Abstract
Background: Trimethylation of histones has been extensively studied, where histone methyltransferases catalyze the transfer of methyl groups from S-adenosyl methionine. Thus far, there have been no researches on the trimethylation of non-histone proteins. The precise mechanisms by which trimethylation affects cell progress and the related protein functions remain unclear.
Purpose: The objective of this study was to identify the Lys-trimethylated proteins in kidney-derived cells and tissues, as well as to better understand the mechanisms underlying Lys-trimethylation-mediated cell metabolism.
Methods: The levels of Lys-trimethylation in kidney-derived cells and tissues were assayed by Western blotting. Additionally, high-resolution mass spectrometry was used to analyze kidney-derived cells and tissues, and the eukaryotic expression vectors that led to the mutations of lysine were constructed and transfected into HEK293T cells. The LDHA activity of HEK293T cells was detected under conditions of Lys-trimethylation inhibition, and the proliferation of HEK293T cells was measured using EdU and Western blotting analyses.
Results: The different proteins in kidney-derived cells and tissues showed different levels of Lys-trimethylation. In particular, lactate dehydrogenase A (LDHA) was Lys-trimethylated on lysine (K5). Inhibition of the Lys-trimethylation in LDHA increased the LDH activity of HEK293T cells and upregulated their proliferation.
Conclusion: We suggested that LDHA affects the metabolism and proliferation of cells via a Lys-trimethylation-mediated mechanism; Lys-trimethylation might be a potential target for therapeutic research or used as a prognostic and treatment biomarker of several diseases.
Keywords: trimethylation, lactate dehydrogenase A, liquid chromatography-tandem mass spectrometry, cell, kidney cancer, proliferation
Introduction
Posttranslational modifications (PTMs) play an important part in regulating cell viability and the interactions between cells.1 Protein methylation is a common type of PTM, which features the addition of one or more methyl groups to proteins. It occurs not only on the nitrogen-containing side-chains of lysine and arginine, but also at the carboxy- and amino-terminals of a few proteins.2 Methyltransferases catalyze the methylation process and are activated primarily by S-adenosyl methionine in cytobiology. Monomethylation may have marginal effects on α-amino nitrogen nucleophilicity and basicity; however, trimethylation (or dimethylation in case of proline) can result in the abolishment of nucleophilicity and a permanent positive charge on the N-terminal amino group; hence, the molecular biological functions of monomethylation may be different from those of trimethylation.3
Trimethylation of histones has been extensively studied, where histone methyltransferases catalyze the transfer of methyl groups from S-adenosyl methionine. Histones that are methylated at certain residues can activate or suppress gene expression epigenetically.4,5 Trimethylation at different lysine sites has different effects on the physical functions of cells. It is well known that dimethylation and trimethylation at H3K27 (H3K27me2/3) can shut down its transcription.6 Trimethylation of histone H3 at lysine 4 (H3K4me3) represents the active chromatin that counters the repressive chromatin milieu suppressed by the methylation of H3K9 and H3K27 in higher eukaryotes.7 H3K4 trimethylation is primarily catalyzed by the vertebrate H3K4 methyltransferases SET1A/SET1B (SET1A/B) and MLL1–MLL4.8 H3K27 histone trimethylation suppresses H3 binding and the function of SET1-like H3K4 methyltransferase complexes.9 Waldmann T et al have focused mainly on post-translational histone modifications since they are one of the major epigenetic mechanisms regulating gene expression, and their imbalance can result in cancer.10 Histone H3 lysine 27 trimethylation (H3K27me3) repression is mediated by AMPK phosphorylation in the antitumor effects of metformin in ovarian cancer.11 The presence of histone H3K36 trimethylation (H3K36me3) in various human tumors is an epigenetic marker of transcription-related histone modification and stem cell regulation. H3K36me3 positivity is related with the expression of biliary markers and is a vital predictor of adverse prognosis in resectable hepatocellular carcinoma (HCC).12
Thus far, there have been no researches on the trimethylation of non-histone proteins. The precise mechanisms by which trimethylation affects cell progress and the related protein functions remain unclear.
In our study, we first discovered that different proteins showed different levels of Lys-trimethylation in HEK293T cells; then, we tried to identify specific Lys-trimethylated proteins and the amino acids in these proteins that were Lys-trimethylated by using high resolution LC-MS/MS and Progenesis LC-MS software. Additionally, we verified the Lys-trimethylated amino acids by point mutation-based analysis. Finally, we analyzed the effect of Lys-trimethylation on the metabolism of HEK293T cells. To the best of our knowledge, the newly identified Lys-trimethylated proteins we have reported here may provide novel, innovative perspectives for understanding the regulatory mechanisms underlying the trimethylation process. Characterization of trimethylation may be important for the interpretation of results from previous biological studies and could reinforce our ability to treat renal diseases.
Materials and methods
Cell culture
The HEK293T and 786-0 ccRCC cell lines were purchased from the cell bank of the Chinese Academy of Sciences (Shanghai, China); they were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen, USA) and RPMI-1640 (Gibco, USA) supplemented with 2 mM glutamine, 100 μg/mL streptomycin, 100 U/mL penicillin and 10% heat-inactivated fetal bovine serum (Gibco, USA), respectively, at 37 °C in a humidified atmosphere composed of 95% air and 5% CO2.
