Abstract
Parturition involves cellular signaling changes driven by the complex interplay between progesterone (P4), inflammation, and the cyclic adenosine monophosphate (cAMP) pathway. To characterize this interplay, we performed comprehensive transcriptomic studies utilizing eight treatment combinations on myometrial cell lines and tissue samples from pregnant women. We performed genome-wide RNA-sequencing on the hTERT-HM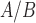 cell line treated with all combinations of P4, forskolin (FSK) (induces cAMP), and interleukin-1
cell line treated with all combinations of P4, forskolin (FSK) (induces cAMP), and interleukin-1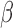 (IL-1
(IL-1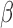 ). We then performed gene set enrichment and regulatory network analyses to identify pathways commonly, differentially, or synergistically regulated by these treatments. Finally, we used tissue similarity index (TSI) to characterize the correspondence between cell lines and tissue phenotypes. We observed that in addition to their individual anti-inflammatory effects, P4 and cAMP synergistically blocked specific inflammatory pathways/regulators including STAT3/6, CEBPA/B, and OCT1/7, but not NF
). We then performed gene set enrichment and regulatory network analyses to identify pathways commonly, differentially, or synergistically regulated by these treatments. Finally, we used tissue similarity index (TSI) to characterize the correspondence between cell lines and tissue phenotypes. We observed that in addition to their individual anti-inflammatory effects, P4 and cAMP synergistically blocked specific inflammatory pathways/regulators including STAT3/6, CEBPA/B, and OCT1/7, but not NF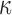 B. TSI analysis indicated that FSK + P4- and IL-1
B. TSI analysis indicated that FSK + P4- and IL-1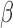 -treated cells exhibit transcriptional signatures highly similar to non-laboring and laboring term myometrium, respectively. Our results identify potential therapeutic targets to prevent preterm birth and show that the hTERT-HM
-treated cells exhibit transcriptional signatures highly similar to non-laboring and laboring term myometrium, respectively. Our results identify potential therapeutic targets to prevent preterm birth and show that the hTERT-HM cell line provides an accurate transcriptional model for term myometrial tissue.
cell line provides an accurate transcriptional model for term myometrial tissue.
Keywords: myometrium, progesterone, cyclic AMP, inflammation, gene expression, cell line, parturition
Introduction
The process and timing of parturition involve transition of the smooth muscle of the uterine wall—the myometrium—from a state of quiescence to the highly contractile laboring state. For most of pregnancy, the myometrial cells are maintained in a relaxed and compliant state and undergo hypertrophy. At parturition, hormonal factors impacting on myometrial cells and intracellular signaling pathways within myometrial cells affect the expression of genes whose products increase excitability and contractility to transition of the uterus to the laboring phenotype wherein rhythmic and forceful myometrial contractions coupled with softening and dilation of the cervix expel the fetus and placenta. The hormonal and intracellular signaling pathways and gene expression networks involved in myometrial cell activation are not clearly understood. This study examined the inter-play of three signaling pathways known to affect the contractile phenotype of the human myometrium during pregnancy: (i) the steroid hormone progesterone (P4) acting via the P4 receptors (PRs); (ii) the pro-labor/pro-inflammatory cytokine interleukin-1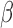 (IL-1
(IL-1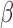 ); and (iii) the 3′,5′-cyclic adenosine monophosphate (cAMP) intracellular signaling cascade.
); and (iii) the 3′,5′-cyclic adenosine monophosphate (cAMP) intracellular signaling cascade.
In all viviparous species examined to date, P4/PR signaling is essential for the establishment and maintenance of pregnancy and P4/PR withdrawal triggers labor. P4/PR signaling maintains pregnancy, in part by inhibiting the response of myometrial cells to pro-labor/pro-inflammatory stimuli (Hardy et al., 2006; Tan et al., 2012; Chen et al., 2014; Amini et al., 2016; Peters et al., 2016). This is important because parturition is an inflammatory process that is triggered by pro-inflammatory stimuli inducing tissue-level inflammation within the myometrium, decidua, and cervix. Desensitization of uterine cells, and especially myometrial cells, to pro-inflammatory stimuli by P4/PR is therefore considered to be a principal mechanism for the P4 block to labor (Mesiano et al., 2002; Mesiano et al., 2011).
The 3′,5′-cAMP intracellular signaling cascade promotes uterine quiescence by inhibiting myometrial cell contractility (Chanrachakul et al., 2004; Price and Bernal, 2001). Typically, cAMP functions as a second messenger, acting through protein kinase A (PKA), a kinase that phosphorylates a diverse set of downstream targets to produce various cellular responses (Skalhegg and Taskén, 1997). PKA suppress myometrial cell contractility by inhibiting myosin light chain kinase activity and decreasing the intracellular concentration of free Ca2+ (Sanborn et al., 2005; Sanborn, 2007; Morgado et al., 2012; Oldenburger et al., 2012; Yulia et al., 2016). Studies in airway smooth muscle cells have shown that cAMP signaling inhibits response to pro-inflammatory stimuli by suppressing the activation of nuclear factor kappa-light-chain-enhancer of activated B cells (NF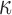 B) and specific mitogen-active protein kinases (MAPKs) (Billington et al., 2013). Thus, hormones whose actions are mediated by the cAMP/PKA pathway may decrease myometrial cell contractility by inhibiting the contractile apparatus and response to pro-inflammatory stimuli.
B) and specific mitogen-active protein kinases (MAPKs) (Billington et al., 2013). Thus, hormones whose actions are mediated by the cAMP/PKA pathway may decrease myometrial cell contractility by inhibiting the contractile apparatus and response to pro-inflammatory stimuli.
We previously found that cAMP/PKA and P4/PR signaling in myometrial cells exerts a potent and synergistic and anti-inflammatory effect (Amini et al., 2019). Those findings were consistent with other studies showing that treatment of myometrial cells derived from term uterus with forskolin (FSK), a natural bicyclic diterpene that increases intracellular cAMP by stimulating adenylyl cyclase, increased the anti-inflammatory actions of P4 (Chen et al., 2014). Thus, anti-inflammatory synergism between cAMP and P4/PR signaling in myometrial cells appears to be a major mechanism by which P4/PR promotes uterine quiescence for most of pregnancy.
In this study, we examined the relationships between P4/PR, cAMP, and IL-1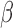 signaling in an immortalized human myometrial cell line. RNA sequencing followed by global, unbiased gene expression analysis was performed to determine the effects of each signaling pathway alone and in combination. Pathway enrichment analysis and regulatory network inference was used to fully characterize P4/PR, cAMP, and IL-1
signaling in an immortalized human myometrial cell line. RNA sequencing followed by global, unbiased gene expression analysis was performed to determine the effects of each signaling pathway alone and in combination. Pathway enrichment analysis and regulatory network inference was used to fully characterize P4/PR, cAMP, and IL-1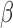 signaling. The same techniques were then used to investigate the anti-inflammatory actions and synergistic effects of P4/PR and cAMP on responsiveness to IL-1
signaling. The same techniques were then used to investigate the anti-inflammatory actions and synergistic effects of P4/PR and cAMP on responsiveness to IL-1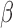 . Results show that the main action of P4/PR in myometrial cells is anti-inflammatory, working through the JUN transcription factor (TF), and suppressing pathways involved in muscle tissue development, leukocyte migration, and MAPK signaling. cAMP also exhibited anti-inflammatory effects and, interestingly, promoted pathways involved in cell hypertrophy and smooth muscle contraction. P4/PR and cAMP acted synergistically to down-regulate genes involved in cytokine production, the STAT signaling cascade, and smooth muscle cell proliferation and up-regulate a set of genes controlled by TFs involved in developmental processes and suppression of inflammation. Our data suggest the P4/PR, cAMP, and IL-1
. Results show that the main action of P4/PR in myometrial cells is anti-inflammatory, working through the JUN transcription factor (TF), and suppressing pathways involved in muscle tissue development, leukocyte migration, and MAPK signaling. cAMP also exhibited anti-inflammatory effects and, interestingly, promoted pathways involved in cell hypertrophy and smooth muscle contraction. P4/PR and cAMP acted synergistically to down-regulate genes involved in cytokine production, the STAT signaling cascade, and smooth muscle cell proliferation and up-regulate a set of genes controlled by TFs involved in developmental processes and suppression of inflammation. Our data suggest the P4/PR, cAMP, and IL-1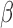 signaling share a subset of regulatory machinery, allowing the observed inter-pathway regulation. Finally, by comparing cell line gene expression in response to various stimuli to transcriptome data from pre-labor and laboring term myometrium, we found that hTERT-HM
signaling share a subset of regulatory machinery, allowing the observed inter-pathway regulation. Finally, by comparing cell line gene expression in response to various stimuli to transcriptome data from pre-labor and laboring term myometrium, we found that hTERT-HM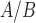 cells exposed to FSK and P4 were an accurate approximation for non-laboring myometrium and IL-1
cells exposed to FSK and P4 were an accurate approximation for non-laboring myometrium and IL-1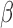 -treated cells best resembled laboring myometrium. Taken together, the data support the concept that labor is induced by inflammatory stimuli in conjunction with the loss of P4/PR, and possibly cAMP signaling.
-treated cells best resembled laboring myometrium. Taken together, the data support the concept that labor is induced by inflammatory stimuli in conjunction with the loss of P4/PR, and possibly cAMP signaling.
Methods
Data
Data were obtained from cell culture studies performed in a telomerase immortalized human myometrial cell line, hTERT-HMA/B, expressing stable and independently inducible PR-A and PR-B as described in Amini et al. (2019). Briefly, cells were made to express equal levels of PR-A and PR-B, and exposed to P4 (100 nM), IL-1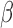 (1 ng/mL), and FSK (to stimulate cyclic AMP, 10 μM) alone and in all combinations (Fig. 1A). All experimental conditions were replicted in triplicate. Stimulation of these pathways was confirmed in Amini et al. (2019) by quantitative reverse transcription polymerase chain reaction (qRT-PCR) showing IL-1
(1 ng/mL), and FSK (to stimulate cyclic AMP, 10 μM) alone and in all combinations (Fig. 1A). All experimental conditions were replicted in triplicate. Stimulation of these pathways was confirmed in Amini et al. (2019) by quantitative reverse transcription polymerase chain reaction (qRT-PCR) showing IL-1 induced expression of IL-8 and that this induced expression of IL-8 was inhibited by P4 in the presence of PR and by FSK. Figure 1b and c depicts the comparisons of various conditions performed throughout this work.
induced expression of IL-8 and that this induced expression of IL-8 was inhibited by P4 in the presence of PR and by FSK. Figure 1b and c depicts the comparisons of various conditions performed throughout this work.
Figure 1.
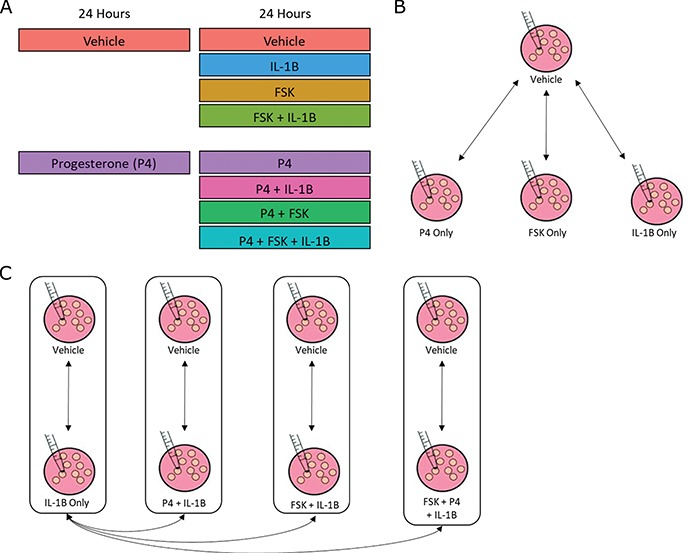
Study design and conceptual illustration of the framework for data analysis.
