Abstract
MicroRNAs (miRs) have been proposed to be involved in the pathological processes of cerebral ischemia/reperfusion (CIR) injury. The present study aimed to investigate the potential role and molecular mechanisms of miR-217 in the regulation of neuronal survival in CIR injury. To perform the investigation, an in vitro cellular model of CIR injury was established by treating neurons with oxygen-glucose deprivation and reoxygenation (OGD/R). miR-217 levels in neurons were detected using reverse transcription-quantitative PCR. The association between miR-217 and sirtuin 1 (SIRT1) was identified using TargetScan and validated in a dual-luciferase reporter assay. Cell viability and apoptosis were measured using a Cell Counting Kit-8 assay and flow cytometry, respectively. The release of lactate dehydrogenase, and the production of proinflammatory factors and oxidative stress biomarkers were analyzed by ELISAs and using specific assay kits. It was revealed that miR-217 was significantly upregulated in OGD/R-treated neurons. SIRT1 was a direct target of miR-217, and was downregulated in neurons following OGD/R treatment. Downregulation of miR-217 significantly ameliorated OGD/R-induced neuronal injury, inflammatory responses and oxidative stress. The effects of miR-217 inhibitor on OGD/R treated neurons were attenuated by SIRT1 knockdown. Additionally, western blotting revealed that the SIRT1/AMP-activated protein kinase-α/NF-κB pathway was partially involved in the regulation of OGD/R-induced neuronal injury by miR-217. In conclusion, the data of the present study indicated that the downregulation of miR-217 protected neurons against OGD/R-induced injury by targeting SIRT1.
Keywords: cerebral ischemia/reperfusion injury, oxygen-glucose deprivation and reoxygenation, microRNA-217, sirtuin 1, neurons
Introduction
Ischemic stroke is one of the leading causes of disability and mortality globally (1,2). Interventions require the recovery of blood flow, which can lead to reperfusion injury. Cerebral ischemia-reperfusion (CIR) injury is a pathological process in which nerve damage induced by ischemia and hypoxia is further aggravated following the short-term recovery of blood perfusion (3). CIR leads to mitochondrial dysfunction, inflammation, massive release of reactive oxygen species (ROS), excessive glutamate excitotoxicity and cell death, ultimately resulting in irreversible brain damage (4). Progress has been made in reperfusion therapy (5); however, the therapeutic outcomes remain unsatisfactory. Therefore, improved understanding of the pathological process of CIR injury and the identification of novel treatment strategies is required.
MicroRNAs (miRNAs/miRs) are a class of small endogenous noncoding RNAs (~22 nucleotides in length) that can regulate the expression of genes at the post-transcriptional level by binding to the 3′-untranslated region (3′-UTR) of target genes (6–8). miRNAs have been reported to serve important roles in the regulation of a variety of biological processes, including proliferation, differentiation and apoptosis, via the regulation of various target genes (7,9–11). Increasing evidence has indicated that miRNAs serve important regulatory roles in normal physiology and disease (12), including in the pathogenesis of CIR injury (13,14). Numerous studies have reported that various miRNAs are dysregulated during CIR injury, and that targeting these miRNAs can effectively relieve neuronal injury in vitro and in vivo (15–17).
miRNA-217 has been studied in various types of cancers, including lung adenocarcinoma, acute myeloid leukemia, liver and gastric cancers, and colorectal carcinoma (18–22). miR-217 has been reported to be downregulated in glioma tissues and cells, and overexpression of miR-217 was observed to reduce the malignancy of glioma cells in vitro and decrease tumor growth in vivo (23); however, to the best of our knowledge, there is no data regarding the expression of miR-217 in other neurological diseases, and whether miR-217 is involved in regulating CIR injury remains unknown.
Research into damage following CIR has increased in previous years, and a large number of studies have demonstrated that the mechanisms underlying the effects of CIR on brain damage are complex (24). At present, there is no treatment strategy to effectively treat CIR injury. Therefore, finding new and effective CIR injury treatment methods has important clinical significance. Novel roles for miRNAs in the pathogenesis of CIR injury have been identified (13–17); however, to the best of our knowledge, the role of miR-217 in CIR injury remains unclear. Therefore, the aims of the present study were to investigate the role of miR-217 in neuronal injury induced by CIR using an in vitro cellular model induced by oxygen-glucose deprivation and reoxygenation (OGD/R).
Materials and methods
Primary neuron culture
Primary rat cerebral cortical neurons were extracted from Sprague-Dawley rat embryos (n=5) at embryonic day 16–18 as previously described (25,26). In brief, brain cortex tissues were extracted and dissected in Hanks' balanced salt solution (HBSS), cut into ~1 mm3 cubes, washed with HBSS, and then digested with 0.25% trypsin (37°C for 15 min). Then, DMEM-F12 (Gibco; Thermo Fisher Scientific, Inc.) with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) was used to stop trypsin digestion. The dissociated cells were seeded in 24-well plates and cultured for 24 h at 37°C. Then, the cells were cultured in neurobasal medium (Gibco; Thermo Fisher Scientific, Inc.) containing 2% B-27, 2 mM glutamine and 50 ug/ml gentamycin, and cells were incubated at 37°C with 5% CO2. Sprague-Dawley rats were obtained from Beijing Vital River Laboratory Animal Technology Co., Ltd., and housed at 25±5°C with 50% humidity under a 12:12-h dark/light cycle. Rats accessed to food and water ad libitum. Animal experiments were conducted according to the the guidelines of the National Institutes of Health for the Care and Use of Laboratory Animals (27). The present study was approved by the Animal Ethics Committee of the First People's Hospital of Wenling (Wenling, China).
OGD/R model establishment
The OGD/R model was established in the present study as previously described (26). In brief, cortical neurons were plated into 6-well plates at a density of 5×105 cells/ml (2 ml) and cultured overnight. Then, the neurons were incubated in glucose-free DMEM in a hypoxic incubator chamber (1% O2, 5% CO2 and 94% N2) at 37°C for 6 h. After three washes with DMEM, cells were incubated in DMEM supplemented with 4.5 g/l glucose at 37°C with 5% CO2 for 24 h.
