Abstract
Background and Objective
Usage of data from electronic health records (EHRs) in clinical research is increasing, but there is little empirical knowledge of the data needed to support multiple types of research these sources support. This study seeks to characterize the types and patterns of data usage from EHRs for clinical research.
Materials and Methods
We analyzed the data requirements of over 100 retrospective studies by mapping the selection criteria and study variables to data elements of two standard data dictionaries, one from the healthcare domain and the other from the clinical research domain. We also contacted study authors to validate our results.
Results
The majority of variables mapped to one or to both of the two dictionaries. Studies used an average of 4.46 (range 1–12) data element types in the selection criteria and 6.44 (range 1–15) in the study variables. The most frequently used items (e.g., Procedure, Condition, Medication) are often available in coded form in EHRs. Study criteria were frequently complex, with 49 of 104 studies involving relationships between data elements and 22 of the studies using aggregate operations for data variables. Author responses supported these findings.
Discussion and Conclusion
The high proportion of mapped data elements demonstrates the significant potential for clinical data warehousing to facilitate clinical research. Unmapped data elements illustrate the difficulty in developing a complete data dictionary.
Keywords: data models, data standards, queries
1. Introduction
Data collected during clinical care can constitute a valuable source of information for secondary use in research studies. Often, this data is used in observational studies; for example, to conduct comparative effectiveness research[1]. Additionally, the data is used to identify patients that might be eligible for prospective studies[2], to populate research data registries[3], and to annotate biospecimens with phenotypic data[4].
The increasing use of electronic health record (EHR) systems and other information systems in clinical practice is increasing the volume of clinical data and provides further opportunities for research. This data, which is in digital form and is codified, also can be much more efficient to use compared to the traditional method of reviewing and abstracting data from patients’ paper medical records or electronic notes (often referred to as chart review). In order to facilitate the use in research of data from clinical information systems, most notably from EHRs, many healthcare organizations are employing clinical data repositories (CDRs).
While CDRs are being increasingly employed to support researchers, there is little empirical knowledge of the data needed from clinical databases to support the types of research studies described above. The study described here aims to address this gap by analyzing the data requirements of retrospective observational studies (also known as “chart reviews”) published within a one-month period. Our objective was to characterize the data needed for performing such studies, by analyzing the selection criteria of the studies and the types of study data collected. This is a follow-up study to our previous pilot work[5] that mapped data elements from eligibility criteria in smaller number of ambulatory care studies. We have broadened this study in sample size and research settings, and have investigated the types of data used during the study. Furthermore, we attempted to validate our results through a survey of the authors of the published studies.
1.1. Background
Many healthcare organizations, primarily academic medical centers, their affiliates, and large health maintenance organizations[6] have implemented CDRs as a tool for researchers. These CDRs draw data mainly from the EHR system, though in many cases, data also are included from other systems such as the billing systems. The data elements that are available in these CDRs are the ones that are commonly recorded as discrete and coded elements in the EHRs such as the patient’s demographics, diagnoses, encounters, laboratory test results, medications, and diagnostic and therapeutic procedures. The structure of the clinical data elements in EHRs is very complex, reflecting the nuances of clinical workflows and the operational needs of healthcare organizations. The data are of high dimensionality and often imprecise.[7] Our institution’s EHR system, a commercially available product, has several hundred tables in its database. This level of breadth and complexity of the database schema is typical of EHR systems. CDR systems tend to use a less complex data schema, typically containing tens of tables. The choices made in the design of CDR database schemas can impact the granularity of the data elements and the relationships amongst them, and can therefore impact the utility and usability of the CDR for research. For example, problem lists in EHRs are used to document clinical problems including admission diagnoses, discharge diagnoses, and differential diagnoses that are to be ruled-in or ruled-out. CDRs may not consider these variations in their diagnoses list, which can potentially lead to incorrect inclusion or exclusion of patients. EHRs might also record preliminary and final results of diagnostic tests. If the CDRs record only the final results, then studies on preliminary results using the CDR might not be possible.
