SUMMARY
How the chromatin regulatory landscape in the inner cell mass cells is established from differentially packaged sperm and egg genomes during preimplantation development is unknown. Here we develop a low-input DNase I sequencing (liDNase-seq) method that allows us to generate maps of DNase I-hypersensitive site (DHS) of mouse preimplantation embryos from 1-cell to morula stage. The DHS landscape is progressively established with a drastic increase at the 8-cell stage. Paternal chromatin accessibility is quickly reprogrammed after fertilization to the level similar to maternal chromatin, while imprinted genes exhibit allelic accessibility bias. We demonstrate that transcription factor Nfya contributes to zygotic genome activation and DHS formation at the 2-cell stage and that Oct4 contributes to the DHSs gained at the 8-cell stage. Our study reveals the dynamic chromatin regulatory landscape during early development and identifies key transcription factors important for DHS establishment in mammalian embryos.
Keywords: Chromatin accessibility, DNase I hypersensitive sites, Transcription factor, Low-input DNase-seq, Zygotic genome activation, Oct4, Nfya
INTRODUCTION
Terminally differentiated sperm and egg genomes are organized in very different ways. While sperm genomes are packaged with protamines, egg genomes are occupied by nucleosomes and are transcriptionally inert (Jenkins and Carrell, 2012). Upon fertilization, the tightly packaged sperm genome undergoes de novo nucleosome assembly before the two parental genomes replicate. This is followed by equal distribution of the replicated chromosomes into the two blastomeres of the 2-cell embryo. After a few round of cleavage divisions, the embryo reaches the morula stage when the first cell lineage specification commences to generate trophectoderm and inner cell mass (ICM) of the blastocyst before implanting to the uterus (Burton and Torres-Padilla, 2014).
Preimplantation development harbors two cell fate transitions. First, the highly differentiated germ cells (sperm and egg) are reprogrammed into a totipotent state characterized by having the highest level of cell fate plasticity (Rossant, 1976). The second cell fate transition takes place when the morula stage cells commit to either the trophectoderm lineage or pluripotent ICM cells (Morgan et al., 2005). Concurrent with the cell fate transitions are dramatic chromatin and transcriptional changes. One of the most notable transcriptional changes taking place during mammalian preimplantation development is zygotic genome activation (ZGA) (Svoboda et al., 2015). In mice, a major ZGA takes place in 2-cell embryos (Hamatani et al., 2004). Despite the fact that ZGA plays an essential role in preimplantation development, no transcription factor (TF) responsible for mammalian major ZGA has been identified. Consequently, the mechanism underlying mammalian ZGA is largely unknown. Recent studies have revealed several TFs, including Zelda, Pou5f1, Nanog, and SoxB1 to be important for ZGA in Drosophila and/or zebrafish (Lee et al., 2013; Liang et al., 2008). These TFs are unlikely to be involved in mammalian ZGA as the mammalian counterpart either does not exist or is not expressed at an appreciable level before ZGA. Mammalian ZGA might be mechanistically different from that of Drosophila and zebrafish as mammalian ZGA takes place early during preimplantation development, while Drosophila/zebrafish ZGA takes place at a much later developmental stage (at cell cycle 14 in Drosophila and cell cycle 10 in zebrafish) (Lee et al., 2014).
Cells at a particular state possess a defined set of cis-regulatory elements that are accessible to trans-acting factors, which underlies the chromatin regulatory network of the cell state (Bell et al., 2011; Gross and Garrard, 1988). Understanding the dynamics of chromatin accessibility during preimplantation development may provide insights into the chromatin and cell fate regulation during the process. DNase I hypersensitivity is one of the best measures of chromatin accessibility (Bell et al., 2011) and has been widely used to map functional elements, including promoters, enhancers, insulators, and locus control regions, as these regions are relatively more accessible (Gross and Garrard, 1988). Recently, DNase I treatment coupled with high-throughput DNA sequencing (DNase-seq) has allowed high-resolution genome-wide mapping of DHSs (Boyle et al., 2008). Using this strategy, millions of regulatory elements in diverse tissue and cell types have been identified in mammalian genome (Thurman et al., 2012; Vierstra et al., 2014). Despite high resolution and robustness of the DNase-seq technique, millions of cells are needed, thereby limiting its application in rare biological samples. Therefore, how the DHS landscape of the pluripotent ICM is initially established during early development is unknown.
In addition to DNase-seq, a technique called ATAC-seq (assay for transposase-accessible chromatin using sequencing) has also been developed and used in studying chromatin accessibility (Buenrostro et al., 2013). Recently, two single-cell ATAC-seq methods have been developed and used in analyzing chromatin heterogeneity among populations of cells (Buenrostro et al., 2015; Cusanovich et al., 2015). However, interpretation of the single cell ATAC-seq data relies on pre-existing chromatin accessibility maps generated using large numbers of cells. DNA loss through the multiple purification steps of traditional DNase-seq is the major reason for its low sensitivity. By minimizing DNA loss, a single-cell DNase sequencing (scDNase-seq) method has been recently developed and used in analyzing chromatin accessibility using as few as one cell (Jin et al., 2015). However, interpretation of the single-cell scDNase-seq data also requires a pre-existing DHS map. Consequently, it remains unknown whether these methods can be used for de novo identification of genome-wide chromatin accessible sites using small number of cells. In this study, we modified scDNase-seq method to achieve de novo genome-wide DHS mapping using as few as 30 cells. Using this low-input DNase-seq (liDNase-seq) method, we have generated DHS maps of mouse preimplantation embryos from 1-cell to the morula stage, revealing the dynamics of chromatin accessibility during mouse preimplantation development process. Using the DHS information, we identified key transcriptional factors involved in the establishment of the regulatory landscape early in mammalian life.
RESULTS
liDNase-seq method for de novo DNase I hypersensitive site identification
Conventional DNase-seq for de novo genome-wide DHS mapping requires millions of cells (Boyle et al., 2008; Thurman et al., 2012; Vierstra et al., 2014), preventing its application to rare samples. To overcome this limitation, we made a few modifications to the scDNase-seq method (Jin et al., 2015) to further prevent DNA loss during sequencing library construction (Figure S1A). The major improvements include: 1) reduction of the purification steps before adaptor ligation and the first amplification; 2) usage of SPRI affinity beads instead of gel purification for size selection. Employing this modified method, we generated DHS maps of 100 and 30 mouse ESCs. We found that both samples generated very similar DHS maps that were highly similar to those of the ENCODE project using 10–20 million ESCs (Figure S1B). Scatterplot analysis revealed a Pearson’s correlation coefficient of 0.93 between the data generated using 30 cells and that of the ENCODE project (Figure S1C). Moreover, the 30-cell and 100-cell data have a Pearson’s correlation coefficient of 0.98, indicating high reproducibility of the method (Figure S1C). Using Hotspot program (John et al., 2011), we identified 49,157 and 77,589 DHSs (FDR < 0.01) from 30-cell and 100-cell data, respectively. Of the 21,330 most reliable DHSs identified by the ENCODE project (FDR < 0.01 and rpm > 2), 93% and 77% were identified by the liDNase-seq using 100 and 30 cells, respectively (Figure S1D). Conversely, of the 44,304 and 29,305 DHSs (FDR < 0.01 and rpm > 1) detected using 30-and 100-cells, 86% and 97% were identified in the ENCODE project (219,515 DHSs, FDR < 0.01), respectively (Figure S1E). These results indicate that the liDNase-seq method allows mapping of de novo DHSs at high resolution using as few as 30 cells without the need of a pre-existing DHS map.
