Abstract
Protein kinases are key players in signal transduction pathways where they are crafted into two functional states. In response to growth factor binding stimulus, epidermal growth factor receptor (EGFR), which is physiologically populated in an autoinhibited inactive state, becomes active. Here, we outline a simple allostery scheme to clarify how an extracellular (ligand-dependent) binding event activates the intracellular EGFR kinase domain via (dimer-dependent) asymmetric dimerization, as well as how pathologically overexpressed EGFR or constitutively active mutants, leads to oncogenic pathway activation. Our underlying allosteric activation mechanism derives from a collection of inactive versus active EGFR structural, biochemical (negatively cooperative ligand binding), and biophysical (weak coupling between extracellular and intracellular kinase dimerization) data. The emerging structural insight reveals that ligand-dependent physiological activation is an outside-in allosteric activation with strong structural coupling across the membrane. In contrast, ligand-independent pathological activation is a weak inside-out activation mediated by intracellular kinase dimerization, which is structurally accommodated by additional extracellular dimers.
Main Text
Members of the epidermal growth factor receptor family (EGFR/ErbB1/HER1, ErbB2/HER2, ErbB3/HER3, and ErbB4/HER4) constitute 1 of the 20 subfamilies, with a total of 58 currently known human receptor tyrosine kinases (1). These receptors play a major role in cell growth, differentiation, and migration (2) and are often dysregulated in human cancers (3). In normal cells, the binding of an epidermal growth factor (EGF) family member to these receptors activates their cytoplasmic kinase domains (KDs), resulting in phosphorylation of tyrosine residues in their C-terminal tails (4) and subsequent recruitment of downstream effectors for initiating various cellular functions (5).
During the last decade, much effort was invested in the structural mechanism of EGFR signaling, seeking to understand how an extracellular binding event propagates via a single transmembrane helix (TM) to accomplish intracellular kinase activation (6, 7, 8, 9, 10). Although the apparent allosteric cause and effect are executed across the plasma membrane at two distally separated sites, the protein-protein interactions involved in the EGFR signal transduction pathway also fall into the general description of allostery (11). The first EGFR signaling event involves the formation of a symmetric homodimer of the extracellular module (EC) upon EGF binding. Because the EGF (ligand) does not bind at the EC dimer interface, this binding event already describes an allosteric scenario that favors EC dimerization. In addition, ligand-bound EC stabilizes an extended conformation, shifting the population of monomeric EC away from a tethered conformation with a large conformational change (12, 13). Finally, the ensuing formation of asymmetric kinase dimer also fits well the description of allosteric activation in which the dimer interface is distal from the kinase active site (14). Although the two allosteric dimerization events are clearly elucidated by structural evidence, the extent of the allosteric coupling between them through a single-pass TM has been described differently (7, 8, 9). Hence, a complete understanding of allostery in EGFR signal transduction may clarify the discrepancy of the coupling as well as the structural mechanism that explains why different ligands (seven members in the EFG family) lead to distinct cellular signaling outcomes, such as promoting cell proliferation versus triggering cell differentiation (15).
In this perspective, we outline a simple allosteric scheme to delineate EGFR signal transduction from available biophysical data (7, 8, 9, 10, 16, 17). The scheme of “how allostery works” is based on population shift among local minima in the free energy landscape, simplified for just two states: the active versus inactive state (11). A protein, either in solution or imbedded in the membrane, populates multiple conformational states. After stimulus, the most populated state typically shifts from an inactive resting state to an active functional state. The population time of each state is determined by its conformational stability denoted in the free energy landscape as a local minimum (Fig. 1). In principle, the barriers between minima are surmountable under physiological conditions for all biomolecules. Thus, the population time of each individual state can be determined by their relative free energy differences among local minima.
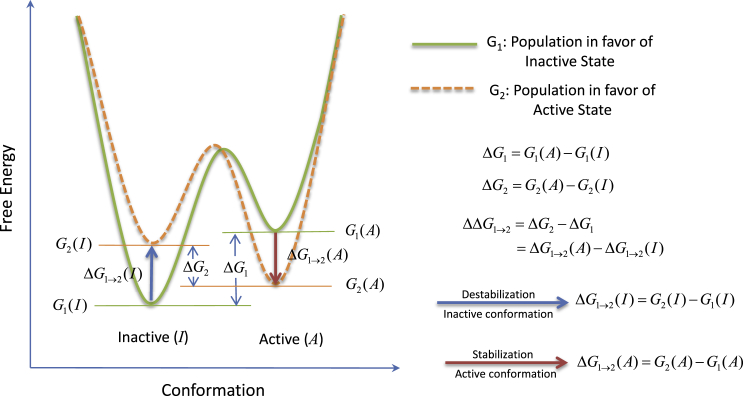
Figure 1.
A simple scheme of allostery depicted as a population shift in the free energy landscape. The allosteric system is represented in conformational space by two structural states: inactive (I) and active (A). G1 stands for the population of a resting system, which favors the inactive state, and the G2 represents the population after stimulation by an allosteric event. This population favors the active state. ΔG gives the population difference in terms of free energy difference between the active and the inactive states. In the figure, we can clearly see the population shift, ΔΔG1→2 = ΔG2 − ΔG1, due to allosteric activation. The population shift can be decomposed into two additive contributions: the destabilization of inactive conformation, ΔG1→2(I), plus the stabilization of active conformation, ΔG1→2(A). The detailed correspondence of the free energy population shift to the thermodynamic view or the structural view of allostery can be found in recent articles (11, 39). To see this figure in color, go online.
As depicted in Fig. 1, allostery is an event of population shift stabilizing the active conformation and/or destabilizing the inactive conformation. Allostery can be described by several key concepts. First, for a representative protein conformation of each local minimum, there are a set of inherent structural stabilization factors responsible for its overall stability. Second, to permit the execution of an allosteric event by shifting the population from one state to the other, these conformational stabilization factors are (to varied extents) structurally coupled. Third, all factors contribute additively to the overall conformational stability. Last, nature crafted a delicate balance between the active and inactive states to execute an allosteric switch through a population shift in favor of the functional state.
