Abstract
The role of genetics in the pathogenesis of necrotizing enterocolitis (NEC) was initially informed by epidemiological data indicating differences in prevalence among different ethnic groups as well as concordance in twins. These early observations, together with major advances in genomic research, paved the way for studies that begin to reveal the contribution of genetics to NEC. Using the candidate gene- or pathway approach, several potential pathogenic variants for NEC in premature infants have already been identified. More recently, exome- and genome-wide association study for NEC has also been completed. These advances, however, are tempered by the lack of adequately powered replication cohorts to validate the accuracy of these discoveries. Despite many challenges, genetic research in NEC is expected to increase, providing new insights into its pathogenesis and bringing the promise of personalized care closer to reality. In this review we provide a summary of genetic studies in NEC along with defining the challenges and possible future approaches.
INTRODUCTION
Necrotizing enterocolitis (NEC) is a devastating inflammatory disease of the preterm intestinal tract that afflicts 5 to 10% of infants weighing less than 1500 grams at birth.1 Despite considerable advances in neonatal medicine, outcomes for NEC have remained stagnant and may even be worsening.2, 3 Many risk factors – such as formula feeds, infection, ischemia, and gut dysbiosis – have been implicated in NEC.4 Clinical risk factors alone, however, cannot explain the inter-individual variability in NEC susceptibility or severity in premature infants, indicating a role for inherited risk factors in disease pathogenesis.5
Epidemiological studies were the first to provide clues regarding a genetic contribution to NEC. Risk for NEC varied widely based on ethnic distribution, occurring less frequently in Japan, Switzerland, and Austria while afflicting preterm infants more often in North America, United Kingdom, and Ireland.4 Although this variation could be due to differences in local neonatal care strategies, studies limited to the United States also demonstrated differences in risk based on race, with African American infants more likely to develop NEC than Caucasians.6, 7 Twin studies by Bhandari et al.5 also demonstrated that more than 50% of variance in NEC is accounted for by genetic and shared environmental factors. These studies triggered investigations by several researchers into the genetic basis of NEC risk. Advances in our understanding of the genetic basis of complex diseases, combined with technological advances in genotyping platforms, are beginning to yield promising, if preliminary results. In this review, we (1) summarize current genetic studies in NEC, (2) discuss various challenges and limitations of these studies, and (3) discuss future approaches for identifying pathogenic loci for NEC.
CANDIDATE GENE APPROACH
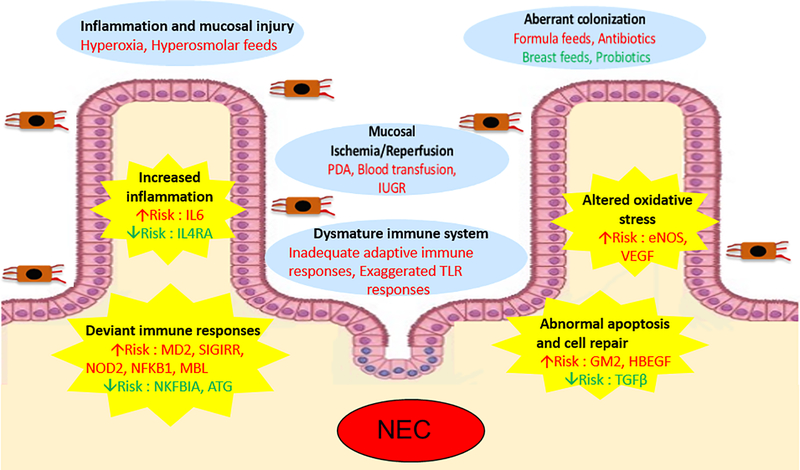
The candidate gene approach has been used in majority of the genetic studies of NEC. In this approach, investigators evaluate single nucleotide polymorphisms (SNPs) of a few, pre-selected genes based on a priori hypothesis of relevance to NEC. The discussion that follows, along with Table 1 and Figure 1, summarize candidate genes for NEC that have been evaluated using this approach, with an emphasis on understanding the biological relevance of the gene or pathway to NEC development.
Table 1:
Summary of genes assessed using the candidate gene approach.
| Gene/Pathway | Increased risk for NEC | Decreased risk for NEC | No evidence of association with NEC |
|---|---|---|---|
| TLRs8–10 | MD2 (C-1625G) | TLR2 (Arg753Gln) TLR4 (Asp299Gly) TLR4 Thr399Ile) TLR4 (Asp299Gly) TLR4 (Thr399Ile) TLR5 (Arg392stop) TLR9 (2849 C>T) CD14 (−260 C>T) IRAK1 (Leu532Ser) TIRAP (Ser180Leu) |
|
| SIGIRR11 | SIGIRR (p.Y168X) SIGIRR (p.S80Y) |
||
| Autophagy8, 12–14 | NOD2 (G908R)** NOD2 (R702W)** NOD2 (1007fs)** |
ATG16L1(Thr300Ala) | NOD2 (Gly908Arg) NOD2 (R702W) NOD2 (1007fs) NOD2 (G908R) NOD2 (rs2066844) NOD1 (37684T>C) NLRP3 (247617036G>T) NLRP3 (247621033G>A) NLRP3 (14384C>A) CARD8 (26498T>A) |
| MBL15 | MBL2 (−221 G>C) | MBL2 (Codon 54 G>A) MBL2 (Codon 57 G>A) | |
| PAF10, 16 | GM2A (g.5220G>A) GM2A (g.19168A>C) |
PAFAH (Ile198Thr) PAFAH (Ala379Val) |
|
| NFKB9 | NFKB1(−24519delATTG) | NFKBIA(−1004A > G) | |
| Cytokines17–22 | IL6(g.4880C>G) | IL4RA(g.54150A>G) | IL1B(−31T>C) IL1B(−511C>T) IL1B(3953 C>T) IL1B(c.−198T>C) IL4RA(c.1902 A>G) IL6(−174G>C) IL6(−596G>A) IL8(−251T>A) IL10(−1082G>A) IL12(g.19532A>C) IL18(−607C>A) IL18(−137G>C) PXR(g.39403G>A) TNF(−238G>A) TNF(8156G>A) TNF(−308G>A) |
| FUT223, 24 | FUT2 (428G>A) | ||
| Antioxidants25, 26 | HMOX1(p.D7H) GCLC(g.−129 C>T) SOD2(p.A16V) GSTP1(p.I1047V) NQO1(p.P187S) NFE2L2(−617C>A) CYBA(C242T) |
||
| Vascular homeostasis22, 27–29 | VEGF(C-2578A) eNOS(894G>T) eNOS(−786T>C) |
VEGF(T-460C) VEGF(+405C) CPS1(p.Thr1406Asn) END-1 (5665G>T) |
|
| HB-EGF30 | HBEGF(140338940G>T) | HBEGF(140333293G>A) HBEGF(140334979C>T) HBEGF(140337403G>C) HBEGF(140347675G>A) |
Genes listed as gene name (SNP or amino acid change).