DHSs are progressively established in mouse preimplantation embryos
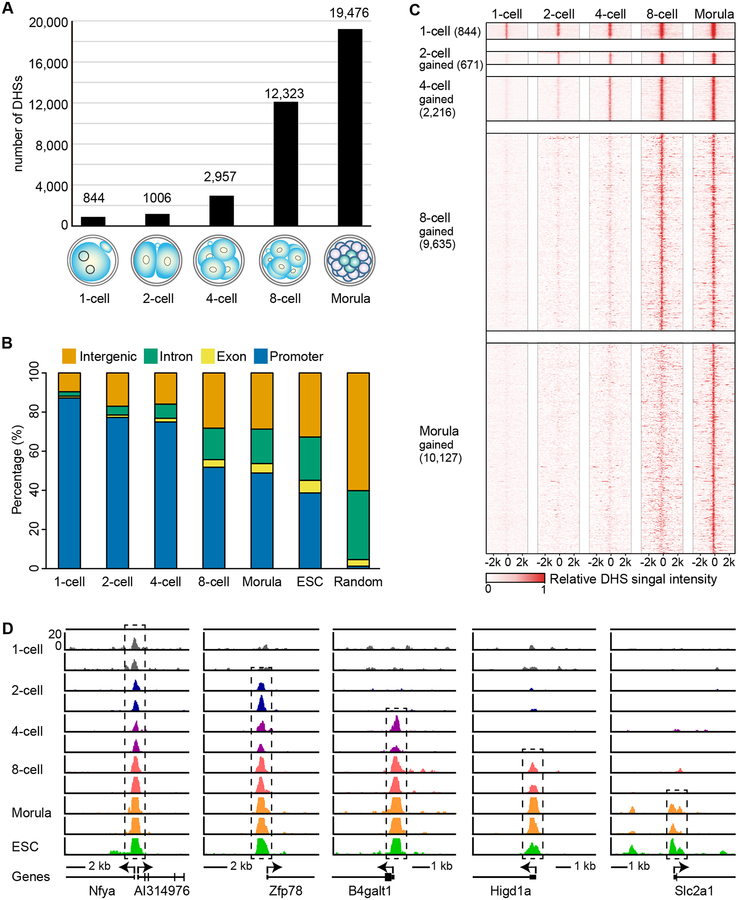
To understand how the chromatin regulatory landscape is established during mouse preimplantation development, we applied the liDNase-seq method to 1-cell, 2-cell, 4-cell, 8-cell and morula stage embryos using 100–200 blastomeres for each sample with two biological replicates in each developmental stage to ascertain generation of high-quality and reproducible data set (Table S1). Comparative analyses indicate that data from the two independent biological replicates in each developmental stage have high reproducibility (Pearson’s correlation coefficient: 0.717–0.895) (Figure S2A). Thus, we combined the data and used the common DHSs from the two replicates for subsequent analysis. As a reference of chromatin regulatory landscape of pluripotent cells, the ESC DHS map generated by liDNase-seq using 100 cells was used. Using stringent criteria (Hotspot FDR < 0.01 and RPM > 1 in both replicates), we identified 844, 1,006, 2,957, 12,323, 19,476, and 23,418 DHSs using the uniquely mapped reads at the 1-cell, 2-cell, 4-cell, 8-cell, morula stage embryos, and ESCs, respectively (Figure 1A and Table S2). The numbers of DHS increase during preimplantation development, suggesting progressive increase of chromatin accessibility at the primary structure level during this process (Figure 1A). To characterize these DHSs, we examined their genomic distribution on annotated genes defined by Refseq (Figure 1B). We found that 87.2% of the DHSs are located at promoter regions at the 1-cell embryos. This ratio decreased in 2-cell embryos (77.2%) and DHSs at distal regions, including distal exons, introns and intergenic regions, are increased (Figure 1B). While there was no big difference in the DHS distribution between 2-cell and 4-cell embryos, a drastic increase was detected in the 8-cell embryos with about half of the DHSs located in the distal regions (Figure 1B). The DHS distribution pattern of morula-stage embryos is similar to that of the 8-cell embryos, but is further shifted to distal distribution in ESCs (Figure 1B). The DHSs are highly enriched in promoter regions in embryos of all the developmental stages and ESCs (Figure 1B), further support the reliability of our DHS mapping results. These DHSs are distributed across the genome without obvious bias toward any specific chromosome or chromosome domains (Figure S2B). To confirm the liDNase-seq results, we performed qPCR analysis of the remaining intact DNA after limited DNase treatment at several DHSs identified by liDNase-seq using 4-cell and morula stage embryos. Consistent with liDNase-seq results, accessible sites had decreased amount of intact DNA remaining while the inaccessible sites were not affected (Figure S3). Taken together, our results suggest that DHS distribution is dynamically changed during preimplantation development, especially at the 2-cell and 8-cell stages when many newly formed DHSs appear in distal regions.
Figure 1. DHSs are gradually established during preimplantation development.
(A) Bar graphs illustrate the number of DHSs detected at each stage of embryo development.
(B) Graph illustrates genomic distribution of the DHSs relative to Refseq annotations. Promoters represent the regions ±0.5 kb around the TSS.
(C) Heat map showing that majority of DHSs persist once generated. Each row represents a DHS centered at its summit. The top block shows all the DHSs detected at the 1-cell stage. The following blocks show DHSs gained at the indicated stages.
(D) Genome browser view of representative DHSs gained in each developmental stage that are maintained during later stages. Black dashed box indicates DHSs.
Next, we examined the dynamics of individual DHS and found that the majority of DHSs are maintained during preimplantation development once established (Figure 1C and 1D). Using DHSs at morula stage as a reference, we found that 96.4% of 1-cell DHSs are maintained during preimplantation development. Similarly, 91.7% of 2-cell DHSs, 94.5% of 4-cell DHSs, and 75.9% of 8-cell DHSs are maintained through the morula stage. Taken together, our liDNase-seq analyses reveal that DHSs are progressively established during preimplantation development and are largely maintained once established.
DHS dynamics during mouse pre-implantation development
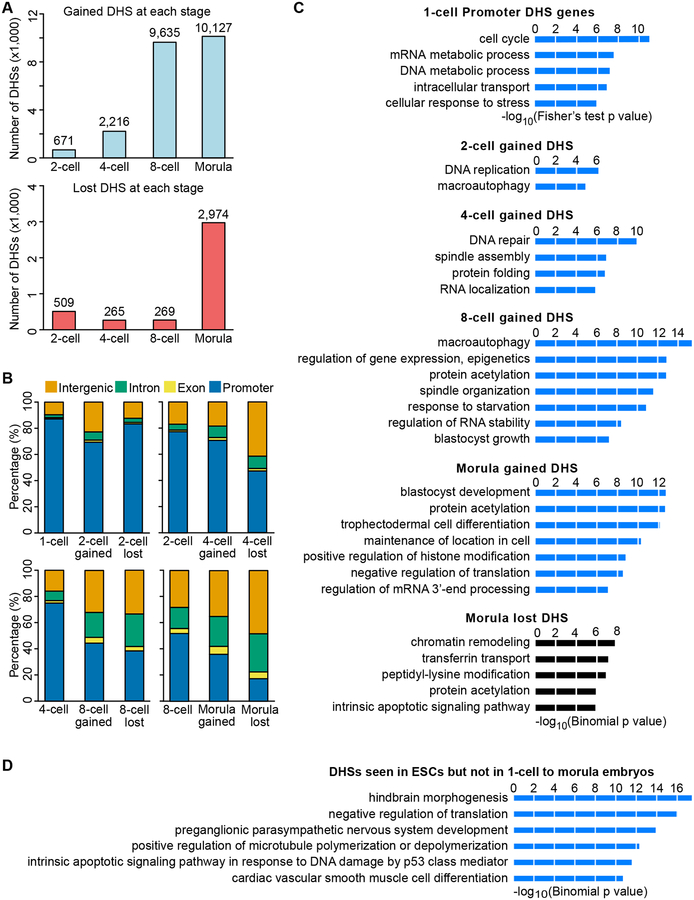
To reveal the detail of DHS dynamics during preimplantation development, we analyzed DHSs gained or lost at each stage. The number of gained DHSs gradually increases during development with the largest increase at the 8-cell and morula stage (Figure 2A). In contrast, the number of lost DHSs is relatively low until the morula stage (Figure 2A). These results are consistent with our observation that the majority of the DHSs are maintained during preimplantation development once they are established. Next, we examined the genomic location of these dynamic DHSs and found that the gained DHSs tend to locate in distal regions compared to all the DHSs at the previous stage (Figures 2B). This is also true for the lost DHSs, especially at the morula stage when more than 80% of the lost DHSs are located in distal regions (Figure 2B). These data indicate that distal DHSs are more dynamic compared to promoter DHSs.
