Abstract
Beta-catenin is well-known as a key effector of Wnt signalling and aberrant expression is associated with several human cancers. Stabilisation of and atypical subcellular localisation of beta-catenin, regulated in part through specific protein-protein interactions has been linked to cancer development, however the mechanisms behind these pathologies is yet to be fully elucidated. Affinity purification and mass spectrometry were used to identify potential β-catenin interacting proteins in SW480 colon cancer cells. Recombinant β-catenin constructs were used to co-isolate interacting proteins from stable isotope labelled cells followed by detection using mass spectrometry. Several known and new putative interactors were observed. In particular, we identified interaction with a set of coatomer complex I subunits implicated in retrograde transport at the Golgi, and confirmed endogenous interaction of β-catenin with coatomer subunit COPB using immunoprecipitation assays and immunofluorescence microscopy. These observations suggest a hitherto unrecognised role for β-catenin in the secretory pathway and warrant further functional studies to unravel its activity at this cellular location.
Keywords: β-catenin, COPB, Golgi, Protein interactions, Mass spectrometry, Cancer
Highlights
-
•
Proteomic screen of beta-catenin in colon cancer cells identifies novel interactors.
-
•
Beta-catenin co-isolates with Coatamer complex I.
-
•
Immunofluorescence demonstrates Golgi localisation of the endogenous interaction.
1. Introduction
β-catenin is a 90 kDa multifunctional, proto-oncogene that is the primary effector of the Wnt signalling pathway [1]. The stabilisation of β-catenin allows it to accumulate in the cell and its translocation into the nucleus results in the transcription of target genes such as T-cell factor (TCF) and lymphoid enhancer-binding factor (LEF) [2]. Downstream outcomes include cell cycle regulation, transmigration [3], cell proliferation [4] and invasion [5] and stem-cell maintenance, differentiation, proliferation and apoptosis [6]. Somatic mutations of β-catenin resulting in stabilisation and nuclear translocation has been reported in a range of cancers [[7], [8], [9], [10], [11], [12]].
β-catenin interacts with a number of proteins governed by various protein structural domains, such as the adhesion facilitating interactions of α-catenin at the N-terminus [13] or WW and PDZ domain-containing protein 1 at the C-terminus [14]. However, the most well described interactions occur at the Arm domains of β-catenin and include interactions with APC [15], axin [16], the cadherins [17] and LEF-1 [18]. The full extent of β-catenin protein interactions is yet to be described, and one approach which has shown utility to discover new interactions is through the use of mass spectrometry [[19], [20], [21]].
In this report we used SILAC based mass spectrometry to identify putative protein interacting partners of β-catenin in SW480 colon cancer cells. We identified interactions with coatamer complex I subunits (COPI), and confirmed this with orthogonal approaches using immunoprecipitation and immunofluorescence targeting the beta-COP subunit (COPB). This observation suggests a potential new role for β-catenin in the secretory pathway.
2. Methods and materials
2.1. Cell culture
Human colon cancer cell line SW480 (American Type Culture Collection) was cultured in Dulbecco's Modified Eagle Medium (DMEM) minus l-Lysine and l-Arginine, supplemented with 10% dialysed fetal bovine serum (FBS), 1% Penicillin/Streptomycin and 10 mM HEPES. Heavy labelled media (for control conditions) was supplemented with 84 mg/L l-Arginine (13C6, 15N4) and 146.2 mg/L l-Lysine (13C6, 15N4) (Novachem, #CNLM539H and #CNLM291H respectively). Light labelled media (for experimental conditions) was supplemented with 84 mg/L l-Arginine (12C6, 14N4) and 146.2 mg/L l-Lysine (12C6, 14N4). Cells were lysed in lysis buffer (pH 7.5) (20 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100) supplemented with protease inhibitor (PI) followed by sonication at 10% output for 5 s and centrifugation at 10,000 rpm.
