Abstract
Polymyxins are important lipopeptide antibiotics that serve as the last-line defense against multidrug-resistant (MDR) Gram-negative bacterial infections. Worryingly, the clinical utility of polymyxins is currently facing a serious threat with the global dissemination of mcr, plasmid-mediated polymyxin resistance. The first plasmid-mediated polymyxin resistance gene, termed as mcr-1 was identified in China in November 2015. Following its discovery, isolates carrying mcr, mainly mcr-1 and less commonly mcr-2 to -7, have been reported across Asia, Africa, Europe, North America, South America and Oceania. This review covers the epidemiological, microbiological and genomics aspects of this emerging threat to global human health. The mcr has been identified in various species of Gram-negative bacteria including Escherichia coli, Klebsiella pneumoniae, Klebsiella oxytoca, Salmonella enterica, Cronobacter sakazakii, Kluyvera ascorbata, Shigella sonnei, Citrobacter freundii, Citrobacter braakii, Raoultella ornithinolytica, Proteus mirabilis, Aeromonas, Moraxella and Enterobacter species from animal, meat, food product, environment and human sources. More alarmingly is the detection of mcr in extended-spectrum-β-lactamases- and carbapenemases-producing bacteria. The mcr can be carried by different plasmids, demonstrating the high diversity of mcr plasmid reservoirs. Our review analyses the current knowledge on the emergence of mcr-mediated polymyxin resistance.
Keywords: mcr, polymyxin resistance, Gram-negative bacteria
Introduction
Polymyxins are cyclic lipopeptide antibiotics that were first discovered in the 1940s (Ainsworth et al. 1947; Benedict and Langlykke 1947; Stansly et al. 1947). Polymyxin B and colistin (polymyxin E) were introduced into clinical practice for the treatment of Gram-negative bacterial infections in 1959 (Ross et al. 1959). Their clinical usage was subsequently withdrawn due to high incidence of nephro- and neuro-toxicity, and also due to the introduction of ‘safer’ antimicrobial agents such as the β-lactams which were equally effective at the time (Fekety et al. 1962; Brown et al. 1970; Koch-Weser et al. 1970). However, over the last two decades the emergence of multidrug-resistant (MDR) Gram-negative bacteria that are resistant to all other antibiotics and paucity of novel antibiotics in the discovery pipeline have led to a resurgence of polymyxin usage in the clinic (Li et al. 2006). Albeit, an increasing incidence of polymyxin-resistant bacterial infections has been reported in both the nosocomial and community settings (Srinivas and Rivard. 2017). The primary mechanism of polymyxin resistance in Gram-negative bacteria involves the modification of lipid A of lipopolysaccharide (LPS), which is a major component of the outer membrane and the initial target of polymyxins. Polymyxin resistance due to modifications of lipid A with positively-charged phosphoethanolamine (pEtN) and/or 4-amino-4-deoxy-L-arabinose (L-Ara4N) was first reported by Vaara et al. (1981); such modifications result in reduced negative charge of the outer membrane and hence reduce the electrostatic interaction with polymyxins (Olaitan et al. 2014; Baron et al. 2016). The modification of lipid A by pEtN and L-Ara4N is mediated by eptA and arnBCADTEF, respectively, which are regulated by two-component systems (TCSs), PhoPQ and PmrAB (Olaitan et al. 2014). The inactivation of mgrB, a negative regulator of PhoPQ system in Klebsiella pneumoniae, can lead to the upregulation of PhoPQ, resulting in polymyxin resistance (Poirel et al. 2015). Secondary polymyxin resistance mechanisms independent of modification of lipid A include production of capsular polysaccharide, expression of efflux pumps and an increased expression of outer membrane proteins (Olaitan et al. 2014). Notably, all of the aforementioned mechanisms of polymyxin resistance are chromosomally-mediated (Olaitan et al. 2014; Baron et al. 2016). Polymyxin resistance in K. pneumoniae and Acinetobacter baumannii is more commonly chromosomal-mediated, and occurs at a higher prevalence in current clinical settings, particularly in Greece and Italy (Giamarellou 2016). On the other hand, mcr is the main polymyxin resistance determinant in Escherichia coli and high prevalence remains in agriculture globally, especially in China. This is coincident with high polymyxin consumption and usage in the aforementioned countries (Giamarellou 2016; Liu YY et al. 2016).
A plasmid-mediated polymyxin resistance gene named mcr-1 was first reported in November 2015 (Liu YY et al. 2016). The emergence of plasmid-mediated polymyxin resistance is a matter of great concern due to the potential for rapid horizontal transfer. The mcr-1 encodes for pEtN transferase enzyme (MCR-1) which catalyzes the addition of pEtN to the phosphate groups in lipid A (Liu YY et al. 2016; Liu YY et al. 2017). The modification of lipid A with pEtN is not a novel mechanism of polymyxin resistance, as this has been frequently associated with the chromosomal gene, eptA (pmrC) (Olaitan et al. 2014; Baron et al. 2016; Huang J et al. 2017). However, the transferability of mcr is of considerable concern due to the potential of MDR Gram-negative bacteria to acquire mcr-harboring plasmids, negating antimicrobial therapy with the important last-line polymyxins.
To date, several other MCR have been identified, including MCR-2, -3, -4, -5, -6 and -7, which share 81%, 32.5%, 34%, 36.1%, 83% and 35% in the amino acid sequence identity, respectively, with MCR-1 (cf. Supplementary information Table S1) (Xavier et al. 2016; AbuOun et al. 2017, Borowiak, Fischer, et al. 2017; Carattoli et al. 2017; Yin et al. 2017; Yang et al. 2018). An additional minor variant was reported for each of MCR-2, -4 and -5, whereas a greater number of minor variants were identified for MCR-1 (11 variants) and MCR-3 (10 variants) (cf. Supplementary information Table S2). It should be noted that mcr-6 (deposited in Genbank in 2018; accession number: MF176240) was described as mcr-2.2 in the study published in 2017 (AbuOun et al. 2017).
Epidemiology
Time-line of mcr discovery
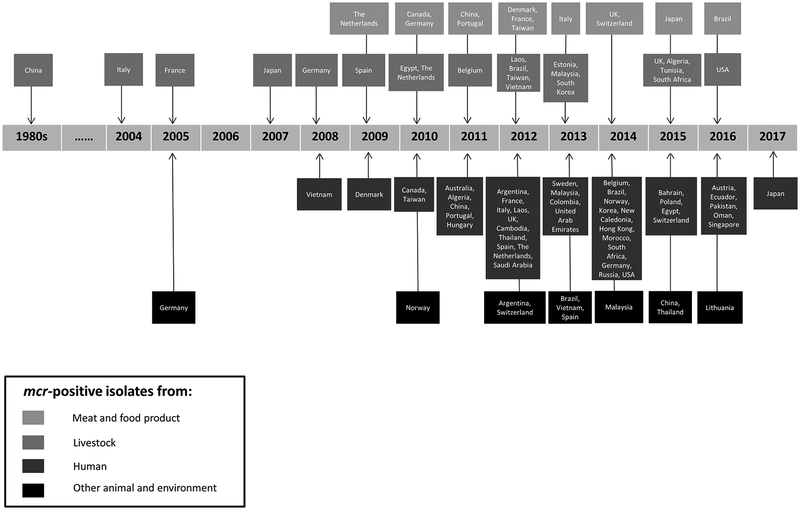
From the time-line diagram (Figure 1), we can clearly see the vast increase in the detection of mcr-positive isolates from various countries dating from 2009 onwards. Retrospective surveillance studies on stored bacterial isolates revealed that the earliest mcr (more specifically mcr-1) was identified from chicken in the 1980s in China, when colistin was started being used for farming purposes (Shen et al. 2016). It was then followed by the identification of mcr from cattle in 2004 (Italy; mcr-1) and veal calves which were raised at local farms in 2005 (France; mcr-1) (Haenni et al. 2016). The earliest mcr-positive bacterial strain from humans was a Shigella sonnei isolated from a hospitalized child in 2008 (Vietnam; mcr-1) (Thanh et al. 2016). It is also important to note that to date, the earliest occurrence of mcr in isolates from wild animals (fish; Aeromonas allosaccharophila; mcr-3.6) and environmental samples (seawater; E. coli; mcr-1) was in 2005 and 2010, respectively (Jørgensen et al. 2017; Eichhorn et al. 2018).
Figure 1.
Time-line of the first identification of mcr in each country.
Among mcr-2 to -7, the earliest mcr-3 (more specifically mcr-3.6) was discovered in 2005 (Eichhorn et al. 2018); whereas mcr-2 (2009), -4 (2013), -5 (2011), -6 (2015) and -7 (2014) were only identified in strains collected over the past decade (Carattoli et al. 2017; Fisher et al. 2017; AbuOun et al. 2018; Borowiak, Eichhorn et al. 2018; Yang et al. 2018). More retrospective studies involving mcr genes should be conducted. Generally, it is evident that mcr has already existed for at least three decades.
Geographical spread of mcr
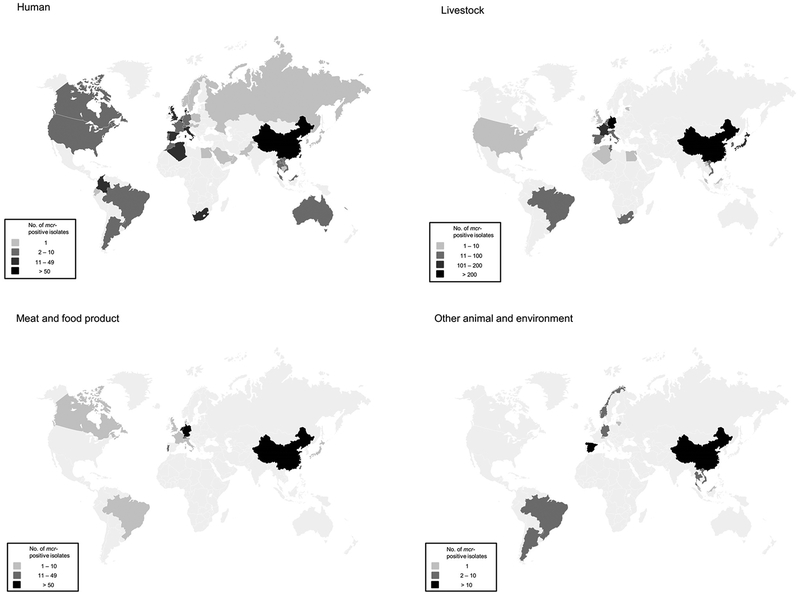
Since the first discovery of the mcr, a number of epidemiological studies have been carried out worldwide (Skov and Monnet 2016). The mcr has been detected in 47 different countries across six continents: Asia (China, Japan, Laos, Vietnam, Malaysia, Singapore, Cambodia, Bahrain, Taiwan, Hong Kong, Thailand, South Korea, Russia, Pakistan, United Arab Emirates, Saudi Arabia and Oman), Europe (Austria, Estonia, UK, The Netherlands, Norway, Spain, Germany, France, Belgium, Denmark, Italy, Poland, Portugal, Russia, Switzerland, Sweden, Lithunia and Hungary), Africa (Algeria, Egypt, Tunisia, Morocco and South Africa), North America (USA and Canada), South America (Colombia, Argentina, Brazil and Ecuador) and Oceania (New Caledonia and Australia) (cf. Table 1 and Figure 2). Among these countries, mcr was identified from human sources in 44 countries, livestock in 21 countries, meat and food products in 13 countries and other sources (including pet/exotic/wild animals and environment) in 11 countries. The only countries in which mcr have been reported from livestock but not humans are Estonia and Tunisia. No trace of mcr has been detected in Antarctica. To date, China, where the first mcr was detected, has the highest prevalence of mcr-positive isolates (cf. Figure 2). This could be due to the fact that polymyxins were heavily used and extensive studies on mcr have been conducted in China.
Table 1.
Characterization of mcr-harboring bacteria.
| Category | Country | Year | Species | Source | No. of isolate | Reference |
|---|---|---|---|---|---|---|
| Human | Algeria | 2011 | E. coli | Urine | 1 | Berrazeg et al. 2016 |
| E. coli | Sperm culture | 1 | Yanat et al. 2016 | |||
| 2013 | E. coli | Rectum | 7 | Leangapichart et al. 2016 | ||
| K. pneumoniae | Rectum | 1 | Leangapichart et al. 2016 | |||
| 2014 | E. coli | Rectum | 1 | Leangapichart et al. 2016 | ||
| Argentina | 2012 | E. coli | Blood (n=1 mcr-1.5) Urine (n=1) |
2 | Rapoport et al. 2016; Tijet et al. 2017 | |
| 2013 | E. coli | Abdominal (n=1 mcr-1.5) Blood (n=1) |
2 | Rapoport et al. 2016; Tijet et al. 2017 | ||
| 2015 | E. coli | Urine (n=2 mcr-1.5) Blood (n=1) Bone (n=1) |
4 | Rapoport et al. 2016; Tijet et al. 2017 | ||
| 2016 | E. coli | Abscess | 1 | Rapoport et al. 2016 | ||
| Australia | 2011 | E. coli | Urine | 1 | Ellem et al. 2017 | |
| 2013 | E. coli | Urine | 1 | Ellem et al. 2017 | ||
| Austria | 2016 | E. coli | Fecal sample | 1 | Hartl et al. 2017 | |
| Bahrain | 2015 | E. coli | Wound (n=1) Urine (n=1) |
2 | Sonnevend et al. 2016 | |
| 2015 | E. coli | Groin and peri-rectal | 4 | Snesrud et al. 2017 | ||
| Belgium | 2014 – 2015 | E. coli | NA | 1 | Castanheira et al. 2016 | |
| 2015 | E. coli | Pus | 1 | Huang TD et al. 2017 | ||
| Brazil | 2014 – 2015 | E. coli | NA | 1 | Castanheira et al. 2016 | |
| 2015 | E. coli | Rectum (n=2) Blood (n=1) |
3 | Conceição-Neto et al. 2017 | ||
| 2016 | K. pneumoniae | Urine | 1 | Aires et al. 2017 | ||
| 2016 | E. coli | Wound | 1 | Fernandes, McCulloch, et al. 2016 | ||
| Cambodia | 2012 | E. coli | Fecal sample | 1 | Stoesser et al. 2016 | |
| Canada | 2010 | E. coli | Blood | 2 | Walkty et al. 2016 | |
| 2011 | E. coli | Gastrostomy tube site, rectum | 1 | Mulvey et al. 2016 | ||
| 2015 – 2016 | E. coli | Urine | 1 | Payne et al. 2016 | ||
| China | 2011 and earlier | NA | Fecal sample (Human microbiome) | 27 | Hu Y et al. 2016; Ruppé et al. 2016 | |
| 2011 | E. coli | Abdominal fluid | 1 | Wang X et al. 2017 | ||
| K. pneumoniae | Wound | 1 | Wang X et al. 2017 | |||
| 2012 | E. coli | Blood (n=8) Urine (n=1) Rectum (n=25) |
34 | Lima Barbieri et al. 2017; Quan et al. 2017; Wang X et al. 2017; Wang Y et al. 2017 | ||
| K. pneumoniae | Blood | 3 | Wang X et al. 2017 | |||
| S. enterica Enteritidis | NA | 2 | Cui et al. 2017 | |||
| S. enterica Typhimurium | NA | 1 | Cui et al. 2017 | |||
| 2013 | E. coli | Blood (n=11) Drainage fluid (n=1) Abdominal fluid (n=4) Sputum (n=1) Urine (n=1) |
18 | Quan et al. 2017; Wang X et al. 2017; Wang Y et al. 2017 | ||
| S. enterica | NA | 1 | Cui et al. 2017 | |||
| S. enterica Typhimurium | NA | 5 | Cui et al. 2017 | |||
| 2014 | E. coli | Urine (n=5) Sputum (n=3) Drainage fluid (n=9) Bile (n=2) Ascites (n=3) Wound (n=1) Blood (n=22) Pus (n=1) |
46 | Du et al. 2016; Liu YY et al. 2016; He QW et al. 2017; Quan et al. 2017; Wang X et al. 2017; Wang Y et al. 2017 | ||
| K. pneumoniae | Urine (n=1) Sputum (n=2) Blood (n=1) |
4 | Liu YY et al. 2016; Quan et al. 2017 | |||
| Enterobacter aerogenes | Vaginal secretion | 1 | Zeng et al. 2016 | |||
| Enterobacter cloacae | Urine | 1 | Zeng et al. 2016 | |||
| S. enterica | Rectum (n=1mcr-1.6) NA (n=3) |
4 | Cui et al. 2017; Lu et al. 2017 | |||
| 2014 – 2015 | E. coli | NA | 2 | Zhang R et al. 2017 | ||
| 2015 | E. coli | Abscess (n=2) Fecal sample (n=63) Blood (n=10) Urine (n=20) Ascites (n=1) Bile (n=8) Catheter (n=2) Drainage fluid (n=7) Pus (n=1) Respiratory tract (n=1) Secretion (n=8) Sputum (n=13) Wound (n=1) |
137 | Du et al. 2016; Gu et al. 2016; Ye et al. 2016; Yu H et al. 2016; Zhang R et al. 2016; Zhang XF et al. 2016; He QW et al. 2017; Tian et al. 2017; Wang Y et al. 2017 | ||
| K. pneumoniae | Fecal sample (n=2) Surgical wound (n=1) Peritoneal fluid (n=1) Sputum (n=1) |
5 | Du et al. 2016; Gu et al. 2016; Tian et al. 2017; Wang Y et al. 2017 | |||
| S. enterica Typhimurium | NA | 16 | Cui et al. 2017 | |||
| 2016 | E. coli | Fecal sample | 34 | Zhong et al. 2016; Hu et al. 2017; Zheng et al. 2017 | ||
| K. pneumoniae | Sputum | 4 | Tian et al. 2017 | |||
| Citrobacter freundii | Fecal sample | 1 | Hu et al. 2017 | |||
| 2017 | E. coli mcr-3.5 | Abdominal abscess | 1 | Liu L et al. 2017 | ||
| NA | E. coli | Blood | 2 | Zheng et al. 2016 | ||
| K. pneumoniae | NA | 13 | Wang Y et al. 2017 | |||
| Enterobacter cloacae | NA | 1 | Wang Y et al. 2017 | |||
| Enterobacter aerogenes | NA | 1 | Wang Y et al. 2017 | |||
| Colombia | 2013 | E. coli | Leg secretion (n=1) Blood (n=1) |
2 | Saavedra et al. 2017 | |
| 2015 | S. enterica Typhimurium | Fecal sample (n=1) Urine (n=1) |
2 | Saavedra et al. 2017 | ||
| 2016 | E. coli | Urine (n=2) Vaginal secretion (n=1) Abdominal abscess (n=1) Toe tissue (n=1) NA (n=1) |
6 | Saavedra et al. 2017 | ||
| K. pneumoniae | Blood | 1 | Saavedra et al. 2017 | |||
| S. enterica Typhimurium | Fecal sample | 1 | Saavedra et al. 2017 | |||
| Denmark | 2009 | S. enterica Typhimurium mcr-3 | NA | 1 | Litrup et al. 2017 | |
| 2010 | S. enterica O:4,5,12;H:i:- mcr-3 | NA | 1 | Litrup et al. 2017 | ||
| 2011 | S. enterica O:4,5,12;H:i:- mcr-3 | NA | 2 | Litrup et al. 2017 | ||
| 2012 | S. enterica Typhimurium mcr-3 | NA | 2 | Litrup et al. 2017 | ||
| S. enterica O:4,5,12;H:i:- mcr-3 | NA | 1 | Litrup et al. 2017 | |||
| 2014 | S. enterica Typhimurium | NA | 2 | Torpdahl et al. 2017 | ||
| E. coli mcr-3 | Blood | 1 | Roer et al. 2017 | |||
| 2015 | E. coli | Blood | 1 | Hasman et al. 2015 | ||
| S. enterica Typhimurium | NA | 2 | Torpdahl et al. 2017 | |||
