Abstract
Introduction
Ischemic stroke (IS) causes severe neurological impairments and physical disabilities and has a high economic burden. Our study aims to identify the key genes and upstream regulators in IS by integrated microarray analysis.
Methods
An integrated analysis of microarray studies of IS was performed to identify the differentially expressed genes (DEGs) in IS compared to normal control. Based on these DEGs, we performed the functional annotation and transcriptional regulatory network constructions. Quantitative real‐time polymerase chain reaction (qRT‐PCR) was performed to verify the expression of DEGs.
Results
From two Gene Expression Omnibus datasets obtained, we obtained 1526 DEGs (534 up‐regulated and 992 down‐regulated genes) between IS and normal control. The results of functional annotation showed that Oxidative phosphorylation and Alzheimer's disease were significantly enriched pathways in IS. Top four transcription factors (TFs) with the most downstream genes including PAX4, POU2F1, ELK1, and NKX2‐5. The expression of six genes (ID3, ICAM2, DCTPP1, ANTXR2, DUSP1, and RGS2) was detected by qRT‐PCR. Except for DUSP1 and RGS2, the other four genes in qRT‐PCR played the same pattern with that in our integrated analysis.
Conclusions
The dysregulation of these six genes may involve with the process of ischemic stroke (IS). Four TFs (PAX4, POU2F1, ELK1 and NKX2‐5) were concluded to play a role in IS. Our finding provided clues for exploring mechanism and developing novel diagnostic and therapeutic strategies for IS.
Keywords: differentially expressed genes, integrated analysis, ischemic stroke, transcription factors
1. INTRODUCTION
Stroke is one of the leading causes of serious long‐term disability and mortality worldwide, which is characterized by symptoms such as inability to move or feel one side of the body, inability to speak, or understand problems and dizziness (Donnan, Fisher, Macleod, & Davis, 2008; Dorrance & Fink, 2015). Ischemic stroke (IS) is one of the main types of stroke, which is characterized by cerebral ischemia. Survival of IS can be lead to severe neurological impairments and physical disabilities with a high economic burden (Evers et al., 2004; Volny, Kasickova, Coufalova, Cimflova, & Novak, 2015). Up to now, thrombolysis is the only efficacious treatment for IS (Chi & Chan, 2017). Despite recent progress, little is known about the underlying pathophysiology mechanisms of IS and much work still needs to be done to fully elucidate the pathophysiology mechanisms.
With high‐throughput genetic analysis, the emergence of gene expression profiles has became an effective method to identify differentially expressed genes (DEGs) in a variety of diseases which help to explore pathogenesis and develop biomarkers(Alieva et al., 2014; Du, Yang, Tian, Wang, & He, 2014; Kong et al., 2018). Due to differences of samples and platforms in multiple microarray studies, integrated analysis of multiple microarray studies can identify more accurate profiles of DEGs with a larger sample size than a single microarray. Exploring the upstream transcription factors (TFs) mediating abnormal gene expression in disease status can help to understand pathophysiological changes in complex diseases (Li, Dani, & Le, 2009; Zhao, Wang, Xu, Li, & Yu, 2017).
Our study aims to make an integrated analysis of multiple IS microarray data to obtain the key DEGs in the pathogenesis of IS. Functional annotation and protein and IS‐specific transcriptional regulatory network construction were performed to explore the biological functions of DEGs, which hopefully provide clues for exploring mechanism and developing novel diagnostic and therapeutic strategies for IS.
2. MATERIALS AND METHODS
2.1. Microarray expression profiling in Gene Expression Omnibus
In this study, we searched datasets from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) with the keywords "ischemic stroke, IS"[MeSH Terms] OR "ischemic stroke, IS" [All Fields]) AND "Homo sapiens"[porgn] AND "gse"[Filter]. The inclusion criteria for this study were: (a) The type of dataset was described as “expression profiling by array. (b) Dataset should be whole‐genome mRNA expression profile by array. (c) Datasets were obtained by blood samples of IS and normal control group (no drug stimulation or transfection). (d) The datasets should be normalized or original, and two sets of mRNA data (GSE22255 and GSE16561) of IS were selected. In GSE22255, Gene expression profiling was performed in peripheral blood mononuclear cells of 20 IS patients and 20 sex‐ and age‐matched controls using Affymetrix microarrays. In GSE16561, total RNA extracted from whole blood in 39 IS patients compared to 24 healthy control subjects.
