Abstract
Polyketides are a large class of bioactive natural products with a wide range of structures and functions. Polyketides are biosynthesized by large, multi-domain enzyme complexes termed polyketide synthases (PKSs). One of the primary challenges when studying PKSs is the high reactivity of their poly-β-ketone substrates. This has hampered structural and mechanistic characterization of PKS–polyketide complexes, and, as a result, little is known about how PKSs position the unstable substrates for proper catalysis while displaying high levels of regio-and stereospecificity. As a first step toward a general plan to use oxetanes as carbonyl isosteres to broadly interrogate PKS chemistry, we describe the development and application of an oxetane-based PKS substrate mimic. This enabled the first structural determination of the acyl-enzyme intermediate of a ketosynthase (KS) in complex with an inert extender unit mimic. The crystal structure, in combination with molecular dynamics simulations, led to a proposed mechanism for the unique activity of DpsC, the priming ketosynthase for daunorubicin biosynthesis. The successful application of an oxetane-based polyketide mimic suggests that this novel class of probes could have wide-ranging applications to the greater biosynthetic community interested in the mechanistic enzymology of iterative PKS.
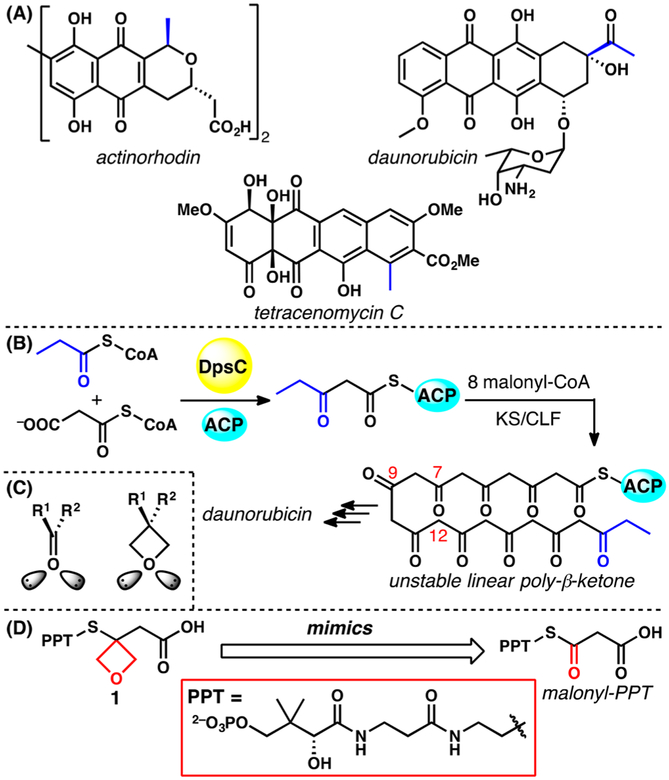
Polyketide natural products are a large and diverse class of secondary metabolites of high impact to human health.1,2 Type II polyketides are biosynthesized by a type II polyketide synthase (PKS) consisting of 5–10 stand-alone enzymes that form complexes in solution.3 Notable examples include actinorhodin, daunorubicin, and tetracenomycin C (Figure 1A).4–6 PKSs have been heavily studied because of their ability to efficiently biosynthesize complex small molecules and their potential to be engineered for combinatorial biosynthesis.1,2
Figure 1.
(A) Examples of type II polyketides with the starter units shown in blue. (B) DpsC catalyzes the transfer of small acyl-CoAs, including propionyl-CoA, to the acyl carrier protein (ACP) and also the initial chain elongation reaction that condenses the propionyl starter unit with malonyl-ACP to afford the growing intermediate. Eight more rounds of chain elongation produce the unstable, linear poly-β-ketone intermediate that is cyclized and tailored to become daunorubicin. (C) Oxetanes are isosteres for the carbonyl group. (D) Probe 1, with the thioester carbonyl group replaced by an oxetane, mimics malonyl-PPT.