Guided by the allostery scheme above, we discuss EGFR transmembrane signaling, focusing on the structural details of exactly how a ligand-dependent extracellular dimerization event physiologically leads to a dimer-dependent allosteric activation of the intracellular tyrosine KD. Under the same structural model description, we next explain how an overexpressed EGFR is pathologically capable of being activated in a ligand-independent way. Thus, based on available experimental data, we first postulate how much of EGFR overexpression level has pathological activity equivalent to that of ligand-stimulated EGFR expression level. Second, we list, bottom-up, the characteristic structural features responsible for the KDs’ activation and their interactions in autoinhibition. We then merge these with the structural model of Shaw and co-workers (7) from molecular dynamics (MD) simulations. Third, we illustrate, top-down, allostery starting from ligand binding to EC in dimerization. Fourth, we identify those key structural species in individual extracellular, transmembrane, and intracellular dimers that participate in the allosteric structural coupling. As implied by experimental data, the bidirectional coupling is not equal. Physiological EC dimerization-dependent signaling shows a strong outside-in allosteric coupling (10), whereas EGFR pathological (mutants) signaling is likely an asymmetric kinase dimerization-dependent weak inside-out coupling (10). Fifth, we postulate that overexpressed EGFR activation follows the same mechanism as activation by cholesterol depletion (18, 19) because of the higher nonmonomer population proximity and the stabilizing coupling between two distinct EC dimers and the intracellular asymmetric kinase dimer. Lastly, we address the observed negatively cooperative ligand binding in EC dimerization in which the first ligand-binding event alters and restricts the conformation of the second binding site (20, 21). Experimental data point to two independent linkages of negative allosteric structural coupling. One refers to a direct coupling between the two ligand-binding sites (20); the other refers to coupling mediated by the tethering arm (domain IV) cascading down to the TM and into the intracellular juxtamembrane segment (JM) (16, 17). The more the negative ligand binding contributes via the latter coupling, the stronger the coupling is between the extracellular EC dimerization and intracellular kinase dimerization.
A mechanistic understanding of allosteric EGFR activation may provide in-depth insight into how ligand-dependent EC dimerization fine tunes EGFR activation: by altering the change in binding affinity in the EC dimer or by adjusting a stronger/weaker allosteric coupling of the asymmetric kinase dimerization. It may also provide clues for pharmaceutical discovery: rather than aiming at the catalytic activity of the oncogenic KD, we suggest to target the protein-protein interface of the identified dimer species that largely account for the EGFR kinase activation in pathological contexts (22).
The structural basis of allosteric activation of receptor tyrosine kinase EGFR
Protein kinases, a large gene superfamily comprising 518 members in the human genome (23), play a dominant role in many cellular signaling pathways. Despite their diversified functionalities, protein kinases have adopted a strikingly similar active conformation in the catalytic domain (24, 25), catalyzing the transfer of the γ-phosphate of ATP to the substrates’ hydroxyl group of Ser, Thr, or Tyr residues. EGFR, a receptor tyrosine kinase, has a sequence layout of an N-terminal EC, a single TM, cytoplasmic JM, intracellular catalytic KD, and a regulatory C-terminus (CT). Cellular signal transduction via EGFR activates a plethora of cellular processes initiated by ligand binding to the EC that leads to intracellular kinase activation and subsequent phosphorylation on the CT.
Activation criteria of EGFR signaling transduction pathway
Cellular network nodes always switch between two states (active and inactive), permitting the network to execute its regulatory function. A cellular node is not represented by a single biomolecule but an ensemble of molecules in the cell. Therefore, the execution of a biological function by a node is determined by the number of active biomolecules, with a threshold of the absolute number of the biological entity existing in the active conformation in a cell. Because EGFR resides on the plasma membrane, the activation criterion of EGFR signaling pathway is best described by the surface density of EGFR with its KD in active conformation. In normal cells, the effective EGFR concentration in terms of surface density is ∼150 molecules per μm−2 (7). In general, the activity of EGFR can be determined by the phosphorylation level of Tyr1068 (pY1068) at its C-terminal tail. According to plots of pY1068 level versus EGFR expression level given by Kuriyan and co-workers (8), a pY1068 level of EGF-bound EGFR at a surface density of 150 molecules per μm−2 matches the ligand-free EGFR at a surface density of 800 molecules per μm−2. This fact indicates that the ligand-free activation threshold for EGFR expression level is ∼800 molecules per μm−2. Together with the definition of activation criteria, the data suggest that the pathological activity of ligand-free EGFR with a fivefold overexpression level will be equivalent to the physiological activity of EGFR stimulated by EGF under a normal expression level.
The key structural features involved in the structural coupling of EGFR activation
Physiological EGFR signaling involves two separate but structurally coupled allosteric activation events across the membrane: an extracellular ligand-dependent EC dimerization (Fig. 2) that leads to an intracellular dimer-dependent kinase activation (Fig. 3). The ligand-binding event stabilizes the extended conformation of the EC. The population shift in favor of the extended conformation from the tethered conformation populates a specific, EC homodimerization-favored state. Then, via domain IV dimerization, the EC homodimer facilitates an alternative transmembrane helical association close to the N-terminal helix, impeding the autoinhibiting C-terminal helical interaction in the preformed ligand-free EC dimer. In turn, the separation of the C-terminal helical interaction allosterically disrupts the interaction of the N-terminal juxtamembrane segment (JM-A) with membrane lipids. The disruption facilitates an antiparallel JM-A helical dimerization, which supports the stabilization of the asymmetric kinase dimerization by the C-terminal juxtamembrane segment (JM-B) of the receiver kinase through binding to the C-lobe of the activator kinase. The key step in physiological EGFR signaling is the release of JM from the membrane (26, 27) that allows the JM-B segment to stabilize the asymmetric kinase dimer (Figs. 3 and 4), leading to allosteric activation of the receiver kinase.
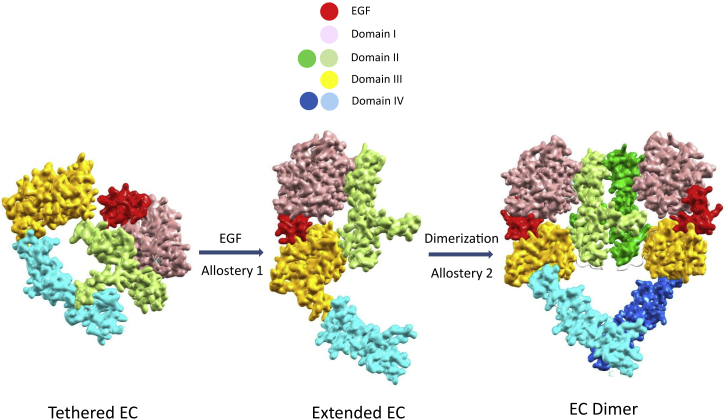
Figure 2.
