Abstract
Combining the anti-angiogenic agent cediranib with the poly(ADP-ribose) polymerase (PARP) inhibitor olaparib improves progression-free survival compared to olaparib alone in ovarian cancer patients through an unknown mechanism. PARP inhibitors are used primarily in the treatment of patients with DNA repair-associated (BRCA1/2) mutated cancers because these mutations cause a deficit in homology-directed DNA repair (HDR) that confers sensitivity to these agents. However, the combination of cediranib and olaparib was effective in patients without BRCA1/2 mutations. We report here that cediranib confers sensitivity to olaparib by down-regulating HDR in tumor cells. This occurs partially as a result of cediranib inducing hypoxia, which suppresses expression of the HDR factors BRCA1/2 and RAD51 recombinase (RAD51). However, we also observed that cediranib has a direct effect on HDR independent of its ability to induce tumor hypoxia. This direct effect occurs through platelet-derived growth factor receptor (PDGFR) inhibition, activation of protein phosphatase 2A (PP2A), and E2F transcription factor 4 (E2F4)/RB transcriptional corepressor like 2 (RB2/p130)-mediated repression of BRCA1/2 and RAD51 gene expression. This down-regulation was seen in mouse tumor xenografts but not in mouse bone marrow, providing a therapeutic window for combining cediranib and olaparib in cancer therapy. Our work reveals a treatment strategy by which DNA repair can be manipulated in human tumors to induce synthetic lethality, broadening the potential therapeutic scope of cediranib based on its activity as a DNA repair inhibitor.
INTRODUCTION
A recent clinical trial in ovarian cancer found improvements in progression-free and overall survival when an anti-angiogenic agent, cediranib (AZD2171), was combined with the poly(adenosine 5’-diphosphate-ribose) polymerase (PARP) inhibitor olaparib, compared to olaparib alone, in patients with recurrent ovarian cancer (1, 2). PARP inhibitors are specifically used in the treatment of BRCA mutation-associated breast and ovarian cancers (3, 4), because the homology-directed DNA repair (HDR) deficit conferred by these mutations renders tumor cells sensitive to this treatment (5, 6). However, the subset of patients in this trial without documented BRCA1/2 mutations showed superior progression-free and overall survival in the combined olaparib/cediranib arm. The basis of this unexpected result remains unclear. We report here that cediranib directly inhibits HDR, thereby conferring synthetic lethality with olaparib.
Cediranib is an inhibitor of the vascular endothelial growth factor receptor (VEGFR) (7), developed with the goal of blocking tumor angiogenesis. Accordingly, acute cediranib treatment decreases tumor perfusion and consequently increases tumor hypoxia (8, 9). Earlier work has demonstrated that hypoxia inhibits HDR in tumors through several mechanisms, including reducing gene expression of the HDR factors BRCA1 and RAD51 (10, 11), epigenetic silencing of the BRCA1 promoter (12), and production of the metabolite S-2-hydroxyglutarate (S-2HG), which functionally inhibits HDR (13). By suppressing HDR, hypoxia may render tumor cells sensitive to PARP inhibitors (14), inducing synthetic lethality in the tumor microenvironment (15).
We initially hypothesized that the clinical success of combining cediranib and olaparib was due to cediranib inducing synthetic lethality in wild-type tumor cells by increasing tumor hypoxia. To test this, we used human tumor models in mice, which allowed us to quantify and separately isolate hypoxic and normoxic tumor cells from stromal cells in tumor xenografts. Although, as expected, cediranib induced a small increase in tumor hypoxia, it strongly suppressed the expression of HDR factors in the overall tumor cell population. Because of this discrepancy, we evaluated for additional effects of cediranib on HDR independent of its ability to cause tumor hypoxia. We found that cediranib reduced the expression of HDR factors even in normoxic tumor cells. Furthermore, in cell culture studies under normoxic conditions, cediranib decreased the expression of several HDR factors, functionally impaired HDR, and increased cellular sensitivity to PARP inhibition. Mechanistically, we present evidence that the impact of cediranib on HDR can be attributed to inhibition of the platelet-derived growth factor receptors (PDGFRs) and occurs via activation of protein phosphatase 2A (PP2A), resulting in suppression of HDR gene expression via promoter occupancy by the repressive transcription regulatory complex E2F transcription factor 4 (E2F4)/p130. These findings identify a pathway by which cediranib can alter the DNA repair capacity of cancer cells that has implications for the design of cancer therapies.
RESULTS
Cediranib induces hypoxia and down-regulates expression of HDR factors in tumors
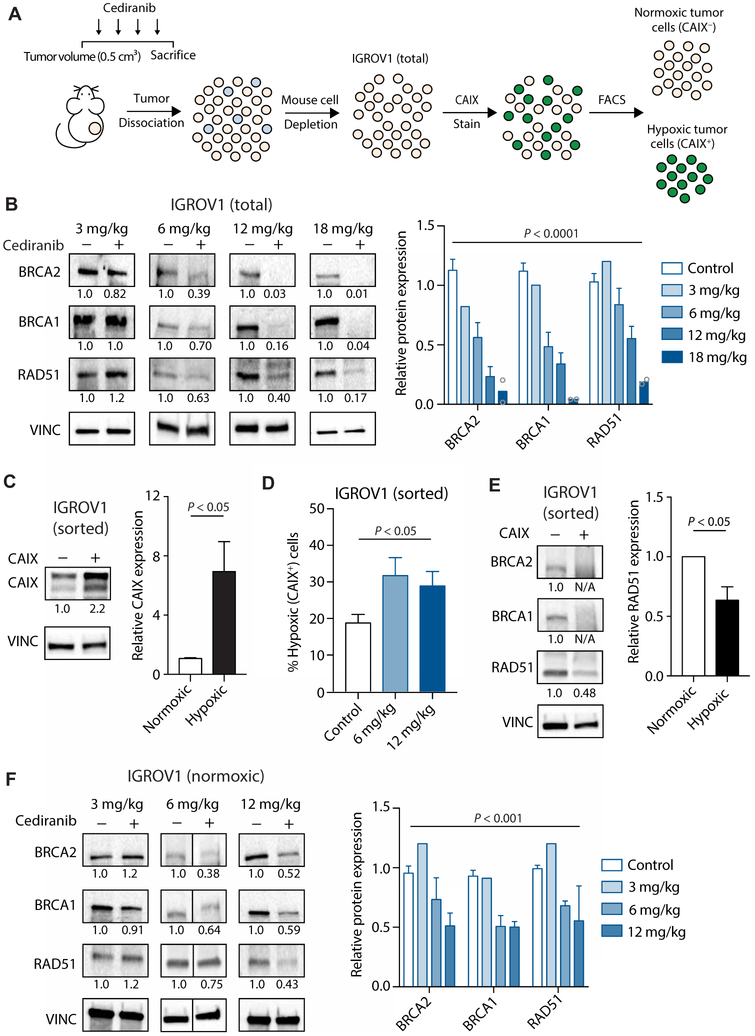
To evaluate the effects of cediranib on DNA repair in tumors in vivo, we established tumor xenografts in mice using the human ovarian cancer cell line IGROV1. The mice were treated or not with cediranib, and tumors were harvested for analysis. We isolated IGROV1 cells from the tumor stroma (Fig. 1A) and assayed for expression of DNA repair factors. We noted a decrease in the expression of HDR factors BRCA1, BRCA2, and RAD51 in the cediranib-treated groups compared to controls (Fig. 1B). To determine whether this down-regulation could be explained by an increase in tumor hypoxia, we isolated hypoxic versus normoxic tumor cells from the IGROV1 xenografts using fluorescence-activated cell sorting (FACS) based on carbonic anhydrase IX (CAIX) expression, a marker of hypoxia (Fig. 1, A and C). We found that cediranib treatment increased the percentage of CAIX+ tumor cells (Fig. 1D). However, the relative increase in hypoxia was mild as compared with the extent of suppression of HDR factor expression. We then assayed HDR factor expression in separately isolated hypoxic and normoxic tumor cells from cediranib-treated and control tumors. As expected, reduced expression of HDR factors was observed in hypoxic cells (CAIX+) compared to the normoxic cells (Fig. 1E). Surprisingly, the normoxic cells from the cediranib-treated tumors showed lower expression of HDR factors compared to the normoxic cells from untreated control tumors (Fig. 1F), raising the possibility that cediranib may affect DNA repair gene expression in a manner independent of its ability to induce hypoxia.
Fig. 1. Cediranib down-regulates HDR factor expression in ovarian tumor xenografts in both hypoxia-dependent and hypoxia-independent pathways.
