Abstract
Mycobacterium tuberculosis (Mtb) is a facultative intracellular pathogen that infects macrophages where it avoids elimination by interfering with host defense mechanisms, including phago-lysosome fusion. Endosomal Toll-like receptors (TLRs) generate Type I Interferons (IFNs), which are associated with active tuberculosis (TB).
We aimed to explore if DNA from different Mtb lineages lead to differences in the inflammatory response of human monocytic/macrophage cells. THP-1 cells which express two inducible reporter constructs for interferons (IFNs) as well as for NF-κB, were stimulated via endosomal delivery of Mtb DNA as a nanocomplex with PEI. DNA from different Mtb phylogenetic lineages elicited differential inflammatory responses in human macrophages. An initial relatively weak IRF-mediated response to DNA from HN878 and H37Rv increased if the cells were pre-treated with Vitamin D (Vit D) for 72 hrs. RNAseq of THP-1 under different transformation conditions showed that pre-treatment with Vit D upregulated several TLR9 variants, as well as genes involved in inflammatory immune response to infection, immune cell activation, Type I IFN regulation, and regulation of inflammation.
Vit D appears to be important in increasing low IRF responses to DNA from certain lineages of Mtb. Variations in the IRF-mediated response to DNA derived from different Mtb genotypes are potentially important in the pathogenesis of tuberculosis since Type I IFN responses are associated with active disease. The role of Vit D in these responses could also translate into future therapeutic approaches.
Keywords: Mycobacterium tuberculosis, Type I IFNs, Vitamin D
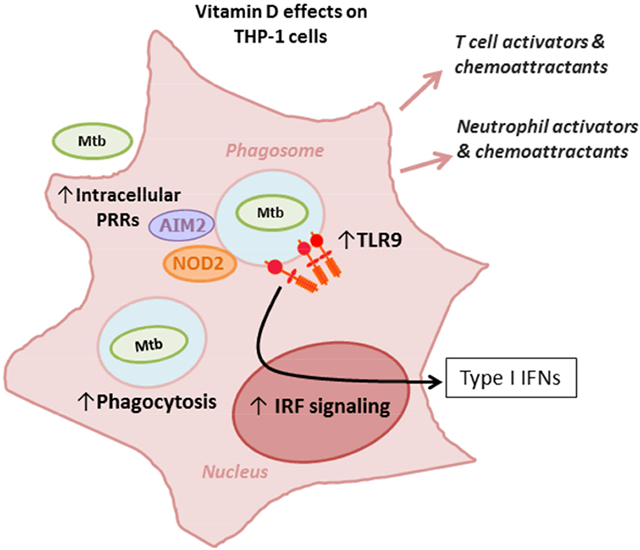
Graphical Abstract
1. Introduction
Tuberculosis (TB) is one of the oldest illnesses in history and still remains a major health problem worldwide, with more than 8 million new cases and more than 1.8 million deaths occurring every year [1]. Mycobacterium tuberculosis (Mtb) mainly infects professional phagocytes in the lungs where it uses strategies such as prevention of phagolysosome maturation and subversion of host cell death pathways, to survive and replicate [2].
In order to mount an appropriate antimicrobial response, the host’s innate immune response first detects pathogens through pattern recognition receptors (PRRs), which recognize conserved pathogen associated molecular patterns (PAMPs). Toll-like receptors (TLRs) expressed on macrophages can recognize PAMPs on Mtb [3, 4] and mediate the production of immune-regulatory cytokines such as tumor necrosis factor (TNF) and type I Interferons (IFNs). A type I IFN gene signature is associated with active TB [5, 6]. Type I IFNs (IFN-α and IFN-β) appear to suppress type II Interferon (IFN-γ)-triggered responses [7, 8], including Mtb killing by macrophages [9]. Although different isolates from Mtb have shown differences in invasiveness [10] and proinflammatory cytokine induction [11, 12], no study has evaluated differences in Type I IFN induction in humans with different Mtb strains. A differential induction of Type I IFNs might explain how TB manifests in some individuals, as type I IFNs have been demonstrated to strongly inhibit monocytes’ and macrophages’ responsiveness to IFN-γ, and are associated with active TB disease [7-9].
The role of Vitamin D (Vit D) in the treatment of tuberculosis has been portrayed in the early 20th century with the use of heliotherapy, and then continued in the form of Vit D supplementation in the pre-antibiotic era [13]. Vit D appears to be crucial for macrophage activation and early Mtb clearance [14-16]. Vit D enhances innate immunity through TLR and IFN-γ, reversal of phagosome maturation arrest, increased expression of various antimicrobial peptides, and induction of autophagy in infected cells, thus restricting Mtb intracellular growth in macrophages [17].
We herein show that Mtb nucleic acid of different lineages elicit differential inflammatory responses in human macrophages. We also investigated the effect of Vit D in these responses and observed that cells which initially showed a weak Interferon regulatory factor (IRF)-mediated response to DNA from certain Mtb strains, could increase it if they were pre-treated with Vit D. Our findings show that both, NF-κB and IRF-activation in macrophages, are differentially affected by the lineage of Mtb, and that Vit D appears to be important in increasing the IRF response when this is low.
2. Material and Methods
2.1. Human cell lines
Human monocytic cell line THP-1 (ATCC TIB-202), and dual THP-1 (Invivogen) cell lines were used for our experiments. The latter expresses two inducible reporter constructs for interferon regulatory factors (IRFs) as well as for NF-κB. The activities of these reporters were measured through a colorimetric assay for the secretory embryonic alkaline phosphatase (SEAP) reporter gene that is linked to NF-κB activation using the Quanti-blue substrate (InvivoGen), and a Luciferase reporter gene linked to IRFs using the QuantiLuc substrate (Invivogen), according to manufacturer’s instructions. Measurement of NF-kB and IRF activation were expressed as a response ratio for each stimulus relative to the reporter activity in unstimulated cells.
THP-1 cells were cultured in RPMI 1640 medium supplemented with 10% FBS, 0.05 mM 2-mercaptoethanol, 100 U/ml Penicillin and 100 μg/ml Streptomycin at 37 °C and 5% CO2. Differentiation and pre-treatment of THP-1 cells were done with 100 ng/mL phorbol ester 12-O-tetradecanoylphorbol-13-acetate (PMA, Sigma-Aldrich) or 0.5μM 1,25-dihydroxyvitamin D3 (Vitamin D3, Sigma-Aldrich) for 72 hours. For some experiments, cells were also activated with 100 ng/mL LPS plus 20 ng/mL IFN-γ for 8 hrs. Reagents were used in working dilutions (1/100) in PBS from stock solutions in DMSO.