Collection of clinical tissue samples
Nine kidney cancer tissue samples (renal clear cell carcinoma, ccRCC) and adjacent morphologically normal renal cortex (adjacent tissues) samples were collected from nine patients with ccRCC who underwent radical or palliative nephrectomy at the Shandong Provincial Hospital Affiliated to the Shandong University between March 2013 and January 2015. Six patients were men and three were women. The patients were aged from 48 to 70. Of these nine patients, three, five, and one patient showed a Fuhrman nuclear grade of G2, G3, and G4, respectively, and two, five, and two patients showed a TNM stage of T1, T2, and T3, respectively. This study was performed in compliance with the Institutional Ethics Review Board’s guidance of Shandong Provincial Hospital, and approved by the board. Written informed consent was collected from each patient involved in the study. The experiments involving human tissues were conducted in compliance with the Declaration of Helsinki. Radiotherapy, chemotherapy, and immunotherapy were not performed before the patients underwent surgery. Cancer tissues were harvested from the core area of their carcinomas, and away from the regions of necrotic and adjacent non-cancerous tissues. Adjacent tissues were harvested from non-cancerous regions at least 3 cm away from the core area of the carcinomas. All samples were verified by two pathologists post operation.
Protein extraction and Western blotting analysis
The culture medium of the cells was removed, and the cells were washed with ice-cold PBS (0.1 M Na2HPO4, 0.15 M NaCl, pH 7.5). They were then lysed in ice-cold RIPA lysis buffer containing 0.5 M Tris-HCl (pH 7.4), 1.5 M NaCl, 2.5% deoxycholic acid, 10% NP-40, 10 mM EDTA, and a protease inhibitor cocktail (Roche). The cells were scraped off by using a curette, and then sucked out using a pipette. The ccRCC and adjacent tissues were first sonicated on ice, and then, the proteins were lysed and extracted in ice-cold RIPA lysis buffer. The lysates were centrifuged at 12,000 g for 20 min at 4 °C. The supernatant was stored at −20 °C until further use. The protein concentrations were determined using a BCA protein assay kit (Biyuntian, Beijing, China). The protein extracts (50 mg) were loaded onto sodium dodecyl sulfate polyacrylamide gel electrophoresis gels (SDS-PAGE, 5% stacking gel and 8% separating gel), and separated at 80 V for 2 hrs; the resulting protein bands were subsequently transferred onto nitrocellulose membranes (Millipore; 0.45 μm). The membranes were blocked and then probed with the following primary antibodies: anti-trimethyl lysine (1:3,000; immunechem, Canada, Catalog # ICP0601) and anti-alpha tubulin antibodies (1:1,000; abcam, USA, Catalog # ab15246). The targeted proteins were stained with fluorescence-conjugated secondary antibodies (IRDye800CW or IRDye680 conjugated IgG, LICOR). After being washed with TBST, the membranes were scanned using the Odyssey infrared imaging system (Li-Cor, Lincoln, NE, USA), or alternatively, the bands were detected with an electrogenerated chemiluminescence reagent (Amersham Pharmacia Biotech, Freiburg, Germany) using an enhanced chemiluminescence system (Clinx, Shanghai, China). The Western blotting results were quantified using the Image J software.
Immunoprecipitation and in-gel tryptic digestion
First, anti-trimethyl lysine antibodies and 40 μL of fully suspended Protein A+G Agarose (beyotime, china, P2012) were added to the protein extracts of the cells and tissues. The samples were slowly shaken for 3 h at 4 °C and centrifuged at 2500 rpm for 5 min; then, the supernatants were carefully removed, and washed and precipitated five times with PBS. The resulting sediments were carefully resuspended in 35 μL of 1×SDS-PAGE electrophoresis buffer. These samples were mixed by vortexing and then centrifuged. After heating at 100 °C for 5 min, 15 μL of each sample was subjected to Western blotting analysis. The IP lysates were separated by SDS-PAGE and stained with Coomassie Blue. The SDS-PAGE gel was cut roughly into 1-mm-thick slices according to the molecular weight markers (at distances representing 25–35, 35–40, 40–50, 50–55, and 55–70 kD) and digested overnight with trypsin, in accordance with the modified in-gel digestion protocol.13
LC-MS/MS analysis
The extracted peptides from each gel band were desalted on a C18 column ZipTip (Millipore, Billerica, MA) and subjected to peptide fractionation using an EASY-nLC II system (Thermo Scientific, Waltham, MA, USA). The gradient-eluted peptides were analyzed by a Velos Pro ion trap mass spectrometer (Thermo Scientific). The liquid chromatography column (150 mm×Ø 0.075 mm) was packed with PepMap C18 (Thermo Scientific; particle size: 3 µm, pore size: 100 Å). Samples were analyzed using a linear gradient of 5–35% acetonitrile in 0.1% formic acid, with a flow rate of 300 nL/min (solvent A: 0.1% formic acid in water, solvent B: 0.1% formic acid in acetonitrile) and a gradient time of 120 min. The mass spectrometer was operated in a data-dependent mode, in which MS/MS fragmentation was performed using the 20 most intense peaks in each full MS scan. MS/MS spectra were searched allowing a maximum deviation of 1 Da for the precursor mass and 0.8 Da for the fragment masses. The MS/MS spectra were searched against human protein databases using MASCOT and SEQUEST. Trypsin (full cleavage) was specified as the cleavage enzyme, allowing up to two missing cleavages. The MS/MS spectra were searched allowing a maximum deviation of 10 ppm for the precursor mass and 0.6 Da for the fragment masses.14 The trimethylation of various amino acid residues was selected as a dynamic modification, and the false discovery rate (FDR) was 1%.
