Summary
During normal mammary gland development, s-SHIP promoter expression marks a distinct type of mammary stem cells, at two different stages, puberty and early mid-pregnancy. To determine whether s-SHIP is a marker of mammary cancer stem cells (CSCs), we generated bitransgenic mice by crossing the C3(1)-SV40 T-antigen transgenic mouse model of breast cancer, and a transgenic mouse (11.5kb-GFP) expressing green fluorescent protein from the s-SHIP promoter. Here we show that in mammary tumors originating in these bitransgenic mice, s-SHIP promoter expression enriches a rare cell population with CSC activity as demonstrated by sphere-forming assays in vitro and limiting dilution transplantation in vivo. These s-SHIP-positive CSCs are characterized by lower expression of Delta-like non-canonical Notch ligand 1 (DLK1), a negative regulator of the Notch pathway. Inactivation of Dlk1 in s-SHIP-negative tumor cells increases their tumorigenic potential, suggesting a role for DLK1 in mammary cancer stemness.
Keywords: breast cancer, CSC, s-SHIP, transgenic mouse, mammary tumor, DLK1, notch, SHIP1
Highlights
-
•
A rare s-SHIP+ cell subset is present in mouse basal-like mammary tumors
-
•
s-SHIP+ tumor cells display higher sphere-forming potential
-
•
s-SHIP+ tumor cells have higher tumorigenicity
-
•
Downregulation of Dlk1 increases tumorigenicity
Bourette and colleagues assess renewal and differentiation potential of s-SHIP-expressing cells in mouse mammary tumors, by in vitro sphere-forming assay and in vivo transplantation. Their data indicate that s-SHIP expression marks a distinct population of mammary cancer stem cells and unveils a potential role of Dlk1, a negative regulator of the Notch pathway, in the regulation of mammary tumor cell stemness.
Introduction
Breast cancer is the leading cause of cancer-related death among females across the world. It is a highly complex disease marked by genetic and clinical heterogeneity (Torre et al., 2015). Intertumoral heterogeneity of breast cancers led to a classification into several subsets with varied patient outcomes and implications for treatment. This heterogeneity is associated with an epithelial hierarchy in tumors resembling that observed in normal mammary gland. On top of this hierarchy that forms the tumor bulk is a cell subpopulation described as cancer stem cells (CSCs) or tumor-initiating cells (TICs) (Al-Hajj et al., 2003, Ginestier et al., 2007). CSCs are defined as cells that retain extensive self-renewal potential through a series of generations and have the ability to recreate the heterogeneity of the original tumor through asymmetric division (Nassar and Blanpain, 2016). The nature of breast CSCs is complex, with the demonstration of their heterogeneity both within and between breast tumors and the discovery of their plasticity (Sreekumar et al., 2015). Strong evidence has suggested that they are responsible for the recurrence, metastasis, and drug resistance of high-grade tumors (Peitzsch et al., 2017). A better understanding of breast CSCs might, therefore, provide prognostic information for relapse and metastasis and lead to the development of more efficient therapies.
Genetically engineered mouse models have been extremely important in helping to elucidate the mechanisms of pathogenesis of breast cancer (Menezes et al., 2014). Different CSC subpopulations have been identified in these transgenic mouse models of mammary carcinogenesis using combinations of cell-surface markers, such as Lin−CD29hiCD24hi (Zhang et al., 2008), Thy1+CD24+ (Cho et al., 2008), CD29hiCD24med (Shafee et al., 2008), and CD61 (Vaillant et al., 2008). However, most of the sorting schemes require multiparameter fluorescence-activated cell sorting (FACS), yield sometimes rather heterogeneous cell populations, and are unclear in their biological meaning (Medema, 2013). Due to the general lack of unique cell-surface markers, the development of new markers to prospectively identify putative CSCs is of the utmost importance (Brooks et al., 2015).
Stem cell-specific expression of s-SHIP (stem-SH2-containing 5′-inositol phosphatase) was initially identified in embryonic and hematopoietic stem cells (Tu et al., 2001). A transgenic mouse (Tg 11.5kb-GFP or s-SHIP-GFP) was generated using the 11.5-kb s-SHIP promoter that specifically expresses enhanced green fluorescent protein (GFP) in several stem cell populations during embryonic development, including skin epidermis, hair follicles, mammary gland, and prostate (Rohrschneider et al., 2005). In the mammary gland, s-SHIP/GFP labels puberty cap cells and pregnancy basal alveolar bud cells, with a stem cell potential (Bai and Rohrschneider, 2010). Notably, s-SHIP expression in cap cells is associated with the expression of Par3-like polarity protein, which is essential for mammary stem cell maintenance (Huo and Macara, 2014).
The C3(1)/SV40 T-antigen transgenic mouse model (Tg C3(1)-Tag mice) develops mammary hyperplasia by 3 months of age with subsequent development of mammary adenocarcinoma by 5–6 months of age (Maroulakou et al., 1994). C3(1)-Tag mouse is a murine model of human basal-like breast cancer, a complex group of breast cancers with both basal and luminal features (Dontu and Ince, 2015, Gusterson and Eaves, 2018). Using a cross-species genomics approach, murine expression profiles were compared with human breast cancers. The vast majority (≈90%) of the C3(1)-Tag mammary tumors displayed characteristics of human basal-like breast cancer, and 5%–10% showed the claudin-low subtype feature (Herschkowitz et al., 2007, Pfefferle et al., 2013, Usary et al., 2016), demonstrating the model's strong resemblance to human disease.
Here we aim to assess whether s-SHIP promoter expression could enable the isolation of a subpopulation of tumor cells with CSC characteristics, i.e., higher ability to self-renew and to form spheres in vitro, and higher ability to form secondary tumors phenotypically similar to the primary tumor, following their transplantation into immunodeficient mice. By crossing Tg 11.5kb-GFP mice with Tg C3(1)-Tag mice, we showed the presence of a subpopulation of GFP+ cells in mammary tumors. These GFP+ mammary cancer cells are CD24+/CD49f+/CD29+, and enriched for sphere-forming activity in vitro. Importantly, they can regenerate heterogeneous tumors that display properties similar to the primary tumor upon subsequent transplantation. Transcriptomic analysis showed that these s-SHIP-positive CSCs were characterized by lower expression of Delta-like non-canonical Notch ligand 1 (DLK1), a negative regulator of the Notch pathway. Inactivation of Dlk1 in s-SHIP-negative tumor cells increased their sphere-forming capacity and their tumorigenic potential, suggesting a role for DLK1 in mammary cancer cell stemness. Altogether, these results demonstrate that s-SHIP promoter expression offers a valuable marker for the isolation and characterization of mammary CSCs.
