Abstract
The epigenetic information present in mammalian gametes and whether it is transmitted to the progeny are relatively unknown. We find that many promoters in mouse sperm are occupied by RNAPII and Mediator. The same promoters are accessible in GV and MII oocytes, and preimplantation embryos. Sperm distal ATAC-seq sites containing motifs for various transcription factors are conserved in monkeys and humans. ChIP-seq analyses confirm that Foxa1, ERα, and AR occupy distal enhancers in sperm. Accessible sperm enhancers containing H3.3/H2A.Z are also accessible in oocytes and pre-implantation embryos. Furthermore, their interactions with promoters in the gametes persist during early development. Sperm- or oocyte-specific interactions mediated by CTCF/cohesin are only present in the paternal or maternal chromosomes, respectively, in the zygote and 2-cell stages. These interactions converge in both chromosomes by the 8-cell stage. Thus, mammalian gametes contain complex patterns of 3D interactions that can be transmitted to the zygote after fertilization.
Keywords: Chromatin, Transcription, Enhancer, Sperm, Oocyte, Foxa1, Androgen, Estrogen, Fertilization, Development
Graphical Abstract
eTOC Blurb
Jung et al. show that sperm and oocyte promoters and enhancers contain ATAC-seq accessible sites suggesting the presence of RNAPII and Foxa1, ERα, and AR. Accessibility in gametes persist in early embryos. Interactions mediated by CTCF in gametes are inherited from each parent and become the same in 8-cell embryos.
INTRODUCTION
Terminally differentiated gametes are the foundational material from which a complete organism is formed. Emerging evidence suggests that phenotypes induced by environmental cues or exposure to toxicants can be inherited by individuals in subsequent generations (Heard and Martienssen, 2014). Environmental factors may affect the epigenome during gametogenesis, since this process provides an opportune window for epigenetic reprogramming of the germline when chromatin undergoes major alterations of epigenetic information. However, it remains unclear how acquired phenotypes can be maintained through the reprograming events taking place in the germline and after fertilization in order to be transferred from parents to offspring. Studies examining sperm chromatin revealed that sperm retain nucleosomes and post-translationally modified histones in different regions of the genome (Brykczynska et al., 2010; Carone et al., 2014; Hammoud et al., 2009). Analyses of nucleosome retention in mouse sperm using ATAC-seq and distribution of covalent histone modifications using native ChIP-seq concluded that approximately 60% of all promoters are in an active epigenetic state, including well positioned nucleosomes with H3K4me3, H3K27ac, H3K9ac, and H3K36me3 (Jung et al., 2017). Sperm also retain nucleosomes containing H3K4me1 and H3K27ac at specific intergenic or intronic regions, suggesting that the sperm genome may contain epigenetically active enhancers despite the absence of transcription (Jung et al., 2017). Furthermore, the presence of regions protected from the insertion of Tn5 transposase suggests that some transcription factors may remain bound to sperm DNA. This has been confirmed by mapping the presence of CTCF and Smc1 using ChIP-seq. Results from these analyses suggest that CTCF and cohesin are bound to the sperm genome in many of the same sites where these proteins are present in the genome of embryonic stem cells (ESCs) (Jung et al., 2017). Analysis of Hi-C data in sperm indicates the presence of CTCF/cohesin loops preferentially formed when CTCF sites are present in convergent orientation, and many of these loops are also present in ESCs and somatic cells. Sperm chromatin is folded into A and B compartmental domains corresponding to regions containing active or repressive histone modifications, respectively (Jung et al., 2017). It is possible that this wealth of epigenetic information present in the sperm is a leftover of prior gene expression during sperm maturation without functional significance during embryonic development. However, it is also possible that proteins bound to the sperm genome play an instructive role in the regulation of transcription in the early embryo.
Here we explore the state of mouse sperm chromatin and find that many sperm promoters contain components of the transcription complex, including RNA polymerase II (RNAPII) phosphorylated in Ser2 and Ser5, and the Mediator complex. Footprint analyses using ATAC-seq data suggest the presence of specific transcription factors, and these factors are conserved in syntenic regions of the macaque and human sperm genomes. In the female germline, based on the presence of ATAC-seq sites containing the CTCF motif, CTCF sites appear to be present in the GV oocyte, where they form loops and, surprisingly, remain in mature MII oocytes without contributing to the 3D organization of metaphase chromosomes at this stage. CTCF-dependent organization is passed on to the zygote in a parent-of-origin-dependent manner until the 8-cell stage, where CTCF loops become the same in both chromosomes. Furthermore, enhancer-promoter interactions mediated by transcription factors in the gametes also persist in the early embryo. These findings suggest that the contribution of information contained in sperm and oocyte chromatin to early embryonic development is greater than was previously recognized, suggesting acquired phenotypes in the parents may be transmitted through the germ line in the form of DNA-bound proteins and their interactions.
RESULTS
Components of the transcription complex are present in sperm promoters
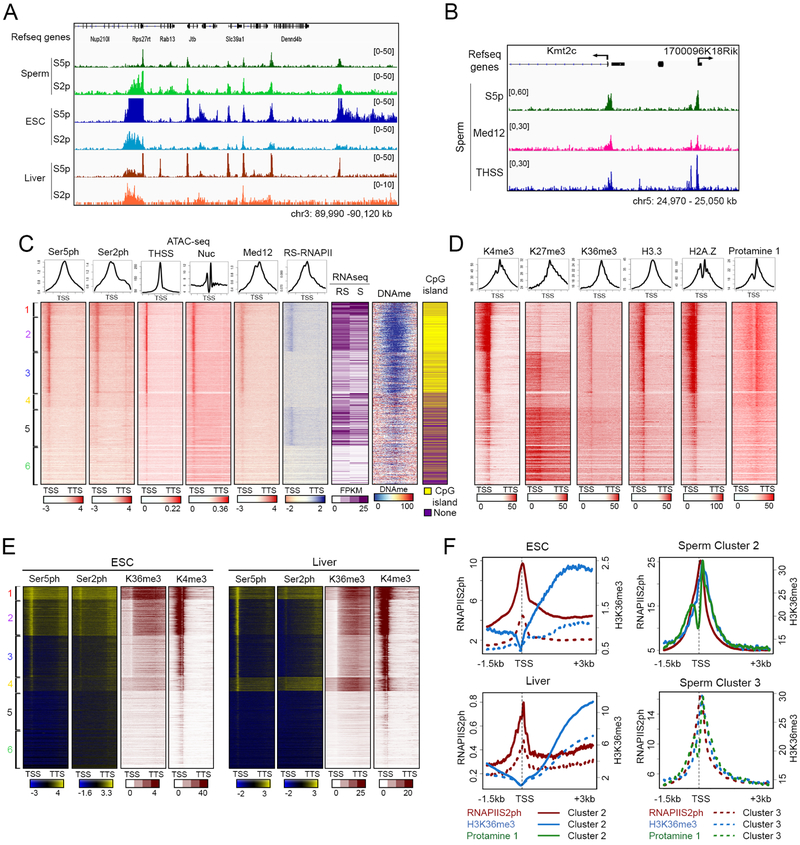
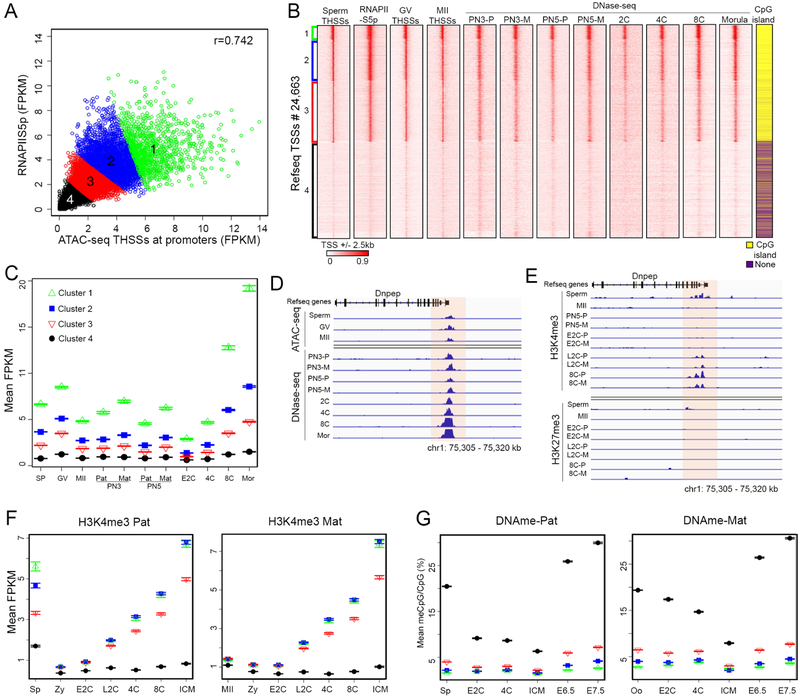
We recently reported that approximately 60% of TSSs of annotated genes in sperm contain a nucleosome-free region flanked by well-positioned nucleosomes marked by active histone modifications (Jung et al., 2017). The presence of a hypersensitive region between the −1 and +1 nucleosomes raises the question of whether TSSs in sperm are occupied by the transcription machinery. To explore this issue, we first investigated the presence of RNAPII in sperm by performing Western analysis using antibodies to RNAPII phosphorylated in Ser5 (RNAPIISer5ph) and RNAPII phosphorylated in Ser2 (RNAPIISer2ph). Sperm was isolated from the cauda epididymis with a purity of less than 1 somatic cells per 1000 sperm. Western analyses of histone H3 and Protamine 1 in sperm and somatic cells are in agreement with a pure population of sperm cells (Figure S1A). Results from Western analyses show that both forms of RNA polymerase are present in sperm (Figure S1B). Furthermore, results from immunofluorescence confocal microscopy also indicate that RNAPIISer5ph and RNAPIISer2ph are present in the sperm nucleus, and the observed staining can be competed with peptides used to prepare the corresponding antibodies (Figure S1C). Staining with a non-specific IgG or with secondary antibodies failed to detect any signal (Figure S1D). Finally, immunofluorescence confocal microscopy of testis cryosections show the presence of RNAPIISer5ph and RNAPIISer2ph in mature sperm present in the seminiferous tubules (Figure S1E). We therefore performed ChIP-seq in sperm with antibodies to these two phosphorylated forms of RNAPII and identified 12,740 and 13,547 peaks of RNAPIISer5ph and RNAPIISer2ph, respectively (Table S1). Analysis of their genome-wide distribution with respect to gene features indicates that a large number of RNAPIISer5ph and RNAPIISer2ph sites are located at TSSs, and their distribution is similar to that in ESCs or somatic cells of the liver (Figure 1A). Both forms of RNAPII are also located at distal intergenic regions (Figures S1F and S1G), where they overlap with H3K4me1 and H3K27ac, suggesting they may represent active enhancers (Figure S1H). To further explore the significance of the distribution of RNAPII in the mouse sperm genome, we performed ATAC-seq using the Omni-ATAC protocol, which allows a higher signal to noise ratio when generating chromatin accessibility profiles (Corces et al., 2017). We obtained two biological replicates (Table S1) and separated ATAC-seq reads into the 50–115 bp range, corresponding to sites where bound proteins protect the DNA from the Tn5 transposase (Tn5 hypersensitive sites, THSSs), and the 180– 247 bp range corresponding to mono-nucleosomes (Schep et al., 2015). We identified a total of 61,238 THSSs. We then used transcription start sites (TSSs) and transcription termination sites (TTSs) of all mouse annotated genes as anchors and carried out k-means clustering using the reads of RNAPIISer5ph and RNAPIISer2ph ChIP-seq (Figure 1C). Results show that approximately 14,756 TSSs are enriched with RNAPIISer5ph and RNAPIISer2ph (clusters 1–4), and the levels of RNAPII at these TSSs correlate with the accessibility to Tn5 transposase and the nucleosome signal obtained by ATAC-seq (Figure 1C). TSSs occupied by RNAPSer5ph and RNAPIISer2ph are hypo-methylated on DNA and are enriched in CpG islands. Using the same TSS order as in Figure 1C, we find that flanking nucleosomes carry the active histone modifications H3K4me3 and H3K36me3 (Figure 1D). We also performed ChIP-seq with the histone variants H3.3 and H2A.Z (Table S1) and found that these nucleosomes also contain H3.3 and H2A.Z, while the +1 nucleosome is enriched in Protamine 1 (Figure 1D). Interestingly, TSSs in cluster 3 are also marked by H3K27me3, suggesting a bivalent state (Figure 1D).