One of the primary challenges associated with investigating type II PKS is the high reactivity of enzymatic substrates and intermediates.7 The poly-β-ketone generated by most type II PKSs is highly susceptible to spontaneous, nonspecific cyclization, which has made structural studies of PKS–substrate complexes extremely difficult (Figure 1B).8 Without this structural knowledge, rational engineering of substrate specificity often leads to inactive enzymes.3 Our group previously synthesized isoxazole-based polyketide isosteres and applied them to the characterization of the interior pocket of an acyl carrier protein.9 Given the substantial structural differences between these first-generation chemical probes and the natural poly-β-ketone substrates, we expect their applications to be rather limited. To interrogate PKS–substrate complexes more broadly, we sought to generate prob molecules that more closely mimicked the natural substrates. Here we present a proof-of-concept study using an oxetane-containing malonate mimic to offer a snapshot of a type II polyketide priming ketosynthase (KS) in action.
The oxetane ring is well recognized as an isostere for the carbonyl group,10 owing primarily to the efforts of Carreira, Müller, and co-workers.11–18 Although slightly larger than the carbonyl, the oxetane orients its oxygen lone pairs along similar vectors to a carbonyl group (Figure 1C). To date, this carbonyl–oxetane replacement strategy has not been used to study questions in polyketide biosynthesis where it is ideally suited for strategic replacement of carbonyl groups in unstable poly-β-ketone intermediates of aromatic polyketides.
As a proof of concept, we present the synthesis of a simple yet representative oxetane-based PKS substrate mimic 1 (Figure 1D) and demonstrate its applicability by cocrystallizing it with the enzyme DpsC from the daunorubicin type II PKS from Streptomyces peucetius.19–22 DpsC is a unique enzyme that has both acyltransferase (AT) and priming ketosynthase (ketosynthase III, KS III) activities.20–22 However, the structural basis for the unique activity of DpsC is unclear because of the lack of high-resolution substrate–DpsC structures. Here we present a cocrystal structure, combined with molecular dynamics (MD) simulations, that provides mechanistic insight into the KS activity of this enzyme.
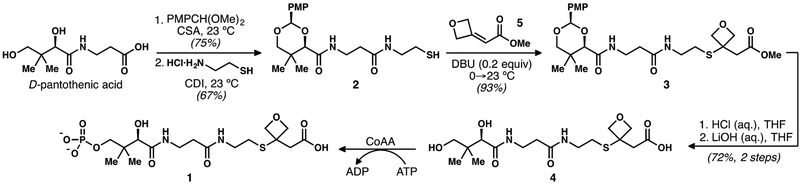
Phosphopantetheine (PPT) malonate mimic 1 was synthesized from commercially available D-pantothenic acid (Scheme 1). Acetal formation, followed by a CDI-mediated amide coupling with cysteamine hydrochloride produced thiol 2. The 1,3-diketone surrogate was installed via base-catalyzed thia-Michael addition of 2 to oxetane-bearing enoate 5, yielding 3.13 Acetal hydrolysis and ester saponification of 3 unveiled the diol and carboxylic acid moieties, respectively, and provided the penultimate intermediate 4. The synthesis of 1 was completed using chemoenzymatic phosphorylation.23
Scheme 1.
Synthesis of Malonyl-PPT Probe 1
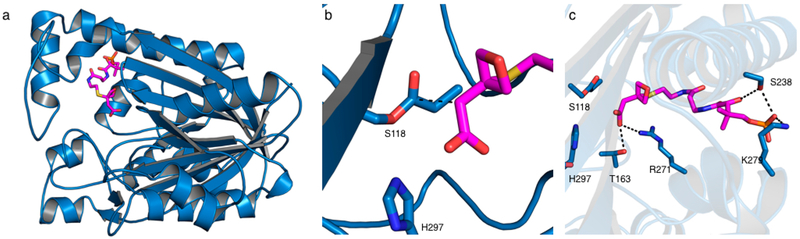
The DpsC–1 cocrystal structure provides new insights into the positioning of substrates just prior to catalysis. As expected, the Asp-His-Ser catalytic triad forms a hydrogen bond network that results in an activated S118 nucleophile (see Figure S4 for details). The resulting propionyl-serine side chain is in close proximity to probe 1. In particular, the two carbon atoms that would normally participate in the Claisen condensation reaction were separated by an appropriate distance (2.9 Å) and were aligned appropriately (Figure 2). The carboxylate of 1 interacts with R271 via a charge–charge interaction and has a hydrogen bond with T163 (Figure 2C). The terminal phosphate shows a charge–charge interaction with K279 as well as a hydrogen bond to S238, which is consistent with many other PKS enzymes that use positively charged surface residues to position the phosphate moiety of the phosphopantetheine prosthetic group.3,8
Figure 2.
