Abstract
Teucrium is one of the most economically and ecologically important genera in the Lamiaceae family; however, it is currently the least well understood at the plastome level. In the current study, we sequenced the complete chloroplast (cp) genomes of T. stocksianum subsp. stenophyllum R.A.King (TSS), T. stocksianum subsp. stocksianum Boiss. (TS) and T. mascatense Boiss. (TM) through next-generation sequencing and compared them with the cp genomes of related species in Lamiaceae (Ajuga reptans L., Caryopteris mongholica Bunge, Lamium album L., Lamium galeobdolon (L.) Crantz, and Stachys byzantina K.Koch). The results revealed that the TSS, TS and TM cp genomes have sizes of 150,087, 150,076 and 150,499 bp, respectively. Similarly, the large single-copy (LSC) regions of TSS, TS and TM had sizes of 81,707, 81,682 and 82,075 bp, respectively. The gene contents and orders of these genomes were similar to those of other angiosperm species. However, various differences were observed at the inverted repeat (IR) junctions, and the extent of the IR expansion into ψrps19 was 58 bp, 23 bp and 61 bp in TSS, TS and TM, respectively. Similarly, in all genomes, the pbsA gene was present in the LSC at varying distances from the JLA (IRa-LSC) junction. Furthermore, 89, 72, and 92 repeats were identified in the TSS, TM and TS cp genomes, respectively. The highest number of simple sequence repeats was found in TSS (128), followed by TS (127) and TM (121). Pairwise alignments of the TSS cp genome with related cp genomes showed a high degree of synteny. However, relatively lower sequence identity was observed when various coding regions were compared to those of related cp genomes. The average pairwise divergence among the complete cp genomes showed that TSS was more divergent from TM (0.018) than from TS (0.006). The current study provides valuable genomic insight into the genus Teucrium and its subspecies that may be applied to a more comprehensive study.
Keywords: Lamiaceae, Chloroplast genomes, Phylogenetic analysis, Comparative analysis, Teucrium species
Introduction
Lamiaceae is one of the largest families in the plant kingdom and comprises 240 genera and almost 72,000 species, which are distributed all over the world (Harley et al., 2004; Salmaki et al., 2016). The genus Teucrium consists of approximately 250 species (Harley et al., 2004) belonging to the family Lamiaceae, the second largest genus of subfamily Ajugoideae and are primarily perennial herbs, shrubs or subshrubs (Navarro & El Oualidi, 1999). The genus Teucrium contains medicinally important and essential-oil-rich plants (Miller & Morris, 1988; Ulubelen, Topu & Sönmez, 2000). Teucrium species have been used in medicines since ancient times, and many species of this genus possess important biological properties, such as antipyretic, anti-inflammatory, anti-ulcerogenic, antiseptic, anthelmintic, antitumour, hypoglycaemic, hypolipidaemic, and hepatoprotective antimicrobial activities (Abdollahi, Karimpour & Monsef-Esfehani, 2003; Barrachina et al., 1995; Sarac & Ugur, 2007). Two taxa of Teucrium (T. stocksianum subsp. stenophyllum and T. mascatense) are endemic to Oman (Ghazanfar, 1994). Furthermore, various taxa of Teucrium are found in the Arabian Peninsula and Middle East (Patzelt, 2015). Despite its considerable variation, this genus can be discriminated from closely related taxa by the combination of characteristics such as a 2-lipped to 5-lobed actinomorphic calyx, 1- (or rarely slightly 2-) lipped corolla, and arched or straight filaments (Salmaki et al., 2016). Furthermore, various factors, such as species richness, high phenotypic plasticity, ploidy variation and widespread distribution, play vital roles in the complexity of Teucrium and make it challenging and attractive for molecular phylogeneticists and systematists (Salmaki et al., 2016).
Chloroplast (cp) DNA is maternally inherited in the majority of angiosperm species but not in all (McCauley et al., 2007). Due to its mode of inheritance, cp DNA plays critical roles in molecular evolution and population genetic studies. Thus, cp DNA can be used not only for species discrimination but also to answer many other unsolved questions related to taxonomy (Liu et al., 2018a; McCauley et al., 2007; Reboud & Zeyl, 1994). Chloroplasts contain their own independent genomes and genetic systems, and DNA replication and transmission to daughter organelles result in the cytoplasmic inheritance of characteristics associated with the primary events in photosynthesis (Allen, 2015; Olmstead & Palmer, 1994). The cp genome is circular in structure, varies in size from 120 kb to 217 kb in angiosperms, and possesses a quadripartite configuration (Chumley et al., 2006; Wicke et al., 2011), being composed of a small single-copy (SSC) region and a large single-copy (LSC) region, which are generally separated by two copies of an inverted repeat region (IRa and IRb) (Wicke et al., 2011). Although the angiosperm cp genome is generally conserved in terms of gene order and gene content, in some angiosperm families, such as Campanulaceae, Fabaceae, Geraniaceae, and Oleaceae, the genome exhibits features such as gene, intron and even inverted repeat (IR) region loss, gene duplications, and large-scale rearrangements (Cai et al., 2008; Frailey et al., 2018; Greiner et al., 2008; Lee et al., 2007). Due to the conserved structure, recombination-free nature, and small size of the cp genome (Barrett et al., 2014), it is widely used in plant phylogenetic studies (Fan et al., 2018). The highly conserved structure of the cp genome facilitates primer design and sequencing, and cp DNA can be used as a barcode for plant identification (Shaw et al., 2005; Shaw et al., 2014).
With the advancement of genomic tools and methods, next-generation technologies have allowed the rapid sequencing of many cp genomes in recent years. These abundant cp genomes have facilitated the verification of evolutionary relationships and have allowed detailed phylogenetic classifications to be conducted at the group, family, and even genus levels in plants (Jansen et al., 2007; Parks, Cronn & Liston, 2009). Therefore, cp genome-scale data have increasingly been used to infer phylogenetic relationships at high taxonomic levels, and even at lower levels, great progress has been made (Barrett et al., 2013; Carbonell-Caballero et al., 2015; Moore et al., 2007; Shaw et al., 2014). Previously, many cp genomes had been sequenced and published from the Lamiaceae family, including Ajuga reptans L., Caryopteris mongholica Bunge (Liu et al., 2018b), Lamium album L., Lamium galeobdolon (L.) Crantz, and Stachys byzantina K.Koch (Liu et al., 2018b). In our current study, we sequenced the cp genomes of two subspecies of T. stocksianum (subspp. stocksianum and stenophyllum) and T. mascatense using a next-generation sequencing platform. These genomes are the first cp genomes to be reported from the genus Teucrium. Because these species possess morphological similarities in their habitats, in the current study, we aimed to sequence and determine the structures of the Teucrium cp genomes, to identify variations in simple sequence repeats (SSRs) and to identify repeat sequences in these eight cp genomes (TM, TS, TSS, Ajuga reptans L., Caryopteris mongholica Bunge, Lamium album L., Lamium galeobdolon (L.) Crantz, and Stachys byzantina K.Koch).
Materials and Methods
Sample collection
Young, fresh photosynthetic leaves from Teucrium stocksianum subsp. stenophyllum (TSS), Teucrium stocksianum subsp. stocksianum (TS) and Teucrium mascatense (TM) were collected from plants in Jabal Al Akhdar in Oman. The Director General of Nature Conservation from the Sultanate of Oman, Ministry of Environment & Climate Affairs issued the collection permit (4/2106). This sampling area is an arid land with a limited amount of rainfall and an average temperature of 25 °C; however, in the summer, temperatures can reach up to 33 °C, with a mean annual rainfall of 40 mm. The collected samples were washed with sterilized water, dried, placed immediately in liquid nitrogen and stored at −80 °C until cp DNA extraction. The specimens were deposited at the University of Nizwa Herbarium Center, Oman, with the voucher numbers UCTM11 (Teucrium mascatense), UCTS32 (Teucrium stocksianum subsp. stenophyllum), and UCTS30 (Teucrium stocksianum subsp. stocksianum).
