Abstract
Local administration of naked self-replicating mRNA (sr-mRNA) in the skin or muscle using electroporation is effective but hampered by low repeatability. In this manuscript, we demonstrated that intradermal electroporation of sr-mRNA in combination with a protein-based RNase inhibitor increased the expression efficiency, success rate, and repeatability of the data. The RNase inhibitor should be added just before administration because storage of the inhibitor together with the sr-mRNA at −80°C resulted in a partial loss of the beneficial effect. Furthermore, the location of intradermal electroporation also had a major effect on the expression of the sr-mRNA, with the highest and longest expression observed at the tail base of the mice. In contrast with previous work, we did not observe a beneficial effect of calcium ions on the efficacy of naked sr-mRNA after intradermal injection. Finally, another important finding was that the traditional representation of in vivo bioluminescence data as means in logarithmic graphs can mask highly variable data. A more truthful representation can be obtained by showing the individual data points or by displaying median values in combination with interquartile ranges. In conclusion, intradermal sr-mRNA electroporation can be improved by adding an RNase inhibitor and injecting at the tail base.
Keywords: self-replicating mRNA, electroporation, mice, RNase inhibitors, Ca2+, intradermal injection
Graphical Abstract
Introduction
In their renowned work of 1990, Wolff et al.1 showed that intramuscular injection of naked synthetic mRNA results in successful expression. However, despite this promising observation, the field of non-viral gene delivery has long been dominated by pDNA-based vectors. mRNA was not considered as an option because the instability of mRNA was perceived as insurmountable. Additionally, the large-scale commercial production and inherent innate immunogenicity of synthetic mRNA were also considered important barriers in the development of mRNA therapeutics.2, 3 However, during the past years most of these issues have been solved, and mRNA is currently widely examined for vaccination, protein (replacement) therapy, gene editing, and stem cell reprogramming applications in preclinical4, 5, 6, 7 and clinical studies.7, 8, 9, 10 The initial preclinical studies with mRNA mainly involved the local administration of naked mRNA. Although this resulted in effective protein expression and immunization, the expression and biological effects were often very variable even when combined with electroporation.11, 12, 13, 14, 15, 16 In more recent studies it has been shown that non-viral carriers can reduce the (biologic) variation and increase the efficacy of mRNA therapeutics.11, 14, 16, 17 However, a drawback of non-viral carriers is that they may cause in vivo toxicity,18, 19, 20, 21 potentiate the inherent innate immunity of synthetic mRNA,22, 23 and make the commercial production and registration of mRNA therapeutics more complicated.
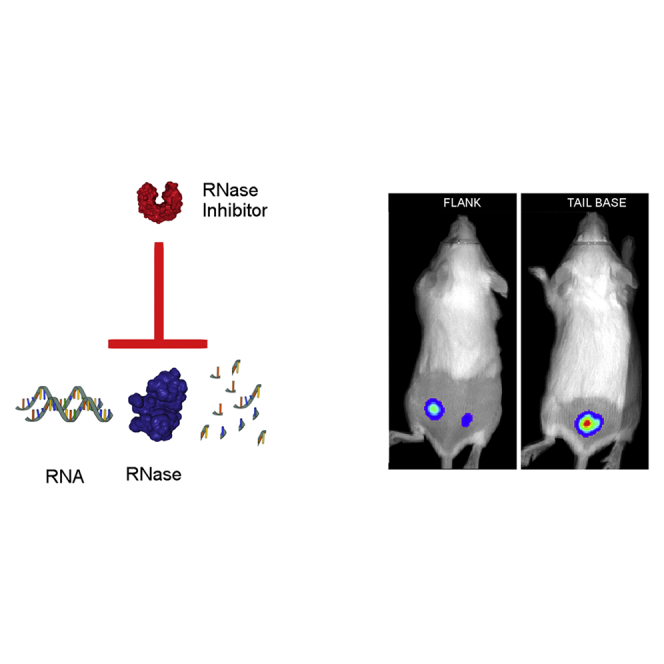
We have recently found that intradermal electroporation of a naked self-replicating mRNA (sr-mRNA) shows, in line with other studies,13, 15 a very variable expression. We hypothesized that degradation of the naked mRNA by skin-resident ribonucleases (RNases) is responsible for this effect, and that RNase inhibitors may increase the efficacy and repeatability of naked mRNA after intradermal electroporation. RNases are present in almost all biological fluids.24 High amounts of RNases are also found on the skin, where they ensure protection of the epithelial barrier against pathogens.25 Several molecules that inhibit RNases have been described.26, 27, 28 A very effective and frequently used RNase inhibitor is the placental RNase inhibitor, which is a natural occurring protein that noncompetitively inhibits a broad spectrum of RNases.29, 30 Here, we evaluated whether this protein-based RNase inhibitor can increase the efficacy and repeatability of sr-mRNA after intradermal electroporation. The protein-based RNase inhibitor was either added just before storage of the sr-mRNA at −80°C or just before injection. Moreover, we injected the sr-mRNA at the flank, as well as at the tail base, because it has been reported that the location of injection can affect the efficacy of mRNA.31, 32 Both areas drain to the same lymph nodes,33 but the skin differs considerably regarding thickness, functionality, and gene expression at these sites.34, 35 In another effort to increase the efficacy of intradermal injected mRNA, we also added calcium ions to the sr-mRNA that was dissolved in a calcium- and magnesium-free phosphate buffer. This maneuver was based on the observation that the calcium ions in Ringer lactate are responsible for the significant increase of luciferase expression after naked mRNA injection in the skin.32
Results
Inhibition of RNases Increases the Repeatability and Efficacy of sr-mRNA after Intradermal Electroporation
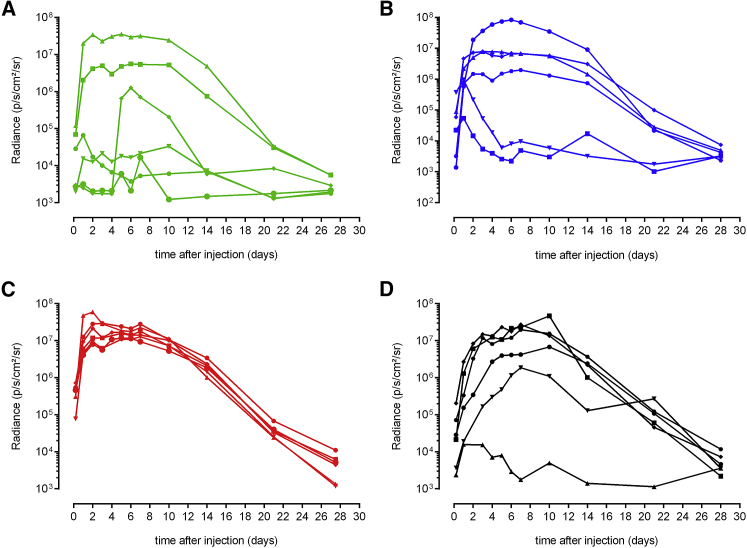
mRNA is a rather unstable molecule that is rapidly degraded by RNases that are abundantly present in the body. Therefore, we hypothesized that the efficacy of naked mRNA therapeutics after intradermal electroporation can be increased by adding an RNase inhibitor. In this study, we used a sr-mRNA based on Venezuelan equine encephalitis virus (VEEV) (Figure 1). Without RNase inhibitor the luciferase expression after intradermal electroporation of this sr-mRNA is extremely variable, and a successful expression was obtained in only two out of six administrations (Figure 2A).