2.2. Expression and purification of human β-catenin-MBP fusion proteins
Full-length, human wild-type β-catenin fused with N-terminal maltose-binding protein (MBP) in a pMALC2 vector (GenScript, NJ, USA) was used to transform competent E. coli (DH5α). An overnight culture of these cells was used to inoculate 2xYT broth at 37 °C at 220 rpm until OD595 = 1.0. Isopropyl β-D-1-thiogalactopyranosid (IPTG) was used to induce protein expression at 25 °C for 4 h. Cells were pelleted, washed and pelleted. Cells were resuspended in H-buffer (TEDM, 0.5 M NaCl, PI) (TEDM = 20 mM Tris (pH 8), 1 mM EDTA, 1 mM DTT, 5 mM MgCl2) and sonicated for 10 s at 10% output (x10) and pelleted. For purification, amylose resin columns were utilised and protein was eluted with 36 mg/ml maltose in TEDM/PI. Dialysis was conducted according to manufacturer instructions using Slide-A-Lyzer dialysis cassettes (Thermo Fisher Scientific, #66380).
2.3. Pull-down using β-catenin-MBP
Amylose beads (120 μg) were incubated with excess fusion-protein (200 μg) for 1 h at 4 °C followed by incubation with excess cell lysate (1 mg) for 1 h 10 min at 4 °C. Samples were centrifuged and pellets washed followed by protein elution with 100 mM maltose/TEDM. Eluates were loaded onto gels for SDS-PAGE. All experiments were independently performed in triplicate. Following SDS-PAGE, gel fixing, staining with Coomassie Brilliant Blue G250 and destaining, the gel was prepared for mass spectrometry by trypsin digestion.
2.4. Mass spectrometry (LC/MS/MS)
Samples were analysed using liquid-chromatography mass spectrometry (LC-MS) instrumentation consisting of an Easy nano-flow HPLC system (Thermo Fisher Scientific) coupled via a nanoelectrospray ion source (Thermo Fisher Scientific) to an Orbitrap Elite mass spectrometer (Thermo Fisher Scientific). Peptide separation was performed on a 20 cm × 100 μm column packed in-house with Halo® 2.7 μm 160 Å ES-C18 (Advanced Materials Technology). Peptides were loaded in buffer A (0.1% (v/v) FA) and eluted with a 97 min linear gradient of buffer B (100% (v/v) ACN/0.1% (v/v) FA) at 300 nL/min (2–85% buffer B in 97 min). Mass spectra were acquired in a data-dependent manner, with an automatic switch between MS and MS/MS using a top 15 method. Mass spectra were acquired in the Orbitrap analyser with a mass range of 350–1800 m/z and 120000 resolution at m/z 400 (Orbitrap Elite) and AGC target of 1e6 ions. High-energy collision dissociation (HCD) peptide fragments, acquired at 35 normalized collision energy, were analysed at high resolution in the Orbitrap with dynamic exclusion of ions for 10 s.
2.5. Data acquisition and statistical analysis
Raw files were processed with Proteome Discoverer 1.3 (Thermo Fisher Scientific). In the spectrum selector nodule, minimum-maximum precursor mass was set to 200–2000 Da. Peptide identification was performed with Mascot against the UniProt human database (20, 121 human entries) and a reversed human decoy database. Searches specified 10 ppm precursor mass tolerance, 0.02 Da fragment mass tolerance, maximum 2 missed cleavages, fixed modifications of carbamidomethylation (C) and variable modifications of oxidation (M), acetylation (N-terminal), 13C6, 15N2 on lysine (K), and 13C6, 15N4 on arginine (R) and peptides filtered at 0.01% false-discovery rate (FDR). SILAC pairs (heavy/light ratio calculation) were identified and quantitated using the built-in SILAC 2-plex quantification method using Arg-10 and Lys-8 labels and mass precision set to 2 ppm for event detection. Utilising this data, candidate lists were produced. This list excluded proteins not present in all 3 replicates at a fold-change of >1.5 for light (β-catenin-MBP):heavy (MBP) and without at least 3 unique peptides detected for it. A t-statistic was generated using the equivalent of a one-sample t-test to test deviance from a fold-change of >1.5. Based on this t-statistic a p-value was generated by using a one-tailed t-distribution with two-degrees of freedom. Proteins with a p-value of >0.05 were excluded from the candidate list.
