Abstract
Emerging evidence suggests that circular RNAs (circRNAs) have crucial roles in various processes, including cancer development and progression. However, the functional roles of circRNAs in breast cancer remain to be elucidated. In this study, we identified a novel circRNA (named circBMPR2) whose expression was lower in breast cancer tissues with metastasis. Moreover, circBMPR2 expression was negatively associated with the motility of breast cancer cells and significantly downregulated in human breast cancer tissues. Functionally, we found that circBMPR2 knockdown effectively enhanced cell proliferation, migration, and invasion. Moreover, circBMPR2 knockdown promoted tamoxifen resistance of breast cancer cells through inhibiting tamoxifen-induced apoptosis, whereas circBMPR2 overexpression led to decreased tamoxifen resistance. Mechanistically, we demonstrated that circBMPR2 could abundantly sponge miR-553 and that miR-553 overexpression could attenuate the inhibitory effects caused by circBMPR2 overexpression. We also found that ubiquitin-specific protease 4 (USP4) was a direct target of miR-553, which functions as a tumor suppressor in breast cancer. Our findings demonstrated that circBMPR2 might function as a miR-553 sponge and then relieve the suppression of USP4 to inhibit the progression and tamoxifen resistance of breast cancer. Targeting this newly identified circRNA may help us to develop potential novel therapies for breast cancer patients.
Keywords: breast cancer, circBMPR2, tumor suppressor, tamoxifen resistance, miR-553, USP4
Introduction
Breast cancer is one of the most frequently diagnosed cancers in women worldwide, and the second major cause of cancer-related mortality in women.1 Although there have been numerous advances in surgical and medical management for breast cancer, the prognosis of breast cancer patients has not been improved significantly,2 due to the high frequency of metastasis, recurrence, and drug resistance.3 Therefore, it is urgent to further understand the molecular mechanism and develop more effective therapeutic strategies to better treat breast cancer.
The initiation and progression of breast cancer is a complex process, including the function of various genetic and epigenetic factors. Accumulating studies suggest that non-coding RNAs (ncRNAs) play important roles in cancer. So far, several studies have revealed that microRNAs (miRNAs)4, 5 and lncRNAs6, 7, 8 participate in metastasis and drug resistance of breast cancer, providing potential targets for cancer treatment. As a new class of ncRNAs, circular RNAs (circRNAs) have been found to be involved in various biological processes,9 including cell proliferation, migration, gene expression, and the regulation of certain responses. circRNAs are characterized by a covalently closed continuous loop where the 3′ and 5′ RNA ends are joined together and have no polyadenylated counterparts,10 which gives them higher stability compared to linear types.11 Based on the different derivation, circRNAs can be classified into several types, such as exonic, intonic, and intergenic circRNAs.12, 13 Recently, circRNAs have been found to be expressed in various cell lines and species, and they are conserved and highly specific to cell type, tissue, or developmental stage.14, 15 Emerging studies have revealed that circRNAs play important roles in the regulation of multiple diseases, especially cancers. Aberrant expression of circRNAs was correlated with progression, drug resistance, and prognosis of cancers.16, 17
circRNAs can participate in gene regulation through various mechanisms, including competing endogenous RNA (ceRNA) mechanisms, sequestering RNA-binding proteins (RBPs), and serving as transcription regulators.18, 19 The most well-known function pattern for circRNAs is acting as a miRNA sponge and functioning through a ceRNA mechanism. circRNA_100290 serves as a sponge for the miR-29 family and participates in oral cancer.20 circ-ABCB10 promotes cell proliferation and progression through competitively binding with miR-1271 and can be a potential therapeutic target for breast cancer.3 Moreover, circular RNA_LARP4 inhibits proliferation and invasion of gastric cancer by attenuating miR-424-5p-mediated suppression on LATS1.21 However, further studies on the function of circRNAs in breast cancer are needed. Tamoxifen (TAM), through competitively binding with estrogen receptor (ER), has proven to be one of the most successful endocrine treatments for ER+ breast cancer patients.22 Unfortunately, the development of TAM resistance limits its clinical application. Many long ncRNAs (lncRNAs)23, 24 and miRNAs25, 26 are associated with TAM resistance, but the functional significance of circRNAs on the acquired resistance remains unclear.
In the present study, we analyzed the expression profiles of circRNAs in breast cancer tissues using RNA sequencing (RNA-seq) and further characterized a novel circRNA derived from the BMPR2 gene, termed circBMPR2. The functions and mechanisms of circBMPR2 in the progression and TAM resistance of breast cancer were explored.
Results
circBMPR2 Is Downregulated in Breast Cancer Tissues with Metastasis
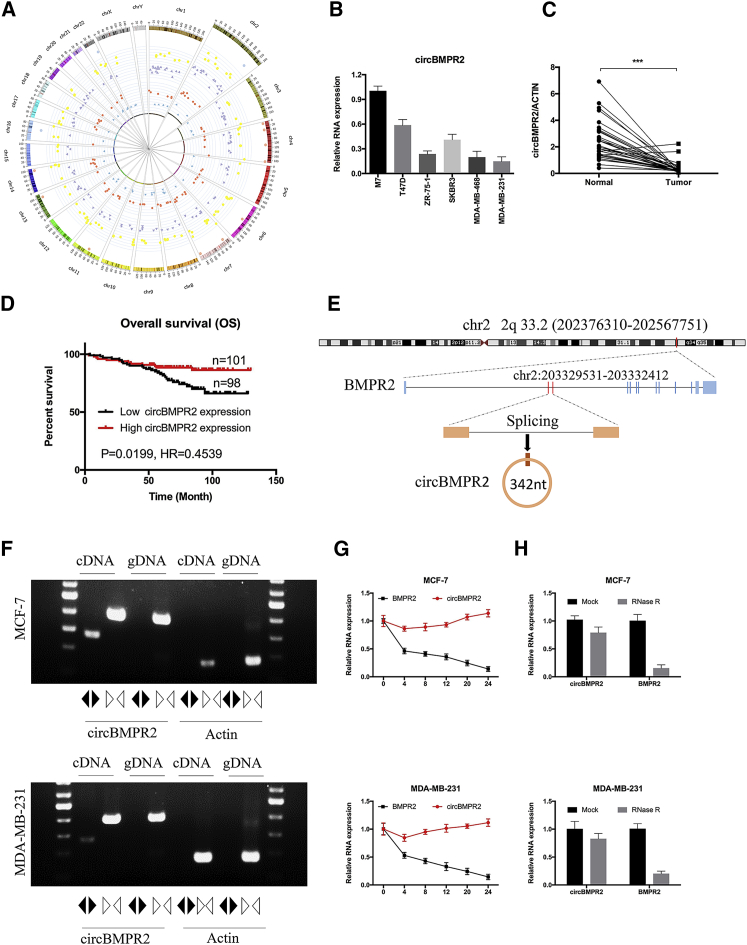
To investigate the role of circRNAs in the progression of breast cancer, we first explored the circRNA expression profiles in breast cancer tissues. According to the GEO database (GEO: GSE77661), 119 circRNAs were upregulated, and 132 circRNAs were downregulated in breast cancer tissue compared with normal tissue. Moreover, our circRNA array assay revealed that 125 circRNAs were differentially expressed in breast cancer tissues with or without metastasis, including 80 circRNAs that increased and 45 circRNAs that decreased in breast cancer tissues with metastasis. Among them, 3 and 10 circRNAs were significantly downregulated and upregulated in the 2 databases (Figure 1A). We then examined the expression of the common differentially expressed circRNAs in breast cancer tissues and cell lines. The quantitative real-time PCR results showed that the expression of hsa_circRNA_0003218 (termed circBMPR2) was decreased in breast cancer cells with higher metastatic ability (such as MDA-MB-231 and MDA-MB-468) compared with cells with lower metastatic potential (such as MCF-7 and T47D) (Figure 1B). Next, 35 pairs of breast cancer tissues and matched adjacent non-tumor tissues were collected and tested for circBMPR2 expression with quantitative real-time PCR. The results showed that the expression of circBMPR2 was downregulated in breast cancer tissues (Figure 1C). Moreover, the decreased expression of circBMPR2 was also confirmed using the GEO (GEO: GSE101123) database. Additionally, a cohort of 199 breast cancer patients with survival data were included to analyze whether circBMPR2 is correlated with the prognosis. The Kaplan-Meier survival curve revealed that lower circBMPR2 expression was associated with a worse overall survival in breast cancer patients (Figure 1D).
Figure 1.
