Abstract
A high diversity of culturable foliar endophytic fungi is known from various mangrove plants, and the core taxa include species from Colletotrichum, Pestalotiopsis, Phoma, Phomopsis, Sporomiella, among others. Since a small fraction of fungi is able to grow in culture, this study investigated the diversity of fungi associated with leaves of Acanthus ilicifolius var. xiamenensis using both isolation and metabarcoding approaches. A total of 203 isolates were cultured from surface-sterilized leaves, representing 47 different fungal species: 30 species from the winter samples (104 isolates), and 26 species from the summer samples (99 isolates). Ascomycota was dominant in both types of leaf samples, while Basidiomycota was isolated only from the summer samples. Drechslera dematioidea (10.58%, percentage of occurrence), Colletotrichum sp. 3 (7.69%) and Alternaria sp. (7.69%) were dominant in the winter samples; Fusarium oxysporum (13.13%), Diaporthe endophytica (10.10%) and Colletotrichum sp. 1 (9.09%) in the summer samples. Overall, Corynespora cassiicola (6.90%), F. oxysporum (6.40%) and Guignardia sp. (6.40%) had the highest overall percentage of occurrence. In the metabarcoding analysis, a total of 111 operational taxonomic units (OTUs) were identified from 17 leaf samples: 96 OTUs from the winter and 70 OTUs from the summer samples. Sequences belonging to Ascomycota and Basidiomycota were detected in both samples but the former phylum was dominant over the latter. Based on read abundance, taxa having the highest percentage of occurrence included Alternaria sp. (3.46%), Cladosporium delicatulum (2.56%) and Pyrenochaetopsis leptospora (1.41%) in the winter leaves, and Aureobasidium sp. (10.72%), Cladosporium sp. (7.90%), C. delicatulum (3.45%) and Hortaea werneckii (3.21%) in the summer leaves. These latter four species also had the highest overall percentage of occurrence. Combining the results from both methods, a high diversity of fungi (at least 110 species) was found associated with leaves of A. ilicifolius var. xiamenensis. Many of the fungi identified were plant pathogens and may eventually cause diseases in the host.
Keywords: High throughput sequencing, Terrestrial fungi, rDNA, Illumina sequencing, Culturomics, Endophytes
Introduction
Mangroves are tropical intertidal forest communities, situated at coastal areas from low to high salinities (Tomlinson, 1986). These communities host both terrestrial and marine fungi: terrestrial fungi, such as endophytic fungi, occur on the aerial parts of the plants, while marine fungi usually grow on the submerged/intertidal dead branches of the trees. Endophytic fungi inhabit plant organs for some time in their life cycle, and they can colonize internal plant tissues without causing apparent harm to the host (Petrini, 1991). Arnold (2007) revised the definition of endophytic fungi as ‘a polyphyletic group of highly diverse, primarily ascomycetous fungi, defined functionally by their occurrence within asymptomatic tissues of plants’.
For the last decade, various mangrove plants were examined for their endophytic fungal assemblages. The Ascomycota was dominant with many asexual species while the Basidiomycota was rare (De Souza Sebastianes et al., 2013; Zhou et al., 2018). Pang et al. (2008) summarized the dominant endophytic fungi of various mangrove plant species and there were several common taxa: Sporormiella minima, Guignardia/Phyllosticta spp., Phoma spp., Diaporthe/Phomopsis spp., Cladosporium spp., Acremonium spp. and Collectotrichum spp. Xylaria spp. and Pestalotiopsis spp. were also common (Suryanarayanan & Kumaresan, 2000; Chaeprasert et al., 2010; Xing & Guo, 2011; De Souza Sebastianes et al., 2013). Abundance and richness of endophytic fungi of mangrove plants are dependent on mangrove plant species and also their tissue types, i.e., stem, leaf or root (Xing et al., 2011; Zhou et al., 2018). Avicennia germinans was found to support the lowest diversity of endophytic fungi compared with Laguncularia racemosa and Rhizophora mangle, and it was concluded to be the effect of salt excreted from leaves of A. germinans, which inhibits spore germination (Gilbert, Mejía-Chang & Rojas, 2002). De Souza Sebastianes et al. (2013) studied endophytic fungi in branches and leaves of Rhizophora mangle, Avicennia schaueriana and Laguncularia racemosa and found that branches had a higher frequency of colonization and diversity than leaves. A higher number of isolates and species richness were also obtained from stems than roots in four species of Rhizophoraceae mangrove plants (Xing & Guo, 2011). Roots of mangrove plants are inhabited with terrestrial, freshwater and marine fungi (Ananda & Sridhar, 2002). Using high throughput sequencing techniques, Arfi et al. (2012) found that different fungal classes/orders were dominant in Avicennia marina and Rhizophora stylosa and between aerial and intertidal parts of the trees. Some endophytic fungi are host-specific and their diversity is seasonally varied (Suryanarayanan, Kumaresan & Johnson, 1998; Costa, Maia & Cavalcanti, 2012). Diversity of endophytic fungi increased with leaf age and some fungi may switch from an endophytic lifestyle to a saprobic one after leaf fall (Kumaresan & Suryanarayanan, 2002).
Acanthus ilicifolius var. xiamenensis is a mangrove plant distributed along the coast of southern China. The only distribution of A. ilicifolius var. xiamenensis in Taiwan is at Liuyu Township, Kinmen County with two small patches. However, their survival is under threat due to construction work for urban development. Previous studies on endophytic fungal assemblages associated with A. ilicifolius found that Colletotrichum spp. and Phomopsis spp. were the dominant species (Suryanarayanan & Kumaresan, 2000; Chaeprasert et al., 2010). This study investigates the cultural diversity of endophytic fungi of surface-sterilized healthy leaves of A. ilicifolius var. xiamenensis and the diversity of fungi of the same leaves using Illumina MiSeq sequencing.
Materials and Methods
Collection of samples
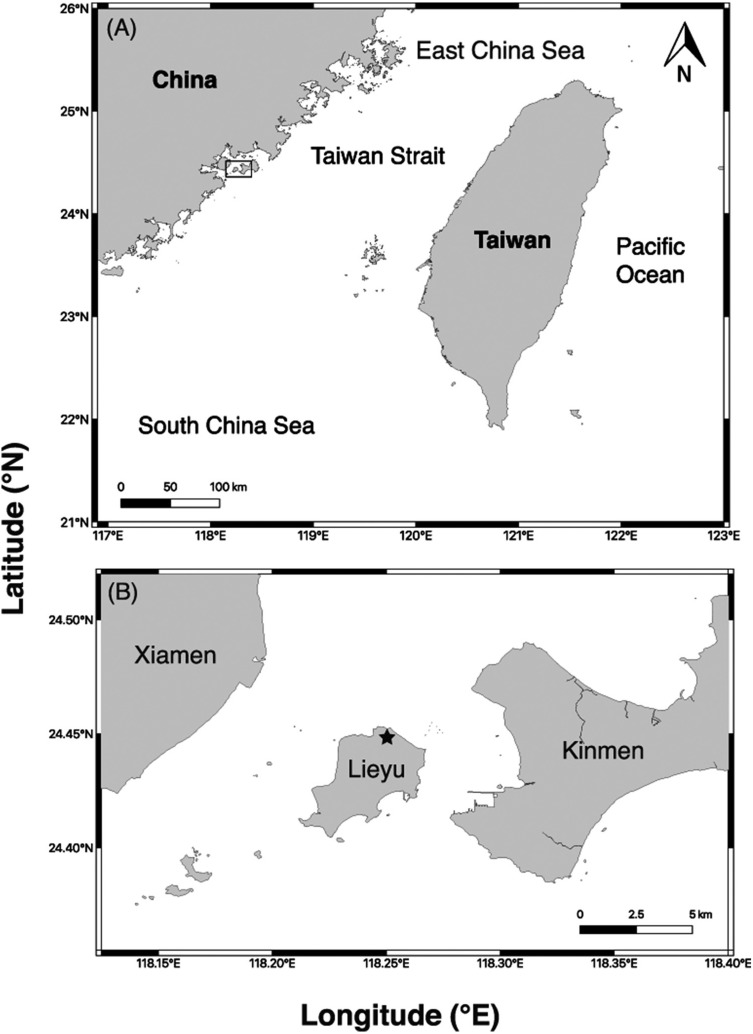
The mangrove plants Aegiceras corniculatum, Acanthus ilicifolius var. xiamenensis and Kandelia obovata are present at Lieyu Township, Kinmen County, Taiwan. A. ilicifolius var. xiamenensis is the only mangrove plant growing at the sampling site at Lieyu Township and it represents the only distribution in Taiwan (Fig. 1). The characteristics of A. ilicifolius var. xiamenensis are shown in Fig. 2. Healthy leaves (i.e., for isolation of endophytic over saprobic/pathogenic fungi) were collected on 16 January (60 leaves) and 11 July (35 leaves) 2014, placed in a cool box and transported to the laboratory at National Taiwan Ocean University for immediate fungal isolation.
Figure 1. Sampling site.
(A) Location of Kinmen County (box), Taiwan; (B) distribution of Acanthus ilicifolius var. xiamenensis at Lieyu Township, where the samples were collected (star).
Figure 2. Morphology ofAcanthus ilicifolius var. xiamenensis.
(A) Trees, (B) healthy leaves, (C) flowers, and (D) fruits surrounded by unhealthy leaves.
Fungal isolation
Leaves were washed with tap water to remove surface dirt. Four discs (6 mm in diameter) were cut out from each leaf, surface-sterilized by immersing in 70% ethanol for 10 s and 4% sodium hypochlorite solution for 30 s, washed twice in sterile distilled water, and plated on 2% malt extract freshwater agar (MEAF, BD Bacto™; BD Biosciences, Sparks, MD, USA), supplemented with 0.5 g/L each of streptomycin sulfate (Sigma-Aldrich, MO, USA) and Penicillin G (Sigma-Aldrich, MO, USA). The inoculated plates were incubated at 25 °C and checked daily to observe fungal growth from the leaf discs for 1 month. Hyphal tips of different mycelial morphotypes from each plate (i.e., from the same leaf) were isolated and subcultured onto fresh MEAF. All cultures were kept at National Taiwan Ocean University.