Results
s-SHIP-GFP/C3(1)-Tag Bitransgenic Mice Develop Mammary Tumors Containing a Rare s-SHIP/GFP+ Cell Subpopulation
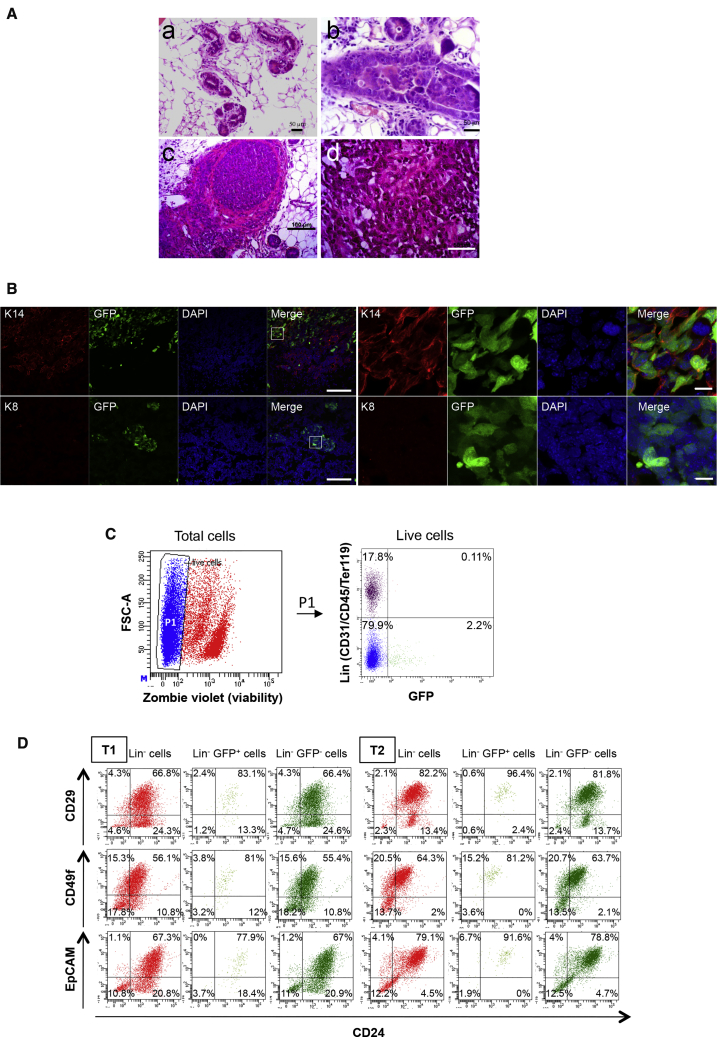
We generated a bitransgenic mouse model by crossing homozygous Tg 11.5kb-GFP mice with hemizygous Tg C3(1)-Tag mice. Progressive mammary gland lesions were observed in female mice that carried the T Ag-containing transgene, from ductal hyperplasia to adenocarcinoma (Figures 1A and S1A). All female mice developed multiple mammary tumors by 4–5 months of age. GFP+ cells were detected on frozen sections (Figure 1B) and by flow cytometry after enzymatic digestion of tumors (Figure 1C) (1.03% ± 0.64% of total cells, n = 10). The vast majority of GFP+ cells were negative for lineage markers (Lin+GFP+ cells = 0.08% ± 0.08% of total cells, n = 5). These results indicated that s-SHIP promoter drives GFP expression specifically in a subpopulation of mammary tumor cells. Moreover, GFP expression correlated with the endogenous s-SHIP mRNA expression, since sorted tumor GFP+ cells expressed higher levels of s-SHIP mRNA compared with sorted tumor GFP− cells (Figure S1C). Analysis of luminal (cytokeratin 8, K8) and basal/myoepithelial (cytokeratin 14, K14) markers showed that few tumors expressed K8 while all tumors displayed expression of K14 (Figure 1B). Importantly, GFP+ tumor cells expressed K14 (Figure 1B). Similarly to s-SHIP/GFP expression at puberty during normal mammary gland development (Bai and Rohrschneider, 2010), some K14+ mammary basal cells of 7-week-old bitransgenic mice also expressed GFP (Figure S1B). We next examined the expression of cell-surface markers historically associated with stem/progenitor cells in the mammary gland. Previous studies using flow cytometry to isolate mouse mammary stem cells have shown the majority of these cells to have a CD49fhiCD29hiCD24+EpCAM+Sca-1neg cell-surface marker phenotype (Shackleton et al., 2006, Shehata et al., 2012, Sleeman et al., 2005, Stingl et al., 2006). Independent tumors were dissociated to single-cell suspensions and stained for CD24, CD29, CD49f, and EpCAM cell-surface markers. Tumors displayed distinct FACS profiles showing heterogeneous expression for different markers but with enrichment for CD24+CD29+ and CD24+EpCAM+ cell subsets (Figures 1D and S1D). The GFP+ cell population was homogeneous, with the majority of cells being located in the Lin−CD24+ cell subset and expressing CD29, CD49f, and EpCAM cell-surface markers (Figure 1D). Moreover, Lin−GFP+ cells showed a higher percentage of double-positive [CD24 CD49f]+ in comparison with total Lin− tumor cells (Figure S1D).
Figure 1.
s-SHIP/GFP Expression Is Detected in Mammary Tumors of s-SHIP-GFP/C3(1)-Tag Bitransgenic Mice
(A) H&E staining of paraffin-embedded sections of mammary tumors illustrating different stages of tumor development: a, ductal hyperplasia; b, ductal carcinoma in situ; c, infiltrative solid carcinoma; d, solid carcinoma. Scale bars, 50 μm (a, b, d) and100 μm (c). See also Figure S1A.