Chloroplast DNA extraction and sequencing
The leaves from TSS, TS and TM were ground into a fine powder in liquid nitrogen, and contamination-free cp DNA (nuclear- and mitochondrial-free DNA) was extracted according to a modified protocol including the addition of several purification steps (Shi et al., 2012). Genomic libraries were prepared according to the manufacturer’s instructions (Life Technologies, Carlsbad, CA, USA). The total cp DNA from each sample was sheared enzymatically into 400 bp fragments using the Ion Shear™ Plus Reagents kit, and libraries were prepared using the Ion Xpress™ Plus gDNA Fragment Library kit. Prepared libraries were quantified and qualified on a Qubit 3.0 fluorometer and bioanalyzer (Agilent 2100 Bioanalyzer system; Life Technologies, Carlsbad, CA, USA). Library preparation was followed by template amplification with the Ion OneTouch™ 2 instrument and the enrichment of the amplified template (Ion OneTouch™ ES enrichment system) using Ion 520 & 530 OT2 Reagents. The sample was loaded onto an Ion S5 Sequencing Chip, and sequencing was performed according to the protocol of the Ion Torrent S5.
Genome assembly
A total of 1,246,225, 1,018,614 and 1,396,422 raw reads were generated for TSS, TS and TM, respectively. The obtained reads of the TSS, TS and TM genomes were mapped to the selected reference genome of Ajuga reptans (NC023102) using Bowtie2 (v.2.2.3) (Langmead & Salzberg, 2012) in Geneious Pro (v.10.2.3) (Kearse et al., 2012) software. The mean coverage of the assemblies for TSS, TS and TM were 186X, 128X and 256X, respectively. The IR junction regions were identified using the already published genome of Ajuga reptans, and an iteration method using MITObim (v.1.8) software (Hahn, Bachmann & Chevreux, 2013) was utilized to adjust the sequence length. After sequencing, FastQC (v0.11.6) (Andrews, 2015) was performed to check the read quality. To reduce biases in analysis, an in-house script was used to filter out reads if less than 90% of the bases that made up the read were below Q20. Trimmomatic (v0.36) (Bolger, Lohse & Usadel, 2014) was used to remove adapter sequences. Only high-quality reads were mapped using Bowtie2 in Geneious Pro (v.10.2.3) (Kearse et al., 2012).
Genome annotation
The cp genomes were annotated with the Dual Organellar Genome Annotator (DOGMA) (Wyman, Jansen & Boore, 2004), BLASTX and BLASTN were used to identify the positions of ribosomal RNAs, tRNA and coding genes, and tRNAscan-SE version 1.21 (Schattner, Brooks & Lowe, 2005) software was used to annotate tRNA genes. Additionally, for manual adjustment, Geneious and tRNAscan-SE (Schattner, Brooks & Lowe, 2005) were used to compare the genomes with the previously reported A. reptans genome. Correspondingly, the start and stop codons and intron boundaries were also manually adjusted by comparison with the published A. reptans cp genome (NC_023102). In addition, the structural features of the Teucrium species cp genomes were illustrated using OGDRAW (Lohse, Drechsel & Bock, 2007). MEGA6 software (Kumar et al., 2008) was used to determine relative synonymous codon usage and deviations in synonymous codon usage while avoiding the influence of amino acid composition. The divergence of the genomes of these three Teucrium species from those of other related species was determined by using mVISTA (Frazer et al., 2004) in Shuffle-LAGAN mode, using A. reptans as the reference genome.
Repeat identification
REPuter software (Kurtz et al., 2001) was used for the identification of palindromic and forward repeats in the genomes. The criterion was a minimum of 15 base pairs with sequence identities of 90%. Furthermore, simple sequence repeats (SSRs) were determined using Phobos version 3.3.12 (Kraemer et al., 2009), with the search parameters set as follows: for mononucleotide repeats, ≥10 repeat units; for dinucleotide repeats, ≥ 8 repeat units; for trinucleotide and tetranucleotide repeats, ≥4 repeat units; and for pentanucleotide and hexanucleotide repeats, ≥3 repeat units. Tandem Repeats Finder version 4.07 b (Benson, 1999), with the default settings, was used to determine tandem repeats.
Sequence distance
The average pairwise sequence distance of the complete cp genomes and the genes shared among Teucrium species and other species were determined. Comparative sequence analyses were used to identify missing and ambiguous gene annotations after comparing gene orders and multiple sequence alignments. MAFFT version 7.222 (Katoh & Standley, 2013), with the default parameters, was used for the alignments of the complete cp genomes, and pairwise sequence distance was calculated using Kimura’s two-parameter (K2P) model (Kimura, 1980). A custom Python script (https://www.biostars.org/p/119214/) and DnaSP 5.10.01 (Librado & Rozas, 2009) were employed to determine single nucleotide polymorphisms (SNPs) and indel polymorphisms, respectively, among the complete genomes.
Results
Organization and general features of chloroplast genomes
The complete cp genomes of the three examined Teucrium species, Teucrium mascatense (TM), (MH325132; Data S1), Teucrium stocksianum subsp. stenophyllum (TSS) (MH325131; Data S2) and Teucrium stocksianum subsp. stocksianum (TS) (MH325133; Data S3) are circular molecules with quadripartite structures, similar to typical angiosperm cp genomes. The sizes of the TSS, TS and TM cp genomes are 150,087, 150,076 and 150,499 bp, respectively (Fig. 1 and Fig. S1). These cp genomes were compared with five related cp genomes with sizes ranging from 149,749 (S. byzantina) (Welch et al., 2016) to 151,707 bp (C. mongholica) (Table 1). The LSC regions of TSS, TS and TM are 81,707, 81,682 and 82,075 bp in length, respectively, while the sizes of the SSC regions are 17,182, 17,372 and 17,193 bp, respectively. The total numbers of genes annotated in these cp genomes are 135 in TSS and TS and 136 in TM, including 89 (TSS), 89 (TM), and 90 (TS) protein-coding genes, which accounted for 66,981, 66,487 and 67,100 bp in TSS, TM, and TS, respectively (Table 1). The total numbers of tRNAs in these genomes are 38 in TSS, 39 in TM and 37 in TS, and these numbers are similar to the numbers found in other cp genomes. The overall GC contents of the TSS, TS and TM genomes are 38.3%, 38.3% and 38.4%, respectively, and the highest GC content was observed in S. byzantina (38.7%) (Welch et al., 2016), while the lowest was 38.2% in C. mongholica (Liu et al., 2018b) (Table 1). There are seventeen intron-containing genes in these three Teucrium species cp genomes, including three genes, ycf3, clpP and rps12, that contain two introns, while the remaining fourteen genes (atpF, ndhA, ndhB, petB, petD, rpoC1, rpl2, rps16, trnA-UGC, trnG-GCC trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC) contain single introns, including eleven protein-coding genes (Table 2). The lengths of the introns vary among these genomes (Table 2). These genomes contain important genes responsible for the photosynthesis and self-replication of chloroplasts, as chloroplasts undergo independent replication (Table S1). These genes encode nine large ribosomal proteins, 12 small ribosomal proteins, 5 genes for photosystem I and 15 genes for photosystem II (Table S1). The total coding sequences in TM, TS and TSS were 66,487, 67,100 and 66,981 bp in length, including 21,190, 20,034 and 20,191 codons, respectively (Table 3).
Figure 1. Genome map of the T. stocksianum. subsp. stenophyllum and T. stocksianum subsp. stocksianum cp genomes.
Thick lines indicate the extent of the inverted repeat regions (IRa and IRb), which separate the genome into small (SSC) and large (LSC) single copy regions. Genes drawn inside the circle are transcribed clockwise, while those outside of the circle are transcribed counter clockwise. Genes belonging to different functional groups are colour coded. The dark grey in the inner circle corresponds to the GC content, while the light grey corresponds to the AT content.