2.2. Identification of DEGs between IS and normal controls
MetaMA, an R package, is used to combine data from multiple microarray datasets, and we obtained the individual p‐values. The Benjamini & Hochberg method were used to obtain multiple comparison correction false discovery rate (FDR). DEGs in IS compared to normal controls were obtained with FDR < 0.05. The heat‐map of top 100 DEGs was obtained by pheatmap package.
2.3. Functional annotation of DEGs
By using GeneCodis3 (http://genecodis.cnb.csic.es/analysis), gene ontology (GO) enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were performed to uncover biological functions of DEGs. FDR < 0.05 was considered as statistically significant.
2.4. Construction of IS‐specific transcriptional regulatory networks
Based on the integrated analysis, the corresponding promoters of the top 20 up‐regulated or down‐regulated DEGs were obtained by UCSC (http://genome.ucsc.edu). The TFs involved in regulating these DEGs were derived from the match tools in TRANSFAC. The IS‐specific transcriptional regulatory network was constructed by Cytoscape software (http://www.cytoscape.org/).
2.5. Validation the expression of DEGs in qRT‐PCR
Five adult patients with IS and five normal controls were enrolled in our study from First Affiliated Hospital of Shantou University Medical College. Patients with neurological diseases, cardiac embolism, transient ischemic attack, hemorrhagic infarction, occult cerebrovascular malformation, traumatic cerebrovascular disease were excluded. Normal control without history of stroke, head trauma and surgery, heart surgery or neurological disease was included in this study. All subjects were first on an empty stomach for 12 hr. Then, we collected the blood samples by venipuncture at 7:00–8:00 of the next morning. The patient demographics, clinical features and risk factors were extracted from medical records as displayed in Table 1. Informed written consent was obtained from all participants, and research protocols were approved by the ethical committee of First Affiliated Hospital of Shantou University Medical College. The criteria of choosing candidate genes as follow: In Top five up/down DEGs, we randomly selected six genes. Therefore, ID3, ICAM2, DCTPP1, ANTXR2, DUSP1, and RGS2 were selected as the validation DEGs.
Table 1.
Subject characteristics
| Case‐1 | Case‐2 | Case‐3 | Case‐4 | Case‐5 | Control‐1 | Control‐2 | Control‐3 | Control‐4 | Control‐5 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Race | Asian | Asian | Asian | Asian | Asian | Asian | Asian | Asian | Asian | Asian |
|
Gender/ Age |
Female/68 | Female/44 | Male/64 | Female/76 | Female/77 | Male/50 | Male/61 | Female/77 | Male/72 | Female/50 |
| GLU (mmol/L) | 12.11 | 4.74 | 4.63 | 8.44 | 6.8 | 4.97 | 4.47 | 5.98 | 6.03 | 5.19 |
| TG (mmol/L) | 2.35 | 0.85 | 2.04 | 1.87 | 4.32 | 1.94 | 0.42 | 1.99 | 1.53 | 1.08 |
| HDL‐C (mmol/L) | 0.67 | 0.95 | 1.09 | 0.92 | 1.98 | 1.4 | 1.29 | 0.94 | 0.89 | 1.57 |
| LDL‐C (mmol/L) | 2.45 | 1.74 | 1.91 | 3.05 | 1.01 | 3.35 | 2.53 | 2.98 | 2.94 | 3.82 |
| Hypertension | Yes | No | Yes | Yes | Yes | No | No | No | No | No |
| Diabetes | Yes | Yes | No | Yes | Yes | Yes | No | No | No | No |
| Hyperlipidemia | Yes | No | Yes | Yes | No | Yes | No | No | No | No |
| Smoker | No | No | No | No | No | No | No | No | Yes | No |
| Drinking | No | No | No | No | No | No | No | No | Yes | No |
Abbreviations: GLU, glucose; HDL‐C, high‐density lipoprotein cholesterol; LDL‐C, low‐density lipoprotein cholesterol; TG, triglyceride.