(B) Representative images (n = 4) of frozen sections of mammary tumors containing GFP+ cells (green) and stained with cytokeratin 14 (K14, red, upper panels), cytokeratin 8 (K8, red, lower panels), and DAPI nuclear stain (blue) Scale bars, 100 μm (left panels) and 10 μm (right panels). Images of 7-week-old mammary gland stained with K14 and K8 are shown in Figure S1B.
(C) Representative flow cytometry analysis (n = 5) of GFP expression in dissociated mammary cancer cells isolated from a 5-month-old bitransgenic mouse. P1 gate is set as to exclude dead cells (Zombie violet+). See also Figure S1C.
(D) Representative flow cytometry analysis (n = 3) of total Lin− cells, Lin−GFP−, and Lin−GFP+ cells from two different tumors: T1 and T2. Cells were analyzed for the expression of CD24, CD29, CD49f, and EpCAM cell-surface markers. See also Figure S1D.
s-SHIP/GFP+ Tumor Cells Are Enriched for Sphere-Forming and Tumorigenic Cells
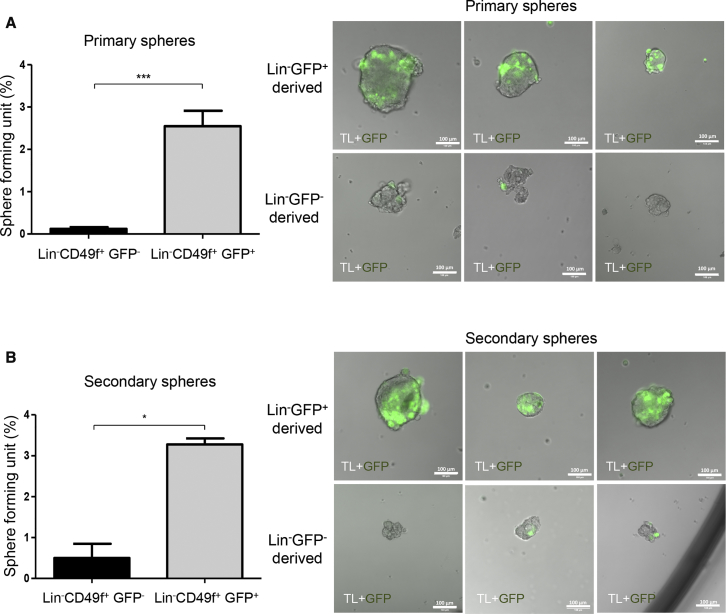
To further characterize GFP+ cells, we used CD49f expression to separate GFP+ epithelial cells (CD49fhigh) from the few GFP+ vascular smooth muscle cells (CD49f−/low) as previously reported (Bai and Rohrschneider, 2010) (Figure S2A). To compare the frequency of TICs in GFP+ versus GFP− cell subsets we isolated cells by FACS, and either cultured them under serum-free suspension conditions to form mammospheres or subcutaneously injected them into SCID mice (Figure S2B). Since the majority of GFP+ cells were located in the Lin− cell fraction, we compared Lin−CD49f+GFP+ cells with Lin−CD49f+GFP− cells (Figure S2B). GFP+ cell populations showed a significant higher sphere-forming potential (2.55% ± 1.92%) compared with the control GFP− cell populations (0.12% ± 0.22%) (Figure 2A, left panel, n = 7). Spheres derived from GFP+ cells were heterogeneous in size but most often larger compared with those derived from GFP− cells (Figure 2A, right panels). To examine the in vitro self-renewal potential of GFP+ cells, we dissociated primary spheres into single-cell suspensions and plated the cells under the same conditions as for primary spheres. Secondary spheres derived from GFP+ subgroups were more numerous and larger as compared with secondary spheres derived from GFP− subgroups (Figure 2B, n = 3). Spheres initially derived from GFP+ cell subsets can be maintained through at least four passages (data not shown). It is noteworthy that few GFP+ cells were always observed in the spheres at all passages (Figures 2 and S3A).
Figure 2.
s-SHIP/GFP+ Cells Have Higher Sphere-Forming Potential and In Vitro Self-Renewal Capacity
(A) Primary mammospheres derived from Lin−CD49f+GFP− and Lin−CD49f+GFP+ cells isolated from bitransgenic mammary tumors. Cells were seeded by limited dilution and grown in suspension for 7–10 days. Data represent the mean ± SEM of seven independent experiments; p values were determined by Student's t test, ∗∗∗p < 0.001. Right panels: representative pictures of primary spheres after 7–10 days of culture.
(B) Primary spheres derived from Lin−CD49f+GFP− or Lin−CD49f+GFP+ cells were dissociated into single cells. Cells were seeded at 200 cells per well in triplicate and grown in suspension for 7–10 days for secondary mammosphere formation. Data represent mean ± SEM of three different experiments; p values were determined by Student's t test, ∗p < 0.05. Right panels: representative pictures of secondary spheres after 7–10 days of culture.
Scale bars, 100 μm. See also Figures S2A and S2B for cell-sorting strategy and experimental design, and Figure S3A.
