Abstract
Background
Myasthenia gravis (MG) is a group of autoimmune disease which could be accompanied by thymoma. Many differences have been observed between thymoma-associated MG (TAMG) and non-MG thymoma (NMG). However, the molecular difference between them remained unknown. This study aimed to explore the differentially expressed genes (DEGs) between the two categories and to elucidate the possible pathogenesis of TAMG further.
Methods
DEGs were calculated using the RNA-Sequencing data from 11 TAMG and 10 NMG in The Cancer Genome Atlas (TCGA) database. GeneAnalytics was performed to characterize the associations between DEGs and tissues and cells, diseases, gene ontology (GO) terms, pathways, phenotypes, and drug/compounds, respectively. Genes related to T cells were sorted out using LifeMapDiscovery Cells and Tissues Database. Genes directly related to the phenotype of autoimmune diseases that were identified by VarElect were validated by reverse transcription-quantitative polymerase chain reaction (RT-qPCR).
Results
The expression level of 169 genes showed a significant difference between the two groups, with 94 up-regulated and 75 down-regulated. Overexpression of six genes (ATM, SFTPB, ANKRD55, BTLA, CCR7, TNFRSF25), which are expressed in T cells and directly related to autoimmune disease through VarElect, was identified. The overexpression of soluble BTLA (sBTLA) (P=0.027), CCR7 (P=0.0018), TNFRSF25 (P=0.0013) and ANKRD55 (P=0.0026) was validated by RT-qPCR in thymoma tissues from our center.
Conclusions
Overexpression of sBTLA, CCR7, TNFRSF25 and ANKRD55 was identified and validated by RT-qPCR, which could partly explain the underlying pathogenesis in TAMG.
Keywords: Thymoma, myasthenia gravis (MG), differentially expressed genes (DEGs), soluble BTLA, CCR7, TNFRSF25, ANKRD55
Introduction
Myasthenia gravis (MG), characterized by T cell involvement and complement dependence, is a group of autoimmune disease with antibodies targeting neuromuscular junctions (1). It presents as fluctuating muscular weakness, mainly affecting extraocular muscles, bulbar muscles and proximal limb muscles. Based on the clinical features and serum antibodies, MG can be classified as early/late-onset, thymoma/nonthymoma, acetylcholine receptor (AChR) antibody/muscle-specific tyrosine kinase (MuSK)/low density lipoprotein receptor related protein 4 (LRP4) antibody positive/antibody-negative and generalized/ocular subgroups (1). In 75–90% cases, MG is combined with abnormalities of thymus, including thymus hyperplasia with germinal centers (85%) and thymoma (15%) (2). Interestingly, thymoma is not the exclusive risk factor of MG, only 15–33% of thymoma patients may develop MG (3,4). With reference to WHO histological classification, type B thymoma is shown to be more frequently associated with MG than type A and AB. Among type B thymoma, type B2 is the most frequently associated with the disease (5).
Compared with early-onset and late-onset MG without thymoma, TAMG usually occurs after 50 years old and a small proportion are diagnosed in their adolescence. The symptoms are usually more severe and with bulbar muscles involved. The gender bias was not significant and its correlation with human leukocyte antigen (HLA) was not strong. Besides anti-AChR antibody, TAMG can also be accompanied by anti-Titin, anti-ryanodine antibodies and other autoimmune diseases (6). Some recent studies indicated that T cells may play a potentially key role in TAMG pathogenesis. Compared with non-MG thymoma (NMG), thymoma in TAMG can generate more mature CD4+CD45RA+T cells exporting to the peripheral while the peripheral AChR-reactive T lymphocytes can be recirculated to the thymus to be activated (2). A higher frequency of circulating IL-17 producing CD4+ T-cell subsets (Th17) and a lower proportion of CD4+CD25highFoxp3+ regulatory T (Treg) cells were also observed (7,8). Besides, our study group verified a higher percentage of T follicular helper (Tfh) cells in MG patients with thymoma (9), which may also demonstrate a difference in pathogenesis between TAMG and NMG. However, the molecular mechanisms behind TAMG still need further investigations.
On a genome-wide scale, gene expression profiling is increasingly used to explore pathogenesis and to find out possible biomarkers in various human disorders. Technological advances in DNA microarrays and subsequent RNA-sequencing (RNA-Seq) are employed to analyze gene expression in a comprehensive and unbiased method (10). Previously, the overexpression of Interferon type I (IFN-I) with abnormal regulation of double-stranded RNA (dsRNA) signaling molecules was demonstrated in TAMG but not in NMG using microarrays (11).
To further explore the pathogenesis of TAMG, we calculated the differentially expressed genes (DEGs) using RNA-seq data from 11 TAMG and 10 NMG patients in The Cancer Genome Atlas (TCGA) database. Based on these genes, we performed gene ontology (GO) classification and functional enrichment analysis using GeneAnalytics (12). We further screened out genes related to T cells and verified these DEGs by reverse transcription-quantitative polymerase chain reaction (RT-qPCR).
Methods
TCGA dataset of thymoma
High throughput RNA-Seq data of the thymoma tissues, including B2 thymoma tissues from 11 TAMG and 10 NMG were downloaded from TCGA database on December 31st, 2017. The summary of the clinical information of these patients from TCGA was shown in Table S1. Considering TCGA data a community resource project, additional approval from the Medical Ethics Committee in our hospital was not necessary. TCGA data access policies and publication guidelines were also obeyed in the present study.
Table S1. Summary of the clinical information of patients from TCGA.
| No. | Case ID | Gender | Age at diagnosis (year) | Pathology | Myasthenia gravis | Race | Case UUID |
|---|---|---|---|---|---|---|---|
| 1 | TCGA-X7-A8DB | Female | 71 | Thymoma; type B2 | No | White | 4165391B-6122-4E87-BA91-CE935E0B3CF4 |
| 2 | TCGA-XU-A92U | Male | 67 | Thymoma; type B2 | No | White | FAC44B50-AA8C-498E-88AF-4EB5C2F5F67A |
| 3 | TCGA-ZC-AAAH | Male | 52 | Thymoma; type B2 | No | White | 3CC2D49B-A3A6-4F55-A355-68CBE84F4056 |
| 4 | TCGA-YT-A95E | Male | 52 | Thymoma; type B2 | No | White | D3189C8E-FFE0-41FF-9C7B-64DA4C83B991 |
| 5 | TCGA-X7-A8M4 | Male | 41 | Thymoma; type B2 | No | White | 23CC4CD3-87DC-4342-9072-BE3695E393FA |
| 6 | TCGA-X7-A8DE | Female | 35 | Thymoma; type B2 | No | White | 15A2BAAE-D15E-4349-9358-74356A2D2AD0 |
| 7 | TCGA-X7-A8M6 | Male | 43 | Thymoma; type B2 | No | White | A6885883-F1A2-48DD-8D72-0038319069A1 |
| 8 | TCGA-XM-A8RC | Male | 67 | Thymoma; type B2 | No | White | B8348ADB-17DD-4849-BD4F-C2E7E15DB1F4 |
| 9 | TCGA-4X-A9FB | Male | 44 | Thymoma; type B2 | No | White | 2C2E169E-9F93-41A7-BFC4-ADE18BB5D7AE |
| 10 | TCGA-XM-A8RD | Female | 69 | Thymoma; type B2 | No | White | 68D2AB61-50FC-41E8-B3EE-75C70F0F4369 |
| 11 | TCGA-4X-A9FD | Female | 43 | Thymoma; type B2 | Yes | White | 0A312064-878F-4B69-8BB7-E7F14E6CD7FE |
| 12 | TCGA-XU-AAXX | Female | 44 | Thymoma; type B2 | Yes | White | 5B7B3E7C-3230-4675-8FDF-3E5119B02CE1 |
| 13 | TCGA-XU-AAXV | Female | 45 | Thymoma; type B2 | Yes | White | 80606B1C-E8C8-4C3D-8475-3D846A84CA98 |
| 14 | TCGA-X7-A8M7 | Male | 26 | Thymoma; type B2 | Yes | White | 56C4F5E8-4E89-4BA0-984D-E17F11A401F5 |
| 15 | TCGA-X7-A8DG | Male | 47 | Thymoma; type B2 | Yes | White | A653B0BB-AACF-4B11-BB11-42E043E49BF6 |
| 16 | TCGA-XU-AAXY | Male | 43 | Thymoma; type B2 | Yes | White | 7933B320-AE8C-4D3D-9468-8ABB135E93D5 |
| 17 | TCGA-4V-A9QS | Female | 53 | Thymoma; type B2 | Yes | White | 88C0AF76-B1BA-478F-A42B-8A50C0568D5E |
| 18 | TCGA-4V-A9QI | Male | 68 | Thymoma; type B2 | Yes | White | EEAA0FA3-408C-4E0F-A18A-095514173FF1 |
| 19 | TCGA-ZC-AAAA | Male | 39 | Thymoma; type B2 | Yes | White | 67FF20BA-1B56-45AE-9310-19DD74401E67 |
| 20 | TCGA-YT-A95G | Female | 61 | Thymoma; type B2 | Yes | White | 7459BEC0-1B54-4FD7-90DB-8375168E6B1A |
| 21 | TCGA-X7-A8D6 | Female | 48 | Thymoma; type B2 | Yes | White | 66C564BA-B42D-4BE9-9E5F-BF93984569EB |
Exploration of DEGs and GeneAnalytics
A simple set of transcript quantification was obtained from the RNA-Seq data of patients with B2 thymoma. RNA-seq relative read counts were measured by counts per million (CPM) to calculate DEGs using edgeR package (13). The value of fold change (FC) was log2 transformed while false discovery rate (FDR) was −log10 transformed for downstream analysis. We further performed differential analysis (|log2FC| >1, log2CPM >1, P value <0.05 and FDR <0.05) with Limma package (14).
Next, we used GeneAnalytics (http://geneanalytics.genecards.org/), a tool empowered by the GeneCards suite, including GeneCards human gene database, MalaCards human disease database, PathCards human biological pathways database and LifeMapDiscovery Cells and Tissues Database to analyze the associations between DEGs and tissues and cells, diseases, GO terms, pathways, phenotypes, and drug/compounds, respectively. Tissues and cells were scored with a matching algorithm that weighs tissue specificity, gene abundance and function. Disease matching scores were originated from the degree of gene-disease relations. Superpathways included the related pathways with high matched genes to reduce redundancy, strengthen inferences and pathway enrichment. Genes related to the T cells were sorted out according to LifeMapDiscovery Cells and Tissues Database (12). Among them, genes directly related to the phenotype of autoimmune diseases were identified by Varlect (15).