Increased risk for NEC reported when more than 2 NOD2 variants are present.
External factors traditionally implicated in the pathogenesis of NEC include mucosal injury, ischemia/reperfusion, aberrant microbial colonization, and immature immune system. In addition, several candidate genes and pathways that may confer an inherent predisposition to NEC are also increasingly being recognized. Red text = increased risk for NEC; green text = protection against NEC.
Toll-like receptors (TLR)
TLRs are pattern recognition receptors (PRR) that recognize conserved structural motifs in pathogens and activate innate immune responses. TLR4, which is important for immunity against Gram-negative bacteria, has been implicated in NEC by studies showing elevated expression in intestinal samples obtained from NEC infants.31 Experimental studies in rodents also demonstrated an obligatory role for TLR4 in NEC, further highlighting its importance in its pathogenesis.32, 33 Preliminary genetic studies in humans, however, have yet to yield strong evidence that genetic variations in TLR pathway increases susceptibility to NEC. In a cohort of 270 preterm infants, Sampath et al.9 did not find an association between NEC and SNPs in TLR receptors and TLR pathway mediators. Szebani et al.8 also did not find any association between NEC and SNPs in TLR4 and cluster of differentiation 14 (CD14, a key co-receptor of TLR4). In another study, Zhou et al.10 investigated myeloid differentiation-2 (MD2, another co-receptor of TLR4) in 42 NEC cases and 83 controls. Overall, there was no difference in the frequency of MD2 SNPs between NEC cases and controls; although analysis based on severity demonstrated a small but significant increased risk of surgical NEC with MD2 variants (OR 1.66, 95% CI 1.09–3.05, p=0.036).
Single immunoglobulin and toll-interleukin 1 receptor (SIGIRR)
Using whole exome sequencing (WES), Sampath et al.11 identified a novel stop variant (p.Y168X) of SIGIRR in an infant with NEC totalis. Additional Sanger sequencing of the SIGIRR gene in 37 other preterm infants (17 with NEC) demonstrated that rare and novel variants of SIGIRR were enriched in preterm infants with NEC compared to controls, further implicating SIGIRR as a candidate gene for NEC. Functional studies in intestinal epithelial cells showed that the identified variants were functional mutations that led to loss of SIGIRR function, dysregulated TLR activation, and excessive inflammation. Additional in-vivo studies using SIGIRR−/− mice confirmed that loss of SIGIRR function results in a phenotype of exaggerated intestinal inflammation and increased NEC susceptibility compared to controls.34 Intriguingly, SIGGIR functions as a “brake” to prevent excessive TLR activation and inflammation in the intestines.35 Thus, although no genes that mediate TLR signaling have been strongly associated with NEC, inhibitors of TLR signaling (such as SIGIRR) may be promising NEC candidate genes.
Nucleotide binding oligomerization domain containing protein 2 (NOD2)
NOD2 is another PRR that recognizes the muramyl dipeptide component of bacterial cell wall and interacts with autophagy pathways to target and degrade invading bacteria.36 Functional polymorphisms in NOD2, which are strongly implicated in inflammatory bowel disease, have been hypothesized to also be important in NEC.37 In a preliminary study, Zouali et al.12 sequenced the entire NOD2 gene of 10 preterm infants with NEC but found no novel NOD2 variants associated with NEC. Szebani et al.8 also found no evidence of association between SNPs of NOD2 and NEC in their study of 118 preterm infants, 41 of whom had NEC. Similar findings of no association was found by Sampath et al.14 between NOD1 or NOD2 variants and NEC in a large, multi-center study involving 1015 preterm infants. In an even larger study, Härtel et al.13 genotyped more than 9,000 preterm infants of European ancestry and found that while no single NOD2 variant was associated with NEC, presence of 2 or more loss of function NOD2 variants was significantly associated with an increased risk for NEC (OR 4.14, 95% CI 1.41–12.12, P=0.009). Thus, while initial studies were negative, the largest study seem to suggest that NOD2 variants do confer increased NEC risk in premature infants.