Table 1. Summary of complete chloroplast genomes.
| T. stenophyllum | T. mascatense | T. stocksianum | A. reptans | C. mongholica | L. album | L. galeobdolon | S. byzantina | |
|---|---|---|---|---|---|---|---|---|
| Size (bp) | 150087 | 150499 | 150076 | 149963 | 151707 | 150505 | 151328 | 149749 |
| Overall GC contents | 38.3 | 38.3 | 38.3 | 38.3 | 38.2 | 38.6 | 38.5 | 38.7 |
| LSC size in bp | 81707 | 82075 | 81682 | 81769 | 83202 | 82444 | 82262 | 81270 |
| SSC size in bp | 17182 | 17193 | 17372 | 17102 | 17226 | 17177 | 17959 | 17679 |
| IR size in bp | 25599 | 25615 | 25511 | 25546 | 25639 | 25442 | 25553 | 25550 |
| Protein coding regions size in bp | 60573 | 56503 | 63572 | 78156 | 77946 | 80187 | 80400 | 80427 |
| tRNA size in bp | 2862 | 2961 | 2794 | 2842 | 2770 | 2793 | 2793 | 2794 |
| rRNA size in bp | 9050 | 9050 | 9048 | 9162 | 9162 | 9052 | 9052 | 9054 |
| Number of genes | 133 | 134 | 133 | 130 | 132 | 133 | 133 | 133 |
| Number of protein coding genes | 87 | 87 | 88 | 84 | 86 | 89 | 89 | 88 |
| Number of rRNA | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 |
| Number of tRNA | 38 | 39 | 37 | 38 | 37 | 37 | 37 | 37 |
| Genes with introns | 14 | 14 | 14 | 15 | 15 | 14 | 15 | 15 |
Notes.
- TM
- Teucrium mascatense
- TS
- Teucrium stocksianum subsp. stocksianum
- TSS
- Teucrium stocksianum subsp. stenophyllum
Table 2. The genes with introns in the Three Teucrium species chloroplast genome and the length of exons and introns.
| Gene | Location | Exon I (bp) | Intron 1 (bp) | Exon II (bp) | Intron II (bp) | Exon III (bp) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TSS | TS | TM | TSS | TS | TM | TSS | TS | TM | TSS | TS | TM | TSS | TS | TMM | ||
| atpF | LSC | 159 | 159 | 159 | 620 | 620 | 621 | 471 | 471 | 471 | ||||||
| petB | LSC | 6 | 6 | 6 | 686 | 693 | 684 | 642 | 654 | 642 | ||||||
| PetD | LSC | 9 | 9 | 9 | 701 | 701 | 695 | 525 | 525 | 525 | ||||||
| rpl2* | IR | 393 | 393 | 393 | 676 | 676 | 661 | 435 | 435 | 435 | ||||||
| rps16 | LSC | 40 | 40 | 40 | 912 | 912 | 908 | 227 | 227 | 227 | ||||||
| rpoC1 | LSC | 456 | 456 | 456 | 803 | 805 | 826 | 1614 | 1614 | 1611 | ||||||
| rps12* | IR/LSC | 114 | 114 | 114 | 232 | 232 | 232 | 539 | 539 | 539 | 26 | 26 | 26 | |||
| clpP | LSC | 69 | 69 | 69 | 732 | 732 | 732 | 291 | 291 | 294 | 626 | 626 | 626 | 228 | 228 | 228 |
| ndhA | SSC | 552 | 552 | 564 | 976 | 979 | 1028 | 540 | 540 | 540 | ||||||
| ndhB* | IR | 777 | 777 | 777 | 679 | 679 | 680 | 756 | 756 | 756 | ||||||
| ycf3 | LSC | 129 | 129 | 129 | 708 | 708 | 708 | 228 | 228 | 228 | 731 | 737 | 736 | 153 | 153 | 153 |
| trnA-UGC* | IR | 38 | 38 | 38 | 806 | 806 | 813 | 35 | 35 | 35 | ||||||
| trnI-GAU* | IR | 42 | 42 | 42 | 947 | 947 | 949 | 35 | 35 | 35 | ||||||
| trnL-UAA | LSC | 37 | 37 | 37 | 480 | 480 | 480 | 50 | 50 | 50 | ||||||
| trnK-UUU | LSC | 37 | 37 | 37 | 2480 | 2480 | 2480 | 26 | 26 | 26 | ||||||
| trnG-GCC | LSC | 35 | 709 | 37 | ||||||||||||
| trnV-UAC | LSC | 38 | 38 | 38 | 578 | 578 | 576 | 37 | 37 | 37 | ||||||
Notes.
- TM
- Teucrium mascatense
- TS
- Teucrium stocksianum subsp. stocksianum
- TSS
- Teucrium stocksianum subsp. stenophyllum
Table 3. Base composition of the Teucrium chloroplast genome.
| T/U | C | A | G | Length (bp) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TSS | TS | TM | TSS | TS | TM | TSS | TS | TM | TSS | TS | TM | TSS | TS | TM | |
| Genome | 31.3 | 31.3 | 31.2 | 19.4 | 19.5 | 19.4 | 30.5 | 30.4 | 30.5 | 18.8 | 18.9 | 18.9 | 150087 | 150499 | 150076 |
| LSC | 32.6 | 32.6 | 32.5 | 18.6 | 18.7 | 18.6 | 31.0 | 30.9 | 31.0 | 17.8 | 17.8 | 17.8 | 81707 | 82075 | 81682 |
| SSC | 33.7 | 33.7 | 33.7 | 16.6 | 17.0 | 16.6 | 34.2 | 33.8 | 34.1 | 15.4 | 15.5 | 15.6 | 17182 | 17193 | 17372 |
| IR | 28.3 | 28.3 | 28.3 | 20.8 | 20.8 | 20.8 | 28.3 | 28.4 | 28.3 | 22.6 | 22.5 | 22.6 | 25599 | 25615 | 25511 |
| tRNA | 25.5 | 25.4 | 25.1 | 23.2 | 23.2 | 23.2 | 22.2 | 22.2 | 22.3 | 29.1 | 29.1 | 29.3 | 2862 | 2961 | 2794 |
| rRNA | 18.8 | 18.7 | 18.6 | 23.8 | 23.8 | 23.9 | 26.1 | 26.1 | 26.1 | 31.4 | 31.5 | 31.4 | 9050 | 9048 | 9048 |
| Protein Coding genes | 31.5 | 31.3 | 31.6 | 18 | 18.1 | 18 | 29.4 | 29.6 | 29.5 | 21.1 | 20.9 | 21.0 | 60073 | 56503 | 63572 |
| 1st position | 22.92 | 25.84 | 21.052 | 18.96 | 19.89 | 16.80 | 29.52 | 21.32 | 26.91 | 28.59 | 31.98 | 24.10 | 20191 | 18834 | 21190 |
| 2nd position | 33.45 | 37.71 | 32.62 | 21.10 | 22.02 | 18.28 | 26.98 | 30.42 | 24.09 | 18.45 | 20.78 | 16.63 | 20191 | 18834 | 21190 |
| 3rd position | 36.35 | 42.95 | 29.87 | 15.03 | 15.77 | 13.24 | 31.62 | 35.78 | 27.91 | 16.19 | 17.98 | 15.092 | 20191 | 18834 | 21190 |
Notes.
- TM
- Teucrium mascatense
- TS
- Teucrium stocksianum subsp. stocksianum
- TSS
- Teucrium stocksianum subsp. stenophyllum
SSR analysis and repeats: an insight into the genome
A total of 89, 72, and 92 repeats were found in the TSS, TM and TS cp genomes, respectively. The TSS cp genome contains 24 palindromic, 26 forward, and 39 tandem repeats; the TM cp genome contains 24 palindromic, 25 forward, and 23 tandem repeats; and the TS cp genome contains 23 palindromic, 27 forward and 42 tandem repeats (Fig. 2). The total numbers of repeats in the cp genomes of related species were also analysed, and 63, 68, 69, 69 and 70 total repeats were detected in the A. reptans, C. mongholica (Liu et al., 2018b), L. album, L. galeobdolon and S. byzantina cp genomes, respectively (Fig. 2). With 31 palindromic repeats, S. byzantina contains the highest number of palindromic repeats (Welch et al., 2016), while TSS contains the highest number of forward repeats at 26, and TS contains 39 tandem repeats, the highest among the compared genomes. We found that C. mongholica (Liu et al., 2018b) contains the lowest number of palindromic repeats (22), S. byzantina (Welch et al., 2016) contains the lowest number of forward repeats (18), and A. reptans contains the lowest number of tandem repeats (13) (Fig. 2).