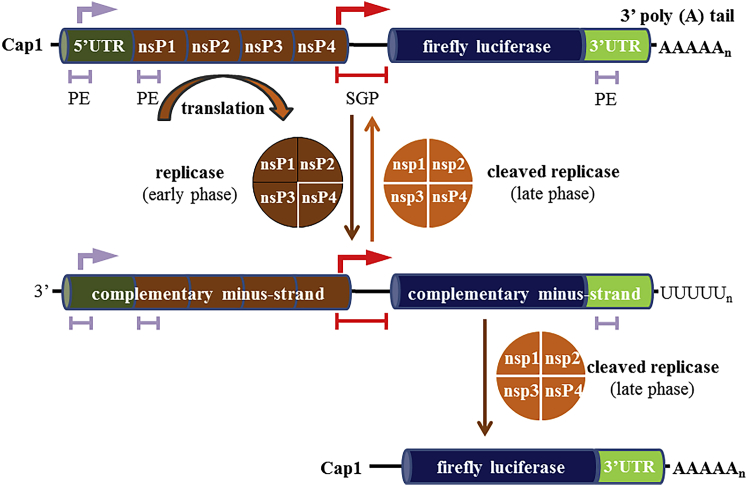
Figure 1.
Schematic Representation of the sr-mRNA and Its Replication
The sr-mRNA starts at its 5′ end with a cap1, a 5′ UTR, and the sequences of the non-structural proteins (nsP1–4) of Venezuelan equine encephalitis virus (VEEV). These non-structural proteins are translated as a polyprotein that forms a replicase (brown). The non-structural proteins are followed by a subgenomic promoter (SGP, red), which starts in nsP4. The sequence of the protein(s) of interest (blue) can be found behind the SGP. In this work, the protein of interest was firefly luciferase. At the 3′ end, an UTR and a polyA tail are present. When the sr-mRNA arrives in the cytosol, the nsP1–4 polyprotein is translated and cleaved by nsP2 to generate the early replication complex (replicase), which consists of nsP1–3 and associated nsP4. In a later phase, nsP1–3 is also cleaved, and the individual nsPs join together to form the cleaved replicase. Three promoter elements (PEs) trigger the replicase and cleaved replicase to generate, respectively, complementary minus-RNA strands and new copies of the original genomic RNA starting from the minus-RNA strands. In addition, the SGP triggers the cleaved replicase to produce an excessive amount of subgenomic RNAs.
Figure 2.
Effect of the RNase Inhibitor on Luciferase Expression after Intradermal Electroporation of sr-mRNA in the Flank of Mice
(A–D) Five micrograms of luciferase encoding sr-mRNA dissolved in 50 μL PBS was supplemented with either 0 (A), 0.33 (B), or 1.0 U/μL (C and D) of RNase inhibitor. In (B) and (C), the RNase inhibitor was added to the sr-mRNA just before administration, whereas in (D), the RNase inhibitor (1.0 U/μL) was added to the sr-mRNA before storage at −80°C. The graphs show the individual expression profiles in each of the six injection spots (n = 6).
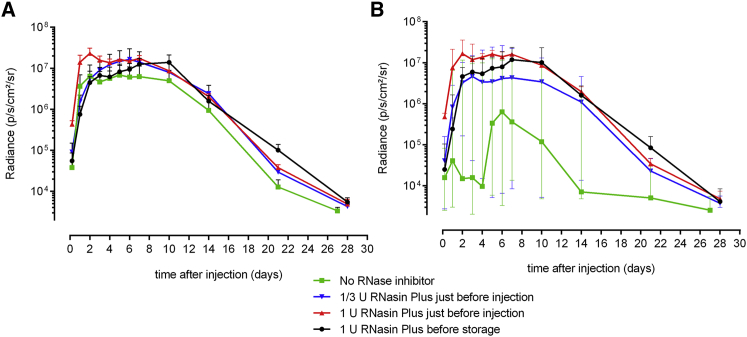
An intradermal electroporation of sr-mRNA was considered successful when the luciferase expression generated a radiance above 4 × 105 photons/s/cm2/steradian (p/s/cm2/sr) during at least four consecutive days as observed previously.23 Supplementation of the sr-mRNA just before injection with 0.33 or 1.0 U RNase inhibitor/μL increased the percentage of successful administrations to, respectively, 67% and 100% (total number of administrations was six; Figures 2B and 2C). Interestingly, the RNase inhibitor not only increased the number of effective administrations, but also drastically reduced the variation of the luciferase expression in the different injection spots (Figure 2C).
Remarkably, a lower success rate (5 out of 6 injections were successful) and more variable results were obtained when the RNase inhibitor (1.0 U/μL) was added to the sr-mRNA before storage at −80°C instead of immediately before injection (Figure 2C versus 2D).