The same treatment was performed in the control group without OGD/R exposure. Briefly, neurons in the control group were cultured in DMEM under standard conditions for 6 h. After three washes with DMEM, cells were incubated in DMEM at 37°C with 5% CO2 for 24 h.
Cell transfection
Cortical neurons were seeded in a 6-well plate (1×106 cells/well) and incubated at 37°C for 24 h. Then, 100 nM miR-217 inhibitor (5′-UACUGCAUCAGGAACUGAUUGGA-3′; Bioneer Corp.), 100 nM inhibitor control (5′-GCCUCCGGCUUCGCACCUCU-3′; Bioneer Corp.), 2 µl control-small interfering RNA (siRNA; cat. no. sc-36869; Santa Cruz Biotechnology, Inc.), 2 µl sirtuin 1 (SIRT1)-siRNA (cat. no. sc-40986; Santa Cruz Biotechnology, Inc.) or 100 nM miR-217 inhibitor + 2 µl SIRT1-siRNA was transfected into the cells using Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocols. At 24 h after transfection, transfection efficiency was measured using reverse transcription-quantitative PCR (RT-qPCR). At 24 h following transfection with miR-217 inhibitor, inhibitor control, or miR-217 inhibitor+SIRT1-siRNA, cells were subjected to OGD/R induction.
Cell Counting Kit-8 (CCK-8) assay
Cell viability was determined at 24 h following OGD/R induction as previously described (26). Cell viability was determined via a CCK-8 assay. In brief, cells were plated into a 96-well plate (5×103 cells/well) and incubated at 37°C overnight. Then, cells were transfected with or without miR-217 inhibitor, inhibitor control or miR-217 inhibitor + SIRT1-siRNA for 24 h (in equal quantities as aforementioned), followed by OGD/R treatment. Subsequently, 10 µl CCK-8 solution (cat. no. C0038; Beyotime Biotechnology) was added to each well and incubated for another 2 h at 37°C. At the end of the experiment, cell viability was calculated by measuring the absorbance at 450 nm using a FLUOstar® Omega microplate reader (BMG Labtech GmbH).
ELISA
The levels of tumor necrosis factor-α (TNF-α; cat. no. PT516), and interleukin (IL)-6 (cat. no. PI326) and IL-1β (cat. no. PI301; all Beyotime Institute of Biotechnology) in the supernatant of cells were measured via sandwich ELISAs according to the manufacturer's protocol.
Lactate dehydrogenase (LDH) assay
An LDH Cytotoxicity Assay kit (cat. no. C0016; Beyotime Institute of Biotechnology) was used to determine cell injury according to the manufacturer's protocols. In brief, cells were lysed and then incubated with NADH and pyruvate at 37°C for 15 min. The absorbance value at a wavelength of 530 nm was determined using a FLUOstar Omega microplate reader.
Cell apoptosis assay
To investigate the effects of miR-217 on the apoptosis of cortical neurons, an Annexin V-FITC/propidium iodide (PI) Apoptosis Detection kit (cat no. 70-AP101-100; MultiSciences Biotech Co., Ltd.) was using according to the manufacturer's protocols. Following the aforementioned treatments, cortical neurons were stained with 5 µl Annexin V-FITC and 5 µl PI for 30 min at room temperature in the dark. Subsequently, a flow cytometer (BD Biosciences) was used to analyze cell apoptosis. The apoptosis rate (early + late) was calculated (percentage of cells in the right quadrants) using WinMDI soft-ware (version 2.5; Purdue University Cytometry Laboratories).
Evaluation of ROS, superoxide dismutase (SOD) and malondialdehyde (MDA) levels
To analyze the levels of ROS in cells, a Reactive Oxygen Species Assay Kit (cat. no. S0033; Beyotime Institute of Biotechnology) was used according to the manufacturer's protocols. A Total Superoxide Dismutase Assay kit with WST-8 (cat. no. S0101; Beyotime Institute of Biotechnology) was used to determine the level of SOD in cortical neurons according to the manufacturer's protocols. A Lipid Peroxidation MDA Assay kit (cat no. S0131; Beyotime Institute of Biotechnology) was used to determine the levels of MDA.
Western blot assay
Proteins were extracted from cells using RIPA lysis buffer (cat no. P0013E; Beyotime Institute of Biotechnology) according to the manufacturer's protocols. A bicinchoninic acid assay kit (Pierce; Thermo Fisher Scientific, Inc.) was used to quantify the protein samples according to the manufacturer's protocols. Equal amounts of protein samples (25 µg/lane) were separated via SDS-PAGE on 12% gels, transferred onto polyvinylidene difluoride membranes (EMD Millipore), and blocked in 5% skim milk at room temperature for 1.5 h. Then, the membranes were incubated with primary antibodies against SIRT1 (1:1,000; cat. no. 9475; Cell Signaling Technology, Inc.), phosphorylated (p)-AMP-activated protein kinase-α (AMPK-α; 1:1,000; cat. no. 50081; Cell Signaling Technology, Inc.), AMPK-α (1:1,000; cat. no. 5831; Cell Signaling Technology, Inc.), NF-κB p65 (1:1,000; cat. no. 8242; Cell Signaling Technology, Inc.), p-p65 (1:1,000; cat. no. 3033; Cell Signaling Technology, Inc.), and β-actin (1:1,000; cat. no. 4970; Cell Signaling Technology, Inc.) overnight at 4°C, followed by incubation with the horseradish peroxidase-conjugated anti-rabbit IgG secondary antibody (1:2,000; cat no. 7074; Cell Signaling Technology, Inc.) at room temperature for 2 h. Finally, protein blots were visualized using chemiluminescent ECL reagent (EMD Millipore) and quantified by densitometry (QuantityOne 4.5.0 software; Bio-Rad Laboratories, Inc.).