Figure 1. RNAPII and Mediator are present at sperm promoters.
(A) Genome browser view showing the distribution of RNAPIISer5ph and RNAPIISer2ph in a specific region of the mouse sperm genome. As comparison, RNAPIISer5ph and RNAPIISer2ph in ESCs and liver are shown.
(B) Genome browser view showing Med12 distribution with respect to that of RNAPIISer5ph and ATAC-seq THSSs in a specific region of the mouse sperm genome.
(C) Heatmaps and average plots showing RNAPIISer5ph and RNAPIISer2ph enrichment at all mouse Refseq annotated genes. Sites are ordered by k-means clustering of RNAPIISer5ph and RNAPIISer2ph signal between TSSs and TTSs. Enrichment of RNAPII in different clusters correlates with enrichment of Med12 and ATAC-seq THSS signal. As comparison, pan-RNAPII signal and RNA-seq in round spermatids and mature sperm are shown. RS, round spermatid; S, mature sperm.
(D) Heatmaps and average plots showing sperm histone modifications H3K4me3, H3K27me3 and H3K36me3, histone variants H3.3 and H2A.Z, and protamine 1. Clusters and gene order are the same as in Figure 1C.
(E) Enrichment of RNAPIISer5ph, RNAPIISer2p, H3K36me3, and H3K4me3 in ESCs and liver at gene promoters clustered in the same order as in Figure 1C.
(F) Average profiles of RNAPIISer2ph, H3K36me3 and protamine 1 for clusters 2 and 3 shown in Figure 1E.
See also Figure S1
Mediator-promoter interactions stabilize the formation of transcription complexes, helping to position the RNAPII complex at the TSS. To test whether Mediator is also present in sperm, we performed Western and immunofluorescence confocal microscopy analyses using antibodies to the Med12 subunit and confirmed the presence of Mediator in sperm nuclei (Figures S1B and S1C). We then performed ChiP-seq with Med12 antibodies (Table S1). Results show that Med12 co-localizes with RNAPIISer5ph and RNAPIISer2ph at specific gene promoters (Figure 1B) and it is present at the same TSSs in the sperm genome (Figure 1C). Taken together, these results suggest that, in addition to nucleosomes, sperm also retain components of the transcription complex at approximately 60% of all annotated promoters despite the lack of transcription.
To test whether the presence of RNAPII in mature sperm correlates with transcriptional activity at earlier stages during spermatogenesis, we compared mature sperm RNAPII ChIP-seq data with published RNAPII ChIP-seq and RNA-seq in round spermatids. Results show that the presence of RNAPII in sperm does not correlate well with RNAPII or RNA levels in round spermatids. For example, TSSs in clusters 1 and 2 also contain RNAPII and are transcribed in round spermatids, but TSSs in clusters 3 and 4 lack RNAPII in round spermatids, and TSSs in cluster 5 are transcribed and contain RNAPII in these cells but not in sperm (Figure 1C). These results suggest that some or all of the accessible promoters enriched with RNAPII in sperm may prepare genes for expression at later stages of development after fertilization. Interestingly, most RNAPIISer2ph and H3K36me3 in sperm appear to be present at TSSs but not extending through gene bodies (Figures 1C and 1D). This is distinct from the canonical patterns of RNAPIISer2ph and H3K36me3 presence throughout coding regions, including the 3’ end of transcribed genes, and correlating with active elongation. To explore differences in the location of RNAPII and H3K36me3 between sperm and cell types undergoing active transcription, we compared their distribution in sperm with that in ESCs and liver cells using published data from these two cell types (Bunch et al., 2014; Ji et al., 2015; Li et al., 2014; Wamstad et al., 2012). In ESCs, RNAPIISer5ph and RNAPIISer2ph are present at TSSs in clusters 1–2 but extend into the gene bodies in ESCs and liver cells, coincident with enrichment of H3K36me3 through the genes (Figures 1E and 1F). Therefore, although sperm promoters are poised in an active chromatin state with RNAPII, Mediator, and active histone modifications, these promoters are not transcribed. One possible explanation for the lack of transcription in sperm despite the apparent similarity of the chromatin state to that of transcriptionally active cell types is the presence of protamines on sperm DNA, specifically enriched just downstream of the TSS and RNAPII (Figures 1D and 1F), raising the possibility that protamines may be involved in preventing poised RNAPII in sperm from elongating into the gene body.
Conservation of the sperm chromatin regulatory landscape in monkeys and humans
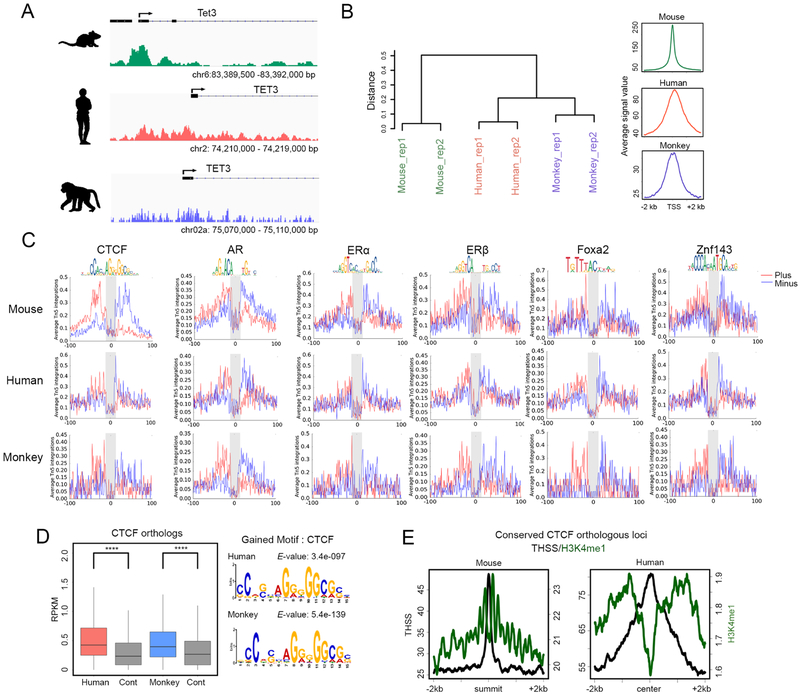
Analysis of accessible regions detected by subnucleosome-sized reads obtained by Omni-ATAC-seq indicates the occurrence of thousands of THSSs at non-TSS sequences, suggesting the presence of DNA binding proteins at promoter-distal regions, including introns and intergenic regions. We identified 50,082 distal THSSs corresponding to putative transcription factor binding sites excluding TSSs in sperm, and searched these sites for DNA footprints of known transcription factor binding motifs using Wellington (Piper et al., 2013). We found significant enrichment of DNA footprints for numerous transcription factors (TFs), including members of the Forkhead family such as Foxa1, Zn finger proteins such as CTCF and Znf143, and nuclear hormone receptors including AR and ER (Figure S2A). To test if this accessibility landscape may have functional significance, we explored whether it is conserved in sperm of other mammalian species. We performed Omni-ATAC-seq with sperm samples from humans and rhesus macaques (M. mulatta). We obtained two independent biological replicates for each species with high correlation coefficients (Table S1). We therefore combined replicates and analyzed reads separately in the 50–115 bp and 180–247 bp ranges, corresponding to the presence of TFs and mono-nucleosomes, respectively. We identified 89,713 THSSs in human and 73,384 THSSs in monkey. We then identified conserved THSSs around TSSs corresponding to the presence of the transcription complex between the +1 and −1 nucleosomes as described above. We considered only genes having syntenic locations in all three species. Tet3 is an example of a conserved gene with a THSS at the promoter in all three-species analyzed (Figure 2A). GO term analysis of genes whose TSS accessibility is maintained among mice, monkeys, and humans are enriched in terms for embryonic development. Hierarchical clustering of homologous genes by accessibility indicates that humans show greater similarity to monkeys than mice (Figure 2B), consistent with the evolutionary distances between the three species. Furthermore, analysis of footprints identified using ATAC-seq reads corresponding to distal THSSs indicates that human and monkey sperm also contain footprints for numerous transcription factors, including CTCF and Znf143, members of the Fox family, and androgen and estrogen receptors (Figure 2C).
Figure 2. Conservation of the sperm chromatin regulatory landscape in mammals.
(A) Genome browser view showing ATAC-seq THSS signal between species at a syntenic region of the genome.
(B) Hierarchical clustering of genes in syntenic regions of the genome by ATAC-seq accessibility between mouse, monkey and human.
(C) Average footprint profile of ATAC-seq THSSs at regions containing the given TF-binding motif.
(D) Boxplots showing the RPKM of ATAC-seq THSSs present at distal accessible regions in mouse sperm containing the CTCF motif in syntenic regions of human and monkey. The motifs found at these syntenic regions in monkey and human are also shown.
(E) Average profiles of ChIP-seq signal for H3K4me1 mouse and human sperm relative to conserved CTCF syntenic loci.
See also Figure S2
To gain further insight into the significance of the conservation of transcription factor binding sites across mammalian species, we first examined mouse CTCF ChIP-seq peaks and determined whether human and monkey sperm have THSSs at CTCF syntenic sites. We first defined syntenic locations of mouse CTCF in human and monkey using LiftOver (mm9 to hg19 and hg19 to rheMac8). Notably, CTCF syntenic locations in both species showed significant chromatin accessibility compared to random control sequences, and consensus CTCF motifs were also found at these conserved sequences (Figure 2D). Furthermore, CTCF syntenic sites are flanked by nucleosomes containing H3K4me1 in mouse and human sperm (Figure 2E). Similar analyses show conservation of Foxa1 and Znf143 sites (Figure S2B). Finally, distal THSSs present in human sperm overlap with previously defined tissue-specific enhancers (Figure S2C) (Gao et al., 2016). Taken together, these observations suggest that putative protein binding sites in mammalian sperm chromatin are evolutionarily conserved among mouse, monkey and human, suggesting that transcription factors may remain bound to the sperm genome and may have functional relevance during embryonic development.