Figure 2. Dynamics of gaining and losing DHSs during preimplantation development.
(A) Bar graph shows the number of gained (upper) and lost (bottom) DHSs at each stage.
(B) Bar graph shows genomic distribution of gained and lost DHSs compared to DHS distribution at the previous stage. Promoters represent the regions ±0.5 kb around the TSS.
(C) Gene ontology analysis of 1-cell DHSs and DHSs gained at each stage, DHSs lost at the morula stage, and DHSs seen in ESCs but not in 1-cell to morula embryos. DAVID was used for the 1-cell Promoter DHS genes, and GREAT was used for the dynamic DHSs.
(D) Gene ontology analysis of DHSs present in ESCs but not in 1-cell to morula embryos. GREAT was used for the GO analysis. The enriched terms are ranked by–log10(p-value).
To gain insight into the potential function of the dynamic DHSs, we performed gene ontology (GO) enrichment analysis on the genes related to 1-cell DHSs and dynamic DHSs at subsequent developmental stages. In 1-cell embryos, the most enriched GO term of the genes with promoter DHSs was cell cycle (Figure 1C), which is consistent with the fact that fertilization triggers the MII-stage arrested eggs to restart the cell cycle. The DHSs gained at the 2-cell stage were enriched for DNA replication and macroautophagy (Figure 2C), which is in line with active DNA replication and autophagy in early preimplantation embryos (Tsukamoto et al., 2008). DHSs gained at the 4-cell stage were enriched for DNA repair, spindle assembly, and protein homeostasis. Interestingly, DHSs gained at the 8-cell stage were enriched for gene expression regulation and protein acetylation, which is consistent with increased expression of various chromatin modifying enzymes at the 8-cell stage (Burton et al., 2013). In addition, genes involved in blastocyst growth begin to become accessible at this stage. The DHSs gained at morula stage were most enriched for blastocyst development and trophectodermal cell differentiation, suggesting that the chromatin regulatory landscape has already prepared for the first cell lineage specification at the morula stage. Unlike the gained DHSs, no GO term enrichment was found in lost DHSs except for the morula stage where chromatin remodeling and covalent modifications were enriched (Figure 2C). The DHSs presented in ESCs, but not in 1-cell to morula embryos, are associated with later differentiation and development (Figure 2D), which is consistent with the expression of genes associated with later developmental processes in serum-cultured ESCs (Marks et al., 2012). These results suggest that the chromatin regulatory landscape is linked to intracellular functions of preimplantation embryos.
DHSs can mark promoters primed for activation
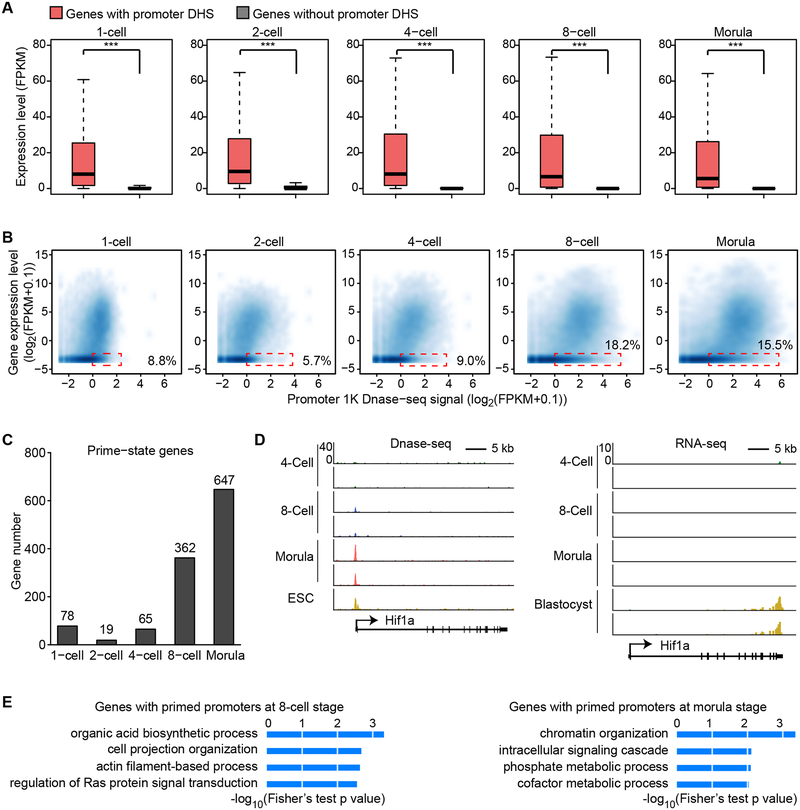
To understand the relationship between DHSs and transcription activity, we integrated the DHS information with the transcriptome of mouse preimplantation embryos (Xue et al., 2013). In general, genes with promoter DHSs are expressed at a higher level compared to those without promoter DHSs in all stages of preimplantation embryos (Figure 3A). Although a majority of promoters with detectable DNase I hypersensitivity signal are active, we noticed a significant portion (5.7–18.2%) of the silenced promoters also harbor detectable DNase I hypersensitivity signal (Figure 3B, dotted boxes). After a close examination, we found that most genes (72–85%) harboring promoter DHSs are expressed in each developmental stage (Figure S4A and S4B), while many of the silent genes with promoter DHSs start transcription at a later developmental stage (FPKM >= 1 and FC > 2), indicating that these promoters are primed for activation (Figures 3C, D and S4A, C). Gene ontology enrichment analysis revealed that genes primed at the 8-cell stage were enriched in organic acid biosynthesis, cell projection organization, and actin filament-based process (Figure 3E), while genes primed at the morula stage are enriched in chromatin organization. Thus, DHSs can mark promoters either actively transcribing or primed for subsequent activation.
Figure 3. DNase accessibility marks promoters that are active or primed for activation in later stages.
(A) Box plot showing the expression level of genes with or without promoter DHSs. Boxes and whiskers represent the 25th/75th and 2.5th/97.5th percentiles, respectively. The difference between the two groups are statistically significant by Wilcoxon rank sum test (*** denotes p < 0.001).
(B) Scatter plot showing the correlation between gene expression level and promoter DNase I hypersensitivity signal. Dashed red rectangles represent non-expressed genes (RNA-seq FPKM < 0.1) harboring promoter DHS signals (DNase-seq RPM > 1).
(C) Bar graph showing number of genes with promoter DHSs that are transcriptionally inactive at the indicated stage but are activated in the following stage (primed-state).
(D) Genome browser view of Hif1a locus as a representative example of primed-state genes. DNase-seq signal is shown on the left. RNA-seq data is shown on the right.
(E) Gene ontology analysis of primed-state genes at the 8-cell and morula stages using DAVID. The enriched terms are ranked by–log10(p-value).
See also Figure S4.