Identification of fungal isolates
All isolated cultures were grouped into different colony morphologies, and identified by comparing their ITS sequences with those deposited in the National Center for Biotechnology Information (NCBI). Mycelia for each morphotype were ground into fine powder in liquid nitrogen using a mortar and pestle. Genomic DNA was extracted using the DNeasy Plant DNA Extraction Kit (Qiagen, Germantown, MD, USA) according to the manufacturer’s instructions. ITS was amplified using the primer pairs ITS1 (or ITS5)/ITS4 (White et al., 1990). PCR reactions were performed in a 25 µL volume containing ca. 20 ng DNA, 0.2 µM of each primer, 0.2 mM of each dNTP, 2.5 mM MgCl2 and 1.25 U of Taq Polymerase (Invitrogen, Sao Paulo, Brazil). The amplification cycle consisted of an initial denaturation step of 95 °C for 2 min followed by 35 cycles of (a) denaturation (95 °C for 1 min), (b) annealing (54 °C for 1 min) and (c) elongation (72 °C for 1.5 min) and a final 10 min elongation step at 72 °C. The PCR products were analysed by agarose gel electrophoresis and sent to Genomics BioSci & Tech (New Taipei City, Taiwan) for sequencing. The sequences returned were checked for ambiguity and the forward/reverse strands were assembled in MEGA7 (Kumar, Stecher & Tamura, 2016). The assembled sequences were submitted to NCBI for a nucleotide BLAST search. The ITS sequences of the fungal isolates were deposited in NCBI with the accession numbers given in Table 1.
Table 1. Fungi isolated from surface-sterilized leaves of Acanthus ilicifolius var. xiamenensis in summer and winter sampling.
Identity was based on BLAST searches in NCBI and percentage of occurrence of fungi was calculated based on number of isolates.
| Isolate number (NTOU) (accession number) | Sequence length (bp) | Phylum | Class | Order | Family | Taxa | Maximum score | Coverage(%) | Similarity(%) | Matched sequence(s) | Occurrence (%) | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Winter | Summer | Total | |||||||||||
| 4398 (MK448262) | 529 | Ascomycota | Dothideomycetes | Capnodiales | Teratosphaeriaceae | Acidiella uranophila | 832 | 100 | 95 | JQ904602 | 0.96 | 0.00 | 0.49 |
| 4330 (MK432953), 4899 (MK432954), 4902 (MK432955), 4904 (MK432956) | 485–544 | Ascomycota | Dothideomycetes | Pleosporales | Pleosporaceae | Alternaria alternata | 896–1005 | 100 | 100 | LC317410, MF422130 | 1.92 | 7.07 | 4.43 |
| 4336 (MK448263), 4368 (same colony morphology as 4336) | 543 | Ascomycota | Dothideomycetes | Pleosporales | Pleosporaceae | Alternaria sp. | 1003 | 100 | 100 | KY190102 | 7.69 | 0.00 | 3.94 |
| 4350 (MK448264) | 556 | Ascomycota | Dothideomycetes | Dothideales | Dothioraceae | Aureobasidium pullulans | 1027 | 100 | 100 | LC277149, LC277150 | 1.92 | 0.00 | 0.99 |
| 4909 (MK432957) | 549 | Ascomycota | Dothideomycetes | Dothideales | Dothioraceae | Aureobasidium sp. | 1014 | 100 | 100 | KF367567 | 0.00 | 1.01 | 0.49 |
| 4875 (MK448265) | 557 | Ascomycota | Dothideomycetes | Botryosphaeriales | Botryosphaeriaceae | Botryosphaeria dothidea | 1029 | 100 | 100 | KU686880 | 0.00 | 3.03 | 1.48 |
| 4340 (MK448266) | 527 | Ascomycota | Dothideomycetes | Capnodiales | Cladosporiaceae | Cladosporium dominicanum | 974 | 100 | 100 | MF472969, MF472970 | 1.92 | 0.00 | 0.99 |
| 4352 (MK432958), 4883 (MK432959) | 524–525 | Ascomycota | Dothideomycetes | Capnodiales | Cladosporiaceae | Cladosporium sp. | 968–970 | 100 | 100 | MG701131, MG572462 | 0.96 | 4.04 | 2.46 |
| 4372 (MK448279), 4386 (MK448280) | 474–492 | Ascomycota | Sordariomycetes | Glomerellales | Glomerellaceae | Colletotrichum boninense | 837–876 | 100 | 99 | FJ981604 | 1.92 | 0.00 | 0.99 |
| 4358 (MK448267), 4402 (MK448268) | 567 | Ascomycota | Sordariomycetes | Glomerellales | Glomerellaceae | Colletotrichum hippeastri | 1001–1011 | 99 | 99 | KR183779 | 4.81 | 0.00 | 2.46 |
| 4370 (MK432992), 4895 (MK432993), 4908 (MK432988) | 515–567 | Ascomycota | Sordariomycetes | Glomerellales | Glomerellaceae | Colletotrichum sp. 1 | 952–1048 | 100 | 100 | MF076596, JN715846 | 2.88 | 9.09 | 5.91 |
| 4326 (MK448269), 4378 (MK448281), 4390 (MK448282) | 536–553 | Ascomycota | Sordariomycetes | Glomerellales | Glomerellaceae | Colletotrichum sp. 2 | 972–1003 | 100 | 99 | HM357614 | 3.85 | 0.00 | 1.97 |
| 4324 (MK432994), 4356 (MK432995), 4364 (MK432989), 4903 (MK432996) | 533–549 | Ascomycota | Sordariomycetes | Glomerellales | Glomerellaceae | Colletotrichum sp. 3 | 985–1013 | 100 | 99–100 | KX620331, KX620330, KY820893 | 7.69 | 1.01 | 4.43 |
| 4346 (MK432960), 4362 (MK432961), 4872 (MK432962), 4889 (MK432963) | 490–533 | Ascomycota | Dothideomycetes | Pleosporales | Corynesporascaceae | Corynespora cassiicola | 898–985 | 99–100 | 99–100 | FJ852578, KF266787, HM535404 | 5.77 | 8.08 | 6.90 |
| 4905 (MK448270), 4907 (MK448271) | 546 | Ascomycota | Sordariomycetes | Xylariales | Hypoxylaceae | Daldinia eschscholtzii | 1003–1009 | 100 | 99–100 | KY792621 | 0.00 | 4.04 | 1.97 |
| 4380 (MK432964), 4869 (same colony morphology as 4380), 4884 (MK432965), 4920 (MK432966) | 550 | Ascomycota | Sordariomycetes | Diaporthales | Diaporthaceae | Diaporthe endophytica | 1011–1016 | 100 | 99–100 | NR_111847 | 0.96 | 10.10 | 5.42 |
| 4886 (MK448272) | 551 | Ascomycota | Sordariomycetes | Diaporthales | Diaporthaceae | Diaporthe longicolla | 1016 | 99 | 100 | JQ754023 | 0.00 | 2.02 | 0.99 |
| 4382 (MK448253) | 552 | Ascomycota | Sordariomycetes | Diaporthales | Diaporthaceae | Diaporthe perseae | 974 | 100 | 98.55 | KC343173 | 0.96 | 0.00 | 0.49 |
| 4376 (MK448273), 4915 (MK448274) | 550–551 | Ascomycota | Sordariomycetes | Diaporthales | Diaporthaceae | Diaporthe phaseolorum | 1002–1011 | 100 | 99 | LN828206, KT964565 | 5.77 | 1.01 | 3.45 |
| 4334 (MK432967), 4354 (MK432968), 4400 (same colony morphology as 4334), 4404 (same colony morphology as 4334), 4878 (MK432998) | 519 | Ascomycota | Dothideomycetes | Pleosporales | Didymellaceae | Didymella sp. | 948 | 100 | 99 | HM012812 | 5.77 | 2.02 | 3.94 |
| 4901 (MK448275) | 533 | Ascomycota | Dothideomycetes | Dothideales | Dothioraceae | Dothioraceae sp. | 894 | 99 | 97 | KU892278 | 0.00 | 1.01 | 0.49 |
| 4410 (MK448276) | 544 | Ascomycota | Dothideomycetes | Pleosporales | Pleosporaceae | Drechslera dematioidea | 1005 | 100 | 100 | KY788112 | 10.58 | 0.00 | 5.42 |
| 4870 (MK448277), 4873 (MK448278), 4879 (MK448283), 4885 (MK448284), 4896 (MK448285), 4898 (MK448286) | 467–518 | Ascomycota | Sordariomycetes | Hypocreales | Nectriaceae | Fusarium oxysporum | 863–957 | 100 | 100 | MG727665, MG722826 | 0.00 | 13.13 | 6.40 |
| 4866 (MK432970), 4876 (MK432971), 4877 (MK432972) | 488–532 | Ascomycota | Sordariomycetes | Hypocreales | Nectriaceae | Fusarium sp. | 902–983 | 100 | 100 | MG562501, MG274294 | 0.00 | 8.08 | 3.94 |
| 4318 (MK432973), 4332 (MK432974), 4388 (MK432975), 4871 (MK432976), 4874 (MK432977) | 587–613 | Ascomycota | Dothideomycetes | Botryosphaeriales | Botryosphaeriaceae | Guignardia sp. | 1085–1133 | 100 | 100 | JQ341114, MF170677, LN828209, JN791605 | 5.77 | 7.07 | 6.40 |
| 4320 (MK432978), 4396 (MK432979) | 432–523 | Ascomycota | Dothideomycetes | Capnodiales | Teratosphaeriaceae | Hortaea werneckii | 798–966 | 100 | 100 | GQ334389, KY434149 | 3.85 | 0.00 | 1.97 |
| 4348 (MK448249) | 525 | Ascomycota | Sordariomycetes | Xylariales | Apiosporaceae | Nigrospora sphaerica | 970 | 100 | 100 | MH028054, MG669225 | 3.85 | 0.00 | 1.97 |
| 4868 (MK432980) | 720 | Ascomycota | Sordariomycetes | Xylariales | Xylariaceae | Nodulisporium sp. | 1297 | 100 | 99 | KR016438 | 0.00 | 2.02 | 0.99 |
| 4408 (MK448250) | 510 | Ascomycota | Dothideomycetes | Pleosporales | Phaeosphaeriaceae | Parastagonospora phoenicicola | 846 | 100 | 97 | KY173428 | 1.92 | 0.00 | 0.99 |
| 4914 (MK432981) | 579 | Ascomycota | Sordariomycetes | Amphisphaeriales | Pestalotiopsidaceae | Pestalotiopsis microspora | 1070 | 100 | 100 | KX755255 | 0.00 | 1.01 | 0.49 |
| 4394 (MK448260) | 550 | Ascomycota | Dothideomycetes | Capnodiales | Mycosphaerellaceae | Phaeophleospora eucalypticola | 1016 | 100 | 100 | NR_145123 | 0.96 | 0.00 | 0.49 |
| 4906 (MK432982) | 618 | Basidiomycota | Agaricomycetes | Polyporales | Phanerochaetaceae | Phanerina mellea | 1136 | 100 | 99 | KX752602 | 0.00 | 1.01 | 0.49 |
| 4917 (MK440618) | 670 | Basidiomycota | Agaricomycetes | Hymenochaetales | Hymenochaetaceae | Phellinus noxius | 1218 | 99 | 99 | KF233592 | 0.00 | 2.02 | 0.99 |
| 4406 (MK448251) | 508 | Ascomycota | Dothideomycetes | Pleosporales | Phoma sp. 1 | 939 | 100 | 100 | KY780194 | 1.92 | 0.00 | 0.99 | |