The pioneer factor Foxa1 co-localizes with nuclear receptors at putative sperm enhancers
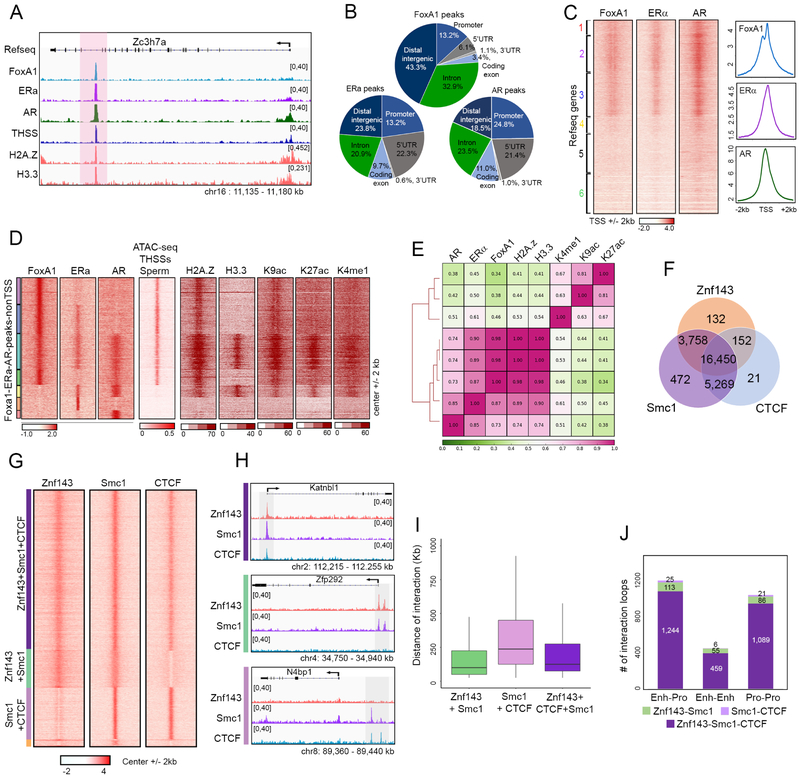
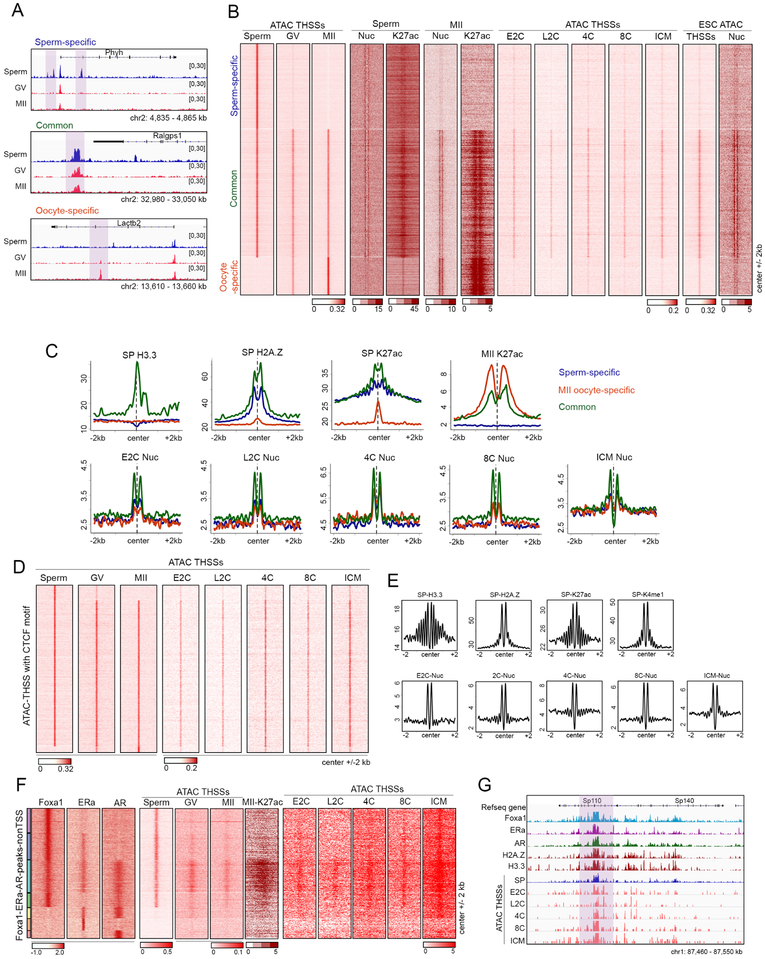
Analysis of footprints generated by ATAC-seq subnucleosome-sized reads suggests the presence of transcription factors bound to the sperm genome, including the pioneer factor Foxa1 and the estrogen and androgen receptors, both of which require Foxa1 for recruitment to chromatin (Hurtado et al., 2011). To directly test the possibility that Foxa1 is bound to the sperm genome, we first confirmed its presence in sperm nuclei using western analysis (Figure S3A). Immunofluorescence confocal microscopy also shows the presence of this protein in sperm nuclei, and the observed staining can be competed with peptides used to prepare the corresponding antibodies (Figure S3B). Furthermore, immunofluorescence confocal microscopy of testis cryosections show the presence of Foxa1 in mature sperm present in the seminiferous tubules (Figure S3C). We then carried out ChIP-seq with antibodies to Foxa1 (Table S1) and identified 7,116 peaks in the mouse sperm genome (Figure 3A). Some of these sites are present at promoters, but most are located in intergenic regions or introns, likely corresponding to putative enhancers (Figure 3B). Analysis of motifs present at Foxa1 binding sites indicates the presence of binding sequences for estrogen, androgen, and glucocorticoid receptors, Grainyhead Like Transcription Factor 1, Nuclear Factor I X (also known as CCAAT-Binding Transcription Factor), and Transcription Factor AP-2 Gamma (Figure S3D). Since Foxa1 has been shown to facilitate binding of nuclear hormone receptors such as ERα and AR, we confirmed that ERα and AR are present in sperm using western analysis (Figure S3A) and immunofluorescence confocal microscopy (Figure S3B and S3C). We then examined the genomic distribution of ERα and AR using ChIP-seq (Figure 3A and Table S1). Like Foxa1, ERα and AR sites are located at promoters, introns and distal intergenic regions (Figure 3B). The three proteins, Foxa1, ERα and AR, co-localize at 52% of all annotated TSSs in the mouse genome (Figure 3C). These proteins can also be found together at a large fraction of TSSs in somatic cells (Figure S3E).
Figure 3. Foxa1, nuclear hormone receptors, and Znf143 are present in sperm chromatin.
(A) Genome browser view showing co-localization of Foxa1, ERα, and AR in a region of the mouse sperm genome.
(B) Genome-wide distribution of Foxa1, ERα, and AR peaks identified by MACS. TSS ± 500 bp are listed as promoters.
(C) Heatmaps and average profiles showing Foxa1, ERα, and AR ChIP-seq signal around TSSs (± 2 kb).
(D) Heatmaps showing Foxa1, ERα, and AR ChIP-seq signal at ATAC-seq distal THSS non-TSS peaks. Clusters derived by k-means clustering of ChIP-seq reads. Histone modifications related to potential enhancers and histone variants at these sites are also shown (center ± 2 kb).
(E) Pearson’s correlation of Foxa1, ERα and AR ChIP-seq read density genome-wide with histone modifications related to potential enhancers and histone variants. Clustering method is hierarchical complete linkage.
(F) Venn diagram showing overlap of non-TSS Znf143 peaks with Smc1 and CTCF.
(G) Heatmaps showing Znf143, Smc1, and CTCF at distal non-TSS peaks from ChIP-seq signal for each protein. Clusters were derived by k-means clustering of ChIP-seq reads.
(H) Genome browser views of specific examples corresponding to the three types of sites shown in Figure 3G.
(I) Box plot showing the distance of interactions mediated by Znf143, Smc1, and CTCF combinations present at anchors. These significant interactions are defined by FitHi-C using sperm Hi-C data.
(J) Number of interaction loops defined by HiCCUPS mediated by combinations of Znf143, Smc1, and/or CTCF. Enhancer-promoter interactions are defined based on the presence of H3K27ac and TSS ± 500 bp.
See also Figure S3
In addition to TSSs, Foxa1, ERα, and AR are also present at a large number of distal sites in the genome. To analyze in detail the relative distribution of these three proteins, we combined the summits of all distal peaks for the three proteins and used them as anchors to perform k-means clustering. Results show that Foxa1 is present at many distal sites containing ERα and AR (Figure 3D). A subset of these sites contains all three proteins, but other sites contain only ERα or AR, others contain Foxa1 alone, and yet others contain ERα and/or AR but lack Foxa1 (Figure 3D). In somatic cells, Foxa1 preferentially binds to lineage-specific enhancers that are defined by H3K27ac, H3K4me1, and histone variant H2A.Z (Jozwik et al., 2016). In sperm, all distal Foxa1 sites are flanked by nucleosomes containing H2A.Z, and those including ERα and AR also contain H3.3, H3K9ac, H3K27ac, and H3K4me1 (Figure 3D). To further analyze the relationship among these proteins and histone modifications, we used ChIP-seq data to perform unsupervised hierarchical cluster analysis and the results are shown in Figure 3E. These results suggest that Foxa1 and associated proteins may remain bound to sperm chromatin to mark enhancer elements involved in the regulation of gene expression in different tissues of the developing embryo. In agreement with this hypothesis, Foxa1 sites in sperm overlap with enhancers defined in embryonic stem cells (ESCs), neural precursor cells (NPCs), heart, lung, kidney and other cell types of the developing embryo (Figure S3F) (Gao et al., 2016).
Znf143 colocalizes with CTCF and cohesin and is involved in loop formation
Analysis of footprints using ATAC-seq subnucleosome-sized reads suggests the presence of multiple Zn finger proteins bound to the mouse sperm genome (Figure S2A). One of these proteins, Znf143, is of special interest because it has been shown to co-localize with CTCF and cohesin, and to mediate interactions between promoters and distal regulatory sequences in somatic cells (Bailey et al., 2015). Western analysis of sperm protein extracts supports the presence of this protein in mouse sperm (Figure S3A). To examine the distribution of Znf143 in mouse sperm, we performed ChIP-seq with antibodies to this protein (Table S1). We observed a total of 20,492 peaks for Znf143, most of which overlap with those of CTCF, Smc1, or both (Figure 3F). Many of these peaks are present at promoters, but also in introns and distal intergenic regions and are enriched in motifs for Znf143 and CTCF. To examine the relative distribution of these three proteins in more detail, we combined all the peaks and used them as anchors to perform k-means clustering of Znf143, Smc1 and CTCF ChIP-seq signal. Most Znf143 peaks overlap with Smc1 and CTCF, and small fractions contain Znf143 and Smc1 or CTCF and Smc1 (Figure 3G). Therefore, most CTCF/cohesin sites in the sperm genome also contain Znf143. Examples of each class are shown in Figure 3H.
To analyze the possible role of Znf143 in CTCF/cohesin-mediated interactions with high resolution, we performed in situ Hi-C with the restriction enzyme DpnII and obtained two biological replicates with a total of 520 million intra-chromosomal contacts (Table S2). We then employed FitHi-C (Ay et al., 2014) to define significant interactions in sperm and we examined the presence of Znf143, CTCF and Smc1 at anchors mediating these interactions. Genomic sites containing the observed combinations of these proteins mediate interactions in the sperm genome, including sites containing Znf143 and Smc1, CTCF and Smc1, and Znf143, CTCF and Smc1 (Figure 3I). Interactions mediated by Znf143/Smc1 take place over shorter distance ranges than those mediated by CTCF/Smc1 or the combination of the three proteins (Figure 3I). Furthermore, most contacts in sperm contain all three proteins at interacting anchors, and interacting sites containing Znf143 are present at both enhancers and promoters (Figure 3J). Examples of the different types of interactions are shown in Figure S3G.
