Abstract
dsDNA Bacteriophages, some dsDNA archaeal viruses and the Herpesviruses share many features including a common capsid assembly pathway and coat protein fold. The coat proteins of these viruses, which have the HK97 fold, co-assemble with a free or attached scaffolding protein and other capsid proteins into a precursor capsid, known as a procapsid or prohead. The procapsid is a metastable state that increases in stability as a result of morphological changes that occur during the dsDNA packaging reaction. We review evidence from several systems indicating that proper contacts acquired in the assembly of the procapsid are critical to forming the correct morphology in the mature capsid.
Graphical Abstract
Introduction
Viruses are thought to be ancient, and likely existed before the division of life into three domains [1]. Although they are made from extremely divergent proteins, often there are detectable lifestyle and structural similarities that suggest distant evolutionary relationships [2]. Viruses use a range of virion designs to protect and deliver their genetic instructions to their host. Some virions have a membrane outer coat, some have a protein coat, while others use a combination of the two. Amongst those with a protein-only coat, some co-assemble the coat and nucleic acid to build a protein capsid, while the tailed dsDNA bacteriophages, some archaeal viruses, and the Herpesviruses build precursor procapsids and then later fill them with DNA using an ATP-driven motor. Curiously, the Herpesviruses, phages and some archaeal viruses appear to be evolutionarily related: they all form icosahedral protein shells or capsids using a shared common assembly pathway and all use a variant of a single protein fold, the HK97 fold (Figure 1A) [3-5].That this fold is found in viruses from all domains of life gives credence to the presence of viruses in evolution for billions of years. Proteins with the HK97 fold are capable of forming particles as small as the T. maritima encapsulins (T=1, 24 nm diameter) [6], to the biggest thus far discovered, jumbophage G (T=52, 150-180 nm diameter) (Figure 1B) [7]. Thus, these particles are composed of 60 to more than 3000 monomers. How can this fold be so amazingly versatile?
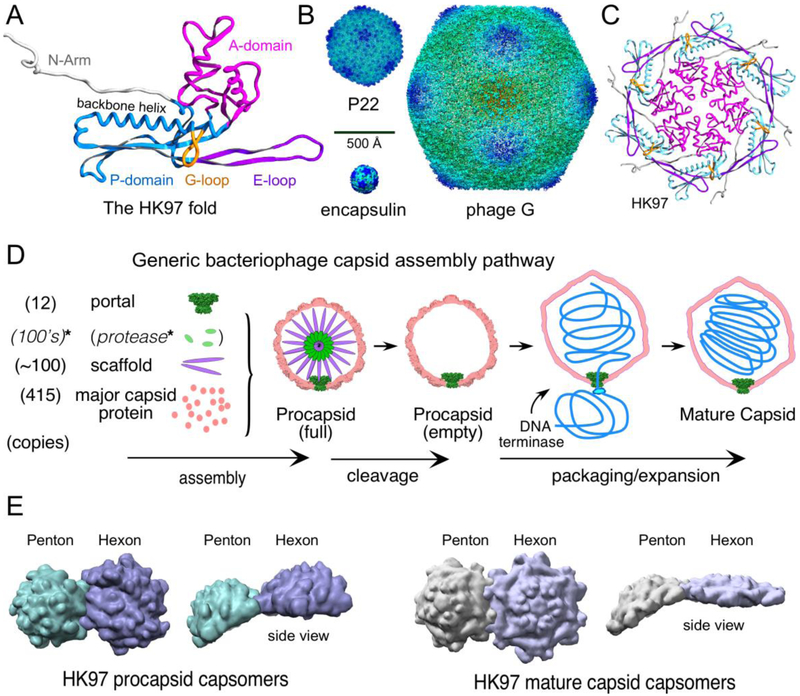
Figure 1. Components and features of capsids.
A. The HK97 fold with important common features labeled and color-coded as indicated in the figure. The ribbon diagram is from the mature HK97 capsid chain A (PDB 1OHG). B. The size range of the HK97 fold. Shown are images of surface rendered cryo-EM density maps from the T=1 encapsulin of Thermotoga maritima [6], the P22 mature capsid [19], and Bacillus megaterium jumbophage G [7]. C. A hexon from the HK97 mature capsid colored as in (A). D. A generic pathway for capsid assembly that applies, in general features, to nearly all double-stranded DNA tailed phages and Herpesviruses. *Note that while a capsid maturation protease is a common feature, there are many bacteriophages that do not utilize one. E. Examples of the shape changes of capsomers that occur when procapsids convert to mature procapsids. Surface rendered images are shown of the hexons and pentons of phage HK97 from X-ray structures (PDB ID 3E8K (procapsid) and 1OGH (mature)). Structural models in figures were visualized using Chimera [47] or SPDBV [48].
Icosahedral virus basics.