2.6. Western blot
Following SDS-PAGE the gel was transferred onto nitrocellulose and the membranes washed and blocked. Membranes were incubated with polyclonal rabbit antibody against human COPB (Thermo Fisher Scientific, #PA1-061) O/N at 4 °C followed by a wash and incubation with rabbit secondary antibody (Millennium Science, #926–32211). Membranes were imaged using the Odyssey Licor 3.0 (Millennium Science) at 700 nm or 800 nm.
2.7. Immunoprecipitation (IP)
2 mg of cell lysate per 1 μg of monoclonal mouse antibody against human β-catenin (BD Biosciences, #610154) was rotated O/N at 4 °C. Protein G beads were added and rotated for 2 h at 4 °C. Samples were centrifuged and pellets washed in lysis buffer and centrifuged. Proteins were eluted and loaded onto a gel.
2.8. Immunofluorescence (IF) microscopy
Cells were fixed, washed, blocked and incubated with primary antibody (the same anti-β-catenin or anti-COPB antibody used for Western blot and IP). Cells were washed and secondary antibody/Hoechst nuclear stain added. Cells were washed and slides mounted using Vectashield. The slides were imaged using the DeltaVision Elite (GE Healthcare) at a magnification of x100 and SoftWoRx Explorer 1.3 (GE Healthcare) was used for analysis. Cells requiring a CSK (100 mM NaCl, 300 mM sucrose, 1 mM MgCl2, 10 mM PIPES, 1 mM EGTA, 1 mM DTT, 1 mM PMSF, 0.1% Triton X-100 and PI) wash were washed on ice before fixation for 1 min.
3. Results
3.1. Mass spectrometry screen identifies novel β-catenin protein interactions in SW480 colon cells
We used a recombinant β-catenin-MBP fusion construct to enrich protein interaction partners from SILAC labelled SW480 colon cancer cells. Bound protein eluates were separated by SDS-PAGE, excised and digested with trypsin, and peptides analysed by high-resolution LC-MS/MS. We independently repeated the enrichments three times and detected a total of 1863 proteins. This list was filtered to focus on those detected in all experiments at the threshold level, resulting in 436 potential interacting proteins (Supplementary Table I). To focus on high confidence interactions the threshold required detection in all three independent purifications at a >1.5 fold difference compared to MBP control (p-value <0.05), and a requirement of a minimum of three unique peptides per protein. Under these stringent conditions several previously reported β-catenin protein interactors were detected including catenin alpha-1 [22], catenin delta-1 [23], four and a half LIM domains protein 2 [24], Na(+)/H(+) exchange regulatory cofactor NHE-RF1 [25], ruvB-like 1 [26] and S-phase kinase-associated protein 1 [27], confirming the reliability of the methodology.