Crystal structure of propionyl-DpsC bound to extender unit mimic 1. (a) Overall structure of propionyl-DpsC in complex with 1 (monomer shown). One DpsC monomer is shown in blue, and 1 is shown in magenta. (b) S118 is shown with its propionylated side chain with the carbon bond that would be formed shown as a black dashed line. (c) Overview of DpsC–1 interactions within the active site and near the enzyme surface.
Canonical KSIIIs use conserved residues in an oxyanion hole to stabilize the buildup of negative charge on the thioester carbonyl.3,8 Residues H244 and N274 in the prototypical KSIII FabH, known to be essential for decarboxylation,24,25 are not conserved in DpsC. In the DpsC–1 structure, the oxetane oxygen atom did not orient itself into a positively charged environment within the DpsC active site (Figure 2C). The structurally equivalent positions in DpsC are P265, which does not contain a suitable side chain for oxy-anion stabilization, and H297 is locked in a hydrogen bond network with D302 and S118. The side chain of H198 is within the active site and could potentially stabilize an oxyanion; however, the oxygen of the oxetane group is pointing away from this side chain in the crystal structure. One possibility is that substrate decarboxylation reorients itself during catalysis of the decarboxylative C–C bond formation to take adfantage of activation by H198 via a simple bond rotation to stabilize the newly formed negative charge (Figure S4). This proposed mechanism is currently under investigation.
To further assess the validity of the carbonyl-oxetane replacement strategy, the atomic coordinates of the cocrystal structure were used to parametrize and generate two types of MD simulations for comparison: DpsC bound to either oxetane-based probe 1 or the more natural, malonate-PPT (Figure 1C). The same atomic coordinates of the cocrystal structure were used to generate the MD simulation for DpsC bound to malonyl-PPT, in which the oxetane substituent was mutated in silico into a carbonyl group. Trajectories of both systems in explicit solvent were collected over a microsecond for comparative analyses of relative binding affinity, backbone fluctuations and low-frequency motions. These simulations demonstrated similar relative binding affinities, overall long-term motion and high-frequency movement of binding site residues (Figures S5–S10). This provides further support that the protein conformation, substrate–DpsC interactions, as well as protein dynamics near the interacting residues between DpsC and probe 1 are self-consistent.
In summary, we report the first design, synthesis and application of an oxetane-based probe as a surrogate for the carbonyl group of an electrophilic thioester. This study clarifies how the substrate is oriented for DpsC-catalyzed decarboxylation/Claisen condensation of malonyl-CoA. More generally, this study provides a proof-of-concept of our use of oxetane isosteres to investigate the biosynthesis catalyzed by iterative PKSs, which have a distinctly different chemical logic from that of modular PKSs. Ongoing efforts include the synthesis of higher-order poly-β-ketone mimics that contain multiple carbonyl to oxetane substitutions, which are currently being applied in mechanistic and structural analyses of other iterative PKSs. These probes can also be attached to ACP via a one-step chemoenzymatic procedure, thereby enabling us to isolate and crystallize ACP-probe-partner complexes for structural and mechanistic studies.9 We are also utilizing these oxetane probes for inhibition enzyme kinetics probing the binding of wild type and mutant PKS domains. These polyketide mimics will enable investigations of important substrate–enzyme and protein–protein interactions that govern the efficiency and selectivity of PKSs, ultimately leading to advances in molecular design and medicinal chemistry.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by NIH GM100305 and GM076330 (to S.-C.T.) and by NSF CHE-1564340 (to C.D.V.). This work was part of the DOE Joint BioEnergy Institute supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, through contract DE-AC02–05CH11231 between Lawrence Berkeley National Laboratory and the U.S. Department of Energy. Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02–76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393). Kostas Vasilakis and Pakorn Wattana-Amorn are acknowledged for their assistance with cloning of the dps PKS.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.7b11793.