Autophagy-related 16-Like 1 (ATG16L1)
ATG16L1 is essential for the proper assembly of lysosomes used in the autophagy of invading pathogens. Induction of autophagy is associated with worse injury in experimental NEC, while diminished autophagy decreases NEC.38 Sampath et al.14 evaluated the key autophagy gene, ATG16L1, and found that a common ATG16L1 variant (Thr300Ala) conferred protection against NEC (OR=0.4, 95% CI 0.19–0.81). This variant, which increases sensitization of ATG16L1 to caspase-3-mediated degradation, results in diminished autophagy and is thus consistent with prior studies showing protective role of deceased autophagy against NEC. An independent replication cohort (260 infants, 23 with NEC) also showed a trend towards decreased NEC among infants who were homozygous for the loss of function variant allele, providing further evidence for the importance of this gene in NEC.14 Further studies are needed to evaluate the role of this and other autophagy-related genes in NEC.
Mannose binding lectin (MBL)
MBL is a circulating PRR that recognizes unique carbohydrate components of invading pathogens and induces their phagocytic clearance by the complement system.39 Elevated MBL levels are found in intestinal tract of preterm infants with NEC, suggesting that excessive MBL activation may play a role in its pathogenesis.40 Prencipe et al.15 evaluated 107 preterm infants (41 with NEC) for SNPs of MBL and found that a promoter SNP (−221 X/Y) of the MBL gene was more common in infants with NEC (OR 4.42, 95% CI 1.11%−17.49%, p=0.03). This polymorphism is associated with increased serum MBL levels, supporting the hypothesis that gain of function variants of MBL is associated with an increased risk for NEC.
Platelet activating factor (PAF)
PAF is an endogenous phospholipid with potent pro-inflammatory actions.41 Infants with NEC have increased plasma levels of PAF, and administration of intravenous PAF in animal models induces NEC.42 Conversely, PAFAH – an enzyme that degrades PAF – has decreased activity and concentration in infants with NEC, and enteral administration of recombinant PAFAH in animal models results in a significant decrease in NEC.43, 44 Sankararaman et al.16 evaluated the role of functional PAFAH polymorphisms in a convenience cohort of 570 preterm infants (36 with stage 1 or 2 NEC, 534 without NEC) and found no evidence of association between PAFAH SNPs and NEC. In contrast, Zhou et al.10 evaluated SNPs of GM2 activator protein (GM2A), which is another PAF antagonist, in a Chinese cohort of 125 infants (42 with NEC) and found a positive association between NEC and 2 variants of GM2A – rs1048719 (OR 1.86, 95% CI 1.04–3.33) and rs2075783 (OR 3.23, 95% CI 1.69–6.21). Inclusion of term infants in this study, however, makes it difficult to determine whether the differences detected were due to prematurity or NEC.
Nuclear factor-kappa B (NFKB)
NFKB is a major transcription factor that regulates the expression of several pro-inflammatory genes.45 NF-κB levels are increased in the intestines following NEC induction in rats, while inhibiting NFKB protects against excessive inflammation.46, 47 Sampath et al.9 studied NFKB in NEC and found that the NFKB1 (g.−24519delATTG) variant was present in all 15 infants with NEC compared to only 166 of 256 without NEC (100% vs 65%, OR ∞, 95% CI 2.1−∞, p=0.003), suggesting that this variant may be necessary, but not sufficient, for developing NEC. On the other hand, the NFKBIA (g.1004A>G) variant was less common in infants with NEC compared to infants without NEC (13.3% vs 49%, OR 0.16, 95% CI 0.04–0.60, P=0.007), suggesting that this variant may confer protection against NEC.
Pro-inflammatory cytokines
As excessive inflammation is a hallmark of NEC, many investigators have examined the relationship of several pro-inflammatory cytokine gene polymorphisms and NEC, with varying results. Treszl et al.17, 18, Heninger et al.19, Henderson et al.20, and Szpecht et al.22 have examined functional variants of several pro-inflammatory cytokines (including tumor necrosis factor-alpha, interleukin (IL)-1β, IL-4 receptor α-chain, IL-6, IL-18, and IL-10) and found no evidence of association of these genes with NEC. In contrast, Franklin et al.21 reported a positive association between IL-6 (rs1800795) variant and NEC in Caucasian neonates; while Tian et al.48 found a positive association between IL17F (rs763780) variant and NEC in a Chinese cohort. Interestingly, the studies showing no association between pro-inflammatory cytokines and NEC comprised of neonates of European descent, suggesting that ethnicity may play an important role on whether certain genetic variants contribute to NEC or not.
Fucosyltransferase 2 (FUT2)
FUT2 mediates the inclusion of fucose sugar units to glycoproteins and glycolipids. Polymorphisms in FUT2 gene, which result in differing phenotypes of secreted fucosylated glycans on mucosal surfaces, have important implications in host-microbiome interactions. Morrow et al.23 investigated non-secretor status (AA) of the FUT2 polymorphism (428G>A) and found no evidence of association between this variant and NEC (OR 1.6, 95% CI 0.7–3.8) or surgical NEC (OR 2.3, 95% CI 0.6–7.4). However, when salivary secretion of H-antigen was used to determine non-secretor status, a positive association with NEC was found. Another study by Demmert et al.24 investigated the same FUT2 polymorphism (248G>A) in a large prospective cohort of 2,406 VLBW infants and also found no association of this polymorphism with NEC. These differential results can be possibly explained by differences in FUT2 genotype and actual secretor phenotype, which has been shown to not always be concordance likely because of epigenetic influences.
Genes regulating the antioxidant response
Anti-oxidants are important in alleviating injury from gut ischemia-reperfusion, and genetic variants that limit anti-oxidant defenses can hypothetically increase risk of NEC in preterm infants.49 Sampath et al.25 investigated loss of function SNPs in 6 anti-oxidant genes belonging to the NF-E2-related factor 2-dependent antioxidant response elements (Nrf2-ARE) pathway and found no evidence of association between the tested SNPs and NEC. Another study by Huizing et al.26 looked at the C242T polymorphism of cytochrome B-245 alpha chain (a gene related to NADPH oxidase family) and also found no association of this variant with NEC in a cohort of 451 preterm infants.
