Abstract
Background
The transcription factor Oct-4 is necessary for maintaining pluripotency and self-renewal of embryonic stem cells, and POU5F1B is a processed pseudogene of Oct-4 with coding capacity. The purpose of this study is to evaluate the expression and clinical implication of POU5F1B in AML.
Material/Methods
The expression of the POU5F1B transcript was evaluated in 175 newly diagnosed AML patients and 39 healthy controls by use of real-time quantitative PCR (RQ-PCR).
Results
POU5F1B was underexpressed in AML compared with controls (P<0.001). The receiver operating characteristic (ROC) curve revealed that the POU5F1B transcript level was able to differentiate AML patients from healthy individuals (AUC=0.682). In non-APL AML patients, the POU5F1Blow group had significantly higher WBC than the POU5F1Bhigh group (20.2×109 vs. 4.6×109 L−1, P=0.021). Among whole-cohort AML, non-APL AML, and intermediate-risk AML, POU5F1Bhigh patients had obviously higher complete remission (CR) rates than POU5F1Blow patients (P=0.012, P=0.012 and P=0.027). In addition, Kaplan-Meier analysis demonstrated better overall survival (OS, P=0.019, P=0.007 and P=0.046, respectively) in POU5F1Bhigh patients compared with POU5F1Blow patients. Furthermore, in multivariate survival analysis, POU5F1B was independently associated with OS in non-APL AML patients and intermediate-risk AML as a favorable prognostic factor.
Conclusions
POU5F1B was frequently underexpressed in AML, and might contribute to the diagnosis and prognosis of AML.
MeSH Keywords: Biological Markers; Diagnosis; Leukemia, Myeloid, Acute; Prognosis; Pseudogenes
Background
Acute myeloid leukemia (AML) is a malignant clonal disorder distinguished by differentiation block of myeloid and accumulation of abnormal myeloid progenitors in the bone marrow (BM) and blood [1]. In adults, AML is the most common form of acute leukemia [2], and the incidence increases with age [3]. Patients may present with bleeding, anemia, and infection complications due to bone marrow failure, but often only have fatigue on initial presentation [3–5]. The evolving molecular genetics facilitate discernment of AML prognostic indicators, especially in cytogenetic results and molecular abnormalities [6].
In the past decade, non-coding RNAs (ncRNAs), involving microRNAs, small interfering RNAs (siRNAs), long non-coding RNAs (lncRNAs), and pseudogenes, have attracted intense attention [7]. There is an increasing focus on the relationship between non-coding RNA and cancer, especially as a diagnostic and prognostic biomarker of cancer from early detection to monitor recurrence or treatment progression [8]. Pseudogenes were defined as ‘junk DNA’ that no longer possess biological functions due to nonsense or frameshift mutations in coding genes, whereas some pseudogene-derived RNAs have been shown to have unique regulatory roles, and some pseudogene fragments can be translated [9]. Increasing evidence indicates that pseudogenes show important biological functions in human cancers [10]. Kong et al. demonstrated that the pseudogene PDIA3P1 is overexpressed in HCC tissues and is associated with tumor size and TNM stage, and knockdown of PDIA3P1 decreases HCC cell proliferation and promotes apoptosis [11].
The POU family possesses a POU DNA-binding domain, and Oct-4 is one of the transcription factors of the family. As a master transcription factor for pluripotent cell self-renewal, Oct-4 plays a critical role in the embryonic development of mammals [12]. Alternative splicing of Oct-4 produces 3 isoforms: Oct-4A, Oct-4B, and Oct-4B1 [13]. Oct-4A (Oct-4) can maintain stem cell self-renewal, while Oct-4B cannot [14]. Two of the 6 highly homologous pseudogenes of human Oct-4 – Oct-4-pg1 and Oct-4-pg5 – were found to be transcribed in somatic cancers [15]. Overexpression of POU5F1B (Oct-4-pg1) is reported in gastric cancer and is associated with unfavorable prognosis in stage IV patients [16]. The aim of this study was to evaluate expression of POU5F1B in patients with an initial diagnosis of AML, as well as to explore the correlation between POU5F1B and AML.
Material and Methods
Patient samples
The study was subject approved by the Institutional Ethics Committee of the Affiliated People’s Hospital of Jiangsu University. Bone marrow samples were collected from 214 adults, including 39 healthy donors and 175 newly diagnosed AML patients before chemotherapy. The patients were diagnosed according to the WHO and French-American-British (FAB) classification [17,18]. All participants signed written informed consent. Previous articles have described treatment protocols for AML patients [19].
RNA isolation, reverse transcription, and RQ-PCR
Lymphocyte Separation Medium (TBD Sciences, Tianjin, China) was used to separate bone marrow mononuclear cells (BMNCs). Trizol reagent (Invitrogen, Carlsbad, CA) was used to separated total RNA. We used 10 mM of dNTPs, 2 μg of total RNA from each sample, 10 μM of random hexamers, 80 U of RNase inhibitor, and 200 U of MMLV reverse transcriptase (MBI Fermentas, Hanover, MA) and reverse-transcribed them into single-stranded complementary DNA (cDNA), and then stored them at −20°C.