Human thymic samples
Eighteen thymoma samples were obtained from patients who underwent thymectomy at the Department of Thoracic Surgery, Huashan and Zhongshan Hospital, Fudan University in 2016–2018. Of them, 9 was with MG and 9 was not. The study was approved by Fudan University Institutional Review Board and written informed consents were granted.
The TAMG group was defined using following criteria: (I) age ≥18 years; (II) definitive diagnosis of generalized MG according to clinical, electrophysiological and antibody status; (III) histopathologic diagnosis of thymoma. The patients were classified by the Myasthenia Gravis Foundation of America (MGFA) Classification (16). Patients who had received corticosteroid or immunosuppressant treatment were excluded.
Validation of RT-qPCR
Total RNA was extracted from the thymoma tissues using Trizol® (Invitrogen, USA) reagent based on the manufacturer’s instructions. The reverse transcription (RT) reactions were conducted with a cDNA synthesis kit (Takara, China). Quantitative polymerase chain reaction (qPCR) was performed with QuantStudio (Applied Biosystems, USA). The results were quantified with the 2−ΔΔCT method against GAPDH for normalization. The data represents the mean value of three experiments. Real-time PCR primers were listed in Table S2.
Table S2. Primers used in RT-PCR.
| Gene | Primers | Direction |
|---|---|---|
| TNFRSF25 | CTTGTGTGTCCCCAAGACAC | F |
| AGTCTAGGCATGGTTGGCAG | R | |
| CCR7 | TCTGCCTGGACTAGAGGGAC | F |
| TATCTTCTGGAGCAGGGGCT | R | |
| ANKRD55 | GGCCGAATGTGTCCAGTCAC | F |
| GTTGAGCACGTTGAACCGTC | R | |
| SFTPB | CTACTTCCAGAACCAGACTGAC | F |
| GCTCGGAGAGATCCTGTGTG | R | |
| BTLA | CATCTTAGCAGGAGATCCCTTTG | F |
| TGACCCATTGTCATTAGGAAGC | R | |
| sBTLA | TGTGACAGGAAAGCAAAATGAAC | F |
| CAGACCCTTCCTGCATCCTG | R | |
| ATM | TGATCTTGTGCCTTGGCTAC | F |
| TATGGTGTACGTTCCCCATG | R | |
| GAPDH | GTGAAGGTCGGAGTCAACG | F |
| GTTCTCAGCCTTGACGGTG | R |
RT-PCR, reverse transcription-polymerase chain reaction.
Statistical analysis
For DEGs analysis, the values of log2CPM were calculated and normalized by quantile normalization. Then the values of FC were calculated with the mean values of log2CPM between the two groups. All data analysis of DEGs was conducted and related figures were generated with R 3.5.1 (www.r-project.org). The results of qPCR were analyzed by Student’s t-test or Mann-Whitney U test using GraphPad software. A P value <0.05 was regarded as statistically significant.
Results
GeneAnalytics
DEGs and GeneAnalytics
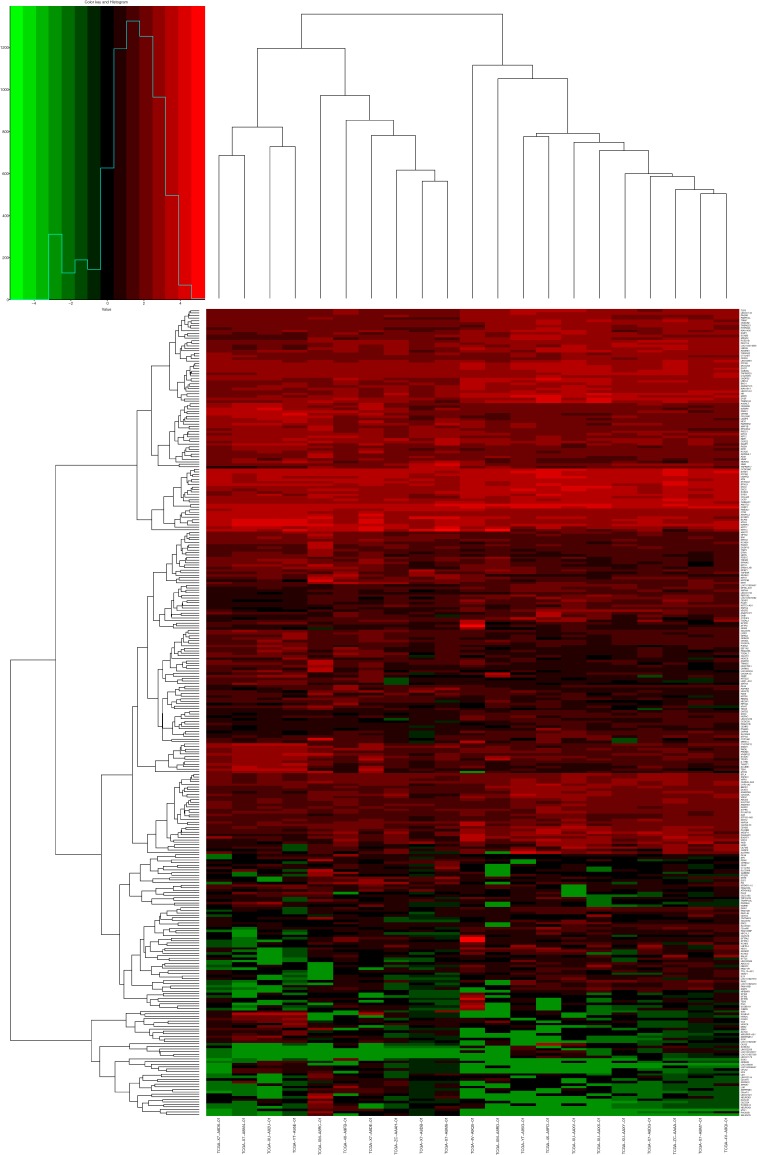
A total of 24,991 transcripts were included and transcripts with CPM value of 0 were excluded. Finally, we screened 16,477 transcripts for analysis. The expression level of 169 genes showed significant difference between the two groups (|log2FC| >1, log2CPM >1, P value <0.05 and FDR <0.05), with 94 up-regulated and 75 down-regulated. The final data was listed as the matrix in Table S3. DEGs calculated by edgeR were shown with volcano plot (Figure S1) and heat map (Figure S2).
Table S3. List of differentially expressed genes derived from edgeR.
| Gene | Log2FC | Log2CPM | P value | FDR |
|---|---|---|---|---|
| SOHLH1 | −8.748095249 | 4.030701899 | 1.36E−06 | 0.001177393 |
| NEUROD1 | −8.29403346 | 1.206443756 | 9.61E−06 | 0.003837044 |
| SLC30A8 | −7.394604588 | 2.46893017 | 5.70E−07 | 0.000758208 |
| KRT14 | −5.961451624 | 8.850437314 | 1.12E−09 | 1.11E−05 |
| CYP19A1 | −5.531242351 | 3.688722857 | 1.51E−05 | 0.004857241 |
| PENK | −5.522444584 | 1.896122199 | 7.44E−05 | 0.013764626 |
| CACNA1G | −4.800224071 | 1.502703602 | 8.66E−08 | 0.000208153 |
| KRT33B | −4.678978991 | 3.516999508 | 1.10E−06 | 0.001051576 |
| SEZ6L2 | −4.394822626 | 2.815492394 | 9.38E−08 | 0.000208153 |
| CHRNG | −4.233245516 | 1.055180283 | 5.61E−06 | 0.002730959 |
| TBX1 | −3.997357832 | 4.27653279 | 0.000715258 | 0.04638443 |
| MYH6 | −3.803434257 | 2.338179784 | 0.000116152 | 0.016448228 |
| SCUBE1 | −3.576521905 | 2.719921949 | 0.000125778 | 0.017320051 |
| TRIM55 | −3.458383411 | 1.634996091 | 0.000109941 | 0.016260644 |
| TFDP3 | −3.348329361 | 3.072405038 | 3.20E−05 | 0.007924085 |
| CRYAB | −3.316888439 | 5.004937173 | 7.62E−06 | 0.003381488 |
| DPEP1 | −3.304235712 | 2.760709455 | 7.47E−06 | 0.003381488 |
| NGEF | −3.259681002 | 1.090315496 | 0.000493254 | 0.038774813 |
| TREM2 | −3.206474566 | 1.610887941 | 5.00E−07 | 0.000712547 |
| TWIST1 | −3.197009539 | 2.286019868 | 0.000357804 | 0.031999183 |
| KIF1A | −3.138683446 | 2.52383897 | 0.000224218 | 0.025293513 |
| EPB41L4B | −3.114798511 | 1.811491458 | 1.51E−05 | 0.004857241 |
| TNFRSF17 | −3.052709294 | 3.52099794 | 9.02E−05 | 0.014635034 |
| ANKFN1 | −3.025104688 | 1.324634834 | 0.00016265 | 0.020685527 |
| MYBPC2 | −2.921742889 | 3.150154799 | 0.000176369 | 0.021342792 |
| SMIM1 | −2.911370466 | 2.541786224 | 1.69E−06 | 0.001347512 |
| KCNQ4 | −2.831610853 | 2.167085633 | 0.000127572 | 0.017411471 |
| C1QTNF12 | −2.825191103 | 3.222153422 | 0.000234636 | 0.02623382 |
| DTNA | −2.811280027 | 1.326718473 | 8.55E−05 | 0.014253189 |
| KRT1 | −2.797637848 | 2.613754179 | 6.92E−05 | 0.013165864 |
| RTN4RL2 | −2.797383855 | 3.565203693 | 3.56E−06 | 0.002151673 |
| NMB | −2.785464392 | 4.769423606 | 0.000144544 | 0.018741018 |
| NKAIN4 | −2.725835329 | 3.32315051 | 0.000175082 | 0.021316194 |
| DUSP15 | −2.712099618 | 1.402249593 | 7.45E−05 | 0.013764626 |
| IGFBP6 | −2.641385346 | 4.705648348 | 0.000357201 | 0.031999183 |
| IRX1 | −2.641371905 | 1.725281185 | 1.65E−06 | 0.001347512 |
| KIF19 | −2.599860383 | 2.634519615 | 0.000756833 | 0.047520993 |
| LAMP5 | −2.521963474 | 3.634943772 | 0.000114638 | 0.016448228 |
| HEY1 | −2.482922047 | 4.131715584 | 0.000115877 | 0.016448228 |
| IL17RD | −2.482847407 | 1.563467996 | 0.00074938 | 0.047350848 |
| NPTX2 | −2.474916128 | 1.944105866 | 0.000114859 | 0.016448228 |
| PRRX1 | −2.418850644 | 5.086227953 | 0.000511045 | 0.039550531 |
| RAMP1 | −2.418519882 | 4.304251887 | 0.000157601 | 0.020171961 |
| SPINK5 | −2.401970407 | 1.419776083 | 0.000392375 | 0.033673167 |
| PRDM6 | −2.376809933 | 2.107794342 | 0.000258927 | 0.027114371 |
| COL26A1 | −2.360436459 | 4.132183181 | 0.000365692 | 0.03234187 |
| PDZD7 | −2.340246855 | 1.531649414 | 0.000262304 | 0.027136948 |
| KRT17 | −2.338793792 | 5.931256513 | 4.11E−05 | 0.009125326 |
| S100A1 | −2.335433447 | 2.256553923 | 8.31E−05 | 0.014253189 |
| TSPEAR | −2.196880683 | 1.24295098 | 0.000641817 | 0.044285573 |
| NNAT | −2.163234419 | 2.316903698 | 0.000406619 | 0.034257196 |
| CPAMD8 | −2.135732976 | 3.744044433 | 0.000728403 | 0.04638443 |
| BCAM | −2.119331059 | 6.855164256 | 9.10E−05 | 0.014649376 |
| KCNJ5 | −2.074088058 | 2.820417316 | 4.19E−05 | 0.009192939 |
| MAST1 | −2.063241259 | 1.728215681 | 0.000334117 | 0.031503459 |
| RARRES2 | −2.050658472 | 4.309808115 | 0.00024593 | 0.026833276 |
| MYL9 | −2.037376153 | 6.269218628 | 3.35E−05 | 0.007924085 |
| RADIL | −2.017851476 | 2.206839739 | 0.000617665 | 0.04373377 |
| MAP1B | −1.983435991 | 3.693569233 | 0.000593846 | 0.042806191 |
| ADPRHL1 | −1.983165118 | 2.371933935 | 0.000411465 | 0.034519841 |