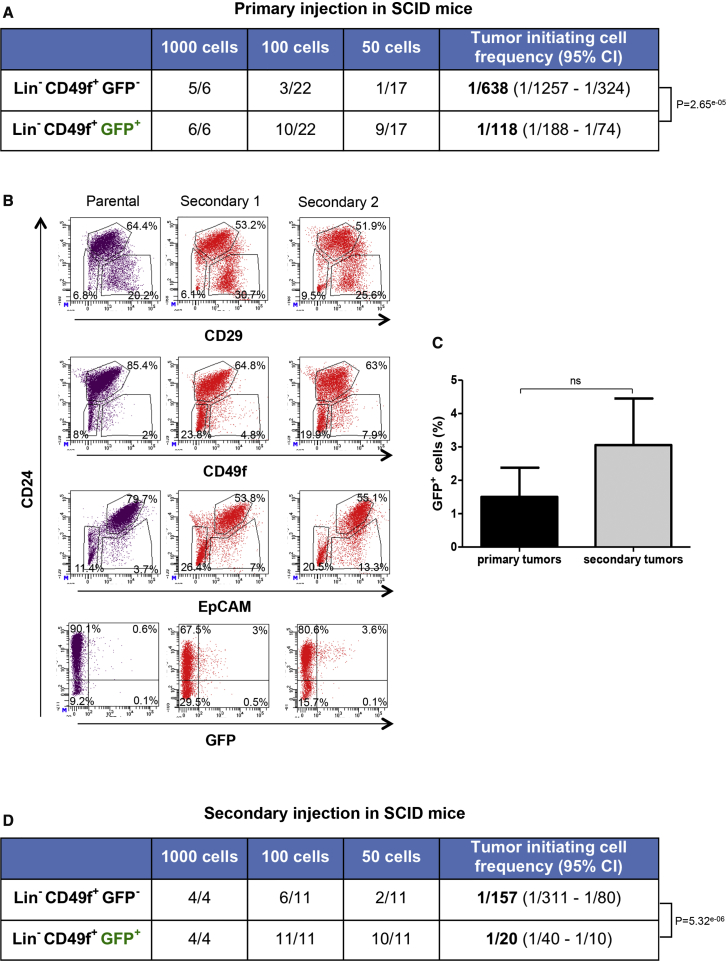
We next performed serial transplantation studies to evaluate the tumorigenic potential of GFP+ cells versus GFP− cells and to determine whether GFP+ cells were able to self-renew in vivo and recapitulate the cell heterogeneity of the original tumors. When injected subcutaneously in the region next to the nipple of the third thoracic gland of immunodeficient SCID mice, GFP+ cells displayed significantly higher tumorigenic potential compared with GFP− cells (Figure 3A). The calculated TIC frequency in the Lin−CD49f+GFP− cell population was 1 in 638 tumor cells whereas it was of 1 in 118 in the Lin−CD49f+GFP+ cell population. No difference was observed in tumor latency (Figure S3B). Similar TIC frequency was observed in CD49f+GFP+ without lineage depletion (Figure S2C). For CD49f+GFP− cells, TIC frequency was lower compared with Lin−CD49f+GFP− cells. This could be explained by a dilution effect of numerous Lin+ cells present in the tumor, but also may indicate an inhibitory effect of some Lin+ cells on tumorigenicity. C3(1)-Tag mammary tumor cells and tumor-derived cell lines cannot be transplanted into immunocompetent FVB mice, since they are rejected likely due to the expression of the SV40 T antigen (Aprelikova et al., 2016). However, we have successfully transplanted primary tumor cells into FVB mice but with a significantly lower engraftment rate as well as longer tumor-formation period (4–7 months) compared with SCID mice (1–3 months, Figure S3B). Nevertheless, Lin−CD49f+GFP+ cells were again capable of generating more tumors than Lin−CD49f+GFP− cells (Figure S2D).
Figure 3.
s-SHIP/GFP+ Cells Are Enriched for Tumor-Initiating Cells and Generate Tumors that Mimic the Parental Tumors
(A) Flow cytometry was used to separate dissociated tumor cells based on cell-surface markers and GFP expression (see Figure S2B for experimental procedures). The collected cell populations were injected subcutaneously into recipient SCID female mice in a limiting dilution manner. Mice were monitored until tumors were observed or up to 7 months if no tumors were detected. See also Figures S2C, S2D, and S3B.
(B) Secondary tumors derived from Lin−CD49f+GFP+ cell fraction have flow-cytometric profiles similar to those of the primary tumors. Representative flow cytometry analysis of dissociated cells from a primary tumor (parental) and two respective secondary tumors (secondary 1 and 2) are presented.
(C) The percentage of Lin−CD49f+GFP+ cells in secondary tumors and corresponding primary parental tumors. Data represent mean values ± SEM of four different experiments; p values were determined by Student's t test, not significant (ns).
(D) Cells from secondary tumors generated by Lin−CD49f+GFP+ fraction were freshly digested and FACS sorted into Lin−CD49f+GFP− or Lin−CD49f+GFP+ subpopulations, and injected into SCID mice as described in (A).
In (A) and (D), tumor-initiating cell frequencies and p values were generated by extreme limiting dilution analysis (ELDA). See also Figures S2A and S2B for cell-sorting strategy and experimental design.
Flow cytometry analysis of secondary tumors generated by Lin−CD49f+GFP+ cells injected into SCID mice showed that the expression patterns of cell-surface markers were similar to those of the original tumor (Figure 3B). Interestingly, GFP+ cells were also present in these secondary tumors (Figure 3B). The percentage of GFP+ cells in secondary tumors was higher but not significantly different (Figure 3C). Transplantation experiments showed that both Lin−CD49f+GFP+ and Lin−CD49f+GFP− cells isolated from these secondary tumors had higher tumor-forming capacities than their corresponding cell populations from primary tumors (Figures 3A and 3D). This suggests that both cell populations changed toward a more aggressive phenotype. Importantly, Lin−CD49f+GFP+ cells still exhibited a higher tumorigenic potential in these secondary injections (1 in 20) compared with Lin−CD49f+GFP− cells (1 in 157) (Figure 3D).
Microarray Analysis Revealed DLK1 as a Negative Regulator of Stemness
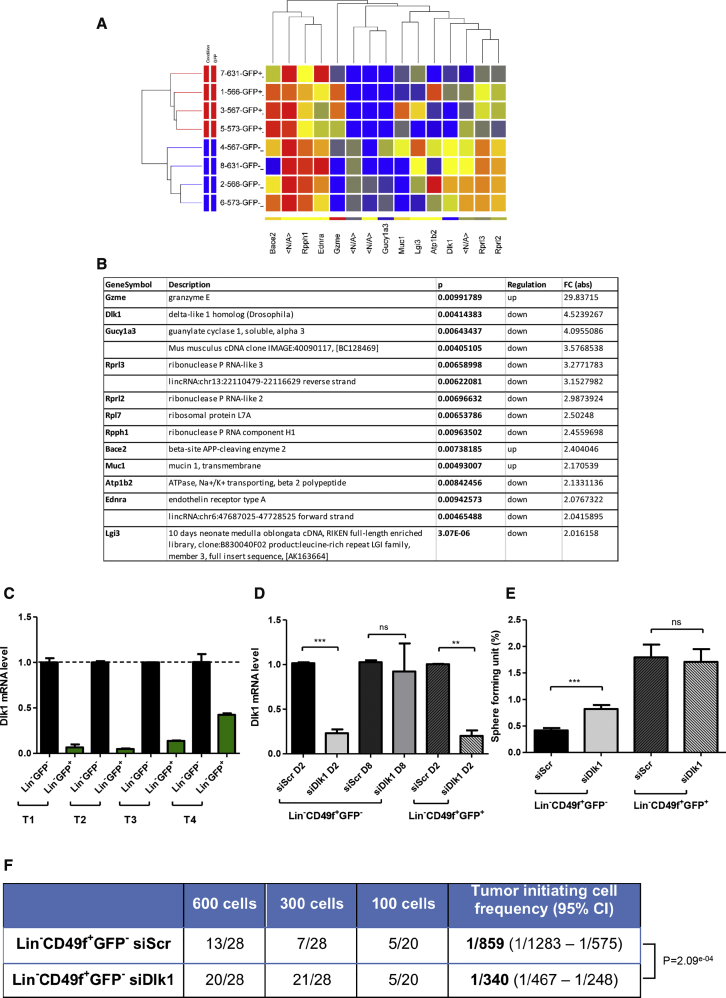
To identify molecular differences between Lin−CD49f+GFP+ and Lin−CD49f+GFP− cells, we performed transcriptional profiling using Agilent microarray technology. As shown in Figure 4A, Lin−CD49f+GFP+ and Lin−CD49f+GFP− cell subpopulations shared a similar gene expression pattern (GEO accession number GEO: GSE108373). Nevertheless, microarray analysis revealed several genes whose expression differs significantly between the two subpopulations (Figure 4B) (Table S1). Granzyme E (GZME) was significantly upregulated in Lin−CD49f+GFP+ cells (fold change of 29.8) (Figure 4B). Granzymes are a family of serine proteases expressed by immune cells (Arias et al., 2017). As a first approach, we delivered small interfering RNA (siRNA) to knock down Gzme expression in Lin−CD49f+GFP+ cells and investigated the outcome of Gzme silencing on the sphere-forming potential of these cells. As shown in Figure S3C, the downregulation of Gzme did not affect the sphere-forming potential of GFP+ cells. We were then interested in the Delta-like non-canonical Notch ligand 1 (Dlk1) gene that was significantly downregulated in Lin−CD49f+GFP+ cells (fold change of 4.5) (Figure 4B). Dlk1 gene encodes for a single-spanning transmembrane protein in the epidermal growth factor-like family, which includes NOTCH receptors and their ligands (Tanimizu et al., 2003). DLK1 is a non-canonical Notch ligand and has been shown to act as an inhibitor of Notch signaling in vitro (Nueda et al., 2017). Microarray analysis revealed the expression of Notch1, Notch3, and Notch4 genes in Lin−CD49f+ cell populations. Chemical inhibition of NOTCH processing with the γ-secretase inhibitor Compound E decreased the sphere-forming potential of Lin−CD49f+GFP+ cells (Figure S3D). Thus, we hypothesized that repression of the Notch-negative regulator DLK1 in Lin−CD49f+GFP+ cells might favor their CSC phenotype. The difference in Dlk1 expression between Lin−CD49f+GFP− and Lin−CD49f+GFP+ subpopulations was first confirmed by qRT-PCR using total RNA isolated from these two subpopulations. Similar to microarray results, we observed a fold change of 11 ± 8 (Figure 4C, n = 4 different tumors). We thus delivered siRNA to knock down Dlk1 expression in Lin−CD49f+GFP− cells and performed sphere-forming assays. Lin−CD49f+GFP− cell subpopulation was transfected with siDlk1 or siScramble (siScr) and then cultured in serum-free sphere medium under non-adherent conditions for 7–10 days. A transient downregulation of Dlk1 expression was obtained with 77% of inhibition after 2 days of culture, and only 10% of inhibition in the generated spheres at day 8 (Figure 4D). We observed a significant increase of the sphere-forming potential of siDlk1-treated cells (0.82% ± 0.21%) compared with siScr-treated cells (0.42% ± 0.12%) (Figure 4E). On the contrary, a downregulation of 80% of Dlk1 expression in Lin−CD49f+GFP+ cells (Figure 4D) did not interfere with their sphere-forming potential (Figure 4E), suggesting that the level of Dlk1 expression in these cells may be too low to have any inhibitory effect on their sphere-forming capacity. Since siDlk1 enhanced the in vitro sphere-forming potential of Lin−CD49f+GFP− cells, we next performed transplantation studies to evaluate the tumorigenic potential of siDlk1- and siScr-transfected Lin−CD49f+GFP− cells. The calculated TIC frequency in the siDlk1-transfected Lin−CD49f+GFP− cell population was 1 in 340 tumor cells whereas it was 1 in 859 in the siScr-transfected Lin−CD49f+GFP− cells (Figure 4F). No difference was observed in the tumor latency (Figure S3E). Thus, downregulation of Dlk1 in Lin−CD49f+GFP− cells significantly increased both their sphere-forming potential in vitro and their tumorigenic potential in vivo compared with control cells. Altogether, these results suggest that DLK1 could act as a negative regulator of stemness of mammary cancer cells.
Figure 4.
Gene Expression Analysis of Lin−CD49f+GFP− versus Lin−CD49f+GFP+ Tumor Cells Reveals the Role of DLK1 in the Regulation of Stemness
(A and B) Fifteen genes (B) were selected from the master list of genes differentially regulated (at least 2-fold with a p value <0.05). A list of 355 deregulated probes (fold change ≥1) is presented in Table S1. For the heatmap (A), each column represents a probe; each row represents various subpopulations from biological replicates. The red color indicates high-level expression while blue indicates a low level of expression.
(C) qRT-PCR confirmed the downregulation of Dlk1 expression in Lin−CD49f+GFP+ cells compared with Lin−CD49f+GFP− cells. Data represent mean values ± SEM from four different tumors (T1 to T4).
(D and E) Inhibition of Dlk1 expression in Lin−CD49f+GFP− cells increased their sphere-forming potential. siDlk1 or control siScramble (siScr) were introduced into Lin−CD49f+GFP− and Lin−CD49f+GFP+ cells isolated from bitransgenic mammary tumors. (D) The level of Dlk1 mRNA was determined by qRT-PCR, 2 days or 8 days after siRNA transfection. (E) Lin−CD49f+GFP− and Lin−CD49f+GFP+ cells were treated by siScr or siDlk1, and plated under sphere conditions. Spheres were counted after 7–10 days in culture. Data represent mean values ± SEM of six independent experiments; p values were determined by Student's t test, ∗∗∗p < 0.001 ∗∗p < 0.01; ns, not significant. See Figures S3C and S3D for additional experiments.
