Abstract
Increased brain α-synuclein (SNCA) protein expression resulting from gene duplication and triplication can cause a familial form of Parkinson’s disease (PD). Dopaminergic neurons exhibit elevated iron levels that can accelerate toxic SNCA fibril formation. Examinations of human post mortem brain have shown that while mRNA levels for SNCA in PD have been shown to be either unchanged or decreased with respect to healthy controls, higher levels of insoluble protein occurs during PD progression. We show evidence that SNCA can be regulated via the 5′untranslated region (5′UTR) of its transcript, which we modeled to fold into a unique RNA stem loop with a CAGUGN apical loop similar to that encoded in the canonical iron-responsive element (IRE) of L- and H-ferritin mRNAs. The SNCA IRE-like stem loop spans the two exons that encode its 5′UTR, whereas, by contrast, the H-ferritin 5′UTR is encoded by a single first exon. We screened a library of 720 natural products (NPs) for their capacity to inhibit SNCA 5′UTR driven luciferase expression. This screen identified several classes of NPs, including the plant cardiac glycosides, mycophenolic acid (an immunosuppressant and Fe chelator), and, additionally, posiphen was identified to repress SNCA 5′UTR conferred translation. Western blotting confirmed that Posiphen and the cardiac glycoside, strophanthidine, selectively blocked SNCA expression (~1 μM IC50) in neural cells. For Posiphen this inhibition was accelerated in the presence of iron, thus providing a known APP-directed lead with potential for use as a SNCA blocker for PD therapy. These are candidate drugs with the potential to limit toxic SNCA expression in the brains of PD patients and animal models in vivo.
Keywords: Parkinson’s disease, Alpha-synuclein, 5′untranslated region, Transfection-based screen, Natural product, Translation blockers, Amyloid precursor protein, Posiphen, Phenserine
Summary
We describe ten clinically relevant compounds, including the AD experimental drugs Posiphen and (−)-phenserine, that we have identified to exhibit specific targeting to the SNCA 5′UTR. In each case, declines in SNCA production in cells lines were achieved at concentrations that are within the clinical realm, and support the future in vivo analysis of these candidates in appropriate rodent models to vigorously define their translational capacity to therapeutically impact SNCA in vivo without toxicity.
Introduction
α-Synuclein (SNCA) is a 104-amino-acid-long protein implicated in the pathogenesis of Parkinson’s disease (PD) among other alpha-synculeinopathies, such as dementia with Lewy bodies (DLB) and multiple system atrophy (MSA) in humans (Ueda et al. 1993). Gene dosage was demonstrated in the etiology of these diseases, as duplication and triplication can cause inheritable forms of PD and DLB in rare families (Singleton et al. 2003). In addition, conformational changes (Lashuel et al. 2002) and oligomerization are also hypothesized to underlie alpha-synucleinopathies, as known point mutations have been shown to accelerate aggregation in vitro to strongly increase the presence of the beta specific fibril-forming conformer that is toxic to dopaminergic neurons and other brain cells (Uversky 2007), in some cases by iron-associated pathways (Ostrerova-Golts et al. 2000). In these rare cases of familial PD several reports have thus far identified three sets of culprit point mutations such as A53T, A30P and E46K (Cabin et al. 2005). These and other rare kindreds express altered or over-expressed SNCA that can cause a toxic gain of function and dopaminergic neural death.
Pathologically, SNCA-related disorders are characterized by the deposition of SNCA aggregates, Lewy Bodies and dystrophic neuritis (Kruger et al. 2000; Baba et al. 1998; Braak et al. 2003, 2006; Forno 1996; Irizarry et al. 1998). In contrast, MSA is characterized by the presence of glial cytoplasmic inclusions (Banati et al. 1998; Campbell et al. 2001).
SNCA is a highly preserved protein (Uversky 2007) normally found in presynaptic terminals (Goedert 2001). In Homo Sapiens there are three confirmed isoforms with the SNCA-140 being the most abundant (Uversky 2007) (Fig. 1). Most of the known mutations of SNCA accumulate at the N-terminus, which also harbors four imperfect repeats (KTKGV). Amino acids 61–96 constitute the so-called non-amyloid component (NAC) of the protein. This region predisposes SNCA to aggregation (Uversky 2007). The C-terminus harbors three of the four tyrosine residues present in SNCA. Truncations of the C-terminus promote aggregation (Li et al. 2005). SNCA is a disordered protein with no structure that can quickly respond to changes in its environment (Uversky 2007). As a natively unfolded protein, SNCA is very sensitive to its environment, and it can undergo a number of conformational switches in response to changes in environmental conditions (Uversky et al. 2001a). The unfolded protein is normally found under physiological conditions in vivo and in vitro. A pre-molten globular state can be induced by changes in temperature and pH or after exposure to pesticides (Uversky et al. 2001b). Shifts from a random coil to alpha-helical structures are observed when SNCA is exposed to lipids and lipidic membranes (Perrin et al. 2000). Shifts in temperature, presence of pro-oxidative conditions (Norris et al. 2003; Souza et al. 2000) and transition metals and divalent cations can promote the formation of dimers and other polymeric forms of SNCA (Conway et al. 1998; Lowe et al. 2004; Yamin et al. 2003). Eventually, SNCA can form large, insoluble aggregates with two predominant morphologies: fibrillar and amorphous aggregates (Uversky et al. 2002).
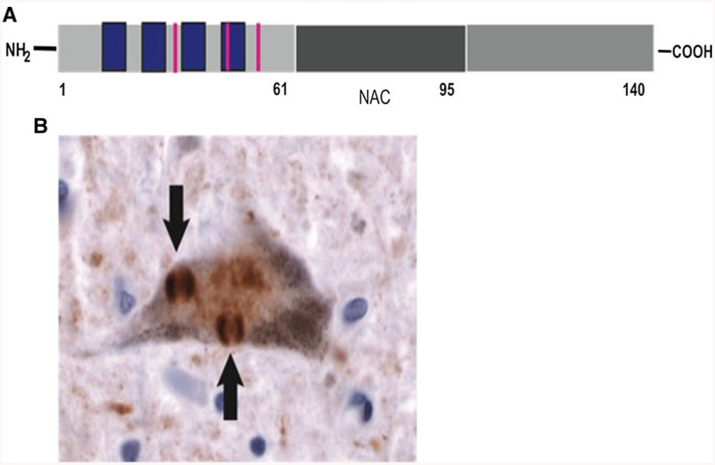
Fig. 1.
a alpha-synuclein (SNCA) sequence and domains. From left to right: the N terminus includes four imperfect repeats (blue boxes) 11-amino-acid long, and harbors three familial mutations associated with familial PD (red lines); the central portion is the non-Aβ (amyloidogenic) component of SNCA (NAC), this region is involved in alpha-synuclein aggregation, the C-terminal domain, the most unstructured part of the protein harbors three of the four tyrosines present in the sequence of alpha-synuclein. b Cortical neuron from a DLB patient stained for alpha-synuclein shows two prominent inclusions known as Lewy bodies (arrows). Lewy bodies can be associated with neuronal loss in the midbrain and other subortical nuclei in PD, while in DLB, Lewy bodies are present also in cortical regions
Events associated with the inflammatory cascade, iron metabolism and translational control of gene expression have been associated with PD and Lewy body dementia (DLB). Disrupted signaling events such as occur in response to inflammatory cytokines, for example mutations to the signaling kinase LRRK-2, may activate inflammatory events in this neurodegenerative disease (Cahill and Rogers 2008; Taylor et al. 2006). Novel signaling pathways may additionally be relevant (Cahill et al. 2009). Certainly, increased iron in the individual dopaminergic neurons of the substantia nigra has been shown to be closely associated with the pathogenesis of PD (Oakley et al. 2007). DLB brains exhibit lowered SNCA mRNA but have higher amounts of insoluble protein (Fig. 1), suggesting mis-regulation of SNCA mRNA translation in addition to its clearance by chaperones (Cantuti-Castelvetri et al. 2005). In this regard, translational control of SNCA is governed, at least in part, by a uniquely configured iron-responsive element (IRE) in the 5′UTR of SNCA mRNA as discussed below (see Cahill et al. (2009) for the relevance of iron in PD).
Iron-responsive element-dependent translational control of iron homeostasis: links to neurodegenerative disease genes
IREs are RNA stem loops that were first described in the untranslated regions (UTRs) of ferritin and transferrin-receptor mRNAs that are critical to iron homeostasis through their modulated interaction with the RNA binding proteins, iron-regulatory proteins (IRP) 1 and 2 (Rogers et al. 2008). In general, IRP2 has been reported to predominate over IRP1 when controlling IRE-dependent regulation of the iron homeostatic pathways for ferritin translation and transferrin receptor mRNA stability (Cho et al. 2010; Wang et al. 2007). However, the IRE stem loop encoded by the 5′UTR of the mRNAs for hypoxia inducible factor 2 alpha (Hif-2a) (Zimmer et al. 2008), ferroportin (IREG-1) (McKie et al. 2000), the erythroid heme biosynthetic aminolevulinate synthase (eALAS) (Wingert et al. 2005) and mitochondrial aconitase (Goforth et al. 2010) each bind more avidly to IRP1 than IRP2. Interestingly, the duodenal divalent metal ion transporter (DMT1) encodes a 3′UTR specific IRE stem loop immediately downstream from its stop codon and preferentially binds IRP1 to mediate iron-dependent message stability, an event that may be critical for setting rates of dietary iron uptake (Gunshin et al. 2001).