A. The human myometrial hTERT-HM cell line, expressing PRs, A and B, was treated with P4, FSK (induces cAMP), and IL-1
cell line, expressing PRs, A and B, was treated with P4, FSK (induces cAMP), and IL-1 individually and in all combinations of these three stimuli. Treatment was performed for 24 hours to establish steady-state cellular transcription. RNA sequencing was performed in triplicate on each condition, resulting in expression measurements for at total of 24 samples. B. Comparisons performed to characterize the signaling carried out by P4, cAMP, and IL-1
individually and in all combinations of these three stimuli. Treatment was performed for 24 hours to establish steady-state cellular transcription. RNA sequencing was performed in triplicate on each condition, resulting in expression measurements for at total of 24 samples. B. Comparisons performed to characterize the signaling carried out by P4, cAMP, and IL-1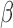 individually. The results of these comparisons are shown in Figs 2–5. C. Comparisons performed to examine the interplay between P4 and cAMP in the context of their anti-inflammatory effects. Samples treated by IL-1
individually. The results of these comparisons are shown in Figs 2–5. C. Comparisons performed to examine the interplay between P4 and cAMP in the context of their anti-inflammatory effects. Samples treated by IL-1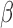 and different combinations of P4 and FSK were first compared to vehicle. Subsequently, the genes, TFs, and processes identified in these comparisons were compared to each other to understand how P4 and cAMP work together or individually to suppress inflammatory processes. The results of these analyses are shown in Figs 6 and 7.
and different combinations of P4 and FSK were first compared to vehicle. Subsequently, the genes, TFs, and processes identified in these comparisons were compared to each other to understand how P4 and cAMP work together or individually to suppress inflammatory processes. The results of these analyses are shown in Figs 6 and 7.
For tissue expression comparison, the raw data files were downloaded from the Gene Expression Omnibus, series accession number GSE50599 (ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE50599) (Chan et al., 2014).
RNA-seq analysis
Cell line and tissue data were processed separately using identical pipelines. FASTQ data files were analyzed using FASTQC (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc). Transcripts were aligned using  against the University of California Santa Cruz hg19 reference transcriptome (Trapnell et al., 2009). Transcripts were assembled and RNA abundance was estimated using
against the University of California Santa Cruz hg19 reference transcriptome (Trapnell et al., 2009). Transcripts were assembled and RNA abundance was estimated using  on the Bam files resulting from
on the Bam files resulting from 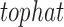 alignment (Trapnell et al., 2010). A single run of the
alignment (Trapnell et al., 2010). A single run of the 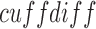 command was run on all samples for each dataset. The resulting output of the cell line data was then further analyzed using
command was run on all samples for each dataset. The resulting output of the cell line data was then further analyzed using 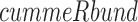 in
in 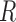 (version 3.5.1) (Goff et al., 2012). Quality control was examined using multiple built-in
(version 3.5.1) (Goff et al., 2012). Quality control was examined using multiple built-in  functions including dispersion, expression box plots, dendograms, and multi-dimensional scaling to verify there were no outlying samples. Differential expression was performed for all sample group comparisons, and a gene was considered to be significant if it had fold change >1.5 and adjusted P-value <0.05 (adjusted using the Benjamini Hochberg procedure for multiple hypothesis test correction). The cell line gene expression matrix was obtained using the repFpkmMatrix() function. The tissue gene expression matrix was built using the genes.read_group_tracking
functions including dispersion, expression box plots, dendograms, and multi-dimensional scaling to verify there were no outlying samples. Differential expression was performed for all sample group comparisons, and a gene was considered to be significant if it had fold change >1.5 and adjusted P-value <0.05 (adjusted using the Benjamini Hochberg procedure for multiple hypothesis test correction). The cell line gene expression matrix was obtained using the repFpkmMatrix() function. The tissue gene expression matrix was built using the genes.read_group_tracking  output file. Genes having very low or zero expression in three or more samples were removed prior to downstream analysis.
output file. Genes having very low or zero expression in three or more samples were removed prior to downstream analysis.
Gene set enrichment
Gene Set Variation Analysis (GSVA) is a publically available tool that is implemented in  to identify the gene sets that are collectively differentially expressed between two different experimental conditions (Hänzelmann et al., 2013). GSVA is a non-parametric method applied to the full expression matrix as opposed to working with a list of differential expression scores for each gene. It transforms the gene by sample matrix to a gene set by sample matrix. This allows for use of expression information about every gene (including non-significant ones) in a given gene set to assess the collective differential expression of the set and calculates an enrichment fold change for each gene set. We used GSVA to identify pathways and TFs that were enriched in differentially expressed genes/targets between treated cell line conditions and the control condition.
to identify the gene sets that are collectively differentially expressed between two different experimental conditions (Hänzelmann et al., 2013). GSVA is a non-parametric method applied to the full expression matrix as opposed to working with a list of differential expression scores for each gene. It transforms the gene by sample matrix to a gene set by sample matrix. This allows for use of expression information about every gene (including non-significant ones) in a given gene set to assess the collective differential expression of the set and calculates an enrichment fold change for each gene set. We used GSVA to identify pathways and TFs that were enriched in differentially expressed genes/targets between treated cell line conditions and the control condition.
The input expression matrix derived from the cell line RNA-seq contained 16 500 genes and 24 samples. The Gene Ontology Biological Processes (GO BPs) and Transcription Factor-Targets gene set collections were obtained from the Molecular Signatures Database (MSigDB) via the Broad Institute website (Subramanian et al., 2005). Gene sets for the FOS and JUN TFs were missing from the Transcription Factor-Targets gene sets. Considering these are of much interest and relevance in inflammatory signaling, and therefore parturition research, sets for FOS and JUN were created using the curated TF network employed in the regulatory network enrichment analysis (RNEA) method (described below; includes data from the TRED, TRRUST, TFACTS, and OREGANNO databases). These gene sets were loaded into  using the
using the 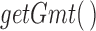 function. The expression matrix was
function. The expression matrix was 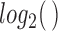 transformed prior to enrichment calculation. IQR filtering was also performed to remove genes with low variance of expression across sample groups. GSVA was applied for each condition against the control sample group. A standard restriction on gene set size, 5 to 500 genes, was applied in the enrichment analysis. Finally, the
transformed prior to enrichment calculation. IQR filtering was also performed to remove genes with low variance of expression across sample groups. GSVA was applied for each condition against the control sample group. A standard restriction on gene set size, 5 to 500 genes, was applied in the enrichment analysis. Finally, the 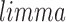 package was used to fit models to each gene set and significance was calculated by applying an empirical Bayes statistical test (Smyth, 2005). P-values were adjusted using the Benjamini Hochberg procedure.
package was used to fit models to each gene set and significance was calculated by applying an empirical Bayes statistical test (Smyth, 2005). P-values were adjusted using the Benjamini Hochberg procedure.
In order to gain insights into how targets of TFs enriched in multiple conditions were regulated, we calculated the overlap of their significantly differentially expressed targets in cells treated with P4, FSK, and IL-1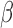 individually as compared to control. For each significant target of a given TF in a given comparison, we identified their direction of fold change (up- or down-regulated) using the original differential expression calculation. The sets of up- and down-regulated targets for each TF were compared across different cell line treatment groups.
individually as compared to control. For each significant target of a given TF in a given comparison, we identified their direction of fold change (up- or down-regulated) using the original differential expression calculation. The sets of up- and down-regulated targets for each TF were compared across different cell line treatment groups.
Regulatory network analysis
RNEA was employed to infer the genomic signaling used by each stimuli, as well as compare the networks of different stimuli (Chouvardas et al., 2016). RNEA uses fold changes and adjusted P-values from differential expression to build a regulatory network specific to an experimental condition, drawing from curated TF target data. Differentially expressed TFs and their targets are included in the network and non-significant TFs along a pathway that have a significant gene both directly upstream and downstream are also included in the final output network. Differential expression statistics were obtained for all genes using the 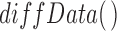 function in the
function in the 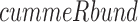 package. These were analyzed using the
package. These were analyzed using the 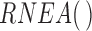 function in
function in 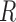 , requiring a fold change >1.5 and an adjusted P-value <0.05 for significance. Networks were visualized using Cytoscape, with fold change values mapped to the included nodes via the same input data that was used for RNEA (Shannon et al., 2003). The networks in the text figures are subnetworks of the full RNEA result achieved using stricter input arguments for fold change and P-value for the sake of visualization. Full networks are available as supplementary Cytoscape files (.cys; view using Cytoscape version 3.3.0 or later; Supplementary Files I–IV). The hierarchical layout was applied for all networks.
, requiring a fold change >1.5 and an adjusted P-value <0.05 for significance. Networks were visualized using Cytoscape, with fold change values mapped to the included nodes via the same input data that was used for RNEA (Shannon et al., 2003). The networks in the text figures are subnetworks of the full RNEA result achieved using stricter input arguments for fold change and P-value for the sake of visualization. Full networks are available as supplementary Cytoscape files (.cys; view using Cytoscape version 3.3.0 or later; Supplementary Files I–IV). The hierarchical layout was applied for all networks.
Comparison of myometrial cell line and tissue
In order to compare the tissue and cell line studies, their gene expression matrices were first 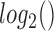 normalized, reduced to the set of genes measured in both studies, and concatenated such that the resulting expression matrix contained 34 samples (10 tissue, 24 cell line) and 15 388 genes (
normalized, reduced to the set of genes measured in both studies, and concatenated such that the resulting expression matrix contained 34 samples (10 tissue, 24 cell line) and 15 388 genes (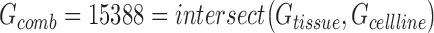 where
where 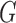 is the set of genes in a gene expression matrix). The
is the set of genes in a gene expression matrix). The 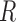 function
function 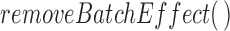 (part of the
(part of the 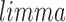 package) was then used to further normalize the expression matrix to make the two datasets comparable. The data was split back into the tissue and cell line matrices, which were then individually row (gene) centered using the function
package) was then used to further normalize the expression matrix to make the two datasets comparable. The data was split back into the tissue and cell line matrices, which were then individually row (gene) centered using the function 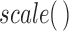 with the input arguments center = TRUE and scale = FALSE.
with the input arguments center = TRUE and scale = FALSE.
To quantify the similarity in expression between the tissue and cell line samples, a method introduced by Sandberg and Ernberg (2005) called tissue similarity index (TSI) was implemented in 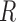 . Briefly, singular value decomposition (SVD) was applied to the tissue gene expression matrix after batch removal and row-centering. A phenotype centroid was calculated for the laboring and non-laboring sample groups by averaging the projected tissue samples using all singular vectors. The cell line samples were then projected into the SVD space, again using all singular vectors derived from the tissue data. Correlations between the projected cell line samples and the phenotype centroids were calculated and median and standard deviation values over treatment conditions were obtained. Visualization of sample clustering was performed using the first two singular vectors.