We used a 7500 Thermal cycler (Applied Biosystems, CA) to perform RQ-PCR. The forward primer used for POU5F1B transcript detection was 5′-GCGATCAAGCAGCGACTA-3′, and the reverse primer was 5′-AGGGAAAGGGACTGAGGAG -3′. POU5F1B transcript level detection was quantified by RT-qPCR as follows: 95°C for 30 s, and then 40 cycles at 95°C for 5 s, 58.1°C for 30 s, 72°C for 30 s, and 80°C for 31 s to collect fluorescence, and finally, the melting program at 95°C for 15 s, 60°C for 60 s, 95°C for 15 s, and 60°C for 15 s. The amount of POU5F1B transcript was calculated by analyzing expression of the housekeeper gene ABL using 2−ΔΔCT method.
Gene mutation detection
Mutations of DNMT3A, N/K-RAS, NPM1, c-KIT, IDH2 R140, IDH1/2, U2AF1, and SRSF2 were detected by PCR and high-resolution melting analysis (HRMA) [20–24]. Mutations of CEBPA and FLT3-ITD genes and all positive samples were detected in genomic DNA by PCR and direct sequencing [25].
Statistical analyses
SPSS22.0 software was used for statistical analysis. P<0.05 was considered statistically significant for all analyses. Continuous variables between the 2 groups were compared using the Mann-Whitney U test. The Fisher exact test or Pearson chi-square analysis was used, as appropriate, to compare categorical variables across groups. The diagnostic accuracy of POU5F1B expression in discriminating AML patients from normal controls was calculated by ROC curve and area under the ROC curve (AUC). Kaplan-Meier curves were used to estimate the effect of POU5F1B expression on survival, and Cox regression models were used to independently assess the prognostic value of POU5F1B expression.
Results
POU5F1B expression in controls and AML patients
Assessment of POU5F1B transcript levels in AML patients (0–246.92, median 0.0536) detected underexpression compared to controls (0–217.99, median 0.7991) (P<0.001, Figure 1). Moreover, the level of POU5F1B expression was lower in non-APL AML patients and in intermediate-risk AML patients (P<0.001 and P<0.001, Figure 1).
Figure 1.
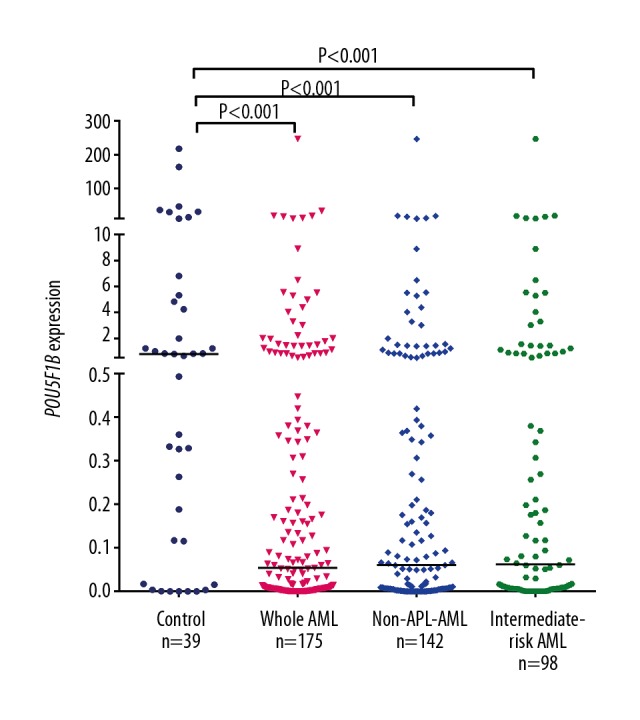
Relative expression levels of POU5F1B in AML patients and controls. The transcript level of POU5F1B in controls, whole-cohort AML patients, non-APL patients, and intermediate-risk AML patients were evaluated by RQ-PCR. Horizontal lines represent the median level of POU5F1B expression in each group.
Diagnostic accuracy of POU5F1B expression
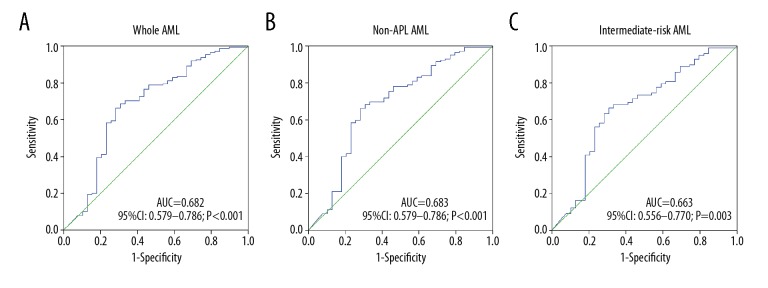
ROC was utilized to analyze the differentiating value of POU5F1B expression. It indicated that POU5F1B can act as a potential marker to distinguish whole-cohort AML patients from controls, with an AUC of 0.682 (95%CI: 0.579–0.786, P<0.001). Meanwhile, the level of POU5F1B expression might be used to segregate non-APL AML (AUC=0.683, 95%CI: 0.579–0.786, P<0.001) from normal controls, and a similar result was found in the intermediate-risk group (AUC=0.663, 95%CI: 0.556–0.770, P=0.003) (Figure 2).