| 4338 (MK432990), 4366 (MK432991) | 465 | Ascomycota | Dothideomycetes | Pleosporales | Phoma sp. 2 | 859 | 100 | 100 | JX157864 | 2.88 | 0.00 | 1.48 | |
| 4384 (MK448252) | 551 | Ascomycota | Sordariomycetes | Diaporthales | Diaporthaceae | Phomopsis asparagi | 1007 | 100 | 99 | JQ613999 | 0.96 | 0.00 | 0.49 |
| 4918 (MK432997) | 554 | Ascomycota | Sordariomycetes | Diaporthales | Diaporthaceae | Phomopsis sp. | 883 | 97 | 96 | AB245060 | 0.00 | 3.03 | 1.48 |
| 4893 (MK432983) | 517 | Ascomycota | Dothideomycetes | Capnodiales | Mycosphaerellaceae | Pseudocercospora nymphaeacea | 955 | 100 | 100 | KY304491 | 0.00 | 1.01 | 0.49 |
| 4374 (MK448254) | 518 | Ascomycota | Dothideomycetes | Capnodiales | Mycosphaerellaceae | Pseudocercospora sp. | 957 | 100 | 100 | KP896027 | 1.92 | 0.00 | 0.99 |
| 4892 (MK448255) | 532 | Ascomycota | Dothideomycetes | Capnodiales | Dissoconiaceae | Ramichloridium punctatum | 839 | 100 | 95 | MF319925 | 0.00 | 2.02 | 0.99 |
| 4890 (MK448256), 4891 (MK448261) | 518–569 | Ascomycota | Dothideomycetes | Pleosporales | Phaeosphaeriaceae | Septoriella hubertusii | 920–970 | 95–100 | 99 | KT827267 | 0.00 | 2.02 | 0.99 |
| 4328 (MK448257) | 603 | Ascomycota | Dothideomycetes | Pleosporales | Lentitheciaceae | Setoseptoria arundinacea | 1075 | 97 | 99 | LC014594 | 0.96 | 0.00 | 0.49 |
| 4360 (MK448258) | 518 | Ascomycota | Dothideomycetes | Pleosporales | Stagonosporopsis cucurbitacearum | 957 | 100 | 100 | KU059901, AB714985, AB714984 | 0.96 | 0.00 | 0.49 | |
| 4392 (MK448259) | 537 | Ascomycota | Dothideomycetes | Capnodiales | Teratosphaeriaceae | Teratosphaeria capensis | 782 | 100 | 93 | JN712501 | 4.81 | 0.00 | 2.46 |
| 4916 (MK432984) | 593 | Basidiomycota | Agaricomycetes | Polyporales | Polyporaceae | Tinctoporellus epimiltinus | 1085 | 100 | 99 | KY948722 | 0.00 | 2.02 | 0.99 |
| 4900 (MK432985) | 564 | Ascomycota | Sordariomycetes | Xylariales | Xylariaceae | Xylaria sp. | 1040 | 99 | 100 | JQ388255 | 0.00 | 2.02 | 0.99 |
| 4322 (MK432986), 4344 (MK432987) | 460–513 | Ascomycota | Dothideomycetes | Capnodiales | Mycosphaerellaceae | Zasmidium citri | 845–937 | 100 | 99 | GU066616 | 2.88 | 0.00 | 1.48 |
Metabarcoding
Seventeen leaves used for the isolation described above were freeze-dried. Total genomic DNA was extracted using QIAGEN DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. A nested PCR approach was used to amplify a region of ITS spanning from 18S to 5.8S rDNA. The first set of primers was NSA3 (5′-AAACTCTGTCGTGCTGGGGATA-3′)/NLC2 (5′-GAGCTGCATTCCCAAACAACTC-3′) (Martin & Rygiewicz, 2005) and the second set was ITS1-F_KYO1 (5′-CTHGGTCATTTAGAGGAASTAA-3′)/ITS2 (5′-GCTGCGTTCTTCATCGATGC-3′) (White et al., 1990; Toju et al., 2012). Adapters were added to the 5′ end of the primers ITS1-F_KYO1 and ITS2. PCR amplification cycle with NSA3/NLC2 primers consisted of an initial denaturation step of 94 °C for 5 min, followed by 35 cycles of 94 °C for 30 s, 55 °C for 30 s and 72 °C for 30 s, and a final 5-min elongation step at 72 °C. For ITS1-F_KYO1/ITS2, the amplification consisted of an initial denaturation step of 95 °C for 10 min, followed by 35 cycles of 95 °C for 30 s, 55 °C for 30 s and 72 °C for 30 s, and a final 72 °C for 7 min. The PCR products were analyzed by agarose gel electrophoresis. For each leaf, five successful PCR products were pooled and purified using EasyPureTM PCR Clean up/Gel Extraction Kit (Bioman, New Taipei City, Taiwan) according to manufacturer’s instructions. The purified product was shipped to Genomics (Taipei, Taiwan) for Illumina MiSeq sequencing.
The raw sequences were filtered with a phred score ≥Q29 (a base call accuracy of ≥99.87%). The raw reads were paired into single reads and adaptors, primers and barcode sequences were removed using the QIIME script “split_library.py” (Caporaso et al., 2010). Clustering was performed using uclust v1.2.22q (Edgar, 2010) in QIIME 1.9.0 (Caporaso et al., 2010). The reads were processed with UCHIME (Edgar et al., 2011) to reject chimeric sequences. Picking of Operation Taxonomic Units (OTUs) and taxonomic assignments were performed with an open-reference OTU picking approach against the UNITE database in QIIME 1.9.0 (Caporaso et al., 2010). A similarity threshold of 97% was adopted. Taxonomic assignment of representative OTUs was run at a 0.97 confidence threshold against the UNITE ITS1 database with UNITE 7.2 reference OTU database (“UNITE+INSD” dataset) using the assignTaxonomy method (Kõljalg et al., 2013).
Statistical analysis
Total number of isolates (total abundance, N), Richness (total number of taxa in the community, S), Species Richness (Margalef), Shannon–Wiener Diversity Index, Pielou’s Evenness and Simpson Diversity Indices (Simpson’s Index, Simpson’s Index of Diversity, Simpson’s Reciprocal Index) were calculated in Microsoft Excel by first computing the variables of the equations and then using the math operators to calculate the different indices.
Rarefaction and extrapolation sampling curves were computed and plotted to estimate sample completeness (sample coverage) in R package iNEXT (iNterpolation/EXTrapolation) with the 95% lower and upper confidence limits for the isolation and metabarcoding data (Hsieh, Ma & Chao, 2016). A principle component analysis (PCA) was calculated by the R software using the R function prcomp() (R Core Team, 2013).
Results
Diversity of culturable fungi
A total of 203 isolates were cultured from leaves of Acanthus ilicifolius var. xiamenensis collected in January and July 2014 at Kinmen Township, Taiwan and ITS of the representative isolate for each morphotype was sequenced (Tables 1–2). The fungi were identified down to species level when the BLAST search results had a high percentage coverage and identity in NCBI; otherwise, they were given an identity at the genus/family level.
Table 2. Diversity indices of fungi associated with leaves of Acanthus ilicifolius var. xiamenensis using culture and metabarcoding analysis.
| Culture | Metabarcoding analysis | |||||
|---|---|---|---|---|---|---|
| Winter | Summer | Total | Winter | Summer | Total | |
| Total No. of isolates/reads (Total Abundance), N | 104 | 99 | 203 | 314692 | 458993 | 773685 |
| Richness (Total number of Taxa in the community), S | 30 | 26 | 47 | 96 | 70 | 111 |
| Species Richness (Margalef): d = (S−1)/ln(N) | 6.24 | 5.44 | 8.66 | 7.50 | 5.29 | 8.11 |
| Shannon-Wiener Diversity Index: H′= −Σ[Pi ln(Pi)] | 3.15 | 2.92 | 3.83 | 1.98 | 2.09 | 2.28 |
| Pielou’s Evenness: J′= H′/ln(S) | 0.93 | 0.90 | 0.99 | 0.44 | 0.49 | 0.49 |
| Simpson Diversity Indices: | ||||||
| Simpson’s Index: D = Σ(Pi2) | 0.05 | 0.07 | 0.29 | 0.29 | 0.23 | 0.18 |
| Simpson’s Index of Diversity: 1−D = 1−Σ(Pi2) | 0.95 | 0.93 | 0.71 | 0.72 | 0.78 | 0.82 |
| Simpson’s Reciprocal Index: 1/D | 19.52 | 14.74 | 3.47 | 3.51 | 4.45 | 5.60 |
A total of 104 and 99 isolates were cultured from the winter (January) and summer (July) samples, representing 30 and 26 fungal species, respectively (Tables 1–2). Nine species were common between the two sampling times, therefore, 47 different fungal species were isolated from leaves of A. ilicifolius var. xiamenensis. The higher percentage of occurrence (Table 1) in the winter samples included Drechslera dematioidea (10.58%), Colletotrichum sp. 3 (7.69%) and Alternaria sp. (7.69%); and in the summer samples, Fusarium oxysporum (13.13%), Diaporthe endophytica (10.10%), Colletotrichum sp. 1 (9.09%), Fusarium sp. (8.08%), Corynespora cassiicola (8.08%), Guignardia sp. (7.07%) and Alternaria alternata (7.07%). Overall, C. cassiicola (6.90%), F. oxysporum (6.40%), Guignardia sp. (6.40%), Colletotrichum sp. 1 (5.91%), D. endophytica (5.42%) and D. dematioidea (5.42%) had the highest percentage of occurrence.
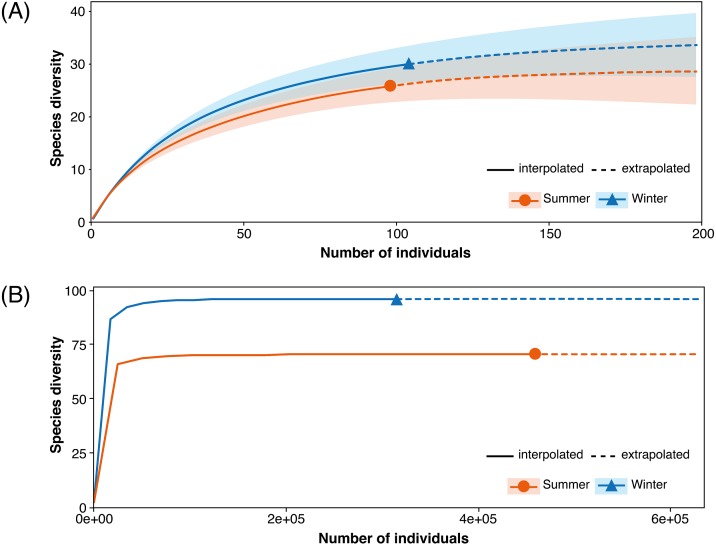
Diversity indices were calculated for the fungal communities in the winter and summer samples (Table 2). The fungal community in the winter samples had a higher species richness of 6.24 (Margalef) and a higher diversity of 3.15 (Shannon–Wiener Diversity Index) than that in the summer samples (5.44 and 2.92, respectively). The Margalef and Shannon–Wiener Diversity indices with the data combining the two seasons were 8.66 and 3.83, respectively. The rarefaction and extrapolation analysis suggested that species diversity was projected to be higher in the winter samples than in the summer samples but both samples did not reach species saturation (Fig. 3A).