. Briefly, singular value decomposition (SVD) was applied to the tissue gene expression matrix after batch removal and row-centering. A phenotype centroid was calculated for the laboring and non-laboring sample groups by averaging the projected tissue samples using all singular vectors. The cell line samples were then projected into the SVD space, again using all singular vectors derived from the tissue data. Correlations between the projected cell line samples and the phenotype centroids were calculated and median and standard deviation values over treatment conditions were obtained. Visualization of sample clustering was performed using the first two singular vectors.
TSI was computed in the same manner on gene expression matrices reduced to sets of genes after batch removal and before row centering. Genes differentially expressed with a 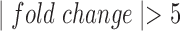 and
and 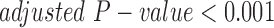 in P4, FSK, and IL-1
in P4, FSK, and IL-1 treated cells versus CTRL and in labor versus non-labor tissue were identified for TSI calculation. These gene sets included 145 and 151 genes, respectively. Key cell line genes were manually selected by identifying highly differentially expressed cell line genes across multiple individual stimuli versus control comparisons that additionally occurred in the visualized RNEA subnetworks. A heatmap of these key genes was generated using the
treated cells versus CTRL and in labor versus non-labor tissue were identified for TSI calculation. These gene sets included 145 and 151 genes, respectively. Key cell line genes were manually selected by identifying highly differentially expressed cell line genes across multiple individual stimuli versus control comparisons that additionally occurred in the visualized RNEA subnetworks. A heatmap of these key genes was generated using the  package in
package in  . Median expression values were obtained for each sample type using the same input matrix as was used with TSI. Rows (genes) were clustered by correlation using the complete clustering method. Sample clustering based on TSI results was visualized using
. Median expression values were obtained for each sample type using the same input matrix as was used with TSI. Rows (genes) were clustered by correlation using the complete clustering method. Sample clustering based on TSI results was visualized using  . Tissue samples, phenotype centroids, and cell line samples were projected onto the first two singular vectors from the SVD calculation using the key cell line genes.
. Tissue samples, phenotype centroids, and cell line samples were projected onto the first two singular vectors from the SVD calculation using the key cell line genes.
Results
Investigation of differential gene expression (comparisons in Fig. 1B) indicates that FSK/cAMP induced a major transcriptional response, altering the expression of 2210 genes, while P4 and IL-1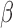 affect fewer genes but a near equal number to each other (932 and 970, respectively) between them (Fig. 2A). cAMP and IL-1
affect fewer genes but a near equal number to each other (932 and 970, respectively) between them (Fig. 2A). cAMP and IL-1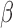 share a substantial number of up-regulated genes (Fig. 2B), cAMP and P4 share a large set of down-regulated genes (Fig. 2C), and P4 down-regulates a number of genes that are up-regulated by IL-1
share a substantial number of up-regulated genes (Fig. 2B), cAMP and P4 share a large set of down-regulated genes (Fig. 2C), and P4 down-regulates a number of genes that are up-regulated by IL-1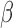 (Fig. 2D). These results suggest that there is inter-gene-regulation between these processes and that P4 is anti-inflammatory while cAMP has a more complicated relationship in that it likely both up and down-regulates different sets of inflammatory-related genes.
(Fig. 2D). These results suggest that there is inter-gene-regulation between these processes and that P4 is anti-inflammatory while cAMP has a more complicated relationship in that it likely both up and down-regulates different sets of inflammatory-related genes.
Figure 2.
Comparison of individual genes that are significantly differentially expressed in response to each individual stimulus. Venn diagrams depicting the overlap of genes that are A. significantly differentially expressed, B. significantly up-regulated, C. significantly down-regulated in hTERT-HM cell lines treated by IL-1
cell lines treated by IL-1 (green circle), P4 (blue circle), and FSK (red circle) as compared to control samples. D. Venn diagram depicting the overlap of genes that are significantly up-regulated in IL-1
(green circle), P4 (blue circle), and FSK (red circle) as compared to control samples. D. Venn diagram depicting the overlap of genes that are significantly up-regulated in IL-1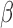 -treated cells as compared to control and significantly down-regulated in P4 and/or FSK treated cells as compared to control. All comparisons are single stimulus versus control and significance is defined as having|fold change| > 1.5 and P-value < 0.05 (after adjustment for multiple hypothesis testing using the Benjamini Hochberg procedure).
-treated cells as compared to control and significantly down-regulated in P4 and/or FSK treated cells as compared to control. All comparisons are single stimulus versus control and significance is defined as having|fold change| > 1.5 and P-value < 0.05 (after adjustment for multiple hypothesis testing using the Benjamini Hochberg procedure).
P4 signaling
Treatment of the hTERT-HM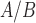 cells made to equally express PR-A and PR-B with P4 significantly (
cells made to equally express PR-A and PR-B with P4 significantly (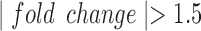 and
and 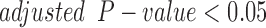 ) altered the expression of 932 genes compared to untreated cells (CTRL). P4 induced more down-regulation (555 genes) than up-regulation (377 genes). At the pathway level, P4 down-regulated the GO BPs muscle tissue development, STAT cascade, chemokine signaling, and leukocyte migration while the TNF signaling pathway and cellular hormone metabolic process were up-regulated (Fig. 3A). As for TFs, P4 up-regulated downstream targets of AR and ISRE and down-regulated the targets of STAT6, RSRFC4 (or MEF2A), CEBP, and HNF1 (Fig. 3B). These results clearly indicate anti-inflammatory actions by P4. Investigation of the inferred transcriptional regulatory machinery underlying the P4-induced gene expression changes (i.e. the TF pathways that P4 uses to alter expression of certain genes) indicates that JUN, a part of the AP-1 complex, is an important component (Fig. 4A). Node colors in this network indicate strong anti-inflammatory action by P4 (the targets MMP1, CCL3, IL-8, and IL-1
) altered the expression of 932 genes compared to untreated cells (CTRL). P4 induced more down-regulation (555 genes) than up-regulation (377 genes). At the pathway level, P4 down-regulated the GO BPs muscle tissue development, STAT cascade, chemokine signaling, and leukocyte migration while the TNF signaling pathway and cellular hormone metabolic process were up-regulated (Fig. 3A). As for TFs, P4 up-regulated downstream targets of AR and ISRE and down-regulated the targets of STAT6, RSRFC4 (or MEF2A), CEBP, and HNF1 (Fig. 3B). These results clearly indicate anti-inflammatory actions by P4. Investigation of the inferred transcriptional regulatory machinery underlying the P4-induced gene expression changes (i.e. the TF pathways that P4 uses to alter expression of certain genes) indicates that JUN, a part of the AP-1 complex, is an important component (Fig. 4A). Node colors in this network indicate strong anti-inflammatory action by P4 (the targets MMP1, CCL3, IL-8, and IL-1 are all down-regulated). Other genes of interest include KLF4, which is up-regulated by P4 and has multiple down-stream targets in our network. KLF4 has been previously linked to P4 and appears to limit cell proliferation [27]. Furthermore, PLAT plays a role in tissue remodeling and cell migration and PTX3 is involved in the regulation of innate resistance to pathogens and inflammatory reactions [10, 21]. Together these results indicate that the majority of signaling by P4 in the hTERT-HM
are all down-regulated). Other genes of interest include KLF4, which is up-regulated by P4 and has multiple down-stream targets in our network. KLF4 has been previously linked to P4 and appears to limit cell proliferation [27]. Furthermore, PLAT plays a role in tissue remodeling and cell migration and PTX3 is involved in the regulation of innate resistance to pathogens and inflammatory reactions [10, 21]. Together these results indicate that the majority of signaling by P4 in the hTERT-HM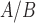 cells is to produce a strong anti-inflammatory action, likely utilizing JUN to achieve this effect.
cells is to produce a strong anti-inflammatory action, likely utilizing JUN to achieve this effect.
Figure 3.
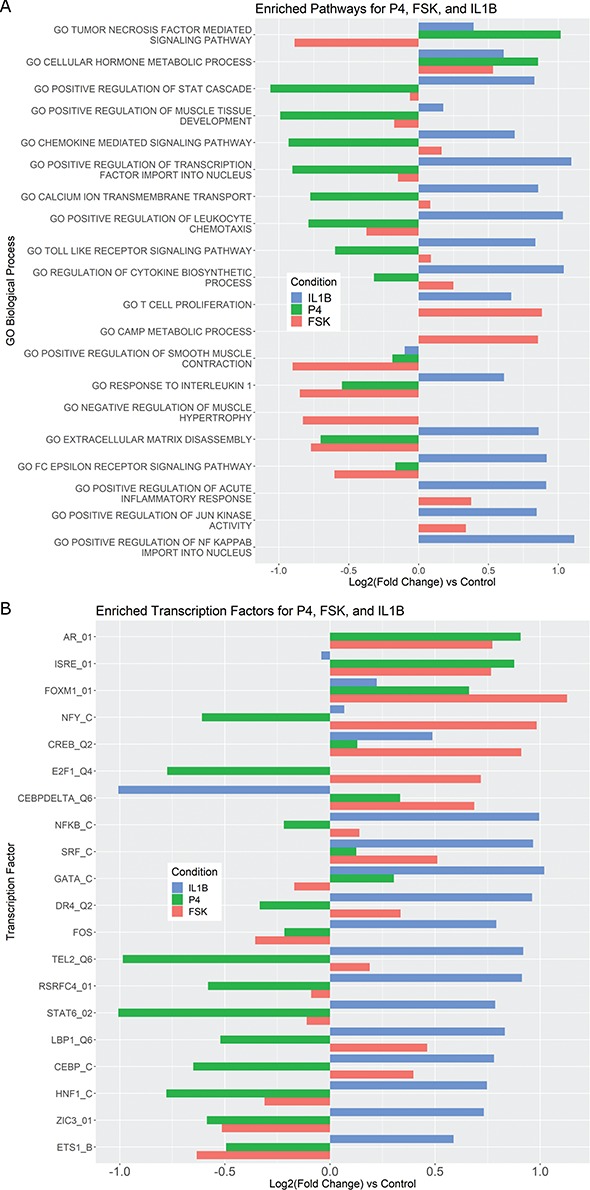
Pathways and TFs that exhibit significant enrichment in differential expression in response to treatment by P4, FSK, and IL-1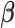 individually. The gene set collections for GO BPs and TF-targets were obtained from the MSigDB. Gene sets significant (|fold change| > 1.5 and adjusted P-value < 0.05) in any single stimulus condition versus control were identified using GSVA. For a subset of these significant A. pathways and B. TFs, the magnitude and direction of the collective fold change of the genes in the corresponding gene set, as quantified by GSVA, is shown using three bars colored according to the stimulus. See Supplementary Tables V and VI for enrichment fold change values for all GOBP and TF terms, respectively, significant in at least one single stimulus condition compared to control.
individually. The gene set collections for GO BPs and TF-targets were obtained from the MSigDB. Gene sets significant (|fold change| > 1.5 and adjusted P-value < 0.05) in any single stimulus condition versus control were identified using GSVA. For a subset of these significant A. pathways and B. TFs, the magnitude and direction of the collective fold change of the genes in the corresponding gene set, as quantified by GSVA, is shown using three bars colored according to the stimulus. See Supplementary Tables V and VI for enrichment fold change values for all GOBP and TF terms, respectively, significant in at least one single stimulus condition compared to control.