Differential Expression Profiles of circRNAs in Human Breast Cancer Tissues
(A) A circos plot was used to show the differentially expressed circRNAs (>1-fold change, p < 0.05). The inner circle shows the downregulated circRNAs in breast cancer tissue with metastasis (blue triangles). The second circle identifies the upregulated circRNAs in breast cancer tissue with metastasis (red dots). The third circle shows the decreased circRNAs in breast cancer tissue (purple triangles), while the fourth circle reveals the increased circRNAs in breast cancer tissue compared with normal tissue (yellow dots). The outside circle represents the overlap of upregulated (pink dot) or downregulated circRNAs (blue dots). (B) The relative expression of circBMPR2 was measured in different breast cancer cell lines using quantitative real-time PCR. (C) The relative expression circBMPR2 in 35 pairs of breast cancer tissues and adjacent non-tumor tissues was measured by quantitative real-time PCR. (D) Kaplan-Meier plotter analysis of the correlation of circBMPR2 expression level with overall survival of breast cancer patients. (E) The schematic diagram indicates the genomic loci of the BMPR2 gene and circBMPR2. (F) Convergent or divergent primers were used to validate the existence of circBMPR2 in MCF-7 and MDA-MB-231 cell lines without RNase R treatment via RT-PCR. circBMPR2 could be amplified by convergent primers in both cDNA and gDNA; however, it could be amplified by divergent primers only in cDNA but not in gDNA. The linear β-actin was used as a negative control, which could be amplified only by convergent primers in both cDNA and gDNA. Black and white triangles represent divergent and convergent primers, respectively. (G) The quantitative real-time PCR indicated the abundance of circBMPR2 and BMPR2 in breast cancer cells after treatment with actinomycin D at the indicated time points. (H) The quantitative real-time PCR indicated the abundance of circBMPR2 and BMPR2 in breast cancer cells after treatment with RNase R. ***p < 0.001, Student’s t test.
According to the circBase database, circBMPR2 is derived from gene BMPR2 on chr2: 203329531–203332412 and ultimately forms the spliced mature sequence with a full length of 342 nt (Figure 1E). We designed two sets of primers for circBMPR2 and actin, and RT-PCR assays were used to verify the head-to-tail splicing of circBMPR2. Using cDNA and genomic DNA (gDNA) templates from breast cancer cells, the convergent primers could amplify both circBMPR2 and actin. However, the divergent primers could only amplify circBMPR2 using cDNA as templates, and no amplifications were observed using gDNA templates (Figure 1F). We next investigated the stability of circBMPR2. After treatment with actinomycin D, an inhibitor of transcription, total RNAs were harvested at the indicated time points from MCF-7 and MDA-MB-231 cells. The quantitative real-time PCR analysis revealed that the transcript half-life of circBMPR2 exceeded 24 h, whereas the associated linear BMPR2 displayed a half-life < 4 h (Figure 1G). Moreover, circBMPR2 displayed resistance to digestion with RNase R exonuclease (Figure 1H), which further confirmed that the circRNA isoforms were highly stable. These results demonstrated that circBMPR2 was consistently expressed in breast cancer cells and might play a significant role in the regulation of breast cancer progression.
Knockdown of circBMPR2 Promotes Proliferation, Migration, and Invasion of Breast Cancer Cells
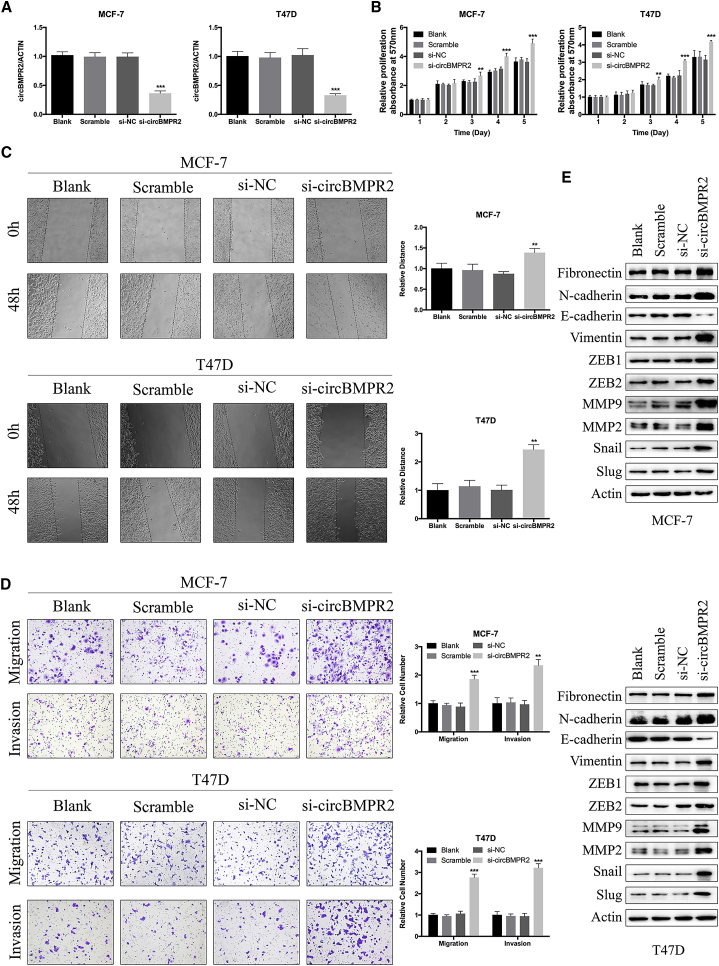
We further investigated the potential functional role of circBMPR2 in breast cancer cells. The breast cancer cells were transfected with small interfering RNA (si)-circBMPR2 or negative control (NC), and the quantitative real-time PCR analysis indicated that the expression levels of circBMPR2 were effectively decreased (Figure 2A; Figure S1A). Subsequent MTT assay showed that downregulation of circBMPR2 significantly promoted the growth of breast cancer cells (Figure 2B; Figure S1B). Flow cytometry analysis was performed to evaluate whether circBMPR2 could affect cell proliferation by altering the cell-cycle profiles. The results showed that more cells were distributed in S phase after circBMPR2 knockdown, indicating that circBMPR2 knockdown promoted cell-cycle progression (Figure S1C). We then evaluated the effect of circBMPR2 on cell motility. Knockdown of circBMPR2 observably increased the wound-healing ability of cells (Figure 2C). Consistently, the Transwell migration and invasion assays indicated that knockdown of circBMPR2 promoted the motility of breast cancer cells (Figure 2D; Figure S1D). Epithelial-mesenchymal transition (EMT) is one of the major mechanisms involved in cancer cell malignant transformation, and we further examined the effect of circBMPR2 on the expression of EMT-related proteins. Western blot (WB) showed that knockdown of circBMPR2 could decrease the levels of epithelial markers and increased the levels of mesenchymal markers (Figure 2E; Figure S1E). We further confirmed these effects by overexpressing circBMPR2 in the MDA-MB-231 and MDA-MB-468 cell lines, and quantitative real-time PCR assays confirmed the overexpression efficiency of circBMPR2 (Figure S1F). In accordance with the aforementioned results, circBMPR2 overexpression led to inhibited proliferation and migration of breast cancer cells (Figures S1G and S1H). These results suggested that circBMPR2 could inhibit the progression of breast cancer cells in vitro.
Figure 2.
Knockdown of circBMPR2 Promotes Proliferation, Migration, and Invasion of Breast Cancer Cells
(A) The knockdown efficiency of circBMPR2-targeting siRNAs on circBMPR2 mRNA in MCF-7 and T47D cells was measured by quantitative real-time PCR. Blank refers to cells just using Lipofectamine 2000; Scramble refers to non-transfected cells; si-NC refers to cells transfected with negative control; si-circBMPR2 refers to cells transfected with siRNAs targeting circBMPR2. (B) Knockdown of circBMPR2 inhibited cell proliferation, as indicated by MTT assay in MCF-7 and T47D cells. (C) Wound healing assay indicated that circBMPR2 knockdown resulted in a faster closing of scratch wounds. (D) Transwell assays were performed to evaluate the cell migration and invasion abilities of MCF-7 and T47D cells transfected with si-circBMPR2 or si-NC. (E) Western blot was used to examine the effect of circBMPR2 knockdown on EMT-related proteins. *p < 0.05; **p < 0.01; ***p < 0.001, Student’s t test.