Structural allostery of ligand-dependent dimerization in EGFR N-terminal EC. The EC is composed of four subdomains, I, II, III, and IV, shown in different colors. At low EGFR levels, monomeric species dominate the population of ligand-free EC, with the most favorable tethered conformation shown on the left. Ligand binding stabilizes the extended conformation as shown in the middle, shifting the monomeric population with a large conformational change for dimerization. The favorable (extended) homodimer conformation then further depletes the population of the monomeric tethered conformation, facilitating the population shift to the dominant back-to-back ligand-bound EC dimer shown on the right. There are two classical allosteric scenarios here. First, a population shift with a large conformational change due to binding is a typical description of allostery. The second scenario is the ligand-dependent dimerization, however, with the ligand not at the dimer interface. To see this figure in color, go online.
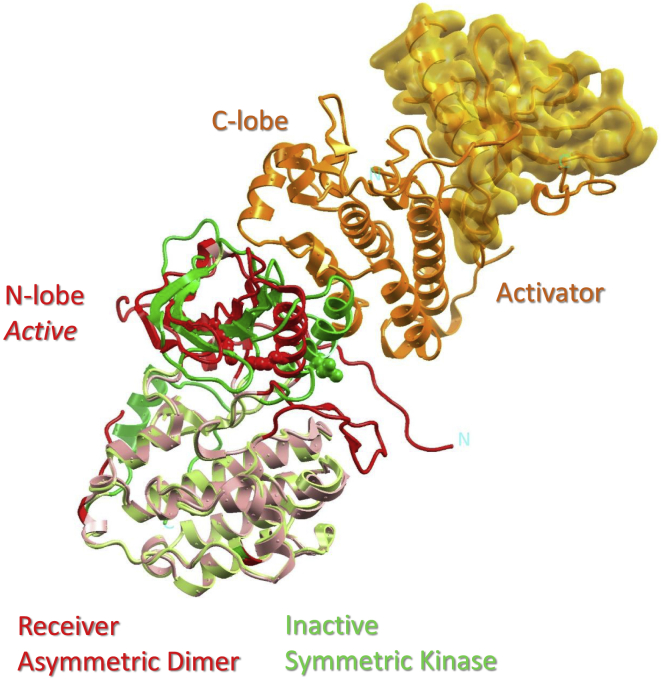
Figure 3.
Allosteric activation of EGFR via an asymmetric KD dimerization. The superposition of two KDs, with the receiver of the asymmetric dimer (red ribbon) and the inactive kinase from the symmetric dimer (green ribbon), clearly shows the inward rotation of the αC-helix (dark color ribbon), which stabilizes a characteristic salt bridge (side chains depicted in space-fill model) in the activated protein kinase. The interaction between the N-lobe of the receiver and the C-lobe of the activator kinase (orange ribbon) is distal from the active site located in between the two lobes. Therefore, the asymmetric dimerization is dubbed a dimer-dependent allosteric kinase activation. The N-lobe of the activator kinase is highlighted by a simulated electron density model in the transparent yellow color. To see this figure in color, go online.
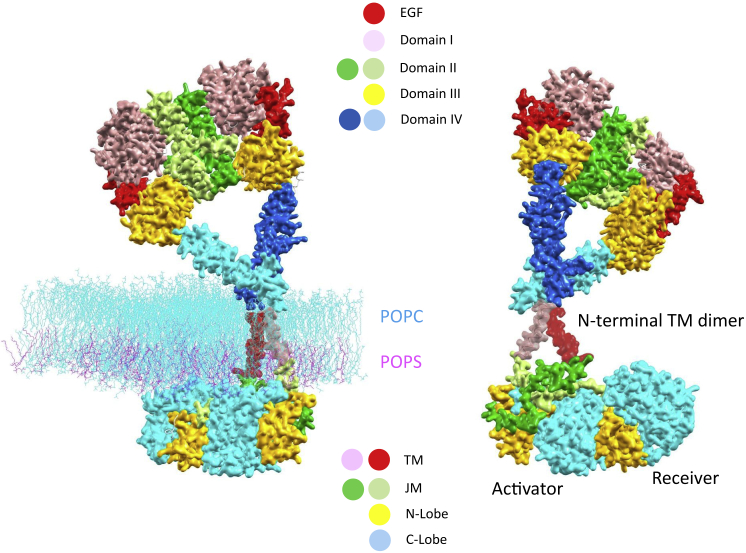
Figure 4.
A near-complete atomic model of the active ligand-bound EGFR dimer in the membrane. Depicted here is a snapshot of the atomic model from MD simulations, which was put together by Shaw and co-workers (7) based on known EGFR segmented structures excluding the disordered part of the C-terminal autophosphorylation CT substrate. On the left, the lipid bilayers included in the simulation (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine in the wire-frame model by the colors cyan and magenta, respectively) specify the position of the membrane with respect to active EGFR. Shown on the right is a view after a 180° rotation along the paper axis, purposely excluding the lipid bilayers to clearly reveal structurally coupled conformations responsible for physiological EGFR activation, including the TM near the N-terminal-cross helical dimer, the antiparallel JM-A helical dimer, and the key interaction of the receiver JM-B with the activator kinase C-lobe. To see this figure in color, go online.
An atomic model of the active EGFR is supported by segmented constructs
A complete atomic model of EGFR (except the C-terminal autophosphorylation substrate CT) has been assembled and simulated by Shaw and co-workers (7). Their model illustrates all the characteristic features of EGF-activated EGFR conformation in physiological signaling. These features are highlighted in Fig. 4. This near-complete model is strongly supported by experimental data of EGFR constructs in different media (7, 8). First, whereas isolated KDs remain predominantly inactive monomers in solution, they become active when JMs are added. However, such a construct loses its activity again when localized to the membrane, indicating that the JM may prefer association with the membrane to interaction with KD. Further addition of the transmembrane segment recovers the kinase activity, indicating that via an N-terminal helical association, the TM may destabilize the interaction between JM and the membrane and facilitate the JM’s availability for stabilizing the asymmetric dimer. Last, further addition of the EC domain autoinhibits ligand-free EGFR at low expression levels by populating isolated monomers and preformed EC dimers that confine the TM dimer in a C-terminal helical association that favors membrane-embedded JM.
Allosteric dimerization of the EC
EGFR allosteric activation is depicted as a population shift (or large conformation change) from an inactive ligand-free tethered monomer (and/or preformed inactive ligand-free dimer) to an active ligand-bound dimer. Unlike many receptor tyrosine kinases with bound ligands at the EC dimer interface (13), as shown in Fig. 2, because the EC ligand-binding site (allosteric site) is distal to the dimerization interface (active site), ligand binding is already a typical allosteric feature. Furthermore, ligand binding not only stabilizes and shifts the population toward the extended EC conformation, but also, through an allosterically coupled conformational change, it tailors the conformation of domain II for EC dimerization (28) and consequent activation via a specific association between two domain IV.