Total RNA was extracted with a RNA simple total RNA kit (Tiangen, China). RNA (2 μg) was reverse‐transcribed using a Fast Quant RT Kit (Tiangen, Beijing, China) according to the manufacturer's instructions. Quantitative real‐time PCR were conducted using the Super Real PreMix Plus SYBR Green (Tiangen, China) on ABI 7500 real‐time PCR system. The amplification process was performed under the following conditions: 15 min at 95°C followed by 40 cycles of 10 s at 95°C, 30 s at 55°C, 32 s at 72°C, and 15 s at 95°C, 60 s at 60°C, 15 s extension at 95°C. Relative quantification of mRNA levels was analyzed by using the 2−∆∆Ct method. Each sample was analyzed in triplicate. The human GAPDH were used as endogenous controls for gene expression in analysis. The PCR primers used are listed in Table 2.
Table 2.
Primer sequences used for qRT‐PCR
| Name | Sequence (5′ to 3′) |
|---|---|
| GAPDH‐F | GGAGCGAGATCCCTCCAAAAT |
| GAPDH‐R | GGCTGTTGTCATACTTCTCATGG |
| ID3‐F | GAGAGGCACTCAGCTTAGCC |
| ID3‐R | TCCTTTTGTCGTTGGAGATGAC |
| ICAM2‐F | CGGATGAGAAGGTATTCGAGGT |
| ICAM2‐R | CACCCACTTCAGGCTGGTTAC |
| DCTPP1‐F | CGCCTCCATGCTGAGTTTG |
| DCTPP1‐R | CCAGGTTCCCCATCGGTTTTC |
| ANTXR2‐F | GATCTCTACTTCGTCCTGGACA |
| ANTXR2‐R | AAATCTCTCCGCAAGTTGCTG |
| DUSP1‐F | ACCACCACCGTGTTCAACTTC |
| DUSP1‐R | TGGGAGAGGTCGTAATGGGG |
| RGS2‐F | AAGATTGGAAGACCCGTTTGAG |
| RGS2‐R | GCAAGACCATATTTGCTGGCT |
Abbreviation: qRT‐PCR, quantitative real‐time polymerase chain reaction.
3. RESULTS
3.1. DEGs in IS
Two datasets (GSE22255 and GSE16561) were downloaded from GEO (Table 3). Samples of GSE22255 and GSE16561 were obtained from participants of Portugal and USA, respectively. Compared with the normal controls, 1526 DEGs in IS were obtained with FDR < 0.05, among which, 534 genes were up‐regulated and 992 genes were down‐regulated. Top 20 DEGs between IS and normal controls were displayed in Table 4. Heat map of top 100 DEGs was displayed in Figure 1.
Table 3.
Gene expression datasets used in this study
| Type | GEO ID | Platform | Sample count (N:P) | Notes | (Refs.) |
|---|---|---|---|---|---|
| mRNA | GSE22255 | GPL570[HG‐U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array | 40 (20:20) | Oliveira SA, Portugal, 2011 | (Krug et al., 2012) |
| mRNA | GSE16561 | GPL6883 Illumina HumanRef‐8 v3.0 expression beadchip | 63 (24:39) | Barr TL, USA, 2010 | (Barr et al., 2010) |
Abbreviation: GEO, Gene Expression Omnibus.
Table 4.
The top 20 DEGs in IS
| ID | Symbol | Combined.ES | P.Value | FDR | Regulation |
|---|---|---|---|---|---|
| 3399 | ID3 | −1.41974 | 8.42E‐10 | 4.33E‐06 | Down |
| 3384 | ICAM2 | −1.32659 | 2.18E‐09 | 4.33E‐06 | Down |
| 79077 | DCTPP1 | −1.36213 | 2.58E‐09 | 4.33E‐06 | Down |
| 8270 | LAGE3 | −1.31394 | 4.31E‐09 | 6.02E‐06 | Down |
| 1677 | DFFB | −1.32119 | 1.14E‐08 | 1.19E‐05 | Down |
| 7693 | ZNF134 | −1.26287 | 2.46E‐08 | 2.05E‐05 | Down |
| 10838 | ZNF275 | −1.22866 | 2.57E‐08 | 2.05E‐05 | Down |
| 10450 | PPIE | −1.22151 | 2.89E‐08 | 2.11E‐05 | Down |
| 353 | APRT | −1.25353 | 4.45E‐08 | 2.96E‐05 | Down |
| 128240 | APOA1BP | −1.23072 | 4.69E‐08 | 2.96E‐05 | Down |
| 118429 | ANTXR2 | 1.573782 | 7.94E‐10 | 4.33E‐06 | Up |
| 1843 | DUSP1 | 1.313089 | 1.71E‐09 | 4.33E‐06 | Up |
| 84898 | PLXDC2 | 1.514479 | 1.82E‐09 | 4.33E‐06 | Up |
| 5997 | RGS2 | 1.518917 | 1.89E‐09 | 4.33E‐06 | Up |
| 728 | C5AR1 | 1.420349 | 2.23E‐09 | 4.33E‐06 | Up |
| 4082 | MARCKS | 1.423355 | 2.42E‐09 | 4.33E‐06 | Up |
| 25909 | AHCTF1 | 1.349131 | 2.55E‐09 | 4.33E‐06 | Up |
| 136319 | MTPN | 1.420982 | 4.21E‐09 | 6.02E‐06 | Up |
| 8349 | HIST2H2BE | 1.307212 | 8.94E‐‐09 | 1.07E‐05 | Up |
| 329 | BIRC2 | 1.287374 | 1.12E‐08 | 1.19E‐05 | Up |
Abbreviation: DEGs, differentially expressed genes.