Figure 2.
ROC curve analysis for distinguishing AML patients from controls. (A) Whole AML patients; (B) Non-APL patients; (C) Intermediate-risk AML.
Association of POU5F1B expression with clinical and laboratory features in AML
We divided the AML patients into 2 groups using a cutoff value of mean minus 2SD obtained in normal controls: POU5F1Bhigh (>1.005) and POU5F1Blow (≤1.005). None of these differences between the 2 groups were statistically significant, such as sex, age, white blood cells (WBC), hemoglobin (HB), platelets (PLT), BM blasts, FAB subtypes, karyotypes, and gene mutations. However, the results, as shown in Table 1, indicated that the CR of POU5F1Bhigh patients was significantly higher than that of POU5F1Blow patients (P=0.012, Table 1). When M3 patients were excluded, the comparison of laboratory characteristics and clinical data between the 2 groups (Table 1), patients with lower expression of POU5F1B had significantly higher WBC than those with higher expression of POU5F1B (20.2×109 vs. 4.6×109 L−1, P=0.021). As with whole AML, in non-APL and intermediate-risk AML patients, the CR was strikingly higher in POU5F1Bhigh patients than in POU5F1Blow (P=0.012 and P=0.027, Tables 1, 2). The intermediate-risk AML patients with lower POU5F1B were older (P=0.038, Table 2).
Table 1.
Comparison of clinical and laboratory features between whole-cohort AML and non-APL AML patients with low and high expression.
| Patient parameters | Whole-cohort AML | Non-APL AML | ||||
|---|---|---|---|---|---|---|
| POU5F1Blow (n=145) | POU5F1Bhigh (n=30) | P value | POU5F1Blow (n=118) | POU5F1Bhigh (n=24) | P value | |
| Sex, Male/Female | 90/55 | 20/10 | 0.683 | 75/43 | 15/9 | 1.000 |
| Median age, years (range) | 57 (20–93) | 54 (21–83) | 0.105 | 61 (20–93) | 57 (28–83) | 0.135 |
| Median WBC, ×109/L (range) | 16.3 (0.4–528.0) | 5.5 (0.3–203.6) | 0.103 | 20.2 (0.4–528.0) | 4.6 (0.8–203.6) | 0.021 |
| Median hemoglobin, g/L (range) | 77.5 (32.0–144.0) | 77.0 (34.0–131.0) | 0.893 | 76.0 (32.0–144.0) | 77.5 (34.0–131.0) | 0.963 |
| Median platelets, ×109/L (range) | 40.0 (3.0–447.0) | 46.5 (4.0–415.0) | 0.937 | 42.0 (3.0–447.0) | 50.0 (4.0–415.0) | 0.714 |
| BM blasts,% (range) | 46.7 (3.0–99.0) | 37.5 (1.0–93.0) | 0.203 | 56.5 (10.5–99.0) | 43.0 (6.0–93.0) | 0.193 |
| CR (+/−) | 50/83 | 18/10 | 0.012 | 29/78 | 13/10 | 0.012 |
| FAB | 0.956 | 0.904 | ||||
| M0 | 2 | 1 | 2 | 1 | ||
| M1 | 10 | 1 | 10 | 1 | ||
| M2 | 56 | 13 | 56 | 13 | ||
| M3 | 27 | 6 | – | – | ||
| M4 | 29 | 5 | 29 | 5 | ||
| M5 | 16 | 3 | 17 | 3 | ||
| M6 | 5 | 1 | 5 | 1 | ||
| Karyotype classification | 0.353 | 0.329 | ||||
| Favorable | 37 | 8 | 12 | 2 | ||
| Intermediate | 78 | 20 | 77 | 20 | ||
| Poor | 21 | 1 | 21 | 1 | ||
| No data | 9 | 1 | 8 | 1 | ||
| Karyotype | 0.320 | 0.209 | ||||
| Normal | 57 | 18 | 56 | 18 | ||
| t(8;21) | 9 | 2 | 9 | 2 | ||
| t(15;17) | 25 | 6 | 0 | 0 | ||
| +8 | 6 | 0 | 6 | 0 | ||
| Others | 23 | 1 | 23 | 1 | ||
| Complex | 16 | 2 | 16 | 2 | ||
| No data | 9 | 1 | 8 | 1 | ||
| Gene mutation | ||||||
| CEBPA (+/−) | 14/106 | 3/23 | 1.000 | 14/85 | 3/18 | 1.000 |
| NPM1 (+/−) | 13/107 | 4/22 | 0.506 | 13/86 | 4/17 | 0.496 |
| FLT3-ITD (+/−) | 16/104 | 2/24 | 0.742 | 13/86 | 2/19 | 1.000 |
| c-KIT (+/−) | 6/114 | 1/25 | 1.000 | 5/94 | 1/20 | 1.000 |
| NRAS or KRAS (+/−) | 7/113 | 3/23 | 0.384 | 7/92 | 3/18 | 0.377 |
| IDH1/2 (+/−) | 6/114 | 1/25 | 1.000 | 6/93 | 1/20 | 1.000 |
| IDH2 R140(+/−) | 4/116 | 1/25 | 1.000 | 4/95 | 1/21 | 1.000 |
| DNMT3A (+/−) | 10/110 | 0/26 | 0.209 | 10/89 | 0/21 | 0.206 |
| U2AF1 (+/−) | 5/115 | 1/25 | 1.000 | 5/94 | 1/20 | 1.000 |
| SRSF2 (+/−) | 7/117 | 0/26 | 0.605 | 7/94 | 0/21 | 0.600 |
Table 2.
