Summary
Myeloid-differentiated hematopoietic stem cells (HSCs) have contributed to a number of novel treatment approaches for lysosomal storage diseases of the central nervous system (CNS), and may also be applied to patients infected with HIV. We quantified hematopoietic stem and progenitor cell (HSPC) trafficking to 20 tissues including lymph nodes, spleen, liver, gastrointestinal tract, CNS, and reproductive tissues. We observed efficient marking of multiple macrophage subsets, including CNS-associated myeloid cells, suggesting that HSPC-derived macrophages are a viable approach to target gene-modified cells to tissues. Gene-marked cells in the CNS were unique from gene-marked cells at any other physiological sites including peripheral blood. This novel finding suggests that these cells were derived from HSPCs, migrated to the brain, were compartmentalized, established myeloid progeny, and could be targeted for lifelong delivery of therapeutic molecules. Our findings have highly relevant implications for the development of novel therapies for genetic and infectious diseases of the CNS.
Keywords: hematopoietic stem cells, cell trafficking, macrophages, microglia, central nervous system, integration site analysis
Highlights
-
•
HSCs give rise to gene-marked, tissue-resident myeloid cells
-
•
HSC-derived myeloid cells in the CNS are unique from any other HSPC-derived lineage
-
•
These subsets are ideal candidates to deliver therapeutic gene cargoes to tissues
Peterson, Adair, and colleagues investigated the trafficking and distribution of autologous hematopoietic stem cells and their progeny to an extensive array of tissues. Gene-marked macrophages were prominent, especially in lymphoid organs. Intriguingly, gene-marked myeloid cells in the CNS were unique from those at other sites. This observation holds great promise to therapeutically deliver disease-relevant transgenes to specific tissue sites.
Introduction
Hematopoietic stem and progenitor cells (HSPCs) give rise to all hematopoietic-origin cells, including lymphoid (T cells, B cells, and natural killer cells), myeloid subsets (monocytes, macrophages, dendritic cells, and neutrophils), and red blood cells. Hence, HSPC gene therapy is an effective means of delivering transgenes of interest to any hematopoietic cell type. Our group and others have extensively investigated the ability to apply HSPC gene therapy for cure of HIV-1 infection, via generation of infection-resistant monocytes, macrophages, and T lymphocytes, and enhancement of virus-specific immune responses (Kiem et al., 2012, Peterson et al., 2016, Peterson et al., 2017, Younan et al., 2013, Younan et al., 2015a, Younan et al., 2015b). In addition, hemoglobinopathies such as sickle cell disease and β-thalassemias can be treated via the expression of corrective transgenes and/or gene-editing approaches to rescue function/expression of hemoglobin molecules (reviewed in Ferrari et al., 2017). Furthermore, other genetic diseases, including pyruvate kinase deficiency and severe combined immunodeficiencies can be similarly treated. Finally, HSPC-based approaches can be used to treat patients with glioblastoma multiforme (GBM) by protecting hematopoietic-origin cells that are otherwise secondarily targeted by chemotherapy regimens directed against the solid tumor. We have shown that the P140K mutant of methylguanine methyltransferase (MGMTP140K) enables resistance to such chemotherapy regimens, namely O6-benzylguanine (O6BG) and either temozolomide or bis-chloroethylnitrosourea (BCNU). Hence, autologous transplantation with MGMTP140K-expressing HSPCs allows for high-dose chemotherapy targeting GBM, and results in improved patient outcomes (Adair et al., 2012, Adair et al., 2014). In short, nearly three decades of clinical and preclinical studies have established that gene-modified HSPCs and their progeny offer therapeutic benefit and protection in a wide range of infectious and genetic disorders of the hematopoietic system.
Lysosomal storage diseases (LSDs) comprise a broad class of over 40 different central nervous system (CNS)-related syndromes that encompass various defects in lysosomal hydrolase genes, leading to the accumulation of undegraded substrates and cell death (reviewed in Biffi, 2017). Likewise, in the setting of suppressed HIV infection, studies in patients and animal models argue strongly that the CNS is a key HIV reservoir site that must be effectively targeted by any curative therapy (Estes et al., 2017, Joseph et al., 2015, Llewellyn et al., 2017, Veenstra et al., 2017). Although allogeneic HSPC transplantation is a first-line therapy for various hematological malignancies, this approach may be less efficacious and more toxic at sites such as the CNS (Biffi, 2017, Muschol et al., 2002), given the risks associated with allogeneic transplantation, including graft-versus-host disease. Genetic modification of an LSD or HIV+ patient's own HSPCs with retroviral/lentiviral vectors expressing ameliorative transgenes provides a lifelong source of therapeutic gene products with lower toxicity. Such autologous approaches for LSDs are currently being evaluated in clinical trials (Biffi et al., 2013, Sessa et al., 2016). In addition, autologous HSCs for the treatment of other CNS disorders such as stroke and multiple sclerosis have also been demonstrated (Borlongan et al., 2011, Boy et al., 2011, Scolding et al., 2017). We are interested in broadening this approach by applying HSPC-based strategies to seed infection-resistant and/or virus-specific cells in various HIV reservoir sites, including the CNS (Zhen et al., 2017).
Several preclinical studies demonstrate the promise of HSPC-based approaches to deliver therapeutic transgenes to solid tissues such as CNS. In a mouse model of LSD, gene-modified, HSPC-derived microglia cells were associated with complete normalization of disease symptoms (Biffi et al., 2006). In nonhuman primates (NHP) transplanted with gene-modified HSPCs, modified HSPC progeny could be detected in macrophage lineages, a highly advantageous HSPC-derived subset for the delivery of genetic cargoes to tissues (Soulas et al., 2009). These findings suggest that HSPC gene therapy is capable of delivering gene-modified progeny to tissue sites. Importantly, while some of the signals regulating HSPC migration to the CNS have been described (reviewed in Lapidot and Kollet, 2010), which cells traffic to the CNS, i.e., whether they are macrophages derived from circulating monocytes or long-term CNS-resident cells such as microglia, remains unclear.
We have previously reported results in a pigtail macaque (M. nemestrina) that was transplanted with autologous HSPCs that were transduced with a lentiviral vector encoding GFP and MGMTP140K. Following engraftment and two subsequent rounds of chemoselection, we observed over 70% of peripheral blood cells expressing GFP (Beard et al., 2010). Here, we tested the hypothesis that high levels of gene marking in the periphery may reveal gene-marked cells in solid tissues, including gene-marked myeloid cells in the CNS. Using flow cytometry, immunohistochemical staining, and lentiviral vector integration site tracking, we describe the systemic localization of gene-marked cells. The goal of this study was to document the cellular subsets and tissue types that are accessible to autologous HSPC gene therapy.
Results
Sustained Engraftment of Lentivirus Gene-Modified Blood Cells over 10 Years Posttransplantation
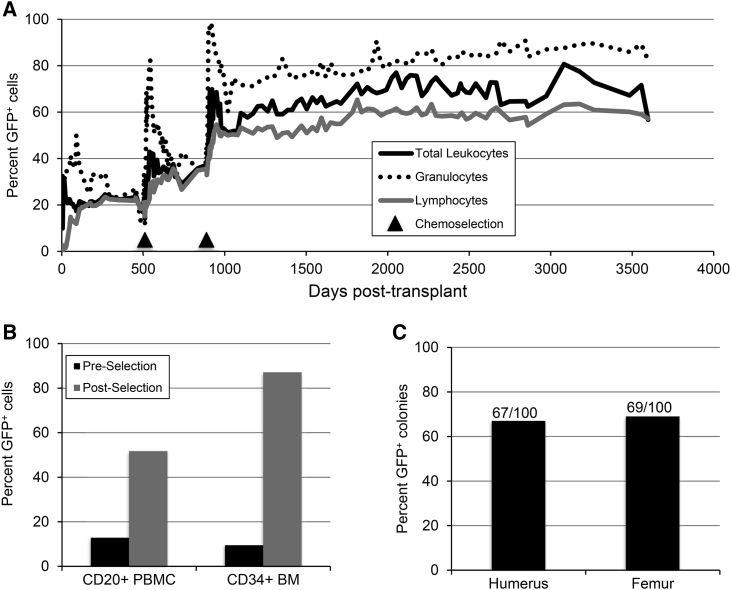
We previously showed that pigtail macaque HSPCs transduced with a lentivirus expressing GFP and the chemoselection marker MGMTP140K exhibited multilineage engraftment in vivo for up to 7 years after transplantation (Beard et al., 2010, Radtke et al., 2017). Here we demonstrate that these levels of lentivirus gene marking are sustained for nearly 10 years (Figure 1A). Two rounds of O6BG and BCNU chemoselection for MGMTP140K-expressing cells led to dramatic increases in transgene marking in peripheral blood lymphocyte (CD20+) subsets, and in CD34+ cells from bone marrow in this animal (Figure 1B). At the latest time point of follow-up, total bone marrow colony-forming cells collected from humerus and femur showed 67% and 69% GFP+ cells in standard methylcellulose assays (Figure 1C).
Figure 1.
Animal J02370 Gene Marking in Peripheral Blood, Subsets, and HSPC Colonies
At the indicated days following transplantation with autologous, lentivirus-transduced CD34+ HSPCs, peripheral blood draws were collected from animal ID J02370.