GV and MII stage oocytes contain accessible sites corresponding to specific transcription factors
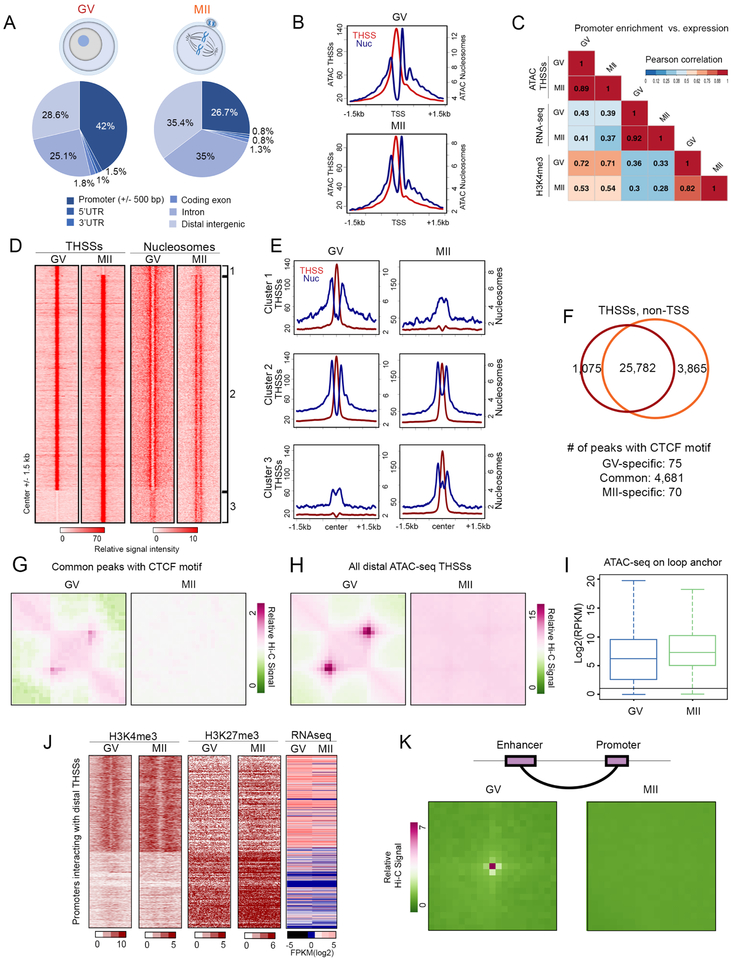
To examine whether female gametes also contain accessible promoters and putative enhancers, we performed Omni-ATAC optimized for a small number of nuclei in mouse oocytes at the GV and MII stages of oogenesis. During fetal development, the mammalian oocyte enters meiosis and is arrested at the diplotene stage of prophase I, termed the germinal vesicle (GV) stage, from birth to puberty. After a hormone surge, the oocyte undergoes meiotic maturation and is arrested again in metaphase of meiosis II (MII), which is the mature stage fertilized by sperm (Von Stetina and Orr-Weaver, 2011). We generated biological replicates of Omni-ATAC-seq libraries using 300–400 oocytes for each sample (Table S1). For subsequent analyses, we used a previously published list of oocyte TSSs that reflect the usage of alternative promoters in this cell type (Veselovska et al., 2015). Using MACS and THSS reads, we identified 28,439 and 31,597 THSS peaks in GV and MII oocytes, respectively. These sites are distributed across various gene features such as promoters, introns, and distal intergenic regions (Figure 4A). Therefore, condensed MII oocyte chromosomes contain a rich map of accessible sites similar to those of GV oocytes. Using nucleosome-size reads to map the location of nucleosomes, we find that oocytes at the GV and MII stages contain accessible TSSs flanked by well-positioned +1 and −1 nucleosomes (Figure 4B). Furthermore, using previously published H3K4me3 ChIP-seq data (Dahl et al., 2016; Zhang et al., 2016)}, we find a good correlation between promoter accessibility and the presence of H3K4me3 (Figure 4C). Specific examples of genes with different degrees of correlation between these three features are shown in Figure S4A. Taken together, these results suggest that a large number of promoters are accessible in GV oocytes, and these promoters remain accessible in MII metaphase chromosomes. Although we have no direct evidence to suggest that accessible promoters in oocytes contain components of the transcription complex, the good correlation observed in sperm between ATAC-seq THSSs and ChIP-seq signal for RNAPII and Med12 can be taken to suggest that these or other components of the transcription complex may also be present at oocyte promoters.
Figure 4. Differences in the distribution of TFs and their interactions in GV and MII oocytes.
(A) Genome-wide distribution of ATAC-seq THSSs from GV and MII oocytes.
(B) Average profiles of ATAC-seq THSSs and nucleosome signals from GV and MII oocytes at TSSs.
(C) Pearson’s correlation of ATAC-seq THSSs with RNA-seq and H3K4me3 ChIP-seq at promoters in GV and MII oocytes.
(D) Heatmaps showing ATAC-seq THSSs and nucleosome signal at distal non-TSS peaks. Clusters derived by k-means clustering of ATAC-seq THSS reads. Cluster 1, GV-specific; Cluster 2, common between GV and MII; Cluster 3, MII-specific.
(E) Average profiles of ATAC-seq THSS and nucleosome signals from GV and MII oocytes at non-TSS peaks corresponding to clusters shown in Figure 4D.
(F) Venn diagram showing overlap of distal ATAC-seq THSS non TSS peaks between GV and MII oocytes. The lower part of the panel shows the number of peaks containing the CTCF motif.
(G) Metaplots of GV and MII oocyte Hi-C significant interactions between anchors occupied by ATAC-seq THSSs (cluster 2 in Figure 4D) containing the CTCF motif.
(H) Metaplots of GV and MII oocyte Hi-C significant interactions between anchors occupied by all distal ATAC-seq THSSs present in GV oocytes.
(I) Boxplot comparing ATAC-seq THSS reads on the loop anchors shown in Figure 4H between GV and MII oocytes. RPKM; Reads Per Kilobase Million
(J) Heatmaps showing RNA-seq, H3K4me3, and H3K27me3 ChIP-seq from GV and MII oocytes at gene promoters interacting with common THSSs (cluster 2 in Figure 4D) analyzed in Figure 4K.
(K) Metaplots of GV and MII oocyte Hi-C significant interactions between enhancers defined by the presence of THSSs and H3K27ac, and active promoter containing H3K4me3.
See also Figure S4
In addition to THSSs present at the promoter, GV and MII oocytes contain a large number of THSSs in distal intergenic regions and introns (Figure 4A). We identified 25,782 distal THSSs shared by the two stages, 1,075 THSSs only present in the GV stage, and 3,865 THSSs newly gained in the MII stage (Figure 4D). These sites are flanked by well positioned nucleosomes in both stages (Figures 4D and 4E), and these nucleosomes lack H3K4me3 but contain H3K27ac in MII oocytes, suggesting they may correspond to enhancers in an active state (Figure S4B). In agreement with this hypothesis, these sites have been identified as enhancers in various tissues of the developing embryo (Figure S4C). To gain insights into the nature of the transcription factors bound at these sites, we performed motif discovery analyses using common, GV-, and MII-specific distal THSSs using MEME (Figure S4D). The most prevalent motifs at GV-specific peaks correspond to the transcription factors Nr5a2, Esrra, Sp1, and Gata3. MII-specific peaks i.e. those appearing de novo as the oocyte goes into metaphase, correspond to the AP-1 factors Fos and JunB, Runx1, and Zfp691, whereas common peaks contain Junb, CTCF, and Nr5a2 (Figure S4D). CTCF, Nr5a2, and Esrra are known to be essential proteins for early development (Wu et al., 2016). Although we lack direct evidence for the presence of these transcription factors at oocyte THSSs, the results raise the intriguing possibility that transcription factors that are important for early embryo development may remain bound to the maternal chromatin through meiosis.
Of the 25,782 THSSs found in both GV and MII oocytes at distal sites, 4,681 contain the CTCF motif (Figure 4F). This is surprising, since CTCF has been shown to be evicted from metaphase chromosomes (Oomen et al., 2019), concomitant to the dramatic reorganization of chromosome structure that accompanies entry into mitosis (Gibcus et al., 2018). Analysis of 3D chromatin organization using Hi-C suggests that GV oocytes show the typical higher-order structures, such as TADs and compartments, whereas MII oocytes lack these structures and, instead, show a similar organization to that observed in mitotic somatic cells (Du et al., 2017). Although it is formally possible that a different protein, rather than CTCF, is bound to THSSs containing the CTCF motif, we asked whether these sites present in GV and MII oocytes are associated with the formation of loops by analyzing published Hi-C data from these stages of oogenesis using HiCCUPS (Durand et al., 2016b). Remarkably, although chromatin accessibility at many putative CTCF binding sites in GV oocytes is preserved in the MII stage, chromatin interactions mediated by these common sites are lost in MII oocytes (Figure 4G). We also examined Hi-C-derived interaction loops anchored by all distal ATAC-seq THSSs. Again, interaction loops present in the GV stage were not detectable in MII oocytes (Figure 4H), even though accessibility at THSSs present at loop anchors defined in GV show no change between the two stages (Figure 4I). To further analyze changes in interactions between the GV and MII oocytes, we defined active promoters as TSSs containing H3K4me3 and lacking H3K27me3, and inactive as those containing opposite histone modifications (Figure 4J). Promoters in an active epigenetic state in GV oocytes maintain this state in MII, and their activity correlates with levels of RNA in these two stages (Figure 4J). Specific examples are shown in Figure S4E. We also defined putative enhancers as the THSSs present in cluster 2 in Figure 4D. We then identified significant interactions in the Hi-C data using FitHi-C and determined enhancer-promoter interactions. We find that putative enhancers contact active promoters in GV oocytes, but these interactions disappear in the MII stage (Figure 4K), although transcription factors and the transcription complex remain bound to these sequences in MII chromosomes. Taken together, these results suggest that compaction of chromosomes in MII oocytes does not affect binding of most proteins present in the GV stage, but interactions mediated by these proteins, including CTCF or a putative different protein that binds to the same motif, as well as other distal sites presumed to be enhancers, are reprogramed during chromosome condensation.
Chromatin accessibility at promoters in the gametes persists in preimplantation embryos
It is possible that promoters in an active state in the gametes play an instructional role in establishing transcription early in embryogenesis. To test this hypothesis, we first examined the correlation between ATAC-seq accessibility of sperm promoters and the levels of RNAPIISer5ph, and found a high degree of correlation, suggesting that the strength of THSSs peaks at promoters may be used as a direct measure of transcription complex occupancy (Figure 5A). We then classified sperm TSSs into four groups based on THSS signal and RNAPIISer5ph levels (Figure 5A). We compared ATAC-seq accessibility in these 4 clusters of sperm promoters with that of GV and MII oocytes. The results suggest that accessible promoters in sperm are also accessible in oocytes (Figure 5B). In order to examine whether these promoters remain accessible during embryogenesis, we made use of DNase-seq obtained at various stages of embryonic development (Lu et al., 2016). Remarkably, we find that promoters with high accessibility in sperm and oocytes, are also accessible in all examined embryonic stages from zygote to morula (Figure 5B). Promoters in cluster 1 consistently have the strongest signal, which becomes progressively weaker in clusters 2–4 with little to no signal in cluster 4, and ATAC-seq accessibility correlates well with density of CPG islands (Figure 5B). A similar result was obtained using ATAC-seq data from preimplantation embryos (Wu et al., 2016) (Figure S5A). We confirmed these qualitative observations on chromatin accessibility in promoter regions in a quantitative manner by calculating RPKM for DNase-seq and FPKM for ATAC-seq data in each stage from the gametes to the ICM. The accessibility of promoters in each cluster remains approximately the same between the gametes, the PN3/PN5 stages of the zygote, 2-cell and 4-cell stages, and increases during the 8 cell and morula stages (Figures 5C and S5B). A detailed quantitative measurement of the degree of correlation of normalized levels of DNase-seq and ATAC-seq at promoters in sperm, oocytes and preimplantation embryos is shown in Figure S5C. An example showing the maintenance of promoter accessibility between the gametes and the early embryonic stages in the Dnpep gene is shown in Figure 5D.
Figure 5. Promoter accessibility is maintained between germ cells and early embryos.
(A) Scatterplot showing the Pearson’s correlation between sperm ATAC-seq THSSs and RNAPIISer5ph ChIP-seq at promoters (TSS ± 500 bp). Four clusters were defined by k-means clustering.
(B) Heatmaps showing sperm and oocyte ATAC-seq THSSs at promoters clustered as shown in Figure 5A. Also shown is DNase-seq signal enrichment at the same promoters in different stages of pre-implantation embryos. GV= GV oocyte; MII= MII oocyte; PN3= zygote pronuclear stage 3; PN5= zygote pronuclear stage 5; 2C= 2-cell stage embryos; 4C= 4-cell stage embryos; 8C= 8-cell stage embryos; P= Paternal; M= Maternal
(C) Plot showing the average ATAC-seq or DNase-seq signal in the different clusters and cell types shown in Figure 5B. Error bars indicate the standard error about the mean. SP= sperm; Mor= Morula; Pat= Paternal; Mat= Maternal. Other symbols as in Figure 5B.
(D) Track view of ATAC-seq and DNase-seq signal at an example gene for the different cell types shown in Figure 5C. E2C= Early 2-cell stage embryo; L2C= Late 2-cell stage embryo. Other symbols as in Figure 5B.
(E) H3K4me3 and H3K27me3 ChIP-seq signal at an example gene for the different cell types shown in Figure 5C. Symbols as in Figure 5B.
(F) Plot showing average allele-specific signal of H3K4me3 ChIP-seq at the TSS clusters defined in Figure 5A. The paternal signal is shown on the left side of each panel, and maternal signal on the right. SP= sperm; Zy= zygote; E2C= Early 2-cell stage embryo; L2C= Late 2-cell stage embryo; 4C= 4-cell stage embryos; 8C= 8-cell stage embryos; ICM= Inner Cell Mass. The number of TSSs present in each cluster are as follows: Cluster 1, 1127; Cluster 2, 3196; Cluster 3, 4809; Cluster 4, 7563.