Comparison of clinical and laboratory features between patients with intermediate-risk AML low and high expression.
| Patient parameters | Intermediate-risk AML | ||
|---|---|---|---|
| POU5F1Blow (n=78) | POU5F1Bhigh (n=20) | P value | |
| Sex, Male/Female | 49/29 | 13/7 | 1.000 |
| Median age, years (range) | 61 (20–93) | 57 (21–83) | 0.038 |
| Median WBC, ×109/L (range) | 21.3 (0.4–528.0) | 7.7 (0.3–203.6) | 0.129 |
| Median hemoglobin, g/L (range) | 81.5 (32.0–144.0) | 77.5 (34.0–131.0) | 0.988 |
| Median platelets, ×109/L (range) | 42.5 (3.0–399.0) | 52.5 (4.0–415.0) | 0.547 |
| BM blasts,% (range) | 55.0 (21.5–99.0) | 48.5 (6.0–93.0) | 0.506 |
| CR (+/−) | 20/52 | 11/8 | 0.027 |
| FAB | 0.883 | ||
| M0 | 1 | 1 | |
| M1 | 6 | 1 | |
| M2 | 39 | 9 | |
| M3 | 1 | 0 | |
| M4 | 17 | 5 | |
| M5 | 11 | 3 | |
| M6 | 3 | 1 | |
| Gene mutation | |||
| CEBPA (+/−) | 11/57 | 3/14 | 1.000 |
| NPM1 (+/−) | 11/57 | 4/13 | 0.487 |
| FLT3-ITD (+/−) | 10/58 | 2/15 | 1.000 |
| c-KIT (+/−) | 1/67 | 1/16 | 0.362 |
| NRAS or KRAS (+/−) | 7/61 | 3/14 | 0.411 |
| IDH1/2 (+/−) | 5/63 | 1/16 | 1.000 |
| IDH2 R140(+/−) | 3/65 | 1/16 | 1.000 |
| DNMT3A (+/−) | 9/59 | 0/17 | 0.194 |
| U2AF1 (+/−) | 4/64 | 1/16 | 1.000 |
| SRSF2 (+/−) | 6/64 | 0/17 | 0.593 |
Prognostic value of POU5F1B in AML
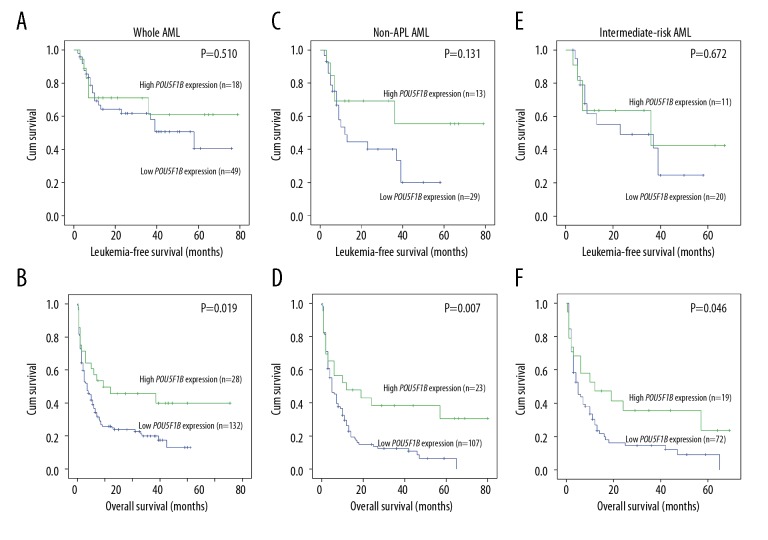
OS and leukemia-free survival (LFS) were estimated according to Kaplan-Meier methods. OS (P=0.019, median 7 vs. 17 months; P=0.007, median 5 vs. 12 months, respectively) was significantly worse in the whole-cohort AML patients and non-APL AML patients with low POU5F1B expression. Kaplan-Meier analysis showed that intermediate-risk AML patients with POU5F1B low expression had significantly shorter OS (P=0.046, median 4.5 vs. 12 months, Figure 3F). There was no significant association between POU5F1B expression and LFS (P=0.510, P=0.131, and P=0.672, respectively) among the 3 AML groups (Figure 3).