Imprinted gene promoter DHSs exhibit parental bias before the onset of allelic expression
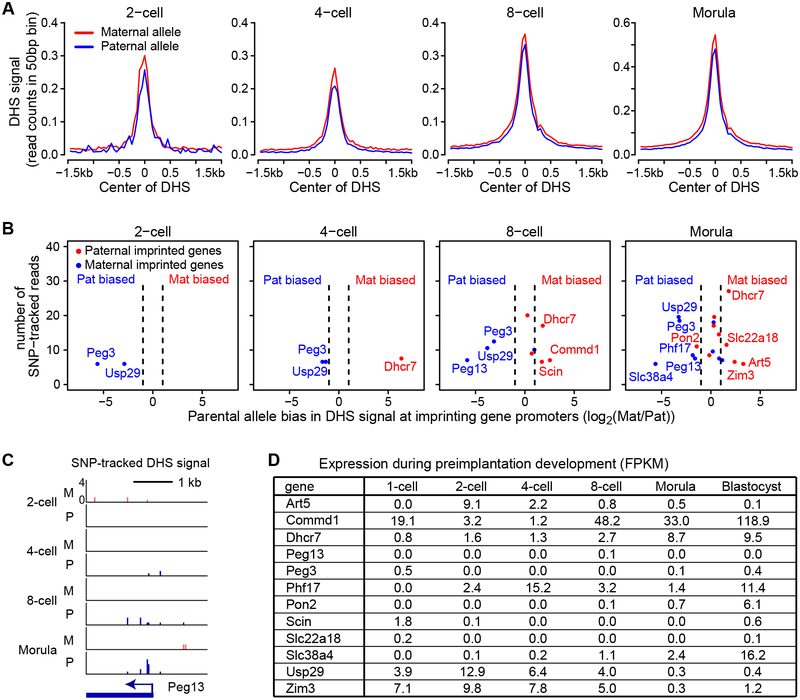
Genomic imprinting confers parental allele-specific regulation of gene expression (Tomizawa and Sasaki, 2012). In addition to differential DNA methylation, differential chromatin accessibility has also been associated with allelic expression of some imprinted genes (Coombes et al., 2003; Feil et al., 1997; Watanabe et al., 2000). However, whether differential chromatin accessibility is a cause or a consequence of mono-allelic expression and when the allelic biased chromatin accessibility is established during development remain unknown. To address these questions, we took advantage of the SNP information between maternal genome (from BDF1 oocytes) and paternal genome (from JF1 sperm). Analysis of the SNP-trackable DHSs revealed that the DHS signal intensity of the two parental alleles are generally similar from 2-cell to morula stage embryos (Figure 4A).
Figure 4. Imprinting gene promoters show allelic-biased accessibility prior to the onset of allelic expression.
(A) SNP-tracking analysis shows that the average DHS signals from the JF1 allele (paternal) and the BDF1 allele (maternal) are similar. The DHS signal was plotted at DHSs detected at each stage.
(B) Promoters of imprinted genes show allelic-biased DNase I accessibility. Shown are the imprinted genes whose promoters harbor SNP-trackable DHSs. The X-axis represents the fold difference of the DHS signal between the two alleles. The Y-axis represents the average number of SNP tracked reads of the two replicates. The dashed lines represent 2-fold difference cutoff for calculating the parental bias. Blue and red indicate paternal and maternal bias, respectively.
(C) Genome browser view of DHS signal at a maternal imprinted gene, Peg13. SNP tracked paternal and maternal DHS signals are shown. M, maternal allele; P, paternal allele.
(D) Expression of the imprinted genes listed in panel B during preimplantation development.
See also Figure S5.
Of the 150 known imprinted genes, 109 were SNP trackable at the promoters in our experimental system (Morison et al., 2005). To examine the allelic DHSs of imprinted gene promoters, we analyzed gene promoters with sufficient coverage (>=4 reads in both biological replicates within ±1.5kb TSS and >=10 reads when both replicates are combined). Using these criteria, we could track 2, 3, 9, and 18 imprinted genes at the 2-cell, 4-cell, 8-cell, and morula stage, respectively (Figure 4B). Among them, 2, 3, 6, and 10 exhibited either paternal or maternal bias at each developmental stage using 2-fold difference as a cutoff, and interestingly the majority of them exhibited weaker DHS signal in the imprinted allele (Figure 4B). For example, Peg13 and Peg3, two maternally-imprinted genes, showed paternal specific promoter DHSs; while a paternally-imprinted gene Dhcr7 showed maternal specific promoter DHSs (Figure 4C and S5). Notably, many of the allelic-biased genes, such as Peg13, Peg3 and Slc22a18, are not expressed in preimplantation embryos (Figure 4D). These results indicate that imprinting information has already been “written” in the form of chromatin accessibility during early preimplantation development before the onset of parental allelic expression.
Rapid reprogramming of paternal chromatin accessibility after fertilization
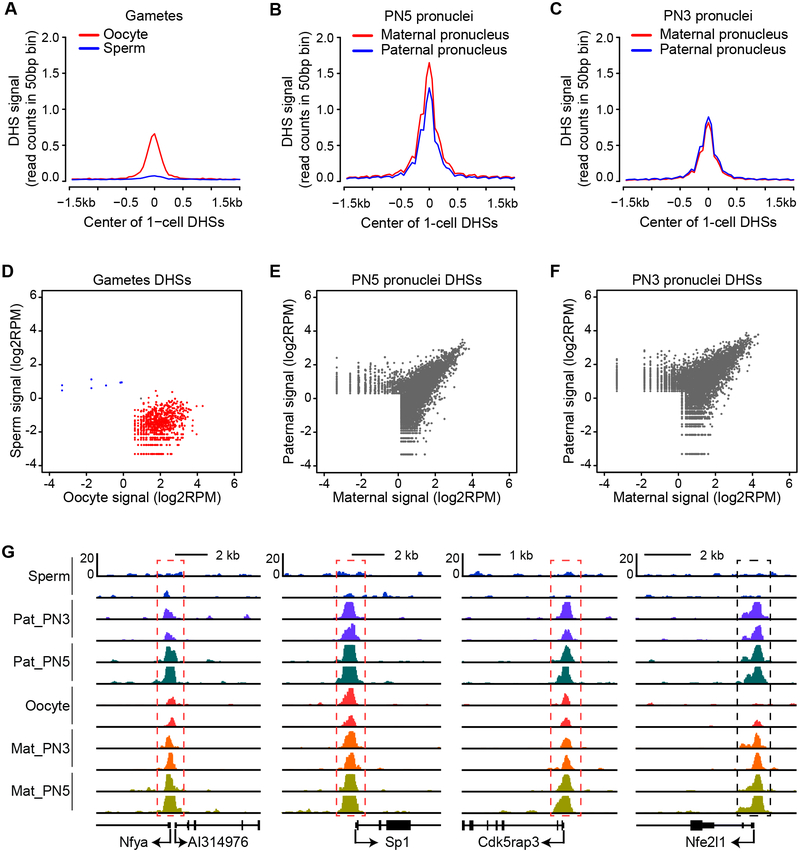
Upon fertilization, the protamine-packaged sperm genome goes through drastic remodeling resulting in de novo assembly of nucleosomes (Inoue and Zhang, 2014). Previous studies have revealed that the newly assembled paternal chromatin harbors distinct histone modifications from those of the maternal chromatin (Burton and Torres-Padilla, 2014; Lu and Zhang, 2015). Due to its tight package, sperm chromatin is largely not accessible compared with that of the oocyte chromatin (Figure 5A and 5D). To determine when the accessibility of the paternal genome becomes similar to that of the maternal genome, we physically isolated paternal and maternal pronuclei from pronucleus stage 5 (PN5) of 1-cell embryos at 12 hours after fertilization and performed liDNase-seq analysis. The DHS maps indicate that the accessibility of the two alleles is quite similar although the maternal genome is slightly more accessible (Figure 5B, 5E and 5G). To further define the time when the paternal genome becomes accessible, we isolated pronuclei from PN3 stage zygotes at 7.5 hours after fertilization, which is technically the earliest time point when the paternal pronuclei can be physically isolated. liDNase-seq analysis revealed that although there is some allelic specific DHSs, the DHS profile of the paternal genome is already very similar to that of the maternal genome (Figure 5C, 5F and 5G). These results indicate that the accessibility of the paternal genome is largely reprogrammed by PN3 stage.
Figure 5: Rapid reprogramming of the paternal chromatin accessibility upon fertilization.
(A) Oocytes and sperm show differential accessibility. The plot shows the DHSs signal detected in sperm and GV-stage oocytes at 1-cell DHSs.
(B) Paternal and maternal genomes exhibit similar accessibility at PN5 stage.