RT-qPCR
Total RNA was extracted from cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocols. A TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) was used for cDNA generation. Reaction conditions for RT were: 50°C for 5 min and 80°C for 2 min. An SYBR Premix Ex Taq™ II (TliRNaseH Plus) kit (Takara Bio, Inc.) was using to analyze the synthesized cDNAs. U6 and GAPDH were used as internal controls for miRNA and mRNA expression, respectively. Primer sequences for PCR were as follows: miR-217, forward, 5′-CGCAGATACTGCATCAGGAA-3′ and reverse, 5′-CTGAAGGCAATGCATTAGGAACT-3′; SIRT1, forward, 5′-AATCCAGTCATTAAACGGTCTACAA-3′ and reverse, 5′-TAGGACCATTACTGCCAGAGG-3′; U6, forward, 5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse, 5′CGCTTCACGAATTTGCGTGTCAT3′; and GAPDH, forward, 5′-CTTTGGTATCGTGGAAGGACTC-3′ and reverse, 5′-GTAGAGGCAGGGATGATGTTCT-3′. The thermocycling conditions were as follows: 95°C for 5 min, followed by 38 cycles of denaturation at 95°C for 15 sec and annealing/elongation at 60°C for 30 sec. The relative gene expression was calculated using the 2−ΔΔCq method (28).
Dual-luciferase reporter assay
TargetScan bioinformatics software (release 7.1; www.targetscan.org/vert_71) was used to predict the targets of miR-217; a putative binding site between the 3′-UTR of SIRT1 and miR-217 was identified. Then, to determine whether miR-217 directly binds to SIRT1, the target sequence (WT-SIRT1; ATGCAGT) and a mutated sequence (MUT-SIRT1; TACGTCA) were synthesized. To point-mutate the miR-217 binding domain on the 3′-UTR of SIRT1, a QuikChange Site-Directed Mutagenesis kit (Stratagene; Agilent Technologies, Inc.) was used according to the manufacturer's protocols. Following digestion with XhoI and NotI restriction enzymes, a pmiR RB Report™ reporter gene plasmid vector (Guangzhou RiboBio Co., Ltd.) were combined with synthesized WT-SIRT1 or MUT-SIRT1 to construct recombinant plasmids, SIRT1-WT or SIRT1-MUT. Neurons (5×104 cells/well) were co-transfected with 50 ng SIRT1-WT or 50 ng SIRT1-MUT, 25 ng pRL-TK (expressing Renilla luciferase as the internal control; Promega Corporation), and 50 nM miR-217 mimic (5′-UACUGCAUCAGGAACUGAUUGGA-3′) or 50 nM mimic control (5′-UUUGUACUACACAAAAGUACUG-3′) using Lipofectamine 2000 according to the manufacturer's protocols. At 24 h following transfection, luciferase activity was determined using a dual-luciferase assay system (Promega Corporation) and normalized to the Renilla luciferase activity.
Statistical analysis
Experiments in the present study were performed three times. Experimental analysis was conducted using SPSS 17.0 software (SPSS, Inc.). Data were presented as the mean ± standard deviation. Differences between groups were determined using Student's t-test or one-way ANOVA with a Bonferroni post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-217 is upregulated in OGD/R-treated neurons
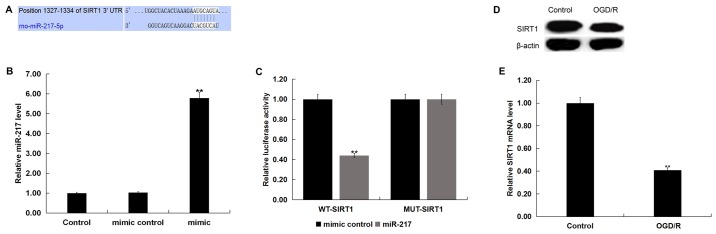
To investigate whether miR-217 was involved in CIR injury in vitro, the levels of miR-217 in neurons following OGD/R or control treatment was determined via RT-qPCR analysis. As presented in Fig. 1, compared with the control, OGD/R treatment significantly upregulated the expression of miR-217 in neurons.
Figure 1.
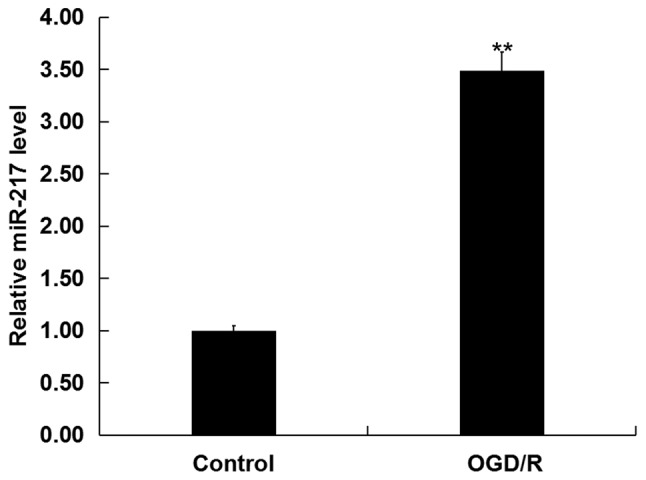
miR-217 expression in OGD/R-treated neurons. miR-217 expression levels in neurons following OGD/R treatment were detected by reverse transcription-quantitative PCR analysis. Data are presented as the mean ± standard deviation. **P<0.01 vs. control. miR-217, microRNA-217; OGD/R, oxygen-glucose deprivation and reoxygenation.
SIRT1 is a target gene of miR-217
TargetScan was used to predict the potential targets of miR-217, and a putative binding site between SIRT1 and miR-217 was identified (Fig. 2A). To demonstrate the association between miR-217 and SIRT1, a luciferase reporter assay was conducted. miR-217 mimic was used to significantly upregulated miR-217 expression in neurons (Fig. 2B). As presented in Fig. 2C, compared with cells co-transfected with WT-SIRT1 and mimic control, luciferase activity was significantly decreased in neurons co-transfected with WT-SIRT1 and miR-217 mimic. The results indicated that miR-217 directly targeted SIRT1.