(A) Schematic of mouse treatment, isolation of human IGROV1 cancer cells from mouse stroma, and FACS-based separation of hypoxic and normoxic cells from IGROV1 tumors. (B) Western blot of HDR factors in IGROV1 ovarian cancer xenograft tumors in mice treated with vehicle or the indicated doses of cediranib [two-way analysis of variance (ANOVA), effect of cediranib: P < 0.0001; control, n = 14; 3 mg/kg, n = 2 (pooled to one sample); 6 mg/kg, n = 9; 12 mg/kg, n = 3; 18 mg/kg, n = 2 tumors]. (C) Western blot of CAIX in sorted normoxic (CAIX−) and hypoxic (CAIX+) cells isolated from IGROV1 tumors (unpaired t test, P < 0.05; n = 5 tumors). (D) Percentage of hypoxic tumor cells after cediranib treatment (one-way ANOVA, P < 0.05; control, n = 14; 6 mg/kg, n = 10; 12 mg/kg, n = 3 tumors). (E) Western blot of HDR factors in sorted normoxic and hypoxic cells isolated from IGROV1 tumors (unpaired t test, P < 0.05; n = 3 tumors). (F) Western blot of HDR factors in sorted normoxic cells isolated from IGROV1 tumors in control [dimethyl sulfoxide (DMSO)] and cediranib-treated mice at the indicated doses [two-way ANOVA, effect of cediranib: P < 0.001; control, n = 9; 3 mg/kg, n = 2 (pooled to one sample); 6 mg/kg, n = 5; 12 mg/kg, n = 3 tumors]. Data are represented as means ± SEM. Numbers below Western blot panels represent relative quantification of the respective bands normalized to loading control by densitometry.
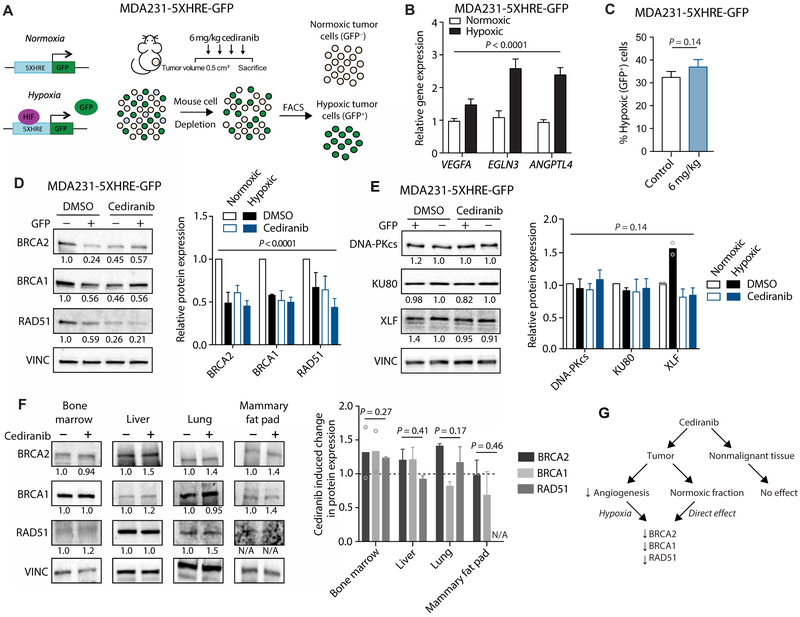
To confirm these results, we used a different tumor xenograft system that allows for FACS isolation of hypoxic and normoxic tumor cells based on hypoxia-induced green fluorescent protein (GFP) expression. The human breast adenocarcinoma cell line MDA-MB-231 was engineered to stably express a construct containing the GFP gene driven by a minimal promoter with five copies of the canonical hypoxia response element (MDA231-5XHRE-GFP). Stabilization of hypoxia-inducible factor–1α (HIF-1 α) by hypoxia induces GFP expression (Fig. 2A). FACS was used to isolate GFP+ (hypoxic) and GFP− (normoxic) MDA231-5XHRE-GFP tumor cells from xenografts (Fig. 2, A and B). Cediranib treatment did not appreciably change the proportions of hypoxic (GFP+) tumor cells (Fig. 2C), but it down-regulated HDR factor expression in normoxic cells (Fig. 2D). This effect appears to be exclusive to the HDR pathway, because the expression of factors necessary for non-homologous end joining (NHEJ), the other major pathway for DNA double-strand break (DSB) repair, remained largely unchanged after cediranib treatment (Fig. 2E).
Fig. 2. Cediranib down-regulates HDR factor expression in breast tumor xenografts in a hypoxia-independent pathway, with no detectable effect on nonmalignant tissue.
(A) Schematic of MDA231-5XHRE-GFP cell xenograft model, cediranib treatment (6 mg/kg), and FACS-based isolation of hypoxic (GFP+) and normoxic (GFP−) cells based on GFP expression. (B) Relative expression of hypoxia-associated genes assayed by qRT-PCR in sorted cell populations isolated from MDA231-5XHRE-GFP tumors (two-way ANOVA, effect of hypoxia: P < 0.0001; n = 7 tumors per group). (C) Percentage of hypoxic tumor cells after cediranib treatment compared to control (DMSO) in MDA231-5XHRE-GFP tumors, as determined by flow cytometry (unpaired t test, P = 0.14; control, n = 11; cediranib, n = 12 tumors). (D) Western blot of HDR factors in sorted cells (normoxic versus hypoxic) isolated from MDA231-5XHRE-GFP tumors in control (DMSO) and cediranib-treated mice (two-way ANOVA, P < 0.0001; DMSO normoxic versus DMSO hypoxic, P < 0.001; DMSO normoxic versus cediranib normoxic, P < 0.001; DMSO, n = 3; cediranib, n = 4 tumors). (E) Western blot of NHEJ factors in tumors from previous panel (two-way ANOVA, P = 0.14; DMSO normoxic versus DMSO hypoxic, P = 0.94; DMSO normoxic versus cediranib normoxic, P = 0.97). (F) Western blot of HDR factors in selected tissues from control and cediranib-treated mice (two-way ANOVA, effect of cediranib: bone marrow, P = 0.27, n = 2 mice; liver, P = 0.41; lung, P = 0.17; mammary fat pad, P = 0.46, n = 3 mice per group). (G) Proposed pathway of dual regulation of tumor HDR gene expression by cediranib compared to a lack of effect on nonmalignant tissue. Data are represented as means ± SEM. Numbers below Western blot panels represent relative quantification of the respective bands normalized to loading control by densitometry.
We then assessed the effect of cediranib treatment on HDR factor expression in normal tissues isolated from cediranib-treated mice. We were concerned about the potential effects of cediranib in the bone marrow, because olaparib is known to cause bone marrow toxicity on its own (16). However, cediranib treatment had no effect on HDR factor expression in bone marrow cells (Fig. 2F) or in other normal tissues, including liver, lung, and mammary fat pad (Fig. 2F). Together, these data suggest that cediranib down-regulates HDR in tumor cells in both a hypoxia-dependent fashion (secondary to reduced angiogenesis) and a hypoxia-independent fashion (a direct effect) but with no corresponding effect on HDR gene expression in nonmalignant, healthy tissue (Fig. 2G).
Cediranib directly suppresses HDR factor expression in cell culture
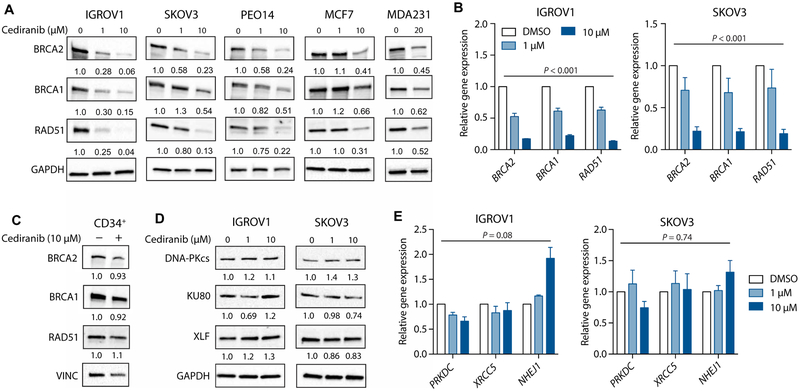
To further evaluate potential hypoxia-independent effects of cediranib on DNA repair, we tested the effects of cediranib on cells in culture. Under standard normoxic culture conditions, we found that cediranib down-regulates the expression of HDR factors in the IGROV1 ovarian cancer cell line in a dose-dependent fashion (Fig. 3A and fig. S1A). These results were confirmed in two additional ovarian cancer cell lines, SKOV3 and PEO14, as well as in breast cancer cell lines, MCF7 and MDA-MB-231 (Fig. 3A and fig. S1A). We observed similar effects at the mRNA level, as assessed by quantitative realtime polymerase chain reaction (qRT-PCR) (Fig. 3B). The effects of cediranib on HDR factor expression are independent of BRCA status, because cediranib reduces BRCA1 and RAD51 expression in both the BRCA2-mutant PEO1 and the BRCA2-restored PEO4 ovarian cancer cell line derived from the same patient (fig. S1B) (17). Cediranib only minimally suppressed HDR factor expression in cultured primary human CD34+ hematopoietic stem cells (Fig. 3C and fig. S1C). In addition, as observed in our in vivo tumor experiments (Fig. 2E), the expression of NHEJ factors remained largely unchanged after cediranib treatment at the protein (Fig. 3D and fig. S1D) and mRNA (Fig. 3E) levels in the IGROV1 and SKOV3 cells. To test for the possibility of a secreted factor causing suppression of HDR factor expression, we treated cells with conditioned medium from control and cediranib-treated cells and found no effect on HDR factor expression in either case (fig. S2).