(C) Paternal and maternal genomes exhibit similar accessibility at PN3 stage. The plot shows the average DHS signals of the parental pronuclei at 1-cell DHSs.
(D) Scatter plot showing that the two gametes have very different DHS signals. DHSs detected in both gametes are combined and plotted. Blue and red dots represent DHSs differentially detected in sperm and oocytes, respectively (FC>2).
(E) Scatter plot showing DHS signals detected in the PN5 pronuclei. DHSs detected in the two parental pronuclei are combined and plotted.
(F) Scatter plot showing DHS signals detected in the PN3 pronuclei.
(G) Genome browser view of representative DHSs in gametes and pronuclei. Red dashed boxes indicate DHSs present in both pronuclei and oocytes. Black dashed box indicates DHS presented in pronuclei but not in gametes.
Oct4 contributes to the DHS increase in 8-cell embryos
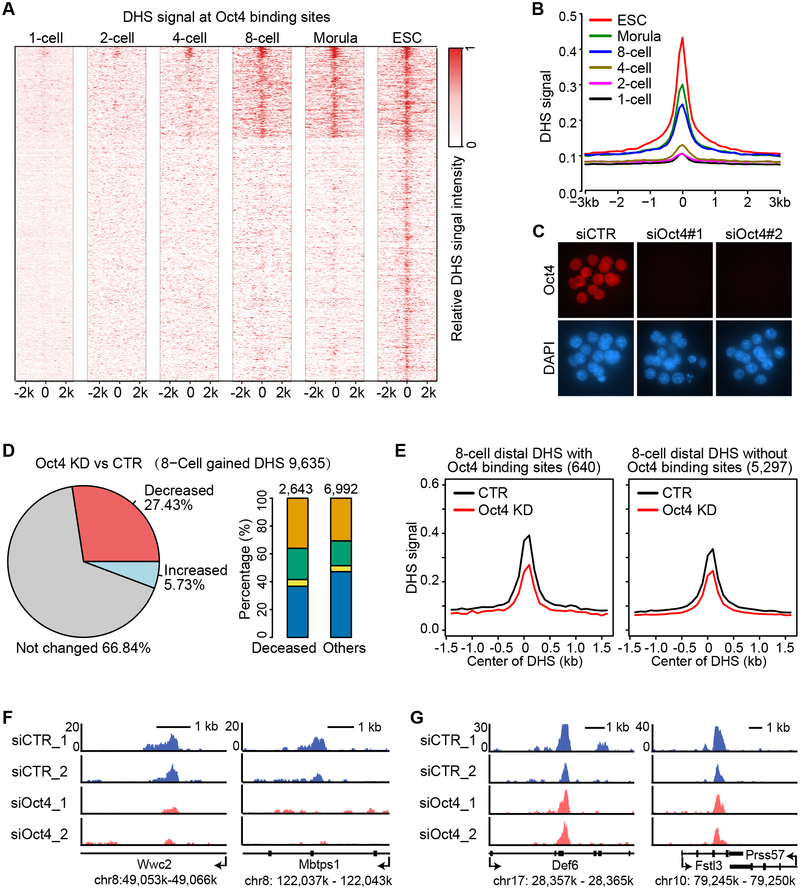
Since 8-cell embryos feature the most dramatic increase in the DHS numbers, we attempted to identify transcription factor (TF) that contribute to this increase. Previous studies have revealed several pluripotency-related TFs called pioneer factors, including Oct4, Sox2 and KLF4, which have the ability to access closed chromatin (Soufi et al., 2012). Since a large proportion (30%) of the DHSs seen in ESCs have already been established in the 8-cell embryos, we reasoned that core pluripotency TFs might contribute to the 8-cell DHS burst. Among the TFs, Oct4 is highly upregulated in the 8-cell embryos (Figure S6A). The fact that a marked increase in DHS signal in 8-cell embryos at Oct4-binding sites in ESCs (Figure 6A and 6B) indicates that Oct4 might be one of the factors involved in the DHS establishment at the 8-cell stage. To test this hypothesis, we depleted Oct4 by injecting two independent siRNAs into fertilized oocytes. After verification of successful Oct4 depletion (Figure 6C), we performed liDNase-seq analysis using the 8-cell stage Oct4 knockdown (KD) embryos of two independent siRNA-injected samples (Figure S6B). Compared to control siRNA-injected embryos (CTR), 27.4% of 8-cell-gained DHSs were lost or decreased more than 2-fold upon Oct4 KD (Figure 6D and Table S3). The decreased DHSs are significantly more enriched in distal regions than in the other regions (Fisher’s exact test p-value < 1.0 × 10−10) (Figure 6D). To examine whether the decreased distal DHSs are enriched for Oct4-binding sites, we separated the distal DHSs into two groups based on the presence or absence of the presumable Oct4 binding sites from Oct4 ChIP-seq in ESCs (Whyte et al., 2013). Comparative analysis revealed that the extent of DHS signal reduction upon Oct4 KD at sites containing Oct4 binding sites is more than that at sites without Oct4 binding sites (Figure 6E). For example, the distal DHSs with Oct4 binding sites in the coding regions of Wwc2 and Mbtps1 show decreased signal upon Oct4 KD (Figure 6F), while the distal DHSs without Oct4 binding sites in the coding regions of Def6 and Fstl3 are not affected (Figure 6G). Taken together, our data support Oct4 as one of the TFs contributing to DHSs gained at the 8-cell stage.
Figure 6. Oct4 contributes to distal DHS establishment at the 8-cell stage.
(A) Heat map showing that presumable Oct4 binding sites become accessible starting from the 8-cell stage. Each row is centered at the mid-point of an Oct4 binding site identified in ESCs.
(B) Average DHS signals at the presumable Oct4 binding sites showing a marked increase at the 8-cell stage.
(C) Representative images of morula embryos stained with anti-Oct4 (red) and DAPI (blue) showing that Oct4 protein is efficiently depleted by siRNA injection. More than 5 embryos per group were examined.
(D) A large proportion of 8-cell gained DHSs are decreased (FC > 2) upon Oct4 KD. Pie-chart (left) shows the proportion of DHSs decreased, increased, or unchanged upon Oct4 KD. Bar graph (right) shows the genomic distribution of either decreased or the rest of DHSs. Orange, green, yellow, and blue represent intergenic, intron, exon, and promoter regions, respectively.
(E) Average DHS signals at 8-cell distal DHSs with or without presumed Oct4 binding sites in control (CTR) and Oct4 KD 8-cell embryos.
(F) Representative loci containing presumed Oct4 binding sites with reduced distal DHS upon Oct4 KD.
(G) Representative loci without presumed Oct4 binding sites with distal DHS unaffected by Oct4 KD.
See also Figure S6.