Figure 2.
SIRT1 is a direct target of miR-217. (A) Interaction between miR-217 and the 3′-UTR of SIRT1 predicted using TargetScan. (B) Level of miR-217 in neurons transfected with miR-217 mimic or mimic control as determined by RT-qPCR. (C) Luciferase activity of reporters containing the WT or MUT SIRT1 3′-UTR following transfection with miR-217 mimic or mimic control. Data are presented as the mean ± standard deviation of three independent experiments. **P<0.01 vs. mimic control. (D) Protein and (E) mRNA expression of SIRT1 in neurons following OGD/R treatment as determined by western blot and RT-qPCR analyses, respectively. Data are presented as the mean ± standard deviation. **P<0.01 vs. Control. RT-qPCR, reverse transcription-quantitative PCR; SIRT1, sirtuin 1; 3′-UTR, 3′-untranslated region; miR-217, microRNA-217; WT, wild-type; MUT, mutated; OGD/R, oxygen-glucose deprivation and reoxygenation.
Subsequently, the protein and mRNA levels of SIRT1 in neurons following control or OGD/R treatment were determined via western blot and RT-qPCR analyses, respectively. As presented in Fig. 2D, OGD/R treatment markedly reduced SIRT1 protein expression in neurons. Additionally, compared with the control group, the mRNA levels of SIRT1 in neurons were significantly downregulated following OGD/R treatment (Fig. 2E).
Downregulation of miR-217 alleviates OGD/R-induced neuronal injury
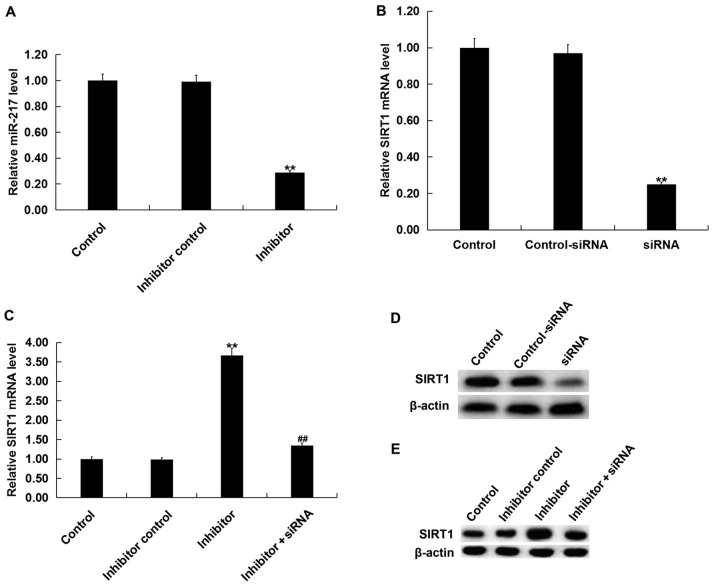
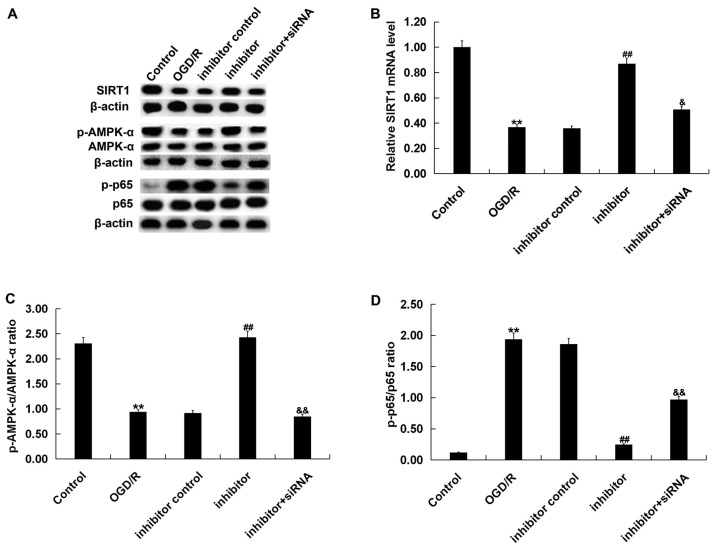
To investigate the role of miR-217 in OGD/R-induced neuronal injury, neurons were transfected with miR-217 inhibitor, inhibitor control, control-siRNA, SIRT1-siRNA or miR-217 inhibitor + SIRT1-siRNA for 24 h, then subjected to OGD/R induction. The transfection efficiency was measured by RT-qPCR and/or western blotting. As presented in Fig. 3A, compared with the transfection control group, miR-217 inhibitor significantly downregulated the expression of miR-217 in neurons, whereas the mRNA levels of SIRT1 in neurons were decreased following SIRT1-siRNA transfection (Fig. 3B). The protein levels of SIRT1 in neurons were also reduced following SIRT1-siRNA transfection (Fig. 3D). Furthermore, it was demonstrated that miR-217 inhibitor significantly increased the mRNA expression of SIRT1 in neurons, and that this upregulation was attenuated by SIRT1-siRNA (Fig. 3C). miR-217 inhibitor also enhanced the protein expression of SIRT1 in neurons, in a manner that was attenuated by SIRT1-siRNA (Fig. 3E).
Figure 3.
Effects of miR-217 inhibitor on SIRT1 expression in neurons. (A) Relative miR-217 expression in neurons following transfection with miR-217 inhibitor or inhibitor control. Relative SIRT1 mRNA expression in neurons following (B) siRNA and/or (C) inhibitor transfection. Protein level of SIRT1 in neurons following (D) siRNA and/or (E) inhibitor transfection. Data are presented as the mean ± standard deviation. **P<0.01 vs. control; ##P<0.01 vs. inhibitor. miR-217, microRNA-217; SIRT1, sirtuin 1; siRNA, small interfering RNA.