Next, we calculated the mean bioluminescence signals of the data in Figure 2. When we plot these means with their SEM on a logarithmic y axis, as is usually done with in vivo bioluminescence signals from reporters, it becomes clear that this way of representing the data masks the strong variability of the data shown in especially Figures 2A, 2B, and 3A. Indeed, based on Figure 3A (green versus red curve), one would conclude that the RNase inhibitor has only a minor and non-significant beneficial effect on the expression efficacy. However, by plotting the median instead of the mean values, a more faithful representation of the data is obtained (Figure 3B).
Figure 3.
The Use of Median Values and Interquartile Ranges Is More Appropriate for Presenting Highly Variable Bioluminescence Data
(A and B) The mean (± SEM) (A) and median (± interquartile range) (B) bioluminescence signals obtained after intradermal electroporation of luciferase encoding sr-mRNA in the absence or presence of RNase inhibitor. Five micrograms of luciferase encoding sr-mRNA dissolved in 50 μL PBS was injected in the flanks of mice and immediately electroporated. The green curves show the luciferase expression of the sr-mRNA without supplementation with RNase inhibitor. The blue and red curves depict the luciferase expression after adding, respectively, 0.33 and 1.0 U/μL RNase inhibitor to the sr-mRNA just before injection. The black curves represent the luciferase expression after adding 1.0 U/μL RNase inhibitor before storage at −80°C. (n = 6). Due to the high variation in luciferase expression, no significant differences were observed between the groups.
The Efficacy of Intradermal sr-mRNA Electroporation Is Location Dependent
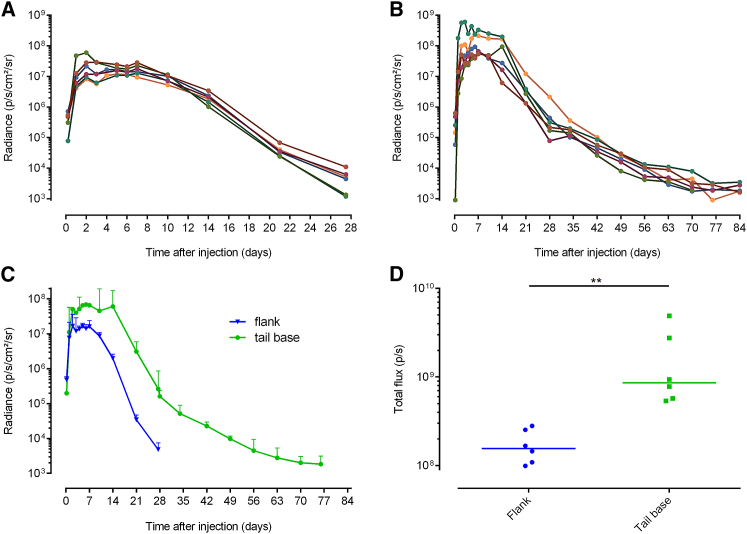
The skin is a large organ with regional variances in thickness, cell-type density, and perfusion that might influence the expression of genetic vectors.36 Because there is a clear difference between the skin characteristics at the flank and the tail base,34, 35 we decided to compare the expression kinetics of sr-mRNA after intradermal electroporation at these two locations. Intradermal electroporation of sr-mRNA in the tail base caused a significantly higher and longer luciferase expression compared with intradermal electroporation in the flank (Figures 4A and 4B).
Figure 4.
Comparison of the Luciferase Expression Kinetics after Intradermal Electroporation of sr-mRNA at the Flank or at the Tail Base
(A–D) Five micrograms of sr-mRNA-encoding luciferase was just before injection supplemented with 1.0 U/μL RNase inhibitor and after intradermal injection immediately electroporated. Each curve in (A) and (B) represents the evolution of the individual bioluminescence signal in the separate injection spots of the flank (A) or the tail base (B) (n = 6). (C) The median bioluminescence signal over time after intradermal electroporation in the flank (blue) and in the tail base (green) is shown (error bars represent interquartile range; n = 6). (D) The area under the curve (AUC) of the curves in (C) and the median AUC (line) are plotted. Note the differences in the scales of the x axes in the graphs. **p = 0.0022.
In more detail, at the plateau, the luciferase expression at the tail base was 4-fold higher and the plateau lasted twice as long (6 versus 12 days) compared with intradermal electroporation in the flank. Additionally, it took about 2 months before the luciferase expression at the tail base disappeared, whereas the expression at the flank vanished already after 1 month. Although the variability of the data after intradermal electroporation in the tail base was slightly higher than after intradermal electroporation in the flank, it stayed within acceptable limits. To estimate the overall protein expression, we calculated the area under the curve (AUC) of the curves shown in Figure 4C and found a 5-fold higher median expression after injection at the tail base (Figure 4D).
Effect of Calcium Ions on the Efficacy of sr-mRNA after Intradermal Administration
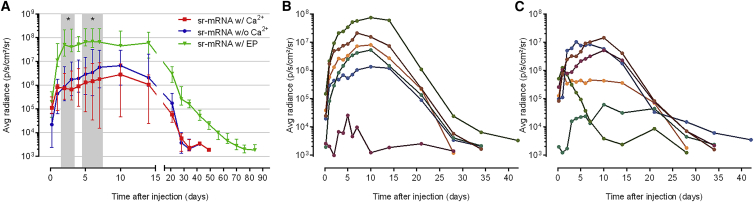
Probst et al.32 reported that the efficacy of naked mRNA after intradermal injection can be drastically increased by using Ringer lactate buffer instead of PBS (without calcium and magnesium). Additionally, they demonstrated that the Ca2+ ions present in the Ringer lactate were responsible for this effect.32 Therefore, we wondered whether addition of Ca2+ ions could improve the transfection of naked sr-mRNA after intradermal injection without using electroporation. To this end, sr-mRNA dissolved in calcium, and magnesium-free PBS was supplemented with 3 mEq/L Ca2+ ions, which is similar to the concentration of this ion in Baxter’s Ringer lactate solution. However, as shown in Figure 5, addition of Ca2+ ions to sr-mRNA did not increase its transfection efficacy after intradermal injection. On the contrary, even a small drop in expression was observed (Figure 5A). Also, the number of successful injections was lower when Ca2+ ions were added (4 out of 6 versus 5 out of 6; Figures 5B and 5C). In contrast, the expression of naked sr-mRNA was significantly increased by electroporation as demonstrated by the green curve in Figure 5A.