Figure 2. Analysis of repeated sequences in T. mascatense, T. stocksianum subsp. stenophyllum and T. stocksianum subsp. stocksianum.
(A) Total numbers of the three repeat types, (B) frequencies of forward repeats by length, (C) frequencies of tandem repeats by length and (D) frequencies of palindromic repeats by length.
SSRs were also identified in the three Teucrium cp genomes and in five other genomes from the Lamiaceae family. The highest number of SSRs was found in TSS (128 SSRs), while the lowest number of SSRs was observed in S. byzantina (121 SSRs) (Welch et al., 2016). Trinucleotide repeats were found to be the most common type of SSRs, comprising 47.67% of all SSRs (Fig. 3). Most SSRs in TSS were trinucleotide repeats (58), followed by di- (31), mono- (28), tetra- (8) and hexanucleotide (3) repeats (Table S2). In TS, most SSRs were trinucleotide repeats (59), followed by di- (29), mono- (28), tetra- (9) and hexanucleotide (3) repeats (Table S3). In TM, the number of trinucleotide SSR repeats was 58, followed by 31 dinucleotide repeats, 28 mononucleotide repeats, 8 tetranucleotide repeats, 3 hexanucleotide repeats and 1 heptanucleotide repeat, which was found in only this genome (Fig. 3, Table S4). Interestingly, pentanucleotide SSRs were found in only the L. album cp genome.
Figure 3. Analysis of simple sequence repeats (SSRs) in the T. mascatense, T. stocksianum subsp. stenophyllum and T. stocksianum subsp. stocksianum plastid genomes.
(A) Number of SSR types in complete genome, (B) Number of SSR types in LSC, SSC and IR regions, and (C) frequency of identified SSR motifs in different repeat class types.
Characteristics of junctions in the cp genomes
One of the aims of our study was to compare the actual positions of junctions within the three Teucrium cp genomes (TSS, TS and TM) and to compare these junction positions with those of three other cp genomes (A. reptans, L. album, and S. byzantina). The overall gene orientations, gene contents, and structures of these Teucrium species were the same, but these genomes possessed obvious differences at the junctions, similar to what has been observed in other typical cp genomes (Fig. 4). At JLB(LSC-IRb), the rps19 gene is present, exceeding the IRb region by 58 bp in TSS, 23 bp in TS and 62 bp in TM. The rpl2 gene is present in the IRb region in all genomes at varying distances from the junction. The JSB(IRb-SSC) junction is located in the ψycf1 gene, a pseudogene in the IRb region with a length equivalent to the length that the IRa is expanded into the ψycf1 gene. Interestingly, the ndhF gene is present in the SSC regions of both TSS and TS, while in TM, it is present at the JSB junction, overlapping the ψycf1 gene (Fig. 4). Moreover, the JLA(IRa-LSC) border is characteristically located upstream of ψrps19 and downstream of the psbA gene. The IRa region was expanded to partially include ψrps19, creating a truncated ψrps19 copy at the JLA border in all the examined Teucrium species. However, in S. byzantina, this pseudogene is missing. The extent of the IR expansion into ψrps19 is 58 bp, 23 bp and 61 bp in TSS, TS and TM, respectively. Similarly, in all genomes, the pbsA gene is present in the LSC at varying distances from the JLAjunction.
Figure 4. Distances between adjacent genes and junctions of the small single-copy (SSC), large single-copy (LSC), and two inverted repeat (IR) regions among eight plastid genomes within the family Lamiaceae.
Boxes above and below the primary line indicate the adjacent border genes. The figure is not to scale with regards to sequence length and only shows relative changes at or near the IR/SC borders.
Comparative analysis of sequence variation
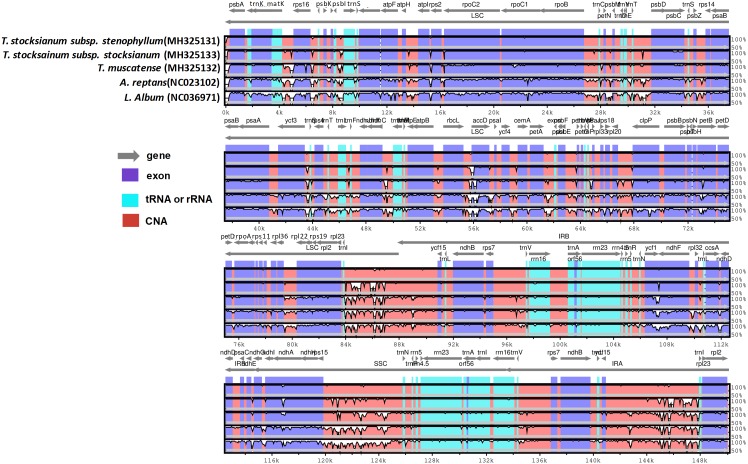
Comparisons among these genomes using mVISTA revealed several regions of sequence variation. The TSS genome was used as the reference genome. Some genes, such as rps16, rpoC1, ycf3, accD, clpP, petB, petD, accD, ycf1, ndhA, petD, and atpF, showed sequence variation among these genomes. In the IRb region, the most divergent regions among the compared genomes was the rps7-trnV region. In the LSC region, the rpoC1 gene showed sequence variation only in A. reptans and L. album, as did the rps16 and petB genes. In the SSC region, the ndhA gene also showed sequence divergence among the compared genomes (Fig. 5).
Figure 5. Visual alignment of plastid genomes from T. mascatense, T. stocksianum subsp. stenophyllum and T. stocksianum subsp. stocksianum with previously reported A. reptans and L. album cp genomes.
VISTA-based identity plot showing sequence identity among eight species, using T. stocksianum subsp. stenophyllum as a reference.
Following these findings, we calculated the average pairwise sequence distances among the complete cp genomes of these eight species (A. reptans, C. mongholica, L. album, L. galeobdolon, S. byzantina, TM, TS, and TSS), and the results revealed that TSS had greater sequence divergence from TM (0.018) than from TS (0.006). We found that the genome sequence distances between TS and TM were smaller than those when these genomes were compared to other genomes (Table S5). Furthermore, when the TSS genome was compared with those of TM, TS, A. reptans, C. mongholica, L. album, L. galeobdolon and S. byzantina with respect to indels and SNPs, 5,458, 6,254, 10,130, 14,106, 19,575, 18,982, and 24,008 SNPs were detected, while 2,275, 2,275, 13,291, 12,991, 18,241, 18,201, and 29,438 indels were detected, respectively (Table S6).
Discussion
The genomic structures and gene orders of the Teucrium cp genomes are highly conserved, and no rearrangement has occurred. The IRs of the Teucrium species are approximately 25.5 kb in length, and this value is within the size range found in most angiosperm cp genomes (20 ± 28 kb) (Chumley et al., 2006). The TSS cp genome is 150,087 bp, the TS genome is 150,076 bp, and the TM genome is 150,499 bp, and the sizes of these reported cp genomes are consistent with the sizes of the previously reported cp genomes of C. mongholica (151,707 bp) (Liu et al., 2018b), Salvia miltiorrhiza (151,328 bp) (Qian et al., 2013), Origanum vulgare L. (151,935 bp) and Mentha spicata (152,132 bp) (Lukas & Novak, 2013). The Teucrium cp genome has a typical quadripartite structure and consists of an SSC and an LSC separated by a pair of IRs. All the sequenced Teucrium species contain higher AT content than GC content. The GC content in TSS, TS, TM and A. reptans is almost identical, and similarly, the GC content in C. mongholica is 38.2% (Abu-Irmaileh & Afifi, 2003), that in L. album is 38.6%, that in L. galeobdolon is 38.5% and that in S. byzantina is 38.7%. The IR region has a higher GC content than the non-coding intergenic regions due to the presence of rRNA genes (Bock, 2007).