Then, the effects of miR-217 inhibitor on OGD/R-induced injury were determined via CCK-8 and LDH assays. Consistent with a previous study (25), OGD/R treatment significantly reduced cell viability and increased LDH release. The effects of OGD/R treatment were inhibited by miR-217 downregulation, and this inhibition was attenuated by SIRT1-siRNA (Fig. 4A and B). Additionally, miR-217 inhibition suppressed the OGD/R treatment-induced apoptosis of neurons (Fig. 4C and D). Collectively, the data indicated that miR-217 downregulation attenuated OGD/R-induced neuronal injury.
Figure 4.
Effects of miR-217 inhibitor on OGD/R-induced neuronal injury. Neurons were transfected with miR-217 inhibitor, inhibitor control or miR-217 inhibitor + sirtuin 1-siRNA for 24 h, followed by OGD/R treatment. (A) Cell viability was detected using a Cell Counting Kit-8 assay. (B) LDH release was measured by an LDH assay. (C and D) Cell apoptosis was determined using flow cytometry. Data are presented as the mean ± standard deviation. *P<0.05, **P<0.01 vs. control; #P<0.05, ##P<0.01 vs. OGD/R; &P<0.05, &&P<0.01 vs. inhibitor. miR-217, microRNA-217; OGD/R, oxygen-glucose deprivation and reoxygenation; LDH, lactate dehydrogenase; siRNA, small interfering RNA; PI, propidium iodide.
Downregulation of miR-217 alleviates OGD/R-induced inflammatory response
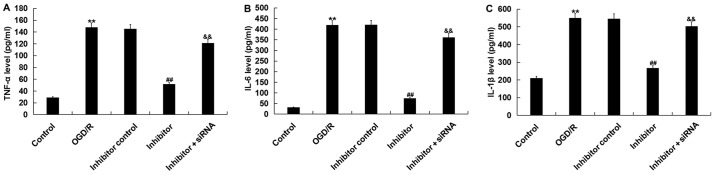
To further investigate the biological function of miR-217 in the regulation of OGD/R injury, the effects of miR-217 inhibitor on OGD/R-induced inflammation were determined. The levels of TNF-α, IL-6 and IL-1β were detected by ELISA. As presented in Fig. 5, the increased levels of TNF-α, IL-6 and IL-1β following OGD/R were significantly reduced by miR-217 inhibitor; however, these reductions were prevented by SIRT1-siRNA.
Figure 5.
Effects of miR-217 inhibitor on proinflammatory factor levels in OGD/R-treated neurons. Neurons were transfected with miR-217 inhibitor, inhibitor control or miR-217 inhibitor + sirtuin 1-siRNA for 24 h, followed by OGD/R treatment. Then, the levels of (A) TNF-α, (B) IL-6 (C) and IL-1β in the supernatant of neurons was measured using ELISA. Data are presented as the mean ± standard deviation. **P<0.01 vs. control; ##P<0.01 vs. OGD/R; &&P<0.01 vs. inhibitor. miR-217, microRNA-217; TNF-α, tumor necrosis factor-α; OGD/R, oxygen-glucose deprivation and reoxygenation; siRNA, small interfering RNA; IL, interleukin.
Downregulation of miR-217 alleviates OGD/R-induced oxidative stress
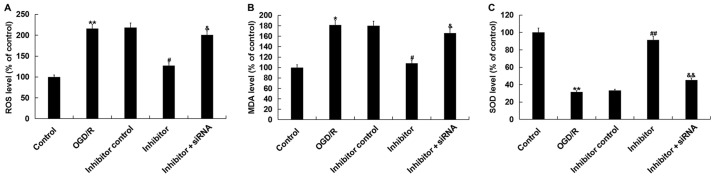
It was then investigated as to whether transfection with miR-217 inhibitor induced an effect on OGD/R-induced oxidative stress; it was observed that compared with the control, OGD/R treatment significantly enhanced the levels of ROS and MDA, and reduced those of SOD in neurons. Downregulation of miR-217 significantly decreased ROS production (Fig. 6A), reduced the levels of MDA (Fig. 6B) and increased those of SOD (Fig. 6C) in OGD/R-treated neurons, whereas these alterations were attenuated by SIRT1 silencing. The results indicated that miR-217 downregulation attenuated OGD/R-induced oxidative stress.
Figure 6.
Effects of miR-217 inhibitor on oxidative stress in OGD/R-treated neurons. Neurons were transfected with miR-217 inhibitor, inhibitor control or miR-217 inhibitor + sirtuin 1-siRNA for 24 h, followed by OGD/R treatment. Then, (A) ROS, (B) MDA and (C) SOD levels were determined. Data are presented as the mean ± standard deviation. *P<0.05, **P<0.01 vs. control; #P<0.05, ##P<0.01 vs. OGD/R; &P<0.05, &&P<0.01 vs. inhibitor. miR-217, microRNA-217; ROS, reactive oxygen species; OGD/R, oxygen-glucose deprivation and reoxygenation; siRNA, small interfering RNA; MDA, malondialdehyde; SOD, superoxide dismutase.
Effects of miR-217 inhibitor on the SIRT1/AMPK-α/NF-κB pathway in OGD/R-treated neurons
Finally, the SIRT1/AMPK-α/NF-κB pathway in OGD/R-treated neurons was analyzed to investigate the molecular mechanisms underlying the effects of miR-217 on OGD/R-induced neuronal injury. As presented in Fig. 7, compared with the control, OGD/R treatment significantly reduced SIRT1 mRNA and protein levels (Fig. 7A and B), decreased AMPK-α protein phosphorylation levels (Fig. 7A and C) and promoted p65 phosphorylation (Fig. 7A and D) in neurons. Compared with the OGD/R treatment group, miR-217 downregulation significantly upregulated SIRT1 and p-AMPK-α expression, and decreased p-p65 levels; these effects were eliminated by SIRT1 downregulation.