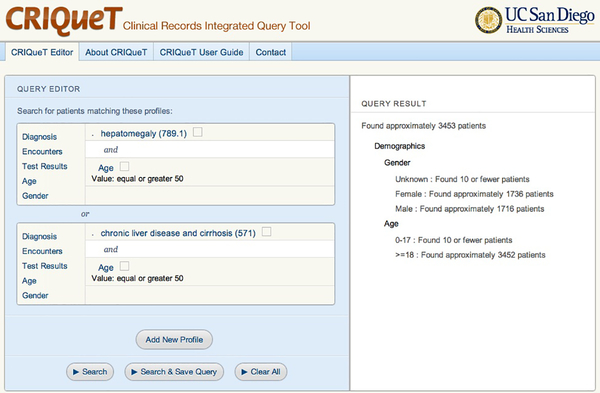
Another important challenge associated with the design of the CDRs and associated tools is usability, enabling researchers to easily obtain study data. Often designers face tradeoffs between usability and database efficiency. Since many biomedical scientists are not trained in writing database queries, graphical query tools are provided with many CDRs[3,8] to assist researchers in specifying the data to be queried. For example, a cohort discovery tool enables the researchers to compose and execute queries that estimate patient counts matching those queries (due to privacy and regulatory concerns, these tools often prevent the user from obtaining more detailed results such as the patient records). The cohort discovery tools allow the researchers to construct cohort specifications in the form of logical combinations of predicates (inclusion criteria). In order to reduce the complexity of the user interface, not all query predicates can be defined in these tools. As illustrated in Figure 1, compared to SQL there are limitations on the logical combinations of predicates. Another significant limitation found in some cases is that the predicates cannot be based on aggregate operations (e.g., all patients who have had two or more visits in the last year). Many cohort discovery tools[3,9], including the CRIQuET system[10] developed at our institution, share these limitations in the user interface. While these user interfaces might make the tools accessible for users without expertise or training in database queries, it is unclear if the queries constructed with these tools have sufficient expressivity for meeting the data needs of the researchers.
Figure 1.
This is a screen image of a cohort discovery tool called CRIQueT used at our institution. The user constructs a query on the left side of the screen. In this case, the user is limited to constructing a query predicate in the form of disjunctions with n nested conjunctions, i.e., (p1 and p2) or (p3 and p4). The results of executing the query, consisting of estimated counts of patients matching the predicate are shown on the right side of the screen.
The study we conducted aims to improve the understanding of the data needed in clinical research studies in order to inform the design of schemas for CDRs, the prioritization of data that are needed for research studies, and the design of query tools that are easy to use and sufficiently expressive.
2. Methods
2.1. Objectives and Overview
The objective of our study was to assess the data requirements for retrospective observational studies. Specifically, we aimed to characterize
The clinical data elements needed in these studies, i.e., the data variables.
The structure of the queries that have to be executed to obtain the data.
We analyzed patient selection criteria and data variables (which formed the study’s data set) for retrospective observational studies. These studies relied upon paper or electronic clinical records to identify patients as the source of the data set. From the full-text manuscripts of a set of observational studies, we extracted the patient selection criteria and the data variables used within the studies. We then mapped the data elements in patient selection criteria and the data variables to data elements in two standards-based data dictionaries. We report the summary statistics of the mappings.
2.2. Selection of Studies
We obtained a convenience sample of studies by performing a PubMed query for retrospective studies in core clinical journals, published in the month of December 2010 (either in print or electronically) and available in English. This query is as follows:
“retrospective studies[MeSH Terms] AND hasabstract[text] AND (“loattrfull text”[sb] AND English[lang] AND jsubsetaim[text] AND (“2010/12/01”[PDAT] : “2010/12/31”[PDAT]))”
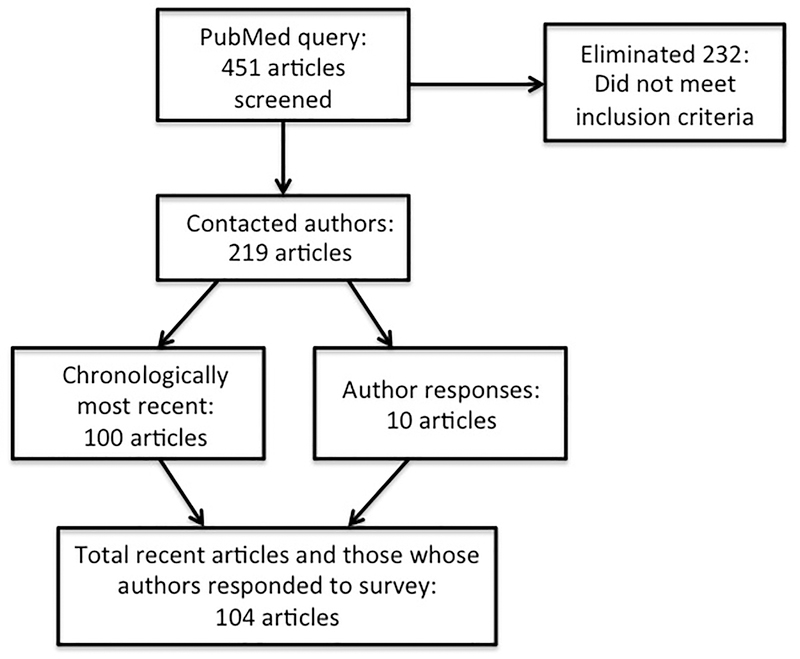
(The search terms “loattrfull text” and “jsubsetaim” refer to full text availability and the subset of core clinical journals, respectively.) This search conducted on July 1, 2011 yielded 451 articles. Of those available publicly or subscribed to by our institution, we only included studies that involved review of medical records. We removed studies that reported use of a database that was designed for a specific and narrow purpose (e.g., a disease registry) since use of these databases would limit queries to the fields in that database. This yielded 219 studies. We contacted the corresponding author of these studies with a questionnaire, inquiring about further information on selection criteria, study data, and the study process and experience.