2.2. Bacterial nucleic acid stimulation
Genomic DNA from different Mtb lineages: Indo-Oceanic T17X strain (NR-44096) (Lineage 1), HN878 strain (bei NR-14867) (Lineage 2), East-African-Indian strain (NR-44095) (Lineage 3), and Mtb strains H37Rv (NR-14865) and H37Ra (ATCC 25177D-5) (Lineage 4), as well as DNA from Mycobacterium bovis (ATCC BAA-935D-2) were acquired from ATCC / bei resources. Strains and correspondent lineages [18] are summarized in Table 1. CpG 2395 (Invivogen) was used as a control ligand.
Table 1.
Mtb Genomic DNA used in this study
| Strain | ATCC | Lineage |
|---|---|---|
| Indo-Oceanic T17X | NR-44096 | Lineage 1 |
| HN878 | NR-14867 | Lineage 2 |
| East-African-Indian | NR-44095 | Lineage 3 |
| H37Rv | NR-14865 | Lineage 4 |
| H37Ra | 25177D-5 | |
| M. bovis | BAA-935D-2 |
Cells, at a density of 2.4×104 cells/well, were stimulated for 24 hours with 100ng of DNA delivered with the use of the cationic polymer polyethilenimine (PEI) according to manufacturer instructions (Polyplus, France). PEI is a transfection reagent that wraps negatively charged RNA and DNA with subsequent transport of the PEI/nucleic acid nanoparticle complex to early and late endosomes [19]. We have previously utilized this endo-lysosomal delivery system for bacterial nucleic acid to evaluate endosomal TLR activation and cytokine induction, including type I IFN in human phagocytic cells [20].
2.3. RNA-Sequencing library preparation
After removing the medium, THP-1 cells were washed twice with cold PBS and centrifugation to get rid of floating cells. Afterwards, total RNA was isolated using RNesay Kit together with QIAshredder (Qiagen). Residual DNA was removed by 30-min DNase treatment (TURBO DNA-free Kit, Ambion) at 37 °C. Subsequently, RNA was purified with RNA Clean and Concentrator (Zymo Research), and its concentration and integrity was checked using ribogreen assay and bioanalyzer (Agilent Technologies 2100 Bioanalyzer). Only RNA samples with RIN number above 8 were further processed. RNA library preparation was carried out using the TruSeq Stranded mRNA Library Prep Kit (Illumina). HiSeq 2500 System Rapid mode (Illumina) was used for RNA-sequencing, giving rise to paired-end reads of 101 bp. A total number of reads per sample was obtained in the range of 123-189×106, with 85.8-91.0% mapped reads.
2.4. RNA-Seq data analysis
The original read quality of our RNA-Seq data was first confirmed by FastQC (Simon Andrews, Babraham Bioinformatics). For gene expression analysis, the paired-end reads were aligned to human genome (UCSC-hg19) using Tophat2 [21] with Bowtie2 [22]. In RNA-Seq data analysis, the default parameters for each tool were used unless otherwise stated. For Tophat2, a maximum of 2 mismatches were allowed and two parameters as --library-type=fr-firststrand and --read-realign-edit-dist=0 were set. Cufflinks, Cuffmerge and Cuffdiff [23] were used for transcripts assembly and quantification of the Tophat2 output. Annotated transfer RNA, ribosomal RNA and mitochondrial transcripts were masked out in Cuffdiff analysis by using the parameter –mask-file with corresponding GTF file. The common FPKM (fragments per kilobase of transcript per million fragments mapped) were used as the expression-levels measure for each gene or transcript.
2.5. Polymerase chain reaction (PCR)
2.5.1. Semi-quantitative PCR
Characterization of expression of TLR-9 and inteferon-1beta was done by semi-quantitative PCR, as described before (Chhabra et al. 2017, JI). In brief, cDNAs were prepared from mRNAs derived from THP-1 cells stimulated with DNA from different MTB strains, using the SuperScript cDNA synthesis kit (Invitrogen Inc.), cDNAs were quantified and 2.5ng cDNA was used for each PCR reaction of 50 ul volume. GAPDH gene amplified as the loading control. PCR conditions included 1 cycle of denaturation step at 94C for 5 min, followed by 40 cycles of amplification step comprised of denaturation at 94C for 30sec, annealing at 56C for 30sec and extension at 72C for 30 sec., followed by one cycle at 72C for 10 min. (PCR Primers: TLR-9 F: 5‘-GAATGCCAGTTGGTTCCGTG-3‘, TLR-9 R: 5‘-TTCAGAGCTGGGAGTGTCCA-3‘, IFN-1b F: 5‘-GCCGCAGTGACCATCTATGA-3‘, IFN-1b R: 5‘-CAGTGACTGTCCTCCTTGGC-3‘, GAPDH F: 5‘-GAAGGTGAAGGTCGGAGTC-3, GAPDH R: 5‘-GAAGATGGTGATGGGATTTC-3‘).
2.5.2. Real-Time quantitative PCR
RT-qPCR analysis was also performed for TLR9 using the previously described cDNA as template. Complementary DNA samples were diluted 1:10 (vol:vol) in deionized water, samples were stored at −20°C. Amplification reactions were performed in triplicate for each tested gene. Each reaction contained 1 μl of cDNA, 5 μl of iQ™ SYBR® Green Supermix (Bio-Rad), 3.7 μl of water and 150 nM of each primer to a final volume of 10 μl. Primer sequences for TLR9, and GAPDH, have been published elsewhere[24, 25]. Non-template controls (NTC) were included in triplicate for each primer pair confirming for the absence of contamination. Amplification reactions were performed on a CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad). PCR conditions were performed as follows, denaturation at 95°C for 3 min, 40 cycles of denaturation at 95°C for 10 sec and annealing/extension at 60°C for 30 sec. To ensure the presence of a single amplicon, a melting curve analysis was performed at the end of each PCR run. Cycle threshold (Ct) values for each gene were used to calculate the transcript levels of TLR9 in reference to the endogenous control GAPDH. The relative changes in gene expression generated were calculated using the 2−ΔΔCt method [26].
