Abstract
A central and causative feature of age-related neurodegenerative disease is the deposition of misfolded proteins in the brain. To devise novel approaches to treatment, regulatory pathways that modulate these aggregation-prone proteins must be defined. One such pathway is the post-translational modification poly(ADP-ribose), which promotes protein localization in a number of cellular contexts. Mounting evidence implicates poly(ADP-ribose) in seeding the abnormal localization and accumulation of proteins causative of neurodegenerative disease. Inhibitors of poly(ADP-ribose) activity have been developed as cancer therapeutics, raising the possibility that they could be applied to treatment of neurodegenerative disease. Here, we focus on pathways regulated by poly(ADP-ribose) in neurodegenerative disease, with emphasis on amyotrophic laterals sclerosis and frontotemporal degeneration.
ADP-ribosylation
Post-translational modifications are fundamental to regulating protein function and stability in all domains of life. One such modification is ADP-ribosylation (Figure 1A), which is the rapid transfer of ADP-ribose (ADPr) onto a target protein via the catalysis of βNAD+ [1–3]. ADPr-transferase activity has independently evolved in 3 protein families: the sirtuins; the bacterial TM1506 family; and the poly(ADPr) polymerases (PARPs), the latter being the principle ADPr-transferase system in humans [4, 5]. The human PARP superfamily consists of 17 proteins with a canonical ADPr-transferase (ART) domain, and each have specific enzymatic activities: 11 are mono(ADP-ribose) transferases (PARP-3, -4, -6, -7, -8, -10, -11, -12, -14, -15, and -16) and link a single ADP-ribose subunit onto the target protein (MARylation); PARP-1, -2, -5a and 5b have the capacity to add sequential ADPr subunits, and generate chains of poly(ADP-ribose) (PAR) (PARylation); and the remaining 2 (PARP-9 and -13) are enzymatically inactive [6]. Amino acids known to be conjugated by ADP-ribsoylated are Arg, Asp, Cys, Glu, Lys, Ser and Tyr [2, 7]. ADP-ribosylation is a labile modification, which is in part due to the activity of the ADPr-hydrolases.
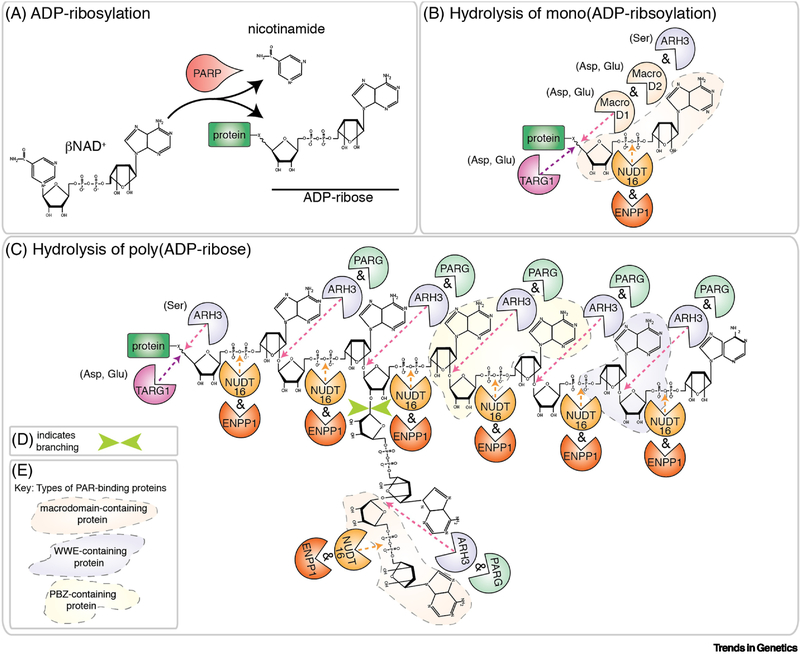
Figure 1: ADP-ribosylation.
A) PARPs catabolize βNAD+ into nicotinamide and ADP-ribose (ADPr), with the resulting ADPr covalently linked onto an acceptor protein. The addition of a single ADPr subunit is called mono(ADP-ribosylation) (MARylation). Amino acids targeted are Arg, Asp, Cys, Glu, Lys, Ser and Tyr [2, 7]. B) Removal of a single ADPr subunit from acidic residues (Asp and Glu) is mediated by MacroD1, MacroD2 and TARG1 [12–14]. ARH3 removes the ADPr from Ser residues [10]. NUDT16 and ENPP1 cleave the 2 phosphate groups in ADPr to generate a phosphoribosylated protein [15–17]. C-D) Poly(ADP-ribosylation) (PARylation) is the sequential addition of ADPr and generates linear and sometimes branched (D, green arrows) polymers of poly(ADP-ribose) (PAR). PARylation is reversed by PARG-mediated hydrolysis of the ribose-ribose bond. ARH3 also cleaves the ribose-ribose bond but unlike PARG, ARH3 can also remove the protein proximal ADPr from Ser residues in the target protein [8–11]. TARG1 removes the entire PAR chain by cleavage of the protein proximal ADPr [14]. The 2 phosphate groups in ADPr are hydrolyzed by NUDT16 and ENPP1 [15–17]. E) Conserved protein domains recognize and bind specific sites of PAR polymers (reviewed in [115]). These domains include the PBZ domain, the WWE domain, the macrodomain and the PAR-binding motif. The physical contacts that are made with the ADPr moiety are known for the PBZ domain, the WWE domain and the macrodomain. Those regions of the PAR chain are shaded in B and C according to the key in E. It is currently unknown what the PBM interacts with in PAR.
In humans there are > 8 ADPr-hydrolases each with specific protein domains and substrates (Figure 1B and C). For example, PAR is cleaved at the ribose-ribose bond by PAR glycohydrolase (PARG) and to a lesser extent by ADP-ribose glycohydrolase 3 (ARH3), but while ARH3 can remove the terminal ADPr conjugated to a serine residue, PARG cannot hydrolyze the terminal ADPr and leaves behind a mono(ADP-ribsoylated) (MARylated) protein [8–11]. MacroD1 and MacroD2 reverse MARylation at acidic residues; similarly, ADP-ribose glycohydrolase OARD1 (TARG1) hydrolyses the ADPr conjugated to acidic residues to remove either a single ADPr subunit or the entire PAR chain [12–14]. ADPr and PAR can also be converted into phosphoribose by the hydrolysis of the 2 phosphate groups in the ADPr subunit by nucleoside diphosphate-linked moiety X-type motif 16 (NUDT16) and ectonucleotide pyrophosphatase/phosphodiesterase 1 (ENPP1) [15–17]. Because of the dynamic and reversible nature of ADP-ribosylation it is often central to stress pathways where there is a need for a rapid response combined with a tightly controlled termination of the signaling pathway.
In mammals, the most widely studied stress-induced function of ADP-ribosylation is in the DNA damage response (DDR). At sites of DNA damage, PARP-1, a nuclear enzyme, modifies itself (automodification) and target proteins (transmodification). DNA-repair enzymes, such as the DNA repair protein XRCC1 (XRCC1), non-covalently bind to the PAR polymer and promote the localization of repair proteins to the sites of damaged DNA [18, 19]. Finally, in order to complete the repair of damaged DNA, PARG is recruited and the PAR polymer is digested [20]. Upon excessive activation of PARP-1, PAR is cleaved and translocated into the cytoplasm where it triggers a cell-death mechanism called parthanatos (see below). Thus, ADP-ribosylation regulates the modified protein, as well as the proteins that bind to PAR, and isolated PAR functions as a signaling molecule. PARylation by the PARP superfamily is now known to regulate many fundamental cellular processes, including chromatin compaction and gene expression, protein stability and cellular signaling, and telomeres and the mitotic spindle (reviewed in [21–26]).
ADP-ribosylation was initially pursued as a cancer therapeutic; however, it is becoming increasingly clear that ADP-ribosylation also regulates many features associated with age-related brain disorders including DNA damage, protein localization and aggregation, and cell death. Hence, it has been proposed that PARP inhibitors developed as cancer treatments could be repurposed for other disorders, including neurological diseases [27]. Here, we discuss recent work that implicates ADP-ribosylation activity as critical for age-related brain function. We will then focus on how ADP-ribosylation relates to amyotrophic lateral sclerosis (ALS) and frontotemporal degeneration (FTD) and we will discuss future considerations.