| ADM | −1.908421627 | 2.164461803 | 0.000166279 | 0.02082272 |
| KIF12 | −1.839197112 | 4.042257933 | 6.80E−05 | 0.013162866 |
| A4GALT | −1.746454553 | 3.356362608 | 0.000628883 | 0.044214475 |
| AFAP1L2 | −1.708529952 | 5.629806254 | 0.000699888 | 0.046273742 |
| EPHX3 | −1.704391406 | 1.161569209 | 0.000131591 | 0.017532068 |
| RYR1 | −1.690586943 | 2.119175707 | 0.000461127 | 0.036829271 |
| KCNIP3 | −1.591561724 | 6.27756293 | 0.000235181 | 0.02623382 |
| RBP1 | −1.396617446 | 4.330992858 | 0.000774057 | 0.048298738 |
| RGS9 | −1.386737859 | 2.212607056 | 0.000317832 | 0.03054823 |
| IGFBP4 | −1.379489998 | 6.898794948 | 0.000192914 | 0.022658341 |
| RAB3IL1 | −1.325468721 | 5.364223263 | 0.000459158 | 0.036827158 |
| COPZ2 | −1.319279321 | 4.042731219 | 0.000626393 | 0.044194982 |
| PXDC1 | −1.284119427 | 3.656120313 | 0.000660007 | 0.044907927 |
| CTSF | −1.231637947 | 4.859352108 | 0.000638457 | 0.044264104 |
| KIFC3 | −1.14093435 | 3.501584415 | 0.000754573 | 0.047520993 |
| CSRP1 | 1.031898606 | 7.236290106 | 0.000196095 | 0.022764126 |
| C1QTNF6 | 1.048423636 | 4.215725309 | 0.000547396 | 0.04108972 |
| PTPN3 | 1.064663452 | 4.79852239 | 0.00063754 | 0.044264104 |
| PTPRS | 1.084992749 | 6.321535934 | 4.01E−05 | 0.008992162 |
| ATM | 1.092312455 | 6.065074206 | 0.000166857 | 0.02082272 |
| CAB39L | 1.114221581 | 4.635029486 | 0.000459256 | 0.036827158 |
| EOGT | 1.13860395 | 5.023332339 | 2.33E−05 | 0.006539235 |
| SYNE1 | 1.15698623 | 6.739375363 | 6.11E−05 | 0.012327227 |
| ST3GAL1 | 1.160699732 | 5.608007883 | 6.86E−06 | 0.003183191 |
| CCDC88C | 1.175757769 | 6.84430386 | 2.17E−05 | 0.006279865 |
| MCOLN3 | 1.176635199 | 4.748109433 | 0.000175053 | 0.021316194 |
| AMOTL1 | 1.224644988 | 7.927186252 | 0.000573374 | 0.042245614 |
| CD226 | 1.251137605 | 2.717868443 | 0.000356512 | 0.031999183 |
| PAQR6 | 1.278244251 | 2.951705012 | 0.000813914 | 0.049547031 |
| C11orf21 | 1.302413542 | 2.972934344 | 0.000586808 | 0.042551936 |
| ADAMTS15 | 1.346155525 | 3.887698093 | 0.000550948 | 0.041201438 |
| COL4A5 | 1.376611342 | 5.435165895 | 0.000658274 | 0.044907927 |
| TSPAN32 | 1.415877755 | 3.247474464 | 0.000352642 | 0.031999183 |
| PCED1B | 1.447718826 | 3.95648763 | 0.000580164 | 0.042551936 |
| PEX11G | 1.450854953 | 3.682162391 | 0.000358722 | 0.031999183 |
| BNC1 | 1.463281485 | 4.041859479 | 0.000164864 | 0.02082272 |
| CABLES1 | 1.466009022 | 5.524096702 | 2.80E−05 | 0.007261196 |
| BTLA | 1.493058148 | 2.296589938 | 0.000661238 | 0.044907927 |
| ZNF831 | 1.521658299 | 1.931796715 | 0.000521141 | 0.040021588 |
| POU5F1B | 1.568868299 | 1.274084132 | 0.000509381 | 0.039550531 |
| AMDHD1 | 1.573918649 | 1.388503093 | 0.000806365 | 0.049388597 |
| PABPC4L | 1.585601776 | 3.026351018 | 0.000604812 | 0.043284181 |
| CCR7 | 1.613695957 | 5.827382462 | 0.000334489 | 0.031503459 |
| SUSD4 | 1.62114103 | 5.625537896 | 9.50E−05 | 0.015059557 |
| CHRM3−AS2 | 1.687459799 | 2.668354407 | 0.000305892 | 0.029793877 |
| DSC2 | 1.713299303 | 5.811930077 | 0.000128245 | 0.017411471 |
| BTNL9 | 1.718941315 | 6.493067076 | 0.000171341 | 0.021118357 |
| OGFRL1 | 1.724670542 | 6.3800536 | 1.27E−07 | 0.000253168 |
| KIAA1614 | 1.724837466 | 4.390292809 | 8.45E−05 | 0.014253189 |
| TMEM221 | 1.747277292 | 3.258767333 | 0.000793345 | 0.048829234 |
| SULT1B1 | 1.751701245 | 1.066856666 | 0.000253265 | 0.027108324 |
| TNFRSF25 | 1.760189383 | 4.648424342 | 1.57E−07 | 0.000285793 |
| SYBU | 1.795524926 | 5.823073573 | 0.000209855 | 0.023943912 |
| S1PR5 | 1.818352037 | 1.726708795 | 0.000513812 | 0.039611173 |
| TLR5 | 1.874988549 | 3.928291175 | 1.61E−05 | 0.005102521 |
| ABCD2 | 1.912033944 | 1.47977496 | 8.64E−05 | 0.014253189 |
| RGPD1 | 1.962107442 | 1.447866581 | 0.000240462 | 0.026426386 |
| VIPR1 | 1.990615221 | 2.346466784 | 4.73E−05 | 0.010050012 |
| DSC3 | 2.060586958 | 6.23696081 | 2.64E−06 | 0.001797271 |
| PITPNM3 | 2.066183086 | 3.19477902 | 0.000279793 | 0.028215335 |
| CADPS2 | 2.072964696 | 5.37328009 | 0.000237964 | 0.026396822 |
| CNKSR2 | 2.118672531 | 3.24874403 | 1.34E−05 | 0.004690262 |
| LRRN3 | 2.156274312 | 4.605360993 | 1.75E−05 | 0.005305741 |
| SCN2B | 2.185969726 | 3.524182011 | 0.000259038 | 0.027114371 |
| TMEM163 | 2.222626461 | 5.546379096 | 0.000443157 | 0.03611641 |
| ADGRB1 | 2.268231032 | 4.599635445 | 0.000354712 | 0.031999183 |
| TRMT9B | 2.270450369 | 4.289344976 | 0.000546679 | 0.04108972 |
| RBFOX3 | 2.281853526 | 1.166636114 | 0.000730959 | 0.04638443 |
| LRRC4 | 2.310897045 | 5.022877443 | 3.72E−06 | 0.002186462 |
| VSIG1 | 2.324453749 | 2.360575948 | 0.000350873 | 0.031999183 |
| SGIP1 | 2.364819235 | 3.463574899 | 8.82E−06 | 0.003621892 |
| ANKRD55 | 2.385252316 | 2.396103806 | 0.000343918 | 0.031939584 |
| HR | 2.392649271 | 5.773120702 | 5.40E−06 | 0.002695495 |
| CYP21A2 | 2.396917721 | 2.816431582 | 0.000123312 | 0.017098387 |
| PLEKHD1 | 2.427148309 | 2.545280033 | 0.000722988 | 0.04638443 |
| DCST2 | 2.469699059 | 1.817009447 | 0.000108736 | 0.016202501 |
| CACNA1H | 2.554062761 | 2.753556499 | 0.000397447 | 0.033771062 |
| MAP2 | 2.595734303 | 5.832143722 | 1.22E−05 | 0.004425089 |
| C2CD4A | 2.621694006 | 3.171878901 | 0.000440538 | 0.036050107 |
| NAPSA | 2.648866778 | 2.812059645 | 0.000111476 | 0.016366482 |
| DYSF | 2.654315409 | 6.120007721 | 4.50E−06 | 0.002430803 |
| BBOX1 | 2.707435961 | 2.624542486 | 2.94E−05 | 0.00744023 |
| MIR452 | 2.726183047 | 4.100742925 | 9.79E−05 | 0.015278367 |
| TRIM7 | 2.749984944 | 3.966026316 | 3.02E−08 | 0.000120607 |
| MEGF11 | 2.768338744 | 3.901573036 | 0.000510894 | 0.039550531 |
| ANAPC1P1 | 2.791896532 | 1.466174301 | 0.000240878 | 0.026426386 |
| CDH26 | 2.817492308 | 3.004824858 | 2.05E−06 | 0.001512784 |
| B3GAT1 | 2.893594108 | 3.129109812 | 7.51E−05 | 0.013764626 |
| NGB | 2.951037162 | 2.930569607 | 0.000684676 | 0.045695125 |
| TCEAL2 | 3.097773295 | 2.284127821 | 0.000392941 | 0.033673167 |
| DAB1 | 3.238208706 | 1.459708099 | 0.000669547 | 0.045110642 |
| CADPS | 3.292163784 | 1.366927466 | 0.000501109 | 0.039237812 |
| VWC2 | 3.392777854 | 3.600284028 | 0.000248537 | 0.026970359 |
| HAS2 | 3.449016578 | 1.627563507 | 5.77E−05 | 0.011869445 |
| PLXNB3 | 3.475439297 | 3.383279965 | 7.05E−07 | 0.000880073 |
| CYP4F3 | 3.787744948 | 1.921251767 | 6.75E−05 | 0.013162866 |
| SFTPD | 3.953404307 | 2.313645037 | 0.000457005 | 0.036827158 |
| MIR614 | 4.203046253 | 1.614918477 | 0.000108146 | 0.016202501 |
| LHFPL4 | 4.340877226 | 1.0676253 | 0.000438816 | 0.036050107 |
| VEGFD | 5.089525645 | 1.953670015 | 7.86E−13 | 1.57E−08 |
| OLFM3 | 5.49125031 | 2.657082163 | 1.86E−08 | 9.29E−05 |
| SALL3 | 5.858632642 | 1.337707021 | 2.36E−05 | 0.006539235 |
| SCGB1A1 | 6.134443217 | 1.230518068 | 0.000617646 | 0.04373377 |
| SLITRK6 | 6.433479385 | 4.049025947 | 1.70E−08 | 9.29E−05 |
| SFTPA2 | 7.881535806 | 5.599887412 | 1.45E−05 | 0.004857241 |
| OLIG2 | 7.883686911 | 2.267494657 | 0.000286309 | 0.028543087 |
| SFTPA1 | 8.457267837 | 5.468790881 | 8.88E−06 | 0.003621892 |
| SFTPC | 9.764859884 | 6.800645422 | 5.66E−08 | 0.00018848 |
| SFTPB | 10.40211808 | 5.172167416 | 3.64E−07 | 0.000605947 |
FC, fold change; CPM, counts per million; FDR, false discovery rate.