The Alzheimer-disease-specific amyloid precursor protein (APP) is up-regulated by iron influx, and it encodes a 5′UTR specific and fully functional IRE RNA stem loop (Cho et al. 2010; Rogers 2002; Rogers et al. 2008). Similar to Hif-2α (Zimmer et al. 2008), IRP1 predominates to control expression of the copper/zinc metalloprotein APP at the level of message translation (Cho et al. 2010). APP also appears to regulate iron homeostasis itself. Recently, APP, like astrocytic bound brain ceruloplasmin, was shown to be a potent ferroxidase via an active iron binding REXXE domain (Rogers 2002) and may thus represent a cofactor with ferroportin to export iron from neurons (Duce et al. 2010) and across brain sub-compartments. In such a scenario, ferroportin may represent a key transporter of iron that has been assimilated across the brain endothelial lining cells. At this stage, the closely bound APP may, for example, directly load incoming iron into brain transferrin (made by oligodendrocytes) in addition to facilitating excess by iron export from neurons, in each case for safe storage of iron.
Similar to APP, SNCA appears to have a role in iron metabolism/homeostasis since this protein has been shown to track with heme biosynthetic genes in its expression profile and is at its highest abundance in developing reticulocytes (Scherzer et al. 2008). Consistent with this finding, this report supports presence of the putative IRE encoded by the 5′UTR of the SNCA transcript (Cahill et al. 2009; Friedlich et al. 2007; Olivares et al. 2009) and demonstrates that FDA approved drugs, including iron chelator mycophenolic acid, can impact to limit SNCA 5′UTR directed translation in neural cell lines and also repress SNCA levels in an iron-dependent manner. These RNA-directed SNCA inhibitors represent a novel neuro-protective strategy for PD.
RNA targeting as a therapeutic strategy to control neurodegenerative diseases
RNA-directed drugs have long been used in the treatment of infectious diseases, as epitomized by antibiotic macrolides and aminoglycosides (Porse et al. 1999). These drugs are capable of controlling reporter gene expression in cell culture models (Thomas and Hergenrother 2008; Werstuck and Green 1998). More recently, RNA-directed therapeutic strategies were employed to control viral gene expression (Hepatitis C) (Malina et al. 2005), HIV (Hamy et al. 1997) and ferritin gene expression (protection from Fe catalyzed oxidative stress) (Tibodeau et al. 2006). For example, the H-ferritin 5′UTR was successfully modulated with an IRE-specific RNA-targeting drug, yohimbine, to increase ferritin translation (Tibodeau et al. 2006).
The role of the APP 5′UTR has been well characterized, which encouraged us to undertake the present study of the SNCA gene (Cahill et al. 2009). Notably, the presence of a ‘CAGA box’ within the 5′UTR mRNA of the APP gene unique to amyloid plaque forming species and absent in all APLP-1/2 genes has prompted us to utilize this unique region as a potential drug target (Cho et al. 2010; Maloney et al. 2004; Lahiri et al. 2005; Shaw et al. 2001). The functional characterization of the APP 5′UTR and its role in cytokine-mediated APP gene expression has obvious implications in Alzheimer’s disease (Lahiri et al. 2003), which led to the proposed concept of a “UTRosome” (Lahiri et al. 2005; Rogers et al. 2008) as the seat of the gene’s post-transcriptional control, the major focus of the present work.
In the case of endogenous neurodegenerative disease genes, we have applied RNA therapeutic strategies to identify drugs that limit the translation of the Alzheimer’s APP mRNA (Rogers 2002). This system provides an excellent precedent for the use of 5′UTR targeting strategies towards other major neurodegenerative diseases, in particular Parkinson’s disease since our alignments revealed a 50% sequences similarity between the APP and SNCA 5′UTRs (Cahill et al. 2009; Friedlich et al. 2007). Based on this, we predicted that there would be overlap in the spectrum of drugs that suppress APP mRNA translation through its 5′UTR with those that suppress SNCA translation. In this regard, we have previously demonstrated that the Alzheimer’s disease anticholinesterase (−)-phenserine (Greig et al. 1995, 2005; Winblad et al. 2010) lowers APP translation via its 5′UTR and, thereby, reduces APP generation in neuronal cell cultures and the brains of animals in vivo (Shaw et al. 2001; Venti et al. 2004; Lahiri et al. 2007a, b). This action is not cholinergically mediated as it is shared by its (+) enantiomeric form, in the experimental Alzheimer’s disease drug, Posiphen. A recent phase 1 clinical trial in subjects with mild cognitive impairment demonstrated that Posiphen safely lowers APP levels in the cerebrospinal fluid and plasma of humans (Maccecchini et al. 2009; Maccecchini 2010). Thus, Posiphen represents a current example of a safe inhibitor of APP translation that can thereby reduce the level of peptides derived from APP processing, such as amyloid-b peptide (Shaw et al. 2001; Lahiri et al. 2007a, b) and other potential toxic N- and C-terminal APP fragments (Maccecchini 2010). It hence provides a lead compound to assess actions to lower SNCA translation.
Success with APP 5′UTR directed translation blockers prompted our current characterization of the SNCA 5′UTR and its usefulness as a drug target for identifying SNCA translation blockers. In this report, we describe a screen of a library of natural products (NPs) to which we added Posiphen and (−)-phenserine as likely leads to impact SNCA translational regulation based on our prior studies. We identified ten specific inhibitors of SNCA translation, including several glycosides, and determined that the cardiac glycoside, strophanthidine, together with Posiphen/(−)-phenserine demonstrated similar high potencies to block SNCA translation and thereby lower its expression in two distinct neuroblastoma cell lines.
Materials and methods
Bioinformatics
RNA sequences from the SNCA gene were located using the NCBI Gene search and the Ensembl database. Since the 5′UTRs were of primary interest, the coding regions were disregarded, apart from the initiating AUG. Thus, in order to study a balanced sequence, 25 nucleotides before the splice junction from the first exon were used to create 50 nucleotide RNA query sequences (mouse and rat had 52 nucleotide sequences due to insertions).
RNA sequences were aligned using the ClustalX2 graphical program to find evolutionary conservation between species. The AUG start region of the CDS was then used as a reference point to align the sequences, allowing a comparison of the sequences in both exons centered around the splice junction. Secondary structure folding of these RNA sequences was generated by the RNAFold webserver at the University of Vienna and was annotated using the RNAFold software package utilities. The RNAFold server provided the most probable RNA secondary structure for SNCA 5′UTR sequences based on minimum free energy calculations (Zuker 1989).
The alignment homology was calculated by comparing a species RNA sequence against the H. sapiens sequence on each side of the splice junction. Only nucleotides that matched respective to the H. sapiens sequence were scored, and the percent homology was calculated by taking the score out of the total nucleotide positions on that side. The two results were then compared to better understand the difference in conservation on each side of the splice junction.
Transfection-based screen of SNCA 5′UTR specific inhibitors
A library of 720 NPs (Microsource Discovery Systems Inc., Gaylordsville, CT, USA) was screened at a concentration of 2 μM in triplicate for inhibition of luciferase expression using (APP 5′UTR-Luc cells) and (SNCA-Luc cells). A compound was scored as a hit if all replicates gave >65% inhibition in this assay, and as contradictory if at least one, but not all replicates gave >65% inhibition.
In order to rule out compounds that reduced luciferase expression due to toxicity, the entire library was also screened in both cell lines for cell viability, using the Alamar blue assay (Invitrogen, Carlsbad, CA, USA) as the readout. Percent inhibition in the Alamar blue assay was calculated and compared with percent inhibition of luciferase activity. Compounds for which the difference was greater than 40%, and that scored as a hit in the APP 5′UTR-luciferase counter-screen luciferase assay, were further evaluated in a dose–response assay.
Luciferase assay
Cells were plated at 2,000 cells/well (APP) or 4,000 cells/well (SNCA) in a 384-well white flat-bottom plate (Greiner) in a volume of 50 μl of media. Following overnight incubation, 50 nl of 2 mM compounds in DMSO was added using a VPN Pintool. Cells were returned to the incubator for 48 h and were then assayed for luciferase activity as follows. Plates were allowed to equilibrate to room temperature. After addition of 25 μl Steady-Glo reagent (Promega, San Luis Obispo, CA, USA), plates were vortexed for 30 s, and 35 min later luminescence was read on an Infinite F2000 plate reader (Tecan, Durham, NC, USA).
Western blot assay
Human H4 neuroglioblastoma and SH-SY5Y and SKNSH neuroblastoma cells were cultured in Dulbecco,s modified essential medium (Invitrogen) supplemented with 10% FBS (Invitrogen) and penicillin/streptomycin (Bio-Whittaker, Walkerville, MD, USA). Cells were exposed to strophanthidine or gitoxigenin at the concentrations indicated and subjected to Western blot analysis as described for Posiphen (legend to Fig. 4b). Cells were exposed to concentrations of Posiphen, as indicated, in the presence and absence of 50 μM iron and ferric ammonium citrate (0, 0.1, 1, 5, 10 micromolar) for 48 h. Cytoplasmic protein lysates were prepared by homogenizing the cells in midRIPA buffer (25 mM Tris pH 7.4, 1% NP40, 0.5% sodium deoxycholate, 15 mM NaCl, protease inhibitors, RNase inhibitor and 10 mM DTT). Western blotting for SNCA was performed using mouse monoclonal anti-SNCA (BD Transduction Laboratories, Lexington, KY, USA), and anti-β-actin (Chemicon, Rosemont, IL, USA). The blots were developed using chemiluminescence (Pierce, Rockford, IL, USA), visualized with a PhosphoImager (BioRad, Hercules, CA, USA), and the bands were quantified using QuantityOne software (BioRad).