Figure 3. Sample-size-based rarefaction and extrapolation sampling curves.
(A) Isolation and (B) metabarcoding studies of endophytic fungi associated with Acanthus ilicifolius var. xiamenensis.
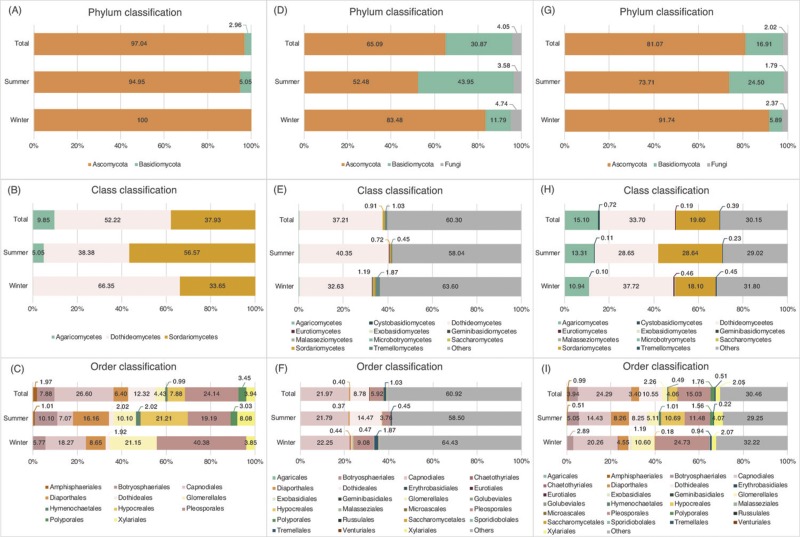
Figures 4A–4C show the taxonomic composition of the cultured fungi at different taxonomic levels. In the winter (January 2014), only Ascomycota was isolated with no Basidiomycota; in the summer, Basidiomycota had a ∼5% occurrence (Fig. 4A). At the class level, Dothideomycetes and Sordariomycetes were the dominant classes in both seasons and Agaricomycetes was only isolated from the summer samples (Fig. 4B). At the ordinal level, the richness of fungi in summer was higher than that in winter. Seven orders Botryosphaeriales, Capnodiales, Diaporthales, Dothideales, Glomerellales, Pleosporales and Xylariales were common between the sampling times but varying in abundance (Fig. 4C). Amphisphaeriales and Hypocreales were not isolated in winter.
Figure 4. Percentage of occurrence of fungi associated with leaves of Acanthus ilicifolius. var. xiamenensis.
Isolation method: (A) phylum, (B) class, and (C) order classification; metabarcoding analysis: (D) phylum, (E) class, and (F) order classification; both isolation and metabarcoding approaches: (G) phylum, (H) class, and (I) order classification.
Metabarcoding analysis
Seventeen samples (leaves) were analyzed by the metabarcoding analysis: 10 for the winter and 7 for the summer samples. A total of 773685 reads were obtained after QIIME analysis, including 314692 reads from the winter samples ranging from 2169 to 54606 reads, and 458993 reads from the summer samples ranging from 39151 to 93295 reads (Table 2). From the set of 17 samples, a total of 111 OTUs were identified, from which 86 could be referred to the generic level and 25 to the family level or above, including 96 OTUs (76 OTUs identified at the genus level) from the winter and 70 OTUs (55 OTUs identified at the genus level) from the summer samples (Table 3). Fifty-five OTUs (41 OTUs identified at the genus level) were common between the two seasons.
Table 3. Fungal diversity associated with leaves of Acanthus ilicifolius var. xiamenensis in summer and winter samples recovered from metabarcoding analysis.
Percentage of occurrence of fungi was calculated based on number of reads.
| Phylum | Class | Order | Family | Taxon | % Occurrence | ||
|---|---|---|---|---|---|---|---|
| Winter | Summer | Total | |||||
| Ascomycota | Sordariomycetes | Hypocreales | Incertae sedis | Acremonium polychromum | 0.040 | 0.031 | 0.035 |
| Basidiomycota | Agaricomycetes | Agaricales | Agaricales | 0.020 | 0.023 | 0.022 | |
| Basidiomycota | Agaricomycetes | Agaricomycetes | 0.166 | 0.136 | 0.148 | ||
| Ascomycota | Dothideomycetes | Pleosporales | Pleosporaceae | Alternaria sp. | 3.463 | 0.122 | 1.481 |
| Ascomycota | Dothideomycetes | Botryosphaeriales | Aplosporellaceae | Aplosporella yalgorensis | 0.009 | 0.000 | 0.004 |
| Ascomycota | Sordariomycetes | Xylariales | Apiosporaceae | Arthrinium sp. | 0.007 | 0.002 | 0.004 |
| Ascomycota | Ascomycota | 49.555 | 11.377 | 26.906 | |||
| Ascomycota | Eurotiomycetes | Eurotiales | Aspergillaceae | Aspergillus penicillioides | 0.008 | 0.000 | 0.003 |
| Ascomycota | Eurotiomycetes | Eurotiales | Aspergillaceae | Aspergillus sp. | 0.093 | 0.016 | 0.047 |
| Ascomycota | Dothideomycetes | Dothideales | Aureobasidiaceae | Aureobasidium sp. | 0.344 | 10.715 | 6.496 |
| Ascomycota | Dothideomycetes | Dothideales | Aureobasidiaceae | Aureobasidium thailandense | 0.000 | 0.064 | 0.038 |
| Basidiomycota | Basidiomycota | 9.308 | 43.089 | 29.349 | |||
| Basidiomycota | Agaricomycetes | Polyporales | Meruliaceae | Bjerkandera adusta | 0.023 | 0.032 | 0.029 |
| Basidiomycota | Tremellomycetes | Tremellales | Bulleraceae | Bullera unica | 0.044 | 0.000 | 0.018 |
| Ascomycota | Dothideomycetes | Capnodiales | Capnodiales | 15.762 | 5.083 | 9.427 | |
| Ascomycota | Dothideomycetes | Chaetothyriales | Herpotrichiellaceae | Capronia semi-immersa | 0.036 | 0.035 | 0.035 |
| Ascomycota | Dothideomycetes | Capnodiales | Cladosporiaceae | Cladosporium delicatulum | 2.561 | 3.449 | 3.088 |
| Ascomycota | Dothideomycetes | Capnodiales | Cladosporiaceae | Cladosporium sp. | 0.189 | 7.901 | 4.764 |
| Ascomycota | Dothideomycetes | Capnodiales | Cladosporiaceae | Cladosporium sphaerospermum | 0.029 | 0.026 | 0.027 |
| Ascomycota | Sordariomycetes | Glomerellales | Glomerellaceae | Colletotrichum brasiliense | 0.011 | 0.063 | 0.042 |
| Ascomycota | Sordariomycetes | Glomerellales | Glomerellaceae | Colletotrichum gloeosporioides | 0.013 | 0.063 | 0.042 |
| Ascomycota | Dothideomycetes | Pleosporales | Coniothyriaceae | Coniothyrium sidae | 0.099 | 0.011 | 0.047 |
| Basidiomycota | Tremellomycetes | Tremellales | Tremellaceae | Cryptococcus dimennae | 0.073 | 0.000 | 0.030 |
| Ascomycota | Dothideomycetes | Chaetothyriales | Cyphellophoraceae | Cyphellophora sessilis | 0.086 | 0.000 | 0.035 |
| Basidiomycota | Tremellomycetes | Tremellales | Bulleribasidiaceae | Derxomyces sp. | 0.003 | 0.000 | 0.001 |
| Ascomycota | Dothideomycetes | Capnodiales | Teratosphaeriaceae | Devriesia sp. | 0.051 | 0.000 | 0.021 |
| Ascomycota | Sordariomycetes | Diaporthales | Diaporthales sp. 1 | 0.332 | 0.157 | 0.228 | |
| Ascomycota | Sordariomycetes | Diaporthales | Diaporthales sp. 2 | 0.099 | 0.167 | 0.139 | |
| Ascomycota | Dothideomycetes | Pleosporales | Didymellaceae | Didymella sp. | 0.000 | 0.014 | 0.009 |
| Ascomycota | Dothideomycetes | Pleosporales | Didymosphaeriaceae | Didymosphaeriaceae | 1.017 | 0.016 | 0.423 |
| Basidiomycota | Tremellomycetes | Tremellales | Bulleribasidiaceae | Dioszegia sp. | 0.367 | 0.002 | 0.150 |
| Basidiomycota | Tremellomycetes | Tremellales | Bulleribasidiaceae | Dioszegia takashimae | 0.113 | 0.032 | 0.065 |
| Ascomycota | Dothideomycetes | Dothideales | Dothideales | 0.000 | 0.491 | 0.291 | |
| Ascomycota | Dothideomycetes | Dothideomycetes | 0.568 | 0.286 | 0.401 | ||
| Basidiomycota | Cystobasidiomycetes | Erythrobasidiales | Erythrobasidiaceae | Erythrobasidium hasegawianum | 0.022 | 0.003 | 0.011 |
| Ascomycota | Dothideomycetes | Chaetothyriales | Herpotrichiellaceae | Exophiala sp. | 0.003 | 0.000 | 0.001 |
| Ascomycota | Dothideomycetes | Chaetothyriales | Herpotrichiellaceae | Exophiala xenobiotica | 0.049 | 0.000 | 0.020 |
| Fungi | 4.735 | 3.577 | 4.048 | ||||
| Ascomycota | Sordariomycetes | Hypocreales | Nectriaceae | Fusarium solani | 0.010 | 0.027 | 0.020 |
| Ascomycota | Sordariomycetes | Hypocreales | Nectriaceae | Fusarium sp. | 0.148 | 0.022 | 0.073 |
| Basidiomycota | Geminibasidiomycetes | Geminibasidiales | Geminibasidiaceae | Geminibasidium sp. | 0.004 | 0.000 | 0.002 |
| Ascomycota | Sordariomycetes | Hypocreales | Nectriaceae | Gibberella intricans | 0.106 | 0.019 | 0.054 |
| Basidiomycota | Exobasidiomycetes | Golubeviales | Golubeviaceae | Golubevia pallescens | 0.092 | 0.000 | 0.037 |
| Ascomycota | Sordariomycetes | Microascales | Halosphaeriaceae | Halosphaeriaceae | 0.010 | 0.000 | 0.004 |
| Basidiomycota | Tremellomycetes | Tremellales | Bulleribasidiaceae | Hannaella oryzae | 0.000 | 0.006 | 0.004 |
| Ascomycota | Dothideomycetes | Dothideales | Incertae sedis | Hortaea werneckii | 0.122 | 3.205 | 1.951 |
| Ascomycota | Sordariomycetes | Hypocreales | Hypocreales | 0.005 | 0.000 | 0.002 | |
| Ascomycota | Sordariomycetes | Xylariales | Xylariaceae | Hypoxylon monticulosum | 0.032 | 0.000 | 0.013 |