Figure S1.
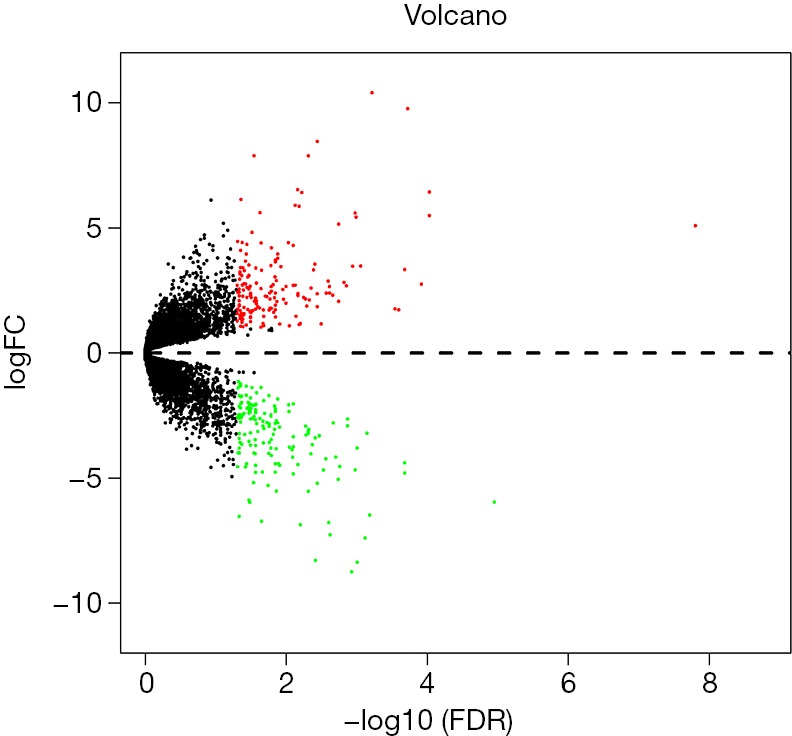
Differentially expressed genes by edgeR. Volcano plot illustrating the differential expression levels of genes of TAMG and NMG group. Genes that are significantly up- and down-regulated in the TAMG group compared to the NMG group are shown in red and green dots, respectively. TAMG, thymoma-associated myasthenia gravis; NMG, non-myasthenia gravis.
Figure S2.
Heat map of the hierarchical clustering based on DEGs (fold change =1, P value <0.05). DEG, differentially expressed gene.
Analysis of tissues and organs (Table S4), diseases (Table S5), biological superpathways (Table S6), GO pathways (Table S7), phenotypes (Table S8) and compounds (Table S9) using GeneAnalytics were listed in Tables S4-S9, respectively. However, we did not see a clearly biological meaning from these results.
Table S4. GeneAnalytics program mapping of systems with 10 highest match scores for 169 DEGs in TAMG and NMG.
| System | Genes matched to system | No. of genes in systems | Score |
|---|---|---|---|
| Nervous system | ABCD2, ADGRB1, ADM, AFAP1L2, AMDHD1, ANKRD55, ATM, B3GAT1, BBOX1, CACNA1G, CACNA1H, CADPS, CCR7, CD226, CNKSR2, COL26A1, COL4A5, COPZ2, CRYAB, CSRP1, CTSF, CYP19A1, CYP4F3, DAB1, DTNA, DUSP15, DYSF, EOGT, EPB41L4B, HAS2, HEY1, IGFBP4, IGFBP6, IL17RD, IRX1, KCNIP3, KCNJ5, KIF1A, KRT14, KRT17, LAMP5, LRRN3, MAP1B, MAP2, MAST1, MCOLN3, MEGF11, MYL9, NAPSA, NEUROD1, NGB, NGEF, NKAIN4, NMB, NNAT, NPTX2, OGFRL1, OLFM3, OLIG2, PABPC4L, PAQR6, PENK, PLEKHD1, PLXNB3, PRRX1, PTPN3, PTPRS, PXDC1, RAB3IL1, RAMP1, RARRES2, RBFOX3, RBP1, RGPD1, RGS9, S100A1, SALL3, SCN2B, SCUBE1, SEZ6L2, SGIP1, SLITRK6, ST3GAL1, SULT1B1, SUSD4, SYBU, SYNE1, TBX1, TCEAL2, TMEM163, TNFRSF25, TRMT9B | 92 | 11.62 |
| Muscoskeletal system | ABCD2, ADAMTS15, ADPRHL1, AMOTL1, ANKFN1, ATM, B3GAT1, BCAM, BNC1, BTLA, BTNL9, C11orf21, CABLES1, CACNA1G, CACNA1H, CADPS, CCR7, CHRNG, COL26A1, COL4A5, COPZ2, CPAMD8, CRYAB, CSRP1, CTSF, CYP4F3, DAB1, DSC2, DTNA, DYSF, EOGT, EPB41L4B, HAS2, HEY1, HR, IGFBP4, IGFBP6, IL17RD, IRX1, KCNIP3, KIF1A, KRT14, KRT17, LAMP5, LRRN3, MAP2, MCOLN3, MIR452, MYBPC2, MYH6, MYL9, NEUROD1, NGEF, NKAIN4, NMB, NNAT, NPTX2, OGFRL1, OLIG2, PENK, PITPNM3, PRRX1, RAB3IL1, RAMP1, RARRES2, RBFOX3, RBP1, RGPD1, RYR1, S100A1, SALL3, SEZ6L2, SFTPA1, SLITRK6, ST3GAL1, SULT1B1, SYBU, SYNE1, TBX1, TCEAL2, TMEM163, TREM2, TRIM55, TRIM7, TSPAN32, TWIST1, VEGFD, VIPR1 | 88 | 10.03 |
| Reproductive system | ADAMTS15, ADM, ANKRD55, ATM, B3GAT1, BNC1, CAB39L, CABLES1, CADPS, CADPS2, CCR7, CD226, COL26A1, COL4A5, COPZ2, CRYAB, CSRP1, CTSF, CYP19A1, CYP4F3, DAB1, DPEP1, DSC2, DSC3, DTNA, DUSP15, DYSF, HAS2, HEY1, HR, IGFBP4, IGFBP6, IL17RD, IRX1, KCNIP3, KCNJ5, KIF1A, KIFC3, KRT1, KRT17, MAP1B, MAP2, MCOLN3, MEGF11, MYL9, NEUROD1, NNAT, NPTX2, PAQR6, PENK, PEX11G, PRRX1, PTPRS, RAB3IL1, RADIL, RAMP1, RARRES2, RBP1, RGPD1, RGS9, S100A1, SOHLH1, SPINK5, ST3GAL1, SUSD4, SYNE1, TCEAL2, TFDP3, TMEM163, TWIST1, VEGFD, VSIG1 | 72 | 7.19 |
| Cardiovascular system | ADAMTS15, ADM, ADPRHL1, ANKRD55, ATM, B3GAT1, BCAM, BNC1, C1QTNF6, CACNA1H, CD226, COPZ2, CRYAB, CSRP1, CTSF, DAB1, DSC2, DSC3, DTNA, DYSF, EOGT, HAS2, HEY1, IGFBP4, IGFBP6, IRX1, KCNIP3, KCNJ5, KIF1A, KRT1, KRT14, LRRN3, MAP1B, MCOLN3, MEGF11, MIR452, MYH6, MYL9, NEUROD1, NKAIN4, OGFRL1, PAQR6, PENK, PLEKHD1, PRRX1, PXDC1, S100A1, SALL3, SPINK5, ST3GAL1, SULT1B1, TBX1, TMEM163, TRIM55, TRIM7, TWIST1 | 56 | 6.39 |
| Hematopoietic system | A4GALT, ADGRB1, ADM, AFAP1L2, AMOTL1, ANKRD55, ATM, BTLA, BTNL9, CABLES1, CACNA1G, CADPS2, CCDC88C, CCR7, CD226, CSRP1, CYP4F3, DSC2, DYSF, EPB41L4B, HAS2, HEY1, KCNIP3, KCNJ5, KRT1, KRT14, KRT17, LAMP5, LRRN3, MYL9, NAPSA, NEUROD1, OGFRL1, OLIG2, PABPC4L, PCED1B, PENK, PITPNM3, PRRX1, RAB3IL1, RAMP1, RBP1, RGPD1, S100A1, SFTPB, SFTPC, SGIP1, SLITRK6, SMIM1, SPINK5, ST3GAL1, SULT1B1, SYNE1, TBX1, TLR5, TNFRSF17, TNFRSF25, TREM2, TSPAN32, VEGFD, VSIG1, ZNF831 | 62 | 6.32 |
| Respiratory system | AMOTL1, ANKFN1, ATM, BCAM, BNC1, BTNL9, CCR7, CD226, CDH26, COL4A5, CRYAB, CSRP1, CYP4F3, DAB1, DSC2, DSC3, EOGT, HR, IRX1, KCNIP3, KCNJ5, KIF1A, KIFC3, KRT14, KRT17, LAMP5, MAP2, MYH6, MYL9, NAPSA, NKAIN4, NMB, NNAT, NPTX2, PAQR6, PENK, PLXNB3, PRRX1, RADIL, RARRES2, RBP1, RYR1, S1PR5, SCGB1A1, SFTPA1, SFTPA2, SFTPB, SFTPC, SFTPD, SLITRK6, SULT1B1, SYNE1, TBX1, TMEM163, TREM2, VEGFD, VIPR1 | 57 | 6.26 |
| Sensory organs | ABCD2, ANKRD55, C1QTNF6, CADPS, CADPS2, CCR7, CHRNG, COL4A5, COPZ2, CPAMD8, CRYAB, CSRP1, CYP4F3, DAB1, DSC2, DTNA, EPHX3, IGFBP6, IRX1, KCNIP3, KCNJ5, KIAA1614, KIF1A, KRT14, LAMP5, MAP1B, MAP2, MEGF11, MYL9, NEUROD1, NKAIN4, NNAT, PABPC4L, PENK, PRRX1, PTPN3, PTPRS, RAB3IL1, RADIL, RARRES2, RBP1, RGPD1, S1PR5, SALL3, SCUBE1, SEZ6L2, SFTPC, SGIP1, SLITRK6, ST3GAL1, SYNE1, TBX1, TNFRSF17, TRMT9B, VWC2 | 55 | 6.05 |
| Integumentary system | A4GALT, ADGRB1, ADM, ATM, BBOX1, BNC1, CABLES1, CACNA1G, CADPS, CCR7, COL4A5, COPZ2, CRYAB, CSRP1, CYP4F3, DAB1, DPEP1, DSC2, DSC3, DYSF, EPHX3, HAS2, HR, IGFBP4, IRX1, KCNIP3, KCNJ5, KIF1A, KRT1, KRT14, KRT17, LAMP5, LRRC4, LRRN3, MAP2, MCOLN3, MYL9, NEUROD1, NKAIN4, NPTX2, PENK, PRRX1, PTPRS, RAMP1, RYR1, S100A1, S1PR5, SCUBE1, SPINK5, ST3GAL1, SYBU, SYNE1, TBX1, TCEAL2, TWIST1 | 55 | 5.81 |