| S. enterica O:4,5,12;H:i:- mcr-3 | NA | 1 | Litrup et al. 2017 | |||
| 2016 | S. enterica O:4,5,12;H:i:- mcr-3 | NA | 1 | Litrup et al. 2017 | ||
| 2017 | S. enterica O:4,5,12;H:i:- mcr-3 | NA | 1 | Litrup et al. 2017 | ||
| Ecuador | 2016 | E. coli | Peritoneal fluid | 1 | Ortega-Paredes et al. 2016 | |
| Egypt | 2015 | E. coli | Sputum | 1 | Elnahriry et al. 2016 | |
| France | 2012 – 2013 | K. pneumoniae | Fecal sample | 2 | Rolain et al. 2016 | |
| 2016 | E. coli | Fecal sample | 1 | Beyrouthy et al. 2017 | ||
| Germany | 2014 | E. coli | Wound | 1 | Falgenhauer, Waezsada, Yao, et al. 2016 | |
| 2014 – 2015 | E. coli | NA | 5 | Castanheira et al. 2016 | ||
| 2016 | E. coli | Urine | 1 | Fritzenwanker et al. 2016 | ||
| Hungary | 2011 | E. coli | Blood | 1 | Juhász et al. 2017 | |
| Hong Kong | 2014 – 2015 | E. coli | NA | 1 | Castanheira et al. 2016 | |
| 2015 – 2016 | E. coli | Blood (n=2) Fecal sample (n=1) Urine (n=1) |
4 | Wong et al. 2016 | ||
| 2015 – 2016 | Enterobacter cloacae | Fecal sample | 1 | Wong et al. 2016 | ||
| Italy | 2012 – 2015 | S. enterica | NA | 10 | Carnevali et al. 2016 | |
| 2013 | E. coli | Urine | 1 | Cannatelli et al. 2016 | ||
| 2014 | E. coli | Urine, surgical wound | 3 | Cannatelli et al. 2016 | ||
| K. pneumoniae mcr-1.2 | Rectum | 1 | Di Pilato et al. 2016 | |||
| 2015 | E. coli | Urine (n=4) Intestinal colonization (n=3) |
7 | Cannatelli et al. 2016; Giufrè et al. 2016 | ||
| 2014 – 2015 | E. coli | NA | 4 | Castanheira et al. 2016 | ||
| 2016 | E. coli | Blood | 2 | Corbella et al. 2017 | ||
| S. enterica Typhimurium mcr-4.2 | Fecal sample | 2 | Carretto et al. 2018 | |||
| 2017 | E. coli | Blood | 1 | Corbella et al. 2017 | ||
| Japan | 2017 | E. coli | Fecal sample | 1 | Tada, Uechi, et al. 2017 | |
| Kingdom of Saudi Arabia | 2012 | E. coli | Blood | 1 | Sonnevend et al. 2016 | |
| Laos | 2012 | E. coli | Fecal sample | 6 | Olaitan et al. 2016 | |
| 2012 | K. pneumoniae | Fecal sample | 4 | Rolain et al. 2016 | ||
| Malaysia | 2013 | E. coli | Urine | 1 | Yu, Ang, Chin, et al. 2016 | |
| 2014 – 2015 | E. coli | NA | 1 | Castanheira et al. 2016 | ||
| Morocco | 2014 | E. coli | Rectum | 2 | Leangapichart et al. 2016 | |
| New Caledonia | 2014 | E. coli | Ascites (n=1) Gastric fluid (n=1) |
2 | Robin et al. 2016 | |
| Norway | 2014 | E. coli | NA | 1 | Solheim et al. 2016 | |
| Oman | 2016 | E. coli | Blood | 1 | Mohsin et al. 2017 | |
| Pakistan | 2016 | E. coli | Wound | 1 | Mohsin et al. 2016 | |
| Poland | 2015 | E. coli | Urine | 1 | Izdebski et al. 2016 | |
| Portugal | 2011 – 2012 | S. enterica 1,4,[5],12:i:- | Blood, fecal sample | 4 | Campos et al. 2016 | |
| 2016 | E. coli | Urine | 1 | Tacão et al. 2017 | ||
| Russia | 2014 – 2015 | E. coli | NA | 1 | Castanheira et al. 2016 | |
| Singapore | 2016 | E. coli | Urine | 2 | Teo et al. 2016 | |
| 2016 | K. pneumoniae | Urine | 1 | Teo et al. 2016 | ||
| South Africa | 2014 – 2016 | E. coli | Blood (n=1) Wound (n=1) Pus (n=1) Urine (n=6) |
9 | Coetzee et al. 2016; Poirel, Kieffer, Brink, et al. 2016 | |
| 2016 | E. coli | Urine (n=9) Superficial abdominal swab (n=1) |
10 | Newton-Foot et al. 2017 | ||
| 2016 | K. pneumoniae | Sputum (n=2) Urine (n=2) |
4 | Newton-Foot et al. 2017 | ||
| 2016 | K. oxytoca | Superficial abdominal swab | 1 | Newton-Foot et al. 2017 | ||
| South Korea | 2014 – 2015 | E. coli | Blood | 1 | Kim et al. 2017 | |
| Spain | 2012 | E. coli | Blood | 1 | Prim et al. 2016 | |
| 2013 | E. coli | Sputum (n=3) Blood (n=4) Urine (n=1) |
8 | Prim et al. 2016 | ||
| 2014 | E. coli | Urine (n=2) Surgical wound (n=1) |
3 | Prim et al. 2016 | ||
| 2015 | E. coli | Urine | 3 | Prim et al. 2016 | ||
| 2014 – 2015 | E. coli | NA | 3 | Castanheira et al. 2016 | ||
| 2016 | E. coli | Peritoneal fluid (n=1) Sputum (n=1) |
2 | Ortiz de la Tabla et al. 2016; Sánchez-Benito et al. 2017 | ||
| 2012 – 2015 | E. coli | Blood (n=5), urine (n=6), sputum (n=3), surgical wound secretion (n=1) | 15 | Prim et al. 2017 | ||
| Sweden | 2013 | E. coli | Fecal sample | 1 | Vading et al. 2016 | |
| Switzerland | 2015 | E. coli | Urine (n=1) Fecal sample (n=3, 1mcr-1.2) Blood (n=2) |
7 | Bernasconi et al. 2016; Nordmann, Lienhard, et al. 2016; Poirel, Kieffer, Liassine, et al. 2016; Donà, Bernasconi, Kasraian, et al. 2017 | |
| 2016 | E. coli | Urine (n=1) Fecal sample (n=2) |
3 | Zurfluh et al. 2017 | ||
| Taiwan | 2010 | E. coli | Sputum | 1 | Kuo et al. 2016 | |
| 2012 | E. coli | Urine | 2 | Kuo et al. 2016 | ||
| 2014 | E. coli | Ascites (n=1) Abscess (n=2) Blood (n=3) Urine (n=5) |
11 | Kuo et al. 2016 | ||
| S. enterica Typhimurium | NA | 5 | Chiou et al. 2017 | |||
| 2015 | S. enterica Typhimurium | NA | 3 | Chiou et al. 2017 | ||
| S. enterica Newport | NA | 1 | Chiou et al. 2017 | |||
| S. enterica Albany | NA | 1 | Chiou et al. 2017 | |||
| NA | E. coli | Urine | 1 | Lai et al. 2017 | ||
| Thailand | 2012 | E. coli | Fecal sample | 2 | Olaitan et al. 2016 | |
| 2014 – 2015 | E. coli | Urine | 1 | Runcharoen et al. 2017 | ||
| 2016 | E. coli | Urine | 1 | Paveenkittiporn et al. 2016 | ||
| The Netherlands | 2012 – 2013 | E. coli | Fecal sample | 6 | Arcilla et al. 2016 | |
| 2014 | E. coli | Fecal sample | 1 | Nijhuis et al. 2016 | ||
| 2015 | E. coli | Fecal sample | 2 | Nijhuis et al. 2016 | ||
| 2014 – 2015 | E. coli | Fecal sample | 1 | Terveer et al. 2017 | ||
| The United States | 2014 | E. coli | Urine | 1 | Mediavilla et al. 2016 | |
| 2015 | E. coli | Urine | 1 | Castanheira et al. 2016 | ||
| 2015 | E. coli | Blood | 1 | Macesic et al. 2017 | ||
| 2016 | E. coli | Urine | 1 | McGann et al. 2016 | ||
| 2016 | E. coli | Fecal sample | 1 | Vasquez et al. 2016 | ||
| The United Kingdom | 2012 | S. enterica Typhimurium | Fecal sample | 1 | Doumith et al. 2016 | |
| 2013 | E. coli | Fecal sample (n=1) Blood (n=1) |
2 | Doumith et al. 2016 | ||
| 2014 | E. coli | Blood | 1 | Doumith et al. 2016 | ||
| 2014 | S. enterica Typhimurium | Fecal sample | 2 | Doumith et al. 2016 | ||
| 2014 | S. enterica Virchow | Fecal sample | 1 | Doumith et al. 2016 | ||
| 2015 | S. enterica Typhimurium | Fecal sample | 5 | Doumith et al. 2016 | ||
| 2015 | S. enterica Paratyphi | Fecal sample | 1 | Doumith et al. 2016 | ||
| United Arab Emirates | 2013 | E. coli | Blood | 1 | Sonnevend et al. 2016 | |
| Vietnam | 2008 | Shigella sonnei | Fecal sample | 1 | Thanh et al. 2016 | |
| 2012 – 2013 | E. coli | Rectum | 3 | Trung et al. 2017 | ||
| 2014 | E. coli | Urine (n=1) Pus (n=1) |
2 | Tada, Nhung, et al. 2017 | ||
| Livestock | Algeria | 2015 | E. coli | Chicken | 1 | Olaitan et al. 2016 |
| Brazil | 2012 | E. coli | Pig | 2 | Fernandes, Moura, et al. 2016 | |
| 2013 | E. coli | Chicken | 14 | Fernandes, Moura, et al. 2016 | ||
| 2015 | E. coli | Chicken | 10 | Lentz et al. 2016 | ||
| Belgium | 2011 – 2012 | E. coli | Calf (n=6) Pig (n=7) Calf (n=1mcr-2) Pig (n=2mcr-2) |
16 | Malhotra-Kumar, Xavier, Das, Lammens, Butaye, et al. 2016; Xavier et al. 2016 | |
| 2013 | E. coli | Cattle (n=1) Pig (n=1) |
2 | El Garch et al. 2016 | ||
| 2015 – 2016 | E. coli mcr-4 | Pig | 2 | Carattoli et al. 2017 | ||
| China | 1980s | E. coli | Chicken | 3 | Shen et al. 2016 | |
| 2004 – 2006 | E. coli | Chicken | 8 | Shen et al. 2016 | ||
| 2007 | S. enterica Typhimurium | Chicken | 1 | Li XP et al. 2016 | ||
| 2008 | S. enterica Typhimurium | Pig | 2 | Li XP et al. 2016 | ||
| 2009 | E. coli | Chicken | 6 | Shen et al. 2016 | ||
| S. enterica Typhimurium | Pig | 1 | Li XP et al. 2016 | |||
| 2010 | E. coli | Chicken | 13 | Shen et al. 2016; Yang YQ et al. 2017 | ||
| S. enterica Typhimurium | Duck | 1 | Li XP et al. 2016 | |||
| 2011 | E. coli | Chicken (n=30) Pig (n=3) |
33 | Li Z et al. 2016; Shen et al. 2016; Yang YQ et al. 2017 | ||
| 2012 | E. coli | Pig (n=33) Chicken (n= 42) |
75 | Li Z et al. 2016; Liu YY et al. 2016; Shen et al. 2016; Lima Barbieri et al. 2017; Yang YQ et al. 2017 | ||
| 2013 | E. coli | Pig (n=69) Chicken (n=39) Chicken (n=1mcr-1.3) |
109 | Li Z et al. 2016; Liu YY et al. 2016; Shen et al. 2016; Lima Barbieri et al. 2017; Wang X et al. 2017; Yang YQ et al. 2017 | ||
| 2014 | E. coli | Pig (n=67) Chicken (n=26) |
93 | Liu YY et al. 2016; Shen et al. 2016; Lima Barbieri et al. 2017; Yang YQ et al. 2017 | ||
| Citrobacter freundii | Pig | 1 | Li XP et al. 2017 | |||
| K. pneumoniae mcr-7 | Chicken | 1 | Yang et al. 2018 | |||
| 2010 – 2015 | K. pneumoniae | Chicken (n=7) Chicken (n=2mcr-7) | 9 | Yang et al. 2018 | ||
| 2014 – 2015 | E.coli | Pig | 35 | Li R et al. 2017 | ||
| S. enterica | Chicken | 4 | Yang YQ et al. 2016 | |||
| 2015 | E. coli | Duck (n=2) Chicken (n=66) Cattle (n=1) Pig (n=1mcr-3) |
70 | Liu BT et al. 2016; Yang RS et al. 2016; He T et al. 2017; Yang YQ et al. 2017; Yi et al. 2017; Yin et al. 2017 | ||
| Aeromonas veronii mcr-3.3 | Chicken | 1 | Ling et al. 2017 | |||
| Cronobacter sakazakii | Chicken (n=2) | 2 | Liu BT et al. 2016 | |||
| 2015 – 2016 | S. enterica | Chicken (n=6) Pig (n=6) |
12 | Ma et al. 2017 | ||
| 2017 | Aeromonas caviae mcr-3.10 | Duck | 1 | Wang et al. 2018 | ||
| E. coli mcr-3.10 | Duck | 1 | Wang et al. 2018 | |||
| Proteus mirabilis mcr-3.10 | Duck | 1 | Wang et al. 2018 | |||
| Egypt | 2010 | E. coli | Chicken | 4 | Lima Barbieri et al. 2017 | |
| 2014 | E. coli | Cattle | 1 | Khalifa et al. 2016 | ||
| Estonia | 2013 | E. coli | Pig | 3 | Brauer et al. 2016 | |
| France | 2005 – 2014 | E. coli | Calf | 106 | Haenni et al. 2016 | |
| 2004 | E. coli | Cattle | 1 | El Garch et al. 2016 | ||
| 2005 | E. coli | Cattle | 1 | El Garch et al. 2016 | ||
| 2006 | E. coli | Pig | 2 | El Garch et al. 2016 | ||
| 2007 | E. coli | Cattle (n=2) Pig (n=1) |
3 | Brennan et al. 2016; El Garch et al. 2016 | ||
| 2008 | E. coli | Cattle (n=1) Pig (n=1) |
2 | El Garch et al. 2016 | ||
| 2009 | E. coli | Pig | 3 | El Garch et al. 2016 | ||
| 2010 | E. coli | Cattle (n=4) Pig (n=5) |
9 | El Garch et al. 2016 | ||
| 2011 | E. coli | Cattle (n=1) Pig (n=2) |
3 | El Garch et al. 2016; Perrin-Guyomard et al. 2016 | ||
| 2012 | E. coli | Pig | 3 | El Garch et al. 2016 | ||
| 2013 | E. coli | Chicken (n=3) Pig (n=4) |
7 | El Garch et al. 2016; Perrin-Guyomard et al. 2016 | ||
| S. enterica 1,4,[5],12:i:- | Chicken | 1 | Webb et al. 2016 | |||
| 2014 | E. coli | Chicken (n=4) Turkey (n=14) Pig (n=1) |
19 | El Garch et al. 2016; Perrin-Guyomard et al. 2016 | ||
| Germany | 2008 | S. enterica Paratyphi B | Chicken | 1 | Borowiak, Hammerl, et al. 2017 | |
| 2010 – 2011 | E. coli | Pig | 3 | Falgenhauer, Waezsada, Yao, et al. 2016 | ||
| S. enterica | Pig | 1 | El Garch et al. 2016 | |||
| 2010 | E. coli | Chicken (n=8) Turkey (n=30) Calf (n=15) Pig (n=1) |
54 | El Garch et al. 2016; Irrgang et al. 2016 | ||
| 2011 | E. coli | Laying hen (n=2) Chicken (n=17) Turkey (n=33) Pig (n=13) |
65 | Irrgang et al. 2016 | ||
| 2011 – 2012 | E. coli | Pig farm (boot swab and fecal sample) | 43 | Roschanski et al. 2017 | ||
| 2012 | Aeromonas media mcr-3.7 | Turkey | 1 | Eichhorn et al. 2018 | ||
| E. coli | Turkey (n=63) Calf (n=5) |
68 | Irrgang et al. 2016 | |||
| S. enterica Paratyphi B mcr-5 | Poultry | 8 | Borowiak, Fischer, et al. 2017 | |||
| 2013 | E. coli | Chicken | 52 | Irrgang et al. 2016 | ||
| 2014 | E. coli | Laying hens(n=1) Chicken (n=22) Turkey (n=37) |
60 | Irrgang et al. 2016 | ||
| 2015 | E. coli | Calf (n=1) Pig (n=11) |
12 | Irrgang et al. 2016 | ||
| 2016 | E. coli | Pig | 11 | Schirmeier et al. 2017 | ||
| Italy | 2004 | E. coli | Cattle | 2 | El Garch et al. 2016 | |
| 2006 | E. coli | Cattle | 1 | El Garch et al. 2016 | ||
| 2007 | E. coli | Pig | 3 | El Garch et al. 2016 | ||
| 2008 | E. coli | Pig | 1 | El Garch et al. 2016 | ||
| 2010 – 2011 | S. enterica | Pig | 2 | El Garch et al. 2016 | ||
| 2011 | E. coli | Pig | 1 | El Garch et al. 2016 | ||
| 2012 | E. coli | Pig | 1 | El Garch et al. 2016 | ||
| 2013 | S. enterica Typhimurium mcr-4 | Pig | 1 | Carattoli et al. 2017 | ||
| 2014 | E. coli | Pig | 1 | El Garch et al. 2016 | ||
| 2012 – 2015 | S. enterica | Poultry (n=2) Pig (n=9) |
11 | Carnevali et al. 2016 | ||
| 2015 – 2016 | E. coli | Pig | 37 | Curcio et al. 2017 | ||
| 2016 | E. coli | Pig | 1 | Pulss et al. 2017 | ||
| Japan | 2007 – 2014 | E. coli | Pig | 90 | Kusumoto et al. 2016 | |
| 2008 | E. coli | Pig | 2 | Kawanishi et al. 2016; Suzuki et al. 2016 | ||
| 2010 | E. coli | Pig | 5 | Kawanishi et al. 2016; Suzuki et al. 2016 | ||
| 2011 | E. coli | Cattle | 1 | Kawanishi et al. 2016 | ||
| 2012 | E. coli | Pig (n=5) Cattle (n=2) Chicken (n=2) |
9 | Kawanishi et al. 2016 | ||
| 2013 | E. coli | Pig (n=3) Cattle (n=1) Chicken (n=2) |
6 | Kawanishi et al. 2016 | ||
| S. enterica Typhimurium | Pig | 1 | Suzuki et al. 2016 | |||
| 2012 – 2013 | E. coli | Cattle | 4 | Suzuki et al. 2016 | ||
| 2014 | E. coli | Pig (n=7) Cattle (n=1) Chicken (n=10) |
18 | Kawanishi et al. 2016 | ||
| Laos | 2012 | E. coli | Pig | 3 | Olaitan et al. 2016 | |
| Malaysia | 2013 | E. coli | Chicken (n=5) Pig (n=1) |
6 | Petrillo et al. 2016; Yu, Ang, Chin, et al. 2016 | |
| South Africa | 2015 | E. coli | Chicken | 19 | Perreten et al. 2016 | |
| South Korea | 2013 | E. coli | Chicken | 1 | Lim et al. 2016 | |
| 2014 | E. coli | Chicken | 6 | Lim et al. 2016 | ||
| 2015 | E. coli | Chicken (n=3) Pig (n=1) |
4 | Lim et al. 2016 | ||
| Spain | 2009 | S. enterica Typhimurium | Pig | 1 | Quesada et al. 2016 | |
| M. pluranimalium mcr-2.2 | Pig | 1 | AbuOun et al. 2017 | |||
| 2010 | S. enterica Typhimurium | Pig | 1 | Quesada et al. 2016 | ||
| S. enterica Rissen | Pig | 1 | Quesada et al. 2016 | |||
| 2011 | E. coli | Pig | 1 | Quesada et al. 2016 | ||
| S. enterica Typhimurium | Pig | 1 | Quesada et al. 2016 | |||
| 2013 | E. coli | Pig | 1 | Quesada et al. 2016 | ||
| 2014 | E. coli | Turkey | 3 | Quesada et al. 2016 | ||
| 2015 | E. coli | Cattle (n=4, 1mcr-3.2) | 5 | Hernández et al. 2017 | ||
| 2015 – 2016 | E. coli mcr-4 | Pig | 9 | Carattoli et al. 2017 | ||
| Taiwan | 2012 | S. enterica Typhimurium | Pig | 1 | Chiou et al. 2017 | |
| 2013 | S. enterica Typhimurium | Pig (n=3) Chicken (n=2) |
5 | Chiou et al. 2017 | ||
| S. enterica Anatum | Pig | 3 | Chiou et al. 2017 | |||
| The Netherlands | 2010 – 2011 | E. coli | Calf (n=15) Chicken (n=2) Turkey (n=1) |
18 | Veldman et al. 2016 | |
| 2012 – 2013 | E. coli | Chicken | 8 | Veldman et al. 2016 | ||
| The United Kingdom | 2014 | M. porci mcr-1.10 | Pig | 1 | AbuOun et al. 2017 | |
| 2015 | E. coli | Pig | 4 | Anjum et al. 2016; Duggett et al. 2016 | ||
| S. enterica Typhimurium | Pig | 1 | Anjum et al. 2016 | |||
| M. pluranimalium mcr-6 | Pig | 1 | AbuOun et al. 2017 | |||
| The United States | 2016 | E. coli | Pig | 3 | Meinersmann, Ladely, Bono, et al. 2016; Meinersmann, Ladely, Plumblee, et al. 2016 | |
| Tunisia | 2015 | E. coli | Chicken | 37 | Grami et al. 2016 | |
| Vietnam | 2012 – 2013 | E. coli | Chicken | 19 | Trung et al. 2017 | |
| 2013 – 2014 | E. coli | Chicken (n=20) Pig (n=17) |