| Ascomycota | Dothideomycetes | Pleosporales | Lentitheciaceae | Keissleriella yonaguniensis | 0.409 | 0.003 | 0.168 |
| Basidiomycota | Tremellomycetes | Tremellales | Cuniculitremaceae | Kockovaella sacchari | 0.262 | 0.000 | 0.106 |
| Ascomycota | Dothideomycetes | Pleosporales | Phaeosphaeriaceae | Leptospora rubella | 0.518 | 0.000 | 0.211 |
| Basidiomycota | Malasseziomycetes | Malasseziales | Malasseziaceae | Malassezia restricta | 0.000 | 0.009 | 0.005 |
| Ascomycota | Dothideomycetes | Pleosporales | Trematosphaeriaceae | Medicopsis romeroi | 0.000 | 0.003 | 0.002 |
| Basidiomycota | Exobasidiomycetes | Exobasidiales | Brachybasidiaceae | Meira argovae | 0.037 | 0.000 | 0.015 |
| Ascomycota | Dothideomycetes | Capnodiales | Mycosphaerellaceae | Mycosphaerella etlingerae | 0.008 | 0.000 | 0.003 |
| Ascomycota | Dothideomycetes | Capnodiales | Mycosphaerellaceae | Mycosphaerella sp. | 0.078 | 0.000 | 0.032 |
| Ascomycota | Dothideomycetes | Capnodiales | Mycosphaerellaceae | Mycosphaerellaceae | 2.517 | 0.556 | 1.354 |
| Ascomycota | Sordariomycetes | Hypocreales | Nectriaceae | Nectriaceae | 0.010 | 0.000 | 0.004 |
| Ascomycota | Sordariomycetes | Xylariales | Apiosporaceae | Nigrospora oryzae | 0.240 | 0.051 | 0.128 |
| Ascomycota | Dothideomycetes | Venturiales | Sympoventuriaceae | Ochroconis musae | 0.009 | 0.000 | 0.004 |
| Basidiomycota | Tremellomycetes | Tremellales | Rhynchogastremataceae | Papiliotrema pseudoalba | 0.840 | 0.212 | 0.467 |
| Basidiomycota | Tremellomycetes | Tremellales | Rhynchogastremataceae | Papiliotrema sp. | 0.000 | 0.194 | 0.115 |
| Ascomycota | Dothideomycetes | Pleosporales | Didymosphaeriaceae | Paraconiothyrium sp. | 0.032 | 0.000 | 0.013 |
| Ascomycota | Eurotiomycetes | Eurotiales | Aspergillaceae | Penicillium sp. | 0.000 | 0.013 | 0.007 |
| Basidiomycota | Agaricomycetes | Russulales | Peniophoraceae | Peniophora sp. | 0.002 | 0.004 | 0.003 |
| Ascomycota | Sordariomycetes | Xylariales | Sporocadaceae | Pestalotiopsis rhododendri | 0.005 | 0.000 | 0.002 |
| Ascomycota | Dothideomycetes | Capnodiales | Mycosphaerellaceae | Phaeophleospora hymenocallidicola | 0.083 | 0.085 | 0.084 |
| Ascomycota | Dothideomycetes | Pleosporales | Phaeosphaeriaceae | Phaeosphaeriaceae | 0.000 | 0.031 | 0.018 |
| Basidiomycota | Agaricomycetes | Polyporales | Meruliaceae | Phanerochaete tuberculata | 0.028 | 0.024 | 0.025 |
| Ascomycota | Sordariomycetes | Xylariales | Incertae sedis | Phialemoniopsis ocularis | 0.007 | 0.000 | 0.003 |
| Ascomycota | Sordariomycetes | Diaporthales | Valsaceae | Phomopsis sp. | 0.012 | 0.042 | 0.030 |
| Ascomycota | Dothideomycetes | Botryosphaeriales | Phyllostictaceae | Phyllosticta capitalensis | 0.000 | 0.007 | 0.004 |
| Ascomycota | Sordariomycetes | Glomerellales | Plectosphaerellaceae | Plectosphaerellaceae | 0.014 | 0.000 | 0.006 |
| Ascomycota | Dothideomycetes | Pleosporales | Pleosporaceae | Pleosporaceae | 0.000 | 0.021 | 0.013 |
| Ascomycota | Dothideomycetes | Pleosporales | Pleosporales sp. 1 | 0.141 | 0.037 | 0.080 | |
| Ascomycota | Dothideomycetes | Pleosporales | Pleosporales sp. 2 | 0.059 | 0.000 | 0.024 | |
| Ascomycota | Dothideomycetes | Pleosporales | Sporomiaceae | Preussia persica | 0.091 | 0.082 | 0.085 |
| Basidiomycota | Agaricomycetes | Agaricales | Strophariaceae | Psilocybe coprophila | 0.003 | 0.000 | 0.001 |
| Basidiomycota | Agaricomycetes | Agaricales | Strophariaceae | Psilocybe sp. | 0.015 | 0.000 | 0.006 |
| Ascomycota | Dothideomycetes | Pleosporales | Cucurbitariaceae | Pyrenochaetopsis leptospora | 1.409 | 1.141 | 1.250 |
| Ascomycota | Dothideomycetes | Pleosporales | Cucurbitariaceae | Pyrenochaetopsis sp. | 1.146 | 0.146 | 0.552 |
| Ascomycota | Dothideomycetes | Capnodiales | Dissoconiaceae | Ramichloridium luteum | 0.048 | 0.000 | 0.020 |
| Basidiomycota | Microbotryomycetes | Sporidiobolales | Sporidiobolaceae | Rhodotorula mucilaginosa | 0.086 | 0.123 | 0.108 |
| Basidiomycota | Agaricomycetes | Polyporales | Meripilaceae | Rigidoporus sp. | 0.013 | 0.019 | 0.017 |
| Ascomycota | Dothideomycetes | Pleosporales | Thyridariaceae | Roussoella solani | 0.000 | 0.007 | 0.004 |
| Basidiomycota | Tremellomycetes | Tremellales | Trimorphomycetaceae | Saitozyma flava | 0.137 | 0.000 | 0.056 |
| Ascomycota | Dothideomycetes | Pleosporales | Phaeosphaeriaceae | Sclerostagonospora ericae | 0.033 | 0.124 | 0.087 |
| Ascomycota | Dothideomycetes | Pleosporales | Phaeosphaeriaceae | Sclerostagonospora phragmiticola | 0.169 | 1.525 | 0.973 |
| Ascomycota | Dothideomycetes | Pleosporales | Lentitheciaceae | Setoseptoria arundinacea | 0.123 | 0.000 | 0.050 |
| Ascomycota | Sordariomycetes | Hypocreales | Cordycipitaceae | Simplicillium obclavatum | 0.010 | 0.008 | 0.009 |
| Ascomycota | Sordariomycetes | Hypocreales | Cordycipitaceae | Simplicillium sp. | 0.001 | 0.000 | 0.001 |
| Ascomycota | Sordariomycetes | Sordariomycetes | 0.035 | 0.010 | 0.020 | ||
| Basidiomycota | Microbotryomycetes | Sporidiobolales | Sporidiobolaceae | Sporobolomyces koalae | 0.029 | 0.000 | 0.012 |
| Ascomycota | Dothideomycetes | Pleosporales | Massarinaceae | Stagonospora neglecta | 0.035 | 0.173 | 0.117 |
| Ascomycota | Dothideomycetes | Pleosporales | Pleosporaceae | Stemphylium vesicarium | 0.000 | 0.030 | 0.018 |
| Ascomycota | Dothideomycetes | Chaetothyriales | Incertae sedis | Strelitziana africana | 0.010 | 0.000 | 0.004 |
| Ascomycota | Dothideomycetes | Chaetothyriales | Incertae sedis | Strelitziana eucalypti | 0.013 | 0.000 | 0.005 |
| Basidiomycota | Cystobasidiomycetes | Symmetrosporaceae | Symmetrospora sp. | 0.067 | 0.029 | 0.044 | |
| Ascomycota | Dothideomycetes | Capnodiales | Teratosphaeriaceae | Teratosphaeria sp. | 0.013 | 0.136 | 0.086 |
| Ascomycota | Dothideomycetes | Capnodiales | Teratosphaeriaceae | Teratosphaeriaceae | 0.798 | 4.551 | 3.024 |
| Basidiomycota | Agaricomycetes | Polyporales | Coriolaceae | Trametes cubensis | 0.004 | 0.006 | 0.005 |
| Basidiomycota | Tremellomycetes | Tremellales | Tremellales | 0.032 | 0.000 | 0.013 | |
| Ascomycota | Sordariomycetes | Hypocreales | Hypocreaceae | Trichoderma lixii | 0.000 | 0.011 | 0.007 |
| Ascomycota | Sordariomycetes | Hypocreales | Hypocreaceae | Trichoderma sp. | 0.035 | 0.044 | 0.040 |
| Ascomycota | Dothideomycetes | Chaetothyriales | Trichomeriaceae | Trichomeriaceae | 0.049 | 0.000 | 0.020 |
| Ascomycota | Dothideomycetes | Capnodiales | Dissoconiaceae | Uwebraunia musae | 0.003 | 0.000 | 0.001 |
| Ascomycota | Dothideomycetes | Chaetothyriales | Herpotrichiellaceae | Veronaea botryosa | 0.004 | 0.000 | 0.002 |
| Ascomycota | Dothideomycetes | Pleosporales | Sporomiaceae | Westerdykella dispersa | 0.332 | 0.277 | 0.299 |
| Ascomycota | Saccharomycetes | Saccharomycetales | Phaffomycetaceae | Wickerhamomyces anomalus | 0.000 | 0.006 | 0.003 |
| Ascomycota | Sordariomycetes | Xylariales | Xylariales | 0.011 | 0.000 | 0.004 | |
| Ascomycota | Dothideomycetes | Capnodiales | Mycosphaerellaceae | Zasmidium sp. | 0.108 | 0.000 | 0.044 |
Figures 4D–4F shows the proportions of the different taxa at the phylum, class and order levels. Both Ascomycota and Basidiomycota were recovered at proportions of 52.5% (percentage of occurrence based on read number) and 44.0% from the summer and 83.5% and 11.8% from the winter samples, respectively, with a higher proportion of basidiomycetous sequences in the summer samples (Fig. 4D). Overall, Ascomycota (65.1%) was dominant over Basidiomycota (30.9%).