Of the 219 studies identified above, we reviewed the chronologically latest 100 articles (first results in PubMed) as well as those studies whose author responded to the questionnaire, for a total of 104 articles. Article selection is depicted in Figure 2.
Figure 2.
Article selection for analysis
2.3. Mapping
After abstracting selection criteria and study analysis variables from the articles, we mapped these first to the data elements of the Health Information Technology Standards Panel (HITSP) dictionary[11], and then for comparison to the Observational Medical Outcomes Partnership Common Data Model, v3 (OMOP).[12] Descriptions of what was considered to be mapped are discussed in the subsection OMOP Comparison and Unmapped Data under Results. The HITSP specification was created for exchange of patient information for healthcare purposes. OMOP is a research model designed for aggregation of health data from multiple organizations for secondary analysis. We chose these models because they represent standard models for EHR data and clinical research data. They are broad enough in domain to cover data in any retrospective study but were sufficiently detailed and concrete to support the analysis. These issues are evidenced by their use in the real-world, where the HITSP dictionary was used for constructing widely used health information exchange document specifications [13] and OMOP has been used to construct research databases [14–16]. By comparison, the HL7 Reference Information Model[17], another very important standard for healthcare information, is very abstract. The Study Data Tabulation Model[18] and the Analysis Data Model [19] used for sharing clinical trials data, specify very generic models and primarily are designed for interventional studies.
The HITSP dictionary is organized into modules, where a module contains one type of data (e.g., Procedures). A module contains data elements for that data type (e.g., Procedure Date/Time). OMOP is a data model for research on the safety and effectiveness of drugs already in clinical use. It is centered on the elements of “Person” and “Provider” and comprises 16 additional data tables as concepts related to them (e.g., Drug_exposure a child of both Person and Provider).
For example, a study[20] that included all vestibular schwannoma resections from May 1990 to May 2009 would be mapped as follows to HITSP (2 selection criteria):
To demonstrate the level of complexity of retrieval of study data, we also calculated the number of join operations (instances in which data elements were dependent on one another) and any aggregate functions that would have to be used in a query to obtain the data elements. For this analysis, we ignored those that simply associate the data element to the patient.
An example of a join operation can be demonstrated in a study of admissions of patients who had a CABG within the 5 days prior to admission; this would be a join between Procedure Date/Time and Encounter Date/Time elements of HITSP. Aggregate functions involved a kind of operation (e.g. count, maximum, last) on the data elements (such as peak bilirubin, which would be a maximum on the data element Result Value). We also noted the data variables that did not map to the dictionary. One author (ECS) performed all the mappings. Another author (AAB) reviewed and verified the mappings performed for a sample of the studies. No significant issues were identified in this second review.
2.4. Corroboration of Analysis
To corroborate the results of our analysis, we contacted the corresponding author of 219 studies with an open-ended questionnaire on study criteria and process (Appendix 1). The questions addressed:
Data sources for finding patients for study inclusion (e.g., EHRs, clinical or research registries, paper charts)
Any further details on selection criteria and analyzed study data, beyond what was covered in the paper
Presence of EHRs, and if they facilitated the study
Approximate number of person-hours to do data acquisition
Researcher responses were collected and summarized, and information on study performance provided by the researchers was compared with information obtained from the articles.
3. Results
3.1. Mapping to Data Elements
Both selection criteria and data variables mapped to 59 types of HITSP data elements in 15 of 20 total different modules in the HITSP dictionary, which corresponded to 17 of 18 types in OMOP. This mapping is summarized in Table 1.
Table 1.