The construction and transfection of plasmids
The eukaryotic expression vectors were constructed by Genechem, Shanghai, China. Lysine (K5) on the N-terminal of the lactate dehydrogenase A (LDHA) protein sequence was mutated to arginine (R) in the purified plasmid DNA, which was abbreviated as LDHA@K5R. Arginine is an alkaline amino acid residue that is similar to lysine. Lysine (K5) on the N-terminal of the LDHA protein sequence was mutated to glutamine (Q) in the purified plasmid DNA, which was abbreviated as LDHA@K5Q. Glutamine is a hydrophilic amino acid residue. The plasmid constructed on the basis of the LDHA wild-type sequence (Genbank NM005566) was transfected into HEK293T cells as the control sample. The structure of the eukaryotic expression vectors was: CMV-MCS-EGFP-SV40-Neomycin. The mutations of lysine (K5) were verified by genetic sequencing.
The plasmids were transfected into H293T cells using Lipofectamine 2000 (Invitrogen), according to the manufacturer’s recommendations. First, the H293T cells (4–6×105 cells at 70–90% confluence) were seeded onto 100-mm dishes. Lipofectamine 2000 was diluted in Opti-MEM Medium, and plasmid diluted in Opti-MEM Medium, adding diluted DNA to diluted Lipofectamine 2000 Reagent (1:1 ratio). The cells were incubated and added DNA-lipid complex with a mixture of 80 mg of purified plasmid and 80 ml of Lipofectamine 2000 for 48 h to visualize/analyze the transfected cells. After anti-LDHA antibodies and Protein A+G Agarose were added to the protein extracts of the H293T cells, immunoprecipitation was performed; additionally, Western blotting was performed using anti-trimethyl lysine (1:3,000; immunechem, Canada, Catalog # ICP0601) and anti-LDHA antibodies (1:500; abcam, USA, Catalog # ab84716) as the primary antibodies.
Lactate dehydrogenase (LDH) activity assay
The LDHA activity of the samples was measured using the LDH Activity Assay Kit (Sigma-Aldrich, St. Louis, MO; Catalog Number: MAK066), strictly following the manufacturer’s instructions. All samples and standards were run in duplicate. The final measurement [(A450) final] for calculating the enzyme activity was the penultimate reading or the reading before the value of the most active sample that was near or exceeded the end of the linear range of the standard curve.
EDU cell proliferation assay
HEK293T cells (2500 cells/well) were seeded in triplicate in 24-well plates (Nunc) and were incubated in DMEM containing 10% bovine serum (Gibco) for 12 h at 37 °C. Four groups of cells (two groups of HEK293T cells; three wells per group) were transfected with either ADAR1 or mutant plasmids for 36 h. Next, 50 μM EdU labeling medium was added to the cell culture, followed by incubation for 2 h at 37 °C under 5% CO2 conditions; the EdU assay was performed using an EdU DNA Cell Proliferation Kit (Guangzhou Ribobio Co., Ltd., China), according to the manufacturer’s instructions. The labeled cells were fixed in 4% paraformaldehyde (pH 7.4) for 30 min and incubated with glycine for 5 min. After the cells were washed with PBS, staining with Apollo working solution was performed at room temperature for 30 min. After the cells were washed with 0.5% Triton X-100 in PBS, they were incubated with the Hoechst 33,342 dye in the dark at room temperature for 30 min; they were then observed under an inverted fluorescence microscope (Olympus, Japan). More than five random fields per well were captured at 200× magnification. Image-Pro Plus 6.0 software (IPP 6.0) was used to calculate the percentage of EdU-positive cells (identified by Apollo® 643 fluorescence) among all cells (identified by Hoechst 33,342 nucleus staining).15
Statistical analysis
Data were presented as the mean ± standard error on the mean (SEM). The SPSS 17.0 software (SPSS Inc., Chicago, IL, USA) was used for statistical analysis. Statistical analyses between two groups were performed using a Student’s t-test, whereas comparisons among multiple groups were performed using two-way ANOVA. P-values less than 0.05 were considered statistically significant.