At the class level, 11 different fungal classes were obtained from both the winter and summer samples (Fig. 4E). Seven classes were common between the samples: Agaricomycetes, Cystobasidiomycetes, Dothideomycetes, Eurotiomycetes, Microbotryomycetes, Sordariomycetes and Tremellomycetes. Dothideomycetes was the dominant class in both winter and summer samples (32.6% and 40.4%, respectively). Other classes only constituted less than 2% of the sequences, excluding those only referred to the phylum level (‘Others’). Exobasidiomycetes and Geminibasidiomycetes were only recovered from the winter samples and likewise, Malasseziomycetes and Saccharomycetes in the summer samples. The proportion of the different major classes overall was similar to that of the individual winter and summer samples.
Twenty-two and nineteen different fungal orders were identified in the winter and summer samples, respectively (Fig. 4F). Agaricales, Botryosphaeriales, Capnodiales, Chaetothyriales, Diaporthales, Dothideales, Erythrobasidiales, Eurotiales, Glomerellales, Hypocreales, Pleosporales, Polyporales, Russulales, Sporidiobolales, Tremellales and Xylariales were recovered from both samples but varying in abundance, excluding the sequences only identified above the order level (‘Others’). The dominant orders in the winter samples were Capnodiales (22.2%), Pleosporales (9.1%) and Tremellales (1.9%). Capnodiales (21.8%) was also the most dominant order in the summer samples, followed by Dothideales (14.5%) and Pleosporales (3.8%). Exobasidiales, Geminibasidiales, Golubeviales, Microascales and Venturiales were only found in the winter samples and Malasseziales and Saccharomycetales in the summer samples; these orders exclusive to their respective sample type only constituted a low sequence abundance (<0.1%). Combining the data from the two seasons, the dominant orders were Capnodiales (22.0%), Dothideales (8.8%) and Pleosporales (5.9%).
At genus and species levels, taxa having the highest percentage of occurrence included Alternaria sp. (3.46%), Cladosporium delicatulum (2.56%) and Pyrenochaetopsis leptospora (1.41%) in the winter samples, and Aureobasidium sp. (10.72%), Cladosporium sp. (7.90%), C. delicatulum (3.45%) and Hortaea werneckii (3.21%) in the summer samples (Table 3). These latter four species also had the highest overall percentage of occurrence (both seasons).
Calculated from the read numbers of the different OTUs, the fungal community in the winter samples had a higher species richness of 7.50 (Margalef) than that in the summer samples (5.29) but the Shannon–Wiener Diversity Index was comparable between the two samples, 1.98 and 2.09 respectively (Table 2). The overall Margalef and Shannon–Wiener Diversity indices were 8.11 and 2.28, respectively. The fungal community in both winter and summer samples had reached species saturation and the winter samples had a higher species diversity (Fig. 3B).
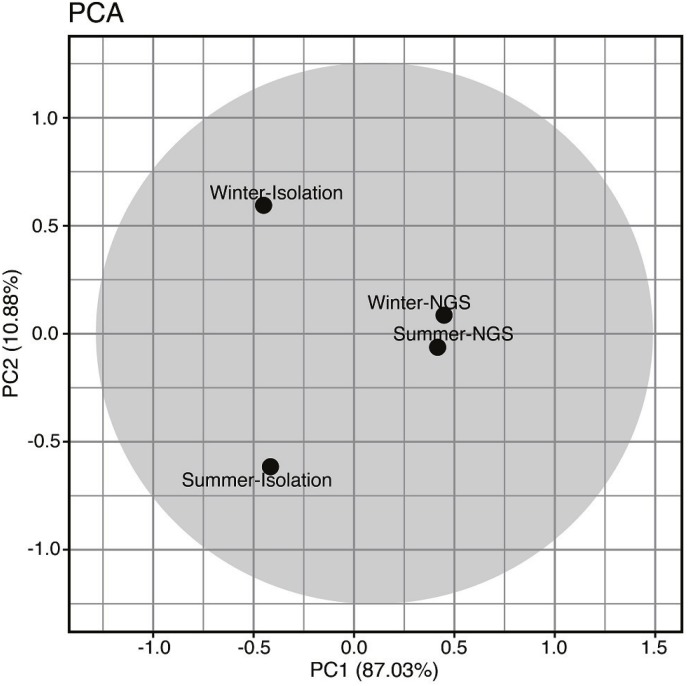
The fungal community among isolation/metabarcoding and winter/summer samples were analyzed by PCA and the result is shown in Fig. 5. A large extent of community variation was found across PC1 (87.03%), and to a lesser extent across PC2 (10.88%). Separation across PC1 was associated with changes in fungal composition between the methods; the fungal communities obtained from the metabarcoding method (Winter-NGS, Summer-NGS) were positively correlated while those obtained from the isolation method (Winter-Isolation, Summer-Isolation) were negatively correlated. For PC2, the community variation was associated with the summer and winter samples; the winter samples (Winter-NGS, Winter-Isolation) and the summer samples (Summer-NGS, Summer Isolation) were positively and negatively correlated, respectively.
Figure 5. Principle component analysis based on percentage of occurrence of foliar endophytic fungal communities of Acanthusilicifolius var. xiamenensis in summer and winter seasons obtained from isolation and metabarcoding (NGS) studies.
Total diversity of fungi on Acanthus ilicifolius var. xiamenensis
Based on the average of the percentage of occurrence in the isolation study (Table 1) and metabarcoding analysis (Table 3), the phylum, class and order classifications of the fungi associated with A. ilicifolius var. xiamenensis were obtained (Figs. 4G–4I). Ascomycota was still dominant, especially in the winter samples (Fig. 4G). The dominant classes in the winter and summer samples were Dothideomycetes and Sordariomycetes (Fig. 4H). Capnodiales, Diaporthales, Glomerellales and Pleosporales were dominant orders in both seasons, although varying in percentage of occurrences (Fig. 4I). The percentages of Dothideales and Hypocreales were much higher in the summer than in the winter.
Table 4 lists the species of fungi identified from the isolation and metabarcoding methods, excluding those taxa at the family level or above. Excluding the composite taxa (i.e., spp.), H. werneckii and Setoseptoria arundinacea were the only fungi recovered from both methods and at least 110 species were identified from leaves of A. ilicifolius var. xiamenensis. The most speciose genus on A. ilicifolius var. xiamenensis was Colletotrichum. Some genera were only obtained with the fungal isolation procedure such as Diaporthe spp., Phoma spp. and Pseudocercospora spp. while some were only recovered with the metabarcoding study, such as Aspergillus spp., Exophiala spp., Trichoderma spp. etc. Species of Alternaria, Aureobasidium, Cladosporium, Colletotrichum, Fusarium, Nigrospora, Pestalotiopsis and Phomopsis were identified with both methods.
Table 4. Fungal taxa associated with leaves of Acanthus ilicifolius var. xiamenensis.
The list was summarized from results of the isolation and metabarcoding analyses.
| ASCOMYCOTA | BASIDIOMYCOTA | |
| Botryosphaeriales | Hypocreales | Agaricales |
| Aplosporella yalgorensis | Acremonium polychromum | Psilocybe coprophila |
| Botryosphaeria dothideaa | Fusarium oxysporuma | Psilocybe sp. |
| Guignardia sp.a | Fusarium solani | Erythrobasidiales |
| Phyllosticta capitalensis | Fusarium spp.b | Erythrobasidium hasegawianum |
| Capnodiales | Gibberella intricans | Exobasidiales |
| Acidiella uranophilaa | Simplicillium obclavatum | Meira argovae |
| Cladosporium delicatulum | Simplicillium sp. | Geminibasidiales |
| Cladosporium spp.b | Trichoderma lixii | Geminibasidium sp. |
| Cladosporium sphaerospermum | Trichoderma sp. | Golubeviales |
| Devriesia sp. | Pleosporales | Golubevia pallescens |
| Mycosphaerella etlingerae | Alternaria alternataa | Hymenochaetales |
| Mycosphaerella sp. | Alternaria spp.b | Phellinus noxiusa |
| Phaeophleospora eucalypticolaa | Coniothyrium sidae | Malasseziales |
| Phaeophleospora hymenocallidicola | Corynespora cassiicolaa | Malassezia restricta |
| Pseudocercospora nymphaeaceaa | Didymella spp.b | Polyporales |
| Pseudocercospora sp.a | Drechslera dematioideaa | Bjerkandera adusta |
| Ramichloridium luteum | Keissleriella yonaguniensis | Phanerina melleaa |
| Ramichloridium punctatuma | Leptospora rubella | Phanerochaete tuberculata |
| Teratosphaeria capensisa | Medicopsis romeroi | Rigidoporus sp. |
| Teratosphaeria sp. | Paraconiothyrium sp. | Tinctoporellus epimiltinusa |
| Uwebraunia musae | Parastagonospora phoenicicolaa | Russulales |
| Zasmidium citria | Phoma spp.a | Peniophora sp. |
| Zasmidium sp. | Preussia persica | Sporidiobolales |
| Chaetothyriales | Pyrenochaetopsis leptospora | Rhodotorula mucilaginosa |
| Capronia semi-immersa | Pyrenochaetopsis sp. | Sporobolomyces koalae |
| Cyphellophora sessilis | Roussoella solani | Tremellales |
| Exophiala sp. | Sclerostagonospora ericae | Bullera unica |
| Exophiala xenobiotica | Sclerostagonospora phragmiticola | Cryptococcus dimennae |
| Strelitziana africana | Septoriella hubertusiia | Derxomyces sp. |
| Strelitziana eucalypti | Setoseptoria arundinaceab | Dioszegia sp. |
| Veronaea botryosa | Stagonospora neglecta | Dioszegia takashimae |
| Diaporthales | Stagonosporopsis cucurbitacearuma | Hannaella oryzae |
| Diaporthe endophyticaa | Stemphylium vesicarium | Kockovaella sacchari |
| Diaporthe longicollaa | Westerdykella dispersa | Papiliotrema pseudoalba |
| Diaporthe phaseoloruma | Saccharomycetales | Papiliotrema sp. |
| Phomopsis asparagi | Wickerhamomyces anomalus | Saitozyma flava |
| Phomopsis spp.b | Venturiales | Trametes cubensis |
| Dothideales | Ochroconis musae | Basidiomycota orderincertae sedis |
| Aureobasidium spp.b | Xylariales | Symmetrospora sp. |
| Aureobasidium thailandense | Arthrinium sp. | |
| Hortaea werneckiib | Daldinia eschscholtziia | |
| Eurotiales | Hypoxylon monticulosum | |
| Aspergillus penicillioides | Nigrospora oryzae | |
| Aspergillus sp. | Nigrospora sp.a | |
| Penicillium sp. | Nodulisporium sp.a | |
| Glomerellales | Pestalotiopsis microsporaa | |
| Colletotrichum boninensea | Pestalotiopsis rhododendri | |
| Colletotrichum brasiliense | Phialemoniopsis ocularis | |
| Colletotrichum gloeosporioides | Xylaria sp.a | |
| Colletotrichum hippeastria | ||
| Colletotrichum spp.a |
Notes.