37 | Nguyen et al. 2016 | ||
| 2014 – 2015 | E. coli | Pig | 9 | Malhotra-Kumar, Xavier, Das, Lammens, Hoang, et al. 2016 | ||
| Meat and food product | Brazil | 2016 | E. coli | Chicken meat | 8 | Monte et al. 2017 |
| Canada | 2010 | E. coli | Beef (Unknown origin) | 2 | Mulvey et al. 2016 | |
| China | 2011 | E. coli | Pork (n=3) Chicken meat(n=10) |
13 | Liu YY et al. 2016 | |
| 2013 | E. coli | Pork (n=11) Chicken meat(n=4) |
15 | Liu YY et al. 2016 | ||
| 2014 | E. coli | Pork (n=29) Chicken meat (n=21) |
50 | Liu YY et al. 2016 | ||
| 2015 – 2016 | E. coli | Retail food sample | 109 | Liu X et al. 2017 | ||
| S. enterica | Chicken meat (n=5) Pork (n=5) |
10 | Ma et al. 2017 | |||
| 2015 | E. coli | Vegetable | 3 | Luo et al. 2017 | ||
| Raoultella ornithinolytica | Vegetable | 2 | Luo et al. 2017 | |||
| 2016 | E. coli | Vegetable | 4 | Luo et al. 2017 | ||
| Denmark | 2012 | E. coli | Chicken meat(imported from Europe) | 3 | Hasman et al. 2015 | |
| 2013 | E. coli | Chicken meat (imported from Europe) | 1 | Hasman et al. 2015 | ||
| 2014 | E. coli | Chicken meat(imported from Europe) | 1 | Hasman et al. 2015 | ||
| France | 2012 | S. enterica Paratyphi B | Chicken breast (n=1) Ready-to-cook guinea fowl pie (n=1) |
2 | Webb et al. 2016 | |
| 2013 | S. enterica Derby | Chipolata sausage | 1 | Webb et al. 2016 | ||
| Italy | 2013 – 2015 | S. enterica | Pork | 4 | Carnevali et al. 2016 | |
| Japan | 2015 | E. coli | Chicken meat | 1 | Ohsaki et al. 2017 | |
| Taiwan | 2012 | E. coli | Beef | 1 | Kuo et al. 2016 | |
| 2013 | E. coli | Chicken meat | 6 | Kuo et al. 2016 | ||
| 2015 | E. coli | Chicken meat (n=9) Pork (n=2) |
11 | Kuo et al. 2016 | ||
| The Netherlands | 2009 | E. coli | Chicken meat(Unknown origin) | 1 | Kluytmans-van den bergh et al. 2016 | |
| 2013 | S. enterica Anatum | Turkey meat (imported) | 1 | Veldman et al. 2016 | ||
| 2014 | E. coli | Chicken meat(imported from Europe) | 2 | Kluytmans-van den bergh et al. 2016 | ||
| 2015 | S. enterica Schwarzengrund | Turkey meat (imported) | 1 | Veldman et al. 2016 | ||
| E. coli | Chicken meat | 33 | Schrauwen et al. 2017 | |||
| K. pneumoniae | Chicken meat | 2 | Schrauwen et al. 2017 | |||
| 2010 – 2015 | S. enterica Java | Chicken meat (local) | 11 | Veldman et al. 2016 | ||
| The United Kingdom | 2014 | S. enterica Paratyphi B | Poultry meat (imported from Europe) | 2 | Doumith et al. 2016 | |
| Germany | 2010 | E. coli | Turkey meat | 17 | Irrgang et al. 2016 | |
| 2011 | E. coli | Chicken meat | 14 | Irrgang et al. 2016 | ||
| S. enterica Paratyphi B mcr-5 | Chicken meat | 1 | Borowiak, Fischer, et al. 2017 | |||
| 2012 | E. coli | Chicken breast (n=1) Turkey hen Schnitzel (n=1) Turkey meat (n=30) Beef (n=2) |
34 | Falgenhauer, Waezsada, Gwozdzinski, et al. 2016; Irrgang et al. 2016 | ||
| S. enterica Paratyphi B mcr-5 | Chicken meat | 1 | Borowiak, Fischer, et al. 2017 | |||
| 2013 | E. coli | Turkey meat (n=1) Chicken meat (n=10) |
11 | Falgenhauer, Waezsada, Gwozdzinski, et al. 2016; Irrgang et al. 2016 | ||
| S. enterica Paratyphi B mcr-5 | Chicken meat | 1 | Borowiak, Fischer, et al. 2017 | |||
| 2014 | E. coli | Chicken meat (n=1) Turkey meat (n=10) |
11 | Irrgang et al. 2016 | ||
| Portugal | 2011 | S. enterica Typhimurium | Food product (originated from swine and poultry) | 3 | Figueiredo et al. 2016; Tse and Yuen 2016 | |
| 2012 | S. enterica Typhimurium | Food product (originated from cattle) | 1 | Figueiredo et al. 2016 | ||
| 2014 – 2015 | S. enterica 1,4,[5],12:i:- | Pork meat/carcass | 5 | Campos et al. 2016 | ||
| S. enterica Rissen | Pork carcass | 2 | Campos et al. 2016 | |||
| Switzerland | 2014 | E. coli | Vegetable (imported from Thailand and Vietnam) | 2 | Zurfuh et al. 2016 | |
| Chicken meat (imported from Germany) | 4 | Donà, Bernasconi, Pires, et al. 2017 | ||||
| 2015 | E. coli | Chicken meat (imported from Germany and Italy) | 2 | Zogg et al. 2016 | ||
| 2016 | E. coli | Chicken meat (imported from Germany and Italy) Turkey meat (imported from Germany and Italy) |
14 | Zurfluh, Buess, et al. 2016; Donà, Bernasconi, Pires, et al. 2017 | ||
| Other animal | Argentina | 2012 | E. coli | Kelp gulls | 5 | Liakopoulos et al. 2016 |
| Brazil | 2013 | E. coli | Magellanic penguins |
1 | Sellera et al. 2016 | |
| China | 2016 | E. coli | Dog (n=4) Cat (n=2) |
6 | Zhang XF et al. 2016 | |
| Germany | 2005 | Aeromonas allosaccharophila mcr-3.6 | Fish | 1 | Eichhorn et al. 2018 | |
| 2006 | Aeromonas hydrophila mcr-3.8, mcr-3.9 | Fish | 1 | Eichhorn et al. 2018 | ||
| 2008 | Aeromonas jandaei mcr-3.8 | Fish | 1 | Eichhorn et al. 2018 | ||
| Lithuania | 2016 | E. coli | European herring gulls | 1 | Ruzauskas and Vaskeviciute 2016 | |
| Vietnam | 2013 – 2014 | E. coli | Asian grass lizard | 2 | Unger et al. 2016 | |
| Environment | Brazil | 2016 | E. coli | Sea water | 3 | Fernandes et al. 2017 |
| China | 2015 | Kluyvera ascorbata | Hospital sewage | 1 | Zhao and Zong 2016 | |
| K. pneumoniae | Hospital sewage | 1 | Zhao, Feng, et al. 2016 | |||
| E. coli | Well water | 2 | Sun et al. 2017 | |||
| 2016 | E. coli | River and lake water | 16 | Zhou et al. 2017 | ||
| Citrobacter freundii | Lake water | 2 | Zhou et al. 2017 | |||
| K. oxytoca | Lake water | 2 | Zhou et al. 2017 | |||
| Citrobacter braakii | Lake water | 2 | Zhou et al. 2017 | |||
| Enterobacter cloacae | River water | 1 | Zhou et al. 2017 | |||
| Germany | 2012 | S. enterica Paratyphi B mcr-5 | NA | 2 | Borowiak, Fischer, et al. 2017 | |
| Malaysia | 2014 | E. coli | Pond water | 1 | Petrillo et al. 2016 | |
| Norway | 2010 | E. coli | Sea water | 2 | Jørgensen et al. 2017 | |
| Spain | 2013 | E. coli | Sewage water | 29 | Ovejero et al. 2017 | |
| K. pneumoniae | Sewage water | 1 | Ovejero et al. 2017 | |||
| Switzerland | 2012 | E. coli | River water | 1 | Zurfuh et al. 2016 | |
| Thailand | 2014 – 2015 | E. coli | Canal water | 2 | Runcharoen et al. 2017 |
NA: not available; Isolates carried mcr-1 unless stated otherwise in superscript.
Figure 2.
Geographical distribution of mcr-carrying bacteria.
To the best of our knowledge, the presence of all mcr except mcr-6 has been detected in samples from China (Yang et al. 2018; Zhang, Chen, Wang, Butaye, et al. 2018; Zhang, Chen, Wang, Yassin, et al. 2018). Thus far, mcr-2 (Belgium and Spain), mcr-3 (Brazil, Denmark, France, Germany, Japan, Spain and Thailand), mcr-4 (Italy and Spain), mcr-5 (Colombia, Japan, Spain and Germany) and mcr-6 (The United Kingdom) have been sparsely detected (Borowiak, Fisher et al. 2017; Carattoli et al. 2017; Liu L et al. 2017; Roer et al. 2017; AbuOun et al. 2018; Eichhorn et al. 2018; Fukuda et al. 2018; García et al. 2018; Haenni et al. 2018; Hammerl et al. 2018; Hernández et al. 2017; Kieffer et al. 2018; Litrup et al. 2017; Wang et al. 2018; Wise et al. 2018; Xavier et al. 2016; Yamaguchi et al. 2018; Yang et al. 2018).
The identification of mcr in sea gulls and migratory penguins is an alarming event due to the possibility for rapid global dissemination, as these flight animals are capable to migrate intercontinentally (Liakopoulos et al. 2016; Ruzauskas and Vaskeviciute 2016; Sellera et al. 2016). Trading of food products such as livestock, meat and vegetables can potentially be another significant force driving the spread of mcr globally (Hasman et al. 2015; Doumith et al. 2016; Grami et al. 2016; Kluytmans-van den bergh et al. 2016; Veldman et al. 2016). In addition, the global trade of exotic animals (Unger et al. 2016) and human travelers may also play a key role in the dissemination of mcr worldwide (Arcilla et al. 2016; Doumith et al. 2016). Fortunately, complete elimination of mcr-carrying isolates from travelers after their return to home country signifies that a biological cost could be conferred by mcr in the absence of polymyxins as the selective pressure, and as such the spread could be mitigated by limiting the use of polymyxins (Arcilla et al. 2016).
Transmission of mcr
By far, livestock is regarded as the main reservoir for mcr due to the heavy usage of polymyxins for prophylaxis, metaphylaxis and therapeutic purposes as well as a growth promoter (Kempf et al. 2016; Liu YY et al. 2016; Nordmann and Poirel 2016). Among livestock, the highest prevalence was observed among poultry, mainly in China and Germany. Approximately one third of the total mcr-positive isolates from livestock were from pigs, mainly attributed by China and Japan. The transferability of the mcr-carrying plasmid from isolates of animal origin to humans was demonstrated by in vitro conjugation and transformation experiments, showing successful transfer of a mcr-1 plasmid (pHNSHP45) from pig into common human pathogenic Enterobacteriaceae and Pseudomonas aeruginosa (Liu YY et al. 2016). Bacteria carrying mcr (mcr-1) have also been identified from pets and the possibility of these bacteria infecting humans is another avenue towards the interspecies spread (Liakopoulos et al. 2016; Zhang XF et al. 2016). The zoonotic potential of mcr-carrying bacteria has been postulated by comparing the genetic determinants of the mcr-carrying isolates from animal and human sources (Elnahriry et al. 2016; Poirel, Kieffer, Liassine, et al. 2016; Poirel and Nordmann 2016). Besides, the widespread of mcr among livestocks/meat/food products and identification of mcr in the human microbiome (mcr-1) suggested the potential transmission via the food chain; however, more definite evidence is required to draw this conclusion (Hu Y et al. 2016). The isolation of a mcr-positive Enterobacteriaceae (mcr-1) from infants who had not started solid diet and had no history of contact with livestock, suggested other possible transmission routes besides food chain and zoonotic transfer (Gu et al. 2016; Zhang R et al. 2016). More worryingly, mcr was identified from water sources, including untreated river wastewater, wastewater treatment plants, seawater, lake, pond, canal and well. This could contribute to the rapid spreading of polymyxin resistance to animals and humans (Petrillo et al. 2016; Zurfuh et al. 2016; Fernandes et al. 2017; Hembach et al. 2017; Jørgensen et al. 2017; Marathe et al. 2017; Runcharoen et al. 2017; Sun et al. 2017).
Epidemiology of mcr in humans and potential clinical impact
The mcr-positive bacteria have been isolated from people of various ages ranging from new-born to elderly (Prim et al. 2016; Zheng et al. 2016). These include patients, elderly residents at long-term aged care facilities in Italy (Giufrè et al. 2016), European travelers who had visited South America, Africa and Asia countries (Arcilla et al. 2016; Bernasconi et al. 2016; Doumith et al. 2016), as well as pilgrims who had traveled to Mecca during Hajj (Leangapichart et al. 2016). The most worrying situation is the detection of mcr in asymptomatic patients (Hu Y et al. 2016; Olaitan et al. 2016; Ruppé et al. 2016), which might further contribute to the silent dissemination. The vast majority of mcr-positive isolates were recovered from fecal samples (cf. Supplementary information Table S3). The Gram-negative species carrying mcr isolated from patients diagnosed with gastrointestinal disorder include E. coli, K. pneumoniae and Salmonella enterica (Doumith et al. 2016; Gu et al. 2016; Ye et al. 2016). The presence of mcr in Shigella sonnei (mcr-1) has been reported only once from a child suffering from dysenteric diarrhea (Thanh et al. 2016). Findings of mcr-positive isolates in the human gut microbiome (mcr-1) of healthy individual is a matter of great concern as gut flora can act as a mixing vessel which facilitates mcr dissemination by horizontal gene transfer (Hu Y et al. 2016; Ruppé et al. 2016). The mcr-harboring bacterial species isolated from patient urine samples were mainly E. coli, and less frequently K. pneumoniae, Enterobacter cloacae and S. enterica (cf. Supplementary information Table S3). Another mcr-carrying Enterobacter species, Enterobacter aerogenes was reported twice from clinical patients in China (Zeng et al. 2016; Wang Y et al. 2017). It is worth noting that the mcr-positive bacterial species isolated from bloodstream were mainly E. coli, with the exception of a few reports on mcr-harboring S. enterica and K. pneumoniae (cf. Supplementary information Table S3). Fortunately, many clinical mcr-harboring isolates were still susceptible to a number of other antimicrobial agents such as carbapenem and tigecycline (Quan et al. 2017; Saavedra et al. 2017). It is debatable whether surveillance cultures should be conducted for mcr when strains are still susceptible to most of antibiotic classes. Nevertheless, the dissemination of mcr-mediated polymyxin resistance should not be dismissed, as plasmids can be easily mobilized to MDR Gram-negatives.
Microbiology
Impact of mcr on polymyxin susceptibility
Transformation and conjugation methods are frequently utilized to study the transferability of the mcr-carrying plasmids and the impact of its acquisition on the polymyxin MIC (cf. Table 2). Broth microdilution is well accepted as the best method for testing polymyxin susceptibility, while other methods (e.g. Etest and disk diffusion) are less reliable but still used in clinical microbiology laboratory worldwide (Poirel, Jayol, et al. 2017; Simar et al. 2017). Generally, an increase in the polymyxin MICs was observed as mcr-carrying plasmid was introduced into polymyxin-susceptible strains (cf. Table 2). The successful conjugation of most mcr-harboring plasmids into the recipient strains led to the formation of transconjugants with comparable polymyxin MICs (4 – 16 mg/L) as the respective mcr donor strains (cf. Table 2). Further increased in colistin resistance in originally resistant E. coli strains (2 mg/L to 8 mg/L; 8 mg/L to 32 mg/L) was observed when mcr-1 plasmid was introduced into these two strains with an existing chromosomal pmrB mutation which is known to confer polymyxin resistance (Jayol et al. 2017). The extent of polymyxin resistance was not affected by the co-existence of multiple mcr-harboring plasmids in a single isolate (Li R et al. 2017; Zurfluh et al. 2017). However, a study demonstrated higher colistin MICs (8 mg/L) in S. enterica carrying multiple copies of mcr-5-harboring plasmid, as compared to the isolates with only one copy of chromosomal mcr-5 (4 mg/L) (Borowiak, Fischer, et al. 2017). Although we know that mcr confers resistance to polymyxins, unexpectedly mcr (more specifically mcr-1) has also been detected in colistin-susceptible E. coli strains (colistin MICs of 0.125 and less than 0.06 mg/L) (Liassine et al. 2016; Quan et al. 2017). The mcr-1 in a susceptible strain was reactivated following exposure to polymyxin, leading to a polymyxin-resistant phenotype (Thanh et al. 2016). This brings about the possibility for silent dissemination of mcr and further reactivation of the gene following polymyxin exposure.
Table 2.