2.6. Endosomal TLR Blocking.
A specific immunoregulatory DNA sequence (IRS869: 5’-TCCTGGAGGGGTTGT-3’) [27] (Integrated DNA Technologies) was used to block TLR9 signaling. IRS869 was used at a concentration of 1.4 μM, as it has shown previously to result in optimal effectiveness and specificity of the inhibitory sequences [27, 28]. The ODN was added 1 hour prior to cell stimulation.
2.7. Statistical Analysis
General statistical analysis was performed using GraphPad Prism 7.01 (GraphPad Software, San Diego, CA). For comparison or response between Untreated vs. Vitamin D-treated for each stimulus a t-test or a non-parametric test (Mann-Whitney) was used, depending on if the data followed a normal distribution or not. p values of <0.05 were considered significant. A two-way ANOVA was performed to assess the interaction of Vit D with the responses upon stimulation with the different Mtb DNAs. For RNAseq, a transcript change consistent in all the three replicates was considered as significant at the statistic level if the q-value was less than 0.05.
3. Results
3.1. DNA from different M.tuberculosis phylogenetic lineages elicits differential inflammatory response in human macrophages
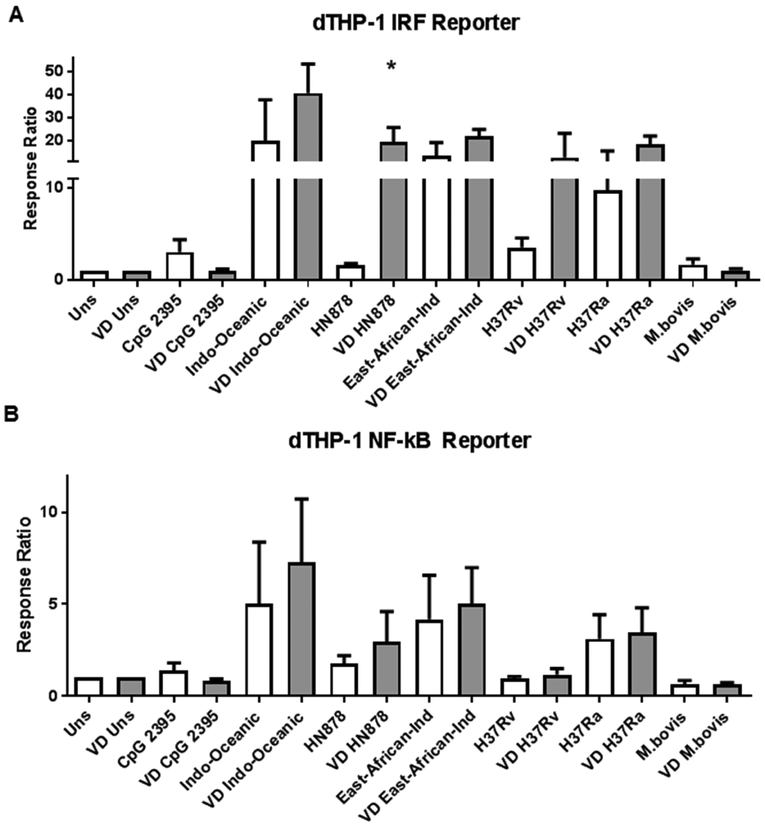
We first aimed to evaluate TLR9 activation by DNA from well characterized Mtb species belonging to different genotypes [18] (available on ATCC) on human macrophages. Utilizing endosomal delivery of Mtb DNA as a nanocomplex with PEI [19, 20], we evaluated differences in pro-inflammatory cytokines and interferon induction in THP-1 cells by nucleic acids belonging to distinct Mtb genotypes. We observed that different genotypes are able to elicit different NF-κB and different IRF-mediated responses after stimulation with the distinct Mtb DNAs (Figure 1, white bars).
Figure 1. DNA from different M.tuberculosis phylogenetic lineages elicits differential inflammatory response in human macrophages.
Dual THP-1 cells were stimulated for 24 hours with 100ng of DNA from several different phylogenetic lineages of Mtb. Response from IRF (A) or an NF-kB (B) reporters are shown. Pretreatment of cells with Vitamin D (gray bars) show increase the response to DNA from HN878 and virulent strain H37Rv (A). * p<0.05 Unpaired t test.
No apparent relationship was observed between genetic distance between lineages and the strength of the elicited NF-kB or IRF-mediated-responses. East-African-Indian (Lineage 3), and Indo-Oceanic (Lineage 1), show the highest response (over 15 and 50 fold, compared to unstimulated). Despite their relative closeness to Lineage 3 [18], DNA from HN878 (Lineage 2), and H37Rv (Lineage 4), showed a relatively weak response (below 5 fold, compared to unstimulated).
3.2. Vitamin D increases the IRF-mediated response to Mtb DNA in cells that were initially poorly reactive
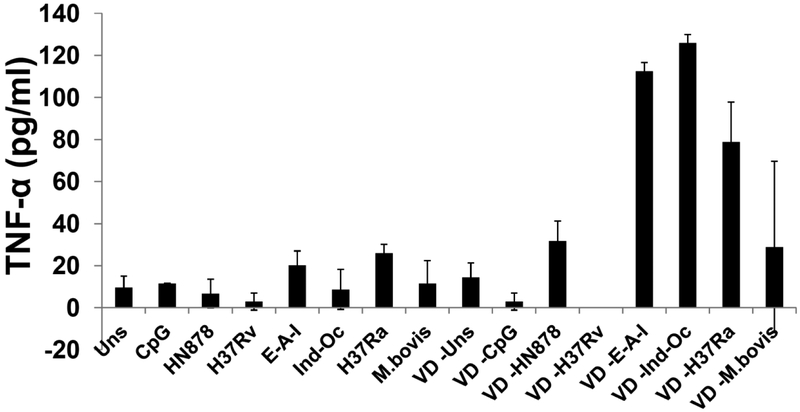
As macrophage activation by Vit D is important in Mtb recognition and clearance [15, 16], we then evaluated the effect of Vit D activation on the response upon Mtb-DNA stimulation. Although pre-treatment of THP-1 cells with Vit D did not substantially affect the overall NF-kB-mediated response (Figure 1B), it did increase the secretion of TNF-α (Figure 2). With respect to the IRF-mediated response, Vit D increased the IRF-mediated response to Mtb-DNA of HN878 and H37Rv (Fig. 1A, gray bars). A two way ANOVA analysis was performed to assess the interaction of Vit D with the responses upon stimulation with the different Mtb DNAs. Vit D treatment constituted a factor that impacted the IRF-mediated responses to Mtb DNA stimuli (p= 0.01). This did not occur in respect to NF-κB-mediated responses (p=0.37). Vit D also increased the IRF-mediated response to other TLR ligands (Figure 3A). In fact, Vit D was the only condition that increased this response. Neither IFN-γ nor IFN-γ + LPS showed any difference compared to untreated or PMA-treated cells (Suppl 1). Overall, treatment of THP-1 cells with Vit D induced upregulation of genes involved in inflammatory immune response to infection, immune cell activation, recruitment of effector cells to site of inflammation, antigen presentation and recognition, Type I IFN regulation, and regulation of inflammation (Suppl 2 and Table 2).