| Endocrine system | AFAP1L2, ATM, B3GAT1, C2CD4A, CAB39L, CACNA1G, CACNA1H, CADPS, CADPS2, CRYAB, CSRP1, CTSF, CYP21A2, CYP4F3, DPEP1, DSC2, EPB41L4B, HEY1, IGFBP6, IRX1, KCNIP3, KCNJ5, KIF1A, KRT14, KRT17, MAP1B, MAP2, MCOLN3, MYH6, MYL9, NEUROD1, NGEF, NNAT, NPTX2, PENK, PRRX1, RAMP1, RARRES2, RBFOX3, RBP1, RGS9, S100A1, SCGB1A1, SEZ6L2, SLC30A8, SLITRK6, SPINK5, ST3GAL1, SYBU, SYNE1, TRMT9B | 51 | 5.12 |
| Gastrointestinal tract | ABCD2, ADM, AFAP1L2, ATM, B3GAT1, BCAM, C2CD4A, CABLES1, CCR7, CD226, CHRNG, COL4A5, CRYAB, CSRP1, CTSF, CYP4F3, DAB1, DPEP1, DSC2, DSC3, DYSF, EPB41L4B, HAS2, HEY1, IGFBP4, IGFBP6, IRX1, KCNIP3, KCNJ5, KIF1A, KRT1, KRT14, KRT17, LAMP5, MAP1B, MAP2, MCOLN3, MYBPC2, MYH6, MYL9, NNAT, PENK, PRRX1, RARRES2, RBP1, S100A1, SEZ6L2, SFTPD, SLITRK6, SPINK5, ST3GAL1, SULT1B1, TBX1, TNFRSF17, TRIM7, VIPR1, VSIG1, ZNF831 | 58 | 4.68 |
DEG, differentially expressed gene; NMG, non-myasthenia gravis thymoma; TAMG, thymoma-associated myasthenia gravis.
Table S5. GeneAnalytics program mapping of diseases with 10 highest match scores for 169 DEGs in TAMG and NMG.
| Disease | Genes matched to disease type | No. of genes in disease type | Score |
|---|---|---|---|
| Lung cancer | KRT14, KRT17, SFTPC, SFTPD, VEGFD, ATM, NAPSA, NMB, SCGB1A1, DSC3, SFTPA2, BNC1, BTNL9, CAB39L, LRRN3, SFTPB | 16 | 9.81 |
| Pulmonary fibrosis, idiopathic | SFTPA2, SFTPA1, SFTPC, CCR7, SCGB1A1, SFTPB, SFTPD | 7 | 8.80 |
| Breast cancer | ATM, KRT14, KRT17, CYP19A1, TWIST1, VEGFD, AMDHD1, BBOX1, DSC3, IRX1, KCNIP3, LAMP5, MAP2, SCUBE1, TCEAL2 | 15 | 8.33 |
| Polycystic kidney disease | BBOX1, CYP4F3, DPEP1, IGFBP6, PRRX1, ADAMTS15, AMDHD1, CNKSR2, LAMP5, MAP1B, MYL9, NAPSA, TWIST1 | 13 | 7.22 |
| Neuroblastoma | CYP21A2, KRT1, CADPS, MAP1B, MAP2, ADM, CADPS2, SNKSR2, EPB41L4B, KIF1A, MYL9, NNAT, RAB3IL1, TCEAL2 | 14 | 7.03 |
| Respiratory distress syndrome in premature infants | SFTPB, SFTPC, SFTPA1 | 3 | 6.83 |
| Dilated cardiomyopathy | MYH6, CRYAB, DSC2, DTNA, MYBPC2, SYNE1, TWIST1 | 7 | 6.50 |
| Lung cancer susceptibility 3 | SCGB1A1, SFTPD, VEGFD, NAPSA, SFTPB, SFTPC, NPTX2, PLXNB3, SFTPA2, VIPR1 | 10 | 6.48 |
| Colorectal cancer | DPEP1, ATM, CACNA1G, SYNE1, VEGFD, NEUROD1, CAB39L, CD226, LRRN3, MYL9, TFDP3 | 11 | 6.02 |
| Tuberous sclerosis complex | CADPS, CNKSR2, DTNA, PRRX1, CYP4F3, EPB41L4B, LAMP5, NEUROD1, NGEF, OLFM3, SCN2B | 11 | 5.97 |
DEG, differentially expressed gene; NMG, non-myasthenia gravis thymoma; TAMG, thymoma-associated myasthenia gravis.
Table S6. GeneAnalytics program mapping of superpathways with 10 highest match scores for 169 DEGs in TAMG and NMG.
| Superpathways | Genes matched to superpathways | No. of genes in superpathways | Score |
|---|---|---|---|
| Defective CSF2RB causes pulmonary surfactant metabolism dysfunction 5 | SFTPA2, SFTPB, SFTPC, SFTPA1, SFTPD | 5 | 26.70 |
| Surfactant metabolism | SFTPA2, SFTPB, SFTPC, SFTPA1, SFTPD, NAPSA | 6 | 22.03 |
| Diseases of metabolism | SFTPA2, SFTPB, SFTPC, SFTPA1, SFTPD | 5 | 14.75 |
| FOXA1 transcription factor network | SCGB1A1, SFTPA2, SFTPA1, SFTPD | 4 | 11.03 |
| Antiarrhythmic pathway, pharmacodynamics | CACNA1G, CACNA1H, SCN2B, KCNJ5 | 4 | 9.72 |
| Circadian entrainment | KRT17, KRT14, KRT33B, COL4A5, VEGFD, CACNA1G, CACNA1H, RYR1, KCNJ5, KCNQ4 | 10 | 9.70 |
| Keratinization | KRT17, KRT14, KRT1, KRT33B, SPINK5, DSC3, DSC2 | 7 | 8.89 |
| Myometrial relaxation and contraction pathways | ADM, RAMP1, RGS9, IGFBP6, IGFBP4, RYR1, KCNJ5 | 7 | 8.74 |
| Cardiac conduction | DYSF, RYR1, SCN2B, KCNIP3, MYBPC2, MYH6, MYL9 | 7 | 8.28 |
| Developmental biology | KRT17, KRT14, KRT1, CNKSR2, KRT33B, COL4A5, NEUROD1, NGEF, PLXNB3, SPINK5, DSC3, DSC2, CACNA1G, CACNA1H, IL17RD, SCN2B, MYL9, TREM2 | 18 | 8.14 |
DEG, differentially expressed gene; NMG, non-myasthenia gravis thymoma; TAMG, thymoma-associated myasthenia gravis.
Table S7. GeneAnalytics program mapping of gene ontology (GO) molecular function and biological processes with 5 highest match scores for 169 DEGs in TAMG and NMG.
| Variable | Genes matched | No. of genes | Score |
|---|---|---|---|
| A. GO-molecular function | |||
| Lipopolysaccharide binding | ADGRB1, SFTPA2, SFTPA1, SRTPD, TREM2 | 5 | 16.93 |
| Microtubule binding | CRYAB, MAP1B, MAP2, SYBU, CCDC88C, KIF12, KIF19, KIF1A, MYH6, KIFC3, SGIP1 | 11 | 15.87 |
| Monosaccharide binding | SFTPA2, SFTPA1, SFTPD | 3 | 12.74 |
| Low voltage-gated calcium channel activity | CACNA1G, CACNA1H | 2 | 10.89 |
| Signaling receptor activity | KRT1, OGFRL1, RAMP1, TNFRSF25, RTN4RL2, TNFRSF17, TREM2 | 7 | 10.49 |
| B. GO-biological process | |||
| Surfactant homeostasis | SFTPA2, SFTPA1, SFTPD, NAPSA | 4 | 19.15 |
| Muscle contraction | CRYAB, DYSF, CACNA1H, RYR1, MYBPC2, MYH6, MYL9, CHRNG | 8 | 17.62 |
| Respiratory gaseous exchange | SFTPA2, SFTPB, SFTPC, SFTPA1, SFTPD | 5 | 15.93 |
| Cornification | KRT17, KRT14, KRT1, KRT33B, SPINK5, DSC3, DSC2 | 7 | 14.34 |
| Cellular protein metabolic process | PENK, IGFBP6, IGFBP4, SFTPA2, SFTPB, SFTPC, SFTPA1, SFTPD, NAPSA | 9 | 13.35 |
DEG, differentially expressed gene; NMG, non-myasthenia gravis thymoma; TAMG, thymoma-associated myasthenia gravis.