Fig. 3. Cediranib suppresses expression of HDR factors in cancer cells in culture.
(A) Western blot of HDR factors in ovarian (IGROV1, SKOV3, and PEO14) and breast (MCF7 and MDA231) cancer cell lines treated with increasing doses of cediranib in culture (n = 3 independent experiments). (B) Relative gene expression assayed by qRT-PCR of HDR factors in IGROV1 and SKOV3 cells treated with cediranib in culture (two-way ANOVA, effect of cediranib: IGROV1, P < 0.001; SKOV3, P < 0.001; n = 3 independent experiments). (C) Western blot of HDR factors in primary human CD34+ hematopoietic stem/progenitor cells treated with cediranib in culture compared to control (n = 3 technical replicates). (D) Western blot of NHEJ factors in IGROV1 and SKOV3 cells treated with increasing doses of cediranib in culture (n = 3 independent experiments). (E) Relative gene expression of NHEJ factors as measured by qRT-PCR in IGROV1 and SKOV3 cells treated with cediranib in culture (two-way ANOVA, effect of cediranib: IGROV1, P = 0.08; SKOV3, P = 0.74; n = 3 independent experiments). Data are represented as means ± SEM. Numbers below Western blot panels represent relative quantification of the respective bands normalized to loading control by densitometry.
Cediranib reduces HDR activity and increases sensitivity to PARP inhibitors
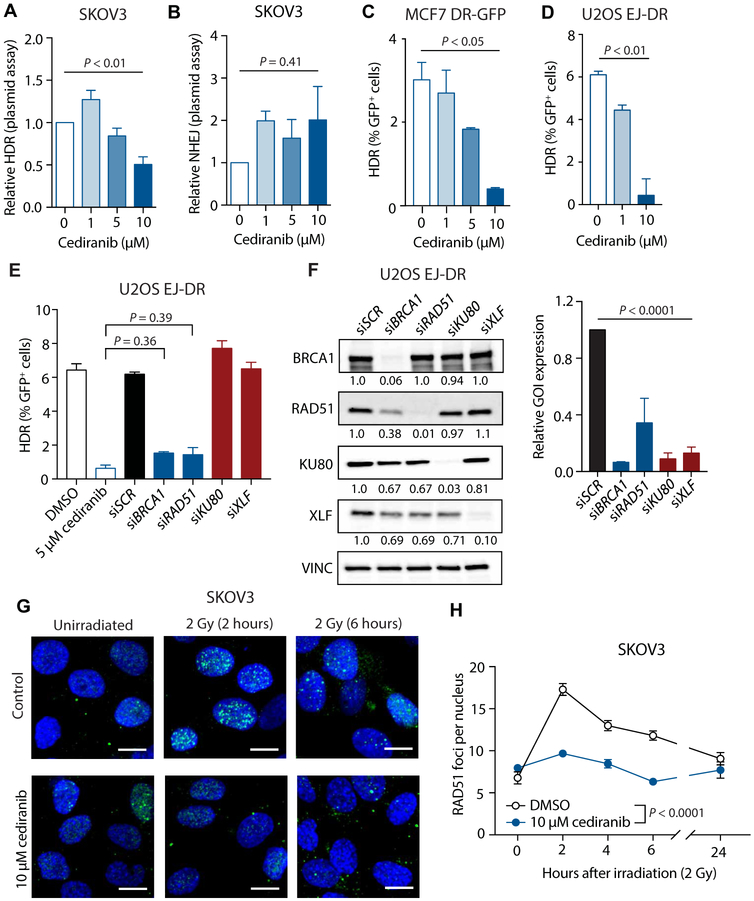
To assess the functional effects of cediranib on DNA repair, we used several DSB repair assays with specificity for either HDR or NHEJ. Using plasmid-based luciferase assays in SKOV3 cells, pretreatment with cediranib suppressed HDR but not NHEJ (Fig. 4, A and B). MCF7 DR-GFP and U2OS EJ-DR cells, two chromosomally based HDR reporter cell lines (10,18,19), were also used to assess HDR function. In both cases, treatment with cediranib markedly reduced HDR capacity (Fig. 4, C and D). Cediranib suppresses HDR to a comparable extent as small interfering RNA (siRNA)–mediated knockdown of BRCA1 and RAD51 (Fig. 4, E and F). We also observed that cediranib blunted the induction of RAD51 foci after irradiation (2 Gy) (Fig. 4, G and H), consistent with our findings that cediranib causes a functional suppression of HDR.
Fig. 4. Cediranib functionally suppresses HDR in cancer cells in culture.
(A) Relative HDR activity after cediranib treatment in SKOV3 cells as determined by luciferase plasmid reactivation assay (one-way ANOVA, P < 0.01; n = 3 independent experiments). (B) Relative NHEJ activity after treatment with increasing doses of cediranib in SKOV3 cells as determined by luciferase reactivation plasmid assay (one-way ANOVA, P = 0.41; n = 4 independent experiments). (C) HDR activity in MCF7 DR-GFP cells after cediranib treatment measured using the chromosomal DR-GFP assay (one-way ANOVA, P < 0.05; n = 3 biological replicates). (D) HDR activity in U2OS EJ-DR cells after cediranib treatment measured using the chromosomal EJ-DR assay (one-way ANOVA, P < 0.01; n = 4 technical replicates). (E) HDR activity in U2OS EJ-DR cells after treatment with DMSO, 5 μM cediranib, or siRNA-mediated knockdown of repair factors (one-way ANOVA, P < 0.0001; DMSO versus 5 μM cediranib, P < 0.0001; siSCR versus siBRCA1, P < 0.0001; siSCR versus siRAD51, P < 0.0001; 5 μM cediranib versus siBRCA1, P = 0.36; 5 μM cediranib versus siRAD51, P = 0.39; n = 3 to 4 technical replicates). (F) Western blot of HDR and NHEJ factor expression in U2OS EJ-DR cells 72 hours after treatment with repair factor siRNAs (one-way ANOVA, P < 0.0001; n = 3 technical replicates). (G) Representative images of RAD51 foci induced by irradiation (2 Gy) in SKOV3 cells treated with or without 10 μM cediranib [green, RAD51; blue, 4’,6-diamidino-2-phenylindole (DAPI)]. Scale bars, 10 μm. (H) Number of RAD51 foci per nucleus induced by irradiation (2 Gy) in SKOV3 cells treated with or without 10 μM cediranib (two-way ANOVA, interaction P < 0.0001; n = 3 technical replicates). Data are represented as means ± SEM. Numbers below Western blot panels represent relative quantification of the respective bands normalized to loading control by densitometry.