The gene orders and gene contents of these genomes are conserved. The number of genes in these Teucrium cp genomes are similar to the numbers that were previously reported in the M. spicata (Wang et al., 2017), Lavandula angustifolia (Ma, 2018), and Perilla frutescens (L.) Britton (Shen et al., 2016) genomes. The number of intron-containing genes (14) in the sequenced genomes (TM, TS, and TSS) was similar in this study. With the exception of the A. reptans and C. mongholica genomes (Liu et al., 2018b), which contain 15 intron-containing genes, the intron contents of cp genomes are conserved; however, in some species such as Lagerstroemia fauriei, structural changes, such as sequence losses or variations (SNP), have been reported (Gu et al., 2016). Some genes, such as atpF (ATP synthase), rpoC2 (RNA polymerase) and ribosomal proteins (rpl12, rps12, and rps16), are known to have structural intron variation (Daniell et al., 2016; He et al., 2017). The cp genome can gain or lose introns during evolution, and this process plays an important role in the regulation of gene expression through the stabilization of the transcript or through alternative splicing (Daniell et al., 2008). Our results reveal that there are 11 protein-coding genes, six tRNA genes (TSS) and five tRNA genes (TS and TM) that contain introns. As in the previously reported cp genomes, both clpP and ycf3 contain double introns. The previously reported O. vulgare cp genome (Lukas & Novak, 2013) shows a similar result, while in the S. miltiorrhiza (Qian et al., 2013) cp genome, there are nine protein-coding genes and six tRNAs that contain introns, and the number of double-intron-containing genes is three (Qian et al., 2013).
In most land plants, the cp genome has a collinear gene order, but it also displays some remarkable changes, such as sequence inversion (Cho et al., 2015), gene loss (Fu et al., 2016), and expansion and contraction at the borders between the LSC, the SSC and the IRs (Choi, Chung & Park, 2016). The expansion and contraction of the IR regions often results in the length variations observed among cp genomes (Cho & Park, 2016; Hu, Woeste & Zhao, 2017). In some genomes, such as Fabaceae (Wang et al., 2018) Erodium and Sarcocaulon (Blazier et al., 2016), the loss of IRs has also been reported. The differences in genome size among the sequenced and compared species can be explained by the variations in the LSC, SSC and IR regions. The sizes of the cp genomes of the three Teucrium species (TSS, TS, and TM) differ, and there are some notable variations in the junction regions. The boundaries between the LSC, the SSC and the IRs were identical in all the cp genomes studied. The LSC/IRb boundary of the three Teucrium cp genomes and of the compared genomes is located in the rps19 gene, and a small fraction of the rps19 gene is also located in the IRb region, similar to the previously reported S. miltiorrhiza cp genome (Qian et al., 2013) and O. vulgare cp genome (Lukas & Novak, 2013) and the cp genomes from seven species from the genus Ilex (Yao et al., 2016). In contrast, there are some cp genomes in which the rps19 gene does not extend into the IR region, such as the Millettia pinnata cp genome (Kazakoff et al., 2012) and Lupinus luteus cp genome (Martin et al., 2014). It has been reported in various studies (Wang et al., 2008) that, mostly in monocots, the rps19 gene occurs inside the IR region, as in the Oryza AA genome (Wambugu et al., 2015). The ycf1 gene extends over the SSC/IRb junction and overlaps with the ndhF gene in most of the compared genomes, including TM, while in TSS and TS, the ycf1 gene does not overlap with the ndhF gene and is located on both sides of the SSC and IRb; a similar result has also been observed in the Petroselinum crispum, Tiedemania filiformis and Panax ginseng cp genomes (Kim & Lee, 2004; Li et al., 2018).
Repetitive sequences, such as tandem repeats and SSRs, play important roles in the stabilization and rearrangement of cp genome sequences (Do Nascimento Vieira et al., 2014) and can affect copy number variation among different and similar species. Such features in cp genomes can be utilized for molecular marker design, which helps in plant identification at the molecular level (Cho et al., 2016) and phylogenetic analyses (Williams et al., 2016). We found that there are more repeats in the intergenic spacer regions than in the coding regions, as expected. Tandem repeats and SSRs can account for recombination in cp genomes, which leads to differences between genomes (Ogihara, Terachi & Sasakuma, 1988). The Teucrium species possess high numbers of repeats in their cp genomes, and it is evident from previous studies that large and complex repeats also play major roles in the rearrangement of sequences within cp genomes and in the evolution of cp genomes (Milligan, Hampton & Palmer, 1989; Cavalier-Smith, 2002; Bausher et al., 2006). Our findings show that TS has the highest number of repeats (92), while TSS has the lowest (53) number of repeats.
In our study, tandem repeats were found to be the most abundant in the Teucrium species genomes, showing similar traits to the previously reported S. miltiorrhiza cp genome (Qian et al., 2013). SSRs in the Teucrium species genomes primarily contain numerous AT subunits, with mononucleotide repeats comprising only A and T repeats. These results are consistent with those for the previously reported cp genomes of angiosperms (Qian et al., 2013; Khan et al., 2017), which have high AT contents (Nie et al., 2012). Furthermore, trinucleotide repeats were more abundant than any other type of nucleotide repeats in these studied genomes, and this finding is consistent with the findings of the previously reported cp genome of Origanum vulgare (Lukas & Novak, 2013).
Our study establishes that a higher level of variation was observed in the following regions of the three Teucrium species and two other compared species: rps16, rpoC1, ycf3, accD, clpP, petB, petD, ycf1, ndhA, and atpF. Therefore, these regions within the genus Teucrium are useful regions for elucidating phylogenetic relationships. These regions contain variation and are suitable for phylogenetic analysis of Teucrium and for evaluation of unresolved phylogenetic relationships. Ycf1, ycf2, rpoC2, and ndhF were confirmed to be the most divergent regions in the previously reported S. miltiorrhiza cp genome within the Lamiaceae family (Qian et al., 2013). Genes such as rpoC1 and ycf1 were also found to be among the most divergent in the six reported cp genomes from Asteraceae species (Nie et al., 2012). Moreover, coding regions such as ndhA, rps16, accD, clpP, ccsA, infA, rpl22, rpl32 and ycf1 were also found to be the most divergent genes in vascular plant cp genomes (Kumar et al., 2009).
Conclusion
This study successfully mapped the first three cp genomes of the genus Teucrium from the family Lamiaceae using next-generation sequencing technology. The genome organizations and gene orders of these three Teucrium cp genomes demonstrated similarity among these three specimens as well as when compared to other genomes, such as S. miltiorrhiza, from the Lamiaceae family. Repetitive sequences, such as SSRs and tandem repeats, were determined within the eight cp genomes. Contraction and expansion as well as sequence divergence inside these genomes were also ascertained. The findings of our study will further facilitate the biological study of this medicinally significant plant genus.
Supplemental Information
Thick lines indicate the extent of the inverted repeat regions (IRa and IRb), which separate the genome into small (SSC) and large (LSC) single copy regions. Genes drawn inside the circle are transcribed clockwise, and those outside are transcribed counter clockwise. Genes belonging to different functional groups are color-coded. The dark grey in the inner circle corresponds to the GC content and the light grey corresponds to the AT content.
Funding Statement
The authors received no funding for this work.
Contributor Information
Abdul Latif Khan, Email: latifepm78@yahoo.co.uk.
Ahmed Al-Harrasi, Email: aharrasi@unizwa.edu.om.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Arif Khan conceived and designed the experiments, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Sajjad Asaf conceived and designed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Abdul Latif Khan conceived and designed the experiments, contributed reagents/materials/analysis tools, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Adil Khan conceived and designed the experiments.
Ahmed Al-Harrasi performed the experiments, contributed reagents/materials/analysis tools, approved the final draft.
Omar Al-Sudairy conceived and designed the experiments, contributed reagents/materials/analysis tools.
Noor Mazin AbdulKareem performed the experiments, contributed reagents/materials/analysis tools.
Nadiya Al-Saady performed the experiments.
Ahmed Al-Rawahi analyzed the data.
Field Study Permissions
The following information was supplied relating to field study approvals (i.e., approving body and any reference numbers):
The Director General of Nature Conversation from the Sultanate of Oman, Ministry of Environment & Climate Affairs provided a permit to collect wild plant specimens (permit no. 4/2106).