Figure 4.
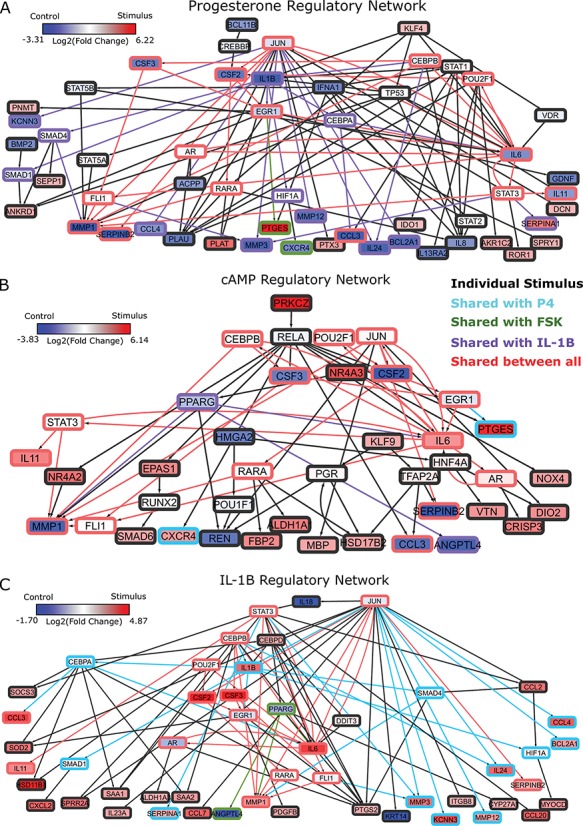
Regulatory networks representing the effects of each individual stimulus. RNEA was performed and significant subnetworks were identified in samples treated by A. P4, B. FSK (cAMP), and C. IL-1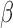 based on gene expression alterations compared to control. Nodes (genes and TFs) are colored based on
based on gene expression alterations compared to control. Nodes (genes and TFs) are colored based on 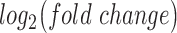 . Edges (regulatory interactions) and node outlines are colored based on occurrence in the three shown networks.
. Edges (regulatory interactions) and node outlines are colored based on occurrence in the three shown networks.  cutoffs used for significance in the RNEA function were 1.5 for P4 and IL-1
cutoffs used for significance in the RNEA function were 1.5 for P4 and IL-1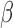 and 1.75 for FSK. Visualization was performed using Cytoscape. See Supplementary Files I–III for full RNEA networks for P4, FSK, and IL-1
and 1.75 for FSK. Visualization was performed using Cytoscape. See Supplementary Files I–III for full RNEA networks for P4, FSK, and IL-1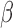 , respectively.
, respectively.
cAMP signaling
Treatment of cells with FSK significantly altered the expression of 2210 genes, the majority of which were up-regulated compared to CTRL (1345 up and 865 down). At the pathway level, partial anti-inflammatory action is observed for cAMP, with response to IL-1 and FC epsilon receptor signaling significantly down-regulated (Fig. 3A). cAMP also induces changes associated with myometrial cell quiescence; genes involved in smooth muscle contraction were down-regulated and those involved in muscle hypertrophy were up-regulated. At the TF level, more anti-inflammatory action was apparent through the down-regulation of FOS and ETS1 targets (Fig. 3B). cAMP up-regulated targets of CREB, expectantly, as well as targets of FOXM1 and NFY, which are involved in regulating cell proliferation. The inferred regulatory network based on these gene expression changes exhibits multiple points of interest (Fig. 4B). Many down-stream genes were regulated by RELA, a sub-unit of the well-known inflammatory TF NF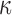 B. RELA was in turn down-stream of the most strongly up-regulated gene after FSK treatment, protein kinase C zeta. There were some similarities with the P4 network, including down-regulation of inflammatory genes MMP1 and CCL3. However, known inflammatory genes such as IL6, IL11, and CXCR4 were up-regulated by cAMP, indicating more diverse and complex action within the cAMP signaling pathway in myometrial cells. Further illustrating this point, some genes in this network correspond to the establishment of the quiescent phenotype, which was also observed from the pathway enrichment (e.g. NR4A2 and NR4A3 participate in cardiac hypertrophy and VTN promotes cell adhesion).
B. RELA was in turn down-stream of the most strongly up-regulated gene after FSK treatment, protein kinase C zeta. There were some similarities with the P4 network, including down-regulation of inflammatory genes MMP1 and CCL3. However, known inflammatory genes such as IL6, IL11, and CXCR4 were up-regulated by cAMP, indicating more diverse and complex action within the cAMP signaling pathway in myometrial cells. Further illustrating this point, some genes in this network correspond to the establishment of the quiescent phenotype, which was also observed from the pathway enrichment (e.g. NR4A2 and NR4A3 participate in cardiac hypertrophy and VTN promotes cell adhesion).
IL-1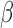 signaling
signaling
IL-1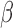 significantly altered the expression of 970 genes in hTERT-HM
significantly altered the expression of 970 genes in hTERT-HM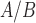 cells, most of which were up-regulated compared to CTRL (586 up and 384 down). At the pathway level, the gene expression alterations corresponded to a classic inflammatory response signal, with pathways related to chemokines, leukocytes, cytokines, and inflammatory molecule signaling (IL-1, FC epsilon receptor, JUN, and NF
cells, most of which were up-regulated compared to CTRL (586 up and 384 down). At the pathway level, the gene expression alterations corresponded to a classic inflammatory response signal, with pathways related to chemokines, leukocytes, cytokines, and inflammatory molecule signaling (IL-1, FC epsilon receptor, JUN, and NF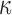 B) all up-regulated (Fig. 3A). Similarly for TFs, IL-1
B) all up-regulated (Fig. 3A). Similarly for TFs, IL-1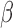 up-regulated targets of GATA, NF
up-regulated targets of GATA, NF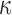 B, SRF, and STAT6 (Fig. 3B). This pattern was also seen in the IL-1
B, SRF, and STAT6 (Fig. 3B). This pattern was also seen in the IL-1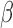 RNEA network with the majority of genes exhibiting increased expression (Fig. 4C). Genes such as CCL3, PTGS2, and IL11 are down-stream of CSF2/3 and CEBPA/B/D TFs as well as JUN, which regulates expression of many genes altered by IL-1
RNEA network with the majority of genes exhibiting increased expression (Fig. 4C). Genes such as CCL3, PTGS2, and IL11 are down-stream of CSF2/3 and CEBPA/B/D TFs as well as JUN, which regulates expression of many genes altered by IL-1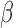 treatment, suggesting an important role for this protein in regulating inflammation.
treatment, suggesting an important role for this protein in regulating inflammation.
Additional insights from single stimulus comparisons
For the most enriched TFs in Fig. 3B, we examined the co-regulation of their target genes by P4, cAMP, and IL-1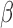 . A substantial portion of the targets of these top TFs were regulated in specific directions (up- or down-regulated) by at least two of the individual stimuli. For example, the TF ISRE was up-regulated by P4 and cAMP (Fig. 3B). Of the 247 known targets of ISRE, 34 were up-regulated by cAMP, 15 by P4, and 7 of these were up-regulated by both (Fig. 5A). cAMP and P4 also altered the same genes of TFs whose activity was down-regulated, such as ETS1 (Fig. 5K). cAMP and IL-1
. A substantial portion of the targets of these top TFs were regulated in specific directions (up- or down-regulated) by at least two of the individual stimuli. For example, the TF ISRE was up-regulated by P4 and cAMP (Fig. 3B). Of the 247 known targets of ISRE, 34 were up-regulated by cAMP, 15 by P4, and 7 of these were up-regulated by both (Fig. 5A). cAMP and P4 also altered the same genes of TFs whose activity was down-regulated, such as ETS1 (Fig. 5K). cAMP and IL-1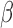 activated many of the same targets of NF
activated many of the same targets of NF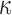 B, whereas STAT6 exhibited that the anti-inflammatory actions of P4 are effective on some of the same targets that were up-regulated by IL-1
B, whereas STAT6 exhibited that the anti-inflammatory actions of P4 are effective on some of the same targets that were up-regulated by IL-1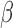 (Fig. 5I). These results indicate that P4, cAMP, and IL-1
(Fig. 5I). These results indicate that P4, cAMP, and IL-1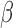 individually regulate the expression of the same genes, with P4 and cAMP typically having the opposite effect as IL-1
individually regulate the expression of the same genes, with P4 and cAMP typically having the opposite effect as IL-1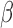 on expression. (See Supplementary Tables I–IV
for shared target information for all TFs in Fig. 3B and Supplementary Tables V–VI
for all GO BPs and TFs significantly enriched in at least one of the comparisons between single stimulus treated cells and control.)
on expression. (See Supplementary Tables I–IV
for shared target information for all TFs in Fig. 3B and Supplementary Tables V–VI
for all GO BPs and TFs significantly enriched in at least one of the comparisons between single stimulus treated cells and control.)
Figure 5.
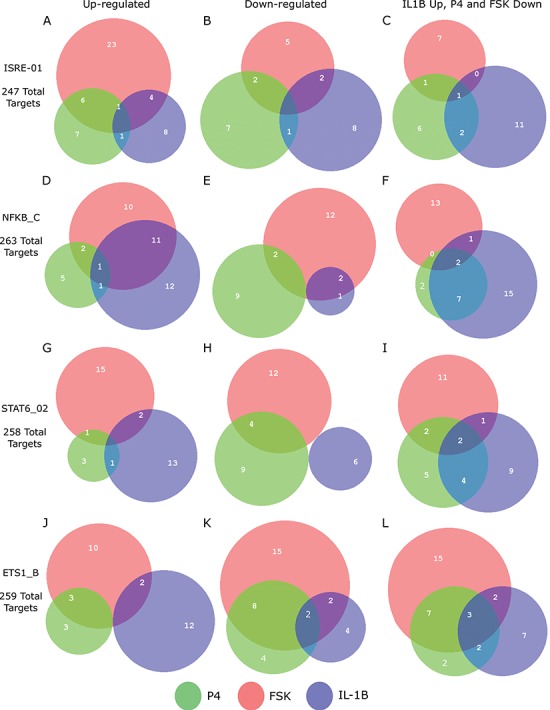
Regulation of shared targets for top enriched TFs in response to individual stimuli. Comparison of how each stimulus alters expression of targets of selected TFs showing strong enrichment from GSVA calculation in at least one of the conditions progesteron (P4), forskoin (FSK), and IL-1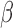 compared to control. For each of these TFs, the number of known targets that are up- or down-regulated by each stimulus and the overlap between these targets are shown, along with the total number of known targets for each TF. See Supplementary Tables I–IV
for statistics on regulation of shared targets for all TFs appearing in Fig. 3B.
compared to control. For each of these TFs, the number of known targets that are up- or down-regulated by each stimulus and the overlap between these targets are shown, along with the total number of known targets for each TF. See Supplementary Tables I–IV
for statistics on regulation of shared targets for all TFs appearing in Fig. 3B.