Table S8. GeneAnalytics program mapping of phenotypes with 10 highest match scores for 169 DEGs in TAMG and NMG.
| Phenotypes | Genes matched to phenotypes | Genes | Score |
|---|---|---|---|
| Abnormal surfactant physiology | SFTPB, SFTPC, SFTPA1, SFTPD | 4 | 18.61 |
| Dehydration | KRT1, NEUROD1, MAP1B, SPINK5, SALL3, CHRNG | 6 | 14.98 |
| Impaired glucose tolerance | COL4A5, SLC30A8, CYP19A1, VIPR1, RARRES2, RGPD1, IGFBP4, CADPS2 | 8 | 11.57 |
| Increased or absent threshold for auditory brainstem response | ADGRB1, SLITRK6, MCOLN3, PDZD7, TBX1, IL17RD, KCNQ4 | 7 | 10.3 |
| Unresponsive to tactile stimuli | MAP1B, RYR1, KIF1A | 3 | 10.2 |
| Blistering | KRT14, KRT1, SPINK5 | 3 | 10.2 |
| No spontaneous movement | OLIG2, RYR1, CHRNG | 3 | 10.02 |
| Abnormal cerebellum morphology | NEUROD1, MAP1B, DAB1, CADPS2, IL17RD | 5 | 9.82 |
| Centrally nucleated skeletal muscle fibers | CRYAB, DYSF, SYNE1, RYR1 | 4 | 9.72 |
| Dystrophic muscle | DTNA, DYSF, SYNE1 | 3 | 9.52 |
DEG, differentially expressed gene; NMG, non-myasthenia gravis thymoma; TAMG, thymoma-associated myasthenia gravis.
Table S9. GeneAnalytics program mapping of compounds with 10 highest match scores for 169 DEGs in TAMG and NMG.
| Compounds | Genes matched to compounds | Genes | Score |
|---|---|---|---|
| Calcium | KRT14, KRT1, NEUROD1, NMB, CRYAB, PITPNM3, MAP1B, MAP2, NPTX2, CYP21A2, DAB1, VIPR1, B3GAT1, DSC3, DSC2, PTPN3, DUSP15, DYSF, RARRES2, PENK, IGFBP6, IGFBP4, CACNA1G, CADPS2, CACNA1H, CADPS, TNFRSF17, RYR1, S100A1, CCR7, CDH26, SCGB1A1, KCNIP3, KCNJ5, KCNQ4, SFTPA2, MYH6, SFTPB, SFTPC, SFTPA1, SFTPD, MYL9 | 42 | 19.79 |
| Progesterone | KRT14, KRT1, ADM, CRYAB, MAP2, CYP21A2, CYP19A1, VEGFD, B3GAT1, RBP1, PENK, IGFBP6, IGFBP4, S100A1, SCGB1A1, SFTPB | 16 | 15.17 |
| Estrogen | KRT14, KRT1, ADM, TWIST1, MAP2, CYP21A2, CYP19A1, VEGFD, ATM, B3GAT1, PTPN3, SULT1B1, RBP1, PENK, IGFBP6, IGFBP4, CACNA1G, S100A1, SCGB1A1, KCNJ5, MYH6 | 21 | 14.51 |
| Infasurf | SFTPB, SFTPC | 2 | 12.88 |
| Calcium citrate | CACNA1G, CADPS2, CACNA1H, CADPS, RYR1 | 5 | 12.78 |
| Calcium phosphate | CACNA1G, CADPS2, CACNA1H, CADPS, RYR1 | 5 | 12.34 |
| Retinoic acid | KRT17, KRT14, KRT1, NEUROD1, ADM, MAP2, OLIG2, VIPR1, HAS2, RARRES2, RBP1, PENK, IGFBP6, IGFBP4, CACNA1G, S100A1, MYH6, SFTPB, SFTPC | 19 | 11.82 |
| Paraffin | KRT17, KRT14, KRT1, ADM, CRYAB, MAP1B, MAP2, ATM, RBP1, S100A1, SFTPB | 11 | 11.76 |
| POPG | SFTPB, SFTPC | 2 | 11.72 |
| ML218 | CACNA1G, CACNA1H | 2 | 11.72 |
DEG, differentially expressed gene; NMG, non-myasthenia gravis thymoma; TAMG, thymoma-associated myasthenia gravis.
DEGs related to T cells
Since thymus is the place where T cells develop, we further selected DEGs in T cells between thymoma tissues of TAMG and NMG by LifeMapDiscovery Cells and Tissues Database. Among 169 DEGs, there were 6 genes mainly expressing in T helper cells (ANKRD55, BTLA, CCR7, LRRN3, TNFRSF25, VSIG1), 3 in T cytotoxic cells (CCR7, LRRN3, TNFRSF25) and 4 in double positive (DP) thymocytes (ATM, CCR7, SFTPB, TNFRSF25) with all 8 genes up-regulated. The genes were exhibited with their log2FC, log2CPM, P value and FDR in Table 1. Further screened by VarElect, 6 genes (ATM, SFTPB, ANKRD55, BTLA, CCR7, TNFRSF25) were directly related while 2 genes (LRRN3, VSIG1) were indirectly related to the phenotype of autoimmune diseases.
Table 1. Eight T-cell associated DEGs screened by LifeMapDiscovery Cells and Tissues Database.
| Gene | Log2FC | Log2CPM | P value | FDR |
|---|---|---|---|---|
| ATM | 1.09 | 6.07 | 0.000167 | 0.0208 |
| SFTPB | 10.4 | 5.17 | 3.64E−07 | 0.000606 |
| ANKRD55 | 2.39 | 2.40 | 0.000344 | 0.0319 |
| BTLA | 1.49 | 2.30 | 0.000661 | 0.0450 |
| CCR7 | 1.61 | 5.83 | 0.000334 | 0.0315 |
| TNFRSF25 | 1.76 | 4.65 | 1.57E−07 | 0.000286 |
| LRRN3 | 2.16 | 4.61 | 1.75E−05 | 0.00531 |
| VSIG1 | 2.32 | 2.36 | 0.000351 | 0.0320 |
DEG, differentially expressed gene; FC, fold change; CPM, counts per million; FDR, false discovery rate.
Validation by RT-qPCR
Patients and samples
Demographic and clinical characteristics were summarized in Table 2. Disease severity was evaluated using the MGFA Clinical Classification (16). There was no statistical difference in sex, age, WHO histologic classification of thymoma and MGFA classification between TAMG and NMG group.
Table 2. Demographic and clinical characteristics of the subjects included in the study.
| Classification/group | NMG | TAMG |
|---|---|---|
| Gender | ||
| Female | 5 | 2 |
| Male | 4 | 7 |
| Age | 18–62 (mean 50.2, median 55) | 30–62 (mean 46, median 42) |
| WHO histologic classification | ||
| A | 1 | 1 |
| AB | 2 | – |
| B1 | 2 | – |
| B2 | 3 | 6 |
| B3 | 1 | 2 |
| MGFA classification | ||
| 1 | – | 1 |
| 2a | – | 2 |
| 2b | – | 3 |
| 3a | – | 3 |
| AChR antibody | ||
| Positive | – | 9 |
| Negative | – | 0 |
TAMG, thymoma-associated MG; NMG, non-MG thymoma; WHO, World Health Organization; MGFA, Myasthenia Gravis Foundation of America; AChR, acetylcholine receptor; MG, myasthenia gravis.
RT-qPCR
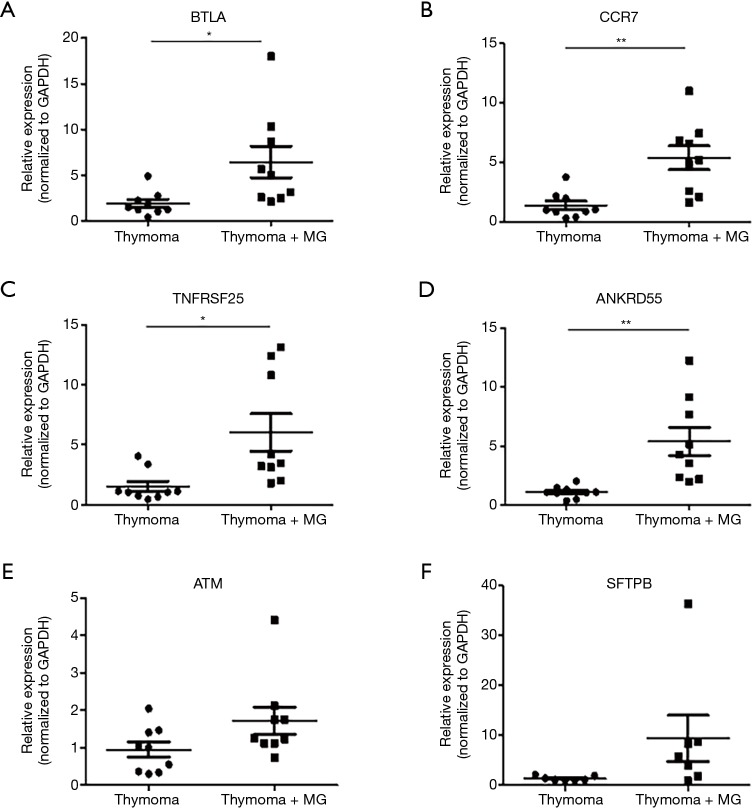
The relative expression level of mRNA in 6 genes (ATM, SFTPB, ANKRD55, BTLA, CCR7, TNFRSF25) that are direct related to autoimmune disease were compared between the thymoma tissues of the TAMG and NMG group. Compared with NMG, significantly higher expression of BTLA (P=0.022), CCR7 (P=0.0018), TNFRSF25 (P=0.013) and ANKRD55 (P=0.0026) were observed in the TAMG group (Figure 1A,B,C,D). However, we did not observe a difference in expression of ATM (P=0.082) and SFTPB (P=0.11) between the two groups (Figure 1E,F). These results may suggest a specific association between the overexpression of BTLA, TNFRSF25, CCR7 or ANKRD55 and the mechanism of TAMG. Since BTLA negatively regulated the immune response, the elevated expression of BTLA could not be explained in the TAMG group. However, soluble BTLA (sBTLA), the isoform of BTLA from alternative splicing, could exhibit a counteracted function from BTLA (17). We further performed RT-qPCR of sBTLA using its specific primers and it was revealed that there was a significant difference (P=0.027) between the two groups (Figure 2).