The population shift from a monomer to proactive EC dimer comes from, first, a greater on rate of dimerization due to the highly populated extended ligand-bound monomer with an exposed dimerization arm as compared to the ligand-free tethered monomer with domain II sequestered by domain IV. And second, binding affinity is further enhanced by a slower dimer off rate due to the coupled conformational change between ligand binding and the dimer interface of active EC.
Population shift: a simple but powerful scheme for EGFR activation
Allosteric activation, depicted in Fig. 1, can be understood as a population shift that stabilizes the active conformation and/or destabilizes the inactive conformation. Such a simple scheme can successfully describe allosteric activation of membrane-anchored EGFR if all essential local minima in both active and inactive states and their corresponding representative structural species are identified. Given the fact that EGFR can be activated by distinct stimulus, population shift in EGFR activation may involve different structural ensembles. These can be dominated by a pair of single active and inactive conformations or involve multiple species with distinct active and inactive conformations. For clarity, we limit our discussion to monomer and dimer species engaged in the population shift.
In the case of ligand-driven EGFR activation, the population shift is predominantly specified by a pair of monomeric ECs, a pair of TMs, and a pair of KDs. Physiological EGFR activation follows a population shift from the inactive tethered EC monomer to the extended ligand-bound EC monomer and the consequent ligand-bound back-to-back EC dimer as described in Fig. 2. Via a strong outside-in structural coupling mediated by domain IV, the back-to-back EC dimer stabilizes the near N-terminal-cross TM helical dimer, which in turn releases JM from the membrane to stabilize the formation of asymmetric KD dimer with an active receiver KD. In the case of an existing inactive ligand-free back-to-back EC dimer species, ligand binding promotes a similar population shift from ligand-free inactive to ligand-bound active EC dimer and a subsequent population shift from the near C-terminal-cross to near N-terminal-cross TM dimer (Fig. 5).
Figure 5.
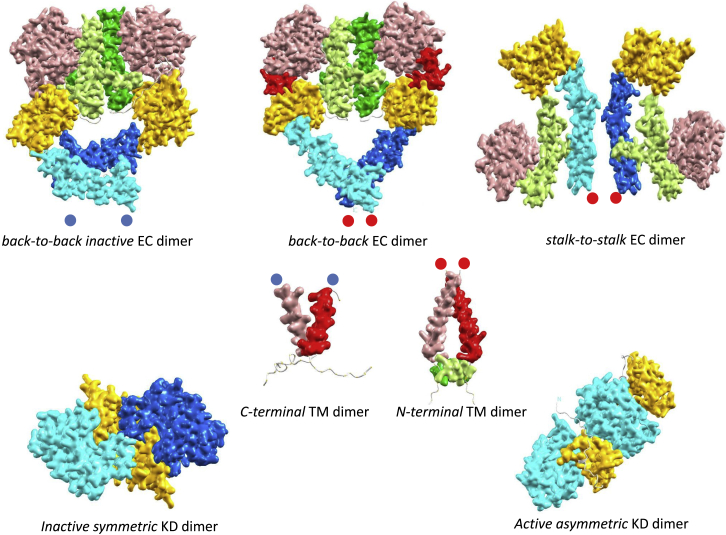
The key structural dimer species involved in EGFR activation. Physiological and pathological (ligand-free) EGFR activation is determined by three independent sources, including ligand-binding-driven EC dimerization, proximity-driven dimerization by clustering on a limited number of lipid rafts, and direct asymmetric KD dimer formation with enhanced affinity. We list important dimer species in three compartments: EC, TM + JM, and KD. In physiological activation, the ligand-binding event allosterically instigates a population shift from the inactive to the active species. Before stimulation by ligands, the dominant ligand-free species include monomer (in inactive tethered conformation (data not shown)) and the back-to-back inactive EC dimer (7) in association with the near C-terminal-cross TM dimer (as indicated by the two separated blue dots) and further down, the associated inactive symmetric KD dimer. On the other hand, the ligand-bound back-to-back EC dimer (PDB: 3NJP) dominates the active species, which via a strong outside-in structural coupling, leads to the stabilization of the near N-terminal-cross TM dimer (indicated by the two adjacent red dots) and the consequent stabilization of the active asymmetric KD dimer through the release of the membrane-embedded JM. In the case of the second activating driver, the force of increasing EGFR proximity either by a drop in the total number of lipid rafts via cholesterol depletion or EGFR overexpression with a fixed number of lipid rafts dominates the nonmonomer species toward their assembly within a lipid raft. Among the pool of ligand-free EC dimers, two leading species accumulate to activate EGFR: the active back-to-back and the stalk-to-stalk EC dimer (29). Both are structurally coupled with the near N-terminal-cross TM dimer and the active asymmetric KD dimer. Last, because oncogenic mutations favor the active KD conformation, the highly populated asymmetric KD dimers are accommodated by the loose inside-out coupling linkage. Through the near N-terminal-cross TM dimer, the active asymmetric KD dimer couples not only with the ligand-free but active back-to-back EC dimer but also the stalk-to-stalk EC dimer. To see this figure in color, go online.
In the case of ligand-free EGFR, although monomeric species dominate at low expression level, imaging technologies reported that at physiological expression levels, there is a mixture of monomers, dimers, and oligomers, in which 65% are populated by nonmonomer species (29). As shown in Fig. 2, crystal structures revealed two highly dynamic monomer conformations, that of tethered and extended. A population of 80–97% has been estimated to be in the inactive tethered conformation (13). Three ligand-free EC dimer models fit experimental data (Fig. 5), including two EC dimers, that of the head-to-head (data not shown) and the back-to-back dimers, both in preferred association with the inactive symmetric KD dimer, and the stalk-to-stalk EC dimer in association with the active asymmetric KD dimer (29). Under pathological conditions, overexpression, or cholesterol-depleted experiments, the increased proximity favors nonmonomer species with a relatively high number of EGFR distributed on a limited number of lipid rafts. Altogether, active species with an asymmetric KD dimer structurally coupled to the N-terminal-cross TM dimer and accommodated by the stalk-to-stalk EC dimer, or the active ligand-free back-to-back EC dimer, exceed the activation threshold.
Last, constitutive KD mutants promote a population shift, stabilizing an active conformation and/or destabilizing inactive conformations. Driven by the mutations, the inside-out structural coupling links the asymmetric KD dimer to the N-terminal-cross TM dimer. However, the experimentally observed loose linkage implies not only active back-to-back EC dimers but also stalk-to-stalk dimers in ligand-independent pathological EGFR activation, which is similar to EGFR overexpression activation, except that one is driven by KD dimerization and the other by increased proximity.