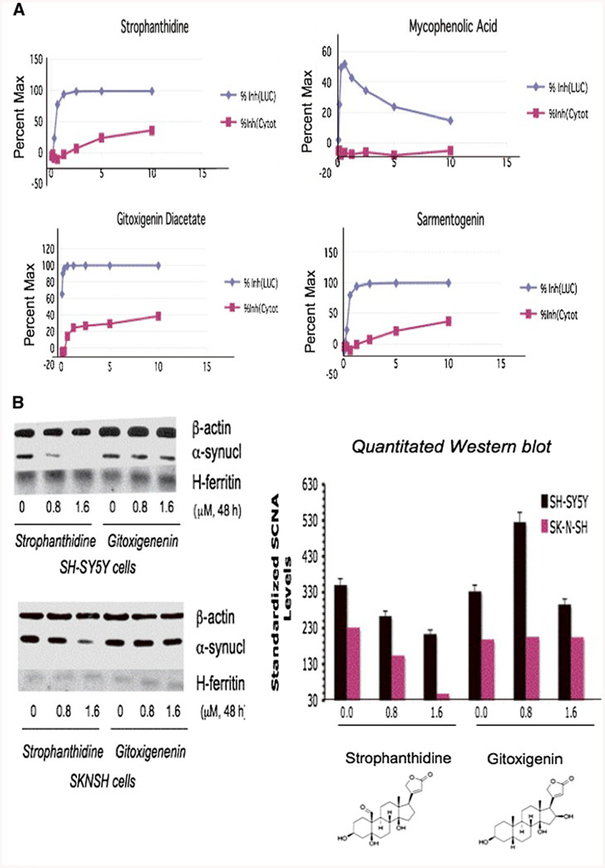
Fig. 4.
Strophanthidine as the confirmed top SNCA translation blocker in three separate neural cell lines. a H4 cells stably transfected with the H4–2a construct in which SNCA 5′UTR drives the translation of a luciferase reporter gene. These cells were treated at incrementally increasing concentrations of the SNCA 5′UTR inhibitor hits (0, 0.08, 0.16, 0.31, 0.63, 1.25, 2.5, 5, 10 μM), and were monitored for luciferase activity and cell viability by Alamar blue staining (48 h). All leads exerted no inhibition when counter-screened in control cell lines expressing the “empty” luciferase vector pGL3.Conclusion: Of 17 hit SNCA 5′UTR directed NP translation inhibitors*, four NPs, the cardiac glycosides strophanthidine, digoxigenin, sarmentogenin plus mycophenolic-acetate, exhibited IC50s at <5 μM in dose–response (without toxicity at initial SNCA inhibitory concentrations). b Western blots confirmed strophanthidine reduced SNCA expression in SK-N-SN/SH-SY5Y cells (see middle band, IC50 <1 μM). Confirming specificity to SNCA, strophanthidine maintained β-actin and H-ferritin levels (IRE encoding mRNA). In contrast, the gitoxigenin hit exerted only a minor inhibition of SNCA levels
Results
The SNCA 5′UTR is encoded by two exons, as illustrated in Fig. 1a. We bioinformatically determined the evolutionary conservation of the SNCA 5′UTR across these two exons. Utilizing the SNCA 5′UTR sequence as a drug target, we then performed a screen of 720 NPs together with Posiphen and (−)-phenserine to identify translation blockers. From the identified lead compounds, we then evaluated the potency of three lead glycosides and mycophenolic acid as bonafide SNCA translation blockers by use of dose-responsive luciferase assays and Western blotting measurements. Finally, we evaluated the iron-dependent mode of action of Posiphen as a known APP 5′UTR directed translation blocker that reduced SNCA levels in SH-SY5Y cells.
Evolutionary characterization of a putative IRE RNA stem loop centered on the single splice junction in the human SNCA mRNA 5′UTR
Using human, chimpanzee, bovine, mouse, rat, dog and chicken 5′UTR sequences deposited in the NCBI database, we confirmed the presence of the exon-1/exon-2 splice junction located 25–28 nucleotides upstream of the start AUG, not only in human and non-human primates, but also in other mammals such as mice (Mus musculus), rats (Rattus norvegicus) cattle (Bovus taurus) and dog (Canis lupus familiaris) (marked in red highlights in Fig. 2b). In humans and chimpanzee, the splice site was found to be 25 bases upstream of the AUG; in rats the splice site is 26 bases upstream of the AUG (single uridine insertion) whereas in mice the SNCA specific AG/UG splice junction is 28 residues upstream of the start codon (5′AUU3′ insertion).
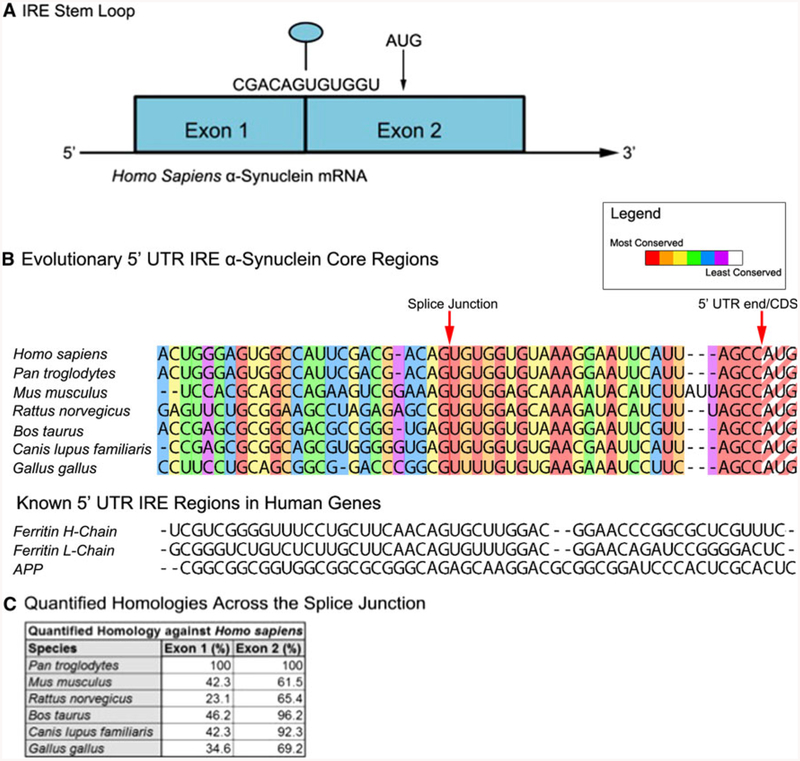
Fig. 2.
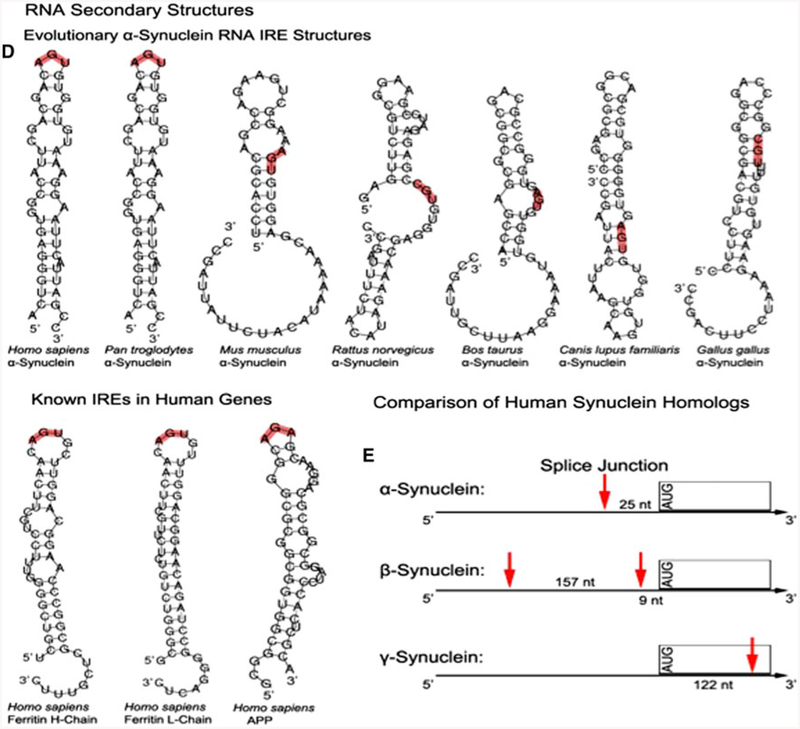
An RNA stem loop is predicted within the 5′untranslated region (5′UTR) of the Parkinson’s disease alpha-synuclein (SNCA) transcript that is homologous to the iron-responsive element (IRE) in H-ferritin mRNA. a The SNCA 5′UTR is encoded by exon-1 and exon-2 of the SNCA gene, which can be alternatively spliced to generate either a shorter exon-1/−2 transcript (Xia et al. 2001). b Evolutionary alignment of the SNCA 5′UTR relative to the human sequence and the CAGUGN loop/splice site sequences (Friedlich et al. 2007). c Quantified homology across the SNCA splice junction. d A SNCA 5′UTR stem loop was predicted by the RNA/FOLD computer program (ΔG = 53 kcal/mol). This SNCA stem loop resembles the classical IRE RNA stem loop (5′CAGUGN3′ loop motif) that controls iron-dependent L- and H-ferritin translation and transferrin receptor (TfR) mRNA stability. Stem loops from the 5′UTRs of several species were predicted to be folded, as described in the materials and methods section, and the pseudotriloop AGU is depicted in red lettering at the apex of the H-ferritin IRE (Goforth et al. 2010) where the analogous AGA from the APP IRE is depicted (Cho et al. 2010). The human SNCA exhibited an AGU triloop whereas in lower vertebrates AGU was located in the stem regions of these transcripts. e Arrangement of splice sites and 5′UTR structures in the SNCA, SNCB, SNCG mRNA
The layout of the 5′UTR of the H. sapiens SNCA transcript was previously reviewed (Olivares et al. 2009). In Fig. 2a of this report the basic features of the SNCA 5′UTR are highlighted, most notably the IRE-specific CAGUGN motif resides at the apex of a predicted hairpin loop, which is also the splice junction within this UTR. In summary, the SNCA mRNA has two exons in the 5′UTR and the splice junction interestingly lies in the very center of the CAGUGN motif. Thus, the splice junctions were key criteria used in evaluating homology between SNCA RNA in different species, as shown in Fig. 2b.