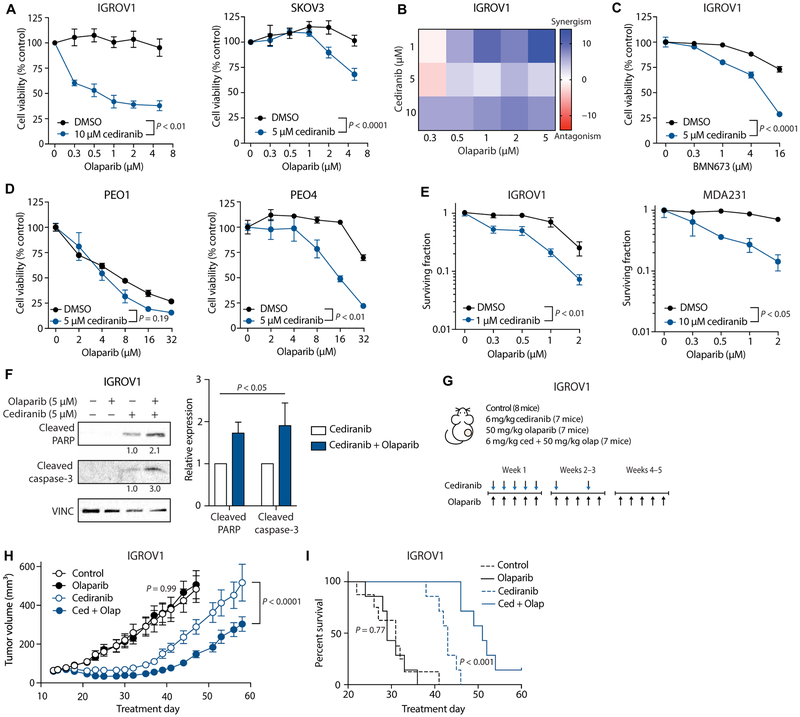
We then tested for cediranib-induced PARP sensitivity in the absence of hypoxia. Olaparib treatment reduced short-term cell viability as measured by an adenosine 5’-triphosphate (ATP)–based assay in cediranib-treated cells but not in control IGROV1 and SKOV3 cells (Fig. 5A). A Bliss analysis of potential synergy found that the combination of cediranib and olaparib treatment was synergistic at several doses (Fig. 5B). These results are not specific to olaparib, because cediranib also sensitized cells to another PARP inhibitor, BMN673 (talazoparib) (Fig. 5C). Whereas cediranib did not further enhance the olaparib sensitivity observed in the already BRCA2-deficient PEO1 cells, it did sensitize the BRCA2-proficient PEO4 cells to olaparib (Fig. 5D), providing evidence that the PARP inhibitor sensitivity observed in cediranib-treated cells is due to a suppression of HDR. Consistent with our short-term cell viability assay, in a long-term clonogenic survival assay, cediranib treatment sensitized IGROV1 and MDA231 cells to increasing doses of olaparib (Fig. 5E). Furthermore, in IGROV1 cells, combined cediranib and olaparib treatment caused an increase in the expression of the apoptotic markers cleaved PARP and cleaved caspase-3 as compared with cediranib alone (Fig. 5F).
Fig. 5. Cediranib increases cancer cell sensitivity to PARP inhibition.
(A) Cell viability (as measured by ATP-based CellTiter-Glo assay) of IGROV1 and SKOV3 cells treated with or without 5 or 10 μM cediranib, as indicated, and increasing doses of olaparib (IGROV1: two-way ANOVA, interaction P < 0.01, n = 3 biological replicates; SKOV3: two-way ANOVA, interaction P < 0.0001, n = 5 biological replicates). (B) Bliss synergism analysis of IGROV1 cells treated with varying doses of cediranib and olaparib. (C) Cell viability of IGROV1 cells treated with or without 5 μM cediranib, as indicated, and increasing doses of BMN673 (two-way ANOVA, interaction P < 0.0001; n = 3 technical replicates). (D) Cell viability of PEO1 (BRCA2-deficient) and PEO4 (BRCA2-proficient) cells treated with or without 5 μM cediranib, as indicated, and increasing doses of olaparib (PEO1: two-way ANOVA, interaction P = 0.19, n = 3 technical replicates; PEO4: two-way ANOVA, interaction P < 0.001, n = 3 technical replicates). (E) Clonogenic survival of IGROV1 and MDA231 cells treated with or without 1 or 10 μM cediranib, as indicated, and increasing doses of olaparib (IGROV1: two-way ANOVA, interaction P < 0.01, n = 6 technical replicates; MDA231: two-way ANOVA, interaction P < 0.05, n = 3 technical replicates). (F) Western blot of apoptotic markers in IGROV1 cells treated with 5 μM cediranib and/or 5 μM olaparib (two-way ANOVA, cediranib versus cediranib + olaparib, P < 0.05; n = 3 independent experiments). (G) Treatment plan for mouse tumor growth and survival study (n = 1 experiment). (H) Growth curves of IGROV1 xenograft tumors in control mice and mice treated with olaparib (50 mg/kg) and/or cediranib (6 mg/kg) (control versus olaparib: two-way ANOVA, interaction P = 0.99, control n = 8, olaparib n = 7 mice; cediranib versus cediranib + olaparib: two-way ANOVA, interaction P < 0.0001, n = 7 mice). (I) Kaplan-Meier plot of survival of mice from tumor growth experiment, with survival cutoff criteria defined as tumor volume greater than three times that at initial treatment [log-rank (Mantel-Cox) test: control versus olaparib, P = 0.77; cediranib versus cediranib + olaparib, P < 0.001]. Data are represented as means ± SEM. Numbers below Western blot panels represent relative quantification of the respective bands normalized to loading control by densitometry.
We next evaluated the effects of combined cediranib and olaparib treatment versus the respective single agents alone on the growth of IGROV1 tumor xenografts in mice (Fig. 5G). We observed that olaparib alone had no effect on either tumor growth or survival (as defined by tumor growth to three times the volume at initial treatment) compared to control (Fig. 5, H and I), whereas cediranib alone did have a detectable effect compared to control for both endpoints (Fig. 5, H and I). However, the combination of cediranib and olaparib was substantially more effective in suppressing tumor growth and improving survival compared to either inhibitor alone (Fig. 5, H and I), demonstrating in vivo sensitization to olaparib by cediranib and matching the effects observed in clinical trials. Together, the above results establish that cediranib increases tumor cell sensitivity to PARP inhibition in a direct manner independent of hypoxia and that this increased sensitivity conferred by cediranib is manifest in vivo in tumors.
Hypoxia-independent effects of cediranib on HDR are mediated by PDGFR inhibition
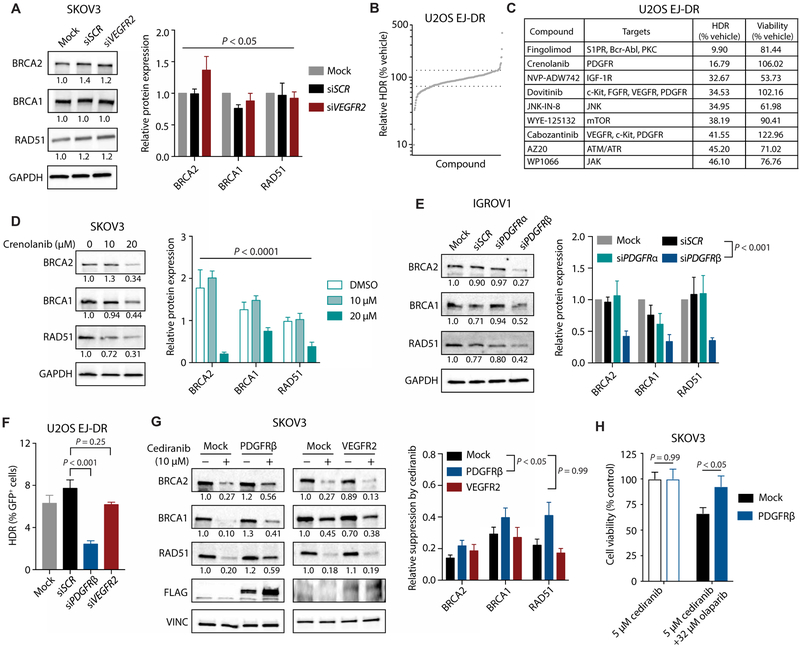
To determine the target mediating HDR inhibition by cediranib, we first evaluated the role of VEGFR2, the receptor through which cediranib inhibits angiogenesis (7). However, siRNA-mediated knockdown of VEGFR2 (fig. S3A) did not alter HDR factor expression (Fig. 6A). Furthermore, inhibition of VEGFR signaling with the small-molecule inhibitors sorafenib and Maz51 did not consistently mimic the effects of cediranib (fig. S3, B and C). Sorafenib did not reduce HDR factor expression in either cell line tested, whereas Maz51 had a modest effect in IGROV1 but not SKOV3 cells. To identify other possible targets that might be mediating this effect of cediranib, we screened more than 400 small-molecule kinase inhibitors from a commercially available kinase inhibitor library for their effects on HDR (Fig. 6, B and C) using the U2OS EJ-DR cell line (18). Several of the most potent inhibitors of HDR identified by this screen were small molecules with known activity against PDGFRs (Fig. 6C). Cediranib also inhibits PDGFRs with nanomolar potency (7). Accordingly, we found that treatment with the PDGFR inhibitors crenolanib and pazopanib suppressed the expression of HDR factors (Fig. 6D and fig. S4, A and B), mimicking the effects of cediranib. The effects of putative PDGFR inhibitors on HDR can be attributed to suppression of PDGFRβ, because siRNA-mediated knockdown of PDGFRβ (fig. S5A) also down-regulates the expression of HDR factors (Fig. 6E and fig. S5B) and functionally impairs HDR (Fig. 6F). Furthermore, overexpression of PDGFRβ partially reverses the effect of cediranib on HDR factor expression (Fig. 6G). These effects are specific to PDGFRβ overexpression, because VEGFR2 overexpression does not reverse the effects of cediranib on HDR gene expression (Fig. 6G). In addition, PDGFRβ overexpression rescues cells from the olaparib sensitivity induced by cediranib (Fig. 6H). These results implicate PDGFRβ in mediating the effects of cediranib on HDR factor expression, HDR capacity, and PARP inhibitor sensitivity.