circBMPR2 Is Negatively Associated with TAM Resistance of Breast Cancer Cells
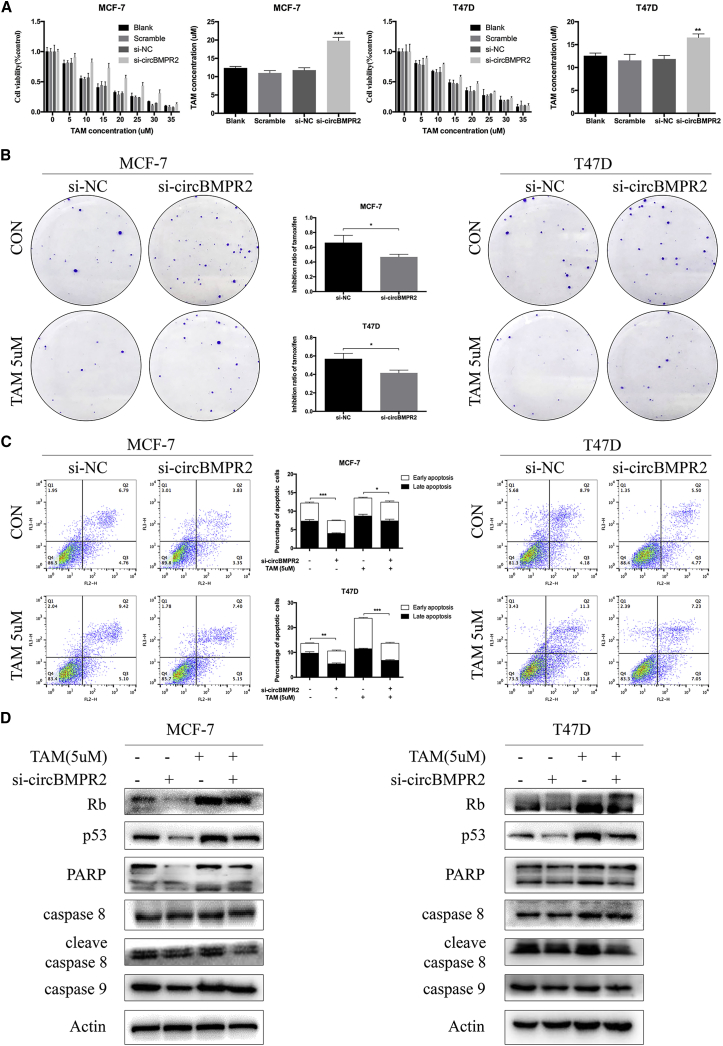
Considering the relatively higher expression of circBMPR2 in ER+ breast cancer cells (Figure 1B), such as MCF-7 and T47D,27 which are more sensitive to TAM, we further investigated the role of circBMPR2 in TAM resistance. After treatment of indicated concentrations of TAM for 48 h,28 the MTT assays showed that knockdown of circBMPR2 led to increased resistance to TAM in MCF-7 and T47D cells (Figure 3A). Then, we chose 5 μM TAM for further clone formation and apoptosis assays, which is approximately half of the 50% inhibitory concentration (IC50) value. The colony formation assay showed that the inhibition ratio of TAM decreased to 49% from 65% in MCF-7 cells and decreased to 42% from 57% in T47D cells after circBMPR2 knockdown, indicating that circBMPR2 knockdown could attenuate the inhibited effect of TAM on colony-forming ability in ER+ cells (Figure 3B). We then performed flow cytometry analysis to evaluate whether circBMPR2 modulated TAM resistance by regulating apoptosis, and the data showed that downregulation of circBMPR2 markedly inhibited the apoptosis caused by TAM treatment (Figure 3C). To verify the mechanism, WB was performed, and the results showed that circBMPR2 knockdown further decreased TAM-induced apoptosis, as determined by the expression levels of apoptosis-related proteins (Figure 3D). We then evaluated the effect of circBMPR2 on TAM-resistant cells (MCF-7/TAMR cells). The circBMPR2 expression of MCF-7/TAMR cells was significantly decreased compared with that of the parental cells (Figure S2A). Consistently, overexpression of circBMPR2 in MCF-7/TAMR cells was found to suppress cell proliferation, migration, invasion, and EMT processes (Figures S2B–S2F). Moreover, circBMPR2 overexpression helped to restore the sensitivity to TAM in MCF-7/TAMR cells by promoting TAM-induced cell apoptosis (Figures S2G and S2H).
Figure 3.
Knockdown of circBMPR2 Promotes Tamoxifen Resistance of Breast Cancer Cells
(A) MTT assays indicated that circBMPR2 knockdown decreased resistance to tamoxifen in MCF-7 and T47D cells. (B) Colony formation assays shows that circBMPR2 knockdown promoted resistance to tamoxifen in MCF-7 and T47D cells. Statistic graphs indicate the inhibition ratio of tamoxifen in the absence and presence of circBMPR2. (C) Knockdown of circBMPR2 attenuated tamoxifen-induced apoptosis in MCF-7 and T47D cells. (D) Western blot was used to evaluate the effect of circBMPR2 knockdown and tamoxifen treatment on apoptosis-related proteins. *p < 0.05; **p < 0.01; and ***p < 0.001, Student’s t test.
We then treated MCF-7 and T47D cells with TAM for 7 days and found that the expression of circBMPR2 was dramatically decreased in accordance with the trend of GREB1 (GenBank: NM_014668.4), a well-known target of ER (Figure S3A). Then, we cultured MCF-7 and T47D cells with estrogen-deprived medium to simulate similar conditions, and quantitative real-time PCR results showed that the expression of circBMPR2 and GREB1 was significantly decreased (Figure S3B). On the contrary, increased concentration of estrogen treatment led to enhanced expression of circBMPR2 and GREB1 in MCF-7 and T47D cells (Figure S3C). Based on the aforementioned results, we wondered whether circBMPR2 could regulate the expression of ER to modulate TAM resistance. The results showed that circBMPR2 overexpression further increased the expression of ER and its target progesterone receptor (PR) in MCF-7 cells and also restored the expression of ER in MCF-7/TAMR cancer cells (Figure S3D). Moreover, circBMPR2 overexpression enhanced the effect of estrogen on its target genes, such as GREB1 and PGR (Figure S3E), which might be mediated by elevated ER expression. All these findings suggested that circBMPR2 could increase TAM sensitivity of breast cancer cells and that mutual regulation might exist between estrogen and circBMPR2.
circBMPR2 Acts as a Sponge of miR-553 in Breast Cancer
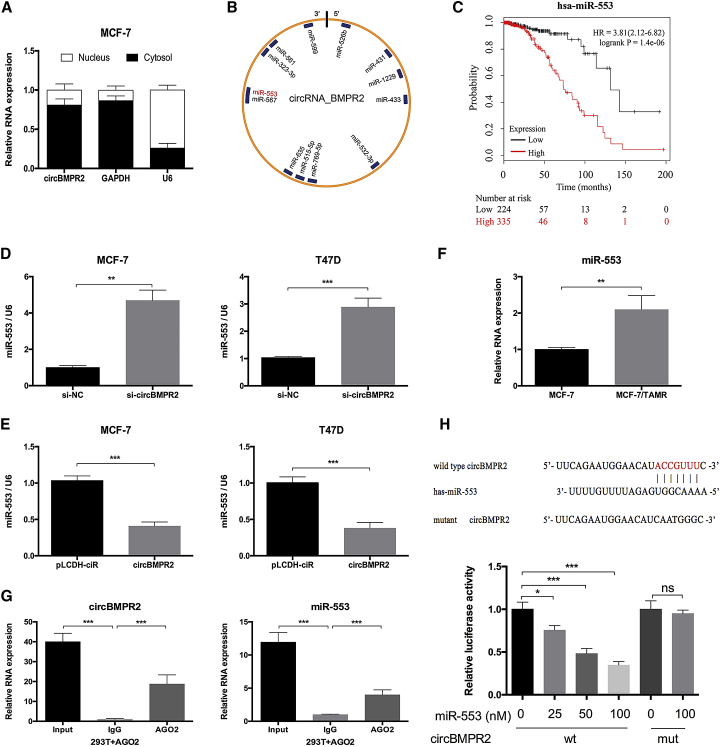
To examine the molecular mechanism of circBMPR2 in breast cancer, we first evaluated the intracellular location of circBMPR2. Nuclear and cytoplasmic fractions were separated from cells, and expression levels of circBMPR2, U6 (nuclear control), and GAPDH (cytoplasmic control) were detected by quantitative real-time PCR. The results revealed that circBMPR2 was predominantly localized in the cytoplasm (Figure 4A), indicating the potential to serve as a miRNA sponge. According to the Circular RNA Interactome database, the potential binding sites of 13 miRNAs were found within the circBMPR2 sequence (Figure 4B; Table S1). Then, the Kaplan-Meier Plotter tool was used to evaluate the association between the expression of miRNAs and overall survival of breast cancer patients. High expression levels of miR-1229, miR-515-5p, miR-635, miR-567, miR-599, and miR-553 were correlated with poor prognosis (Figure 4C; Figure S4), indicating that they may play oncogenic roles and could be regulated by circBMPR2 in breast cancer. The quantitative real-time PCR results revealed increased expression of these miRNAs after circBMPR2 knockdown (Figure S5A). Among them, the change of miR-553 was the most significant, and overexpression of circBMPR2 exhibited the opposite roles (Figures 4D and 4E). Moreover, given the potential role of circBMPR2 in regulating TAM resistance, we also examined the expression of these miRNAs in MCF-7 and MCF-7/TAMR cells and found that the expression of miR-553 was increased in MCF-7/TAMR cells (Figure 4F; Figure S5B). Therefore, we chose miR-553 as a regulatory target of circBMPR2 and further determined the binding potential between them. RNA immunoprecipitation (RIP) assay was then performed in HEK293T cells to determine the association between cirBMPR2 and miR-553. The quantitative real-time PCR results showed that the expression of circBMPR2 and miR-553 pulled down with anti-AGO2 antibodies was significantly higher compared to the anti-IgG (immunoglobulin G) group (Figure 4G), suggesting that miR-553 could directly target circBMPR2 in an AGO2 manner. In addition, luciferase reporter assays demonstrated that overexpression of miR-553 could remarkably reduce the luciferase activity of the vector containing the wild-type circBMPR2 sequence but not the luciferase activity of the vector with mutant binding sites (Figure 4H). On the other hand, miR-553 overexpression could cause decreased expression of circBMPR2 (Figure S5C), indicating that there existed mutual regulation. These results implied that circBMPR2 could function as a ceRNA for miR-553 through direct binding between them.