Caspar and Klug [8] showed how coat proteins in pentameric and hexameric arrangements (pentons and hexons, or capsomers, Figure 1C shows the arrangement of subunits in a phage HK97 hexamer) can build icosahedral capsids of different sizes. The twenty faces that define an icosahedron are composed of ~flat protein hexamers, while the 12 vertices (corners) are pyramids made of five copies of the same or similar protein. The smallest capsid is composed of 60 subunits in 12 pentamers. Larger capsids are formed by moving the pentons apart and connecting them with a group of hexons that form the icosahedral faces. These concepts are easy to understand when equilateral triangles of paper or plastic are used, but the real viruses we wish to discuss are composed of proteins subunits made using one of the many variants of the unusually shaped HK97 fold (Figure 2).
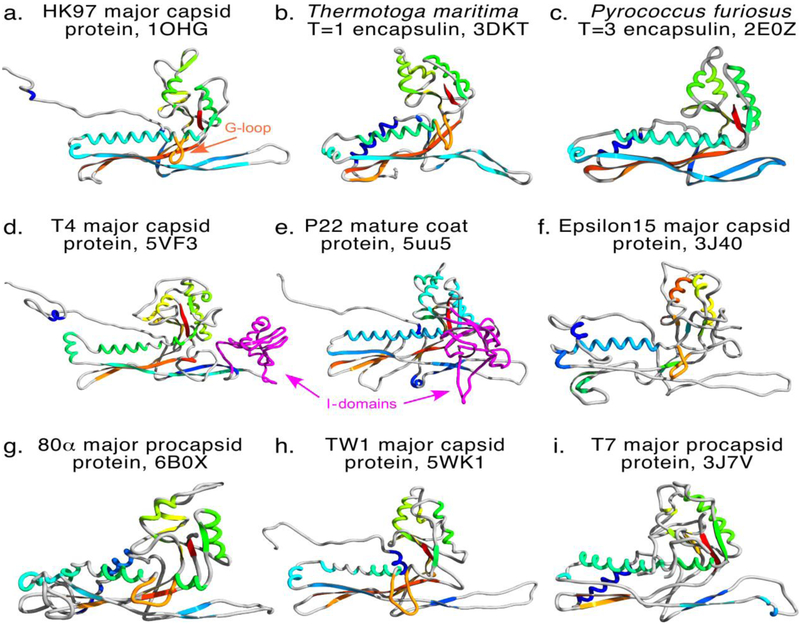
Figure 2. HK97 fold gallery.
Nine examples of the HK97 fold determined by X-ray crystallography or cryo-electron microscopy with secondary structure colored from N- to C-termini in blue to red hues. Protruding G-loops are colored orange, I-domains (Insertion domains) are colored magenta. Panels, in order: a, phage HK97 mature capsid protein [43]; b, Thermotoga maritima encapsulin [6]; c, Pyrococcus furiosus encapsulin (previously “PFV” for Pyrococcus fuiosus virus-like particle [39]); d, phage T4 mature major capsid protein gp23* [49]; e, P22 coat protein [19]; f, epsilon 15 mature capsid protein [50]; g, 80alpha procapsid major capsid protein [42]; h, phage TW1 major capsid protein [18]; i, phage T7 major capsid protein from the procapsid [41].
Features of the HK97 fold (Figure 1A).
The conserved elements are 1) A-domain (axial domain), 2) the P-domain (peripheral domain), E-loop (extended Loop), and N-arm (N-terminal arm), but there are additional “bells and whistles” which appear in some, but not all capsid proteins (Figure 2). These include the I-domain (insertion domain, in P22 this is grafted onto the A-domain and contains the D-loop, Figure 2, D and E), and the G-loop (gycine-rich loop) in HK97 and many others. The triangular A-domain makes up the central wedge-shaped sectors of hexons and pentons and has a β-sheet-rich core surrounded by prominent α-helices along the edges that interact with adjacent A-domains. The P-domain forms the periphery of hexons and pentons and is composed of a long α-helix (backbone or spine helix) nestled against a multi-stranded β-sheet with prominent loops at one end. The flexible E-loop is a long 2-strand β-sheet hairpin that protrudes from the P-domain and makes contacts with other subunits. In an isolated subunit, the N-arm appears to protrude from the subunit body, but it associates intimately with other domains and changes its associations during maturation.
Procapsids are essential.
