Abstract
Intrinsically disordered proteins (IDPs) as well as intrinsically disordered regions (IDRs) of complex protein machineries have recently been recognized as key players in many cellular functions. NMR represents a unique tool to access atomic resolution structural and dynamic information on highly flexible IDPs/IDRs. Improvements in instrumental sensitivity made heteronuclear direct detection possible for biomolecular NMR applications. The CON experiment has become one of the most useful NMR experiments to get a snapshot of an IDP/IDR in conditions approaching physiological ones. The availability of NMR spectrometers equipped with multiple receivers now enables the acquisition of several experiments simultaneously instead of one after the other. Here, we propose several variants of the CON experiment in which, during the recovery delay, a second two-dimensional experiment is acquired, either based on 1H detection (CON//HN) or on 15N detection (CON//btNH, CON//(H)CAN). The possibility to collect simultaneous snapshots of an IDP/IDR through different two-dimensional spectra provides a novel tool to follow chemical reactions, such as the occurrence of posttranslational modifications, as well as to study samples of limited lifetime such as cell lysates or whole cells.
Significance
The exploitation of multiple receivers available in most of the newly designed NMR spectrometers enables the simultaneous collection of several snapshots of the very same event from different atomic perspectives.
Introduction
Proteins or protein regions that lack a well-defined three-dimensional (3D) structure and that are characterized by high flexibility and disorder are widespread in living organisms and essential for protein function (1, 2, 3). Their contributions to biological processes are highly complementary to those typical of folded proteins. Indeed, intrinsically disordered proteins (IDPs) or intrinsically disordered protein regions (IDRs) are often involved in key regulatory processes for which the adaptability of the protein structure and dynamics represents a clear functional advantage (4). Their important functional role also becomes evident from the strong link that has been found between misfunction in IDPs/IDRs and many diseases such as cancer and neurodegenerative diseases. The atomic-level characterization of IDPs/IDRs has thus become a topic of central importance also in light of the development of new drugs capable of interfering with them, a completely novel area for which the traditional approach of drug design based on the identification of well-defined binding pockets in folded proteins is obviously bound to fail (5, 6, 7, 8).
NMR plays a central role in this area of research being the only method able to provide atomic resolution information on the structural and dynamic properties of highly flexible macromolecules (9, 10, 11, 12, 13). NMR observables are, however, strongly influenced by the peculiar properties of IDPs, and thus the NMR approaches should be optimized to overcome critical points (14). For example, it is well known that 1H-detected experiments, usually the first choice on the grounds of instrumental sensitivity, do have some drawbacks for the study of IDPs, in particular when approaching physiological pH and temperature conditions. In fact, the largely solvent-exposed backbones typical of highly flexible IDPs leave amide protons accessible to the solvent and are responsible for efficient chemical exchange processes that may lead to extreme broadening of HN resonances, reducing the sensitivity of 1H-detected experiments. In addition, 1H nuclear spins are characterized by a moderate chemical shift dispersion. These are two features that have stimulated the development of exclusively heteronuclear NMR experiments based on 13C direct detection for the study of IDPs. The two-dimensional (2D) CON experiment (15, 16) is now widely used to characterize highly flexible IDPs, thanks to the excellent chemical shift dispersion of the crosspeaks observed in this experiment and the possibility to reveal atomic resolution information on IDPs also in experimental conditions in which HN resonances are not observable (17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28). The wide use of exclusively heteronuclear NMR experiments for protein investigations has been stimulated by improvements in instrumental sensitivity (29). Technological advances provided a jump of a factor of more than 10 in 13C instrumental sensitivity, and lately, similar efforts have even enabled direct detection of 15N (30, 31, 32, 33), a nucleus characterized by a lower gyromagnetic ratio with respect to 13C and thus by a very low intrinsic sensitivity.
In parallel to significant increase in instrumental sensitivity, new technologies have also become easily accessible such as the possibility to exploit NMR spectrometers with multiple receivers, enabling the design of multinuclear pulse sequences for the acquisition of more than one free induction decay (FID) for each repetition. Several different strategies exploiting multiple receivers have been proposed in the literature (34, 35, 36, 37, 38, 39) to acquire more experiments in the time needed for a single one. Applications of multiple receivers for biomolecular NMR experiments were also proposed either in solution or in the solid state (36, 37, 38, 39, 40, 41). One strategy consists of the acquisition of different FIDs within one main coherence transfer pathway to recover signals that would otherwise be lost or suppressed (36, 37). The other strategy consists in exploiting polarization that remains unused at the end of one experiment to collect more than one spectrum in the time needed for the longest one of them (38).
The development of more sensitive instruments and the possibility to use multiple receivers stimulates the design of novel experimental strategies to investigate IDPs, and we will show some examples of the useful information that can be gathered combining the CON with other experimental schemes.
Materials and Methods
Samples preparation
The 13C, 15N-labeled α-synuclein sample was prepared as previously described in the literature (42), with a final concentration of 1.0 mM in 100 mM NaCl, 50 μM ethylenediaminetetraacetic acid (EDTA), and 20 mM phosphate buffer at different pH between 6.5 and 7.4 with the addition of 2–10% D2O for the lock. Each figure reports the details about sample conditions. The 13C, 15N-labeled α-synuclein samples used for in-cell experiments and for experiments on cell lysate were prepared as previously described (43) with an estimated protein concentration of 250 μM, using the same buffer as for the purified samples at pH 7.4.
For the phosphorylation reaction, 700 U of tyrosine kinase Fyn (Sigma Aldrich, St. Louis, MO) were added to a 200 μM sample of 13C, 15N-labeled α-synuclein in 20 mM phosphate buffer, 100 mM NaCl, 50 μM EDTA, 2 mM dithiothreitol, 6 mM MgCl2, and 3 mM ATP buffer at pH 7.0 for a final volume of 300 μL, with 5% D2O added for the lock.
NMR experiments
The NMR experiments were acquired at different temperatures on a Bruker AVANCE NEO spectrometer (Billerica, MA) operating at 700.06 MHz 1H, 176.05 MHz 13C, and 70.97 MHz 15N frequencies equipped with a cryogenically cooled probehead optimized for 13C-direct detection (TXO). Standard radio frequency pulses and carrier frequencies for triple resonance experiments were used and are summarized hereafter. Q5- and Q3-shaped pulses (44) of durations of 300 and 231 μs, respectively, were used for 13C band-selective π/2 and π flip angle pulses except for the π pulses that should be band selective on the Cα region (Q3, 1200 μs) and for the adiabatic π pulse to invert both C′ and Cα (smoothed chirp 500 μs, 20% smoothing, 80 kHz sweep width, 11.3 kHz radio frequency field strength) (45). An Eburp2 shape of duration of 1.768 ms and a Reburp shape of duration of 2.076 ms were used, respectively, for 1H band-selective π/2 and π flip angle pulses. In the CON//HN experiment, solvent suppression was achieved through the 3:9:19 pulse scheme (46).
The 13C band-selective pulses on Cα and C′ were applied at the center of each region, respectively, and the 1H-band-selective pulses were applied at the center of the HN region at 8.13 ppm. Decoupling of 13Cα and 15N was achieved with p5m4sp180 (Q3, 900 μs) and Waltz65 (100 μs) decoupling sequences, respectively (44, 47). All gradients employed had a smoothed square shape.
All the spectra were acquired, processed, and analyzed by using Bruker TopSpin 4.0.1 software. Calibration of the spectra was achieved using 4,4-dimethyl-4-silapentane-1-sulfonic acid as a standard for 1H and 13C; 15N shifts were calibrated indirectly (48).
The CON//HN was acquired with a CON interscan delay of 1.7 s; the HN was acquired within this delay. For each increment of the CON experiment, the in-phase (IP) and antiphase (AP) components were acquired and properly combined to achieve IPAP virtual decoupling (49) as described in Fig. S1. The CON spectrum was acquired with two scans, with sweep widths of 5555 Hz (13C) × 2500 Hz (15N) and 1024 × 512 real points in the two dimensions, respectively. The HN spectrum was acquired with 4 scans (2 scans as used for the CON × 2 additional scans because no IPAP decoupling is necessary for the HN), with sweep widths of 10,869 Hz (1H) × 2000 Hz (15N) and 1536 × 512 real points in the two dimensions, respectively. The total duration of the experiment was 1 h and 7 min. For comparison purposes, the two independent experiments with the same parameters were also acquired.
Similar parameters were used also for CON//HN experiments acquired for cell lysates and for in-cell samples as well as to follow the phosphorylation reaction, with the main difference being the use of four scans in the latter case because of lower sample concentration.
The CON//btNH was acquired with a CON interscan delay of 2.5 s; during this time, the btNH was repeated four times (recovery delay of 400 ms). For each increment of each experiment, the IP and AP components were acquired and properly combined to achieve IPAP virtual decoupling (49) as described in Fig. S2. The CON spectrum was acquired with 4 scans, with sweep widths of 5263 Hz (13C′) × 2403 Hz (15N) and 1024 × 1024 real points in the 2 dimensions, respectively. The btNH spectrum was acquired with 128 scans (4 scans as used for the CON × 4 repetitions of the btNH experiment × 8 because of the different number of increments needed in the indirect dimension), with sweep widths of 5263 Hz (15N) × 2403 Hz (1H) and 1024 × 128 real points in the two dimensions, respectively. The total duration of the experiment was 6 h and 19 min. For comparison purposes, the two independent experiments with the same parameters were also acquired.
The CON//(H)CAN was acquired with a CON interscan delay of 2.3 s; during this time, the (H)CAN was repeated three times (recovery delay of 700 ms). For each increment of each experiment, the IP and AP components were acquired and properly combined to achieve IPAP virtual decoupling (49) as described in Fig. S3. The CON spectrum was acquired with 8 scans, with sweep widths of 5555 (13C′) × 2554 Hz (15N) and 1024 × 2048 real points in the two dimensions, respectively. The (H)CAN spectrum was acquired with 192 scans (8 scans as used for the CON × 3 repetitions of the (H)CAN experiment × 8 because of the different number of increments needed in the indirect dimension), with sweep widths of 5555 Hz (15N) × 4000 Hz (13C) and 1024 × 256 real points in the 2 dimensions, respectively. The total duration of the experiment was 60 h and 10 min. For comparison purposes, the two independent experiments with the same parameters were also acquired.