The regulatory network analysis also indicates clear similarities between the three individual stimuli. All three networks shared 15 nodes and 25 edges, indicated in red (edges and node borders in Fig. 4). A majority of the shared genes included upstream TFs, suggesting that each stimulus induced its cellular response through common regulatory machinery. The anti-inflammatory action by P4 is clearly represented in the shared sub-network between P4 and IL-1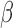 (purple in Fig. 4A and blue in Fig. 4C). These two networks shared 12 nodes and 20 edges (in addition to the nodes and edges shared by all three conditions), where most genes were up-regulated in the IL-1
(purple in Fig. 4A and blue in Fig. 4C). These two networks shared 12 nodes and 20 edges (in addition to the nodes and edges shared by all three conditions), where most genes were up-regulated in the IL-1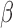 network and down-regulated in the P4 network. This indicates that the main action of P4 is anti-inflammatory, directly down-regulating many of the same genes that IL-1
network and down-regulated in the P4 network. This indicates that the main action of P4 is anti-inflammatory, directly down-regulating many of the same genes that IL-1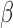 stimulates. Although P4 and cAMP had anti-inflammatory action and equally acted on many of the same TF targets, little was shared between top-enriched P4 and cAMP regulatory networks that is not also shared with the IL-1
stimulates. Although P4 and cAMP had anti-inflammatory action and equally acted on many of the same TF targets, little was shared between top-enriched P4 and cAMP regulatory networks that is not also shared with the IL-1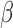 network. Taken together, it is clear that P4, cAMP, and IL-1
network. Taken together, it is clear that P4, cAMP, and IL-1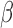 are not only regulating the same genes, but also using similar transcriptional pathways to achieve the regulation.
are not only regulating the same genes, but also using similar transcriptional pathways to achieve the regulation.
Anti-inflammatory actions of P4 and cAMP
Terms significantly enriched by IL-1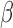 treatment were examined to better characterize the anti-inflammatory effects of P4 and cAMP in the presence of inflammation (performing comparisons in Fig. 1C; see Supplementary Tables VII–VIII
for all GO BP and TF terms enriched in IL-1β versus Ctrl and their enrichment in all IL-1β treatment conditions). Certain pathways up- (chromatin access and organization) and down-regulated (inflammatory-related pathways including glucose starvation and type 1 interferon) by IL-1
treatment were examined to better characterize the anti-inflammatory effects of P4 and cAMP in the presence of inflammation (performing comparisons in Fig. 1C; see Supplementary Tables VII–VIII
for all GO BP and TF terms enriched in IL-1β versus Ctrl and their enrichment in all IL-1β treatment conditions). Certain pathways up- (chromatin access and organization) and down-regulated (inflammatory-related pathways including glucose starvation and type 1 interferon) by IL-1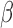 were observed to be unaffected by the addition of P4 and/or cAMP (Fig. 6A, top panel). The addition of P4 and cAMP individually decreased specific IL-1
were observed to be unaffected by the addition of P4 and/or cAMP (Fig. 6A, top panel). The addition of P4 and cAMP individually decreased specific IL-1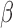 responsive pathways. P4 strongly inhibited GO terms related to cell dynamics. cAMP again exhibits a dual action of promoting relaxation (reversal of pathways relating to muscle and calcium transport) and inhibition of inflammation (IL-1 signaling and monocyte chemotaxis). Interestingly, multiple pathways significantly up-regulated by IL-1
responsive pathways. P4 strongly inhibited GO terms related to cell dynamics. cAMP again exhibits a dual action of promoting relaxation (reversal of pathways relating to muscle and calcium transport) and inhibition of inflammation (IL-1 signaling and monocyte chemotaxis). Interestingly, multiple pathways significantly up-regulated by IL-1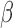 were inhibited only when both cAMP and P4 were present, consisting of processes related to cytokine production and signaling cascades involved in inflammatory response (STAT, IL12, and LPS). As for TFs, we found that the activity of NF
were inhibited only when both cAMP and P4 were present, consisting of processes related to cytokine production and signaling cascades involved in inflammatory response (STAT, IL12, and LPS). As for TFs, we found that the activity of NF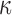 B, in the context of inflammatory signaling, was not inhibited by P4 and/or cAMP (Fig. 6B, top panel). The extent to which P4 inhibited inflammation at the level of TF activity was clear, almost completely reversing the expression of downstream targets. For most of these cases, cAMP had relatively little or no effect suggesting anti-inflammatory action specific to P4 signaling. cAMP again seemed to oppose down-regulation by IL-1
B, in the context of inflammatory signaling, was not inhibited by P4 and/or cAMP (Fig. 6B, top panel). The extent to which P4 inhibited inflammation at the level of TF activity was clear, almost completely reversing the expression of downstream targets. For most of these cases, cAMP had relatively little or no effect suggesting anti-inflammatory action specific to P4 signaling. cAMP again seemed to oppose down-regulation by IL-1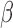 , particularly on TFs related to relaxatory actions (GR and PR). Similar to results seen at the pathway level, there were cases where the combination of cAMP and P4 had a stronger effect on IL-1
, particularly on TFs related to relaxatory actions (GR and PR). Similar to results seen at the pathway level, there were cases where the combination of cAMP and P4 had a stronger effect on IL-1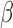 -altered gene sets than the two individually. Inhibited inflammatory TFs included GATA, SRF (part of the MAPK pathway), and STAT6, which was also a synergistically inhibited pathway.
-altered gene sets than the two individually. Inhibited inflammatory TFs included GATA, SRF (part of the MAPK pathway), and STAT6, which was also a synergistically inhibited pathway.
Figure 6.
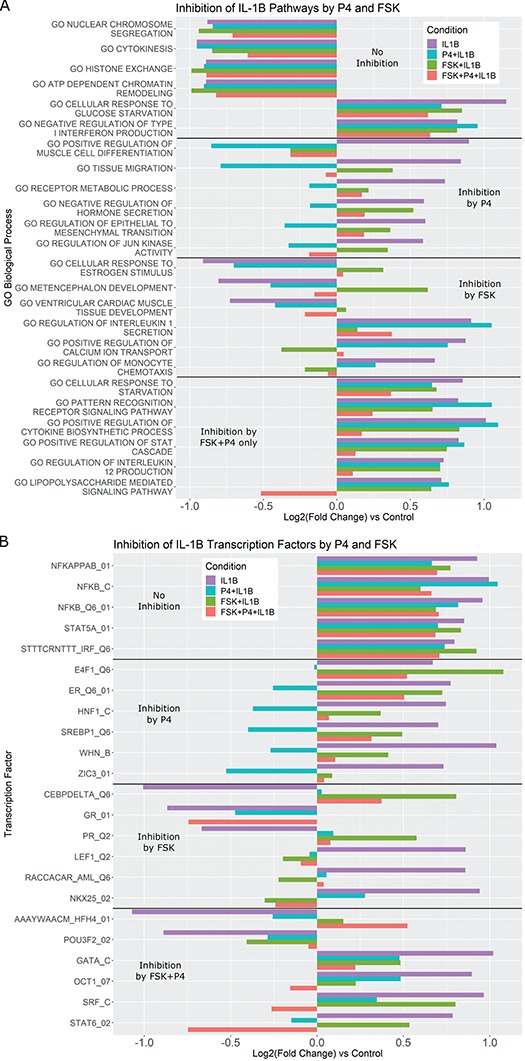
Pathways that are activated/inhibited by IL-1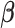 are inhibited/activated by different combinations of P4 and FSK. The gene set collections of GO BPs and Transcription Factor-Targets were obtained from the MSigDB. Gene sets significantly altered (|fold change| > 1.5 and adjusted P-value < 0.05) from control to IL-1
are inhibited/activated by different combinations of P4 and FSK. The gene set collections of GO BPs and Transcription Factor-Targets were obtained from the MSigDB. Gene sets significantly altered (|fold change| > 1.5 and adjusted P-value < 0.05) from control to IL-1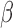 were identified using GSVA. These gene sets were then re-evaluated by GSVA for the conditions of P4 + IL-1
were identified using GSVA. These gene sets were then re-evaluated by GSVA for the conditions of P4 + IL-1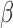 , FSK + IL-1
, FSK + IL-1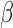 , and FSK + P4 + IL-1
, and FSK + P4 + IL-1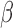 compared with control. These gene sets were classified into four categories according to their regulation pattern: (i) no inhibition (activated by IL-1
compared with control. These gene sets were classified into four categories according to their regulation pattern: (i) no inhibition (activated by IL-1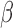 and not inhibited by the addition of P4 and/or FSK), (ii) inhibition by P4 (activated by IL-1
and not inhibited by the addition of P4 and/or FSK), (ii) inhibition by P4 (activated by IL-1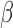 , but this activation was reversed by the addition of P4), (iii) inhibition by FSK (activated/inhibited by IL-1
, but this activation was reversed by the addition of P4), (iii) inhibition by FSK (activated/inhibited by IL-1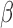 , but this activation/inhibition was reversed by the addition of FSK), (iv) inhibition by P4 + FSK (activated/inhibited by IL-1
, but this activation/inhibition was reversed by the addition of FSK), (iv) inhibition by P4 + FSK (activated/inhibited by IL-1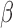 , this regulation was not reversed by the addition of P4 or FSK individually, but it was reversed by the addition of P4 and FSK together). For each of these four categories, the fold change of sample gene sets (representing A. GO BPs, B. Transcription Factor-Targets) in response to different combinations of stimuli are shown. See Supplementary Tables VII and VIII
for enrichment fold change values for all GO BP and TF terms, respectively, significant in IL-1
, this regulation was not reversed by the addition of P4 or FSK individually, but it was reversed by the addition of P4 and FSK together). For each of these four categories, the fold change of sample gene sets (representing A. GO BPs, B. Transcription Factor-Targets) in response to different combinations of stimuli are shown. See Supplementary Tables VII and VIII
for enrichment fold change values for all GO BP and TF terms, respectively, significant in IL-1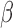 compared to control. These include enrichment fold changes for all IL-1
compared to control. These include enrichment fold changes for all IL-1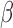 conditions.
conditions.
We further investigated the anti-inflammatory effects through annotation of the previously calculated IL-1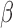 RNEA network (Fig. 4C). Figure 7A depicts how genes from the IL-1
RNEA network (Fig. 4C). Figure 7A depicts how genes from the IL-1 RNEA network are influenced, in terms of their expression, by addition of cAMP and/or P4. A sizable portion of the network includes IL-1
RNEA network are influenced, in terms of their expression, by addition of cAMP and/or P4. A sizable portion of the network includes IL-1 genes that were inhibited in the presence of P4 alone, cAMP alone, and cAMP + P4 (indicated in yellow). Note that all of these genes were up-regulated by IL-1
genes that were inhibited in the presence of P4 alone, cAMP alone, and cAMP + P4 (indicated in yellow). Note that all of these genes were up-regulated by IL-1 (boxed nodes), indicating the pro-inflammatory genes and regulatory interactions that P4 and cAMP inhibit or inactivate individually and in combination. There were additional cases in blue, green, and orange where only one of P4 or cAMP was needed to achieve inhibition. Genes in red indicate those where inhibition was only achieved in the presence of P4 and cAMP, representing the GSVA-observed synergistic inhibition. Finally, this network also highlights genes and regulatory interactions that are unaffected by P4 and/or cAMP (in purple), meaning they remain active in the presence of anti-inflammatory stimuli.