(G) Plot showing average allele-specific signal of BS-seq data at the TSS clusters defined in Figure 5A. The paternal signal is shown on the left side of each panel, and maternal signal on the right. Oo= oocytes. Other symbols as in Figure 5F. The number of TSSs present in each cluster are as follows: Cluster 1, 513; Cluster 2, 1498; Cluster 3, 2290; Cluster 4, 3325.Other symbols as in Figure 5F.
See also Figure S5
Widespread transcription in the mouse embryo does not occur until the 2-cell stage, suggesting that the existence of accessible promoters containing components of the transcription complex is not sufficient for gene expression. To understand the relationship between epigenetic modifications and accessibility at promoters, we re-analyzed published H3K4me3 and H3K27me3 data obtained in preimplantation embryos and gametes (Zhang et al., 2016; Zheng et al., 2016). For allele-specific comparisons, we distinguished between the paternal and maternal allele reads using single-nucleotide polymorphism (SNP) information and processed only normalized SNP-trackable reads in each allele (see STAR*Methods). Results show that the majority of H3K4me3 signal present in sperm accessible promoters (clusters 1–3) is lost in the zygote and begins reestablishment in the late 2 cell stage, perhaps as a consequence of the start of major zygotic genome activation (ZGA) (Figure 5F). An example of the distribution of H3K4me3 during embryonic development at the promoter of the Dnpep gene is shown in Figure 5E. H3K4me3 is present at low levels in the promoters of MII oocytes, the maternal chromosomes of the PN5 zygote, and, as is the case for the paternal chromosome, increases in the late 2 cell stage (Figure 5F). In contrast, H3K27me3 shows a negative correlation with accessibility at gene promoters in sperm, with those in cluster 4 showing highest levels of this modification (Figure S5D). Interestingly, accessible sperm promoters also loose this histone modification in the zygote, and appreciable levels are only restored in the ICM (Figure S5D). On the other hand, H3K27me3 is retained in the metaphase chromosomes of the MII oocyte and persists during all stages of early embryonic development in the maternal chromosome (Figure S5D). To further explore the relationship between promoter accessibility in the gametes and preimplantation embryos and epigenetic modifications, we examined previously published GWBS data (Jung et al., 2017; Wang et al., 2014). We find that most accessible promoters in sperm have low DNA methylation levels (clusters 1–3) and remain unmethylated until E6.5, at which time methylation levels increase slightly. Interestingly, most methylation changes in the early embryo take place in non-accessible promoters (cluster 4), which become demethylated up to the blastocyst stage and then rapidly remethylated in both maternal and paternal chromosomes (Figure 5G).
Comparison of chromatin accessibility at regulatory regions between sperm and oocytes
A large fraction of THSS ATAC-seq peaks in sperm and oocytes display enrichment in distal regions or introns, which are presumed to be regulatory elements (Figures 2C and 4A). We therefore asked whether these distal sites, which are bound by various transcription factors in sperm, are different between male and female gametes. Visual inspection of ATAC-seq data suggests the existence of common and gamete-specific distal sites (Figure 6A). Therefore, we combined the distal ATAC-seq sites in sperm, GV, and MII oocytes and used them as anchors to perform k-means clustering. We find 23,168 sperm-specific, 10,058 oocyte-specific, and 32,465 common distal THSSs (Figure 6B). All sites are flanked by well positioned nucleosomes containing H3K27ac in the corresponding cell type. Interestingly, MII oocyte-specific sites are flanked by better positioned nucleosomes containing higher levels of H3K27ac (Figure 6B and 6C). To examine whether there are other chromatin features that distinguish sperm-specific versus common sites, we examined the distribution of histone variants around these sites. Results show that sperm distal sites that are also present in oocytes contain H3.3 and their flanking nucleosomes contain H2A.Z and high levels of H3K27ac in sperm, whereas those that are sperm- or oocyte-specific lack histone H3.3 (Figure 6C).
Figure 6. Chromatin accessibility at regulatory regions in gametes and pre-implantation embryos.
(A) Track view of distal ATAC-seq THSSs in sperm, GV, and MII oocytes at allele-specific or common accessible distal regulatory regions (highlighted).
(B) Heatmaps comparing distal ATAC-seq THSSs and nucleosomes between sperm, oocytes, and pre-implantation embryos. Also shown is the distribution of H3K27ac ChIP-seq signal in sperm and MII oocytes, and ATAC-seq THSSs in preimplantation embryos and ESCs. Clusters were derived by k-means clustering of sperm and oocytes ATAC-seq THSS reads.
(C) Average profiles of ChIP-seq signal for histone variants H3.3 and H2A.Z in sperm, and H3K27ac in sperm and MII oocytes at the same distal regulatory regions as in Figure 6B. Lower panels show average plots of ATAC-seq nucleosome signals from preimplantation embryos.
(D) Heatmap showing ATAC-seq THSSs from sperm, GV and MII oocytes, and preimplantation embryos at all distal regulatory regions containing the CTCF motif.
(E) Average profiles of ChIP-seq signal for histone variants H3.3 and H2A.Z in sperm, and H327ac in sperm and MII oocytes, at the same distal regulatory regions as in Figure 6D. Average plots of ATAC-seq nucleosome signals from preimplantation embryos are also shown.
(F) K-means clustering of Foxa1, ERα, and AR ChIP-seq signal at distal THSSs overlapping a peak for at least one of these three proteins. ATAC-seq THSS signal from sperm, GV and MII oocytes, and preimplantation embryos is shown at the same clusters.
(G) Track view of a specific genomic region showing maintenance of accessibility at Foxa1 sites in sperm during embryo development.
See also Figure S6
We next used ATAC-seq data obtained in preimplantation embryos (Wu et al., 2016) to ask whether distal sites accessible to Tn5 in sperm and oocytes are also accessible during early embryonic development. We find that sperm- and oocyte-specific distal sites are not maintained in preimplantation embryos. However, common sites present in both sperm and oocytes show robust and similar accessibility at all embryonic stages beginning 7.5 hr after fertilization (Figure 6B). Using nucleosome-sized reads from ATAC-seq data, we find that these sites are flanked by well positioned nucleosomes in all preimplantation stages (Figures 6C and 6SA). Interestingly, gamete-specific distal sites that lack ATAC-seq THSSs in the embryo, suggesting the absence of bound transcription factors at these sites, still contain nucleosome-sized reads suggestive of positioned nucleosomes, although the intensity of this signal is lower than at common sites (Figures 6C and S6A). These findings suggest that distal THSSs present in both gametes, which presumably represent regulatory sequences, might be transmitted from germ cells to preimplantation embryos, where they maintain an open chromatin state throughout early embryonic development. In sperm, where this information is available, sites that persist in the embryo are marked by H3.3 and flanking nucleosomes containing H2A.Z (Figure 6C). To explore the possibility of additional epigenetic differences between distal THSS sites in the gametes that persist in the embryo and those that do not, we examined the DNA methylation state at these sites. Sperm- and oocyte-specific sites are methylated at intermediate levels in the gametes, become demethylated up to the blastocyst stage, and then remethylated again to levels similar to those present in the gametes (Figure S6B). However, accessible regions common to the sperm and oocyte, which remain accessible in the embryo, have methylation levels below 20% at all stages from gamete through epiblast (Figure S6B).
To determine which transcription factors are present at THSS sites that persist between the gametes and the early embryo, we performed a motif analysis with MEME-ChIP on these sites. We find a number of transcription factor motifs known to be essential for early development, including CTCF, ZIC4, NFYA, NFYB, AR and ESR2 (Figure S6C). To further analyze the possible persistence of these proteins, we first examined the maintenance of binding sites for CTCF. We combined all ATAC-seq THSSs present in sperm or oocytes containing a CTCF motif and used them as anchors to perform k-means clustering. Only a small fraction of CTCF sites are gamete-specific, and those present in both sperm and oocytes are maintained in preimplantation embryos (Figure 6D). CTCF sites maintained in the early embryo are flanked by H3.3 and H2A.Z, contain H3K27ac and H3K4me1, and are flanked by well positioned nucleosomes in all stages or early embryonic development (Figure 6E). In addition to CTCF, distal THSSs common to sperm and oocytes and maintained in the early embryo contain motifs for androgen and estrogen receptors. Many of these sites also contain Foxa1 in sperm. Analysis of ATAC-seq subnucleosome-sized reads suggests that ChIP-seq sites containing Foxa1, AR, and ERα in sperm are also present in GV and MII oocytes (Figure 6F). Furthermore, these sites co-occupied by Foxa1, AR, and ERα in both sperm and oocytes, are maintained in preimplantation embryos based on the presence of THSSs observed in ATAC-seq signal (Figure 6F). An example of a specific region of the genome showing this maintenance of accessibility is shown in Figure 6G.
Conservation of CTCF- and TF-mediated DNA loops from gametes through preimplantation embryos
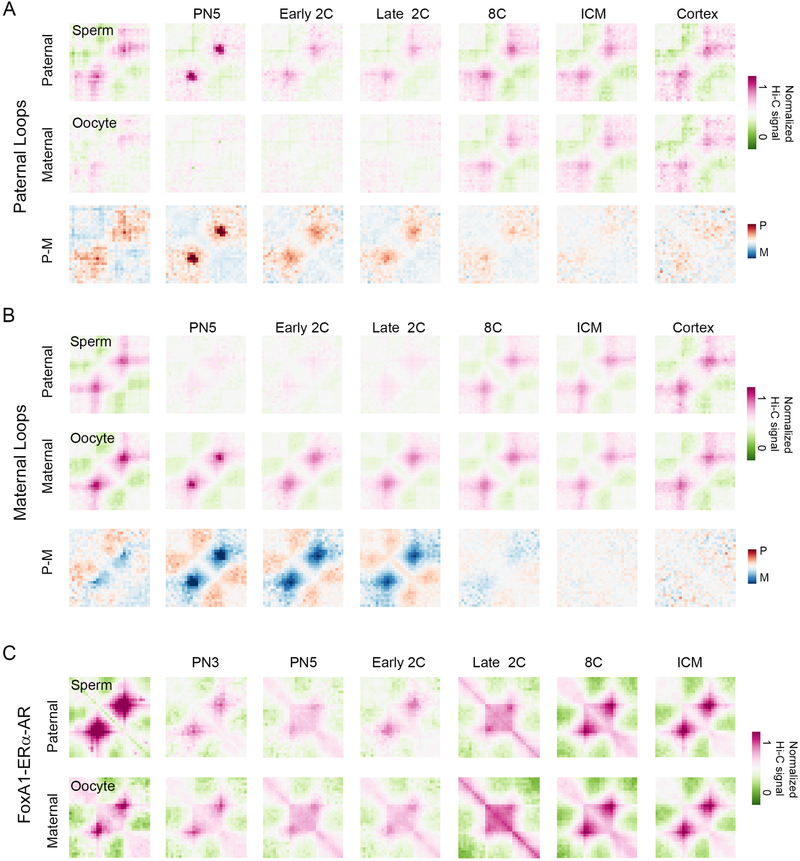
It is possible that interactions mediated by CTCF and other transcription factors in sperm are maintained in preimplantation embryos and contribute to the establishment of specific transcription patterns during early embryogenesis. To test this hypothesis, we examined publicly available Hi-C data obtained with oocytes, preimplantation embryos (Du et al., 2017; Flyamer et al., 2017), sperm (this study), and mouse brain cortex (Du et al., 2017). Hi-C data obtained in mouse embryos was separated into interactions present in the paternal or maternal chromosomes using SNP information. First, we determined significant interactions in the different Hi-C datasets using FitHi-C (Ay et al., 2014) and then we analyzed CTCF-mediated interactions using CTCF motif-containing THSSs common to sperm, GV, and MII oocytes. These sites mediate interactions in both sperm and GV oocytes (Figure S7A). Furthermore, some or all of the sperm or oocyte interactions are also present in the paternal or maternal chromosomes of preimplantation embryos, respectively (Figure S7A). Based on ChIP-seq experiments, we have identified three types of cohesin-containing sites, those containing CTCF, Znf143, or CTCF plus Znf143. All of these three types of cohesin-containing sites mediate specific interactions in sperm and are maintained in the paternal chromosomes from the one-cell stage to the ICM of the blastocyst (Figure S7B). To examine the persistence of interactions between the gametes and the early embryo in a different manner, we called paternal chromosome-specific loops at the PN5 zygote stage using a derivative of the HiCCUPS method (Cubenas-Potts et al., 2017) and examined the presence of these paternal-specific loops in sperm and embryos. We find that paternal-specific loops present in the zygote are also present in the sperm and in the paternal-chromosomes up to the blastocyst stage, as well as in the brain cortex (Figure 7A). These loops are not present in the GV oocyte or in the maternal chromosomes of one- and two-cell embryos. However, by the 8-cell stage the maternal chromosomes acquire the same loops as those present in the paternal alleles (Figure 7A). Similarly, CTCF loops present in the PN5 maternal chromosome are present in the GV oocyte and are conserved in the maternal chromosomes throughout early embryonic development. These loops are not present in the paternal chromosomes until the 8-cell stage, when both chromosomes appear to acquire the same 3D organization.