Polymyxin B and colistin MICs of mcr-carrying strains and their respective transformants and/or transconjugants.
| Reference | Bacterial strain | Description | mcr | Polymyxin B | Colistin | ||
|---|---|---|---|---|---|---|---|
| MIC (mg/L) | MIC fold-change | MIC (mg/L) | MIC fold-change | ||||
| Liu YY et al. 2016 | E. coli SHP45 | Original mcr-1 positive isolate from pig | + | 4 | 8 | ||
| E. coli C600 | Recipient | − | 0.5 | 8 | 0.5 | 16 | |
| E. coli C600 transconjugant of E. coli SHP45 | Transconjugant | + | 4 | 8 | |||
| E. coli E11 | Recipient | − | 0.5 | 4 | 0.5 | 8 | |
| E. coli E11 + pHNSHP45 | Transformant | + | 2 | 4 | |||
| K. pneumoniae MPC11 | Recipient | − | 0.5 | 8 | 0.5 | 16 | |
| K. pneumoniae MPC11 + pHNSHP45 | Transformant | + | 4 | 8 | |||
| K. pneumoniae 1202 | Recipient | − | 0.5 | 8 | 0.5 | 8 | |
| K. pneumoniae 1202 + pHNSHP45 | Transformant | + | 4 | 4 | |||
| P. aeruginosa HE26 | Recipient | − | 0.5 | 8 | 0.5 | 16 | |
| P. aeruginosa HE26 + pHNSHP45 | Transformant | + | 4 | 8 | |||
| E. coli W3110 + pUC18 | Recipient (lab strain) | − | 0.5 | 4 | 0.5 | 4 | |
| E. coli W3110 + pUC18mcr-1 | Transformant | + | 2 | 2 | |||
| Gu et al. 2016 | K. pneumoniae 15451–1 | Original mcr-1 positive isolate from human | + | NA | 16 | ||
| E.coli C600 | Recipient | − | NA | NA | ≤1 | >16 | |
| E. coli C600 transconjugant of K. pneumoniae 15451–1 | Transconjugant | + | NA | 16 | |||
| E. coli 15451–2 | Original mcr-1 positive isolate from human | + | NA | 16 | |||
| E.coli C600 | Recipient | − | NA | NA | ≤1 | >16 | |
| E. coli C600 transconjugant of E. coli 15451–2 | Transconjugant | + | NA | 16 | |||
| Yang YQ et al. 2016 | S. enterica SC23 | Original mcr-1 positive isolate from chicken | + | 8 | 8 | ||
| E. coli J53 | Recipient | − | <0.25 | >32 | <0.25 | >32 | |
| E. coli J53 transconjugant of S. enterica SC23 | Transconjugant | + | 8 | 8 | |||
| Zeng et al. 2016 | Enterobacter aerogenes GB68 | Original mcr-1 positive isolate from human | + | 16 | 16 | ||
| E.coli C600 | Recipient | − | <0.25 | >64 | <0.25 | >64 | |
| E.coli C600 transconjugant of Enterobacter aerogenes GB68 | Transconjugant | + | 16 | 16 | |||
| Enterobacter cloacae GB38 | Original mcr-1 positive isolate from human | + | >32 | >32 | |||
| E.coli C600 | Recipient | − | <0.25 | >64 | <0.25 | >64 | |
| E.coli C600 transconjugant of Enterobacter cloacae GB38 | Transconjugant | + | 16 | 16 | |||
| Sonnevend et al. 2016 | E. coli BA76 | Original mcr-1 positive isolate from human | + | NA | 16 | ||
| E. coli J53RAZ | Recipient | − | NA | NA | 0.25 | 16 | |
| E. coli J53RAZ transconjugant of E. coli BA76 | Transconjugant | + | NA | 4 | |||
| E. coli BA77 | Original mcr-1 positive isolate from human | + | NA | 16 | |||
| E. coli J53RAZ | Recipient | − | NA | NA | 0.25 | 16 | |
| E. coli J53RAZ transconjugant of E. coli BA77 | Transconjugant | + | NA | 4 | |||
| E. coli SA26 | Original mcr-1 positive isolate from human | + | NA | 16 | |||
| E. coli J53RAZ | Recipient | − | NA | NA | 0.25 | 16 | |
| E. coli J53RAZ transconjugant of E. coli SA26 | Transconjugant | + | NA | 4 | |||
| E. coli ABC149 | Original mcr-1 positive isolate from human | + | NA | 16 | |||
| E. coli DH5α | Recipient | − | NA | NA | 0.25 | 32 | |
| E. coli DH5α + pABC149 | Transformant | + | NA | 8 | |||
| Berrazeg et al. 2016 | E. coli SE65 | Original mcr-1 positive isolate from human | + | NA | 4 | ||
| E. coli J53 | Recipient | − | NA | NA | 0.125 | 32 | |
| E. coli J53 transconjugant of E. coli SE65 | Transconjugant | + | NA | 4 | |||
| Li XP et al. 2016 | S. enterica GDS78, GDS79, GDS82, GDS141 | Original mcr-1 positive isolate from animal | + | NA | 16 | ||
| E. coli C600 | Recipient | − | NA | NA | 0.125 | 32 | |
| E. coli C600 T(GDS78T, GDS79T, GDS82T, GDS141T) | Transconjugant | + | NA | 4 | |||
| Zheng et al. 2016 | E. coli 1002 | Original mcr-1 positive isolate from human | + | 4 | 4 | ||
| E. coli J53 | Recipient | − | 0.25 | 8 | 0.5 | 8 | |
| E. coli J53 transconjugant of E. coli 1002 | Transconjugant | + | 2 | 4 | |||
| E. coli 2474 | Original mcr-1 positive isolate from human | + | 4 | 4 | |||
| E. coli J53 | Recipient | − | 0.25 | 16 | 0.5 | 8 | |
| E. coli J53 transconjugant of E. coli 2474 | Transconjugant | + | 4 | 4 | |||
| Zhong et al. 2016 | E. coli GB049 | Original mcr-1 positive isolate form human | + | 16 | 8 | ||
| E.coli EC600 | Recipient | − | 0.5 | 32 | 0.25 | 64 | |
| E.coli EC600 transconjugant of E. coli GB049 | Transconjugant | + | 16 | 16 | |||
| E. coli GB090 | Original mcr-1 positive isolate form human | + | 16 | 16 | |||
| E.coli EC600 | Recipient | − | 0.5 | 32 | 0.25 | 64 | |
| E.coli EC600 transconjugant of E. coli GB090 | Transconjugant | + | 16 | 16 | |||
| Liu BT et al. 2016 | E. coli WF5–19 | Original mcr-1 positive isolate from chicken | + | NA | 4 | ||
| E. coli C600 | Recipient | − | NA | NA | 0.25 | 16 | |
| E. coli C600 transconjugant of E. coli WF5–19 | Transconjugant | + | NA | 4 | |||
| Cronobacter sakazakii WF5–19C | Original mcr-1 positive isolate from chicken | + | NA | 4 | |||
| E. coli C600 | Recipient | − | NA | NA | 0.25 | 16 | |
| E. coli C600 transconjugant of Cronobacter sakazakii WF5–19C | Transconjugant | + | NA | 4 | |||
| Cronobacter sakazakii WF5–21C | Original mcr-1 positive isolate from chicken | + | NA | 4 | |||
| E. coli C600 | Recipient | − | NA | NA | 0.25 | 16 | |
| E. coli C600 transconjugant of Cronobacter sakazakii WF5–21C | Transconjugant | + | NA | 4 | |||
| Ortiz de la Tabla et al. 2016 | E. coli (unnamed) | Original mcr-1 positive isolate from human | + | NA | 4 | ||
| E. coli Hb101 | Recipient | − | NA | NA | 0.5 | 8 | |
| E. coli Hb101 transconjugant of E. coli (unnamed) | Transconjugant | + | NA | 4 | |||
| Lu et al. 2017 | S. enterica Typhimurium YL14P053 | Original mcr-1.6 positive isolate from human | + | NA | 4 | ||
| S. enterica Typhi CT18 | Recipient | − | NA | NA | 0.125 | 32 | |
| S. enterica Typhi CT18 transconjugant of S. enterica Typhimurium YL14P053 | Transconjugant | + | NA | 4 | |||
| E. coli J53 AziR | Recipient | − | NA | NA | 0.125 | 32 | |
| E. coli J53 AziR transconjugant of S. enterica Typhimurium YL14P053 | Transconjugant | + | NA | 4 | |||
| K. pneumoniae BJ1988 | Recipient | − | NA | NA | 0.125 | 32 | |
| K. pneumoniae BJ1988 transconjugant of S. enterica Typhimurium YL14P053 | Transconjugant | + | NA | 4 | |||
| Conceição-Neto et al. 2017 | E. coli CCBH20178 | Original mcr-1 positive isolate from human | + | NA | 8 | ||
| E. coli J53 | Recipient | − | NA | NA | <0.125 | >32 | |
| E. coli J53 transconjugant of E. coli CCBH20178 | Transconjugant | + | NA | 4 | |||
| E. coli CCBH20607 | Original mcr-1 positive isolate from human | + | NA | 8 | |||
| E. coli J53 | Recipient | − | NA | NA | <0.125 | >32 | |
| E. coli J53 transconjugant of E. coli CCBH20607 | Transconjugant | + | NA | 4 | |||
| E. coli CCBH20180 | Original mcr-1 positive isolate from human | + | NA | 4 | |||
| E. coli J53 | Recipient | − | NA | NA | <0.125 | >64 | |
| E. coli J53 transconjugant of E. coli CCBH20180 | Transconjugant | + | NA | 8 | |||
| Aires et al. 2017 | K. pneumoniae CCBH24080 | Original mcr-1 positive isolate from human | + | NA | 16 | ||
| E. coli J53 | Recipient | − | NA | NA | <0.125 | >64 | |
| E. coli J53 transconjugant of K. pneumoniae CCBH24080 | Transconjugant | + | NA | 8 | |||
| Kim et al. 2017 | E. coli 28 | Original mcr-1 positive isolate from human | + | NA | 8 | ||
| E. coli J53 | Recipient | − | NA | NA | 0.5 | 16 | |
| E. coli J53 transconjugant of E. coli 28 | Transconjugant | + | NA | 8 | |||
| Yin et al. 2017 | E. coli WJ1 | Original mcr-3 positive isolate from porcine | + | NA | 8 | ||
| E. coli C600 | Recipient | − | NA | NA | 0.5 | 8 | |
| E. coli C600 transconjugant of E. coli WJ1 | Transconjugant | + | NA | 4 | |||
| Liu L et al. 2017 | E. coli WCHECLL123 | Original mcr-3.5 and mcr-1 positive isolate from human | + | NA | 8 | ||
| E. coli J53 | Recipient | − | NA | NA | 1 | 4 | |
| E. coli J53 transconjugant of E. coli WCHECLL123 (mcr-1) | Transconjugant | + | NA | 4 | |||
| E. coli J53 transconjugant of E. coli WCHECLL123 (mcr-3.5) | Transconjugant | + | NA | NA | 4 | 4 | |
| Carattoli et al. 2017 | S. enterica Typhimurium R3445 | Original mcr-4 positive isolate from pig | + | NA | 8 | ||
| E. coli DH5α | Recipient | − | NA | NA | 0.25 | 8 | |
| E. coli DH5α + pMCR_R3445 | Transformant | + | NA | 2 | |||
| E. coli R4287 | Original mcr-4 positive isolate from pig | + | NA | 8 | |||
| E. coli CSH26 RifR | Recipient | − | NA | NA | 0.25 | 16 | |
| E. coli CSH26 RifR transconjugant of E. coli R4287 | Transconjugant | + | NA | 4 | |||
NA: not available; Original mcr-positive isolates are highlighted in grey.
Prevalence of mcr in Gram-negative species
E. coli is the most prevalent species among the mcr-positive isolates reported so far, accounting for approximately 91% of the total mcr-carrying isolates, followed by S. enterica (~7%) and K. pneumoniae (~2%). It is noteworthy that the total number of S. enterica screened for mcr was at least 12-fold greater than K. pneumoniae. This is likely due to the fact that S. enterica is one of the major food-borne pathogens and mcr is very likely to be disseminated via food chain (Zurfluh et al. 2017). The mcr has been detected on very rare occasion in Klebsiella oxytoca, Citrobacter freundii, Citrobacter braakii, Cronobacter sakazakii, Kluyvera ascorbata, Shigella sonnei, Raoultella ornithinolytica, Proteus mirabilis, Moraxella, Aeromonas and Enterobacter species with a total prevalence rate of approximately 0.2%. Among the bacterial species which have been tested, mcr has not been detected in wild-type isolates of Klebsiella ozaenae, Morganella morganii, Providencia rettgeri, Pseudomonas aeruginosa, Campylobacter, Serratia and Acinetobacter species. Although mcr has yet to be found in wildtype Pseudomonas and Acinetobacter species, it has been demonstrated that, after mcr-1 was introduced into Acinetobacter baumannii and Pseudomonas aeruginosa, their lipid A was modified by pEtN; interestingly, greater colistin resistance was observed in Acinetobacter baumannii (64- to >128-fold increase in colistin MICs) as compared to only modest changes in colistin susceptibility in Pseudomonas aeruginosa (2- to 4-fold increase in colistin MICs) (Liu YY et al. 2017). Overall, the true prevalence of mcr has yet to be fully understood due to the limits of many studies which only screened for the presence of mcr in extended-spectrum-β-lactamase (ESBL)-producing isolates and polymyxin-resistant isolates. Such limitations could lead to the underestimation of the true prevalence for mcr isolates.
Co-occurrence with β-lactamases and carbapenemases
Carbapenem is often the treatment option for ESBL-associated bacterial infection and unfortunately increasing emergence of carbapenemase-producing bacteria has been reported (Meletis 2016; Thomson 2010). This situation has brought back polymyxins as a last-resort against carbapenemase-producing MDR Gram-negative bacteria (Trecarichi and Tumbarello 2017). Hence, the co-occurrence of mcr with carbapenemases may herald the rise of a post-antibiotic era. The mcr was found to be frequently associated with β-lactamase-producing Enterobacteriaceae carrying blaCTX-M and blaTEM of various variants as well as carbapenem-resistant Enterobacteriaceae harboring blaOXA-48, blaOXA-181, blaKPC-2, blaKPC-3, blaNDM-1, blaNDM-4, blaNDM-5 and blaNDM-9 (cf. Supplementary information Table S4). Importantly, the discovery of β-lactamase and carbapenemase genes co-localizing with mcr on the same conjugative plasmid is the most worrisome, as Gram-negative pathogens can acquire both types of antibiotic resistance genes via horizontal transmission. The co-localization of the β-lactamase and carbapenemase genes (blaTEM-1, blaCTX-M-1, blaCTX-M-55, blaNDM-5) with mcr most commonly occurs on the IncHI2 (Grami et al. 2016; Sonnevend et al. 2016; Zurfluh, Klumpp, et al. 2016; Yi et al. 2017) and IncI2 (Sonnevend et al. 2016; Yang YQ et al. 2016; Yi et al. 2017) plasmids, and less commonly on IncFI (Zeng et al. 2016; Zhong et al. 2016), IncK2 (Donà, Bernasconi, Pires, et al. 2017) and IncX3-IncX4 hybrid plasmids (Sun et al. 2016).
Potential origins of mcr
Thus far, mcr-1 to -7 have been reported. MCR-1 and MCR-2 share the highest percentage of amino acid sequence identity (81%), are believed to be originated from Moraxella species, common animal pathogens. MCR-1 and MCR-2 share 59 – 64% amino acid similarities with those found in M. porci, M. osloensis, M. lincolnii and M. catarrhalis (Kieffer et al. 2017). MCR-3 and MCR-7, which share 70% amino acid similarity, might be of Aeromonas origin (Yang et al. 2018; Yin et al. 2017). MCR-4, sharing only 34% amino acid sequence identity with MCR-1 might have been originated from Shewanella frigidimarina (Carattoli et al. 2017). MCR-5 has distinct amino acid sequences from the all the others (34 – 37% similarities) and its origin is still unknown (Borowiak, Fischer, et al. 2017). The heavy usage of colistin in animals is very likely a major selective factor facilitating the mobilization of these mcr genes from natural source to Enterobacteriaceae.
Genetic organization of mcr-harboring plasmids
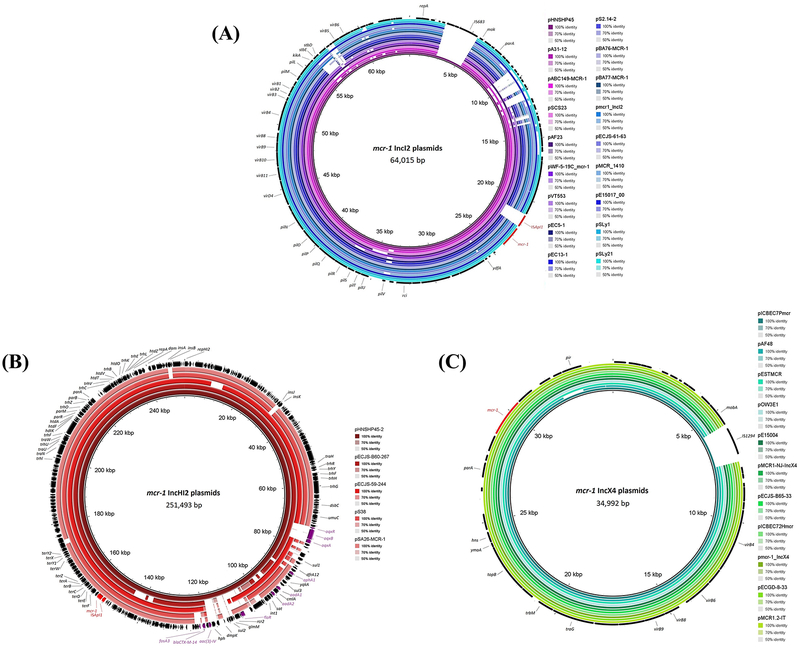
The detection of mcr on different classes of plasmids indicated a high diversity of mcr plasmid reservoirs (cf. Table 3). Numerous studies have confirmed mcr on three major types of plasmids: IncI2, IncHI2 and IncX4. There are also several other types of plasmids carrying mcr, including IncHI1, IncF, IncFI, IncFIB, IncFII, IncP, IncP-1, IncK2 and phage-like IncY (cf. Supplementary information Table S5). To date, only mcr-4 and mcr-5 have been identified on small ColE-type plasmid (Borowiak, Fischer, et al. 2017; Carattoli et al. 2017). Of all the different plasmid reservoirs, the IncHI2-type plasmids are often associated with various antimicrobials resistance (cf. Figure 3). As compared to IncI2 and IncHI2-type plasmids, the genetic contexts are more conserved in IncX4-type plasmids except pICBEC7Pmcr which has an additional IS1294 resulting in truncated mobA (Sellera et al. 2016) (cf. Figure 3). Although mcr was initially reported as a plasmid-mediated polymyxin resistance gene, mcr-1 has been identified in the chromosome of E. coli strains and mcr-5 was identified in the chromosome of a S. enterica (cf. Supplementary information Table S5). The presence of ISApl1-mcr-1 region in the chromosome postulates the importance of ISApl1 for the chromosomal integration of mcr-1 and represents another transmissibility factor for the vertical transmission. Remarkably, a triplicated ISApl1-mcr-1-Δpap in a tandem arrangement in the chromosome was described in an E. coli isolated from humans, indicating a more diverse genetic context of mcr (Yu, Ang, Chong, et al. 2016).
Table 3.