Figure 2. Pre-treatment of human macrophages with Vitamin D increases secretion of TNF-α after stimulation with DNA from different M.tuberculosis phylogenetic lineages.
Dual THP-1 cells were pre-treated (or not) with Vitamin (VD) for 72 hours, and then stimulated for 24 hours with DNA from several different phylogenetic lineages of Mtb. Two way ANOVA p<0.05
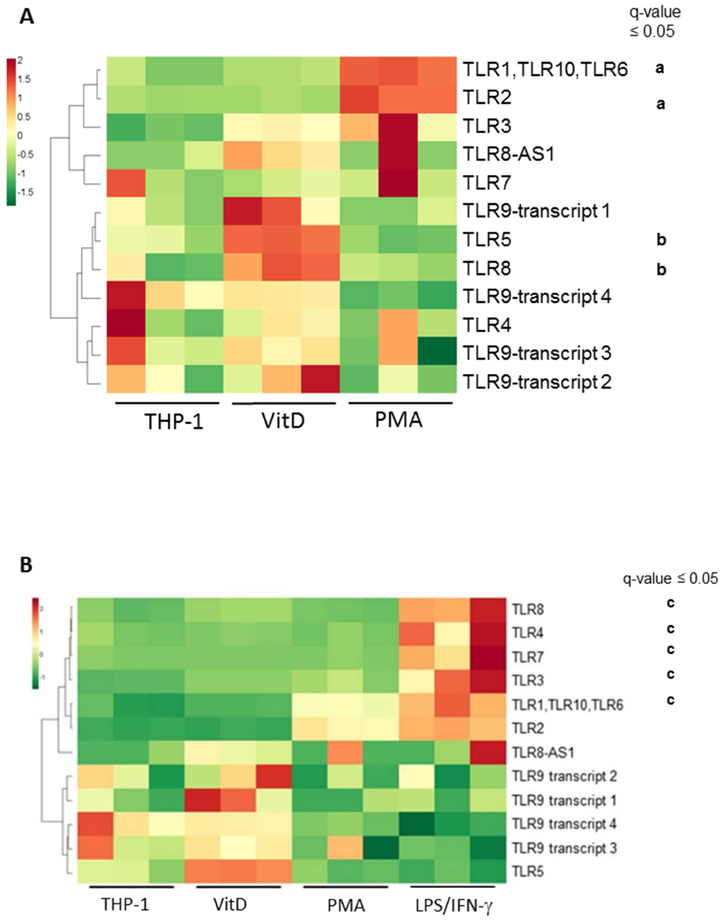
Figure 3. Vitamin D promotes upregulation of TLR9 splicing variants in THP-1 cells THP-1 cells were pre-treated with Vit D or PMA for 72 hrs, this latter with or without LPS plus IFN-γ for 8 hrs.
Heatmap shows upregulation of a set of TLR genes by Vit D that includes specific TLR9 splicing variants. Data for A and B excludes and includes, respectively, the data with PMA followed by LPS and IFN-γ treatment. Each of the three columns per condition indicate a biological replicate (different cell preparation). PMA and LPS/IFN-γ treatments induced upregulation of other TLRs that do not include TLR9.
a Comparison between THP-1 vs. PMA
b Comparison between THP-1 vs. Vit D
c Comparison between PMA vs. LPS/IFN-γ
Table 2.
Genes upregulated by Vit D treatment of THP-1 cells
| Function | Gene name | Role in TB |
|---|---|---|
| CXCL8 | Stimulator of phagocytosis | |
| Mediators of inflammatory response | IL1B | Caspase-1 activation. Stimulator of MIP-I alpha production |
| CCL22 | Lung inflammation through CCR4 activation | |
| Transcription Factor for Type I IFNs | IRF5 | Necessary for Type I IFN in response to Mtb |
| Recruitment of effector immune cells to site of inflammation | CXCL8 | Neutrophil chemoattractant |
| CCR1 | Receptor for CCL-3, −4, −5, and MIP-1 alpha | |
| CD38 | M1 polarization inflammatory marker in Mtb infection | |
| T cell chemoattractant | IL16 | Increased in TB pleural effusion |
| CCL22 | Lung inflammation through CCR4 activation | |
| CD86 | M1 polarization marker in Mtb infection | |
| T cell activation | CD84 | Increased in B cell precursors in Mtb infection |
| IL27RA | Involved in resolution of inflammation. Induced by Type I IFNs in Mtb infection | |
| Neutrophil and NK-mediated bacterial killing | CXCR1 | Oxidative defense in active TB |
| CD244 | Increased in lymphocytes of TB patients | |
| Antigen presentation | HLA-B,HLA-C | Restricting alleles for TB CD8 antigens |
| Pathogen Recognition Receptor | TLR8 | Enhances the innate and adaptive immune response |
| TLR5 | Expressed on myeloid cells in TB granulomas | |
| P2RX7 | Induces cell death and subsequent loss of intracellular bacterial viability in human macrophages infected with mycobacteria | |
| CD14 | Facilitates the uptake of non-opsonized Mtb | |
| Intracellular PRR | NOD2 | Enhances the innate response of alveolar macrophages to Mtb |
| AIM1 | IL-1β and IL-18 production and Th1 responses to Mtb | |
| TLR adapter | TIRAP | Influences disease susceptibility by modulating the inflammatory response |
| Regulated by Type I IFNs and MyD88 | IL10RA | Anti-inflammatory cytokine expressed in chronic Mtb granulomas |
| Serine protease inhibitors | SERPINB9 | Inhibits apoptosis and promotes survival of Mtb-infected macrophages |
| SERPING1 | Component of active TB signature | |
| SERPINB1 | Critical for neutrophil survival | |
| Complement degradation | CD46 | Bactericidal activity of macrophages and granuloma formation |
| Regulation of Inflammation | SIGLEC10 | Increased in CD14+CD16++ monocytes upon Mtb infection |
| SIGLEC14,SIGLEC5 | Involved in Mtb replication in monocytes |
3.3. Vitamin D promotes upregulation of TLR9 splicing variants in THP-1 cells
To address the question if the effect of Vit D was due to upregulation of TLR9 in THP-1 cells, RNAseq was carried on these cells, comparing different activation conditions. Transcriptome analysis showed that Vit D treatment of cells lead to upregulation of a cluster of TLR genes, including TLR5, TLR8, and certain splicing variants of TLR9, although the latter where not statistically significant (Fig. 3). This set of TLR genes were observed to be downregulated with the other cell treatment conditions such as PMA or IFN-γ + LPS (Figure 3B). PMA led to the known differential expression (DE) of TLR2 [29] while IFN-γ + LPS-treatment lead to upregulation of a different set of TLRs, namely TLR3, 4, 7, and 8.
