Abstract
Background
The pathology of juvenile dermatomyositis (JDM) is characterized by prominent vessel wall and perivascular inflammation. This feature of the disease has remained unexplained and under-investigated. We have hypothesized that plasma exosomes, which play an important role in inter-cellular communication, may play a role in the vascular injury associated with JDM.
Objective
To characterize the circulating exosomes of children with JDM and determine whether the small RNA cargoes within those exosomes are capable of altering transcriptional programs within endothelial cells.
Design/methods
We purified exosomes from plasma samples of children with active, untreated JDM (n = 6) and healthy controls (n = 9). We characterized the small RNA cargoes in JDM and control exosomes by RNA sequencing using the Illumina HiSeq 2500 platform. We then incubated isolated exosomes from healthy controls and children with JDM with cultured human aortic endothelial cells (HAEC) for 24 h. Fluorescence microscopy was used to confirm that both control and JDM exosomes were taken up by HAEC. RNA was then purified from HAEC that had been incubated with either control or JDM exosomes and sequenced on the Illumina platform. Differential expression of mRNAs from HAEC incubated with control or JDM exosomes was ascertained using standard computational methods. Finally, we assessed the degree to which differential gene expression in HAEC could be attributed to the different small RNA cargoes in JDM vs control exosomes using conventional and novel analytic methods.
Results
We identified 10 small RNA molecules that showed differential abundance when we compared JDM and healthy control exosomes. Fluorescence microscopy of labeled exosomes confirmed that both JDM and control exosomes were taken up by HAEC. Differential gene expression analysis revealed 59 genes that showed differential expression between HAEC incubated with JDM exosomes vs HAEC incubated with exosomes from controls. Statistical analysis of gene expression data demonstrated that multiple miRNAs exerted transcriptional control on multiple genes with HAEC.
Conclusions
Plasma exosomes from children with active, untreated JDM are taken up by HAEC and are associated with alterations in gene expression in those cells. These findings provide new insight into potential mechanisms leading to the targeting of vascular tissue by the immune system in JDM.
Electronic supplementary material
The online version of this article (10.1186/s12969-019-0347-0) contains supplementary material, which is available to authorized users.
Keywords: Juvenile dermatomyositis, Plasma exosomes, Endothelial cells, Gene expression, RNA-seq, miRNA-seq
Background
Prominent vascular changes and perivascular inflammation are distinct clinical [1] and pathological [2, 3] features of juvenile dermatomyosits (JDM) and underlie much of the morbidity associated with the disease [3]. The mechanisms through which the blood vessels are targeted by the immune system in this disease remain incompletely understood. A prominent type 1 interferon-induced gene expression signature is seen in both affected muscle tissue and peripheral blood of children with JDM [4], and there is accumulating evidence that type 1 interferons can directly harm the vasculature [5], but the specific relationship between vascular damage and the interferon signature in dermatomyositis remains uncertain [6]. We have recently reported that endothelial cells in JDM are targeted by a brisk autoantibody response [7], but we were unable to ascertain whether such autoantibodies represent a primary driver of the pathology or a response to immunologically-injured endothelium. Thus, the question of how the vasculature becomes targeted or injured in JDM remains unanswered.
Exosomes are one of several families of microparticles that are released from cells either after specific stimuli or during cellular apoptosis [8]. We are now coming to understand that these subcellular components contain small, non-coding RNA molecules that form a previously unrecognized level of transcriptional control in mammals and possibly in simpler organisms [9]. The RNA molecules contained within exosomes and other microparticles are, among other things, a mechanism through which cells of the immune system communicate with one-another [10]. These microparticles are abundant in serum and plasma and have been a source of considerable interest as biomarkers in a broad range of diseases [11–13]. Since the endothelium is in intimate contact with plasma, and therefore circulating microparticles such as exosomes, we believe that understanding more about the composition and biological behavior of circulating exosomes may yield new insights into basic disease mechanisms in JDM.
In this paper, we describe the small RNA contents of exosomes purified from the plasma of children with JDM and describe their interactions with endothelial cells in an in vitro model. We believe these studies provide new insights into the complex mechanisms that appear to underlie JDM and very likely other forms of vasculitis.
Methods
Patients and patient specimens
Plasma was obtained from 6 children with newly diagnosed, untreated JDM who presented at the University of Oklahoma Pediatric Rheumatology Clinic between 2007 and 2011. All children had weakness, rash, elevated serum concentrations of CPK, AST, and ALT and altered nailfold capillaries. There were 4 girls (age range 3–10 years) and 2 boys (ages 14 and 15 years) whose plasma samples were used for these studies. Plasma was also obtained from healthy children (HC; n = 9) who were patients at Hodge Pediatrics, a general pediatrics clinic operated by the Women and Children’s Hospital of Buffalo Hodge Pediatrics Clinic. Among the HC, there were 3 boys (ages 8–13 years) and 6 girls (ages 6–14 years). HC were excluded if they were taking antibiotics or systemic steroids, had an underlying autoimmune condition such as type 1 diabetes or autoimmune thyroiditis, had asthma and on a leukotriene receptor antagonist or a medium or high dose inhaled corticosteroid, had invasive surgery in the previous 3 weeks, were obese with a BMI at greater than the 95th percentile for their age, or had temperature greater than 38.0 within the previous 36 h.