(A) Percent of granulocytes and lymphocytes expressing GFP. Arrows indicate time points at which O6BG/BCNU chemotherapy was applied.
(B) GFP expression in peripheral blood CD20+ and bone marrow CD34+ subsets. Preselection samples were collected on day 174 posttransplant, while postselection samples were collected on days 3,380 (CD20+ PBMC) or 3,424 (CD34+ BM) posttransplant.
(C) Bone marrow-derived total leukocytes from the indicated long bones were plated in colony-forming assays. Gene marking was measured by counting the proportion of GFP+ colonies out of 100 total.
Lentiviral HSPC Gene Therapy Leads to Robust Marking in Tissues
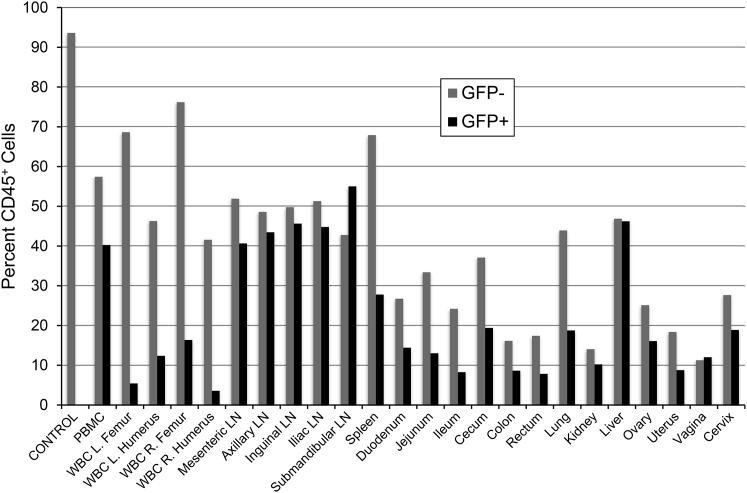
Following 10 years of longitudinal study, an extensive necropsy was performed on animal J02370 in order to assess the level of lentivirus gene marking in tissues, most of which were not accessible by survival surgeries. Our list of necropsy tissues comprises well-characterized secondary lymphoid sites (lymph nodes, spleen, and gut), reproductive organs (ovary, uterus, vagina, and cervix), and other tissues of particular interest to the gene therapy field (lung, kidney, and liver, CNS) (Figure 2). A similar tissue set has been investigated in our HIV gene therapy studies (Peterson et al., 2017). To focus on HSPC-derived cells, we gated on the pan-hematopoietic marker, CD45. Single-cell suspensions were generated from each tissue by mechanical and enzymatic dissociation, and the fractions of CD45+GFP+ and CD45+GFP– cells were quantified by flow cytometry using peripheral blood from an untransplanted animal (“CONTROL”) for gating. We observed transgene marking above background levels in all tissues tested. Unsurprisingly, CD45+GFP+ cells were most readily detected in lymphocyte-rich tissues including lymph nodes, spleen, gut, lung, and liver (Figure 2). These findings suggest that transgene-marked HSPCs and their progeny traffic to a broad range of secondary lymphoid and tertiary tissue sites in vivo.
Figure 2.
Identification of GFP+ Cells in Tissues
A panel of 20 tissues, peripheral blood, and 4-limb bone marrow draws were collected at necropsy from animal ID J02370. Tissues were dissociated enzymatically and/or mechanically, and single-cell suspensions were analyzed by flow cytometry. Following gating for live singlets, the percentage of CD45+GFP− (gray bars) and CD45+GFP+ (black bars) were measured from the total cell populations from each tissue. Control: PBMC from an untransplanted (GFP−) animal.
All Peripheral Blood T Cell and Innate Immune Subsets Contain Transgene-Marked Cells
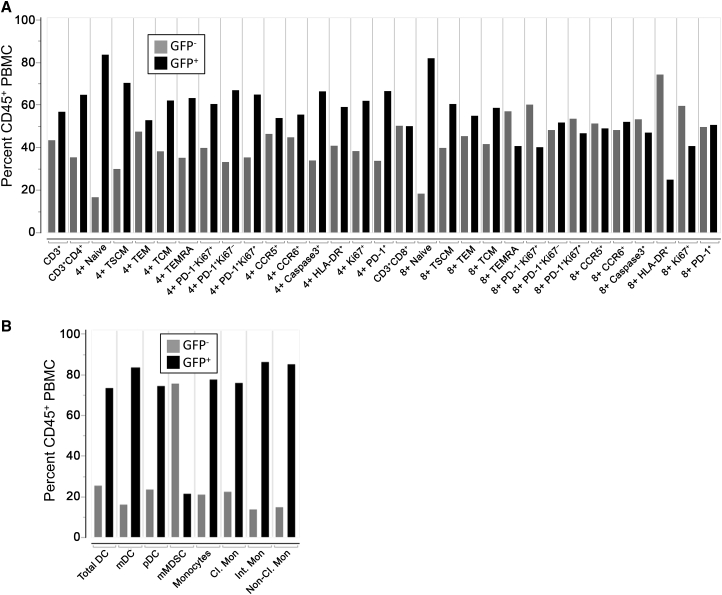
Fifty-two percent of CD20+ lymphocytes and 87% of CD34+ bone marrow cells, respectively, were marked with our lentiviral vector following MGMTP140K chemoselection (Figure 1B). To examine the phenotype of transgene-expressing T lymphocytes and innate immune cells, we developed a pair of multicolor flow panels and applied them to peripheral blood samples from animal J02370. As shown in Figure S1, the T cell panel enumerated CD45+CD3+CD4+ and CD45+CD3+CD8+ T cells, and naive and memory subsets on the basis of CD28, CD95, CD45RA, and CCR7 staining. The percentage of CD4+ and CD8+ T cells expressing the proliferation/exhaustion markers Ki-67 and PD-1, respectively, chemokine receptors CCR5 and CCR6, the apoptosis marker caspase-3, and the activation marker HLA-DR were also quantified. For the innate immune panel, CD45+SSCHiLin– cells were separated based on CD14 and HLA-DR staining into monocytic myeloid-derived suppressor cell (mMDSC), dendritic cell, and monocyte subsets, on the basis of staining for markers including CD11b, CD33, CD11c, and CD16 (Figure S1). Both the T cell and innate immune panels were constructed such that the proportion of transgene-positive (GFP+) cells could be measured in each subset (Figure 3), and, importantly, almost all of the immune cell subsets analyzed were greater than 50% GFP+. Unsurprisingly, naive T cells were the highest marked subset in the T cell panel, with upward of 80% expressing GFP. Interestingly, only 20% of activated CD8+ T cells (CD45+CD3+CD8+HLA-DR+) expressed GFP (Figure 3A). Transgene marking was high in subsets from the innate panel, with 75%–80% GFP+ cells found in dendritic cell and monocyte subsets (Figure 3B). Similar to activated CD8+ T cells, only 20% of mMDSCs expressed GFP. In summary, lentivirus-marked HSPCs gave rise to every lymphoid and myeloid subset measured, at proportions that were frequently comparable with those found in total peripheral leukocytes (Figure 1A).
Figure 3.
GFP Marking in Peripheral Blood T Cell and Innate Immune Subsets
Peripheral blood was drawn prior to necropsy of animal ID J02370 and analyzed by flow cytometry.
(A) T cell subsets in peripheral blood mononuclear cells. Shown is the proportion of CD45+GFP– (gray bars) and CD45+GFP+ cells (black bars) from the indicated CD3+, CD3+CD4+, or CD3+CD8+ T cell subsets. Subset definitions and gating are described in Figure S1.
(B) Innate immune subsets from hemolyzed total leukocytes. CD45+ GFP+ proportions were calculated as in (A) from mMDSC, DC, and monocyte subsets as outlined in Figure S1.