Fig. 6. Inhibition of PDGFRβ, but not VEGFR2, suppresses HDR.
(A) Western blot of HDR factors after siRNA-mediated knockdown of VEGFR2 in SKOV3 cells (two-way ANOVA, effect of siRNA, P < 0.05; n = 3 independent experiments). (B) Relative HDR capacity in U2OS EJ-DR cells treated with a library of small-molecule kinase inhibitors. (C) List of top hits from small-molecule inhibitor screen, with relative HDR capacity and cell viability as compared with vehicle control–treated cells. (D) Western blot of HDR factors in SKOV3 cells treated with increasing doses of the PDGFR inhibitor crenolanib (two-way ANOVA, effect of crenolanib, P < 0.0001; n = 3 independent experiments). (E) Western blot of HDR factors in IGROV1 cells after siRNA-mediated knockdown of PDGFRα and PDGFRβ (two-way ANOVA, effect of siRNA, P < 0.0001; siSCR versus siPDGFRβ, P < 0.001; n = 3 technical replicates). (F) HDR activity in U2OS EJ-DR cells after siRNA-mediated knockdown of PDGFRβ and VEGFR2 (one-way ANOVA, P < 0.001; siSCR versus siPDGFRβ, P < 0.001; siSCR versus siVEGFR2, P = 0.25; n = 3 to 5 technical replicates). (G) Western blot of HDR factors in control (mock) and PDGFRβ- and VEGFR2-overexpressing SKOV3 cells treated with cediranib or not (two-way ANOVA, effect of overexpression, P < 0.01; mock versus PDGFRβ, P < 0.05; mock versus VEGFR2, P = 0.99; mock, n = 14; PDGFRβ, n = 7; VEGFR2, n = 5 biological replicates). (H) Cell viability (as measured by ATP-based CellTiter-Glo assay) of control and PDGFRβ-overexpressing SKOV3 cells treated with 5 μM cediranib and with or without 32 μM olaparib, as indicated (t test mock versus PDGFRβ: cediranib, P = 0.99; cediranib + olaparib, P < 0.05; n = 9 technical replicates). Data are represented as means ± SEM. Numbers below Western blot panels represent relative quantification of the respective bands normalized to loading control by densitometry.
Because HDR gene expression can vary by cell cycle phase, we also examined the impact of cediranib on cell cycle phase distribution as compared to other small-molecule inhibitors of PDGFR and VEGFR, and to siRNA knockdown of PDGFRs and VEGFR2 (fig. S6, A and B). However, no clear correlation emerged between the ability to suppress HDR and effect on cell cycle. For example, we found that cediranib causes a shift to G1, but crenolanib, which also suppresses HDR function and HDR gene expression, does the opposite. Furthermore, siRNA-mediated knockdown of PDGFRβ, which strongly suppresses HDR function, has no effect on the cell cycle. Hence, the effects on HDR through PDGFRs cannot be simply attributed to cell cycle perturbations.
Cediranib suppresses HDR factor gene expression through an E2F4-dependent mechanism
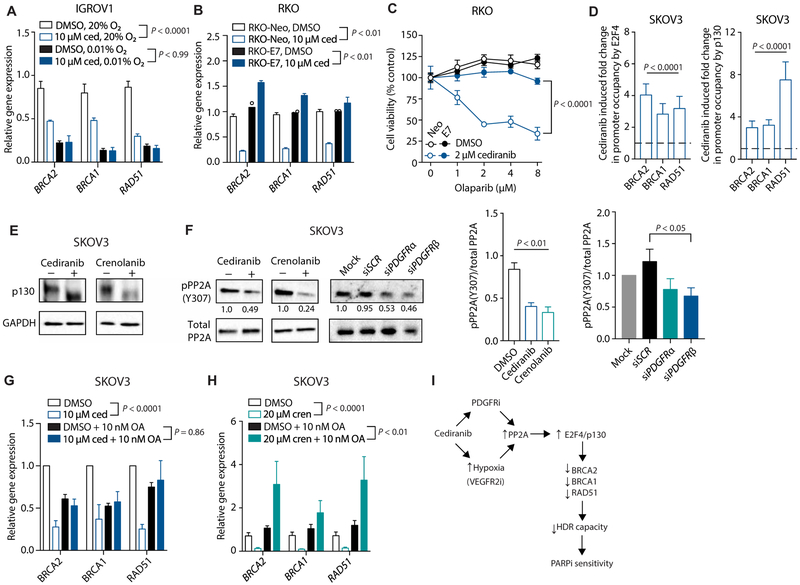
To further probe the mechanism by which cediranib suppresses HDR, we investigated the interactions between cediranib and hypoxia in regulating HDR factor expression. Whereas cediranib and hypoxia both individually suppressed HDR factor expression, the combination of cediranib with hypoxia did not cause a further suppression than hypoxia alone (Fig. 7A). This epistasis suggests that cediranib and hypoxia suppress HDR factor expression through the same mechanism.
Fig. 7. Cediranib suppresses HDR factor expression through activation of PP2A and E2F4/p130.
(A) Relative gene expression of HDR factors in IGROV1 cells treated with DMSO or 10 μM cediranib under normoxic (20% O2) or hypoxic (0.01% O2) conditions (two-way ANOVA, effect of cediranib: normoxia, P < 0.0001; hypoxia, P > 0.99; n = 3 biological replicates). (B) Relative gene expression of HDR factors in control and E7-expressing RKO cells treated with 5 μM cediranib compared to DMSO control (two-way ANOVA, effect of cediranib: RKO-Neo, P < 0.01; RKO-E7, P < 0.01; n = 2 to 3 biological replicates). (C) Cell viability (as measured by ATP-based CellTiter-Glo assay) of control and E7-expressing RKO cells treated with or without 2 μM cediranib, as indicated, and increasing doses of olaparib (two-way ANOVA, RKO-Neo 2 μM cediranib versus RKO-E7 2 μM cediranib, P < 0.0001; n = 3 technical replicates). (D) ChIP analysis of E2F4 and p130 occupancy at HDR promoters in SKOV3 cells after cediranib treatment compared to controls (two-way ANOVA, effect of cediranib: E2F4, P < 0.0001; p130, P < 0.0001; n = 5 biological replicates). (E) Western blot of p130 in SKOV3 cells after treatment with cediranib and crenolanib compared to controls (n = 3 technical replicates). (F) Western blot of phosphorylated and total PP2A in SKOV3 cells after treatment with cediranib, crenolanib, or siRNA against PDGFRα or PDGFRβ, as indicated (one-way ANOVA, inhibitors: P < 0.01; DMSO versus cediranib, P < 0.001; DMSO versus crenolanib, P < 0.001; DMSO, n = 5; crenolanib and cediranib, n = 3 technical replicates; siRNAs, P = 0.11; siSCR versus siPDGFRβ, P < 0.05; n = 3 technical replicates). (G) Relative gene expression of HDR factors as measured by qRT-PCR in SKOV3 cells treated with cediranib and/or the PP2A inhibitor okadaic acid (OA) (two-way ANOVA, effect of cediranib: DMSO, P < 0.0001; okadaic acid, P = 0.86; n = 3 biological replicates). (H) Relative gene expression of HDR factors as measured by qRT-PCR in SKOV3 cells treated with crenolanib and/or okadaic acid (two-way ANOVA, effect of cediranib: DMSO, P < 0.0001; okadaic acid, P < 0.01; n = 5 to 6 biological replicates). (I) Dual mechanisms underlying cediranib-mediated suppression of HDR in vivo in tumor cells: an indirect effect via VEGFR inhibition and induction of hypoxia and a direct effect via inhibition of PDGFR signaling, both causing phosphatase PP2A activation, formation of repressive E2F4/p130 complexes, and down-regulation of HDR gene expression, as indicated. Data are represented as means ± SEM. Numbers below Western blot panels represent relative quantification of the respective bands normalized to loading control by densitometry.
Hypoxia induces p130/E2F4 complexes that bind to E2F consensus sequences in the promoters of HDR genes, reducing gene expression (10, 11). Disruption of these complexes by the expression of human papillomavirus (HPV) E7 protein prevents this down-regulation (10). To test whether cediranib also acts through E2F4/p130 complexes, we evaluated the effects of cediranib on HDR factor expression in the presence or absence of E7. E7 expression blocked the effects of cediranib on HDR factor mRNA expression (Fig. 7B) and also blocked the synergistic effect of cediranib and olaparib on tumor cell killing (Fig. 7C).