DNA Deposition
Data Availability
The following information was supplied regarding data availability:
The specimens are accessible at the University of Nizwa Herbarium Center, Oman, with the voucher numbers UCTM11 (Teucrium mascatense), UCTS32 (Teucrium stocksianum subsp. stenophyllum), and UCTS30 (Teucrium stocksianum subsp. stocksianum). http://sweetgum.nybg.org/science/ih/herbarium-list/?NamOrganisationAcronym=NMSRC.
References
- Abdollahi, Karimpour & Monsef-Esfehani (2003).Abdollahi M, Karimpour H, Monsef-Esfehani HR. Antinociceptive effects of Teucrium polium L. total extract and essential oil in mouse writhing test. Pharmacological Research. 2003;48:31–35. [PubMed] [Google Scholar]
- Abu-Irmaileh & Afifi (2003).Abu-Irmaileh BE, Afifi FU. Herbal medicine in Jordan with special emphasis on commonly used herbs. Journal of Ethnopharmacology. 2003;89:193–197. doi: 10.1016/S0378-8741(03)00283-6. [DOI] [PubMed] [Google Scholar]
- Allen (2015).Allen JF. Why chloroplasts and mitochondria retain their own genomes and genetic systems: colocation for redox regulation of gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(33):10231–10238. doi: 10.1073/pnas.1500012112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews (2015).Andrews S. Babraham bioinformatics-FastQC a quality control tool for high throughput sequence data. 2015. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [06 December 2018]. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- Barrachina et al. (1995).Barrachina M, Bello R, Martínez-Cuesta M, Esplugues J, Primo-Yúfera E. Antiinflammatory activity and effects on isolated smooth muscle of extracts from different Teucrium species. Phytotherapy Research. 1995;9:368–371. doi: 10.1002/ptr.2650090512. [DOI] [Google Scholar]
- Barrett et al. (2013).Barrett CF, Davis JI, Leebens-Mack J, Conran JG, Stevenson DW. Plastid genomes and deep relationships among the commelinid monocot angiosperms. Cladistics. 2013;29:65–87. doi: 10.1111/j.1096-0031.2012.00418.x. [DOI] [PubMed] [Google Scholar]
- Barrett et al. (2014).Barrett CF, Freudenstein JV, Li J, Mayfield-Jones DR, Perez L, Pires JC, Santos C. Investigating the path of plastid genome degradation in an early-transitional clade of heterotrophic orchids, and implications for heterotrophic angiosperms. Molecular Biology and Evolution. 2014;31:3095–3112. doi: 10.1093/molbev/msu252. [DOI] [PubMed] [Google Scholar]
- Bausher et al. (2006).Bausher MG, Singh ND, Lee S-B, Jansen RK, Daniell H. The complete chloroplast genome sequence of Citrus sinensis (L.) Osbeck var’Ridge Pineapple’: organization and phylogenetic relationships to other angiosperms. BMC Plant Biology. 2006;6:21. doi: 10.1186/1471-2229-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson (1999).Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Research. 1999;27(2):573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazier et al. (2016).Blazier JC, Jansen RK, Mower JP, Govindu M, Zhang J, Weng M-L, Ruhlman TA. Variable presence of the inverted repeat and plastome stability in Erodium. Annals of Botany. 2016;117:1209–1220. doi: 10.1093/aob/mcw065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock (2007).Bock R. Cell and molecular biology of plastids. Springer; Berlin, Heidelberg: 2007. Structure, function, and inheritance of plastid genomes; pp. 29–63. [Google Scholar]
- Bolger, Lohse & Usadel (2014).Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai et al. (2008).Cai Z, Guisinger M, Kim H-G, Ruck E, Blazier JC, McMurtry V, Kuehl JV, Boore J, Jansen RK. Extensive reorganization of the plastid genome of Trifolium subterraneum (Fabaceae) is associated with numerous repeated sequences and novel DNA insertions. Journal of Molecular Evolution. 2008;67:696–704. doi: 10.1007/s00239-008-9180-7. [DOI] [PubMed] [Google Scholar]
- Carbonell-Caballero et al. (2015).Carbonell-Caballero J, Alonso R, Ibañez V, Terol J, Talon M, Dopazo J. A phylogenetic analysis of 34 chloroplast genomes elucidates the relationships between wild and domestic species within the genus Citrus. Molecular Biology and Evolution. 2015;32:2015–2035. doi: 10.1093/molbev/msv082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith (2002).Cavalier-Smith T. Chloroplast evolution: secondary symbiogenesis and multiple losses. Current Biology. 2002;12:R62–R64. doi: 10.1016/S0960-9822(01)00675-3. [DOI] [PubMed] [Google Scholar]
- Cho et al. (2016).Cho K-S, Cheon K-S, Hong S-Y, Cho J-H, Im J-S, Mekapogu M, Yu Y-S, Park T-H. Complete chloroplast genome sequences of Solanum commersonii and its application to chloroplast genotype in somatic hybrids with Solanum tuberosum. Plant Cell Reports. 2016;35:2113–2123. doi: 10.1007/s00299-016-2022-y. [DOI] [PubMed] [Google Scholar]
- Cho & Park (2016).Cho K-S, Park T-H. Complete chloroplast genome sequence of Solanum nigrum and development of markers for the discrimination of S. nigrum. Horticulture, Environment, and Biotechnology. 2016;57:69–78. doi: 10.1007/s13580-016-0003-2. [DOI] [Google Scholar]
- Cho et al. (2015).Cho K-S, Yun B-K, Yoon Y-H, Hong S-Y, Mekapogu M, Kim K-H, Yang T-J. Complete chloroplast genome sequence of tartary buckwheat (Fagopyrum tataricum) and comparative analysis with common buckwheat (F. esculentum) PLOS ONE. 2015;10:e0125332. doi: 10.1371/journal.pone.0125332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, Chung & Park (2016).Choi KS, Chung MG, Park SJ. The complete chloroplast genome sequences of three Veroniceae species (Plantaginaceae): comparative analysis and highly divergent regions. Frontiers in Plant Science. 2016;7:355–362. doi: 10.3389/fpls.2016.00355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chumley et al. (2006).Chumley TW, Palmer JD, Mower JP, Fourcade HM, Calie PJ, Boore JL, Jansen RK. The complete chloroplast genome sequence of Pelargonium × hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Molecular Biology and Evolution. 2006;23:2175–2190. doi: 10.1093/molbev/msl089. [DOI] [PubMed] [Google Scholar]
- Daniell et al. (2016).Daniell H, Lin CS, Yu M, Chang WJ. Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biology. 2016;17:134–162. doi: 10.1186/s13059-016-1004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell et al. (2008).Daniell H, Wurdack KJ, Kanagaraj A, Lee S-B, Saski C, Jansen RK. The complete nucleotide sequence of the cassava (Manihot esculenta) chloroplast genome and the evolution of atpF in Malpighiales: RNA editing and multiple losses of a group II intron. Theoretical and Applied Genetics. 2008;116:723–737. doi: 10.1007/s00122-007-0706-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do Nascimento Vieira et al. (2014).Do Nascimento Vieira L, Faoro H, Rogalski M, De Freitas Fraga HP, Cardoso RLA, De Souza EM, De Oliveira Pedrosa F, Nodari RO, Guerra MP. The complete chloroplast genome sequence of Podocarpus lambertii: genome structure, evolutionary aspects, gene content and SSR detection. PLOS ONE. 2014;9:e90618. doi: 10.1371/journal.pone.0090618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan et al. (2018).Fan W-B, Wu Y, Yang J, Shahzad K, Li Z-H. Comparative chloroplast genomics of dipsacales species: insights into sequence variation, adaptive evolution, and phylogenetic relationships. Frontiers in Plant Science. 2018;9:689–692. doi: 10.3389/fpls.2018.00689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frailey et al. (2018).Frailey DC, Chaluvadi SR, Vaughn JN, Coatney CG, Bennetzen JL. Gene loss and genome rearrangement in the plastids of five Hemiparasites in the family Orobanchaceae. BMC Plant Biology. 2018;18:30. doi: 10.1186/s12870-018-1249-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer et al. (2004).Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Research. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu et al. (2016).Fu P-C, Zhang Y-Z, Geng H-M, Chen S-L. The complete chloroplast genome sequence of Gentiana lawrencei var. farreri (Gentianaceae) and comparative analysis with its congeneric species. PeerJ. 2016;4:e2540. doi: 10.7717/peerj.2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazanfar (1994).Ghazanfar SA. Handbook of Arabian medicinal plants. CRC Press Inc.; Boca Raton: 1994. p. 265. [Google Scholar]