Figure 4.
circBMPR2 Acts as a miRNA Sponge for miR-553 in Breast Cancer Cells
(A) Subcellular fractionation location assay determined that circBMPR2 was mainly located in cytoplasm. (B) A schematic model showed the putative binding sites for miRNAs and the 3′ UTR of circBMPR2. (C) Kaplan-Meier Plotter tool was used to detect the association between the expression of miR-553 and the overall survival of breast cancer patients. (D) Knockdown of circBMPR2 promoted the expression of miR-553 in MCF-7 and T47D cells. (E) Overexpression of circBMPR2 inhibited the expression of miR-553 in MCF-7 and T47D cells. (F) The expression of miR-553 was increased in MCF-7/TAMR cells, compared to their parental cells. (G) RIP experiments were performed in HEK293T cells, and the co-precipitated RNA was subjected to quantitative real-time PCR for circBMPR2 and miR-553. (H) Schematic representation showed the wild-type and mutant potential binding sites of miR-553 in circBMPR2 (top). Luciferase assays was performed in HEK293T cells transfected wild-type or mutant pmirGLO-circBMPR2 and miR-553 or NC (bottom). **p < 0.01; ***p < 0.001, Student’s t test.
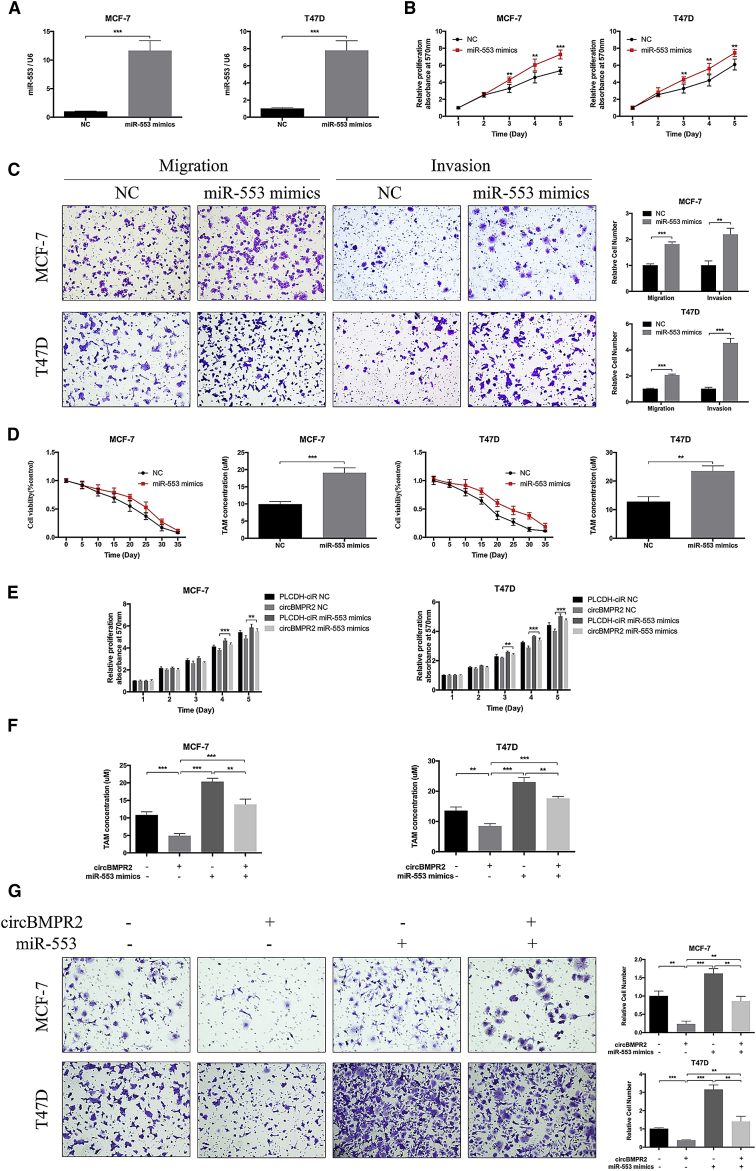
miR-553 Plays an Oncogenic Role and Partly Reverses the Effects Caused by circBMPR2 in Breast Cancer Cells
We then determined the biological function of miR-553 in breast cancer. The quantitative real-time PCR assay was used to confirm the overexpression efficiency of miR-553 (Figure 5A). MTT assay showed that upregulation of miR-553 significantly enhanced the proliferation and migration of breast cancer cells (Figures 5B and 5C). Moreover, increased resistance to TAM was observed in MCF-7 and T47D cells transfected with miR-553 mimics compared to the control group (Figure 5D). TAM treatment and estrogen deprivation could increase the expression of miR-553, whereas estrogen treatment caused opposite trends (Figures S6A–S6C), which may be due to elevated expression of circBMPR2. To address whether circBMPR2 inhibited proliferation, migration, and TAM resistance of breast cancer cells via interacting with miR-553, we co-transfected miR-553 mimics and circBMPR2-overexpressed vectors into breast cancer cells. The rescue experiments showed that overexpression of miR-553 could attenuate the function of circBMPR2 in breast cancer (Figures 5E–5G). Taken together, these results suggested that circBMPR2 suppressed proliferation, migration, and TAM resistance through sponging miR-553 in breast cancer cells.
Figure 5.
miR-553 Acts as an Oncogene and Eliminated the Repression Function of circBMPR2 in Breast Cancer Cells
(A) The expression levels of miR-553 in MCF-7 and T47D cells after transfection of miR-553 mimics or negative control (NC) were detected by quantitative real-time PCR. (B) MTT assay indicated that miR-553 promoted the vitality of MCF-7 and T47D cells. (C) Overexpression of miR-553 promoted the migration and invasion of breast cancer cells. (D) Overexpression of miR-553 increased the tamoxifen resistance of breast cancer cells. (E–G) MTT and Transwell migration assays were used to determine the cell proliferation (E), tamoxifen resistance (F), and migration (G) of MCF-7 and T47D cells co-transfected with pLCDH-circBMPR2 and miR-553 mimics. **p < 0.01; ***p < 0.001, Student’s t test.