Figure 5.
The Effect of Calcium Ions on the Expression of Naked sr-mRNA after Intradermal Administration without Electroporation
(A) Median luciferase expression with interquartile range after intradermal injection of sr-mRNA (5 μg) dissolved in PBS (Ca/Mg free) without (blue curve) or with 3 mEq/L CaCl2 (red curve). The naked sr-mRNA was supplemented with 1.0 U/μL RNasin just before intradermal injection (six mice per group). The luciferase expression after intradermal electroporation of 5 μg sr-mRNA (dissolved in Ca/Mg-free PBS, green curve) in the tail base is also shown as a reference. The gray zones display significant differences between intradermally electroporated sr-mRNA and intradermally injected naked sr-mRNA with or without calcium. *p < 0.05. (B and C) Luciferase expression in the individual intradermal injection spots of 5 μg sr-mRNA dissolved in PBS (B) or in PBS supplemented with 3 mEq/L CaCl2 (C) (6 mice per group). All intradermal injections in these graphs were performed at the tail base.
Size and zeta potential measurements indicated that addition of Ca2+ ions to sr-mRNA resulted in the formation of nanoparticles. In more detail, the zeta potential of sr-mRNA decreased from −7 ± 4 to −29 ± 1 mV when PBS was supplemented with Ca2+ ions. Additionally, the size of the sr-mRNA slightly increased upon addition of the Ca2+ ions (Table S1).
Discussion
Our data reveal that supplementation of naked sr-mRNA with an RNase inhibitor just before intradermal electroporation resulted in a 70-fold increase of the median total luciferase expression (Figure S1). The latter was obtained by calculating the median of the AUCs of the individual curves in Figures 2A and 2C. The addition of the RNase inhibitor also improved the success rates of the administration procedure (from 2 out of 6 to 6 out of 6 successful expressions; Figures 2A and 2C) and the repeatability of the data (Figure 2C). These beneficial effects of the RNase inhibitor were less pronounced after intradermal injection of sr-mRNA in the absence of electroporation (Figure S2), where a better repeatability was seen in only the first 4 days. To inhibit endogenous RNases, we used a protein-based RNase inhibitor derived from human placenta. In the past, Phua et al.31 also evaluated this RNase inhibitor, but found that addition of this inhibitor to naked mRNA did not increase nor decrease its efficacy. This discrepancy between both studies is likely due to the difference in the pH of the buffers. In the work of Phua et al.,31 the naked RNA and the RNase inhibitor were dissolved in an acidic buffer with pH 5, whereas we used PBS at pH 7.4. This difference is very important because the inhibitor capacity of the human placental RNase inhibitor is maximal between pH 7 and 8.37 Moreover, the inhibitor starts to lose its RNase blocking capacities at a pH below 6.38 The pH-dependent efficacy of this protein-based RNase inhibitor is not surprising because electrostatic interactions are involved in the binding of the RNase inhibitor (negatively charged at physiologic pH) with RNases (positively charged at physiologic pH).39 Based on the sequence of the human placental RNase inhibitor used in the work of Phua et al.,31 we calculated that it has an isoelectric point of 4.8 (UniProt: P13489).40 Consequently, at the pH used by Phua et al.31 (i.e., pH 5), this protein-based RNase inhibitor will be almost neutral and hence largely lose its RNase binding and blocking capacities. This most likely explains why they did not observe any beneficial effect of the RNase inhibitor in their paper.
In Figure 2A, we noticed that one unsuccessful injection suddenly gave rise to a moderate luciferase expression after day 4. Although the exact reason is not known, we speculate that in this specific case many of the sr-mRNAs were degraded by extracellular RNases, and that hence only a limited amount of sr-mRNAs entered the cytoplasm. We assume that this higher amount of degraded mRNAs triggered a stronger innate immune response than usual. This intense innate response caused, probably 6–12 h after sr-mRNA administration, a premature arrest of the translation of the replicase. Nevertheless, over the next hours and days the replicase succeeded: (1) to replicate the few intact copies of the sr-mRNA and (2) to generate many copies of the luciferase encoding subgenomic RNA. However, this did not directly result in a luciferase expression because the elicited innate immune response still inhibited the translation of the subgenomic RNAs. To explain the sudden rise of the luciferase, we speculate that the elicited factors causing translation block steadily dropped, and that after day 4 they became below the “translation inhibition threshold.” This allowed a massive translation of the generated subgenomic RNA.
Surprisingly, we observed that the beneficial effect of the protein-based RNase inhibitor was partly lost when it was added to the sr-mRNA prior to storage at −80°C. This brings us to another important pitfall when using human placental RNase inhibitor to protect mRNA therapeutics against RNases. In laboratory protocols involving the manipulation of isolated RNA, it is often stated that, e.g., human placental RNase inhibitor can be added to the isolated RNA before freezing. The stock solution of the protein-based RNase inhibitor (40 U/μL) is stored at −20°C and contains 50% glycerol, which prevents freezing of the stock solution at −20°C. However, by adding the advised amount of 1.0 U of RNase inhibitor per microliter RNA, a 40-fold dilution of the RNase inhibitor occurs. Consequently, this mixture will freeze below 0°C and hence the protein-based RNase inhibitor will be exposed to freeze denaturation. This most likely explains the lower success rate and higher variation of the data in Figure 2D when we used RNase inhibitor-supplemented sr-mRNA solutions that had been stored at −80°C. Therefore, to protect naked mRNA against RNases in the body, it is advised to add the human placental RNase inhibitor just before administration and not store it together with the mRNA in the freezer. Our study also shows that the beneficial effect of the RNase inhibitor on RNA expression was dose dependent (Figures 2B and 2C). The optimal dose was 1 U/μL, because increasing the dose to 3 U/μL did not affect RNA expression (Figure S3). A unit of this inhibitor is defined as the amount that is necessary to inhibit the activity of 5 ng of RNase A by 50%. When using 50 U in 50 μL of injection solution, as in our experiments, this comes down to the potential of inhibiting 250 ng of RNase A by 50%.