- Greiner et al. (2008).Greiner S, Wang X, Herrmann RG, Rauwolf U, Mayer K, Haberer G, Meurer J. The complete nucleotide sequences of the 5 genetically distinct plastid genomes of Oenothera, subsection Oenothera: II. A microevolutionary view using bioinformatics and formal genetic data. Molecular Biology and Evolution. 2008;25:2019–2030. doi: 10.1093/molbev/msn149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu et al. (2016).Gu C, Tembrock LR, Johnson NG, Simmons MP, Wu Z. The complete plastid genome of Lagerstroemia fauriei and loss of rpl2 intron from Lagerstroemia (Lythraceae) PLOS ONE. 2016;11:e0150752. doi: 10.1371/journal.pone.0150752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn, Bachmann & Chevreux (2013).Hahn C, Bachmann L, Chevreux B. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Research. 2013;41:e129–e129. doi: 10.1093/nar/gkt371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley et al. (2004).Harley RM, Atkins S, Budantsev AL, Cantino PD, Conn BJ, Grayer R, Harley MM, De Kok RD, Krestovskaja TD, Morales R, Paton AJ. Flowering plants. Dicotyledons. Springer; Berlin, Heidelberg: 2004. Labiatae; pp. 167–275. [Google Scholar]
- He et al. (2017).He L, Qian J, Li X, Sun Z, Xu X, Chen S. Complete chloroplast genome of medicinal plant Lonicera japonica: genome rearrangement, intron gain and loss, and implications for phylogenetic studies. Molecules. 2017;22(2):249–261. doi: 10.3390/molecules22020249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Woeste & Zhao (2017).Hu Y, Woeste KE, Zhao P. Completion of the chloroplast genomes of five Chinese Juglans and their contribution to chloroplast phylogeny. Frontiers in Plant Science. 2017;7:1955–1970. doi: 10.3389/fpls.2016.01955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen et al. (2007).Jansen RK, Cai Z, Raubeson LA, Daniell H, Leebens-Mack J, Müller KF, Guisinger-Bellian M, Haberle RC, Hansen AK, Chumley TW. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:19369–19374. doi: 10.1073/pnas.0709121104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh & Standley (2013).Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazakoff et al. (2012).Kazakoff SH, Imelfort M, Edwards D, Koehorst J, Biswas B, Batley J, Scott PT, Gresshoff PM. Capturing the biofuel wellhead and powerhouse: the chloroplast and mitochondrial genomes of the leguminous feedstock tree Pongamia pinnata. PLOS ONE. 2012;7(12):e51687. doi: 10.1371/journal.pone.0051687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse et al. (2012).Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan et al. (2017).Khan AL, Al-Harrasi A, Asaf S, Park CE, Park G-S, Khan AR, Lee I-J, Al-Rawahi A, Shin J-H. The first chloroplast genome sequence of Boswellia sacra, a resin-producing plant in Oman. PLOS ONE. 2017;12(1):e0169794. doi: 10.1371/journal.pone.0169794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim & Lee (2004).Kim K-J, Lee H-L. Complete chloroplast genome sequences from Korean ginseng (Panax schinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Research. 2004;11:247–261. doi: 10.1093/dnares/11.4.247. [DOI] [PubMed] [Google Scholar]
- Kimura (1980).Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Kraemer et al. (2009).Kraemer L, Beszteri B, Gäbler-Schwarz S, Held C, Leese F, Mayer C, Pöhlmann K, Frickenhaus S. S TAMP: Extensions to the S TADEN sequence analysis package for high throughput interactive microsatellite marker design. BMC Bioinformatics. 2009;10:41. doi: 10.1186/1471-2105-10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar et al. (2009).Kumar S, Hahn FM, McMahan CM, Cornish K, Whalen MC. Comparative analysis of the complete sequence of the plastid genome of Parthenium argentatum and identification of DNA barcodes to differentiate Parthenium species and lines. BMC Plant Biology. 2009;9:131. doi: 10.1186/1471-2229-9-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar et al. (2008).Kumar S, Nei M, Dudley J, Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Briefings in Bioinformatics. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz et al. (2001).Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Research. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead & Salzberg (2012).Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee et al. (2007).Lee H-L, Jansen RK, Chumley TW, Kim K-J. Gene relocations within chloroplast genomes of Jasminum and Menodora (Oleaceae) are due to multiple, overlapping inversions. Molecular Biology and Evolution. 2007;24:1161–1180. doi: 10.1093/molbev/msm036. [DOI] [PubMed] [Google Scholar]
- Li et al. (2018).Li Y, Zhang Z, Yang J, Lv G. Complete chloroplast genome of seven Fritillaria species, variable DNA markers identification and phylogenetic relationships within the genus. PLOS ONE. 2018;13(3):e0194613. doi: 10.1371/journal.pone.0194613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado & Rozas (2009).Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Liu et al. (2018a).Liu H-J, Ding C-H, He J, Cheng J, Pei LY, Xie L. Complete chloroplast genomes of Archiclematis, Naravelia and Clematis (Ranunculaceae), and their phylogenetic implications. Phytotaxa. 2018a;343:214–226. doi: 10.11646/phytotaxa.343.3.2. [DOI] [Google Scholar]
- Liu et al. (2018b).Liu L, Zhang C, Wang Y, Dong M, Shang F, Li P. The complete chloroplast genome of Caryopteris mongholica and phylogenetic implications in Lamiaceae. Conservation Genetics Resources. 2018b;10(3):281–285. doi: 10.1007/s12686-017-0802-5. [DOI] [Google Scholar]
- Lohse, Drechsel & Bock (2007).Lohse M, Drechsel O, Bock R. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Current Genetics. 2007;52:267–274. doi: 10.1007/s00294-007-0161-y. [DOI] [PubMed] [Google Scholar]
- Lukas & Novak (2013).Lukas B, Novak J. The complete chloroplast genome of Origanum vulgare L.(Lamiaceae) Gene. 2013;528:163–169. doi: 10.1016/j.gene.2013.07.026. [DOI] [PubMed] [Google Scholar]
- Ma (2018).Ma L. The complete chloroplast genome sequence of the fragrant plant Lavandula angustifolia (Lamiaceae) Mitochondrial DNA Part B. 2018;3:135–136. doi: 10.1080/23802359.2018.1431067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin et al. (2014).Martin GE, Rousseau-Gueutin M, Cordonnier S, Lima O, Michon-Coudouel S, Naquin D, De Carvalho JF, Aïnouche M, Salmon A, Aïnouche A. The first complete chloroplast genome of the Genistoid legume Lupinus luteus: evidence for a novel major lineage-specific rearrangement and new insights regarding plastome evolution in the legume family. Annals of Botany. 2014;113:1197–1210. doi: 10.1093/aob/mcu050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCauley et al. (2007).McCauley DE, Sundby AK, Bailey MF, Welch ME. Inheritance of chloroplast DNA is not strictly maternal in Silene vulgaris (Caryophyllaceae): evidence from experimental crosses and natural populations. American Journal of Botany. 2007;94:1333–1337. doi: 10.3732/ajb.94.8.1333. [DOI] [PubMed] [Google Scholar]
- Miller & Morris (1988).Miller AG, Morris M. Plants of Dhofar: the southern region of Oman, traditional, economic and medicinal uses. Office of the Adviser for Conservation of the Environment, Diwan of Royal Court Sultanate of Oman; Muscat: 1988. xxvii, 361p-col illus. [Google Scholar]