(F) siScr or siDlk1 transfected Lin−CD49f+GFP− cells were injected subcutaneously into recipient SCID female mice in a limiting dilution manner. Mice were monitored until tumors were observed or up to 3 months if no tumors were detected. Tumor-initiating cell frequencies and p values were generated by ELDA. See also Figure S3E.
Discussion
Stem cell-specific expression of s-SHIP was initially identified in hematopoietic and embryonic stem cells (Rohrschneider et al., 2005, Tu et al., 2001). Using s-SHIP-GFP promoter reporter, its stem cell-specific expression has been demonstrated in prostate tissues (Al Shareef et al., 2018, Bauderlique-Le Roy et al., 2015, Brocqueville et al., 2016). In postnatal mammary gland development, GFP+ cap cells in puberty and basal alveolar bud cells in pregnancy each exhibit stem cell properties (Bai and Rohrschneider, 2010). In the same study, the authors described the presence of GFP+ tumor cells in heterogeneous luminal/basal-like mammary tumors originating in bitransgenic s-SHIP-GFP/MMTV-Wnt1 mice. Subsequent studies have demonstrated the value of s-SHIP promoter to investigate normal mammary stem cells (Huo and Macara, 2014, Kogata et al., 2013, Roarty et al., 2015). However, s-SHIP/GFP usefulness in isolating mammary CSCs remains to be determined. Here we used a mouse model of basal-like breast cancer to show that s-SHIP is a marker of mammary CSCs. In our s-SHIP-GFP/C3(1)-Tag bitransgenic mice, s-SHIP/GFP+ cells expressed commonly used mammary CSC cell-surface markers, exhibited a higher sphere-forming potential in vitro, and a higher ability to reform secondary tumors when transplanted into recipient mice. Interestingly, transplanted s-SHIP/GFP+ cells generated heterogeneous tumors that displayed properties similar to those of the primary tumors, including the presence of a subset of s-SHIP/GFP+ cells with CSC properties. Bai and Rohrschneider (2010) did not observe s-SHIP/GFP+ cells in tumors of bitransgenic s-SHIP-GFP/MMTV-ErbB2/neu/HER2 mice exhibiting luminal features. Considering breast CSC heterogeneity, it remains to be determined whether s-SHIP expression is restricted or not to CSCs in basal-like breast cancer. To address this question, we aim to characterize the exact nature of s-SHIP/GFP+ tumor cells that have been observed in heterogeneous mammary tumors developed by bitransgenic s-SHIP-GFP/MMTV-Wnt1 mice (Bai and Rohrschneider, 2010), together with an in-depth analysis of data for breast cancer from The Cancer Genome Atlas.
Transcriptional profiling showed few genes differentially expressed between Lin−CD49f+GFP+ and Lin−CD49f+GFP− cells. Among them, the significant decrease of Dlk1 expression in GFP+ cells has attracted our attention. DLK1 acts as a non-canonical ligand of the Notch signaling pathway and is expressed at a high frequency in various human tumors including breast carcinoma (Yanai et al., 2010). Several studies have demonstrated that DLK1 can act as an inhibitor of Notch signaling in vitro, including in breast cancer cells (Nueda et al., 2017). Our results strongly suggest that DLK1 acts as a negative regulator of stemness in breast cancer, which agrees with several studies pointing to a role of Notch pathway in breast cancer, especially in subtypes with stem-like features and endothelial-to-mesenchymal transition (Harrison et al., 2010). Based on our results, we can hypothesize that DLK1 interacts with Notch receptor expressed by tumor cells and can act as a negative regulator of Notch activation and signaling in these tumors cells. It is likewise possible that DLK1 action implicates cells of the tumor microenvironment. Interestingly, a recent study showed that Notch ligand Dll1 mediates crosstalk between mammary stem cells and tissue-resident macrophages (Chakrabarti et al., 2018). This implies that DLK1 may also be involved in the modulation of the oncogenic potential of mammary tumor cells through the inhibition of Notch signaling in surrounding macrophages. To address this issue, it would be important to determine the relationship between breast CSCs, macrophages, DLK1, and Notch signaling.
Taken together, our results strongly support the relationship between mammary CSCs and s-SHIP expression. This is in accordance with high s-SHIP expression in murine claudin-low tumors that are enriched for CSC signature (Herschkowitz et al., 2012). s-SHIP protein is a shorter isoform of SHIP1 protein that lacks the SH2 domain in its N-terminal region (Tu et al., 2001). However, the function of s-SHIP protein remains unknown. SHIP2, the ubiquitous homolog of SHIP1, is crucial for maintaining the ER-negative breast CSCs through activation of Akt and JNK (Fu et al., 2014). Since our data indicate s-SHIP as a potential clinical target for breast cancer therapy, the expression and role of s-SHIP in human breast cancers will be the subject of our future investigation.
Experimental Procedures
All animal procedures were performed under a protocol (#01989.02) approved by the Animal Protocol Review Committees of the Institut Pasteur de Lille (France) in accordance with European regulation.
Statistics
Data are expressed as means ± SEM of at least three independent experiments. The statistical analysis was done by using two-way ANOVA or Student's t test with GraphPad Prism5, and a p value of <0.05 was considered significant. For in vivo tumor-initiating potential, the results were analyzed by extreme limiting dilution analysis (ELDA) (Hu and Smyth, 2009).
Author Contributions
M-J. T. and C.L. contributed equally to this work. L.T., M.-J.T., E.B., H.B.-L.R., M.F., and R.P.B. did the experiments L.T., C.L., E.A., X.L.B., and R.P.B. designed the study. L.T. and R.P.B. wrote the main manuscript text, and all authors reviewed the manuscript.
Acknowledgments
We thank S. Sebda and F. Leprêtre (Functional Genomic Platform, Lille) for microarrays hybridization and for GEO repository assistance, Q. Pascal (OCR, Loos) for anatomopathological analysis, and S. Salomé-Desnoulez (BICel), M.-C. Bouchez (CNRS), and T. Chassat (IPL) for their help. This study was supported by grants from the SIRIC ONCOLille (INCa-DGOS-Inserm 6041), the Ligue Contre le Cancer (comités du Nord et du Pas-de-Calais), and the Cancéropole Nord-Ouest. L.T. was supported by fellowships from the Région Hauts-de-France and the Fondation ARC pour la Recherche sur le Cancer.