(boxed nodes), indicating the pro-inflammatory genes and regulatory interactions that P4 and cAMP inhibit or inactivate individually and in combination. There were additional cases in blue, green, and orange where only one of P4 or cAMP was needed to achieve inhibition. Genes in red indicate those where inhibition was only achieved in the presence of P4 and cAMP, representing the GSVA-observed synergistic inhibition. Finally, this network also highlights genes and regulatory interactions that are unaffected by P4 and/or cAMP (in purple), meaning they remain active in the presence of anti-inflammatory stimuli.
Figure 7.
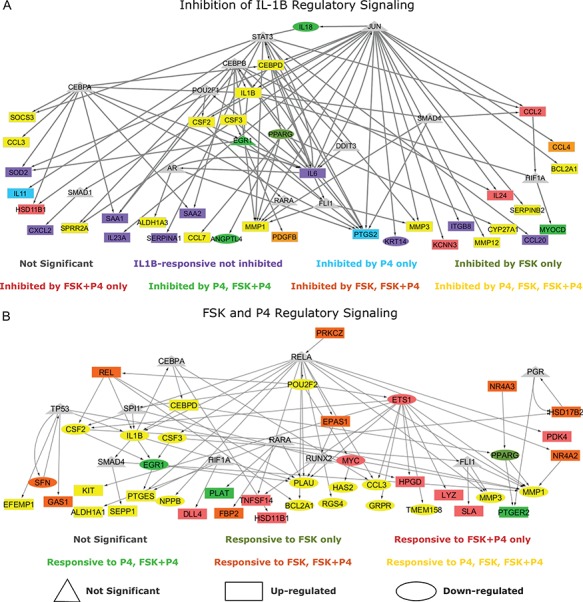
Regulation interplay of genomic signaling by IL-1
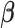 , FSK, and P4.
A. Inhibition of the regulatory network induced by IL-1
, FSK, and P4.
A. Inhibition of the regulatory network induced by IL-1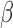 (same network and inputs used for Fig. 5C) by different combinations of P4 and FSK (e.g. an orange node is significant in IL-1
(same network and inputs used for Fig. 5C) by different combinations of P4 and FSK (e.g. an orange node is significant in IL-1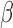 versus Ctrl and remains significant in the same direction in P4 + IL-1
versus Ctrl and remains significant in the same direction in P4 + IL-1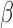 versus Ctrl but becomes insignificant or significant in the opposite direction in FSK + IL-1
versus Ctrl but becomes insignificant or significant in the opposite direction in FSK + IL-1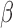 versus Ctrl and FSK + P4 + IL-1
versus Ctrl and FSK + P4 + IL-1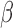 versus Ctrl). B. Regulatory patterns of P4 and FSK. The network is obtained by performing RNEA for FSK + P4 versus Ctrl with a
versus Ctrl). B. Regulatory patterns of P4 and FSK. The network is obtained by performing RNEA for FSK + P4 versus Ctrl with a 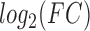 cutoff of 2.4 and P-value cutoff of 0.05. Node colors are based on P4 versus Ctrl, FSK versus Ctrl, and FSK + P4 versus Ctrl expression comparisons. Node shape indicates differential expression of each node, including direction of regulation. See Supplementary File IV
for full RNEA networks for FSK + P4.
cutoff of 2.4 and P-value cutoff of 0.05. Node colors are based on P4 versus Ctrl, FSK versus Ctrl, and FSK + P4 versus Ctrl expression comparisons. Node shape indicates differential expression of each node, including direction of regulation. See Supplementary File IV
for full RNEA networks for FSK + P4.
Synergy between P4 and cAMP
Much of our previous results indicate a synergistic effect between cAMP and P4. Such a relationship is important in the context of establishing and maintaining pregnancy. cAMP and P4 each had anti-inflammatory actions and relaxatory actions (Figs 2–5), but these did not fully overlap, meaning that they work similarly in certain biological contexts and complimentarily in others. We examine this further by annotating a regulatory enrichment network calculated from the FSK + P4 versus CTRL comparison of expression (Fig. 7B). A sizable portion of this network (indicated in red) shows the activated regulatory interactions present only under the combination treatment of FSK and P4. Many of the genes were up-regulated, implicating them as important molecules for pro-relaxtory signaling as opposed to the dominant anti-inflammatory signaling seen in Fig. 7A. Again, many of the nodes in this network are co-regulated by P4 and cAMP (indicated in yellow). These genes are mostly down-regulated (ovular nodes), which is consistent with the idea that cAMP and P4 are anti-inflammatory. It is important to understand how these two processes work individually and in concert to elucidate their role in maintaining quiescence. The results in this work identify pathways and regulatory mechanisms that may be important for fulfilling this role.
Cell line and tissue similarity
Tissue similarity analysis was used to determine the extent to which hTERT-HM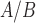 cells in response to various stimuli mimic term myometrium. To do this, we collected and compared an RNA-seq dataset of term laboring and non-laboring myometrial samples (two phenotypes, N = 5 for each) to our treated cell line using a method introduced by Sandberg and Ernberg (2005) called TSI. TSI uses SVD, an unsupervised dimensionality reduction method, to find an average profile for all samples of a given type (e.g. a specific cancer or tissue) based on the variability within the gene expression matrix. The cell line samples are then projected (using the calculated singular vectors) into the reduced SVD space and their correlations to the different average tissue profiles are calculated. The correlations produce the TSI values, which range from −1 (low similarity) to 1 (high similarity).
cells in response to various stimuli mimic term myometrium. To do this, we collected and compared an RNA-seq dataset of term laboring and non-laboring myometrial samples (two phenotypes, N = 5 for each) to our treated cell line using a method introduced by Sandberg and Ernberg (2005) called TSI. TSI uses SVD, an unsupervised dimensionality reduction method, to find an average profile for all samples of a given type (e.g. a specific cancer or tissue) based on the variability within the gene expression matrix. The cell line samples are then projected (using the calculated singular vectors) into the reduced SVD space and their correlations to the different average tissue profiles are calculated. The correlations produce the TSI values, which range from −1 (low similarity) to 1 (high similarity).
Correlation for each hTERT-HM cell line sample was calculated with the averaged non-labor profile and labor profile and were symmetric (i.e. samples with a correlation of 1 to the non-labor representative had a correlation of −1 to the labor representative). Based on our current understanding of P4, cAMP, and inflammatory signaling in parturition, as well as the results presented in this work, we expect a given trend for these cell line sample groups. IL-1
cell line sample was calculated with the averaged non-labor profile and labor profile and were symmetric (i.e. samples with a correlation of 1 to the non-labor representative had a correlation of −1 to the labor representative). Based on our current understanding of P4, cAMP, and inflammatory signaling in parturition, as well as the results presented in this work, we expect a given trend for these cell line sample groups. IL-1 should likely be most labor-like, addition of P4 and/or FSK to IL-1
should likely be most labor-like, addition of P4 and/or FSK to IL-1 -treated cells should make them less labor-like, untreated cells should be roughly in a middle-ground between labor and non-labor, while FSK-, P4-, and FSK + P4-treated cells should be more non-labor-like. We performed TSI on four separate gene expression matrices containing (i) all genes, (ii) highly significant (|fold change| > 5 and adjusted P-value < 0.001) genes from P4, FSK, and IL-1
-treated cells should make them less labor-like, untreated cells should be roughly in a middle-ground between labor and non-labor, while FSK-, P4-, and FSK + P4-treated cells should be more non-labor-like. We performed TSI on four separate gene expression matrices containing (i) all genes, (ii) highly significant (|fold change| > 5 and adjusted P-value < 0.001) genes from P4, FSK, and IL-1 individual treatment (145 genes), (iii) a set of key cell line genes selected based on the results presented in Figs 2–7 (17 genes), and (iv) highly significant genes from the tissue dataset (non-labor versus labor; 151) (Fig. 8A, see Methods for a detailed description of the four gene sets).
individual treatment (145 genes), (iii) a set of key cell line genes selected based on the results presented in Figs 2–7 (17 genes), and (iv) highly significant genes from the tissue dataset (non-labor versus labor; 151) (Fig. 8A, see Methods for a detailed description of the four gene sets).
Figure 8.
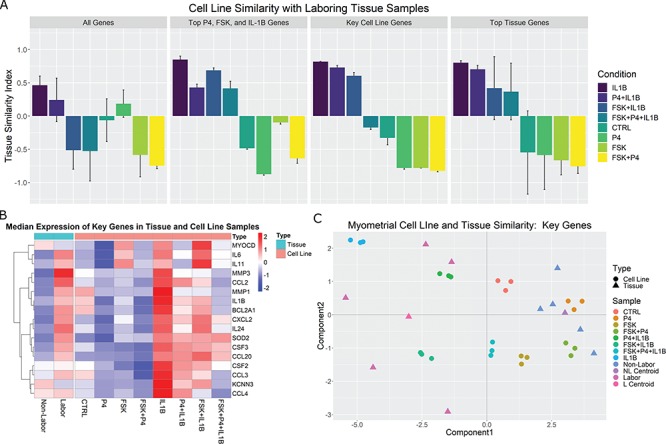
Comparison of cell line and tissue samples using TSI. SVD was performed on the tissue gene expression matrix and phenotype centroids were calculated in the reduced SVD space. Cell line samples were then projected to this space and their correlations to the labor centroid were calculated. A. Median correlations and standard deviations per sample group are shown for all genes shared between the two studies, highly significant (|fold change| > 5 and adjusted P-value < 0.001) genes from the individual stimulus treated cells compared to control, a set of key genes selected from the comprehensive analyses of cell lines reported in Figs 2–7, and highly significant genes from the tissue study by comparing in-labor and not-in-labor samples. B. A heatmap of the key genes with median expression across all sample groups. C. Visualization of samples and tissue centroids using the first two singular vectors (or components) from the SVD analysis on the key genes selected based on the cell line analysis.
Using all genes shared between the two expression studies (gene set 1) in the TSI calculation produces fair TSI values (i.e. similar to our hypothesized pattern of similarity), with IL-1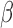 -treated cells having the highest correlation, ~0.5, to the averaged labor profile while FSK + P4-treated cells are highly correlated, around −0.75, with the averaged non-labor profile. Using gene set 2, the correlation to labor increased to 0.85 for IL-1
-treated cells having the highest correlation, ~0.5, to the averaged labor profile while FSK + P4-treated cells are highly correlated, around −0.75, with the averaged non-labor profile. Using gene set 2, the correlation to labor increased to 0.85 for IL-1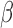 and all IL-1
and all IL-1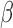 conditions were more labor-like. In contrast, P4-treated cells achieved the highest correlation to the non-laboring phenotype. This pattern indicates that the strong anti-inflammatory action of P4 is captured well by this gene set, but the relaxatory action of cAMP is less represented. Use of gene set 3 produced a result highly similar to our prior expected cell condition-tissue phenotype similarity. Additionally, the synergistic effect of FSK and P4 seen in the GSVA results were replicated, with FSK + P4 being more highly correlated with the non-laboring phenotype and FSK + P4 + IL-1
conditions were more labor-like. In contrast, P4-treated cells achieved the highest correlation to the non-laboring phenotype. This pattern indicates that the strong anti-inflammatory action of P4 is captured well by this gene set, but the relaxatory action of cAMP is less represented. Use of gene set 3 produced a result highly similar to our prior expected cell condition-tissue phenotype similarity. Additionally, the synergistic effect of FSK and P4 seen in the GSVA results were replicated, with FSK + P4 being more highly correlated with the non-laboring phenotype and FSK + P4 + IL-1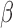 correlated with non-labor, almost equivalent to the untreated cells. cAMP and P4 were also clearly relaxatory stimuli in this result, seen by the substantial increase in correlation to non-labor compared with control. These patterns were also clearly visible in the gene expression matrix (Fig. 8B). Finally, gene set 4 (tissue-derived) produced the same ordering as the key cell line genes, albeit with more variability.
correlated with non-labor, almost equivalent to the untreated cells. cAMP and P4 were also clearly relaxatory stimuli in this result, seen by the substantial increase in correlation to non-labor compared with control. These patterns were also clearly visible in the gene expression matrix (Fig. 8B). Finally, gene set 4 (tissue-derived) produced the same ordering as the key cell line genes, albeit with more variability.