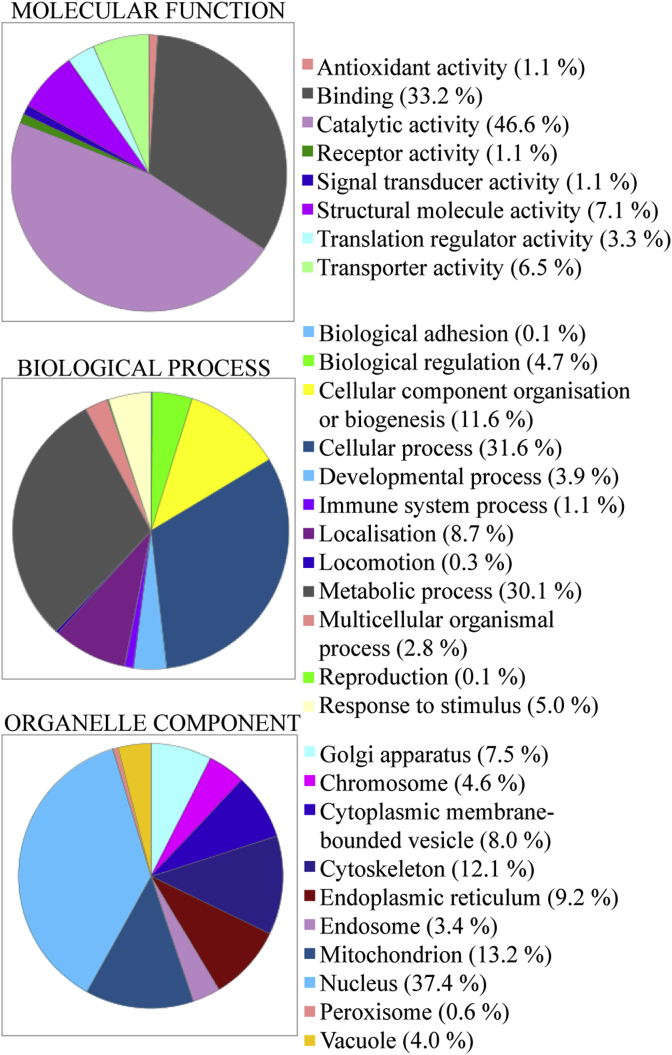
Gene ontology analysis was used to ascribe likely molecular functions, biological processes and reported subcellular locations for each of the putative β-catenin interacting proteins (Fig. 1). Of the proteins annotated to localise to organelles, this analysis showed that 7.5% are located at the Golgi, 9.2% at the endoplasmic reticulum (ER) and 3.4% at endosomes. The remainder of the proteins were at the chromosomes (4.6%), peroxisomes (0.6%), vacuoles (4.0%), membrane-bound (8.0%), cytoskeletal (12.1%), mitochondrial (13.2%) or nuclear (37.4%).
Fig. 1.
Distribution of subcellular component, molecular function and biological processes of candidate binders of β-catenin. Gene ontology analysis of candidate novel binding partners of β-catenin in colon cancer cell line SW480.
3.2. Interaction of β-catenin with coatomer complex (COPI)
Detection of β-catenin in the secretory pathway is not well established, so to explore this further we focused on the coatomer complex (COPI) as a putative novel interactor with β-catenin. COPI is a protein complex essential for retrograde vesicular transport and budding from the Golgi membrane [28]. COPI consists of seven subunits, each of which were detected by mass spectrometry in the β-catenin pulldown experiments (Table 1), with five subunits satisfying our stringent reporting threshold. To confirm this interaction we carried out a separate pull-down experiment using β-catenin-MBP fusion protein, and separately, an in vivo immunoprecipitation using anti-β-catenin antibody to bind endogenous β-catenin. These experiments demonstrated that β-catenin and endogenously expressed COPB subunit could be co-isolated in whole cell lysates of SW480 cells (Fig. 2). Interestingly, analysis of β-catenin primary sequence showed the presence of three dilysine motifs; KK (amino acids 180–181 and 671–672) and KxK (433–435) which are also known as coatomer complex binding sites in Type I transmembrane proteins [29].
Table 1.
Interactors of β-catenin detected by MS.