We also demonstrated that the interpretation of highly variable in vivo bioluminescence data is influenced by the presentation of the data. In the literature, in vivo bioluminescence data of vectors encoding, e.g., luciferase are frequently displayed as means accompanied by error bars representing the SEM.4, 15, 17, 31, 41 The value of in vivo bioluminescence signals ranges in general between 104 and 109. Therefore, a few high data points in a series of bioluminescence data can drastically increase the mean, whereas low signals will have almost no effect on the value of the mean. The error bars on graphs that represent bioluminescence data should depict this variability. However, in vivo bioluminescence data are represented on graphs with a logarithmic y-axis. Consequently, the upper error bar in such graphs is always quite small compared with the mean, and the opposite applies for the lower error bar. Additionally, in many papers the error bar in these graphs represents the SEM, which makes it even more difficult for the reader to get an idea of the variability of the data. Therefore, a more reliable way of presenting in vivo bioluminescence data is showing all of the individual data points.11, 14, 32 However, when using large group samples, graphs often become cluttered, resulting in loss of clarity. Another good alternative in this case is the use of medians and interquartile ranges for the representation of the data.
The site where the sr-mRNA is injected in the skin of the mice (tail base versus flank) had a drastic effect on the expression efficacy and duration. Intradermal electroporation at the tail base resulted in a significant, 5-fold higher overall expression and lasted twice as long compared with administration at the flank. Also, Phua et al.31 found that subcutaneous injection at the tail base resulted in a higher expression of the mRNA compared with injection at the base of the ear pinna. To exclude that the higher luciferase expression at the tail base was due to a better tissue penetration of the bioluminescent light at the tail base, we intradermally injected at the flank and tail base an equal amount of 4T1 cells that stably express luciferase. The bioluminescence signals at both locations were compared, and no significant differences were observed (Figure S4). Therefore, we can conclude that the differences in luciferase expression at the flank and tail base are not due to a better tissue penetration of the bioluminescent light at the tail base. Hence the differences are really due to an increased luciferase expression at the tail base.
It has been reported that the efficacy of intradermal or subcutaneous injected naked mRNA can also be improved by dissolving the mRNA in Ringer lactate buffer. Probst et al.32 found that the Ca2+ ions in Ringer lactate are causing this increase in expression. However, our study did not find any beneficial effect of Ca2+ ions on the transfection efficacy of naked sr-mRNA after intradermal injection without electroporation. The reason for this discrepancy is not clear, but a possible explanation is that the Ca2+ ions partly protect the mRNA against RNases by forming a complex (particle) with mRNA. In our study, the sr-mRNA was already protected because an RNase inhibitor was used, and therefore we probably could not detect the beneficial effects of the Ca2+ ions. The hypothesis that Ca2+ ions increase the efficacy of naked mRNA by reducing its degradation by RNase is further supported by the fact that Ca2+ ions have been reported to inhibit human pancreatic RNase activity.42, 43
Conclusions
In conclusion, we found that addition of a protein-based RNase inhibitor to sr-mRNA just before intradermal electroporation improved its repeatability and increased the efficacy and success rate of the sr-mRNA expression. The beneficial effect of the RNase inhibitors was partly lost when it was added beforehand and thus stored together with the sr-mRNA at −80°C. This can be attributed to a partial degradation of the protein-based RNase inhibitor by freeze denaturation. In this paper, we also demonstrated that the traditional representation of in vivo bioluminescence data as mean ± SEM in logarithmic graphs can mask highly variable data. A more truthful representation can be obtained by displaying median values in combination with interquartile ranges. Furthermore, in an attempt to increase the translation efficiency of the mRNA, we demonstrated that the expression of intradermally electroporated sr-mRNA in the tail base was significantly superior compared with its expression in the flank. This vast difference emphasizes that the location of an intradermal mRNA injection should be carefully chosen in human clinical trials. Finally, it has been reported that Ca2+ can increase the efficacy of naked mRNA.32 However, using RNase inhibitor-supplemented sr-mRNA, we could not confirm this effect. We speculate that the beneficial effect of calcium in previous work is due to its capacity to block RNases and to condense mRNA into nanoparticles.
Materials and Methods
Mice
Female wild-type BALB/cJRj mice were purchased from Janvier (France) or bred in-house in a climate-controlled facility under a 14/10-h light/dark cycle. The mice were kept in individually ventilated cages with ad libitum access to feed and water. The described experiments with mice in this paper were conducted with the approval of the ethical committee of the Faculty of Veterinary Medicine, Ghent University (EC 2016/17). At the start of the experiments the mice were between 7 and 10 weeks old.
Plasmids and mRNA Synthesis
The plasmid pTK160 (11,519 bp) used for in vitro transcription (IVT) of firefly luciferase encoding sr-mRNA (Figure 1) was a kind gift of Dr. Tasuku Kitada and Prof. Ron Weiss (Massachusetts Institute of Technology, Cambridge, MA, USA). The plasmid is derived from VEEV strain TC-83 containing a substitution in the 5′ UTR (r.3a>g) and in nsP2 (p.Q739L). The sequence coding for the structural proteins was replaced by the reporter gene luc2. This plasmid was constructed using standard Gateway cloning procedures and contains, besides the reporter genes, also VEEV-derived 5′ and 3′ UTRs. E. coli bacteria containing the plasmids were cultivated in lysogeny broth (LB; Invitrogen, MA, USA), and the plasmids were subsequently isolated using the Plasmid Plus midi kit (QIAGEN, Germany). mRNA was produced by IVT of I-SceI (NEB, MA, USA) linearized pTK160 using the MEGAscript T7 Transcription kit (Life Technologies, MA, USA). Following IVT, the RNA was treated with DNase I (Invitrogen) and purified using the RNeasy Mini Kit (QIAGEN), denatured at 65°C, and enzymatically (cap1) capped using the ScriptCap m7G Capping System (Cellscript, Madison, WI, USA) and ScriptCap 2′-O-Methyltransferase Kit (Cellscript). All mRNAs were purified with the RNeasy Mini kit (QIAGEN) between two modifications and before in vivo application. After the final purification, the sr-mRNA was stored at −80°C. The mean hydrodynamic diameter and zeta potential were determined using a Nano ZS90 (Malvern Pananalytical, Worcestershire, UK).