Results and Discussion
The 2D CON has by now become one of the key experiments to characterize IDPs. Many experimental variants of the basic CON pulse sequence have been developed (43, 50). Important features include the nuclear spins used as starting polarization source as well as the approach employed for homonuclear decoupling. When working with sample conditions approaching the “physiological” ones, like in the presence of high ionic strength, pH above 7, and relatively high temperature, the most preferred scheme is the simplest one, which starts on 13C and actively exploits heteronuclear spins only. However, it requires a relaxation delay of ∼2 s to allow the C′ magnetization to relax back to equilibrium. Can we profitably use this delay to collect additional experiments?
The CON//HN experiment
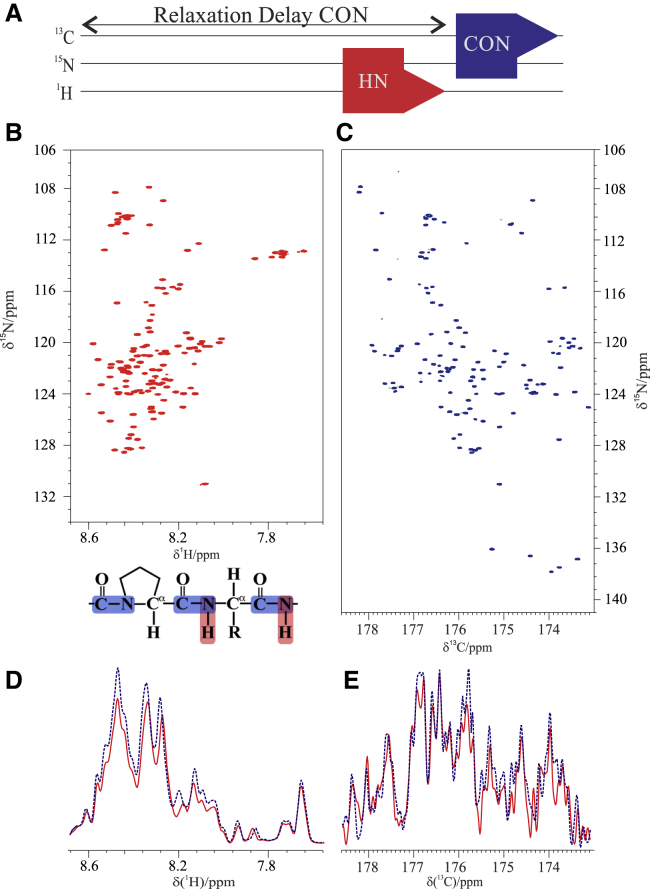
The first two spectra generally collected on an IDP are the 2D HN and 2D CON. The experimental variant designed to acquire them simultaneously is here referred to as the CON//HN, and the spectra obtained on α-synuclein are shown in Fig. 1. The pulse sequence is reported in Fig. S1 and sketched in Fig. 1 A. In brief, a very simple logic was followed in the design of the pulse sequence. The CON experiment essentially exploits only two backbone nuclear spins (C′ and N). During the relaxation delay, an additional experiment can be acquired provided that different nuclear spins are perturbed and that it does not interfere with the CON itself. The 1H-15N heteronuclear single-quantum coherence (HSQC) exploits the amide proton as a starting polarization source as well as for acquisition of the FID; 15N chemical shifts are sampled in the indirect dimension, just like in the CON experiment, and this does not cause any interference. Thus, it can be profitably acquired during the 2-s recycle delay of the CON. The only compromise to combine the two experiments derives from the need to decouple 15N from 1H and 13C during the chemical shift evolution period. For the CON experiment, it is very important to use composite pulse decoupling of 1H throughout the whole pulse sequence to minimize possible influence of exchange processes of amide protons with the solvent protons, which reduce signal intensities in particular when approaching physiological conditions. This does not constitute a problem because the 1H-15N HSQC pulse sequence precedes the CON one. Decoupling of 15N from 13C during the 15N evolution period of the 1H-15N HSQC could be achieved by a combination of two 180° pulses in appropriate positions to restore C′ magnetization to equilibrium as a starting polarization source for the CON experiment. Thanks to this solution, the two spectra can be acquired simultaneously in the optimal conditions required for each of them as one can see by inspecting the quality of the spectra reported in Fig. 1. These were acquired on α-synuclein at 285 K and pH 6.5, which are conditions useful to evaluate whether all the expected resonances could be observed through the CON//HN experiment. Indeed, more than 99% of the backbone correlations could be detected in both experiments (133 out of 134 in the HN and 138 out of 139 in the CON, respectively).
Figure 1.
(A) Scheme of the CON//HN experiment. (B) HN and (C) CON 2D spectra acquired through the CON//HN experiment on 13C, 15N-labeled α-synuclein 1 mM at 285 K in 100 mM NaCl, 50 μM EDTA, and 20 mM phosphate buffer (pH 6.5) are shown. (D) A projection of the HN and the (E) CON spectra acquired with the CON//HN experiment (red solid traces) and with the corresponding experiments acquired independently (blue dotted traces) is shown. To see this figure in color, go online.
Comparison of the intensities of the spectra collected through the CON//HN experiment with those acquired with exactly the same parameters but in an independent fashion shows that reduction in signal to noise ratio (S/N) resulting from the combination of the two experiments is minimal for the CON (5%) and more significant for the HN (15%) which, however, has an intrinsically higher sensitivity and provides the same number of peaks as the independent spectrum (Fig. 1, D and E).
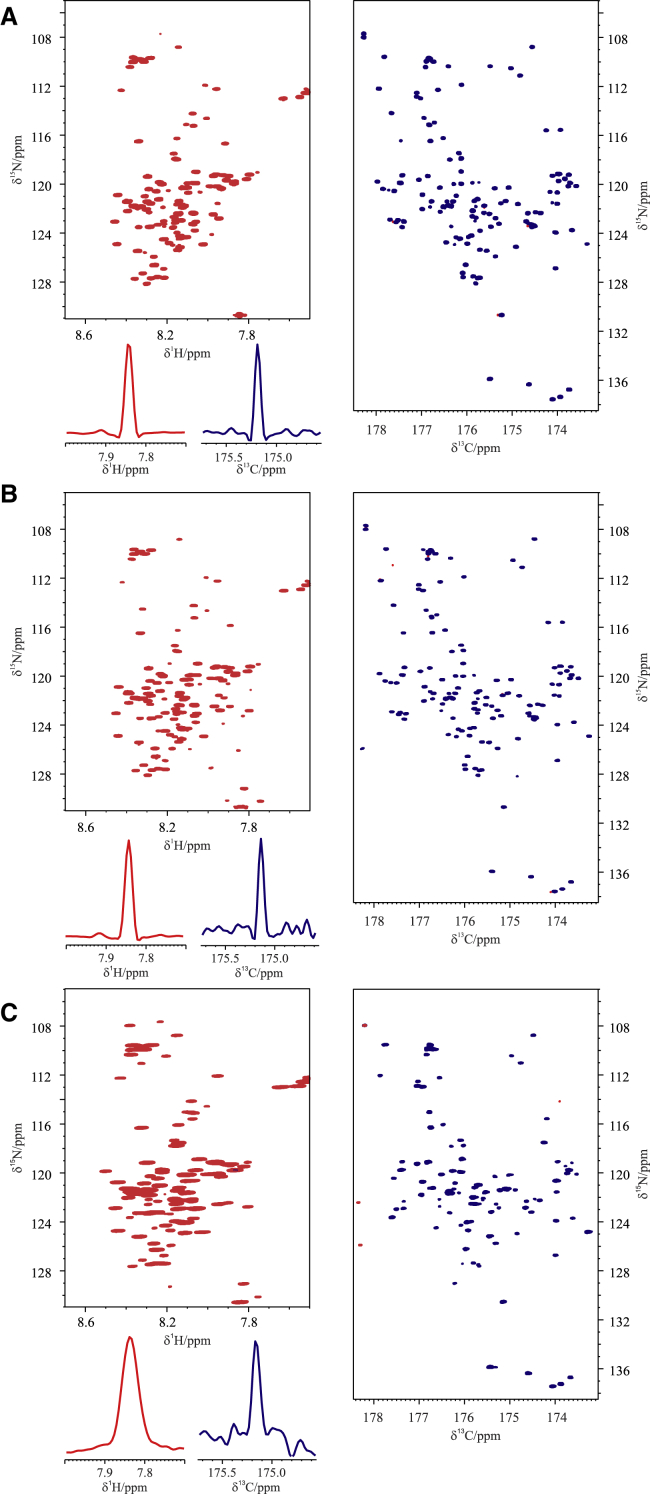
When going to more demanding samples of limited lifetime, the advantages of recording the CON//HN are even more striking. The spectra acquired on α-synuclein in cell are shown in Fig. 2 C and are compared with those acquired on α-synuclein on cell lysates (Fig. 2 B) as well as on purified α-synuclein (Fig. 2 A) in conditions approaching physiological ones (310 K; pH 7.4). First of all, it is interesting to observe the spectra acquired on purified α-synuclein in these conditions and compare them with the analogous one acquired at lower pH and temperature (Fig. 1). The CON results are particularly useful to access complete information (138 detected backbone correlations; 99.3% of the expected ones); the quality of the HSQC is instead reduced because of increased solvent exchange broadening at higher pH and temperature (108 detected backbone correlations, 80.6% of the expected ones). Still, the two spectra provide interesting complementary information, including qualitative information about exchange processes.
Figure 2.
Comparison of the 2D spectra (HN left; CON right) acquired through the CON//HN experiment on 13C, 15N-labeled α-synuclein at 310 K in different experimental conditions: (A) purified sample in 100 mM NaCl, 50 μM EDTA, and 20 mM phosphate buffer at pH 7.4 is shown; (B) in Escherichia coli cells lysate resuspended in the same buffer as in (A) and (C) in-cell. The traces of a representative signal extracted from the HN (red; left) and the CON (blue; right) spectra are also reported. To see this figure in color, go online.
The same spectra acquired on cell lysate show that most of the signals detected on the purified sample can be observed also in cell lysates. The possibility to acquire the two spectra simultaneously is very important, in particular for cell lysates, which have limited lifetimes. It is worth noting that the linewidths of the observed crosspeaks in the two spectra are very similar to those observed for purified α-synuclein, a nontrivial observation considering the complexity of cell lysate composition. When moving to in-cell experiments, the two spectra still show that a vast majority of the expected signals can be detected, providing atomic resolution information to characterize IDPs inside whole cells. However, the increase in linewidth when going to in-cell spectra is much more pronounced for the 1H signals with respect to 13C signals. Simultaneous acquisition of the two spectra is important to confirm that it is a real property of in-cell spectra and that it does not depend on possible changes in the experimental conditions or in-cell sample quality. Therefore, the CON//HN provides at the same time information derived from the exclusively heteronuclear spectra, characterized by narrow linewidths, as well as from proton-detected spectra, characterized by higher sensitivity but more influenced by line broadening deriving from both the inhomogeneous environment as well as from exchange processes.
The CON//15N-detected experiments
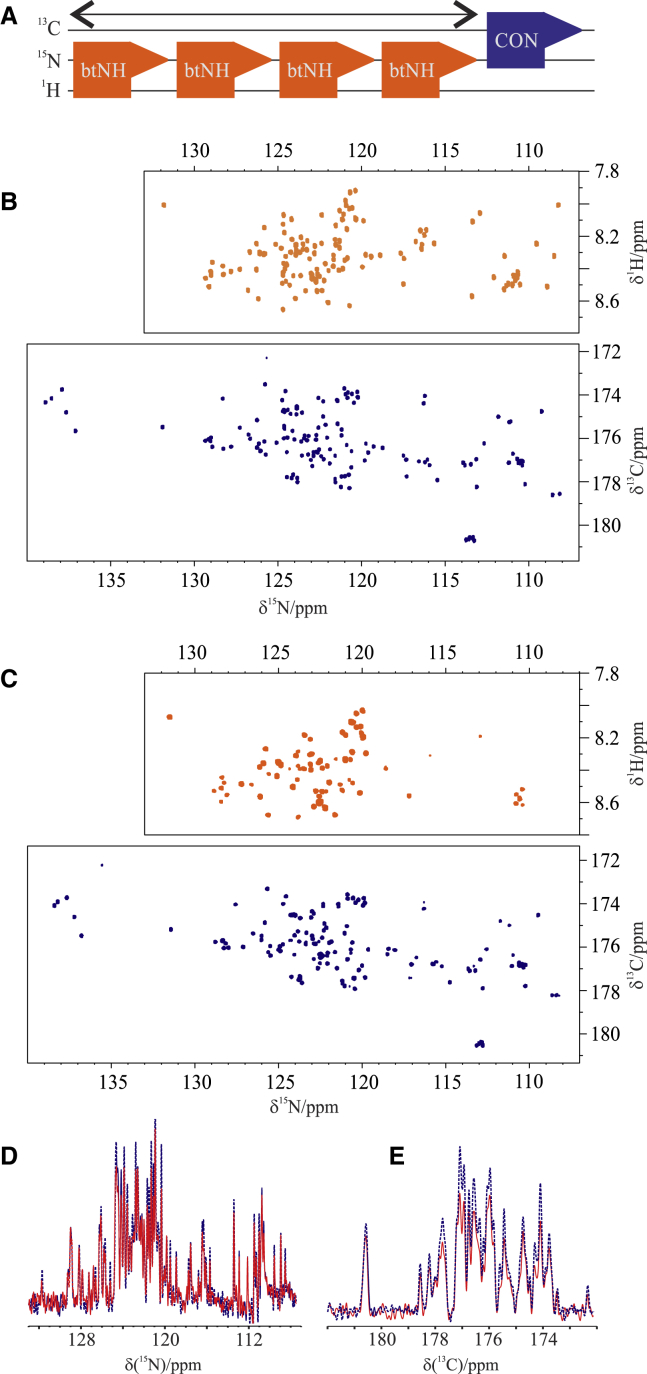
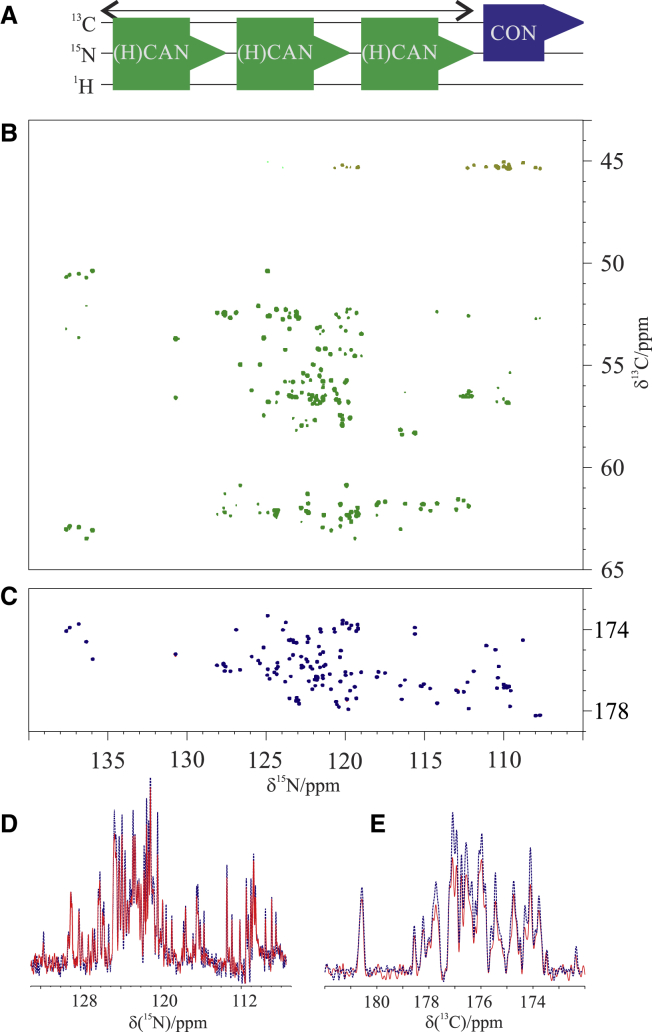
15N detection has interesting features for the study of IDPs because of the narrow linewidths and large dispersion of chemical shifts, provided one is in the appropriate sensitivity regime, which nowadays still requires highly concentrated samples (30, 31, 32, 33). The CON experiment was first combined with the 15N-detected experiment to collect 2D HN spectra (CON//btNH) (51). The BEST-TROSY variant (52), which uses band-selective pulses for the amide protons, was selected to exploit the very efficient longitudinal relaxation enhancement of amide protons in IDPs (53, 54, 55) as well as to avoid 1H decoupling during acquisition of the 15N FID, two features that enable the acquisition of 3–4 15N FIDs during the CON relaxation delay to overcome the very low sensitivity of 15N. Decoupling of 1H during the 15N evolution delay of the CON was achieved by pairs of 180° HN band-selective pulses to restore the starting polarization source for the BEST-TROSY experiments. In addition, there is no proton decoupling during the acquisition of the CON. These two features reintroduce some dependence on solvent exchange processes as well as C′-HN couplings in the direct acquisition dimension. The major modification included in the BEST-TROSY part of the experiment was to avoid composite pulse decoupling of carbonyl carbon nuclear spins during the acquisition of the 15N FID to preserve C′ polarization for the following CON experiment. To this end, virtual decoupling of C′ during the acquisition of the 15N FID was implemented through the IPAP approach (56) in combination with band-selective decoupling of the Cα spins. The pulse sequence is reported in Fig. S2 and schematically illustrated in Fig. 3 A. The results of the CON//btNH experiments recorded on α-synuclein at different temperatures are reported in Fig. 3, B and C. The good performance of the experiment is shown by the well-resolved spectra that can be acquired; essentially all expected correlations can be detected at 285 K and pH 6.5 (138 detected backbone correlations for the CON and 133 for the btNH; 99.3% of the expected ones for each spectrum). The modifications introduced to acquire the two spectra simultaneously slightly reduce the sensitivity of the CON spectrum, which is, however, the more sensitive of the two and thus only has a minor impact on the final outcome (Fig. 3, D and E). The 15N-detected BEST-TROSY HN spectrum, although less influenced by exchange broadening of amide protons than its 1H-detected counterpart, still relies on amide protons in the initial coherence transfer step as well as in the 1H chemical shift evolution achieved in the indirect dimension. As expected, with increasing temperature, the quality of the CON is essentially maintained, whereas the intensity of crosspeaks in the 15N-detected BEST-TROSY spectrum is reduced. Still, it can be observed that even at higher temperature than physiological one (315 K), a few additional crosspeaks can be observed in the 15N-detected BEST-TROSY HN spectrum that were not identified in these conditions through 1H-detected experiments (Biological Magnetic Resonance Bank [BMRB]: 27348) (57). In particular, in the 1H-detected experiment, 93/134 peaks were visible, whereas when performing the 15N-detected variant, seven additional peaks are visible (thus 100/134), mostly due to polar residues (Thr-22, -64, -44, -33, Ser-9, Lys-43, Ala-76). Even if the signal intensity is lower in the 15N-detected experiment because of the lower gyromagnetic ratio of 15N, it is possible to collect a few more signals because the proton magnetization is transverse for a shorter amount of time, reducing losses. This shows that the method has some potential, in particular using optimized hardware (high fields and optimized probes). Another experiment based on 15N detection that was combined with the CON is the (H)CAN (58). The CON//(H)CAN acquired at 310 K and pH 7.4 is shown in Fig. 4. The pulse sequence is reported in Fig. S3 and schematically illustrated in Fig. 4 A. As for the previous experiment, this magnetization transfer pathway starts from a proton, the Hα, and is then transferred to the Cα. The Cα dimension evolves as the indirect one. The magnetization can now be transferred to N of the same amino acid, through the 1J constant, and to the previous one, through the 2J constant. The two constants are similar and it is thus difficult to distinguish one pathway from the other. However, we decided to prioritize the intraresidue correlations, which are more sensitive and provide complementary information, with respect to the one available through the CON experiment. When exploiting Hα as starting polarization source, the results are optimal at any temperature because of the nonexchangeable property of this atom. Longitudinal relaxation enhancements are expected to be modest for Hα in IDPs (55), and thus nonselective 1H pulses can be used. More than one repetition of this experiment during the CON relaxation delay can be collected thanks to the relatively short time needed for Hα to recover to the steady state (hundreds of milliseconds). Technical compromises needed in this case are very similar to those discussed for the previous experiment. They are responsible for a small reduction in the sensitivity of the CON (Fig. 4, D and E), which, however, is the more sensitive between the two. Decoupling of 1H and 13C was achieved by pairs of 180° pulses to restore the magnetization to equilibrium for subsequent experiments. For the same reason, a virtual CO decoupling scheme is required for the acquisition of 15N FIDs. To this end, we implemented the IPAP scheme, in combination with band-selective decoupling of the Cα spins.
Figure 3.
(A) Scheme of the CON//btNH experiment. The 2D spectra (btNH top; CON bottom) acquired with the CON//btNH experiment on 13C,15N-labeled α-synuclein 1 mM at (B) 285 K and (C) 315 K at pH 6.5 in the same buffer reported in Fig. 1 are shown. (D) and (E) report the comparison of the S/N of the projection of the spectra acquired with the CON//btNH experiment (red solid traces) with the analogous spectra acquired independently (blue dotted traces). To see this figure in color, go online.
Figure 4.
(A) Scheme of the CON//(H)CAN experiment. (B) (H)CAN and (C) CON spectra recorded on 13C, 15N-labeled α-synuclein at 310 K (pH 7.4) are shown. (D) and (E) report the comparison of the S/N of the projection of the spectra acquired with the CON//(H)CAN experiment (red solid traces) with the analogous spectra acquired independently (blue dotted traces). To see this figure in color, go online.
In this spectrum, all the correlations of the intraresidue crosspeaks are observed as well as part of the interresidue ones (113/139; 81.0%). However, significant overlap still occurs, and only a fraction of them are resolved. It is worth noting however that with increasing magnetic fields and with tailored probes, the CON//(H)CAN becomes a promising tool for the study of IDPs. Finally, should long experimental times be necessary to compensate for a lower 15N sensitivity, the CON//(H)CAN experiment can reveal weak crosspeaks present in the CON such as those deriving from the cis-trans proline isomerization, which has not been extensively studied for IDPs and has been found to be important in many regulatory processes.
Comparison with other methods
Here, we have chosen to opt for the most straightforward approach to combine two experiments: instead of simply waiting for the magnetization to recover before the next transient, we use the recovery delay of one particularly useful experiment, which would be acquired anyway, to collect an additional one providing complementary information for free. This method follows the idea of activating unexploited magnetization (UTOPIA) (34) but shifts the central interest to the 13C-detected experiment, as schematically indicated in Fig. 1. The simplest variant of the CON experiment was selected because it represents the most robust one to ensure detection of all signals of an IDP near physiological conditions, including those that are invisible in HN spectra (residues experiencing fast solvent exchange and proline residues). This was combined either with 1H-detected or with 15N-detected 2D experiments.
In principle, a 3D HNCO (or a 3D HNCA), which can nowadays be collected in a very quick time thanks to projection spectroscopy (59) or nonuniform acquisition and processing strategies (60, 61), could provide very similar information. However, the results of these 3D spectra depend on the observability of amide protons. Therefore, the approach presented here is more suitable to study IDPs approaching physiological conditions (310 K; pH 7.4; ionic strength value around 0.25 M; in-cell environment) because the CON is not affected by fast solvent exchange processes.
Similar arguments hold when comparing the current approach with an elegant experimental variant of the HNCO proposed for parallel acquisition of the HN and (HN)CON experiments (33). In this case, the two spectra (HN and (HN)CON) are obtained by recovering magnetization from two different pathways that however both originate from HN polarization. As a result, the two spectra are influenced by solvent exchange broadening, and the (HN)CON does not reveal proline signals.
The relative sensitivity of the two experiments to be combined is an important issue to be considered. There is no doubt that the intrinsic sensitivity of 13C is lower than that of 1H because of the lower gyromagnetic ratio. However, thanks to instrumental improvements, the sensitivity of the CON experiments acquired on highly flexible IDPs is sufficient to collect the experiment with just a few scans per increment. Therefore, it is worth combining it with the other experiment that is generally collected on an IDP, the HN correlation spectrum. The relative sensitivity of the two experiments is inverted when combining the CON with 15N-detected experiments. This difference can be partly compensated by exploiting 1H as a starting polarization source, exploiting longitudinal relaxation enhancement (when possible) (49, 51, 52, 55), or accumulating more 15N FIDs. A leap in instrumental sensitivity exploiting dedicated probe heads will provide the necessary push to make this combination of experiments really amenable for every application.
From this analysis, it is clear that more than for time savings, which are generally moderate when considering combinations of simple 2D experiments, the simultaneous acquisition of two spectra is important for accessing a snapshot of an IDP in cases in which the sample either has limited stability in time or it changes because of chemical reactions as it happens when studying posttranslational modifications (PTMs). In these cases, to investigate the IDP of interest, in addition to the CON one can access complementary information through the multiple receiver CON variants proposed here.
A case study for PTMs
PTMs represent the most efficient way to modulate the biological activity of proteins after their biosynthesis through chemical reactions, catalyzed by enzymes. Indeed, it is possible to modulate the chemo-physical properties of a protein and thus its function by adding or removing specific chemical groups or through the cleavage of chemical bonds. The diversity of PTMs found in proteins arises from the very different processes that they can modulate in a cell such as protein folding, signaling, molecular recognition, interactions, and degradation. In this way, the same protein can be handled for different aims, ranging from cell fate control to regulation of metabolism (62).
IDPs are frequently involved in regulatory and signaling functions, which can be modulated by PTMs (63, 64, 65). PTMs can trigger both local and long-range changes as well as intermolecular interactions. It has been supposed that structural flexibility could also be a biological strategy to overcome the classical problem of the “one lock/one key” model. The lack of a stable 3D structure guarantees adaptability for the enzymatic site and allows high specificity of the process, maintaining, however, low affinity (64, 66).
Among a variety of possible modifications, one of the most common is phosphorylation. It was estimated that at least 2% of the human genome is constituted by kinases and more than 30% of the eukaryotic proteins are involved in phosphorylation, with 700,000 sites that can potentially act as substrates (67). Investigating this mechanism is fundamental to understand functions of these important enzymes. PTMs usually involve only a few residues, and NMR provides a unique tool to study these processes, giving access to residue-specific atomic resolution information.
It is well known that the role of some IDPs or IDRs and their link to the onset of several diseases is strictly dependent on PTMs, as phosphorylation (64, 65, 66, 68). A clear example is the supposed key role of phosphorylation of specific α-synuclein residues in the modulation of Lewy bodies formation. In this widely studied process, serine 129 is selectively and extensively phosphorylated, promoting fibrillation in vitro (69, 70, 71). It has emerged that this process occurs together with phosphorylation of tyrosine 125, a priming event in the efficient modification of serine 129 by CK1 kinase, both in vitro under physiological conditions and in vivo (72, 73, 74).
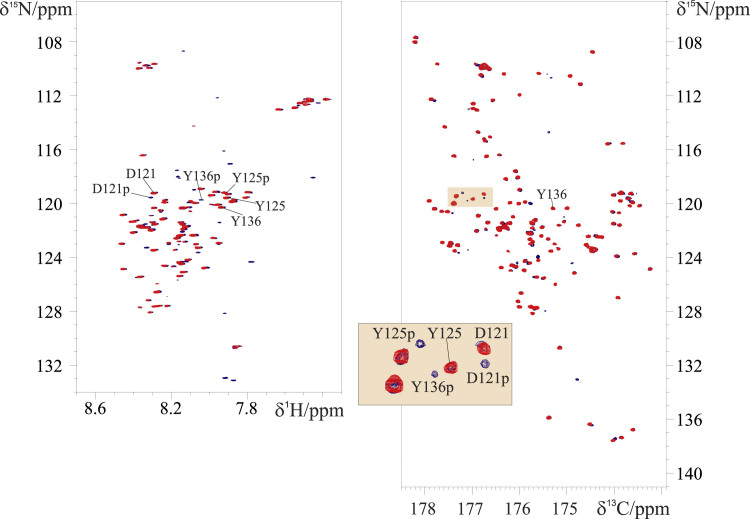
To provide an example of the utility of the CON//HN experiment, we show here the phosphorylation reaction of tyrosine residues by Fyn near physiological conditions. Fyn is a nonreceptor tyrosine kinase of the Src family that has been shown to react with all the four tyrosine residues present in α-synuclein (Y39, Y125, Y133, Y136) with high-percentage levels but preferentially targeting Y125. The spectra acquired with the CON//HN experiment during the phosphorylation reaction are shown in Fig. 5. It can be observed that the changes in HN crosspeaks agree with previous observations (73) and that additional details are monitored through the CON spectrum acquired simultaneously (Fig. 5). In particular, the CON spectrum, which exploits the 13C in the direct dimension, has a higher resolution with respect to the HN spectrum. Thus, we can also monitor the shift of those peaks, which, in the HN spectrum, are found in crowded regions such as those of Tyr125, which is well resolved in the CON (Fig. 5).
Figure 5.
