion injury by inhibiting myocardial apoptosis through ROS/Mapks pathway. Cell Physiol Biochem. 2017;44:1011–1023. doi: 10.1159/000485401. [DOI] [PubMed] [Google Scholar]
- 31.Liu K, Chen H, You QS, Ye Q, Wang F, Wang S, Zhang SL, Yu KJ, Lu Q. Curcumin attenuates myocardial ischemia-reperfusion injury. Oncotarget. 2017;8:112051–112059. doi: 10.18632/oncotarget.23002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang R, Zhang JY, Zhang M, Zhai MG, Di SY, Han QH, Jia YP, Sun M, Liang HL. Curcumin attenuates IR-induced myocardial injury by activating SIRT3. Eur Rev Med Pharmacol Sci. 2018;22:1150–1160. doi: 10.26355/eurrev_201802_14404. [DOI] [PubMed] [Google Scholar]
- 33.Yang Y, Duan W, Lin Y, Yi W, Liang Z, Yan J, Wang N, Deng C, Zhang S, Li Y, et al. SIRT1 activation by curcumin pretreatment attenuates mitochondrial oxidative damage induced by myocardial ischemia reperfusion injury. Free Radic Biol Med. 2013;65:667–679. doi: 10.1016/j.freeradbiomed.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 34.Zhang L, Wei S, Tang JM, Guo LY, Zheng F, Yang JY, Kong X, Huang YZ, Chen SY, Wang JN. PEP-1-CAT protects hypoxia/reoxygenation-induced cardiomyocyte apoptosis through multiple sigaling pathways. J Transl Med. 2013;11:113. doi: 10.1186/1479-5876-11-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duan W, Yang Y, Yan J, Yu S, Liu J, Zhou J, Zhang J, Jin Z, Yi D. The effects of curcumin post-treatment against myocardial ischemia and reperfusion by activation of the JAK2/STAT3 signaling pathway. Basic Res Cardiol. 2012;107:263. doi: 10.1007/s00395-012-0263-7. [DOI] [PubMed] [Google Scholar]
- 36.Bagheri F, Khori V, Alizadeh AM, Khalighfard S, Khodayari S, Khodayari H. Reactive oxygen species-mediated cardiac-reperfusion injury: Mechanisms and therapies. Life Sci. 2016;165:43–55. doi: 10.1016/j.lfs.2016.09.013. [DOI] [PubMed] [Google Scholar]
- 37.Jeong CW, Yoo KY, Lee SH, Jeong HJ, Lee CS, Kim SJ. Curcumin protects against regional myocardial ischemia/reperfusion injury through activation of RISK/GSK-3β and inhibition of p38 MAPK and JNK. J Cardiovasc Pharmacol Ther. 2012;17:387–394. doi: 10.1177/1074248412438102. [DOI] [PubMed] [Google Scholar]
- 38.Zhou XL, Wan L, Liu JC. Activated Notch1 reduces myocardial ischemia reperfusion injury in vitro during ischemic postconditioning by crosstalk with the RISK signaling pathway. Chin Med J (Engl) 2013;126:4545–4551. [PubMed] [Google Scholar]
- 39.Zhang S, Zhang R, Wu F, Li X. MicroRNA-208a regulates H9c2 cells simulated ischemia-reperfusion myocardial injury via targeting CHD9 through Notch/NF-kappa B signal pathways. Int Heart J. 2018;59:580–588. doi: 10.1536/ihj.17-147. [DOI] [PubMed] [Google Scholar]
- 40.Jing R, Zhou Z, Kuang F, Huang L, Li C. microRNA-99a reduces lipopolysaccharide-induced oxidative injury by activating Notch pathway in H9c2 cells. Int Heart J. 2017;58:422–427. doi: 10.1536/ihj.16-261. [DOI] [PubMed] [Google Scholar]