The alignment of the SNCA sequence of these species was conducted with the ClustalX2 program, and we observed and highlighted a clear difference in evolutionary conservation between exon-1 and exon-2 around the splice junction (see the homology Table in Fig. 2c). Across the vertebrate species shown, the sequence of the SNCA exon-2 is much more conserved than the sequence in exon-1 preceding the splice junction.
The primates (H. sapiens and Pan troglodytes) showed complete conservation within the entire SNCA 5′UTR sequence of the selected RNA regions (100% homology for both exon 1 and exon 2). Bos taurus and Canis lupus familiaris also showed high level (~96 and 92%, respectively) of conservation skewed towards exon 2, compared with a limited (46 and 42%) homology within the human exon 1 sequences (see the homology Table in Fig. 2). However, M. musculus and R. norvegicus each demonstrated stronger evolutionary divergence from H. sapiens in the overall sequence, such that exon 2 retained greater homology (i.e., 65% homology between exon 2 of the mouse and human SNCA 5′UTRs but only 42% homology between exon 1 for the human and mouse SNCA 5′UTRs). Both species had two nucleotide insertions and extended similarities in the exon-2 sequence (expected due to the related nature of the two species). Gallus gallus was found to be the most distantly related to H. sapiens, with least conservation at the splice junction site and in exon-2, as tabulated in Fig. 2c.
The 5′UTRs of the SNCA transcript in humans and in non-human primates were predicted to fold into RNA stem loops with an apex sequence that is related to the canonical ferritin L- and H-chain IREs (Fig. 2d). This incorporates an apical CAGUGN motif (Fig. 2d). Indeed, the prediction for the secondary structure of the SNCA 5′UTR has been refined to demonstrate a RNA stem loop that can fold to the same AGU psuedotriloop observed at the apex of the H-ferritin IREs (Goforth et al. 2010). This is related to a recently reported AGA tri-loop that we showed was significant for iron-dependent binding of IRP1 to APP IRE sequences as a means to control APP expression (Cho et al. 2010). In sum, L- and H-Ferritin IREs encode an AGU triloop sequence at the apex of their hairpin loops, similar to the RNA structure of the 5′UTRs of human and chimpanzee SNCA transcripts (Fig. 2d). We observed that the central “AGU” motifs from mouse, bovine and canine SNCA mRNA sources were predicted to be within the stem, not apex of their 5′UTR specific RNA stem loops. This motif was a CGU tri-nucleotide in the case of the rat and chicken genes.
One of the key features of the SNCA 5′UTR was the splice junction that overlapped with the CAGUGN motif that is trypical of IREs (Xia et al. 2001). In contrast, the γ- and β-homologs of SNCA encoded dissimilar 5′UTRs as shown in Fig. 2e. First, β-synuclein has two splice junctions in its 5′UTR, at 9 nucleotides and 166 nucleotides from the AUG. Second, the entire 5′UTR of γ-synuclein is encoded by a single exon; the first splice junction actually appears 122 nucleotides into the coding sequence. This lack of homology is consistent with our rationale for the use of the uniquely folded SNCA IRE as a drug target. Any specific inhibitor would be preferred to not reduce β- and γ-synuclein levels and, thereby, allow them to maintain their compensatory genetic function.
SNCA translation blockers screened from a library of natural products
To identify SNCA 5′UTR directed translation blockers of SNCA expression in H4 neuroblastoma cells, we employed a transfection-based screen similar to that used to identify several APP 5′UTR directed modulators (Bandyopadhyay et al. 2006b) (Fig. 3). Specifically, derived from our library screen, Table 1 summarizes the best ten candidate drugs identified as SNCA mRNA translation blockers. Compounds were screened in triplicate for inhibition of luciferase expression using SNCA 5UTR cells as the primary screening target, and APP 5′UTR cells were employed in the parallel counter-screen. A compound was scored as a hit if all replicates gave >65% inhibition in this assay, and as contradictory if at least one, but not all replicates gave >65% inhibition. The cutoff for specificity was less than 20% inhibitory effect on APP 5′UTR directed translation.
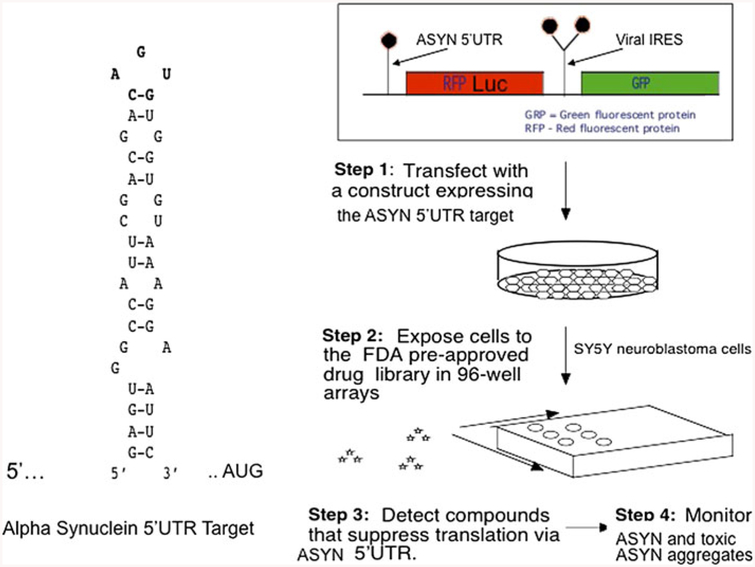
Fig. 3.
Dicistronic construct designed to screen small molecule inhibitors of SNCA translation: Left panel the unique RNA stem-loop target in the 5′UTR of the transcript for SNCA (RNA predicted by MULTIFOLD (Cahill and Rogers 2008; Friedlich et al. 2007; Olivares et al. 2009; Zuker 1989). Right panel transfection-based screen with our dicistronic pIRES(SNCA) construct to identify SNCA 5′UTR directed inhibitors of luciferase reporter translation, but which selectively maintain GFP translation from the downstream IRES RNA structure. This RNA targeting technology has already identified translation inhibitors of APP mRNA (as a precedent for AD therapy (Rogers et al. 2002b; Bandyopadhyay et al. 2006b). Chemical structures of HTS inhibitors of the APP 5′UTR can be downloaded from PUBCHEM at the NCBI website as AID: 1285; we are now also screening for SNCA 5′UTR directed translation blockers (Broad Inst. Cambridge, MA, USA)
Table 1.
Alpha synuclein translation blockers; Tabulated results of top ten alpha-synuclein 5′UTR directed hits
| Natural product | % inhibition (2 μM) |
Toxicity (%) (Alamar blue)/(2 μM) |
Class/source/citation |
|---|---|---|---|
| Digoxin | 100 | 40 | Glycoside |
| Strophantidine | 100 | 35 | Glycoside |
| Sarmentogenin | 100 | 10 | Strophanthus sarmentosis |
| Mycophenolic acid | 80 | 50 | Immunosuppressant/Fe chelator (Mudge et al. 2004) |
| Kinetine riboside | 40 | Non toxic | Glyoside |
| Harmine | 60 | 10 | PD drug/Peganium harmala |
| Kinetine Riboside | 40 | Non toxic | Anti cancer |
| Helenine | 12 | Non toxic | Natural product |
| Posiphen | 80 | 10% | Experimental Alzheimer drug, amyloid precursor protein synthesis inhibitor (Shaw et al. 2001; Lahiri et al. 2007a, b) |
| Phenserine (PS) | 50 | 10 | Alzheimer experimental drug, anticholinesterase (Greig et al. 2005; Winblad et al. 2010) |
Ten NPs were listed as SNCA 5′UTR directed inhibitors by the criterion that these had <20% inhibitory effect on APP 5′UTR directed translation (counter-screen). Included for further study were all replicates that gave >65% inhibition in this assay, and as contradictory if at least one, but not all replicates gave >65%. The first four of these compounds adhered to the selectivity criterion for which the difference between SNCA and APP 5′UTR inhibition was >40%. Since these were NPs of clinical interest they were evaluated in a dose–response assay and by western blot (as for the APP and SNCA 5′UTR blocker Posiphen)
Interestingly, one hit was the drug, harmine, which is already in use for the treatment of PD. It induced a 60% inhibition of SNCA translation coupled with low toxicity (10%). Harmine is a reversible inhibitor of monoamine oxidase and is a fluorescent harmala analog. Kinetine riboside inhibited SNCA translation by 45% and proved not to be toxic (less than 10%). It has been reported to possess anticancer activity in a number of human, mouse and plant tumor cell lines (Griffaut et al. 2004) and had only a modest inhibitory effect of APP translation (10%).
We conducted dose response experiments with four SNCA 5′UTR inhibitors: the glycosides, sarmentogenin, strophanthidine and gitoxigenin acetate and the immunosuppressant mycophenolic acid (Fig. 4a). The three gylcosides all significantly reduced SNCA 5′UTR activity (IC50 ~0.08 μM), and mycophenolic acid exhibited similar inhibition, although this declined at higher concentrations. In a concurrent Alamar blue assay to assess drug-induced toxicity, mycophenolic acid proved 25% more toxic to H4 cells than the three glycosides (n = 3), as was the case in SH-SY5Y cells (not shown).
To define whether translation inhibition determined in the luciferase assay translated to lowered protein expression, specific compounds were screened by Western blot analyses. Strophanthidine proved to be one of the three glycosidic SNCA 5′UTR directed translation blockers that likewise reduced SNCA protein expression (Fig. 4b). In SK-N-SH cells, strophanthidine almost completely repressed SNCA expression at 1.6 μM, and possessed an IC50 of 0.8 μM. In SH-SY5Y cells SNCA levels were reduced substantially but to a lesser extent, approaching 50% at 1.6 μM strophanithidine. We observed no change to the steady-state levels of either ferritin or β-actin in these experiments (n = 4). Hence, by this criterion, strophanthidine proved to be a selective SNCA translation blocker in two distinct neuroblastoma cells lines (Fig. 4b). To illustrate the importance of the secondary Western blot screen, another SNCA 5′UTR inhibitor lead, gitoxigenin, did not alter SNCA protein expression in either SK-N-SH or SH-SY5Y cells, in which 0.8 and 1.6 mM gitoxigenin was without affect (Fig. 4b).