All research procedures were reviewed and approved by the Institutional Review Boards (IRB) of the University of Oklahoma Health Sciences Center and the University at Buffalo Jacobs School of Medicine & Biomedical Sciences. All research was performed in compliance with the IRB-approved protocol. Written informed consent was obtained from the parents or guardians of all subjects, and, where appropriate, assent was obtained from children in accordance with IRB guidelines at each institution.
Blood was drawn into CPT tubes (Becton Dickinson, Cat # 362761, Franklin Lakes, NJ) and brought to the laboratory for processing within an hour of being obtained. Plasma was separated from cells by centrifugation and stored at -80 °C until used.
Purification of exosomes from plasma
Exosomes were isolated from 500 μL of plasma using the ExoQuick exosome precipitation solution (System Biosciences, Mountain View, CA, USA) according to the manufacturer’s methods. Briefly, the plasma was incubated with thrombin liquid suspension (500 U/mL) (System Biosciences) for 5 min while mixing at room temperature. After centrifugation at 10,000 rpm for 5 min, the supernatant was then filtered through a 0.2 μm Spin X column filtration system (Sigma) to remove any cell debris and particles larger than 200 nm. The filtrate was mixed with 120 μL of ExoQuick solution and RNase A (Sigma, St. Louis, MO, USA) to a final concentration of 10 μg/mL. The mixture was kept at 4 °C overnight and then further mixed with 100 units/mL of murine RNase inhibitor (NEB) before centrifugation at 13,000 rpm for 2 min. The pellet was washed in phosphate-buffered saline (PBS), then the PBS was removed.
Exosome identification
To verify that the material prepared as described above represented plasma exosomes, Western blot assay was performed for a common exosome marker, CD63. Protein was loaded on 10% SDS-PAGE gels. After electrophoretic separation, proteins were transferred to a nitrocellulose membrane (Bio-Rad). The membranes were blocked with 5% milk in 1× TBS with 0.05% Tween 20 (TBST) for 1 h at room temperature. Primary antibodies (rabbit anti-human CD63 antibody, System Biosciences, Mountain View, CA, USA) were added to the membrane and incubated overnight at 4 °C with gentle agitation. The membranes were further washed with TBST and incubated for 1 h with horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (System Biosciences). The immunoreactive proteins were detected using an enhanced chemiluminescence substrate kit (SuperSignal, Pierce, Rockford, IL) according to the manufacturer’s instructions.
Further verification that our isolation procedures had resulted in purification of plasma exosomes was undertaken using transmission electron microscopy. Exosome enriched fractions from thrombin treated plasma were fixed using 2% glutaraldehyde at 4 °C overnight. The following day, the fixed exosomes were deposited on the surface of a 400-mesh carbon-coated grids, air-dried, stained with uranyl acetate, and transmission election microscope (TEM) images were recorded with a JEM-100CX II transmission electron microscope (JEOL, Peabody, MA, USA).
Purification of RNA from isolated exosomes
Exosomal RNA was extracted using a miRNeasy Micro Kit (QIAGEN, Valencia, CA, USA). Exosome pellets was mixed with 700 μL QIAzol lysis buffer, and the mixture was processed according to the manufacturer’s standard protocol. To eliminate potential co-precipitated DNA, the column-bound RNA was treated with DNase I for 15 min at room temperature. The RNA was then eluted with 14 μl of DNase- and RNase-free water. RNA quality and quantity were estimated by Agilent Bioanalyzer 2100 using a Small RNA Chip (Agilent, Santa Clara, CA).
Uptake of exosomes by cultured human aortic endothelial cells (HAEC) and purification of total RNA from HAEC
HAECs were cultured in 4-well Lab-Tek chamber coated with collagen in Medium 200 plus Low Serum Supplement (Life Technologies) for 24 h. Exosomes from plasma were labeled with PKH67 (Sigma) for 30 min followed by PBS washes. HAECs were incubated with PKH67 labeled exosomes for 24 h. HAECs were fixed by 4% formaldehyde and the slide was mounted using ProLong Gold antifade reagent with DAPI (Life Technologies). The chamber slides and exosomes were visualized with a fluorescent imaging microscope (Zeiss).
In order to determiner the biological effects of JDM exosomes after their uptake by HAEC, we cultured HAECs in 6-well plates coated with collagen in Medium 200 plus Low Serum Supplement. When HAEC cells had approximate 85% confluence, they were incubated with exosomes from JDM (n = 4) or healthy control plasma (n = 4) for 24 h. HAEC total RNA was isolated using Trizol and RNeasy Mini Kit (Qiagen) as described previously [14].
RNA sequencing
Small RNA libraries were constructed following the manufacturer’s instructions using the TruSeq Small RNA Library Prep Set from Illumina. RNA libraries were amplified by 11 PCR cycles using Illumina compatible index primers. Size selection of the amplified libraries was done using Blue Pippin 3% gel cassettes (Sage Sciences). cDNA fragments from 140 to 160 bp (the length of miRNA inserts plus the 3′ and 5′ adaptors) were eluted. cDNA libraries were quantified using Picogreen Assay (Invitrogen) and Library Quantification kit (Kapa Biosystems, Wilmington, MA). Agilent Bioanalyzer 2100 Hi Sensitivity DNA chip was used to confirm the sizes of the cDNA libraries. The cDNA products were then sequenced using the Illumina HiSeq2500 at the UB Genomics and Bioinformatics Core Facility (Buffalo, NY). Ten sequencing libraries were pooled into a single sequencing lane.
mRNA libraries were constructed at the UB Genomics and Bioinformatics Core Facility (Buffalo, NY) using the TruSeq RNA Library Preparation Kit. All samples were subjected to 50-cycle, single-read sequencing in the HiSeq 2500 (Illumina).