Vascular endothelial growth factor (VEGF)
Studies demonstrate that decreased intestinal expression of VEGF is seen in human NEC compared to controls, suggesting that dysregulated VEGF signaling contributes to NEC pathogenesis. Banyasz et al.29 evaluated VEGF polymorphisms in NEC and discovered that a carrier state for VEGF-2578 mutant allele, which predisposes to low VEGF production, is associated with an increased risk for NEC (OR 2.77, 95% CI 1.00–7.65, p=0.049). This is consistent with studies in mice that demonstrate decreased VEGF signaling increases susceptibility to NEC, while increased VEGF levels through subcutaneous administration decreases the extent of mucosal injury in NEC.50, 51
Arginine and nitric oxide
Arginine is an important substrate for nitric oxide, which is a potent vasodilator that regulates mucosal blood flow and helps maintain mucosal integrity. Low arginine levels have been implicated in NEC based on observational studies demonstrating hypoarginemia in preterm infants with NEC, and clinical trials showing decreased NEC in infants following arginine supplementation.52, 53 Moonen et al.27 evaluated C-to-A nucleotide transversion (T1405N) of carbamoyl-phosphate synthase (CPS), which is functionally correlated with low plasma arginine levels, in a small study of 51 preterm infants (17 with NEC). They found that the CC genotype was associated with increased NEC (unadjusted OR 3.43, 95% CI 1.01–11.49), although this association was not significant once adjusted for gestational age and birth weight. As a follow-up to this study, Moonen et al.28 prospectively recruited 477 preterm infants from 4 different centers to investigate the same CPS1 polymorphism in NEC. They again found no significant association between CC genotype and NEC. Instead, a significant negative association was found between the minor A-allele and the combined outcome of NEC or death, suggesting that the A-allele variant may confer protection against NEC.
In another study, Szpecht et al.22 more directly evaluated nitric oxide pathways by measuring polymorphisms of eNOS in 100 preterm infants, 22 of whom had NEC. They found that eNOS 894G>T polymorphism was more common in infants with NEC (OR 20, 95% CI 3.71–208.7, p=0.0004), and the eNOS 786T>C was increased in infants with surgical NEC (OR 4.88, 95% CI 1.33–21.99, p=0.013). Both variants have been associated in other studies with impaired eNOS enzymatic activity, providing biological plausibility that these variants are functional and potentially significant in NEC.54
Heparin-binding epidermal growth factor-like growth factor (HB-EGF)
HB-EGF has been shown in animal studies to promote repair of intestinal epithelial cells resulting from various forms in injury in vitro, including in NEC.55 The protective role of HB-EGF is thought to be mediated by increased intestinal blood flow, decreased apoptosis, and increased enterocyte migration and proliferation.56 Ma et al.30 measured polymorphisms of HB-EGF in 30 NEC patients and 80 controls and found that rs4912711 SNP was associated with increased NEC. They also measured plasma levels of HB-EGF and found it to be decreased in NEC infants with the rs4912711 SNP, providing additional evidence that the identified SNP is potentially important.
GENOME-WIDE APPROACH
In genome-wide approach, millions of SNPs are evaluated in an unbiased manner, allowing for discovery of previously unknown associations between genes and disease. Jilling et al.57 performed the first genome-wide association study (GWAS) in NEC in 751 extremely preterm infants (30 with surgical NEC) and identified 35 SNPs significantly associated with NEC. A cluster of SNPs in chromosome 8 (8q23.3) had the strongest association for NEC, with odds ratio of 4.72 (95% CI 2.51–8.88). Other clusters of SNPs identified in chromosome 14 and 11 also demonstrated significant association. A validation cohort did not have sufficient power to replicate these findings, except for a single SNP found in chromosome 8. Additional studies are required to confirm functional significance of the identified SNPs. Further fine mapping or sequencing studies may also be necessary to identify pathogenic genes or variants in identified clusters.
CHALLENGES AND LIMITATIONS
Several challenges and limitations can confound the results of genetic association studies, especially for complex diseases such as NEC.58 The following discussion summarizes these challenges as well as important considerations for performing robust genetic studies in NEC. Table 2 characterizes the different genetic studies included in this review based on these criteria.
Table 2:
Characteristics of included studies.