Figure 1.
Validation of the differences of six directly immune-related DEGs using RT-qPCR. (A-D) Compared with NMG, significantly higher expression of BTLA (P=0.022), CCR7 (P=0.0018), TNFRSF25 (P=0.013) and ANKRD55 (P=0.0026) was observed in TAMG. (E,F) The difference in expression of ATM (P=0.082) and SFTPB (P=0.11) was not significant between the two groups. *, P<0.05; **, P<0.01. DEG, differentially expressed gene; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; NMG, non-myasthenia gravis thymoma; TAMG, thymoma-associated myasthenia gravis.
Figure 2.
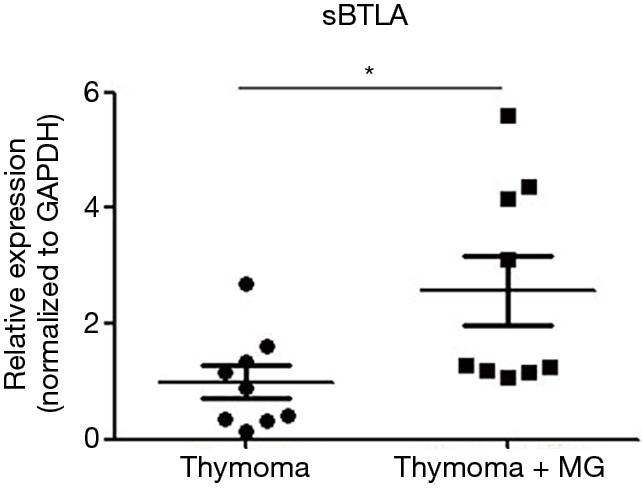
Validation of the difference of soluble BTLA using RT-qPCR. A significant difference (P=0.027) was observed between the two groups. *, P<0.05. RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Discussion
In the present study, with high throughput data from 11 TAMG and 10 NMG from TCGA database, we tried to find the possible molecular changes in the pathogenesis of TAMG. With the edgeR analysis tool, we found that among 24,991 transcripts, 169 genes showed a significant difference between the TAMG and NMG group. Further screening using LifeMapDiscovery Cells and Tissues Database and VarElect, overexpression of six genes (ATM, SFTPB, ANKRD55, BTLA, CCR7, TNFRSF25) directly related to autoimmune diseases expressed on T cells was found and four (ANKRD55, BTLA, CCR7, TNFRSF25) of them were validated in thymoma tissues in our center by RT-qPCR.
C-C chemokine receptor-7 (CCR7), a member of the C-C chemokine receptor subfamily, can combine with its ligand (CCL19 or CCL21). Mainly expressed on naïve and central memory CD4+ T cells, naïve B cells and mature dendritic cells (DCs), this receptor activates them to travel to their target tissues selectively and initiates the autoimmune response (18,19). It also mediates the settling of T-cell precursors in the thymus to initiate thymopoiesis (20). CCR7-deficient mice and mice lacking the expression of CCL19 and CCL21 exhibited defective migration of DCs and lymphocytes toward the T-cell zones (21). When under antigenic stimulation, some activated T cells lost their CCR7 expression and upregulated CXCR5, the receptor for CXCL13 expressed on B cells. CXCR5+ T cells then migrated towards the B cell-rich follicles to interact with B cells (22). Moschovakis et al. demonstrated that CCR7-deficient mice showed complete resistance to collagen-induced arthritis, and the antibody response to collagen II was severely impaired (23). Belikan et al. demonstrated that the induction of experimental autoimmune encephalomyelitis (EAE) was halted with the specific constitutive deletion of CCR7 on CD4+ T cells (24). When compared with normal subjects, the frequency of CD4+CCR7+ and CD8+CCR7+ T cells increased significantly in patients with primary Sjögren’s syndrome. The expression level of CCR7 on CD4+ T cells was associated with disease activity (25). In MG thymus, Li Qi et al. showed overexpression of CCR7 may block the differentiation of CD4+CD8+ DP thymocytes from the CD4-CD8- double negative (DN) stage, leading to the production of AChR autoreactive T cells (26). Our results also showed a significant higher expression of CCR7 in TAMG (P=0.0018), which further validated the positive correlation between CCR7 and TAMG.
B and T lymphocyte attenuator (BTLA), the ligand of herpesvirus entry mediator (HVEM), is expressed on DCs, natural killer (NK) cells, T and B lymphocytes (27). It was detected on double positive (DP) thymocytes, CD4+ or CD8+ single positive but not DN thymocytes (28). Belonging to the immunoglobulin superfamily and with similarities to cytotoxic T lymphocyte antigen 4 (CTLA-4) and programmed death 1 (PD-1), BTLA is another coinhibitory receptor that negatively regulate the immune response. It was demonstrated that BTLA-deficient mice have enhanced specific antibody reactions and improved sensitivity to EAE, with earlier disease onset, prolonged disease duration and increased clinical score (29). DCs require the functions of BTLA to actively adjust tolerizing T cell responses in CD5 dependent mechanisms (30). BTLA could also inhibit γδ T cell-dependent dermatitis through negatively regulating the production of IL-17 and tumor necrosis factor (TNF) (31). In multiple sclerosis, CD19+BTLA+ cells were significantly reduced compared with health controls while the disease remission was related to the increase in CD19+BTLA+ cells, suggesting a potentially pathogenic role (32). In our study, we first found the relationship between the higher expression of BTLA and TAMG. It could be explained by the possibility of reactive responses following antigen specific activation of T cells. Since we did not see such responses in other autoimmune diseases in literature, we further quantified and confirmed the elevation of sBTLA by RT-qPCR. In collagen-induced arthritis, soluble PD-1 aggravates the disease progression through Th1 and Th17 pathway (33). Like PD-1, the production of soluble form of BTLA may also interfere the normal regulatory function of HVEM/BTLA pathway and lead to the phenotype we observed.
Tumor necrosis factor receptor superfamily member 25 (TNFRSF25), also called death receptor 3 (DR3), is expressed on T cells and combines with the ligand of TNF-like protein 1A (TL1A) (34). In addition to mediating necroptotic cell death, this receptor has been shown to stimulate NF-κB activity through promoting the activation of phosphoinositide 3-kinase (PI3K) and Akt (protein kinase B, PKB), leading to the increased production of pro-inflammatory cytokines, enhanced differentiation and clonal expansion of T cells (35,36). It was demonstrated that the contact of TNFRSF25 and TL1A is required for the immunopathological process in EAE with IL-9 producing Th9 cells (37,38). This may explain the effect of elevated expression of TNFRSF25 in TAMG samples.
Although the precise function of ankyrin repeat domain containing protein 55 (ANKRD55) is still unknown, it functions almost exclusively to mediate protein-protein interactions (39). Single-nucleotide polymorphism (SNP) of this gene was reported to be related to several autoimmune diseases, such as rheumatoid arthritis (40) and multiple sclerosis (41). ANKRD55 is also overexpressed in EAE with neuroinflammation (42). These demonstrations in autoimmune diseases were in accordance with our results. However, the detailed mechanisms underlying TAMG need to be further clarified.
ATM, the ataxia telangiectasia-mutated gene, is required for DNA damage response and genome stability (43). The mutations in ATM have been reported in patients with brain and breast tumor (44). Surfactant Protein B (SFTPB) could influence lung function and innate immunity (45). Altered SFTPB is associated with several lung diseases including acute respiratory distress syndrome (46) and chronic obstructive pulmonary disease (47). In this study two genes mentioned above did not show significant difference between the two groups, and this may partly be due to the lower number of thymoma samples.
Combined with the findings above, the overexpression of 4 T-cell associated genes could be a characteristic of MG-associated thymoma with potential pathogenic importance. The induction and maintenance of MG may be related with the overexpression of some functional genes from T cells and other immune cells in the thymoma microenvironment. In other words, whether or not thymoma develops MG may depend on the heterogeneity of the thymoma microenvironment. As for how this heterogeneity was formed, further research should be conducted.
Of course, there are some limitations in this study. First, the conclusion may be limited by the small sample size. Second, the heterogeneity of individual patients including disease severity and WHO thymoma classification would also influence the reliability of the study. Third, tissue validation could not limit its results to T cells like bioinformatics analysis. Although aberrantly expressed genes were validated using RT-qPCR, they may originate in various cells from thymic tissues, so the results should be interpreted with caution. This was the first study to use TCGA database for investigating the possible molecular changes and pathogenesis in TAMG and in the study we find the correlation of TAMG with higher expression of CCR7, BTLA, TNFRSF25 as well as ANKRD55. The expression and SNPs of these genes may be promising biomarkers in TAMG and need to be further investigated.
Conclusions
In this study, we first identified 6 genes that were directly related to autoimmune diseases through GeneAnalytics from 169 DEGs calculated in TCGA. Then, overexpression of sBTLA, CCR7, TNFRSF25 and ANKRD55 was validated in thymoma tissues from TAMG in our center, which could partly explain the underlying pathogenesis of TAMG.
Acknowledgments
Funding: This work was supported by The National Key Research and Development Program of China (No. 2016YFC0901504), the National Natural Science Foundation of China (No. 81870988) and Shanghai Municipal Commission of Health and Family Planning (No. 201540044).