Further visualization of the TSI results was achieved using the first two singular vectors, or components, of the SVD calculation. Figure 8C depicts this sample clustering for gene set 3 (selected key cell line genes). Tissue phenotype ‘centroids’ indicate the representative profile for labor and non-labor. The top two SVD components indicate that the most dominant patterns in the gene expression data are first the IL-1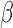 /inflammatory signaling and cAMP signaling second. The former, component 1, represents the TSI result in that the FSK/P4 and non-labor samples are separated from the IL-1
/inflammatory signaling and cAMP signaling second. The former, component 1, represents the TSI result in that the FSK/P4 and non-labor samples are separated from the IL-1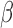 and labor samples with opposite values. The latter, component 2, identifies cAMP signaling, separating FSK-treated samples from the rest. These results are interesting because they were clearly observed in the cell line data, but the components were identified from the tissue data only. The genes came from the observed expression in the cell line, but no direct information from the cells is included in the SVD calculation. This indicates a clear role for cAMP in parturition. These genes also capture the inherent diversity in the laboring phenotype, clearly exhibiting more variability in expression than the non-labor samples. These high correlations and the ordering of the cell line sample groups indicates that this experimental model of human myometrium accurately captures the transcriptional landscapes of myometrium at term and during parturition.
and labor samples with opposite values. The latter, component 2, identifies cAMP signaling, separating FSK-treated samples from the rest. These results are interesting because they were clearly observed in the cell line data, but the components were identified from the tissue data only. The genes came from the observed expression in the cell line, but no direct information from the cells is included in the SVD calculation. This indicates a clear role for cAMP in parturition. These genes also capture the inherent diversity in the laboring phenotype, clearly exhibiting more variability in expression than the non-labor samples. These high correlations and the ordering of the cell line sample groups indicates that this experimental model of human myometrium accurately captures the transcriptional landscapes of myometrium at term and during parturition.
Discussion
RNA-seq data derived from an immortalized human myometrial cell line, hTERT-HM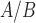 induced to express PR-A and PR-B, treated with combinations of IL-1
induced to express PR-A and PR-B, treated with combinations of IL-1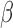 , P4, and FSK (activates cAMP signaling) and vehicle were used to address three questions: (i) what genes and regulatory pathways are affected by IL-1
, P4, and FSK (activates cAMP signaling) and vehicle were used to address three questions: (i) what genes and regulatory pathways are affected by IL-1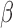 , P4, and cAMP in myometrial cells, (ii) how do P4 and cAMP affect responsiveness of myometrial cells to IL-1
, P4, and cAMP in myometrial cells, (ii) how do P4 and cAMP affect responsiveness of myometrial cells to IL-1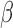 , and (iii) how do the gene expression patterns in differently treated hTERT-HM
, and (iii) how do the gene expression patterns in differently treated hTERT-HM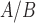 cells compare with expression patterns in laboring and non-laboring term myometrium. These questions were answered using global differential gene expression analysis, gene set enrichment, regulatory network enrichment, and the TSI. Those analyses suggest that (i) P4/PR signaling inhibits responsiveness to IL-1
cells compare with expression patterns in laboring and non-laboring term myometrium. These questions were answered using global differential gene expression analysis, gene set enrichment, regulatory network enrichment, and the TSI. Those analyses suggest that (i) P4/PR signaling inhibits responsiveness to IL-1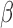 primarily through AP-1 (JUN), (ii) cAMP signaling exerts anti-inflammatory effects alone while also helping to establish a relaxed and hypertrophic state, and (iii) that P4 and cAMP work together in a synergistic manner to further inhibit certain inflammatory processes. Specific regulatory molecules/interactions were observed to have altered activity under individual treatment with the three stimuli, providing context to the inter-process regulation observed from gene set enrichment. Lastly, comparison between a gene expression study of clinical myometrial tissue and the treated hTERT-HM
primarily through AP-1 (JUN), (ii) cAMP signaling exerts anti-inflammatory effects alone while also helping to establish a relaxed and hypertrophic state, and (iii) that P4 and cAMP work together in a synergistic manner to further inhibit certain inflammatory processes. Specific regulatory molecules/interactions were observed to have altered activity under individual treatment with the three stimuli, providing context to the inter-process regulation observed from gene set enrichment. Lastly, comparison between a gene expression study of clinical myometrial tissue and the treated hTERT-HM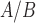 cells suggested that exposure to IL-1
cells suggested that exposure to IL-1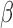 mimics the laboring phenotype, whereas exposure to P4 and FSK mimics the pre-labor phenotype.
mimics the laboring phenotype, whereas exposure to P4 and FSK mimics the pre-labor phenotype.
Human parturition is associated with an increase in the PR-A:PR-B ratio that is thought to cause functional P4 withdrawal. Our recent studies suggest that the PR-A:PR-B ratio is affected by P4 and pro-inflammatory stimuli due to differential effects of serine phosphorylation on the stability of PR-B (steady-state level decreases) and PR-A (steady-state level increases). We proposed that the labor-associated increase in the myometrial PR-A:PR-B ratio occurs in response to pro-inflammatory stimuli inducing increased stability of PR-A (Amini et al., 2019). Using TSI analyses, we found that hTERT-HM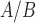 cells exposed to P4 and FSK mimic quiescent myometrium, whereas cells exposed to IL-1
cells exposed to P4 and FSK mimic quiescent myometrium, whereas cells exposed to IL-1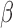 mimic laboring myometrium. This suggests that transition to the labor phenotype (at least based on transcriptome analyses) could be caused by a loss of cAMP/PKA signaling and/or gain of pro-inflammatory stimuli that alters P4/PR activity due to changes in PR phosphorylation. In this context, the PR-A:PR-B ratio in myometrial cells reflects the balance between the cAMP/PKA and inflammatory load stimuli.
mimic laboring myometrium. This suggests that transition to the labor phenotype (at least based on transcriptome analyses) could be caused by a loss of cAMP/PKA signaling and/or gain of pro-inflammatory stimuli that alters P4/PR activity due to changes in PR phosphorylation. In this context, the PR-A:PR-B ratio in myometrial cells reflects the balance between the cAMP/PKA and inflammatory load stimuli.
Analysis of gene expression changes using gene set enrichment allows for better interpretation of the altered expression of hundreds of genes by grouping them in biologically meaningful sets (e.g. pathways or TFs targets). While this analysis is highly useful, there are caveats to consider. One is that the top enriched sets are not always the definitive cause/effect of the observed expression changes. This is especially the case for TF enrichment. While many targets of a specific TF may exhibit altered expression, we cannot be certain that these alterations were due to increased or decreased activity of a specific TF in question. Furthermore, when many genes are changing, it is even less likely that a specific TF is solely responsible but rather a combination of some of the significantly enriched TFs. As an example, targets of P4/PR were significantly down-regulated in the IL-1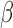 versus CTRL comparison (Fig. 6B). By experimental design, these cells express PR; however, PR is only expected to be active when bound to its ligand P4. Therefore, it is likely that other factors are responsible for changes in these genes (or potentially ligand-independent actions of PRs), but the enrichment shows PR as a top candidate.
versus CTRL comparison (Fig. 6B). By experimental design, these cells express PR; however, PR is only expected to be active when bound to its ligand P4. Therefore, it is likely that other factors are responsible for changes in these genes (or potentially ligand-independent actions of PRs), but the enrichment shows PR as a top candidate.
GSVA was used to calculate enrichment partly because it provides directionality for each gene set in the form of an enrichment fold change and our initial hypotheses suggested opposite regulation of similar pathways in different conditions (e.g. pro-inflammatory signaling by IL-1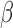 and anti-inflammatory action of P4). GSVA takes into account the direction of expression change in each gene in the gene set, while other enrichment approaches calculate differential expression for each gene first, followed by tallying the number of significant genes in the set, regardless of up or down-regulation. Both analyses have their advantages and shortcomings and the approach used should be based on the study design and research questions. For RNEA, TFs that undergo significant changes in expression are included in the network. However, it is known that this is not solely dependent on gene expression as most TFs are activated by post-translational modifications. RNEA covers this case by allowing TFs exhibiting minimal or no change in expression in the network if it is up- and down-stream of two differentially expressed genes. TFs having no differentially expressed up-stream regulators may play a role in the observed down-stream changes in target expression but will not be incorporated into the network. GSVA and RNEA were combined to circumvent these shortcomings. RNEA gives the regulatory pathway detail with targets and the directionality of their changes, while GSVA requires no information about the transcriptional alteration of the TFs themselves. Combining the techniques allows for identification of pathways, TFs, and targets of those TFs that fit specific relationships between P4, cAMP, and IL-1
and anti-inflammatory action of P4). GSVA takes into account the direction of expression change in each gene in the gene set, while other enrichment approaches calculate differential expression for each gene first, followed by tallying the number of significant genes in the set, regardless of up or down-regulation. Both analyses have their advantages and shortcomings and the approach used should be based on the study design and research questions. For RNEA, TFs that undergo significant changes in expression are included in the network. However, it is known that this is not solely dependent on gene expression as most TFs are activated by post-translational modifications. RNEA covers this case by allowing TFs exhibiting minimal or no change in expression in the network if it is up- and down-stream of two differentially expressed genes. TFs having no differentially expressed up-stream regulators may play a role in the observed down-stream changes in target expression but will not be incorporated into the network. GSVA and RNEA were combined to circumvent these shortcomings. RNEA gives the regulatory pathway detail with targets and the directionality of their changes, while GSVA requires no information about the transcriptional alteration of the TFs themselves. Combining the techniques allows for identification of pathways, TFs, and targets of those TFs that fit specific relationships between P4, cAMP, and IL-1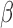 signaling.
signaling.