| Study | Country | Number of cases/controls | Definition of NEC cases | Functional studies | Validation cohort |
|---|---|---|---|---|---|
| Trezl 200118 | Hungary | 46/90 | Included Bell Stage I | No | No |
| Heninger 200219 | Hungary | 46/90 | Included Bell Stage I | No | No |
| Treszl 200317 | Hungary | 46/90 | Included Bell Stage I | No | No |
| Zouali 200512 | France | 10/103 | Unclear | No | No |
| Szebani 20068 | Hungary | 41/77 | Included Bell Stage I | No | No |
| Banyasz 200629 | Hungary | 49/79 | Included Bell Stage I | No | No |
| Moonen 200727 | Netherlands | 17/34 | ≥ Bell Stage II | No | No |
| Henderson 200720 | Australia | 50/50 | Included Bell Stage I | No | No |
| Sampath 20119 | USA | 15/255 | Included Bell Stage I | No | No |
| Morrow 201123 | USA | 30/410 | ≥ Bell Stage II | Yes | No |
| Prencipe 201215 | Italy | 41/66 | ≥ Bell Stage II | Yes | No |
| Sankararaman 201316 | USA | 36/534 | Included Bell Stage I | No | No |
| Zhou 201510 | China | 42/83 | ≥ Bell Stage II | No | No |
| Franklin 201521 | USA | 66/118 | ≥ Bell Stage 2 | No | No |
| Sampath 201511 | USA | 17/20 | ≥ Bell Stage II | Yes | No |
| Sampath 201525 | USA | 52/585 | ≥ Bell Stage II | No | No |
| Demmert 201524 | Germany | 108/2406 | Included SIP and Surgical NEC | No | No |
| Hartel 201613 | Germany | 262/6680 | Included SIP and ≥ Bell Stage II | No | No |
| Moonen 201628 | Netherlands | 36/441 | ≥ Bell Stage II | No | Yes |
| Tian 201748 | China | 102/120 | ≥ Bell Stage II | No | No |
| Huizing 201726 | Netherlands | 28/392 | ≥ Bell Stage II | No | No |
| Sampath 201714 | USA | 86/929 | Included Bell Stage I | No | Yes |
| Ma 201830 | China | 30/80 | ≥ Bell Stage II | Yes | No |
| Szpecht 201822 | Poland | 22/78 | Included Bell Stage I | No | No |
| Jilling 201857 | USA | 30/721 | Surgical NEC | No | Yes |
Studies should begin with precise phenotypic definition of NEC cases and controls. Only confirmed cases of NEC in preterm infants (i.e. Bell’s Stage ≥ II) should be included. Inclusion of term infants, spontaneous intestinal perforations (SIP), or unconfirmed NEC (Bell Stage I) as NEC cases can limit the study’s power. Further confining to infants who developed surgical NEC may confer additional precision to the phenotype. Ethnic composition of the cohort should also be accurately described. Variant allele frequencies can vary significantly by ethnicity, and false associations may arise because of population admixture. In cohorts with mixed ethnicity, additional analysis of variants stratified by race should be completed.
An adequate sample size is also key. Ideally, candidate genome approaches should include hundreds of infants, while genome-wide approaches often need thousands of infants to attain sufficient power. Such numbers are difficult to achieve for any disease, and is especially difficult in NEC which variably afflicts about 5–10% of infants born preterm. Compounding this challenge is the need to correct for multiple testing across the number of SNPs tested. Given the challenges with sample size, only a handful of studies have been able to adjust for multiple testing.
Separate validation cohorts are also important. In general, genetic association studies should be treated as preliminary, hypothesis-generating studies until further validation has been provided by subsequent replication cohorts. Given the challenge of obtaining adequate sample size for discovery cohorts, few genetic studies in NEC have been replicated in validation cohorts. Lastly, functional studies that explain the mechanisms underlying the association of the gene with the disease are also important. Functional studies are particularly important for genome-wide approaches such as GWAS and exome/whole genome sequencing, where identified variants are likely of unknown significance to NEC pathogenesis.
FUTURE CONSIDERATIONS
Advances in genetic research have allowed identification of many promising genes associated with increased or decreased risk of NEC. Most of these studies, however, have been limited by small sample sizes as well as lack of validation and functional studies. Future efforts should be made to validate these candidate genes using independent replication cohorts and functional studies. Although challenging to complete, such studies are necessary to establish the importance of these genes and decrease risk of false discovery.
Future studies should also utilize unbiased genome-wide approaches, such as GWAS, WES, or whole genome sequencing (WGS), to discover new genes and pathways. Linkage-based approaches used in GWAS typically identify common variants (present in >1% of the population) in intronic regions. Because of this, additional fine mapping/sequencing as well as functional studies are typically needed to identify pathogenic genes and variants. In contrast, WES directly identifies variants in the coding or exonic regions that are more likely to have a functional impact on the gene product. WES is thus increasingly emerging as the primary modality to query the genetic basis of complex diseases.59–61 WGS probes both coding and non-coding regions of the genome.62 However, WGS encumbers very large samples sizes; and functional evaluation of regulatory variants found in non-coding regions (i.e. introns, promoters, etc.) is still not fully developed.63 Thus, combining GWAS with fine mapping, or alternatively WES directly, are suitable approaches to identify pathogenic loci for NEC susceptibility.
Contribution of other “-omic” fields, which can be helpful in identifying genetic signatures underlying NEC, should also be pursued. For example, Chan et al.64 performed transcriptome (mRNA) sequencing of intestinal tissues from infants with surgical NEC compared to controls and identified several dysregulated genes and pathways in NEC. Using a similar strategy, Ng et al. 65 probed microRNAs (miR), a subset of small RNAs which are not protein-coding but regulate other mRNAs, to identify regulatory pathways of interest in NEC. Interestingly, both of these studies identified the TLR pathway as a key pathway involved in NEC pathogenesis. This discovery, which further highlights the importance of TLR in NEC, also provides evidence supporting the usefulness of this approach to identify potential genetic pathways in NEC.
Finally, functional genomics approaches that combine genomics with transcriptome and proteomics approaches may provide even deeper insights into understanding how pathogenic genes direct molecular phenotypes that program NEC vulnerability.66–68 Major advances in bioinformatics and network analysis provide platforms to interrogate and define the key genetic pathways as well as molecular signatures that prime NEC vulnerability.
CONCLUSION
It is expected that genetic studies in NEC will continue to increase as technological advances allow for more robust and less expensive exploration of the genome. Although a number of potential gene variants have been identified, these discoveries have to be interpreted with caution until further validation studies are completed. Future directions include use of whole exome/genome sequencing to uncover novel and rare genes/variants in NEC. Complimentary functional genomic approaches may also allow NEC prediction in premature infants as well as defining pathways that may be targeted to prevent NEC. While these exciting approaches hold significant promise for defining the molecular pathogenesis of NEC and informing preventive approaches, they require collaborative approaches between clinicians, researchers, and bioinformaticians for their success.