Nfya promotes 2-cell promoter DHS establishment and ZGA
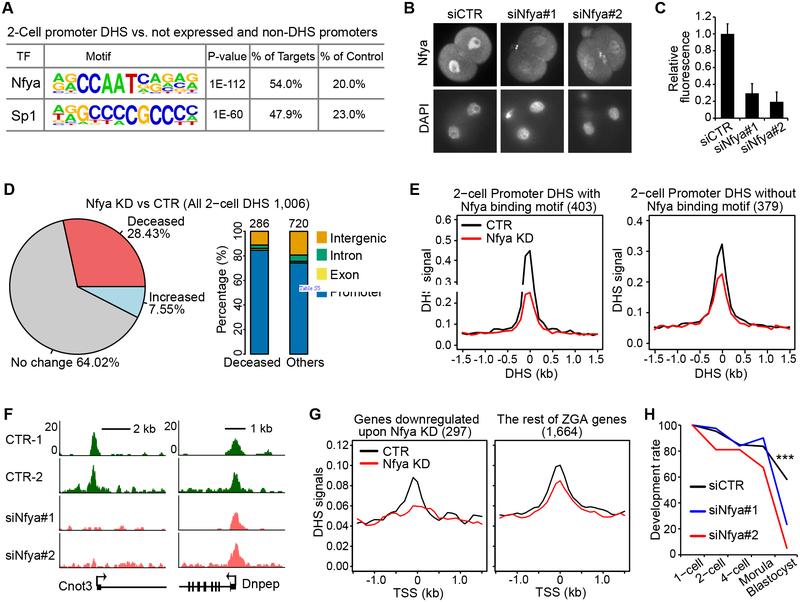
As DHSs are often occupied by TFs, our DHS mapping results may facilitate identification of TFs involved in early development. To this end, we performed TF-binding motif enrichment analysis and identified Nfya binding motif (CCAAT) as the most enriched motif in the 2-cell promoter DHSs (Figure 7A). Nfya is the sequence-specific DNA binding subunit of an Nfy complex which forms a histone-like structure binding to DNA and promotes chromatin accessibility (Nardini et al., 2013; Oldfield et al., 2014). To determine whether Nfya contributes to DHS formation in 2-cell embryos, we attempted to deplete maternally deposited Nfya protein. We injected siRNAs targeting Nfya (siNfya) into GV-stage oocytes followed by meiotic maturation and in vitro fertilization. Immunostaining with anti-Nfya antibody revealed that Nfya protein level is significantly reduced in 2-cell embryos by siRNA injection (Figure 7B and 7C). We then performed liDNase-seq on control and Nfya KD 2-cell embryos using two biological replicates. Although the Nfya KD samples exhibit a relatively large variation, we believe that the variation is likely contributed by the in vitro oocyte/embryo culturing system and micromanipulations that we used to achieve Nfya depletion because parallel control KD samples also exhibited a similar variation (Figure S7). To avoid bias, we used the average signal of the two biological replicates in our analysis. Compared to the control embryos, we found that 28.4% of 2-cell DHSs were decreased more than 2-fold, and these decreased DHSs are enriched in the promoter regions (Figure 7D and Table S4). Moreover, the reduction of the average DHS signal in Nfya motif-containing promoter DHSs is greater than that in promoter DHSs without the Nfya motif (Figure 7E). For example, DHSs are lost at the promoters of the Cnot3, Ppm1b and Osbp genes that contain Nfya binding motifs in Nfya KD embryos, while DHSs persist at the Dnpep, Fbrsl1 and Hdgfrp2 promoters that do not harbor the Nfya motif (Figure 7F and S7B).
Figure 7. Nfya is involved in shaping promoter DHSs and ZGA at 2-cell embryos.
(A) Nfya binding motif is the most enriched DNA motif at 2-cell promoter DHSs. The background (control) used are promoters of genes that are neither expressed nor accessible.
(B) Representative images of 2-cell embryos stained with anti-Nfya (top) and DAPI (bottom) showing that Nfya protein is efficiently reduced by siRNA injection.
(C) Quantification of the fluorescent signal of Nfya staining in control (siCTR) and Nfya KD 2-cell embryos. The total numbers of embryos quantified were 10, 11, and 7, respectively. Error bars indicate s.d.
(D) Pie-chart (left) shows a large proportion of 2-cell promoter DHSs is decreased (FC > 2) upon Nfya KD. Bar graph (right) shows the genomic distribution of decreased DHSs.
(E) Average DHS signals at 2-cell promoter DHSs with (left) or without (right) the Nfya binding motif. The promoter DHS peaks are centered.
(F) A representative locus of promoters with the Nfya binding motif (Cnot3 gene promoter) shows loss of DHS upon Nfya KD (left). A representative example of promoters without Nfya binding motif (Dnpep gene promoter) shows little change upon Nfya KD (right).
(G) Downregulated genes upon Nfya KD show decreased promoter DHSs. Genes activated at the 2-cell stage were divided into 2 groups; those downregulated upon Nfya KD (FC >1.5) and the rest of the genes (Figure S7C and S7D). Average promoter DHS signals were plotted for each group. The plots are centered at TSSs.
(H) Nfya is required for preimplantation development. Graph shows the percentage of embryos reaching the indicated stages. The numbers of embryos examined were 86 (siCTR), 81 (siNfya#1), and 58 (siNfya#2). *** denotes p < 0.001 (Fisher’s chi-squire test).
See also Figure S7.
Due to the broad effect of Nfya KD on 2-cell promoter DHSs, we asked whether ZGA is affected by Nfya KD. By comparing transcriptomes of 1-cell and 2-cell embryos, we defined 1,961 genes that are significantly up-regulated in 2-cell embryos (FC>5.0) as ZGA genes (Figure S7C and Table S5). Among these genes, 297 (15.1%) were downregulated while 83 (4.2%) were upregulated in Nfya KD embryos (FC>1.5, Figure S7D and Table S5). Interestingly, the downregulated genes showed large decreases in promoter DHS signal upon Nfya KD, while the rest of ZGA genes did not (Figure 7G). Consistent with the defects in DHS formation and ZGA, the majority of the Nfya KD embryos arrested at the morula stage (Figure 7H). These results demonstrate that Nfya contributes to promoter DHS formation and ZGA at the 2-cell stage.
DISCUSSION
DHS dynamics during preimplantation development
We show that DHSs are gradually established during preimplantation development to reach the highest number at morula stage immediately prior to formation of the pluripotent ICM. A previous study comparing DHS maps of human ESCs, differentiated progenitors, and terminally differentiated cells indicate that chromatin accessibility is positively correlated to pluripotency and cell plasticity (Stergachis et al., 2013). Our work extends the previous study by revealing how the chromatin regulatory landscape in pluripotent cells is established during preimplantation development. One of our most surprising results is that the chromatin of earlier stage embryos contains fewer DHSs compared to that of the later stage embryos; previous studies using DAPI staining and electro-microscopy had revealed that chromatin of zygotes have little densely packaged chromatin (Ahmed et al., 2010; Burton and Torres-Padilla, 2014). These observations may be due to the detection of chromatin structure at different levels by the two methods. Electronic or fluorescent microscopy images may mainly reflect the higher order chromatin structure, while DNase I hypersensitivity mainly reflect nucleosome and TF occupancy at the primary chromatin structure level. Thus, the relatively loose chromatin observed in zygotes by microscopes may indicate reduced higher-order chromatin structure rather than reduced density of nucleosome occupancy. Notably, not only the number of DHSs but also the width and height of each DHS peak are increased during preimplantation development (Figure 1C and 1D), suggesting that the DHSs become more stable during this process. It is intriguing to speculate that when higher-order chromatin structure is established along preimplantation development, maintenance of a locally accessible chromatin environment might be important for permissive cis-regulation. In contrast, less dense chromatin in the earlier stage of preimplantation embryos might facilitate the access of transcriptional machineries to unstable DHSs.
It is important to point out that while we observed an increase in the number of DHSs during mouse preimplantation development, the DHS numbers are not significantly changed in the Drosophila embryos from the cellular blastoderm (stage 5) to the beginning of head involution (stage 14) (Thomas et al., 2011). However, since no embryos earlier than the stage 5, when 13 nuclear divisions have already taken place, were analyzed, it remains to be determined how DHSs are established from zygote to stage 5 embryos in Drosophila. Profiling DHSs during early embryonic development in other organisms will reveal whether progressive establishment of chromatin accessibility that we observed in mice is evolutionarily conserved. The liDNase-seq technique will certainly facilitate this endeavor.
In addition to the increase in DHS numbers, the genomic distribution of accessible chromatin is also developmentally regulated. While promoters are generally more accessible than the other genomic regions during the entire preimplantation development, it appears that there are two phases of DHS increase in the distal regions. The first phase is at the 2-cell stage. While only about 10% of the 1-cell DHSs are located in the distal regulatory regions, this proportion increased to 25% in the 2-cell DHSs (Figure 1B). This observation is consistent with a previous study demonstrating that an enhancer is required for efficient transcription starting from 2-cell embryos using a reporter assay (Schultz, 2002; Wiekowski et al., 1991). Further studies are needed to determine which of the gained DHSs indeed function as enhancer elements in the 2- cell embryos. The second phase of increase in distal DHS takes place in 8-cell embryos when about 55% of the newly formed DHSs are located in the distal regions (Figure 2B). This might be caused by the fact that a large number of TFs, such as Oct4, Tead4 and Gata3 implicated in ICM or trophectoderm cell lineage specification, commence expression at the 8-cell stage (Hamatani et al., 2004). Indeed, we demonstrate that Oct4 is involved in the establishment of 8- cell distal DHSs (Figure 6D and 6E). In addition to TFs, many chromatin modifying enzymes are upregulated at the 8-cell stage (Burton et al., 2013), which might also contribute to the 8-cell DHS burst. Our study should contribute to the understanding of the TF networks in preimplantation development.