Figure 1.

The heat‐map of top 100 DEGs in IS compared to normal control. Row and column represented DEGS and GEO data, respectively. The color scale represented the expression levels. DEGs, differentially expressed genes; GEO, Gene Expression Omnibus; IS, ischemic stroke
3.2. Functional annotation of DEGs
According to the GO enrichment analysis with FDR < 0.05, apoptotic process (FDR = 1.25E‐12), respiratory electron transport chain (FDR = 2.58E‐10) and mitochondrion (FDR = 1.28E‐66) were significantly enriched GO terms. The top 15 GO terms of DEGs in IS were displayed in Figure 2. After the KEGG pathway enrichment analysis (FDR < 0.05), we found that Oxidative phosphorylation (FDR = 4.56E‐09) and Alzheimer's disease (AD) (FDR = 4.56E‐09) were significantly enriched pathways in IS. Top 15 most significantly enriched KEGG pathways of DEGs in IS were shown in Figure 3.
Figure 2.
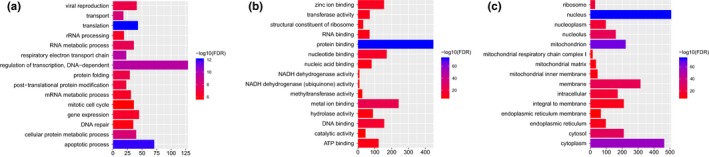
The top 15 most significantly enriched GO terms of DEGs in IS compared to normal control. The x‐axis shows −log FDR and y‐axis shows GO terms (a) Biological process. (b) Molecular function. (c) Cellular component. DEGs, differentially expressed genes; FDR, false discovery rate; GO, gene ontology; IS, ischemic stroke
Figure 3.
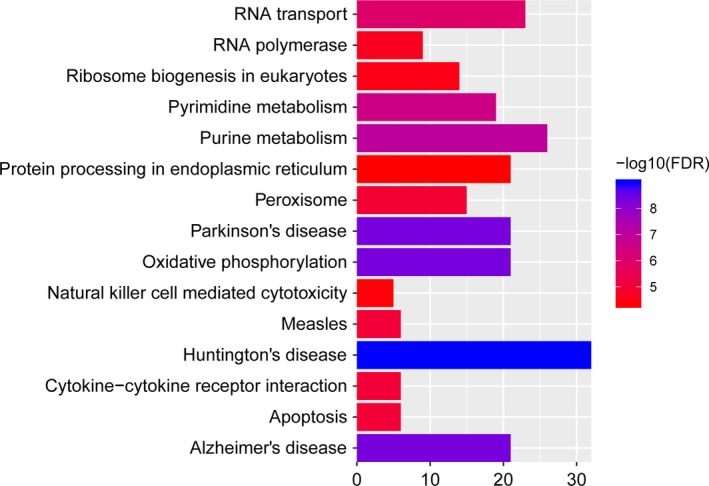
Top 15 significantly enriched pathways of DEGs in IS compared to normal control. The x‐axis shows −log FDR and y‐axis shows KEGG pathways. DEGs, differentially expressed genes; FDR, false discovery rate; IS, ischemic stroke; KEGG, Kyoto Encyclopedia of Genes and Genomes
3.3. Construction of IS‐specific transcriptional regulatory networks
According to TRANSFAC, 35 TFs targeting 20 DEGs (top 10 up‐regulated or down‐regulated genes) were identified. Top four TFs with the most downstream genes (top 20 DEGs) including PAX4, POU2F1, ELK1, and NKX2‐5 (Table 5). Top four TFs and their target gene regulatory network maps were built, which consisted of 22 nodes and 26 edges (Figure 4).