Furthermore, after treatment with cediranib, the BRCA1 and RAD51 promoters displayed enhanced occupancy by E2F4 and p130, as measured in chromatin immunoprecipitation (ChIP) assays (Fig. 7D). Because the down-regulation of BRCA2 expression is also seen under hypoxic conditions (20) and E2F binding sites exist within the BRCA2 promoter (21), we similarly evaluated the role of E2F4/p130 in cediranib-mediated suppression of BRCA2. We found that E7 expression blocks the effect of cediranib on BRCA2 expression (Fig. 7B). Likewise, cediranib treatment enhanced BRCA2 promoter occupancy by E2F4 and p130 (Fig. 7D). Cediranib also down-regulates FANCD2 (fig. S7), whose expression is also regulated by E2F4/p130 complexes (22).
Hypoxia-induced suppression of HDR factor expression involves hypophosphorylation of p130 (11), which mediates its activation. We observed p130 hypophosphorylation, as determined by increased migration of the p130 band in SDS–polyacrylamide gel electrophoresis, in cells treated with cediranib and crenolanib (Fig. 7E and fig. S8), providing evidence of a common mechanism linking PDGFR inhibition and activation of p130/E2F4-mediated suppression of HDR gene expression.
Next, we hypothesized that cediranib may act on p130 through activation of the phosphatase PP2A. Hypoxia activates PP2A (23, 24), and the hypophosphorylation and activation of p130 that occur under hypoxic conditions are likely mediated through enhanced activity of PP2A. PP2A activity is modulated by phosphorylation of tyrosine-307 (Y307), which inactivates the enzyme (25, 26). PP2A also physically interacts with p130 (27) and plays a role in p130 activation (28). We therefore evaluated the role of PP2A activation in cediranib-mediated suppression of HDR factor expression. Treatment with cediranib reduced PP2A phosphorylation at Y307, indicating that cediranib increases PP2A activity (Fig. 7F). This dephosphorylation likely occurs due to cediranib-mediated inhibition of PDGFR signaling, because inhibition of PDGFRs through the small-molecule inhibitor crenolanib or siRNA-mediated knockdown also reduces phosphorylation of PP2A at Y307 (Fig. 7F). Furthermore, incubation of SKOV3 cells with okadaic acid, a small-molecule inhibitor of PP2A, blocks the effects of cediranib and crenolanib on HDR factor expression (Fig. 7, G and H), providing further evidence linking PDGFR inhibition, PP2A activation, and suppression of HDR factor expression. These data indicate that, by inhibiting PDGFR signaling, cediranib enhances PP2A activity and that the activation of PP2A is required for the suppression of HDR factor expression caused by cediranib.
DISCUSSION
In this study, we show that cediranib exerts a direct effect on HDR in tumor cells in vivo and in culture, independent of its ability to promote hypoxia. We present evidence that this direct effect on HDR occurs by suppressing PDGFR signaling, activating PP2A, and inducing E2F4/p130-mediated suppression of HDR factor gene expression. The effects of cediranib on DNA repair partially mimic the effects of hypoxia in that both activate PP2A and suppress gene expression through E2F4/p130 complexes. In tumor xenografts, we show that, as expected, cediranib treatment increases hypoxia, and further document that HDR gene expression is reduced in hypoxic cells isolated from tumors. We therefore propose two mechanisms by which cediranib impairs HDR in tumors: (i) a direct effect via PDGFR inhibition that suppresses DNA repair gene expression through PP2A activation and E2F4/p130 occupancy of the HDR gene promoters and (ii) an indirect effect via VEGFR2-mediated inhibition of angiogenesis, causing decreased perfusion and increased hypoxia, which secondarily suppresses HDR gene expression (Fig. 7I).
We present several lines of evidence supporting a role for PDGFR in mediating the effects of cediranib on DNA repair. We identified a group of kinase inhibitors that have a suppressive effect on HDR similar to cediranib, many of which are PDGFR inhibitors. Cediranib inhibits PDGFRα and PDGFRβ with IC50’s (median inhibitory concentrations) of 36 and 5 nM, respectively (7). Hence, PDGFR is a plausible target for cediranib at the doses used in the ovarian cancer clinical trial discussed above as well as in the work reported here. We show that siRNA knockdown of PDGFRβ, but not of VEGFR2, decreases HDR activity and expression of HDR factors, matching the effects of cediranib. We further show that the forced overexpression of PDGFRβ partially reverses the effects of cediranib. We also show that the activation of PP2A is a required step in the pathway by which cediranib down-regulates HDR factors. Together, these findings place PDGFRs in the pathway by which cediranib suppresses HDR. Although cediranib has been advanced to the clinic as an anti-angiogenic agent inhibiting VEGFR, its additional activity against PDGFR may further contribute to its clinical utility by establishing cediranib as a DNA repair inhibitor.
In keeping with the suppression of HDR, we also find that cediranib increases sensitivity to PARP inhibition, providing the basis for combining cediranib with olaparib even in the setting of patients with BRCA1/2 wild-type tumors. The dual effects of cediranib on HDR that we have elucidated also provide a mechanistic explanation for the improved outcomes reported in patients with recurrent ovarian cancer treated with cediranib plus olaparib versus olaparib alone (1). Furthermore, our findings broaden the implications of this clinical trial and expand the potential clinical scope of cediranib.
The ability of cediranib to inhibit DNA repair appears to be specific to tumor cells, because no effect of cediranib on DNA repair gene expression was seen in several normal mouse tissues in vivo (including bone marrow). This differential effect may allow for a therapeutic window for the clinical use of the combination of cediranib and olaparib, because the dose-limiting toxicity of PARP inhibitors is bone marrow suppression (16).
One potential limitation to this study is that it did not explore how variations in PDGFR expression or mutations in PDGFRs, PP2A, pocket proteins, or E2Fs may impact the effects of cediranib on HDR in cancer cells. Further, in vitro synergism was observed at micromolar concentrations of olaparib and not at the nanomolar concentrations that induce synthetic lethality in BRCA mutant cells (5, 6). However, clinical dosing of olaparib yields plasma concentrations in the micromolar range (29), supporting the relevance of these findings for cancer therapeutics.
There is growing evidence of therapeutic and other exogenous compounds regulating DNA repair, including the ability of androgen receptor inhibitors to inhibit HDR in castration-resistant prostate cancer (30), which has recently shown promising results in a clinical trial (31). HDR can also be suppressed by environmental manipulations, including hyperthermia (32). These findings shed light on the diverse mechanisms through which DNA repair can be manipulated, and they have potential implications for the development of combination therapies designed to exploit induced DNA repair deficiencies in cancer cells.
MATERIALS AND METHODS
Study design
The objective of this study was to define the effects of cediranib on the HDR pathway of DNA repair. In vitro experiments were performed in biological triplicate unless otherwise stated. Sample sizes were determined on the basis of previous experience. Tumor growth studies were performed with n = 7 to 8 mice, which have a power of about 80% to detect a difference of 1.25 SDs between two groups at a 5% significance level. For in vivo experiments, mice were randomly assigned to treatment groups. Experiments were not blinded.
Mouse tumor xenograft studies
All studies were approved by the Yale University Institutional Animal Care and Use Committee. Human IGROV1 (5 × 106) or MDA2315XHRE-GFP (2 × 106) cells were implanted subcutaneously in 100 μl of RPMI medium unilaterally in the flanks of athymic nu/nu mice (Harlan). For MDA231-5XHRE-GFP xenografts, medium contained 30% Matrigel. Three times a week, tumors were measured using calipers to determine tumor volume by the following formula: V = 1/2(4π/3)(length/2)(width/2)(height).
Cell culture
All cell lines were validated by short tandem repeat analysis and were regularly tested for mycoplasma contamination. Reported mutations in DNA repair genes for all available cell lines are listed in table S1. Sources and culture conditions can be found in Supplementary Materials and Methods.
Drugs and reagents
For in vivo gene expression experiments, cediranib (3 to 18 mg/kg; Selleckchem) in DMSO/1% Tween 80 was administered daily for 3 to 4 days via oral gavage once tumors reached an average size of 500 mm3. DMSO/1% Tween 80 was used as a vehicle control. For tumor growth studies, mice were treated once tumors reached an average size of 100 mm3. Cediranib (6 mg/kg) was administered by oral gavage daily for 1 week, followed by twice weekly for 2 weeks. Olaparib (50 mg/kg in DMSO/10% 2-hydroxy-propyl-β-cyclodextrin; Sigma) was administered by intraperitoneal injection five times a week for 5 weeks. For cell culture experiments, cediranib, olaparib, sorafenib, crenolanib, and pazopanib (all from Selleckchem) and Maz51 (Cayman Chemical) were dissolved in DMSO and used at final concentrations ranging from 0.25 to 32 μM. The final percentage of DMSO in the medium did not exceed 0.1%.