Results and discussion
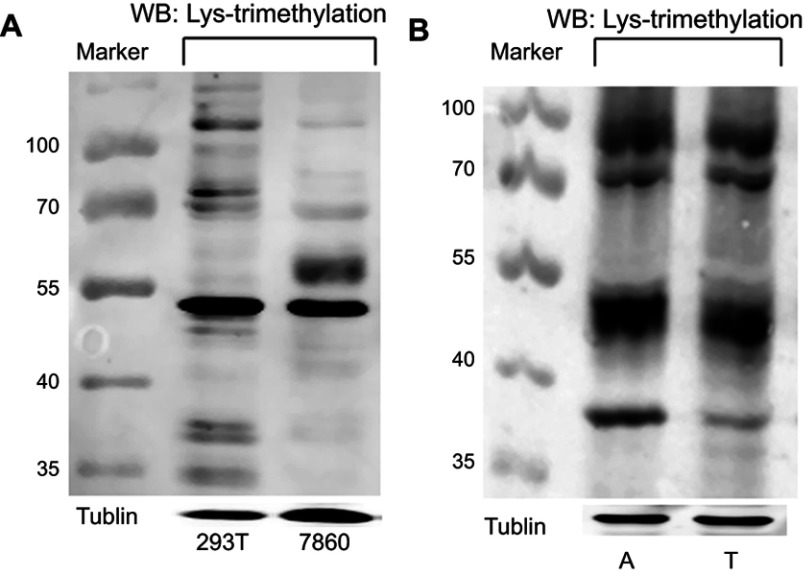
Quantification of the levels of Lys-trimethylation in different kidney-derived cells and tissues by Western blotting
The levels of Lys-trimethylation in HEK293T and 786–0 cells, kidney cancer tissues, and adjacent normal renal cortex tissues were quantified by Western blotting, using anti-trimethyl lysine antibodies as the primary antibodies. The results of the Western blotting analysis are shown in Figure 1. Our results revealed that the HEK293T and 786-0 cells, kidney cancer tissues, and adjacent normal renal cortex tissues showed different levels of Lys-trimethylation, and that Lys-trimethylation was seen in different proteins. HEK293T cells are derived from human embryonic kidney cells, and 786-0 cells are derived from human kidney clear cell adenocarcinoma cells. Overall, the different kidney-derived cells and tissues showed different levels of Lys-trimethylation. In this study, we detected the levels of Lys-trimethylation in different kidney-derived cells and tissues by Western blotting analysis; for the first time, it was found that different proteins in kidney-derived cells and tissues showed different levels of Lys-trimethylation expression. Previous research about trimethylation has focused on trimethylation in the histones or DNA.
Figure 1.
Lys-trimethylation was observed in different proteins in kidney-derived cells and tissues.
Notes: (A) The levels of Lys-trimethylation in HEK293T and 786-0 cells were quantified by Western blotting; (B) The levels of Lys-trimethylation in kidney cancer tissues and adjacent normal renal cortex tissues were quantified by Western blotting. Tubulin served as the loading control. Marker: protein marker (Fermentas, USA, Catalog # 26616-ladder-002); The targeted proteins were stained with fluorescence-conjugated secondary antibodies (IRDye800CW or IRDye680 conjugated IgG, LICOR).
Abbreviations: 293T, HEK293T cells; 7860, 786-0 cells; A, adjacent normal renal cortex tissues; T, kidney cancer tissues.
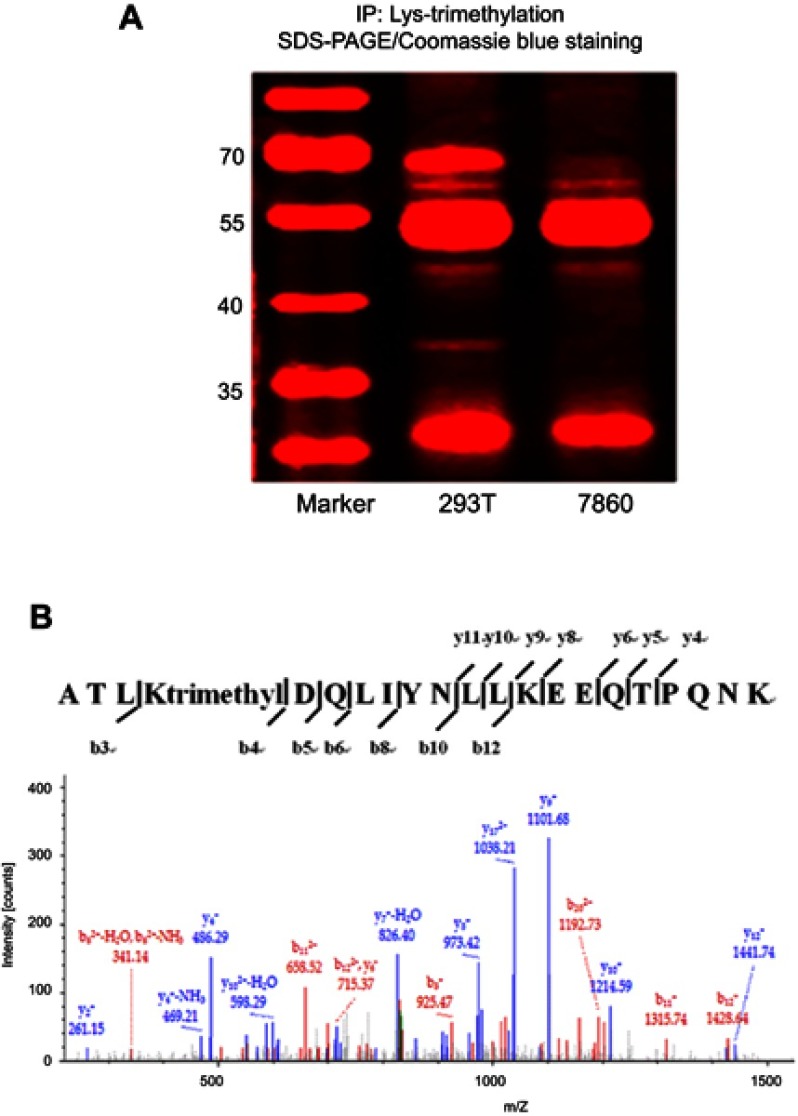
Identification of the Lys-trimethylated proteins by LC-MS/MS analysis
To confirm which proteins were Lys-trimethylated, the cell and tissue lysates were immunoprecipitated with anti-trimethyl lysine antibodies, and the proteins in these samples were separated by SDS-PAGE and stained with Coomassie Blue (Figure 2A). The bands on the SDS-PAGE gel stained markedly with Coomassie Blue were cut off based on the molecular weight markers used and analyzed by LC-MS/MS; the results of this analysis are shown in Figure 2B. The proteins identified by LC-MS/MS must meet the following requirements. The molecular weight of the proteins identified by LC-MS/MS must be approximately the same as that of the proteins on the SDS-PAGE gel (obtained as bands) identified according to the molecular weight markers. LC-MS/MS analysis proved that proteins had higher abundance ratios and were Lys-trimethylated, and that the modifications showed high credibility. Although the MASCOT and SEQUEST analyses showed that several proteins were Lys-trimethylated, only L-lactate dehydrogenase A chain (LDHA) was found to conform to all the above requirements simultaneously, because of the limitations of our experimental operations. The molecular weight of LDHA was 36.7 kDa, and its Σ coverage was 57.83–73.21%. It was Lys-trimethylated on the peptides comprising the sequence ATLKDQLIYNLLKEEQTPQNK, and the peptides coverage A2 was high. LDHA was Lys-trimethylated on the fifth amino acid, lysine (abbreviated as K5), of its protein sequence. The immunoprecipitation and LC-MS/MS analyses were repeated thrice. In conclusion, LDHA was found to be Lys-trimethylated in kidney-derived cells and tissues.
Figure 2.