REFERENCES
- 1.Neu J, Walker WA. Necrotizing enterocolitis. The New England journal of medicine. 2011;364:255–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patel RM, Kandefer S, Walsh MC, et al. Causes and timing of death in extremely premature infants from 2000 through 2011. The New England journal of medicine. 2015;372:331–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blakely ML, Lally KP, McDonald S, et al. Postoperative outcomes of extremely low birth-weight infants with necrotizing enterocolitis or isolated intestinal perforation: a prospective cohort study by the NICHD Neonatal Research Network. Annals of surgery. 2005;241:984–989; discussion 989–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zani A, Pierro A. Necrotizing enterocolitis: controversies and challenges. F1000Research. 2015;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhandari V, Bizzarro MJ, Shetty A, et al. Familial and genetic susceptibility to major neonatal morbidities in preterm twins. Pediatrics. 2006;117:1901–1906. [DOI] [PubMed] [Google Scholar]
- 6.Llanos AR, Moss ME, Pinzon MC, Dye T, Sinkin RA, Kendig JW. Epidemiology of neonatal necrotising enterocolitis: a population-based study. Paediatric and perinatal epidemiology. 2002;16:342–349. [DOI] [PubMed] [Google Scholar]
- 7.Uauy RD, Fanaroff AA, Korones SB, Phillips EA, Phillips JB, Wright LL. Necrotizing enterocolitis in very low birth weight infants: biodemographic and clinical correlates. National Institute of Child Health and Human Development Neonatal Research Network. The Journal of pediatrics. 1991;119:630–638. [DOI] [PubMed] [Google Scholar]
- 8.Szebeni B, Szekeres R, Rusai K, et al. Genetic polymorphisms of CD14, toll-like receptor 4, and caspase-recruitment domain 15 are not associated with necrotizing enterocolitis in very low birth weight infants. Journal of pediatric gastroenterology and nutrition. 2006;42:27–31. [DOI] [PubMed] [Google Scholar]
- 9.Sampath V, Le M, Lane L, et al. The NFKB1 (g.−24519delATTG) variant is associated with necrotizing enterocolitis (NEC) in premature infants. The Journal of surgical research. 2011;169:e51–57. [DOI] [PubMed] [Google Scholar]
- 10.Zhou W, Yuan W, Huang L, Wang P, Rong X, Tang J. Association of neonatal necrotizing enterocolitis with myeloid differentiation-2 and GM2 activator protein genetic polymorphisms. Molecular medicine reports. 2015;12:974–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sampath V, Menden H, Helbling D, et al. SIGIRR genetic variants in premature infants with necrotizing enterocolitis. Pediatrics. 2015;135:e1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zouali H, Bonnard A, De Lagausie DL, et al. CARD15/NOD2 is not a predisposing factor for necrotizing enterocolitis. Digestive diseases and sciences. 2005;50:1684–1687. [DOI] [PubMed] [Google Scholar]
- 13.Hartel C, Hartz A, Pagel J, et al. NOD2 Loss-of-Function Mutations and Risks of Necrotizing Enterocolitis or Focal Intestinal Perforation in Very Low-birth-weight Infants. Inflammatory bowel diseases. 2016;22:249–256. [DOI] [PubMed] [Google Scholar]
- 14.Sampath V, Bhandari V, Berger J, et al. A functional ATG16L1 (T300A) variant is associated with necrotizing enterocolitis in premature infants. Pediatric research. 2017;81:582–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prencipe G, Azzari C, Moriondo M, et al. Association between mannose-binding lectin gene polymorphisms and necrotizing enterocolitis in preterm infants. Journal of pediatric gastroenterology and nutrition. 2012;55:160–165. [DOI] [PubMed] [Google Scholar]
- 16.Sankararaman S, Yanamandra K, Napper D, Caldito G, Dhanireddy R. The prevalence of platelet activating factor acetylhydrolase single nucleotide polymorphisms in relationship to necrotizing enterocolitis in Northwest Louisiana infants. SpringerPlus. 2013;2:294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Treszl A, Heninger E, Kalman A, Schuler A, Tulassay T, Vasarhelyi B. Lower prevalence of IL-4 receptor alpha-chain gene G variant in very-low-birth-weight infants with necrotizing enterocolitis. Journal of pediatric surgery. 2003;38:1374–1378. [DOI] [PubMed] [Google Scholar]
- 18.Treszl A, Kocsis I, Szathmari M, Schuler A, Tulassay T, Vasarhelyi B. Genetic variants of the tumour necrosis factor-alpha promoter gene do not influence the development of necrotizing enterocolitis. Acta paediatrica. 2001;90:1182–1185. [DOI] [PubMed] [Google Scholar]
- 19.Heninger E, Treszl A, Kocsis I, Derfalvi B, Tulassay T, Vasarhelyi B. Genetic variants of the interleukin-18 promoter region (−607) influence the course of necrotising enterocolitis in very low birth weight neonates. European journal of pediatrics. 2002;161:410–411. [DOI] [PubMed] [Google Scholar]
- 20.Henderson G, Craig S, Baier RJ, Helps N, Brocklehurst P, McGuire W. Cytokine gene polymorphisms in preterm infants with necrotising enterocolitis: genetic association study. Archives of disease in childhood. Fetal and neonatal edition. 2009;94:F124–128. [DOI] [PubMed] [Google Scholar]