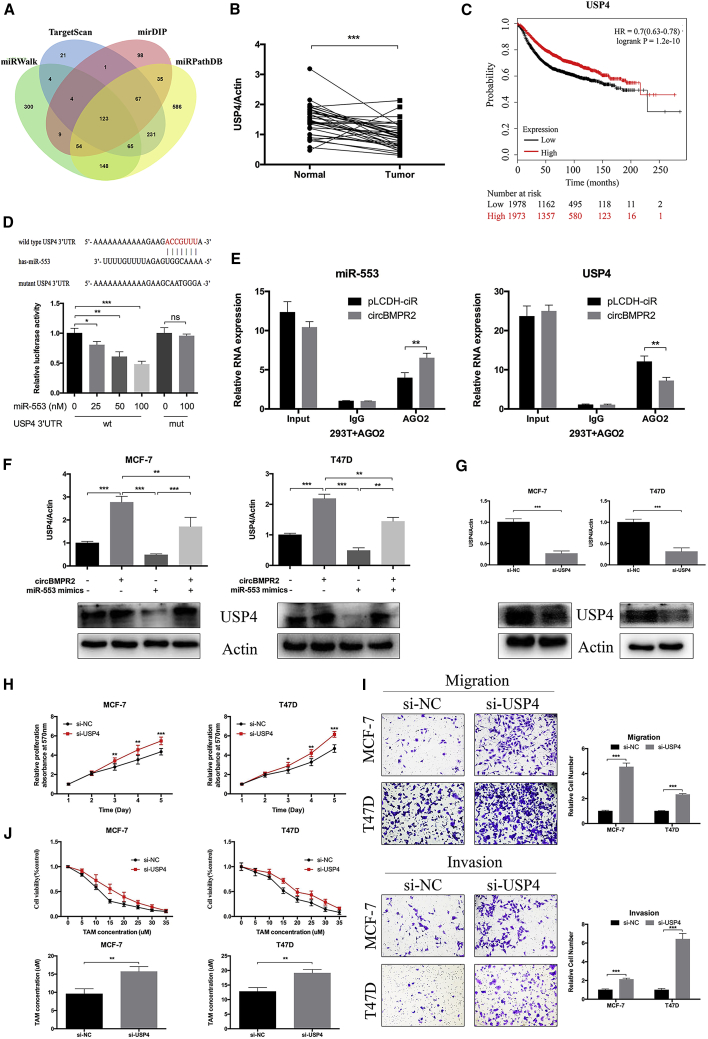
circBMPR2 Upregulates USP4 Expression via Inhibition of miR-553 in Breast Cancer Cells
Then, we utilized the prediction software (miRWalk, TargetScan, mirDIP, and miRPathDB) to forecast the potential target genes of miR-553 in breast cancer, and 123 genes were identified with very high stringency (Figure 6A). Among them, USP4, an enzyme that can cleave ubiquitin from various protein substrates, ranked relatively higher and was one of the most likely predicted targets. The quantitative real-time PCR results indicated that the expression of USP4 was lower in breast cancer tissues, compared with normal tissues (Figure 6B). The Cancer Genome Atlas (TCGA) database also determined lower expression of USP4 in breast cancer tissues, especially triple-negative breast cancer tissues (Figure S7A). The Kaplan-Meier Plotter tool revealed that higher expression of USP4 was associated with better prognosis of breast cancer patients (Figure 6C). Moreover, the expression of USP4 was positively associated with prognosis of lung cancer and renal cancer patients according to the Human Protein Atlas database (Figure S7B). Luciferase reporter assays demonstrated that overexpression of miR-553 led to decreased luciferase activity of vectors containing wild-type 3′ UTR of USP4 but did not affect the luciferase activity of mutant types (Figure 6D). Moreover, a RIP assay was performed to determine the effect of circBMPR2 on the association between USP4 and miR-553. The quantitative real-time PCR results showed that the expression of miR-553 or USP4 pulled down with anti-AGO2 antibodies in the circBMPR2-overexpressing group was significantly higher or lower compared to the control group, respectively (Figure 6E), suggesting that circBMPR2 could inhibit the association between USP4 and miR-553 in an AGO2 manner. Overexpression of miR-553 or circBMPR2 knockdown could decreased the protein and RNA expression of USP4 in MCF-7 and T47D cells (Figure S7C). Moreover, the increased expression of USP4 induced by circBMPR2 overexpression could be partly abrogated by ectopic expression of miR-553 (Figure 6F). Next, we used RNAi to knock down the expression of USP4 to evaluate its biological functions in breast cancer. The quantitative real-time PCR and WB assays were used to demonstrate that knockdown efficiency (Figure 6G). The results of an MTT assay showed that USP4 knockdown significantly promoted the proliferation, migration, and invasion of breast cancer cells (Figures 6H and 6I). The protein and RNA levels of USP4 in MCF-7/TAMR cells were lower compared to those of the parental cells (Figure S7D), and knockdown of USP4 increased the resistance to TAM in MCF-7 and T47D cells (Figure 6J). Moreover, USP4 knockdown could lead to decreased ER protein expression levels, although no significant changes on ER mRNA levels were found (Figure S7E). Altogether, these results indicated that USP4 was a direct target of miR-553 and played a similar role to that of circBMPR2 in breast cancer.
Figure 6.
miR-553 Acts as an Oncogene and Eliminated the Repression Function of circBMPR2 in Breast Cancer Cells
(A) Schematic illustration showing overlapping of the target genes of miR-553 predicted by TargetScan, miRWalk, mirDIP, and miRPathDB. (B) The expression of USP4 was decreased in breast cancer cells, compared to the adjacent normal tissues. (C) Kaplan-Meier Plotter tool showed that the expression of miR-553 was associated with better overall survival of breast cancer patients. (D) The binding sites of wild-type or mutant USP4 3′ UTR with miR-553 (top). The luciferase activity of wild-type or mutant USP4 3′ UTR after transfection with miR-55 mimic or NC in HEK293T cells (bottom). (E) circBMPR2 overexpression leads to decreased association between mir-553 and USP4. (F) The quantitative real-time PCR and western blot assays were used to determine the USP4 expression level after co-transfection with pLCDH-circBMPR2 and miR-553 mimics in MCF-7 and T47D cells. (G) The quantitative real-time PCR and western blot assays revealed reduced USP4 expression after transfection with si-USP4 in MCF-7 and T47D cells. (H) Knockdown of USP4 promoted proliferation of MCF-7 and T47D cells as detected by MTT assay. (I) Transwell assay showed that knockdown of USP4 promoted migration and invasion in MCF-7 and T47D cells. (J) Knockdown of USP4 led to increased resistance to tamoxifen in MCF-7 and T47D cells. **p < 0.01; ***p < 0.001, Student’s t test.
Discussion
Emerging evidence demonstrates that circRNAs play crucial roles in carcinogenesis and cancer progression and have attracted much attention recently. Aberrantly expressed circRNAs have been reported in diverse cancer types, such as pancreatic cancer,29 colorectal cancer,30 hepatocellular carcinoma,31 and bladder cancer,32 indicating that circRNAs may serve as regulators and diagnostic markers for cancer. As the relationship between expression and function of circRNAs and breast cancer progression is not well established, we screened the differentially expressed circRNAs in breast cancer tissues and focused on the function and potential mechanism of circBMPR2 in breast cancer.
Using RNA-sequencing analysis and the GEO database, we identified a novel circRNA, circBMPR2, which is downregulated in breast cancer tissue, especially tissue with metastasis. Moreover, the expression of circBMPR2 is also downregulated in gastric cancer and hepatocellular cancer tissues based on the GEO (GEO: GSE77661) database. The downregulation of circBMPR2 in breast cancer tissues was further confirmed using 35 pairs of breast cancer tissues and matched adjacent non-tumor tissues. These results suggested that circBMPR2 might act as a tumor suppressor in breast cancer. circRNAs are promising potential biomarkers because of their unique structure, high stability, and specific expression patterns.33 Using a cohort of 199 breast cancer patients, we found that patients with lower levels of circBMPR2 had a worse overall survival than those with higher circBMPR2 expression, indicating that circBMPR2 might act as a prognostic factor for survival in patients with breast cancer.
The past few decades have witnessed outstanding advances in breast cancer treatment. However, tumor metastasis and the development of resistance remain great threats to human health, leading to a poor prognosis for breast cancer patients.34 Through our in vitro experiments, we found that knockdown of circBMPR2 could promote cell-cycle progression and enhance cell proliferation. Moreover, circBMPR2 was able to decrease the metastatic ability of breast cancer cells through interfering with the EMT process, and the expression levels of EMT-related proteins showed corresponding changes. TAM is one of the most successful agents for ER+ breast cancer; however, the developed resistance during treatment or intrinsic resistance to TAM limits its therapeutic benefits. Accumulating evidence suggests that tumor metastasis and TAM resistance may be closely associated processes, which can be regulated by common factors.35, 36, 37 Our results demonstrated that circBMPR2 overexpression could markedly inhibit the migration and invasion ability of MCF-7/TAMR cells, which was in accordance with the WB results. Moreover, circBMPR2 knockdown could promote the resistance to TAM in breast cancer cells, whereas circBMPR2 overexpression significantly inhibited TAM resistance. These results revealed the regulatory role of circBMPR2 in the two processes and preliminarily suggested the relevance between metastasis and TAM resistance. Previous studies showed that TAM could suppress cell proliferation through inducing apoptosis,38, 39 and our results found that circBMPR2 knockdown could significantly inhibit TAM-induced apoptosis, while circBMPR2 knockdown alone could also inhibit cell apoptosis. It was plausible that circBMPR2 attenuated TAM resistance through increasing TAM-induced apoptosis. The aforementioned results suggested a tumor-suppressor role of circBMPR2 in breast cancer, revealing the potential of circBMPR2 as a therapeutic target and prognosis predictor.
Recently, circRNAs have been reported to regulate the expression of oncogenic or tumor-suppressive genes through competing for binding with miRNA,18 constructing a complex posttranscriptional regulatory network. The interaction between circRNAs and miRNAs has already been found to perform significant roles in multiple cancers.40 Our results demonstrated that circBMPR2 was predominantly localized in the cytoplasm, served as a miR-553 sponge, and decreased the abundance of miR-553 in the cytoplasm. We first revealed that miR-553 promoted proliferation, metastasis, and TAM resistance in breast cancer, and miR-553 overexpression could partly reverse the effects of circBMPR2 on breast cancer cells. Moreover, circBMPR2 and miR-553 could regulate the expression of each other, forming a negative-feedback loop.