| Protein name |
Σ# Unique |
Heavy/light SILAC ratio |
p-value | |||
|---|---|---|---|---|---|---|
| Accession | Peptides | Replicate 1 | Replicate 2 | Replicate 3 | ||
| Known interactors | ||||||
| P35221 | Catenin alpha-1 | 33 | 0.003 | 0.003 | 0.001 | 6.71E-07 |
| O60716 | Catenin delta-1 | 4 | 0.002 | 0.002 | 0.001 | 1.07E-07 |
| Q14192 | Four and a half LIM domains protein 2 | 13 | 0.001 | 0.001 | 0.001 | 7.27E-10 |
| O14745 | Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | 5 | 0.025 | 0.001 | 0.001 | 7.15E-05 |
| Q9Y265 | RuvB-like 1 | 19 | 0.004 | 0.002 | 0.001 | 8.86E-07 |
| P63208 | S-phase kinase-associated protein 1 | 3 | 0.001 | 0.020 | 0.015 | 3.81E-05 |
| COPI subunits | ||||||
| P53621 | Coatomer subunit alpha | 19 | 0.002 | 0.297 | 0.001 | 1.44E-02 |
| P53618 | Coatomer subunit beta | 7 | 0.001 | 0.472 | 0.001 | 4.17E-02 |
| P35606 | Coatomer subunit beta' | 7 | 0.001 | 0.001 | 0.001 | 1.25E-09 |
| P48444 | Coatomer subunit delta | 6 | 0.628 | 0.035 | 0.260 | 8.66E-02 |
| O14579 | Coatomer subunit epsilon | 3 | 0.001 | 0.444 | 0.001 | 3.62E-02 |
| Q9Y678 | Coatomer subunit gamma-1 | 11 | 0.727 | 0.436 | 0.001 | 1.58E-01 |
| P61923 | Coatomer subunit zeta-1 | 3 | 0.460 | 0.462 | 0.423 | 1.67E-03 |
Known interactors (accounting for < 2% of the candidate list) and novel candidate interactor COPI of which all subunits were detected.
β-catenin at the Golgi.
Fig. 2.
Detection of β-catenin–COPB protein interactions in SW480 total cell extracts following enrichment.
(A) Pull-down with β-catenin-MBP construct, followed by immunoblot with COPB specific antibody. * indicates location of COPB protein. (B) Co-immunoprecipitation of COPB using anti-β-catenin antibody. Detection using anti-COPB antibody. * indicates location of COPB protein.
Having demonstrated that β-catenin and COPB co-isolate, we next performed immunofluorescence microscopy experiments in SW480 colon cancer cells. As shown in Fig. 3, we observed co-localisation of COPB with the trans-Golgi-marker TGN46 [30], confirming COPB is located in the secretory pathway including at the Golgi in these cells. Normally β-catenin is overexpressed in SW480 cells due to an APC mutation, and hence the high background of β-catenin makes it difficult to pinpoint it to any specific organelle. Furthermore, the oversaturation of β-catenin signal limits the ability to confidently assign points of co-localisation. We therefore performed a mild detergent extraction to remove soluble proteins, leaving behind only those proteins retained at specific structures (Fig. 4A). This immunofluorescence microscopy study showed Golgi co-localisation of endogenous β-catenin and COPB in SW480 colon cancer cells (Fig. 4B).
Fig. 3.
Co-localisation of COPB and Golgi-marker (TGN46) in SW480. Cells were stained with α-COPB (green) and α-TGN46 (red) antibody (scalebar = 5 μm). The zoom image underneath each box demonstrates that each arrow points to a spot of co-localisation, indicating that COPB can sometimes be found at the Golgi although it cannot be considered as a Golgi-marker.
Fig. 4.
Co-localisation of β-catenin and COPB in SW480
A) Cells were stained for IF pre- and post-CSK wash (contains Triton X-100) using β-catenin (red) and COPB (green) (scalebar = 20 μm). With the overexpression of β-catenin in this cell line, no sub-structures can be detected without a mild detergent extraction which removes cytoplasmic β-catenin and COPB, retaining only β-catenin and COPB that is anchored at specific sites. Subsequently, any co-localisation of β-catenin with COPB indicates it is affixed, potentially at the Golgi and in complex with other COP proteins. B) The SW480 pre-zoom image is post-CSK buffer wash (from panel A). Due to the CSK wash, any co-localisation of β-catenin and COPB would be fastened to a structure, such as the Golgi. Arrows in the inset zooms point to sites of co-localisation.