Fluorescence-activated cell sorting
Tumors were dissected and dissociated to single-cell suspension using dispase/collagenase followed by trypsinization. Red blood cells were removed by ammonium chloride lysis. Human tumor cells were isolated using a Mouse Cell Depletion kit (Miltenyi Biotec). For CAIX staining, cells were incubated in fluorescein-conjugated CAIX antibody (R&D MAB2188) for 30min. Cells were sorted into fluorescein isothiocyanate (FITC)/GFP+ and FITC/GFP− fractions using a FACSAria cell sorter (BD Biosciences).
Western blotting
Cells were lysed in AZ lysis buffer (50 mM tris, 250 mM NaCl, 1% Igepal, 0.1% SDS, 5 mM EDTA, 10 mM Na2P2O7, 10 mM NaF) supplemented with Protease Inhibitor Cocktail (Roche) and PhosSTOP (Sigma). Band intensities were quantified using ImageJ64 software. Antibodies can be found in Supplementary Materials and Methods. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and vinculin were used as endogenous loading controls.
qRT-PCR analysis
RNA was isolated using an RNA Miniprep kit (Qiagen) and used to synthesize complementary DNA (cDNA) using a High-Capacity cDNA Reverse Transcription kit (Applied Biosystems). The resulting cDNA was used in qRT-PCR assays using TaqMan Universal PCR Master Mix and Gene Expression Assays (Applied Biosystems; information in Supplementary Materials and Methods). ACTB and 18S expression were used as endogenous normalization controls. A StepOnePlus RT-PCR system (Thermo Fisher Scientific) was used to measure fluorescence intensity in real time and to calculate cycle thresholds.
Cell-based HDR assays
HDR assays were performed using MCF7 DR-GFP and U2OS EJ-DR cells as previously described (10,19,33). Briefly, cells were seeded and pretreated with cediranib for 6 to 24 hours. For MCF7 DR-GFP cells, site-specific DSBs were introduced using electroporation with Amaxa Nucleofector II and Nucleofector Kit V (Lonza) to deliver 4 μg of a plasmid encoding the restriction enzyme I-Sce I per 1 × 106 cells. For U2OS EJ-DR cells, DSBs were induced by the addition of triamcinolone (0.5 μM; Sigma) and Shield1 (100 nM; Clontech) to cell culture medium for 24 hours to activate the endogenously expressed I-Sce I enzyme. MCF7 DR-GFP and U2OS EJ-DR cells were analyzed by flow cytometry to quantify GFP+ cells 72 hours after DSB induction.
Cell viability and clonogenic assays
Cell viability assays were performed using CellTiter-Glo (Promega). Briefly, 2 to 20,000 cells per well were seeded in a 96-well plate and then treated with cediranib and/or olaparib for 48 to 72 hours. Luminescence was quantified using a Synergy HT plate reader (BioTek). Synergism analysis was calculated by the Bliss model using Combenefit software (34). For clonogenic survival assays, 250 to 1,000 cells per well were seeded in six-well plates and treated with cediranib (1 to 10 μM) and/or olaparib (0.25 to 5 μM) for 48 hours. About 1.5 weeks later, cells were permeabilized in 0.9% saline solution and stained with crystal violet in 80% methanol. Colonies were counted manually.
siRNA-mediated knockdown
Cells were treated with siRNA for BRCA1, RAD51, KU80, XLF, VEGFR2, PDGFRα, or PDGFRβ (Dharmacon SMARTpool) at final concentrations of 10 to 100 nM for 72 to 96 hours. A nontargeting scrambled siRNA (Dharmacon SMARTpool) was used as a control. All knockdowns were confirmed using Western blot or qRT-PCR.
Chromatin immunoprecipitation
ChIP was performed as previously described (10). Primer information can be found in Supplementary Materials and Methods.
Statistical analysis
Statistical analysis was performed in GraphPad Prism. The following methods were used to determine significance: unpaired t test, one-way ANOVA, and two-way ANOVA. All testing was two sided with a testing level (α) of 0.05. Unless otherwise stated, data are presented as means ± SEM, with n = 3 replicates. Original data are provided in data file S1.
Supplementary Material
Data file S1. Individual data points.
Fig. S1. Cediranib suppresses expression of HDR factors in cancer cells in culture.
Fig. S2. Effects of cediranib on HDR factor expression are not mediated through a trans-acting factor.
Fig. S3. Inhibition of VEGFRs does not suppress HDR factor expression.
Fig. S4. Inhibition of PDGFRs suppresses HDR factor expression.
Fig. S5. siRNA-mediated targeting of PDGFRβ suppresses HDR factor expression.
Fig. S6. No consistent changes were observed in cell cycle profiles after various kinase inhibitor and siRNA treatments.
Fig. S7. Cediranib suppresses expression of FANCD2 in cancer cells in culture.
Fig. S8. PDGFR inhibition shifts migration of p130.
Table S1. Reported mutations in DNA repair genes.
Acknowledgments:
We thank N. Fons, D. Hegan, and A. Dhawan for assistance. We thank E. Ratner, T. Taniguchi, K. Cho, and R. Bindra for reagents. Funding: P.M.G. was supported by NIH grants R01ES005775 and R35CA197574. Z.Y. was supported by NIH grant R01CA178254. A.R.K. was supported by NIH grant F30CA221065. Support for this research was also provided by NIH Medical Scientist Training Program Training grant T32GM007205.
Footnotes
SUPPLEMENTARY MATERIALS
Competing interests: P.M.G. is a consultant to and has equity interests in Trucode Gene Repair Inc. of San Francisco, CA, and Cybrexa Therapeutics Inc. of New Haven, CT, and has an equity interest in Patrys Ltd. of Melbourne, Australia. None are related to the topic of this manuscript.
Data and materials availability: All data associated with this study are present in the paper or Supplementary Materials.
REFERENCES AND NOTES
- 1.Liu JF, Barry WT, Birrer M, Lee J-M, Buckanovich RJ, Fleming GF, Rimel BJ, Buss MK, Nattam S, Hurteau J, Luo W, Quy P, Whalen C, Obermayer L, Lee H, Winer EP, Kohn EC, Ivy SP, Matulonis UA, Combination cediranib and olaparib versus olaparib alone for women with recurrent platinum-sensitive ovarian cancer: A randomised phase 2 study. Lancet Oncol. 15, 1207–1214 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu JF, Barry WT, Birrer M, Lee J-M, Buckanovich RJ, Fleming GF, Rimel BJ, Buss MK, Nattam SR, Hurteau J, Luo W, Curtis J, Whalen C, Kohn EC, Ivy SP, Matulonis UA, Overall survival and updated progression-free survival outcomes in a randomized phase II study of combination cediranib and olaparib versus olaparib in relapsed platinum-sensitive ovarian cancer. Ann. Oncol, mdz018 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robson M, Im S-A, Senkus E, Xu B, Domchek SM, Masuda N, Delaloge S, Li W, Tung N, Armstrong A, Wu W, Goessl C, Runswick S, Conte P, Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N. Engl. J. Med 377, 523–533 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Pujade-Lauraine E, Ledermann JA, Selle F, Gebski V, Penson RT, Oza AM, Korach J, Huzarski T, Poveda A, Pignata S, Friedlander M, Colombo N, Harter P, Fujiwara K, Ray-Coquard I, Banerjee S, Liu J, Lowe ES, Bloomfield R, Pautier P; SOLO2/ENGOT-Ov21 Investigators, Olaparib tablets as maintenance therapy in patients with platinum-sensitive, relapsed ovarian cancer and a BRCA1/2 mutation (SOLO2/ENGOT-Ov21): A double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. 18, 1274–1284 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Farmer H, McCabe N, Lord CJ, Tutt ANJ, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NMB, Jackson SP, Smith GCM, Ashworth A, Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T, Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434, 913–917 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Wedge SR, Kendrew J, Hennequin LF, Valentine PJ, Barry ST, Brave SR, Smith NR, James NH, Dukes M, Curwen JO, Chester R, Jackson JA, Boffey SJ, Kilburn LL, Barnett S, Richmond GHP, Wadsworth PF, Walker M, Bigley AL, Taylor ST, Cooper L, Beck S, Jurgensmeier JM, Ogilvie DJ, AZD2171: A highly potent, orally bioavailable, vascular endothelial growth factor receptor-2 tyrosine kinase inhibitor for the treatment of cancer. Cancer Res. 65, 4389–4400 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Grkovski M, Emmas S-A, Carlin SD, 18F-Fluoromisonidazole kinetic modeling for characterization of tumor perfusion and hypoxia in response to antiangiogenic therapy. J. Nucl. Med 58, 1567–1573 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang Y, Allen D, Kersemans V, Devery AM, Bokobza SM, Smart S, Ryan AJ, Acute vascular response to cediranib treatment in human non-small-cell lung cancer xenografts with different tumour stromal architecture. Lung Cancer 90, 191–198 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bindra RS, Gibson SL, Meng A, Westermark U, Jasin M, Pierce AJ, Bristow RG, Classon MK, Glazer PM, Hypoxia-induced down-regulation of BRCA1 expression by E2Fs. Cancer Res. 65, 11597–11604 (2005) [DOI] [PubMed] [Google Scholar]