Figure 7. Conservation of CTCF- and TF-mediated DNA loops from gametes through preimplantation embryos.
(A) Metaplot showing median, distance-normalized Hi-C of sperm, GV oocytes, and allele-specific preimplantation embryos at loops defined in paternal PN5 chromosomes using a derivative of the HiCCUPS method. The bottom row shows the difference between paternal and maternal median, distance-normalized Hi-C interactions.
(B) Metaplot showing median, distance-normalized Hi-C of sperm, GV oocytes, and allele-specific preimplantation embryos at loops defined in maternal PN5 chromosomes using a derivative of the HiCCUPS method. The bottom row gives the difference between paternal and maternal median, distance-normalized Hi-C interactions.
(C) Metaplot showing median, distance-normalized Hi-C of sperm, GV oocytes, and allele-specific preimplantation embryos at Foxa1-mediated enhancer-promoter significant interactions determined by FitHi-C.
See also Figure S7
In addition to CTCF, a subset of distal Foxa1 sites containing the AR and ERα receptors are present in the gametes and persist in different stages of preimplantation embryos (Figure 6F). Using significant interactions determined by FitHi-C, we examined contacts between these putative enhancers and promoters. These Foxa1 sites mediate interactions with promoters in chromatin of sperm and GV oocytes (Figure 7C). Furthermore, these contacts appear to be maintained in the paternal and maternal chromosomes during embryonic development between the zygote and the ICM of the blastocyst (Figure 7C). Taken together, these results suggest that chromosomal interactions mediated by architectural proteins and transcription factors in the germline persist in the chromosomes of the embryo and could play a significant role in the expression of the genome during early embryogenesis and in the inheritance of transcriptional states between the gametes and the early embryo.
DISCUSSION
Here we present evidence for persistence of transcription factor occupancy between the gametes and the early mouse embryo. The molecular mechanisms responsible for the transmission of acquired epigenetic information between males and their offspring via sperm remain uncertain, despite increasing reports of environment-altering exposures producing heritable phenotypes in the next generation. Current models to explain transgenerational inheritance emphasize DNA methylation, non-coding RNAs, and histone modifications, which undergo extensive reprogramming during early development. Results described here expand the repertoire of information present in the gametes to include transcription factors, architectural proteins, and the long-range contacts they mediate, and show that this information persists in preimplantation embryos, making DNA-bound proteins ideal candidates to explain the inheritance of acquired epiphenotypes.
The chromatin state of the mammalian gametes has been difficult to explore in detail due to the high compaction of their chromatin. However, using Omni-ATAC we have been able to map thousands of sites in the mouse sperm genome where the occurrence of Tn5 transposase hypersensitive sites suggests the presence of bound proteins. A subset of these sites is located at TSSs of approximately 60% of all mapped genes. Surprisingly, given the transcriptionally inactive state of sperm, these promoters contain Mediator and RNAPII phosphorylated in Ser5 and Ser2 i.e. at the initiation and elongation stages of transcription, respectively. Oocytes at the prophase GV stage also contain accessible promoters, which remain accessible in the MII metaphase stage. Furthermore, the accessibility state is quantitatively maintained from the gametes to various stages of the preimplantation embryo i.e. those promoters with the highest subnucleosome-sized ATAC-seq reads in embryos correspond to those with the highest levels of ATAC-seq and RNAPII in sperm and oocytes. It is possible that the apparent maintenance of the transcription complex at these promoters is related to a requirement for their expression in preimplantation embryos. Alternatively, the persistence of promoter accessibility between gametes and the early embryo may be related to the reprograming of DNA methylation of the paternal and maternal genomes after fertilization. This hypothesis is supported by the low methylation levels of these promoters in the gametes, and the maintenance of this state in preimplantation embryos, which is in contrast to the high methylation and dramatic reprograming of methylation in non-accessible promoters.
In addition to promoters, Omni-ATAC-seq allows the identification of thousands of sites located in distal regions of the sperm and oocyte genomes. These sites may correspond to regulatory sequences such as enhancers, and their conservation in syntenic regions of the rhesus macaque and human genomes suggests a functional role for these sequences. In agreement with this, analysis of DNA binding motifs at these regions suggest the presence of transcription factors that remain bound to the sperm and oocyte genomes, including MII oocytes. A subset of these sites persists in the genome of the embryo at different stages up to the time of implantation. Interestingly, the sites that are maintained correspond to those present in both sperm and oocytes. In sperm, where this information is known, sites that persist in the embryo are flanked by nucleosomes containing H3.3, H2A.Z, and high levels of H3K27ac, whereas those that are not maintained lack H3.3 and have lower levels of H2A.Z and H3K27ac. The results suggest a regulated and determined assembly of sperm chromatin with the possible objective of controlling information passed on to the embryo. These observations agree with results obtained in zebrafish, where H2A.Z-containing placeholder nucleosomes correlate with the persistence of epigenetic information between sperm and the embryo (Murphy et al., 2018). Interestingly, sperm-specific sites that fail to persist still contain nucleosomes around these sites in the embryo, but their positioning becomes less marked as development proceeds. These results raise the interesting question of whether the presence of the transcription factor determines the presence of the H2A.Z nucleosomes or conversely.
As proof of concept to show that transcription factors are actually present in sperm chromatin at ATAC-seq accessible regions, we used ChIP-seq to analyze the distribution of Foxa1 and nuclear hormone receptors. We find that indeed these proteins are present at thousands of TSSs and distal sites in the sperm genome flanked by nucleosomes containing H2A.Z and active histone modifications. A subset of these sites is also present in GV and MII oocytes, and these common sites persist in preimplantation embryos. Importantly, persistent sites contain H3.3 and higher levels of H2A.Z and active histone modifications, including H3K4me1, in sperm. These inheritable distal sites also contain AR and ERα, supporting the idea that they constitute regulatory elements that may play a role in controlling gene expression during early embryo development. In agreement with this, Foxa1-mediated interactions between these sites and gene promoters can be observed in Hi-C data obtained in preimplantation embryo stages. Importantly, these interactions are already present in the gametes, suggesting the transmission of 3D chromatin organization between the gametes and the embryo.
The organization of the chromatin fiber in the nuclear space arises as a consequence of interactions between compartmental domains in the same transcriptional state but point-to-point contacts mediated by CTCF and cohesin are also important contributors to the establishment of this organization. Other proteins with less clear roles, such as YY1 and Znf143, are also present at CTCF sites and may regulate the specificity or frequency of these interactions. In agreement with the finding of the Znf143 binding motif at accessible sites, ChIP-seq experiments demonstrate the presence of this protein at thousands of sites in the genome where it colocalizes with CTCF and/or cohesin. Some Znf143 sites lack CTCF but contain cohesin, and these sites appear to mediate shorter-range interactions among enhancers and promoters than classical CTCF/cohesin loops. Interestingly, most CTCF sites present in sperm are accessible to Tn5 transposase in both GV and MII oocytes, and these sites shared by the gametes persist during early embryogenesis up to the blastocyst stage. In spite of their persistence in MII oocytes, putative CTCF sites present at this stage do not mediate interactions in MII metaphase chromosomes. However, CTCF loops present in the GV oocyte have been re-established in the maternal chromosome of the zygote by the PN5 stage, persist during early embryonic development, and can be observed in adult brain cortex. Interestingly, these interactions are absent from the paternal chromosome in the zygote and 2-cell stage but are established by the 8-cell stage. Similarly, paternal chromosome-specific interactions present in sperm and early embryo are absent in the maternal chromosomes, which become the same as the sperm by the 8-cell stage. The mechanisms by which parent-of-origin specific interactions are established in the sperm or the zygote and converge in the two parental chromosomes by the 8-cell stage are unknown and an important issue for future discovery.
In summary, we have shown that sperm and oocytes contain far more information with the potential to encode epigenetic memory than was previously recognized. Specific sites on gamete chromatin are poised with transcription factors despite lack of transcriptional inactivity, and chromatin accessibility at these sites and at distal regulatory elements is maintained in the embryo until at least the ICM stage. Since global remethylation occurs after the ICM stage, persistent sites with bound transcription factors inherited from gametes and retained through early embryogenesis may inhibit remethylation at their binding motifs. These sites may then remain accessible to the transcription machinery later in development as differentiation ensues. These observations open the possibility that transcription factors, whose distribution in the genome may be altered by environmental effects, are the basis for the transmission of epiphenotypes between generations.
STAR METHODS
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed and will be fulfilled by the Lead Contact Victor Corces, vgcorces@gmail.com, Phone: 404–727-4250, Fax: 404–727-2880.
Experimental Model and Subject Details
Experiments presented in this study make use of sperm isolated from mice, monkeys, and humans. All experiments were conducted according to the animal research guidelines from NIH and all protocols for animal usage were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) or the Institutional Review Board (IRB) of Emory University. Human sperm was obtained from anonymous volunteers and no informed consent was required in the protocol approved by the IRB. Mice were maintained and handled in accordance with the Institutional Animal Care and Use policies at Emory University. Mice were housed in standard cages on a 12: 12 h light:dark cycle and given ad lib access to food and water. Healthy 8-week old CD1 mice (Charles River Labs) not involved in previous procedures were used for sperm isolation. No genotyping was performed.
Method Details
Isolation of mouse sperm and oocytes
Euthanasia was performed by CO2 asphyxiation and the epididymes were removed. Mature sperm were collected from the dissected cauda epididymis of 8–10-week-old CD1 mice (Charles River Labs). After dissection to eliminate blood vessels and fat, the cauda epididymis was rinsed with PBS, deposited in Donners medium in a cell culture plate, and punctured with a needle. Sperm were then transferred to a tube and allowed to swim up for 1 hr (Hisano et al., 2013). Purity of sperm was determined by examination under a microscope after DAPI staining. After counting 1000 sperm, purity was determined to be at least 99.9% if no contaminating cells were observed.
For the preparation of GV-stage oocytes, the ovaries were harvested from 16-day-old CD1 female pups. GV oocytes were harvested from the ovaries. The zona pellucida was removed with hyaluronidase to avoid any residual cumulus cells. For MII oocyte collection, CD1 females at 6–8 weeks of age were hormonally stimulated by injection of 7.5 IU of pregnant mare serum gonadotropin (PMSG) followed by 5 IU of human chorionic gonadotropin (hCG) at 48 h post-PMSG injection. MII oocytes were harvested at 13–14 h post-hCG injection. Zona pellucida and polar bodies were removed with hyaluronidase.
Preparation of monkey semen
Ejaculates were collected from two male wild-type rhesus monkeys (Macaca mulatta) via electro-ejaculation. Each male was chair trained (Primate Product Inc.) using the “pole and collar” technique (Bliss-Moreau et al., 2013; Moran et al., 2016). The monkey was then lightly sedated with Ketamine (~0.4 mg/kg body weight) administered via intramuscular injection. One pre-sized defibrillator gel electrode was wrapped around the base of the penis and connected to the negative electrode lead. The second gel electrode was positioned immediately behind the glans and connected to the positive lead. The electro-ejaculator device was slowly raised up to 32 V or until ejaculation. Ejaculates were kept at room temperature for 25 min to liquefy. The liquid sample was transferred into a fresh 15 ml conical tube and washed once with TALP-HEPES medium supplemented with 4 mg/ml BSA (Moran et al., 2016; Putkhao et al., 2013), then centrifuged at 400 x g at room temperature for 5 min. Supernatant was removed, and sample was resuspended into 1 ml total volume with TALP-HEPES + BSA. Density gradient centrifugation was carried out using PureSperm® (Nidacon). Briefly, samples were layered on a double density gradient of 2 ml of PureSperm®40 (top) and 2 ml PureSperm®80 (bottom) and centrifuged at 300 x g for 20 min at room temperature. Supernatant was removed and the pellets were washed with 1 ml PureSperm® Wash and centrifuged at 500 x g for 10 min. Samples were then resuspended in 1 ml TALP-HEPES + BSA and an aliquot was examined for sperm concentration and purity.