- 21.Franklin AL, Said M, Cappiello CD, et al. Are Immune Modulating Single Nucleotide Polymorphisms Associated with Necrotizing Enterocolitis? Scientific reports. 2015;5:18369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szpecht D, Neumann-Klimasinska N, Blaszczynski M, et al. Candidate gene analysis in pathogenesis of surgically and non-surgically treated necrotizing enterocolitis in preterm infants. Molecular and cellular biochemistry. 2018;439:53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morrow AL, Meinzen-Derr J, Huang P, et al. Fucosyltransferase 2 non-secretor and low secretor status predicts severe outcomes in premature infants. The Journal of pediatrics. 2011;158:745–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Demmert M, Schaper A, Pagel J, et al. FUT 2 polymorphism and outcome in very-low-birth-weight infants. Pediatric research. 2015;77:586–590. [DOI] [PubMed] [Google Scholar]
- 25.Sampath V, Garland JS, Helbling D, et al. Antioxidant response genes sequence variants and BPD susceptibility in VLBW infants. Pediatric research. 2015;77:477–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huizing MJ, Cavallaro GH, 2017 #396}, Moonen RM, et al. Is the C242T Polymorphism of the CYBA Gene Linked with Oxidative Stress-Associated Complications of Prematurity? Antioxidants & redox signaling. 2017;27:1432–1438. [DOI] [PubMed] [Google Scholar]
- 27.Moonen RM, Paulussen AD, Souren NY, Kessels AG, Rubio-Gozalbo ME, Villamor E. Carbamoyl phosphate synthetase polymorphisms as a risk factor for necrotizing enterocolitis. Pediatric research. 2007;62:188–190. [DOI] [PubMed] [Google Scholar]
- 28.Moonen RM, Cavallaro G, Huizing MJ, Gonzalez-Luis GE, Mosca F, Villamor E. Association between the p.Thr1406Asn polymorphism of the carbamoyl-phosphate synthetase 1 gene and necrotizing enterocolitis: A prospective multicenter study. Scientific reports. 2016;6:36999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Banyasz I, Bokodi G, Vasarhelyi B, et al. Genetic polymorphisms for vascular endothelial growth factor in perinatal complications. European cytokine network. 2006;17:266–270. [PubMed] [Google Scholar]
- 30.Ma F, Li S, Hao H, et al. Association of Heparin-binding EGF-like Growth Factor Polymorphisms with Necrotizing Enterocolitis in Preterm Infants. Journal of pediatric gastroenterology and nutrition. 2017. [DOI] [PubMed] [Google Scholar]
- 31.Leaphart CL, Cavallo J, Gribar SC, et al. A critical role for TLR4 in the pathogenesis of necrotizing enterocolitis by modulating intestinal injury and repair. Journal of immunology. 2007;179:4808–4820. [DOI] [PubMed] [Google Scholar]
- 32.Jilling T, Simon D, Lu J, et al. The roles of bacteria and TLR4 in rat and murine models of necrotizing enterocolitis. Journal of immunology. 2006;177:3273–3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu J, Jilling T, Li D, Caplan MS. Polyunsaturated fatty acid supplementation alters proinflammatory gene expression and reduces the incidence of necrotizing enterocolitis in a neonatal rat model. Pediatric research. 2007;61:427–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fawley J, Cuna A, Menden HL, et al. Single-Immunoglobulin Interleukin-1-Related Receptor regulates vulnerability to TLR4-mediated necrotizing enterocolitis in a mouse model. Pediatric research. 2018;83:164–174. [DOI] [PubMed] [Google Scholar]
- 35.Wald D, Qin J, Zhao Z, et al. SIGIRR, a negative regulator of Toll-like receptor-interleukin 1 receptor signaling. Nature immunology. 2003;4:920–927. [DOI] [PubMed] [Google Scholar]
- 36.Chen G, Shaw MH, Kim YG, Nunez G. NOD-like receptors: role in innate immunity and inflammatory disease. Annual review of pathology. 2009;4:365–398. [DOI] [PubMed] [Google Scholar]
- 37.Cho JH, Abraham C. Inflammatory bowel disease genetics: Nod2. Annual review of medicine. 2007;58:401–416. [DOI] [PubMed] [Google Scholar]
- 38.Neal MD, Sodhi CP, Dyer M, et al. A critical role for TLR4 induction of autophagy in the regulation of enterocyte migration and the pathogenesis of necrotizing enterocolitis. Journal of immunology. 2013;190:3541–3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gadjeva M, Thiel S, Jensenius JC. The mannan-binding-lectin pathway of the innate immune response. Current opinion in immunology. 2001;13:74–78. [DOI] [PubMed] [Google Scholar]
- 40.Schlapbach LJ, Aebi C, Fisch U, et al. Higher cord blood levels of mannose-binding lectin-associated serine protease-2 in infants with necrotising enterocolitis. Pediatric research. 2008;64:562–566. [DOI] [PubMed] [Google Scholar]
- 41.Caplan MS, Sun XM, Hsueh W. Hypoxia causes ischemic bowel necrosis in rats: the role of platelet-activating factor (PAF-acether). Gastroenterology. 1990;99:979–986. [DOI] [PubMed] [Google Scholar]
- 42.Caplan MS, Sun XM, Hseuh W, Hageman JR. Role of platelet activating factor and tumor necrosis factor-alpha in neonatal necrotizing enterocolitis. The Journal of pediatrics. 1990;116:960–964. [DOI] [PubMed] [Google Scholar]
- 43.Caplan MS, Hedlund E, Adler L, Lickerman M, Hsueh W. The platelet-activating factor receptor antagonist WEB 2170 prevents neonatal necrotizing enterocolitis in rats. Journal of pediatric gastroenterology and nutrition. 1997;24:296–301. [DOI] [PubMed] [Google Scholar]
- 44.Caplan MS, Lickerman M, Adler L, Dietsch GN, Yu A. The role of recombinant platelet-activating factor acetylhydrolase in a neonatal rat model of necrotizing enterocolitis. Pediatric research. 1997;42:779–783. [DOI] [PubMed] [Google Scholar]