Summary of mappings of study data to distinct modules and data elements in HITSP and OMOP.
| HITSP Module | Number of Types from Studies | OMOP | ||||
|---|---|---|---|---|---|---|
| Section Number | Name | All Data Types | Selection Criteria | Data Variables | Section Number | Corresponding Table Name |
| 1 | Personal Information | 8 | 5 | 7 | 1 | Person |
| 4 | Healthcare Provider | 1 | 1 | 14 | Provider | |
| 5 | Insurance Provider | 1 | 1 | 17 | Payer Plan Period | |
| 6 | Allergy/Drug Sensitivity | 3 | 1 | 2 | 4 & 5 | Condition Occurrence, Condition Era |
| 7 | Condition | 8 | 4 | 8 | 4, 5, 10 | Condition Occurrence, Condition Era, Death |
| 8 | Medication | 10 | 9 | 8 | 2, 3, 11 | Drug Exposure, Drug Era, Drug Cost |
| 9 | Pregnancy | 1 | 1 | 1 | 8 | Observation |
| 13 | Immunization | 2 | 2 | 2 | Drug Exposure | |
| 14 | Vital Sign | 3 | 3 | 2 | 8, 9 | Observation, Observation Period |
| 15 | Result | 5 | 5 | 4 | 8, 9 | Observation, Observation Period |
| 16 | Encounter | 10 | 8 | 8 | 6, 13, 15, 16 | Visit Occurrence, Location, Organization, Care Site |
| 17 | Procedure | 4 | 3 | 3 | 7, 12 | Procedure Occurrence, Procedure Cost |
| 18 | Family History | 1 | 1 | n/a | Observation | |
| 19 | Social History | 2 | 2 | n/a | Observation | |
| 20 | Medical Equipment | (module under development) | n/a | n/a | ||
The results and descriptive statistics, as in Table 2, provided hereafter are for HITSP mappings only; details on how the two dictionaries compared are in Table 1 and Table 3. We use the terms “selection criteria” (i.e., inclusion and exclusion criteria) and “data variables” to refer to types specified in the studies whereas we use the term “data element” to refer to the corresponding components of the dictionaries themselves.
Table 2.
A summary of the mappings of data variables, joins, and aggregate functions to HITSP data elements within queries. Unless specified otherwise, the results are for statistics across studies.
| Statistic | Selection Criteria | Data Variables |
|---|---|---|
| Average number of data elements per study | 4.46 | 6.44 |
| Range of number of data elements | 1–12 | 1–15 |
| Median number of data elements | 4 | 6 |
| Number of distinct data elements | 39 | 50 |
| Unmapped data elements | 25 | 60 |
| Number (percent) of studies with at least one unmapped variable | 19 (18.3%) | 35 (33.7%) |
| Number of joins across studies | 36 | 86 |
| Number (percent) of studies requiring joins | 25 (24.0%) | 49 (47.1%) |
| Joins per study | range 1–3 | range 1–9 |
| Modules in joins | 6 | 8 |
| Data elements in joins | 20 | 19 |
| Number of aggregates | 5 in 5 studies | 27 in 22 studies |
| Aggregate functions used | Count and Last | Count, last, minimum, maximum, average, first |
| Number (percent) of studies including both join and aggregate | 2 (1.9%) | 8 (7.7%) |
Table 3.
A summary of the most common HITSP components from the studies’ selection criteria.
| Selection Criteria Query | Three Most Frequent HITSP Components (Number, Percent) |
|---|---|
| Modules | Procedure (128, 27.6%), Condition (103, 22.2%), Result (79, 17.0%) |
| Data elements | Problem Code (95, 20.5%), Procedure Type (65, 14.0%), Procedure Date/Time (59, 12.7%) |
| Modules in joins | Procedure (19, 52.8%), Result (18, 50.0%), Encounter (14, 38.9%) |
| Elements in joins | Procedure Date/Time (16, 44.4%), Encounter Date/Time (12, 33.3%), Result Date/Time (11, 30.6%) |
The middle column of Table 2 shows the results for the mappings of data elements within the selection criteria of the studies. The selection criteria across all 104 studies totaled 464 elements, averaging 4.46 per study. The minimum number of data elements per study was 1 (a study that included all cases of a specific procedure, so the only selection criterion was Procedure Type). There were 25 selection criteria in 19 studies that did not map (more details of unmapped data are in Table 4). The most frequent HITSP components among study selection criteria are summarized in Table 3.
Table 4.