In addition to the ability of HK97-fold capsid proteins to adapt to generate many particle sizes, the first product of the assembly reaction is not the final virion but a metastable precursor capsid, known as a procapsid, prehead, or prohead depending on the system. A generic capsid assembly pathway is shown in Figure 1D. Procapsids convert to mature capsids via large scale conformational changes during DNA packaging (Figure 1D) [9]. Assembly of procapsids is driven by a scaffolding protein or an equivalent scaffold domain (“delta domain”) that is covalently attached to the major capsid protein. Scaffolding proteins are discarded after assembly is complete and leave through ports in the capsid, either intact or after proteolysis [10,11]. Procapsids are round, lumpy and thick-shelled (Figure 3), while usually mature capsids are angular, thin-shelled and usually larger, which is why maturation is often referred to as “expansion”. All capsomers in procapsids have a dome-like shapes that gives procapsids the lumpy appearance that is lost during maturation. The domed hexons in proheads are asymmetric (elongated, “skewed,” or oval) and morph into a nearly flat regular hexagons in mature capsids (Figure 1E). Pentons also change conformation, but retain the pyramid shape needed at vertices. All procapsids examined to date have asymmetric hexons, suggesting that they are an intrinsic and essential feature (Figure 3). The asymmetric hexon shape is a result of the breaking of the normal contacts found in the mature capsids between adjacent A-domains to allow a shift or twist of the subunits along that boundary [12]. We propose that assembly is controlled at the prohead stage, and it is at this stage that interactions determine capsid size and shape - interactions that may be missing, and therefore not discoverable, in the mature capsid [12,13]. Below we highlight some interactions that we deem important to formation of proper capsids, concentrating on the A-domain and E-loop.
Figure 3. Procapsid gallery.
Ten examples of procapsid structures from the Protein Data Base (PDB) or Electron microscopy database (EMDB) rendered at low resolution using Chimera [47] so that the shapes of the procapsid hexons can be discerned. Procapsids are round (not angular) and have lumpy capsomers with a domed and asymmetric shape, as noted in the text. The procapsids are radially cued with yellow being the furthest from and blue being closest to the center of each particle. The scale bar represents 20 nm. Shown are the procapsids of phages HK97 (PDB 3E8K, [17]); T7 (EMD-1321 ,[51]); Gifsy-2 (EMD-1691, [52]); Lambda (EMD-1507, [53]); 80alpha (EMD-7030, PDB 6B0X;[42]); P2 (EMD-5406, [54]); P22 (EMD-5149, [15]); Sf6 (EMD-5724, [55]); SIO2 (EMD-5383, [56]); and T5 (not deposited, [57]).
A-domain
The A-domain fills the centers of the hexons and pentons, and undergoes important changes during capsid maturation [14,15]. The skewed and dome-shaped hexons become flat and hexagonal when the subunits tilt and slide along the A-domain:A-domain interfaces so that all of those interactions are about the same [12,16]. There are other changes as well that involve the A-domain’s pointed tip or A-loop [17] and an attached segment that is tucked under the tip and usually contains an alpha-helix, which is very short in HK97, and longer in phage TW1 [18], P22 [19] and others. In P22 this segment is like a flap because it unfurls from its location in the interior of the procapsid to attain a fully formed and stable secondary structure that fills the centers of the hexons [20-22]. In P22, this A-domain refolding is much larger than seen for other phages because the procapsid is assembled with holes in the center of the hexons that are used for scaffolding protein exit during DNA packaging and maturation. Prevelige’s group found that this part of P22’s A-domain is highly sensitive to amino acid insertions and could tolerate the addition of three, but not four alanines [23]. The thermophilic phage P74-26, which infects Thermus thermophilus, has evolved the largest T=7 capsid thus far identified to contain its 83.3 kb genome. Encapsidating this larger genome (a typical T=7 phage has a ~48 kb genome) is accomplished by increasing the size of the A-domain, and lengthening the E-loop (see below for the importance of the E-loop), along with a number of other changes [24]. In phage P74-26, the A-loop is extended by 6 Å by the addition of a new helix and the A-domain is widened as well.
Mutants in the A-domain can lead to alternate capsid forms in many phages, indicating that the A-domain also is important for proper assembly. The A-domain has a conserved feature in its core that we call the β-hinge, which is a 5 stranded β-sheet that connects all of the other domains in the protein [25,26]. In P22, the β-hinge is important for coat protein folding, in addition to proper capsid morphology. One of the 18 P22 coat protein temperature-sensitive-folding (tsf) mutants is in the β-hinge, as well as all three of the tsf global suppressor substitutions D163G, T166I and F170L that alleviate folding defects from tsf mutations all over coat protein, signifying the importance of the β-hinge in P22 coat protein folding [25,27]. The global suppressor substitutions alone have no phenotype, but double and triple combinations are cold-sensitive [28]. F170L coat protein, even though it is able to support phage growth, makes short tubes of coat protein in vivo and in vitro [28]. When this residue is changed to alanine or lysine, a ts phenotype results and long regular tubes of hexameric coat protein are assembled in vitro, highlighting the importance of the β-hinge in P22 assembly as well as folding [29,30]. The F170L tubes have the skewed hexons seen in procapsids. F170A tubes have the symmetric hexons of mature capsids, but likely assemble with skewed procapsid hexons and become symmetric during purification [30]. The F170 substitutions appear to decrease conformational flexibility in the A-domain, as probed by accessibility to protease digestion in procapsids. We hypothesize this leads to the assembly tubes of only hexons because penton formation is inhibited by steric crowding in the center of pentons, but not in hexons [29,30].