Semi-quantitative PCR on Mtb-DNA stimulated cells, showed a minimal increase in the transcription of IFN-β, after stimulation with DNA of certain Mtb-strains, and an increase in TLR9 transcript in THP-1 cells pre-treated with Vit D, for all Mtb-DNA, except Indo-Oceanic (Suppl 3A-C). Even though the RT-PCR could not confirm these results, it did show a prominent increase in TLR9 expression on Vit D-treated cells after stimulation with DNA from M. bovis (Suppl 3D). To further examine the involvement of TLR-9 in the responses to Mtb DNA, we performed TLR9 inhibition utilizing a specific immunoregulatory sequence (IRS). Although a reduction was observed for the NF-kB mediated response, and not for the IRF-mediated one, this was only observed for certain Mtb lineages (Suppl 4).
4. Discussion
Little is known about nucleic acid signaling in tuberculosis or its role in susceptibility to the disease. It has been reported that stimulation of human alveolar macrophages with DNA from attenuated mycobacteria (H37Ra or BCG) vs. generates a greater TNF-α response, compared to stimulation with DNA from a virulent mycobacteria (H37Rv or M.bovis) [30]. In this study, we observed that DNA from hypervirulent HN878 strain (Lineage 2) showed both low NF-kB and IRF-mediated responses. HN878 is genetically close to virulent Mtb Beijing strain [31], which induces a diminished inflammatory production [12]. Our findings demonstrate that DNA from different Mtb phylogenetic lineages elicits differential IRF-mediated responses in human macrophages. This suggests that variations in Type I IFN responses correlate with differences in Mtb nucleic acids. This is of outmost importance as type I IFNs are mediators implicated in host resistance to TB [32].
Mtb is fundamentally a bacterium that remains in the phagosome [33], inhibiting phagosomal acidification and recruitment of lysosomal proteins in order to maintain a hospitable niche within macrophages [2]. Reports of escape of Mtb from its vacuole into the cytosol [34, 35] appear to have been generated by the experimental technique used [36, 37], and occurred in a limited percentage of cells when they are infected ex vivo [38] but not in vivo [39]. Furthermore, cytosolic Mtb-strain dependent DNA-recognizing receptor activation observed in mice, is associated with host’s DNA release, and not with that one from mycobacteria [40]. The phagosome could then be considered the platform from where components of Mtb are recognized by PRRs present at this site, as it has been shown to occur with other pathogens [41-43].
The importance of TLR signaling in the recognition of Mtb components is underlined by the fact that TLR-adaptor molecule MyD88 pathway in macrophages is needed for an adequate innate and adaptive immune response, mycobacterial clearance [44-46], and confining Mtb within phagolysosomes [38]. The subcellular localization of endosomal TLRs guarantees release of ligands, like nucleic acids, from the pathogen in a natural occurring immunity-promoting manner [47]. Endosomal TLR signaling may play an important role in the outcome of infection, as these receptors are involved in the generation of Type I IFNs. TLR9 is the endosomal TLR receptor able to recognize mycobacterial DNA in humans [4], and mice [48]. Several studies have demonstrated an association between specific endosomal TLR polymorphisms of TLR9, with susceptibility to tuberculosis [49, 50].
It is well known that TLR9-mediated recognition of the unmethylated CpG motifs can lead to distinct signaling responses depending on the sequence of oligodeoxynucleotide (ODN)s that express CpG motifs [51]. Newer and more reliable phylogenetic analysis of MTBC strains through the use of Whole Genome Sequencing (WGS) has shown that there exists more genetic diversity between strains, with genetic distance sometimes comparable to that one existing between Mtb and M. bovis [52]. So it is possible that genetic differences between Mtb strains could be sufficient to elicit different responses after PRR recognition.
We here observed that Vit D rescued low IRF-mediated response to stimulation with DNA from certain Mtb genotypes. That fact that Vit D increases the response to DNA from some of the strains that were initially “weak”, rules out the possibility that the initial differential response is due to differences in DNA preparation, as the same amount of the same DNA preparations was able to elicit an increase. In fact, Mtb DNA preparations were fairly pure, with 260/280 ratios in the range of 1.7-188 (data not shown). Also, we have previously observed that the degree of bacterial DNA fragmentation did not alter the degree of cell activation, nor the size or PEI-DNA complex particle [20].
Pretreatment of THP-1 cell with Vit D also increased secretion of TNF-α after stimulation with Mtb DNA. This is concordant with a previous study showing increased secretion of TNF-α and other cytokines upon Mtb infection of Vit D-treated THP-1 cells [53]. Vit D has also been shown to rescue impaired TNF-α response from U937 and alveolar macrophages after infection with Mtb and BCG [54]. Our transcriptome findings on Vit D activation of THP-1 cells are concordant with previous studies showing that Vit D-stimulated top-ranking genomic regions and genes related to the anti-microbial response and other immunity-related pathways [55, 56]. Only Vit D pretreatment induced upregulation of TLR9, while other forms of cell activation upregulated other TLRs. An increase in TLR 9 expression was observed upon Mtb-DNA stimulation in Vit D pre-treated cells. Besides being useful model to study Mtb infection and persistence [57], using THP-1 cell line helped to avoid the potential bias of the presence of TLR variants which could affect the amount of TLR which they generate [58].