Characterization of mcr-harboring plasmids with complete sequences in Genbank.
| Year | Country | Source | Species | Plasmid | Type | Length (bp) | ISApI1 | Accession number |
|---|---|---|---|---|---|---|---|---|
| 2011 – 2012 | Belgium | Pig | E. coli | pKP37-BE | IncX4 | 35,104 | − | LT598652 |
| 2012 | Switzerland | River water | E. coli | pOW3E1 | IncX4 | 34,640 | − | KX129783 |
| 2013 | Estonia | Pig | E. coli | pESTMCR | IncX4 | 33,311 | − | KU743383 |
| 2013 | Brazil | Magellanic penguin | E. coli | pICBEC7Pmcr | IncX4 | 34,992 | − | CP017246 |
| 2014 | China | Pig | E. coli | pECGD-8–33 | IncX4 | 33,307 | − | KX254343 |
| 2014 | Italy | Human | K. pneumoniae | pMCR1.2-IT mcr-1.2 | IncX4 | 33,303 | − | KX236309 |
| 2014 | The United States | Human | E. coli | pMCR1-NJ-IncX4 | IncX4 | 33,395 | − | KX447768 |
| 2015 | China | Human | K. pneumoniae | pmcr1_IncX4 | IncX4 | 33,287 | − | KU761327 |
| 2015 | China | Human | E. coli | pE15004 | IncX4 | 33,309 | − | KX772777 |
| 2015 | China | Pig | E. coli | pECJS-B65–33 | IncX4 | 33,298 | − | KX084392 |
| 2015 | China | Sewage | E. coli | pMCR_WCHEC1618 mcr-1.4 | IncX4 | 33,309 | − | KY463454 |
| 2015 | Portugal | Pig | E. coli | pLV23529-MCR-1.9 mcr-1.9 | IncX4 | 33,303 | − | KY964067 |
| 2015 | South Africa | Human | E. coli | pAF48 | IncX4 | 31,808 | − | KX032520 |
| 2016 | Brazil | Human | E. coli | pICBEC72Hmcr | IncX4 | 33,304 | − | CP015977 |
| 2011 | Australia | Human | E. coli | pJIE2288–1 | IncI2 | 60,733 | − | KY795977 |
| 2013 | Australia | Human | E. coli | pJIE3685–1 | IncI2 | 60,960 | − | KP795978 |
| 2013 | Malaysia | Chicken | E. coli | pEC5–1 | IncI2 | 61,735 | − | CP016185 |
| 2013 | Malaysia | Pond water | E. coli | pEC13–1 | IncI2 | 60,218 | − | CP016186 |
| 2013 | Malaysia | Chicken | E. coli | pS2.14–2 | IncI2 | 60,950 | − | CP016187 |
| 2013 | China | Chicken | E. coli | pHeN867 mcr-1.3 | IncI2 | 60,757 | − | KU934208 |
| 2014 | China | Chicken | K. pneumoniae | pSC20141012 mcr-7 | IncI2 | 65,631 | − | MG267386 |
| 2015 | Bahrain | Human | E. coli | pBA76-MCR-1 | IncI2 | 64,942 | − | KX013540 |
| 2015 | Bahrain | Human | E. coli | pBA77-MCR-1 | IncI2 | 62,661 | − | KX013539 |
| 2015 | China | Human | E. coli | pmcr1_IncI2 | IncI2 | 64,964 | − | KU761326 |
| 2015 | China | Pig | E. coli | pECJS-61–63 | IncI2 | 63,656 | − | KX084393 |
| 2015 | China | Hospital sewage | Kluyvera ascorbata | pMCR_1410 | IncI2 | 57,059 | − | KU922754 |
| 2015 | China | Human | E. coli | pE15017_00 | IncI2 | 65,375 | − | KX772778 |
| 2016 | The United States | Pig | E. coli | pSLy1 | IncI2 | 65,888 | − | CP015913 |
| 2016 | The United States | Pig | E. coli | pSLy21 | IncI2 | 63,329 | − | CP016405 |
| 2013 – 2015 | Argentina | Human | E. coli | pMCR-M15049 mcr-1.5 | IncI2 | 61,198 | + | KY471308 |
| 2013 – 2015 | Argentina | Human | E. coli | pMCR-M17059 mcr-1.5 | IncI2 | 61,531 | + | KY471310 |
| 2013 – 2015 | Argentina | Human | E. coli | pMCR-M19241 mcr-1.5 | IncI2 | 61,584 | + | KP471311 |
| 2012 | China | Chicken | E. coli | pA31–12 | IncI2 | 67,134 | + | KX034083 |
| 2013 | China | Pig | E. coli | pHNSHP45 | IncI2 | 64,015 | + | KP347127 |
| 2013 | United Arab Emirates | Human | E. coli | pABC149-MCR-1 | IncI2 | 61,228 | + | KX013538 |
| 2014 | China | Chicken | S. enterica | pSCS23 | IncI2 | 65,419 | + | KU934209 |
| 2014 | South Africa | Human | E. coli | pAf23 | IncI2 | 61,177 | + | KX032519 |
| 2015 | China | Chicken | Cronobacter sakazakii | pWF-5–19C_mcr-1 | IncI2 | 65,203 | + | KX505142 |
| 2015 | China | Sewage | E. coli | pMCR_WCHEC1604-IncI2 mcr-1.7 | IncI2 | 62,098 | + | KY829117 |
| 2015 | South Africa | Chicken | E. coli | pVT553 | IncI2 | 62,219 | + | KU870627 |
| 2011 – 2012 | Belgium | Cattle | E. coli | pKH457–3-BE | IncP | 79,798 | − | KU353730 |
| 2014 | China | Human | S. enterica Typhimurium | pMCR16_P053 mcr-1.6 | IncP | 47,824 | − | KY352406 |
| 2015 | China | Hospital sewage | K. pneumoniae | pMCR_1511 | IncP | 57,278 | + | KX377410 |
| 2017 | China | Human | E. coli | pMCR3_LL123 mcr-3.5 | IncP | 52,208 | − | MF489760 |
| 2011 – 2012 | Belgium | Pig | E. coli | pKP81-BE | IncFII | 91,041 | + | KU994859 |
| 2016 | The United States | Human | E. coli | pMR0516mcr | IncF | 225,069 | + | KX276657 |
| 2014 | Switzerland | Vegetables (imported from Thailand) | E. coli | pH226B | IncHI1 | 209,401 | − | KX129784 |
| 2013 | Malaysia | Chicken | E. coli | pEC2_1–4 | IncHI1 | 230,278 | + | CP016183 |
| 2013 | Malaysia | Pig | E. coli | pEC2–4 | IncHI1 | 235,403 | + | CP016184 |
| 2008 | Germany | Chicken | S. enterica Paratyphi B | pSE08-00436-1 | IncHI2 | 264,914 | + | CP020493 |
| 2012 | Saudi Arabia | Human | E. coli | pSA26-MCR-1 | IncHI2 | 240,367 | + | KU743384 |
| 2013 | China | Pig | E. coli | pHNSHP45–2 | IncHI2 | 251,493 | + | KU341381 |
| 2015 | China | Pig | E. coli | pECJS-59–244 | IncHI2 | 243,572 | + | KX084394 |
| 2015 | China | Pig | E. coli | pECJS-B60–267 | IncHI2 | 267,486 | + | KX254341 |
| 2015 | Switzerland | Chicken meat (imported from Italy) | E. coli | pS38 | IncHI2 | 247,885 | + | KX129782 |
| 2015 | China | Pig | E. coli | pWJ1 mcr-3 | IncHI2 | 261,119 | − | KY924928 |
| 2015 | China | Pig | E. coli | pHYEC7-mcr1 | IncY | 97,559 | + | KX518745 |
| 2016 | China | Pig | E. coli | pMCR-1-P3 | IncY | 97,386 | + | KX880944 |
| 2012 | Germany | Poultry | S. enterica Paratyphi B | pSE12–02541 mcr-5 | ColE | 17,156 | − | KY807920 |
| 2013 | Germany | Chicken meat | S. enterica Paratyphi B | pSE13-SA01718 mcr-5 | ColE | 12,201 | − | KY807921 |
| 2013 | Italy | Pig | S. enterica Typhimurium | pMCR_R3445 mcr-4 | ColE10 | 8,749 | − | MF543359 |
Plasmids carried mcr-1 unless stated otherwise in superscript.
Figure 3.
Comparison of the genetic context of mcr-1 harboring (A) IncI2 plasmids with pHNSHP45 as the reference sequence; (B) IncHI2 plasmids with pHNSHP45–2 as the reference sequence; (C) IncX4 plasmids with pICBEC7Pmcr as the reference sequence, using BRIG (Alikhan et al. 2011). The mcr-1 and ISApl1 transposase are illustrated in red, whereas other antimicrobial resistance genes are illustrated in purple at the outermost circle containing the CDS annotations.
It has been demonstrated that the transposon Tn6330 element (ISApl1-mcr-1-orf-ISApl1) could be excised from the plasmid, forming a circular intermediate of ISApl1-mcr-1-orf, which might be integrated into other ISApl1-carrying plasmids (Li R et al. 2016). The mobilization of mcr-1 by ISApl1-mediated transposition was demonstrated in vitro (Poirel, Kieffer and Nordmann 2017). Plasmid analysis from various studies revealed that the ISApl1 family transposase does not always co-present with mcr in the plasmid. ISApl1 is usually present in IncHI2-type plasmids (~200 kb), can be either present or absent in IncI2-type plasmids (~60 kb), and completely absent in IncX4-type plasmids (~30 kb) (cf. Figure 3). A possible explanation for this observation is that mcr may have originated from plasmid and their co-evolution occurred via acquisition of ISApl1, leading to rapid mobilization onto other plasmids (Kuo et al. 2016; Petrillo et al. 2016). Another possibility is that ISApl1 transposase is lost after translocation of the ISApl1-mcr-1 element into the plasmid with the purpose to strengthen the stability of mcr per se in the plasmid (Li A et al. 2016; Snesrud et al. 2016). The mcr-3 was found to be associated with TnAs2 (Yin et al. 2017) and TnAs3 (Liu L et al. 2017), which are the Tn3-family transposon and were found only in Aeromonas salmonicida. This fortifies the possible transfer of mcr-3 from Aeromonas species to Enterobacteriaceae. The mcr-5 gene was found to be located on a Tn3-family transposon which has been identified in Cupriavidus gilardii (Borowiak, Fischer, et al. 2017).
The transferability of mcr-carrying plasmids between bacteria depends on the conjugative properties of the plasmid backbone. The defective conjugation potential can be due to the disrupted or lacking of tra region (encodes for conjugal transfer protein) (Bernasconi et al. 2016; Cui et al. 2017). The mcr-1 carrying plasmid, pMCR_1410 from Kluyvera ascorbata was found to possess the ability to transfer between different Gram-negative species, unlike the first mcr-1-harboring plasmid identified, pHNSHP45. Comparison of pMCR_1410 and pHNSHP45 revealed that the absence of traC in pHNSHP45 could cause its inability for interspecies conjugation, suggested that traC gene could possibly be responsible for interspecies transfer of the plasmid (Zhao and Zong 2016). This finding is alarming due to the potential for the spread of mcr to a more diverse bacterial species pool, which highlights the need for further investigations into the genes involved in the conjugative transfer of mcr.
Future perspective
Since the discovery of mcr, the number of mcr-harboring isolates has been increasingly reported worldwide at an alarming rate. Notwithstanding, the prevalence of mcr remains much higher in livestock as compared to humans, which is in line with its purported agricultural origins. Luckily, the use of colistin has been recently banned for agriculture purposes in China and Brazil (Walsh and Wu 2016; Monte et al. 2017). The increasing usage of polymyxins clinically may increase the potential for wide dissemination of mcr in the nosocomial setting. As polymyxins are the last therapeutic option for life-threatening infections caused by Gram-negative ‘superbugs’, every effort must be made to minimize the emergence of resistance, in particular due to mcr.
Supplementary Material
Acknowledgments
Disclosure statement
J.L. and T.V. are supported by grants from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (R01 AI111965 and AI132154). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health. J.L. is an Australian National Health and Medical Research Council (NHMRC) Senior Research Fellow. T.V. is an Australian NHMRC Career Development Research Fellow.
Footnotes
Declaration of interest
The authors report no declarations of interest.
References
- AbuOun M, Stubberfield EJ, Duggett NA, Kirchner M, Dormer L, Nunez-Garcia J, Randall LP, Lemma F, Crook DW, Teale C et al. 2017. mcr-1 and mcr-2 variant genes identified in Moraxella species isolated from pigs in Great Britain from 2014 to 2015. 72(10):2745–2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainsworth GC, Brown AM, Brownlee G. 1947. ‘Aerosporin’, an antibiotic produced by Bacillus aerosporus Greer. Nature. 160:263. [DOI] [PubMed] [Google Scholar]
- Aires CAM, da Conceicao-Neto OC, Oliveira T, Dias CF, Montezzi LF, Picão RC, Albano RM, Asensi MD, Carvalho-Assef APD. 2017. Emergence of plasmid-mediated mcr-1 gene in clinical KPC-2-producing Klebsiella pneumoniae ST392 in Brazil. Antimicrob Agents Chemother. 61(7):pii=e00317–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST Ring Image Genarator (BRIG): simple prokaryote genome comparisons. BMC Genom. 12:402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anjum MF, Duggett NA, AbuOun M, Randall L, Nunez-Garcia J, Ellis RJ, Rogers J, Horton R, Brena C, Williamson S et al. 2016. Colistin resistance in Salmonella and Escherichia coli isolates from a pig farm in Great Britain. J Antimicrob Chemother. 71(8):2306–2313. [DOI] [PubMed] [Google Scholar]
- Arcilla MS, van Hattem JM, Matamoros S, Melles DC, Penders J, de Jong MD, Schultsz C. 2016. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis. 16(2):147–149. [DOI] [PubMed] [Google Scholar]
- Baron S, Hadjadj L, Rolain JM, Olaitan AO. 2016. Molecular mechanisms of polymyxin resistance: knowns and unknowns. Int J Antimicrob Agents. 48(6):583–591. [DOI] [PubMed] [Google Scholar]
- Benedict RG, Langlykke AF. 1947. Antibiotic activity of Bacillus polymyxa. J Bacteriol. 54(1):24. [PubMed] [Google Scholar]
- Bernasconi OJ, Kuenzli E, Pires J, Tinguely R, Carattoli A, Hatz C, Perreten V, Endimiani A. 2016. Travelers can import colistin-resistant Enterobacteriaceae including those possessing the plasmid-mediated mcr-1 gene. Antimicrob Agents Chemother. 60(8):5080–5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrazeg M, Hadjadj L, Ayad A, Drissi M, Rolain JM. 2016. First detected human case in Algeria of mcr-1 plasmid mediated colistin resistance: a 2011 Escherichia coli isolate. Antimicrob Agents Chemother. 60(11):6996–6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyrouthy R, Robin F, Lessene A, Lacombat I, Dortet L, Naas T, Ponties V, Bonnet R. 2017. MCR-1 and OXA-48 in vivo acquisition in KPC-producing Escherichia coli after colistin treatment. Antimicrob Agents Chemother. 61(8):pii=e02540–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowiak M, Fischer J, Hammerl JA, Hendriksen RS, Szabo I, Malorny B. 2017. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J Antimicrob Chemother. 72(12):3317–3324. [DOI] [PubMed] [Google Scholar]
- Borowiak M, Hammerl JA, Fischer J, Szabo I, Malorny B. 2017. Complete genome sequence of Salmonella enterica subsp. enterica Serovar Paratyphi B sequence type 28 harboring mcr-1. Genome Announc. 5(37):pii=e00991–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer A, Telling K, Laht M, Kalmus P, Lutsar I, Remm M, Kisand V, Tenson T. 2016. Plasmid with colistin resistance gene mcr-1 in extended-spectrum-β-lactamase-producing Escherichia coli strains isolated from pig slurry in Estonia. Antimicrob Agents Chemother. 60(11):6933–6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan E, Martins M, McCusker MP, Wang J, Alves BM, Hurley D, El Garch F, Woehrlé F, Miossec C, McGrath L et al. 2016. Multidrug-resistant Escherichia coli in bovine animals, Europe. Emerg Infect Dis. 22(9):1650–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JM, Dorman DC, Roy LP. 1970. Acute renal failure due to overdosage of colistin. Med J Aust. 2(20):923–924. [DOI] [PubMed] [Google Scholar]
- Campos J, Cristino L, Peixe L, Antunes P. 2016. MCR-1 in multidrug-resistant and copper-tolerant clinically relevant Salmonella 1,4,[5],12:i:- and S. Rissen clones in Portugal, 2011 to 2015. Euro Surveill. 21(26):pii=30270. [DOI] [PubMed] [Google Scholar]
- Cannatelli A, Giani T, Antonelli A, Principe L, Luzzaro F, Rossolini GM. 2016. First detection of the mcr-1 colistin resistance gene in Escherichia coli in Italy. Antimicrob Agents Chemother. 60(5):3257–3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A, Villa L, Feudi C, Curcio L, Orsini S, Luppi A, Pezzotti G, Magistrali CF. 2017. Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro Surveill. 22(31):pii=30589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnevali C, Morganti M, Scaltriti E, Bolzoni L, Pongolini S, Casadei G. 2016. Occurrence of mcr-1 colistin-resistant Salmonella isolates recovered from human and animals in Italy, 2012–2015. Antimicrob Agents Chemother. 60(12):7532–7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carretto E, Brovarone F, Nardini P, Russello G, Barbarini D, Pongolini S, Gagliotti C, Carattoli A, Sarti M. 2018. Detection of mcr-4 positive Salmonella enterica serovar Typhimurium in clinical isolates of human origin, Italy, October to November 2016. Euro Surveill. 23(2): pii=17–00821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanheira M, Griffin MA, Deshpande LM, Mendes RE, Jones RN, Flamm RK. 2016. Detection of mcr-1 among Escherichia coli clinical isolates collected worldwide as part of the SENTRY Antimicrobial Surveillance Program during 2014–2015. Antimicrob Agents Chemother. 60(9):5623–5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou CS, Chen YT, Wang YW, Liu YY, Kuo HC, Tu YH, Lin AC, Liao YS, Hong YP. 2017. Dissemination of mcr-1-carrying plasmids among colistin-resistant Salmonella strains from humans and food-producing animals, Taiwan. Antimicrob Agents Chemother. 61(7):pii=e00338–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coetzee J, Corcoran C, Prentice E, Moodley M, Mendelson M, Poirel L, Nordmann P, Brink AJ. 2016. Emergence of plasmid-mediated colistin resistance (MCR-1) among Escherichia coli isolated from South African patients. S Afr Med J. 106(5):449–450. [DOI] [PubMed] [Google Scholar]
- Conceição-Neto OC, Aires CAM, Pereira NF, da Silva LHJ, Picão RC, Siqueira BN, Albano RM, Asensi MD, Carvalho-Assef APD. 2017. Detection of the plasmid-mediated mcr-1 gene in clinical KPC-2-producing Escherichia coli isolates in Brazil. Int J Antimicrob Agents. 50(2):282–284. [DOI] [PubMed] [Google Scholar]
- Corbella M, Mariani B, Ferrari C, Comandatore F, Scaltriti E, Marone P, Cambieri P. 2017. Three cases of mcr-1-positive colistin-resistant Escherichia coli bloodstream infections in Italy, August 2016 to January 2017. Euro Surveill. 22(16):pii=30517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui M, Zhang J, Gu Z, Li R, Chan EW, Yan M, Xu X, Chen S, Wu C. 2017. Prevalence and molecular characterization of mcr-1-positive Salmonella strains recovered from clinical specimens in China. Antimicrob Agents Chemother. 61(5):pii=e02471–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcio L, Luppi A, Bonilauri P, Gherpelli Y, Pezzotti G, Pesciaroli M, Magistrali CF. 2017. Detection of the colistin resistance gene mcr-1 in pathogenic Escherichia coli from pigs affected by post-weaning diarrhoea in Italy. J Glob Antimicrob Resist. 10:80–83. [DOI] [PubMed] [Google Scholar]
- Di Pilato V, Arena F, Tascini C, Cannatelli A, Henrici De Angelis L, Fortunato S, Giani T, Menichetti F, Rossolini GM. 2016. mcr-1.2: a new mcr variant encoded by a transferable plasmid from a colistin-resistant KPC carbapenemase-producing Klebsiella pneumoniae of sequence type 512. Antimicrob Agents Chemother. 60(9):5612–5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donà V, Bernasconi OJ, Kasraian S, Tinguely R, Endimiani A. 2017. A SYBR green-based real-time PCR method for improved detection of mcr-1-mediated colistin resistance in human stool samples. J Glob Antimicrob Resist. 9:57–60. [DOI] [PubMed] [Google Scholar]