Putting it all together
EGFR activation is not equivalent to cell transformation
EGFR activation, physiological or pathological, can lead to several cellular outcomes, including cell proliferation, transformation, and differentiation. Activation via asymmetric KD dimerization induces tyrosine autophosphorylation in the C-terminal tail with the signaling outcome distributed among several pathways. Phosphorylation of each of the nine tail tyrosines creates docking sites for signaling effectors (30). Although the distinct signaling kinetics of EGFR ligands, such as weak sustained versus strong transient activation, can induce different cellular results (15), it is believed that distinct tyrosine phosphorylation patterns via direct effector recruitment shape the signaling outputs (30). Considering this signaling complexity, the contradictory literature reports as to whether EGFR can be activated via overexpression alone are not surprising. A 3-decade-old report (31) indicated that a more than 500-fold EGFR overexpression cannot transform NIH 3T3 cells as compared to ligand-mediated stimulation, implying that EGFR overexpression alone may not lead to KD activation. This contradicts our premise that a fivefold EGFR overexpression can lead to the same level of ligand-mediated EGFR activation at physiological expression levels for which the activation level is determined by a single 1068Y phosphorylation (8).
The multimer model proposed by Kuriyan’s laboratory (32) and the EGFR activation scheme in Fig. 5 can resolve this discrepancy. EGFR-stimulated cell transformation requires both the MAPK and PI3K pathways (33). Considering only direct effectors recruitment, both distal 1068Y and proximal 992Y (4) (or 920Y in the kinase C-lobe, which contains the pYMXM binding motif for the p85α nSH2 domain (34)) need to be fully phosphorylated to recruit effectors through their Src homology 2 or phosphotyrosine binding domains. Binding these phosphorylated tyrosines promotes the activation of MAPK and PI3K pathways, respectively. Receiver or activator 1068Y, distal to the KD, can reach the active site of the receiver kinase (35). However, this is not the case for the proximal 992Y. Restricted by length, only the cis-phosphorylation is feasible for the receiver proximal site, but not trans-phosphorylation for the activator (35). Thus, compared to the distal tyrosine in MAPK activation, PI3K activation, which relies only on the proximal site, can accomplish half of the activation strength because only cis-phosphorylation is achievable, but not trans-phosphorylation in the asymmetric kinase dimer.
In the multimer (or oligomer) model constructed by Kuriyan’s laboratory (32), with a linear side-by-side stacking of an active back-to-back EC dimer (Fig. 5), the receiver of the asymmetric KD dimer becomes the activator of the interasymmetric KD dimer in the multimer, thus activating the activator of the intraasymmetric KD dimer. In the multimer of n back-to-back EC dimers, there is only one inactive kinase. Thus, the phosphorylation strength of the proximal site is close to that of the distal site with a fraction of 2n-1/2n, instead of 1/2 of the isolated asymmetric dimers. This may explain why overexpression of HER2 (36), but not of EGFR (31), can transform NIH 3T3 cells. Activated by EGFR overexpression, the most populated active asymmetric dimer could be the stalk-to-stalk EC dimer (Fig. 5), which is not compatible with the linear side-by-side stacking. Thus, EGFR overexpression activates PI3K at half strength. In the case of HER2, the monomer is already populated in extended conformation even in a ligand-free state. The low population of unfavorable stalk-to-stalk EC dimers with the monomer in closed conformation in HER2 does not constrain PI3K activation, thus cell transformation.
Structural coupling in EGFR signaling
Structural coupling is a fundamental principle of allostery. Coupling between the extracellular and intracellular EGFR components is mediated by the TM, which alters its conformation to promote ligand-mediated dimerization. Thus, ligand-mediated EC signaling propagates through the EC domain IV (17), resulting in an apparent conformational change in the TM dimer that in turn shifts the most populated state from inactive autoinhibited conformations (monomer + preformed symmetric KD dimer) to an active asymmetric KD dimer.
Negative cooperativity in ligand binding
Under physiological conditions, ligand-mediated EC dimerization populates the dominant back-to-back assembly (Fig. 5). Allosteric coupling through domain IV reflects the negative ligand cooperativity (17). Domain IV association in the EC dimer favors the N-terminal TM dimer conformation (Protein Data Bank [PDB]: 2M20) over the C-terminal (MD simulation coordinates (7)). The favorable TM change propagates, perturbing the autoinhibited association of the intracellular membrane-proximal JM-A (residues 645–665) segment (16) with membrane. Consequently, the N-terminal TM dimer populates its associated active asymmetric KD dimer and shifts the favorable intracellular population from autoinhibited to active states.
Strong structural outside-in coupling and weak inside-out coupling
The scenario above reveals that in physiological ligand-mediated outside-in activation, only one dominant back-to-back EC dimer is structurally coupled to the active asymmetric KD dimer. However, in pathological overexpression, the active asymmetric KD dimer is associated with two different EC dimers, that of stalk-to-stalk and back-to-back dimers. The one-to-one linkage between the back-to-back EC dimer and the asymmetric KD dimer displays direct coupling, making the outside-in coupling strong. By contrast, the inside-out coupling with one intracellular structure corresponding to two extracellular structures is weak. However, although the outside-in one-to-one coupling is strong, disulfide cross-linking and systematic mutagenesis show that the interactions of two IV domains as well as two N-terminal TMs are nonspecific (9). Thus, interactions resulting from the structural coupling through the back-to-back EC dimer do not increase the stability of the active conformation; they only release the autoinhibition (or destabilize the inactive state) of the closed EC conformation.
Our description of bidirectional allosteric coupling, as inspired by EGFR signaling, proposes that a forward structural coupling between two distal sites (the outside-in coupling) is not always equivalent to a backward structural coupling (the inside-out coupling) if those interactions participating in the coupling do not create a relative stabilization energy advantage to sustain the allosteric propagation. In the outside-in coupling, the ligand-bound EC dimer promotes an arrangement of domain IVs that propagates the allosteric coupling to the TM and a favorable asymmetric KD dimer. By contrast, the inside-out coupling stops at the membrane with a near N-terminal-cross TM dimer, which provides no energy advantage for domain IV to propagate back to the ligand-bound EC dimer conformation.
Conclusions
Based on a core principle of how allostery works (11), we unified the canonical view of ligand-mediated EGFR activation at physiological EGFR expression and sufficient agonist levels and pathological ligand-independent activation due to oncogenic mutations or overexpression. Constructs of domain combinations and negative ligand-binding data all point to EC domain IV and JM-mediated structural coupling across the membrane, establishing a population shift between an active N-terminal-cross TM dimer and an inactive, C-terminal-cross TM dimer. To clarify the role of allostery in EGFR activation, we ask how a stimulus executes this population shift via stabilizing the active conformation and/or destabilizing the inactive conformation. Within this framework, physiological EGFR activation can be described as ligand-mediated EC dimerization that stabilizes the N-terminal-cross TM dimer. In turn, this disrupts the autoinhibiting interaction between the membrane and JM, allowing the released JM-B segment to stabilize the asymmetric KD dimer with the active receiver kinase representing the resulting EGFR signaling.