Intradermal Delivery of sr-mRNA
Five micrograms of sr-mRNA dissolved in 50 μL PBS (Ca2+ and Mg2+ free; Ambion; Thermo Fisher Scientific, MA, USA) was supplemented with different concentrations of RNase inhibitor (RNasin Plus; Promega, WI, USA) before storage at −80°C or just before injection. RNasin Plus is a recombinant variant of the human placental RNase inhibitor that is more resistant toward oxidation than RNase inhibitor due to the substitution of two cysteine residues (Cys328 and Cys329) by alanine residues.44 RNasin Plus is a noncompetitive inhibitor of RNase A, RNase B, and placental RNase.45 Where indicated in the Results, RNase-free CaCl2 (Sigma-Aldrich, Germany) was also added to the sr-mRNA at a final concentration of 3 mEq/L Ca2+ (as present in Baxter’s Ringer lactate buffer). Prior to intradermal administration, the mice were shaved to allow a better exit of the photons produced during the luciferase-mediated oxidation of luciferin. The sr-mRNA (5 μg in 50 μL PBS) was intradermally injected at the flanks or at the tail base of the mice using a 29G insulin needle (VWR, the Netherlands). Electroporation, when used, was executed immediately after each injection with a needle array electrode consisting of two rows of four 2-mm-long needles. The gap between the rows was 4 mm, and within the rows the needles were 1.5 mm apart from each other (AgilePulse; BTX Harvard Apparatus, MA, USA). Needles were completely inserted in the skin covering the injection site. The electroporation has been previously described and involved two short 0.05-ms high-voltage pulses of 450 V with an interval of 300 ms, followed by eight long low-voltage pulses of 100 V with a duration of 10 ms and an interval of 300 ms.46 In experiments where sr-mRNA was supplemented with Ca2+ ions, no electroporation was used, because it is well known that the combination of Ca2+ ions with electroporation leads to cell death.47, 48, 49
Bioluminescence Imaging
To monitor the luciferase expression, we intraperitoneally injected the mice with 200 μL D-luciferin (15 mg/mL; Gold Biotechnology, MO, USA), and in vivo bioluminescent imaging was performed 15 min later using an IVIS Lumina II (Perkin-Elmer, Zaventem, Belgium). The average radiance in the region of interest was determined using the Living ImageSoftware 4.3.1. In vivo bioluminescence imaging was repeated at different time points to study the expression kinetics of the sr-mRNA. Time point 0 in the graphs indicates the time of administration, and the first imaging time point was 5 h (0.2 day) after injection. The intradermal injections and bioluminescence imaging were performed under gas anesthesia (isoflurane: 5% induction and 2% maintenance).
Statistical Analysis
Statistical analysis was performed with the software GraphPad Prism 6 (GraphPad Software, CA, USA). Longitudinal experiments were analyzed with repeated-measures two-way ANOVA, followed by Sidak’s or Tukey’s multiple comparisons test. Differences between medians were analyzed using the non-parametric Mann-Whitney U test. Differences are found to be significant when the p value is <0.05.
Author Contributions
Experiments were designed by H.H., J.D.T., and N.N.S. H.H. and J.D.T. conducted the experiments with the support of Z.Z., S.M.C., and F.C. The manuscript was written by H.H. and J.D.T. with input from all authors. F.H. provided critical feedback and assisted with the supervision. N.N.S. supervised the project and corrected the drafts of the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
The plasmid pTK160 was a kind gift from Tasuku Kitada and Ron Weiss (Massachusetts Institute of Technology, USA). This work was financially supported by the concerted research action (GOA) fund of Ghent University (Project Code BOF15/GOA/013) and by the China Scholarship Council (grant 201607650018).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.06.011.
Supplemental Information
References
- 1.Wolff J.A., Malone R.W., Williams P., Chong W., Acsadi G., Jani A., Felgner P.L. Direct gene transfer into mouse muscle in vivo. Science. 1990;247:1465–1468. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- 2.Sahin U., Karikó K., Türeci Ö. mRNA-based therapeutics—developing a new class of drugs. Nat. Rev. Drug Discov. 2014;13:759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 3.Kwon H., Kim M., Seo Y., Moon Y.S., Lee H.J., Lee K., Lee H. Emergence of synthetic mRNA: In vitro synthesis of mRNA and its applications in regenerative medicine. Biomaterials. 2018;156:172–193. doi: 10.1016/j.biomaterials.2017.11.034. [DOI] [PubMed] [Google Scholar]
- 4.Vogel A.B., Lambert L., Kinnear E., Busse D., Erbar S., Reuter K.C., Wicke L., Perkovic M., Beissert T., Haas H. Self-Amplifying RNA Vaccines Give Equivalent Protection against Influenza to mRNA Vaccines but at Much Lower Doses. Mol. Ther. 2018;26:446–455. doi: 10.1016/j.ymthe.2017.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sehgal A., Barros S., Ivanciu L., Cooley B., Qin J., Racie T., Hettinger J., Carioto M., Jiang Y., Brodsky J. An RNAi therapeutic targeting antithrombin to rebalance the coagulation system and promote hemostasis in hemophilia. Nat. Med. 2015;21:492–497. doi: 10.1038/nm.3847. [DOI] [PubMed] [Google Scholar]
- 6.Zhong X., Zhang D., Xiong M., Zhang L. Noncoding RNA for Cancer Gene Therapy. Recent Results Cancer Res. 2016;209:51–60. doi: 10.1007/978-3-319-42934-2_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhong Z., Mc Cafferty S., Combes F., Huysmans H., De Temmerman J., Gitsels A., Vanrompay D., Catani J.P., Sanders N.N. mRNA therapeutics deliver a hopeful message. Nano Today. 2018;23:16–39. [Google Scholar]