Figure 3.
Differences in leukemia-free survival and overall survival between the POU5F1Bhigh group and the POU5F1Blow group were estimated using to Kaplan-Meier method. (A) LFS for whole AML patients; (B) OS for whole AML patients; (C) LFS for non-APL patients; (D) OS for non-APL patients; (E) LFS for intermediate-risk patients; (F) OS for intermediate-risk patients.
In multivariate analyses, POU5F1B overexpression remained a significant favorable prognostic factor for OS (P=0.014 and P=0.023) in non-APL and intermediate-risk AML patients. However, improved OS was not observed among whole-cohort AML patients (Table 3).
Table 3.
Multivariate analyses of prognostic factors for overall survival in non-APL and intermediate-risk AML cases.
| Prognostic factors | Non-APL AML | Intermediate-risk AML | ||
|---|---|---|---|---|
| Hazard ratio (95% CI) | P value | Hazard ratio (95% CI) | P value | |
| Sex (Male vs. Female) | 1.136 (0.713–1.812) | 0.591 | 0.876 (0.512–1.498) | 0.629 |
| Age (≤60 vs. >60 years) | 1.384 (0.905–2.117) | 0.134 | 1.116 (0.642–1.939) | 0.698 |
| WBC (≥30×109/L vs. <30×109/L) | 1.209 (0.776–1.884) | 0.401 | 1.499 (0.882–2.548) | 0.134 |
| PLT (100×109/L vs. 100×109/L) | 1.340 (0.766–2.343) | 0.305 | 1.074 (0.528–2.184) | 0.845 |
| Karyotypic classifications (favorable vs. intermediate vs. poor) | 1.683 (1.251–2.266) | 0.001 | – | – |
| POU5F1B expression (high vs. low) | 0.453 (0.241–0.852) | 0.014 | 0.448 (0.225–0.896) | 0.023 |
| IDH1/2 mutation (mutant vs. wild-type) | 4.470 (1.860–10.740) | 0.001 | 5.732 (2.197–14.960) | 0.000 |
| IDH2 R140 mutation (mutant vs. wild-type) | 0.650 (0.118–3.585) | 0.621 | 0.636 (0.102–3.948) | 0.627 |
| U2AF1 mutation (mutant vs. wild-type) | 2.452 (1.013–5.931) | 0.047 | 2.206 (0.831–5.858) | 0.112 |
| SRSF2 mutation (mutant vs. wild-type) | 1.339 (0.540–3.321) | 0.529 | 2.032 (0.766–5.390) | 0.154 |
Discussion
Oct-4 was reported to be overexpressed in several types of cancers, such as bladder cancer [26], hepatocellular carcinoma [27], primary endometrioid endometrial and ovarian carcinomas [28], and non-small-cell lung cancer [29]. Oct-4 high expression in AML was a common molecular event, and AML patients with high expression of Oct-4 showed a shorter overall survival rate [30]. POU5F1B was reported as a susceptibility gene in breast cancer [31], prostate cancer [32], and gastric cancer [33]. Hayashi et al. reported that overexpression of POU5F1B induces overexpression of GC cell growth factors, which in turn promotes cell proliferation and inhibits apoptosis [16]. POU5F1B promotes HCC proliferation by activating AKT, and patients with high POU5F1B level have shorter survival times [34]. HPV integrated in POU5F1B in cervical tumor cells survives during radiotherapy and may lead to resistance to radiation therapy [35]. To date, abnormal expression of a few pseudogenes in AML have been reported, including Vim2p, DUSP5P1, and BMI1P1 [36–38]. However, there has been no research focused on pseudogene POU5F1B in AML. In this study, POU5F1B transcript was expressed at lower levels in AML patients compared with the control group. ROC analysis revealed that low POU5F1B expression is a prospective biomarker for use in discriminating AML patients, including non-APL AML and intermediate-risk AML patients, from healthy controls. AML patients with high WBC count have a particularly poor prognosis [39]. Similarly, our finding suggested that non-APL AML patients with lower POU5F1B expression had higher WBC counts and worse survival.
Furthermore, CR rates and OS were significantly better in the POU5F1Bhigh group than in the POU5F1Blow group in total AML, non-APL, and intermediate-risk AML patients. Multivariate analyses demonstrated that POU5F1B was an independent prognostic factor for OS in non-APL and intermediate-risk AML patients. Collectively, these results showed that detecting the expression of the POU5F1B transcript in AML patients, especially non-APL AML and intermediate-risk patients, might have important prognostic and curative implications. POU5F1B expression has no significant correlation with LFS, and we considered that it was caused by the differences in consolidation and intensification therapy. A more comprehensive study including all the subgroups is needed, such as different treatment and age groups, CR and non-CR groups, and the cases need to be expanded.