Detailed experimental procedures, protein structural analysis, molecular dynamic simulations, and compound spectra (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Staunton J; Weissman KJ Nat. Prod. Rep 2001, 18 (4), 380–416. [DOI] [PubMed] [Google Scholar]
- (2).Borchardt JK Modern Drug Discovery 1999, 2 (4), 22–29. [Google Scholar]
- (3).Das A; Khosla C Acc. Chem. Res 2009, 42 (5), 631–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Kim ES; Bibb MJ; Butler MJ; Hopwood DA; Sherman DH Gene 1994, 141 (1), 141–2. [DOI] [PubMed] [Google Scholar]
- (5).Motamedi H; Hutchinson CR Proc. Natl. Acad. Sci. U. S. A 1987, 84 (13), 4445–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Malpartida F; Hopwood DA Nature 1984, 309 (5967), 462–4. [DOI] [PubMed] [Google Scholar]
- (7).Harris TM; Harris CM; Hindley KB Fortschr Chem. Org. Naturst 1974, 31 (0), 217–82. [DOI] [PubMed] [Google Scholar]
- (8).Tsai SC; Ames BD Methods Enzymol. 2009, 459, 17–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Shakya G; Rivera H Jr.; Lee DJ; Jaremko MJ; La Clair JJ; Fox DT; Haushalter RW; Schaub AJ; Bruegger J; Barajas JF; White AR; Kaur P; Gwozdziowski ER; Wong F; Tsai SC; Burkart MD J. Am. Chem. Soc 2014, 136 (48), 16792–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Bull JA; Croft RA; Davis OA; Doran R; Morgan KF Chem. Rev 2016, 116 (19), 12150–12233. [DOI] [PubMed] [Google Scholar]
- (11).Burkhard JA; Tchitchanov BH; Carreira EM Angew. Chem., Int. Ed 2011, 50 (23), 5379–82. [DOI] [PubMed] [Google Scholar]
- (12).Burkhard JA; Wuitschik G; Plancher JM; Rogers-Evans M; Carreira EM Org. Lett 2013, 15 (17), 4312–5. [DOI] [PubMed] [Google Scholar]
- (13).Burkhard JA; Wuitschik G; Rogers-Evans M; Muller K; Carreira EM Angew. Chem., Int. Ed 2010, 49 (48), 9052–67. [DOI] [PubMed] [Google Scholar]
- (14).McLaughlin M; Yazaki R; Fessard TC; Carreira EM Org. Lett 2014, 16 (16), 4070–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Rogers-Evans M; Knust H; Plancher JM; Carreira EM; Wuitschik G; Burkhard J; Li DB; Guerot C Chimia 2014, 68 (7–8), 492–9. [DOI] [PubMed] [Google Scholar]
- (16).Ruider SA; Muller S; Carreira EM Angew. Chem., Int. Ed 2013, 52 (45), 11908–11. [DOI] [PubMed] [Google Scholar]
- (17).Wuitschik G; Rogers-Evans M; Buckl A; Bernasconi M; Marki M; Godel T; Fischer H; Wagner B; Parrilla I; Schuler F; Schneider J; Alker A; Schweizer WB; Muller K; Carreira EM Angew. Chem., Int. Ed 2008, 47 (24), 4512–5. [DOI] [PubMed] [Google Scholar]
- (18).Wuitschik G; Rogers-Evans M; Muller K; Fischer H; Wagner B; Schuler F; Polonchuk L; Carreira EM Angew. Chem., Int. Ed 2006, 45 (46), 7736–9. [DOI] [PubMed] [Google Scholar]
- (19).Grimm A; Madduri K; Ali A; Hutchinson CR Gene 1994, 151 (1–2), 1–10. [DOI] [PubMed] [Google Scholar]
- (20).Bao W; Sheldon PJ; Hutchinson CR Biochemistry 1999, 38(30), 9752–7. [DOI] [PubMed] [Google Scholar]
- (21).Bao W; Sheldon PJ; Wendt-Pienkowski E; Hutchinson CR J. Bacteriol 1999, 181 (15), 4690–4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Rajgarhia VB; Priestley ND; Strohl WR Metab. Eng 2001, 3 (1), 49–63. [DOI] [PubMed] [Google Scholar]
- (23).Yang K; Eyobo Y; Brand LA; Martynowski D; Tomchick D; Strauss E; Zhang HJ Bacteriol. 2006, 188 (15), 5532–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Davies C; Heath RJ; White SW; Rock CO Structure 2000, 8 (2), 185–95. [DOI] [PubMed] [Google Scholar]
- (25).Sachdeva S; Musayev F; Alhamadsheh MM; Neel Scarsdale J; Tonie Wright H; Reynolds KA Bioorg. Chem 2008, 36 (2), 85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.