From isolation.
from both methods.
Discussion
This study investigated the diversity of fungi associated with leaves of the mangrove plant Acanthus ilicifolius var. xiamenensis using the traditional isolation technique and the metabarcoding approach. In the isolation study, most of the isolates did not fruit on the agar plates and sequence analysis of the internal transcribed spacer regions of the rDNA including the 5.8S rDNA (ITS) was used to identify the cultures. ITS is easily amplifiable by PCR and has the highest probability of successful identification for the broadest range of fungi as compared to other rDNA regions and protein genes (Schoch et al., 2012). In the metabarcoding analysis, many OTUs were only identified to the phylum or kingdom levels (Table 3) and the UNITE database was not extensive enough to identify these sequences down to genus/species level (Nilsson et al., 2019). However, the metabarcoding approach offers the advantages of finding signatures of unculturable fungi and potential cryptic species not identifiable with other methods. The nested PCR approach used in this study was found to be able to specifically amplify fungal sequences in the samples.
The leaves were surface-sterilized before isolation and therefore the diversity of fungi recovered from isolation represented the endophytic fungal diversity. On the other hand, the diversity obtained from the metabarcoding analysis represented predominantly endophytic fungi and might represent partial diversity of the epiphytic fungi as surface sterilization of leaves by sodium hypochlorite and ethanol does not completely eliminate all fungal DNA on the surface of the leaves (Burgdorf et al., 2014). This might have resulted in the differences in fungal richness (Margalef species richness, total richness) between the two methods, i.e., generally higher in the metabarcoding analysis (winter: 7.50 (Margalef), 96 species, summer: 5.29, 70) than in the isolation study (winter: 6.24, 30, summer: 5.44, 26) although the Shannon–Wiener diversity index of the two samples was comparable.
The winter samples had a higher fungal species diversity. The weather conditions of Kinmen, Taiwan in January 2014, when the winter samples were collected, were much colder and drier (13.7 °C, 0 mm rainfall, 65% relative humidity) than July 2014 (29.8 °C, 106.9 mm rainfall, 81% relative humidity) for the summer samples. Generally, higher richness and abundance of endophytic fungi were found in hotter and wetter seasons (Pang et al., 2008).
Only nine out of a total of 47 fungal species isolated from A. ilicifolius var. xiamenensis were common between the two sampling times, showing a seasonal variation of fungal diversity using the culture method. However from the metabarcoding analysis, 55 taxa were found to be common between the winter and summer samples (41 and 15 exclusive fungi in the winter and summer samples, respectively), suggesting there was an overall similarity in fungal diversity between the samples. These results show the weakness of using isolation techniques as the sole methods to study diversity of endophytic fungi of mangrove plants (Abdelfattah et al., 2015) Inoculation of leaf discs on a nutritious medium always favors fast-growing fungi to be isolated. In addition, the isolation medium (MEAF) used in this study only recovered a fraction of culturable fungal diversity and it is advisable to use multiple media to widen the number of fungal isolates (Rosa et al., 2011; Potshangbam et al., 2017). Three basidiomycetes Phellinus noxius, Phanerina mellea and Tinctoporellus epimiltinus were isolated from the summer samples, but a number of basidiomycetous OTUs were recovered from both seasons from the metabarcoding analysis and this further confirms the importance of culture-independent techniques in studying diversity of fungi.
A core group of culturable endophytic fungi was found to be associated with mangrove plants, including species of the genera Acremonium, Cladosporium, Colletotrichum, Fusarium, Pestalotiopsis, Phyllosticta (sexual morph Guignardia), Phoma, Phomopsis (sexual morph Diaporthe) and Sporomiella (Pang et al., 2008). Many of these genera, such as Acremonium, Cladosporium, Phomopsis, Phyllosticta, among others, were isolated from leaves of A. ilicifolius var. xiamenensis in this study, confirming their prevalence in mangrove plants. However, Sporomiella, a universal endophytic taxon of mangrove plants, was not found in this study (Pang et al., 2008). The number of species isolated from leaves of A. ilicifolius var. xiamenensis (47) was much higher than those found in related studies in this species: 11 species from roots in Udupi, India (Ananda & Sridhar, 2002), 10 species from leaves in Ranong, Thailand (Chaeprasert et al., 2010), eight species from leaves in Tamil Nadu, India (Suryanarayanan & Kumaresan, 2000) and 14 species from leaves in Muthupet, India (Priyadharshini, Ambikapathy & Panneerselvam, 2015). However, the fungal community obtained from the metabarcoding analysis was different from that of the isolation study. The dominant fungi included Cladosporium spp. and other common terrestrial fungi, such as Hortaea werneckii. H. werneckii is a cosmopolitan halophilic fungus and can potentially cause human diseases (Marchetta et al., 2018). Together with Setoseptoria arundinacea, H. werneckii was also cultured from leaves of A. ilicifolius var. xiamenensis and it was previously reported from surface-sterilized roots and stems of the mangrove plant Aegiceras corniculatum (Chen et al., 2012).
At least 110 species (excluding the composite genera) were obtained from both isolation and metabarcoding studies suggesting a much higher fungal diversity associated with leaves of A. ilicifolius var. xiamenensis. Ascomycota was dominant with a small proportion of Basidiomycota from both methods, agreeing with similar studies using the traditional culture methods (Hamzah et al., 2018; Zhou et al., 2018) and with Arfi et al. (2012) who used a culture-independent approach. As expected, Basidiomycetes were not commonly cultured as endophytes (Chaeprasert et al., 2010; Xing & Guo, 2011; Costa, Maia & Cavalcanti, 2012).
Dothideomycetes was found to be the most dominant class in both seasons from both methods. Dothideomycetes were also found to be the dominant class of fungi on the aerial parts (trunk, bark and leaf) of the mangrove plants Avicennia marina and Rhizophora stylosa and Lecanoromycetes in R. stylosa using 454 pyrosequencing of 18S and ITS rDNA genes (Arfi et al., 2012). Lecanoromycetes is a group of lichenized fungi; it was not found in this study, probably because tree trunk and bark, where this group of fungi normally inhabits, were not analyzed. Also according to Arfi et al. (2012), Capnodiales, Diaporthales, Dothideales and Pleosporales were dominant on the emerged plant parts, especially in A. marina. This result generally agrees with this study, with variations related to the abundance.
A number of fungi recovered from A. ilicifolius var. xiamenensis are well known pathogens such as Cladosporium, Colletotrichum, and Fusarium, which might ultimately cause plant diseases. The Botryosphaeriales was reported to potentially cause diseases of mangrove plants (Osorio et al., 2017). In this study, Aplosporella yalgorensis, Botryosphaeria dothidea, Guignardia sp. and Phyllosticta capitalensis of the Botryosphaeriales were recovered. Whether these fungi cause diseases in A. ilicifolius var. xiamenensis is not known and requires further research. A Purpureocillium sp. isolated endophytically from roots of Kandelia candel was found to protect growth of the plant from copper(II) stress when this fungus was added to the growth pots (Gong et al., 2017). Whether endophytic fungi help to relief metal stress imposed on mangrove plants also requires further studies. A high quantity of RNA transcripts of fungi from surface-sterilized leaves of A. marina was found (Huang et al., 2014) and it may suggest that endophytic fungi live in a close symbiotic relationship with the mangrove plant.
In conclusion, this study discovered a high diversity of fungi associated with leaves of A. ilicifolius var. xiamenensis with a total of 110 taxa recovered from the isolation and metabarcoding methods. From the isolation study, Ascomycota was dominant, with Basidiomycota isolated only in the summer samples. C. cassiicola (6.90%), F. oxysporum (6.40%) and Guignardia sp. (6.40%) had the highest overall percentage of occurrence. In the metabarcoding analysis, Ascomycota was also dominant over the Basidiomycota. Based on reads, Aureobasidium sp. (10.72%), Cladosporium sp. (7.90%), C. delicatulum (3.45%) and H. werneckii (3.21%) had the highest percentage of occurrence. The use of both methods discovered a much higher diversity of endophytic fungi associated with A. ilicifolius var. xiamenensis. The association of these fungi with the plant is not known and future studies should focus on the ecological roles of these fungi. However, a chemical analysis of the spent culture liquid of the fungal isolates in this study suggests that 28 isolates produced antimicrobial substances against some Gram-positive and Gram-negative bacteria and fungi and thus might protect the plant from microbial diseases (Chi et al., 2019).
Supplemental Information
The fungal isolates were cultured from leaves of Acanthus ilicifolius var. xiamenensis collected at Lieyu Township, Kinmen County, Taiwan on malt extract freshwater agar supplemented with antibiotics.
Acknowledgments
We would like to thank Kuang-Yao Chen for assistance with plant collection. We thank journal editor and many anonymous referees for their editing and suggestions that substantially improved the quality of the manuscript.
Funding Statement
This work was supported by grants from the Center of Excellence for the Oceans (National Taiwan Ocean University), which is financially supported by The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education in Taiwan. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Wei-Chiung Chi conceived and designed the experiments, performed the experiments, analyzed the data, contributed reagents/materials/analysis tools, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft, collect samples in field.
Weiling Chen, Chih-Chiao He, Hyo-Jung Cha and Tsz Wai Ho performed the experiments, approved the final draft.
Sheng-Yu Guo performed the experiments, analyzed the data, prepared figures and/or tables, approved the final draft.
Ling Ming Tsang performed the experiments, analyzed the data, contributed reagents/materials/analysis tools, approved the final draft.
Ka-Lai Pang conceived and designed the experiments, analyzed the data, contributed reagents/materials/analysis tools, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
DNA Deposition
The following information was supplied regarding the deposition of DNA sequences:
All identified fungal sequences are accessible in GenBank. The accession numbers are available in Table 1.