Through bioinformatics analysis, we chose USP4 as a potential target of miR-553, which was confirmed by further luciferase reporter assay, RIP assays, and functional studies. Moreover, overexpression of circBMPR2 led to increased expression of USP4, as expected in our hypothesis, implying a circBMPR2/miR-553/USP4 axis in breast cancer. Ubiquitin-specific protease 4 (USP4), a member of the USPs, mediates the removal and processing of ubiquitin. USP4 has been reported to participate in various human tumors with diverse biological functions. Previous studies have shown that USP4 expression was significantly decreased in breast cancer tissue and that USP4 inhibited breast cancer cell growth through inhibiting PDCD4 degradation.41 USP4 was downregulated in lung adenocarcinoma and served as an independent predictor.42 Some other studies claimed that USP4 might have oncogenic properties. USP4 could inhibit p53 through deubiquitinating and stabilizing ARF-BP143 or HDAC244 and was upregulated in several cancer tissues. The WNT/β-catenin pathway was reported to be regulated by USP4 through deubiquitination and facilitating nuclear localization of β-catenin in colorectal cancer.45 Moreover, another study claimed that USP4 could promote migration and invasion of breast cancer cells.46 It is controversial to define the exact role of USP4 in cancer, especially breast cancer. It is plausible that the functions of USP4 may depend on different types of cancer, different stages, or different regulators. Here, we showed that USP4 was decreased in breast cancer tissues compared with adjacent normal tissues. Knockdown of USP4 significantly promoted proliferation, metastasis, and TAM resistance in breast cancer, indicating a tumor-suppressive role for USP4. In addition, USP4 knockdown led to significantly decreased ER protein expression levels, which might result in decreased sensitivity to TAM. Remarkably, the ER mRNA levels exhibited no significant change after USP4 knockdown, indicating that the regulatory effects of USP4 on ER expression is at the post-transcriptional level. Considering that USP4 promotes the removal of ubiquitins from proteins, it may also protect ER from ubiquitin-mediated degradation. However, further study is needed to reveal the specific regulation mechanism.
In summary, circRNAs can influence various biological behaviors of tumor cells and act as promising potential biomarkers for cancer diagnosis and prognosis prediction. The present study provides novel insights for future studies on the role of circRNAs in tumor progression and TAM resistance. It is reasonable for us to conclude that circBMPR2 inhibits breast cancer cell proliferation, migration, invasion, and TAM resistance by functioning as a sponge of miR-553 to upregulate USP4 expression. Therefore, circBMPR2 may serve as a potential therapeutic target and prognostic predictor for breast cancer patients.
Materials and Methods
Patient Samples
Tumor tissues and paired adjacent non-tumorous tissues were collected from the breast cancer patients who received treatment at Qilu Hospital from February 2009 to January 2015. The median follow-up was 69 months. All the tissue specimens were confirmed by histological and pathological diagnoses. Written informed consent was obtained from all participants. This study was approved by the Ethical Committee of Shandong University.
Microarray Analysis
Three pairs of breast cancer tissues with or without metastasis were collected at Qilu Hospital of Shandong University in 2017. Tissue specimens were obtained during operation and immediately frozen at −80°C until further use. Total RNA was extracted using TRIzol reagent (Takara Bio, Shiga, Japan). The concentration and quality of each RNA sample was evaluated with the Nanodrop ND-1000, and the quality control was based on a ratio of optical density 260 (OD260) to optical density 280 OD280 (1.8–2.0). The sample preparation and circRNA sequencing were performed using Cloud-Seq Biotech (Shanghai, China). Briefly, total RNAs were digested with Rnase R (Epicenter Technologies, Madison, WI, USA) to remove linear RNAs and enrich circRNAs. The enriched circRNAs were used to construct the RNA libraries with the TruSeq Stranded Total RNA Library Prep Kit (Illumina, San Diego, CA, USA), and the quality of libraries was controlled using the BioAnalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA). The denatured single-stranded DNA molecules were sequenced on an Illumina HiSeq Sequencer, and the paired-end reads were further analyzed by bowtie2 and find_circ software. The raw data were quantile normalized, and further data analysis was performed with the R software package, such as the impute package and the limma package. The statistical significance of differentially regulated circRNAs was identified through p value and fold change. Significantly differentially expressed circRNAs were retained by screening for fold change ≥ 2.0 and p < 0.05. Hierarchical clustering was performed to generate an overview of the characteristics of expression profiles based on values of all significantly differentially expressed transcripts.
Gene Expression Profiles
The gene expression data matrix of normal tissue and breast cancer tissue was obtained from the National Center for Biotechnology GEO database (https://www.ncbi.nlm.nih.gov/geo/), which is accessible through the GEO platforms GPL11154 (GEO: GSE77661) and GPL19978 (GEO: GSE101123). As reported, the matrix contained a total of 26 samples, including various cancer types.47 In the present study, we selected the data of normal breast tissue and breast cancer tissue for further study. The data analysis was performed with R software using the DEGseq package. Significantly differentially expressed circRNAs were identified with fold change ≥ 2.0 and p < 0.05.
Cell Lines and Transfection
All the cells were purchased from the American Type Culture Collection (Manassas, VA, USA). MCF-7, MDA-MB-231, MDA-MB-468, and HEK293T cells were cultured in DMEM. T47D, SKBR3, and ZR-75-1 cells were cultured in RPMI 1640 medium. The medium was supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 μg/mL streptomycin. Cells in the medium were incubated in a humidified atmosphere containing 5% CO2 at 37°C.
We constructed circBMPR2-overexpressed vector as previously reported.48 Briefly, the sequences of exons 2 and 3 of BMPR2 with a full length of 342 bp was subcloned into a pLCDH-ciR vector (RiboBio, Guangzhou, China) to generate pLCDH-circBMPR2 constructs. The subcloned sequence containing a front circular frame (SA), back circular frame (SD) of circRNA biogenesis and full length of circBMPR2, and 5′-TGAAATATGCTATCTTACAG-circBMPR2-GTGAATATATTTTTTCTTGA-3′ was directly synthesized. The pLCDH-ciR empty vector was used as an NC. For the knockdown experiments, the small interfering RNA (siRNA) target sequences for circBMPR2 were as follows: sense: 5′-CACCACUCACUUCGCAGAATT-3′; antisense: 5′-UUCUGCGAAGUGAGUGGUGTT-3′. The sequences for NC siRNA were as follows: sense: 5′-UUCUCCGAACGUGUCACGUTT-3′; antisense: 5′-ACGUGACACGUUCGGAGAATT-3′. The procedure of transfection was performed using Lipofectamine 2000 (Invitrogen) as previously reported.48
Development of a TAMR Cell Line
The MCF-7 cells were continuously exposed to 10 μM TAM (Sigma-Aldrich, St. Louis, MO, USA) in phenol red-free DMEM culture supplied with 10% charcoal-stripped FBS for at least 1 year,49 termed as MCF-7/TAMR cells. The medium was replaced every 4 days, and the cells were passaged by trypsinization when 70% confluence was reached. The MCF-7/TAMR cells were routinely cultured in medium with 1 μM TAM, and dead cells were rarely observed. Corresponding parental cells (WT) were cultured in parallel to resistant ones without the addition of TAM.
RNA Extraction and Real-Time qPCR
Total RNA from cells or tissues was isolated using TRIzol Reagent (Takara Bio, Shiga, Japan). cDNA was synthesized using the PrimeScript RT Reagent Kit (Takara, Shiga, Japan), and the PrimeScript miRNA cDNA Synthesis Kit (Takara, Shiga, Japan) was used for miRNA reverse transcription. Then, real-time qPCR was performed using SYBR Premix Ex Taq (Takara, Japan). The primers used in this study are shown in Table S2. β-actin was used as the endogenous control for the relative expression of mRNA, while U6 was used as the internal control for miRNA expression. The relative expression was calculated by the 2−ΔΔCT method.
Actinomycin D and RNase R Treatment
Transcription was prevented by the addition of 2 μg/mL actinomycin D or DMSO (Sigma-Aldrich, St. Louis, MO, USA) as the NC to the cell culture medium. After treatment in the indicated time, the RNA expression levels of circBMPR2 and BMPR2 were detected by quantitative real-time PCR. For RNase R treatment, the original RNAs extracted from MCF-7 or MDA-MB-231 cells were divided into two equal parts, respectively, one for RNase R treatment (RNase R) and the other for non-treatment (mock). 2 μg total RNA was incubated for 30 min at 37°C with or without 3 U/μg RNase R (Epicenter Technologies, Madison, WI, USA). The internal reference (actin) in the mock group was used as the calculation standard.50
MTT Assay
Transfected cells were seeded into a 96-well plate at a density of 1,500 cells per well and allowed to incubate up to 6 days. To evaluate the sensitivity to TAM, the cells were cultured in medium with indicated concentrations of TAM for 48 h. 20 μL MTT (5 mg/mL) was added to each well, and the cells were incubated for another 4 h. 100 μL DMSO was used to resolve the crystal, and the absorbance at 570 nm was measured using a microplate reader (Bio-Rad, Hercules, CA, USA).
Colony Formation Assay
Transfected MCF-7 and T47D cells were seeded into a 6-cm plate at a density of 500 cells per well and cultured for 2–3 weeks. 5 μM TAM was used to examine the effect of circBMPR2 on TAM resistance. The colonies were fixed with methanol for 15 min and then stained with 0.1% crystal violet for 20 min. The colonies were then counted and photographed.
Cell-Cycle Assay
Cell cycle assays were performed at 48 h after transfection. The transfected breast cancer cells were stained with 500 μL cell-cycle staining buffer (MultiSciences (Lianke), Hangzhou, Zhejiang, China) for 30 min in a dark place and then measured by flow cytometry (Becton Dickinson, Franklin Lakes, NJ, USA). The data were analyzed with ModFit LT V4.0 software.