Interaction of different TLRs appears to have important regulatory and cooperative effects against Mtb infection [48, 59]. TLR9-TLR2 crosstalk could be important in promoting vaccine enhancing responses, as the evidence of the benefits of using endosomal agonists as adjuvants for TB vaccines is increasing [60, 61].
Tissue destruction and pathogenesis during Mtb infection is not mediated by pathogens alone but induced by an immunopathological inflammatory response of the host. Differences in the inflammatory cytokine induction, especially type I and type II IFNs after recognition of Mtb nucleic acids could account for differences in the severity of the disease manifestation. Further studies could elucidate the role of the different PRRs in the response after Mtb-DNA recognition.
Supplementary Material
Suppl 1. Vitamin D pre-treatment activates THP-1 cells. Vitamin D increased IRF-mediated response to TLR 2 ligand stimulation. Dual THP-1 cells were pre-treated with PMA. Vit D, IFN-γ for 72 hrs., or IFN-γ for 72 hrs. followed by LPS for 8 hrs. Bars show the response ratio (compared to Unstimulated) after stimulation with TLR-2 ligand Pam3CSSNA for 24 hours.
Suppl 2. Genes differentially expressed in THP-1 cells by Vitamin D. Heatmap of transcriptome analysis of THP-1 cells pre-treated with PMA, or Vit D for 72 hrs. Each of the three columns per condition indicate a biological replicate (different cell preparation). Blue rectangle shows DE genes involved in inflammatory immune response to infection, cell adhesion, signal transduction, Type I IFN regulation, macrophage activation, pathogen recognition, pathogen DNA recognition, and recruitment of effector cells to site of inflammation.
Suppl 3. Vitamin D increases transcription of IFN-β in THP-1 cells PCR for IFN-1beta, and TLR9 of THP-1 cells untreated and pre-treated with Vit D for 72 hrs, followed by 24 hr. stimulation with genomic Mtb DNA from different lineages. A Bands show an overall increase in IFN-1beta transcription after Vit D treatment. B and C Adjusted band density values show an overall increase in TLR9 transcription in cells pretreated with Vit D (black bars), with the exception of cells stimulated with Indo-Oceaninc Mtb DNA. GAPDH is used as a housekeeping gene. Gray bars: Untreated cells. D RT-PCR for TLR9 showing an increase in the TLR9 transcript for Vit D-treated cells after stimulation with CpG or M. bovis DNA.
Suppl 4. TLR9 transcription inhibition partially affects NF-kB response to DNA from different M.tuberculosis phylogenetic lineages THP-1 cells were pre-treated with a specific immunoregulatory DNA sequence (IRS869: 5’-TCCTGGAGGGGTTGT-3’) [27], to block TLR9 signaling. IRS869 was used at a concentration of 1.4 μM, added 1 hour prior to cell stimulation. A NF-kB response was decreased after stimulation with DNA from some strains, such as Indo-Oceanic, or H37Rv, but it was increased in the case of East-Africa-Indian strain. B No change was observed in the IRF-mediated response.
Acknowledgements
Special thanks to Dr. Wendy Walker, and Olga Estrada (TTUHSC), for their assistance with RT-PCR. This work was presented in part at the Texas Tuberculosis Research Symposium (TTRS) 2018, El Paso, Texas, USA, sponsored by the Texas Tech University Health Sciences Center El Paso.
Funding
This work was supported by NIAID grants AI0901656 (JS) and CCMC Arrison and Burr Curtis Research funds (JS). XR acknowledges funding from Academic Research Fund Tier 2 MOE2013-T2-1-101 (ARC 45/13) from Singapore’s Ministry of Education. The funders played no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interest: None
Each article needs to have a financial disclosure line, usually within the author bio section. Publication of this supplement was supported by The University of Texas Health Science Center at Houston.
References:
- 1.Matteelli A, et al. , Tuberculosis elimination and the challenge of latent tuberculosis. Presse Med, 2017. 46(2 Pt 2): p. e13–e21. [DOI] [PubMed] [Google Scholar]
- 2.Soldati T and Neyrolles O, Mycobacteria and the intraphagosomal environment: take it with a pinch of salt(s)! Traffic, 2012. 13(8): p. 1042–52. [DOI] [PubMed] [Google Scholar]
- 3.Basu J, Shin DM, and Jo EK, Mycobacterial signaling through toll-like receptors. Front Cell Infect Microbiol, 2012. 2: p. 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hossain MM and Norazmi MN, Pattern recognition receptors and cytokines in Mycobacterium tuberculosis infection--the double-edged sword? Biomed Res Int, 2013. 2013: p. 179174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mayer-Barber KD, et al. , Host-directed therapy of tuberculosis based on interleukin-1 and type I interferon crosstalk. Nature, 2014. 511(7507): p. 99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berry MP, et al. , An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature, 2010. 466(7309): p. 973–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Teles RM, et al. , Type I interferon suppresses type II interferon-triggered human anti-mycobacterial responses. Science, 2013. 339(6126): p. 1448–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Paus RA, et al. , Inhibition of the type I immune responses of human monocytes by IFN-alpha and IFN-beta. Cytokine, 2013. 61(2): p. 645–55. [DOI] [PubMed] [Google Scholar]
- 9.McNab FW, et al. , Type I IFN induces IL-10 production in an IL-27-independent manner and blocks responsiveness to IFN-gamma for production of IL-12 and bacterial killing in Mycobacterium tuberculosis-infected macrophages. J Immunol, 2014. 193(7): p. 3600–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashiru OT, Pillay M, and Sturm AW, Mycobacterium tuberculosis isolates grown under oxygen deprivation invade pulmonary epithelial cells. Anaerobe, 2012. 18(4): p. 471–4. [DOI] [PubMed] [Google Scholar]