In pathological EGFR activation, oncogenic mutations in KD, the stimulus, lead to stabilization of the active KD conformation, which in turn stabilizes the asymmetric KD dimerization. The inside-out coupling is not as strong as the outside-in physiological coupling. This suggests that EC dimer species are coupled to the oncogenic asymmetric KD dimer through the N-terminal-cross TM dimer. Indeed, a recently verified stalk-to-stalk dimer EC dimer species (29), in addition to the active ligand-free back-to-back dimer, fits well this loose-linkage inside-out coupling scenario (9).
In the case of the third stimulus, that of increasing EGFR proximity via EGFR overexpression or cholesterol depletion, the assembled and activated species resemble those in the second stimulus. The driving force is purely the proximity-mediated dimerization in the extracellular, transmembrane, and intracellular components.
The complete description of EGFR activation with structural models of known or newly identified dimers in the extracellular, transmembrane, and intracellular modules given here further provides a robust structural basis for verification of oncogenic mutations as “drivers” or “latent drivers” (37, 38). These mutations can be defined as offering increased stabilization to the active conformation of EGFR, albeit to different extents.
Acknowledgments
This project was funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract number HHSN261200800001E. This research was supported (in part) by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
Editor: Brian Salzberg.
References
- 1.Lemmon M.A., Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lemmon, M. A., and J. Schlessinger. 2010. Cell signaling by receptor tyrosine kinases. Cell. 141:1117-1134. [DOI] [PMC free article] [PubMed]
- 2.Yarden Y., Sliwkowski M.X. Untangling the ErbB signalling network. Nat. Rev. Mol. Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]; Yarden, Y., and M. X. Sliwkowski. 2001. Untangling the ErbB signalling network. Nat. Rev. Mol. Cell Biol. 2:127-137. [DOI] [PubMed]
- 3.Hynes N.E., MacDonald G. ErbB receptors and signaling pathways in cancer. Curr. Opin. Cell Biol. 2009;21:177–184. doi: 10.1016/j.ceb.2008.12.010. [DOI] [PubMed] [Google Scholar]; Hynes, N. E., and G. MacDonald. 2009. ErbB receptors and signaling pathways in cancer. Curr. Opin. Cell Biol. 21:177-184. [DOI] [PubMed]
- 4.Jones R.B., Gordus A., MacBeath G. A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature. 2006;439:168–174. doi: 10.1038/nature04177. [DOI] [PubMed] [Google Scholar]; Jones, R. B., A. Gordus, …, G. MacBeath. 2006. A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature. 439:168-174. [DOI] [PubMed]
- 5.Morandell S., Stasyk T., Huber L.A. Quantitative proteomics and phosphoproteomics reveal novel insights into complexity and dynamics of the EGFR signaling network. Proteomics. 2008;8:4383–4401. doi: 10.1002/pmic.200800204. [DOI] [PubMed] [Google Scholar]; Morandell, S., T. Stasyk, …, L. A. Huber. 2008. Quantitative proteomics and phosphoproteomics reveal novel insights into complexity and dynamics of the EGFR signaling network. Proteomics. 8:4383-4401. [DOI] [PubMed]
- 6.Maeda R., Sato T., Sako Y. Lipid-protein interplay in dimerization of juxtamembrane domains of epidermal growth factor receptor. Biophys. J. 2018;114:893–903. doi: 10.1016/j.bpj.2017.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]; Maeda, R., T. Sato, …, Y. Sako. 2018. Lipid-protein interplay in dimerization of juxtamembrane domains of epidermal growth factor receptor. Biophys. J. 114:893-903. [DOI] [PMC free article] [PubMed]
- 7.Arkhipov A., Shan Y., Shaw D.E. Architecture and membrane interactions of the EGF receptor. Cell. 2013;152:557–569. doi: 10.1016/j.cell.2012.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]; Arkhipov, A., Y. Shan, …, D. E. Shaw. 2013. Architecture and membrane interactions of the EGF receptor. Cell. 152:557-569. [DOI] [PMC free article] [PubMed]
- 8.Endres N.F., Das R., Kuriyan J. Conformational coupling across the plasma membrane in activation of the EGF receptor. Cell. 2013;152:543–556. doi: 10.1016/j.cell.2012.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]; Endres, N. F., R. Das, …, J. Kuriyan. 2013. Conformational coupling across the plasma membrane in activation of the EGF receptor. Cell. 152:543-556. [DOI] [PMC free article] [PubMed]
- 9.Lu C., Mi L.Z., Springer T.A. Structural evidence for loose linkage between ligand binding and kinase activation in the epidermal growth factor receptor. Mol. Cell. Biol. 2010;30:5432–5443. doi: 10.1128/MCB.00742-10. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lu, C., L. Z. Mi, …, T. A. Springer. 2010. Structural evidence for loose linkage between ligand binding and kinase activation in the epidermal growth factor receptor. Mol. Cell. Biol. 30:5432-5443. [DOI] [PMC free article] [PubMed]
- 10.Lu C., Mi L.Z., Springer T.A. Mechanisms for kinase-mediated dimerization of the epidermal growth factor receptor. J. Biol. Chem. 2012;287:38244–38253. doi: 10.1074/jbc.M112.414391. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lu, C., L. Z. Mi, …, T. A. Springer. 2012. Mechanisms for kinase-mediated dimerization of the epidermal growth factor receptor. J. Biol. Chem. 287:38244-38253. [DOI] [PMC free article] [PubMed]
- 11.Tsai C.J., Nussinov R. A unified view of “how allostery works”. PLoS Comput. Biol. 2014;10:e1003394. doi: 10.1371/journal.pcbi.1003394. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tsai, C. J., and R. Nussinov. 2014. A unified view of “how allostery works”. PLoS Comput. Biol. 10:e1003394. [DOI] [PMC free article] [PubMed]