Published: June 13, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2019.05.013.
Accession Numbers
The accession number for the microarray dataset reported in this paper is GEO: GSE108373.
Supplemental Information
The p value was calculated with a paired t test.
References
- Al-Hajj M., Wicha M.S., Benito-Hernandez A., Morrison S.J., Clarke M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Shareef Z., Kardooni H., Murillo-Garzón V., Domenici G., Stylianakis E., Steel J.H., Rabano M., Gorroño-Etxebarria I., Zabalza I., Vivanco M.D. Protective effect of stromal dickkopf-3 in prostate cancer: opposing roles for TGFBI and ECM-1. Oncogene. 2018;37:5305–5324. doi: 10.1038/s41388-018-0294-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aprelikova O., Tomlinson C.C., Hoenerhoff M., Hixon J.A., Durum S.K., Qiu T., He S., Burkett S., Liu Z.-Y., Swanson S.M. Development and preclinical application of an immunocompetent transplant model of basal breast cancer with lung, liver and brain metastases. PLoS One. 2016;11:e0155262. doi: 10.1371/journal.pone.0155262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias M., Martinez-Lostao L., Santiago L., Ferrandez A., Granville D.J., Pardo J. The untold story of granzymes in oncoimmunology: novel opportunities with old acquaintances. Trends Cancer. 2017;3:407–422. doi: 10.1016/j.trecan.2017.04.001. [DOI] [PubMed] [Google Scholar]
- Bai L., Rohrschneider L.R. s-SHIP promoter expression marks activated stem cells in developing mouse mammary tissue. Genes Dev. 2010;24:1882–1892. doi: 10.1101/gad.1932810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauderlique-Le Roy H., Vennin C., Brocqueville G., Spruyt N., Adriaenssens E., Bourette R.P. Enrichment of human stem-like prostate cells with s-SHIP promoter activity uncovers a role in stemness for the long noncoding RNA H19. Stem Cells Dev. 2015;24:1252–1262. doi: 10.1089/scd.2014.0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocqueville G., Chmelar R.S., Bauderlique-Le Roy H., Deruy E., Tian L., Vessella R.L., Greenberg N.M., Rohrschneider L.R., Bourette R.P. s-SHIP expression identifies a subset of murine basal prostate cells as neonatal stem cells. Oncotarget. 2016;7:29228–29244. doi: 10.18632/oncotarget.8709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks M.D., Burness M.L., Wicha M.S. Therapeutic implications of cellular heterogeneity and plasticity in breast cancer. Cell Stem Cell. 2015;17:260–271. doi: 10.1016/j.stem.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti R., Celià-Terrassa T., Kumar S., Hang X., Wei Y., Choudhury A., Hwang J., Peng J., Nixon B., Grady J.J. Notch ligand Dll1 mediates cross-talk between mammary stem cells and the macrophageal niche. Science. 2018;360:eaan4153. doi: 10.1126/science.aan4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho R.W., Wang X., Diehn M., Shedden K., Chen G.Y., Sherlock G., Gurney A., Lewicki J., Clarke M.F. Isolation and molecular characterization of cancer stem cells in MMTV-Wnt-1 murine breast tumors. Stem Cells. 2008;26:364–371. doi: 10.1634/stemcells.2007-0440. [DOI] [PubMed] [Google Scholar]
- Dontu G., Ince T.A. Of mice and women: a comparative tissue biology perspective of breast stem cells and differentiation. J. Mammary Gland Biol. Neoplasia. 2015;20:51–62. doi: 10.1007/s10911-015-9341-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C.-H., Lin R.-J., Yu J., Chang W.-W., Liao G.-S., Chang W.-Y., Tseng L.-M., Tsai Y.-F., Yu J.-C., Yu A.L. A novel oncogenic role of inositol phosphatase SHIP2 in ER-negative breast cancer stem cells: involvement of JNK/vimentin activation. Stem Cells. 2014;32:2048–2060. doi: 10.1002/stem.1735. [DOI] [PubMed] [Google Scholar]
- Ginestier C., Hur M.H., Charafe-Jauffret E., Monville F., Dutcher J., Brown M., Jacquemier J., Viens P., Kleer C.G., Liu S. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusterson B., Eaves C.J. Basal-like breast cancers: from pathology to biology and back again. Stem Cell Reports. 2018;10:1676–1686. doi: 10.1016/j.stemcr.2018.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison H., Farnie G., Brennan K.R., Clarke R.B. Breast cancer stem cells: something out of notching? Cancer Res. 2010;70:8973–8976. doi: 10.1158/0008-5472.CAN-10-1559. [DOI] [PubMed] [Google Scholar]
- Herschkowitz J.I., Simin K., Weigman V.J., Mikaelian I., Usary J., Hu Z., Rasmussen K.E., Jones L.P., Assefnia S., Chandrasekharan S. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8:R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschkowitz J.I., Zhao W., Zhang M., Usary J., Murrow G., Edwards D., Knezevic J., Greene S.B., Darr D., Troester M.A. Comparative oncogenomics identifies breast tumors enriched in functional tumor-initiating cells. Proc. Natl. Acad. Sci. U S A. 2012;109:2778–2783. doi: 10.1073/pnas.1018862108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Smyth G.K. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- Huo Y., Macara I.G. The Par3-like polarity protein Par3L is essential for mammary stem cell maintenance. Nat. Cell Biol. 2014;16:526–534. doi: 10.1038/ncb2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogata N., Zvelebil M., Howard B.A. Neuregulin 3 and Erbb signalling networks in embryonic mammary gland development. J. Mammary Gland Biol. Neoplasia. 2013;18:149–154. doi: 10.1007/s10911-013-9286-4. [DOI] [PubMed] [Google Scholar]