- 11.Portevin D, et al. , Human macrophage responses to clinical isolates from the Mycobacterium tuberculosis complex discriminate between ancient and modern lineages. PLoS Pathog, 2011. 7(3): p. e1001307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen YY, et al. , The pattern of cytokine production in vitro induced by ancient and modern Beijing Mycobacterium tuberculosis strains. PLoS One, 2014. 9(4): p. e94296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jarrett P and Scragg R, A short history of phototherapy, vitamin D and skin disease. Photochem Photobiol Sci, 2017. 16(3): p. 283–290. [DOI] [PubMed] [Google Scholar]
- 14.Liu PT, et al. , Toll-like receptor triggering of a vitamin D-mediated human antimicrobial response. Science, 2006. 311(5768): p. 1770–3. [DOI] [PubMed] [Google Scholar]
- 15.Verrall AJ, et al. , Early clearance of Mycobacterium tuberculosis: a new frontier in prevention. Immunology, 2014. 141(4): p. 506–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martineau AR, et al. , Vitamin D in the treatment of pulmonary tuberculosis. J Steroid Biochem Mol Biol, 2007. 103(3–5): p. 793–8. [DOI] [PubMed] [Google Scholar]
- 17.Ralph AP, Lucas RM, and Norval M, Vitamin D and solar ultraviolet radiation in the risk and treatment of tuberculosis. Lancet Infect Dis, 2013. 13(1): p. 77–88. [DOI] [PubMed] [Google Scholar]
- 18.Coscolla M and Gagneux S, Consequences of genomic diversity in Mycobacterium tuberculosis. Semin Immunol, 2014. 26(6): p. 431–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suh J, et al. , Real-time gene delivery vector tracking in the endo-lysosomal pathway of live cells. Microsc Res Tech, 2012. 75(5): p. 691–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cervantes JL, et al. , Human TLR8 is activated upon recognition of Borrelia burgdorferi RNA in the phagosome of human monocytes. J Leukoc Biol, 2013. 94(6): p. 1231–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim D, et al. , TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol, 2013. 14(4): p. R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langmead B and Salzberg SL, Fast gapped-read alignment with Bowtie 2. Nat Methods, 2012. 9(4): p. 357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trapnell C, et al. , Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol, 2013. 31(1): p. 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Durand SH, et al. , Lipoteichoic acid increases TLR and functional chemokine expression while reducing dentin formation in in vitro differentiated human odontoblasts. J Immunol, 2006. 176(5): p. 2880–7. [DOI] [PubMed] [Google Scholar]
- 25.Cantaert T, et al. , Type I interferons have no major influence on humoral autoimmunity in rheumatoid arthritis. Rheumatology (Oxford), 2010. 49(1): p. 156–66. [DOI] [PubMed] [Google Scholar]
- 26.Livak KJ and Schmittgen TD, Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods, 2001. 25(4): p. 402–8. [DOI] [PubMed] [Google Scholar]
- 27.Barrat FJ, et al. , Nucleic acids of mammalian origin can act as endogenous ligands for Toll-like receptors and may promote systemic lupus erythematosus. J Exp Med, 2005. 202(8): p. 1131–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petzke MM, et al. , Recognition of Borrelia burgdorferi, the Lyme disease spirochete, by TLR7 and TLR9 induces a type I IFN response by human immune cells. J Immunol, 2009. 183(8): p. 5279–92. [DOI] [PubMed] [Google Scholar]
- 29.Daigneault M, et al. , The identification of markers of macrophage differentiation in PMA-stimulated THP-1 cells and monocyte-derived macrophages. PLoS One, 2010. 5(1): p. e8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiemer AK, et al. , Attenuated activation of macrophage TLR9 by DNA from virulent mycobacteria. J Innate Immun, 2009. 1(1): p. 29–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fallow A, Domenech P, and Reed MB, Strains of the East Asian (W/Beijing) lineage of Mycobacterium tuberculosis are DosS/DosT-DosR two-component regulatory system natural mutants. J Bacteriol, 2010. 192(8): p. 2228–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Friedland JS, Targeting the inflammatory response in tuberculosis. N Engl J Med, 2014. 371(14): p. 1354–6. [DOI] [PubMed] [Google Scholar]
- 33.Wong KW and Jacobs WR Jr., Critical role for NLRP3 in necrotic death triggered by Mycobacterium tuberculosis. Cell Microbiol, 2011. 13(9): p. 1371–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Wel N, et al. , M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell, 2007. 129(7): p. 1287–98. [DOI] [PubMed] [Google Scholar]
- 35.Houben D, et al. , ESX-1-mediated translocation to the cytosol controls virulence of mycobacteria. Cell Microbiol, 2012. 14(8): p. 1287–98. [DOI] [PubMed] [Google Scholar]
- 36.Welin A and Lerm M, Inside or outside the phagosome? The controversy of the intracellular localization of Mycobacterium tuberculosis. Tuberculosis (Edinb), 2012. 92(2): p. 113–20. [DOI] [PubMed] [Google Scholar]
- 37.Simeone R, et al. , Cytosolic access of Mycobacterium tuberculosis: critical impact of phagosomal acidification control and demonstration of occurrence in vivo. PLoS Pathog, 2015. 11(2): p. e1004650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rahman A, et al. , Mycobacterium tuberculosis subverts the TLR-2-MyD88 pathway to facilitate its translocation into the cytosol. PLoS One, 2014. 9(1): p. e86886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Zon M, et al. , Subcellular localization of M. tuberculosis in vivo and effect of the adaptive immunity. Ultrastruct Pathol, 2017. 41(1): p. 133. [Google Scholar]
- 40.Wiens KE and Ernst JD, The Mechanism for Type I Interferon Induction by Mycobacterium tuberculosis is Bacterial Strain-Dependent. PLoS Pathog, 2016. 12(8): p. e1005809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cervantes JL, et al. , Phagosomal signaling by Borrelia burgdorferi in human monocytes involves Toll-like receptor (TLR) 2 and TLR8 cooperativity and TLR8-mediated induction of IFN-beta. Proc Natl Acad Sci U S A, 2011. 108(9): p. 3683–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bonham KS and Kagan JC, Endosomes as platforms for NOD-like receptor signaling. Cell Host Microbe, 2014. 15(5): p. 523–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sokolovska A, et al. , Activation of caspase-1 by the NLRP3 inflammasome regulates the NADPH oxidase NOX2 to control phagosome function. Nat Immunol, 2013. 14(6): p. 543–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berod L, et al. , MyD88 signalling in myeloid cells is sufficient to prevent chronic mycobacterial infection. Eur J Immunol, 2014. 44(5): p. 1399–409. [DOI] [PubMed] [Google Scholar]