- 12.Burgess A.W., Cho H.S., Yokoyama S. An open-and-shut case? Recent insights into the activation of EGF/ErbB receptors. Mol. Cell. 2003;12:541–552. doi: 10.1016/s1097-2765(03)00350-2. [DOI] [PubMed] [Google Scholar]; Burgess, A. W., H. S. Cho, …, S. Yokoyama. 2003. An open-and-shut case? Recent insights into the activation of EGF/ErbB receptors. Mol. Cell. 12:541-552. [DOI] [PubMed]
- 13.Ferguson K.M., Berger M.B., Lemmon M.A. EGF activates its receptor by removing interactions that autoinhibit ectodomain dimerization. Mol. Cell. 2003;11:507–517. doi: 10.1016/s1097-2765(03)00047-9. [DOI] [PubMed] [Google Scholar]; Ferguson, K. M., M. B. Berger, …, M. A. Lemmon. 2003. EGF activates its receptor by removing interactions that autoinhibit ectodomain dimerization. Mol. Cell. 11:507-517. [DOI] [PubMed]
- 14.Zhang X., Gureasko J., Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]; Zhang, X., J. Gureasko, …, J. Kuriyan. 2006. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 125:1137-1149. [DOI] [PubMed]
- 15.Freed D.M., Bessman N.J., Lemmon M.A. EGFR ligands differentially stabilize receptor dimers to specify signaling kinetics. Cell. 2017;171:683–695.e18. doi: 10.1016/j.cell.2017.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]; Freed, D. M., N. J. Bessman, …, M. A. Lemmon. 2017. EGFR ligands differentially stabilize receptor dimers to specify signaling kinetics. Cell. 171:683-695.e18. [DOI] [PMC free article] [PubMed]
- 16.Adak S., Yang K.S., Pike L.J. The membrane-proximal intracellular domain of the epidermal growth factor receptor underlies negative cooperativity in ligand binding. J. Biol. Chem. 2011;286:45146–45155. doi: 10.1074/jbc.M111.274175. [DOI] [PMC free article] [PubMed] [Google Scholar]; Adak, S., K. S. Yang, …, L. J. Pike. 2011. The membrane-proximal intracellular domain of the epidermal growth factor receptor underlies negative cooperativity in ligand binding. J. Biol. Chem. 286:45146-45155. [DOI] [PMC free article] [PubMed]
- 17.Adak S., DeAndrade D., Pike L.J. The tethering arm of the EGF receptor is required for negative cooperativity and signal transduction. J. Biol. Chem. 2011;286:1545–1555. doi: 10.1074/jbc.M110.182899. [DOI] [PMC free article] [PubMed] [Google Scholar]; Adak, S., D. DeAndrade, and L. J. Pike. 2011. The tethering arm of the EGF receptor is required for negative cooperativity and signal transduction. J. Biol. Chem. 286:1545-1555. [DOI] [PMC free article] [PubMed]
- 18.Chen X., Resh M.D. Cholesterol depletion from the plasma membrane triggers ligand-independent activation of the epidermal growth factor receptor. J. Biol. Chem. 2002;277:49631–49637. doi: 10.1074/jbc.M208327200. [DOI] [PubMed] [Google Scholar]; Chen, X., and M. D. Resh. 2002. Cholesterol depletion from the plasma membrane triggers ligand-independent activation of the epidermal growth factor receptor. J. Biol. Chem. 277:49631-49637. [DOI] [PubMed]
- 19.Lambert S., Vind-Kezunovic D., Gniadecki R. Ligand-independent activation of the EGFR by lipid raft disruption. J. Invest. Dermatol. 2006;126:954–962. doi: 10.1038/sj.jid.5700168. [DOI] [PubMed] [Google Scholar]; Lambert, S., D. Vind-Kezunovic, …, R. Gniadecki. 2006. Ligand-independent activation of the EGFR by lipid raft disruption. J. Invest. Dermatol. 126:954-962. [DOI] [PubMed]
- 20.Alvarado D., Klein D.E., Lemmon M.A. Structural basis for negative cooperativity in growth factor binding to an EGF receptor. Cell. 2010;142:568–579. doi: 10.1016/j.cell.2010.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]; Alvarado, D., D. E. Klein, and M. A. Lemmon. 2010. Structural basis for negative cooperativity in growth factor binding to an EGF receptor. Cell. 142:568-579. [DOI] [PMC free article] [PubMed]
- 21.Lemmon M.A., Freed D.M., Kiyatkin A. The dark side of cell signaling: positive roles for negative regulators. Cell. 2016;164:1172–1184. doi: 10.1016/j.cell.2016.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lemmon, M. A., D. M. Freed, …, A. Kiyatkin. 2016. The dark side of cell signaling: positive roles for negative regulators. Cell. 164:1172-1184. [DOI] [PMC free article] [PubMed]
- 22.Herrero A., Pinto A., Crespo P. Small molecule inhibition of ERK dimerization prevents tumorigenesis by RAS-ERK pathway oncogenes. Cancer Cell. 2015;28:170–182. doi: 10.1016/j.ccell.2015.07.001. [DOI] [PubMed] [Google Scholar]; Herrero, A., A. Pinto, …, P. Crespo. 2015. Small molecule inhibition of ERK dimerization prevents tumorigenesis by RAS-ERK pathway oncogenes. Cancer Cell. 28:170-182. [DOI] [PubMed]
- 23.Manning G., Whyte D.B., Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]; Manning, G., D. B. Whyte, …, S. Sudarsanam. 2002. The protein kinase complement of the human genome. Science. 298:1912-1934. [DOI] [PubMed]
- 24.Nolen B., Taylor S., Ghosh G. Regulation of protein kinases; controlling activity through activation segment conformation. Mol. Cell. 2004;15:661–675. doi: 10.1016/j.molcel.2004.08.024. [DOI] [PubMed] [Google Scholar]; Nolen, B., S. Taylor, and G. Ghosh. 2004. Regulation of protein kinases; controlling activity through activation segment conformation. Mol. Cell. 15:661-675. [DOI] [PubMed]
- 25.Huse M., Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]; Huse, M., and J. Kuriyan. 2002. The conformational plasticity of protein kinases. Cell. 109:275-282. [DOI] [PubMed]
- 26.Thiel K.W., Carpenter G. Epidermal growth factor receptor juxtamembrane region regulates allosteric tyrosine kinase activation. Proc. Natl. Acad. Sci. USA. 2007;104:19238–19243. doi: 10.1073/pnas.0703854104. [DOI] [PMC free article] [PubMed] [Google Scholar]; Thiel, K. W., and G. Carpenter. 2007. Epidermal growth factor receptor juxtamembrane region regulates allosteric tyrosine kinase activation. Proc. Natl. Acad. Sci. USA. 104:19238-19243. [DOI] [PMC free article] [PubMed]