Other interesting phenotypes are associated with A-domain mutations in phages T4 and lambda. Phage T4 has two coat proteins that each have the HK97 fold: protein gp23 forms hexons, while gp24 makes the pentons need to form the vertices [31]. There are mutants in gp23 that bypass the need for gp24 at the 5-fold vertices, with the mutant gp23 taking the penton position [32]. Many of these mutants are found in gp23’s A-domain [31]. In phage lambda, three of the five of the mutants that lead to assembly of thick tubes are found in the A-domain [33,34]. These observations indicate the A-domain is critical for proper assembly of coat proteins with the HK97 fold.
A-domain embellishments
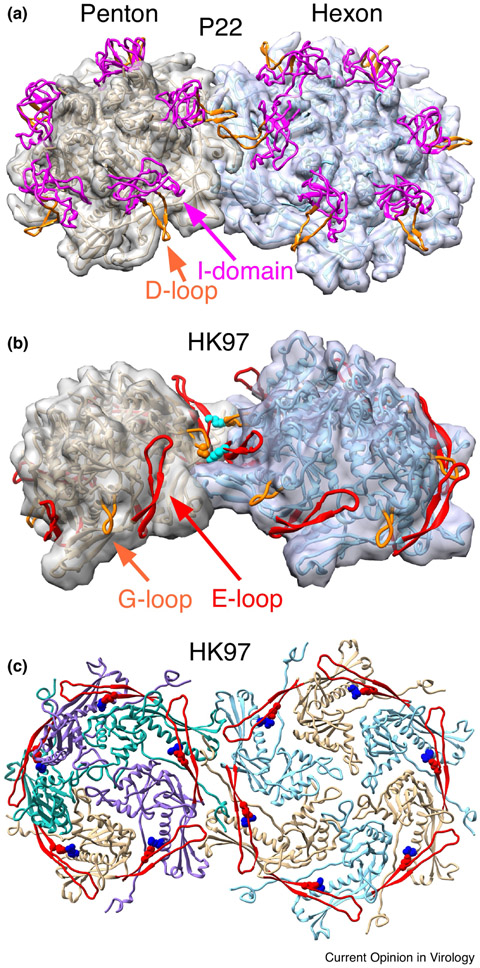
In the P22-like phages, an extra domain is inserted between sheets β1 and β3 of the β-hinge called the Insertion domain (I-domain) [15,22] (Figure 2e). The 123 residue I-domain is a six-stranded β-barrel that forms P22 coat protein’s folding nucleus and contributes half of its overall thermodynamic stability [35,36]. Loops connecting some of the I-domain β-strands have functionally significant roles. Mutations in the I-domain’s S-loop (for size determination) at position 285 lead to the assembly of smaller than normal (petite) capsids with T=4 rather than the normal T=7 geometry. The S-loop is also involved in incorporation of the portal protein complex [22,37]. The petite capsids get smaller with increasing bulkiness of the residue 285 side chain, suggesting a mechanism involving contacts with another domain. Another loop of P22’s I-domain, the D-loop, also appears to be involved in regulating assembly (Figure 4A). The D-loop makes crucial salt bridges across the two-fold axes of symmetry in both in capsids and procapsids. Mutations of critical residues at the tip of the D-loop lead to aberrant partial procapsids or irregular tubes, showing the importance of these contacts for proper capsid assembly [38]. Though not an A-domain embellishment, phage HK97 G-loop makes similar important interactions across the two fold axes of symmetry or the HK97 procapsid and mutations in the G-loop result in similar mis-assembly products (Figure 4B)[13]. P22’s I-domain D-loop and HK97’s coat protein G-loop occupy spatially analogous positions on the exteriors of their respective coat protein subunits (Figure 2A and E, Figure 4) although the G-loop is inserted into the spine helix rather than the A-domain, suggesting that they mediate potentially analogous inter-capsomer interactions that result in similar phenotypes when perturbed. Though little is known about the function of the elongated variant of the G-loop (or β-tongue) that is seen in lambda [33] and CFT073 coat proteins (PDB:3BQW), we can speculate that it may have a similar function in proper capsid assembly. These observations show that interactions that reach across 2-fold axes of symmetry between adjacent capsomers that are mediated by loops protruding from the outer surface of the coat protein subunits are an important and common feature of the HK97 fold.
Figure 4. Capsid protein interactions that are important for assembly.