- 41.Boccalini G, Sassoli C, Formigli L, Bani D, Nistri S. Relaxin protects cardiac muscle cells from hypoxia/reoxygenation injury: Involvement of the Notch-1 pathway. FASEB J. 2015;29:239–249. doi: 10.1096/fj.14-254854. [DOI] [PubMed] [Google Scholar]
- 42.Wang K, Ding R, Ha Y, Jia Y, Liao X, Wang S, Li R, Shen Z, Xiong H, Guo J, Jie W. Hypoxia-stressed cardiomyocytes promote early cardiac differentiation of cardiac stem cells through HIF-1α/Jagged1/Notch1 signaling. Acta Pharm Sin B. 2018;8:795–804. doi: 10.1016/j.apsb.2018.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kamei N, Kwon SM, Ishikawa M, Ii M, Nakanishi K, Yamada K, Hozumi K, Kawamoto A, Ochi M, Asahara T. Endothelial progenitor cells promote astrogliosis following spinal cord injury through Jagged1-dependent Notch signaling. J Neurotrauma. 2012;29:1758–1769. doi: 10.1089/neu.2011.2139. [DOI] [PubMed] [Google Scholar]
- 44.Wu X, Zou Y, Zhou Q, Huang L, Gong H, Sun A, Tateno K, Katsube K, Radtke F, Ge J, et al. Role of Jagged1 in arterial lesions after vascular injury. Arterioscler Thromb Vasc Biol. 2011;31:2000–2006. doi: 10.1161/ATVBAHA.111.225144. [DOI] [PubMed] [Google Scholar]
- 45.Hu Y, Su H, Li X, Guo G, Cheng L, Qin R, Qing G, Liu H. The NOTCH ligand JAGGED2 promotes pancreatic cancer metastasis independent of NOTCH signaling activation. Mol Cancer Ther. 2015;14:289–297. doi: 10.1158/1535-7163.MCT-14-0501. [DOI] [PubMed] [Google Scholar]
- 46.Francis JC, Radtke F, Logan MP. Notch1 signals through Jagged2 to regulate apoptosis in the apical ectodermal ridge of the developing limb bud. Dev Dyn. 2005;234:1006–1015. doi: 10.1002/dvdy.20590. [DOI] [PubMed] [Google Scholar]
- 47.Zhang H, Ye Y, Bai Z, Wang S. The COX-2 selective inhibitor-independent COX-2 effect on colon carcinoma cells is associated with the Delta1/Notch1 pathway. Dig Dis Sci. 2008;53:2195–2203. doi: 10.1007/s10620-007-0139-0. [DOI] [PubMed] [Google Scholar]
- 48.Pelullo M, Quaranta R, Talora C, Checquolo S, Cialfi S, Felli MP, te Kronnie G, Borga C, Besharat ZM, Palermo R, et al. Notch3/Jagged1 circuitry reinforces notch signaling and sustains T-ALL. Neoplasia. 2014;16:1007–1017. doi: 10.1016/j.neo.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weerateerangkul P, Chattipakorn S, Chattipakorn N. Roles of the nitric oxide signaling pathway in cardiac ischemic preconditioning against myocardial ischemia-reperfusion injury. Med Sci Monit. 2011;17:RA44–RA52. doi: 10.12659/MSM.881385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu L, Ye T, Tang Q, Wang Y, Wu X, Li H, Jiang Y. Exercise preconditioning regulates the toll-like receptor 4/nuclear Factor-κB signaling pathway and reduces cerebral ischemia/reperfusion inflammatory injury: A study in rats. J Stroke Cerebrovasc Diseases. 2016;25:2770–2779. doi: 10.1016/j.jstrokecerebrovasdis.2016.07.033. [DOI] [PubMed] [Google Scholar]
- 51.Zheng Y, Bu J, Yu L, Chen J, Liu H. Nobiletin improves propofol-induced neuroprotection via regulating Akt/mTOR and TLR 4/NF-κB signaling in ischemic brain injury in rats. Biomed Pharmacother. 2017;91:494–503. doi: 10.1016/j.biopha.2017.04.048. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.