4. Discussion
4.1. Novel localisation and interaction of β-catenin
We used mass spectrometry in a discovery screening mode to identify peptides derived from proteins isolated from interactions of full-length β-catenin-MBP fusion protein in lysates of SW480 colon cancer cells. Strict filtering of the resultant data revealed 436 putative β-catenin interactors, including several with previously confirmed in vivo interactions. β-catenin appears to facilitate multiple cellular activities, with particularly well characterised roles at the plasma membrane with cadherins (cell adhesion) and in the cytoplasm and nucleus as an effector of Wnt signalling. However, less is known about β-catenin and its putative role in the secretory pathway. In this paper we have shown that in SW480 colon cancer cells: (i) recombinant β-catenin precipitates the COPI complex (Table 1), (ii) endogenous β-catenin and COPB can be co-isolated by immunoprecipitation (Fig. 2), and (iii) endogenous β-catenin and COPB colocalise to structures that stain positive with Golgi-marker TGN46 (Figs. 3 and 4), suggesting localisation at the Golgi. These findings are consistent with a role for β-catenin in transport between the ER and Golgi and implicate the requirement of COPI in this process. The biochemical properties and functional consequences underlying this interaction remain to be explored.
4.2. β-catenin at the Golgi and its interaction with the coatomer complex
There is little documented in the literature regarding β-catenin and the Golgi. Experiments here identified a number of novel candidate binding partners that normally reside at specific cellular compartments in the secretory pathway, including the Golgi, endoplasmic reticulum and endosomes (7.5%, 9.2% and 3.4% of the candidate interactor list respectively). While our discovery approach cannot exclude artefactual binders due to the requirement to use soluble lysates for protein capture, it is particularly noteworthy that we detected all seven subunits of the COPI coatomer complex subunits [31] to co-isolate with β-catenin, providing solid evidence for in vivo interaction, which was subsequently confirmed by microscopy. The coatomer complex is essential for budding from the Golgi membrane [28] and coatomer subunit beta (COPB) in particular mediates traffic between Golgi compartments and is necessary for protein transport from the ER to the cis-Golgi [32]. Follow-up experiments are required to determine the precise subcellular structures containing β-catenin and the functional consequences of its interactions.
Previously, a large pool of cytoplasmic β-catenin was discovered using IF to accumulate in recycling endosomes and the Golgi in MCF-7 breast cancer cells [33]. Additionally, IF demonstrated that β-catenin and COPB co-localise in MCF-7 breast cancer cells, but attempts to validate this observation using immunoprecipitation were inconclusive [33]. Other studies have also shown the potential for β-catenin to function in transport at the Golgi/ER in addition to its normal roles in the nucleus. For example, Brunner et al. (2006) analysed the subcellular location of β-catenin in meningiomas and found that β-catenin was mostly localised at the Golgi and the ER/Golgi intermediate compartment (ERGIC) [34]. A hallmark of meningioma tumorigenesis is loss of the neurofibromatosis type 2 (NF2) gene which controls cell-cell adhesion mediated by E-cadherin/β-catenin. They showed that loss of β-catenin at the membrane in the majority of meningiomas (74%) correlated with relocation to the Golgi apparatus/ERGIC.
Furthermore, Du et al. (2012) demonstrated the importance of β-catenin phosphorylation in directing its localisation [35] in prostate cancer cells. They demonstrated that in normal prostate tissue β-catenin was predominantly (73%) localised to the TGN due to phosphorylation at threonine 120 and that this was altered and diminished to only 7% in prostate cancer, concluding that phosphorylation at threonine 120 could alter subcellular localisation and transcriptional activity of β-catenin and may be a driver for its Golgi localisation.
Though the above literature supports our observations of localisation of β-catenin at the Golgi, to date evidence for the Golgi proteins responsible for this interaction have been missing. In lieu of this, our proteomics screen suggests that COPI is a site of β-catenin interaction in structures which stain with TGN46, such as the Golgi. While it remains to be investigated, the interaction of β-catenin and COPI may function in the formation of the COPI vesicular coat or transport of the vesicles between the ER and Golgi [32], implicating the Golgi as a site of vesicle transport for β-catenin.
Funding
BRH and MPM acknowledge funding support from the National Health and Medical Research Council (NHMRC) of Australia (APP1046767). Aspects of this research where facilitated by access to the Australian Proteome Analysis Facility supported by the Australian Government’s National Collaborative Research Infrastructure Scheme.
Conflicts of interest
The authors have reviewed the manuscript and approved its submission. We have no competing interests to disclose. The article is not under consideration elsewhere.