LDHA is Lys-trimethylated in HEK293T cells.
Notes: (A) HEK293T and 786–0 cell lysates were immunoprecipitated with anti-trimethyl lysine antibodies, and the proteins in these lysates were separated by SDS-PAGE and stained using Coomassie Blue; (B) The protein bands on the SDS-PAGE gel stained notably with Coomassie Blue were cut off based on the molecular weight markers used and analyzed by LC-MS/MS. The results of the MASCOT and SEQUEST analyses indicated that LDHA was Lys-trimethylated on the peptides comprising the ATLKDQLIYNLLKEEQTPQNK sequence.
Abbreviations: 293T, HEK293T cells; 7860, 786–0 cells.
Verification of Lys-trimethylation on the K5 of LDHA by point mutation and Western blotting analyses
To further verify that LDHA was Lys-trimethylated on the K5 of its protein sequence, two kinds of eukaryotic expression vectors were constructed. Lysine (K5) on the N-terminal of LDHA protein sequence was mutated to arginine (R) in the purified plasmid DNA, which was abbreviated as LDHA@K5R, and lysine (K5) on the N-terminal of the LDHA protein sequence was mutated to glutamine (Q) in the purified plasmid DNA, which was abbreviated as LDHA@K5Q. The mutations of lysine in the eukaryotic expression vectors were verified by genetic sequencing, and the results are shown in Figure 3A and B. Both arginine and lysine are alkaline amino acids, and glutamine is a hydrophilic amino acid; thus, when the eukaryotic expression vectors were transfected into HEK293T cells, it was assumed that if LDHA was Lys-trimethylated on the K5, the mutation of lysine (K5) will lead to the reduction or disappearance of trimethylation in LDHA, but the expression of LDHA remained unchanged. The HEK293T cell lysates transfected using Lipofectamine 2000 were immunoprecipitated with anti-LDHA antibodies, and Western blotting was performed using anti-trimethyl lysine and anti-LDHA antibodies as primary antibodies. The results of the Western blotting are shown in Figure 3C and D, and E, and they prove that our hypothesis is true and that LDHA was indeed Lys-trimethylated on lysine (K5) in HEK293T cells. We confirmed that LDHA was the protein that was Lys-trimethylated in HEK293T cells by using LC-MS/MS analysis; the molecular weight of this protein was in the range of 35–40 kD. However, we could not confirm the existence of other Lys-trimethylated proteins because of the limitations of our experimental operations. LDHA is an enzyme encoded by the LDHA gene in humans.16 It is a monomer of lactate dehydrogenase, which exists as a tetramer. LDHA catalyzes the inter-conversion of pyruvate and L-lactate, with the concomitant, mutual transformation of NADH and NAD+. LDHA is found predominantly in not only muscle tissues and tumors, but also in most somatic tissues.17 It has long been known that many types of human cancer cells show higher LDHA levels compared to their normal counterparts. LDHA plays an important role in the invasion and metastasis of malignancies.18 Once again, we verified that LDHA was Lys-trimethylated on the K5 of its protein sequence by constructing eukaryotic expression vectors that led to the mutations of lysine (K5) on LDHA. The results of the Western blotting analysis showed that the mutation of lysine (K5) decreased the level of trimethylation in LDHA, while the expression of LDHA remained unchanged. Fibroblast growth factor receptor 1 (FGFR1) reduced LDHB expression by promoting the methylation of its promoter, and regulated the cell metabolism, causing a shift from oxidative phosphorylation to aerobic glycolysis.19 The relationship between LDH and methylation has been researched previously, and these researches have focused on the methylation of the LDH promoter. Thus far, the relationship between Lys-trimethylation and LDHA remains unclear.