- Donà V, Bernasconi OJ, Pires J, Collaud A, Overesch G, Ramette A, Perreten V, Endimiani A. 2017. Heterogeneous genetic location of mcr-1 in colistin-resistant Escherichia coli isolated from humans and retail chicken meat in Switzerland: emergence of mcr-1-carrying IncK2 plasmids. Antimicrob Agents Chemother. 61(11):pii=e01245–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doumith M, Godbole G, Ashton P, Larkin L, Dallman T, Day M, Day M, Muller-Pebody B, Ellington MJ, de Pinna E et al. 2016. Detection of the plasmid-mediated mcr-1 gene conferring colistin resistance in human and food isolates of Salmonella enterica and Escherichia coli in England and Wales. J Antimicrob Chemother. 71(8):2300–2305. [DOI] [PubMed] [Google Scholar]
- Du H, Chen L, Tang Y-W, Kreiswirth BN. 2016. Emergence of the mcr-1 colistin resistance gene in carbapenem-resistant Enterobacteriaceae. Lancet Infect Dis. 16(3):287–288. [DOI] [PubMed] [Google Scholar]
- Duggett NA, Sayers E, AbuOun M, Ellis RJ, Nunez-Garcia J, Randall L, Horton R, Rogers J, Martelli F, Smith RP et al. 2016. Occurrence and characterization of mcr-1-harboring Escherichia coli isolated from pigs in Great Britain from 2013 to 2015. J Antimicrob Chemother. 72(3):691–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichhorn I, Feudi C, Wang Y, Kaspar H, Feßler AT, Lübke-Becker A, Michael GB, Shen J, Schwarz S. 2018. Identification of novel variants of the colistin resistance gene mcr-3 in Aeromonas spp. from the national resistance monitoring programme GERM-Vet and from diagnostic submissions. J Antimicrob Chemother. 73(5): 1217–1221. [DOI] [PubMed] [Google Scholar]
- El Garch F, Sauget M, Hocquet D, Lechaudee D, Woehrle F, Bertrand X. 2016. mcr-1 is borne by highly diverse Escherichia coli isolates since 2004 in food-producing animals in Europe. Clin Microbiol Infect. 23(1):51. [DOI] [PubMed] [Google Scholar]
- Ellem JA, Ginn AN, Chen SC, Ferguson J, Partridge SR, Iredell JR. 2017. Locally Acquired mcr-1 in Escherichia coli, Australia, 2011 and 2013. Emerg Infect Dis. 23(7):1160–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elnahriry SS, Khalifa HO, Soliman AM, Ahmed AM, Hussein AM, Shimamoto T, Shimamoto T. 2016. Emergence of plasmid-mediated colistin resistance gene mcr-1 in a clinical Escherichia coli isolate from Egypt. Antimicrob Agents Chemother. 60(5):3249–3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falgenhauer L, Waezsada S-E, Yao Y, Imirzalioglu C, Käsbohrer A, Roesler U, Michael GB, Schwarz S, Werner G, Kreienbrock L et al. 2016. Colistin resistance gene mcr-1 in extended-spectrum β-lactamase-producing and carbapenemase-producing Gram-negative bacteria in Germany. Lancet Infect Dis. 16(3):282–283. [DOI] [PubMed] [Google Scholar]
- Falgenhauer L, Waezsada SE, Gwozdzinski K, Ghosh H, Doijad S, Bunk B, Spröer C, Imirzalioglu C, Seifert H, Irrgang A et al. 2016. Chromosomal locations of mcr-1 and blaCTX-M-15 in fluoroquinolone-resistant Escherichia coli ST410. Emerg Infect Dis. 22(9):1689–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekety JFR, Norman PS, Cluff LE. 1962. The treatment of Gram-negative bacillary infections with colistin: the toxicity and efficacy of large doses in forty-eight patients. Ann Intern Med. 57:214–229. [DOI] [PubMed] [Google Scholar]
- Fernandes MR, McCulloch JA, Vianello MA, Moura Q, Pérez-Chaparro PJ, Esposito F, Sartori L, Dropa M, Matté MH, Lira DP et al. 2016. First report of the globally disseminated IncX4 plasmid carrying the mcr-1 gene in a colistin-resistant Escherichia coli sequence type 101 isolate from a human infection in Brazil. Antimicrob Agents Chemother. 60(10):6415–6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes MR, Moura Q, Sartori L, Silva KC, Cunha MP, Esposito F, Lopes R, Otutumi LK, Goncalves DD, M. D et al. 2016. Silent dissemination of colistin-resistant Escherichia coli in South America could contribute to the global spread of the mcr-1 gene. Euro Surveill. 21(17):pii=30214. [DOI] [PubMed] [Google Scholar]
- Fernandes MR, Sellera FP, Esposito F, Sabino CP, Cerdeira L, Lincopan N. 2017. Colistin-resistant mcr-1-positive Escherichia coli in public beaches, an infectious threat emerging in recreational waters. Antimicrob Agents Chemother. 61(7):pii=e00234–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo R, Card RM, Nunez J, Pomba C, Mendonca N, Anjum MF, Da Silva GJ. 2016. Detection of an mcr-1-encoding plasmid mediating colistin resistance in Salmonella enterica from retail meat in Portugal. J Antimicrob Chemother. 71(8):2338–2340. [DOI] [PubMed] [Google Scholar]
- Fritzenwanker M, Imirzalioglu C, Gentil K, Falgenhauer L, Wagenlehner FM, Chakraborty T. 2016. Incidental detection of a urinary Escherichia coli isolate harboring mcr-1 of a patient with no history of colistin treatment. Clin Microbiol Infect. 22(11):954–955. [DOI] [PubMed] [Google Scholar]
- Fukuda A, Sato T, Shinagawa M, Takahashi S, Asai T, Yokota SI, Usui M, Tamura Y. 2018. High prevalence of mcr-1, mcr-3 and mcr-5 in Escherichia coli derived from diseased pigs in Japan. Int J Antimicrob Agents. 51(1):163–164. [DOI] [PubMed] [Google Scholar]
- García V, García-Meniño I, Mora A, Flament-Simon SC, Díaz-Jiménez D, Blanco JE, Alonso MP, Blanco J. 2018. Co-occurrence of mcr-1, mcr-4 and mcr-5 genes in multidrug-resistant ST10 Enterotoxigenic and Shiga toxin-producing Escherichia coli in Spain (2006–2017). Int J Antimicrob Agents. [accessed 2018 May 16]. 10.1016/j.ijantimicag.2018.03.022. [DOI] [PubMed] [Google Scholar]
- Giamarellou H 2016. Epidemiology of infections caused by polymyxin-resistant pathogens. Int J Antimicrob Agents. 48(6):614–621. [DOI] [PubMed] [Google Scholar]
- Giufrè M, Monaco M, Accogli M, Pantosti A, Cerquetti M. 2016. Emergence of the colistin resistance mcr-1 determinant in commensal Escherichia coli from residents of long-term-care facilities in Italy. J Antimicrob Chemother. 71(8):2329–2331. [DOI] [PubMed] [Google Scholar]
- Grami R, Mansour W, Mehri W, Bouallègue O, Boujaâfar N, Madec JY, Haenni M. 2016. Impact of food animal trade on the spread of mcr-1-mediated colistin resistance, Tunisia, July 2015. Euro Surveill. 21(8):pii=30144. [DOI] [PubMed] [Google Scholar]
- Gu DX, Huang YL, Ma JH, Zhou HW, Fang Y, Cai JC, Hu YY, Zhang R. 2016. Detection of colistin resistance gene mcr-1 in hypervirulent Klebsiella pneumoniae and Escherichia coli isolates from an infant with diarrhea in China. Antimicrob Agents Chemother. 60(8):5099–5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haenni M, Beyrouthy R, Lupo A, Châtre P, Madec JY, Bonnet R. 2018. Epidemic spread of Escherichia coli ST744 isolates carrying mcr-3 and blaCTX-M-55 in cattle in France. J Antimicrob Chemother. 73(2):533–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haenni M, Poirel L, Kieffer N, Châtre P, Saras E, Métayer V, Dumoulin R, Nordmann P, Madec J-Y. 2016. Co-occurrence of extended spectrum β lactamase and MCR-1 encoding genes on plasmids. Lancet Infect Dis. 16(3):281–282. [DOI] [PubMed] [Google Scholar]
- Hammerl JA, Borowiak M, Schmoger S, Shamoun D, Grobbel M, Malorny B, Tenhagen BA, Käsbohrer A. 2018. mcr-5 and a novel mcr-5.2 variant in Escherichia coli isolates from food and food-producing animals, Germany, 2010 to 2017. J Antimicrob Chemother. [accessed 2018 May 16]. 10.1093/jac/dky020. [DOI] [PubMed] [Google Scholar]
- Hartl R, Kerschner H, Lepuschitz S, Ruppitsch W, Allerberger F, Apfalter P. 2017. Detection of the mcr-1 gene in a multidrug-resistant Escherichia coli isolate from an Austrian patient. Antimicrob Agents Chemother. 61(4):pii=e02623–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasman H, Hammerum AM, Hansen F, Hendriksen RS, Olesen B, Agerø Y, Zankari E, Leekitcharoenphon P, Stegger M, Kaas RS et al. 2015. Detection of mcr-1 encoding plasmid-mediated colistin-resistant Escherichia coli isolates from human bloodstream infection and imported chicken meat, Denmark 2015. Euro Surveill. 20(49):pii=30085. [DOI] [PubMed] [Google Scholar]
- He QW, Xu XH, Lan FJ, Zhao ZC, Wu ZY, Cao YP, Li B. 2017. Molecular characteristic of mcr-1 producing Escherichia coli in a Chinese university hospital. Ann Clin Microbiol Antimicrob. 16(1):32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He T, Wei R, Zhang L, Sun L, Pang M, Wang R, Wang Y. 2017. Characterization of NDM-5-positive extensively resistant Escherichia coli isolates from dairy cows. Vet Microbiol. 207:153–158. [DOI] [PubMed] [Google Scholar]
- Hembach N, Schmid F, Alexander J, Hiller C, Rogall ET, Schwartz T. 2017. Occurrence of the mcr-1 colistin resistance gene and other clinically relevant antibiotic resistance genes in microbial populations at different municipal wastewater treatment plants in Germany. Front Microbiol. 8:1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández M, Iglesias MR, Rodríguez-Lázaro D, Gallardo A, Quijada N, Miguela-Villoldo P, Campos MJ, Píriz S, López-Orozco G, de Frutos C et al. 2017. Co-occurrence of colistin-resistance genes mcr-1 and mcr-3 among multidrug-resistant Escherichia coli isolated from cattle, Spain, September 2015. Euro Surveill. 22(31):pii=30586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Liu F, Lin IYC, Gao GF, Zhu B. 2016. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis. 16(2):146–147. [DOI] [PubMed] [Google Scholar]
- Hu YY, Wang YL, Sun QL, Huang ZX, Wang HY, Zhang R, Chen GX. 2017. Colistin-resistance gene mcr-1 in children’s gut flora. Int J Antimicrob Agents. 50(4):593–597. [DOI] [PubMed] [Google Scholar]
- Huang J, Zhu Y, Han M, Li M, Song J, Velkov T, Li C, Li J. 2017. Comparative analysis of phoephoethanolamine transferases involved in polymyxin resistance across ten clinically relevant Gram-negative bacteria. Int J Antimicrob Agents. 51(4):586–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang TD, Bogaerts P, Berhin C, Hoebeke M, Bauraing C, Glupczynski Y. 2017. Increasing proportion of carbapenemase-producing Enterobacteriaceae and emergence of a MCR-1 producer through a multicentric study among hospital-based and private laboratories in Belgium from September to November 2015. Euro Surveill. 22(19):pii=30530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irrgang A, Roschanski N, Tenhagen BA, Grobbel M, Skladnikiewicz-Ziemer T, Thomas K, Roesler U, Käsbohrer A. 2016. Prevalence of mcr-1 in E. coli from livestock and food in Germany, 2010–2015. PLoS One. 11(7):e0159863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izdebski R, Baraniak A, Bojarska K, Urbanowicz P, Fiett J, Pomorska-Wesolowska M, Hryniewicz W, Gniadkowski M, Żabicka D. 2016. Mobile MCR-1-associated resistance to colistin in Poland. J Antimicrob Chemother. 71(8):2331–2333. [DOI] [PubMed] [Google Scholar]
- Jayol A, Nordmann P, André C, Dubois V, Poirel L. 2017. Increased colistin resistance upon acquisition of the plasmid-mediated mcr-1 gene in Escherichia coli isolates with chromosomally encoded reduced susceptibility to polymyxins. Int J Antimicrob Agents. 50(3):503–504. [DOI] [PubMed] [Google Scholar]
- Jørgensen SB, Søraas A, Arnesen LS, Leegaard T, Sundsfjord A, Jenum PA. 2017. First environmental sample containing plasmid-mediated colistin-resistant ESBL-producing Escherichia coli detected in Norway. APMIS. 125(9):822–825. [DOI] [PubMed] [Google Scholar]
- Juhász E, Iván M, Pintér E, Pongrácz J, Kristóf K. 2017. Colistin resistance among blood culture isolates at a tertiary care centre in Hungary. J Glob Antimicrob Resist. 11:167–170. [DOI] [PubMed] [Google Scholar]
- Kawanishi M, Abo H, Ozawa M, Uchiyama M, Shirakawa T, Suzuki S, Shima A, Yamashita A, Sekizuka T, Kato K et al. 2016. Prevalence of colistin-resistance gene mcr-1 and absence of mcr-2 in Escherichia coli isolated from healthy food producing animals in Japan. Antimicrob Agents Chemother. 61(1):pii=e02057–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempf I, Jouy E, Chauvin C. 2016. Colistin use and colistin resistance in bacteria from animals. Int J Antimicrob Agents. 48(6):598–606. [DOI] [PubMed] [Google Scholar]
- Khalifa HO, Ahmed AM, Oreiby AF, Eid AM, Shimamoto T, Shimamoto T. 2016. Characterisation of the plasmid-mediated colistin resistance gene mcr-1 in Escherichia coli isolated from animals in Egypt. Int J Antimicrob Agents. 47(5):413–414. [DOI] [PubMed] [Google Scholar]
- Kieffer N, Nordmann P, Laurent P. 2017. Moraxella species as potential sources of MCR-like polymyxin resistance determinants. Antimicrob Agents Chemother. 61(6):e00129–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer N, Nordmann P, Moreno AM, Moreno LZ, Chaby R, Breton A, Tissières P, Poirel L. 2018. Genetic and functional characterization of an MCR-3-like producing Escherichia coli recovered from swine, Brazil. Antimicrob Agents Chemother. [accessed 2018 May 16]. 10.1128/AAC.00278-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim ES, Chong YP, Park SJ, Kim MN, Kim SH, Lee SO, Choi SH, Woo JH, Jeong JY, Kim YS. 2017. Detection and genetic features of MCR-1-producing plasmid in human Escherichia coli infection in South Korea. Diagn Microbiol Infect Dis. 89(2):158–160. [DOI] [PubMed] [Google Scholar]
- Kluytmans-van den bergh M, Huizinga P, Bonten MJ, Bos M, De Bruyne K, Friedrich AW, Rossen JW, Savelkoul PH, Kluytmans JA. 2016. Presence of mcr-1-positive Enterobacteriaceae in retail chicken meat but not in humans in the Netherlands since 2009. Euro Surveill. 21(9):pii=30149. [DOI] [PubMed] [Google Scholar]
- Koch-Weser J, Sidel VW, Federman EB, Kanarek P, Finer DC, Eaton AE. 1970. Adverse effects of sodium colistimethate. manifestations and specific reaction rates during 317 courses of therapy. Ann Intern Med. 72(6):857–868. [DOI] [PubMed] [Google Scholar]
- Kuo SC, Huang WC, Wang HY, Shiau YR, Cheng MF, Lauderdale TL. 2016. Colistin resistance gene mcr-1 in Escherichia coli isolates from humans and retail meats, Taiwan. J Antimicrob Chemother. 71(8):2327–2329. [DOI] [PubMed] [Google Scholar]
- Kusumoto M, Ogura Y, Gotoh Y, Iwata T, Hayashi T, Akiba M. 2016. Colistin-resistant mcr-1-positive pathogenic Escherichia coli in swine, Japan, 2007–2014. Emerg Infect Dis. 22(7):1315–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CC, Chuang YC, Chen CC, Tang HJ. 2017. Coexistence of MCR-1 and NDM-9 in a clinical carbapenem-resistant Escherichia coli isolate. Int J Antimicrob Agents. 49(4):517–518. [DOI] [PubMed] [Google Scholar]
- Leangapichart T, Gautret P, Brouqui P, Memish ZA, Raoult D, Rolain JM. 2016. Acquisition of mcr-1 plasmid-mediated colistin resistance in Escherichia coli and Klebsiella pneumoniae during Hajj 2013 and 2014. Antimicrob Agents Chemother. 60(11):6998–6999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentz SA, de Lima-Morales D, Cuppertino VM, Nunes Lde S, da Motta AS, Zavascki AP, Barth AL, Martins AF. 2016. Letter to the editor: Escherichia coli harboring mcr-1 gene isolated from poultry not exposed to polymyxins in Brazil. Euro Surveill. 21(26):pii=30267. [DOI] [PubMed] [Google Scholar]
- Li A, Yang Y, Miao M, Kalyan DC, Mediavilla JR, Xie X, Feng P, Tang YW, Kreiswirth BN, Chen L et al. 2016. Complete sequences of mcr-1 harboring plasmids from extended-spectrum-β-lactamase- and carbapenemase-producing Enterobacteriaceae. Antimicrob Agents Chemother. 60(7):4351–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Nation RL, Turnidge JD, Milne RW, Coulthard K, Rayner CR, Paterson DL. 2006. Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect Dis. 6(9):589–601. [DOI] [PubMed] [Google Scholar]
- Li R, Xie M, Lv J, Chan E, Chen S. 2016. Complete genetic analysis of plasmids carrying mcr-1 and other resistance genes in an Escherichia coli isolate of animal origin. J Antimicrob Chemother. 72(3):696–699. [DOI] [PubMed] [Google Scholar]
- Li R, Xie M, Zhang J, Yang Z, Liu L, Liu X, Zheng Z, Chan EW, Chen S. 2017. Genetic characterization of mcr-1-bearing plasmids to depict molecular mechanisms underlying dissemination of the colistin resistance determinant. J Antimicrob Chemother. 72(2):393–401. [DOI] [PubMed] [Google Scholar]
- Li XP, Fang LX, Song JQ, Xia J, Huo W, Fang JT, Liao XP, Liu YH, Feng Y, Sun J. 2016. Clonal spread of mcr-1 in PMQR-carrying ST34 Salmonella isolates from animals in China. Sci Rep. 6:38511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XP, Fang LX, Jiang P, Pan D, Xia J, Liao XP, Liu YH, Sun J. 2017. Emergence of the colistin resistance gene mcr-1 in Citrobacter freundii. Int J Antimicrob Agents. 49(6):786–787. [DOI] [PubMed] [Google Scholar]
- Li Z, Tan C, Lin J, Feng Y. 2016. Diversified variants of the mcr-1-carrying plasmid reservoir in the swine lung microbiota. Sci China Life Sci. 59(9):971–973. [DOI] [PubMed] [Google Scholar]
- Liakopoulos A, Mevius DJ, Olsen B, Bonnedahl J. 2016. The colistin resistance mcr-1 gene is going wild. J Antimicrob Chemother. 71(8):2335–2336. [DOI] [PubMed] [Google Scholar]
- Liassine N, Assouvie L, Descombes MC, Tendon VD, Kieffer N, Poirel L, Nordmann P. 2016. Very low prevalence of MCR-1/MCR-2 plasmid-mediated colistin resistance in urinary tract Enterobacteriaceae in Switzerland. Int J Infect Dis. 51:4–5. [DOI] [PubMed] [Google Scholar]