Observing results from these two approaches in the context of this work suggests two potential avenues for treating preterm birth. The TF complex AP-1, specifically the JUN sub-unit, was seen to be a key part of the inflammatory signaling induced by IL-1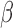 . This was distinctly inhibited with co-treatment of P4 and up-regulated targets of JUN were also strongly inhibited by P4 alone, suggesting that this is one of the key mechanisms influenced by P4 to maintain quiescence throughout pregnancy. On the other hand, NF
. This was distinctly inhibited with co-treatment of P4 and up-regulated targets of JUN were also strongly inhibited by P4 alone, suggesting that this is one of the key mechanisms influenced by P4 to maintain quiescence throughout pregnancy. On the other hand, NF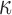 B was clearly a major part of the IL-1
B was clearly a major part of the IL-1 -induced inflammatory signaling and this activity remained significant under co-treatment with P4 and/or cAMP. Therefore, molecular targeting of JUN or NF
-induced inflammatory signaling and this activity remained significant under co-treatment with P4 and/or cAMP. Therefore, molecular targeting of JUN or NF B may reduce the likelihood of preterm delivery. Inhibition of NF
B may reduce the likelihood of preterm delivery. Inhibition of NF B would provide a second anti-inflammatory action separate from that carried out by P4 and cAMP, while inhibition of JUN would act alongside P4 and cAMP in preventing signaling through this TF. Treatment with a JUN inhibitor during late stages of pregnancy may work most effectively, as it could work in place of P4 after its functional withdrawal. NF
B would provide a second anti-inflammatory action separate from that carried out by P4 and cAMP, while inhibition of JUN would act alongside P4 and cAMP in preventing signaling through this TF. Treatment with a JUN inhibitor during late stages of pregnancy may work most effectively, as it could work in place of P4 after its functional withdrawal. NF B is thought to cooperate with AP-1 and CEBP to activate labor genes due to the presence of both AP-1 and NF
B is thought to cooperate with AP-1 and CEBP to activate labor genes due to the presence of both AP-1 and NF B binding sites on the promoters of these genes (Nadeau-Vallée et al., 2015; Sakowicz, 2018). Therefore, an inhibitor of just one of AP-1 or NF
B binding sites on the promoters of these genes (Nadeau-Vallée et al., 2015; Sakowicz, 2018). Therefore, an inhibitor of just one of AP-1 or NF B could be successful in reducing the chance of preterm birth. Studies suggest that full inhibition of NF
B could be successful in reducing the chance of preterm birth. Studies suggest that full inhibition of NF B signaling would be undesirable for a number of reasons and suggest partial inhibition through a small noncompetitive IL-1R biased ligand or compounds targeting the non-canonical NF
B signaling would be undesirable for a number of reasons and suggest partial inhibition through a small noncompetitive IL-1R biased ligand or compounds targeting the non-canonical NF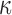 B signaling pathways (Nadeau-Vallée et al., 2015; Wang et al., 2018).
B signaling pathways (Nadeau-Vallée et al., 2015; Wang et al., 2018).
The use of cell line models as an approximation of in vivo systems provides alternative and useful avenues of research to address various biological questions. The setup in this work of using treated cell lines allows for isolated investigation of processes that are interconnected in a complex manner throughout parturition. Through the comparisons in this work, we identified synergistic effects of P4 and cAMP and how cAMP inhibits signaling by IL-1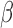 . These insights are made meaningful when the in vitro model was shown to effectively approximate term myometrium. Importantly, as expected, IL-1
. These insights are made meaningful when the in vitro model was shown to effectively approximate term myometrium. Importantly, as expected, IL-1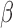 - and FSK + P4-treated cells had similar gene expression profiles to laboring and non-laboring myometrial samples, respectively. The similarity—at least at the gene expression level—suggests that the hTERT-HM
- and FSK + P4-treated cells had similar gene expression profiles to laboring and non-laboring myometrial samples, respectively. The similarity—at least at the gene expression level—suggests that the hTERT-HM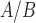 cell line mimics the gravid myometrium in response to specific treatments. Interestingly, under basal conditions, hTERT-HM
cell line mimics the gravid myometrium in response to specific treatments. Interestingly, under basal conditions, hTERT-HM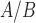 cells appear less relaxed than non-laboring myometrium but can be transformed to this phenotype by exposure to P4 and FSK.
cells appear less relaxed than non-laboring myometrium but can be transformed to this phenotype by exposure to P4 and FSK.
This study is, to our knowledge, the first to examine the relationship between P4, cAMP, and IL-1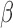 signaling at the genome-wide level in myometrium. Each of these signaling processes are believed to play a major role in pregnancy and parturition. Through our analysis, we generated testable hypotheses on how these processes interact to guide future research efforts. This includes using the results presented in this work to find new ways to increase the anti-inflammatory actions of P4 and cAMP to promote uterine quiescence as a means for treating preterm birth.
signaling at the genome-wide level in myometrium. Each of these signaling processes are believed to play a major role in pregnancy and parturition. Through our analysis, we generated testable hypotheses on how these processes interact to guide future research efforts. This includes using the results presented in this work to find new ways to increase the anti-inflammatory actions of P4 and cAMP to promote uterine quiescence as a means for treating preterm birth.
Supplementary Material
Authors’ roles
Research pipeline was conceived by Z.S., M.K., and S.M. M.R.C. provided early insights for study direction. L.Y. and H.T. performed early molecular biology experiments to establish cell line system. P.A. performed cell cultures and RNA extraction. J.W. carried out RNA sequencing. Z.S. performed all data analysis. M.K. supervised application of computational methods and S.M. supervised biological interpretation and conceptualization of results. Z.S. drafted the manuscript. M.K. and S.M. edited and commented on earlier drafts. All authors read and approved the final manuscript.
Funding
March of Dimes Ohio Prematurity Research Collaborative; The Eunice Kennedy Shriver National Institute of Child Health and Human Development; National Institutes of Health (HD069818, U01-CA198941).
Conflict of interest
The authors declare they have no conflicts of interest.
References
- Amini P, Michniuk D, Kuo K, Yi L, Skomorovska-Prokvolit Y, Peters GA, Tan H, Wang J, Malemud CJ, Mesiano S. Human parturition involves phosphorylation of progesterone receptor-A at serine-345 in myometrial cells. Endocrinology 2016;157:4434 –4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amini P, Wilson R, Koeblitz W, Wang J, Tan H, Yi L, Stanfield Z, Romani A, Malemud C, Mesiano S. Mechanism by which progesterone and camp synergize to maintain uterine quiescence during pregnancy. Mol Cell Endocrinol 2019;479:1–11. [DOI] [PubMed] [Google Scholar]
- Billington CK, Ojo OO, Penn RB, Ito S. Camp regulation of airway smooth muscle function. Pulm Pharmacol Ther 2013;26:112–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y-W, van den Berg HA, Moore JD, Quenby S, Blanks AM. Assessment of myometrial transcriptome changes associated with spontaneous human labour by high-throughput RNA-seq. Exp Physiol 2014;99:510–524. [DOI] [PubMed] [Google Scholar]
- Chanrachakul B, Broughton Pipkin F, Khan RN. Contribution of coupling between human myometrial 2-adrenoreceptor and the bkca channel to uterine quiescence. Am J Physiol Cell Physiol 2004;287:C1747–C1752. [DOI] [PubMed] [Google Scholar]
- Chen L, Lei K, Malawana J, Yulia A, Sooranna SR, Bennett PR, Liang Z, Grammatopoulos D, Johnson MR. Cyclic amp enhances progesterone action in human myometrial cells. Mol Cell Endocrinol 2014;382:334–343. [DOI] [PubMed] [Google Scholar]
- Chouvardas P, Kollias G, Nikolaou C. Inferring active regulatory networks from gene expression data using a combination of prior knowledge and enrichment analysis. BMC Bioinform 2016;17:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff L.A., Trapnell C., and Kelley D.. Cummerbund: Visualization and Exploration of Cufflinks High-Throughput Sequencing Data. R package version 2. 2012.
- Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform 2013;14:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy DB, Janowski BA, Corey DR, Mendelson CR. Progesterone receptor plays a major antiinflammatory role in human myometrial cells by antagonism of nuclear factor-b activation of cyclooxygenase 2 expression. Mol Endocrinol 2006;20:2724–2733. [DOI] [PubMed] [Google Scholar]
- Mesiano S, Chan E-C, Fitter JT, Kwek K, Yeo G, Smith R. Progesterone withdrawal and estrogen activation in human parturition are coordinated by progesterone receptor A expression in the myometrium. J Clin Endocrinol Metabol 2002;87:2924–2930. [DOI] [PubMed] [Google Scholar]
- Mesiano S, Wang Y, Norwitz ER. Progesterone receptors in the human pregnancy uterus: do they hold the key to birth timing? Reprod Sci 2011;18:6–19. [DOI] [PubMed] [Google Scholar]
- Morgado M, Cairrão E, Santos-Silva AJ, Verde I. Cyclic nucleotide-dependent relaxation pathways in vascular smooth muscle. Cell Mol Life Sci 2012;69:247–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau-Vallée M, Quiniou C, Palacios J, Hou X, Erfani A, Madaan A, Sanchez M, Leimert K, Boudreault A, Duhamel F et al. . Novel noncompetitive IL-1 receptor-biased ligand prevents infection-and inflammation-induced preterm birth. J Immunol 2015; 1500758. [DOI] [PubMed] [Google Scholar]
- Oldenburger A, Roscioni SS, Jansen E, Menzen MH, Halayko AJ, Timens W, Meurs H, Maarsingh H, Schmidt M. Anti-inflammatory role of the cAMP effectors EPAC and PKA: implications in chronic obstructive pulmonary disease. PLoS One 2012;7: 31574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters GA, Yi L, Skomorovska-Prokvolit Y, Patel B, Amini P, Tan H, Mesiano S. Inflammatory stimuli increase progesterone receptor-A stability and transrepressive activity in myometrial cells. Endocrinology 2016;158:158–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price SA, Bernal AL. Uterine quiescence: the role of cyclic amp. Exp Physiol 2001;86:265–272. [DOI] [PubMed] [Google Scholar]
- Sakowicz A. The role of NFKB in the three stages of pregnancy: implantation, maintenance and labour; a review article. BJOG 2018. [DOI] [PubMed] [Google Scholar]
- Sanborn BM. Hormonal signaling and signal pathway crosstalk in the control of myometrial calcium dynamics. Semin Cell Dev Biol 2007;18:305–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanborn BM, Ku C-Y, Shlykov S, Babich L. Molecular signaling through g-protein-coupled receptors and the control of intracellular calcium in myometrium. J Soc Gynecol Investig 2005;12:479–487. [DOI] [PubMed] [Google Scholar]
- Sandberg R, Ernberg I. Assessment of tumor characteristic gene expression in cell lines using a tissue similarity index (TSI). Proc Natl Acad Sci USA 2005;102:2052–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalhegg B, Taskén K. Specificity in the cAMP/PKA signaling pathway: differential expression, regulation, and subcellular localization of subunits of PKA. Front Biosci 1997;2:d331–d342. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Limma: linear models for microarray data Bioinformatics and Computational Biology Solutions Using r and Bioconductor. New York, NY: Springer, 2005, 397–420. [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES et al. . Gene set enrichment analysis: a knowledge based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005;102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan H, Yi L, Rote NS, Hurd WW, Mesiano S. Progesterone receptor-A and-B have opposite effects on pro-inflammatory gene expression in human myometrial cells: implications for progesterone actions in human pregnancy and parturition. J Clin Endocrinol 2012;97:E719–E730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. Tophat: discovering splice junctions with RNA-seq. Bioinformatics 2009;25:1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, Van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 2010;28:511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Parobchak N, Martin A, Rosen M, Yu LJ, Nguyen M, Gololobova K, Rosen T. Screening a small molecule library to identify inhibitors of NF-KB inducing kinase and pro-labor genes in human placenta. Sci Rep 2018;8:1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yulia A, Singh N, Lei K, Sooranna SR, Johnson MR. Cyclic amp effectors regulate myometrial oxytocin receptor expression. Endocrinology 2016;157:4411–4422. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