Lentivirus Gene-Marked HSPCs Give Rise to GFP-Expressing Macrophages
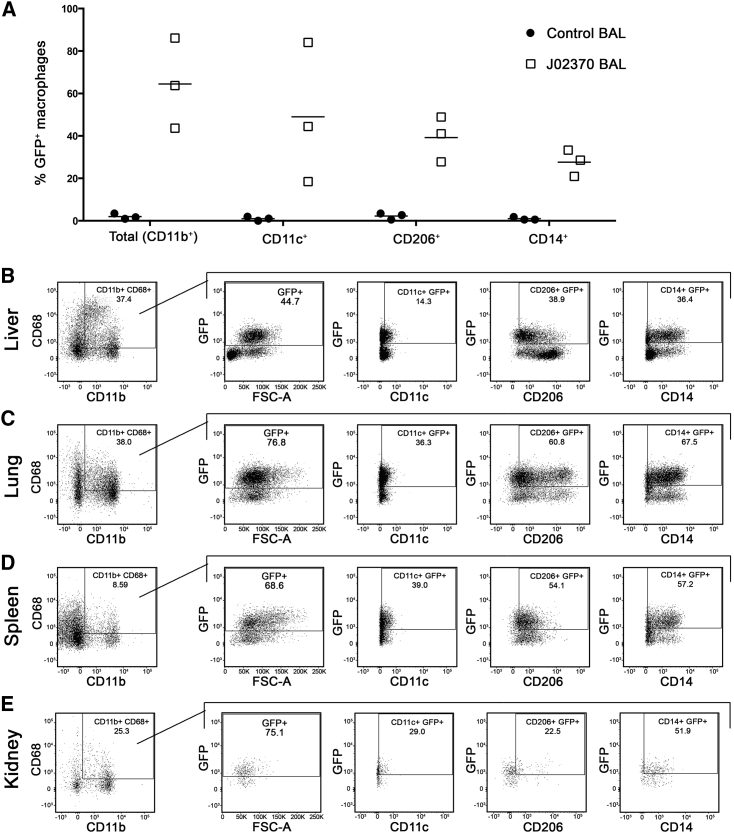
To investigate the potential of gene-modified HSPCs and their progeny to engraft in tissues, we first quantified gene-modified macrophages. Based on past reports (Soulas et al., 2009), we reasoned that macrophages' preferential residence in tissue sites relative to peripheral blood made them an ideal marker for tissue engraftment of HSPC-derived cells. Prior to necropsy, a series of bronchioalveolar lavage (BAL) samples were collected from animal J02370. BAL products are known to be enriched for alveolar macrophages and can be repeatedly collected during longitudinal survival surgeries. We developed a macrophage panel to assay these cells by flow cytometry, building on methods reported in a recent publication (Ortiz et al., 2015). Our objective in phenotyping macrophage subsets was to assess several surface marker definitions that were consistent with a macrophage phenotype, but rely primarily on a functional readout, phagocytosis of fluorescent beads, to define these cells. As shown in Figure S2, BAL macrophages were gated based on viability dye exclusion and fluorescent bead uptake in CD45+Lin–HLA-DR+ cells. We refer to the CD11b+CD68+ population as “Total (CD11b+)” macrophages, and measured the percentage of GFP-expressing cells in this subset, as well as in CD11b+CD68+CD11c+ (“CD11c+”), CD11b+CD68+CD206+ (“CD206+”), and CD11b+CD68+CD14+ (“CD14+”) subsets. Unstained (Figure S2A) and stained BAL samples (Figure S2B) from a nontransplanted control animal were used to set gates for animal J02370 (Figure S2C). In three separate BAL collections from animal J02370, we observed an average of 62% GFP+ total (CD11b+) macrophages, 48% CD11c+, 40% CD206+, and 32% CD14+ macrophages (Figure 4A). Next, we applied the same macrophage panel to single-cell suspensions derived from tissues collected at necropsy. Up to 44.7% of liver, 76.8% of lung, 68.6% of spleen, and 75.1% of kidney-associated total macrophages were GFP+ (Figures 4B–4E). Transgene marking in bone marrow macrophages varied between the four long bones assayed (Figures S3A–S3D). Comparable levels of marking were found in lymph node and reproductive tissue-associated macrophages, although the frequency of these cells was low (Figures S3E–S3I). In gut-associated lymphoid tissues, macrophage marking was proportional to the number of CD11b+CD68+ macrophages that were detected (Figure S3J–S3N). In subsets that were negative for uptake of fluorescent beads (A-Gonzalez et al., 2017, Ayata et al., 2018), we detected comparable levels of gene marking (Table 1). These results demonstrate that lentivirus-marked HSPCs give rise to macrophage progeny with similar efficiency to other HSPC-derived lymphoid and myeloid subsets.
Figure 4.
Gene Marking in Macrophages following HSC Transplantation
Macrophage subsets were delineated from bronchoalveolar lavage (BAL) (A) and tissue-derived macrophages (B–E) as exemplified in Figure S2. (A) GFP+ percentages of total macrophages, and from CD11c+, CD206+, and CD14+ macrophage subsets from three untransplanted control animals (circles) and from animal ID J02370 (squares). Three independent BAL collections were performed from J02370. Macrophage subsets from (B) liver, (C) lung, (D) spleen, and (E) kidney were analyzed identically to BAL samples. See also Figures S2 and S3.
Table 1.
Percentage of Gene-Modified, Phagocytic, and Nonphagocytic Macrophage Subsets
| Site | % GFP+ of HLA-DR+ Phago+ | % GFP+ of HLA-DR+ Phago− |
|---|---|---|
| BAL no. 1 | 43.6 | 16.1 |
| BAL no. 2 | 63.7 | 3.1 |
| BAL no. 3 | 86.1 | 42.8 |
| Left femur | 52.6 | 33.0 |
| Left humerus | 22.8 | 15.3 |
| Right femur | 36.1 | 26.6 |
| Right humerus | 34.8 | 26.0 |
| Mesenteric lymph node | 57.6 | 34.3 |
| Inguinal lymph node | 45.7 | 32.5 |
| Iliac lymph node | 60.5 | 28.7 |
| Spleen | 68.6 | 36.6 |
| Duodenum | 58.9 | 43.3 |
| Jejunum | 74.9 | 17.0 |
| Ileum | 57.3 | 20.0 |
| Cecum | 70.5 | 12.1 |
| Colon | 70.7 | 18.3 |
| Lung | 76.8 | 56.8 |
| Liver | 44.7 | 52.8 |
| Kidney | 75.1 | 3.6 |
| Cervix | 68.1 | 47.2 |
| Vagina | 51.9 | 33.6 |
See also Figures S2 and S3.
Gene-Marked HSPCs Give Rise to CNS-Resident Myeloid Subsets
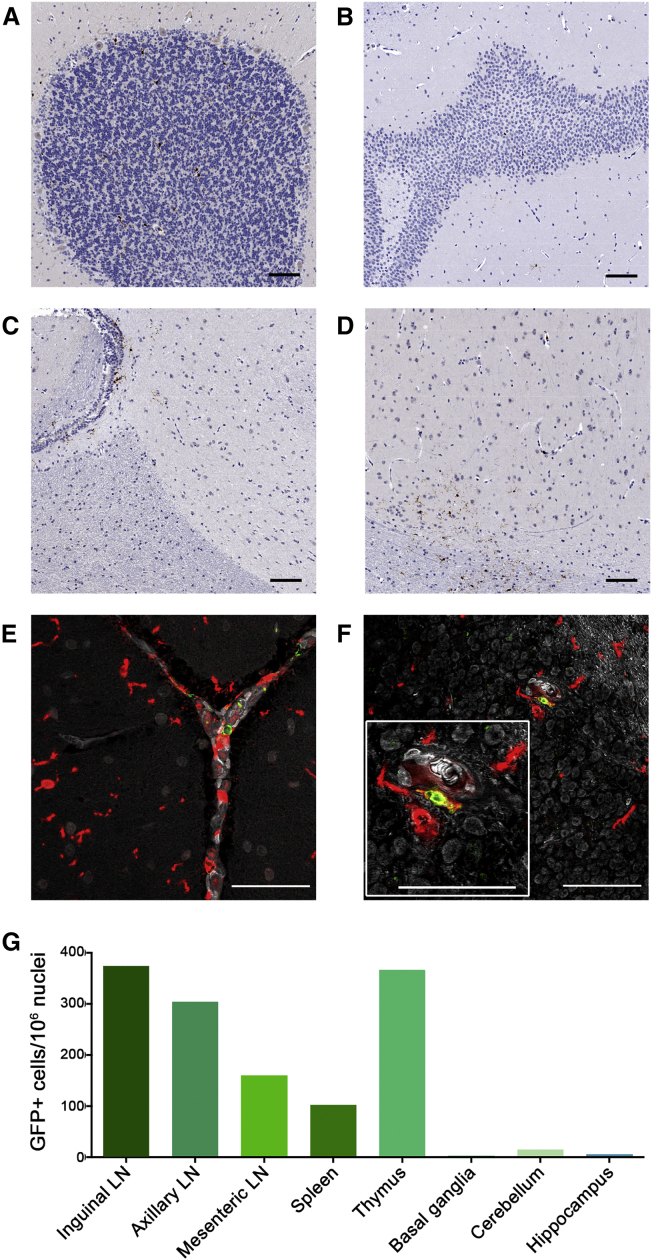
Although the bulk of microglia and perivascular macrophages are of yolk sac origin (Prinz et al., 2017), HSPC-derived cells have also been described (Biffi et al., 2006, Rocca et al., 2017). Previously, HSPC-derived perivascular macrophages have been shown to engraft in the CNS in transplanted NHP (Biffi et al., 2006, Soulas et al., 2009). We adapted our macrophage panel to detect CNS-resident myeloid cells in necropsy tissues from animal J02370 and quantify the proportion of microglia cells that expressed our lentiviral GFP transgene. Total cerebral tissue was dissociated and separated on a Percoll gradient before antibody staining. CD68+Lin– cells were identified, and the GFP+ proportion was quantified by flow cytometry. As with the macrophage panel, CNS samples from an untransplanted control animal, with and without antibody staining, were used to set gates. Flow cytometry measurements demonstrated that over 20% of myeloid-lineage, phagocytic cells in the CNS expressed GFP (Figure S4). We next performed immunohistochemistry on tissue sections using an anti-GFP antibody to confirm that these cells were indeed CNS resident. This approach was validated in GFP+ Jurkat cells (Figure S5A) and thymus tissue from animal J02370 (Figure S5B); the background was assessed in spleen sections from a nontransplanted animal, compared with animal J02370 (Figures S5C and S5D). Consistent with our flow cytometry data (Figure 2), lymph node sections contained a comparatively high level of transgene marking; gene-marked cells localized to the T cell zone, as well as within B cell follicles (Figures S5E–S5H and S6). When we applied this staining protocol to sections of cerebellum, hippocampus, parietal cortex, and basal ganglia, we detected GFP+ foci at each CNS site (Figures 5A–5D). Importantly, we detected cells that costained with anti-GFP antibodies and the macrophage/microglia marker Iba1, consistent with the labeling of a gene-modified microglial cell (Figures 5E, 5F, and S7). Unsurprisingly, quantification of our imaging data demonstrated that GFP+ cells in the CNS were less frequent than those in secondary lymphoid tissues (Figure 5G). Notably, Iba1 marks both macrophages and microglia in the CNS; CD45 staining was not available to distinguish whether these cells were macrophages or microglia (Martin et al., 2017). These data clearly show that gene-modified HSPC traffic to the brain, and give rise to transgene-expressing myeloid subsets.