Inter and intra-capsomer interactions discussed in the text are illustrated in close-up views. A. The site of inter-capsomer interactions between phage P22 coat protein D-loops. This shows the interaction in the mature capsid [19], but similar interactions occur in procapsids during assembly [22,38,58]. B. Interactions between the E-loops (in red) and G-loops (in orange) of adjacent capsomers in HK97 procapsids (PDB ID 3e8k). These interactions (mediated by residues K178 (in cyan) on the E-loop and D231 (orange) on the G-loop) have been shown to be important for controlling the assembly of procapsids, but are not present in the mature capsid [13]. C. Interactions made by HK97 E-loops (shown as red ribbons) within capsomers. Residue E153 (in red) on the E-loop interacts with R210 (in blue) on the backbone helix of the adjacent subunit. These interactions are essential for assembly of the HK97 major capsid protein into procapsids [12]. The figure shows the mature capsid (PDB ID 1OHG), but the interactions are present at all stages of HK97 assembly.
What does the E-loop do?
The E-loop is shown as an isolated feature in subunit illustrations, but it is never actually without binding partners. In procapsids and capsids it always has multiple interactions that may include parts of 1) the same subunit, 2) the adjacent subunit in the same capsomer, and 3) subunits in adjacent capsomers. We give examples below and describe how some have demonstrated roles in regulating assembly.
In the non-viral T=1 and T=3 encapsulins from T. maritima [6] and P. furiosus [39] and in all instances of the HK97 fold examined so far in viruses (all T=4 or larger), the E-loops make intra-capsomer connections to the adjacent subunits’ P- or A-domain, as can be seen in HK97 (Figure 1C, Figure 4C), T4, phi29 [40], P22 [15], T7 [41], HSV-1 [4] 80alpha [42], and many others. Given the prevalence of this interaction, we would suggest that it is universal. In HK97, a salt bridge between the E-loop and the spine helix of the adjacent subunit is a key part of that essential connection: breaking it leads to the formation of sheets and tubes of flattened hexons similar to those in mature capsids (Figure 4C) [12]. This suggests that these E-loop connections tether adjacent subunits to each so that hexons can take on the domed, asymmetric shape needed to build procapsids. The exact nature of these intra-capsomer connections may not be important: salt bridges may mediate some connections, as tested in HK97 and suggested for others [12,19], Indeed, T4 uses an extra domain inserted end of the E-loop’s end to make analogous intra-capsomer connections (Figure 2D) [31].
E-loops also make many inter-capsomer contacts which are likely to play roles in assembly. In HK97 procapsids, E-loops from each capsomer contact the G-loop on the “hillside” of the adjacent capsomer (Figure 4B). Those interactions occur only in procapsids, but are essential and appear to set the dihedral angle between adjacent capsomers [13] since disrupting them either prevents normal assembly or leads to the formation of tubes or small incomplete shells. The all-penton T=1 encapsulins from T. maritima [6] use E-loop:E-loop interactions exclusively to bind adjacent capsomers to each other. In other procapsids, including T7’s [41], the E-loops on adjacent capsomers contact each other across the canyon of the inter-capsomer interface, or around the local-3-fold symmetric junctions. We suggest that some of these interactions play roles in setting inter-capsomer angles (as they do in HK97 [13]) and thus are likely to have a role in determining capsid size. The E-loop often extends far enough to make distant interactions with adjacent capsomers near its tip, whose roles are generally unknown, including in HK97 [43], T7 [41] and P22 [19]. One example is a lysine residue near the tip of the HK97 E-loop that inserts into a socket between subunits during expansion and forms a covalent link to the interior of an adjacent capsomer [5,44-46], An unusual E-loop connection is found in P22, where it appears to be bound to the underside of the I-domain of the same subunit, but we do not yet know if that interaction is important.
Thus, what the E-loop does is make connections, some of which we can tentatively assign functions: intra-capsomer connections that appear needed for holding capsomers in the correct conformation for procapsid assembly, and inter-capsomer connections that participate in and can regulate the higher order assembly into capsid shells.
Conclusion
Here we have highlighted some of the features and interactions that appear to be widely conserved and play important roles in assembling viral capsids from subunits having the HK97 fold. We propose that the size and shape of capsid particles are determined at the procapsid stage through inter- and intra-capsomer interactions that we have begun to reveal by studying the locations and phenotypes of mutants that divert capsid assembly into abnormal pathways.
Highlights.
Mature capsid size and shape is determined during the assembly of procapsids.
Intra- and inter-capsomer contacts are critical for regulating procapsid assembly.
Coat protein subunits often have extra loops and domains with roles in assembly.