- Lim SK, Kang HY, Lee K, Moon DC, Lee HS, Jung SC. 2016. First detection of the mcr-1 gene in Escherichia coli isolated from livestock between 2013 and 2015 in South Korea. Antimicrob Agents Chemother. 60(11):6991–6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima Barbieri N, Nielsen DW, Wannemuehler Y, Cavender T, Hussein A, Yan SG, Nolan LK, Logue CM. 2017. mcr-1 identified in Avian Pathogenic Escherichia coli (APEC). PLoS One. 12(3):e0172997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling Z, Yin W, Li H, Zhang Q, Wang X, Wang Z, Ke Y, Wang Y, Shen J. 2017. Chromosome-mediated mcr-3 variants in Aeromonas veronii from chicken meat. Antimicrob Agents Chemother. 61(11): pii: e01272–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litrup E, Kiil K, Hammerum AM, Roer L, Nielsen EM, Torpdahl M. 2017. Plasmid-borne colistin resistance gene mcr-3 in Salmonella isolates from human infections, Denmark, 2009–17. Euro Surveill. 22(31):pii=30587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu BT, Song FJ, Zou M, Hao ZH, Shan H. 2016. Emergence of colistin resistance gene mcr-1 in Cronobacter sakazakii producing NDM-9 and Escherichia coli from the same animal. Antimicrob Agents Chemother. 61(2):pii=e01444–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Feng Y, Zhang X, McNally A, Zong Z. 2017. New variant of mcr-3 in an extensively drug-resistant Escherichia coli clinical isolate carrying mcr-1 and blaNDM-5. Antimicrob Agents Chemother. 61(12):pii=e01757–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Li R, Zheng Z, Chen K, Xie M, Chan EW, Geng S, Chen S. 2017. Molecular characterization of Escherichia coli isolates carrying mcr-1, fosA3, and extended-spectrum-beta-lactamase genes from food samples in China. Antimicrob Agents Chemother. 61(6):pii=e00064–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YY, Chandler CE, Leung LM, McElheny CL, Mettus RT, Shanks RMQ, Liu JH, Goodlett DR, Ernst RK, Doi Y. 2017. Structural modification of lipopolysaccharide conferred by mcr-1 in Gram-negative ESKAPE pathogens. Antimicrob Agents Chemother. 61(6):pii=e00580–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X et al. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 16(2):161–168. [DOI] [PubMed] [Google Scholar]
- Lu X, Hu Y, Luo M, Zhou H, Wang X, Du Y, Li Z, Xu J, Zhu B, Xu X et al. 2017. MCR-1.6: a new MCR variant carried by an IncP plasmid in a colistin-resistant Salmonella enterica serovar Typhimurium isolated from a healthy individual. Antimicrob Agents Chemother. 61(5):pii=e02632–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Yao X, Lv L, Doi Y, Huang X, Huang S, Liu JH. 2017. Emergence of mcr-1 in Raoultella ornithinolytica and Escherichia coli from retail vegetables, China. Antimicrob Agents Chemother. 61(10):pii=e01139–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Lei C, Kong L, Jiang W, Liu B, Men S, Yang Y, Cheng G, Chen Y, Wang H. 2017. Prevalence, antimicrobial resistance, and relatedness of Salmonella isolated from chickens and pigs on farms, abattoirs, and markets in Sichuan Province, China. Foodborne Pathog Dis. 14(11):667–677. [DOI] [PubMed] [Google Scholar]
- Macesic N, Green D, Wang Z, Sullivan SB, Shim K, Park S, Whittier S, Furuya EY, Gomez-Simmonds A, Uhlemann AC. 2017. Detection of mcr-1-carrying Escherichia coli causing bloodstream infection in a New York city hospital: Avian origins, human concerns? Open Forum Infect Dis. 4(3):ofx115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra-Kumar S, Xavier BB, Das AJ, Lammens C, Butaye P, Goossens H. 2016. Colistin resistance gene mcr-1 harboured on a multidrug resistant plasmid. Lancet Infect Dis. 16(3):283–284. [DOI] [PubMed] [Google Scholar]
- Malhotra-Kumar S, Xavier BB, Das AJ, Lammens C, Hoang HTT, Pham NT, Goossens H. 2016. Colistin-resistant Escherichia coli harboring mcr-1 isolated from food animals in Hanoi, Vietnam. Lancet Infect Dis. 16(3):286–287. [DOI] [PubMed] [Google Scholar]
- Marathe NP, Pal C, Gaikwad SS, Jonsson V, Kristiansson E, Larsson DGJ. 2017. Untreated urban waste contaminates Indian river sediments with resistance genes to last resort antibiotics. Water Res. 124:388–397. [DOI] [PubMed] [Google Scholar]
- McGann P, Snesrud E, Maybank R, Corey B, Ong AC, Clifford R, Hinkle M, Whitman T, Lesho E, Schaecher KE. 2016. Escherichia coli harboring mcr-1 and blaCTX-M on a novel IncF-plasmid: first report of mcr-1 in the USA. Antimicrob Agents Chemother. 60(7):4420–4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mediavilla JR, Patrawalla A, Chen L, Chavda KD, Mathema B, Vinnard C, Dever LL, Kreiswirth BN. 2016. Colistin- and carbapenem-resistant Escherichia coli harboring mcr-1 and blaNDM-5, causing a complicated urinary tract infection in a patient from the United States. mBio. 7(4):pii=e01191–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinersmann RJ, Ladely SR, Bono JL, Plumblee JR, Hall MC, Genzlinger LL, Cook KL. 2016. Complete genome sequence of a colistin resistance gene (mcr-1)-bearing isolate of Escherichia coli from the United States. Genome Announc. 4(6):pii=e01283–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinersmann RJ, Ladely SR, Plumblee JR, Cook KL, Thacker E. 2016. Prevalence of mcr-1 in the cecal contents of food-animals in the United States. Antimicrob Agents Chemother. 61(2):pii=e02244–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meletis G 2016. Carbapenem resistance: overview of the problem and future perspectives. Ther Adv Infect Dis. 3(1): 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohsin J, Pál T, Petersen JE, Darwish D, Ghazawi A, Ashraf T, Sonnevend A. 2017. Plasmid-mediated colistin resistance gene mcr-1 in an Escherichia coli ST10 bloodstream isolate in the Sultanate of Oman. Microb Drug Resist. 24(3):278–282. [DOI] [PubMed] [Google Scholar]
- Mohsin M, Raza S, Roschanski N, Guenther S, Ali A, Schierack P. 2016. Description of the first Escherichia coli clinical isolate harboring colistin-resistance mcr-1 gene from the Indian subcontinent. Antimicrob Agents Chemother. 61(1):pii=e01945–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monte DF, Mem A, Fernandes MR, Cerdeira L, Esposito F, Galvão JA, Franco BD, Lincopan N, Landgraf M. 2017. Chicken meat as reservoir of colistin-resistant Escherichia coli carrying mcr-1 genes in South America. Antimicrob Agents Chemother. 61(5):pii=e02718–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvey MR, Mataseje LF, Robertson J, Nash JHE, Boerlin P, Toye B, Irwin R, Melano RG. 2016. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis. 16(3):289–290. [DOI] [PubMed] [Google Scholar]
- Newton-Foot M, Snyman Y, Maloba MRB, Whitelaw AC. 2017. Plasmid-mediated mcr-1 colistin resistance in Escherichia coli and Klebsiella spp. clinical isolates from the Western Cape region of South Africa. Antimicrob Resist Infect Control. 6:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen NT, Nguyen HM, Nguyen CV, Nguyen TV, Nguyen MT, Thai HQ, Ho MH, Thwaites G, Ngo HT, Baker S et al. 2016. The use of colistin and other critical antimicrobials on pig and chicken farms in southern Vietnam and their association with resistance in commensal Escherichia coli. Appl Environ Microbiol. 82(13):3727–3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijhuis RH, Veldman KT, Schelfaut J, Van Essen-Zandbergen A, Wessels E, Claas EC, Gooskens J. 2016. Detection of the plasmid-mediated colistin-resistance gene mcr-1 in clinical isolates and stool specimens obtained from hospitalized patients using a newly developed real-time PCR assay. J Antimicrob Chemother. 71(8):2344–2346. [DOI] [PubMed] [Google Scholar]
- Nordmann P, Lienhard R, Kieffer N, Clerc O, Poirel L. 2016. Plasmid-mediated colistin-resistant Escherichia coli in bacteremia in Switzerland. Clin Infect Dis. 62(10):1322–1323. [DOI] [PubMed] [Google Scholar]
- Nordmann P, Poirel L. 2016. Plasmid-mediated colistin resistance: an additional antibiotic resistance menace. Clin Microbiol Infect. 22(5):398–400. [DOI] [PubMed] [Google Scholar]
- Ohsaki Y, Hayashi W, Saito S, Osaka S, Taniguchi Y, Koide S, Kawamura K, Nagano Y, Arakawa Y, Nagano N. 2017. First detection of Escherichia coli harboring mcr-1 gene from retail domestic chicken meat in Japan. Jpn J Infect Dis. 70(5):590–592. [DOI] [PubMed] [Google Scholar]
- Olaitan AO, Chabou S, Okdah L, Morand S, Rolain J-M. 2016. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis. 16(2):147. [DOI] [PubMed] [Google Scholar]
- Olaitan AO, Morand S, Rolain JM. 2014. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol. 5:643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega-Paredes D, Barba P, Zurita J. 2016. Colistin-resistant Escherichia coli clinical isolate harboring the mcr-1 gene in Ecuador. Epidemiol Infect. 144(14):2967–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz de la Tabla V, Ortega A, Buñuel F, Pérez-Vázquez M, Marcos B, Oteo J. 2016. Detection of the high-risk clone ST131 of Escherichia coli carrying the colistin resistance gene mcr-1 and causing acute peritonitis. Int J Antimicrob Agents. 49(1):115–116. [DOI] [PubMed] [Google Scholar]
- Ovejero CM, Delgado-Blas JF, Calero-Caceres W, Muniesa M, Gonzalez-Zorn B. 2017. Spread of mcr-1-carrying Enterobacteriaceae in sewage water from Spain. J Antimicrob Chemother. 72(4):1050–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paveenkittiporn W, Kerdsin A, Chokngam S, Bunthi C, Sangkitporn S, Gregory CJ. 2016. Emergence of plasmid-mediated colistin resistance and New Delhi metallo-beta-lactamase genes in extensively drug-resistant Escherichia coli isolated from a patient in Thailand. Diagn Microbiol Infect Dis. 87(2):157–159. [DOI] [PubMed] [Google Scholar]
- Payne M, Croxen MA, Lee TD, Mayson B, Champagne S, Leung V, Bariso S, Hoang L, Lowe C. 2016. mcr-1-positive colistin-resistant Escherichia coli in traveler returning to Canada from China. Emerg Infect Dis. 22(9):1673–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perreten V, Strauss C, Collaud A, Gerber D. 2016. Colistin resistance gene mcr-1 in avian-pathogenic Escherichia coli in South Africa. Antimicrob Agents Chemother. 60(7):4414–4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin-Guyomard A, Bruneau M, Houée P, Deleurme K, Legrandois P, Poirier C, Soumet C, Sanders P. 2016. Prevalence of mcr-1 in commensal Escherichia coli from French livestock, 2007 to 2014. Euro Surveill. 21(6):pii=30135. [DOI] [PubMed] [Google Scholar]
- Petrillo M, Angers-Loustau A, Kreysa J. 2016. Possible genetic events producing colistin resistance gene mcr-1. Lancet Infect Dis. 16(3):280. [DOI] [PubMed] [Google Scholar]
- Poirel L, Jayol A, Nordmann P. 2017. Polymyxins: Antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev. 30(2):557–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L, Jayol A, Bontron S, Villegas MV, Ozdamar M, Tükoglu S, Nordmann P. 2015. The mgrB gene as a key target for acquired resistance to colistin in Klebsiella pneumoniae. J Antimicrob Chemother. 70(1):75–80. [DOI] [PubMed] [Google Scholar]
- Poirel L, Kieffer N, Brink A, Coetze J, Jayol A, Nordmann P. 2016. Genetic features of MCR-1-producing colistin-resistant Escherichia coli isolates in South Africa. Antimicrob Agents Chemother. 60(7):4394–4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L, Kieffer N, Liassine N, Thanh D, Nordmann P. 2016. Plasmid-mediated carbapenem and colistin resistance in a clinical isolate of Escherichia coli. Lancet Infect Dis. 16(3):281. [DOI] [PubMed] [Google Scholar]
- Poirel L, Kieffer N, Nordmann P. 2017. In vitro study of ISApl1-mediated mobilization of the colistin resistance gene mcr-1. Antimicrob Agents Chemother. 61(7):pii=e00127–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L, Nordmann P. 2016. Emerging plasmid-encoded colistin resistance: the animal world as the culprit? J Antimicrob Chemother. 71(8):2326–2327. [DOI] [PubMed] [Google Scholar]
- Prim N, Rivera A, Rodriguez-Navarro J, Español M, Turbau M, Coll P, Mirelis B. 2016. Detection of MCR-1 colistin resistance gene in polyclonal Escherichia coli isolates in Barcelona, Spain, 2012 to 2015. Euro Surveill. 21(13):pii=30183. [DOI] [PubMed] [Google Scholar]
- Prim N, Turbau M, Rivera A, Rodriguez-Navarro J, Coll P, Mirelis B. 2017. Prevalence of colistin resistance in clinical isolates of Enterobacteriaceae: A four-year cross-sectional study. J Infect. 75(6):493–498. [DOI] [PubMed] [Google Scholar]
- Pulss S, Semmler T, Prenger-Berninghoff E, Bauerfeind R, Ewers C. 2017. First report of an Escherichia coli strain from swine carrying an OXA-181 carbapenemase and the colistin resistance determinant MCR-1. Int J Antimicrob Agents. 50(2):232–236. [DOI] [PubMed] [Google Scholar]
- Quan J, Li X, Chen Y, Jiang Y, Zhou Z, Zhang H, Sun L, Ruan Z, Feng Y, Akova M et al. 2017. Prevalence of mcr-1 in Escherichia coli and Klebsiella pneumoniae recovered from bloodstream infections in China: a multicentre longitudinal study. Lancet Infect Dis. 17(4):400–410. [DOI] [PubMed] [Google Scholar]
- Quesada A, Ugarte-Ruiz M, Iglesias MR, Porrero MC, Martínez R, Florez-Cuadrado D, Campos MJ, Garcia M, Píriz S, Sáez JL et al. 2016. Detection of plasmid mediated colistin resistance (MCR-1) in Escherichia coli and Salmonella enterica isolated from poultry and swine in Spain. Res Vet Sci. 105:134–135. [DOI] [PubMed] [Google Scholar]
- Rapoport M, Faccone D, Pasteran F, Ceriana P, Albornoz E, Petroni A, Corso A. 2016. First description of mcr-1-mediated colistin resistance in human infections caused by Escherichia coli in Latin America. Antimicrob Agents Chemother. 60(7):4412–4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin F, Beyrouthy R, Colot J, Saint-Sardos P, Berger-Carbonne A, Dalmasso G, Delmas J, Bonnet R. 2016. MCR-1 in ESBL-producing Escherichia coli responsible for human infections in New Caledonia. J Antimicrob Chemother. 72(3):946–947. [DOI] [PubMed] [Google Scholar]
- Roer L, Hansen F, Stegger M, Sönksen UW, Hasman H, Hammerum AM. 2017. Novel mcr-3 variant, encoding mobile colistin resistance, in an ST131 Escherichia coli isolate from bloodstream infection, Denmark, 2014. Euro Surveill. 22(31):pii=30584. [PubMed] [Google Scholar]
- Rolain JM, Kempf M, Leangapichart T, Chabou S, Olaitan AO, Le Page S, Morand S, Raoult D. 2016. Plasmid-mediated mcr-1 gene in colistin-resistant clinical isolates of Klebsiella pneumoniae in France and Laos. Antimicrob Agents Chemother. 60(11):6994–6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roschanski N, Falgenhauer L, Grobbel M, Guenther S, Kreienbrock L, Imirzalioglu C, Roesler U. 2017. Retrospective survey of mcr-1 and mcr-2 in German pig-fattening farms, 2011–2012. Int J Antimicrob Agents. 50(2):266–271. [DOI] [PubMed] [Google Scholar]
- Ross S, Puig JR, Zaremba EA. 1959. Colistin: some preliminary laboratory and clinical observations in specific gastroenteritis in infants and children. Antibiot Annu. 7:89–100. [PubMed] [Google Scholar]
- Runcharoen C, Raven KE, Reuter S, Kallonen T, Paksanont S, Thammachote J, Anun S, Blane B, Parkhill J, Peacock SJ et al. 2017. Whole genome sequencing of ESBL-producing Escherichia coli isolated from patients, farm waste and canals in Thailand. Genome Med. 9(1):81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruppé E, Chatelier EL, Pons N, Andremont A, Ehrlich SD. 2016. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis. 16(3):290–291. [DOI] [PubMed] [Google Scholar]
- Ruzauskas M, Vaskeviciute L. 2016. Detection of the mcr-1 gene in Escherichia coli prevalent in the migratory bird species Larus argentatus. J Antimicrob Chemother. 71(8):2333–2334. [DOI] [PubMed] [Google Scholar]
- Saavedra SY, Diaz L, Wiesner M, Correa A, Arévalo SA, Reyes J, Hidalgo AM, de la Cadena E, Perenguez M, Montaño LA et al. 2017. Genomic and molecular characterization of clinical isolates of Enterobacteriaceae harboring mcr-1 in Colombia, 2002 to 2016. Antimicrob Agents Chemother. 61(12):pii=e00841–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Benito R, Iglesias MR, Quijada NM, Campos MJ, Ugarte-Ruiz M, Hernández M, Pazos C, Rodríguez-Lázaro D, Garduño E, Domínguez L, et al. 2017. Escherichia coli ST167 carrying plasmid mobilisable mcr-1 and blaCTX-M-15 resistance determinants isolated from a human respiratory infection. Int J Antimicrob Agents. 50(2):285–286. [DOI] [PubMed] [Google Scholar]
- Schirmeier E, Zimmermann P, Hofmann V, Biebl M, Gerstmans H, Maervoet VE, Briers Y. 2017. Inhibitory and bactericidal effect of Artilysin(R) Art-175 against colistin-resistant mcr-1-positive Escherichia coli isolates. Int J Antimicrob Agents. 51(3):528–529. [DOI] [PubMed] [Google Scholar]
- Schrauwen EJA, Huizinga P, van Spreuwel N, Verhulst C, Kluytmans-van den Bergh MFQ, Kluytmans J. 2017. High prevalence of the mcr-1 gene in retail chicken meat in the Netherlands in 2015. Antimicrob Resist Infect Control. 6:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellera FP, Fernandes MR, Sartori L, Carvalho MP, Esposito F, Nascimento CL, Dutra GH, Mamizuka EM, Pérez-Chaparro PJ, McCulloch JA et al. 2016. Escherichia coli carrying IncX4 plasmid-mediated mcr-1 and blaCTX-M genes in infected migratory Magellanic penguins (Spheniscus magellanicus). J Antimicrob Chemother. 72(4):1255–1256. [DOI] [PubMed] [Google Scholar]
- Shen Z, Wang Y, Shen Y, Shen J, Wu C. 2016. Early emergence of mcr-1 in Escherichia coli from food-producing animals. Lancet Infect Dis. 16(3):293. [DOI] [PubMed] [Google Scholar]
- Simar S, Sibley D, Ashcraft D, Pankey G. 2017. Colistin and polymyxin B minimal inhibitory concentrations determined by Etest found unreliable for Gram-negative Bacilli. Ochsner J. 17(3):239–242. [PMC free article] [PubMed] [Google Scholar]
- Skov RL, Monnet DL. 2016. Plasmid-mediated colistin resistance (mcr-1 gene): three months later, the story unfolds. Euro Surveill. 21(9):pii=30155. [DOI] [PubMed] [Google Scholar]
- Snesrud E, He S, Chandler M, Dekker JP, Hickman AB, McGann P, Dyda F. 2016. A model for transposition of the colistin resistance gene mcr-1 by ISApl1. Antimicrob Agents Chemother. 60(11):6973–6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snesrud E, Ong AC, Corey B, Kwak YI, Clifford R, Gleeson T, Wood S, Whitman TJ, Lesho EP, Hinkle M et al. 2017. Analysis of serial isolates of mcr-1-positive Escherichia coli reveals a highly active ISApl1 transposon. Antimicrob Agents Chemother. 61(5):pii=e00056–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solheim M, Bohlin J, Ulstad CR, Schau Slettemeås J, Naseer U, Dahle UR, Wester AL. 2016. Plasmid-mediated colistin-resistant Escherichia coli detected from 2014 in Norway. Int J Antimicrob Agents. 48(2):227–228. [DOI] [PubMed] [Google Scholar]