Figure 5.
Gene-Marked Cells Are Detected in CNS Tissues
(A–D) Anti-GFP antibody staining was used to identify lentivirus gene-marked cells in situ from tissue sections from animal ID J02370. Shown are sections from the brain, including (A) cerebellum, (B) hippocampus, (C) parietal cortex, and (D) basal ganglia, imaged with chromogenic staining.
(E and F) Cerebellum images following immuno-fluorescent staining with antibodies against GFP (green) and the microglia marker Iba1 (red); double-positive cells are in yellow, and DAPI staining for nuclei are gray; (E) meninges, (F) granular layer, including higher-magnification inset image.
(G) Quantification of GFP+ cells in CNS and non-CNS tissues, normalized to nuclei number via segmentation of hematoxylin staining.
Scale bars, = 100 μm. See also Figures S4 through S7.
CNS-Derived Progeny Are Distinct from Other Gene-Marked Clones
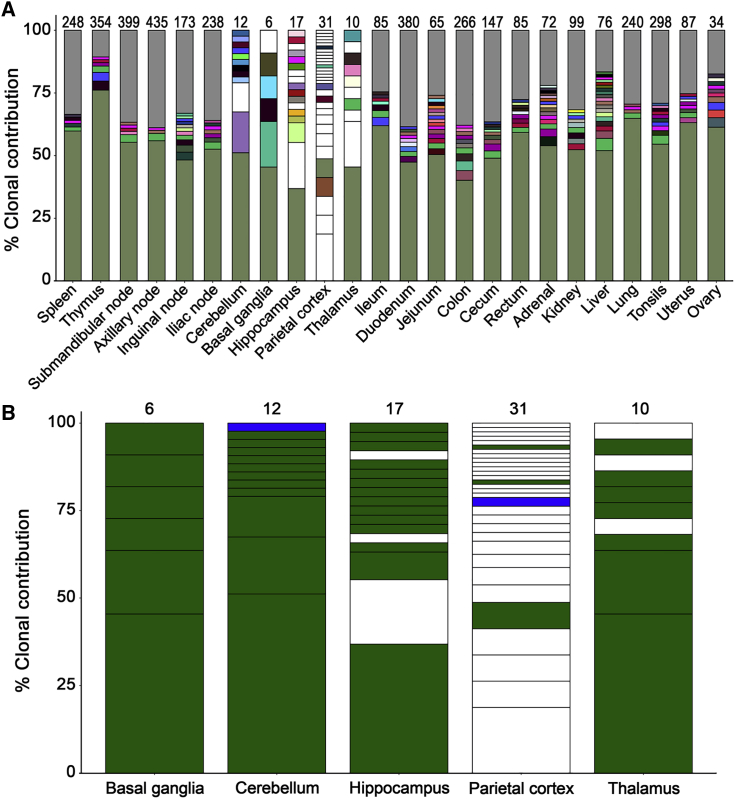
Two nonmutually exclusive models could explain the findings above. First, gene-marked myeloid cells in the CNS could be derived from circulating monocytes that trafficked to the brain and differentiated into macrophages. Second, myeloablative conditioning, which was administered to the animal immediately before infusion of lentivirus-transduced HSPCs and induces temporary breakdown of the blood-brain barrier (BBB), could have facilitated direct trafficking of HSPCs to the CNS. Following BBB repair, gene-marked cells in the CNS would become compartmentalized and distinct from gene-marked cells in peripheral blood and other tissues. To distinguish between these models, we applied our well-established lentivirus integration site (IS) analysis platform (Beard et al., 2014, Hocum et al., 2015) to peripheral blood and tissues from animal J02370. Unique IS in the CNS would reinforce our hypothesis that CNS clones are unique and compartmentalized. Notably, a dominant clone was identified in peripheral blood cells, which emerged during two rounds of chemotherapy that the animal received approximately 2 years posttransplant (Figure S8). Although this clone grew to comprise more than 70% of the gene-marked cells in peripheral blood, we did not find any evidence during the subsequent 8 years of follow-up that it negatively impacted the animal's health. Next, we assayed IS in tissues collected at necropsy. Genomic DNA was extracted from total tissue homogenates that were flash-frozen immediately on collection, and processed identically to IS samples from peripheral blood. The dominant clone detected in peripheral blood was also observed in multiple tissues, both CNS and non-CNS, at frequencies as high as 75% (Figure 6A). Multiple other clones were observed in non-CNS tissues and peripheral blood; these could represent either blood contamination in tissues or cells that are derived from a clone that is present in both compartments.
Figure 6.
CNS Tissues Display a Unique Clonal Repertoire Relative to Other Tissues at Necropsy
(A) IS profiles in the indicated tissues collected at necropsy from animal ID J02370. Methods and color schemes are matched to those used for IS analyses in peripheral blood (Figure S8).
(B) IS in the CNS with a contribution >1.0%. Colors indicate in which additional samples each IS was detected: only detected in a single CNS tissue (white), multiple tissues within the CNS (blue); multiple tissues, including CNS, non-CNS, and/or peripheral blood (green).
The number of IS detected in each tissue is denoted at the top of each bar. See also Figure S8.
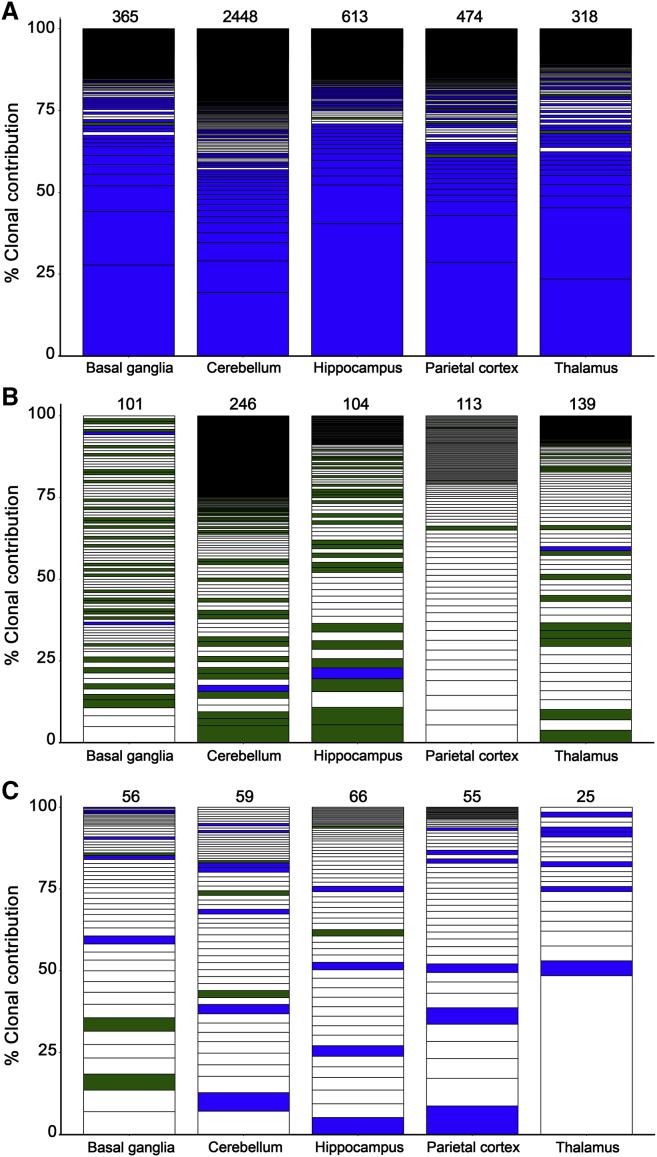
Interestingly, we observed a distinct pattern of IS in CNS tissues relative to all other tissues and all peripheral blood clones detected over the follow-up term. IS detected in CNS sites were often not found in any non-CNS tissues. This was particularly striking in the parietal cortex, where only 5 of 31 IS were found outside the CNS at any time after transplant (Figure 6B). CNS-restricted IS were also found in the cerebellum, hippocampus, and thalamus. We did not detect CNS-restricted IS in the basal ganglia, although this tissue also yielded the lowest number of total IS (Figure 6B). To reinforce these findings, we interrogated IS in CNS tissues from an additional cohort of three macaques that underwent similar transplantation protocols with more than 1,000 days of posttransplant follow-up. Across target CNS tissues from these three animals, between 25 and 2,448 unique IS were identified, and the profiles of these unique CNS-associated IS were even more striking (Figure 7). In each of these animals, the vast majority of IS detected were unique to either one or multiple CNS tissues and were not found outside of the CNS, strongly suggesting that the identification of unique CNS clones was not specific to animal J02370 (Figure 7). Our analysis demonstrates that up to 90% of detected gene-marked HSPC progeny in the brain are unique from those found in the blood or other tissues.