The importance in regulating ADP-ribosylation in the brain
Mounting evidence implicates ADP-ribosylation as a key pathway required for normal brain function. Copy number variants (CNVs) and intronic single nucleotide polymorphisms (SNPs) at the ADPr-hydrolase MacroD2 locus have been identified in epilepsy, autism spectrum disorder (ASD), ischemic stroke, multiple sclerosis and schizophrenia [28–39]. Non-coding SNPs can regulate local genes, long-range genes and genes positioned on different chromosomes, and intronic SNPs in MacroD2 have been linked to transcriptional clusters on several other chromosomes [40]. Thus, it remains to be elucidated whether the intronic SNPs and CNVs contribute to disease via misregulation of MacroD2 or genes elsewhere in the genome that are associated with the MacroD2 locus. However, in 6 independent families, recessive and inactivating mutations in the ADPr-hydrolase ARH3 were identified in patients that suffer from early-onset neurodegeneration [41]. Affected individuals experience stress-induced epilepsy, ataxia and death within the first decade of life [41]. Recessive and inactivating mutations have also been identified in the gene encoding for TARG1 [14]. Affected individuals from an extended family experienced severe neurodevelopmental delay, seizures and motor deficits [14]. TARG1 removes ADPr from Glu and Asp residues while ARH3 hydrolyzes PAR and ADPr from Ser residues [9, 10, 12–14] (Figure 1). The clinical presentation of patients harboring inactivating mutations in TARG1 and ARH3 suggests that a failure to reverse ADP-ribosylation at Ser, Glu and Asp residues is catastrophic for human brain function and results in rapid and severe neurological dysfunction.
ADP-ribosylation is implicated in neurodegenerative disease characterized by abnormal protein aggregation. For example, proteins linked to the motor neuron disease amyotrophic lateral sclerosis (ALS), frontotemporal degeneration (FTD) and Parkinson’s disease (PD) (TDP-43, FUS and α-synuclein, respectively) are PAR-binding proteins, and PAR regulates their cellular localization, aggregation and associated neurotoxicity [42–46]. Inhibiting the activity of PARP-1 and PARP-2 (collectively referred to as PARP-1/2) and PARP-5a and PARP-5b (PARP-5a/5b) is beneficial in a variety of cellular, neuronal and rodent models of ALS, FTD and PD [43, 45–47]. Additionally, in cellular and animal models of ischemic stroke, downregulation or small-molecule inhibition of PARP-1 profoundly mitigates neural loss [48–50]. Collectively, these studies indicate that ADP-ribosylation facilitates disease progression. Indeed, inactivating mutations in XRCC1 lead to ataxia in the fifth decade of life [51]. XRCC1 is localized to damaged DNA by binding the PAR scaffold generated by PARP-1 [18, 19]. Loss of XRCC1 leads to sustained PARylation, impaired repair of single-stranded DNA breaks (SSB) and neuronal loss in mice that is rescued by deletion of PARP-1 [51], suggesting that XRCC1 mutations lead to ataxia via the activation of PARP-1. Thus, ADP-ribosylation regulation of brain disease could be considered a spectrum, where mutations in the ADPr-hydrolase enzymes cause extreme and early onset neurological disease, whereas low-level, but perhaps long-term PARP activation by disease-causing factors, promotes age-related brain dysfunction.
Seminal findings of the role of PARP-1 in parthanatos have emerged from studies in disease models of ischemic stroke and PD (see below). The role of ADP-ribosylation in promoting protein aggregation in diseases such as ALS, FTD and PD is a more recent concept. Many of the disease pathways linked in ALS/FTD (reviewed in [52, 53]) are regulated by ADP-ribosylation including: 1) DNA damage; 2) protein localization; 3) protein aggregation; and 4) cell death. In the following sections we discuss the pathways regulated by ADP-ribosylation in relation to ALS/FTD and PD. We will consider the benefits and challenges in deciphering the potential of targeting ADP-ribosylation activity as a therapeutic strategy.
ALS/FTD and DNA damage
Brain disorders share remarkable overlap in genetics and affected pathways. Two exemplary diseases are ALS and FTD, which are collectively known as ALS/FTD (Figure 2 and Box 1). In >95% of ALS and ~45% of FTD the normally nuclear protein TDP-43 is mislocalized to the cytoplasm where it accumulates as phosphorylated protein in ubiquitin-positive inclusions (Figure 3 and Box 1). A second notable gene is FUS (Fused in sarcoma), which is mislocalized to the cytoplasm of affcted brain regions in <4% ALS and ~5% of FTD (Box 1). FUS is an RNA-binding protein that localizes to sites of DNA damage in a manner dependent upon PARP-1 (reviewed in [54]). More recently, TDP-43 was shown to facilitate the repair of damaged DNA [55, 56]. Thus, the loss of TDP-43 and FUS from the nucleus in ALS/FTD could impact the repair of damaged DNA in disease. Analysis of post-mortem tissue indicates that some of the early signs of DNA damage are present in the ALS spinal cord [46, 57, 58], suggesting that at the endpoint of ALS, motor neurons may be undergoing increased levels of DNA damage and/or their ability to repair damaged DNA is impaired. Increased DNA damage has also been detected in human iPSC-derived motor neurons derived from a variety of ALS genetic backgrounds [59–62]. However, it is unclear if DNA damage plays a vital role in disease progression.
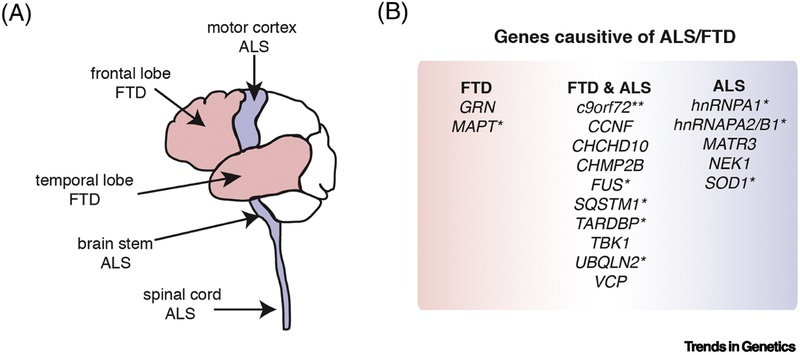
Figure 2. The ALS/FTD disease spectrum.
A) Upon autopsy many regions of the brain can present with neuronal loss and pathology in ALS and FTD. However, the most consistently affected regions in ALS are the motor cortex, brain stem and spinal cord, and for FTD the frontal and temporal cerebral lobes (reviewed in [98, 111]). B) Genes that when mutated give rise to either ALS, FTD or both ALS and FTD (reviewed in [52]). *Indicates genes whose protein products are known to aggregate in disease tissue. ** The mutation in c9orf72 is an intronic G4C2-hexanucleotide expansion that gives rise to abnormal accumulation of G4C2-hexanucleotide containing RNA and dipeptide-repeat proteins that are translated from the intronic repeat.
Box 1: The pathophysiology of ALS and FTD.
ALS and FTD (Figure 2) are fatal disorders for which there are few treatments and no cures. ALS is a motor neuron disease that commonly occurs in midlife, patients lose all motor control and typically die within 2–5 years of symptomatic onset (reviewed in [98]). In >95% of ALS, the normally nuclear protein TDP-43 is cleared from the nucleus (Figure 3) and instead accumulates as phosphorylated protein in ubiquitin positive inclusions in the cytoplasm reviewed in [98]. For the most part ALS is apparently sporadic, however, ~10% of cases are the result of inherited genetic mutations (reviewed in [98]). SOD1 (superoxide dismutase) was the first gene found to be mutated in ALS, and along with patients that harbor mutations in FUS (Fused in sarcoma), these patients lack TDP-43 pathology [106–108]. TDP-43 pathology is the hallmark of all other disease backgrounds, suggesting that disease-causing factors converge on TDP-43.