Figure 3.
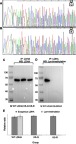
LDHA is Lys-trimethylated on K5 in HEK293T cells.
Notes: (A) The genetic sequencing results of LDHA@K5Q showed that lysine (K5) on the peptides comprising the ATLKDQLIYNLLKEEQTPQNK sequence was mutated into glutamine (Q); (B) The genetic sequencing results of LDHA@K5R showed that lysine (K5) on the peptides comprising the ATLKDQLIYNLLKEEQTPQNK sequence was mutated into arginine (R); (C, D) After LDHA@K5Q and LDHA@K5R were transfected into HEK293T cells, the HEK293T cell lysates were immunoprecipitated with anti-LDHA antibodies, and Western blotting was performed using anti-trimethyl lysine and anti-LDHA antibodies as the primary antibodies. The molecular weight of LDHA is 36.7 kDa, however, because of its connection with GFP in the plasmid sequence, theoretically, the molecular weight of exogenous LDHA was approximately 64 kDa. (E) The results of Western blotting indicated that the mutation of lysine (K5) led to the disappearance of trimethylation on Lysine (K5), while the expression of LDHA remained unchanged (p<0.01, n=3).
Abbreviations: M, marker; K5-Q, lysine (K5) on LDHA was mutated into glutamine (Q) in HEK293T cells; K5-R, lysine (K5) on LDHA was mutated into arginine (R) in HEK293T cells; WT LDHA, The plasmid constructed on the basis of LDHA wild-type sequence (Genbank NM005566) was transfected into HEK293T cells as the control sample.
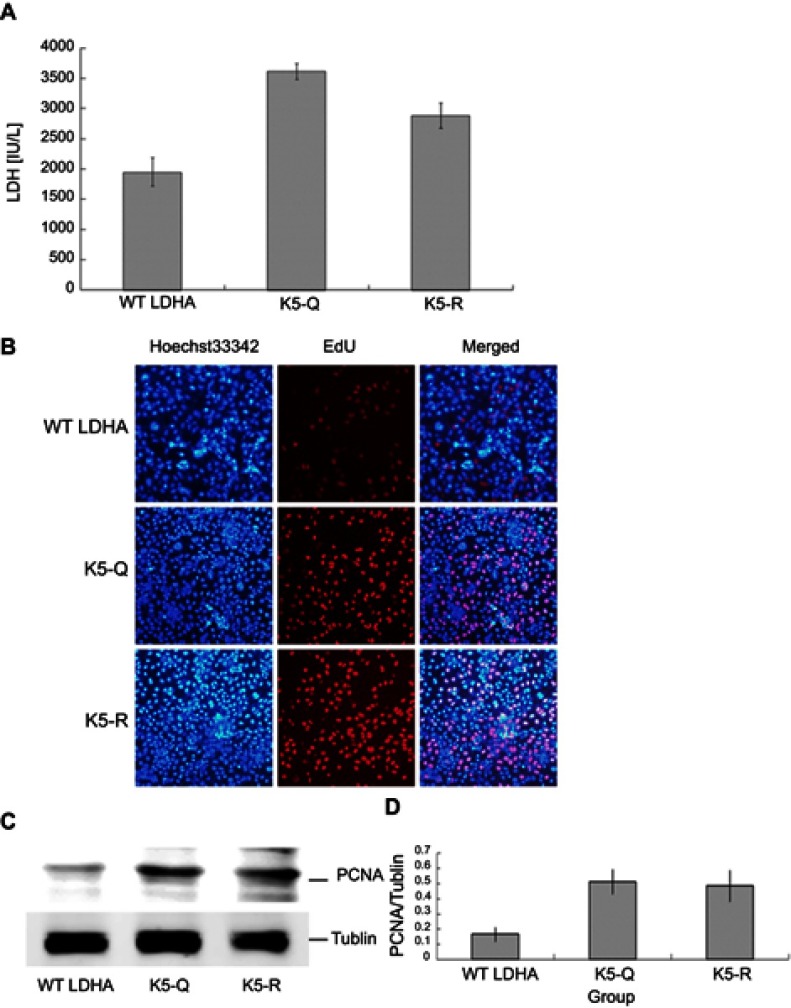
The effect of Lys-trimethylation of LDHA on the metabolism of HEK293T cells
To examine the effect of the Lys-trimethylation of LDHA on the metabolism of HEK293T cells, first, the LDH activity of HEK293T cells transfected with LDHA@K5R and LDHA@K5Q was measured by using the LDH Activity Assay Kit, strictly following the manufacturer’s instructions. This assay was quick, convenient, and sensitive. In this kit, LDH reduced NAD to NADH, which was specifically detected by colorimetric measurements (at 450 nm). The result of the LDH activity assay indicated that the reduction of the level of Lys-trimethylation in LDHA increased the LDH activity of HEK293T cells accordingly by 1.65-fold (P<0.01, n=3). These results are shown in Figure 4A. Next, 5-ethynyl-2ʹdeoxyuridine (EdU) was used to measure the proliferation of HEK293T cells with or without Lys-trimethylation inhibition. EdU has previously been shown to label the newly synthesized DNA with red fluorescence and has been used to quantitatively measure cell proliferation. The total DNA was also determined by staining with Hoechst 33,342 to normalize the fluorescence of HEK293T cells with and without Lys-trimethylation inhibition. The ratio of HEK293T cells showing EdU and Hoechst fluorescent signals was 0.22±0.03 without and 0.41±0.06 with Lys-trimethylation inhibition (Figure 4B), and the inhibition of Lys-trimethylation in LDHA upregulated the proliferation of HEK293T cells by 1.87-fold (p<0.01, n=3). We further measured the proliferation of HEK293T cells with or without Lys-trimethylation inhibition by Western blotting. To this end, total lysates of HEK293T cells transfected with LDHA@K5R and LDHA@K5Q were resolved by SDS-PAGE and transferred onto nitrocellulose membranes. The membranes were probed with anti-PCNA (proliferating cell nuclear antigen; 1:1,000; CST, USA), and antibodies against tubulin were used as the control. Consistent with the label-free quantification, PCNA expression increased by 2.91-fold (p<0.01, n=3) (Figure 4C and D) when Lys-trimethylation was inhibited.