Acknowledgements
Not applicable.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2019.100662.
Contributor Information
Crystal Semaan, Email: C.semaan@centenary.org.au.
Beric R. Henderson, Email: beric.henderson@sydney.edu.au.
Mark P. Molloy, Email: m.molloy@sydney.edu.au.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Saifo M.S. Targeting the oncogenic protein beta-catenin to enhance chemotherapy outcome against solid human cancers. Mol. Cancer. 2010;9:310. doi: 10.1186/1476-4598-9-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li V.S. Wnt signaling through inhibition of beta-catenin degradation in an intact Axin1 complex. Cell. 2012;149(6):1245–1256. doi: 10.1016/j.cell.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 3.Freese J.L., Pino D., Pleasure S.J. Wnt signaling in development and disease. Neurobiol. Dis. 2010;38(2):148–153. doi: 10.1016/j.nbd.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernard S., Eilers M. Control of cell proliferation and growth by Myc proteins. Results Probl. Cell Differ. 2006;42:329–342. doi: 10.1007/400_004. [DOI] [PubMed] [Google Scholar]
- 5.Stamenkovic I. Matrix metalloproteinases in tumor invasion and metastasis. Semin. Canc. Biol. 2000;10(6):415–433. doi: 10.1006/scbi.2000.0379. [DOI] [PubMed] [Google Scholar]
- 6.Cadigan K.M., Peifer M. Wnt signaling from development to disease: insights from model systems. Cold Spring Harb Perspect Biol. 2009;1(2):a002881. doi: 10.1101/cshperspect.a002881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Korinek V. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma. Science. 1997;275(5307):1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 8.Morin P.J. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275(5307):1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 9.Yokoya F. beta-catenin can be transported into the nucleus in a Ran-unassisted manner. Mol. Biol. Cell. 1999;10(4):1119–1131. doi: 10.1091/mbc.10.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miyoshi Y. Activation of the beta-catenin gene in primary hepatocellular carcinomas by somatic alterations involving exon 3. Cancer Res. 1998;58(12):2524–2527. [PubMed] [Google Scholar]
- 11.Lee J.M. beta-Catenin signaling in hepatocellular cancer: Implications in inflammation, fibrosis, and proliferation. Cancer Lett. 2014;343(1):90–97. doi: 10.1016/j.canlet.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rubinfeld B. Stabilization of beta-catenin by genetic defects in melanoma cell lines. Science. 1997;275(5307):1790–1792. doi: 10.1126/science.275.5307.1790. [DOI] [PubMed] [Google Scholar]
- 13.Aberle H. Single amino acid substitutions in proteins of the armadillo gene family abolish their binding to alpha-catenin. J. Biol. Chem. 1996;271(3):1520–1526. doi: 10.1074/jbc.271.3.1520. [DOI] [PubMed] [Google Scholar]
- 14.Dobrosotskaya I.Y., James G.L. MAGI-1 interacts with beta-catenin and is associated with cell-cell adhesion structures. Biochem. Biophys. Res. Commun. 2000;270(3):903–909. doi: 10.1006/bbrc.2000.2471. [DOI] [PubMed] [Google Scholar]
- 15.Rubinfeld B. Association of the APC gene product with beta-catenin. Science. 1993;262(5140):1731–1734. doi: 10.1126/science.8259518. [DOI] [PubMed] [Google Scholar]
- 16.Behrens J. Functional interaction of an axin homolog, conductin, with beta-catenin, APC, and GSK3beta. Science. 1998;280(5363):596–599. doi: 10.1126/science.280.5363.596. [DOI] [PubMed] [Google Scholar]
- 17.Kemler R. From cadherins to catenins: cytoplasmic protein interactions and regulation of cell adhesion. Trends Genet. 1993;9(9):317–321. doi: 10.1016/0168-9525(93)90250-l. [DOI] [PubMed] [Google Scholar]