TDP-43 pathology is also observed in ~45% of FTD [106, 109, 110]. FTD, the second most common presenile dementia, is characterized by degeneration of the frontal and temporal lobes (Figure 2A). FTD leads to changes in behavior, language, personality and death within ~10 years of diagnosis. Up to 50% of FTD is the result of inherited mutations (reviewed in [111]). Intrigingly, at least 10 genes are shared by ALS and FTD (Figure 2B), including the most frequently affected gene, c9orf72 [52, 112, 113]. The clinical, genetic and pathological overlap between ALS and FTD has led to the understanding that these two diseases are on opposing ends of a shared ALS/FTD-disease spectrum [52]. TDP-43 pathology can also be observed in Alzheimer’s, Pick’s, Alexander’s disease and hippocampal sclerosis (reviewed in [114]), suggesting that dysfunction in multiple situations impact pathways that converge on TDP-43.
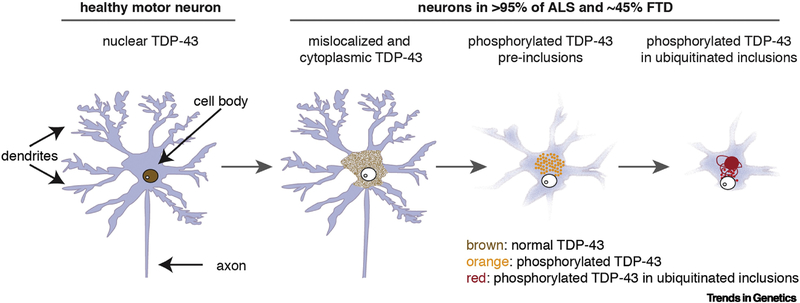
Figure 3: TDP-43 pathology in ALS/FTD.
In healthy neurons TDP-43 (brown) is nuclear. In > 95% of ALS, and ~45% of FTD, TDP-43 is cleared from the nucleus and accumulates in the cytoplasm. Cytoplasmic TDP-43 concentrates into phosphorylated foci (orange) that mature into ubiquitinated inclusions (red) [106, 109, 110].
ALS-associated mutations that occur in the nuclear localization sequence (NLS) of FUS reduce the recruitment of the protein to sites of DNA damage to varying degrees [58, 60, 63–65], they alter key protein interactions required for the repair of DNA [58, 65, 66], and they lower the efficiency of DDR in proliferating cells and human iPSC-derived motor neurons [58, 61, 63, 64]. However, while the mutations in the NLS of FUS cause neurodegenerative phenotypes in the mouse, DNA damage is only detected in ALS-FUS model with the most aggressive neurodegenerative symptoms [66, 67]. It is important to note that it is the RGG domain of FUS that binds to PAR and localizes the protein to sites of DNA damage [42, 63]. It is possible that mutations in the PAR-binding region (the RGG domain) of FUS may have a greater capacity to misregulate the function of the protein in the repair of damaged DNA.
It is still to be determined whether targeting DDR could be a potential therapeutic strategy in ALS/FTD. PARP-1/2 inhibitors mitigate the neurotoxicity of TDP-43 [46, 47], suggesting that downregulation of a response to DNA damage may be beneficial, however PARP-1/2 regulates many pathways. As discussed below, excessive DNA damage and activation of PARP-1 promotes PAR-mediated protein aggregation and cell loss. In the following sections, we discuss abnormal protein localization and cell death in ALS/FTD and the role that PAR may play in these processes.
Protein mislocalization in ALS/FTD
The disease-associated pathology of TDP-43 and FUS suggest that nucleocytoplasmic transport is affected in disease, thus, agents that can return TDP-43 and FUS back to the nucleus are of therapeutic interest (reviewed in [68]). Studies that launched from Drosophila uncovered that a reduction of cytoplasmic (PARP-5a/5b) ADP-ribosylation activity decreases the accumulation of TDP-43 in the cytoplasm and mitigates TDP-43 neurotoxicity [43]. Additionally, inhibition of nuclear ADP-ribosylation (PARP-1/2) mitigates neurotoxicity in rodent neurons and reduces arsenite-induced accumulation of TDP-43 in the cytoplasm of mammalian cells [44, 47]. Furthermore, upon treatment with hydrogen peroxide, inhibition of PARP-1/2 inhibits cytoplasmic accumulation of FUS [69]. Collectively, these data indicate that PARP-1/2 and PARP-5a/5b can regulate the nuclear and cytoplasmic levels of TDP-43 and FUS. PARP-1 regulates nucleocytoplasmic localization of several proteins (reviewed in [70]). It remains to be seen how PARP-1/2 promotes TDP-43, and FUS, accumulation in the cytoplasm, whether it involves a nuclear export mechanism, and whether PARP-1/2 works together with PARP-5a/5b.
There is, however, evidence to suggest that stress is involved in TDP-43 pathology in the ALS spinal cord [71–73]. ADP-ribosylation activity is widely recognized as a stress-induced protein modification (see above). In the following section we discuss the role of stress and the ways in which stress-activated ADP-ribosylation may regulate cytoplasmic protein aggregation in ALS/FTD.
Protein aggregation in ALS/FTD
Some proteins have an intrinsic capacity to undergo fibrilization and these proteins, including TDP-43 and FUS, often harbor an intrinsically disordered region (IDR) (reviewed in [74]). Abnormally aggregated proteins differ from functional high-molecular weight fibrils in that they have precipitated into insoluble clumps of dysfunctional protein; however, similar mechanisms can induce both types of fibrilization (reviewed in [75]). Elucidation of these biophysical properties is uncovering critical insight into disease progression. A thermodynamic phenomenon linked to normal and abnormal protein fibrilization is liquid-liquid phase separation (LLPS). LLPS occurs when inter- and intra-molecular protein interactions are stronger than the interactions the protein has with the surrounding liquid, such that protein solubility is reduced and LLPS is promoted (reviewed in [76]). Multivalent interactions determine the phase separation properties of a protein, influencing factors include protein domains, e.g. an IDR (reviewed in [77]) and extrinsic factors like osmolarity and interactions with charged molecules, such as nucleic acids and PAR (Figure 4A). The role of PAR in localizing proteins is well established; however, more recently it has been proposed (see below) that PAR may localize proteins by promoting LLPS.
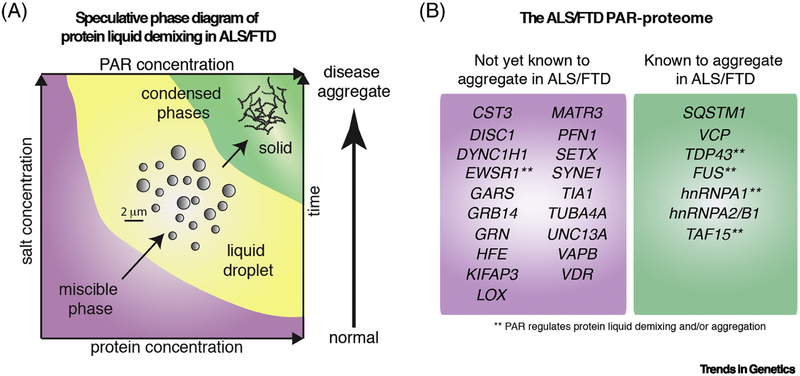
Figure 4: ALS/FTD-associated genes and their relationship with PARP activity.
A) A speculative phase diagram representing the conditions that may regulate protein phase separation. PAR promotes liquid-liquid phase separation of TDP-43, FUS and hnRNAPA1 [43, 47, 85]; however, long-term incubation of PAR with FUS, TAF15 and EWSR1 promotes insoluble protein aggregation [42]. Thus, PAR-mediated phase separation may initially promote phase separation, but with time disease-like protein fibrils are formed. B) Proteins of ALS/FTD that are known to be PARylated or interact with PAR. The 128 genes and corresponding proteins from the amyotrophic lateral sclerosis online genetics database ((http://alsod.iop.kcl.ac.uk/) genes associated with ALS, and causative for ALS and FTD), plus the FTD genes TIA-1 and KIF5A, were cross-referenced to the PAR proteome [84, 116–119]. Proteins identified as directly PARylated or non-covalent binders of PAR are listed and grouped into proteins that are not yet known to aggregate in disease and those that are known to aggregate in disease.
In the cell, LLPS governs the formation of non-membrane bound, RNA-rich organelles, including P bodies and stress granules (reviewed in [78]), the latter of which are enriched for PAR [43, 79, 80]. Stress granules inhibit global translation by sequestering translation pre-initiation complexes in the cytoplasm during times of stress (reviewed in [81]). Several mechanisms regulate stress granules such as interactions mediated by IDR domains (reviewed in [78]). Bioinformatic analyses has revealed overlap between PAR-binding and ADP-ribsoylated proteins with IDR-containing proteins and proteins that localize to stress granules [42, 82, 83], suggesting that PAR may influence LLPS of IDR-containing proteins. Indeed, stress-granule proteins have been shown to be PARylated in cells exposed to chemical stressors [47, 79]. It is not known which PARP promotes PARylation of stress-granule proteins or where the PARylation happens in the cell. It is known that 5 PARPs (PARP-5a, -12, -13.1, -13.2 and -14) localize to stress granules [43, 79, 80], that PARP-1 activity promotes stress-granule formation [46, 47, 80, 84], and that PARP-12 translocates from the Golgi to stress granules in a manner dependent upon PARP-1 activity and its WWE domain (a PAR-binding module (Figure 1)) [80]. Moreover, after the removal of stress, stress granules resolve, however, upon inhibition of PARG, stress granules persist [47, 79]. This suggests that PARG promotes stress-granule dissolution. In the in vitro setting, PAR promotes LLPS of TDP-43, FUS and hnRNPA1 [42, 43, 47, 85], and PARG can dissolve FUS aggregates localized to DNA damage [69] suggesting that in the cell, PAR promotes localized phase separation in stress granules, as well as sites of DNA damage.
Many of the proteins that cause ALS/FTD, including TDP-43, localize to stress granules (reviewed in [86]), thus it was proposed that stress granules may be the initial seed for the abnormal TDP-43 aggregates. TDP-43 harbors a PAR-binding motif (PBM), which when mutated prevents PAR binding and TDP-43 recruitment to stress granules [43]. TDP-43 unable to bind to PAR forms protein aggregates excluded from stress granules that are aberrantly phosphorylated [43], suggesting that PAR-dependent recruitment of TDP-43 to stress granules protects the protein from disease-associated phosphorylation. The finding that TDP-43 aggregates that form outside of stress granules are pathological has been confirmed in independent studies that also demonstrate that phosphorylated TDP-43 aggregates are neurotoxic [87–89]. Collectively, these studies indicate that PAR-mediated aggregation of TDP-43 is important for preventing disease-associated aggregation, and they suggest that in conditions where βNAD+ and PAR may be limiting, stress may give rise to neurotoxic aggregation of TDP-43.
An increased concentration of ALS/FTD-associated proteins in liquid droplets can promote pathological fibrilization [85, 90–92]. Thus, LLPS provides an environment for aggregation-prone proteins to fibrilize and mature into insoluble precipitates over time. In the cell, exposure to chronic stress leads to a dissolution of stress granules and the formation of phosphorylated TDP-43 aggregates akin to those observed in disease [43]. Thus, although PAR-recruitment to stress granules is initially protective (see above), PAR is an early seeding agent of protein-rich liquid droplets that with time can transition into insoluble disease-like aggregates (Figure 4A). Indeed, long-term incubation of FUS, TAF15 and EWSR1 with PAR leads to the formation of disease-like aggregates in vitro [42]. ADP-ribosylation in ALS/FTD could be considered a catch-22: it facilitates the localization of protein into soluble and protective environments, that unfortunately also have the capacity to transition the proteins into toxic species. A therapeutic strategy could be to ensure proper stress-granule localization and the timely resolution of stress granules [43]. Long-term activation of stress and ADP-ribosylation activity is also known to lead to cell death. In the following section we discuss PAR-mediated cell death and ALS/FTD.
Hyperactivation of PARP-1 and parthanatos
PARP-1 aids the response to stress by assisting in cell survival or cell death. The defining factor correlates with the extent to which PARP-1 is automodified. Upon hyper-automodification of PARP-1, PAR is cleaved and released into the cytoplasm where it elicits a caspase-independent cell-death mechanism called parthanatos (reviewed in [93]). Parthanatos has been studied for > 2 decades, and much of our understanding has come from cellular and animal models of PD and ischemic stroke, the latter of which is associated with glutamate excitotoxicity (reviewed in [94]). Glutamate excitotoxicity lead to a hyper-activation of PARP-1, the subsequent translocation of PAR from the nucleus the cytoplasm, where it binds to Apoptosis Inducing Factor (AIF) releasing the protein from the mitochondria and into the cytoplasm [95]. In the cytoplasm, AIF binds to the nuclease Macrophage Migration Inhibitory Factor (MIF), both proteins translocate into the nucleus and MIF digests the genomic DNA, triggering cell death [96]. The protein(s) that mediate transport of PAR into the cytoplasm and to the mitochondria during parthanatos remain enigmatic. Reducing PARP-1 in cellular and rodent models of stroke mitigates neuronal toxicity [45, 48–50], indicating that PARP-1 activation is important for disease progression.
In a mouse model of PD, preformed fibrils of α-synuclein (α-syn) injected into the cortex leads to excitotoxicity, PAR-induced aggregation of α-syn, and a neurotoxicity that is mitigated by inhibition of PARP-1/2 [45]. These studies suggested a feed-forward mechanism whereby the excitotoxicity induced by the preformed α-syn fibrils hyperactivates PARP-1, which leads to the release of PAR into the cytoplasm where it interacts with α-syn, promoting more aggregation and toxicity, which in turn further activates PARP-1. Intriguingly, measurements of the cerebral spinal fluid of a cohort of PD patients revealed elevated levels of PAR compared to controls [45], suggesting that PAR poses as a potential biomarker for PD. Elevated levels of PAR have also been detected in the nuclei of spinal cord motor neurons in ALS patients [46]; is it possible that a similar mechanism regulates TDP-43 and that PAR is a potential biomarker in the cerebral spinal fluid ALS/FTD?
Neuronal excitotoxic mechanisms known to activate PARP-1 in parthanatos, such as glutamate excitotoxicity, ER stress, mitochondrial stress and increased ROS [93, 97] have been widely implicated in ALS/FTD [98]. For example, glutamate excitotoxicity was one of the first pathways implicated in ALS (reviewed in [98]). FTD- and ALS-causative mutations occur in genes involved in ER homeostasis (Valosin-Containing Protein (VCP) (reviewed in [52]) and in mitochondrial ROS production (superoxide dismutase (SOD1) (reviewed in [98]). Could PARP activation link the stress pathways activated in disease to TDP-43 nuclear export, TDP-43 cytoplasmic aggregation and neuronal death (Figure 5, key figure), and if so how? For example, PARP-1/2 and PARP-5a/5b promote TDP-43 accumulation in the cytoplasm; is PAR the direct facilitator or is it acting via an intermediary protein or proteins? Once in the cytoplasm, PAR incorporates TDP-43 into stress granules, but under prolonged stress, stress granules resolve, leaving behind phosphorylated TDP-43; does the resolution of chronic stress granules also release PAR into the cytoplasm to initiate parthanatos? Inhibition of PARP-1/2 and PARP-5a/5b mitigate toxicity in neuronal models, will the identification of pathways that activate ADP-ribosylation in disease provide insight into earlier intervention strategies? Finally, PAR is upregulated in motor neuron post-mortem spinal cord tissue, is it also elevated in patient cerebral spinal fluid? Addressing these questions may help to elucidate the potential of targeting ADP-ribosylation activity in ALS/FTD.
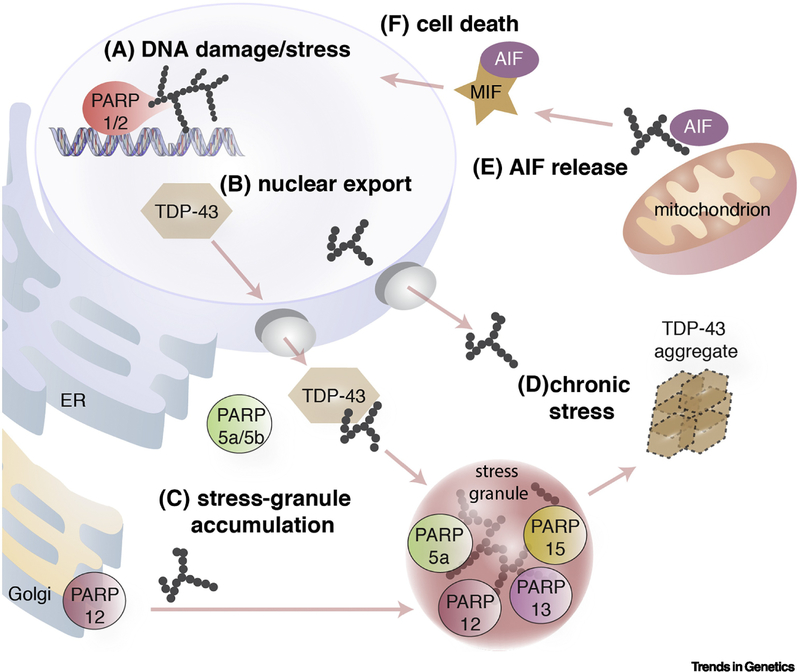
Figure 5 (Key Figure). Model for ADP-ribosylation in ALS/FTD.
A) Upon activation of stress pathways PARP-1 becomes auto-modified with PAR.
B) Stress-induced accumulation of TDP-43 in the cytoplasm is promoted by PARP-1/2 and PARP-5a/5b activity.
C) The activity of PARP-1 promotes cytoplasmic stress-granule formation. PARP-12 tranlsocates from the Golgi to stress granules where it accumulates alongside PARP-5a, -12, -13 and -15. TDP-43 is recruited to stress granules by binding to PAR.
D) Under conditions of chronic stress, TDP-43 aggregates form that lack stress granule proteins and are phosphorylated.
E) Chronic stress also induces PAR-mediated release of Apoptosis Inducing Factor (AIF) from mitochondria. It is unknown whether the chronic stress that leads to TDP-43 aggregation also leads to AIF release from mitochondria.
F) AIF in the cytoplasm binds to and forms a protein complex with Macrophage migration inhibitory factor (MIF), which translocates to the nucleus where MIF digests the genomic DNA leading to death of the cell.
Concluding Remarks and Future Perspectives
Inhibition of ADP-ribosylation activity remarkably mitigates the neurodegenerative features in a number of animal disease models (Table 1), enforcing the suggestion that PARP inhibitors, developed as cancer therapeutics, may be an option for the treatment of neurological disease [27]. We know very little of the role of ADP-ribosylation in the brain, such as which proteins are modified and how these modified or unmodified proteins regulate brain development, function and integrity. Would long-term inhibition of PARP activity have a detrimental effect on the normal function of the brain, and how would long-term PARP inhibition impact severe neurological disease? Perhaps the identification of ADP-ribsoylated proteins in disease could lead to the generation of pathway-specific inhibitors with minimal side effects on other cellular pathways regulated by ADP-ribosylation.
Table 1:
Rodent models of neurodegenerative disease that shown benefit from therapeutic inhibition of PARP-1/2.
| Disease | Model | Inhibitors | Treatment | Sacrifice | Effect of PARP-1/2 inhibitor | ref. |
|---|---|---|---|---|---|---|
| AD | rat - Injection of Aβ(1–42) into hippocampal C3 region | niacin | daily i.p.i. for 7 d | 7 d post Aβ(1–42) injection | reduced lipid peroxidation & ROS | [120] |
| HD | mouse – transgenic for exon 1 of Htt with 150 CAG repeats driven by human Htt promoter | daily i.p.i. from 28 d onward | / | improved lifespan & motor deficits | [121] | |
| Ischemic stroke | mouse –MCAO & ACAO | INH2BP | i.p.i. 2 hr before occlusion. | 25 hr/75 hr post MCAO & ACAO | inhibition of PARP-1/2 activity & reduced infarct size | [122] |
| mouse – MCAO | PJ34 | i.p.i. 1 hr and 2hr before occlusion | 24 hr & 72 hr post MCAO | reduced iNOS expression & reduced infarct size | [123] | |
| mouse – MCAO | DPQ | i.p.i. 1 hr and 2hr before occlusion | 24 hr & 72 hr post MCAO | reduced infarct size | [123] | |
| mouse – MCAO | DPQ | i.p.i. 2 hr before and 2 hr after occlusion | 24 hr post MCAO | reduced infarct size | [124] | |
| PD | mouse – i.p.i. of MPTP 4 times at 1hr intervals | benzamide | i.p.i. 30 min before and 90 min after first MPTP injection | 5 d post MPTP injection | mitigated loss of tyrosine hydroxylase | [125] |
| mouse – unilateral injection of α-syn PFF into striatum | ABT-888 | Mice were fed ABT-888 | 3 & 6 mo post injection of α-syn PFF | improved motor deficits (3 mo post α-syn PFF injection) & mitigated loss of DA neurons (6 mo post α-syn PFF injection) | [45] | |
Abbreviations: AD: Alzheimer’s disease; HD: Huntington’s disease; PD: Parkinson’s disease; MCAO: middle cerebral artery occlusion; ACAO anterior cerebral artery occlusion; i.p.i.: intraperitoneal injection; PFF: preformed fibril; MPTP: 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine; iNOS: induced nitric oxide synthase; INH2BP: 5-iodo-6-amino-1,2-benzopyrone; DPQ: 4-dihydro-5-[4(1-piperidinl)butoxyl]-1-(2H)-isoquinolinone; mo: month; DA: dopaminergic neurons.
In the past, uncovering the pathways regulated by ADP-ribosylation has been limited by the detection of the ADP-ribsoylated substrates (either proteins directly modified or that bind the ADPr or PAR modification), including the conditions that maintain the modification and the conditions that allow for the identification of the substrates by mass spectrometry. Recent advances have been made in the development of methods that identify ADP-ribsoylated targets [99–102] and that measure the levels of ADP-ribose [100]. Could these state-of-the-art techniques assist in identifying ADP-ribsoylated targets in the aging brain? Could it be that PARP-1, which is the most active and highly modified substrate known to date, is the substrate central to neurological disease characterized by abnormal ADP-ribosylation? If not, and rather that PARP-1 is simply one of many proteins abnormally modified in disease, how will the ADP-ribsoylated substrates critical for brain function or specific to disease be identified?
Despite the need for greater understanding of ADP-ribosylation function in the brain, initial studies in cells and animal models indicate that inhibition of PARP activity is a promising therapeutic for neurological disease, including for ALS/FTD. Important advances have been made in developing rodent models of ALS/FTD that recapitulate features of the human disease [103–105]. Given the efficacy of PARP inhibition in rodent models of PD and ischemic stroke, and the reported benefit of PARP inhibition in neuronal models of ALS/FTD, PARP inhibitors may be of therapeutic benefit in ALS/FTD rodent models.
Outstanding Questions Box.
Which proteins are substrates of ADP-ribosylation in the brain, how does their ADP-ribosylation modulate brain function and how might their misregulation by ADP-ribosylation contribute to neurodegeneration?
Is increased ADP-ribosylation driving disease progression in ALS? And does this directly promote nuclear export and aggregation of TDP-43?
Is disease-like aggregation of TDP-43 linked to parthanatos and is parthanatos occurring in ALS/FTD?
Will PARP inhibitors be beneficial in ALS/FTD rodent models? How will potential negative effects of PARP inhibitors be prioritized and balanced against potential side effects from long term PARP inhibition?
Highlights.
ADP-ribose activity is essential for brain development, function and integrity.
Poly(ADP-ribose) promotes disease-like aggregation of proteins causative of ALS and FTD.
Inhibition of PARP activity mitigates neurotoxicity of ALS- and FTD-causing proteins.
GLOSSARY
- CNV
Copy number variants, are either duplications or deletions of sections of the genome. There is variation in CNVs in the general population and some are positively associated with disease.
- DDR
The DNA-damage response (DDR) is initiated upon detection of a DNA lesion, such as a singlestrand break (SSB), double-strand break (DSB), mismatches, insertion/deletion loops, and abnormal DNA bases.
- DSB
Double-strand breaks (DSBs) are lesions in DNA that affect both strands of DNA.
- MacroD1 and MacroD2
MacroD1 and MacroD2 are mono(ADP-ribose) hydrolases that cleave and remove the terminal ADP-ribose from the target protein.
- MARylation
Mono(ADP-ribosylation) is the addition of a single ADP-ribose subunit on to a target protein via the hydrolysis of βNAD+.
- PAR
Poly(ADP-ribose) is a post-translational modification comprised of multiple ADP-ribose molecules in a single chain or in branched forms.
- PAR-binding motif
A consensus motif consisting of hydrophobic and basic residues that bind to the negatively charged PAR polymer.
- PAR interactome
The PAR interactome is comprised of proteins that bind to PAR non-covalently or are direct targets of PARylation.
- PAR-reader protein
PAR reader proteins are proteins that bind to PAR non-covalently.
- Parthanatos
A caspase-independent cell-death mechanism that is triggered by the release of PAR into the cytoplasm. The name is derived from Greek mythology: Thanatos is the personification of death.
- PARP
Poly(ADP-ribose) polymerases, or PARPs, are an enzyme family characterized by the presence of an ADP-ribose transferase catalytic domain. Within the family, the 17 PARPs have structural, enzymatic and functional differences.
- PARG
Poly(ADP-ribose) glycohydrolase, or PARG, is an enzyme that hydrolyzes PAR at glycosidic linkages to form free ADP-ribose molecules.
- PARylation
Poly(ADP-ribosylation) is the covalent addition of sequential subunits of ADP-ribose onto an acceptor protein to generate linear and sometimes branched polymers of poly(ADP-ribose).
- TARG1
Terminal ADP-ribose protein glycohydrolase, or TARG1, is an enzyme that cleaves the ester bond between the terminal ADP-ribose molecule and the target protein.
- SNP
A single nucleotide polymorphism is a nucleotide in the DNA sequence that differs from that of the general population. Some SNPs are significantly correlated with disease.
- SSB
Single-strand breaks (SSBs) are lesions in DNA that affect one strand of DNA that can elicit the DNA-damage response.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gupte R et al. (2017) PARPs and ADP-ribosylation: recent advances linking molecular functions to biological outcomes. Genes Dev 31 (2), 101–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crawford K et al. (2018) Specificity of reversible ADP-ribosylation and regulation of cellular processes. Crit Rev Biochem Mol Biol 53 (1), 64–82. [DOI] [PubMed] [Google Scholar]
- 3.Cohen MS and Chang P (2018) Insights into the biogenesis, function, and regulation of ADP-ribosylation. Nat Chem Biol 14 (3), 236–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perina D et al. (2014) Distribution of protein poly(ADP-ribosyl)ation systems across all domains of life. DNA Repair (Amst) 23, 4–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aravind L et al. (2015) The natural history of ADP-ribosyltransferases and the ADP-ribosylation system. Curr Top Microbiol Immunol 384, 3–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vyas S et al. (2014) Family-wide analysis of poly(ADP-ribose) polymerase activity. Nat Commun 5, 4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leslie Pedrioli DM et al. (2018) Comprehensive ADP‐ribosylome analysis identifies tyrosine as an ADP‐ribose acceptor site. EMBO reports 19 (8), e45310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin W et al. (1997) Isolation and characterization of the cDNA encoding bovine poly(ADP-ribose) glycohydrolase. J Biol Chem 272 (18), 11895–901. [DOI] [PubMed] [Google Scholar]
- 9.Slade D et al. (2011) The structure and catalytic mechanism of a poly(ADP-ribose) glycohydrolase. Nature 477 (7366), 616–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fontana P et al. (2017) Serine ADP-ribosylation reversal by the hydrolase ARH3. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oka S et al. (2006) Identification and characterization of a mammalian 39-kDa poly(ADP-ribose) glycohydrolase. J Biol Chem 281 (2), 705–13. [DOI] [PubMed] [Google Scholar]
- 12.Rosenthal F et al. (2013) Macrodomain-containing proteins are new mono-ADP-ribosylhydrolases. Nat Struct Mol Biol 20 (4), 502–7. [DOI] [PubMed] [Google Scholar]
- 13.Jankevicius G et al. (2013) A family of macrodomain proteins reverses cellular mono-ADP-ribosylation. Nat Struct Mol Biol 20 (4), 508–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharifi R et al. (2013) Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease. EMBO J 32 (9), 1225–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palazzo L et al. (2016) ENPP1 processes protein ADP-ribosylation in vitro. FEBS J 283 (18), 3371–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palazzo L et al. (2015) Processing of protein ADP-ribosylation by Nudix hydrolases. Biochem J 468 (2), 293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daniels CM et al. (2015) Nudix hydrolases degrade protein-conjugated ADP-ribose. Sci Rep 5, 18271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Breslin C et al. (2015) The XRCC1 phosphate-binding pocket binds poly (ADP-ribose) and is required for XRCC1 function. Nucleic Acids Res 43 (14), 6934–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li M et al. (2013) The FHA and BRCT domains recognize ADP-ribosylation during DNA damage response. Genes Dev 27 (16), 1752–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fisher AE et al. (2007) Poly(ADP-ribose) polymerase 1 accelerates single-strand break repair in concert with poly(ADP-ribose) glycohydrolase. Mol Cell Biol 27 (15), 5597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abplanalp J and Hottiger MO (2017) Cell fate regulation by chromatin ADP-ribosylation. Semin Cell Dev Biol 63, 114–122. [DOI] [PubMed] [Google Scholar]
- 22.Posavec Marjanovic M et al. (2017) PARP, transcription and chromatin modeling. Semin Cell Dev Biol 63, 102–113. [DOI] [PubMed] [Google Scholar]
- 23.Wright RH et al. (2016) Insight into the machinery that oils chromatin dynamics. Nucleus 7 (6), 532–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mariotti L et al. (2017) Regulation of Wnt/beta-catenin signalling by tankyrase-dependent poly(ADP-ribosyl)ation and scaffolding. Br J Pharmacol 174 (24), 4611–4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Lange T (2018) Shelterin-Mediated Telomere Protection. Annu Rev Genet 52, 223–247. [DOI] [PubMed] [Google Scholar]
- 26.Langelier MF et al. (2018) PARP family enzymes: regulation and catalysis of the poly(ADP-ribose) posttranslational modification. Curr Opin Struct Biol 53, 187–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berger NA et al. (2018) Opportunities for the repurposing of PARP inhibitors for the therapy of non-oncological diseases. Br J Pharmacol 175 (2), 192–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu B et al. (2009) Elucidating the genetic architecture of familial schizophrenia using rare copy number variant and linkage scans. Proc Natl Acad Sci U S A 106 (39), 16746–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lesca G et al. (2012) Epileptic encephalopathies of the Landau-Kleffner and continuous spike and waves during slow-wave sleep types: genomic dissection makes the link with autism. Epilepsia 53 (9), 1526–38. [DOI] [PubMed] [Google Scholar]
- 30.Egger G et al. (2014) Identification of risk genes for autism spectrum disorder through copy number variation analysis in Austrian families. Neurogenetics 15 (2), 117–27. [DOI] [PubMed] [Google Scholar]
- 31.Autism Spectrum Disorders Working Group of The Psychiatric Genomics, C. (2017) Meta-analysis of GWAS of over 16,000 individuals with autism spectrum disorder highlights a novel locus at 10q24.32 and a significant overlap with schizophrenia. Mol Autism 8, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Debette S et al. (2010) Genome-wide association studies of MRI-defined brain infarcts: meta-analysis from the CHARGE Consortium. Stroke 41 (2), 210–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baranzini SE et al. (2009) Genome-wide association analysis of susceptibility and clinical phenotype in multiple sclerosis. Hum Mol Genet 18 (4), 767–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anney R et al. (2010) A genome-wide scan for common alleles affecting risk for autism. Hum Mol Genet 19 (20), 4072–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Curran S et al. (2011) No association between a common single nucleotide polymorphism, rs4141463, in the MACROD2 gene and autism spectrum disorder. Am J Med Genet B Neuropsychiatr Genet 156B (6), 633–9. [DOI] [PubMed] [Google Scholar]
- 36.Grove J et al. (2019) Identification of common genetic risk variants for autism spectrum disorder. Nat Genet 51 (3), 431–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones RM et al. (2014) MACROD2 gene associated with autistic-like traits in a general population sample. Psychiatr Genet 24 (6), 241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuo PH et al. (2015) Genome-Wide Association Study for Autism Spectrum Disorder in Taiwanese Han Population. PLoS One 10 (9), e0138695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Namjou B et al. (2014) Phenome-wide association study (PheWAS) in EMR-linked pediatric cohorts, genetically links PLCL1 to speech language development and IL5-IL13 to Eosinophilic Esophagitis. Front Genet 5, 401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duan S et al. (2008) Genetic architecture of transcript-level variation in humans. Am J Hum Genet 82 (5), 1101–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ghosh SG et al. (2018) Biallelic Mutations in ADPRHL2, Encoding ADP-Ribosylhydrolase 3, Lead to a Degenerative Pediatric Stress-Induced Epileptic Ataxia Syndrome. Am J Hum Genet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Altmeyer M et al. (2015) Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose). Nat Commun 6, 8088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McGurk L et al. (2018) Poly(ADP-Ribose) Prevents Pathological Phase Separation of TDP-43 by Promoting Liquid Demixing and Stress Granule Localization. Mol Cell 71 (5), 703–717 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McGurk L et al. (2018) Poly(ADP-ribose) engages the TDP-43 nuclear-localization sequence to regulate granulo-filamentous aggregation. Biochemistry 57 (51), 6923–6926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kam TI et al. (2018) Poly(ADP-ribose) drives pathologic alpha-synuclein neurodegeneration in Parkinson’s disease. Science 362 (6414). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McGurk L et al. (2018) Nuclear poly(ADP-ribose) activity is a therapeutic target in amyotrophic lateral sclerosis. Acta Neuropathol Commun 6 (1), 84–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duan Y et al. (2019) PARylation regulates stress granule dynamics, phase separation, and neurotoxicity of disease-related RNA-binding proteins. Cell Res 29 (3), 233–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu SW et al. (2002) Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science 297 (5579), 259–63. [DOI] [PubMed] [Google Scholar]
- 49.Goto S et al. (2002) Poly(ADP-ribose) polymerase impairs early and long-term experimental stroke recovery. Stroke 33 (4), 1101–6. [DOI] [PubMed] [Google Scholar]
- 50.Li X et al. (2010) Contributions of poly(ADP-ribose) polymerase-1 and -2 to nuclear translocation of apoptosis-inducing factor and injury from focal cerebral ischemia. J Neurochem 113 (4), 1012–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoch NC et al. (2017) XRCC1 mutation is associated with PARP1 hyperactivation and cerebellar ataxia. Nature 541 (7635), 87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao FB et al. (2017) Dysregulated molecular pathways in amyotrophic lateral sclerosis-frontotemporal dementia spectrum disorder. EMBO J 36 (20), 2931–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Balendra R and Isaacs AM (2018) C9orf72-mediated ALS and FTD: multiple pathways to disease. Nat Rev Neurol 14 (9), 544–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sama RR et al. (2014) Functions of FUS/TLS from DNA repair to stress response: implications for ALS. ASN Neuro 6 (4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hill SJ et al. (2016) Two familial ALS proteins function in prevention/repair of transcription-associated DNA damage. Proc Natl Acad Sci U S A 113 (48), E7701–E7709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mitra J et al. (2019) Motor neuron disease-associated loss of nuclear TDP-43 is linked to DNA double-strand break repair defects. Proc Natl Acad Sci U S A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Farg MA et al. (2017) The DNA damage response (DDR) is induced by the C9orf72 repeat expansion in amyotrophic lateral sclerosis. Hum Mol Genet 26 (15), 2882–2896. [DOI] [PubMed] [Google Scholar]
- 58.Wang WY et al. (2013) Interaction of FUS and HDAC1 regulates DNA damage response and repair in neurons. Nat Neurosci 16 (10), 1383–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lopez-Gonzalez R et al. (2016) Poly(GR) in C9ORF72-Related ALS/FTD Compromises Mitochondrial Function and Increases Oxidative Stress and DNA Damage in iPSC-Derived Motor Neurons. Neuron 92 (2), 383–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Naumann M et al. (2018) Impaired DNA damage response signaling by FUS-NLS mutations leads to neurodegeneration and FUS aggregate formation. Nat Commun 9 (1), 335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Higelin J et al. (2016) FUS Mislocalization and Vulnerability to DNA Damage in ALS Patients Derived hiPSCs and Aging Motoneurons. Front Cell Neurosci 10, 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Higelin J et al. (2018) NEK1 loss-of-function mutation induces DNA damage accumulation in ALS patient-derived motoneurons. Stem Cell Res 30, 150–162. [DOI] [PubMed] [Google Scholar]
- 63.Mastrocola AS et al. (2013) The RNA-binding protein fused in sarcoma (FUS) functions downstream of poly(ADP-ribose) polymerase (PARP) in response to DNA damage. J Biol Chem 288 (34), 24731–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rulten SL et al. (2014) PARP-1 dependent recruitment of the amyotrophic lateral sclerosis-associated protein FUS/TLS to sites of oxidative DNA damage. Nucleic Acids Res 42 (1), 307–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang H et al. (2018) Mutant FUS causes DNA ligation defects to inhibit oxidative damage repair in Amyotrophic Lateral Sclerosis. Nat Commun 9 (1), 3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Qiu H et al. (2014) ALS-associated mutation FUS-R521C causes DNA damage and RNA splicing defects. J Clin Invest 124 (3), 981–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lopez-Erauskin J et al. (2018) ALS/FTD-Linked Mutation in FUS Suppresses Intra-axonal Protein Synthesis and Drives Disease Without Nuclear Loss-of-Function of FUS. Neuron 100 (4), 816–830 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guo L et al. (2019) Therapeutic Dissolution of Aberrant Phases by Nuclear-Import Receptors. Trends Cell Biol 29 (4), 308–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Singatulina AS et al. (2019) PARP-1 Activation Directs FUS to DNA Damage Sites to Form PARG-Reversible Compartments Enriched in Damaged DNA. Cell Rep 27 (6), 1809–1821 e5. [DOI] [PubMed] [Google Scholar]
- 70.Grimaldi G and Corda D (2019) ADP-ribosylation and intracellular traffic: an emerging role for PARP enzymes. Biochem Soc Trans 47 (1), 357–370. [DOI] [PubMed] [Google Scholar]
- 71.McGurk L et al. (2014) Poly-A binding protein-1 localization to a subset of TDP-43 inclusions in amyotrophic lateral sclerosis occurs more frequently in patients harboring an expansion in C9orf72. J Neuropathol Exp Neurol 73 (9), 837–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bentmann E et al. (2012) Requirements for stress granule recruitment of fused in sarcoma (FUS) and TAR DNA-binding protein of 43 kDa (TDP-43). J Biol Chem 287 (27), 23079–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu-Yesucevitz L et al. (2010) Tar DNA binding protein-43 (TDP-43) associates with stress granules: analysis of cultured cells and pathological brain tissue. PLoS One 5 (10), e13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Harrison AF and Shorter J (2017) RNA-binding proteins with prion-like domains in health and disease. Biochem J 474 (8), 1417–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Franzmann TM and Alberti S (2019) Protein Phase Separation as a Stress Survival Strategy. Cold Spring Harb Perspect Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Banani SF et al. (2017) Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 18 (5), 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Boeynaems S et al. (2018) Protein Phase Separation: A New Phase in Cell Biology. Trends Cell Biol 28 (6), 420–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shin Y and Brangwynne CP (2017) Liquid phase condensation in cell physiology and disease. Science 357 (6357), doi: 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- 79.Leung AK et al. (2011) Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol Cell 42 (4), 489–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Catara G et al. (2017) PARP1-produced poly-ADP-ribose causes the PARP12 translocation to stress granules and impairment of Golgi complex functions. Sci Rep 7 (1), 14035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ivanov P et al. (2018) Stress Granules and Processing Bodies in Translational Control. Cold Spring Harb Perspect Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Leung AK (2014) Poly(ADP-ribose): an organizer of cellular architecture. J Cell Biol 205 (5), 613–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fischbach A et al. (2018) The C-terminal domain of p53 orchestrates the interplay between non-covalent and covalent poly(ADP-ribosyl)ation of p53 by PARP1. Nucleic Acids Res 46 (2), 804–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Isabelle M et al. (2012) Quantitative proteomics and dynamic imaging reveal that G3BP-mediated stress granule assembly is poly(ADP-ribose)-dependent following exposure to MNNG-induced DNA alkylation. J Cell Sci 125 (Pt 19), 4555–66. [DOI] [PubMed] [Google Scholar]
- 85.Patel A et al. (2015) A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell 162 (5), 1066–77. [DOI] [PubMed] [Google Scholar]
- 86.Li YR et al. (2013) Stress granules as crucibles of ALS pathogenesis. J Cell Biol 201 (3), 361–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mann JR et al. (2019) RNA Binding Antagonizes Neurotoxic Phase Transitions of TDP-43. Neuron. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen Y and Cohen TJ (2019) Aggregation of the nucleic acid-binding protein TDP-43 occurs via distinct routes that are coordinated with stress granule formation. J Biol Chem 294 (10), 3696–3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gasset-Rosa F et al. (2019) Cytoplasmic TDP-43 De-mixing Independent of Stress Granules Drives Inhibition of Nuclear Import, Loss of Nuclear TDP-43, and Cell Death. Neuron. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xiang S et al. (2015) The LC Domain of hnRNPA2 Adopts Similar Conformations in Hydrogel Polymers, Liquid-like Droplets, and Nuclei. Cell 163 (4), 829–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lin Y et al. (2015) Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol Cell 60 (2), 208–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Molliex A et al. (2015) Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163 (1), 123–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dawson TM and Dawson VL (2017) Mitochondrial Mechanisms of Neuronal Cell Death: Potential Therapeutics. Annu Rev Pharmacol Toxicol 57, 437–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fatokun AA et al. (2014) Parthanatos: mitochondrial-linked mechanisms and therapeutic opportunities. Br J Pharmacol 171 (8), 2000–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang Y et al. (2011) Poly(ADP-ribose) (PAR) binding to apoptosis-inducing factor is critical for PAR polymerase-1-dependent cell death (parthanatos). Sci Signal 4 (167), ra20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang Y et al. (2016) A nuclease that mediates cell death induced by DNA damage and poly(ADP-ribose) polymerase-1. Science 354 (6308). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kaus A and Sareen D (2015) ALS Patient Stem Cells for Unveiling Disease Signatures of Motoneuron Susceptibility: Perspectives on the Deadly Mitochondria, ER Stress and Calcium Triad. Front Cell Neurosci 9, 448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Taylor JP et al. (2016) Decoding ALS: from genes to mechanism. Nature 539 (7628), 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bonfiglio JJ et al. (2017) Mass spectrometry for serine ADP-ribosylation? Think o-glycosylation! Nucleic Acids Res 45 (11), 6259–6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ando Y et al. (2019) ELTA: Enzymatic Labeling of Terminal ADP-Ribose. Mol Cell 73 (4), 845–856 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Daniels Casey M. et al. (2015) The Promise of Proteomics for the Study of ADP-Ribosylation. Molecular Cell 58 (6), 911–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Vivelo CA and Leung AK (2015) Proteomics approaches to identify mono-(ADP-ribosyl)ated and poly(ADP-ribosyl)ated proteins. Proteomics 15 (2–3), 203–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Porta S et al. (2018) Patient-derived frontotemporal lobar degeneration brain extracts induce formation and spreading of TDP-43 pathology in vivo. Nat Commun 9 (1), 4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liu Y et al. (2016) C9orf72 BAC Mouse Model with Motor Deficits and Neurodegenerative Features of ALS/FTD. Neuron 90 (3), 521–34. [DOI] [PubMed] [Google Scholar]
- 105.Chew J et al. (2015) C9ORF72 repeat expansions in mice cause TDP-43 pathology, neuronal loss, and behavioral deficits. Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mackenzie IR et al. (2007) Pathological TDP-43 distinguishes sporadic amyotrophic lateral sclerosis from amyotrophic lateral sclerosis with SOD1 mutations. Ann Neurol 61 (5), 427–34. [DOI] [PubMed] [Google Scholar]
- 107.Kwiatkowski TJ Jr. et al. (2009) Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 323 (5918), 1205–8. [DOI] [PubMed] [Google Scholar]
- 108.Vance C et al. (2009) Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 323 (5918), 1208–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Neumann M et al. (2007) TDP-43-positive white matter pathology in frontotemporal lobar degeneration with ubiquitin-positive inclusions. J Neuropathol Exp Neurol 66 (3), 177–83. [DOI] [PubMed] [Google Scholar]
- 110.Arai T et al. (2006) TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun 351 (3), 602–11. [DOI] [PubMed] [Google Scholar]
- 111.Mackenzie IR and Neumann M (2016) Molecular neuropathology of frontotemporal dementia: insights into disease mechanisms from postmortem studies. J Neurochem 138 Suppl 1, 54–70. [DOI] [PubMed] [Google Scholar]
- 112.DeJesus-Hernandez M et al. (2011) Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72 (2), 245–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Renton AE et al. (2011) A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72 (2), 257–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chornenkyy Y et al. (2019) Tau and TDP-43 proteinopathies: kindred pathologic cascades and genetic pleiotropy. Lab Invest. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Krietsch J et al. (2013) Reprogramming cellular events by poly(ADP-ribose)-binding proteins. Mol Aspects Med 34 (6), 1066–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gagne JP et al. (2012) Quantitative proteomics profiling of the poly(ADP-ribose)-related response to genotoxic stress. Nucleic Acids Res 40 (16), 7788–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gagne JP et al. (2008) Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res 36 (22), 6959–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jungmichel S et al. (2013) Proteome-wide identification of poly(ADP-Ribosyl)ation targets in different genotoxic stress responses. Mol Cell 52 (2), 272–85. [DOI] [PubMed] [Google Scholar]
- 119.Gagne JP et al. (2003) A proteomic approach to the identification of heterogeneous nuclear ribonucleoproteins as a new family of poly(ADP-ribose)-binding proteins. Biochem J 371 (Pt 2), 331–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Turunc Bayrakdar E et al. (2014) Nicotinamide treatment reduces the levels of oxidative stress, apoptosis, and PARP-1 activity in Abeta(1–42)-induced rat model of Alzheimer’s disease. Free Radic Res 48 (2), 146–58. [DOI] [PubMed] [Google Scholar]
- 121.Cardinale A et al. (2015) PARP-1 Inhibition Is Neuroprotective in the R6/2 Mouse Model of Huntington’s Disease. PLoS One 10 (8), e0134482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Endres M et al. (1998) Protective effects of 5-iodo-6-amino-1,2-benzopyrone, an inhibitor of poly(ADP-ribose) synthetase against peroxynitrite-induced glial damage and stroke development. Eur J Pharmacol 351 (3), 377–82. [DOI] [PubMed] [Google Scholar]
- 123.Park EM et al. (2004) Interaction between inducible nitric oxide synthase and poly(ADP-ribose) polymerase in focal ischemic brain injury. Stroke 35 (12), 2896–901. [DOI] [PubMed] [Google Scholar]
- 124.Takahashi K et al. (1997) Neuroprotective effects of inhibiting poly(ADP-ribose) synthetase on focal cerebral ischemia in rats. J Cereb Blood Flow Metab 17 (11), 1137–42. [DOI] [PubMed] [Google Scholar]
- 125.Yokoyama H et al. (2010) Poly(ADP-ribose)polymerase inhibitor can attenuate the neuronal death after 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine-induced neurotoxicity in mice. J Neurosci Res 88 (7), 1522–36. [DOI] [PubMed] [Google Scholar]