Data Availability
References
- Abdelfattah et al. (2015).Abdelfattah A, Nicosia MGLD, Cacciola SO, Droby S, Schena L. Metabarcoding analysis of fungal diversity in the phyllosphere and carposphere of olive (Olea europaea) PLOS ONE. 2015;10(7):e0131069. doi: 10.1371/journal.pone.0131069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananda & Sridhar (2002).Ananda K, Sridhar KR. Diversity of endophytic fungi in the roots of mangrove species on the west coast of India. Canadian Journal of Microbiology. 2002;48:871–878. doi: 10.1139/w02-080. [DOI] [PubMed] [Google Scholar]
- Arfi et al. (2012).Arfi Y, Buée M, Marchand C, Levasseur A, Record E. Multiple markers pyrosequencing reveals highly diverse and host-specific fungal communities on the mangrove trees Avicennia marina and Rhizophora stylosa. FEMS Microbiology Ecology. 2012;79:433–444. doi: 10.1111/j.1574-6941.2011.01236.x. [DOI] [PubMed] [Google Scholar]
- Arnold (2007).Arnold AE. Understanding the diversity of foliar endophytic fungi: progress, challenges, and frontiers. Fungal Biology Review. 2007;21:51–66. doi: 10.1016/j.fbr.2007.05.003. [DOI] [Google Scholar]
- Burgdorf et al. (2014).Burgdorf RJ, Laing MD, Morris CD, Jamal-Ally SF. A procedure to evaluate the efficiency of surface sterilization methods in culture-independent fungal endophyte studies. Brazilian Journal of Microbiology. 2014;45:977–983. doi: 10.1590/S1517-83822014000300030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso et al. (2010).Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK. QIIME allows analysis of high-throughput community sequencing data. Nature Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaeprasert et al. (2010).Chaeprasert S, Piapukiew J, Whalley AJS, Sihanonth P. Endophytic fungi from mangrove plant species of Thailand: their antimicrobial and anticancer potentials. Botanica Marina. 2010;53(6):555–564. doi: 10.1515/bot.2010.074. [DOI] [Google Scholar]
- Chen et al. (2012).Chen JX, Xing K, Zhang LC, Xing YM, Guo SX. Identification of Hortaea werneckii isolated from mangrove plant Aegiceras corniculatum based on morphology and rDNA sequences. Mycopathologia. 2012;174:457–466. doi: 10.1007/s11046-012-9568-1. [DOI] [PubMed] [Google Scholar]
- Chi et al. (2019).Chi WC, Pang KL, Chen WL, Wang GJ, Lee TH. Antimicrobial and iNOS inhibitory activities of the endophytic fungi isolated from the mangrove plant Acanthus ilicifolius var. xiamenensis. Botanical Studies. 2019;60 doi: 10.1186/s40529-019-0252-3. Article 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa, Maia & Cavalcanti (2012).Costa I, Maia LC, Cavalcanti MA. Diversity of leaf endophytic fungi in mangrove plants of northeast Brazil. Brazilian Journal of Microbiology. 2012;43:1165–1173. doi: 10.1590/S1517-83822012000300044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza Sebastianes et al. (2013).De Souza Sebastianes FL, Romão Dumaresq AS, Lacava PT, Harakava R, Azevedo JL, De Melo IS, Pizzirani-Kleiner AA. Species diversity of culturable endophytic fungi from Brazilian mangrove forests. Current Genetics. 2013;59:153–166. doi: 10.1007/s00294-013-0396-8. [DOI] [PubMed] [Google Scholar]
- Edgar (2010).Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Edgar et al. (2011).Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, Mejía-Chang & Rojas (2002).Gilbert GS, Mejía-Chang M, Rojas E. Fungal diversity and plant disease in mangrove forests: salt excretion as a possible defense mechanism. Oecologia. 2002;132:278–285. doi: 10.1007/s00442-002-0966-9. [DOI] [PubMed] [Google Scholar]
- Gong et al. (2017).Gong B, Liu G, Liao R, Song J, Zhang H. Endophytic fungus Purpureocillium sp. A5 protect mangrove plant Kandelia candel under copper stress. Brazilian Journal of Microbiology. 2017;48(3):530–536. doi: 10.1016/j.bjm.2016.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamzah et al. (2018).Hamzah TNT, Lee SY, Hidayat A, Terhem R, Faridah-Hanum I, Mohamed R. Diversity and characterization of endophytic fungi isolated from the tropical mangrove species, Rhizophora mucronata, and identification of potential antagonists against the soil-borne fungus, Fusarium solani. Frontiers in Microbiology. 2018;9 doi: 10.3389/fmicb.2018.01707. Article 1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh, Ma & Chao (2016).Hsieh TC, Ma KH, Chao A. iNEXT: an R package for rarefaction and extrapolation of species diversity (Hill numbers) Methods in Ecology and Evolution. 2016;7:1451–1456. doi: 10.1111/2041-210X.12613. [DOI] [Google Scholar]
- Huang et al. (2014).Huang J, Lu X, Zhang W, Huang R, Chen S, Zheng Y. Transcriptome sequencing and analysis of leaf tissue of Avicennia marina using the Illumina platform. PLOS ONE. 2014;9:e108785. doi: 10.1371/journal.pone.0108785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kõljalg et al. (2013).Kõljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AFS, Bahram M, Bates ST, Bruns TD, Bengtsson-Palme J, Callaghan TM, Douglas B, Drenkhan T, Eberhardt U, Dueñas M, Grebenc T, Griffith GW, Hartmann M, Kirk PM, Kohout P, Larsson E, Lindahl BD, Lücking R, Martín MP, Matheny PB, Nguyen NH, Niskanen T, Oja J, Peay KG, Peintner U, Peterson M, Põldmaa K, Saag L, Saar I, Schüßler A, Scott JA, Senés C, Smith ME, Suija A, Taylor DL, Telleria MT, Weiß M, Larsson KH. Towards a unified paradigm for sequence-based identification of Fungi. Molecular Ecology. 2013;22:5271–5277. doi: 10.1111/mec.12481. [DOI] [PubMed] [Google Scholar]
- Kumar, Stecher & Tamura (2016).Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaresan & Suryanarayanan (2002).Kumaresan V, Suryanarayanan TS. Endophytic assemblages in young, mature and senescent leaves of Rhizophora apiculata: evidence for the role of endophytes in mangrove litter degradation. Fungal Diversity. 2002;9:81–91. [Google Scholar]
- Marchetta et al. (2018).Marchetta A, Van den Ende BG, Al-Hatmi AMS, Hagen F, Zalar P, Sudhadham M, Gunde-Cimerman N, Urzì C, De Hoog S, De Leo F. Global molecular diversity of the halotolerant fungus Hortaea werneckii. Life. 2018;8(3) doi: 10.3390/life8030031. Article 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin & Rygiewicz (2005).Martin KJ, Rygiewicz PT. Fungal-specific PCR primers developed for analysis of the ITS region of environmental DNA extracts. BMC Microbiology. 2005;5:28. doi: 10.1186/1471-2180-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson et al. (2019).Nilsson RH, Larsson KH, Taylor AFS, Bengtsson-Palme J, Jeppesen TS, Schigel D, Kennedy P, Picard K, Glöckner FO, Tedersoo L, Saar I, Kõljalg U, Abarenkov K. The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Research. 2019;47:D259–D264. doi: 10.1093/nar/gky1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio et al. (2017).Osorio JA, Crous CJ, De Beer ZW, Wingfield MJ, Roux J. Endophytic Botryosphaeriaceae, including five new species, associated with mangrove trees in South Africa. Fungal Biology. 2017;121(4):361–393. doi: 10.1016/j.funbio.2016.09.004. [DOI] [PubMed] [Google Scholar]
- Pang et al. (2008).Pang KL, Vrijmoed LLP, Goh TK, Plaingam N, Jones EBG. Fungal endophytes associated with Kandeliacandel (Rhizophoraceae) in Mai Po Nature Reserve, Hong Kong. Botanica Marina. 2008;51(3):171–178. doi: 10.1515/BOT.2008.012. [DOI] [Google Scholar]
- Petrini (1991).Petrini O. Fungal endophytes of tree leaves. In: Andrews JH, Hirano SS, editors. Microbial ecology of leaves. Springer-Verlag; New York: 1991. pp. 179–197. [DOI] [Google Scholar]
- Potshangbam et al. (2017).Potshangbam M, Devi SI, Sahoo D, Strobel GA. Functional characterization of endophytic fungal community associated with Oryza sativa L., and Zea mays L. Frontiers in Microbiology. 2017;8 doi: 10.3389/fmicb.2017.00325. Article 325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priyadharshini, Ambikapathy & Panneerselvam (2015).Priyadharshini R, Ambikapathy V, Panneerselvam A. Isolation and identification of endophytic fungi from Acanthus ilicifolius in Muthupet Mangroves. International Journal of Scientific Research. 2015;4:169–171. [Google Scholar]
- R Core Team (2013).R Core Team . R Foundation for Statistical Computing; Vienna: 2013. [Google Scholar]
- Rosa et al. (2011).Rosa LH, Vieira MLA, Cota BB, Johann S, Alves TMA, Zani CL, Rosa CA. Endophytic fungi of tropical forests a promising source of bioactive prototype molecules for the treatment of neglected diseases. In: Rundfeldt C, editor. Drug development—a case study based insight into modern strategies. IntechOpen; London: 2011. pp. 469–486. [Google Scholar]
- Schoch et al. (2012).Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W, Fungal Barcoding Consortium Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryanarayanan & Kumaresan (2000).Suryanarayanan TS, Kumaresan V. Endophytic fungi of some halophytes from an estuarine mangrove forest. Mycological Research. 2000;104:1465–1467. doi: 10.1017/S0953756200002859. [DOI] [Google Scholar]
- Suryanarayanan, Kumaresan & Johnson (1998).Suryanarayanan TS, Kumaresan V, Johnson JA. Foliar fungal endophytes from two species of the mangrove Rhizophora. Canadian Journal of Microbiology. 1998;44:1003–1006. doi: 10.1139/w98-087. [DOI] [Google Scholar]
- Toju et al. (2012).Toju H, Tanabe AS, Yamamoto S, Sato H. High-coverage ITS primers for the DNA-based identification of ascomycetes and basidiomycetes in environmental samples. PLOS ONE. 2012;7(7):e40863. doi: 10.1371/journal.pone.0040863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson (1986).Tomlinson PB. The botany of mangroves. Cambridge University Press; New York: 1986. [Google Scholar]
- White et al. (1990).White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and application. Academic Press; San Diego: 1990. pp. 315–322. [Google Scholar]
- Xing et al. (2011).Xing XK, Chen J, Xu MJ, Lin WH, Guo SX. Fungal endophytes associated with Sonneratia (Sonneratiaceae) mangrove plants on the south coast of China. Forest Pathology. 2011;41:334–340. doi: 10.1111/j.1439-0329.2010.00683.x. [DOI] [Google Scholar]
- Xing & Guo (2011).Xing XK, Guo SX. Fungal endophyte communities in four Rhizophoraceae mangrove species on the south coast of China. Ecological Research. 2011;26:403–409. doi: 10.1007/s11284-010-0795-y. [DOI] [Google Scholar]
- Zhou et al. (2018).Zhou J, Diao X, Wang T, Chen G, Lin Q, Yang X, Xu J. Phylogenetic diversity and antioxidant activities of culturable fungal endophytes associated with the mangrove species Rhizophora stylosa and R. mucronata in the South China Sea. PLOS ONE. 2018;13(6):e0197359. doi: 10.1371/journal.pone.0197359. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The fungal isolates were cultured from leaves of Acanthus ilicifolius var. xiamenensis collected at Lieyu Township, Kinmen County, Taiwan on malt extract freshwater agar supplemented with antibiotics.
Data Availability Statement
The following information was supplied regarding data availability: