Abstract
The naturally occurring amino acid cysteine has often been implicated with a crucial role in maintaining protein structure and stability. An intriguing duality in the intrinsic hydrophobicity of the cysteine side chain is that it exhibits both polar as well as hydrophobic characteristics. Here, we have utilized a cysteine-scanning mutational strategy on the transmembrane β-barrel PagP to examine the membrane depth-dependent energetic contribution of the free cysteine side chain (thiolate) versus the parent residue at an experimental pH of 9.5 in phosphatidylcholine vesicles. We find that introduction of cysteine causes destabilization at several of the 26 lipid-facing sites of PagP that we mutated in this study. The destabilization is minimal (0.5–1.5 kcal/mol) when the mutation is toward the bilayer midplane, whereas it is higher in magnitude (3.0–5.0 kcal/mol) near the bilayer interface. These observations suggest that cysteine forms more favorable interactions with the hydrophobic lipid core as compared to the amphiphilic water-lipid interface. The destabilizing effect is more pronounced when cysteine replaces the interfacial aromatics, which are known to participate in tertiary interaction networks in transmembrane β-barrels. Our observations from experiments involving the introduction of cysteine at the bilayer midplane further strengthen previous views that the free cysteine side chain does possess strongly apolar characteristics. Additionally, the free energy changes observed upon cysteine incorporation show a depth-dependent correlation with the estimated energetic cost of partitioning derived from reported hydrophobicity scales. Our results and observations from the thermodynamic analysis of the PagP barrel may explain why cysteine, despite possessing a polar sulfhydryl group, tends to behave as a hydrophobic (rather than polar) residue in folded protein structures.
Significance
Cysteine displays an enigmatic duality in its physicochemical properties. Here, we show that in membrane proteins, cysteine destabilizes a membrane protein irrespective of its positioning at most of the lipid-facing sites of a model transmembrane β-barrel. However, cysteine energetically favors hydrophobicity over its polar nature. Our findings provide a thermodynamic basis for why cysteine residues are largely infrequent in membrane proteins.
Introduction
Membrane protein folding is dictated by intra- and intermolecular interactions established between the protein and lipid moieties participating in the folding event. The energetics associated with the transfer of an amino acid side chain from an aqueous solvent into the lipid bilayer is the key contributor that drives the folding process. This parameter, also referred to as the partitioning free energy, has been derived for the 20 naturally occurring amino acids using a multitude of methods in the past several decades (1, 2, 3, 4, 5, 6, 7, 8, 9). Most amino acids can be unequivocally classified as hydrophobic, polar, or hydrophilic, based on the chemical properties of the side chain, which are indicated by the relative position occupied by the particular amino acid on the hydrophobicity scale. For a small subset of amino acid residues, however, the position on the free energy scale alters when the experimental or theoretical technique employed to generate the scale is varied. Cysteine is one such amino acid residue whose side-chain hydrophobicity is a matter of ambiguity (10).
In general, the free cysteine side chain plays a variety of roles in protein structure and function, including dimerization, metal coordination, redox regulation, and thermal stability (10, 11, 12, 13). Previously reported hydrophobicity scales support both a hydrophilic (3, 5) (because of the polar sulfhydryl group in its side chain) as well as a hydrophobic (1, 2) (because of its occurrence in hydrophobic regions in native protein structures) nature for this residue. Existing hydrophobicity scales have mostly been derived either from theoretical predictions of amino acid propensities (3, 7) or from experimental calculations of side-chain transfer from an aqueous environment to apolar solvents (4, 5). In the latter case, it has been noted that the chemical nature of hydrophobic solvents and the solubility of water within them can change the rank order of transfer free energies (14, 15). In interesting contrast, studies on the amino acid distributions in membrane protein structures indicate that cysteine is preferentially located toward the membrane midplane (16, 17, 18) and would therefore possess a hydrophobic nature. There is, however, a constraint associated with statistical analyses of small sample size (fewer available structures), particularly while dealing with an infrequently occurring amino acid such as cysteine. It is, therefore, becoming increasingly important to carry out experimental derivations using actual protein systems (8, 19), to deduce the context-dependent hydrophobicity of a particular side chain.
In terms of the chemically apolar nature of the side chain, the sulfhydryl group is mostly inactive toward water molecules. More specifically, unlike the hydroxyl (−OH) group, the sulfhydryl (−SH) group has a limited ability to form a hydrogen bond with water. There is the added complexity of the highly reactive sulfhydryl (−SH) group, which, upon participation in covalent bond formation, might modify the side-chain properties. Therefore, it is imperative to precisely estimate the magnitude of hydrophobicity presented by the free cysteine side chain. Assessment of both the hydrophobic and polar nature of cysteine requires the formation of the thiolate ion, which has a pKa ≈ 8.4. Therefore, we examined the thermodynamics of the cysteine side chain in an alkaline environment. Here, we carried out folding analysis of the eight-stranded β-barrel membrane protein PagP (PhoPQ-activated gene P) in phosphatidylcholine vesicles at an experimental pH of 9.5. Our aim was to introduce context-dependent effects (the context here being local structural variations in the global scaffold of a transmembrane β-barrel protein) on the apparent hydrophobicity of cysteine in a model membrane protein. To this end, we employed a mutational strategy that involved introducing cysteines in place of a total of 27 lipid-facing residues located toward the midplane as well as near the interface region of each of the eight β-strands. Along similar lines, we used a lipid bilayer system consisting of phosphatidylcholine vesicles to create a near-native environment to accommodate PagP, as opposed to our previous studies with membrane mimetics such as detergent micelles (20, 21). This membrane system now recreates the chemical environment of the biological membrane by providing a polarity gradient across the bilayer and, at the same time, possesses the physical properties of membrane lipids such as lateral bilayer pressure. Phosphatidylcholine vesicles thereby serve as an ideal system to study the energetics of membrane protein folding. Our results establish that the introduction of cysteine causes varying degrees of destabilization at several host sites of PagP. Furthermore, we find a strong depth-dependent correlation between the free energy changes observed upon cysteine incorporation and the estimated energetic cost of partitioning derived from the reported hydrophobicity scales. Our observations from the thermodynamic analyses of the PagP barrel support the previous findings that the free cysteine side chain, when present in folded proteins, exhibits substantial hydrophobic character.
Materials and Methods
Protein preparation
The wild-type PagP construct was generated by PCR amplification of the pagP gene (NCBI accession number NC_000913) from the genome of Escherichia coli K12 MG1655 without the signal sequence and cloning into pET3a vector (Novagen/MilliporeSigma, Burlington, MA). A total of 27 single-cysteine mutants (at lipid-facing host sites) described in this study were generated by site-directed mutagenesis and expressed without affinity tags. E. coli BL21(DE3) cells were transformed with these plasmid constructs and used for protein production. Proteins were expressed in the form of inclusion bodies and processed to ∼95% purity using reported methods (20).
Equilibrium folding experiments using steady-state fluorescence
Measurements of guanidine hydrochloride (GdnHCl)-mediated folding titrations were carried out by monitoring the change in tryptophan fluorescence on a SpectraMax M5 Microplate Reader (Molecular Devices, San Jose, CA). PagP possesses 12 tryptophans that are distributed throughout the protein. We and others have observed that PagP exhibits hysteresis or incomplete unfolding in alkaline conditions in lipid vesicles (20, 22). Hence, we examined only the folding titration of the equilibrium reaction in small unilamellar vesicles (SUVs) of 1,2-dilauroyl-sn-glycero-3-phosphocholine (DLPC), as reported previously (20). Briefly, we added 5 μg/μL (0.26 mM) unfolded PagP in a 1:9 ratio to 8.9 mM DLPC SUVs to generate a folding stock containing 26 μM protein and 8 mM lipid dissolved in 4.5 M GdnHCl prepared in 20 mM Tris-Cl (pH 9.5) and 20 mM 1,4-dithiothreitol (DTT). The reaction was centrifuged for 1 h at 16,600 × g, and the supernatant was collected to avoid traces of aggregated protein. This stock was further diluted 10-fold into the GdnHCl gradient, ranging from concentrations of 1.0 to 4.5 M, and the final samples were incubated at 25°C to monitor the progress of the reaction. The final lipid-to-protein ratio in all the titrations has been maintained at ∼300:1.
Data analysis
For equilibrium folding experiments, we recorded the tryptophan fluorescence emission spectra between 320 and 400 nm using a λex of 280 nm. Unfolded fractions were calculated from fluorescence data at 340 nm using the following equation (Eq. 1) (23):
| (1) |
Here, yO is the observed fluorescence at GdnHCl concentration [D], whereas yN, mN, yU, and mU are intercepts and slopes of the pre- and post-transition baselines, respectively.
The thermodynamic parameters (ΔG0, the apparent equilibrium free energy of folding; m value, the apparent change in solvent-accessible surface area) were derived by globally fitting the data to the two-state equation (Eq. 2) (23):
| (2) |
According to this model, the protein folds from the unfolded (U) to the native (N) state, in a cooperative manner, without a detectable intermediate. By means of the global analysis of the folding curves, we derived a common m value, which describes the apparent change in accessible surface area (ASA) between the U and N states for all mutants. The midpoint of chemical denaturation (Cm) was derived as Cm = ΔG0/m.
Enzymatic assay
We measured the activity of PagP folded in DLPC SUVs by performing a modified version of the previously reported lipase assay using the substrate analog p-nitrophenyl palmitate (pNPP) (24). Briefly, we added 0.02 μg/μL (1.0 μM) protein, refolded in 1.2 mM lipid, to 1.0 mM of the substrate analog pNPP prepared in 50 mM phosphate buffer (pH 8.0) containing 2% Triton X-100 and 2 mM DTT. We measured the activity by monitoring the release of p-nitrophenol spectrophotometrically at 405 nm. All the reactions were carried out at 25°C. The rate of the reaction was monitored for 60 min, and enzymatic activity was calculated using the initial linear segment of the kinetics plot (25).
Circular dichroism measurements
We monitored the far ultraviolet (far-UV) circular dichroism (CD) of samples containing 0.22 μg/μL (11.0 μM) protein, folded in 4.0 mM DLPC. We recorded wavelength scans using a quartz cuvette of 1 mm path length, between 210 and 260 nm at a temperature of 25°C, as described previously (25). Data were averaged over three accumulations, subtracted for buffer contributions and smoothened, and represented as ellipticity values (25).
Results and Discussion
Global structural features are largely retained in the PagP cysteine variants
The eight-stranded β-barrel, PagP, has been widely used as a model for membrane protein folding studies owing to its ability to spontaneously fold into an array of membrane mimetics ranging from detergent micelles to lipidic vesicles (20, 22, 26). Here, we generated a library of single-cysteine mutants (native PagP from E. coli is devoid of cysteine residues) by strategically choosing 27 lipid-facing host sites near the water-lipid interface and toward the midplane on each strand of the eight-stranded transmembrane barrel (Fig. 1). We utilized a membrane system involving SUVs of DLPC to fold PagP and its mutants (20). DLPC is ideally suited for folding and stability analysis of PagP because it provides a membrane thickness of ∼20 Å, which matches the lateral dimensions of the PagP structure (27). DLPC has also been utilized in other studies examining PagP thermodynamics (19, 22). Thereafter, we probed into the structural details of the PagP barrel and the effect of cysteine incorporation on the global structural properties. Accordingly, we first performed spectroscopic analysis of a subset of the single-Cys variants using far-UV CD measurements to examine whether the PagP structure is perturbed by the introduction of cysteine at strategic host sites (Fig. S1). Here, we chose sufficient representatives from the midplane as well as the interface variants spanning the length of the protein to exhaustively determine the folding efficiency of the mutants. In addition to the signature negative maximum at 215 nm, natively folded PagP exhibits a characteristic tertiary CD Cotton effect (28, 29) with an observable positive maximum at ∼231 nm and a negative maximum at ∼225 nm. The molecular basis of this CD exciton couplet has been discussed extensively in earlier reports (28, 30). We find that the folded single-Cys variants exhibit negligible variation in the secondary structure content (θ215 value), ranging from −30 to −20 mdeg. These ellipticity values are representative of the native state of PagP. On the other hand, the θ231 value, which is an indicator of the extent of tertiary interactions between Y26 and W66 in folded PagP (28, 30), carries a negligible variation across the mutants and ranges from ∼+7 mdeg for PagP-R59C to ∼+2 mdeg for PagP-P127C. A remarkable feature of the tertiary interaction data is that all the single-Cys variants examined here show positive θ231 values, which indicates that strong tertiary interactions leading to the formation of a compact folded barrel is retained in all the mutants. Nevertheless, we note here that the specific geometry of the aromatic interaction between Y26 and W66 could be abolished without severely affecting the overall compaction of the barrel (21). Based on the structural profiling, we deduce that all of the single-Cys variants examined here exhibit comparable ellipticities (for secondary structure as well as tertiary packing interactions) when refolded into DLPC vesicles.
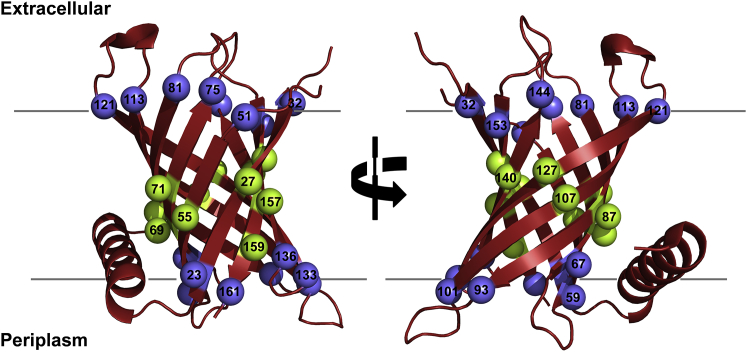
Figure 1.
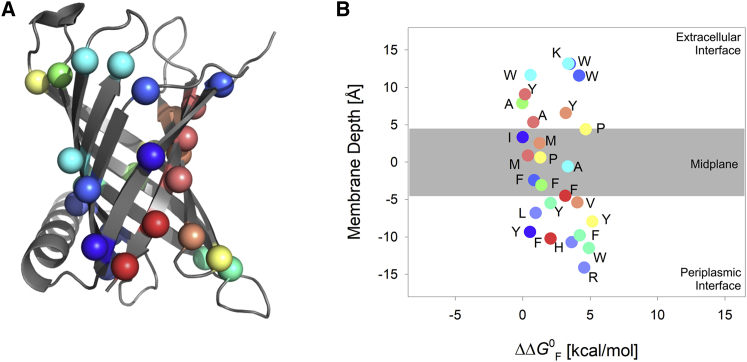
Incorporation of cysteine at strategic sites of the transmembrane barrel PagP. Schematic representation of PagP from E. coli (red) was generated using PyMOL (40) from the crystal structure (PDB: 3GP6 (34)). The structure of PagP is tilted by ∼25° with respect to the membrane normal and consists of eight transmembrane β-strands and a periplasmic α-helix. Host sites at which cysteine was introduced are highlighted as spheres along with the residue number. A total of 27 lipid-facing sites, including nine midplane residues (green spheres; note that strand 3 has two lipid-facing midplane residues), two lipid-facing residues on the terminal strand (residues 155 and 159; green spheres), and 16 residues lining the water-lipid interface (blue spheres) were chosen from across the eight β-strands of PagP for mutation to cysteine. To retain visual clarity, not all the spheres are numbered. To see this figure in color, go online.
PagP and its variants show highly cooperative two-state transitions
Next, we followed the chemical denaturation titrations for each PagP variant using fluorescence measurements of the 12 intrinsic tryptophans (11 for W32C, W51C, W81C, and W93C). The protonated state of cysteine is largely apolar. Assessment of both the hydrophobic and polar nature of cysteine requires the formation of the thiolate ion, which has a pKa ≈ 8.4. Although large unilamellar vesicles (LUVs) are considered a more thermodynamically stable membrane mimetic as compared to SUVs, we found that unlike SUVs, PagP demonstrated lower folding efficiencies in LUVs at an alkaline pH and showed a tendency to aggregate in solution (data not shown). Therefore, we performed equilibrium titration experiments on PagP refolded in DLPC SUVs, using GdnHCl as the denaturant (20).
Full-length PagP exhibits hysteresis in DLPC SUVs despite prolonged incubation at the alkaline conditions (pH > 8.4) required for our experiments and in acidic buffer conditions (Figs. S2 and S3). In other words, the PagP-DLPC system is not under equilibrium and exhibits path dependence for the folding or the unfolding titration. Therefore, the measured free energy in our experiments must be considered an apparent value. However, prolonged incubation used in our experiments (up to 72 h incubation) allows for an appreciable level of equilibration to be achieved with SUVs (Fig. S2) (31). Additionally, we have previously observed that energetics derived from the folding titrations in DLPC SUVs correlate well with data from phosphocholine micelle systems that are under thermodynamic equilibrium (20). Furthermore, the folding cooperativity that we derive from the folding titrations is in excellent agreement with previously reported values for the equilibrium measurements of PagP folded into LUVs (19, 32). We reason that this observation is one of the several factors that validate the use of apparent free energy values in our thermodynamic analysis of PagP. Based on this result and previous studies (32), we conclude that the unfolding measurements are the likely source of the hysteresis (i.e., a kinetically unattained two-state system), and the energetics derived from the folding titrations could be considered as near equilibrium values. Finally, by uniformly subtracting the folding free energy of wild-type PagP from each mutant to derive the partitioning free energy values, we believe we can potentially minimize the light-scattering artifacts associated with the use of SUVs (31).
We obtained the thermodynamic parameters (ΔG0, free energy of folding), m value (change in accessible surface area between unfolded and folded states; also represents the cooperativity of folding), and Cm (midpoint of chemical denaturation) by fitting data from the fluorescence profiles to the two-state mechanism (23) using a common m value of 5.07 ± 0.04 kcal/mol/M (Fig. 2). This value is in good agreement with previously reported m values for PagP in SUVs (m = 4.80 kcal/mol/M) (20) and LUVs (m = 4.97 kcal/mol/M) (19, 32). It is interesting to note here that the physical heterogeneity and nonequilibrium nature of SUVs when compared with LUVs (33) does not significantly affect the folding cooperativity of the transitions observed for PagP. The overall ΔG0, however, is lower in SUVs (difference of ∼6.0 kcal/mol for wild-type PagP in LUVs in pH 3.8 (Cm = 4.57 M) (19) versus SUVs in pH 9.5 (Cm = 3.28 M)). We can deduce from the considerable differences in Cm that the higher stability of wild-type PagP in LUVs results from the ability of the folded protein to resist solvation by GdnHCl. However, the ability to demonstrate highly cooperative two-state transitions is a property afforded by both lipidic systems. We validate this further from the result obtained from the far-UV CD wavelength scans (Fig. S1). As mentioned above, the equilibrium folding profiles for all the mutants characterized in this study have been analyzed by assuming a common m value. The m value represents the difference between the solvent-accessible surface areas (ΔASA) associated with the U and N states (32). Assuming that the denatured state ensemble does not carry significant residual structure, the ΔASA (and therefore, the m value) would strongly depend on the structural content of the native state of PagP. We have previously observed that micelles of dodecyl phosphocholine allow structural heterogeneity in the folded state of PagP mutants and consequently alter the m value in a residue-specific manner (20). For the same reason, in our previous analyses of PagP energetics (20, 21), we computed individual m values for each mutant to derive the thermodynamic parameters. However, in this study with single-Cys mutants in SUVs, we do not observe any significant variation in the structural content of the native state across mutants. Accordingly, we infer that the ΔASA of PagP folded in SUVs remains the same, irrespective of point mutations, thereby validating the use of a common m value.
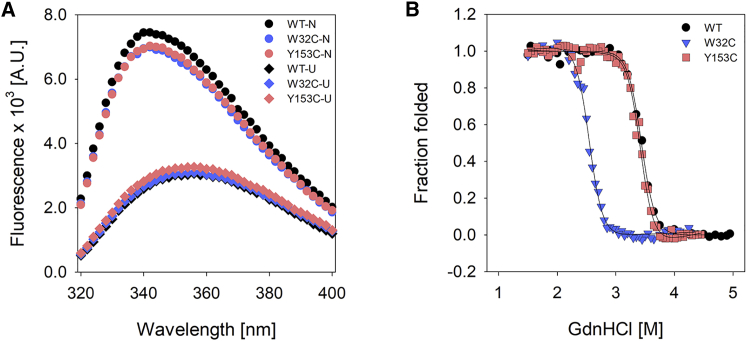
Figure 2.
Equilibrium folding profiles of PagP cysteine variants. (A) Shown are the fluorescence emission profiles of PagP wild-type (black) and representative mutants W32C (blue) and Y153C (red) derived from GdnHCl-induced chemical denaturation. Emission spectra are displayed for low (circles) and high (diamonds) denaturant concentrations, indicating the native (N) and unfolded (U) states of the protein. A.U., arbitrary units. (B) Shown are folding titrations displayed as folded fraction (fF) for representative mutants W32C and Y153C (see Fig. S5 for the complete data set). Folding profiles were acquired by monitoring the change in fluorescence emission intensity at a λem of 340 nm, corresponding to the λem-max of the folded protein. Data sets were fitted to the two-state equation (23) (fits are shown as solid lines). Data for wild-type PagP are shown in black. To see this figure in color, go online.
The ΔG0 obtained for all the single-cysteine mutants of PagP in SUVs is summarized in Table 1. Inspection of the values immediately reveals that replacing most of the intrinsic PagP residues with cysteine destabilizes the barrel by ∼2.0–4.0 kcal/mol. Mutating I27, A113, or Y153 to cysteine does not affect PagP stability. Two of these mutations (I27C and M157C) are at the midplane, whereas the other mutation is at the interface (extracellular side). The lack of a direct correlation between the observed stabilization and the physicochemical nature of the parent residue being replaced or the membrane depth suggests a likely stabilizing contribution of the cysteine in these positions as arising from favorable local interactions with spatially vicinal residues. Despite this stabilizing effect on the barrel, the θ231 is marginally lowered in these mutants (see Fig. S8). To further assess whether the introduction of cysteine affected the PagP scaffold, we checked the enzymatic activity of wild-type and its mutants.
Table 1.
Thermodynamic Parameters for the PagP Single-Cysteine Mutant Library Derived from Equilibrium Folding Titrations
| Strand Number | PagP Variantsa | ΔG0 (kcal/mol)b | Cm (M) | ΔΔG0 (kcal/mol) |
|---|---|---|---|---|
| – | Wild-type | −16.65 ± 0.98 | 3.28 ± 0.19 | – |
| Interface Mutants | ||||
| 1 | Y23C | −16.10 ± 0.47 | 3.18 ± 0.09 | 0.54 ± 1.13 |
| 1 | W32C | −13.14 ± 0.35 | 2.59 ± 0.07 | 3.51 ± 1.09 |
| 2 | W51C | −12.46 ± 0.57 | 2.46 ± 0.11 | 4.19 ± 1.18 |
| 2 | R59C | −12.08 ± 0.93 | 2.38 ± 0.18 | 4.57 ± 1.03 |
| 3 | H67C | −13.02 ± 0.11 | 2.57 ± 0.02 | 3.63 ± 1.04 |
| 3 | K75C | −13.25 ± 0.78 | 2.61 ± 0.15 | 3.40 ± 1.29 |
| 4 | W81C | −16.06 ± 0.68 | 3.17 ± 0.13 | 0.59 ± 1.23 |
| 4 | W93C | −11.01 ± 0.20 | 2.17 ± 0.04 | 5.64 ± 1.05 |
| 5 | F101C | −12.41 ± 0.76 | 2.45 ± 0.15 | 4.24 ± 1.28 |
| 5 | A113C | −16.67 ± 0.45 | 3.29 ± 0.09 | 0.03 ± 1.13 |
| 6 | P121C | −11.97 ± 0.71 | 2.36 ± 0.14 | 4.68 ± 1.25 |
| 6 | Y133C | −11.48 ± 0.76 | 2.26 ± 0.15 | 5.17 ± 1.28 |
| 7 | V136C | −12.59 ± 0.58 | 2.48 ± 0.11 | 4.06 ± 1.18 |
| 7 | Y142C | −13.45 ± 0.91 | 2.65 ± 0.18 | 3.20 ± 1.38 |
| 8 | Y153C | −16.47 ± 1.26 | 3.25 ± 0.25 | 0.18 ± 1.63 |
| 8 | F161C | −14.58 ± 0.01 | 2.87 ± 0.01 | 2.07 ± 1.03 |
| Midplane Mutants | ||||
| 1 | I27C | −16.65 ± 0.52 | 3.28 ± 0.10 | 0.01 ± 1.15 |
| 2 | F55C | −15.80 ± 0.33 | 3.12 ± 0.06 | 0.84 ± 1.08 |
| 3 | L69C | −15.68 ± 0.17 | 3.09 ± 0.03 | 0.97 ± 1.04 |
| 3 | A71C | −13.29 ± 0.07 | 2.62 ± 0.01 | 3.36 ± 1.03 |
| 4 | Y87C | −14.58 ± 0.83 | 2.87 ± 0.16 | 2.07 ± 1.32 |
| 5 | F107C | −15.26 ± 0.83 | 3.01 ± 0.16 | 1.39 ± 1.32 |
| 6 | P127C | −15.34 ± 0.59 | 3.02 ± 0.12 | 1.31 ± 1.19 |
| 7 | M140C | −15.39 ± 0.24 | 3.04 ± 0.05 | 1.26 ± 1.06 |
| 8 | M157C | −16.26 ± 1.17 | 3.21 ± 0.23 | 0.39 ± 1.56 |
| Additional Terminal Strand Mutantsc | ||||
| 8 | A155C | −15.86 ± 1.01 | 3.13 ± 0.20 | 0.79 ± 1.44 |
| 8 | F159C | −13.49 ± 0.28 | 2.66 ± 0.05 | 3.16 ± 1.07 |
Mutants are labeled based on the residue mutations using the single letter code of the amino acid, along with the residue number at which the substitution has been carried out.
All mutants were analyzed for two-state folding behavior and were fitted using a common m value of 5.07 ± 0.04 kcal/mol/M. Errors are SDs from n = 2 independent measurements.
The five terminal strand mutants (lipid-facing residues) are Y153C, A155C, M157C, F159C, and F161C. The two mutants—A155C and F159C—can neither be categorized as interface mutants nor as midplane mutants as they are situated midway between the bilayer midplane and the interface. Therefore, they have been listed separately as additional terminal strand mutants.
Protein activity does not correlate with the thermodynamic stability of single-Cys mutants
PagP is an outer membrane enzyme with the ability to transfer palmitoyl lipid chains from glycerophospholipids in the outer membrane to the lipopolysaccharide moiety (26). The first step of this catalytic reaction can be monitored by measuring the rate of p-nitrophenol release from the lipase substrate pNPP in vitro, as reported earlier (24). The active site residues are located in the extracellular loop 2 of PagP. By making the reasonable assumption that PagP activity is not detrimentally affected when the crucial active site residues remain unperturbed (25, 34), we can deduce that the catalytic ability of the single-Cys mutant library also represents a marker for the ability of the particular mutant to attain its native structure. Based on earlier studies on PagP activity (35, 36), we reasoned that the introduction of a cysteine in place of a hydrophobic residue in the lipid-facing host site might alter the structural compaction of the barrel, which in turn, can affect activity. Hence, we examined the catalytic ability of all the eight midplane mutants. Additionally, we carried out the activity assay on all the terminal strand mutants (a total of five mutants including the midplane mutant of strand 8) to assess whether there was depth dependence to the same contribution and to examine whether the mutation does not inadvertently affect the active site of the protein. We used a pH of 8.0 for the lipase assay, as reported earlier (30). Accordingly, we find that all of the mutants that we examined in this study retain the ability to hydrolyze pNPP and exhibit lipase activity (Fig. S4).
Interestingly, the catalytic efficiency measured for each of the single-Cys mutants shows no correlation with the folding free energy of the mutant. The lowest activity, of 0.023 ± 0.004 nmol min−1 μM−1, in comparison with that of wild-type PagP (0.059 ± 0.009 nmol min−1 μM−1), was observed for the PagP-I27C mutant. This result is particularly intriguing because the thermodynamic stability of the PagP-I27C mutant (ΔG0 = −16.65 ± 0.52 kcal/mol) is nearly identical to that of wild-type PagP. On the other hand, the highest activity among the variants was observed for PagP-Y87C (ΔG0 = −14.58 ± 0.83 kcal/mol), a mutant which is less stable than wild-type PagP. We have previously observed that the outer membrane enzyme PagP has the tendency to demonstrate an activity-stability trade-off in its enzymatic behavior (25). The observation that all of the mutants examined here show lower activity than wild-type PagP might be taken to represent global structural perturbation. However, for the cysteine mutants, we infer that the lowering of protein activity is largely insignificant when we consider the effects of single-residue substitutions on PagP activity from our earlier studies (25, 36). The reduced activity for this set of mutants is within the same order of magnitude (∼0.02 − ∼0.06 nmol min−1 μM−1), thereby indicating that the introduction of cysteine in PagP does not impair the ability of the mutant protein to hydrolyze the substrate analog.
It is important to note the pH difference between the experimental conditions for the lipase activity assay (pH 8.0) and our folding experiments (pH 9.5). Essentially, it is an experimental limitation that one cannot avoid. As reported in an earlier work on PagP activity using native substrates (30), the activity profile of PagP shows optimum levels around pH 7.0–8.0, with little to no activity at pH 9.5. Furthermore, based on our earlier studies on PagP activity (25, 36), it is reasonable to argue that protonation states of cysteine residues introduced at lipid-facing host sites along the transmembrane region of PagP do not drastically affect the active site of PagP, which is located toward the periplasmic loops involving a catalytic triad. Therefore, we use the lipase assay as an overall measure of the folded state of the particular cysteine variant and restrict the analysis to a qualitative correlation of the activity with the folding free energy values.
Near universal destabilization of PagP barrel by cysteine incorporation
To further delve into the effect of Cys incorporation on PagP folding energetics, we measured the folding free energy for the single Cys-containing mutant series with respect to wild-type PagP (ΔΔG0 = − ) (Fig. 2; Table 1). This analysis enables us to examine the compounded effect of the loss of the hydrophobic residue (present in wild-type PagP) as well as the introduction of cysteine at the lipid-facing host site on the overall thermodynamic stability of PagP. We find that the majority of single-cysteine mutants are destabilized in comparison with wild-type PagP, evidenced directly by a shift in the equilibrium folding curves toward lower denaturant concentrations (Figs. S5–S7). This destabilization is not strongly linked with the host residue, which has been replaced by cysteine. For example, the three variants, W32C, K75C, and V136C, show comparable extents of destabilization, despite the diametrically opposite side-chain properties of each host residue. In other words, the introduction of Cys in place of an aromatic, charged, or aliphatic residue leads to a comparable reduction in the Cm by ∼0.7 M and a corresponding lowering of the free energy by ∼3 kcal/mol. Furthermore, we did not find any strong correlation between the magnitude of change in the free energy and the position of the host site in the primary sequence of the protein (see Fig. S6). Essentially, the extent of stabilization or destabilization does not depend on whether the host site is situated toward the N-terminus or the C-terminus of the eight-stranded barrel.
Another interesting aspect that merits discussion is the possible accumulation of disulfide-bonded dimeric species when the single-Cys variants are analyzed using sodium dodecyl sulfate polyacrylamide gel electrophoresis (Fig. S7). It is known that wild-type PagP (devoid of cysteine residues) shows a significant amount of dimer population when folded into DLPC vesicles, as we and several other groups have reported. In our analysis, we do not observe any significant variation in the dimer population upon introducing a cysteine, allowing us to conclude that disulfide-mediated PagP dimers (if any) are present only in trace amounts. In addition, we have retained 2.0 mM DTT in all our experiments to ensure that the formation of transient disulfides is greatly reduced. Thereafter, we compared the folding free energies with the tertiary interaction data (θ231) derived from far-UV CD wavelength scans (see Fig. S1). However, we find no significant correlation between the magnitude of tertiary interaction (which is an indicator of barrel compaction) and the thermodynamic stability of the single-Cys variant (Fig. S8).
Comparison of the normalized free energy values we obtain with those in an earlier study on the ϕ-value analysis of PagP in lipid vesicles (22) shows a strong linear correlation between the levels of destabilization in both cases (R = 0.98) (Fig. S9). Interestingly, the magnitude of destabilization we obtain for the mutation of the host site to cysteine is similar to corresponding mutations to alanine (22) (slope = 0.8, Fig. S9), suggesting that cysteine has a similar hydrophobic character as alanine when accommodated in the lipid-facing sites of the folded protein scaffold. As noted earlier, conventional hydrophobicity scales annotate both hydrophobic as well as hydrophilic behavior to the cysteine side chain (1, 2, 3, 5), depending on the method employed to calculate the free energy of side-chain transfer. However, in our experimental system using a membrane protein model system, we find that the cysteine side chain is largely hydrophobic in nature.
Side-chain partitioning free energy of cysteine demonstrates strong dependence on membrane depth
A major driving force in membrane protein folding is the energy associated with the transfer of hydrophobic residues from water to the lipid bilayer. Based on a multitude of theoretical and experimental studies, we can correlate the hydrophobicity of an amino acid side chain with the free energy of its transfer from water to an apolar environment (3, 5, 6, 8, 9). An interesting ambiguity exists in the case of cysteine, in which the intrinsic nature of the side chain can be considered either polar or hydrophobic, depending on the method used to derive the hydrophobicity (1, 2, 3, 5). Nine of the 27 lipid-facing host sites where we have incorporated cysteine represent the true apolar environment corresponding to the membrane midplane (green spheres in Fig. 1) and are ideally suited for the measurement of side-chain hydrophobicity. Analysis of the side-chain partitioning free energy of cysteine at these sites reveals unfavorable partitioning energies (Table 1). In other words, the calculation of normalized folding free energy (ΔΔG0) for the subset of mutants returned positive values, indicating that cysteine is intrinsically destabilizing at all 27 positions.
To assess the true hydrophobicity of cysteine at any particular host site, we took into account the side-chain properties of the natively present residue in wild-type PagP, which was being replaced by cysteine in this analysis. Nearly all of the residues natively occupying these sites are highly hydrophobic in nature (Ile, Phe, Leu, Ala, Tyr, Pro, and Met). Accordingly, when we correlated the side-chain partitioning free energy derived at these host sites with the estimated energetic cost of cysteine incorporation based on reported hydrophobicity scales (Fig. 3), we found reasonably strong correlations (r = ∼0.8) with the Moon-Fleming scale (8) and the translocon (Hessa) scale (6). The better correlation (r = 0.82) and higher slope (0.76) observed with the Moon-Fleming scale indicates that the whole protein system utilized for hydrophobicity calculation is a better match with the magnitude of side-chain transfer free energies we obtain here. The hydrophobicity values derived for the translocon (Hessa) scale have been examined previously, and it has been observed that the magnitude of the partitioning free energy in this scale is underestimated because of the experimental system employed (14, 37). In both comparisons, the observed correlations highlight the importance of the natively present residue being displaced—in addition to the incoming amino acid side chain—in hydrophobicity calculations. The reasonably strong correlations also indicate that the hydrophobicity of the host residue is mimicked by the lipid-facing cysteine to acceptable levels, irrespective of the aliphatic or aromatic nature of the host residue. We therefore surmise, based on this correlation, that the magnitude of free energy change we measure for cysteine supports the hydrophobic nature of the thiolate ion (when referenced to a typical anion). In other words, this result is particularly interesting because it brings to light the hydrophobicity of a negatively charged functional group. In comparison to a typical anion, the thiolate is relatively more hydrophobic because of the higher electron density of the sulfur atom. This property allows the thiolate to establish van der Waals interactions and makes the cysteine side chain more suited to hydrophobic environments when compared with other residues with charged functional groups.
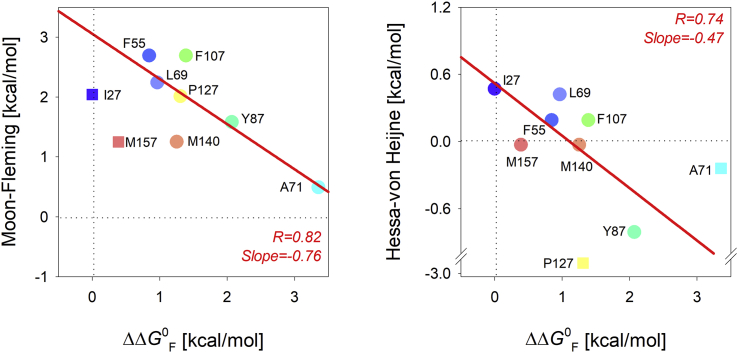
Figure 3.
Correlation of the energetic cost of cysteine incorporation at the membrane midplane with reported hydrophobicity scales. Correlation plot was derived by mapping the partitioning free energy for only the midplane substituents with the estimated energetic cost of substitution (Xxx→Cys) calculated from the previously reported whole protein (Moon-Fleming) hydrophobicity scale (8) (left panel) and the biological translocon (Hessa) scale (6) (right panel). Regression coefficients for the correlation are indicated toward the bottom right-hand corner, and linear fits to the correlation are shown as solid red lines. In the left panel, we have omitted I27 and M157 from the fit to make the correlation more obvious. Even with the inclusion of the data, the correlation still holds true, except with a lower regression coefficient (R = 0.6). In the right panel, the exclusion of A71 and P127 is necessary because they are clear outliers to the data. Points excluded from the fit are shown as square symbols. Mutants are labeled using the single letter code for each amino acid followed by the residue position at which the substitution has been carried out. A rainbow color scheme (N-→C-terminus) is used for the scatter plots. To see this figure in color, go online.
Along similar lines, we investigated the depth-dependent effect of cysteine incorporation using the free energy values obtained for the lipid-facing terminal strand mutants Y153C, A155C, M157C, F159C, and F161C (see Fig. 1). When we mapped the side-chain partitioning free energy of cysteine at these sites against their respective membrane depths, we found an asymmetric distribution of free energy values along the membrane normal (Fig. 4). We observe that cysteine incorporation is more favorable as we move toward the extracellular interface as opposed to the periplasmic interface of PagP. Similar to the analysis in Fig. 3, we compared the partitioning free energy calculated at each host site with the estimated energetic cost of cysteine incorporation (horizontal gray bars in Fig. 4) based on two reported hydrophobicity scales derived as a function of membrane depth, namely 1) the Liang scale (38), a computational scale derived using a model β-barrel protein accommodated in the native outer membrane using molecular dynamics simulations; and 2) the DeGrado scale (7), a depth-dependent empirical scale derived using amino acid propensities in known structures of helical membrane proteins. Here, we find a better correlation with the Liang scale (Fig. 4, left panel), which reflects the asymmetry inherent in the model OMP as well as the membrane system utilized in the computational analysis. Although our experimental setup involves the use of DLPC vesicles, which represents a homogenous and symmetrical lipid system, the asymmetry observed in the free energy values along the membrane normal may be an intrinsic property of the membrane protein and the host site at which the mutation has been carried out. As anticipated, the correlation observed with the DeGrado scale was poorer (Fig. 4, right panel). In the DeGrado scale, the estimated energetic cost of cysteine incorporation showed a linear decrease toward the membrane midplane and reaches a maximum at the interface region, irrespective of whether the interface host site is located near the extracellular face or the periplasmic face. On the other hand, we find an asymmetric distribution of partitioning free energies across the bilayer normal, with the periplasmic interface site showing higher destabilization compared to the extracellular interface host site. This asymmetry correlates well with the computational transfer free energy scale (left panel) (38), which was derived using an outer membrane protein assembled in an asymmetric bacterial outer membrane by molecular dynamics simulations.
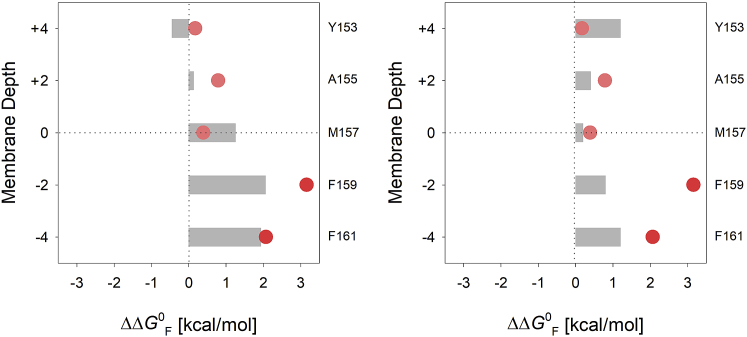
Figure 4.
Comparison of the partitioning free energies of the terminal strand mutants with reported hydrophobicity scales. Partitioning free energies derived for only the terminal strand substituents have been plotted as a function of depth of lipid-facing host sites at which cysteine has been incorporated. The estimated energetic cost of substitution (Xxx→Cys) calculated from a previously reported depth-dependent computational transfer free energy scale (Liang scale) (38) (left panel) and an empirical potential for amino acid insertion into the membrane (DeGrado scale) (7) (right panel) are depicted as gray histograms. To see this figure in color, go online.
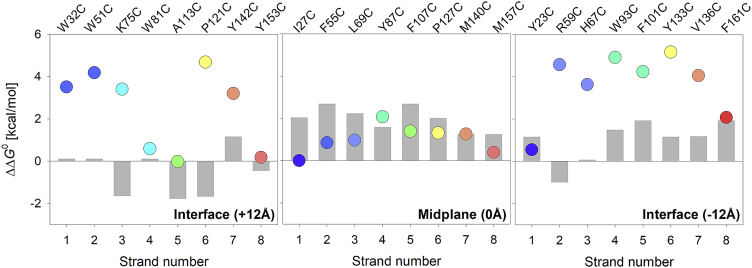
Next, we performed a comprehensive strand-wise analysis of the interface and midplane substituents to examine whether cysteine preferentially partitions toward the hydrophobic core at the middle of the membrane or near the amphipathic chemical milieu presented at the water-lipid interface (Fig. 5). As discussed earlier, cysteine shows unfavorable partitioning energies at a majority of the interface and midplane host sites (scatter plots in Fig. 5, also see Table 1). The magnitude of destabilization is higher in the interface substituents (top and bottom panels in Fig. 5) as compared to the midplane substituents (middle panel in Fig. 5). To examine whether this behavior is due to displacement of the natively present residue from the lipid-facing host site, we further mapped the estimated energetic cost of cysteine incorporation (vertical gray bars in Fig. 5) derived from two reported hydrophobic scales, namely 1) the Moon-Fleming scale (8) for the midplane (membrane depth of 0 Å) substituents, and 2) the Wimley-White interface scale (5) for the interface (membrane depth of ±12 Å) substituents. In this case, we find that the modest destabilization observed for the midplane substituents (∼0.5–1.5 kcal/mol) is indeed a consequence of the removal of the natively present hydrophobic residue from the host site (also see Fig. 3). On the other hand, the large destabilization observed for the interface substituents (∼3.0–5.0 kcal/mol) does not correlate with the estimated energetic cost of substitution (top and bottom panels in Fig. 5). In a majority of cases, the magnitude of destabilization at the midplane is lesser than the estimated energetic cost for the corresponding substitution. Additionally, we performed a statistical significance test to verify this result. We compared the free energy values derived for all of the midplane mutants taken together with those of the interface mutants using the Mann-Whitney U test. Indeed, the U-value derived using this statistical test suggests that the result is significant at p < 0.5. Furthermore, the Mann-Whitney U test delivers a p-value of 0.0251, indicating a statistically significant difference between the ΔΔG values of the interface and the midplane mutants. Put simply, whereas the incorporation of cysteine itself is less favored than hydrophobic residues, cysteine is favorably accommodated at the midplane region in comparison with the water-lipid interface.
Figure 5.
Correlation of measured hydrophobicity of cysteine with the intrinsic hydrophobicity of the residue. Scatter plots representing partitioning free energies derived for the midplane and interface host site substituents are plotted as a function of the β-strand (from one to eight) in which the host site is located. A rainbow color scheme (N→C-terminus) is used for the scatter plots. The estimated energetic cost of substitution (Xxx→Cys) calculated from the whole protein hydrophobicity scale for midplane mutants (8) and the Wimley-White interface scale derived from peptide insertion into lipid membranes for the interface mutants (5) are depicted as gray histograms. We find that the estimated energetic costs correlate reasonably well with the ΔΔG0 for the midplane mutants (center panel). However, in the case of interface mutants (left and right panels), there is a significant difference between the estimated energetic cost of cysteine incorporation and the observed destabilization computed in the form of ΔΔG0. Additionally, the Mann-Whitney U test delivers a p-value of 0.0251, indicating a statistically significant difference between the ΔΔG values of the interface and the midplane mutants. This deviation could be explained by the difference in the model systems used to generate the two respective hydrophobicity scales. The whole protein hydrophobicity scale used mutations at a host site in a β-barrel membrane protein (used here for comparison with midplane mutants), whereas the Wimley-White interface scale measured the insertion of an array of helical peptides into phospholipid bilayers (used here for comparison with interface mutants). To see this figure in color, go online.
A similar conclusion can be deduced from our depth-dependent analysis of the entire single-Cys mutant library (Fig. 6). Here, we calculated the membrane depths of various lipid-facing host sites with reference to a center of mass, using the crystal structure of PagP from E. coli (Protein Data Bank [PDB]: 3GP6 (34)). When we mapped the normalized folding free energy of the cysteine variants against the membrane depth of the particular host site at which the mutation has been introduced, we observed that the side-chain partitioning free energy of cysteine increases toward the midplane and decreases as we move upwards or downwards along the membrane normal. The magnitude of destabilization is heavily influenced by the nature of the host residue (predominantly hydrophobic, see Fig. 6 B) that was displaced to accommodate cysteine and the associated local interactions that were lost as a result of the mutation. These observations indicate that the cysteine side chain can possess the ability to demonstrate hydrophobic characteristics in folded protein structures.
Figure 6.
Depth-dependent effect of cysteine substitution on PagP energetics. Correlation plot was derived by mapping the partitioning free energies derived for the single-cysteine mutants characterized in this study against the membrane depth of the host site at which the substitution was carried out. The membrane depth was derived using the crystal structure of PagP from E. coli (PDB: 3GP6 (34); shown in (A)). The scatter plot is labeled using the single letter code of the host residue that has been mutated to cysteine (B). The transmembrane region corresponding to the midplane has been depicted as a gray histogram in the background in (B). A rainbow color scheme (N-→C-terminus) is used for the scatter plot. To see this figure in color, go online.
Conclusions
In this study, we have performed a detailed analysis of free energy contributions of the cysteine side chain at strategic lipid-facing host sites, ranging from the N-terminal β-strand 1 to the C-terminal β-strand 8 of the outer membrane enzyme PagP. We find that cysteine partitions favorably toward the hydrophobic core at the membrane midplane rather than the amphiphilic interface region. The key reason for this could be the chemically apolar nature of the sulfhydryl group, which may form stabilizing hydrophobic interactions with the lipid hydrocarbon tail (11). Indeed, there have been precedents for the formation of a thiolate anion in the interior of β-barrel structures, giving rise to hydrophilic behavior for the cysteine side chain (21, 30). However, because the thermodynamic analysis carried out in this study involves lipid-facing host sites, it is reasonable to assume that the chemical environment provided to cysteine in the membrane promotes the hydrophobic behavior of the side chain. It is worth noting that complete reliance on the fluorescence signal of PagP to derive the global picture of protein folding and stability could come with certain limitations. However, we faced major challenges in validating these results using an independent technique—such as CD or cold sodium dodecyl sulfate polyacrylamide gel electrophoresis—for the PagP-DLPC system that uses GdnHCl as the perturbant. Therefore, the need for an independent investigative technique is only overstated, and not resolved, by the complex system that we are dealing with. Another important feature of this study is the utilization of lipid vesicles to determine the hydrophobicity of an amino acid, which serves as a robust working system ideally suited to 1) provide a near native environment to accommodate a membrane protein and 2) compute side-chain transfer free energies with high accuracy (8, 14, 15). The only limitation of this study is the occurrence of hysteresis, which hampers the precise measurement of side-chain transfer free energies. Nevertheless, this study provides important insight on the important contribution of protein-lipid interactions in determining the thermodynamic fate of the thiolate ion in native membrane protein structures. It also raises the open-ended question of the evolutionary significance as to why cysteines are infrequent (or absent) in OMPs of bacterial origin (e.g., PagP, OmpX, tOmpA, OmpG, OmpLA, OmpW, OmpF, FadL from E. coli), as opposed to proteins from more complex organisms (39). Finally, interesting observations made from this thermodynamic investigation could prove very useful for a variety of applications in the analysis and the ab initio design of membrane proteins.
Author Contributions
R.M. designed the research. B.R.I. performed the research. Both authors analyzed the data, wrote the article, and have approved the final version of the manuscript.
Acknowledgments
B.R.I. thanks the University Grants Commission, Government of India for a senior research fellowship. R.M. is a Wellcome Trust – Department of Biotechnology India Alliance Intermediate Fellow. This work was supported by funds from the Science and Engineering Research Board award SB/WEA-13/2016 and the Wellcome Trust – Department of Biotechnology India Alliance award IA/I/14/1/501305 to R.M.
Editor: Charles Deber.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2019.05.024.
Supporting Material
References
- 1.Rose G.D. Prediction of chain turns in globular proteins on a hydrophobic basis. Nature. 1978;272:586–590. doi: 10.1038/272586a0. [DOI] [PubMed] [Google Scholar]; Rose, G. D. 1978. Prediction of chain turns in globular proteins on a hydrophobic basis. Nature. 272:586-590. [DOI] [PubMed]
- 2.Janin J. Surface and inside volumes in globular proteins. Nature. 1979;277:491–492. doi: 10.1038/277491a0. [DOI] [PubMed] [Google Scholar]; Janin, J. 1979. Surface and inside volumes in globular proteins. Nature. 277:491-492. [DOI] [PubMed]
- 3.Kyte J., Doolittle R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]; Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed]
- 4.Radzicka A., Wolfenden R. Comparing the polarities of the amino-acids - side-chain distribution coefficients between the vapor-phase, cyclohexane, 1-octanol, and neutral aqueous-solution. Biochemistry. 1988;27:1664–1670. [Google Scholar]; Radzicka, A., and R. Wolfenden. 1988. Comparing the polarities of the amino-acids - side-chain distribution coefficients between the vapor-phase, cyclohexane, 1-octanol, and neutral aqueous-solution. Biochemistry. 27:1664-1670.
- 5.Wimley W.C., White S.H. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat. Struct. Biol. 1996;3:842–848. doi: 10.1038/nsb1096-842. [DOI] [PubMed] [Google Scholar]; Wimley, W. C., and S. H. White. 1996. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat. Struct. Biol. 3:842-848. [DOI] [PubMed]
- 6.Hessa T., Kim H., von Heijne G. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature. 2005;433:377–381. doi: 10.1038/nature03216. [DOI] [PubMed] [Google Scholar]; Hessa, T., H. Kim, …, G. von Heijne. 2005. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature. 433:377-381. [DOI] [PubMed]
- 7.Senes A., Chadi D.C., Degrado W.F. E(z), a depth-dependent potential for assessing the energies of insertion of amino acid side-chains into membranes: derivation and applications to determining the orientation of transmembrane and interfacial helices. J. Mol. Biol. 2007;366:436–448. doi: 10.1016/j.jmb.2006.09.020. [DOI] [PubMed] [Google Scholar]; Senes, A., D. C. Chadi, …, W. F. Degrado. 2007. E(z), a depth-dependent potential for assessing the energies of insertion of amino acid side-chains into membranes: derivation and applications to determining the orientation of transmembrane and interfacial helices. J. Mol. Biol. 366:436-448. [DOI] [PubMed]
- 8.Moon C.P., Fleming K.G. Side-chain hydrophobicity scale derived from transmembrane protein folding into lipid bilayers. Proc. Natl. Acad. Sci. USA. 2011;108:10174–10177. doi: 10.1073/pnas.1103979108. [DOI] [PMC free article] [PubMed] [Google Scholar]; Moon, C. P., and K. G. Fleming. 2011. Side-chain hydrophobicity scale derived from transmembrane protein folding into lipid bilayers. Proc. Natl. Acad. Sci. USA. 108:10174-10177. [DOI] [PMC free article] [PubMed]
- 9.Elazar A., Weinstein J., Fleishman S.J. Mutational scanning reveals the determinants of protein insertion and association energetics in the plasma membrane. eLife. 2016;5:e12125. doi: 10.7554/eLife.12125. [DOI] [PMC free article] [PubMed] [Google Scholar]; Elazar, A., J. Weinstein, …, S. J. Fleishman. 2016. Mutational scanning reveals the determinants of protein insertion and association energetics in the plasma membrane. eLife. 5:e12125. [DOI] [PMC free article] [PubMed]
- 10.Marino S.M., Gladyshev V.N. Cysteine function governs its conservation and degeneration and restricts its utilization on protein surfaces. J. Mol. Biol. 2010;404:902–916. doi: 10.1016/j.jmb.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]; Marino, S. M., and V. N. Gladyshev. 2010. Cysteine function governs its conservation and degeneration and restricts its utilization on protein surfaces. J. Mol. Biol. 404:902-916. [DOI] [PMC free article] [PubMed]
- 11.Nagano N., Ota M., Nishikawa K. Strong hydrophobic nature of cysteine residues in proteins. FEBS Lett. 1999;458:69–71. doi: 10.1016/s0014-5793(99)01122-9. [DOI] [PubMed] [Google Scholar]; Nagano, N., M. Ota, and K. Nishikawa. 1999. Strong hydrophobic nature of cysteine residues in proteins. FEBS Lett. 458:69-71. [DOI] [PubMed]
- 12.Barford D. The role of cysteine residues as redox-sensitive regulatory switches. Curr. Opin. Struct. Biol. 2004;14:679–686. doi: 10.1016/j.sbi.2004.09.012. [DOI] [PubMed] [Google Scholar]; Barford, D. 2004. The role of cysteine residues as redox-sensitive regulatory switches. Curr. Opin. Struct. Biol. 14:679-686. [DOI] [PubMed]
- 13.Poole L.B. The basics of thiols and cysteines in redox biology and chemistry. Free Radic. Biol. Med. 2015;80:148–157. doi: 10.1016/j.freeradbiomed.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]; Poole, L. B. 2015. The basics of thiols and cysteines in redox biology and chemistry. Free Radic. Biol. Med. 80:148-157. [DOI] [PMC free article] [PubMed]
- 14.MacCallum J.L., Tieleman D.P. Hydrophobicity scales: a thermodynamic looking glass into lipid-protein interactions. Trends Biochem. Sci. 2011;36:653–662. doi: 10.1016/j.tibs.2011.08.003. [DOI] [PubMed] [Google Scholar]; MacCallum, J. L., and D. P. Tieleman. 2011. Hydrophobicity scales: a thermodynamic looking glass into lipid-protein interactions. Trends Biochem. Sci. 36:653-662. [DOI] [PubMed]
- 15.McDonald S.K., Fleming K.G. Aromatic side chain water-to-lipid transfer free energies show a depth dependence across the membrane normal. J. Am. Chem. Soc. 2016;138:7946–7950. doi: 10.1021/jacs.6b03460. [DOI] [PMC free article] [PubMed] [Google Scholar]; McDonald, S. K., and K. G. Fleming. 2016. Aromatic side chain water-to-lipid transfer free energies show a depth dependence across the membrane normal. J. Am. Chem. Soc. 138:7946-7950. [DOI] [PMC free article] [PubMed]
- 16.Ulmschneider M.B., Sansom M.S. Amino acid distributions in integral membrane protein structures. Biochim. Biophys. Acta. 2001;1512:1–14. doi: 10.1016/s0005-2736(01)00299-1. [DOI] [PubMed] [Google Scholar]; Ulmschneider, M. B., and M. S. Sansom. 2001. Amino acid distributions in integral membrane protein structures. Biochim. Biophys. Acta. 1512:1-14. [DOI] [PubMed]
- 17.Ulmschneider M.B., Sansom M.S., Di Nola A. Properties of integral membrane protein structures: derivation of an implicit membrane potential. Proteins. 2005;59:252–265. doi: 10.1002/prot.20334. [DOI] [PubMed] [Google Scholar]; Ulmschneider, M. B., M. S. Sansom, and A. Di Nola. 2005. Properties of integral membrane protein structures: derivation of an implicit membrane potential. Proteins. 59:252-265. [DOI] [PubMed]
- 18.Koehler Leman J., Bonneau R., Ulmschneider M.B. Statistically derived asymmetric membrane potentials from α-helical and β-barrel membrane proteins. Sci. Rep. 2018;8:4446. doi: 10.1038/s41598-018-22476-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; Koehler Leman, J., R. Bonneau, and M. B. Ulmschneider. 2018. Statistically derived asymmetric membrane potentials from α-helical and β-barrel membrane proteins. Sci. Rep. 8:4446. [DOI] [PMC free article] [PubMed]
- 19.Marx D.C., Fleming K.G. Influence of protein scaffold on side-chain transfer free energies. Biophys. J. 2017;113:597–604. doi: 10.1016/j.bpj.2017.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]; Marx, D. C., and K. G. Fleming. 2017. Influence of protein scaffold on side-chain transfer free energies. Biophys. J. 113:597-604. [DOI] [PMC free article] [PubMed]
- 20.Iyer B.R., Zadafiya P., Mahalakshmi R. Energetics of side-chain partitioning of β-signal residues in unassisted folding of a transmembrane β-barrel protein. J. Biol. Chem. 2017;292:12351–12365. doi: 10.1074/jbc.M117.789446. [DOI] [PMC free article] [PubMed] [Google Scholar]; Iyer, B. R., P. Zadafiya, …, R. Mahalakshmi. 2017. Energetics of side-chain partitioning of β-signal residues in unassisted folding of a transmembrane β-barrel protein. J. Biol. Chem. 292:12351-12365. [DOI] [PMC free article] [PubMed]
- 21.Iyer B.R., Vetal P.V., Mahalakshmi R. Salvaging the thermodynamic destabilization of interface histidine in transmembrane β-barrels. Biochemistry. 2018;57:6669–6678. doi: 10.1021/acs.biochem.8b00805. [DOI] [PMC free article] [PubMed] [Google Scholar]; Iyer, B. R., P. V. Vetal, …, R. Mahalakshmi. 2018. Salvaging the thermodynamic destabilization of interface histidine in transmembrane β-barrels. Biochemistry. 57:6669-6678. [DOI] [PMC free article] [PubMed]
- 22.Huysmans G.H., Baldwin S.A., Radford S.E. The transition state for folding of an outer membrane protein. Proc. Natl. Acad. Sci. USA. 2010;107:4099–4104. doi: 10.1073/pnas.0911904107. [DOI] [PMC free article] [PubMed] [Google Scholar]; Huysmans, G. H., S. A. Baldwin, …, S. E. Radford. 2010. The transition state for folding of an outer membrane protein. Proc. Natl. Acad. Sci. USA. 107:4099-4104. [DOI] [PMC free article] [PubMed]
- 23.Moon C.P., Fleming K.G. Using tryptophan fluorescence to measure the stability of membrane proteins folded in liposomes. Methods Enzymol. 2011;492:189–211. doi: 10.1016/B978-0-12-381268-1.00018-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; Moon, C. P., and K. G. Fleming. 2011. Using tryptophan fluorescence to measure the stability of membrane proteins folded in liposomes. Methods Enzymol. 492:189-211. [DOI] [PMC free article] [PubMed]
- 24.Huysmans G.H., Radford S.E., Baldwin S.A. The N-terminal helix is a post-assembly clamp in the bacterial outer membrane protein PagP. J. Mol. Biol. 2007;373:529–540. doi: 10.1016/j.jmb.2007.07.072. [DOI] [PMC free article] [PubMed] [Google Scholar]; Huysmans, G. H., S. E. Radford, …, S. A. Baldwin. 2007. The N-terminal helix is a post-assembly clamp in the bacterial outer membrane protein PagP. J. Mol. Biol. 373:529-540. [DOI] [PMC free article] [PubMed]
- 25.Iyer B.R., Mahalakshmi R. Residue-dependent thermodynamic cost and barrel plasticity balances activity in the PhoPQ-activated enzyme PagP of Salmonella typhimurium. Biochemistry. 2015;54:5712–5722. doi: 10.1021/acs.biochem.5b00543. [DOI] [PubMed] [Google Scholar]; Iyer, B. R., and R. Mahalakshmi. 2015. Residue-dependent thermodynamic cost and barrel plasticity balances activity in the PhoPQ-activated enzyme PagP of Salmonella typhimurium. Biochemistry. 54:5712-5722. [DOI] [PubMed]
- 26.Hwang P.M., Choy W.Y., Kay L.E. Solution structure and dynamics of the outer membrane enzyme PagP by NMR. Proc. Natl. Acad. Sci. USA. 2002;99:13560–13565. doi: 10.1073/pnas.212344499. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hwang, P. M., W. Y. Choy, …, L. E. Kay. 2002. Solution structure and dynamics of the outer membrane enzyme PagP by NMR. Proc. Natl. Acad. Sci. USA. 99:13560-13565. [DOI] [PMC free article] [PubMed]
- 27.Scott K.A., Bond P.J., Sansom M.S. Coarse-grained MD simulations of membrane protein-bilayer self-assembly. Structure. 2008;16:621–630. doi: 10.1016/j.str.2008.01.014. [DOI] [PubMed] [Google Scholar]; Scott, K. A., P. J. Bond, …, M. S. Sansom. 2008. Coarse-grained MD simulations of membrane protein-bilayer self-assembly. Structure. 16:621-630. [DOI] [PubMed]
- 28.Khan M.A., Neale C., Bishop R.E. Gauging a hydrocarbon ruler by an intrinsic exciton probe. Biochemistry. 2007;46:4565–4579. doi: 10.1021/bi602526k. [DOI] [PMC free article] [PubMed] [Google Scholar]; Khan, M. A., C. Neale, …, R. E. Bishop. 2007. Gauging a hydrocarbon ruler by an intrinsic exciton probe. Biochemistry. 46:4565-4579. [DOI] [PMC free article] [PubMed]
- 29.Khan M.A., Moktar J., Bishop R.E. Inscribing the perimeter of the PagP hydrocarbon ruler by site-specific chemical alkylation. Biochemistry. 2010;49:9046–9057. doi: 10.1021/bi1011496. [DOI] [PMC free article] [PubMed] [Google Scholar]; Khan, M. A., J. Moktar, …, R. E. Bishop. 2010. Inscribing the perimeter of the PagP hydrocarbon ruler by site-specific chemical alkylation. Biochemistry. 49:9046-9057. [DOI] [PMC free article] [PubMed]
- 30.Khan M.A., Moktar J., Bishop R.E. A thiolate anion buried within the hydrocarbon ruler perturbs PagP lipid acyl chain selection. Biochemistry. 2010;49:2368–2379. doi: 10.1021/bi901669q. [DOI] [PMC free article] [PubMed] [Google Scholar]; Khan, M. A., J. Moktar, …, R. E. Bishop. 2010. A thiolate anion buried within the hydrocarbon ruler perturbs PagP lipid acyl chain selection. Biochemistry. 49:2368-2379. [DOI] [PMC free article] [PubMed]
- 31.Ladokhin A.S., Jayasinghe S., White S.H. How to measure and analyze tryptophan fluorescence in membranes properly, and why bother? Anal. Biochem. 2000;285:235–245. doi: 10.1006/abio.2000.4773. [DOI] [PubMed] [Google Scholar]; Ladokhin, A. S., S. Jayasinghe, and S. H. White. 2000. How to measure and analyze tryptophan fluorescence in membranes properly, and why bother? Anal. Biochem. 285:235-245. [DOI] [PubMed]
- 32.Moon C.P., Zaccai N.R., Fleming K.G. Membrane protein thermodynamic stability may serve as the energy sink for sorting in the periplasm. Proc. Natl. Acad. Sci. USA. 2013;110:4285–4290. doi: 10.1073/pnas.1212527110. [DOI] [PMC free article] [PubMed] [Google Scholar]; Moon, C. P., N. R. Zaccai, …, K. G. Fleming. 2013. Membrane protein thermodynamic stability may serve as the energy sink for sorting in the periplasm. Proc. Natl. Acad. Sci. USA. 110:4285-4290. [DOI] [PMC free article] [PubMed]
- 33.Schmidt C.F., Lichtenberg D., Thompson T.E. Vesicle- vesicle interactions in sonicated dispersions of dipalmitoylphosphatidylcholine. Biochemistry. 1981;20:4792–4797. doi: 10.1021/bi00519a041. [DOI] [PubMed] [Google Scholar]; Schmidt, C. F., D. Lichtenberg, and T. E. Thompson. 1981. Vesicle- vesicle interactions in sonicated dispersions of dipalmitoylphosphatidylcholine. Biochemistry. 20:4792-4797. [DOI] [PubMed]
- 34.Cuesta-Seijo J.A., Neale C., Privé G.G. PagP crystallized from SDS/cosolvent reveals the route for phospholipid access to the hydrocarbon ruler. Structure. 2010;18:1210–1219. doi: 10.1016/j.str.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cuesta-Seijo, J. A., C. Neale, …, G. G. Prive. 2010. PagP crystallized from SDS/cosolvent reveals the route for phospholipid access to the hydrocarbon ruler. Structure. 18:1210-1219. [DOI] [PMC free article] [PubMed]
- 35.Ahn V.E., Lo E.I., Privé G.G. A hydrocarbon ruler measures palmitate in the enzymatic acylation of endotoxin. EMBO J. 2004;23:2931–2941. doi: 10.1038/sj.emboj.7600320. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ahn, V. E., E. I. Lo, …, G. G. Prive. 2004. A hydrocarbon ruler measures palmitate in the enzymatic acylation of endotoxin. EMBO J. 23:2931-2941. [DOI] [PMC free article] [PubMed]
- 36.Iyer B.R., Mahalakshmi R. Distinct structural elements govern the folding, stability, and catalysis in the outer membrane enzyme PagP. Biochemistry. 2016;55:4960–4970. doi: 10.1021/acs.biochem.6b00678. [DOI] [PubMed] [Google Scholar]; Iyer, B. R., and R. Mahalakshmi. 2016. Distinct structural elements govern the folding, stability, and catalysis in the outer membrane enzyme PagP. Biochemistry. 55:4960-4970. [DOI] [PubMed]
- 37.Öjemalm K., Higuchi T., von Heijne G. Apolar surface area determines the efficiency of translocon-mediated membrane-protein integration into the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 2011;108:E359–E364. doi: 10.1073/pnas.1100120108. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ojemalm, K., T. Higuchi, …, G. von Heijne. 2011. Apolar surface area determines the efficiency of translocon-mediated membrane-protein integration into the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 108:E359-E364. [DOI] [PMC free article] [PubMed]
- 38.Lin M., Gessmann D., Liang J. Outer membrane protein folding and topology from a computational transfer free energy scale. J. Am. Chem. Soc. 2016;138:2592–2601. doi: 10.1021/jacs.5b10307. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lin, M., D. Gessmann, …, J. Liang. 2016. Outer membrane protein folding and topology from a computational transfer free energy scale. J. Am. Chem. Soc. 138:2592-2601. [DOI] [PMC free article] [PubMed]
- 39.Landeta C., Boyd D., Beckwith J. Disulfide bond formation in prokaryotes. Nat. Microbiol. 2018;3:270–280. doi: 10.1038/s41564-017-0106-2. [DOI] [PubMed] [Google Scholar]; Landeta, C., D. Boyd, and J. Beckwith. 2018. Disulfide bond formation in prokaryotes. Nat. Microbiol. 3:270-280. [DOI] [PubMed]
- 40.Schrödinger. Schrödinger, LLC; 2008. The PyMOL Molecular Graphics System, Version 2.0. [Google Scholar]; Schrodinger. (2008). The PyMOL Molecular Graphics System, Version 2.0 (Schrodinger, LLC).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.