Figure 7.
Effects of miR-217 inhibitor on SIRT1/AMPK-α/NF-κB pathway in OGD/R-treated neurons. Neurons were transfected with miR-217 inhibitor, inhibitor control or miR-217 inhibitor + SIRT1-siRNA for 24 h, followed by OGD/R treatment. Then, the (A) protein and (B) mRNA expression levels of SIRT1 were determined by western blot and reverse transcription-quantitative PCR analyses, respectively. Additionally, the protein levels of p-AMPK-α, AMPK-α, p65, and p-p65 were measured by western blotting. The ratios of (C) p-AMPK-α/AMPK-α and (D) p-p65/p65 normalized to β-actin were calculated and presented. Data are presented as the mean ± standard deviation. **P<0.01 vs. control; ##P<0.01 vs. OGD/R; &P<0.05, &&P<0.01 vs. inhibitor. miR-217, microRNA-217; OGD/R, oxygen-glucose deprivation and reoxygenation; siRNA, small interfering RNA; SIRT1, sirtuin 1; p, phosphorylated; AMPK-α, AMP-activated protein kinase-α.
Discussion
Recent studies have reported that miR-217 serves an important role in the development of tumors (18–22). miR-217 is involved in the apoptosis of human podocyte cells by targeting TNF ligand superfamily member 11 in membranous nephropathy (29). Another study demonstrated that miR-217 promotes fibroblast senescence (30). Additionally, miR-217 was identified as a key regulator in ethanol-induced hepatic inflammation (31); however, the role of miR-217 in neuronal injury induced by CIR injury remains unclear. In the present study, the results indicated that miR-217 was upregulated in OGD/R-treated neurons, and its downregulation relieved OGD/R treatment-induced neuronal injury, as revealed by increased cell viability, reduced LDH release and decreased cell apoptosis. Additionally, it was observed that miR-217 downregulation notably inhibited OGD/R-induced inflammatory responses and oxidative stress. Furthermore, the findings of the present study revealed that SIRT1 was a direct target of miR-217, and that the effects of miR-217 inhibitor on OGD/R-treated neurons were eliminated by SIRT1 silencing.
A number of experimental studies have revealed that the mechanisms underlying the effects of CIR on brain damage are complex (24,25). At present, there is no strategy to effectively treat CIR injury. Therefore, finding novel and effective methods has important clinical relevance. The emerging roles of miRNAs in the pathogenesis of CIR injury have been identified in numerous studies (12–16). In the present study, the role of miR-217 in neuron injury induced by OGD/R was investigated.
An in vitro cellular model of CIR injury was established by treating neurons with OGD/R. The OGD/R model is a cell injury model widely used to study CIR injury in vitro (24,26,32). Then, the levels of miR-217 in neurons subjected to control or OGD/R treatment were determined, and the results revealed that compared with the control group, miR-217 was significantly upregulated in OGD/R-treated neurons. Subsequently, it was revealed that SIRT1 was a direct target of miR-217 that was downregulated in neurons treated with OGD/R. SIRT1 is a NAD+-dependent deacetylase important for apoptosis, cell cycle, mitochondrial function and metabolism (33), and serves an important role in the regulation of inflammatory responses and oxidative stress (33–37). To explore the potential function of miR-217 in the regulation of OGD/R-induced neuronal injury, loss-of-function experiments were performed by transfection with miR-217 inhibitor. The results suggested that miR-217 downregulation significantly mitigated OGD/R-induced neuronal injury, as determined by increased cell viability, reduced LDH release and decreased cell apoptosis. Additionally, miR-217 downregulation notably suppressed inflammatory responses and oxidative stress enhanced by OGD/R treatment in neurons.
The activation of the AMPK-α-SIRT1 pathway can inhibit NF-κB inflammation (38). Activation of AMPK/SIRT1 signaling attenuated TNF-α-induced MMP-3 expression in human nucleus pulposus cells (39). Additionally, activation of the AMPK/SIRT1 pathway protected PC12 cells against cisplatin-induced neurotoxicity (40). The pathway has also been reported to relieve oxidized low-density lipoprotein-induced endothelial cell injury (41). These results indicated that AMPK/SIRT1 signaling serves important roles in cell injury and inflammation regulation. Furthermore, a recent study revealed that AMPK/SIRT1 pathway activation was involved in the protective effects of lncRNA SNHG12 on CIR injury (42). Therefore, to investigate the molecular mechanisms underlying the effects of miR-217 inhibitor on OGD/R-induced neuronal injury, the activity of the SIRT1/AMPK-α/NF-κB pathway was analyzed in the present study. Results revealed that OGD/R treatment significantly reduced SIRT1 mRNA and protein levels, decreased AMPK-α phosphorylation, and increased p65 phosphorylation level in neurons; these effects were reversed by miR-217 inhibitor. Of note, all the observed effects of miR-217 inhibitor on OGD/R-treated neurons were attenuated by SIRT1 silencing.
In conclusion, the results of the present study demonstrated that miR-217 was upregulated in OGD/R-treated neurons, and that its inhibition attenuated OGD/R-induced neuronal injury via the upregulation of SIRT1 and the SIRT1/AMPK-α/NF-κB pathway. Of note, this is a preliminary study of the role of miR-217 in CIR injury, and further research is required to more comprehensively and convincingly determine the precise roles and mechanisms of miR-217 in CIR injury. For example, the effects of miR-217 on CIR injury in vivo should be investigated, in addition to the effects of SIRT1 manipulation alone on CIR injury. Furthermore, in the present study, experiments were performed at only one time point (24 h following OGD/R treatment); thus, additional time points should be investigated in the future. Finally, an investigation into the association between miR-217 expression and the clinical characteristics and prognosis of patients with CIR injury is required to confirm the clinical relevance of miR-217 in CIR injury. In-depth research into these issues will be conducted in the future.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Medical and Health Science and Technology Planning Project of Zhejiang Province (grant no. 2018KY918).