Figure 7.
Unique HSPC-Derived CNS Clones in Three Additional Transplanted Animals
IS profiles in the indicated CNS tissues collected at necropsy from animal IDs Z13105 (A), Z09132 (B), and Z08103 (C). Methods and color schemes are matched to those used in Figure 6. IS in the CNS with a contribution >1.0% are shown, and the number of IS detected in each tissue is denoted at the top of each bar. See also Figure S8.
Discussion
HSPC gene therapy has shown great promise in patients with infectious and genetic diseases of the hematopoietic system. However, the extent to which this approach can be applied to pathologies in solid tissues, namely targeting of CNS-localized HIV reservoirs, remains unclear. We have previously shown that an MGMTP140K-based in vivo chemoselection approach enables high levels of gene marking in a large animal model and significant clinical benefit in GBM patients. Here, we extended the study of our highest gene-marked macaque, ID J02370, to investigate engraftment and clonality of gene-modified HSPC progeny in tissues. We show that HSPC-derived progeny in the brain were not the result of ongoing trafficking of peripheral subsets such as monocytes but, rather, were bona fide CNS-resident cells that were likely compartmentalized following immigration to the brain after total body irradiation (TBI) and autologous HSPC transplantation. In a total of four animals that underwent autologous transplantation with GFP-modified HSPCs, we were able to detect highly unique CNS clones that contrasted starkly with the clonal repertoire found in 20 other peripheral anatomic sites.
Following an in-depth analysis of peripheral blood subset gene marking, we observe robust levels of GFP+ cells in every subset tested. Most subsets expressed GFP in equal proportion to our observations in total peripheral blood (60%–80% GFP+), with the exception of memory T cell and mMDSC subsets. Over 80% of naive CD4+ and CD8+ T cells in peripheral blood expressed GFP, whereas 50%–60% of central and effector memory T cells were GFP+. This may reflect memory T cells that persisted through the myeloablative TBI conditioning regimen preceding infusion of gene-modified cells. Past reports suggest that TBI only partially depletes T cell subsets in tissues (Donahue et al., 2015). As such, the slightly reduced frequency of GFP+ memory T cells may reflect dilution by persistent pretransplant immune cells that have since undergone homeostatic proliferation in response to cognate antigen. We have followed the nomenclature and phenotypic definition of MDSC subsets of Bronte et al. (2016), in which the former granulocytic MDSC are instead referred to as polymorphonuclear MDSC and are distinct from the mMDSC measured here. In general, the half-life of MDSC populations has been measured in days (Parker et al., 2015), suggesting that no pretransplant MDSCs should have persisted through this 10-year study. Instead, it is formally possible that the integrated MGMTP140K/GFP transgene has been transcriptionally silenced in this subset. Collectively, our flow-based analyses suggest that gene-modified HSPC differentiation is highly multilineage, with little or no subset bias.
In contrast to the even distribution of gene-modified HSPC progeny in peripheral blood, the percentage of gene-modified CD45+ cells in tissues varied drastically between sites, as various tissues may contain different levels of CD45+ cells that persisted through myeloablative TBI and 10 years of follow-up (e.g., memory T cells). We therefore focused on macrophages and microglia, which we hypothesized would be the most advantageous HSPC progeny for targeting of latent HIV+ cells in the CNS. In quantifying the percentage of gene-modified macrophages, our goal was to investigate multiple subset definitions, consistent with past reports (Ortiz et al., 2015). Macrophage marking in BAL from animal J02370 ranged between 30% and 60%, depending on the macrophage subset analyzed. We observed over 70% GFP+ macrophages in select subsets, including CD11b+CD68+ subsets from kidney and lung. Importantly, by quantifying uptake of a fluorescently labeled phagocytosis substrate, we were able to restrict our analysis to functionally phagocytic cells. An intriguing future direction is the consideration of how best to apply these tissue-resident cells in a therapeutic setting. Both microglia and macrophages are characterized primarily by their phagocytic functions but are also known to secrete cytokines and cell-signaling molecules (Lopez-Atalaya et al., 2018, Pranzatelli, 2018). Our data clearly demonstrate that HSPC gene therapy strategies are well positioned to provide therapeutic benefit, for example, by secreting broadly neutralizing antibodies to target and destroy HIV-infected cells at various reservoir sites.
Finally, we leveraged the high percentage of peripheral gene marking in animal J02370 to investigate the engraftment of HSPC-derived progeny in the CNS. We found over 20% of phagocytic, CD68+ myeloid cells in the CNS were GFP+, and observed GFP+ Iba1+ cells by IHC in multiple CNS sites including cerebellum, hippocampus, parietal cortex, and basal ganglia. Previous studies have identified gene-modified perivascular macrophages at these sites (Soulas et al., 2009). We were not able to directly distinguish CNS-associated perivascular macrophages from microglia by costaining with purinergic receptor P2Y, G-protein coupled, 12 (P2RY12) in this study (Butovsky et al., 2014), although we observed cells that localized both proximal (perivascular macrophages) and distal to blood vessels (microglia). Instead, we used IS analysis to compare the individual HSPC-derived clones that were identified in CNS tissue versus peripheral blood and other tissue sites. Consistent with our hypothesis that gene-modified clones in the CNS were distinct and compartmentalized from the rest of the body, unique IS were identified in CNS tissues that were not found in any other peripheral or non-CNS tissue sites. We found no evidence in our IS dataset that lentiviral vector-induced insertional mutagenesis conferred selective advantage to CNS-resident, gene-modified cells. Instead, these results strongly suggest that the HSPC-derived progeny in the brain are not the result of ongoing trafficking of peripheral subsets such as monocytes, but, rather, are bona fide CNS-resident cells that could have migrated through a permeabilized BBB following myeloablative TBI and were compartmentalized following BBB repair. Notably, we were able to detect even more unique CNS clones in an additional three NHPs that underwent analogous autologous transplantation procedures with GFP-modified HSPCs, suggesting that, across a range of peripheral gene marking levels, unique clones are consistently identified in this compartment. While additional studies are required to determine whether a subset(s) of CD34+ cells with CNS potential can be isolated, these results raise the exciting possibility that, in addition to perivascular macrophages, gene-modified microglia could also be exploited for HSPC-based gene therapies for CNS pathologies. Such an approach would have immediate implications in LSDs such as adrenoleukodystrophy or metachromatic leukodystrophy, and possibly other CNS disorders including stroke (Borlongan et al., 2011, Boy et al., 2011), multiple sclerosis (Scolding et al., 2017), and autism (Dawson et al., 2017, Sharma et al., 2013, Sun and Kurtzberg, 2018).
Notably, the developmental origin of tissue-associated macrophages and microglia remains controversial (reviewed in Ginhoux et al., 2013). In mouse embryos, early stages of hematopoiesis initiate in the yolk sac, including generation of “fetal macrophage populations” that bypass monocyte differentiation stages (Naito et al., 1990). These are thought to be among the first hematopoietic cells established during prenatal development, followed by hematopoietic progenitor infiltration of fetal liver, and eventually bone marrow (Naito et al., 1990, Stremmel et al., 2018). From these sites, more traditional pathways of HSPC-monocyte-macrophage differentiation are initiated; transcriptional profiling suggests that HSPC-derived macrophages and yolk sac-derived macrophages are distinct lineages (Hoeffel and Ginhoux, 2015, Schulz et al., 2012). Importantly, recent in vivo studies suggest that the yolk sac may not be the only site from which tissue macrophages are derived (Fehrenbach et al., 2018, Xu et al., 2015). We show that, although gene-modified microglia/macrophage populations in the CNS constitute a relatively small proportion relative to gene-modified macrophages in other tissues, they are bona fide macrophage/microglia cells that are distinct from gene-marked cells found in any other compartment. Our findings support a model in which HSPCs contribute to microglia/macrophage homeostasis in the CNS, especially following treatments such as TBI that permeabilize the BBB and facilitate HSPC migration. This approach could ideally be coupled with CNS-targeted, chemotherapy-based conditioning regimens that enhance the niche for gene-modified HSPCs in the brain (Capotondo et al., 2012).
In summary, we report the system-wide trafficking of gene-modified HSPC and progeny cells in a macaque with high-level gene marking in peripheral blood. The detection of gene-marked tissue macrophages and microglia unlock new applications for HSPC gene therapy in a number of tissue-based pathologies and for latent HIV infection. Our findings support the use of CNS-engrafting HSPCs and their progeny for a wide range of therapeutic applications, including expression of enhanced, virus-specific immune effector proteins, or as “delivery vehicles” capable of secreting peptides/proteins designed to target a given disease.