Data element mapped to one dictionary but not to the other (explicit OMOP mappings only).
| General Category | Mapped to HITSP (not to OMOP) | Mapped to OMOP (not to HITSP) |
|---|---|---|
| Personal Information (HITSP)/Person (OMOP) | Marital status | |
| Encounter (HITSP)/Visit (OMOP) | Reason for visit Associated provider | |
| Medication (HITSP)/Drug (OMOP) | Route, dose Dispensing pharmacy Response or reaction | Reason for discontinuation Costs |
| Procedure | Procedure time Body location/site | Associated condition |
| Medical Equipment | (module under development) | Devices implanted |
| History | Family History Social History Allergies |
There were five aggregates in the selection criteria in five studies. The following functions were used in the selection criteria: count (occurring four times, e.g., at least 2 vancomycin troughs, at least 2 CAD risk factors), and last (once, follow-up duration >6 months). Elements involved were Result Type (2), Encounter Date/Time (2), and Problem Code (1). The instance of a selection criterion consisting of both a join and an aggregate occurred twice. One study included patients with at least 2 (aggregate count) post-operative radiographs (join Result Type with Procedure Date/Time); another included those with a minimum of 2 years follow-up (aggregate last) from a procedure (join Encounter and Procedure Date/Time).
The right column of Table 2 shows the results for the mappings of the studies’ data variables to the data elements. They encompassed 50 distinct types of data elements (the breakdown per module is in Table 1). This sample of retrospective studies evaluated an average of 6.44 different types of data elements, with as few as one (Result Value only), a maximum of 15, and a median of 6. There were 60 data variables in 35 studies that did not map (not including Medical Equipment, which has a designated module but no assigned data elements in the HITSP dictionary, more details of unmapped data are in Table 4).
There were 27 aggregate functions in 22 studies. Functions were last (11 instances, e.g., duration of follow-up), count (9, e.g., number of procedures, UTIs, completed medication regimens), minimum (1, e.g., lowest heart rate), maximum (4, e.g., peak bilirubin), average (1, e.g., average result during a procedure), and first (1, e.g., baseline laboratory result value). Data in aggregates were mostly Encounter Date/Time (11 instances) and Result Value (10). Some study data involved both a join and an aggregate; this phenomenon occurred 8 times in our sample of studies. Five were duration of follow-up (join of Encounter Date/Time with Procedure Date/Time, with aggregate function of “last” on Encounter Date/Time). One was the average result during a procedure (join of Result Date/Time with Procedure Date/Time, with aggregate average of Result Value); another was number of UTIs per year during follow-up (join of Encounter Date/Time with count of Problem Code).
3.2. OMOP Comparison and Unmapped Data
Subsequent to the analysis of HITSP elements, we then re-mapped inclusion criteria and data variables to the OMOP dictionary for comparison. The OMOP specification includes an “Observation” table described as containing “General catch-all categories from various data sources that cannot be otherwise categorized within the entities provided.” This approach of using a generic table requires precoordinated terms. This accommodates any element to functionally map to this table, although many data types explicitly described in HITSP (such as family history, allergies) are covered but incompletely so by this table. For example, encoding a family history observation would require a term for “family history of colon cancer in first-degree relative.” Even so, the model cannot accommodate all details. For example, HITSP has a separate family history field for age at onset, but this cannot be modeled in the OMOP Observation table without extending the data model. Across both selection criteria and study data, there were 138 elements that did not map explicitly to OMOP-provided entities, compared with 75 elements unmapped to HITSP. Thirty-nine of the 104 studies contained at least one inclusion criterion or data variable that did not map to either dictionary. These data types that did not map to HITSP, OMOP, or either, are summarized by general categories in Table 4 and Table 5.
Table 5.
Data element unmapped to either dictionary.
| Category | Example |
|---|---|
| Details at the point of diagnosis | Date diagnosis made, method and circumstance (whether by tests or history, because of symptoms versus by screening), provider and specialty or institution that made diagnosis |
| Patient thought | Reason for delay of care, patient’s preference, influences on patient decision, satisfaction |
| Clinician thought | Recommendation of treatment, prior suspicion of a condition, provider satisfaction |
| Procedural characteristics | Urgency of procedure, equipment utilized (not implanted), details of process |
| Success of treatment | Failed prior procedure or medication |
| Clinical status | Listing for transplant |
| Documentation | Availability of video recordings |
| Administrative/ managerial elements | Costs of hospitalization, specialty of referring physician, appropriateness of management, experience of surgeon, presentation at a teaching conference |
| Obstetric details | Lactation, pregnancy planned or unplanned |
| Social factors (not part of the standard social history) | Current education plan, size of family or household, vaccination of family members, Worker’s Compensation, age of partner, partner’s prior pregnancy, age at menarche/voice break, quality of housing conditions |
Other difficulties in mapping were encountered in some studies that required the joining of two charts. One common occasion of this was in obstetric studies, in which data on both the mother and infant were needed. Another found recipients of a particular treatment and examined clinical courses of their household contacts. These are complex studies not easily described by a data query.