The 2D spectra acquired with the CON//HN experiment to monitor phosphorylation of 13C, 15N-labeled α-synuclein 200 μM with Fyn tyrosine kinase as a function of time (t = 0 h, red spectra; t = 50 h blue spectra). The HN spectra are reported on the left and the CON on the right. The highlighted region of the CON spectra is enlarged to show, as an example, a subset of α-synuclein signals that are influenced by phosphorylation. The reaction was performed at 303 K in 20 mM phosphate buffer, 100 mM NaCl, 50 μM EDTA, 2 mM dithiothreitol, 6 mM MgCl2, and 3 mM ATP (pH 7.0). To see this figure in color, go online.
A final comment is necessary on the specific conditions needed to follow PTMs. Most enzymes indeed fulfill their roles at physiological conditions. Therefore, the CON//HN experiment offers a particularly useful tool because in addition to the HN, often used to study PTMs, it exploits the CON, which is not influenced by exchange broadening because of the chemical properties of the nuclei participating in the magnetization transfer pathway. The fairly low substrate concentration generally required for these reactions is still sufficient to acquire good quality 13C-detected spectra, demonstrating the general applicability of this approach.
Conclusion
Research in the area of IDPs is continuously expanding. Partly or completely disordered proteins of increasing size and complexity and with high biomedical relevance are being discovered at fast pace, adding a novel layer to structural biology. In this frame, NMR spectroscopy has a central role, enabling the investigation of systems of increasing complexity in an environment that resembles more and more the physiological one. Heteronuclear direct detection has contributed to establishing the importance of NMR in this area of research. The possibility to collect two spectra simultaneously is very attractive to study IDPs. Several experimental variants are presented here in which during the recovery delay needed to acquire the CON, an additional 2D spectrum is collected either based on 1H detection (CON//HN) or on 15N detection (CON//btNH, CON//(H)CAN). This, to our knowledge, novel approach is particularly useful to study samples of limited lifetime, such as IDPs inside whole cells or cell lysates, as well as to follow chemical reactions such as PTMs of IDPs in conditions approaching physiological ones (pH, temperature, ionic strength, inhomogeneous in-cell environment). The availability of cryogenically cooled probeheads optimized for heteronuclear direct detection is important to alleviate the problem of the intrinsic lower sensitivity of heteronuclei. The usefulness of these experiments is bound to increase with increasing magnetic fields to take maximal advantage of the narrow linewidths of C′ and N of highly flexible IDPs, enabling the study of IDPs of increasing complexity.
Author Contributions
I.C.F. and R.P. designed the research. All the authors performed research, analyzed the data, discussed the results, and wrote the manuscript.
Acknowledgments
The support and the use of resources of the CERM/CIRMMP center of Instruct-ERIC, a Landmark European Strategy Forum on Research Infrastructures project, is gratefully acknowledged. Dr. F. X. Theillet, and Dr. A. Binolfi are gratefully acknowledged for the stimulating discussions.
This work has been supported in part by a grant from the Fondazione CR Firenze and by a grant from the Italian Ministry of Foreign Affairs and International Cooperation (MAE0057283) to R.P.
Editor: David Eliezer.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2019.05.017.
Contributor Information
Roberta Pierattelli, Email: roberta.pierattelli@unifi.it.
Isabella C. Felli, Email: felli@cerm.unifi.it.
Supporting Material
References
- 1.Habchi J., Tompa P., Uversky V.N. Introducing protein intrinsic disorder. Chem. Rev. 2014;114:6561–6588. doi: 10.1021/cr400514h. [DOI] [PubMed] [Google Scholar]; Habchi, J., P. Tompa, …, V. N. Uversky. 2014. Introducing protein intrinsic disorder. Chem. Rev. 114:6561-6588. [DOI] [PubMed]
- 2.Wright P.E., Dyson H.J. Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 2015;16:18–29. doi: 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wright, P. E., and H. J. Dyson. 2015. Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 16:18-29. [DOI] [PMC free article] [PubMed]
- 3.van der Lee R., Buljan M., Babu M.M. Classification of intrinsically disordered regions and proteins. Chem. Rev. 2014;114:6589–6631. doi: 10.1021/cr400525m. [DOI] [PMC free article] [PubMed] [Google Scholar]; van der Lee, R., M. Buljan, …, M. M. Babu. 2014. Classification of intrinsically disordered regions and proteins. Chem. Rev. 114:6589-6631. [DOI] [PMC free article] [PubMed]
- 4.Uversky V.N., Oldfield C.J., Dunker A.K. Intrinsically disordered proteins in human diseases: introducing the D2 concept. Annu. Rev. Biophys. 2008;37:215–246. doi: 10.1146/annurev.biophys.37.032807.125924. [DOI] [PubMed] [Google Scholar]; Uversky, V. N., C. J. Oldfield, and A. K. Dunker. 2008. Intrinsically disordered proteins in human diseases: introducing the D2 concept. Annu. Rev. Biophys. 37:215-246. [DOI] [PubMed]
- 5.Ambadipudi S., Zweckstetter M. Targeting intrinsically disordered proteins in rational drug discovery. Expert Opin. Drug Discov. 2016;11:65–77. doi: 10.1517/17460441.2016.1107041. [DOI] [PubMed] [Google Scholar]; Ambadipudi, S., and M. Zweckstetter. 2016. Targeting intrinsically disordered proteins in rational drug discovery. Expert Opin. Drug Discov. 11:65-77. [DOI] [PubMed]
- 6.Joshi P., Chia S., Vendruscolo M. A fragment-based method of creating small-molecule libraries to target the aggregation of intrinsically disordered proteins. ACS Comb. Sci. 2016;18:144–153. doi: 10.1021/acscombsci.5b00129. [DOI] [PubMed] [Google Scholar]; Joshi, P., S. Chia, …, M. Vendruscolo. 2016. A fragment-based method of creating small-molecule libraries to target the aggregation of intrinsically disordered proteins. ACS Comb. Sci. 18:144-153. [DOI] [PubMed]
- 7.Heller G.T., Aprile F.A., Vendruscolo M. Methods of probing the interactions between small molecules and disordered proteins. Cell. Mol. Life Sci. 2017;74:3225–3243. doi: 10.1007/s00018-017-2563-4. [DOI] [PMC free article] [PubMed] [Google Scholar]; Heller, G. T., F. A. Aprile, and M. Vendruscolo. 2017. Methods of probing the interactions between small molecules and disordered proteins. Cell. Mol. Life Sci. 74:3225-3243. [DOI] [PMC free article] [PubMed]
- 8.Tsafou K., Tiwari P.B., Toretsky J.A. Targeting intrinsically disordered transcription factors: changing the paradigm. J. Mol. Biol. 2018;430:2321–2341. doi: 10.1016/j.jmb.2018.04.008. [DOI] [PubMed] [Google Scholar]; Tsafou, K., P. B. Tiwari, …, J. A. Toretsky. 2018. Targeting intrinsically disordered transcription factors: changing the paradigm. J. Mol. Biol. 430:2321-2341. [DOI] [PubMed]
- 9.Eliezer D. Biophysical characterization of intrinsically disordered proteins. Curr. Opin. Struct. Biol. 2009;19:23–30. doi: 10.1016/j.sbi.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; Eliezer, D. 2009. Biophysical characterization of intrinsically disordered proteins. Curr. Opin. Struct. Biol. 19:23-30. [DOI] [PMC free article] [PubMed]
- 10.Felli I.C., Pierattelli R., editors. Intrinsically Disordered Proteins Studied by NMR Spectroscopy. Springer; Basel, Switzerland: 2015. [Google Scholar]; Felli, I. C., and R. Pierattelli, eds. 2015. Intrinsically Disordered Proteins Studied by NMR Spectroscopy; Springer, Basel, Switzerland.
- 11.Gil S., Hošek T., Felli I.C. NMR spectroscopic studies of intrinsically disordered proteins at near-physiological conditions. Angew. Chem. Int. Ed. Engl. 2013;52:11808–11812. doi: 10.1002/anie.201304272. [DOI] [PubMed] [Google Scholar]; Gil, S., T. Hošek, …, I. C. Felli. 2013. NMR spectroscopic studies of intrinsically disordered proteins at near-physiological conditions. Angew. Chem. Int. Ed. Engl. 52:11808-11812. [DOI] [PubMed]
- 12.Dyson H.J., Wright P.E. Nuclear magnetic resonance methods for elucidation of structure and dynamics in disordered states. Methods Enzymol. 2001;339:258–270. doi: 10.1016/s0076-6879(01)39317-5. [DOI] [PubMed] [Google Scholar]; Dyson, H. J., and P. E. Wright. 2001. Nuclear magnetic resonance methods for elucidation of structure and dynamics in disordered states. Methods Enzymol. 339:258-270. [DOI] [PubMed]
- 13.Konrat R. NMR contributions to structural dynamics studies of intrinsically disordered proteins. J. Magn. Reson. 2014;241:74–85. doi: 10.1016/j.jmr.2013.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]; Konrat, R. 2014. NMR contributions to structural dynamics studies of intrinsically disordered proteins. J. Magn. Reson. 241:74-85. [DOI] [PMC free article] [PubMed]
- 14.Brutscher B., Felli I.C., Sólyom Z. NMR methods for the study of instrinsically disordered proteins structure, dynamics, and interactions: general overview and practical guidelines. Adv. Exp. Med. Biol. 2015;870:49–122. doi: 10.1007/978-3-319-20164-1_3. [DOI] [PubMed] [Google Scholar]; Brutscher, B., I. C. Felli, …, Z. Solyom. 2015. NMR methods for the study of instrinsically disordered proteins structure, dynamics, and interactions: general overview and practical guidelines. Adv. Exp. Med. Biol. 870:49-122. [DOI] [PubMed]
- 15.Bermel W., Bertini I., Pierattelli R. Protonless NMR experiments for sequence-specific assignment of backbone nuclei in unfolded proteins. J. Am. Chem. Soc. 2006;128:3918–3919. doi: 10.1021/ja0582206. [DOI] [PubMed] [Google Scholar]; Bermel, W., I. Bertini, …, R. Pierattelli. 2006. Protonless NMR experiments for sequence-specific assignment of backbone nuclei in unfolded proteins. J. Am. Chem. Soc. 128:3918-3919. [DOI] [PubMed]
- 16.Bermel W., Bertini I., Pierattelli R. Novel 13C direct detection experiments, including extension to the third dimension, to perform the complete assignment of proteins. J. Magn. Reson. 2006;178:56–64. doi: 10.1016/j.jmr.2005.08.011. [DOI] [PubMed] [Google Scholar]; Bermel, W., I. Bertini, …, R. Pierattelli. 2006. Novel 13C direct detection experiments, including extension to the third dimension, to perform the complete assignment of proteins. J. Magn. Reson. 178:56-64. [DOI] [PubMed]
- 17.Pérez Y., Gairí M., Bernadó P. Structural characterization of the natively unfolded N-terminal domain of human c-Src kinase: insights into the role of phosphorylation of the unique domain. J. Mol. Biol. 2009;391:136–148. doi: 10.1016/j.jmb.2009.06.018. [DOI] [PubMed] [Google Scholar]; Perez, Y., M. Gairi, …, P. Bernado. 2009. Structural characterization of the natively unfolded N-terminal domain of human c-Src kinase: insights into the role of phosphorylation of the unique domain. J. Mol. Biol. 391:136-148. [DOI] [PubMed]
- 18.Knoblich K., Whittaker S., Günther U. Backbone assignment of the N-terminal polyomavirus large T antigen. Biomol. NMR Assign. 2009;3:119–123. doi: 10.1007/s12104-009-9155-7. [DOI] [PubMed] [Google Scholar]; Knoblich, K., S. Whittaker, …, U. Gunther. 2009. Backbone assignment of the N-terminal polyomavirus large T antigen. Biomol. NMR Assign. 3:119-123. [DOI] [PubMed]
- 19.Contreras-Martos S., Piai A., Tompa P. Linking functions: an additional role for an intrinsically disordered linker domain in the transcriptional coactivator CBP. Sci. Rep. 2017;7:4676. doi: 10.1038/s41598-017-04611-x. [DOI] [PMC free article] [PubMed] [Google Scholar]; Contreras-Martos, S., A. Piai, …, P. Tompa. 2017. Linking functions: an additional role for an intrinsically disordered linker domain in the transcriptional coactivator CBP. Sci. Rep. 7:4676. [DOI] [PMC free article] [PubMed]
- 20.Hošek T., Calçada E.O., Pierattelli R. Structural and dynamic characterization of the molecular hub early region 1A (E1A) from human adenovirus. Chemistry. 2016;22:13010–13013. doi: 10.1002/chem.201602510. [DOI] [PubMed] [Google Scholar]; Hošek, T., E. O. Calçada, …, R. Pierattelli. 2016. Structural and dynamic characterization of the molecular hub early region 1A (E1A) from human adenovirus. Chemistry. 22:13010-13013. [DOI] [PubMed]
- 21.Motáčková V., Nováček J., Sklenář V. Strategy for complete NMR assignment of disordered proteins with highly repetitive sequences based on resolution-enhanced 5D experiments. J. Biomol. NMR. 2010;48:169–177. doi: 10.1007/s10858-010-9447-3. [DOI] [PMC free article] [PubMed] [Google Scholar]; Motačkova, V., J. Novaček, …, V. Sklenař. 2010. Strategy for complete NMR assignment of disordered proteins with highly repetitive sequences based on resolution-enhanced 5D experiments. J. Biomol. NMR. 48:169-177. [DOI] [PMC free article] [PubMed]
- 22.Haba N.Y., Gross R., Chill J.H. NMR determines transient structure and dynamics in the disordered C-terminal domain of WASp interacting protein. Biophys. J. 2013;105:481–493. doi: 10.1016/j.bpj.2013.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]; Haba, N. Y., R. Gross, …, J. H. Chill. 2013. NMR determines transient structure and dynamics in the disordered C-terminal domain of WASp interacting protein. Biophys. J. 105:481-493. [DOI] [PMC free article] [PubMed]
- 23.Nováček J., Haba N.Y., Sklenář V. 4D non-uniformly sampled HCBCACON and 1J(NCα)-selective HCBCANCO experiments for the sequential assignment and chemical shift analysis of intrinsically disordered proteins. J. Biomol. NMR. 2012;53:139–148. doi: 10.1007/s10858-012-9631-8. [DOI] [PubMed] [Google Scholar]; Novaček, J., N. Y. Haba, …, V. Sklenař. 2012. 4D non-uniformly sampled HCBCACON and 1J(NCα)-selective HCBCANCO experiments for the sequential assignment and chemical shift analysis of intrinsically disordered proteins. J. Biomol. NMR. 53:139-148. [DOI] [PubMed]
- 24.Lawrence C.W., Showalter S.A. Carbon-detected 15N NMR spin relaxation of an intrinsically disordered protein: FCP1 dynamics unbound and in complex with RAP74. J. Phys. Chem. Lett. 2012;3:1409–1413. doi: 10.1021/jz300432e. [DOI] [PubMed] [Google Scholar]; Lawrence, C. W., and S. A. Showalter. 2012. Carbon-detected (15)N NMR spin relaxation of an intrinsically disordered protein: FCP1 dynamics unbound and in complex with RAP74. J. Phys. Chem. Lett. 3:1409-1413. [DOI] [PubMed]
- 25.Sahu D., Bastidas M., Showalter S.A. Generating NMR chemical shift assignments of intrinsically disordered proteins using carbon-detected NMR methods. Anal. Biochem. 2014;449:17–25. doi: 10.1016/j.ab.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sahu, D., M. Bastidas, and S. A. Showalter. 2014. Generating NMR chemical shift assignments of intrinsically disordered proteins using carbon-detected NMR methods. Anal. Biochem. 449:17-25. [DOI] [PMC free article] [PubMed]
- 26.Lopez J., Schneider R., Lippens G. Studying intrinsically disordered proteins under true in vivo conditions by combined cross-polarization and carbonyl-detection NMR spectroscopy. Angew. Chem. Int.Engl. 2016;55:7418–7422. doi: 10.1002/anie.201601850. [DOI] [PubMed] [Google Scholar]; Lopez, J., R. Schneider, …, G. Lippens. 2016. Studying intrinsically disordered proteins under true in vivo conditions by combined cross-polarization and carbonyl-detection NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 55:7418-7422. [DOI] [PubMed]
- 27.Piai A., Calçada E.O., Pierattelli R. Just a flexible linker? The structural and dynamic properties of CBP-ID4 revealed by NMR spectroscopy. Biophys. J. 2016;110:372–381. doi: 10.1016/j.bpj.2015.11.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]; Piai, A., E. O. Calçada, …, R. Pierattelli. 2016. Just a flexible linker? The structural and dynamic properties of CBP-ID4 revealed by NMR spectroscopy. Biophys. J. 110:372-381. [DOI] [PMC free article] [PubMed]
- 28.Eftekharzadeh B., Piai A., Salvatella X. Sequence context influences the structure and aggregation behavior of a PolyQ tract. Biophys. J. 2016;110:2361–2366. doi: 10.1016/j.bpj.2016.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]; Eftekharzadeh, B., A. Piai, …, X. Salvatella. 2016. Sequence context influences the structure and aggregation behavior of a PolyQ tract. Biophys. J. 110:2361-2366. [DOI] [PMC free article] [PubMed]
- 29.Kovacs H., Moskau D., Spraul M. Cryogenically cooled probes - a leap in NMR technology. Prog. Nucl. Mag. Res. Sp. 2005;46:131–155. [Google Scholar]; Kovacs, H., D. Moskau, and M. Spraul. 2005. Cryogenically cooled probes - a leap in NMR technology. Prog. Nucl. Mag. Res. Sp. 46:131-155.
- 30.Takeuchi K., Heffron G., Wagner G. Nitrogen-detected CAN and CON experiments as alternative experiments for main chain NMR resonance assignments. J. Biomol. NMR. 2010;47:271–282. doi: 10.1007/s10858-010-9430-z. [DOI] [PMC free article] [PubMed] [Google Scholar]; Takeuchi, K., G. Heffron, …, G. Wagner. 2010. Nitrogen-detected CAN and CON experiments as alternative experiments for main chain NMR resonance assignments. J. Biomol. NMR. 47:271-282. [DOI] [PMC free article] [PubMed]
- 31.Takeuchi K., Arthanari H., Wagner G. Perspective: revisiting the field dependence of TROSY sensitivity. J. Biomol. NMR. 2016;66:221–225. doi: 10.1007/s10858-016-0075-4. [DOI] [PMC free article] [PubMed] [Google Scholar]; Takeuchi, K., H. Arthanari, and G. Wagner. 2016. Perspective: revisiting the field dependence of TROSY sensitivity. J. Biomol. NMR. 66:221-225. [DOI] [PMC free article] [PubMed]
- 32.Chhabra S., Fischer P., Arthanari H. 15N detection harnesses the slow relaxation property of nitrogen: delivering enhanced resolution for intrinsically disordered proteins. Proc. Nat. Acad. Sci. USA. 2018;115:1710–1719. doi: 10.1073/pnas.1717560115. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chhabra, S., P. Fischer, …, H. Arthanari. 2018. 15N detection harnesses the slow relaxation property of nitrogen: delivering enhanced resolution for intrinsically disordered proteins. Proc. Nat. Acad. Sci. USA. 115:1710-1719. [DOI] [PMC free article] [PubMed]
- 33.Gibbs E.B., Kriwacki R.W. Direct detection of carbon and nitrogen nuclei for high-resolution analysis of intrinsically disordered proteins using NMR spectroscopy. Methods. 2018;138-139:39–46. doi: 10.1016/j.ymeth.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gibbs, E. B., and R. W. Kriwacki. 2018. Direct detection of carbon and nitrogen nuclei for high-resolution analysis of intrinsically disordered proteins using NMR spectroscopy. Methods. 138-139:39-46. [DOI] [PMC free article] [PubMed]
- 34.Kupče E., Freeman R., John B.K. Parallel acquisition of two-dimensional NMR spectra of several nuclear species. J. Am. Chem. Soc. 2006;128:9606–9607. doi: 10.1021/ja0634876. [DOI] [PubMed] [Google Scholar]; Kupče, E., R. Freeman, and B. K. John. 2006. Parallel acquisition of two-dimensional NMR spectra of several nuclear species. J. Am. Chem. Soc. 128:9606-9607. [DOI] [PubMed]
- 35.Kupče E., Freeman R. Molecular structure from a single NMR sequence (fast-PANACEA) J. Magn. Reson. 2010;206:147–153. doi: 10.1016/j.jmr.2010.06.018. [DOI] [PubMed] [Google Scholar]; Kupče, E., and R. Freeman. 2010. Molecular structure from a single NMR sequence (fast-PANACEA). J. Magn. Reson. 206:147-153. [DOI] [PubMed]
- 36.Chakraborty S., Paul S., Hosur R.V. Simultaneous acquisition of 13Cα-15N and 1H-15N-15N sequential correlations in proteins: application of dual receivers in 3D HNN. J. Biomol. NMR. 2012;52:5–10. doi: 10.1007/s10858-011-9596-z. [DOI] [PubMed] [Google Scholar]; Chakraborty, S., S. Paul, and R. V. Hosur. 2012. Simultaneous acquisition of 13Cα-15N and 1H-15N-15N sequential correlations in proteins: application of dual receivers in 3D HNN. J. Biomol. NMR. 52:5-10. [DOI] [PubMed]
- 37.Kupče E., Kay L.E. Parallel acquisition of multi-dimensional spectra in protein NMR. J. Biomol. NMR. 2012;54:1–7. doi: 10.1007/s10858-012-9646-1. [DOI] [PubMed] [Google Scholar]; Kupče, E., and L. E. Kay. 2012. Parallel acquisition of multi-dimensional spectra in protein NMR. J. Biomol. NMR. 54:1-7. [DOI] [PubMed]
- 38.Viegas A., Viennet T., Etzkorn M. UTOPIA NMR: activating unexploited magnetization using interleaved low-gamma detection. J. Biomol. NMR. 2016;64:9–15. doi: 10.1007/s10858-015-0008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; Viegas, A., T. Viennet, …, M. Etzkorn. 2016. UTOPIA NMR: activating unexploited magnetization using interleaved low-gamma detection. J. Biomol. NMR. 64:9-15. [DOI] [PMC free article] [PubMed]
- 39.Kupče E. NMR with multiple receivers. Top. Curr. Chem. 2013;335:71–96. doi: 10.1007/128_2011_226. [DOI] [PubMed] [Google Scholar]; Kupče, E. 2013. NMR with multiple receivers. Top. Curr. Chem. 335:71-96. [DOI] [PubMed]
- 40.Sharma K., Madhu P.K., Mote K.R. A suite of pulse sequences based on multiple sequential acquisitions at one and two radiofrequency channels for solid-state magic-angle spinning NMR studies of proteins. J. Biomol. NMR. 2016;65:127–141. doi: 10.1007/s10858-016-0043-z. [DOI] [PubMed] [Google Scholar]; Sharma, K., P. K. Madhu, and K. R. Mote. 2016. A suite of pulse sequences based on multiple sequential acquisitions at one and two radiofrequency channels for solid-state magic-angle spinning NMR studies of proteins. J. Biomol. NMR. 65:127-141. [DOI] [PubMed]
- 41.Kupče Ē., Kay L.E., Freeman R. Detecting the “afterglow” of 13C NMR in proteins using multiple receivers. J. Am. Chem. Soc. 2010;132:18008–18011. doi: 10.1021/ja1080025. [DOI] [PubMed] [Google Scholar]; Kupče, Ē., L. E. Kay, and R. Freeman. 2010. Detecting the “afterglow” of 13C NMR in proteins using multiple receivers. J. Am. Chem. Soc. 132:18008-18011. [DOI] [PubMed]
- 42.Huang C., Ren G., Wang C.C. A new method for purification of recombinant human α-synuclein in Escherichia coli. Protein Expr. Purif. 2005;42:173–177. doi: 10.1016/j.pep.2005.02.014. [DOI] [PubMed] [Google Scholar]; Huang, C., G. Ren, …, C. C. Wang. 2005. A new method for purification of recombinant human α-synuclein in Escherichia coli. Protein Expr. Purif. 42:173-177. [DOI] [PubMed]
- 43.Felli I.C., Gonnelli L., Pierattelli R. In-cell 13C NMR spectroscopy for the study of intrinsically disordered proteins. Nat. Protoc. 2014;9:2005–2016. doi: 10.1038/nprot.2014.124. [DOI] [PubMed] [Google Scholar]; Felli, I. C., L. Gonnelli, and R. Pierattelli. 2014. In-cell 13C NMR spectroscopy for the study of intrinsically disordered proteins. Nat. Protoc. 9:2005-2016. [DOI] [PubMed]
- 44.Emsley L., Bodenhausen G. Optimization of shaped selective pulses for NMR using a quaternion description of their overall propagators. J. Magn. Reson. 1992;97:135–148. [Google Scholar]; Emsley, L., and G. Bodenhausen. 1992. Optimization of shaped selective pulses for NMR using a quaternion description of their overall propagators. J. Magn. Reson. 97:135-148.
- 45.Böhlen J.M., Bodenhausen G. Experimental aspects of Chirp NMR spectroscopy. J. Magn. Reson. 1993;102:293–301. [Google Scholar]; Bohlen, J. M., and G. Bodenhausen. 1993. Experimental aspects of Chirp NMR spectroscopy. J. Magn. Reson. 102:293-301.
- 46.Piotto M., Saudek V., Sklenár V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]; Piotto, M., V. Saudek, and V. Sklenar. 1992. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 2:661-665. [DOI] [PubMed]
- 47.Freeman R., Geen H. Band-selective radiofrequency pulses. J. Magn. Reson. 1991;93:93–141. [Google Scholar]; Freeman, R., and H. Geen. 1991. Band-selective radiofrequency pulses. J. Magn. Reson. 93:93-141.
- 48.Markley J.L., Bax A., Wüthrich K. Recommendations for the presentation of NMR structures of proteins and nucleic acids. Pure Appl. Chem. 1998;70:117–142. doi: 10.1006/jmbi.1998.1852. [DOI] [PubMed] [Google Scholar]; Markley, J. L., A. Bax, …, K. Wuethrich. 1998. Recommendations for the presentation of NMR structures of proteins and nucleic acids. Pure Appl. Chem. 70:117-142. [DOI] [PubMed]
- 49.Felli I.C., Pierattelli R. Spin-state-selective methods in solution- and solid-state biomolecular 13C NMR. Prog. Nucl. Magn. Reson. Spectrosc. 2015;84–85:1–13. doi: 10.1016/j.pnmrs.2014.10.001. [DOI] [PubMed] [Google Scholar]; Felli, I. C., and R. Pierattelli. 2015. Spin-state-selective methods in solution- and solid-state biomolecular 13C NMR. Prog. Nucl. Magn. Reson. Spectrosc. 84-85:1-13. [DOI] [PubMed]
- 50.Felli I.C., Pierattelli R. Novel methods based on 13C detection to study intrinsically disordered proteins. J. Magn. Reson. 2014;241:115–125. doi: 10.1016/j.jmr.2013.10.020. [DOI] [PubMed] [Google Scholar]; Felli, I. C., and R. Pierattelli. 2014. Novel methods based on (13)C detection to study intrinsically disordered proteins. J. Magn. Reson. 241:115-125. [DOI] [PubMed]
- 51.Takeuchi K., Arthanari H., Wagner G. Nitrogen detected TROSY at high field yields high resolution and sensitivity for protein NMR. J. Biomol. NMR. 2015;63:323–331. doi: 10.1007/s10858-015-9991-y. [DOI] [PMC free article] [PubMed] [Google Scholar]; Takeuchi, K., H. Arthanari, …, G. Wagner. 2015. Nitrogen detected TROSY at high field yields high resolution and sensitivity for protein NMR. J. Biomol. NMR. 63:323-331. [DOI] [PMC free article] [PubMed]
- 52.Solyom Z., Schwarten M., Brutscher B. BEST-TROSY experiments for time-efficient sequential resonance assignment of large disordered proteins. J. Biomol. NMR. 2013;55:311–321. doi: 10.1007/s10858-013-9715-0. [DOI] [PubMed] [Google Scholar]; Solyom, Z., M. Schwarten, …, B. Brutscher. 2013. BEST-TROSY experiments for time-efficient sequential resonance assignment of large disordered proteins. J. Biomol. NMR. 55:311-321. [DOI] [PubMed]
- 53.Schanda P., Forge V., Brutscher B. HET-SOFAST NMR for fast detection of structural compactness and heterogeneity along polypeptide chains. Magn. Reson. Chem. 2006;44:S177–S184. doi: 10.1002/mrc.1825. [DOI] [PubMed] [Google Scholar]; Schanda, P., V. Forge, and B. Brutscher. 2006. HET-SOFAST NMR for fast detection of structural compactness and heterogeneity along polypeptide chains. Magn. Reson. Chem. 44:S177-S184. [DOI] [PubMed]
- 54.Schanda P., Van Melckebeke H., Brutscher B. Speeding up three-dimensional protein NMR experiments to a few minutes. J. Am. Chem. Soc. 2006;128:9042–9043. doi: 10.1021/ja062025p. [DOI] [PubMed] [Google Scholar]; Schanda, P., H. Van Melckebeke, and B. Brutscher. 2006. Speeding up three-dimensional protein NMR experiments to a few minutes. J. Am. Chem. Soc. 128:9042-9043. [DOI] [PubMed]
- 55.Hošek T., Gil-Caballero S., Felli I.C. Longitudinal relaxation properties of 1HN and 1Hα determined by direct-detected 13C NMR experiments to study intrinsically disordered proteins (IDPs) J. Magn. Reson. 2015;254:19–26. doi: 10.1016/j.jmr.2015.01.017. [DOI] [PubMed] [Google Scholar]; Hošek, T., S. Gil-Caballero, …, I. C. Felli. 2015. Longitudinal relaxation properties of (1)H(N) and (1)H(α) determined by direct-detected (13)C NMR experiments to study intrinsically disordered proteins (IDPs). J. Magn. Reson. 254:19-26. [DOI] [PubMed]
- 56.Bermel W., Bertini I., Pierattelli R. 13C-detected protonless NMR spectroscopy of proteins in solution. Prog. Nucl. Mag. Res. Sp. 2006;48:25–45. [Google Scholar]; Bermel, W., I. Bertini, …, R. Pierattelli. 2006. 13C-detected protonless NMR spectroscopy of proteins in solution. Prog. Nucl. Mag. Res. Sp. 48:25-45.