Acknowledgements:
This work was supported by NIH grant GM076661 to CMT, and GM47795 to RLD. We thank Alexis Huet for his assistance in making the figures.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Hendrix RW: Evolution: the long evolutionary reach of viruses. Curr Biol 1999, 9:R914–917. [DOI] [PubMed] [Google Scholar]
- **2.Abrescia NG, Bamford DH, Grimes JM, Stuart DI: Structure unifies the viral universe. Annual Review of Biochemistry 2012, 81:795–822. [DOI] [PubMed] [Google Scholar]
- *3.Pietila MK, Laurinmaki P, Russell DA, Ko CC, Jacobs-Sera D, Hendrix RW, Bamford DH, Butcher SJ: Structure of the archaeal head-tailed virus HSTV-1 completes the HK97 fold story. Proc Natl Acad Sci U S A 2013, 110:10604–10609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *4.Huet A, Makhov AM, Huffman JB, Vos M, Homa FL, Conway JF: Extensive subunit contacts underpin herpesvirus capsid stability and interior-to-exterior allostery. Nat Struct Mol Biol 2016, 23:531–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wikoff WR, Liljas L, Duda RL, Tsuruta H, Hendrix RW, Johnson JE: Topologically linked protein rings in the bacteriophage HK97 capsid. Science 2000, 289:2129–2133. [DOI] [PubMed] [Google Scholar]
- 6.Sutter M, Boehringer D, Gutmann S, Gunther S, Prangishvili D, Loessner MJ, Stetter KO, Weber-Ban E, Ban N: Structural basis of enzyme encapsulation into a bacterial nanocompartment. Nat Struct Mol Biol 2008, 15:939–947. [DOI] [PubMed] [Google Scholar]
- *7.Hua J, Huet A, Lopez CA, Toropova K, Pope WH, Duda RL, Hendrix RW, Conway JF: Capsids and Genomes of Jumbo-Sized Bacteriophages Reveal the Evolutionary Reach of the HK97 Fold. MBio 2017, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caspar DLD, Klug A: Physical principles in the construction of regular viruses. Cold Spring Harbor Symposia on Quantitative Biology 1962, 27:1–24. [DOI] [PubMed] [Google Scholar]
- **9.Steven AC, Heymann JB, Cheng N, Trus BL, Conway JF: Virus maturation: dynamics and mechanism of a stabilizing structural transition that leads to infectivity. Curr Opin Struct Biol 2005, 15:227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dokland T: Scaffolding proteins and their role in viral assembly. Cell Mol Life Sci 1999, 56:580–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prevelige PE, Fane BA: Building the machines: scaffolding protein functions during bacteriophage morphogenesis. Adv Exp Med Biol 2012, 726:325–350. [DOI] [PubMed] [Google Scholar]
- *12.Hasek ML, Maurer JB, Hendrix RW, Duda RL: Flexible Connectors between Capsomer Subunits that Regulate Capsid Assembly. J Mol Biol 2017, 429:2474–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *13.Tso DJ, Hendrix RW, Duda RL: Transient contacts on the exterior of the HK97 procapsid that are essential for capsid assembly. J Mol Biol 2014, 426:2112–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hendrix RW, Johnson JE: Bacteriophage HK97 capsid assembly and maturation. Adv Exp Med Biol 2012, 726:351–363. [DOI] [PubMed] [Google Scholar]
- 15.Parent KN, Khayat R, Tu LH, Suhanovsky MM, Cortines JR, Teschke CM, Johnson JE, Baker TS: P22 coat protein structures reveal a novel mechanism for capsid maturation: Stability without auxiliary proteins or chemical cross-links. Structure 2010, 18:1568–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wikoff WR, Conway JF, Tang J, Lee KK, Gan L, Cheng N, Duda RL, Hendrix RW, Steven AC, Johnson JE: Time-resolved molecular dynamics of bacteriophage HK97 capsid maturation interpreted by electron cryo-microscopy and X-ray crystallography. J Struct Biol 2006, 153:300–30 [DOI] [PubMed] [Google Scholar]
- 17.Gertsman I, Komives EA, Johnson JE: HK97 maturation studied by crystallography and H/2H exchange reveals the structural basis for exothermic particle transitions. J Mol Biol 2010, 397:560–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Z, Hardies SC, Fokine A, Klose T, Jiang W, Cho BC, Rossmann MG: Structure of the Marine Siphovirus TW1: Evolution of Capsid-Stabilizing Proteins and Tail Spikes. Structure 2018, 26:238–248 e233. [DOI] [PubMed] [Google Scholar]
- 19.Hryc CF, Chen DH, Afonine PV, Jakana J, Wang Z, Haase-Pettingell C, Jiang W, Adams PD, King JA, Schmid MF, et al. : Accurate model annotation of a near-atomic resolution cryo-EM map. Proc Natl Acad Sci U S A 2017, 114:3103–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang S, Prevelige PE Jr.: Domain study of bacteriophage p22 coat protein and characterization of the capsid lattice transformation by hydrogen/deuterium exchange. J Mol Biol 2005, 347:935–948. [DOI] [PubMed] [Google Scholar]