Analysis of small RNA cargoes in exosomes from RNAseq data
For miRNA sequence analysis, read counts for individual miRNAs were obtained for each sample using miRDeep* [15], which directly takes fastq sequences as input. The resulting read counts were used to build an expression matrix, which was then subjected to edgeR analysis. Differentially expressed miRNAs were defined as those miRNA with FDR < 0.05 between two groups of compared samples.
Analysis of transcriptional changes after incubation of exosomes with cultured HAEC
We generated strand-specific RNA libraries using TruSeq Stranded Total RNA Plus Ribo-zero kits (Illumina). Sequencing was performed at the Genomics and Bioinformatics Core of the State University of New York at Buffalo. Single-end reads per sample were obtained using the HiSeq 2500 platform from Illumina. For mRNA, reads were first trimmed using Cutadap [16] to remove the 3′ end adapters and trailing sequences, followed by aligning to human RefSeq mRNAs (hg19) using TopHat2 [17]. Transcript counts were estimated using HTSeq [18]. Differences in gene expression levels between samples were assessed with edgeR [19].
Identifying miRNAs associating with individual differentially expressed genes (DEGs)
We first used the miRNA target gene dataset predicted from the algorithm of TargetScanS [20] to find miRNAs that target individual DEGs resulted from RNA profiling analysis. We then searched for the corresponding miRNAs in the miRNA expression dataset, as not all miRNAs from TargetScanS are included in the miRNA expression dataset. The association between individual DEGs and miRNAs from the expression dataset were assessed by plotting their expression changes between JDM and HC.
Results
Characterization of exosomes isolated from plasma of children with JDM and controls
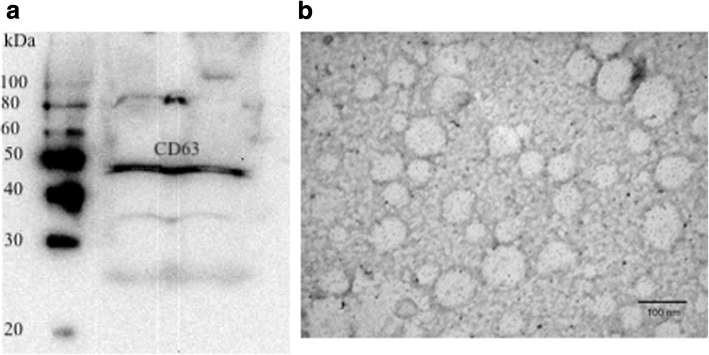
Figure 1a shows results from the western blotting procedure. All samples were CD63+ by western blotting. Furthermore, exosomes showed the appropriate size and morphology under electron microscopy, as shown in Fig. 1b.
Fig. 1.
Results of western blotting for CD63 (a) and electron micrograph showing size and morphology of exosomes purified from plasma of a child with juvenile dermatomyositis (b). Molecular weight markers (100-20 kD) are shown in the first lane of the left panel. The scale for the electron micrograph (100 nm) is shown in the lower right side of the right panel. Exosomes were prepared with the ExoQuick™ reagent from System Biosciences
The small RNA cargoes of JDM-derived exosomes differ from those found in healthy controls
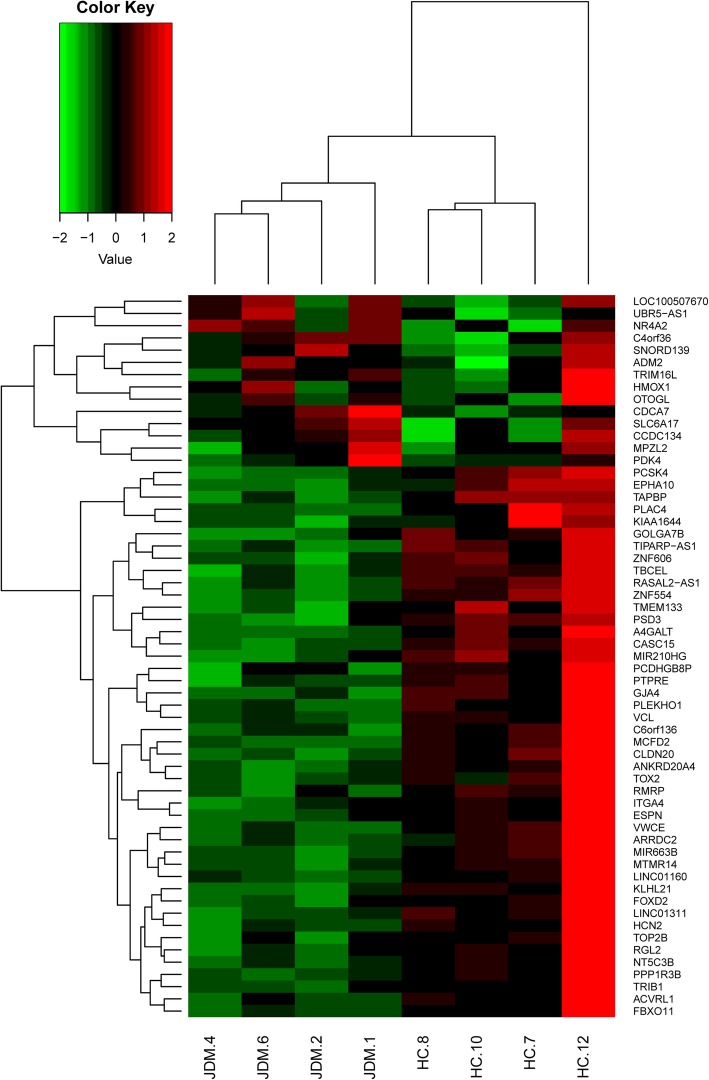
We used small RNA library RNA-seq to compare the differential expression of miRNAs within the exosomes purified from children with JDM and HC. For this analysis, we used 6 JDM and 9 HC exosome samples. Setting the false discovery rate (FDR) at < 0.05, we identified 10 differentially expressed miRNAs in the comparison between JDM and HC, of which 2 show down-regulation, and 8 show up-regulation (Table 1). Hierarchical cluster analysis classified the JDM and HC samples into distinct clusters (Fig. 2).