Competing endogenous RNA (ceRNAs) are RNA transcripts, including long non-coding RNAs, pseudogenes, and circular RNAs [40], and they regulate each other by competing to bind to shared microRNA (miRNA) recognition elements (MREs) [41]. Pseudogenes show sequence similarity to their parental genes, with many identical MREs in its sequence [42]. Pseudogenes regulate their parental transcripts by competing for shared miRNAs [43]. A number of studies have found that ceRNAs abundance and activity are underexpressed in cancer; therefore, these ceRNAs may be potential diagnostic biomarkers in cancer [44]. Previous studies have reported that 2 pseudogenes of HMGA1 – HMGA1P6 and HMGA1P7 – act as competitive endogenous RNA decoys for carcinogenesis genes HMGA1, H19, and Igf2, and pseudogene overexpression increases oncogenes levels, inhibiting their mRNA suppression by miRNAs that target HMGA1P7 gene [45]. Scarola et al. showed that the lncRNA produced by Oct-4-pg4 (X-linked Oct-4 pseudogene) during transcription is overexpressed during mESC differentiation and forms a complex with SUV39H1 HMTase to regulate the ancestral Oct-4 gene promoter, resulting in Oct-4 gene silence and reduced mESC self-renewal [46]. In hepatocellular carcinoma cells (HCC) and endometrial carcinoma, Oct-4-pg4 and pg5 function as miRNA sponges protecting Oct-4 transcript by competing with miR-145 [47]. To date, the function of the pseudogene POU5F1B in AML remains largely unknown. Combined with the expression of Oct-4 in AML, an interesting pattern emerges: the transcript levels of Oct-4 and POU5F1B showed the exact opposite trend. We anticipate that the pseudogene-derived lncRNAs may regulate the promoter of the parental gene Oct-4, resulting in reduced Oct-4 gene silencing. POU5F1B was found to have 95% homology with Oct-4 [48]; therefore, POU5F1B may serve as ceRNA, allowing Oct-4 to evade miRNA inhibition. More research on this topic is needed, including in vivo and in vitro functional assays, correlation analysis with Oct-4 and POU5F1B expression levels, prediction and detection of pseudogene-derived lncRNAs and POU5F1B-targeted miRNAs, and stemness potential assays.
Conclusions
Expression of the POU5F1B transcript is significantly decreased in AML patients and is associated with unfavorable clinical variables and poor prognosis. POU5F1B has promise as a potential novel biomarker and target for future therapy.
Footnotes
Source of support: This study was supported by National Natural Science Foundation of China (20120016), the Medical Innovation Team of Jiangsu Province (CXTDB2017002), the Zhenjiang Clinical Research Center of Hematology (SS2018009), and the Jiangsu Provincial “Innovative & Entrepreneurial Talent Team” Program
Conflicts of interest
None.
References
- 1.Grove CS, Vassiliou GS. Acute myeloid leukaemia: A paradigm for the clonal evolution of cancer? Dis Model Mech. 2014;7:941–51. doi: 10.1242/dmm.015974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perna F, Berman SH, Soni RK, et al. Integrating proteomics and transcriptomics for systematic combinatorial chimeric antigen receptor therapy of AML. Cancer Cell. 2017;32:506–19. doi: 10.1016/j.ccell.2017.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abelson S, Collord G, Ng SWK, et al. Prediction of acute myeloid leukaemia risk in healthy individuals. Nature. 2018;559(7714):400–4. doi: 10.1038/s41586-018-0317-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyd AL, Reid JC, Salci KR, et al. Acute myeloid leukaemia disrupts endogenous myelo-erythropoiesis by compromising the adipocyte bone marrow niche. Nat Cell Biol. 2017;19(11):1336–47. doi: 10.1038/ncb3625. [DOI] [PubMed] [Google Scholar]
- 5.Lacourt TE, Kavelaars A, Ohanian M, et al. Patient-reported fatigue prior to treatment is prognostic of survival in patients with acute myeloid leukemia. Oncotarget. 2018;9:31244–52. doi: 10.18632/oncotarget.25787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li W, Zhong C, Jiao J, et al. Characterization of hsa_circ_0004277 as a new biomarker for acute myeloid leukemia via circular RNA profile and bioinformatics analysis. Int J Mol Sci. 2017;18(3):597–610. doi: 10.3390/ijms18030597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Groen JN, Capraro D, Morris KV. The emerging role of pseudogene expressed non-coding RNAs in cellular functions. Int J Biochem Cell Biol. 2014;54:350–55. doi: 10.1016/j.biocel.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anfossi S, Babayan A, Pantel K, Calin GA. Clinical utility of circulating non-coding RNAs-an update. Nat Rev Clin Oncol. 2018;15(9):541–63. doi: 10.1038/s41571-018-0035-x. [DOI] [PubMed] [Google Scholar]