Cell Apoptosis Analysis
The transfected cells were treated with or without 5 μM TAM for 48 h. Cells were stained using an Annexin V fluorescein isothiocyanate (FITC)/propidium iodide (PI) apoptosis detection kit (BD Biosciences, San Jose, CA, USA) according to the manufacturer’s instructions and analyzed by flow cytometry. The data were analyzed with FlowJo software.
Wound-Healing Assay
Breast cancer cells were seeded onto a 24-well plate and scraped with the fine end of 200-μL pipette tips (time 0 h). Cell migration was photographed using 10 high-power fields at indicated hours post-wounding. Remodeling was measured as diminishing distance across the induced injury and was normalized to the 0-h control. Then, relative distance was calculated.
Transwell Migration and Invasion Assays
The cell migration and invasion assays were performed using Transwell chambers (Corning, Corning, NY, USA), which were coated with (invasion assay) or without (migration assay) Matrigel (BD Falcon, Bedford, MA, USA). A total of 1 × 105 transfected cells were suspended in 200 μL serum-free medium and then added to the upper chambers. The lower chambers contained culture medium with 20% FBS as a chemoattractant. After 24–48 h, the cells located on the lower surfaces were fixed with methanol and stained with 0.1% crystal violet. The stained cells were photographed, and relative cell number was calculated.
Protein Isolation and WB
Total proteins were extracted, separated by 10% SDS-PAGE gel, and then transferred to 0.22 μm polyvinylidine difluoride (PVDF) membranes (Millipore, Billerica, MA, USA). The PVDF membranes were incubated with 5% skim milk to block nonspecific binding at room temperature for 1 h. The membranes were incubated with primary antibodies overnight at 4°C and secondary antibodies for 2 h at room temperature. The antibodies used in this study are presented in Table S3. The protein levels were detected by chemiluminescence (Millipore, Billerica, MA, USA). β-actin was used as endogenous control.
Subcellular Fractionation Location
The nuclear and cytoplasmic fractions were isolated using the PARIS Kit (Life Technologies, Carlsbad, CA, USA) following the manufacturer’s instruction.
Kaplan-Meier Plotter Tool Analysis
The Kaplan-Meier Plotter tool (http://kmplot.com/analysis/) was used to detect the association between miRNAs or USP4 and the overall survival of breast cancer patients.
RNA Immunoprecipitation Assay
An RNA immunoprecipitation (RIP) assay was conducted in HEK293T cells using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA, USA) according to manufacturer’s instructions. Anti-AGO2 (AGO2) antibody or NC mouse IgG (Millipore, Billerica, MA, USA) were used. The extracted RNAs were analyzed by quantitative real-time PCR.
Luciferase Reporter Assay
The full sequence of circBMPR2 and the mutant version were constructed into pmirGLO vector (Promega, Madison, WI, USA). Luciferase reporter vector with the wild-type or mutant sequence of the 3′ UTR of USP4 was constructed. HEK293T cells were seeded in 96-well plates and transfected with luciferase reporter vector and miR-553 or control miRNA using Lipofectamine 2000. After 48 h of incubation, luciferase reporter assays were conducted using a dual-luciferase reporter assay system (Promega, Madison, WI, USA) according to the manufacturer’s instructions. The subtracted difference of firefly and Renilla luciferase activities was calculated as relative luciferase activity.
Statistical Analysis
The statistical analysis was performed using SPSS software (v18.0). Data were expressed as mean ± SD from 3 independent experiments. Comparisons between groups were analyzed with the Student’s t test. p < 0.05 was considered statistically significant. Survival analysis was performed by Kaplan-Meier curves and log-rank test for significance.
Author Contributions
Y. Liang and Q.Y. conceived the study; Y. Liang, X.S., Y. Li, H.Z., Y.S., Y. Liu, and Y.D. performed the experiments; T.M., P.S., R.G., N.Z., and X.L. collected clinical samples; B.C., W.Z., and L.W. analyzed the data; Y. Liang and X.S. wrote the paper; Y. Liang and Q.Y. revised the paper. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (nos. 81272903, 81672613, 81502285, and 81602329), the Natural Science Foundation of Shandong Province (ZR2014HQ078), the China Postdoctoral Science Foundation (2018M630787 to N.Z.), the Key Research and Development Program of Shandong Province (nos. 2015GSF118093 and 2016GSF201119), the Shandong Science and Technology Development Plan (2016CYJS01A02), and the Special Support Plan for National High Level Talents (“Ten Thousand Talents Program” to Q.Y.).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.05.005.
Supplemental Information
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2017. CA Cancer J. Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Rodgers R.J., Reid G.D., Koch J., Deans R., Ledger W.L., Friedlander M., Gilchrist R.B., Walters K.A., Abbott J.A. The safety and efficacy of controlled ovarian hyperstimulation for fertility preservation in women with early breast cancer: a systematic review. Hum. Reprod. 2017;32:1033–1045. doi: 10.1093/humrep/dex027. [DOI] [PubMed] [Google Scholar]
- 3.Liang H.F., Zhang X.Z., Liu B.G., Jia G.T., Li W.L. Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am. J. Cancer Res. 2017;7:1566–1576. [PMC free article] [PubMed] [Google Scholar]
- 4.di Gennaro A., Damiano V., Brisotto G., Armellin M., Perin T., Zucchetto A., Guardascione M., Spaink H.P., Doglioni C., Snaar-Jagalska B.E. A p53/miR-30a/ZEB2 axis controls triple negative breast cancer aggressiveness. Cell Death Differ. 2018;25:2165–2180. doi: 10.1038/s41418-018-0103-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guarnieri A.L., Towers C.G., Drasin D.J., Oliphant M.U.J., Andrysik Z., Hotz T.J., Vartuli R.L., Linklater E.S., Pandey A., Khanal S. The miR-106b-25 cluster mediates breast tumor initiation through activation of NOTCH1 via direct repression of NEDD4L. Oncogene. 2018;37:3879–3893. doi: 10.1038/s41388-018-0239-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu W., Chen F., Cui X., Yang L., Chen J., Zhao J., Huang D., Liu J., Yang L., Zeng J. LncRNA NKILA suppresses TGF-β-induced epithelial-mesenchymal transition by blocking NF-κB signaling in breast cancer. Int. J. Cancer. 2018;143:2213–2224. doi: 10.1002/ijc.31605. [DOI] [PubMed] [Google Scholar]
- 7.Tang J., Li Y., Sang Y., Yu B., Lv D., Zhang W., Feng H. LncRNA PVT1 regulates triple-negative breast cancer through KLF5/beta-catenin signaling. Oncogene. 2018;37:4723–4734. doi: 10.1038/s41388-018-0310-4. [DOI] [PubMed] [Google Scholar]
- 8.Liang Y., Song X., Li Y., Sang Y., Zhang N., Zhang H., Liu Y., Duan Y., Chen B., Guo R. A novel long non-coding RNA-PRLB acts as a tumor promoter through regulating miR-4766-5p/SIRT1 axis in breast cancer. Cell Death Dis. 2018;9:563. doi: 10.1038/s41419-018-0582-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lasda E., Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20:1829–1842. doi: 10.1261/rna.047126.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilusz J.E., Sharp P.A. Molecular biology. A circuitous route to noncoding RNA. Science. 2013;340:440–441. doi: 10.1126/science.1238522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y., Zheng Q., Bao C., Li S., Guo W., Zhao J., Chen D., Gu J., He X., Huang S. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qu S., Yang X., Li X., Wang J., Gao Y., Shang R., Sun W., Dou K., Li H. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365:141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 13.Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., Zhong G., Yu B., Hu W., Dai L. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 14.Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., Marzluff W.F., Sharpless N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enuka Y., Lauriola M., Feldman M.E., Sas-Chen A., Ulitsky I., Yarden Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 2016;44:1370–1383. doi: 10.1093/nar/gkv1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J., Li Y., Zheng Q., Bao C., He J., Chen B., Lyu D., Zheng B., Xu Y., Long Z. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388:208–219. doi: 10.1016/j.canlet.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 17.Kun-Peng Z., Xiao-Long M., Chun-Lin Z. Overexpressed circPVT1, a potential new circular RNA biomarker, contributes to doxorubicin and cisplatin resistance of osteosarcoma cells by regulating ABCB1. Int. J. Biol. Sci. 2018;14:321–330. doi: 10.7150/ijbs.24360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 19.Zhong Z., Huang M., Lv M., He Y., Duan C., Zhang L., Chen J. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305–317. doi: 10.1016/j.canlet.2017.06.027. [DOI] [PubMed] [Google Scholar]