Preparation of human semen
Human semen samples were obtained from 20–25 year-old heathy donors. After seminal liquefaction, sperm was transferred to sterile 10 ml centrifuge tubes and washed twice with Human Tubal Fluid medium (Irvine Scientific) supplemented with human serum albumin. After washing, isolation and purification of human spermatozoa was carried out as described (Hisano et al., 2013).
Assay for transposase-accessible chromatin using sequencing (ATAC-seq)
ATAC-seq was carried out using the Omni-ATAC protocol (Corces et al., 2017). After sperm cells were counted, the nuclei from 100,000 sperm were isolated with Lysis Buffer (10 mM Tris-HCl pH 7.4, 10 mM NaCl, 3 mM MgCl2) containing 0.1% NP40, 0.1% Tween-20, and 0.01% digitonin. The purified sperm nuclei pellet was then resuspended in the transposase reaction mix containing 0.05% digitonin and incubated for 30 min at 37°C. Following incubation, sperm were treated with Proteinase K at 55°C for 2 hr, an d gDNA was isolated by phenol:chloroform:isoamyl alcohol and EtOH precipitation. Library amplification was done with 2x KAPA HiFi mix (Kapa Biosystems) and 1.25 μM indexed primers using the following PCR conditions: 72°C for 5 min; 98°C for 30 s; and 10–1 1 cycles at 98°C for 10 s, 63°C for 30 s, and 72°C for 1 min.
ChIP-seq in sperm cells
Chromatin immunoprecipitation to detect the localization of DNA-binding proteins in sperm was performed using the standard ChIP-seq protocol with some modifications. Briefly, 10–20 million sperm cells were crosslinked with 1% formaldehyde in 1x PBS for 10 min at RT, and the reaction was quenched with 125 mM glycine for 10 min at RT. After washing with PBS, crosslinked sperm were lysed with 5 mM PIPES, 85 mM KCl, 0.5% NP40 and 1x protease inhibitors (P8107S, NEB) on ice for 15 min. After centrifugation, sperm cells were resuspendedin RIPA buffer (1x PBS; 1% NP40; 0.5% sodium deoxycholate; 0.1% SDS; 1x protease inhibitors) and incubated on ice for 20 min. The purified sperm chromatin was sonicated to 300–1000 bp using a Diagenode Bioruptor. After 25 cycles (30 seconds on and 60 seconds off) the supernatant containing sheared chromatin was collected. Immunoprecipitation was performed overnight at 4°C with antibodies against Med12 (A30 0–774A, Bethyl laboratories), RNAPIISer5ph (ab5131, Abcam), RNAPIISer2ph (ab5095, Abcam), Foxa1(ab5089, Abcam), AR (sc-816, Santa Cruz), Znf143 (16618–1-AP, Proteintech) and ERα (sc-543, Santa Cruz). Libraries for Illumina sequencing were constructed using the following standard protocol. Fragment ends were repaired using the NEBNext End Repair Module and adenosine was added at the 3’ ends using Klenow fragment (3’ to 5’ exo minus, New England Biolabs). Precipitated DNA and input DNA were incubated with adaptors at room temperature for 1 hr with T4 DNA ligase (New England Biolabs) and amplified with Illumina primers. Chromatin immunoprecipitation to detect histone variants was carried out as described (Hisano et al., 2013) using antibodies to H2A.Z (ab4174, Abcam) and H3.3 (ab176840, Abcam).
Western analysis
For Western analysis of chromatin proteins, an equal number of sperm and J1 mESCs were resuspended in 1x Laemmli sample buffer (5% 2-mercaptoethanol, 0.002% bromophenol blue, 10% Glycerol, 2% SDS, 62.5 mM Tris-HCl pH 6.8). Samples were boiled for 5 min and supernatant was loaded onto SDS-PAGE 4–15% gels at a ratio of 12:1 sperm:mESCs. Because sperm are haploid, sperm lanes contain 6-fold more genome equivalents than lanes with mESCs. Membranes were blocked in TBST (20 mM Tris, pH7.4, 150 mM NaCl, 0.05% Tween 20) with 5% nonfat milk powder and incubated overnight with the antibodies described above. Membranes were washed 3 times with TBST and incubated with secondary antibodies-conjugated to HRP (1:5000, Jackson ImmunoResearch Laboratories) for 1 hr. After three more washes, the presence of different proteins was detected using SuperSignal West Pico/Dura Chemiluminescent substrate (Thermo Scientific).
In-situ Hi-C
in-situ Hi-C libraries were prepared using DpnII restriction enzyme as previously described (Rao et al., 2014). Briefly, 10 million sperm were crosslinked with 1% formaldehyde, quenched with glycine, washed with PBS, and permeabilized to obtain intact nuclei. Nuclear DNA was then digested with DpnII, the 5’-overhangs were filled with biotinylated dCTPs and dA/dT/dGTPs to make blunt-end fragments, which were then ligated, reverse-crosslinked, and purified by standard DNA ethanol precipitation. Purified DNA was sonicated to 200–500 bp small fragments and captured with streptavidin beads. Standard Illumina TruSeq library preparation steps, including end-repairing, A-tailing, and ligation with universal adaptors were performed on beads, washing twice in Tween Washing Buffer (5mM Tris-HCl pH 7.5, 0.5mM EDTA, 1M NaCl, 0.05% Tween 20) between each step. DNA on the beads was PCR amplified with barcoded primers using KAPA SYBR FAST qPCR Master Mix (Kapa Biosystems) for 5~12 PCR cycles to obtain enough DNA for sequencing. Generated libraries were paired-end sequenced on Illumina HiSeq2500 v4 or NovaSeq 6000 instruments. Two biological replicates were generated, and replicates were combined for all analyses after ensuring high correlation.
Data Processing
Analysis of ATAC-seq data
All libraries were sequenced using an Illumina Hiseq2500 v4 sequencer and 50 bp paired-end format. Paired reads were aligned to the mouse reference genome mm9, human reference genome hg19 and monkey reference genomes rheMac8 and MacaM using Bowtie2. ATAC-seq reads were aligned using default parameters except -X 2000 -m 1. PCR duplicates were removed using Picard Tools. To adjust for fragment size, we aligned all reads as + strands offset by +4 bp and – strands offset by −5 bp (Buenrostro et al., 2013). For all ATAC-seq datasets except the pre-implantation embryo data, the THSS (Tn5 hypersensitive sites) and mono-nucleosome fractions were separated by choosing fragments 50–115 bp and 180–247 bp in length, respectively. For the pre-implantation embryo ATAC-seq data, THSS and mono-nucleosome fractions were separated by choosing fragments 50–125 bp and 171–256 bp in length, respectively. Mono-nucleosome reads were analyzed using DANPOS2 (Chen et al., 2013). MACS2 was used for peak calling for THSSs (Liu, 2014).
ChIP-Seq data processing
All reads were mapped to unique genomic regions using Bowtie2 (Langmead and Salzberg, 2012) and the mm9 genome. PCR duplicates were removed using Picard Tools. MACS2 was used to call peaks using default parameters with IgG ChIP-seq data as a control.
Transcription factor footprint analysis
To analyze the footprints of TFs in ATAC-seq data, motifs on a set of peaks were used as anchors for running dnase_average_profile.py scripts of the Wellington program in ATAC-seq mode. The footprint p values of all motifs on a set of peaks were derived using the wellington_footprints.py scripts of the Wellington program in ATAC-Seq mode on read-normalized ATAC-seq THSSs (<115 bp) fragments.
Analysis of publicly available SNP-trackable ATAC-seq, DNase-seq, and ChIP-seq of gametes and embryos
All data were aligned to the mm9 mouse reference genome using Bowtie2 using the --no-mixed–no-discordant flags. Additionally, the -X 2000 flag was used for the ATAC-seq and H3K4me3 ChIP-seq data. All data were aligned in paired-end mode except for DNAse-seq, which were aligned in single-end mode. Uniquely mapped reads were extracted and converted to bam format for all downstream analyses. Uniquely mapped reads were inputted into the SNPsplit software in order to determine the parent-of-origin for the data. SNP tables for the crosses for each dataset were downloaded from the Sanger Institute mouse genome project. DNase-seq data were not split according to SNPs.
Analysis of publicly available DNA methylation data
Publicly available bed files containing counts of CpG methylation vs. unmethlyated CpGs were separated by parent-of-origin according to SNPs based on methylC-seq data (Wang et al., 2014). Average methylation values were calculated as a weighted average, with the weight at each CpG being equal to the number of reads covering that CpG.
Normalization of SNP-trackable sequencing data
After separating reads by strain according to SNPs, regions were normalized with a modified form of either RPKM or FPKM. The typical definitions for RPKM/FPKM were used, except instead of dividing by the total number of reads for each sample, we divided by the total number of SNP-containing reads. For example, if N_P were the number of SNP-containing reads in the paternal allele, and N_M were the corresponding number of reads on the maternal allele, the number of reads (or fragments) per kilobase in a region of interest were divided by N_P+N_M in millions of reads. For visualization of tracks in IGV, reads were normalized by RPM using the same N_P+N_M factor. This normalization was used for all strain-specific analyses using SNP reads coming from the same sample.
Hi-C data processing
Paired-end reads from Hi-C experiments were aligned to the mouse mm9 reference genome using Juicer (Rao et al., 2014). After PCR duplicates and low-quality reads were removed, high-quality reads were assigned to DpnII restriction fragments, and Hi-C interaction contacts where mapped in a binned matrix to create a hic file. A derivative of the HiCCUPS method (Cubenas-Potts et al., 2017; Rao et al., 2014) was used to call significant peaks in the Hi-C interaction matrix. All statistically significant peaks were post-filtered for observed values greater than 12 Hi-C contacts, observed over expected values greater than 2 Hi-C contacts, and interactions that are less than 5 Mb apart. The WashU Epigenome browser was used to obtain arc views of significant loops. FitHi-C (Ay et al., 2014) was used to call significant interaction at 25 kb resolution from the second pass with a q-value threshold of q > 0.001.
Data and Software Availability
ChIP-seq, ATAC-seq, and Hi-C data are available from NCBI’s Gene Expression Omnibus (GEO). The accession number for all the datasets reported in this paper is GSE116857. Reviewers can access these data using token whqtysowzriddkp. Custom scripts were used to separate ATAC-seq reads into subnucleosomal and nucleosome-size ranges; to obtain the line plots shown in Figure 5 and Figure S5; and to obtain metaplots of Hi-C data shown in Figure 7 and Figure S7. These scripts are available without restrictions upon request.
Supplementary Material
HIGLIGHTS.