- 9.Prieto-Godino LL, Rytz R, Bargeton B, et al. Olfactory receptor pseudo-pseudogenes. Nature. 2016;539(7627):93–97. doi: 10.1038/nature19824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi X, Nie F, Wang Z, Sun M. Pseudogene-expressed RNAs: A new frontier in cancers. Tumour Biol. 2016;37(2):1471–78. doi: 10.1007/s13277-015-4482-z. [DOI] [PubMed] [Google Scholar]
- 11.Kong Y, Zhang L, Huang Y, et al. Pseudogene PDIA3P1 promotes cell proliferation, migration and invasion, and suppresses apoptosis in hepatocellular carcinoma by regulating the p53 pathway. Cancer Lett. 2017;407:76–83. doi: 10.1016/j.canlet.2017.07.031. [DOI] [PubMed] [Google Scholar]
- 12.Radzisheuskaya A, Chia Gle B, dos Santos RL, et al. A defined Oct4 level governs cell state transitions of pluripotency entry and differentiation into all embryonic lineages. Nat Cell Biol. 2013;15(6):579–90. doi: 10.1038/ncb2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao Y, Wei J, Han J, et al. The novel function of OCT4B isoform-265 in genotoxic stress. Stem Cells. 2012;30(4):665–72. doi: 10.1002/stem.1034. [DOI] [PubMed] [Google Scholar]
- 14.Wang X, Dai J. Concise review: Isoforms of OCT4 contribute to the confusing diversity in stem cell biology. Stem Cells. 2010;28(5):885–93. doi: 10.1002/stem.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao S, Yuan Q, Hao H, et al. Expression of OCT4 pseudogenes in human tumours: Lessons from glioma and breast carcinoma. J Pathol. 2011;223(5):672–82. doi: 10.1002/path.2827. [DOI] [PubMed] [Google Scholar]
- 16.Hayashi H, Arao T, Togashi Y, et al. The OCT4 pseudogene POU5F1B is amplified and promotes an aggressive phenotype in gastric cancer. Oncogene. 2015;34(2):199–208. doi: 10.1038/onc.2013.547. [DOI] [PubMed] [Google Scholar]
- 17.Giagounidis AA, Hildebrandt B, Heinsch M, et al. Acute basophilic leukemia. Eur J Haematol. 2001;67(2):72–76. doi: 10.1034/j.1600-0609.2001.t01-1-00487.x. [DOI] [PubMed] [Google Scholar]
- 18.Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391–405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, Lin J, Yang J, et al. Overexpressed let-7a-3 is associated with poor outcome in acute myeloid leukemia. Leuk Res. 2013;37(12):1642–47. doi: 10.1016/j.leukres.2013.09.022. [DOI] [PubMed] [Google Scholar]
- 20.Lin J, Yao DM, Qian J, et al. Recurrent DNMT3A R882 mutations in Chinese patients with acute myeloid leukemia and myelodysplastic syndrome. PLoS One. 2011;6(10):e26906. doi: 10.1371/journal.pone.0026906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin J, Yao DM, Qian J, et al. IDH1 and IDH2 mutation analysis in Chinese patients with acute myeloid leukemia and myelodysplastic syndrome. Ann Hematol. 2012;91(4):519–25. doi: 10.1007/s00277-011-1352-7. [DOI] [PubMed] [Google Scholar]
- 22.Yang X, Qian J, Sun A, et al. RAS mutation analysis in a large cohort of Chinese patients with acute myeloid leukemia. Clin Biochem. 2013;46(7–8):579–83. doi: 10.1016/j.clinbiochem.2012.12.022. [DOI] [PubMed] [Google Scholar]
- 23.Qian J, Yao DM, Lin J, et al. U2AF1 mutations in Chinese patients with acute myeloid leukemia and myelodysplastic syndrome. PLoS One. 2012;7(9):e45760. doi: 10.1371/journal.pone.0045760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skrdlant L, Lin RJ. Characterization of RNA-protein interactions: lessons from two RNA-binding proteins, SRSF1 and SRSF2. Methods Mol Biol. 2016;1421:1–13. doi: 10.1007/978-1-4939-3591-8_1. [DOI] [PubMed] [Google Scholar]
- 25.Wen XM, Lin J, Yang J, et al. Double CEBPA mutations are prognostically favorable in non-M3 acute myeloid leukemia patients with wild-type NPM1 and FLT3-ITD. Int J Clin Exp Pathol. 2014;7(10):6832–40. [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou J, Dong D, Cheng R, et al. Aberrant expression of KPNA2 is associated with a poor prognosis and contributes to OCT4 nuclear transportation in bladder cancer. Oncotarget. 2016;7(45):72767–76. doi: 10.18632/oncotarget.11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang PZ, Lu CL, Li BK, et al. OCT4 expression in hepatocellular carcinoma and its clinical significance. Chin J Cancer. 2010;29(1):111–16. doi: 10.5732/cjc.009.10232. [DOI] [PubMed] [Google Scholar]
- 28.Zhang R, Jiao J, Chu H, et al. Expression of microRNA-145, OCT4, and SOX2 in double primary endometrioid endometrial and ovarian carcinomas. Histol Histopathol. 2018;33(8):859–70. doi: 10.14670/HH-11-986. [DOI] [PubMed] [Google Scholar]