Figure 4.
Lys-trimethylation of LDHA has some effects on the metabolism of HEK293T cells.
Notes: (A) LDH activity of HEK293T cells that were transfected with LDHA@K5R and LDHA@K5Q was detected by using the LDH Activity Assay Kit. The results indicated that the reduction of Lys-trimethylation on LDHA increased the lactate dehydrogenase activity accordingly by 1.65-fold (p<0.01, n=3). (B) The proliferation of transfected HEK293T cells was detected by using 5-ethynyl-2ʹdeoxyuridine (EdU). The ratio of HEK293T cells showing EdU and Hoechst fluorescent signals was 0.22±0.03 without and 0.41±0.06 with Lys-trimethylation inhibition. Inhibition of Lys-trimethylation in LDHA upregulated the proliferation of HEK293T cells by 1.87-fold (p<0.01, n=3). (C) The proliferation of HEK293T cells was detected by Western blotting using anti-PCNA antibodies, and antibodies against tubulin were used as a control. (D) PCNA expression increased by 2.91-fold (p<0.01, n=3) (C, D) when Lys-trimethylation was inhibited.
We detected the LDH activity of HEK293T cells following Lys-trimethylation inhibition by using the LDH Activity Assay Kit, and the results showed that when the expression of Lys-trimethylation in LDHA was suppressed, the LDH activity increased accordingly, while the LDHA expression remained unchanged. We measured the proliferation of HEK293T cells following Lys-trimethylation inhibition by using EdU and Western blotting analyses, and found that the inhibition of Lys-trimethylation in LDHA upregulated the proliferation of HEK293T cells. There is a close relationship between LDHA and cell metabolism. Overexpression of LDHA in a pituitary adenoma cell line (GH3) has been shown to induce cellular invasion via the upregulation of matrix metalloproteinase2 (MMP2), and promote glucose uptake via the upregulation of lactate secretion and glucose transporter-1 (Glut1).20 Overexpression of miR-383 significantly suppressed cell proliferation and invasion as well as glycolysis. Furthermore, LDHA is a target gene of miR-383; the expression of LDHA has been shown to be inversely related with that of miR-383 in hepatocellular cancer.21 LDHA activation mediated by phosphorylation promotes tumor cell invasion and tumor metastasis.22 In lung cancer cells and leukemia cells harboring dysregulated fibroblast growth factor receptor 1 (FGFR1), the phosphorylation of LDHA at tyrosine has been shown to promote tumor cell metabolism and the growth of tumors by regulating NADH/NAD+ redox homeostasis.23 LDHA K5 may be also acetylated and succinylated,24,25 and the LDHA K5 substitutions will similarly affect all other potential lysine modifications at this residue. Hence, whether the reduction of Lys-trimethylation in LDHA directly or indirectly affects the metabolism of cells should be further studied. In this study, for the first time, we reported that LDHA affects the metabolism and proliferation of cells via a Lys-trimethylation-mediated mechanism; however, this mechanism is yet to be investigated.
Conclusion
LDHA is Lys-trimethylated on lysine (K5) in kidney-derived cells and tissues. Lys-trimethylation in LDHA plays an important role in the metabolism and proliferation of cells and may be useful as a potential target for therapy or a prognostic and treatment biomarker of several diseases.