Posiphen inhibition of neural SNCA expression is accelerated by cellular iron
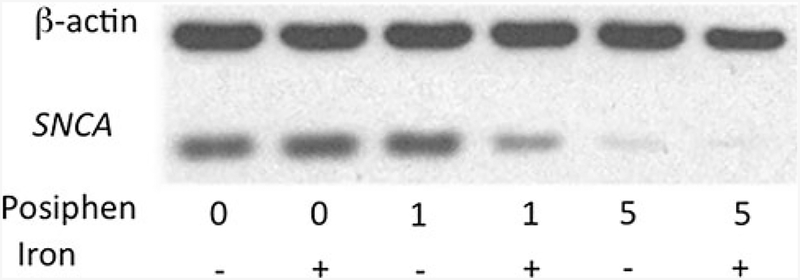
Based on the 50% sequence similarity between the APP and SNCA 5′UTRs (Fig. 2), we predicted that drugs that would suppress APP mRNA translation via its 5′UTR may usefully impact SNCA mRNA translation, and utilized the Alzheimer experimental drugs Posiphen and (−)-phenserine to explore this. Both agents demonstrated activity in the primary screen to lower SNCA translation in the luciferase H4 cell screen and, likewise, lowered SNCA expression in SK-N-SH cells (not shown). Interestingly and as illustrated with Posiphen in Fig. 5, not only was SNCA expression in SH-SY5Y neural cells suppressed, but the potency with which this occurred increased in the presence of iron, which is known to be elevated in PD brain (Olivares et al. 2009). In this representative experiment, Posiphen inhibited SNCA with the same potency as (−)-phenserine IC50 > 5 μM (not shown); however, in the presence of iron, Posiphen lowered SNCA expression with an IC50 < 1 μM (Fig. 5).
Fig. 5.
Posiphen decreased SNCA levels dose-dependently in dopaminergic SH-SY5Y cells, as was reported for APP: The 5′UTRs of both APP and SNCA share 50% homology with the IRE H-ferritin mRNA (Friedlich et al. 2007). SH-SY5Y cells were treated with concentrations ranging from 0 to 10 μM phenserine and Posiphen for 48 h. Harvested cell lysates were prepared [see (Bandyopadhyay et al. 2006a)]. Quantitative Western blotting established the anti-SNCA efficacy of Posiphen and phenserine (IC-50 < 5 M); after standardization for β-actin [Densitometry of multiple lanes (n = 8) by ImageQuant]. Cell viability was unaffected [measured by standardized ATP levels/cell (Tm, Cell-Titer-glo™, Promega, Inc.)]
Discussion
Protein–RNA interactions have been successfully targeted by small drug-like molecules. As a recent example, the natural alkaloid yohimbine increased the endogenous expression of iron storage protein ferritin by 40% in a cell-free expression system (rabbit reticulocyte lysate) by activating the IRE RNA stem loop in the 5′UTR of its transcript (Tibodeau et al. 2006). Such RNA protein types of interactions are critically important to support the continuous endogenous regulation of protein levels in cells to allow them to maintain and optimize their function with changes in activity and environment. Pharmacological interventions in this process have been sought to prevent viral infections (Howe et al. 2004). We recently provided proof-of-concept that it is possible to reduce APP mRNA translation and in vivo amyloidosis through the use of FDA pre-approved translation blockers that target the APP 5′UTR (Payton et al. 2003; Tucker et al. 2006; Rogers et al. 2002a, b) as well as by the experimental Alzheimer drugs, Posiphen and (−)-phenserine (Shaw et al. 2001; Venti et al. 2004; Lahiri et al. 2007a, b).
Given the direct association between SNCA and PD, we sought to discover novel drugs to reduce SNCA translation by targeting its 5′UTR stem-loop. If successful, this approach would provide small molecule probes to elucidate the mechanism of SNCA translation as well as provide a new therapeutic strategy for treating PD. To achieve this goal, we initially defined conserved sequence elements within the SNCA 5′UTR in order to predict secondary RNA structures that may control SNCA translation expression and improve understanding of the RNA alignment and folding patterns of this therapeutic RNA target.
The homology percentages listed in Fig. 2c underpin a strong contrast in the pattern of evolutionary conservation between exon-1 and exon-2 of non-coding sequences in the SNCA genes. The conservation between H. sapiens and P. troglodytes is 100% for both exons, as would be predicted from the close evolutionary relationship between these two primates. However, the percent homology of exon-1 across all species was found to be significantly lower than that observed for exon-2. This suggests that non-coding sequences in exon-2 are significantly more evolutionarily conserved across species than exon-1.
Conspicuously, the 5′UTR of the SNCA transcript folds into a unique RNA stem loop that resembles an IRE RNA structure that is related to (but distinct from) the H-ferritin and APP 5′UTR specific IREs (Cho et al. 2010; Bandyopadhyay et al. 2006a; Rogers 2002). As Fig. 2e shows, the SNCA IRE is formed at the splice junction of the first two exons in the SNCA gene (Olivares et al. 2009). By contrast, the H-ferritin and APP IREs are transcribed from the single first exon of their genes, confirming the uniqueness of SNCA translational repression via its 5′UTR. Interestingly, the predicted RNA stem loop in the SNCA 5′UTR is not found in β- and γ-synuclein mRNAs, as is apparent in Fig. 2e. In this scenario, SNCA 5′UTR inhibitors would be predicted to leave β- and γ-synuclein unchanged, and such an action could prove of value for a proposed drug as these proteins appear to compensate for diminished or absent SNCA expression in vivo (Dauer et al. 2002). Although we characterized (−)-phenserine and Posiphen as SNCA 5′UTR directed leads, based on their known ability to lower translation of APP through related 5′UTR sequences (Shaw et al. 2001; Lahiri et al. 2007a, b; Utsuki et al. 2007), we nevertheless co-screened for actions on APP translation to further define selectivity. From a clinical perspective, however, compounds that lack significant differential action between SNCA and APP could retain great value, as there remains significant overlap between PD and Alzheimer’s disease beyond DLB (Strobel 2009).
From our transfection-based initial screen of a luciferase reporter gene expression when driven by the full length 5′UTR of SNCA mRNA in a neural cell line, the major drug class of SNCA 5′UTR inhibitors deriving from our 720 NP library proved to be plant glycosides. Although these drugs are well known as effective in the treatment of congestive heart failure and primarily act to inhibit Na+/K+-ATPase, an enzyme located in the cell membrane that promotes the outward transport of Na+ and the inward transport of K+, they were unexpected lead anti-SNCA inhibitors. The most potent inhibitors of Na+/K+-ATPase are cardenolides and include digoxin, digitoxin, digitoxigenin and strophathidine.
We reproducibly found that strophanthidine’s action as a SNCA 5′UTR inhibitor carried through to dopaminergic SK-N-SH and SH-SY5Y cells, where SNCA expression was reduced without toxicity (that occurred at 10 μM or greater) (Fig. 4). Others have reported an alternative indication in which cardiac glycosides can provide neuroprotection against ischemic stroke in a brain slice-based compound screening platform (Wang et al. 2006). Strophanthidine derives from the seeds of Strophanthus kombé that has, likewise been reported to impact Na/K-ATPase activity and to increase the tone, excitability and contractility of cardiac muscles (Wang et al. 2006). In contrast, the actions of the cardenolide, gitoxigenin, which is a close analog of strophanthidine and was similarly identified as a lead SNCA 5′UTR directed translation blocker from our transfection based screen, did not translate to the key secondary assay focused on quantitative changes in SNCA expression by Western blot. Whether this relates to a difference between the cell lines used in the primary (H4 cells) and secondary (SK-N-SH and SH-SY5Y cells) assays or to the different position of hydroxyl groups present on the rings that differentiates the two glycosides, remains to be determined. It is clearly apparent that a hierarchy of supporting primary and secondary assays is essential for effective screening of agents prior to assessment in animal models of PD. Also evident is that SNCA lowering activity is not a property shared by all cardiac glycosides but this activity, nevertheless, provides insight into potential mechanisms underpinning SNCA activity. Digoxin and related cardiac glycosides repress HIF-1alpha synthesis and block tumor growth (Zhang et al. 2008). Such inhibition of Hif-1-alpha is an iron-related activity and provides a means for its inhibition of the IRE-dependent pathways of gene expression (i.e., suppression of the SNCA IRE activity) (Toth et al. 1999). Indeed, IREs are responsive to hypoxic induction of Hif-1-alpha dependent activation of IRP1-dependent gene expression (Li et al. 2006).
A structurally unrelated SNCA 5′UTR directed translation blocker proved to be mycophenolic acid, an immunosuppressant drug that has been widely used to prevent rejection of organ transplantation, in particular renal transplant rejection (Villarroel et al. 2009). In addition to being an immunosuppressant, mycophenolic acid possesses iron chelator activity, thus potentially explaining the mechanism via which it may have generated activity against the IRE encoded by the SNCA 5′UTR sequences (Mudge et al. 2004).
Unlike strophanthidine, mycophenolic acid’s concentration-dependent action on luciferase activity followed an inverted U-shaped curve. Nevertheless, it represents an interesting lead agent that warrants further study.
Similarly, the structurally unrelated compounds Posiphen and (−)-phenserine effectively and reproducibly lowered SNCA translation as well as expression across assays and neural cell types. Both agents possess the classic hexahydropyrroloindole backbone, present in the natural alkaloid physostigmine, but Posiphen exists in the unnatural (+) enantiomeric form that lacks anticholinesterase action (Greig et al. 1995, 2005). Posiphen and phenserine, both readily enter the brain with a brain/plasma ratio of 7:1 (Greig et al. 2000, 2005). However, due to its cholinomimetic effects phenserine cannot be dosed high enough in vivo to achieve APP inhibition, whereas Posiphen has been shown to effectively lower APP as well as Aβ levels in the brains of a number of rodent models (Lahiri et al. 2007b; Marutle et al. 2007). Posiphen, therefore, represents an interesting compound to translate into animal models of PD involving elevated SNCA levels. The translation of Posiphen, in particular, into animal models is additionally supported by the recent demonstration of the ability of Posiphen (60 mg QID) to safely lower plasma and CSF levels of APP in a phase 1 clinical trial of subjects with mild cognitive impairment and achieve target concentrations commensurate with those described herein to lower SNCA expression (Maccecchini et al. 2009; Maccecchini 2010).