Table 5.
Top 4 TFs that covered the most target genes
| Transcription factor | ID | Count | Genes |
|---|---|---|---|
| PAX4 | 5078 | 9 | PPIE, ZNF275, ANTXR2, DFFB, AHCTF1, BIRC2, ICAM2, C5AR1, DCTPP1 |
| POU2F1 | 5451 | 8 | PPIE, ZNF275, APOA1BP, DUSP1, MARCKS, RGS2, LAGE3, HIST2H2BE |
| ELK1 | 2002 | 5 | ANTXR2, DUSP1, ICAM2, APRT, ZNF134 |
| NKX2‐5 | 1482 | 4 | MTPN, ICAM2, C5AR1, LAGE3 |
Abbreviation: TFs, transcription factors.
Figure 4.
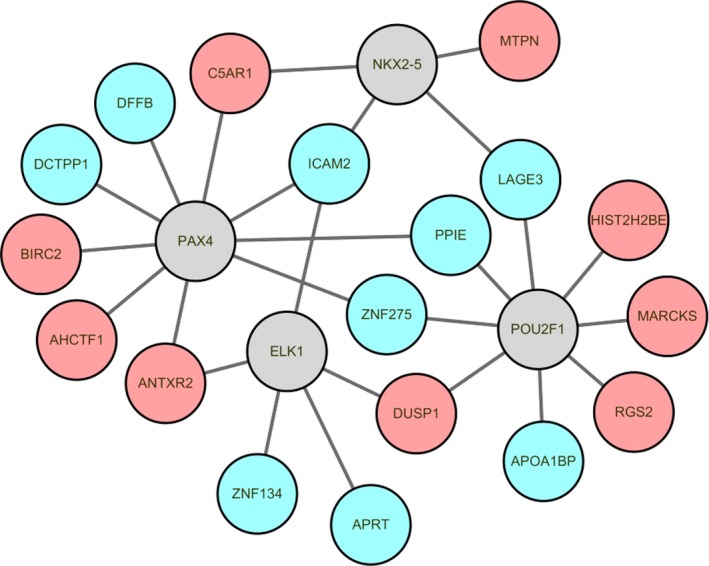
The IS‐specific transcriptional regulatory network. Red‐ and green‐color nodes represent the up‐ and down‐regulated DEGs targeted by TFs, respectively. Gray nodes denote the TFs which predicted to interact with the corresponding DEGs. DEGs, differentially expressed genes; IS, ischemic stroke; TFs, transcription factors
3.4. Validation the expression of DEGs in qRT‐PCR
According to our integrated microarray analysis based on GEO, six DEGs including ID3, ICAM2, DCTPP1, ANTXR2, DUSP1, and RGS2 were selected to perform the quantitative real‐time polymerase chain reaction (qRT‐PCR) confirmation (Figure 5). ID3, ICAM2, DCTPP1, ANTXR2, DUSP1, and RGS2 were top 5 DEGs. Compared to normal control, ID3, ICAM2 and DCTPP1 were down‐regulated while ANTXR2 was up‐regulated in IS in the qRT‐PCR confirmation which was consistent with that in integrated microarray analysis. Compared to normal control, DUSP1 and RGS2 were down‐regulated in IS in qRT‐PCR confirmation while up‐regulated in IS in integrated microarray analysis results. We hypothesized that this inconsistence might be influenced by the small sample size of qRT‐PCR.
Figure 5.

The validation of the expression levels of selected DEGs in IS. The x‐axis shows DEGs and y‐axis shows log2 (fold change) between IS and normal controls. DEGs, differentially expressed genes; IS, ischemic stroke
4. DISCUSSION
To better uncover the pathogenesis and develop novel diagnostic and therapeutic strategies for IS, we performed this integrated analysis between IS patients and normal controls. A total of 1526 genes across the studies were consistently differentially expressed in IS (534 up‐regulated and 992 down‐regulated) with FDR < 0.05. PAX4, POU2F1, ELK1, and NKX2‐5 were top four TFs with the most downstream genes. ID3, ICAM2, DCTPP1, ANTXR2, DUSP1, and RGS2, which were top 10 up‐regulated or down‐regulated DEGs, were selected candidate gene to verify their expression in IS. Except for DUSP1 and RGS2, the other four genes in qRT‐PCR played the same pattern with that in our integrated analysis, suggesting the results of our integration analysis are reliable.