- 8.Weide B., Carralot J.P., Reese A., Scheel B., Eigentler T.K., Hoerr I., Rammensee H.G., Garbe C., Pascolo S. Results of the first phase I/II clinical vaccination trial with direct injection of mRNA. J. Immunother. 2008;31:180–188. doi: 10.1097/CJI.0b013e31815ce501. [DOI] [PubMed] [Google Scholar]
- 9.Rittig S.M., Haentschel M., Weimer K.J., Heine A., Muller M.R., Brugger W., Horger M.S., Maksimovic O., Stenzl A., Hoerr I. Intradermal vaccinations with RNA coding for TAA generate CD8+ and CD4+ immune responses and induce clinical benefit in vaccinated patients. Mol. Ther. 2011;19:990–999. doi: 10.1038/mt.2010.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sahin U., Derhovanessian E., Miller M., Kloke B.P., Simon P., Löwer M., Bukur V., Tadmor A.D., Luxemburger U., Schrörs B. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature. 2017;547:222–226. doi: 10.1038/nature23003. [DOI] [PubMed] [Google Scholar]
- 11.Lutz J., Lazzaro S., Habbeddine M., Schmidt K.E., Baumhof P., Mui B.L., Tam Y.K., Madden T.D., Hope M.J., Heidenreich R., Fotin-Mleczek M. Unmodified mRNA in LNPs constitutes a competitive technology for prophylactic vaccines. NPJ Vaccines. 2017;2:29. doi: 10.1038/s41541-017-0032-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leyman B., Huysmans H., Mc Cafferty S., Combes F., Cox E., Devriendt B., Sanders N.N. Comparison of the expression kinetics and immunostimulatory activity of replicating mRNA, nonreplicating mRNA, and pDNA after intradermal electroporation in pigs. Mol. Pharm. 2018;15:377–384. doi: 10.1021/acs.molpharmaceut.7b00722. [DOI] [PubMed] [Google Scholar]
- 13.Cu Y., Broderick K.E., Banerjee K., Hickman J., Otten G., Barnett S., Kichaev G., Sardesai N.Y., Ulmer J.B., Geall A. Enhanced Delivery and Potency of Self-Amplifying mRNA Vaccines by Electroporation in Situ. Vaccines (Basel) 2013;1:367–383. doi: 10.3390/vaccines1030367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geall A.J., Verma A., Otten G.R., Shaw C.A., Hekele A., Banerjee K., Cu Y., Beard C.W., Brito L.A., Krucker T. Nonviral delivery of self-amplifying RNA vaccines. Proc. Natl. Acad. Sci. USA. 2012;109:14604–14609. doi: 10.1073/pnas.1209367109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johansson D.X., Ljungberg K., Kakoulidou M., Liljeström P. Intradermal electroporation of naked replicon RNA elicits strong immune responses. PLoS ONE. 2012;7:e29732. doi: 10.1371/journal.pone.0029732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brito L.A., Chan M., Shaw C.A., Hekele A., Carsillo T., Schaefer M., Archer J., Seubert A., Otten G.R., Beard C.W. A cationic nanoemulsion for the delivery of next-generation RNA vaccines. Mol. Ther. 2014;22:2118–2129. doi: 10.1038/mt.2014.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pardi N., Tuyishime S., Muramatsu H., Kariko K., Mui B.L., Tam Y.K., Madden T.D., Hope M.J., Weissman D. Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J. Control. Release. 2015;217:345–351. doi: 10.1016/j.jconrel.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reichmuth A.M., Oberli M.A., Jaklenec A., Langer R., Blankschtein D. mRNA vaccine delivery using lipid nanoparticles. Ther. Deliv. 2016;7:319–334. doi: 10.4155/tde-2016-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aramaki Y., Takano S., Tsuchiya S. Induction of apoptosis in macrophages by cationic liposomes. FEBS Lett. 1999;460:472–476. doi: 10.1016/s0014-5793(99)01386-1. [DOI] [PubMed] [Google Scholar]
- 20.Dokka S., Toledo D., Shi X., Castranova V., Rojanasakul Y. Oxygen radical-mediated pulmonary toxicity induced by some cationic liposomes. Pharm. Res. 2000;17:521–525. doi: 10.1023/a:1007504613351. [DOI] [PubMed] [Google Scholar]
- 21.Yun C.H., Bae C.S., Ahn T. Cargo-Free Nanoparticles Containing Cationic Lipids Induce Reactive Oxygen Species and Cell Death in HepG2 Cells. Biol. Pharm. Bull. 2016;39:1338–1346. doi: 10.1248/bpb.b16-00264. [DOI] [PubMed] [Google Scholar]
- 22.Fotin-Mleczek M., Duchardt K.M., Lorenz C., Pfeiffer R., Ojkić-Zrna S., Probst J., Kallen K.J. Messenger RNA-based vaccines with dual activity induce balanced TLR-7 dependent adaptive immune responses and provide antitumor activity. J. Immunother. 2011;34:1–15. doi: 10.1097/CJI.0b013e3181f7dbe8. [DOI] [PubMed] [Google Scholar]
- 23.Huysmans H., Zhong Z., De Temmerman J., Mui B.L., Tam Y.K., Mc Cafferty S. Expression kinetics and innate immune response after electroporation and lipid nanoparticle mediated delivery of a self-amplifying mRNA in the skin of mice. bioRxiv. 2019 doi: 10.1016/j.omtn.2019.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu L., Li J., Moussaoui M., Boix E. Immune Modulation by Human Secreted RNases at the Extracellular Space. Front. Immunol. 2018;9:1012. doi: 10.3389/fimmu.2018.01012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Probst J., Brechtel S., Scheel B., Hoerr I., Jung G., Rammensee H.G., Pascolo S. Characterization of the ribonuclease activity on the skin surface. Genet. Vaccines Ther. 2006;4:4. doi: 10.1186/1479-0556-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kime L., Vincent H.A., Gendoo D.M., Jourdan S.S., Fishwick C.W., Callaghan A.J., McDowall K.J. The first small-molecule inhibitors of members of the ribonuclease E family. Sci. Rep. 2015;5:8028. doi: 10.1038/srep08028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith B.D., Soellner M.B., Raines R.T. Potent inhibition of ribonuclease A by oligo(vinylsulfonic acid) J. Biol. Chem. 2003;278:20934–20938. doi: 10.1074/jbc.M301852200. [DOI] [PubMed] [Google Scholar]