In summary, several recent reports have shown SNCA duplication to be a genetic cause of familial early onset PD, implicating simple SNCA dose as one of the causative factors in disease progression in the striatal neurons of PD patients (Nishioka et al. 2006). A reduction in SNCA expression hence represents a rational approach to slow disease progression. In this regard, brain infusion of SiRNA by canula and related approaches have successfully limited SNCA production in vivo in rodents (Lewis 2009). Our identification of compounds that suppress SNCA expression, described in Table 1, may provide a less invasive approach to potentially achieve the same goal if target efficacy concentrations can be safely achieved. As several of the agents are in clinic, they are amenable to rapid translation into animal models and, if effective, to human PD clinical trials.
Acknowledgments
This research was supported by the Michael J. Fox Foundation Novel Drug Discoveries Award, National Institute of Aging R01 AG20181 (JTR), Alzheimer’s Association Zenith Award 09–131352 (JTR), ISOA (JTR), QR Pharma Inc. (MLM), the National Institutes of Health Grants [AG18379 and AG18884 (DKL)] and the Intramural Research Program, National Institute on Aging, National Institutes of Health (NHG). ICC was supported by the MGH/MIT Morris Udall Center of Excellence in PD Research (NIH NS38372), and the APDA Advanced Center for Parkinson Research at MGH. We are indebted for the input and support of Professor Moussa Youdim, a pioneer in the fields of Parkinson’s disease research, and an innovative drug discoverer and developer.
Contributor Information
Jack T. Rogers, Neurochemistry Laboratory, Psychiatry-Neuroscience, Massachusetts General Hospital, Charlestown, MA 02129, USA
Sohan Mikkilineni, Neurochemistry Laboratory, Psychiatry-Neuroscience, Massachusetts General Hospital, Charlestown, MA 02129, USA.
Ippolita Cantuti Castelvetri, Neurochemistry Laboratory, Psychiatry-Neuroscience, Massachusetts General Hospital, Charlestown, MA 02129, USA; MassGeneral Institute for Neurodegenerative Disease (MIND), Massachusetts General Hospital, Charlestown, MA, USA.
Deborah H. Smith, Yale University, New Haven, CT, USA
Xudong Huang, Neurochemistry Laboratory, Psychiatry-Neuroscience, Massachusetts General Hospital, Charlestown, MA 02129, USA.
Sanghamitra Bandyopadhyay, Indian Institute of Toxicology Research (CSIR), Lucknow, India.
Catherine M. Cahill, Neurochemistry Laboratory, Psychiatry-Neuroscience, Massachusetts General Hospital, Charlestown, MA 02129, USA
Maria L. Maccecchini, QR Pharma Inc., Radnor, PA 19087, USA
Debomoy K. Lahiri, Laboratory of Molecular Neurogenetics, Department of Psychiatry, Institute of Psychiatric Research, Indiana University School of Medicine, IN, USA
Nigel H. Greig, Drug Design and Development Section, Laboratory of Neurosciences, Intramural Research Program, National Institute on Aging, Baltimore, MD 21224, USA
References
- Baba M, Nakajo S, Tu PH, Tomita T, Nakaya K, Lee VM, Trojanowski JQ, Iwatsubo T (1998) Aggregation of alpha-synuclein in Lewy bodies of sporadic Parkinson’s disease and dementia with Lewy bodies. Am J Pathol 152(4):879–884 [PMC free article] [PubMed] [Google Scholar]
- Banati RB, Daniel SE, Blunt SB (1998) Glial pathology but absence of apoptotic nigral neurons in long-standing Parkinson’s disease [see comments]. Mov Disord 13(2):221–227 [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay S, Huang X, Cho H, Greig NH, Youdim MB, Rogers JT (2006a) Metal specificity of an iron-responsive element in Alzheimer’s APP mRNA 5′untranslated region, tolerance of SH-SY5Y and H4 neural cells to desferrioxamine, clioquinol, VK-28, and a piperazine chelator. J Neural Transm Suppl 71: 237–247 [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay S, Ni J, Ruggiero A, Walshe K, Rogers MS, Chattopadhyay N, Glicksman MA, Rogers JT (2006b) A high-throughput drug screen targeted to the 5′untranslated region of Alzheimer amyloid precursor protein mRNA. J Biomol Screen 11(5):469–480 [DOI] [PubMed] [Google Scholar]
- Braak H, Rub U, Del Tredici K (2003) Involvement of precerebellar nuclei in multiple system atrophy. Neuropathol Appl Neurobiol 29(1):60–76 [DOI] [PubMed] [Google Scholar]
- Braak H, Rub U, Del Tredici K (2006) Cognitive decline correlates with neuropathological stage in Parkinson’s disease. J Neurol Sci 248:255–258 [DOI] [PubMed] [Google Scholar]
- Cabin DE, Gispert-Sanchez S, Murphy D, Auburger G, Myers RR, Nussbaum RL (2005) Exacerbated synucleinopathy in mice expressing A53T SNCA on a Snca null background. Neurobiol Aging 26(1):25–35 [DOI] [PubMed] [Google Scholar]
- Cahill CM, Rogers JT (2008) Interleukin-1beta induction of IL-6 is mediated by a novel phosphatidylinositol 3-kinase dependent AKT/Ikappa B kinase alpha pathway targeting activator protein-1. J Biol Chem 283:212–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill CM, Lahiri DK, Huang X, Rogers JT (2009) Amyloid precursor protein and alpha synuclein translation, implications for iron and inflammation in neurodegenerative diseases. Biochim Biophys Acta 1790(7):615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell BC, McLean CA, Culvenor JG, Gai WP, Blumbergs PC, Jakala P, Beyreuther K, Masters CL, Li QX (2001) The solubility of alpha-synuclein in multiple system atrophy differs from that of dementia with Lewy bodies and Parkinson’s disease. J Neurochem 76(1):87–96 [DOI] [PubMed] [Google Scholar]
- Cantuti-Castelvetri I, Klucken J, Ingelsson M, Ramasamy K, McLean PJ, Frosch MP, Hyman BT, Standaert DG (2005) Alpha-synuclein and chaperones in dementia with Lewy bodies. J Neuropathol Exp Neurol 64(12):1058–1066 [DOI] [PubMed] [Google Scholar]
- Cho HH, Cahill CM, Vanderburg CR, Scherzer CR, Wang B, Huang X, Rogers JT (2010) Selective translational control of the Alzheimer amyloid precursor protein transcript by iron regulatory protein-1. J Biol Chem 285:31217–31232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway KA, Harper JD, Lansbury PT (1998) Accelerated in vitro fibril formation by a mutant alpha-synuclein linked to early-onset Parkinson disease. Nat Med 4(11):1318–1320 [DOI] [PubMed] [Google Scholar]
- Dauer W, Kholodilov N, Vila M, Trillat AC, Goodchild R, Larsen KE, Staal R, Tieu K, Schmitz Y, Yuan CA, Rocha M, Jackson-Lewis V, Hersch S, Sulzer D, Przedborski S, Burke R, Hen R (2002) Resistance of alpha-synuclein null mice to the parkinsonian neurotoxin MPTP. Proc Natl Acad Sci USA 99:14524–14529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duce JA, Tsatsanis A, Cater MA, James SA, Robb E, Wikhe K, Leong SL, Perez K, Johanssen T, Greenough MA, Cho HH, Galatis D, Moir RD, Masters CL, McLean C, Tanzi RE, Cappai R, Barnham KJ, Ciccotosto GD, Rogers JT, Bush AI (2010) Iron-export ferroxidase activity of beta-amyloid precursor protein is inhibited by zinc in Alzheimer’s disease. Cell 142(6):857–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forno LS (1996) Neuropathology of Parkinson’s disease. J Neuropathol Exp Neurol 55(3):259–272 [DOI] [PubMed] [Google Scholar]
- Friedlich AL, Tanzi RE, Rogers JT (2007) The 5′-untranslated region of Parkinson’s disease alpha-synuclein messengerRNA contains a predicted iron responsive element. Mol Psychiatry 12(3):222–223 [DOI] [PubMed] [Google Scholar]
- Goedert M (2001) Alpha-synuclein and neurodegenerative diseases. Nat Rev Neurosci 2(7):492–501 [DOI] [PubMed] [Google Scholar]
- Goforth JB, Anderson SA, Nizzi CP, Eisenstein RS (2010) Multiple determinants within iron-responsive elements dictate iron regulatory protein binding and regulatory hierarchy. RNA 16(1):154–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig NH, Pei X, Soncrant TT, Ingram DK, Brossi A (1995) Phenserine and ring C hetero-analogues: drug candidates for the treatment of Alzheimer’s disease. Med Res Rev 15(1):3–31 [DOI] [PubMed] [Google Scholar]
- Greig NH, DeMicheli E, Utsuki T, Holloway HW, Yu QS, Perry T, Brossi A, Deutsch J, Ingram DK, Lahiri DK, Soncrant T (2000) The experimental Alzheimer drug phenserine: pharmacodynamics and kinetics in the rat. Acta Neurol Scand 102:74–84 [DOI] [PubMed] [Google Scholar]
- Greig NH, Sambamurti K, Yu QS, Brossi A, Bruinsma GB, Lahiri DK (2005) An overview of phenserine tartrate, a novel acetylcholinesterase inhibitor for the treatment of Alzheimer’s disease. Curr Alzheimer Res 2(3):281–290 [DOI] [PubMed] [Google Scholar]