As one of Inhibitor of DNA binding family, ID3 have been known to regulate cell growth, self‐renewal, senescence, angiogenesis, and neurogenesis (Doke, Avecilla, & Felty, 2018). The ID3 is biologically relevant to neurological and behavior research because of its involvement in the stress response, neural plasticity, and neural circuitry (Avecilla, Doke, & Felty, 2017). ID3 was down‐regulated in peripheral blood of IS patient, and identified candidate gene that can accurately detect IS (O'Connell et al., 2016). The diagnostic robustness of the identified 10 candidate genes (ID3 is one of 10 candidate genes) in an independent patient population, and further suggest that it is temporally stable over the first 24 hr of stroke pathology (O'Connell, Chantler, & Barr, 2017). In this study, the expression of ID3 was down‐regulated in the blood of patients with IS in the results of bioinformatics analysis and qRT‐PCR validation. Our results provide further evidence that ID3 may be an important biomarker for the diagnosis of IS.
Intercellular adhesion molecule‐2 (ICAM‐2) belongs to the ICAM family of adhesion proteins. ICAM2 is expressed in vascular endothelial cells and blood cells, and plays a key role in cell‐cell interactions during humoral immunity (Lyck & Enzmann, 2015). ICAM2 also promotes neutrophil binding to and migration through vascular endothelium as a component of immune reactions (Huang et al., 2006). Platelet‐leukocyte aggregation and platelet activation are found to be on the higher side in IS patients, and ICAM2 is an important gene in regulating interaction of platelets with leukocytes. Herein, ICAM2 was down‐regulated in the blood of patients with IS in the results of bioinformatics analysis and qRT‐PCR validation. Transcriptional regulatory networks results showed that ICAM2 was co‐expression with PAX4, ELK1 and NKX2‐5. Therefore, we speculated that ICAM2 may be involved in the occurrence of IS.
ANTXR cell adhesion molecule 2 (ANTXR2), also known as CMG2, encodes a 55‐kDa type I transmembrane protein serves as capillary morphogenesis protein 2 (Youssefian et al., 2017). ANTXR2 also known as the main receptor of the anthrax toxin (Scobie, Rainey, Bradley, & Young, 2003). ANTXR2 was up‐regulated in peripheral blood of IS patient, and identified candidate gene that can accurately detect IS (O'Connell et al., 2016). In this study, the results of bioinformatics analysis and qRT‐PCR validation showed that ANTXR2 was up‐regulated in IS patient. ANTXR2 was co‐expression with PAX4 and ELK1 in transcriptional regulatory networks. These finds indicated ANTXR2 may play a pivotal role in IS.
Base on functional annotation, apoptotic process, respiratory electron transport chain and mitochondrion were significantly enriched GO terms, and Oxidative phosphorylation and AD were significantly enriched pathways in IS. In mitochondrial genome‐wide association study of IS, a genetic score comprised of the sum of all common variants in the mitochondrial genome showed association with IS (Anderson et al., 2011). The associations for small vessel stroke and deep intracerebral hemorrhage suggest that genetic variation in oxidative phosphorylation influences small vessel pathobiology (Anderson et al., 2013). Oxidative phosphorylation plays an important role in neuronal response to oxidative damage (Nicholls, 2008). Is and AD, despite being distinct disease entities, share numerous pathophysiological mechanisms such as those mediated by inflammation, immune exhaustion, and neurovascular unit compromise (Lucke‐Wold et al., 2015). Therefore, we speculated that Oxidative phosphorylation and AD pathway may play an important role in pathogenesis of IS.
In conclusion, our study identified several DEGs, TFs, and pathways in IS which provides clues to understand the pathology and develop diagnostic and therapeutic targets for the IS. The new DEGs, TFs, and pathways of IS obtained in our integrated analysis suggested that integrated microarray analysis is a good way to uncover the molecular mechanism of diseases. However, there are limitations in this study. The number of samples for qRT‐PCR confirmation was small. Further research with larger sample size was performed to confirm our finding and explore the precise role of key DEGs in IS.