Table 1.
Differentially expressed miRNAs between JDM and HC
| miRNA | logFC | Fold change | p value | FDR |
|---|---|---|---|---|
| hsa-mir-21 | −6.34 | −80.84 | 5.28E-27 | 7.18E-25 |
| hsa-mir-221 | 2.55 | 5.86 | 4.59E-09 | 3.12E-07 |
| hsa-mir-206 | 3.77 | 13.64 | 3.90E-04 | 1.77E-02 |
| hsa-mir-122 | 2.47 | 5.56 | 6.85E-04 | 1.96E-02 |
| hsa-mir-222 | 1.37 | 2.59 | 7.21E-04 | 1.96E-02 |
| hsa-let-7b | −3.53 | −11.56 | 9.86E-04 | 2.23E-02 |
| hsa-mir-501 | 2.37 | 5.15 | 1.32E-03 | 2.51E-02 |
| hsa-mir-744 | 2.60 | 6.05 | 1.48E-03 | 2.51E-02 |
| hsa-mir-193a | 1.45 | 2.74 | 2.64E-03 | 3.99E-02 |
| hsa-mir-484 | 1.21 | 2.32 | 3.57E-03 | 4.85E-02 |
Fig. 2.
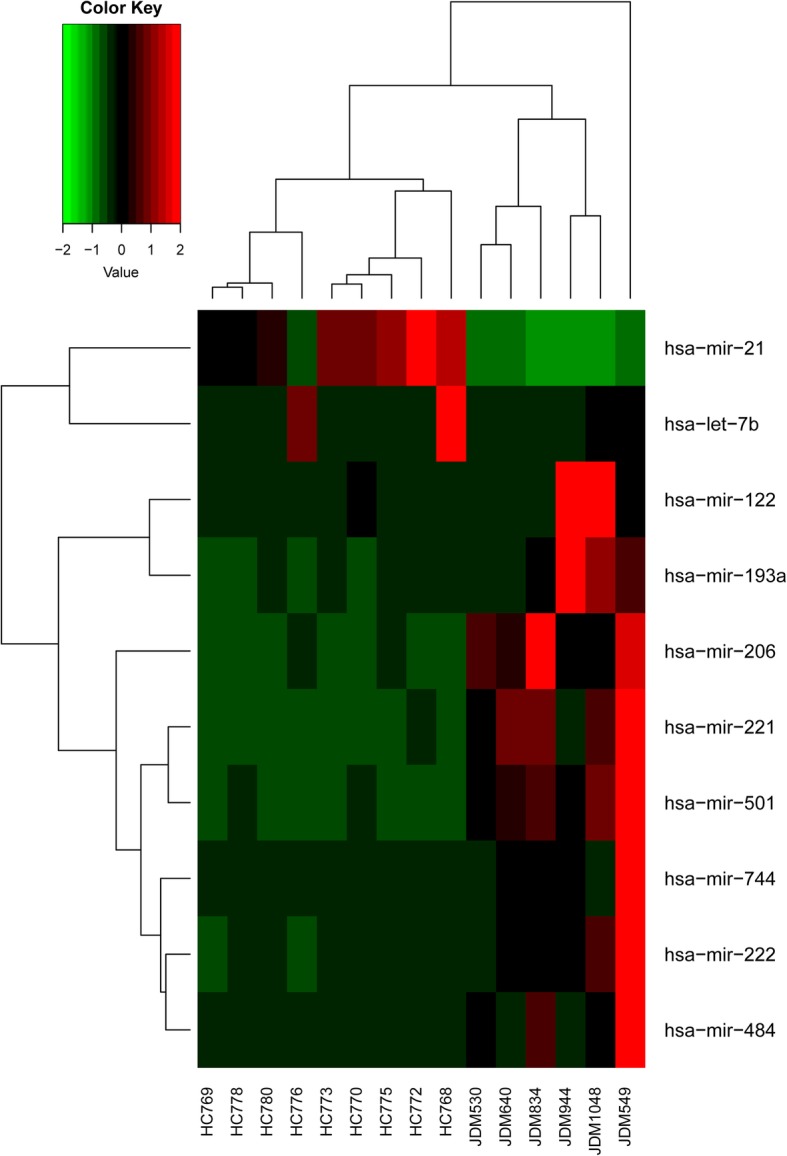
Unsupervised hierarchical clustering analysis of gene expression for the 10 differentially expressed miRNA genes between JDM and HC. The heatmap shows the median-normalized expression of individual miRNA genes across all samples. Heatmap colors represent relative miRNA expression as indicated in the color key
The RNA cargoes that we identified as showing higher abundance in JDM exosome samples (compared to controls) showed no overlap with small RNAs that show higher abundance in plasma exosomes of children with untreated polyarticular JIA. We show results from 40 children with untreated polyarticular JIA and 20 healthy controls in Additional file 1: Table S1. This work is part of an ongoing investigation and will be published in more detail separately.