- Sonnevend Á, Ghazawi A, Alqahtani M, Shibl A, Jamal W, Hashmey R, Pal T. 2016. Plasmid-mediated colistin resistance in Escherichia coli from the Arabian Peninsula. Int J Infect Dis. 50:85–90. [DOI] [PubMed] [Google Scholar]
- Srinivas P, Rivard K. 2017. Polymyxin resistance in Gram-negative pathogens. Curr Infect Dis Rep. 19(11): 38. [DOI] [PubMed] [Google Scholar]
- Stansly PG, Shepherd RG, White HJ. 1947. Polymyxin: a new chemotherapeutic agent. Bull Johns Hopkins Hosp. 81(1):43–54. [PubMed] [Google Scholar]
- Stoesser N, Mathers AJ, Moore CE, Day NPJ, Crook DW. 2016. Colistin resistance gene mcr-1 and pHNSHP45 plasmid in human isolates of Escherichia coli and Klebsiella pneumoniae. Lancet Infect Dis. 16(3):285–286. [DOI] [PubMed] [Google Scholar]
- Sun J, Yang RS, Zhang Q, Feng Y, Fang LX, Xia J, Li L, Lv XY, Duan JH, Liao XP, et al. 2016. Co-transfer of blaNDM-5 and mcr-1 by an IncX3–X4 hybrid plasmid in Escherichia coli. Nat Microbiol. 1:16176. [DOI] [PubMed] [Google Scholar]
- Sun P, Bi Z, Nilsson M, Zheng B, Berglund B, Stålsby Lundborg C, Börjesson S, Li X, Chen B, Yin H, et al. 2017. Occurrence of blaKPC-2, blaCTX-M and mcr-1 in Enterobacteriaceae from well water in rural China. Antimicrob Agents Chemother. 61(4):pii=e02569–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S, Ohnishi M, Kawanishi M, Akiba M, Kuroda M. 2016. Investigation of a plasmid genome database for colistin-resistance gene mcr-1. Lancet Infect Dis. 16(3):284–285. [DOI] [PubMed] [Google Scholar]
- Tacão M, Tavares RDS, Teixeira P, Roxo I, Ramalheira E, Ferreira S, Henriques I. 2017. mcr-1 and blaKPC-3 in Escherichia coli sequence type 744 after meropenem and colistin therapy, Portugal. Emerg Infect Dis. 23(8):1419–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada T, Nhung PH, Shimada K, Tsuchiya M, Phuong DM, Anh NQ, Ohmagari N, Kirikae T. 2017. Emergence of colistin-resistant Escherichia coli clinical isolates harboring mcr-1 in Vietnam. Int J Infect Dis. 63:72–73. [DOI] [PubMed] [Google Scholar]
- Tada T, Uechi K, Nakasone I, Shimada K, Nakamatsu M, Kirikae T, Fujita J. 2017. Emergence of a colistin-resistant Escherichia coli clinical isolate harboring mcr-1 in Japan. Int J Infect Dis. 63:21–22. [DOI] [PubMed] [Google Scholar]
- Teo JW, Chew KL, Lin RT. 2016. Transmissible colistin resistance encoded by mcr-1 detected in clinical Enterobacteriaceae isolates in Singapore. Emerg Microbes Infect. 5(8):e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terveer EM, Nijhuis RHT, Crobach MJT, Knetsch CW, Veldkamp KE, Gooskens J, Kuijper EJ, Claas ECJ. 2017. Prevalence of colistin resistance gene (mcr-1) containing Enterobacteriaceae in feces of patients attending a tertiary care hospital and detection of a mcr-1 containing, colistin susceptible E. coli. PLoS One. 12(6):e0178598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanh DP, Tuyen HT, Nguyen TNT, The HC, Wick RR, Thwaites GE, Baker S, Holt KE. 2016. Inducible colistin resistance via a disrupted plasmid-borne mcr-1 gene in a 2008 Vietnamese Shigella sonnei isolate. J Antimicrob Chemother. 71(8):2314–2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson KS. 2010. Extended-spectrum-β-lactamase, AmpC, and carbapenemase issues. J Clin Microbiol. 48(4): 1019–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian GB, Doi Y, Shen J, Walsh TR, Wang Y, Zhang R, Huang X. 2017. MCR-1-producing Klebsiella pneumoniae outbreak in China. Lancet Infect Dis. 17(6):577. [DOI] [PubMed] [Google Scholar]
- Tijet N, Faccone D, Rapoport M, Seah C, Pasterán F, Ceriana P, Albornoz E, Corso A, Petroni A, Melano RG. 2017. Molecular characteristics of mcr-1-carrying plasmids and new mcr-1 variant recovered from polyclonal clinical Escherichia coli from Argentina and Canada. PLoS One. 12(7):e0180347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torpdahl M, Hasman H, Litrup E, Skov RL, Nielsen EM, Hammerum AM. 2017. Detection of mcr-1-encoding plasmid-mediated colistin-resistant Salmonella isolates from human infection in Denmark. Int J Antimicrob Agents. 49(2):261–262. [DOI] [PubMed] [Google Scholar]
- Trecarichi EM, Tumbarello M. 2017. Therapeutic options for carbapenem-resistant Enterobacteriaceae infections. Virulence. 8(4):470–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trung NV, Matamoros S, Carrique-Mas JJ, Nghia NH, Nhung NT, Chieu TT, Mai HH, van Rooijen W, Campbell J, Wagenaar JA, et al. 2017. Zoonotic transmission of mcr-1 colistin resistance gene from small-scale poultry farms, Vietnam. Emerg Infect Dis. 23(3):529–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse H, Yuen K-Y. 2016. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis. 16(2):145–146. [DOI] [PubMed] [Google Scholar]
- Unger F, Eisenberg T, Prenger-Berninghoff E, Leidner U, Ludwig ML, Rothe M, Semmler T, Ewers C. 2016. Imported reptiles as a risk factor for the global distribution of Escherichia coli harboring the colistin resistance gene mcr-1. Int J Antimicrob Agents. 49(1):122–123. [DOI] [PubMed] [Google Scholar]
- Vaara M, Vaara T, Jensen M, Helander I, Nurminen M, Rietschel ET, Mäkelä PH. 1981. Characterization of the lipopolysaccharide from the polymyxin-resistant pmrA mutants of Salmonella typhimurium. FEBS Lett. 129(1):145–149. [DOI] [PubMed] [Google Scholar]
- Vading M, Kabir MH, Kalin M, Iversen A, Wiklund S, Nauclér P, Giske CG. 2016. Frequent acquisition of low-virulence strains of ESBL-producing Escherichia coli in travellers. J Antimicrob Chemother. 71(12):3548–3555. [DOI] [PubMed] [Google Scholar]
- Vasquez AM, Montero N, Laughlin M, Dancy E, Melmed R, Sosa L, Watkins LF, Folster JP, Strockbine N, Moulton-Meissner H, et al. 2016. Investigation of Escherichia coli harboring the mcr-1 resistance gene - Connecticut, 2016. Morb Mortal Wkly Rep. 65(36):979–980. [DOI] [PubMed] [Google Scholar]
- Veldman K, van Essen-Zandbergen A, Rapallini M, Wit B, Heymans R, van Pelt W, Mevius D. 2016. Location of colistin resistance gene mcr-1 in Enterobacteriaceae from livestock and meat. J Antimicrob Chemother. 71(8):2340–2342. [DOI] [PubMed] [Google Scholar]
- Walkty A, Karlowsky JA, Adam HJ, Lagacé-Wiens P, Baxter M, Mulvey MR, McCracken M, Poutanen SM, Roscoe D, Zhanel GG. 2016. Frequency of MCR-1-mediated colistin resistance among Escherichia coli clinical isolates obtained from patients in Canadian hospitals (CANWARD 2008–2015). CMAJ Open. 4(4):E641–E645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh TR, Wu Y. 2016. China bans colistin as a feed additive for animals. Lancet Infect Dis. 16(10):1102–1103. [DOI] [PubMed] [Google Scholar]
- Wang Q, Sun J, Li J, Ding Y, Li XP, Lin J, Hassan B, Feng Y. 2017. Expanding landscapes of the diversified mcr-1-bearing plasmid reservoirs. Microbiome. 5(1):70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Liu Y, Qi X, Wang R, Jin L, Zhao M, Zhang Y, Wang Q, Chen H, Wang H. 2017. Molecular epidemiology of colistin-resistant Enterobacteriaceae in inpatients and avian from China: high prevalence of mcr-negative Klebsiella pneumoniae. Int J Antimicrob Agents. 50(4):536–541. [DOI] [PubMed] [Google Scholar]
- Wang X, Zhai W, Li J, Liu D, Zhang Q, Shen Z, Wang S, Wang Y. 2018. Presence of a mcr-3 variant in Aeromonas caviae, Proteus mirabilis, and Escherichia coli from one domestic duck. Antimicrob Agents Chemother. 62(2):pii=e02106–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Tian GB, Zhang R, Shen Y, Tyrrell JM, Huang X, Zhou H, Lei L, Li HY, Doi Y, et al. 2017. Prevalence, risk factors, outcomes, and molecular epidemiology of mcr-1-positive Enterobacteriaceae in patients and healthy adults from China: an epidemiological and clinical study. Lancet Infect Dis. 17(4):390–399. [DOI] [PubMed] [Google Scholar]
- Webb HE, Granier SA, Marault M, Millemann Y, den Bakker HC, Nightingale KK, Bugarel M, Ison SA, Scott HM, Loneragan GH. 2016. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis. 16(2):144–145. [DOI] [PubMed] [Google Scholar]
- Wise MG, Estabrook MA, Sahm DF, Stone GG, Kazmierczak KM. 2018. Prevalence of mcr-type genes among colistin-resistant Enterobacteriaceae collected in 2014–2016 as part of the INFORM global surveillance program. PLoS One. 13(4):e0195281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong SC, Tse H, Chen JH, Cheng VC, Ho PL, Yuen KY. 2016. Colistin-resistant Enterobacteriaceae carrying the mcr-1 gene among patients in Hong Kong. Emerg Infect Dis. 22(9):1667–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier BB, Lammens C, Ruhal R, Kumar-Singh S, Butaye P, Goossens H, Malhotra-Kumar S. 2016. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill. 21(27):pii=30280. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Kawahara R, Harada K, Teruya S, Nakayama T, Motooka D, Nakamura S, Nguyen PD, Kumeda Y, Van Dang C, et al. 2018. The presence of colistin resistance gene mcr-1 and mcr-3 in ESBL producing Escherichia coli isolated from food in Ho Chi Minh City, Vietnam. FEMS Microbiol Lett. [accessed 2018 May 16]. 10.1093/femsle/fny100. [DOI] [PubMed] [Google Scholar]
- Yanat B, Machuca J, Yahia RD, Touati A, Pascual Á, Rodríguez-Martínez JM. 2016. First report of the plasmid-mediated colistin resistance gene mcr-1 in a clinical Escherichia coli isolate in Algeria. Int J Antimicrob Agents. 48(6):760–761. [DOI] [PubMed] [Google Scholar]
- Yang RS, Feng Y, Lv XY, Duan JH, Chen J, Fang LX, Xia J, Liao XP, Sun J, Liu YH. 2016. Emergence of NDM-5 and MCR-1 producing Escherichia coli clone ST648 and ST156 from a single muscovy duck (Cairina moschata). Antimicrob Agents Chemother. 60(11):6899–6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YQ, Li YX, Song T, Yang YX, Jiang W, Zhang AY, Guo XY, Liu BH, Wang YX, Lei CW et al. 2017. Colistin resistance gene mcr-1 and its variant in Escherichia coli isolates from chickens in China. Antimicrob Agents Chemother. 61(5):pii=e01204–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YQ, Li YX, Lei CW, Zhang AY, Wang HN. 2018. Novel plasmid-mediated colistin resistance gene mcr-7.1 in K. pneumoniae. J Antimicrob Chemother. [accessed 2018 May 14]. 10.1093/jac/dky111. [DOI] [PubMed] [Google Scholar]
- Yang YQ, Zhang AY, Ma SZ, Kong LH, Li YX, Liu JX, Davis MA, Guo XY, Liu BH, Lei CW, et al. 2016. Co-occurrence of mcr-1 and ESBL on a single plasmid in Salmonella enterica. J Antimicrob Chemother. 71(8):2336–2338. [DOI] [PubMed] [Google Scholar]
- Yao X, Doi Y, Zeng L, Lv L, Liu J-H. 2016. Carbapenem-resistant and colistin-resistant Escherichia coli co-producing NDM-9 and MCR-1. Lancet Infect Dis. 16(3):288–289. [DOI] [PubMed] [Google Scholar]
- Ye H, Li Y, Li Z, Gao R, Zhang H, Wen R, Gao GF, Hu Q, Feng Y. 2016. Diversified mcr-1-harboring plasmid reservoirs confer resistance to colistin in human gut microbiota. mBio. 7(2):e00177–00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi L, Wang J, Gao Y, Liu Y, Doi Y, Wu R, Zeng Z, Liang Z, Liu JH. 2017. mcr-1-harboring Salmonella enterica serovar Typhimurium sequence type 34 in pigs, China. Emerg Infect Dis. 23(2):291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W, Li H, Shen Y, Liu Z, Wang S, Shen Z, Zhang R, Walsh TR, Shen J, Wang Y. 2017. Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. mBio. 8(3):pii=e00543–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CY, Ang GY, Chin PS, Ngeow YF, Yin WF, Chan KG. 2016. Emergence of mcr-1-mediated colistin resistance in Escherichia coli in Malaysia. Int J Antimicrob Agents. 47(6):504–505. [DOI] [PubMed] [Google Scholar]
- Yu CY, Ang GY, Chong TM, Chin PS, Ngeow YF, Yin WF, Chan KG. 2016. Complete genome sequencing revealed novel genetic contexts of the mcr-1 gene in Escherichia coli strains. J Antimicrob Chemother. 72(4):1253–1255. [DOI] [PubMed] [Google Scholar]
- Yu H, Qu F, Shan B, Huang B, Jia W, Chen C, Li A, Miao M, Zhang X, Bao C, et al. 2016. Detection of mcr-1 colistin resistance gene in carbapenem-resistant Enterobacteriaceae (CRE) from different hospitals in China. Antimicrob Agents Chemother. 60(8):5033–5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng KJ, Doi Y, Patil S, Huang X, Tian GB. 2016. Emergence of the plasmid-mediated mcr-1 gene in colistin-resistant Enterobacter aerogenes and Enterobacter cloacae. Antimicrob Agents Chemother. 60(6):3862–3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Feng Y, Liu F, Jiang H, Qu Z, Lei M, Wang J, Zhang B, Hu Y, Ding J, et al. 2016. A phage-like IncY plasmid carrying the mcr-1 gene in Escherichia coli from a pig farm in China. Antimicrob Agents Chemother. 61(3):pii=e02035–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Seward CH, Wu Z, Ye H, Feng Y. 2016. Genomic insights into the ESBL and MCR-1-producing ST648 Escherichia coli with multi-drug resistance. Sci Bull (Beijing). 61:875–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Chen L, Wang J, Butaye P, Huang K, Qiu H, Zhang X, Gong W, Wang C. 2018. Molecular detection of colistin resistance genes (mcr-1 to mcr-5) in human vaginal swabs. BMC Res Notes. 11(1):143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Chen L, Wang J, Yassin AK, Butaye P, Kelly P, Gong J, Guo W, Li J, Li M, et al. 2018. Molecular detection of colistin resistance genes (mcr-1, mcr-2 and mcr-3) in nasal/oropharyngeal and anal/cloacal swabs from pigs and poultry. Sci Rep. 8(1):3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Huang Y, Chan E, Zhou H, Chen S. 2016. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis. 16(3):291–292. [DOI] [PubMed] [Google Scholar]
- Zhang R, Liu L, Zhou H, Chan EW, Li J, Fang Y, Li Y, Liao K, Chen S. 2017. Nationwide surveillance of clinical carbapenem-resistant Enterobacteriaceae (CRE) strains in China. EBioMedicine. 19:98–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XF, Doi Y, Huang X, Li HY, Zhong LL, Zeng KJ, Zhang YF, Patil S, Tian GB. 2016. Possible transmission of mcr-1–harboring Escherichia coli between companion animals and human. Emerg Infect Dis. 22(9):1679–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liao K, Gao H, Wang Q, Wang X, Li H, Wang R, Wang H. 2017. Decreased fitness and virulence in ST10 Escherichia coli harboring blaNDM-5 and mcr-1 against a ST4981 strain with blaNDM-5. Front Cell Infect Microbiol. 7:242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F, Feng Y, Lu X, McNally A, Zong Z. 2016. IncP plasmid carrying the colistin resistance gene mcr-1 in Klebsiella pneumoniae from hospital sewage. Antimicrob Agents Chemother. 61(2):pii=e02229–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F, Zong Z. 2016. Kluyvera ascorbata carrying the mcr-1 colistin resistance gene from hospital sewage. Antimicrob Agents Chemother. 60(12):7498–7501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Dong H, Xu H, Lv J, Zhang J, Jiang X, Du Y, Xiao Y, Li L. 2016. Coexistence of MCR-1 and NDM-1 in clinical Escherichia coli isolates. Clin Infect Dis. 63(10):1393–1395. [DOI] [PubMed] [Google Scholar]
- Zheng B, Lv T, Xu H, Yu X, Chen Y, Li J, Huang C, Guo L, Zhang J, Jiang X, et al. 2017. Discovery and characterization of an Escherichia coli ST206 strain producing NDM-5 and MCR-1 from a patient with acute diarrhea. Int J Antimicrob Agents. 51(2):273–275. [DOI] [PubMed] [Google Scholar]
- Zhi C, Lv L, Yu L-F, Doi Y, Liu J-H. 2016. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis. 16(3):292–293. [DOI] [PubMed] [Google Scholar]
- Zhong LL, Zhang YF, Doi Y, Huang X, Zhang XF, Zeng KJ, Shen C, Patil S, Xing Y, Zou Y, et al. 2016. Co-production of MCR-1 and NDM-1 by colistin-resistant Escherichia coli isolated from a healthy individual. Antimicrob Agents Chemother. 61(1):pii=e01962–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HW, Zhang T, Ma JH, Fang Y, Wang HY, Huang ZX, Wang Y, Wu C, Chen GX. 2017. Occurrence of plasmid- and chromosome-encoded mcr-1 in water-borne Enterobacteriaceae in China. Antimicrob Agents Chemother. 61(8):pii=e00017–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zogg AL, Zurfluh K, Nüesch-Inderbinen M, Stephan R. 2016. Characteristics of ESBL-producing Enterobacteriaceae and methicillin resistant Staphylococcus aureus (MRSA) isolated from Swiss and imported raw poultry meat collected at retail level. Schweiz Arch Tierheilkd. 158(6):451–456. [DOI] [PubMed] [Google Scholar]
- Zurfluh K, Buess S, Stephan R, Nüesch-Inderbinen M. 2016. Assessment of the occurrence of MCR producing Enterobacteriaceae in Swiss and imported poultry meat. SDRP J Food Sci Technol. 1(4). [Google Scholar]
- Zurfluh K, Klumpp J, Nüesch-Inderbinen M, Stephan R. 2016. Full-length nucleotide sequences of mcr-1 harboring plasmids isolated from extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli of different origins. Antimicrob Agents Chemother. 60(9):5589–5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurfluh K, Nüesch-Inderbinen M, Klumpp J, Poirel L, Nordmann P, Stephan R. 2017. Key features of mcr-1-bearing plasmids from Escherichia coli isolated from humans and food. Antimicrob Resist Infect Control. 6:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurfluh K, Tasara T, Poirel L, Nordmann P, Stephan R. 2016. Draft genome sequence of Escherichia coli S51, a chicken isolate harboring a chromosomally encoded mcr-1 gene. Genome Announc. 4(4):e00796–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurfuh K, Poirel L, Nordmann P, Nüesch-Inderbinen M, Hächler H, Stephan R. 2016. Occurrence of the plasmid-borne mcr-1 colistin resistance gene in extended-spectrum-beta-lactamase-producing Enterobacteriaceae in river water and imported vegetable samples in Switzerland. Antimicrob Agents Chemother. 60(4):2594–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.