CONFLICT OF INTEREST
None.
DATA AVAILABILITY STATEMENT
The dataset supporting the conclusions of this article is included within the article.
Supporting information
ACKNOWLEDGMENTS
We thank Beijing Medintell Bioinformatic Technology Co., LTD for assistance in data analysis.
Zhang Q, Chen W, Chen S, Li S, Wei D, He W. Identification of key genes and upstream regulators in ischemic stroke. Brain Behav. 2019;9:e01319 10.1002/brb3.1319
Qian Zhang and Wenjie Chen contributed equally to this work and should be considered as co‐first authors.
Data Availability Statement: The dataset supporting the conclusions of this article is included within the article.
REFERENCES
- Alieva, A. , Shadrina, M. I. , Filatova, E. V. , Karabanov, A. V. , Illarioshkin, S. N. , Limborska, S. A. , & Slominsky, P. A. (2014). Involvement of endocytosis and alternative splicing in the formation of the pathological process in the early stages of Parkinson's disease. BioMed Research International, 2014, 718732 10.1155/2014/718732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, C. D. , Biffi, A. , Nalls, M. A. , Devan, W. J. , Schwab, K. , Ayres, A. M. , … Rosand, J. (2013). Common variants within oxidative phosphorylation genes influence risk of ischemic stroke and intracerebral hemorrhage. Stroke, 44, 612–619. 10.1161/STROKEAHA.112.672089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, C. D. , Biffi, A. , Rahman, R. , Ross, O. A. , Jagiella, J. M. , Kissela, B. , … Rosand, J. (2011). Common mitochondrial sequence variants in ischemic stroke. Annals of Neurology, 69, 471–480. 10.1002/ana.22108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avecilla, V. , Doke, M. , & Felty, Q. (2017). Contribution of inhibitor of DNA binding/differentiation‐3 and endocrine disrupting chemicals to pathophysiological aspects of chronic disease. BioMed Research International, 2017, 6307109 10.1155/2017/6307109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr, T. L. , Conley, Y. , Ding, J. , Dillman, A. , Warach, S. , Singleton, A. , & Matarin, M. (2010). Genomic biomarkers and cellular pathways of ischemic stroke by RNA gene expression profiling. Neurology, 75, 1009–1014. 10.1212/WNL.0b013e3181f2b37f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi, M. S. , & Chan, L. Y. (2017). Thrombolytic therapy in acute ischemic stroke in patients not fulfilling conventional criteria. The Neurologist, 22, 219–226. 10.1097/NRL.0000000000000149 [DOI] [PubMed] [Google Scholar]
- Doke, M. , Avecilla, V. , & Felty, Q. (2018). Inhibitor of differentiation‐3 and estrogenic endocrine disruptors: Implications for susceptibility to obesity and metabolic disorders. BioMed Research International, 2018, 6821601 10.1155/2018/6821601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnan, G. A. , Fisher, M. , Macleod, M. , & Davis, S. M. (2008). Stroke. The Lancet, 371, 1612–1623. 10.1016/S0140-6736(08)60694-7 [DOI] [PubMed] [Google Scholar]
- Dorrance, A. M. , & Fink, G. (2015). Effects of stroke on the autonomic nervous system. Comprehensive Physiology, 5, 1241–1263. 10.1002/cphy.c140016 [DOI] [PubMed] [Google Scholar]
- Du, J. Y. , Yang, H. , Tian, D. R. , Wang, Q. M. , & He, L. (2014). Identification and functional analysis of differentially expressed genes related to obesity using DNA microarray. Genetics and Molecular Research, 13, 64–72. 10.4238/2014.January.8.5 [DOI] [PubMed] [Google Scholar]
- Evers, S. M. , Struijs, J. N. , Ament, A. J. , van Genugten, M. L. , Jager, J. H. , & van den Bos, G. A. (2004). International comparison of stroke cost studies. Stroke, 35, 1209–1215. 10.1161/01.STR.0000125860.48180.48 [DOI] [PubMed] [Google Scholar]