Availability of data and materials
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.
Authors' contributions
GR and WZ contributed to study design, data collection, statistical analysis, data interpretation and manuscript preparation. SS contributed to data collection and interpretation. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Animal Ethics Committee of The First People's Hospital of Wenling.
Patient consent for publication
Not applicable.
Competing interests
All authors declare that they have no competing interests.
References
- 1.Feigin VL. Stroke epidemiology in the developing world. Lancet. 2005;365:2160–2161. doi: 10.1016/S0140-6736(05)66755-4. [DOI] [PubMed] [Google Scholar]
- 2.Goldstein LB, Adams R, Becker K, Furberg CD, Gorelick PB, Hademenos G, Hill M, Howard G, Howard VJ, Jacobs B, et al. Primary prevention of ischemic stroke A statement for healthcare professionals from the stroke council of the American heart association. Circulation. 2001;103:163–182. doi: 10.1161/01.CIR.103.1.163. [DOI] [PubMed] [Google Scholar]
- 3.Jean WC, Spellman SR, Nussbaum ES, Low WC. Reperfusion injury after focal cerebral ischemia: The role of inflammation and the therapeutic horizon. Neurosurgery. 1998;43:1382–1397. doi: 10.1227/00006123-199812000-00076. [DOI] [PubMed] [Google Scholar]
- 4.Schrepfer E, Scorrano L. Mitofusins, from mitochondria to metabolism. Mol Cell. 2016;61:683–694. doi: 10.1016/j.molcel.2016.02.022. [DOI] [PubMed] [Google Scholar]
- 5.Bhaskar S, Stanwell P, Cordato D, Attia J, Levi C. Reperfusion therapy in acute ischemic stroke: Dawn of a new era? BMC Neurol. 2018;18:8. doi: 10.1186/s12883-017-1007-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Hammond SM. An overview of microRNAs. Adv Drug Deliv Rev. 2015;87:3–14. doi: 10.1016/j.addr.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghildiyal M, Zamore PD. Small silencing RNAs: An expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soifer HS, Rossi JJ, Saetrom P. MicroRNAs in disease and potential therapeutic applications. Mol Ther. 2017;15:2070–2079. doi: 10.1038/sj.mt.6300311. [DOI] [PubMed] [Google Scholar]
- 10.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 11.O'Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10:111–122. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 12.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148:1172–1187. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Di Y, Lei Y, Yu F, Changfeng F, Song W, Xuming M. MicroRNAs expression and function in cerebral ischemia reperfusion injury. J Mol Neurosci. 2014;53:242–250. doi: 10.1007/s12031-014-0293-8. [DOI] [PubMed] [Google Scholar]
- 14.Hu Y, Deng H, Xu S, Zhang J. MicroRNAs regulate mitochondrial function in cerebral ischemia-reperfusion injury. Int J Mol Sci. 2015;16:24895–24917. doi: 10.3390/ijms161024895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang P, Liang X, Lu Y, Zhao X, Liang J. MicroRNA-93 downregulation ameliorates cerebral ischemic injury through the Nrf2/HO-1 defense pathway. Neurochem Res. 2016;41:2627–2635. doi: 10.1007/s11064-016-1975-0. [DOI] [PubMed] [Google Scholar]
- 16.Wang N, Zhang L, Lu Y, Zhang M, Zhang Z, Wang K, Lv J. Down-regulation of microRNA-142-5p attenuates oxygen-glucose deprivation and reoxygenation-induced neuron injury through up-regulating Nrf2/ARE signaling pathway. Biomed Pharmacother. 2017;89:1187–1195. doi: 10.1016/j.biopha.2017.03.011. [DOI] [PubMed] [Google Scholar]
- 17.Shi F, Dong Z, Li H, Liu X, Liu H, Dong R. MicroRNA-137 protects neurons against ischemia/reperfusion injury through regulation of the notch signaling pathway. Exp Cell Res. 2017;352:1–8. doi: 10.1016/j.yexcr.2017.01.015. [DOI] [PubMed] [Google Scholar]
- 18.Liu AN, Qu HJ, Yu CY, Sun P. Knockdown of LINC01614 inhibits lung adenocarcinoma cell progression by up-regulating miR-217 and down-regulating FOXP1. J Cell Mol Med. 2018;22:4034–4044. doi: 10.1111/jcmm.13483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang LP, Wang JP, Wang XP. HOTAIR contributes to the growth of liver cancer via targeting miR-217. Oncol Lett. 2018;15:7963–7972. doi: 10.3892/ol.2018.8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan J, Wu G, Chen J, Xiong L, Chen G, Li P. Downregulated miR-217 expression predicts a poor outcome in acute myeloid leukemia. Cancer Biomark. 2018;22:73–78. doi: 10.3233/CBM-170936. [DOI] [PubMed] [Google Scholar]
- 21.Safaralizadeh R, Ajami N, Nemati M, Hosseinpourfeizi M, Azimzadeh Isfanjani A, Moaddab SY. Disregulation of miR-216a and miR-217 in gastric cancer and their clinical significance. J Gastrointest Cancer. 2019;50:78–83. doi: 10.1007/s12029-017-0019-6. [DOI] [PubMed] [Google Scholar]