- 11.Bindra RS, Glazer PM, Repression of RAD51 gene expression by E2F4/p130 complexes in hypoxia. Oncogene 26, 2048–2057 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Lu Y, Chu A, Turker MS, Glazer PM, Hypoxia-induced epigenetic regulation and silencing of the BRCA1 promoter. Mol. Cell. Biol 31, 3339–3350 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sulkowski PL, Corso CD, Robinson ND, Scanlon SE, Purshouse KR, Bai H, Liu Y, Sundaram RK, Hegan DC, Fons NR, Breuer GA, Song Y, Mishra-Gorur K, De Feyter HM, de Graaf RA, Surovtseva YV, Kachman M, Halene S, Gunel M, Glazer PM, Bindra RS, 2-Hydroxyglutarate produced by neomorphic IDH mutations suppresses homologous recombination and induces PARP inhibitor sensitivity. Sci. Transl. Med 9, eaal2463 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hegan DC, Lu Y, Stachelek GC, Crosby ME, Bindra RS, Glazer PM, Inhibition of poly(ADP-ribose) polymerase down-regulates BRCA1 and RAD51 in a pathway mediated by E2F4 and p130. Proc. Natl. Acad. Sci. U.S.A 107, 2201–2206 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan N, Pires IM, Bencokova Z, Coackley C, Luoto KR, Bhogal N, Lakshman M, Gottipati P, Oliver FJ, Helleday T, Hammond EM, Bristow RG, Contextual synthetic lethality of cancer cell kill based on the tumor microenvironment. Cancer Res. 70, 8045–8054 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo XX, Wu HL, Shi HY, Su L, Zhang X, The efficacy and safety of olaparib in the treatment of cancers: A meta-analysis of randomized controlled trials. Cancer Manag. Res 10, 2553–2562 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakai W, Swisher EM, Jacquemont C, Chandramohan KV, Couch FJ, Langdon SP, Wurz K, Higgins J, Villegas E, Taniguchi T, Functional restoration of BRCA2 protein by secondary BRCA2 mutations in BRCA2-mutated ovarian carcinoma. Cancer Res. 69, 6381–6386 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bindra RS, Goglia AG, Jasin M, Powell SN, Development of an assay to measure mutagenic non-homologous end-joining repair activity in mammalian cells. Nucleic Acids Res. 41, e115 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pierce AJ, Johnson RD, Thompson LH, Jasin M, XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 13, 2633–2638 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan N, Koritzinsky M, Zhao H, Bindra R, Glazer PM, Powell S, Belmaaza A, Wouters B, Bristow RG, Chronic hypoxia decreases synthesis of homologous recombination proteins to offset chemoresistance and radioresistance. Cancer Res. 68, 605–614 (2008) [DOI] [PubMed] [Google Scholar]
- 21.Davis PL, Miron A, Andersen LM, Iglehart JD, Marks JR, Isolation and initial characterization of the BRCA2 promoter. Oncogene 18, 6000–6012 (1999). [DOI] [PubMed] [Google Scholar]
- 22.Scanlon SE, Glazer PM, Hypoxic stress facilitates acute activation and chronic downregulation of fanconi anemia proteins. Mol. Cancer Res 12, 1016–1028 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen T-I, Chiu H-W, Pan Y-C, Hsu S-T, Lin J-H, Yang K-T, Intermittent hypoxiainduced protein phosphatase 2A activation reduces PC12 cell proliferation and differentiation. J. Biomed. Sci 21, 46 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Conza G, Trusso Cafarello S, Loroch S, Mennerich D, Deschoemaeker S, Di Matteo M, Ehling M, Gevaert K, Prenen H, Zahedi RP, Sickmann A, Kietzmann T, Moretti F, Mazzone M, The mTOR and PP2A pathways regulate PHD2 phosphorylation to fine-tune HIF1α levels and colorectal cancer cell survival under hypoxia. Cell Rep. 18, 1699–1712 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen J, Martin BL, Brautigan DL, Regulation of protein serine-threonine phosphatase type-2A by tyrosine phosphorylation. Science 257, 1261–1264 (1992). [DOI] [PubMed] [Google Scholar]
- 26.Guo H, Damuni Z, Autophosphorylation-activated protein kinase phosphorylates and inactivates protein phosphatase 2A. Proc. Natl. Acad. Sci. U.S.A 90, 2500–2504 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vuocolo S, Purev E, Zhang D, Bartek J, Hansen K, Soprano DR, Soprano KJ, Protein phosphatase 2A associates with Rb2/p130 and mediates retinoic acid-induced growth suppression of ovarian carcinoma cells. J. Biol. Chem 278, 41881–41889 [DOI] [PubMed] [Google Scholar]
- 28.Cicchillitti L, Fasanaro P, Biglioli P, Capogrossi MC, Martelli F, Oxidative stress induces protein phosphatase 2A-dependent dephosphorylation of the pocket proteins pRb, p107, and p130. J. Biol. Chem 278, 19509–19517 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O’Connor MJ, Ashworth A, Carmichael J, Kaye SB, Schellens JHM, de Bono JS, Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med 361, 123–134 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Li L, Karanika S, Yang G, Wang J, Park S, Broom BM, Manyam GC, Wu W, Luo Y, Basourakos S, Song JH, Gallick GE, Karantanos T, Korentzelos D, Azad AK, Kim J, Corn PG, Aparicio AM, Logothetis CJ, Troncoso P, Heffernan T, Toniatti C, Lee H-S, Lee J-S, Zuo X, Chang W, Yin J, Thompson TC, Androgen receptor inhibitor-induced “BRCAness” and PARP inhibition are synthetically lethal for castration-resistant prostate cancer. Sci. Signal 10, eaam7479 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clarke N, Wiechno P, Alekseev B, Sala N, Jones R, Kocak I, Chiuri VE, Jassem J, Flechon A, Redfern C, Goessl C, Burgents J, Kozarski R, Hodgson D, Learoyd M, Saad F, Olaparib combined with abiraterone in patients with metastatic castration-resistant prostate cancer: A randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Oncol. 19, 975–986 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Krawczyk PM, Eppink B, Essers J, Stap J, Rodermond H, Odijk H, Zelensky A, van Bree C, Stalpers LJ, Buist MR, Soullie T, Rens J, Verhagen HJM, O’Connor MJ, Franken NAP, ten Hagen TLM, Kanaar R, Aten JA, Mild hyperthermia inhibits homologous recombination, induces BRCA2 degradation, and sensitizes cancer cells to poly (ADP-ribose) polymerase-1 inhibition. Proc. Natl. Acad. Sci. U.S.A 108, 9851–9856 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Surovtseva YV, Jairam V, Salem AF, Sundaram RK, Bindra RS, Herzon SB, Characterization of cardiac glycoside natural products as potent inhibitors of DNA double-strand break repair by a whole-cell double immunofluorescence assay. J. Am. Chem. Soc 138, 3844–3855 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Di Veroli GY, Fornari C, Wang D, Mollard S, Bramhall JL, Richards FM, Jodrell DI, Combenefit: An interactive platform for the analysis and visualization of drug combinations. Bioinformatics 32, 2866–2868 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data file S1. Individual data points.
Fig. S1. Cediranib suppresses expression of HDR factors in cancer cells in culture.
Fig. S2. Effects of cediranib on HDR factor expression are not mediated through a trans-acting factor.
Fig. S3. Inhibition of VEGFRs does not suppress HDR factor expression.
Fig. S4. Inhibition of PDGFRs suppresses HDR factor expression.
Fig. S5. siRNA-mediated targeting of PDGFRβ suppresses HDR factor expression.
Fig. S6. No consistent changes were observed in cell cycle profiles after various kinase inhibitor and siRNA treatments.
Fig. S7. Cediranib suppresses expression of FANCD2 in cancer cells in culture.
Fig. S8. PDGFR inhibition shifts migration of p130.
Table S1. Reported mutations in DNA repair genes.