3.3. Researcher Responses
Ten of the 219 researchers we contacted responded to our questionnaire on the study details and process, for a response rate of only ~5%. Six reported using EHRs for their studies; five used clinical databases (not mentioned in their manuscripts). Three used research registries and only two used paper charts. To our inquiry for further details on study selection criteria and data examined, researchers responded with no major deviation from what could be obtained from the article, with one exception: one researcher reported examining seven data elements that were not mentioned in the publication. All others referred to their paper as containing all relevant information. This was helpful with our mapping, allowing us to conclude that our analysis of elements as we obtained them from publications was closely correlated with the actual study process.
Eight investigators provided estimates on the time spent gathering and analyzing study data; these varied widely from only “a few” hours to 1700 person-hours (two estimated 60 hours, while the other responses were 30, 120, 160, and 250). Researchers expressed desire for improvements in EHRs for performing retrospective studies, mentioning specific needs such as data warehousing, easy exporting to databases, enhanced searching features, and a nationally standardized EHR.
4. DISCUSSION
4.1. Significance
We attempted to identify data needs for performing retrospective observational studies by analyzing the data that were used in studies published in the literature. By mapping data elements to standard definitions to be used in HIEs and research data repositories, we can perform preliminary assessment of the feasibility of obtaining data for research studies from clinical databases. We found, similarly to our previous findings, that the most frequently used data elements are Encounter/Visit, Procedure, Condition, Result/Observation, Personal Information, and Medication/Drug. This is an encouraging result since many of these data are often available in coded form due to billing and reporting needs.
We also were able to gain some insight into the nature and complexity of queries. We found that the median number of data elements used in study selection criteria was about four and in study variables was six. We also observed that in about a quarter of the studies, the selection criteria required a join operation, and in about half the studies, the query for data variables required one. The data elements that were joined were usually temporal data elements. Five studies needed aggregate functions in the selection criteria and 22 studies needed those for the data variable queries. Features to perform aggregation or to join data often are not available in the graphical query tools for research data warehouses. These results also indicate, as we expected, that queries for obtaining the data variables are more complex than those for selection criteria.
More than one-third of the studies involved at least one element that could not map explicitly to either dictionary, demonstrating limits in the current standards to adequately encompass the breadth and granularity of information that is included in clinical data and examined in clinical research. Some of the unmapped elements were very specific, uncommonly sought information in clinical histories (e.g., quality of housing conditions, influences on a patient’s decision). Others were elements generally known at the time of care and likely documented (e.g., prior treatment failure, clinician’s recommended course), whereas others would require a great deal of expert interpretation (e.g., appropriateness of prior management) or are not reasonably considered priority to store in a patient’s EHR (e.g., experience of surgeon). The incorporation of this type of information in study data, as well as the knowledge that future research will also assuredly include nontraditional and unforeseen variables in studies, demonstrates not only the current needs of research queries, but also the difficulty in building a sufficiently thorough and timeless data dictionary. While many retrospective studies may be completed with simple queries, additional effort may often be needed to retrieve all desired data elements, whether by natural language processing, manual individual chart review, or extending queries to other databases. Clinical research studies rarely, if ever, refer to standard data dictionaries when describing study design and outcomes assessed. This may indicate a possible area for future collaborative efforts with data modelers, as improved standards may greatly facilitate retrospective research.
The differences in successful mappings between the two data element dictionaries also demonstrate their different purposes. The HITSP model was created for patient care and strived for explicit comprehensiveness, increasing its complexity. The OMOP model was created for medication surveillance, and therefore the model was not as comprehensive for certain data elements. Analysis of studies alongside two data element dictionaries suggests the difficulty and perhaps limitations of the delineation of clinical data into discrete modules. Several very important data elements are not discrete entities but rather exist as a function or linkage of two or more other types, such as Allergy (medication and condition or symptom), Reason for visit (encounter and condition), but with various relations between these entities (cause, effect, etc.).
The methodology of this study provides an approach to identify requirements of data needs for clinical research from published studies. It is complementary to requirements we are gathering from traditional methods used in software engineering such as by engaging users, reviewing existing tools and methods they use, analyzing the sources of data, and reviewing similar tools. The results of this study will be helpful to designers, developers, and operators of CDRs for research. First, the results validate that much of the data needed for clinical research is available in EHRs which may justify the investments made in the development of CDRs. Second, it provides insights into the data needs, which can guide the schedule and prioritization of data availability in the CDR. Finally, the structure and complexity of queries can provide insights into the design of the graphical user interface. Our analysis indicates the need to perform joins using temporal operators in about a quarter of the studies for selection criteria and in almost half the studies for study variables. Yet many of the cohort discovery tools do not provide the ability to join data elements.