AEC take up JDM exosomes; transcriptional profiles differ between cells incubated with JDM exosomes and exosomes from healthy children
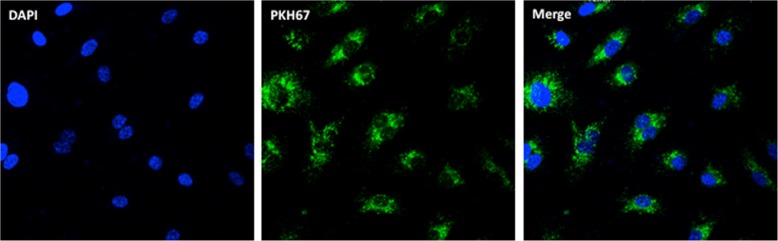
We next sought to determine whether plasma exosomes might be taken up by HAEC, and whether we could identify transcriptional alterations within HAEC that could be attributed to the RNA cargoes within the JDM exosomes. Figure 3 shows the results of the uptake experiments. Exosomes isolated from both JDM and healthy control plasma were seen within the cytoplasm of HAEC within 24 h hours of exposure.
Fig. 3.
Uptake of exosomes by cultured human aortic endothelial cells (HAEC). HAECs were incubated with PKH67 labeled exosomes (Green) for 24 h. HAECs were fixed by 4% formaldehyde and the slide was mounted using ProLong Gold antifade reagent with DAPI (Blue). The chamber slides and exosomes were visualized with a fluorescent imaging microscope
We next used RNA-seq to compare the expression of genes in HAEC incubated with exosomes from children with untreated JDM (n = 4) JDM and HAEC incubated with exosomes from HC (n = 4). Setting the cutoff off p < 0.01 and fold change > 1.3, we identified 59 differentially expressed genes (DEGs) when we compared HAEC incubated with JDM exosomes with HAEC incubated with exosomes from HC. Of these 59 genes, 45 show higher expression in HAEC incubated with HC exosomes, and 14 show lower expression when compared to HAEC incubated with JDM exosomes (Table 2). Hierarchical cluster analysis using the expression data of the 59 genes from both JDM and HC was able to classify the HAEC incubated with exosomes from JDM samples into a distinct cluster from those incubated with HC exosomes. The later formed one major cluster with three of the 4 samples, as shown in the heatmap (Fig. 4).
Table 2.
Differentially expressed genes after incubation with HAEC
| Gene smybol | Fold change | p value | miRNA targeta |
|---|---|---|---|
| TBCEL | −2.225083292 | 0.000232193 | Yes |
| TOX2 | −2.043385398 | 0.001375425 | Yes |
| PCDHGB8P | −2.019271064 | 0.008283468 | no |
| EPHA10 | − 1.937909029 | 0.003578788 | Yes |
| TMEM133 | −1.913578811 | 0.006586193 | Yes |
| A4GALT | − 1.893256114 | 0.003843604 | no |
| ZNF606 | −1.892954092 | 0.003339793 | Yes |
| C6orf136 | −1.884378467 | 0.002680209 | Yes |
| MIR663B | −1.880005391 | 0.006120389 | no |
| CLDN20 | −1.854101267 | 0.006351915 | no |
| PTPRE | −1.853862355 | 0.006160045 | Yes |
| TAPBP | −1.764985627 | 0.006569879 | Yes |
| TOP2B | −1.731232787 | 0.001904384 | Yes |
| PCSK4 | −1.731028601 | 0.000191937 | no |
| KIAA1644 | −1.713535532 | 0.006150785 | Yes |
| ARRDC2 | −1.708274585 | 0.005457507 | Yes |
| RMRP | −1.599489386 | 0.003469351 | Yes |
| PLAC4 | −1.590352236 | 0.001600752 | no |
| MTMR14 | −1.566438827 | 0.004540765 | Yes |
| RGL2 | −1.562608346 | 0.003102877 | Yes |
| TIPARP-AS1 | −1.532284267 | 0.000747686 | no |
| PLEKHO1 | −1.527310688 | 0.00386381 | Yes |
| ITGA4 | −1.502769183 | 0.002712255 | Yes |
| ESPN | −1.489929407 | 0.000536184 | Yes |
| PPP1R3B | −1.484443928 | 0.00593945 | Yes |
| MCFD2 | −1.463147761 | 0.009630371 | Yes |
| GOLGA7B | −1.460005241 | 0.009801722 | Yes |
| VCL | −1.442300805 | 0.001415881 | Yes |
| NT5C3B | −1.436375137 | 0.008060946 | Yes |
| ANKRD20A4 | −1.431959897 | 0.006739324 | no |
| ACVRL1 | −1.424058578 | 0.003299645 | Yes |
| GJA4 | −1.413440473 | 0.005108807 | no |
| FBXO11 | −1.411318412 | 0.003194483 | Yes |
| CASC15 | −1.407311311 | 0.000150042 | no |
| FOXD2 | −1.396058124 | 0.001731055 | Yes |
| LINC01311 | −1.387030428 | 0.003652969 | no |
| KLHL21 | −1.376156001 | 0.001415717 | Yes |
| PSD3 | −1.365960862 | 0.003093348 | Yes |
| RASAL2-AS1 | −1.34551208 | 0.00966025 | no |
| TRIB1 | −1.336306596 | 0.008051487 | Yes |
| LINC01160 | −1.319909279 | 0.0099622 | no |
| VWCE | −1.31119374 | 0.005042249 | no |
| ZNF554 | −1.310679787 | 0.002771376 | no |
| HCN2 | −1.300708485 | 0.00084889 | Yes |
| MIR210HG | −1.30045409 | 0.006892233 | no |
| TRIM16L | 1.302986687 | 0.000129035 | no |
| HMOX1 | 1.309164138 | 0.00026656 | Yes |