- 28.Earl C.C., Smith M.T., Lease R.A., Bundy B.C. Polyvinylsulfonic acid: A Low-cost RNase inhibitor for enhanced RNA preservation and cell-free protein translation. Bioengineered. 2018;9:90–97. doi: 10.1080/21655979.2017.1313648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee F.S., Fox E.A., Zhou H.M., Strydom D.J., Vallee B.L. Primary structure of human placental ribonuclease inhibitor. Biochemistry. 1988;27:8545–8553. doi: 10.1021/bi00423a007. [DOI] [PubMed] [Google Scholar]
- 30.Dickson K.A., Haigis M.C., Raines R.T. Ribonuclease inhibitor: structure and function. Prog. Nucleic Acid Res. Mol. Biol. 2005;80:349–374. doi: 10.1016/S0079-6603(05)80009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phua K.K., Leong K.W., Nair S.K. Transfection efficiency and transgene expression kinetics of mRNA delivered in naked and nanoparticle format. J. Control. Release. 2013;166:227–233. doi: 10.1016/j.jconrel.2012.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Probst J., Weide B., Scheel B., Pichler B.J., Hoerr I., Rammensee H.G., Pascolo S. Spontaneous cellular uptake of exogenous messenger RNA in vivo is nucleic acid-specific, saturable and ion dependent. Gene Ther. 2007;14:1175–1180. doi: 10.1038/sj.gt.3302964. [DOI] [PubMed] [Google Scholar]
- 33.Harrell M.I., Iritani B.M., Ruddell A. Lymph node mapping in the mouse. J. Immunol. Methods. 2008;332:170–174. doi: 10.1016/j.jim.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Azzi L., El-Alfy M., Martel C., Labrie F. Gender differences in mouse skin morphology and specific effects of sex steroids and dehydroepiandrosterone. J. Invest. Dermatol. 2005;124:22–27. doi: 10.1111/j.0022-202X.2004.23545.x. [DOI] [PubMed] [Google Scholar]
- 35.Rinn J.L., Wang J.K., Liu H., Montgomery K., van de Rijn M., Chang H.Y. A systems biology approach to anatomic diversity of skin. J. Invest. Dermatol. 2008;128:776–782. doi: 10.1038/sj.jid.5700986. [DOI] [PubMed] [Google Scholar]
- 36.Larsson B.A., Norman M., Bjerring P., Egekvist H., Lagercrantz H., Olsson G.L. Regional variations in skin perfusion and skin thickness may contribute to varying efficacy of topical, local anaesthetics in neonates. Paediatr. Anaesth. 1996;6:107–110. doi: 10.1111/j.1460-9592.1996.tb00370.x. [DOI] [PubMed] [Google Scholar]
- 37.Blackburn P., Wilson G., Moore S. Ribonuclease inhibitor from human placenta. Purification and properties. J. Biol. Chem. 1977;252:5904–5910. [PubMed] [Google Scholar]
- 38.Shultz J., Hurst R., Betz N. RNasin® Ribonuclease Inhibitor Part I: Characterization of the Protein. Promega Notes. 2001;77:8–11. https://pdfs.semanticscholar.org/059b/f1cd98540b9256466e9b51705831ef3ecbf8.pdf [Google Scholar]
- 39.Johnson R.J., McCoy J.G., Bingman C.A., Phillips G.N., Jr., Raines R.T. Inhibition of human pancreatic ribonuclease by the human ribonuclease inhibitor protein. J. Mol. Biol. 2007;368:434–449. doi: 10.1016/j.jmb.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stothard P. The Sequence Manipulation Suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques. 2000;28:1102–1104. doi: 10.2144/00286ir01. [DOI] [PubMed] [Google Scholar]
- 41.Pepini T., Pulichino A.-M., Carsillo T., Carlson A.L., Sari-Sarraf F., Ramsauer K., Debasitis J.C., Maruggi G., Otten G.R., Geall A.J. Induction of an IFN-Mediated Antiviral Response by a Self-Amplifying RNA Vaccine: Implications for Vaccine Design. J. Immunol. 2017;198:4012–4024. doi: 10.4049/jimmunol.1601877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roth J.S. Ribonuclease. III. Ribonuclease activity in rat liver and kidney. J. Biol. Chem. 1954;208:181–194. [PubMed] [Google Scholar]
- 43.Morrill G.A., Reiss M.M. Inhibition of enzymatic degradation of RNA by bound calcium and magnesium. Biochim. Biophys. Acta. 1969;179:43–49. doi: 10.1016/0005-2787(69)90120-8. [DOI] [PubMed] [Google Scholar]
- 44.Kim B.M., Schultz L.W., Raines R.T. Variants of ribonuclease inhibitor that resist oxidation. Protein Sci. 1999;8:430–434. doi: 10.1110/ps.8.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Promega. (2018). RNasin® Plus RNase Inhibitor: usage information. https://be.promega.com/-/media/files/resources/protocols/product-information-sheets/n/rnasin-plus-rnase-inhibitor-protocol.pdf?la=en.
- 46.Denies S., Cicchelero L., Van Audenhove I., Sanders N.N. Combination of interleukin-12 gene therapy, metronomic cyclophosphamide and DNA cancer vaccination directs all arms of the immune system towards tumor eradication. J. Control. Release. 2014;187:175–182. doi: 10.1016/j.jconrel.2014.05.045. [DOI] [PubMed] [Google Scholar]
- 47.Staresinic B., Jesenko T., Kamensek U., Krog Frandsen S., Sersa G., Gehl J., Cemazar M. Effect of calcium electroporation on tumour vasculature. Sci. Rep. 2018;8:9412. doi: 10.1038/s41598-018-27728-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Falk H., Forde P.F., Bay M.L., Mangalanathan U.M., Hojman P., Soden D.M., Gehl J. Calcium electroporation induces tumor eradication, long-lasting immunity and cytokine responses in the CT26 colon cancer mouse model. OncoImmunology. 2017;6:e1301332. doi: 10.1080/2162402X.2017.1301332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hansen E.L., Sozer E.B., Romeo S., Frandsen S.K., Vernier P.T., Gehl J. Dose-dependent ATP depletion and cancer cell death following calcium electroporation, relative effect of calcium concentration and electric field strength. PLoS ONE. 2015;10:e0122973. doi: 10.1371/journal.pone.0122973. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.