4.2. Limitations
We limited the mapping to data elements and attributes to two dictionaries: HITSP/C154 and OMOP v3. Our study aims to provide empirical support for CDR schema design by characterizing the data elements used in published studies. Our desire was to use definitions of data elements that would have broad agreement and be well understood. Hence, we leveraged the Health Information Technology Standards Panel Data Dictionary (formally referred to as construct C154).[11] The data dictionary is used as the basis of data standards in health information interchange. The dictionary defines types of data elements such as Encounter, Result, Condition, Medication, and Procedure. These each have comparable types within the OMOP dictionary. For each type of element, both list attributes and the definitions and constraints on values of those attributes.
Our dictionary selection was influenced by the fact that HITSP is designed to represent a broad set of EHR data. The common data model was created by the Observational Medical Outcomes Partnership (OMOP) was originally designed to support the narrower use case of medication surveillance from clinical data [21], and later expanded for CER applications. Other data standards being used in research are designed for reporting results of or data sets associated with clinical trials[22,23], not necessarily those obtained from EHRs.
We performed the mappings on version 3 of OMOP as that was the published version at the time of our analysis. Subsequent to this, however, a newer version of OMOP was released. A comparison of the two versions indicated that use of the later version would not have changed our results since the changes to the data schema are insignificant.
A limitation of our research we analyzed only 104 studies due to the significant effort required to map the data needs of each study. This volume of studies may not represent the full range of observational studies. While this may impact the quantitative result values, the overall findings are consistent with our experience in providing data to researchers from our institution’s CDR.
Another limitation of the analysis is that the data was mapped primarily by one researcher. This was in part, mitigated by the review by the other author. While this review did not identify any significant issues in mapping, it was a simple verification and not a blinded check, and we did not quantify the results of the second review. Additionally, we had a low response rate from the study authors. This may be explained by the fact that we offered no incentives for the respondents to participate, and we did not send reminders. The responses we received corroborated our findings. However, a larger response rate would provide more confidence in the study results.
The study indicates that EHRs contain the types of data needed for research studies. However, effective use of data from EHRs must also consider issues of data quality[24,25], but we did not address that in this study.
4.3. Conclusions
This study characterized the types of EHR data that are needed in clinical research studies and the complexity of queries required to obtain such data. The results should be helpful in the implementation of data repositories for research and the design of cohort discovery and other data access tools. Our results confirm that a majority of the data elements needed for research are mappable to standard dictionaries and typically available in an EHR system in a coded format.
Supplementary Material
| Data examined | Element | Module | Identifier |
|---|---|---|---|
| vestibular schwannoma resection | Procedure Type | 17. Procedure | 17.02 |
| May 1990 to May 2009 | Procedure Date/Time | 17. Procedure | 17.04 |
Acknowledgments
This research was supported by the National Library of Medicine Training Grant T15 LM011271-01, and by the Agency for Healthcare Research and Quality (AHRQ) through the American Recovery & Reinvestment Act of 2009, Grant R01 HS019913
References
- 1.Sox HC, Greenfield S. Comparative effectiveness research: a report from the Institute of Medicine. Ann Intern Med 2009;151:203–5. [DOI] [PubMed] [Google Scholar]
- 2.Wilke RA, Berg RL, Peissig P, et al. Use of an electronic medical record for the identification of research subjects with diabetes mellitus. Clin Med Res 2007;5:1–7. doi: 10.3121/cmr.2007.726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murphy SN, Weber G, Mendis M, et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2). J Am Med Inform Assoc JAMIA 2010;17:124–30. doi: 10.1136/jamia.2009.000893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCarty CA, Chisholm RL, Chute CG, et al. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics 2011;4:13. doi: 10.1186/1755-8794-4-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boxwala A, Kim H, Choi J, et al. Understanding data and query requirements for cohort identification in clinical research studies. Proc AMIA Clin Res Inform Summit 2011. [Google Scholar]
- 6.Vogt TM, Elston-Lafata J, Tolsma D, et al. The role of research in integrated healthcare systems: the HMO Research Network. Am J Manag Care 2004;10:643–8. [PubMed] [Google Scholar]
- 7.Harrison JH Jr. Introduction to the mining of clinical data. Clin Lab Med 2008;28:1–7. doi: 10.1016/j.cll.2007.10.001 [DOI] [PubMed] [Google Scholar]
- 8.caBIG Strategic Planning Workspace. The Cancer Biomedical Informatics Grid (caBIG): infrastructure and applications for a worldwide research community. Stud Health Technol Inform 2007;129:330–4. [PubMed] [Google Scholar]
- 9.Lowe HJ, Ferris TA, Hernandez PM, et al. STRIDE--An integrated standards-based translational research informatics platform. AMIA Annu Symp Proc AMIA Symp AMIA Symp 2009;2009:391–5. [PMC free article] [PubMed] [Google Scholar]
- 10.Clinical Records Integrated Query Tool (CRIQueT). http://dbmi.ucsd.edu/pages/viewpage.action?pageId=524610 (accessed 25 Jul2013).