- Milligan, Hampton & Palmer (1989).Milligan BG, Hampton JN, Palmer JD. Dispersed repeats and structural reorganization in subclover chloroplast DNA. Molecular Biology and Evolution. 1989;6:355–368. doi: 10.1093/oxfordjournals.molbev.a040558. [DOI] [PubMed] [Google Scholar]
- Moore et al. (2007).Moore MJ, Bell CD, Soltis PS, Soltis DE. Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:19363–19368. doi: 10.1073/pnas.0708072104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro & El Oualidi (1999).Navarro T, El Oualidi J. Trichome morphology in Teucrium L.(Labiatae). A taxonomic review. Anales del Jardín Botánico de Madrid. 1999;57(2):277–297. [Google Scholar]
- Nie et al. (2012).Nie X, Lv S, Zhang Y, Du X, Wang L, Biradar SS, Tan X, Wan F, Weining S. Complete chloroplast genome sequence of a major invasive species, crofton weed (Ageratina adenophora) PLOS ONE. 2012;7(5):e36869. doi: 10.1371/journal.pone.0036869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogihara, Terachi & Sasakuma (1988).Ogihara Y, Terachi T, Sasakuma T. Intramolecular recombination of chloroplast genome mediated by short direct-repeat sequences in wheat species. Proceedings of the National Academy of Sciences of the United States of America. 1988;85:8573–8577. doi: 10.1073/pnas.85.22.8573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmstead & Palmer (1994).Olmstead RG, Palmer JD. Chloroplast DNA systematics: a review of methods and data analysis. American Journal of Botany. 1994;81(9):1205–1224. [Google Scholar]
- Parks, Cronn & Liston (2009).Parks M, Cronn R, Liston A. Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biology. 2009;7:84. doi: 10.1186/1741-7007-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patzelt (2015).Patzelt A. Oman plant red data book. Oman botanic Garden: Office of the Adviser for Conservation of the Environment, Diwan of Royal Court Sultanate of Oman; Muscat: 2015. [Google Scholar]
- Qian et al. (2013).Qian J, Song J, Gao H, Zhu Y, Xu J, Pang X, Yao H, Sun C, Xe Li, Li C. The complete chloroplast genome sequence of the medicinal plant Salvia miltiorrhiza. PLOS ONE. 2013;8(2):e57607. doi: 10.1371/journal.pone.0057607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboud & Zeyl (1994).Reboud X, Zeyl C. Organelle inheritance in plants. Heredity. 1994;72:132–140. doi: 10.1038/hdy.1994.19. [DOI] [Google Scholar]
- Salmaki et al. (2016).Salmaki Y, Kattari S, Heubl G, Bräuchler C. Phylogeny of non-monophyletic Teucrium (Lamiaceae: Ajugoideae): implications for character evolution and taxonomy. Taxon. 2016;65:805–822. doi: 10.12705/654.8. [DOI] [Google Scholar]
- Sarac & Ugur (2007).Sarac N, Ugur A. Antimicrobial activities and usage in folkloric medicine of some Lamiaceae species growing in Mugla, Turkey. EurAsian Journal of BioSciences. 2007;4:28–37. [Google Scholar]
- Schattner, Brooks & Lowe (2005).Schattner P, Brooks AN, Lowe TM. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Research. 2005;33:W686–W689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw et al. (2005).Shaw J, Lickey EB, Beck JT, Farmer SB, Liu W, Miller J, Siripun KC, Winder CT, Schilling EE, Small RL. The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. American Journal of Botany. 2005;92:142–166. doi: 10.3732/ajb.92.1.142. [DOI] [PubMed] [Google Scholar]
- Shaw et al. (2014).Shaw J, Shafer HL, Leonard OR, Kovach MJ, Schorr M, Morris AB. Chloroplast DNA sequence utility for the lowest phylogenetic and phylogeographic inferences in angiosperms: the tortoise and the hare IV. American Journal of Botany. 2014;101:1987–2004. doi: 10.3732/ajb.1400398. [DOI] [PubMed] [Google Scholar]
- Shen et al. (2016).Shen Q, Yang J, Lu C, Wang B, Song C. The complete chloroplast genome sequence of Perilla frutescens (L.) Mitochondrial DNA Part A. 2016;27:3306–3307. doi: 10.3109/19401736.2015.1015015. [DOI] [PubMed] [Google Scholar]
- Shi et al. (2012).Shi C, Hu N, Huang H, Gao J, Zhao Y-J, Gao L-Z. An improved chloroplast DNA extraction procedure for whole plastid genome sequencing. PLOS ONE. 2012;7(2):e31468. doi: 10.1371/journal.pone.0031468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulubelen, Topu & Sönmez (2000).Ulubelen A, Topu G, Sönmez U. Chemical and biological evaluation of genus Teucrium. In: Atta-ur Rahman, editor. Studies in natural products chemistry. Elsevier Science; Amsterdam: 2000. pp. 591–648. [Google Scholar]
- Wambugu et al. (2015).Wambugu PW, Brozynska M, Furtado A, Waters DL, Henry RJ. Relationships of wild and domesticated rices (Oryza AA genome species) based upon whole chloroplast genome sequences. Scientific Reports. 2015;5:13957. doi: 10.1038/srep13957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2017).Wang K, Li L, Hua Y, Zhao M, Li S, Sun H, Lv Y, Wang Y. The complete chloroplast genome of Mentha spicata, an endangered species native to South Europe. Mitochondrial DNA Part B. 2017;2:907–909. doi: 10.1080/23802359.2017.1413311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2008).Wang R-J, Cheng C-L, Chang C-C, Wu C-L, Su T-M, Chaw S-M. Dynamics and evolution of the inverted repeat-large single copy junctions in the chloroplast genomes of monocots. BMC Evolutionary Biology. 2008;8:36. doi: 10.1186/1471-2148-8-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2018).Wang Y-H, Wicke S, Wang H, Jin J-J, Chen S-Y, Zhang S-D, Li D-Z, Yi T-S. Plastid genome evolution in the early-diverging Legume subfamily Cercidoideae (Fabaceae) Frontiers in Plant Science. 2018;9:138–150. doi: 10.3389/fpls.2018.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch et al. (2016).Welch AJ, Collins K, Ratan A, Drautz-Moses DI, Schuster SC, Lindqvist C. Data characterizing the chloroplast genomes of extinct and endangered Hawaiian endemic mints (Lamiaceae) and their close relatives. Data in Brief. 2016;7:900–922. doi: 10.1016/j.dib.2016.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicke et al. (2011).Wicke S, Schneeweiss GM, Müller KF, Quandt D. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Molecular Biology. 2011;76:273–297. doi: 10.1007/s11103-011-9762-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams et al. (2016).Williams AV, Miller JT, Small I, Nevill PG, Boykin LM. Integration of complete chloroplast genome sequences with small amplicon datasets improves phylogenetic resolution in Acacia. Molecular Phylogenetics and Evolution. 2016;96:1–8. doi: 10.1016/j.ympev.2015.11.021. [DOI] [PubMed] [Google Scholar]
- Wyman, Jansen & Boore (2004).Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20:3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- Yao et al. (2016).Yao X, Tan Y-H, Liu Y-Y, Song Y, Yang J-B, Corlett RT. Chloroplast genome structure in Ilex (Aquifoliaceae) Scientific Reports. 2016;6:28559. doi: 10.1038/srep28559. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Thick lines indicate the extent of the inverted repeat regions (IRa and IRb), which separate the genome into small (SSC) and large (LSC) single copy regions. Genes drawn inside the circle are transcribed clockwise, and those outside are transcribed counter clockwise. Genes belonging to different functional groups are color-coded. The dark grey in the inner circle corresponds to the GC content and the light grey corresponds to the AT content.
Data Availability Statement
The following information was supplied regarding data availability:
The specimens are accessible at the University of Nizwa Herbarium Center, Oman, with the voucher numbers UCTM11 (Teucrium mascatense), UCTS32 (Teucrium stocksianum subsp. stenophyllum), and UCTS30 (Teucrium stocksianum subsp. stocksianum). http://sweetgum.nybg.org/science/ih/herbarium-list/?NamOrganisationAcronym=NMSRC.