- Griffaut B, Bos R, Maurizis JC, Madelmont JC, Ledoigt G (2004) Cytotoxic effects of kinetin riboside on mouse, human and plant tumour cells. Int J Biol Macromol 34(4):271–275 [DOI] [PubMed] [Google Scholar]
- Gunshin H, Allerson CR, Polycarpou-Schwarz M, Rofts A, Rogers JT, Kishi F, Hentze MW, Rouault TA, Andrews NC, Hediger MA (2001) Iron-dependent regulation of the divalent metal ion transporter. FEBS Lett 509(2):309–316 [DOI] [PubMed] [Google Scholar]
- Hamy F, Felder ER, Heizmann G, Lazdins J, Aboul-ela F, Varani G, Karn J, Klimkait T (1997) An inhibitor of the Tat/TAR RNA interaction that effectively suppresses HIV-1 replication. Proc Natl Acad Sci USA 94(8):3548–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe AY, Bloom J, Baldick CJ, Benetatos CA, Cheng H, Christensen JS, Chunduru SK, Coburn GA, Feld B, Gopalsamy A, Gorczyca WP, Herrmann S, Johann S, Jiang X, Kimberland ML, Krisnamurthy G, Olson M, Orlowski M, Swanberg S, Thompson I, Thorn M, Del Vecchio A, Young DC, van Zeijl M, Ellingboe JW, Upeslacis J, Collett M, Mansour TS, O’Connell JF (2004) Novel nonnucleoside inhibitor of hepatiti6s C virus RNA-dependent RNA polymerase. Antimicrob Agents Chemother 48(12):4813–4821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry MC, Growdon W, Gomez-Isla T, Newell K, George JM, Clayton DF, Hyman BT (1998) Nigral and cortical Lewy bodies and dystrophic nigral neurites in Parkinson’s disease and cortical Lewy body disease contain alpha-synuclein immunoreactivity. J Neuropathol Exp Neurol 57(4):334–337 [DOI] [PubMed] [Google Scholar]
- Kruger R, Muller T, Riess O (2000) Involvement of alpha-synuclein in Parkinson’s disease and other neurodegenerative disorders. J Neural Transm 107(1):31–40 [DOI] [PubMed] [Google Scholar]
- Lahiri DK, Chen D, Vivien D, Ge Y-W, Greig NH, Rogers JT (2003) Role of cytokines in the gene expression of amyloid ß-protein precursor: identification of a 5′-UTR-binding nuclear factor and its implications in Alzheimer’s disease. J Alzheimer’s Dis 5(2):81–90 [DOI] [PubMed] [Google Scholar]
- Lahiri DK, Ge Y-W, Maloney B (2005) Characterization of the APP proximal promoter and 5′-untranslated regions: Identification of cell type specific domains and implications in APP gene expression and Alzheimer’s disease. FASEB J 19(6):653–655 [DOI] [PubMed] [Google Scholar]
- Lahiri DK, Alley GM, Tweedie D, Chen D, Greig NH (2007a) Differential effects of two hexahydropyrroloindole carbamate-based anticholinesterase drugs on the amyloid beta protein pathway involved in Alzheimer’s disease. Neuromolecular Med 9(2):157–168 [DOI] [PubMed] [Google Scholar]
- Lahiri DK, Chen D, Maloney B, Holloway HW, Yu QS, Utsuki T, Giordano T, Sambamurti K, Greig NH (2007b) The experimental Alzheimer’s disease drug Posiphen [(+)-phenserine] lowers amyloid-beta peptide levels in cell culture and mice. J Pharmacol Exp Ther 320(1):386–396 [DOI] [PubMed] [Google Scholar]
- Lashuel HA, Hartley D, Petre BM, Walz T, Lansbury PT Jr (2002) Neurodegenerative disease: amyloid pores from pathogenic mutations. Nature 418(6895):291. [DOI] [PubMed] [Google Scholar]
- Lewis JA (2009) Digoxin blocks tumor growth through HIF-1alpha inhibition. Curr Top Med Chem 9(1):117. [DOI] [PubMed] [Google Scholar]
- Li W, West N, Colla E, Pletnikova O, Troncoso JC, Marsh L, Dawson TM, Jakala P, Hartmann T, Price DL, Lee MK (2005) Aggregation promoting C-terminal truncation of alpha-synuclein is a normal cellular process and is enhanced by the familial Parkinson’s disease-linked mutations. Proc Natl Acad Sci USA 102(6):2162–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Chen H, Huang X, Costa M (2006) Effects of 12 metal ions on iron regulatory protein 1 (IRP-1) and hypoxia-inducible factor-1 alpha (HIF-1alpha) and HIF-regulated genes. Toxicol Appl Pharmacol 213(3):245–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe R, Pountney DL, Jensen PH, Gai WP, Voelcker NH (2004) Calcium(II) selectively induces alpha-synuclein annular oligomers via interaction with the C-terminal domain. Protein Sci 13(12):3245–3252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccecchini ML (2010) Posiphen’s pharmacokinetics and mechanism of action in mild cognitive impaired patients. Alzheimer’s & Dementia 6(4S1):e54 [Google Scholar]
- Maccecchini ML, Roffman M, Greig NH (2009) Posiphen lowers amyloid precursor protein and amyloid β as well as acetylcholinesterase levels in culture, animals and humans. Alzheimer’s & Dementia 5(4S):47–48 [Google Scholar]
- Malina A, Khan S, Carlson CB, Svitkin Y, Harvey I, Sonenberg N, Beal PA, Pelletier J (2005) Inhibitory properties of nucleic acid-binding ligands on protein synthesis. FEBS Lett 579(1):79–89 [DOI] [PubMed] [Google Scholar]
- Maloney B, Ge Y-W, Greig N, Lahiri DK (2004) Presence of a ‘CAGA box’ in the APP gene unique to amyloid plaque forming species and absent in all APLP-1/2 genes: Implications in Alzheimer’s disease. FASEB J 18(11):1288–1290 [DOI] [PubMed] [Google Scholar]
- Marutle A, Ohmitsu M, Nilbratt M, Greig NH, Nordberg A, Sugaya K (2007) Modulation of human neural stem cell differentiation in APP23 transgenic mice by phenserine treatment. PNAS 104:12506–12511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKie AT, Marciani P, Rolfs A, Brennan K, Wehr K, Barrow D, Miret S, Bomford A, Peters TJ, Farzaneh F, Hediger MA, Hentze MW, Simpson RJ (2000) A novel duodenal iron-regulated transporter, IREG1, implicated in the basolateral transfer of iron to the circulation. Mol Cell 5(2):299–309 [DOI] [PubMed] [Google Scholar]
- Mudge DW, Atcheson B, Taylor PJ, Sturtevant JM, Hawley CM, Campbell SB, Isbel NM, Nicol DL, Pillans PI, Johnson DW (2004) The effect of oral iron admiinistration on mycophenolate mofetil absorption in renal transplant recipients: a randomized, controlled trial. Transplantation 77(2):206–209 [DOI] [PubMed] [Google Scholar]
- Nishioka K, Hayashi S, Farrer MJ, Singleton AB, Yoshino H, Imai H, Kitami T, Sato K, Kuroda R, Tomiyama H, Mizoguchi K, Murata M, Toda T, Imoto I, Inazawa J, Mizuno Y, Hattori N (2006) Clinical heterogeneity of alpha-synuclein gene duplication in Parkinson’s disease. Ann Neurol 59(2):298–309 [DOI] [PubMed] [Google Scholar]
- Norris EH, Giasson BI, Ischiropoulos H, Lee VM (2003) Effects of oxidative and nitrative challenges on alpha-synuclein fibrillo-genesis involve distinct mechanisms of protein modifications. J Biol Chem 278(29):27230–27240 [DOI] [PubMed] [Google Scholar]
- Oakley AE, Collingwood JF, Dobson J, Love G, Perrott HR, Edwardson JA, Elstner M, Morris CM (2007) Individual dopaminergic neurons show raised iron levels in Parkinson disease. Neurology 68(21):1820–1825 [DOI] [PubMed] [Google Scholar]
- Olivares D, Huang X, Branden L, Greig NH, Rogers JT (2009) Physiological and pathological role of alpha-synuclein in Parkinson’s disease through iron mediated oxidative stress; the role of a putative iron-responsive element. Int J Mol Sci 10(3):1226–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrerova-Golts N, Petrucelli L, Hardy J, Lee JM, Farer M, Wolozin B (2000) The A53T alpha-synuclein mutation increases iron-dependent aggregation and toxicity. J Neurosci 20(16):6048–6054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payton S, Cahill CM, Randall JD, Gullans SR, Rogers JT (2003) Drug discovery targeted to the Alzheimer’s APP mRNA 5′-untranslated region: the action of paroxetine and dimercaptopropanol. J Mol Neurosci 20(3):267–275 [DOI] [PubMed] [Google Scholar]
- Perrin RJ, Woods WS, Clayton DF, George JM (2000) Interaction of human alpha-Synuclein and Parkinson’s disease variants with phospholipids. Structural analysis using site-directed mutagenesis. J Biol Chem 275(44):34393–34398 [DOI] [PubMed] [Google Scholar]
- Porse BT, Kirillov SV, Awayez MJ, Garrett RA (1999) UV-induced modifications in the peptidyl transferase loop of 23S rRNA dependent on binding of the streptogramin B antibiotic, pristinamycin IA. RNA 5(4):585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J (2002) Pepides derived from the human amyloid precursor protein used to protect cells form iron catalyzed oxidative damage Issued US Patent WO/2002/034766
- Rogers JT, Lahiri DK (2004) Metal andinflammatory targets for Alzheimer’s disease. Curr Drug Targets 5(6):535–551 [DOI] [PubMed] [Google Scholar]