Experimental Procedures
Transplantation and Gene Marking in Peripheral Blood and Bone Marrow
All experimental procedures performed were reviewed and approved by the Institutional Animal Care and Use Committee of the Fred Hutchinson Cancer Research Center and University of Washington (Protocol no. 3235-01). This study was carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals (“The Guide”) of the NIH. The autologous transplantation protocol for pigtail macaques (M. nemestrina), chemoselection for MGMTP140K-expressing cells, and methods to track lentiviral gene marking in peripheral blood total leukocytes, granulocytes, lymphocytes, and bone marrow have been described previously (Beard et al., 2010, Radtke et al., 2017). Peripheral blood mononuclear cells (PBMCs) were collected by Ficoll density centrifugation, and total leukocytes were collected by ammonium chloride hemolysis. GFP marking in CD20+ PBMC was assessed with anti-CD20 antibody (clone L27) and gating on lymphocytes (SSCLoFSCLo). GFP marking in CD34+ bone marrow total leukocytes was assessed with anti-CD34 antibody (clone 12.8). Two rounds of chemoselection were administered to animal J02370, at 510 and 888 days posttransplant, whereas animals Z09132 and Z08103 received one round of chemoselection each at day 365 and 400 posttransplant, respectively. Animal Z13105 did not receive any chemotherapy. Follow-up included peripheral blood and bone marrow samples both pre- and postchemotherapy treatment for up to 3,598 days posttransplant (Animal J02370). The shortest follow-up period was 1,030 days (Animal Z13105). Necropsy was performed at the final time point of follow-up for each animal.
Gene Marking in Tissues
Collection of tissues at necropsy followed our previously described protocol (Peterson et al., 2017). Following mechanical and enzymatic dissociation, cell suspensions were analyzed by flow cytometry using anti-CD45 antibody (clone D058-1,283) to identify the proportion of CD45+GFP+ hematopoietic-origin cells from each tissue site.
Flow Cytometry Panels
Four multicolor flow panels were applied to samples from animal J02370; in each, the FITC channel was left open in order to quantify GFP fluorescence from transgene-marked cells. The T cell and innate immune panels (Figure S1) have been described previously (Peterson et al., 2017). The macrophage panel (Figure S2) was adapted from Ortiz et al. (2015) and included the following antibodies: CD3-PerCP clone SP34-2, and CD45 clone D058-1283 (Thermo Fisher, Waltham, MA); CD11c-APC clone S-HCL, CB11b-APC-Cy7 clone ICRF44, HLA-DR-PE-CF594 clone G46-6, CD206-PE-Cy5 clone 19.2, CD14-PE-Cy7 clone M5E2, and CD8-Pacific Blue clone RPA-T8 (Becton Dickinson, Franklin Lakes, NJ); CD68-PE clone Y182A (Miltenyi Biotec, Cologne, Germany); and CD20-BV605 clone 2H7 (BioLegend, San Diego, CA). 0.2 μm FluoSpheres “Dark Red” (APC Cy5.5) and Aqua Live/Dead stain (both from Life Technologies, Carlsbad, CA) were also used. Intracellular staining was required to identify CD68+ cells. The microglia panel was adapted from the macrophage panel and included CD3-PerCP clone SP34-2 (Thermo Fisher), CD8-Pacific Blue clone RPA-T8 (Becton Dickinson), CD68-PE clone Y182A (Miltenyi Biotec), and CD20-BV605 clone 2H7 (BioLegend). 0.2 μm FluoSpheres Dark Red (APC Cy5.5) was used to positively identify phagocytic cells.
BAL
Animals were intubated before introduction of a 5-mm video bronchoscope into the trachea. Aliquots of sterile saline were lavaged into the terminal bronchioles, after which the infusate was aspirated. BAL samples were counted with a Countess automated cytometer (Life Technologies), and either cryopreserved or stained fresh. Cryopreserved samples were thawed in the presence of Dornase media (1 vial Dornase per 100 mL DMEM + 10% fetal bovine serum [FBS] + 1% penicillin/streptomycin [PenStrep], “DMEM 10/1”). Fresh or thawed BAL samples were filtered through 70-μm cell strainers to remove mucous and debris. Before Aqua Live/Dead and antibody staining, samples were brought to 2 × 106 cells/mL in DMEM 10/1 and incubated for 1–2 h in a 37°C, 5% CO2 incubator with FluoSpheres diluted 1:72 in PBS (5 × 106 beads per 10 μL working volume). Bead uptake was stopped by adding 1 mL of cold 0.5 mM EDTA in 1× PBS to the cell suspension, followed by washing. Cryopreserved single-cell suspensions from necropsy tissue collections were thawed and stained in batch, identically to BAL samples.
Microglia Isolation
Total cerebellum tissue was collected at necropsy from animal J02370 in R10 media (RPMI 1640 + 10% FBS, 2 mM L-glutamine, and 1% PenStrep). Tissue was transferred to a biosafety cabinet where it was rinsed with medium and minced in a tissue culture dish with isolation medium (DMEM/F12 + 10% FBS, 2 mM L-glutamine, and 1% PenStrep). Minced tissue was washed with PBS and further processed by trypsin-mediated enzymatic digestion (DMEM + 4.5 g/L glucose + 0.25% trypsin + 3 U/mL Dornase) at 37°C for 30 min followed by vigorously pipetting to create a homogeneous mix. The trypsinization was stopped by addition of DMEM with 20% FBS. The trypsin mixture was filtered with a 100-μm nylon membrane and the filtrate was pelleted using a tabletop centrifuge spinning for 10 min at 1,200 rpm. The supernatant was aspirated, the pellet was resuspended in a 30% Percoll/PBS solution (GE Healthcare Life Sciences), and centrifuged for 1.5 h at 4,700 rpm with no brake. The microglia-enriched phase, which migrated between myelin debris and blood vessel phases, was extracted using a syringe, filtered through a 40-μm mesh to remove small capillaries, and washed with isolation medium. The cells were pelleted by centrifugation at 2,000 rpm for 10 min, resuspended, and processed for immunostaining and flow cytometry.
Immunohistochemistry and Immunofluorescent Confocal Analysis
Lymph node, spleen, thymus, and CNS tissues (cerebellum, hippocampus, parietal cortex, thalamus, and basal ganglia) were collected in 4% paraformaldehyde (PFA) (Electron Microscopy Sciences, Hatfield, PA) in 1× PBS, and fixed overnight at room temperature. Samples were then transferred to 80% ethanol for 24–48 h. In parallel, samples were collected in optical coherence tomography media and stored at −80°C.
Immunohistochemistry was performed using a biotin-free polymer approach (Golden Bridge International) on 5-μm tissue sections mounted on glass slides, which were dewaxed and rehydrated with double-distilled H2O. Heat-induced epitope retrieval was performed by heating sections in 0.01% citraconic anhydride containing 0.05% Tween 20 in a pressure cooker (Biocare Medical) set at 122°C–125°C for 30 s. Slides were incubated with blocking buffer (Tris-buffered saline [TBS] with 0.05% Tween 20 and 0.25% casein) for 10 min followed by rabbit anti-GFP (1:100; Invitrogen-A11122) diluted in blocking buffer overnight at 4°C. Slides were washed in 1× TBS with 0.05% Tween 20 and endogenous peroxidases blocked using 1.5% (v/v) H2O2 in TBS (pH 7.4) for 10 min. Slides were incubated with the Rabbit Polink-2 HRP staining system (Golden Bridge International) according to the manufacturer's recommendations and subsequently developed with Impact 3,3′-diaminobenzidine. All slides were scanned at high magnification (200–400×) using the Aperio AT-2 System (Leica Microsystems) yielding high-resolution data for the entire tissue section. Immunofluorescent staining was performed as described previously (Osuna et al., 2016) using the following primary antibodies: myeloid cells (CD163, Novo Castra-NCL-CD163 and CD68, Biocare-CM33C), microglia cells (Iba1, Biocare CP290 A, B), and rabbit anti-GFP (1:100; Invitrogen-A11122). Slides were cover slipped with no. 1.5 GOLD SEAL cover glass (EMS) using Prolong Gold reagent (Invitrogen) and imaged on an Olympus FV10i confocal microscope using a 60× phase-contrast oil-immersion objective (NA 1.35) in sequential mode to separately capture the fluorescence from the different fluorochromes at an image resolution of 1,024 × 1,024 pixels.
Lentivirus IS Analyses
Total genomic DNA was isolated from peripheral blood cells or homogenized tissue (Peterson et al., 2017) by QIAGEN DNA Blood Mini kit (QIAGEN, Waltham, MA), and analyzed via our previously described IS methods (Beard et al., 2014). Analysis of resulting sequencing reads was conducted as previously described using custom scripts (Radtke et al., 2017). The clonal contribution for any given IS was calculated as the proportion of genomically aligned reads attributable to that specific IS relative to all genomically aligned reads in the sample, which sum to 100%. Color codes are matched in Figures 6A and S8 to indicate clones that were found both in peripheral blood and tissues, respectively.