- 21.Chen D, Baker M, Hryc C, DiMaio F, Jakana J, Wu W, Dougherty M, Haase-Pettingell C, Schmid M, Jiang W, et al. : Structural basis for scaffolding-mediated assembly and maturation of a dsDNA virus. Proc Natl Acad Sci U S A 2011, 108:1355–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *22.Rizzo AA, Suhanovsky MM, Baker ML, Fraser LC, Jones LM, Rempel DL, Gross ML, Chiu W, Alexandrescu AT, Teschke CM: Multiple functional roles of the accessory I-domain of bacteriophage P22 coat protein revealed by NMR structure and CryoEM modeling. Structure 2014, 22:830–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morris DS, Prevelige PE: The role of the coat protein A-domain in p22 bacteriophage maturation. Viruses 2014, 6:2708–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stone NP, Demo G, Agnello E, Kelch BA: Principles for enhancing virus capsid capacity and stability from a thermophilic virus capsid structure. BioRxiv.org 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teschke CM, Parent KN: ‘Let the phage do the work’: using the phage P22 coat protein structures as a framework to understand its folding and assembly mutants. Virology 2010, 401:119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *26.Suhanovsky MM, Teschke CM: Nature's favorite building block: Deciphering folding and capsid assembly of proteins with the HK97-fold. Virology 2015, 479-480:487–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aramli LA, Teschke CM: Single amino acid substitutions globally suppress the folding defects of temperature-sensitive folding mutants of phage P22 coat protein. J. Biol. Chem 1999, 274:22217–22224. [DOI] [PubMed] [Google Scholar]
- 28.Parent KN, Suhanovsky MM, Teschke CM: Polyhead formation in phage P22 pinpoints a region in coat protein required for conformational switching. Mol. Microbiol 2007, 65:1300–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suhanovsky MM, Parent KN, Dunn SE, Baker TS, Teschke CM: Determinants of bacteriophage P22 polyhead formation: the role of coat protein flexibility in conformational switching. Mol. Microbiol 2010, 77:1568–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parent KN, Sinkovits RS, Suhanovsky MM, Teschke CM, Egelman EH, Baker TS: Cryo-reconstructions of P22 polyheads suggest that phage assembly is nucleated by trimeric interactions among coat proteins. . Phys. Biol 2010, 7:045004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fokine A, Leiman PG, Shneider MM, Ahvazi B, Boeshans KM, Steven AC, Black LW, Mesyanzhinov VV, Rossmann MG: Structural and functional similarities between the capsid proteins of bacteriophages T4 and HK97 point to a common ancestry. Proc Natl Acad Sci USA 2005, 102:7163–7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson K, Condie B, Mooney DT, Doermann AH: Mutations that eliminate the requirement for the vertex protein in bacteriophage T4 capsid assembly. J Mol Biol 1992, 224:601–611. [DOI] [PubMed] [Google Scholar]
- 33.Singh P, Nakatani E, Goodlett DR, Catalano CE: A pseudo-atomic model for the capsid shell of bacteriophage lambda using chemical cross-linking/mass spectrometry and molecular modeling. J Mol Biol 2013, 425:3378–3388. [DOI] [PubMed] [Google Scholar]
- 34.Katsura I, Kobayashi H: Structure and inherent properties of the bacteriophage lambda head shell. VII. Molecular design of the form-determining major capsid protein. J Mol Biol 1990, 213:503–511. [DOI] [PubMed] [Google Scholar]
- 35.Suhanovsky MM, Teschke CM: An Intramolecular Chaperone Inserted in Bacteriophage P22 Coat Protein Mediates Its Chaperonin-independent Folding. J Biol Chem 2013, 288:33772–33783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harprecht C, Okifo O, Robbins KJ, Motwani T, Alexandrescu AT, Teschke CM: Contextual Role of a Salt Bridge in the Phage P22 Coat Protein I-Domain. J Biol Chem 2016, 291:11359–11372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *37.Suhanovsky MM, Teschke CM: Bacteriophage P22 capsid size determination: Roles for the coat protein telokin-like domain and the scaffolding protein amino-terminus. Virology 2011, 417:418–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *38.D'Lima NG, Teschke CM: A Molecular Staple: D-Loops in the I Domain of Bacteriophage P22 Coat Protein Make Important Intercapsomer Contacts Required for Procapsid Assembly. J Virol 2015, 89:10569–10579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Akita F, Chong KT, Tanaka H, Yamashita E, Miyazaki N, Nakaishi Y, Suzuki M, Namba K, Ono Y, Tsukihara T, et al. : The crystal structure of a virus-like particle from the hyperthermophilic archaeon Pyrococcus furiosus provides insight into the evolution of viruses. J Mol Biol 2007, 368:1469–1483. [DOI] [PubMed] [Google Scholar]
- 40.Morais MC, Choi KH, Koti JS, Chipman PR, Anderson DL, Rossmann MG: Conservation of the capsid structure in tailed dsDNA bacteriophages: the pseudoatomic structure of phi29. Mol Cell 2005, 18:149–159. [DOI] [PubMed] [Google Scholar]