Role of Nfya in 2-cell DHS generation and zygotic genome activation
ZGA is one of the most important molecular events that take place during preimplantation development. Our DHS maps make systematic identification of TFs involved in regulation of preimplantation development possible. Through TF binding motif enrichment analysis using the 2-cell DHS map followed by functional validation, we identified Nfya as a TF involved in 2-cell DHS, particularly promoter DHS, generation. This suggests that Nfya may function as a pioneer TF in opening and binding to the local chromatin in 2-cell embryos. This idea is supported by previous observations in cultured cells that the NF-Y is required for opening local chromatin and recruiting other TFs (Oldfield et al., 2014; Sherwood et al., 2014). RNA-seq for the Nyfa KD 2-cell embryos revealed that Nfya is required for activation of ~15% of 2-cell activated genes, suggesting that Nfya is one of the TFs responsible for ZGA. Several factors may cause a possible underestimation of the role of Nfya in ZGA. First, our siRNA-mediated knockdown only achieved 70–80% of Nfya depletion (Figure 7C). The remaining Nfya may still be partially functional. Second, Nfya may function together with other TFs in ZGA. Loss function of Nfya alone may not have a significant effect on some of the ZGA genes. Nevertheless, our study has provided genetic evidence demonstrating Nfya’s involvement in mammalian ZGA.
Allele-specific chromatin accessibility and imprinting gene regulation
It is of great interest to find out when the paternal and maternal genomes become equally accessible after fertilization. To our surprise, the global levels of DHS signal in the parental chromatins have already become similar at PN3 stage (Figure 5C, F), indicating quick reprogramming of sperm chromatin after fertilization. Since many histone modifications are asymmetric in the two parental alleles in PN3 zygotes (Burton and Torres-Padilla, 2010), our results suggest that chromatin accessibility might be uncoupled from the asymmetric histone modifications.
Although only a small number of imprinted gene promoters are with SNP trackable DHS information, many of them showed allelic biased DHSs in every stage of preimplantation development (Figure 4B, 4C, and S5). This suggests that accessibility of the two parental alleles of the imprinted genes is already different in embryos. In addition, we found that some of the imprinted promoters show an allelic bias in chromatin accessibility even when not expressed from either alleles (Figure 4B–D, and S5). Therefore, our data suggest that the allelic differences in chromatin accessibility of imprinted genes are established and maintained prior to the onset of parental allelic gene expression, raising the possibility that allelic difference of chromatin accessibility might be one mechanism contributing to allelic gene expression.
Collectively, our study provides new insights into dynamics of the chromatin accessibility landscape during preimplantation development. Analyses of the datasets enable the identification of TFs involved in shaping chromatin structure and regulating transcription during mammalian preimplantation development. The described technique and the study itself have opened a new window for understanding the chromatin basis of transition from totipotent state to pluripotent state.
EXPERIMENTAL PROCEDURES
Preparation of mouse preimplantation embryos and oocytes
All animal studies were performed in accordance with guidelines of the Institutional Animal Care and Use Committee at Harvard Medical School. MII-stage oocytes were collected from 8 week-old superovulated B6D2F1/J (BDF1) females. For in vitro fertilization (IVF), MII oocytes were inseminated with activated spermatozoa obtained from the caudal epididymides of adult JF1/MsJ male mice (Jackson laboratory, 003720). The 1-cell, 2-cell, 4-cell, 8-cell, and morula stage embryos were harvested at 12, 30, 46, 60, 78 hpi, respectively. The numbers of the embryos collected for DNase-seq are 100, 100, 50, 25, and 10, respectively. For preparation of the 1-cell stage zygotes, BDF1 sperm was used. For preparation of oocytes, the ovaries were harvested from 14 day-old BDF1 female mice, punctured with a 30-gauge needle; 150 oocytes were collected for DNase-seq. Just before sampling the preimplantation embryos for DNase-seq, the polar bodies were removed to avoid genomic contamination.
Isolation of maternal and paternal pronuclei at PN3 and PN5 stages
BDF1 zygotes were obtained by in vitro fertilization. For PN3-stage pronuclei isolation, zygotes were transferred into M2 media containing 10 μM cytochalasin B (Sigma-Aldrich) at 5 hpi. Zona pellucidae were cut by a Piezo impact-driven micromanipulator and the pronuclei were isolated from the zygotes. At 7.5 hpi (PN3-stage), they were washed with 0.2% BSA/PBS and treated with DNase I. 140 PN3 pronuclei were collected for each experiment. For PN5-stage pronuclei isolation, zygotes were transferred into M2 media containing 10 μM cytochalasin B at 10 hpi. At 12 hpi (PN5-stage), they were washed and treated with DNase I. 170 PN5 pronuclei were collected for each experiment. The parental pronuclei were distinguished by (1) the distance from the second polar body and (2) the size of the pronucleus.
Low-input DNase-seq
Fresh cells were collected and resuspended in 36 μl lysis buffer (10 mM Tris-HCl, pH 7.5, 10 mM NaCl, 3 mM MgCl2, 0.1% Triton X-100). DNase I (Roche) was added to final concentration of 80 U/ml (for 1-cell embryo and oocyte samples) or 40 U/ml (for all the other samples in this study) and incubated at 37 °C for exactly 5 minutes. The reaction was stopped by adding 80 μl Stop Buffer (10 mM Tris-HCl, pH 7.5, 10 mM NaCl, 0.15% SDS, 10 mM EDTA) containing 40 μg Proteinase K (Life technologies) and 20 ng circular carrier DNA. After incubation at 50 °C for 1 hour, DNA was purified by phenol-chloroform extraction and ethanol precipitated. The DNA were end repaired and adaptor ligated. Size selection for insertion between 50 bp and 200 bp were performed after 8 cycles of first PCR amplification and again after 2nd PCR amplification. The detailed procedures can be found in supplemental experiment procedures. The libraries were sequenced on a Hiseq2500 with single-end 100 bp reads (Illumina).
For qPCR validation of DHSs, samples (32 4-cell embryos or 10 morula embryos) were treated as described in liDNase-seq with or without DNase I until resuspending the ethanol precipitated DNA in 15 μl TE. The DNA was then used for multiplexed amplification for 14 cycles in 50 μl volume with SsoFast EvaGreen Supermix (Bio-Rad) in the presence of 4% DMSO. The pre-amplified product was used for qPCR quantification with technical replicates. The relative amount of intact DNA at DHSs amplifiable after DNase I treatment was quantified against DNase I free control samples. The primers used here are included in Table S6.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Shinpei Yamaguchi for helpful discussion and Dr. Luis Tuesta for critical reading of the manuscript. This project is supported by HHMI. F.L. is supported by a Charles A. King Trust Postdoctoral Research Fellowship. Y.Z. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
The Gene Expression Omnibus (GEO) accession number for the datasets reported in this paper is GSE76642.