- 27.Red Brewer M., Choi S.H., Carpenter G. The juxtamembrane region of the EGF receptor functions as an activation domain. Mol. Cell. 2009;34:641–651. doi: 10.1016/j.molcel.2009.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]; Red Brewer, M., S. H. Choi, …, G. Carpenter. 2009. The juxtamembrane region of the EGF receptor functions as an activation domain. Mol. Cell. 34:641-651. [DOI] [PMC free article] [PubMed]
- 28.Freed D.M., Alvarado D., Lemmon M.A. Ligand regulation of a constitutively dimeric EGF receptor. Nat. Commun. 2015;6:7380. doi: 10.1038/ncomms8380. [DOI] [PMC free article] [PubMed] [Google Scholar]; Freed, D. M., D. Alvarado, and M. A. Lemmon. 2015. Ligand regulation of a constitutively dimeric EGF receptor. Nat. Commun. 6:7380. [DOI] [PMC free article] [PubMed]
- 29.Zanetti-Domingues L.C., Korovesis D., Martin-Fernandez M.L. The architecture of EGFR’s basal complexes reveals autoinhibition mechanisms in dimers and oligomers. Nat. Commun. 2018;9:4325. doi: 10.1038/s41467-018-06632-0. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zanetti-Domingues, L. C., D. Korovesis, …, M. L. Martin-Fernandez. 2018. The architecture of EGFR’s basal complexes reveals autoinhibition mechanisms in dimers and oligomers. Nat. Commun. 9:4325. [DOI] [PMC free article] [PubMed]
- 30.Wilson K.J., Gilmore J.L., Riese D.J., II Functional selectivity of EGF family peptide growth factors: implications for cancer. Pharmacol. Ther. 2009;122:1–8. doi: 10.1016/j.pharmthera.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wilson, K. J., J. L. Gilmore, …, D. J. Riese II. 2009. Functional selectivity of EGF family peptide growth factors: implications for cancer. Pharmacol. Ther. 122:1-8. [DOI] [PMC free article] [PubMed]
- 31.Di Fiore P.P., Pierce J.H., Aaronson S.A. Overexpression of the human EGF receptor confers an EGF-dependent transformed phenotype to NIH 3T3 cells. Cell. 1987;51:1063–1070. doi: 10.1016/0092-8674(87)90592-7. [DOI] [PubMed] [Google Scholar]; Di Fiore, P. P., J. H. Pierce, …, S. A. Aaronson. 1987. Overexpression of the human EGF receptor confers an EGF-dependent transformed phenotype to NIH 3T3 cells. Cell. 51:1063-1070. [DOI] [PubMed]
- 32.Huang Y., Bharill S., Kuriyan J. Molecular basis for multimerization in the activation of the epidermal growth factor receptor. eLife. 2016;5:e14107. doi: 10.7554/eLife.14107. [DOI] [PMC free article] [PubMed] [Google Scholar]; Huang, Y., S. Bharill, …, J. Kuriyan. 2016. Molecular basis for multimerization in the activation of the epidermal growth factor receptor. eLife. 5:e14107. [DOI] [PMC free article] [PubMed]
- 33.Nussinov R., Tsai C.J., Keskin O. Principles of K-Ras effector organization and the role of oncogenic K-Ras in cancer initiation through G1 cell cycle deregulation. Expert Rev. Proteomics. 2015;12:669–682. doi: 10.1586/14789450.2015.1100079. [DOI] [PubMed] [Google Scholar]; Nussinov, R., C. J. Tsai, …, O. Keskin. 2015. Principles of K-Ras effector organization and the role of oncogenic K-Ras in cancer initiation through G1 cell cycle deregulation. Expert Rev. Proteomics. 12:669-682. [DOI] [PubMed]
- 34.Nolte R.T., Eck M.J., Harrison S.C. Crystal structure of the PI 3-kinase p85 amino-terminal SH2 domain and its phosphopeptide complexes. Nat. Struct. Biol. 1996;3:364–374. doi: 10.1038/nsb0496-364. [DOI] [PubMed] [Google Scholar]; Nolte, R. T., M. J. Eck, …, S. C. Harrison. 1996. Crystal structure of the PI 3-kinase p85 amino-terminal SH2 domain and its phosphopeptide complexes. Nat. Struct. Biol. 3:364-374. [DOI] [PubMed]
- 35.Koland J.G. Coarse-grained molecular simulation of epidermal growth factor receptor protein tyrosine kinase multi-site self-phosphorylation. PLoS Comput. Biol. 2014;10:e1003435. doi: 10.1371/journal.pcbi.1003435. [DOI] [PMC free article] [PubMed] [Google Scholar]; Koland, J. G. 2014. Coarse-grained molecular simulation of epidermal growth factor receptor protein tyrosine kinase multi-site self-phosphorylation. PLoS Comput. Biol. 10:e1003435. [DOI] [PMC free article] [PubMed]
- 36.Di Fiore P.P., Pierce J.H., Aaronson S.A. erbB-2 is a potent oncogene when overexpressed in NIH/3T3 cells. Science. 1987;237:178–182. doi: 10.1126/science.2885917. [DOI] [PubMed] [Google Scholar]; Di Fiore, P. P., J. H. Pierce, …, S. A. Aaronson. 1987. erbB-2 is a potent oncogene when overexpressed in NIH/3T3 cells. Science. 237:178-182. [DOI] [PubMed]
- 37.Nussinov R., Jang H., Cheng F. Precision medicine review: rare driver mutations and their biophysical classification. Biophys Rev. 2019;11:5–19. doi: 10.1007/s12551-018-0496-2. [DOI] [PMC free article] [PubMed] [Google Scholar]; Nussinov, R., H. Jang, …, F. Cheng. 2019. Precision medicine review: rare driver mutations and their biophysical classification. Biophys Rev. 11:5-19. [DOI] [PMC free article] [PubMed]
- 38.Nussinov R., Tsai C.J. ‘Latent drivers’ expand the cancer mutational landscape. Curr. Opin. Struct. Biol. 2015;32:25–32. doi: 10.1016/j.sbi.2015.01.004. [DOI] [PubMed] [Google Scholar]; Nussinov, R., and C. J. Tsai. 2015. ‘Latent drivers’ expand the cancer mutational landscape. Curr. Opin. Struct. Biol. 32:25-32. [DOI] [PubMed]
- 39.Nussinov R., Tsai C.J. Unraveling structural mechanisms of allosteric drug action. Trends Pharmacol. Sci. 2014;35:256–264. doi: 10.1016/j.tips.2014.03.006. [DOI] [PubMed] [Google Scholar]; Nussinov, R., and C. J. Tsai. 2014. Unraveling structural mechanisms of allosteric drug action. Trends Pharmacol. Sci. 35:256-264. [DOI] [PubMed]