- 41.Guo F, Liu Z, Fang PA, Zhang Q, Wright ET, Wu W, Zhang C, Vago F, Ren Y, Jakana J, et al. : Capsid expansion mechanism of bacteriophage T7 revealed by multistate atomic models derived from cryo-EM reconstructions. Proc Natl Acad Sci U S A 2014, 111:E4606–4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dearborn AD, Wall EA, Kizziah JL, Klenow L, Parker LK, Manning KA, Spilman MS, Spear JM, Christie GE, Dokland T: Competing scaffolding proteins determine capsid size during mobilization of Staphylococcus aureus pathogenicity islands. Elife 2017, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Helgstrand C, Wikoff WR, Duda RL, Hendrix RW, Johnson JE, Liljas L: The refined structure of a protein catenane: the HK97 bacteriophage capsid at 3.44 A resolution. J Mol Biol 2003, 334:885–899. [DOI] [PubMed] [Google Scholar]
- 44.Dierkes LE, Peebles CL, Firek BA, Hendrix RW, Duda RL: Mutational analysis of a conserved glutamic acid required for self-catalyzed cross-linking of bacteriophage HK97 capsids. J Virol 2009, 83:2088–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gan L, Conway JF, Firek BA, Cheng N, Hendrix RW, Steven AC, Johnson JE, Duda RL: Control of crosslinking by quaternary structure changes during bacteriophage HK97 maturation. Mol Cell 2004, 14:559–569. [DOI] [PubMed] [Google Scholar]
- 46.Ross PD, Cheng N, Conway JF, Firek BA, Hendrix RW, Duda RL, Steven AC: Crosslinking renders bacteriophage HK97 capsid maturation irreversible and effects an essential stabilization. Embo J 2005, 24:1352–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE: UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 2004, 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 48.Guex N, Peitsch MC: SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 1997, 18:2714–2723. [DOI] [PubMed] [Google Scholar]
- 49.Chen Z, Sun L, Zhang Z, Fokine A, Padilla-Sanchez V, Hanein D, Jiang W, Rossmann MG, Rao VB: Cryo-EM structure of the bacteriophage T4 isometric head at 3.3-A resolution and its relevance to the assembly of icosahedral viruses. Proc Natl Acad Sci U S A 2017, 114:E8184–E8193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baker ML, Hryc CF, Zhang Q, Wu W, Jakana J, Haase-Pettingell C, Afonine PV, Adams PD, King JA, Jiang W, et al. : Validated near-atomic resolution structure of bacteriophage epsilon15 derived from cryo-EM and modeling. Proceedings of the National Academy of Sciences of the United States of America 2013, 110:12301–12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Agirrezabala X, Velazquez-Muriel JA, Gomez-Puertas P, Scheres SH, Carazo JM, Carrascosa JL: Quasi-atomic model of bacteriophage t7 procapsid shell: insights into the structure and evolution of a basic fold. Structure 2007, 15:461–472. [DOI] [PubMed] [Google Scholar]
- 52.Effantin G, Figueroa-Bossi N, Schoehn G, Bossi L, Conway JF: The tripartite capsid gene of Salmonella phage Gifsy-2 yields a capsid assembly pathway engaging features from HK97 and lambda. Virology 2010, 402:355–365. [DOI] [PubMed] [Google Scholar]
- 53.Lander GC, Evilevitch A, Jeembaeva M, Potter CS, Carragher B, Johnson JE: Bacteriophage lambda stabilization by auxiliary protein gpD: timing, location, and mechanism of attachment determined by cryo-EM. Structure 2008, 16:1399–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dearborn AD, Laurinmaki P, Chandramouli P, Rodenburg CM, Wang S, Butcher SJ, Dokland T: Structure and size determination of bacteriophage P2 and P4 procapsids: function of size responsiveness mutations. J Struct Biol 2012, 178:215–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parent KN, Gilcrease EB, Casjens SR, Baker TS: Structural evolution of the P22-like phages: comparison of Sf6 and P22 procapsid and virion architectures. Virology 2012, 427:177–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lander GC, Baudoux AC, Azam F, Potter CS, Carragher B, Johnson JE: Capsomer dynamics and stabilization in the T = 12 marine bacteriophage SIO-2 and its procapsid studied by CryoEM. Structure 2012, 20:498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Preux O, Durand D, Huet A, Conway JF, Bertin A, Boulogne C, Drouin-Wahbi J, Trevarin D, Perez J, Vachette P, et al. : A two-state cooperative expansion converts the procapsid shell of bacteriophage T5 into a highly stable capsid isomorphous to the final virion head. J Mol Biol 2013, 425:1999–2014. [DOI] [PubMed] [Google Scholar]
- 58.Chen DH, Baker ML, Hryc CF, DiMaio F, Jakana J, Wu W, Dougherty M, Haase-Pettingell C, Schmid MF, Jiang W, et al. : Structural basis for scaffolding-mediated assembly and maturation of a dsDNA virus. Proc Natl Acad Sci U S A 2011, 108:1355–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]