REFERENCES
- Ahmed K, Dehghani H, Rugg-Gunn P, Fussner E, Rossant J, and Bazett-Jones DP (2010). Global chromatin architecture reflects pluripotency and lineage commitment in the early mouse embryo. PloS one 5, e10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell O, Tiwari VK, Thoma NH, and Schubeler D (2011). Determinants and dynamics of genome accessibility. Nature reviews Genetics 12, 554–564. [DOI] [PubMed] [Google Scholar]
- Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, and Crawford GE (2008). High-resolution mapping and characterization of open chromatin across the genome. Cell 132, 311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, and Greenleaf WJ (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nature methods 10, 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, Chang HY, and Greenleaf WJ (2015). Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton A, Muller J, Tu S, Padilla-Longoria P, Guccione E, and Torres-Padilla M-E (2013). Single-Cell Profiling of Epigenetic Modifiers Identifies PRDM14 as an Inducer of Cell Fate in the Mammalian Embryo. Cell Reports 5, 687–701. [DOI] [PubMed] [Google Scholar]
- Burton A, and Torres-Padilla ME (2010). Epigenetic reprogramming and development: a unique heterochromatin organization in the preimplantation mouse embryo. Briefings in functional genomics 9, 444–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton A, and Torres-Padilla ME (2014). Chromatin dynamics in the regulation of cell fate allocation during early embryogenesis. Nature reviews Molecular cell biology 15, 723–734. [DOI] [PubMed] [Google Scholar]
- Coombes C, Arnaud P, Gordon E, Dean W, Coar EA, Williamson CM, Feil R, Peters J, and Kelsey G (2003). Epigenetic properties and identification of an imprint mark in the Nesp-Gnasxl domain of the mouse Gnas imprinted locus. Molecular and cellular biology 23, 5475–5488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusanovich DA, Daza R, Adey A, Pliner HA, Christiansen L, Gunderson KL, Steemers FJ, Trapnell C, and Shendure J (2015). Multiplex single-cell profiling of chromatin accessibility by combinatorial cellular indexing. Science 348, 910–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil R, Boyano MD, Allen ND, and Kelsey G (1997). Parental chromosome-specific chromatin conformation in the imprinted U2af1-rs1 gene in the mouse. The Journal of biological chemistry 272, 20893–20900. [DOI] [PubMed] [Google Scholar]
- Gross DS, and Garrard WT (1988). Nuclease hypersensitive sites in chromatin. Annual review of biochemistry 57, 159–197. [DOI] [PubMed] [Google Scholar]
- Hamatani T, Carter MG, Sharov AA, and Ko MS (2004). Dynamics of global gene expression changes during mouse preimplantation development. Developmental cell 6, 117–131. [DOI] [PubMed] [Google Scholar]
- Inoue A, and Zhang Y (2014). Nucleosome assembly is required for nuclear pore complex assembly in mouse zygotes. Nat Struct Mol Biol 21, 609–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins TG, and Carrell DT (2012). Dynamic alterations in the paternal epigenetic landscape following fertilization. Frontiers in genetics 3, 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W, Tang Q, Wan M, Cui K, Zhang Y, Ren G, Ni B, Sklar J, Przytycka TM, Childs R, et al. (2015). Genome-wide detection of DNase I hypersensitive sites in single cells and FFPE tissue samples. Nature 528, 142–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S, Sabo PJ, Thurman RE, Sung MH, Biddie SC, Johnson TA, Hager GL, and Stamatoyannopoulos JA (2011). Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nature genetics 43, 264–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MT, Bonneau AR, and Giraldez AJ (2014). Zygotic genome activation during the maternal-to-zygotic transition. Annual review of cell and developmental biology 30, 581–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MT, Bonneau AR, Takacs CM, Bazzini AA, DiVito KR, Fleming ES, and Giraldez AJ (2013). Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature 503, 360–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang HL, Nien CY, Liu HY, Metzstein MM, Kirov N, and Rushlow C (2008). The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature 456, 400–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F, and Zhang Y (2015). Cell totipotency: molecular features, induction, and maintenance. National Science Review 2, 217–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H, Kalkan T, Menafra R, Denissov S, Jones K, Hofemeister H, Nichols J, Kranz A, Stewart AF, Smith A, et al. (2012). The transcriptional and epigenomic foundations of ground state pluripotency. Cell 149, 590–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan HD, Santos F, Green K, Dean W, and Reik W (2005). Epigenetic reprogramming in mammals. Human molecular genetics 14 Spec No 1, R47–58. [DOI] [PubMed] [Google Scholar]
- Morison IM, Ramsay JP, and Spencer HG (2005). A census of mammalian imprinting. Trends in genetics: TIG 21, 457–465. [DOI] [PubMed] [Google Scholar]
- Nardini M, Gnesutta N, Donati G, Gatta R, Forni C, Fossati A, Vonrhein C, Moras D, Romier C, Bolognesi M, et al. (2013). Sequence-specific transcription factor NF-Y displays histone-like DNA binding and H2B-like ubiquitination. Cell 152, 132–143. [DOI] [PubMed] [Google Scholar]
- Oldfield AJ, Yang P, Conway AE, Cinghu S, Freudenberg JM, Yellaboina S, and Jothi R (2014). Histone-fold domain protein NF-Y promotes chromatin accessibility for cell type-specific master transcription factors. Mol Cell 55, 708–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossant J (1976). Postimplantation development of blastomeres isolated from 4-and 8-cell mouse eggs. Journal of embryology and experimental morphology 36, 283–290. [PubMed] [Google Scholar]
- Schultz RM (2002). The molecular foundations of the maternal to zygotic transition in the preimplantation embryo. Human reproduction update 8, 323–331. [DOI] [PubMed] [Google Scholar]
- Sherwood RI, Hashimoto T, O’Donnell CW, Lewis S, Barkal AA, van Hoff JP, Karun V, Jaakkola T, and Gifford DK (2014). Discovery of directional and nondirectional pioneer transcription factors by modeling DNase profile magnitude and shape. Nature biotechnology 32, 171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A, Donahue G, and Zaret KS (2012). Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell 151, 994–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stergachis AB, Neph S, Reynolds A, Humbert R, Miller B, Paige SL, Vernot B, Cheng JB, Thurman RE, Sandstrom R, et al. (2013). Developmental fate and cellular maturity encoded in human regulatory DNA landscapes. Cell 154, 888–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda P, Franke V, and Schultz RM (2015). Sculpting the Transcriptome During the Oocyte-to-Embryo Transition in Mouse. Current topics in developmental biology 113, 305–349. [DOI] [PubMed] [Google Scholar]
- Thomas S, Li XY, Sabo PJ, Sandstrom R, Thurman RE, Canfield TK, Giste E, Fisher W, Hammonds A, Celniker SE, et al. (2011). Dynamic reprogramming of chromatin accessibility during Drosophila embryo development. Genome biology 12, R43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, et al. (2012). The accessible chromatin landscape of the human genome. Nature 489, 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa S, and Sasaki H (2012). Genomic imprinting and its relevance to congenital disease, infertility, molar pregnancy and induced pluripotent stem cell. Journal of human genetics 57, 84–91. [DOI] [PubMed] [Google Scholar]
- Tsukamoto S, Kuma A, Murakami M, Kishi C, Yamamoto A, and Mizushima N (2008). Autophagy Is Essential for Preimplantation Development of Mouse Embryos. Science 321, 117–120. [DOI] [PubMed] [Google Scholar]
- Vierstra J, Rynes E, Sandstrom R, Zhang M, Canfield T, Hansen RS, Stehling-Sun S, Sabo PJ, Byron R, Humbert R, et al. (2014). Mouse regulatory DNA landscapes reveal global principles of cis-regulatory evolution. Science 346, 1007–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Yoshimura A, Mishima Y, Endo Y, Shiroishi T, Koide T, Sasaki H, Asakura H, and Kominami R (2000). Differential chromatin packaging of genomic imprinted regions between expressed and non-expressed alleles. Human molecular genetics 9, 3029–3035. [DOI] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, and Young RA (2013). Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiekowski M, Miranda M, and DePamphilis ML (1991). Regulation of gene expression in preimplantation mouse embryos: Effects of the zygotic clock and the first mitosis on promoter and enhancer activities. Developmental biology 147, 403–414. [DOI] [PubMed] [Google Scholar]
- Xue Z, Huang K, Cai C, Cai L, Jiang CY, Feng Y, Liu Z, Zeng Q, Cheng L, Sun YE, et al. (2013). Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature 500, 593–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.