- 57.Murrali M.G., Schiavina M., Felli I.C. 13C APSY-NMR for sequential assignment of intrinsically disordered proteins. J. Biomol. NMR. 2018;70:167–175. doi: 10.1007/s10858-018-0167-4. [DOI] [PubMed] [Google Scholar]; Murrali, M. G., M. Schiavina, …, I. C. Felli. 2018. 13C APSY-NMR for sequential assignment of intrinsically disordered proteins. J. Biomol. NMR. 70:167-175. [DOI] [PubMed]
- 58.Gal M., Edmonds K.A., Wagner G. Speeding up direct 15N detection: hCaN 2D NMR experiment. J. Biomol. NMR. 2011;51:497–504. doi: 10.1007/s10858-011-9580-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gal, M., K. A. Edmonds, …, G. Wagner. 2011. Speeding up direct (15)N detection: hCaN 2D NMR experiment. J. Biomol. NMR. 51:497-504. [DOI] [PMC free article] [PubMed]
- 59.Hiller S., Fiorito F., Wider G. Automated projection spectroscopy (APSY) Proc. Natl. Acad. Sci. USA. 2005;102:10876–10881. doi: 10.1073/pnas.0504818102. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hiller, S., F. Fiorito, …, G. Wider. 2005. Automated projection spectroscopy (APSY). Proc. Natl. Acad. Sci. USA. 102:10876-10881. [DOI] [PMC free article] [PubMed]
- 60.Robson S., Arthanari H., Wagner G. Nonuniform sampling for NMR spectroscopy. Methods Enzymol. 2019;614:263–291. doi: 10.1016/bs.mie.2018.09.009. [DOI] [PubMed] [Google Scholar]; Robson, S., H. Arthanari, …, G. Wagner. 2019. Nonuniform sampling for NMR spectroscopy. Methods Enzymol. 614:263-291. [DOI] [PubMed]
- 61.Kazimierczuk K., Stanek J., Koźmiński W. Random sampling in multidimensional NMR spectroscopy. Prog. Nucl. Magn. Reson. Spectrosc. 2010;57:420–434. doi: 10.1016/j.pnmrs.2010.07.002. [DOI] [PubMed] [Google Scholar]; Kazimierczuk, K., J. Stanek, …, W. Koźmiński. 2010. Random sampling in multidimensional NMR spectroscopy. Prog. Nucl. Magn. Reson. Spectrosc. 57:420-434. [DOI] [PubMed]
- 62.Cheng H.C., Qi R.Z., Zhu H.J. Regulation and function of protein kinases and phosphatases. Enzyme Res. 2011;2011:794089. doi: 10.4061/2011/794089. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cheng, H. C., R. Z. Qi, …, H. J. Zhu. 2011. Regulation and function of protein kinases and phosphatases. Enzyme Res. 2011:794089. [DOI] [PMC free article] [PubMed]
- 63.Zhou J., Zhao S., Dunker A.K. Intrinsically disordered proteins link alternative splicing and post-translational modifications to complex cell signaling and regulation. J. Mol. Biol. 2018;430:2342–2359. doi: 10.1016/j.jmb.2018.03.028. [DOI] [PubMed] [Google Scholar]; Zhou, J., S. Zhao, and A. K. Dunker. 2018. Intrinsically disordered proteins link alternative splicing and post-translational modifications to complex cell signaling and regulation. J. Mol. Biol. 430:2342-2359. [DOI] [PubMed]
- 64.Darling A.L., Uversky V.N. Intrinsic disorder and posttranslational modifications: the darker side of the biological dark matter. Front. Genet. 2018;9:158. doi: 10.3389/fgene.2018.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]; Darling, A. L., and V. N. Uversky. 2018. Intrinsic disorder and posttranslational modifications: the darker side of the biological dark matter. Front. Genet. 9:158. [DOI] [PMC free article] [PubMed]
- 65.Bah A., Forman-Kay J.D. Modulation of intrinsically disordered protein function by post-translational modifications. J. Biol. Chem. 2016;291:6696–6705. doi: 10.1074/jbc.R115.695056. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bah, A., and J. D. Forman-Kay. 2016. Modulation of intrinsically disordered protein function by post-translational modifications. J. Biol. Chem. 291:6696-6705. [DOI] [PMC free article] [PubMed]
- 66.Uversky V.N. Wrecked regulation of intrinsically disordered proteins in diseases: pathogenicity of deregulated regulators. Front. Mol. Biosci. 2014;1:6. doi: 10.3389/fmolb.2014.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]; Uversky, V. N. 2014. Wrecked regulation of intrinsically disordered proteins in diseases: pathogenicity of deregulated regulators. Front. Mol. Biosci. 1:6. [DOI] [PMC free article] [PubMed]
- 67.Ubersax J.A., Ferrell J.E., Jr. Mechanisms of specificity in protein phosphorylation. Nat. Rev. Mol. Cell Biol. 2007;8:530–541. doi: 10.1038/nrm2203. [DOI] [PubMed] [Google Scholar]; Ubersax, J. A., and J. E. Ferrell, Jr. 2007. Mechanisms of specificity in protein phosphorylation. Nat. Rev. Mol. Cell Biol. 8:530-541. [DOI] [PubMed]
- 68.Fujiwara H., Hasegawa M., Iwatsubo T. α-synuclein is phosphorylated in synucleinopathy lesions. Nat. Cell Biol. 2002;4:160–164. doi: 10.1038/ncb748. [DOI] [PubMed] [Google Scholar]; Fujiwara, H., M. Hasegawa, …, T. Iwatsubo. 2002. Alpha-synuclein is phosphorylated in synucleinopathy lesions. Nat. Cell Biol. 4:160-164. [DOI] [PubMed]
- 69.Choi H.S., Liew H., Suh Y.H. Phosphorylation of α-synuclein is crucial in compensating for proteasomal dysfunction. Biochem. Biophys. Res. Commun. 2012;424:597–603. doi: 10.1016/j.bbrc.2012.06.159. [DOI] [PubMed] [Google Scholar]; Choi, H. S., H. Liew, …, Y. H. Suh. 2012. Phosphorylation of α-synuclein is crucial in compensating for proteasomal dysfunction. Biochem. Biophys. Res. Commun. 424:597-603. [DOI] [PubMed]
- 70.Mahul-Mellier A.L., Fauvet B., Lashuel H.A. c-Abl phosphorylates α-synuclein and regulates its degradation: implication for α-synuclein clearance and contribution to the pathogenesis of Parkinson’s disease. Hum. Mol. Genet. 2014;23:2858–2879. doi: 10.1093/hmg/ddt674. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mahul-Mellier, A. L., B. Fauvet, …, H. A. Lashuel. 2014. c-Abl phosphorylates α-synuclein and regulates its degradation: implication for α-synuclein clearance and contribution to the pathogenesis of Parkinson’s disease. Hum. Mol. Genet. 23:2858-2879. [DOI] [PMC free article] [PubMed]
- 71.Schmid A.W., Fauvet B., Lashuel H.A. α-synuclein post-translational modifications as potential biomarkers for Parkinson disease and other synucleinopathies. Mol. Cell. Proteomics. 2013;12:3543–3558. doi: 10.1074/mcp.R113.032730. [DOI] [PMC free article] [PubMed] [Google Scholar]; Schmid, A. W., B. Fauvet, …, H. A. Lashuel. 2013. Alpha-synuclein post-translational modifications as potential biomarkers for Parkinson disease and other synucleinopathies. Mol. Cell. Proteomics. 12:3543-3558. [DOI] [PMC free article] [PubMed]
- 72.Nakamura T., Yamashita H., Nakamura S. Activated Fyn phosphorylates α-synuclein at tyrosine residue 125. Biochem. Biophys. Res. Commun. 2001;280:1085–1092. doi: 10.1006/bbrc.2000.4253. [DOI] [PubMed] [Google Scholar]; Nakamura, T., H. Yamashita, …, S. Nakamura. 2001. Activated Fyn phosphorylates α-synuclein at tyrosine residue 125. Biochem. Biophys. Res. Commun. 280:1085-1092. [DOI] [PubMed]
- 73.Kosten J., Binolfi A., Selenko P. Efficient modification of α-synuclein serine 129 by protein kinase CK1 requires phosphorylation of tyrosine 125 as a priming event. ACS Chem. Neurosci. 2014;5:1203–1208. doi: 10.1021/cn5002254. [DOI] [PubMed] [Google Scholar]; Kosten, J., A. Binolfi, …, P. Selenko. 2014. Efficient modification of alpha-synuclein serine 129 by protein kinase CK1 requires phosphorylation of tyrosine 125 as a priming event. ACS Chem. Neurosci. 5:1203-1208. [DOI] [PubMed]
- 74.Paleologou K.E., Schmid A.W., Lashuel H.A. Phosphorylation at Ser-129 but not the phosphomimics S129E/D inhibits the fibrillation of α-synuclein. J. Biol. Chem. 2008;283:16895–16905. doi: 10.1074/jbc.M800747200. [DOI] [PMC free article] [PubMed] [Google Scholar]; Paleologou, K. E., A. W. Schmid, …, H. A. Lashuel. 2008. Phosphorylation at Ser-129 but not the phosphomimics S129E/D inhibits the fibrillation of alpha-synuclein. J. Biol. Chem. 283:16895-16905. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.