| OTOGL | 1.314890882 | 0.003869475 | Yes |
| MPZL2 | 1.316672807 | 8.40E-06 | Yes |
| ADM2 | 1.436182568 | 0.003847478 | Yes |
| CCDC134 | 1.465832907 | 0.001426697 | Yes |
| PDK4 | 1.545392888 | 0.002047088 | Yes |
| SNORD139 | 1.584940011 | 0.002580409 | no |
| LOC100507670 | 1.670751157 | 0.004775702 | no |
| UBR5-AS1 | 1.744471642 | 0.004954936 | no |
| SLC6A17 | 1.751169685 | 0.008800857 | Yes |
| NR4A2 | 1.969010915 | 0.001416847 | Yes |
| CDCA7 | 2.099733467 | 0.003238882 | Yes |
| C4orf36 | 2.128247182 | 0.00275716 | Yes |
abased on the prediction of Target Scan
Fig. 4.
Unsupervised hierarchical clustering analysis of gene expression for the 59 differentially expressed genes between JDM and HC. The heatmap shows the median-normalized expression of individual genes across all samples. Heatmap colors represent relative mRNA expression as indicated in the color key
We next asked if the genes that showed differential expression in HAEC after incubation with exosomes could be grouped by identifiable biological functions. We used gene ontology features of Biological Process Ontology from GSEA (http://software.broadinstitute.org/gsea/msigdb/index.jsp) to identify specific biological pathways that are altered in HAEC incubated with JDM exosomes. The results indicated that, although no biological functions were linked to the DEGs that showed higher expression in HAEC incubated with JDM exosomes, DEGs with that showed lower expression were associated with a broad range of physiologic functions. Those include cell migration, intercellular junction assembly, cytoplasm organization and biogenesis, and cell-cell adhesion (Additional file 2: Table S2).
Correlation between miRNA cargoes and patterns of gene expression in HAEC
We next looked for evidence that the miRNAs cargoes of JDM exosomes have direct impact on gene expression within HAEC cells. We first undertook an analysis to determine whether the DEGs identified in the HAEC RNAseq experiments were enriched for target genes of the differentially expressed miRNAs. No significant enrichment was observed, indicating that individual miRNAs have no significant impact on the expression of these DEGs.
However, individual miRNAs can mildly down-regulate hundreds of targets by direct or indirect effects, providing a mechanism of fine-tuning for endothelial gene expression. Furthermore, individual genes can be the targets for multiple miRNAs. Our previous studies have shown that miRNA binding sites have an additive effect on mRNA stability [21]. In our previous studies we demonstrated that the number of miRNAs binding to specific transcripts influences the mRNA decay rate in those miRNA target genes. We found mRNA degradation was enhanced through an additive effect from multiple miRNA targeting [17].
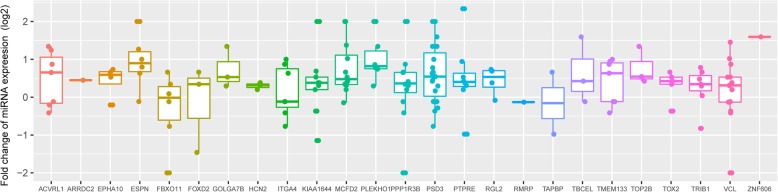
We therefore undertook a second analysis to determine whether the pattern of DEG expression identified in HAEC incubated with JDM vs HC exosomes might reflect gene regulation by multiple miRNAs instead of the effects of individual miRNAs on individual genes. As shown in Fig. 5, for genes that showed lower expression in HAEC incubated with exosomes from children with JDM (compared to exosomes from HC), this is indeed the case. The majority (22 out of 25) of the DEGs have corresponding up-regulated miRNA abundance in JDM exosomes compared to exosomes from HC.
Fig. 5.
Distribution of fold changes between JDM and HC for miRNAs (each dot represents one miRNA) targeting the corresponding DEGs, which are listed on the x axis. Fold change above zero stands for up-regulation of miRNAs in JDM, when compared to their expression in HC
We did not observe negative association between up-regulated DEGs and corresponding miRNAs (data not shown). Since the majority of the DEGs exhibit lower expression and control a broader range of physiologic functions, the findings indicate that miRNAs from the exosomes of children with with JDM, when taken up by HAEC, impact gene expression within those cells in an additive manner.