- 22.Yu B, Du Q, Li H, Liu HY, Ye X, Zhu B, Zhai Q, Li XX. Diagnostic potential of serum exosomal colorectal neoplasia differentially expressed long non-coding RNA (CRNDE-p) and microRNA-217 expression in colorectal carcinoma. Oncotarget. 2017;8:83745–83753. doi: 10.18632/oncotarget.19407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng J, Liu X, Xue Y, Gong W, Ma J, Xi Z, Que Z, Liu Y. TTBK2 circular RNA promotes glioma malignancy by regulating miR-217/HNF1β/Derlin-1 pathway. J Hematol Oncol. 2017;10:52. doi: 10.1186/s13045-017-0422-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sahu S, Nag DS, Swain A, Samaddar DP. Biochemical changes in the injured brain. World J Biol Chem. 2017;8:21–31. doi: 10.4331/wjbc.v8.i1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J, Zhao F, Zhao Y, Wang J, Pei L, Sun N, Shi J. Hypoxia induces an increase in intracellular magnesium via transient receptor potential melastatin 7 (TRPM7) channels in rat hippocampal neurons in vitro. J Biol Chem. 2011;286:20194–20207. doi: 10.1074/jbc.M110.148494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu X, Li M, Hou M, Huang W, Song J. MicroRNA-135a alleviates oxygen-glucose deprivation and reoxygenation-induced injury in neurons through regulation of GSK-3β/Nrf2 signaling. J Biochem Mol Toxicol. 2018;2:e22159. doi: 10.1002/jbt.22159. [DOI] [PubMed] [Google Scholar]
- 27.Bayne K. Revised guide for the care and use of laboratory animals available. American physiological society. Physiologist. 1996;39:199208–199211. [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Li J, Liu B, Xue H, Zhou QQ, Peng L. miR-217 Is a useful diagnostic biomarker and regulates human podocyte cells apoptosis via targeting TNFSF11 in membranous nephropathy. Biomed Res Int. 2017;2017:2168767. doi: 10.1155/2017/2168767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang B, Du R, Xiao X, Deng ZL, Jian D, Xie HF, Li J. Microrna-217 modulates human skin fibroblast senescence by directly targeting DNA methyltransferase 1. Oncotarget. 2017;8:33475–33486. doi: 10.18632/oncotarget.16509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yin H, Liang X, Jogasuria A, Davidson NO, You M. miR-217 regulates ethanol-induced hepatic inflammation by disrupting sirtuin 1-lipin-1 signaling. Am J Pathol. 2015;185:1286–1296. doi: 10.1016/j.ajpath.2015.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xin L, Junhua W, Long L, Jun Y, Yang X. Exogenous hydrogen sulfide protects SH-SY5Y cells from OGD/rinduced injury. Curr Mol Med. 2017;17:563–567. doi: 10.2174/1566524018666180222121643. [DOI] [PubMed] [Google Scholar]
- 33.Nogueiras R, Habegger KM, Chaudhary N, Finan B, Banks AS, Dietrich MO, Horvath TL, Sinclair DA, Pfluger PT, Tschöp MH. Sirtuin 1 and sirtuin 3: Physiological modulators of metabolism. Physiol Rev. 2012;92:1479–1514. doi: 10.1152/physrev.00022.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.da Cunha MSB, Arruda SF. Tucum-do-cerrado (Bactris setosa Mart.) may promote anti-aging effect by Upregulating SIRT1-Nrf2 pathway and attenuating oxidative stress and inflammation. Nutrients. 2017;9:E1243. doi: 10.3390/nu9111243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rada P, Pardo V, Mobasher MA, García-Martínez I, Ruiz L, González-Rodríguez Á, Sanchez-Ramos C, Muntané J, Alemany S, James LP, et al. SIRT1 controls acetaminophen hepatotoxicity by modulating inflammation and oxidative stress. Antioxid Redox Signal. 2018;28:1187–1208. doi: 10.1089/ars.2017.7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan SH, Hung CH, Shih JY, Chu PM, Cheng YH, Lin HC, Tsai KL. SIRT1 inhibition causes oxidative stress and inflammation in patients with coronary artery disease. Redox Biol. 2017;13:301–309. doi: 10.1016/j.redox.2017.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng YY, Kao CL, Ma HI, Hung CH, Wang CT, Liu DH, Chen PY, Tsai KL. SIRT1-related inhibition of pro-inflammatory responses and oxidative stress are involved in the mechanism of nonspecific low back pain relief after exercise through modulation of Toll-like receptor 4. J Biochem. 2015;158:299–308. doi: 10.1093/jb/mvv041. [DOI] [PubMed] [Google Scholar]
- 38.Tian Y, Ma J, Wang W, Zhang L, Xu J, Wang K, Li D. Resveratrol supplement inhibited the NF-κB inflammation pathway through activating AMPKα-SIRT1 pathway in mice with fatty liver. Mol Cell Biochem. 2016;422:75–84. doi: 10.1007/s11010-016-2807-x. [DOI] [PubMed] [Google Scholar]
- 39.Wang XH, Zhu L, Hong X, Wang YT, Wang F, Bao JP, Xie XH, Liu L, Wu XT. Resveratrol attenuated TNF-α-induced MMP-3 expression in humannucleus pulposus cells by activating autophagy via AMPK/SIRT1 signaling pathway. Exp Biol Med (Maywood) 2016;241:848–853. doi: 10.1177/1535370216637940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferreira RS, Dos Santos NAG, Bernardes CP, Sisti FM, Amaral L, Fontana ACK, Dos Santos AC. Caffeic acid phenethyl ester (CAPE) protects PC12 cells against cisplatin-induced neurotoxicity by activating the AMPK/SIRT1, MAPK/Erk, and PI3k/Akt signaling pathways. Neurotox Res. 2019 Apr 23; doi: 10.1007/s12640-019-00042-w. (Epub ahead of print). doi: 10.1007/s12640-019-00042-w. [DOI] [PubMed] [Google Scholar]
- 41.Zhao D, Sun X, Lv S, Sun M, Guo H, Zhai Y, Wang Z, Dai P, Zheng L, Ye M, Wang X. Salidroside attenuates oxidized low-density lipoprotein-induced endothelial cell injury via promotion of the AMPK/SIRT1 pathway. Int J Mol Med. 2019;43:2279–2290. doi: 10.3892/ijmm.2019.4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yin WL, Yin WG, Huang BS, Wu LX. LncRNA SNHG12 inhibits miR-199a to upregulate SIRT1 to attenuate cerebral ischemia/reperfusion injury through activating AMPK signaling pathway. Neurosci Lett. 2019;690:188–195. doi: 10.1016/j.neulet.2018.08.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.