- 18.Behrens J. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature. 1996;382(6592):638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 19.Selbach M., Mann M. Protein interaction screening by quantitative immunoprecipitation combined with knockdown (QUICK) Nat. Methods. 2006;3(12):981–983. doi: 10.1038/nmeth972. [DOI] [PubMed] [Google Scholar]
- 20.Hwang J.R. Identification of beta-catenin-interacting proteins in nuclear fractions of native rat collecting duct cells. Am. J. Physiol. Renal. Physiol. 2017;313(1):F30–F46. doi: 10.1152/ajprenal.00054.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yakulov T. Wnt3a-dependent and -independent protein interaction networks of chromatin-bound beta-catenin in mouse embryonic stem cells. Mol. Cell. Proteom. 2013;12(7):1980–1994. doi: 10.1074/mcp.M112.026914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drees F. Alpha-catenin is a molecular switch that binds E-cadherin-beta-catenin and regulates actin-filament assembly. Cell. 2005;123(5):903–915. doi: 10.1016/j.cell.2005.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kucerova D. Expression and interaction of different catenins in colorectal carcinoma cells. Int. J. Mol. Med. 2001;8(6):695–698. [PubMed] [Google Scholar]
- 24.Wei Y. Identification of the LIM protein FHL2 as a coactivator of beta-catenin. J. Biol. Chem. 2003;278(7):5188–5194. doi: 10.1074/jbc.M207216200. [DOI] [PubMed] [Google Scholar]
- 25.Shibata T. EBP50, a beta-catenin-associating protein, enhances Wnt signaling and is over-expressed in hepatocellular carcinoma. Hepatology. 2003;38(1):178–186. doi: 10.1053/jhep.2003.50270. [DOI] [PubMed] [Google Scholar]
- 26.Baron B.W., Baron R.M., Baron J.M. The relationship between RUVBL1 (Pontin, TIP49, NMP238) and BCL6 in benign and malignant human lymphoid tissues. Biochem Biophys Rep. 2016;6:1–8. doi: 10.1016/j.bbrep.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu G. Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity of the SCF beta-TrCP1 ubiquitin ligase. Mol. Cell. 2003;11(6):1445–1456. doi: 10.1016/s1097-2765(03)00234-x. [DOI] [PubMed] [Google Scholar]
- 28.Orci L. Budding from Golgi membranes requires the coatomer complex of non-clathrin coat proteins. Nature. 1993;362(6421):648–652. doi: 10.1038/362648a0. [DOI] [PubMed] [Google Scholar]
- 29.Ma W., Goldberg J. Rules for the recognition of dilysine retrieval motifs by coatomer. EMBO J. 2013;32(7):926–937. doi: 10.1038/emboj.2013.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang J.S. COPI acts in both vesicular and tubular transport. Nat. Cell Biol. 2011;13(8):996–1003. doi: 10.1038/ncb2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Futatsumori M. Identification and characterization of novel isoforms of COP I subunits. J. Biochem. 2000;128(5):793–801. doi: 10.1093/oxfordjournals.jbchem.a022817. [DOI] [PubMed] [Google Scholar]
- 32.Jackson L.P. Structure and mechanism of COPI vesicle biogenesis. Curr. Opin. Cell Biol. 2014;29:67–73. doi: 10.1016/j.ceb.2014.04.009. [DOI] [PubMed] [Google Scholar]
- 33.Jamieson C. Characterization of a beta-catenin nuclear localization defect in MCF-7 breast cancer cells. Exp. Cell Res. 2016;341(2):196–206. doi: 10.1016/j.yexcr.2016.01.020. [DOI] [PubMed] [Google Scholar]
- 34.Brunner E.C. Altered expression of beta-catenin/E-cadherin in meningiomas. Histopathology. 2006;49(2):178–187. doi: 10.1111/j.1365-2559.2006.02440.x. [DOI] [PubMed] [Google Scholar]
- 35.Du C. Beta-catenin phosphorylated at threonine 120 antagonizes generation of active beta-catenin by spatial localization in trans-Golgi network. PLoS One. 2012;7(4) doi: 10.1371/journal.pone.0033830. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.