- Rogers JT, Randall JD, Cahill CM, Eder PS, Huang X, Gunshin H, Leiter L, McPhee J, Sarang SS, Utsuki T, Greig NH, Lahiri DK, Tanzi RE, Bush AI, Giordano T, Gullans SR (2002a) An iron-responsive element type II in the 5′-untranslated region of the Alzheimer’s amyloid precursor protein transcript. J Biol Chem 277(47):45518–45528 [DOI] [PubMed] [Google Scholar]
- Rogers JT, Randall JD, Eder PS, Huang X, Bush AI, Tanzi RE, Venti A, Payton SM, Giordano T, Nagano S, Cahill CM, Moir R, Lahiri DK, Greig N, Sarang SS, Gullans SR (2002b) Alzheimer’s disease drug discovery targeted to the APP mRNA 5′untranslated region. J Mol Neurosci 19:77–82 [DOI] [PubMed] [Google Scholar]
- Rogers JT, Bush AI, Cho HH, Smith DH, Thomson AM, Friedlich AL, Lahiri DK, Leedman PJ, Huang X, Cahill CM (2008) Iron and the translation of the amyloid precursor protein (APP) and ferritin mRNAs: riboregulation against neural oxidative damage in Alzheimer’s disease. Biochem Soc Trans 36(Pt 6):1282–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherzer CR, Grass JA, Liao Z, Pepivani I, Zheng B, Eklund AC, Ney PA, Ng J, McGoldrick M, Mollenhauer B, Bresnick EH, Schlossmacher MG (2008) GATA transcription factors directly regulate the Parkinson’s disease-linked gene alpha-synuclein. Proc Natl Acad Sci USA 105(31):10907–10912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw KT, Utsuki T, Rogers J, Yu QS, Sambamurti K, Brossi A, Ge YW, Lahiri DK, Greig NH (2001) Phenserine regulates translation of beta-amyloid precursor protein mRNA by a putative interleukin-1 responsive element, a target for drug development. Proc Natl Acad Sci USA 98(13):7605–7610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, Hulihan M, Peuralinna T, Dutra A, Nussbaum R, Lincoln S, Crawley A, Hanson M, Maraganore D, Adler C, Cookson MR, Muenter M, Baptista M, Miller D, Blancato J, Hardy J, Gwinn-Hardy K (2003) alpha-Synuclein locus triplication causes Parkinson’s disease. Science 302(5646):841. [DOI] [PubMed] [Google Scholar]
- Souza JM, Giasson BI, Chen Q, Lee VM, Ischiropoulos H (2000) Dityrosine cross-linking promotes formation of stable alpha-synuclein polymers. Implication of nitrative and oxidative stress in the pathogenesis of neurodegenerative synucleinopathies. J Biol Chem 275(24):18344–18349 [DOI] [PubMed] [Google Scholar]
- Strobel G (2009) The spectrum series: grappling with the overlap between Alzheimer’s and Parkinson’s diseases. 9th International Conference on Alzheimer’s and Parkinson’s Diseases, 11–15 March 2009, Prague, Czech Republic. J Alzheimers Dis 18(3):625–640 [DOI] [PubMed] [Google Scholar]
- Taylor JP, Mata IF, Farrer MJ (2006) LRRK2: a common pathway for parkinsonism, pathogenesis and prevention? Trends Mol Med 12(2):76–82 [DOI] [PubMed] [Google Scholar]
- Thomas JR, Hergenrother PJ (2008) Targeting RNA with small molecules. Chem Rev 108(4):1171–1224 [DOI] [PubMed] [Google Scholar]
- Tibodeau JD, Fox PM, Ropp PA, Theil EC, Thorp HH (2006) The up-regulation of ferritin expression using a small-molecule ligand to the native mRNA. Proc Natl Acad Sci USA 103(2):253–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth I, Yuan L, Rogers JT, Boyce H, Bridges KR (1999) Hypoxia alters iron-regulatory protein-1 binding capacity and modulates cellular iron homeostasis in human hepatoma and erythroleukemia cells. J Biol Chem 274:4467–4473 [DOI] [PubMed] [Google Scholar]
- Tucker S, Ahl M, Cho HH, Bandyopadhyay S, Cuny GD, Bush AI, Goldstein LE, Westaway D, Huang X, Rogers JT (2006) RNA therapeutics directed to the non coding regions of APP mRNA, in vivo anti-amyloid efficacy of paroxetine, erythromycin, and N-acetyl cysteine. Curr Alzheimer Res 3(3):221–227 [DOI] [PubMed] [Google Scholar]
- Ueda K, Fukushima H, Masliah E, Xia Y, Iwai A, Yoshimoto M, Otero DA, Kondo J, Ihara Y, Saitoh T (1993) Molecular cloning of cDNA encoding an unrecognized component of amyloid in Alzheimer disease. Proc Natl Acad Sci USA 90(23):11282–11286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsuki T, Uchimura N, Irikura M, Moriuchi H, Holloway HW, Yu QS, Spangler EL, Mamczarz J, Ingram DK, Irie T, Greig NH (2007) Preclinical investigation of the topical administration of phenserine: transdermal flux, cholinesterase inhibition, and cognitive efficacy. J Pharmacol Exp Ther 321(1):353–361 [DOI] [PubMed] [Google Scholar]
- Uversky VN (2007) Neuropathology, biochemistry, and biophysics of alpha-synuclein aggregation. J Neurochem 103(1):17–37 [DOI] [PubMed] [Google Scholar]
- Uversky VN, Li J, Fink AL (2001a) Metal-triggered structural transformations, aggregation, and fibrillation of human alpha-synuclein. A possible molecular NK between Parkinson’s disease and heavy metal exposure. J Biol Chem 276(47): 44284–44296 [DOI] [PubMed] [Google Scholar]
- Uversky VN, Li J, Fink AL (2001b) Pesticides directly accelerate the rate of alpha-synuclein fibril formation: a possible factor in Parkinson’s disease. FEBS Lett 500(3):105–108 [DOI] [PubMed] [Google Scholar]
- Uversky VN, Li J, Souillac P, Millett IS, Doniach S, Jakes R, Goedert M, Fink AL (2002) Biophysical properties of the synucleins and their propensities to fibrillate: inhibition of alpha-synuclein assembly by beta- and gamma-synucleins. J Biol Chem 277(14):11970–11978 [DOI] [PubMed] [Google Scholar]
- Venti A, Giordano T, Eder P, Bush AI, Lahiri DK, Greig NH, Rogers JT (2004) The integrated role of desferrioxamine and phenserine targeted to an iron-responsive element in the APP-mRNA 5′-untranslated region. Ann NY Acad Sci 1035:34–48 [DOI] [PubMed] [Google Scholar]
- Villarroel MC, Hidalgo M, Jimeno A (2009) Mycophenolate mofetil: an update. Brugs Today (Barc) 45:521–532 [DOI] [PubMed] [Google Scholar]
- Wang JK, Portbury S, Thomas MB, Barney S, Ricca DJ, Morris DL, Warner DS, Lo DC (2006) Cardiac glycosides provide neuro-protection against ischemic stroke: discovery by a brain slice-based compound screening platform. Proc Natl Acad Sci USA 103(27):10461–10466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Di X, D’Agostino RB Jr, Torti SV, Torti FM (2007) Excess capacity of the iron regulatory protein system. J Biol Chem 282(34):24650–24659 [DOI] [PubMed] [Google Scholar]
- Wang WW, Khajavi M, Patel BJ, Beach J, Jankovic J, Ashizawa T (1998) The G209A mutation in the alpha-synuclein gene is not detected in familial cases of Parkinson disease in non-Greek and/or Italian populations. Arch Neurol 55(12):1521–1523 [DOI] [PubMed] [Google Scholar]
- Werstuck G, Green MR (1998) Controlling gene expression in living cells through small molecule-RNA interactions. Science 282:296–298 [DOI] [PubMed] [Google Scholar]
- Winblad B, Giacobini E, Frölich L, Friedhoff L, Bruinsma G, Becker RE, Greig NH (2010) Phenserine efficacy in Alzheimer’s disease. J Alzheimer’s Dis. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingert RA, Galloway JL, Barut B, Foott H, Fraenkel P, Axe JL, Weber GJ, Dooley K, Davidson AJ, Schmid B, Paw BH, Shaw GC, Kingsley P, Palis J, Schubert H, Chen O, Kaplan J, Zon LI (2005) Deficiency of glutaredoxin 5 reveals Fe-S clusters are required for vertebrate haem synthesis. Nature 436(7053):1035–1039 [DOI] [PubMed] [Google Scholar]
- Xia Y, Saitoh T, Ueda K, Tanaka S, Chen X, Hashimoto M, Hsu L, Conrad C, Sundsmo M, Yoshimoto M, Thal L, Katzman R, Masliah E (2001) Characterization of the human alpha-synuclein gene: genomic structure, transcription start site, promoter region and polymorphisms. J Alzheimers Dis 3(5):485–494 [DOI] [PubMed] [Google Scholar]
- Yamin G, Glaser CB, Uversky VN, Fink AL (2003) Certain metals trigger fibrillation of methionine-oxidized alpha-synuclein. J Biol Chem 278(30):27630–27635 [DOI] [PubMed] [Google Scholar]
- Zhang H, Qian DZ, Tan YS, Lee K, Gao P, Ren YR, Rey S, Hammers H, Chang D, Pili R, Dang CV, Liu JO, Semenza GL (2008) Digoxin and other cardiac glycosides inhibit HIF-1alpha synthesis and block tumor growth. Proc Natl Acad Sci USA 105(50):19579–19586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer M, Ebert BL, Neil C, Brenner K, Papaioannou I, Melas A, Tolliday N, Lamb J, Pantopoulos K, Golub T, Iliopoulos O (2008) Small-molecule inhibitors of HIF-2a translation link its 5′UTR iron-responsive element to oxygen sensing. Mol Cell 32(6):838–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M (1989) Computer prediction of RNA structure. Methods Enzymol 180:262–288 [DOI] [PubMed] [Google Scholar]