- 11.HITSP Data Dictionary Component (HITSP/C154). 2010http://www.hitsp.org/ConstructSet_Details.aspx?&PrefixAlpha=4&PrefixNumeric=154 (accessed 24 Jul2013).
- 12.Common Data Model Version 3 Specifications. 2011http://omop.fnih.org/CDMV3newestversionhttp://omop.org/CDMvocabV4 (accessed 24 Jul2013).
- 13.Health Information Technology Standards Panel. HITSP Summary Documents Using HL7 Continuity of Care Document (CCD) Component (HITSP/C32). 2009http://www.hitsp.org/ConstructSet_Details.aspx?&PrefixAlpha=4&PrefixNumeric=32 (accessed 25 Sep2014).
- 14.Harpaz R, DuMouchel W, LePendu P, et al. Performance of pharmacovigilance signal-detection algorithms for the FDA adverse event reporting system. Clin Pharmacol Ther 2013;93:539–46. doi: 10.1038/clpt.2013.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou X, Murugesan S, Bhullar H, et al. An evaluation of the THIN database in the OMOP Common Data Model for active drug safety surveillance. Drug Saf Int J Med Toxicol Drug Exp 2013;36:119–34. doi: 10.1007/s40264-012-0009-3 [DOI] [PubMed] [Google Scholar]
- 16.Schuemie MJ, Gini R, Coloma PM, et al. Replication of the OMOP experiment in Europe: evaluating methods for risk identification in electronic health record databases. Drug Saf Int J Med Toxicol Drug Exp 2013;36 Suppl 1:S159–169. doi: 10.1007/s40264-013-0109-8 [DOI] [PubMed] [Google Scholar]
- 17.Schadow G, Mead CN, Walker DM. The HL7 reference information model under scrutiny. Stud Health Technol Inform 2006;124:151–6. [PubMed] [Google Scholar]
- 18.CDISC Submission Data Standards Team. Study Data Tabulation Model, version 1.4. Austin, TX: : Clinical Data Interchange Standards Consortium, Inc. 2013. [Google Scholar]
- 19.CDISC Analysis Data Model Team. Analysis Data Model, Version 2.1. Austin, TX: : Clinical Data Interchange Standards Consortium, Inc. 2009. [Google Scholar]
- 20.Wind JJ, Leonetti JP, Raffin MJM, et al. Hearing preservation in the resection of vestibular schwannomas: patterns of hearing preservation and patient-assessed hearing function. J Neurosurg 2011;114:1232–40. doi: 10.3171/2010.11.JNS091752 [DOI] [PubMed] [Google Scholar]
- 21.Overhage JM, Ryan PB, Reich CG, et al. Validation of a common data model for active safety surveillance research. J Am Med Inform Assoc JAMIA 2012;19:54–60. doi: 10.1136/amiajnl-2011-000376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fridsma DB, Evans J, Hastak S, et al. The BRIDG project: a technical report. J Am Med Inform Assoc JAMIA 2008;15:130–7. doi: 10.1197/jamia.M2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuchinke W, Aerts J, Semler SC, et al. CDISC standard-based electronic archiving of clinical trials. Methods Inf Med 2009;48:408–13. doi: 10.3414/ME9236 [DOI] [PubMed] [Google Scholar]
- 24.Kahn MG, Raebel MA, Glanz JM, et al. A pragmatic framework for single-site and multisite data quality assessment in electronic health record-based clinical research. Med Care 2012;50 Suppl:S21–29. doi: 10.1097/MLR.0b013e318257dd67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bayley KB, Belnap T, Savitz L, et al. Challenges in using electronic health record data for CER: experience of 4 learning organizations and solutions applied. Med Care 2013;51:S80–86. doi: 10.1097/MLR.0b013e31829b1d48 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.