- Maroulakou I.G., Anver M., Garrett L., Green J.E. Prostate and mammary adenocarcinoma in transgenic mice carrying a rat C3(1) simian virus 40 large tumor antigen fusion gene. Proc. Natl. Acad. Sci. U S A. 1994;91:11236–11240. doi: 10.1073/pnas.91.23.11236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema J.P. Cancer stem cells: the challenges ahead. Nat. Cell Biol. 2013;15:338–344. doi: 10.1038/ncb2717. [DOI] [PubMed] [Google Scholar]
- Menezes M.E., Das S.K., Emdad L., Windle J.J., Wang X.Y., Sarkar D., Fisher P.B. Genetically engineered mice as experimental tools to dissect the critical events in breast cancer. Adv. Cancer Res. 2014;121:331–382. doi: 10.1016/B978-0-12-800249-0.00008-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassar D., Blanpain C. Cancer stem cells: basic concepts and therapeutic implications. Annu. Rev. Pathol. 2016;11:47–76. doi: 10.1146/annurev-pathol-012615-044438. [DOI] [PubMed] [Google Scholar]
- Nueda M.L., Naranjo A.I., Baladrón V., Laborda J. Different expression levels of DLK1 inversely modulate the oncogenic potential of human MDA-MB-231 breast cancer cells through inhibition of NOTCH1 signaling. FASEB J. 2017;31:3484–3496. doi: 10.1096/fj.201601341RRR. [DOI] [PubMed] [Google Scholar]
- Peitzsch C., Tyutyunnykova A., Pantel K., Dubrovska A. Cancer stem cells: the root of tumor recurrence and metastases. Semin. Cancer Biol. 2017;44:10–24. doi: 10.1016/j.semcancer.2017.02.011. [DOI] [PubMed] [Google Scholar]
- Pfefferle A.D., Herschkowitz J.I., Usary J., Harrell J.C., Spike B.T., Adams J.R., Torres-Arzayus M.I., Brown M., Egan S.E., Wahl G.M. Transcriptomic classification of genetically engineered mouse models of breast cancer identifies human subtype counterparts. Genome Biol. 2013;14:R125. doi: 10.1186/gb-2013-14-11-r125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roarty K., Shore A.N., Creighton C.J., Rosen J.M. Ror2 regulates branching, differentiation, and actin-cytoskeletal dynamics within the mammary epithelium. J. Cell Biol. 2015;208:351–366. doi: 10.1083/jcb.201408058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrschneider L.R., Custodio J.M., Anderson T.A., Miller C.P., Gu H. The intron 5/6 promoter region of the ship1 gene regulates expression in stem/progenitor cells of the mouse embryo. Dev. Biol. 2005;283:503–521. doi: 10.1016/j.ydbio.2005.04.032. [DOI] [PubMed] [Google Scholar]
- Shackleton M., Vaillant F., Simpson K.J., Stingl J., Smyth G.K., Asselin-Labat M.L., Wu L., Lindeman G.J., Visvader J.E. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- Shafee N., Smith C.R., Wei S., Kim Y., Mills G.B., Hortobagyi G.N., Stanbridge E.J., Lee E.Y. Cancer stem cells contribute to cisplatin resistance in Brca1/p53–mediated mouse mammary tumors. Cancer Res. 2008;68:3243–3250. doi: 10.1158/0008-5472.CAN-07-5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehata M., Teschendorff A., Sharp G., Novcic N., Russell I.A., Avril S., Prater M., Eirew P., Caldas C., Watson C.J. Phenotypic and functional characterisation of the luminal cell hierarchy of the mammary gland. Breast Cancer Res. 2012;14:R134. doi: 10.1186/bcr3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleeman K.E., Kendrick H., Ashworth A., Isacke C.M., Smalley M.J. CD24 staining of mouse mammary gland cells defines luminal epithelial, myoepithelial/basal and non-epithelial cells. Breast Cancer Res. 2005;8:R7. doi: 10.1186/bcr1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreekumar A., Roarty K., Rosen J.M. The mammary stem cell hierarchy: a looking glass into heterogeneous breast cancer landscapes. Endocr. Relat. Cancer. 2015;22:T161–T176. doi: 10.1530/ERC-15-0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingl J., Eirew P., Ricketson I., Shackleton M., Vaillant F., Choi D., Li H.I., Eaves C.J. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- Tanimizu N., Nishikawa M., Saito H., Tsujimura T., Miyajima A. Isolation of hepatoblasts based on the expression of Dlk/Pref-1. J. Cell Sci. 2003;116:1775–1786. doi: 10.1242/jcs.00388. [DOI] [PubMed] [Google Scholar]
- Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- Tu Z., Ninos J.M., Ma Z., Wang J.W., Lemos M.P., Desponts C., Ghansah T., Howson J.M., Kerr W.G. Embryonic and hematopoietic stem cells express a novel SH2–containing inositol 5′-phosphatase isoform that partners with the Grb2 adapter protein. Blood. 2001;98:2028–2038. doi: 10.1182/blood.v98.7.2028. [DOI] [PubMed] [Google Scholar]
- Usary J., Darr D.B., Pfefferle A.D., Perou C.M. Overview of genetically engineered mouse model of distinct breast cancer subtypes. Curr. Protoc. Pharmacol. 2016;72:14.38.1–14.38.11. doi: 10.1002/0471141755.ph1438s72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillant F., Asselin-Labat M.-L., Shackleton M., Forrest N.C., Lindeman G.J., Visvader J.E. The mammary progenitor marker CD61/β3 integrin identifies cancer stem cells in mouse models of mammary tumorigenesis. Cancer Res. 2008;68:7711–7717. doi: 10.1158/0008-5472.CAN-08-1949. [DOI] [PubMed] [Google Scholar]
- Yanai H., Nakamura K., Hijioka S., Kamei A., Ikari T., Ishikawa Y., Shinozaki E., Mizunuma N., Hatake K., Miyajima A. Dlk-1, a cell surface antigen on foetal hepatic stem/progenitor cells, is expressed in hepatocellular, colon, pancreas and breast carcinomas at a high frequency. J. Biochem. 2010;148:85–92. doi: 10.1093/jb/mvq034. [DOI] [PubMed] [Google Scholar]
- Zhang M., Behbod F., Atkinson R.L., Landis M.D., Kittrell F., Edwards D., Medina D., Tsimelzon A., Hilsenbeck S., Green J.E. Identification of tumor-initiating cells in a p53 null mouse model of breast cancer. Cancer Res. 2008;68:4674–4682. doi: 10.1158/0008-5472.CAN-07-6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The p value was calculated with a paired t test.