Author Contributions
C.W.P. assembled the final data and oversaw necropsy sample collections and processing. J.E.A. oversaw the animal studies, generated flow cytometry data, and analyzed integration site data, which was generated by Z.K.N., L.E.S., and G.M.S.-H. C.D. and J.D.E. generated and analyzed immunohistochemistry data. M.E.W., S.R., and K.K.N. generated flow data from CNS samples. K.K.N. and A.R. generated the flow data from non-CNS samples, which was analyzed by K.K.N., A.R., and C.W.P. A.B. and R.D.M. performed necropsy tissue sample collections and generated histopathological reports. H-.P.K. is the principle investigator of the study. C.W.P., J.E.A., and H.-P.K. wrote the manuscript, which was reviewed by all authors.
Acknowledgments
We thank Helen Crawford for preparing this manuscript; Veronica Nelson, Erica Curry, and Kelvin Sze for outstanding support in our pigtail macaque studies; Willi Obenza and Sowmya Reddy for processing of macaque samples; and Alexandra Ortiz, Nikki Klatt and Steve De Rosa for helpful advice regarding flow cytometry panels. This study was supported by grants from the NIH National Institute of Allergy and Infectious Diseases (U19 AI096111 and UM1 AI126623 to H.-P.K. and K.R.J.) and the National Heart, Lung, and Blood Institute (R01 HL116217 and U19 HL129902 to H.-P.K.), as well as funds from the Fred Hutchinson Cancer Research Center (to J.E.A.). This study was also supported by NIH P51 OD010425 and UW/FHCRC CFAR AI027757, and in part through the NIH/NCI Cancer Center Support Grant P30 CA015704. In addition, this project has been funded in part by the Oregon National Primate Research Center NIH grant award P51OD011092, and in part with Federal funds from the National Cancer Institute, NIH, under contract no. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government. H.-P.K. is a Markey Molecular Medicine Investigator and received support as the inaugural recipient of the José Carreras/E. Donnall Thomas Endowed Chair for Cancer Research and the Fred Hutch Endowed Chair for Cell and Gene Therapy.
Published: June 13, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2019.05.016.
Supplemental Information
References
- A-Gonzalez N., Quintana J.A., Garcia-Silva S., Mazariegos M., Gonzalez de la Aleja A., Nicolas-Avila J.A., Walter W., Adrover J.M., Crainiciuc G., Kuchroo V.K. Phagocytosis imprints heterogeneity in tissue-resident macrophages. J. Exp. Med. 2017;214:1281–1296. doi: 10.1084/jem.20161375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adair J.E., Beard B.C., Trobridge G.D., Neff T., Rockhill J.K., Silbergeld D.L., Mrugala M., Kiem H.P. Extended survival of glioblastoma patients after chemoprotective HSC gene therapy. Sci. Transl. Med. 2012;4:133ra157. doi: 10.1126/scitranslmed.3003425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adair J.E., Johnston S.K., Mrugala M.M., Beard B.C., Guyman L.A., Baldock A.L., Bridge C.A., Hawkins-Daarud A., Gori J.L., Born D.E. Gene therapy enhances chemotherapy tolerance and efficacy in glioblastoma patients. J. Clin. Invest. 2014;124:4082–4092. doi: 10.1172/JCI76739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayata P., Badimon A., Strasburger H.J., Duff M.K., Montgomery S.E., Loh Y.E., Ebert A., Pimenova A.A., Ramirez B.R., Chan A.T. Epigenetic regulation of brain region-specific microglia clearance activity. Nat. Neurosci. 2018;21:1049–1060. doi: 10.1038/s41593-018-0192-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beard B.C., Adair J.E., Trobridge G.D., Kiem H.P. High-throughput genomic mapping of vector integration sites in gene therapy studies. Methods Mol. Biol. 2014;1185:321–344. doi: 10.1007/978-1-4939-1133-2_22. [DOI] [PubMed] [Google Scholar]
- Beard B.C., Trobridge G.D., Ironside C., McCune J.S., Adair J.E., Kiem H.P. Efficient and stable MGMT-mediated selection of long-term repopulating stem cells in nonhuman primates. J. Clin. Invest. 2010;120:2345–2354. doi: 10.1172/JCI40767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biffi A. Hematopoietic stem cell gene therapy for storage disease: current and new indications. Mol. Ther. 2017;25:1155–1162. doi: 10.1016/j.ymthe.2017.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biffi A., Capotondo A., Fasano S., del C.U., Marchesini S., Azuma H., Malaguti M.C., Amadio S., Brambilla R., Grompe M. Gene therapy of metachromatic leukodystrophy reverses neurological damage and deficits in mice. J. Clin. Invest. 2006;116:3070–3082. doi: 10.1172/JCI28873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biffi A., Montini E., Lorioli L., Cesani M., Fumagalli F., Plati T., Baldoli C., Martino S., Calabria A., Canale S. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science. 2013;341:1233158. doi: 10.1126/science.1233158. [DOI] [PubMed] [Google Scholar]
- Borlongan C.V., Glover L.E., Tajiri N., Kaneko Y., Freeman T.B. The great migration of bone marrow-derived stem cells toward the ischemic brain: therapeutic implications for stroke and other neurological disorders. Prog. Neurobiol. 2011;95:213–228. doi: 10.1016/j.pneurobio.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boy S., Sauerbruch S., Kraemer M., Schormann T., Schlachetzki F., Schuierer G., Luerding R., Hennemann B., Orso E., Dabringhaus A. Mobilisation of hematopoietic CD34+ precursor cells in patients with acute stroke is safe – results of an open-labeled non randomized phase I/II trial. PLoS One. 2011;6:e23099. doi: 10.1371/journal.pone.0023099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronte V., Brandau S., Chen S.H., Colombo M.P., Frey A.B., Greten T.F., Mandruzzato S., Murray P.J., Ochoa A., Ostrand-Rosenberg S. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat. Commun. 2016;7:12150. doi: 10.1038/ncomms12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butovsky O., Jedrychowski M.P., Moore C.S., Cialic R., Lanser A.J., Gabriely G., Koeglsperger T., Dake B., Wu P.M., Doykan C.E. Identification of a unique TGF-beta-dependent molecular and functional signature in microglia. Nat. Neurosci. 2014;17:131–143. doi: 10.1038/nn.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capotondo A., Milazzo R., Politi L.S., Quattrini A., Palini A., Plati T., Merella S., Nonis A., di Serio C., Montini E. Brain conditioning is instrumental for successful microglia reconstitution following hematopoietic stem cell transplantation. Proc. Natl. Acad. Sci. U S A. 2012;109:15018–15023. doi: 10.1073/pnas.1205858109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson G., Sun J.M., Davlantis K.S., Murias M., Franz L., Troy J., Simmons R., Sabatos-DeVito M., Durham R., Kurtzberg J. Autologous cord blood infusions are safe and feasible in young children with autism spectrum disorder: results of a single-center phase I open-label trial. Stem Cells Transl. Med. 2017;6:1332–1339. doi: 10.1002/sctm.16-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue R.E., Srinivasula S., Uchida N., Kim I., St Claire A., Duralde G., DeGrange P., St Claire M., Reba R.C., Bonifacino A.C. Discordance in lymphoid tissue recovery following stem cell transplantation in rhesus macaques: an in vivo imaging study. Blood. 2015;126:2632–2641. doi: 10.1182/blood-2015-07-657346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes J.D., Kityo C., Ssali F., Swainson L., Makamdop K.N., Del Prete G.Q., Deeks S.G., Luciw P.A., Chipman J.G., Beilman G.J. Defining total-body AIDS-virus burden with implications for curative strategies. Nat. Med. 2017;23:1271–1276. doi: 10.1038/nm.4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehrenbach M.K., Tjwa M., Bechmann I., Krueger M. Decreased microglial numbers in Vav1-Cre(+):dicer knock-out mice suggest a second source of microglia beyond yolk sac macrophages. Ann. Anat. 2018;218:190–198. doi: 10.1016/j.aanat.2018.03.004. [DOI] [PubMed] [Google Scholar]
- Ferrari G., Cavazzana M., Mavilio F. Gene therapy approaches to hemoglobinopathies. Hematol. Oncol. Clin. North Am. 2017;31:835–852. doi: 10.1016/j.hoc.2017.06.010. [DOI] [PubMed] [Google Scholar]
- Ginhoux F., Lim S., Hoeffel G., Low D., Huber T. Origin and differentiation of microglia. Front. Cell. Neurosci. 2013;7:45. doi: 10.3389/fncel.2013.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocum J.D., Battrell L.R., Maynard R., Adair J.E., Beard B.C., Rawlings D.J., Kiem H.P., Miller D.G., Trobridge G.D. VISA–Vector Integration Site Analysis server: a web-based server to rapidly identify retroviral integration sites from next-generation sequencing. BMC Bioinformatics. 2015;16:212. doi: 10.1186/s12859-015-0653-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffel G., Ginhoux F. Ontogeny of tissue-resident macrophages. Front. Immunol. 2015;6:486. doi: 10.3389/fimmu.2015.00486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph S.B., Arrildt K.T., Sturdevant C.B., Swanstrom R. HIV-1 target cells in the CNS. J. Neurovirol. 2015;21:276–289. doi: 10.1007/s13365-014-0287-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiem H.P., Jerome K.R., Deeks S.G., McCune J.M. Hematopoietic-stem-cell-based gene therapy for HIV disease (Review) Cell Stem Cell. 2012;10:137–147. doi: 10.1016/j.stem.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapidot T., Kollet O. The brain-bone-blood triad: traffic lights for stem-cell homing and mobilization. Hematology Am. Soc. Hematol. Educ. Program. 2010;2010:1–6. doi: 10.1182/asheducation-2010.1.1. [DOI] [PubMed] [Google Scholar]