Discussion
Vascular and perivascular inflammation of the skin and muscles are the most prominent features of the pathology of JDM and are like to be directly related to the tissue injury and dysfunction that characterize the clinical disease [22]. Indeed, one of the first comprehensive English language descriptions of the clinical features and natural history of the entity we call “juvenile dermatomyositis” referred to the illness as “systemic angiopathy” of childhood. How and why the vasculature of the skin and muscle (and other organs) become targets of the brisk immune/inflammatory response remains one of the most important unanswered questions with regard to this disease.
Exosomes are one of several families of microparticles that are released from cells either after specific stimuli or during cellular apoptosis. We are now coming to understand that these subcellular components contain small, non-coding RNA molecules that form a previously unrecognized level of transcriptional control in mammals and possibly in simpler organisms [9]. The RNA molecules contained within exosomes and other microparticles are, among other things, a mechanism through which cells of the immune system communicate with one-another [10]. These microparticles are abundant in serum and plasma and have been a source of considerable interest as biomarkers in a broad range of diseases [11–13].
In this paper, we show that exosomes purified from the plasma of children with untreated JDM carry different small RNA cargoes compared to those purified from healthy control children’s plasma. These exosomes can be taken up by HAEC and are associated with altered gene expression in those cells. Furthermore, endothelial cell genes whose expression is altered by these exosomes regulate basic and essential functions for endothelial cells, including include cell migration, intercellular junction assembly, cell-cell adhesion, and cytoplasm organization and biogenesis. Furthermore, computational analysis that incorporates the additive effects of multiple miRNA on gene expression [17] support the idea that the transcriptional effects are due to the RNA cargoes within the JDM exosomes.
There are several limitations to this study that need to be considered in interpreting the data. The first is the small number of samples studied. While there was considerable homogeneity in the RNA cargoes found in the plasma exosomes of healthy children (Fig. 2), there was considerable inter-subject variability within the JDM exosome samples. This finding is consistent with the broad range of clinical phenotypes subsumed within the single disease entity, JDM. This limitation prevents us from making broad generalizations regarding the full range of miRNA cargoes within JDM exosomes or associating specific miRNA with specific clinical phenotypes. However, we believe that despite the small numbers, these findings are sufficient to establish that there are real differences in the miRNA cargoes of exosomes derived from children with with JDM. Furthermore, the biological effects of these exosomes, as reflected in the transcriptomes of HAEC after they have taken up the exosomes, are quite uniform (Fig. 4) and demonstrate cogent biological effects. We believe that further efforts to categorize the small RNA content of JDM exosomes and to relate specific small RNAs to particular clinical features could provide useful clinical and pathological insights.
A second limitation of the study is the cells that we used. We chose AEC because we had used them in our earlier work examining anti-endothelial cell antibodies (AECA) in JDM [7]. However, JDM pathology is most prominent in small arterial vessels [22], although larger vessels can be involved. It is possible that the patterns of gene expression would have differed if we had used a different cell line.
Despite these limitations, we believe that these simple experiments provide the basis for further inquiries into the mechanisms leading to the targeting of blood vessels by immune/inflammatory cells in JDM. Understanding this process better can be expected to provide the foundation for new therapeutic strategies.
Conclusions
The RNA cargoes carried by plasma exosomes of children with JDM differ from those found in healthy children. These exosomes are taken up by endothelial cells and influence the transcriptional program in these cells. These findings suggest a previously unsuspected mechanism underlying the targeting of the vasculature in JDM.
Additional files
Table S1. Small RNAs Showing Differential Abundance in Plasma Exosomes of Children With Polyarticular JIA. (DOCX 20 kb)
Table S2. Ontology analysis for down- and up regulated DEGs. (DOCX 18 kb)
Acknowledgements
None
Abbreviations
- DEGs
Differentially expressed genes
- HAEC
Human aortic endothelial cells
- HC
Healthy controls
- JDM
Juvenile dermatomyositis
- RNAseq
RNA sequencing
Authors’ contributions
KJ: Assisted in preparing and characterizing exosomes. Performed the experiments with human aortic endothelial cells. RK: Assisted in developing the concept for these experiments. Assisted in purifying and characterizing exosomes. Provided technical advice in culturing endothelial cells. ZH: Performed computational analysis of miRNA and mRNA sequencing data. YC: Assisted in performance of the endothelial cell experiments. LH: Assisted in acquiring samples from healthy controls and establishing appropriate inclusion and exclusion criteria for control samples. KOM: Assisted in acquiring patient samples and establishing clinical phenotype. JNJ: Designed the study in collaboration with RK, directed the study and assisted with data analysis and interpretation. All authors read and approved the final manuscript.
Funding
This work was supported by a grant from the Cure JM Foundation (to JNJ). This work was also the National Center for Advancing Translational Sciences of the National Institutes of Health under award number UL1TR001412 to the University at Buffalo. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. This work was also supported by JSPS KAKENHI Grant Number JP 16 K10045 (to RK).
Availability of data and materials
We are now preparing both the mRNAseq and small RNAseq data for public availability on the Gene Expression Omnibus (GEO) website.