- 45.Lawrence T The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harbor perspectives in biology. 2009;1:a001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chung DH, Ethridge RT, Kim S, et al. Molecular mechanisms contributing to necrotizing enterocolitis. Annals of surgery. 2001;233:835–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Plaen IG, Liu SX, Tian R, et al. Inhibition of nuclear factor-kappaB ameliorates bowel injury and prolongs survival in a neonatal rat model of necrotizing enterocolitis. Pediatric research. 2007;61:716–721. [DOI] [PubMed] [Google Scholar]
- 48.Tian J, Liu Y, Jiang Y, et al. Association of single nucleotide polymorphisms of IL23R and IL17 with necrotizing enterocolitis in premature infants. Molecular and cellular biochemistry. 2017;430:201–209. [DOI] [PubMed] [Google Scholar]
- 49.Marseglia L, D’Angelo G, Manti S, et al. Oxidative Stress-Mediated Damage in Newborns with Necrotizing Enterocolitis: A Possible Role of Melatonin. American journal of perinatology. 2015;32:905–909. [DOI] [PubMed] [Google Scholar]
- 50.Yan X, Managlia E, Liu SX, et al. Lack of VEGFR2 signaling causes maldevelopment of the intestinal microvasculature and facilitates necrotizing enterocolitis in neonatal mice. American journal of physiology. Gastrointestinal and liver physiology 2016:ajpgi 00273 02015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karatepe HO, Kilincaslan H, Berber M, et al. The effect of vascular endothelial growth factor overexpression in experimental necrotizing enterocolitis. Pediatric surgery international. 2014;30:327–332. [DOI] [PubMed] [Google Scholar]
- 52.Zamora SA, Amin HJ, McMillan DD, et al. Plasma L-arginine concentrations in premature infants with necrotizing enterocolitis. The Journal of pediatrics. 1997;131:226–232. [DOI] [PubMed] [Google Scholar]
- 53.Polycarpou E, Zachaki S, Tsolia M, et al. Enteral L-arginine supplementation for prevention of necrotizing enterocolitis in very low birth weight neonates: a double-blind randomized pilot study of efficacy and safety. JPEN. Journal of parenteral and enteral nutrition 2013;37:617–622. [DOI] [PubMed] [Google Scholar]
- 54.Veldman BA, Spiering W, Doevendans PA, et al. The Glu298Asp polymorphism of the NOS 3 gene as a determinant of the baseline production of nitric oxide. Journal of hypertension. 2002;20:2023–2027. [DOI] [PubMed] [Google Scholar]
- 55.Radulescu A, Zhang HY, Yu X, et al. Heparin-binding epidermal growth factor-like growth factor overexpression in transgenic mice increases resistance to necrotizing enterocolitis. Journal of pediatric surgery. 2010;45:1933–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feng J, Besner GE. Heparin-binding epidermal growth factor-like growth factor promotes enterocyte migration and proliferation in neonatal rats with necrotizing enterocolitis. Journal of pediatric surgery. 2007;42:214–220. [DOI] [PubMed] [Google Scholar]
- 57.Jilling T, Ambalavanan N, Cotten CM, et al. Surgical necrotizing enterocolitis in extremely premature neonates is associated with genetic variations in an intergenic region of chromosome eight. Pediatric research. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marian AJ. Molecular genetic studies of complex phenotypes. Translational research : the journal of laboratory and clinical medicine. 2012;159:64–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bamshad MJ, Ng SB, Bigham AW, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nature reviews. Genetics 2011;12:745–755. [DOI] [PubMed] [Google Scholar]
- 60.Tennessen JA, Bigham AW, O’Connor TD, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kiezun A, Garimella K, Do R, et al. Exome sequencing and the genetic basis of complex traits. Nature genetics. 2012;44:623–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bick D, Dimmock D. Whole exome and whole genome sequencing. Current opinion in pediatrics. 2011;23:594–600. [DOI] [PubMed] [Google Scholar]
- 63.Panoutsopoulou K, Tachmazidou I, Zeggini E. In search of low-frequency and rare variants affecting complex traits. Human molecular genetics. 2013;22:R16–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chan KY, Leung KT, Tam YH, et al. Genome-wide expression profiles of necrotizing enterocolitis versus spontaneous intestinal perforation in human intestinal tissues: dysregulation of functional pathways. Annals of surgery. 2014;260:1128–1137. [DOI] [PubMed] [Google Scholar]
- 65.Ng PC, Chan KY, Leung KT, et al. Comparative MiRNA Expressional Profiles and Molecular Networks in Human Small Bowel Tissues of Necrotizing Enterocolitis and Spontaneous Intestinal Perforation. PloS one. 2015;10:e0135737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pastinen T Genome-wide allele-specific analysis: insights into regulatory variation. Nature reviews. Genetics 2010;11:533–538. [DOI] [PubMed] [Google Scholar]
- 67.Albert FW, Kruglyak L. The role of regulatory variation in complex traits and disease. Nature reviews. Genetics 2015;16:197–212. [DOI] [PubMed] [Google Scholar]
- 68.Stunnenberg HG, Hubner NC. Genomics meets proteomics: identifying the culprits in disease. Human genetics. 2014;133:689–700. [DOI] [PMC free article] [PubMed] [Google Scholar]