- 29.Li X, Wang J, Xu Z, et al. Expression of Sox2 and Oct4 and their clinical significance in human non-small-cell lung cancer. Int J Mol Sci. 2012;13(6):7663–75. doi: 10.3390/ijms13067663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yin JY, Tang Q, Zhai LL, et al. High expression of OCT4 is frequent and may cause undesirable treatment outcomes in patients with acute myeloid leukemia. Tumour Biol. 2015;36(12):9711–16. doi: 10.1007/s13277-015-3731-5. [DOI] [PubMed] [Google Scholar]
- 31.Wei W, Jiang M, Luo L, et al. Colorectal cancer susceptibility variants alter risk of breast cancer in a Chinese Han population. Genet Mol Res. 2013;12(4):6268–74. doi: 10.4238/2013.December.4.14. [DOI] [PubMed] [Google Scholar]
- 32.Breyer JP, Dorset DC, Clark TA, et al. An expressed retrogene of the master embryonic stem cell gene POU5F1 is associated with prostate cancer susceptibility. Am J Hum Genet. 2014;94(3):395–404. doi: 10.1016/j.ajhg.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen L, Du M, Wang C, et al. Clinical significance of POU5F1P1 rs10505477 polymorphism in Chinese gastric cancer patients receiving cisplatin-based chemotherapy after surgical resection. Int J Mol Sci. 2014;15(7):12764–77. doi: 10.3390/ijms150712764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karreth FA, Reschke M, Ruocco A, et al. The BRAF pseudogene functions as a competitive endogenous RNA and induces lymphoma in vivo. Cell. 2015;161(2):319–32. doi: 10.1016/j.cell.2015.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang D, Zhang W, Liu Y, et al. Single-cell whole-genome sequencing identifies human papillomavirus integration in cervical tumour cells prior to and following radiotherapy. Oncol Lett. 2018;15(6):9633–40. doi: 10.3892/ol.2018.8567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhai LL, Zhou J, Zhang J, et al. Down-regulation of pseudogene Vimentin 2p is associated with poor outcome in de novo acute myeloid leukemia. Cancer Biomark. 2017;18(3):305–12. doi: 10.3233/CBM-160247. [DOI] [PubMed] [Google Scholar]
- 37.Zhou LY, Yin JY, Tang Q, et al. High expression of dual-specificity phosphatase 5 pseudogene 1 (DUSP5P1) is associated with poor prognosis in acute myeloid leukemia. Int J Clin Exp Pathol. 2015;8(12):16073–80. [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou LY, Zhai LL, Yin JY, et al. Pseudogene BMI1P1 expression as a novel predictor for acute myeloid leukemia development and prognosis. Oncotarget. 2016;7(30):47376–86. doi: 10.18632/oncotarget.10156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ganzel C, Manola J, Douer D, et al. Extramedullary disease in adult acute myeloid leukemia is common but lacks independent significance: Analysis of patients in ECOG-ACRIN Cancer Research Group Trials, 1980–2008. J Clin Oncol. 2016;34(29):3544–53. doi: 10.1200/JCO.2016.67.5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomson DW, Dinger ME. Endogenous microRNA sponges: Evidence and controversy. Nat Rev Genet. 2016;17(5):272–83. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- 41.Li LJ, Zhao W, Tao SS, et al. Competitive endogenous RNA network: Potential implication for systemic lupus erythematosus. Expert Opin Ther Targets. 2017;21(6):639–48. doi: 10.1080/14728222.2017.1319938. [DOI] [PubMed] [Google Scholar]
- 42.Karreth FA, Reschke M, Ruocco A, et al. The BRAF pseudogene functions as a competitive endogenous RNA and induces lymphoma in vivo. Cell. 2015;161(2):319–32. doi: 10.1016/j.cell.2015.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chan JJ, Kwok ZH, Chew XH, et al. A FTH1 gene: Pseudogene: MicroRNA network regulates tumorigenesis in prostate cancer. Nucleic Acids Res. 2018;46(4):1998–2011. doi: 10.1093/nar/gkx1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qi X, Zhang DH, Wu N, et al. ceRNA in cancer: Possible functions and clinical implications. J Med Genet. 2015;52(10):710–18. doi: 10.1136/jmedgenet-2015-103334. [DOI] [PubMed] [Google Scholar]
- 45.De Martino M, Forzati F, Marfella M, et al. HMGA1P7-pseudogene regulates H19 and Igf2 expression by a competitive endogenous RNA mechanism. Sci Rep. 2016;6:37622. doi: 10.1038/srep37622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scarola M, Comisso E, Pascolo R, et al. Epigenetic silencing of Oct4 by a complex containing SUV39H1 and Oct4 pseudogene lncRNA. Nat Commun. 2015;6:7631. doi: 10.1038/ncomms8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Villodre ES, Kipper FC, Pereira MB, Lenz G. Roles of OCT4 in tumorigenesis, cancer therapy resistance and prognosis. Cancer Treat Rev. 2016;51:1–9. doi: 10.1016/j.ctrv.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 48.Wezel F, Pearson J, Kirkwood L, Southgate J. Differential expression of Oct4 variants and pseudogenes in normal urothelium and urothelial cancer. Am J Pathol. 2013;183(4):1128–36. doi: 10.1016/j.ajpath.2013.06.025. [DOI] [PubMed] [Google Scholar]