Ethics approval and consent to participate
Patient samples were acquired under the auspices of Institutional Review Board (IRB)-approved protocols at the University of Oklahoma Health Sciences Center and the University at Buffalo Jacobs School of Medicine and Biomedical Sciences. Informed consent was obtained from the parents of the participating subjects and, where required, assent was also obtained from the subjects themselves.
Consent for publication
All patients/parents consented to publication of data in de-identified form.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Kaiyu Jiang and Rie Karasawa contributed equally to this work.
Contributor Information
Kaiyu Jiang, Email: kaiyujia@buffalo.edu.
Rie Karasawa, Email: r2karasawa@marianna-u.ac.jp.
Zihua Hu, Email: zihuahu@buffalo.edu.
Yanmin Chen, Email: yanminch@buffalo.edu.
Lucy Holmes, Email: luholmes@buffalo.edu.
Kathleen M. O’Neil, Email: kmoneil@iu.edu
James N. Jarvis, Email: jamesjar@buffalo.edu
References
- 1.Spencer-Green G, Crowe WE, Levinson JE. Nailfold capillary abnormalities and clinical outcome in childhood dermatomyositis. Arthritis Rheum. 1982;25(8):954–958. doi: 10.1002/art.1780250807. [DOI] [PubMed] [Google Scholar]
- 2.Crowe WE, Bove KE, Levinson JE, Hilton PK. Clinical and pathogenetic implications of histopathology in childhood polydermatomyositis. Arthritis Rheum. 1982;25(2):126–139. doi: 10.1002/art.1780250203. [DOI] [PubMed] [Google Scholar]
- 3.Banker BQ, Victor M. Dermatomyositis (systemic angiopathy) of childhood. Medicine (Baltimore) 1966;45(4):261–289. doi: 10.1097/00005792-196607000-00001. [DOI] [PubMed] [Google Scholar]
- 4.Greenberg SA. Dermatomyositis and type 1 interferons. Curr Rheumatol Rep. 2010;12(3):198–203. doi: 10.1007/s11926-010-0101-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaplan MJ, Salmon JE. How does interferon-alpha insult the vasculature? Let me count the ways. Arthritis Rheum. 2011;63(2):334–336. doi: 10.1002/art.30161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Paepe B. Vascular changes and perifascicular muscle fiber damage in dermatomyositis: another question of the chicken or the egg that is on our mind. Ann Transl Med. 2017;5(1):22. doi: 10.21037/atm.2016.12.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karasawa R, Tamaki M, Sato T, Tanaka M, Nawa M, Yudoh K, Jarvis JN. Multiple target autoantigens on endothelial cells identified in juvenile dermatomyositis using proteomics. Rheumatology (Oxford) 2018;57(4):671–676. doi: 10.1093/rheumatology/kex468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Distler JH, Pisetsky DS, Huber LC, Kalden JR, Gay S, Distler O. Microparticles as regulators of inflammation: novel players of cellular crosstalk in the rheumatic diseases. Arthritis Rheum. 2005;52(11):3337–3348. doi: 10.1002/art.21350. [DOI] [PubMed] [Google Scholar]
- 9.Tewari M. A functional extracellular transcriptome in animals? Implications for biology, disease and medicine. Genome Biol. 2015;16:47. doi: 10.1186/s13059-015-0613-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robbins PD, Morelli AE. Regulation of immune responses by extracellular vesicles. Nat Rev Immunol. 2014;14(3):195–208. doi: 10.1038/nri3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jia S, Zocco D, Samuels ML, Chou MF, Chammas R, Skog J, Zarovni N, Momen-Heravi F, Kuo WP. Emerging technologies in extracellular vesicle-based molecular diagnostics. Expert Rev Mol Diagn. 2014;14(3):307–321. doi: 10.1586/14737159.2014.893828. [DOI] [PubMed] [Google Scholar]
- 12.Properzi F, Logozzi M, Fais S. Exosomes: the future of biomarkers in medicine. Biomark Med. 2013;7(5):769–778. doi: 10.2217/bmm.13.63. [DOI] [PubMed] [Google Scholar]
- 13.Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Jr, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10(12):1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang K, Sun X, Chen Y, Shen Y, Jarvis JN. RNA sequencing from human neutrophils reveals distinct transcriptional differences associated with chronic inflammatory states. BMC Med Genet. 2015;8:55. doi: 10.1186/s12920-015-0128-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.An J, Lai J, Lehman ML, Nelson CC. miRDeep*: an integrated application tool for miRNA identification from RNA sequencing data. Nucleic Acids Res. 2013;41(2):727–737. doi: 10.1093/nar/gks1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:3. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 17.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 21.Hu Z. Insight into microRNA regulation by analyzing the characteristics of their targets in humans. BMC Genomics. 2009;10:594. doi: 10.1186/1471-2164-10-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wargula JC, Lovell DJ, Passo MH, Bove KE, Santangelo JD, Levinson JE. What more can we learn from muscle histopathology in children with dermatomyositis/polymyositis? Clin Exp Rheumatol. 2006;24(3):333–343. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Small RNAs Showing Differential Abundance in Plasma Exosomes of Children With Polyarticular JIA. (DOCX 20 kb)
Table S2. Ontology analysis for down- and up regulated DEGs. (DOCX 18 kb)
Data Availability Statement
We are now preparing both the mRNAseq and small RNAseq data for public availability on the Gene Expression Omnibus (GEO) website.