- 20.Chen L., Zhang S., Wu J., Cui J., Zhong L., Zeng L., Ge S. circRNA_100290 plays a role in oral cancer by functioning as a sponge of the miR-29 family. Oncogene. 2017;36:4551–4561. doi: 10.1038/onc.2017.89. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Zhang J., Liu H., Hou L., Wang G., Zhang R., Huang Y., Chen X., Zhu J. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol. Cancer. 2017;16:151. doi: 10.1186/s12943-017-0719-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burstein H.J., Lacchetti C., Anderson H., Buchholz T.A., Davidson N.E., Gelmon K.E., Giordano S.H., Hudis C.A., Solky A.J., Stearns V. Adjuvant endocrine therapy for women with hormone receptor-positive breast cancer: American Society of Clinical Oncology clinical practice guideline update on ovarian suppression. J. Clin. Oncol. 2016;34:1689–1701. doi: 10.1200/JCO.2015.65.9573. [DOI] [PubMed] [Google Scholar]
- 23.Xue X., Yang Y.A., Zhang A., Fong K.W., Kim J., Song B., Li S., Zhao J.C., Yu J. LncRNA HOTAIR enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogene. 2016;35:2746–2755. doi: 10.1038/onc.2015.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng W.X., Huang J.G., Yang L., Gong A.H., Mo Y.Y. Linc-RoR promotes MAPK/ERK signaling and confers estrogen-independent growth of breast cancer. Mol. Cancer. 2017;16:161. doi: 10.1186/s12943-017-0727-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayes E.L., Lewis-Wambi J.S. Mechanisms of endocrine resistance in breast cancer: an overview of the proposed roles of noncoding RNA. Breast Cancer Res. 2015;17:40. doi: 10.1186/s13058-015-0542-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Q., Yao Y., Eades G., Liu Z., Zhang Y., Zhou Q. Downregulation of miR-140 promotes cancer stem cell formation in basal-like early stage breast cancer. Oncogene. 2014;33:2589–2600. doi: 10.1038/onc.2013.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gavgiotaki E., Filippidis G., Markomanolaki H., Kenanakis G., Agelaki S., Georgoulias V., Athanassakis I. Distinction between breast cancer cell subtypes using third harmonic generation microscopy. J. Biophotonics. 2017;10:1152–1162. doi: 10.1002/jbio.201600173. [DOI] [PubMed] [Google Scholar]
- 28.Zhu Y., Liu Y., Zhang C., Chu J., Wu Y., Li Y., Liu J., Li Q., Li S., Shi Q. Tamoxifen-resistant breast cancer cells are resistant to DNA-damaging chemotherapy because of upregulated BARD1 and BRCA1. Nat. Commun. 2018;9:1595. doi: 10.1038/s41467-018-03951-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang W.J., Wang Y., Liu S., Yang J., Guo S.X., Wang L., Wang H., Fan Y.F. Silencing circular RNA hsa_circ_0000977 suppresses pancreatic ductal adenocarcinoma progression by stimulating miR-874-3p and inhibiting PLK1 expression. Cancer Lett. 2018;422:70–80. doi: 10.1016/j.canlet.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 30.Zeng K., Chen X., Xu M., Liu X., Hu X., Xu T., Sun H., Pan Y., He B., Wang S. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9:417. doi: 10.1038/s41419-018-0454-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Huang X.Y., Huang Z.L., Xu Y.H., Zheng Q., Chen Z., Song W., Zhou J., Tang Z.Y., Huang X.Y. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-100338/miR-141-3p pathway in hepatitis B-related hepatocellular carcinoma. Sci. Rep. 2017;7:5428. doi: 10.1038/s41598-017-05432-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang C., Yuan W., Yang X., Li P., Wang J., Han J., Tao J., Li P., Yang H., Lv Q., Zhang W. Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21, PTEN expression. Mol. Cancer. 2018;17:19. doi: 10.1186/s12943-018-0771-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhong Z., Lv M., Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci. Rep. 2016;6:30919. doi: 10.1038/srep30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Greenberg P.A., Hortobagyi G.N., Smith T.L., Ziegler L.D., Frye D.K., Buzdar A.U. Long-term follow-up of patients with complete remission following combination chemotherapy for metastatic breast cancer. J. Clin. Oncol. 1996;14:2197–2205. doi: 10.1200/JCO.1996.14.8.2197. [DOI] [PubMed] [Google Scholar]
- 35.Liang Y.K., Zeng D., Xiao Y.S., Wu Y., Ouyang Y.X., Chen M., Li Y.C., Lin H.Y., Wei X.L., Zhang Y.Q. MCAM/CD146 promotes tamoxifen resistance in breast cancer cells through induction of epithelial-mesenchymal transition, decreased ERα expression and AKT activation. Cancer Lett. 2017;386:65–76. doi: 10.1016/j.canlet.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 36.Shah N., Jin K., Cruz L.A., Park S., Sadik H., Cho S., Goswami C.P., Nakshatri H., Gupta R., Chang H.Y. HOXB13 mediates tamoxifen resistance and invasiveness in human breast cancer by suppressing ERα and inducing IL-6 expression. Cancer Res. 2013;73:5449–5458. doi: 10.1158/0008-5472.CAN-13-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ward A., Balwierz A., Zhang J.D., Küblbeck M., Pawitan Y., Hielscher T., Wiemann S., Sahin Ö. Re-expression of microRNA-375 reverses both tamoxifen resistance and accompanying EMT-like properties in breast cancer. Oncogene. 2013;32:1173–1182. doi: 10.1038/onc.2012.128. [DOI] [PubMed] [Google Scholar]
- 38.Shah K.N., Mehta K.R., Peterson D., Evangelista M., Livesey J.C., Faridi J.S. AKT-induced tamoxifen resistance is overturned by RRM2 inhibition. Mol. Cancer Res. 2014;12:394–407. doi: 10.1158/1541-7786.MCR-13-0219. [DOI] [PubMed] [Google Scholar]
- 39.Bergamaschi A., Katzenellenbogen B.S. Tamoxifen downregulation of miR-451 increases 14-3-3ζ and promotes breast cancer cell survival and endocrine resistance. Oncogene. 2012;31:39–47. doi: 10.1038/onc.2011.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ebert M.S., Neilson J.R., Sharp P.A. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat. Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y., Jiang D., Zhang Q., Liu X., Cai Z. Ubiquitin-specific protease 4 inhibits breast cancer cell growth through the upregulation of PDCD4. Int. J. Mol. Med. 2016;38:803–811. doi: 10.3892/ijmm.2016.2685. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Zhong M., Jiang Q., Jin R. USP4 expression independently predicts favorable survival in lung adenocarcinoma. IUBMB Life. 2018;70:670–677. doi: 10.1002/iub.1755. [DOI] [PubMed] [Google Scholar]
- 43.Zhang X., Berger F.G., Yang J., Lu X. USP4 inhibits p53 through deubiquitinating and stabilizing ARF-BP1. EMBO J. 2011;30:2177–2189. doi: 10.1038/emboj.2011.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Z., Hao Q., Luo J., Xiong J., Zhang S., Wang T., Bai L., Wang W., Chen M., Wang W. USP4 inhibits p53 and NF-κB through deubiquitinating and stabilizing HDAC2. Oncogene. 2016;35:2902–2912. doi: 10.1038/onc.2015.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yun S.I., Kim H.H., Yoon J.H., Park W.S., Hahn M.J., Kim H.C., Chung C.H., Kim K.K. Ubiquitin specific protease 4 positively regulates the WNT/β-catenin signaling in colorectal cancer. Mol. Oncol. 2015;9:1834–1851. doi: 10.1016/j.molonc.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cao W.H., Liu X.P., Meng S.L., Gao Y.W., Wang Y., Ma Z.L., Wang X.G., Wang H.B. USP4 promotes invasion of breast cancer cells via Relaxin/TGF-β1/Smad2/MMP-9 signal. Eur. Rev. Med. Pharmacol. Sci. 2016;20:1115–1122. [PubMed] [Google Scholar]
- 47.Zheng Q., Bao C., Guo W., Li S., Chen J., Chen B., Luo Y., Lyu D., Li Y., Shi G. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang L., Tong X., Zhou Z., Wang S., Lei Z., Zhang T., Liu Z., Zeng Y., Li C., Zhao J. Circular RNA hsa_circ_0008305 (circPTK2) inhibits TGF-β-induced epithelial-mesenchymal transition and metastasis by controlling TIF1γ in non-small cell lung cancer. Mol. Cancer. 2018;17:140. doi: 10.1186/s12943-018-0889-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao S., Li X., Ding X., Jiang L., Yang Q. Huaier extract restrains the proliferative potential of endocrine-resistant breast cancer cells through increased ATM by suppressing miR-203. Sci. Rep. 2017;7:7313. doi: 10.1038/s41598-017-07550-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang S., Li X., Zheng H., Si X., Li B., Wei G., Li C., Chen Y., Chen Y., Liao W. Loss of super-enhancer-regulated circRNA Nfix induces cardiac regeneration after myocardial infarction in adult mice. Circulation. 2019;139:2857–2876. doi: 10.1161/CIRCULATIONAHA.118.038361. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.