Acknowledgments
The authors are grateful for the support from National Natural Science Foundation of China (#31570352 and 31170321).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Dhar SK, St Clair DK. Manganese superoxide dismutase regulation and cancer. Free Radic Biol Med. 2012;52:2209–2222. doi: 10.1016/j.freeradbiomed.2012.03.009 [DOI] [PubMed] [Google Scholar]
- 2.Schubert HL, Blumenthal RM, Cheng X. 1 Protein methyltransferases: their distribution among the five structural classes of adoMet-dependent methyltransferases. Enzymes. 2006;24:3–28. doi: 10.1016/S1874-6047(06)80003-X [DOI] [PubMed] [Google Scholar]
- 3.Varland S, Osberg C, Arnesen T. N-terminal modifications of cellular proteins: the enzymes involved, their substrate specificities and biological effects. Proteomics. 2015;15:2385–2401. doi: 10.1002/pmic.201400619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grewal SI, Rice JC. Regulation of heterochromatin by histone methylation and small RNAs. Curr Opin Cell Biol. 2004;16:230–238. doi: 10.1016/j.ceb.2004.04.002 [DOI] [PubMed] [Google Scholar]
- 5.Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118 [DOI] [PubMed] [Google Scholar]
- 6.Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991 [DOI] [PubMed] [Google Scholar]
- 7.Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422 [DOI] [PubMed] [Google Scholar]
- 8.Lee S, Roeder RG, Lee JW. Roles of histone H3-lysine 4 methyltransferase complexes in NR-mediated gene transcription. Prog Mol Biol Transl Sci. 2009;87:343–382. doi: 10.1016/S1877-1173(09)87010-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim DH, Tang Z, Shimada M, et al. Histone H3K27 trimethylation inhibits H3 binding and function of SET1-like H3K4 methyltransferase complexes. Mol Cell Biol. 2013;33:4936–4946. doi: 10.1128/MCB.00601-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waldmann T, Schneider R. Targeting histone modifications–epigenetics in cancer. Curr Opin Cell Biol. 2013;25:184–189. doi: 10.1016/j.ceb.2013.01.001 [DOI] [PubMed] [Google Scholar]
- 11.Tang G, Guo J, Zhu Y, et al. Metformin inhibits ovarian cancer via decreasing H3K27 trimethylation. Int J Oncol. 2018;52:1899–1911. doi: 10.3892/ijo.2018.4343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lien HC, Jeng YM, Jhuang YL, Yuan RH. Increased Trimethylation of histone H3K36 associates with biliary differentiation and predicts poor prognosis in resectable hepatocellular carcinoma. PLoS One. 2018;13:e0206261. doi: 10.1371/journal.pone.0206261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li C, Wang D, Lv X, et al. Yersinia pestis acetyltransferase-mediated dual acetylation at the serine and lysine residues enhances the auto-ubiquitination of ubiquitin ligase MARCH8 in human cells. Cell Cycle. 2017;16:649–659. doi: 10.1080/15384101.2017.1281481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Z, Azadzoi KM, Choi HP, et al. LC-MS/MS analysis unravels deep oxidation of manganese superoxide dismutase in kidney cancer. Int J Mol Sci. 2017;18:E319. doi: 10.3390/ijms18020319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo J, Wang X, Lü X, et al. Unraveling molecular effects of ADAR1 overexpression in HEK293T cells by label-free quantitative proteomics. Cell Cycle. 2016;15:1591–1601. doi: 10.1080/15384101.2016.1176657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chung FZ, Tsujibo H, Bhattacharyya U, Sharief FS, SS LI. Genomic organization of human lactate dehydrogenase-A gene. Biochem J. 1985;231:537–541. doi: 10.1042/bj2310537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mack N, Mazzio EA, Bauer D, Flores-Rozas H, Soliman KF. Stable shRNA silencing of lactate dehydrogenase A (LDHA) in human MDA-MB-231 breast cancer cells fails to alter lactic acid production, glycolytic activity, ATP or survival. Anticancer Res. 2017;37:1205–1212. doi: 10.21873/anticanres.11435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, Xu T, Wang Y, et al. Prognostic role of lactate dehydrogenase expression in urologic cancers: a systematic review and meta-analysis. Oncol Res Treat. 2016;39:592–604. doi: 10.1159/000449138 [DOI] [PubMed] [Google Scholar]
- 19.Liu J, Chen G, Liu Z, et al. Aberrant FGFR tyrosine kinase signaling enhances the warburg effect by reprogramming LDH isoform expression and activity in prostate cancer. Cancer Res. 2018;78:4459–4470. doi: 10.1158/0008-5472.CAN-17-3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.An J, Zhang Y, He J, et al. Lactate dehydrogenase A promotes the invasion and proliferation of pituitary adenoma. Sci Rep. 2017;7:4734. doi: 10.1038/s41598-017-04366-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fang Z, He L, Jia H, Huang Q, Chen D, Zhang Z. The miR-383-LDHA axis regulates cell proliferation, invasion and glycolysis in hepatocellular cancer. Iran J Basic Med Sci. 2017;20:187–192. doi: 10.22038/ijbms.2017.8246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin L, Chun J, Pan C, et al. Phosphorylation-mediated activation of LDHA promotes cancer cell invasion and tumour metastasis. Oncogene. 2017;36:3797–3806. doi: 10.1038/onc.2017.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan J, Hitosugi T, Chung TW, et al. Tyrosine phosphorylation of lactate dehydrogenase A is important for NADH/NAD(+) redox homeostasis in cancer cells. Mol Cell Biol. 2011;31:4938–4950. doi: 10.1128/MCB.06120-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abdo AA (1), Ackermann M, Ajello M, et al. Detection of 16 gamma-ray pulsars through blind frequency searches using the fermi LAT. Science. 2009;325:840–844. doi: 10.1126/science.1175558 [DOI] [PubMed] [Google Scholar]
- 25.Park J, Chen Y, Tishkoff DX, et al. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol Cell. 2013;50:919–930. doi: 10.1016/j.molcel.2013.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]