- Huang, M. T. , Larbi, K. Y. , Scheiermann, C. , Woodfin, A. , Gerwin, N. , Haskard, D. O. , & Nourshargh, S. (2006). ICAM‐2 mediates neutrophil transmigration in vivo: Evidence for stimulus specificity and a role in PECAM‐1‐independent transmigration. Blood, 107, 4721–4727. 10.1182/blood-2005-11-4683 [DOI] [PubMed] [Google Scholar]
- Kong, P. , Lei, P. , Zhang, S. , Li, D. , Zhao, J. , & Zhang, B. (2018). Integrated microarray analysis provided a new insight of the pathogenesis of Parkinson's disease. Neuroscience Letters, 662, 51–58. 10.1016/j.neulet.2017.09.051 [DOI] [PubMed] [Google Scholar]
- Krug, T. , Gabriel, J. P. , Taipa, R. , Fonseca, B. V. , Domingues‐Montanari, S. , Fernandez‐Cadenas, I. , … Oliveira, S. A. (2012). TTC7B emerges as a novel risk factor for ischemic stroke through the convergence of several genome‐wide approaches. Journal of Cerebral Blood Flow and Metabolism, 32, 1061–1072. 10.1038/jcbfm.2012.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Dani, J. A. , & Le, W. (2009). The role of transcription factor Pitx3 in dopamine neuron development and Parkinson's disease. Current Topics in Medicinal Chemistry, 9, 855–859. [PMC free article] [PubMed] [Google Scholar]
- Lucke‐Wold, B. P. , Turner, R. C. , Logsdon, A. F. , Simpkins, J. W. , Alkon, D. L. , Smith, K. E. , … Rosen, C. L. (2015). Common mechanisms of Alzheimer's disease and ischemic stroke: The role of protein kinase C in the progression of age‐related neurodegeneration. Journal of Alzheimer's Disease, 43, 711–724. 10.3233/JAD-141422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyck, R. , & Enzmann, G. (2015). The physiological roles of ICAM‐1 and ICAM‐2 in neutrophil migration into tissues. Current Opinion in Hematology, 22, 53–59. 10.1097/MOH.0000000000000103 [DOI] [PubMed] [Google Scholar]
- Nicholls, D. G. (2008). Oxidative stress and energy crises in neuronal dysfunction. Annals of the New York Academy of Sciences, 1147, 53–60. 10.1196/annals.1427.002 [DOI] [PubMed] [Google Scholar]
- O'Connell, G. C. , Chantler, P. D. , & Barr, T. L. (2017). Stroke‐associated pattern of gene expression previously identified by machine‐learning is diagnostically robust in an independent patient population. Genomics Data, 14, 47–52. 10.1016/j.gdata.2017.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell, G. C. , Petrone, A. B. , Treadway, M. B. , Tennant, C. S. , Lucke‐Wold, N. , Chantler, P. D. , & Barr, T. L. (2016). Machine‐learning approach identifies a pattern of gene expression in peripheral blood that can accurately detect ischaemic stroke. NPJ Genomic Medicine, 1, 16038 10.1038/npjgenmed.2016.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scobie, H. M. , Rainey, G. J. , Bradley, K. A. , & Young, J. A. (2003). Human capillary morphogenesis protein 2 functions as an anthrax toxin receptor. Proceedings of the National Academy of Sciences, 100, 5170–5174. 10.1073/pnas.0431098100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volny, O. , Kasickova, L. , Coufalova, D. , Cimflova, P. , & Novak, J. (2015). MicroRNAs in cerebrovascular disease. Advances in Experimental Medicine and Biology, 888, 155–195. 10.1007/978-3-319-22671-2_9 [DOI] [PubMed] [Google Scholar]
- Youssefian, L. , Vahidnezhad, H. , Aghighi, Y. , Ziaee, V. , Zeinali, S. , Abiri, M. , & Uitto, J. (2017). Hyaline fibromatosis syndrome: A novel mutation and recurrent founder mutation in the CMG2/ANTXR2 gene. Acta Dermato Venereologica, 97, 108–109. 10.2340/00015555-2459 [DOI] [PubMed] [Google Scholar]
- Zhao, B. , Wang, M. , Xu, J. , Li, M. , & Yu, Y. (2017). Identification of pathogenic genes and upstream regulators in age‐related macular degeneration. BMC Ophthalmology, 17, 102 10.1186/s12886-017-0498-z [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The dataset supporting the conclusions of this article is included within the article.