- Llewellyn G.N., Alvarez-Carbonell D., Chateau M., Karn J., Cannon P.M. HIV-1 infection of microglial cells in a reconstituted humanized mouse model and identification of compounds that selectively reverse HIV latency. J. Neurovirol. 2017;24:192–203. doi: 10.1007/s13365-017-0604-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Atalaya J.P., Askew K.E., Sierra A., Gomez-Nicola D. Development and maintenance of the brain's immune toolkit: microglia and non-parenchymal brain macrophages. Dev. Neurobiol. 2018;78:561–579. doi: 10.1002/dneu.22545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin E., El-Behi M., Fontaine B., Delarasse C. Analysis of microglia and monocyte-derived macrophages from the central nervous system by flow cytometry. J. Vis. Exp. 2017;22 doi: 10.3791/55781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muschol N., Matzner U., Tiede S., Gieselmann V., Ullrich K., Braulke T. Secretion of phosphomannosyl-deficient arylsulphatase A and cathepsin D from isolated human macrophages. Biochem. J. 2002;368:845–853. doi: 10.1042/BJ20020249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito M., Takahashi K., Nishikawa S. Development, differentiation, and maturation of macrophages in the fetal mouse liver. J. Leukoc. Biol. 1990;48:27–37. doi: 10.1002/jlb.48.1.27. [DOI] [PubMed] [Google Scholar]
- Ortiz A.M., DiNapoli S.R., Brenchley J.M. Macrophages are phenotypically and functionally diverse across tissues in simian immunodeficiency virus-infected and uninfected Asian macaques. J. Virol. 2015;89:5883–5894. doi: 10.1128/JVI.00005-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osuna C.E., Lim S.Y., Deleage C., Griffin B.D., Stein D., Schroeder L.T., Omange R.W., Best K., Luo M., Hraber P.T. Zika viral dynamics and shedding in rhesus and cynomolgus macaques. Nat. Med. 2016;22:1448–1455. doi: 10.1038/nm.4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker K.H., Beury D.W., Ostrand-Rosenberg S. Myeloid-derived suppressor cells: critical cells driving immune suppression in the tumor microenvironment. Adv. Cancer Res. 2015;128:95–139. doi: 10.1016/bs.acr.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson C.W., Benne C., Polacino P., Kaur J., McAllister C.E., Filali-Mouhim A., Obenza W., Pecor T.A., Huang M.L., Baldessari A. Loss of immune homeostasis dictates SHIV rebound after stem-cell transplantation. JCI Insight. 2017;2:e91230. doi: 10.1172/jci.insight.91230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson C.W., Haworth K.G., Burke B.P., Polacino P., Norman K.K., Adair J.E., Hu S.L., Bartlett J.S., Symonds G.P., Kiem H.P. Multilineage polyclonal engraftment of Cal-1 gene-modified cells and in vivo selection after SHIV infection in a nonhuman primate model of AIDS. Mol. Ther. Methods Clin. Dev. 2016;3:16007. doi: 10.1038/mtm.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pranzatelli M.R. Advances in biomarker-guided therapy for pediatric- and adult-onset neuroinflammatory disorders: targeting chemokines/cytokines. Front. Immunol. 2018;9:557. doi: 10.3389/fimmu.2018.00557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz M., Erny D., Hagemeyer N. Ontogeny and homeostasis of CNS myeloid cells. Nat. Immunol. 2017;18:385–392. doi: 10.1038/ni.3703. [DOI] [PubMed] [Google Scholar]
- Radtke S., Adair J.E., Giese M.A., Chan Y.Y., Norgaard Z.K., Enstrom M., Haworth K.G., Schefter L.E., Kiem H.P. A distinct hematopoietic stem cell population for rapid multilineage engraftment in nonhuman primates. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aan1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocca C.J., Goodman S.M., Dulin J.N., Haquang J.H., Gertsman I., Blondelle J., Smith J.L.M., Heyser C.J., Cherqui S. Transplantation of wild-type mouse hematopoietic stem and progenitor cells ameliorates deficits in a mouse model of Friedreich's ataxia. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aaj2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz C., Gomez Perdiguero E., Chorro L., Szabo-Rogers H., Cagnard N., Kierdorf K., Prinz M., Wu B., Jacobsen S.E., Pollard J.W. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 2012;336:86–90. doi: 10.1126/science.1219179. [DOI] [PubMed] [Google Scholar]
- Scolding N.J., Pasquini M., Reingold S.C., Cohen J.A., International Conference on Cell-Based Therapies for Multiple Sclerosis. International Conference on Cell-Based Therapies for Multiple Sclerosis. International Conference on Cell-Based Therapies for Multiple Sclerosis Cell-based therapeutic strategies for multiple sclerosis. Brain. 2017;140:2776–2796. doi: 10.1093/brain/awx154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa M., Lorioli L., Fumagalli F., Acquati S., Redaelli D., Baldoli C., Canale S., Lopez I.D., Morena F., Calabria A. Lentiviral haemopoietic stem-cell gene therapy in early-onset metachromatic leukodystrophy: an ad-hoc analysis of a non-randomised, open-label, phase 1/2 trial. Lancet. 2016;388:476–487. doi: 10.1016/S0140-6736(16)30374-9. [DOI] [PubMed] [Google Scholar]
- Sharma A., Gokulchandran N., Sane H., Nagrajan A., Paranjape A., Kulkarni P., Shetty A., Mishra P., Kali M., Biju H. Autologous bone marrow mononuclear cell therapy for autism: an open label proof of concept study. Stem Cells Int. 2013;2013:623875. doi: 10.1155/2013/623875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulas C., Donahue R.E., Dunbar C.E., Persons D.A., Alvarez X., Williams K.C. Genetically modified CD34+ hematopoietic stem cells contribute to turnover of brain perivascular macrophages in long-term repopulated primates. Am. J. Pathol. 2009;174:1808–1817. doi: 10.2353/ajpath.2009.081010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stremmel C., Schuchert R., Wagner F., Thaler R., Weinberger T., Pick R., Mass E., Ishikawa-Ankerhold H.C., Margraf A., Hutter S. Yolk sac macrophage progenitors traffic to the embryo during defined stages of development. Nat. Commun. 2018;9:75. doi: 10.1038/s41467-017-02492-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J.M., Kurtzberg J. Cell therapy for diverse central nervous system disorders: inherited metabolic diseases and autism. Pediatr. Res. 2018;83:364–371. doi: 10.1038/pr.2017.254. [DOI] [PubMed] [Google Scholar]
- Veenstra M., Leon-Rivera R., Li M., Gama L., Clements J.E., Berman J.W. Mechanisms of CNS viral seeding by HIV(+) CD14(+) CD16(+) monocytes: establishment and reseeding of viral reservoirs contributing to HIV-associated neurocognitive disorders. mBio. 2017;8 doi: 10.1128/mBio.01280-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Zhu L., He S., Wu Y., Jin W., Yu T., Qu J.Y., Wen Z. Temporal-spatial resolution fate mapping reveals distinct origins for embryonic and adult microglia in zebrafish. Dev. Cell. 2015;34:632–641. doi: 10.1016/j.devcel.2015.08.018. [DOI] [PubMed] [Google Scholar]
- Younan P.M., Peterson C.W., Polacino P., Kowalski J.P., Obenza W., Miller H.W., Milless B.P., Gafken P., DeRosa S.C., Hu S.L. Lentivirus-mediated gene transfer in hematopoietic stem cells is impaired in SHIV-infected, ART-treated nonhuman primates. Mol. Ther. 2015;23:943–951. doi: 10.1038/mt.2015.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younan P.M., Polacino P., Kowalski J.P., Hu S.L., Kiem H.P. Combinatorial hematopoietic stem cell transplantation and vaccination reduces viral pathogenesis following SHIV89.6P-challenge. Gene Ther. 2015;22:1007–1012. doi: 10.1038/gt.2015.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younan P.M., Polacino P., Kowalski J.P., Peterson C.W., Maurice N.J., Williams N.P., Ho O., Trobridge G.D., von Laer D., Prlic M. Positive selection of mC46-expressing CD4+ T cells and maintenance of virus specific immunity in a primate AIDS model. Blood. 2013;122:179–187. doi: 10.1182/blood-2013-01-482224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen A., Peterson C.W., Carrillo M.A., Reddy S.S., Youn C.S., Lam B.B., Chang N.Y., Martin H.A., Rick J.W., Kim J. Long-term persistence and function of hematopoietic stem cell-derived chimeric antigen receptor T cells in a nonhuman primate model of HIV/AIDS. PLoS Pathog. 2017;13:e1006753. doi: 10.1371/journal.ppat.1006753. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.