Ethical Statement: The study was approved by Fudan University Institutional Review Board and written informed consents were granted.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Gilhus NE, Verschuuren JJ. Myasthenia gravis: subgroup classification and therapeutic strategies. Lancet Neurol 2015;14:1023-36. 10.1016/S1474-4422(15)00145-3 [DOI] [PubMed] [Google Scholar]
- 2.Marx A, Willcox N, Leite MI, et al. Thymoma and paraneoplastic myasthenia gravis. Autoimmunity 2010;43:413-27. 10.3109/08916930903555935 [DOI] [PubMed] [Google Scholar]
- 3.Nakajima J, Murakawa T, Fukami T, et al. Postthymectomy myasthenia gravis: relationship with thymoma and antiacetylcholine receptor antibody. Ann Thorac Surg 2008;86:941-5. 10.1016/j.athoracsur.2008.04.070 [DOI] [PubMed] [Google Scholar]
- 4.Thomas CR, Wright CD, Loehrer PJ. Thymoma: state of the art. J Clin Oncol 1999;17:2280-9. 10.1200/JCO.1999.17.7.2280 [DOI] [PubMed] [Google Scholar]
- 5.Okumura M, Fujii Y, Shiono H, et al. Immunological function of thymoma and pathogenesis of paraneoplastic myasthenia gravis. Gen Thorac Cardiovasc Surg 2008;56:143-50. 10.1007/s11748-007-0185-8 [DOI] [PubMed] [Google Scholar]
- 6.Marx A, Porubsky S, Belharazem D, et al. Thymoma related myasthenia gravis in humans and potential animal models. Exp Neurol 2015;270:55-65. 10.1016/j.expneurol.2015.02.010 [DOI] [PubMed] [Google Scholar]
- 7.Masuda M, Matsumoto M, Tanaka S, et al. Clinical implication of peripheral CD4+CD25+ regulatory T cells and Th17 cells in myasthenia gravis patients. J Neuroimmunol 2010;225:123-31. 10.1016/j.jneuroim.2010.03.016 [DOI] [PubMed] [Google Scholar]
- 8.Nishi T, Yokoyama S, Takamori S, et al. Thymoma in Patient with Myasthenia Gravis Has Significantly Fewer Forkhead Box P3 Positive Lymphocytes than that without One. Kurume Med J 2015;61:65-71. 10.2739/kurumemedj.MS65002 [DOI] [PubMed] [Google Scholar]
- 9.Song Y, Zhou L, Miao F, et al. Increased frequency of thymic T follicular helper cells in myasthenia gravis patients with thymoma. J Thorac Dis 2016;8:314-22. 10.21037/jtd.2016.03.03 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaussabel D, Pascual V, Banchereau J. Assessing the human immune system through blood transcriptomics. BMC Biol 2010;8:84. 10.1186/1741-7007-8-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cufi P, Soussan P, Truffault F, et al. Thymoma-associated myasthenia gravis: On the search for a pathogen signature. J Autoimmun 2014;52:29-35. 10.1016/j.jaut.2013.12.018 [DOI] [PubMed] [Google Scholar]
- 12.Ben-Ari Fuchs S, Lieder I, Stelzer G, et al. GeneAnalytics: An Integrative Gene Set Analysis Tool for Next Generation Sequencing, RNAseq and Microarray Data. Omics 2016;20:139-51. 10.1089/omi.2015.0168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139-40. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stelzer G, Plaschkes I, Oz-Levi D, et al. VarElect: the phenotype-based variation prioritizer of the GeneCards Suite. BMC Genomics 2016;17 Suppl 2:444. 10.1186/s12864-016-2722-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaretzki A, 3rd, Barohn RJ, Ernstoff RM, et al. Myasthenia gravis: recommendations for clinical research standards. Task Force of the Medical Scientific Advisory Board of the Myasthenia Gravis Foundation of America. Neurology 2000;55:16-23. 10.1212/WNL.55.1.16 [DOI] [PubMed] [Google Scholar]
- 17.Han L, Wang W, Lu J, et al. AAV-sBTLA facilitates HSP70 vaccine-triggered prophylactic antitumor immunity against a murine melanoma pulmonary metastasis model in vivo. Cancer Lett 2014;354:398-406. 10.1016/j.canlet.2014.08.006 [DOI] [PubMed] [Google Scholar]
- 18.Ramirez PW, Famiglietti M, Sowrirajan B, et al. Downmodulation of CCR7 by HIV-1 Vpu results in impaired migration and chemotactic signaling within CD4(+) T cells. Cell Rep 2014;7:2019-30. 10.1016/j.celrep.2014.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xin H, Zhu J, Miao H, et al. Adenovirus-Mediated CCR7 and BTLA Overexpression Enhances Immune Tolerance and Migration in Immature Dendritic Cells 2017;2017:3519745. [DOI] [PMC free article] [PubMed]
- 20.Reinhardt A, Ravens S, Fleige H, et al. CCR7-mediated migration in the thymus controls gammadelta T-cell development. Eur J Immunol 2014;44:1320-9. 10.1002/eji.201344330 [DOI] [PubMed] [Google Scholar]
- 21.Gunn MD, Kyuwa S, Tam C, et al. Mice lacking expression of secondary lymphoid organ chemokine have defects in lymphocyte homing and dendritic cell localization. J Exp Med 1999;189:451-60. 10.1084/jem.189.3.451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vaccari M, Franchini G. T Cell Subsets in the Germinal Center: Lessons from the Macaque Model. Front Immunol 2018;9:348. 10.3389/fimmu.2018.00348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moschovakis GL, Bubke A, Friedrichsen M, et al. The chemokine receptor CCR7 is a promising target for rheumatoid arthritis therapy. Cell Mol Immunol 2018. [Epub ahead of print]. 10.1038/s41423-018-0056-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belikan P, Buhler U, Wolf C, et al. CCR7 on CD4(+) T Cells Plays a Crucial Role in the Induction of Experimental Autoimmune Encephalomyelitis. J Immunol 2018;200:2554-62. 10.4049/jimmunol.1701419 [DOI] [PubMed] [Google Scholar]
- 25.Wu C, Yang P, Liu H, et al. Increased frequency of CCR7(+)CD4(+) T cells from patients with primary Sjogren's syndrome: An indicator of disease activity rather than of damage severity. Cytokine 2018;110:9-17. 10.1016/j.cyto.2018.04.015 [DOI] [PubMed] [Google Scholar]
- 26.Li Q, Liu P, Xuan X, et al. CCR9 AND CCR7 are overexpressed in CD4(-) CD8(-) thymocytes of myasthenia gravis patients. Muscle Nerve 2017;55:84-90. 10.1002/mus.24999 [DOI] [PubMed] [Google Scholar]
- 27.Sedy JR, Gavrieli M, Potter KG, et al. B and T lymphocyte attenuator regulates T cell activation through interaction with herpesvirus entry mediator. Nat Immunol 2005;6:90-8. 10.1038/ni1144 [DOI] [PubMed] [Google Scholar]
- 28.Han P, Goularte OD, Rufner K, et al. An inhibitory Ig superfamily protein expressed by lymphocytes and APCs is also an early marker of thymocyte positive selection. J Immunol 2004;172:5931-9. 10.4049/jimmunol.172.10.5931 [DOI] [PubMed] [Google Scholar]
- 29.Watanabe N, Gavrieli M, Sedy JR, et al. BTLA is a lymphocyte inhibitory receptor with similarities to CTLA-4 and PD-1. Nat Immunol 2003;4:670-9. 10.1038/ni944 [DOI] [PubMed] [Google Scholar]
- 30.Jones A, Bourque J, Kuehm L, et al. Immunomodulatory Functions of BTLA and HVEM Govern Induction of Extrathymic Regulatory T Cells and Tolerance by Dendritic Cells. Immunity 2016;45:1066-77. 10.1016/j.immuni.2016.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bekiaris V, Sedy JR, Macauley MG, et al. The inhibitory receptor BTLA controls gammadelta T cell homeostasis and inflammatory responses. Immunity 2013;39:1082-94. 10.1016/j.immuni.2013.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piancone F, Saresella M, Marventano I, et al. B Lymphocytes in Multiple Sclerosis: Bregs and BTLA/CD272 Expressing-CD19+ Lymphocytes Modulate Disease Severity. Sci Rep 2016;6:29699. 10.1038/srep29699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu C, Jiang J, Gao L, et al. Soluble PD-1 aggravates progression of collagen-induced arthritis through Th1 and Th17 pathways. Arthritis Res Ther 2015;17:340. 10.1186/s13075-015-0859-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meylan F, Richard AC, Siegel RM. TL1A and DR3, a TNF family ligand-receptor pair that promotes lymphocyte costimulation, mucosal hyperplasia, and autoimmune inflammation. Immunol Rev 2011;244:188-96. 10.1111/j.1600-065X.2011.01068.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bittner S, Knoll G, Ehrenschwender M. Death receptor 3 mediates necroptotic cell death. Cell Mol Life Sci 2017;74:543-54. 10.1007/s00018-016-2355-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.So T, Croft M. Regulation of PI-3-Kinase and Akt Signaling in T Lymphocytes and Other Cells by TNFR Family Molecules. Front Immunol 2013;4:139. 10.3389/fimmu.2013.00139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meylan F, Davidson TS, Kahle E, et al. The TNF-family receptor DR3 is essential for diverse T cell-mediated inflammatory diseases. Immunity 2008;29:79-89. 10.1016/j.immuni.2008.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richard AC, Tan C, Hawley ET, et al. Correction: The TNF-Family Ligand TL1A and Its Receptor DR3 Promote T Cell-Mediated Allergic Immunopathology by Enhancing Differentiation and Pathogenicity of IL-9-Producing T Cells. J Immunol 2015;195:5839-40. 10.4049/jimmunol.1502026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li J, Mahajan A, Tsai MD. Ankyrin repeat: a unique motif mediating protein-protein interactions. Biochemistry 2006;45:15168-78. 10.1021/bi062188q [DOI] [PubMed] [Google Scholar]
- 40.Stahl EA, Raychaudhuri S, Remmers EF, et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet 2010;42:508-14. 10.1038/ng.582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alloza I, Otaegui D, de Lapuente AL, et al. ANKRD55 and DHCR7 are novel multiple sclerosis risk loci. Genes Immun 2012;13:253-7. 10.1038/gene.2011.81 [DOI] [PubMed] [Google Scholar]
- 42.Lopez de Lapuente A, Feliu A, Ugidos N, et al. Novel Insights into the Multiple Sclerosis Risk Gene ANKRD55. J Immunol 2016;196:4553-65. 10.4049/jimmunol.1501205 [DOI] [PubMed] [Google Scholar]
- 43.Pandita TK. 14th International Workshop on Ataxia-Telangiectasia ATW2012. DNA Repair (Amst) 2012;11:853-6. 10.1016/j.dnarep.2012.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Estiar MA, Mehdipour P. ATM in breast and brain tumors: a comprehensive review. Cancer Biol Med 2018;15:210-27. 10.20892/j.issn.2095-3941.2018.0022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin Z, Thorenoor N, Wu R, et al. Genetic Association of Pulmonary Surfactant Protein Genes, SFTPA1, SFTPA2, SFTPB, SFTPC, and SFTPD With Cystic Fibrosis. Front Immunol 2018;9:2256. 10.3389/fimmu.2018.02256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin Z, Pearson C, Chinchilli V, et al. Polymorphisms of human SP-A, SP-B, and SP-D genes: association of SP-B Thr131Ile with ARDS. Clin Genet 2000;58:181-91. 10.1034/j.1399-0004.2000.580305.x [DOI] [PubMed] [Google Scholar]
- 47.Guo X, Lin HM, Lin Z, et al. Surfactant protein gene A, B, and D marker alleles in chronic obstructive pulmonary disease of a Mexican population. Eur Respir J 2001;18:482-90. 10.1183/09031936.01.00043401 [DOI] [PubMed] [Google Scholar]