- 45.Fremond CM, et al. , Fatal Mycobacterium tuberculosis infection despite adaptive immune response in the absence of MyD88. J Clin Invest, 2004. 114(12): p. 1790–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reiling N, Ehlers S, and Holscher C, MyDths and un-TOLLed truths: sensor, instructive and effector immunity to tuberculosis. Immunol Lett, 2008. 116(1): p. 15–23. [DOI] [PubMed] [Google Scholar]
- 47.Sander LE, et al. , Detection of prokaryotic mRNA signifies microbial viability and promotes immunity. Nature, 2011. 474(7351): p. 385–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simmons DP, et al. , Mycobacterium tuberculosis and TLR2 agonists inhibit induction of type I IFN and class I MHC antigen cross processing by TLR9. J Immunol, 2010. 185(4): p. 2405–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bharti D, et al. , The role of TLR9 polymorphism in susceptibility to pulmonary tuberculosis. Immunogenetics, 2014. 66(12): p. 675–81. [DOI] [PubMed] [Google Scholar]
- 50.Velez DR, et al. , Variants in toll-like receptors 2 and 9 influence susceptibility to pulmonary tuberculosis in Caucasians, African-Americans, and West Africans. Hum Genet, 2010. 127(1): p. 65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Steinhagen F, et al. , Activation of type I interferon-dependent genes characterizes the "core response" induced by CpG DNA. J Leukoc Biol, 2012. 92(4): p. 775–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Galagan JE, Genomic insights into tuberculosis. Nat Rev Genet, 2014. 15(5): p. 307–20. [DOI] [PubMed] [Google Scholar]
- 53.Verway M, et al. , Vitamin D induces interleukin-1beta expression: paracrine macrophage epithelial signaling controls M. tuberculosis infection. PLoS Pathog, 2013. 9(6): p. e1003407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anandaiah A, et al. , Vitamin D rescues impaired Mycobacterium tuberculosis-mediated tumor necrosis factor release in macrophages of HIV-seropositive individuals through an enhanced Toll-like receptor signaling pathway in vitro. Infect Immun, 2013. 81(1): p. 2–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Neme A, et al. , The vitamin D-dependent transcriptome of human monocytes. J Steroid Biochem Mol Biol, 2016. 164: p. 180–187. [DOI] [PubMed] [Google Scholar]
- 56.Liu H, et al. , Alternative splicing analysis in human monocytes and macrophages reveals MBNL1 as major regulator. Nucleic Acids Res, 2018. 46(12): p. 6069–6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Estrella JL, et al. , A Novel in vitro Human Macrophage Model to Study the Persistence of Mycobacterium tuberculosis Using Vitamin D(3) and Retinoic Acid Activated THP-1 Macrophages. Front Microbiol, 2011. 2: p. 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gantier MP, et al. , Genetic modulation of TLR8 response following bacterial phagocytosis. Hum Mutat, 2010. 31(9): p. 1069–79. [DOI] [PubMed] [Google Scholar]
- 59.Bafica A, et al. , TLR9 regulates Th1 responses and cooperates with TLR2 in mediating optimal resistance to Mycobacterium tuberculosis. J Exp Med, 2005. 202(12): p. 1715–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Commandeur S, et al. , The in vivo expressed Mycobacterium tuberculosis (IVE-TB) antigen Rv2034 induces CD4(+) T-cells that protect against pulmonary infection in HLA-DR transgenic mice and guinea pigs. Vaccine, 2014. 32(29): p. 3580–8. [DOI] [PubMed] [Google Scholar]
- 61.Baldwin SL, et al. , Intradermal immunization improves protective efficacy of a novel TB vaccine candidate. Vaccine, 2009. 27(23): p. 3063–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Suppl 1. Vitamin D pre-treatment activates THP-1 cells. Vitamin D increased IRF-mediated response to TLR 2 ligand stimulation. Dual THP-1 cells were pre-treated with PMA. Vit D, IFN-γ for 72 hrs., or IFN-γ for 72 hrs. followed by LPS for 8 hrs. Bars show the response ratio (compared to Unstimulated) after stimulation with TLR-2 ligand Pam3CSSNA for 24 hours.
Suppl 2. Genes differentially expressed in THP-1 cells by Vitamin D. Heatmap of transcriptome analysis of THP-1 cells pre-treated with PMA, or Vit D for 72 hrs. Each of the three columns per condition indicate a biological replicate (different cell preparation). Blue rectangle shows DE genes involved in inflammatory immune response to infection, cell adhesion, signal transduction, Type I IFN regulation, macrophage activation, pathogen recognition, pathogen DNA recognition, and recruitment of effector cells to site of inflammation.
Suppl 3. Vitamin D increases transcription of IFN-β in THP-1 cells PCR for IFN-1beta, and TLR9 of THP-1 cells untreated and pre-treated with Vit D for 72 hrs, followed by 24 hr. stimulation with genomic Mtb DNA from different lineages. A Bands show an overall increase in IFN-1beta transcription after Vit D treatment. B and C Adjusted band density values show an overall increase in TLR9 transcription in cells pretreated with Vit D (black bars), with the exception of cells stimulated with Indo-Oceaninc Mtb DNA. GAPDH is used as a housekeeping gene. Gray bars: Untreated cells. D RT-PCR for TLR9 showing an increase in the TLR9 transcript for Vit D-treated cells after stimulation with CpG or M. bovis DNA.
Suppl 4. TLR9 transcription inhibition partially affects NF-kB response to DNA from different M.tuberculosis phylogenetic lineages THP-1 cells were pre-treated with a specific immunoregulatory DNA sequence (IRS869: 5’-TCCTGGAGGGGTTGT-3’) [27], to block TLR9 signaling. IRS869 was used at a concentration of 1.4 μM, added 1 hour prior to cell stimulation. A NF-kB response was decreased after stimulation with DNA from some strains, such as Indo-Oceanic, or H37Rv, but it was increased in the case of East-Africa-Indian strain. B No change was observed in the IRF-mediated response.