ATAC-seq accessibility at sperm and oocyte promoters is maintained in the embryo
Sperm enhancers containing transcription factors are conserved in mammals
Accessible sperm enhancers are also open in oocytes and pre-implantation embryos
Interactions mediated by FoxA1 and CTCF/cohesin persist from gametes to embryos
ACKNOWLEGMENTS
We would like to thank the Genomic Services Lab at the HudsonAlpha Institute for Biotechnology, and specially Drs. Angela Jones and Braden Boone, for their help in performing Illumina sequencing of samples. This work was supported by U.S. Public Health Service Award R01 ES027859 from the National Institutes of Health to VGC. Rhesus macaque sperm samples were provided by the Transgenic Huntington’s Disease Monkey Resource (THDMR; OD010930) sponsored by the Office of Research and Infrastructure Programs (ORIP) at the National Institute of Health. M.J.R was supported by NIH Pathway to Independence Award NIGMS K99GM127671. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests
REFERENCES
- Ay F, Bailey TL, and Noble WS (2014). Statistical confidence estimation for Hi-C data reveals regulatory chromatin contacts. Genome Res 24, 999–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae WK, Kang K, Yu JH, Yoo KH, Factor VM, Kaji K, Matter M, Thorgeirsson S, and Hennighausen L (2015). The methyltransferases enhancer of zeste homolog (EZH) 1 and EZH2 control hepatocyte homeostasis and regeneration. FASEB journal : official publication of the Federation of American Societies for Experimental Biology 29, 1653–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey SD, Zhang X, Desai K, Aid M, Corradin O, Cowper-Sal Lari R, Akhtar-Zaidi B, Scacheri PC, Haibe-Kains B, and Lupien M (2015). ZNF143 provides sequence specificity to secure chromatin interactions at gene promoters. Nat Commun 2, 6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Williams N, Misleh C, and Li WW (2006). MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34, W369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliss-Moreau E, Theil JH, and Moadab G (2013). Efficient cooperative restraint training with rhesus macaques. Journal of applied animal welfare science : JAAWS 16, 98–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brykczynska U, Hisano M, Erkek S, Ramos L, Oakeley EJ, Roloff TC, Beisel C, Schubeler D, Stadler MB, and Peters AH (2010). Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nature structural & molecular biology 17, 679–687. [DOI] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, and Greenleaf WJ (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nature methods 10, 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunch H, Zheng X, Burkholder A, Dillon ST, Motola S, Birrane G, Ebmeier CC, Levine S, Fargo D, Hu G, et al. (2014). TRIM28 regulates RNA polymerase II promoter-proximal pausing and pause release. 21, 876–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carone BR, Hung JH, Hainer SJ, Chou MT, Carone DM, Weng Z, Fazzio TG, and Rando OJ (2014). High-resolution mapping of chromatin packaging in mouse embryonic stem cells and sperm. Dev Cell 30, 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Xi Y, Pan X, Li Z, Kaestner K, Tyler J, Dent S, He X, and Li W (2013). DANPOS: dynamic analysis of nucleosome position and occupancy by sequencing. Genome Res 23, 341–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces MR, Trevino AE, Hamilton EG, Greenside PG, and Sinnott-Armstrong NA (2017). An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. 14, 959–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubenas-Potts C, Rowley MJ, Lyu X, Li G, Lei EP, and Corces VG (2017). Different enhancer classes in Drosophila bind distinct architectural proteins and mediate unique chromatin interactions and 3D architecture. Nucleic Acids Res 45, 1714–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl JA, Jung I, Aanes H, Greggains GD, Manaf A, Lerdrup M, Li G, Kuan S, Li B, Lee AY, et al. (2016). Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature 537, 548–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zheng H, Huang B, Ma R, Wu J, Zhang X, He J, Xiang Y, Wang Q, Li Y, et al. (2017). Allelic reprogramming of 3D chromatin architecture during early mammalian development. Nature 547, 232–235. [DOI] [PubMed] [Google Scholar]
- Durand NC, Robinson JT, Shamim MS, Machol I, Mesirov JP, Lander ES, and Aiden EL (2016a). Juicebox Provides a Visualization System for Hi-C Contact Maps with Unlimited Zoom. Cell Systems 3, 99–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand NC, Shamim MS, Machol I, Rao SS, Huntley MH, Lander ES, and Aiden EL (2016b). Juicer Provides a One-Click System for Analyzing Loop-Resolution Hi-C Experiments. Cell Syst 3, 95–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flyamer IM, Gassler J, Imakaev M, Brandao HB, Ulianov SV, Abdennur N, Razin SV, Mirny LA, and Tachibana-Konwalski K (2017). Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature 544, 110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao T, He B, Liu S, Zhu H, Tan K, and Qian J (2016). EnhancerAtlas: a resource for enhancer annotation and analysis in 105 human cell/tissue types. Bioinformatics 32, 3543–3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibcus JH, Samejima K, Goloborodko A, Samejima I, Naumova N, Nuebler J, Kanemaki MT, Xie L, Paulson JR, Earnshaw WC, et al. (2018). A pathway for mitotic chromosome formation. Science 359, eaao6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, and Noble WS (2011). FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoud SS, Low DH, Yi C, Carrell DT, Guccione E, and Cairns BR (2014). Chromatin and transcription transitions of mammalian adult germline stem cells and spermatogenesis. Cell Stem Cell 15, 239–253. [DOI] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, and Cairns BR (2009). Distinctive chromatin in human sperm packages genes for embryo development. Nature 460, 473–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa K, Sin HS, Maezawa S, Broering TJ, Kartashov AV, Alavattam KG, Ichijima Y, Zhang F, Bacon WC, Greis KD, et al. (2015). SCML2 establishes the male germline epigenome through regulation of histone H2A ubiquitination. Dev Cell 32, 574–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard E, and Martienssen RA (2014). Transgenerational epigenetic inheritance: myths and mechanisms. Cell 157, 95–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt SC, Li L, Grimm SA, Chen Y, Liu L, Li Y, Bushel PR, Fargo D, and Korach KS (2012). Research resource: whole-genome estrogen receptor alpha binding in mouse uterine tissue revealed by ChIP-seq. Mol Endocrinol 26, 887–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichs AS, Karolchik D, Baertsch R, Barber GP, Bejerano G, Clawson H, Diekhans M, Furey TS, Harte RA, Hsu F, et al. (2006). The UCSC Genome Browser Database: update 2006. Nucleic Acids Res 34, D590–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisano M, Erkek S, Dessus-Babus S, Ramos L, Stadler MB, and Peters AH (2013). Genome-wide chromatin analysis in mature mouse and human spermatozoa. Nature protocols 8, 2449–2470. [DOI] [PubMed] [Google Scholar]
- Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, and Carroll JS (2011). FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet 43, 27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Dadon DB, Abraham BJ, Lee TI, Jaenisch R, Bradner JE, and Young RA (2015). Chromatin proteomic profiling reveals novel proteins associated with histone-marked genomic regions. Proc Natl Acad Sci U S A 112, 3841–3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jozwik KM, Chernukhin I, Serandour AA, Nagarajan S, and Carroll JS (2016). FOXA1 Directs H3K4 Monomethylation at Enhancers via Recruitment of the Methyltransferase MLL3. Cell Rep 17, 2715–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung YH, Sauria ME, Lyu X, Cheema MS, Ausio J, Taylor J, and Corces VG (2017). Chromatin States in Mouse Sperm Correlate with Embryonic and Adult Regulatory Landscapes. Cell Rep 18, 1366–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, and Salzberg SL (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nature methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, and Salzberg SL (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, and Genome Project Data Processing, S. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Mav D, Grimm SA, Jothi R, Shah R, and Wade PA (2014). Fine-tuning of epigenetic regulation with respect to promoter CpG content in a cell type-specific manner. Epigenetics 9, 747–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T (2014). Use model-based Analysis of ChIP-Seq (MACS) to analyze short reads generated by sequencing protein-DNA interactions in embryonic stem cells. Methods Mol Biol 1150, 81–95. [DOI] [PubMed] [Google Scholar]
- Lu F, Liu Y, Inoue A, Suzuki T, Zhao K, and Zhang Y (2016). Establishing Chromatin Regulatory Landscape during Mouse Preimplantation Development. Cell 165, 1375–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maza I, Caspi I, Zviran A, Chomsky E, Rais Y, Viukov S, Geula S, Buenrostro JD, Weinberger L, Krupalnik V, et al. (2015). Transient acquisition of pluripotency during somatic cell transdifferentiation with iPSC reprogramming factors. 33, 769–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran SP, Chi T, Prucha MS, Agca Y, and Chan AW (2016). Cryotolerance of Sperm from Transgenic Rhesus Macaques (Macaca mulatta). Journal of the American Association for Laboratory Animal Science : JAALAS 55, 520–524. [PMC free article] [PubMed] [Google Scholar]
- Murphy PJ, Wu SF, James CR, Wike CL, and Cairns BR (2018). Placeholder Nucleosomes Underlie Germline-to-Embryo DNA Methylation Reprogramming. Cell 172, 993–1006.e1013. [DOI] [PubMed] [Google Scholar]
- Oomen ME, Hansen AS, Liu Y, Darzacq X, and Dekker J (2019). CTCF sites display cell cycle-dependent dynamics in factor binding and nucleosome positioning. Genome Res 29, 236–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pihlajamaa P, Sahu B, Lyly L, Aittomaki V, Hautaniemi S, and Janne OA (2014). Tissue-specific pioneer factors associate with androgen receptor cistromes and transcription programs. EMBO J 33, 312–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper J, Elze MC, Cauchy P, Cockerill PN, Bonifer C, and Ott S (2013). Wellington: a novel method for the accurate identification of digital genomic footprints from DNase-seq data. Nucleic Acids Res 41, e201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putkhao K, Chan AW, Agca Y, and Parnpai R (2013). Cryopreservation of transgenic Huntington’s disease rhesus macaque sperm-A Case Report. Cloning & transgenesis 2. Quinlan, A.R. (2014). BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Current protocols in bioinformatics 47, 11 12 11–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. (2014). A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu B, Pihlajamaa P, Dubois V, Kerkhofs S, Claessens F, and Janne OA (2014). Androgen receptor uses relaxed response element stringency for selective chromatin binding and transcriptional regulation in vivo. Nucleic Acids Res 42, 4230–4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schep AN, Buenrostro JD, Denny SK, Schwartz K, Sherlock G, and Greenleaf WJ (2015). Structured nucleosome fingerprints enable high-resolution mapping of chromatin architecture within regulatory regions. Genome Res 25, 1757–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z, Zhang Y, Yuan GC, Orkin SH, and Waxman DJ (2012). MAnorm: a robust model for quantitative comparison of ChIP-Seq data sets. Genome Biol 13, R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Shao N, Liu X, and Nestler E (2014). ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics 15, 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, and Pachter L (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology 28, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veselovska L, Smallwood SA, Saadeh H, Stewart KR, Krueger F, Maupetit-Mehouas S, Arnaud P, Tomizawa S, Andrews S, and Kelsey G (2015). Deep sequencing and de novo assembly of the mouse oocyte transcriptome define the contribution of transcription to the DNA methylation landscape. Genome Biol 16, 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Stetina JR, and Orr-Weaver TL (2011). Developmental control of oocyte maturation and egg activation in metazoan models. Cold Spring Harbor perspectives in biology 3, a005553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wamstad JA, Alexander JM, Truty RM, Shrikumar A, Li F, Eilertson KE, Ding H, Wylie JN, Pico AR, Capra JA, et al. (2012). Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell 151, 206–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhang J, Duan J, Gao X, Zhu W, Lu X, Yang L, Zhang J, Li G, Ci W, et al. (2014). Programming and inheritance of parental DNA methylomes in mammals. Cell 157, 979–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Huang B, Chen H, Yin Q, Liu Y, Xiang Y, Zhang B, Liu B, Wang Q, Xia W, et al. (2016). The landscape of accessible chromatin in mammalian preimplantation embryos. Nature 534, 652–657. [DOI] [PubMed] [Google Scholar]
- Zhang B, Zheng H, Huang B, Li W, Xiang Y, Peng X, Ming J, Wu X, Zhang Y, Xu Q et al. (2016). Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature 537, 553–557. [DOI] [PubMed] [Google Scholar]
- Zheng H, Huang B, Zhang B, Xiang Y, Du Z, Xu Q, Li Y, Wang Q, Ma J, Peng X, et al. (2016). Resetting Epigenetic Memory by Reprogramming of Histone Modifications in Mammals. Mol Cell 63, 1066–1079. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ChIP-seq, ATAC-seq, and Hi-C data are available from NCBI’s Gene Expression Omnibus (GEO). The accession number for all the datasets reported in this paper is GSE116857. Reviewers can access these data using token whqtysowzriddkp. Custom scripts were used to separate ATAC-seq reads into subnucleosomal and nucleosome-size ranges; to obtain the line plots shown in Figure 5 and Figure S5; and to obtain metaplots of Hi-C data shown in Figure 7 and Figure S7. These scripts are available without restrictions upon request.