Abstract
Purpose
To establish a gemcitabine-resistant lung adenocarcinoma cell line, A549/G+, and to screen the differences of miRNA expression in exosomes from A549 and A549/G+ cells.
Methods
A549 cells were exposed in gemcitabine until they were resistant to gemcitabine, and extracted exosomes from A549 and A549/G+. The RNAs from exosomes were subjected to miRNA expression microarray experiments.
Results
After 39 weeks of continuous induction, we induced drug resistance in A549 cells. The resistance index was 6. Via GeneChip miRNA 4.0 analysis, there were 446 differential miRNAs between A549 and A549/G+. Target gene prediction and pathway analysis discovered the microRNAs in the intersections may participate in drug resistance.
Conclusion
These differential miRNAs help to do in-depth research to elucidate the mechanism of resistance to gemcitabine in non-small cell lung cancer.
Keywords: non-small cell lung cancer, NSCLC, exosomes, miRNA, microarray, drug resistance, gemcitabine
Introduction
Lung cancer is one of the most prevalent malignant tumors in China. In the past three decades, the mortality rate of lung cancer patients in China has increased by 464.84%, and about 100,000 people die of lung cancer every year.1 It is estimated that there were 1.8 million new cases of lung cancer worldwide in 2012, accounting for 13% of all cancer cases.2 In lung cancer, the incidence of non-small cell lung cancer (NSCLC) which includes squamous cell carcinoma, adenocarcinoma, and large cell carcinoma is the highest, accounting for 85%. Therefore, it is necessary to expand the research on the treatment of NSCLC.
Gemcitabine is a nucleoside analog that primarily effects on the synthesis of DNA. And it is a first-line chemotherapy drug in the treating guidelines of NSCLC established by National Comprehensive Cancer Network and National Institute for Health and Care Excellence . However, drug resistance always affects the efficacy of gemcitabine.3 It is reported that the objective response rate of NSCLC patients was only 18.4–22.2% when gemcitabine was treated as a first-line drug alone.4 Many patients have no response to the treatment of gemcitabine.5 At present, there was no conclusive theory to reasonably explain the mechanism of NSCLC resistance to gemcitabine. Current research indicated that multiple aspects including drug target, membrane transporter, and drug metabolism mainly involved in gemcitabine-resistance. Davidson et al, found that the expression of ribonucleotide reductase subunit 1, one of the gemcitabine targets, was higher in gemcitabine-resistant cell line H358-G200 and H460-G400 compared with the gemcitabine-sensitive cell line.6 Oguri et al, reported the expression level of ATP-binding cassette C5 which mediates the transport of intracellular metabolites and exogenous drugs was negatively correlated with the sensitivity of gemcitabine.7 In a study by Qu et al, immunohistochemistry showed that Sorcin expression was positive in 83.3% of the gemcitabine-resistant patients, which was significantly higher than 42.3% of the gemcitabine-sensitive patients.8
Thus, it can be seen that the production of resistance to gemcitabine in NSCLC cells is a complex process. In recent years, exosomes as a “natural mini-map” about the origin, physiology and pathological of cells was gradually being widely recognized by researchers.9 Exosomes are small nano-vesicles released by cells with diameters between 30 and 120 nm. They could carry cellular contents including lipids, proteins, DNAs, and RNAs, and pass on these biological information from cells to cells.10 Various studies proved that exosomes play an essential role in cancer drug resistance via carrying various biomolecules,and were effective in transmitting drug resistance via the delivery of particular miRNAs.11,12 Due to the characteristics of exosomes, we wanted to analyze the exosomes isolated from A549/G+ which had been induced to resist gemcitabine. Through the miRNA microarray technique, we found the differences in miRNA contained in exosomes between resistant cell line and sensitive cell line. Then, we try to identify some miRNAs involved in drug resistance by bioinformatics analysis for providing individualized medication guidance in clinical practice.
Materials and methods
Cell culture
Human lung adenocarcinoma cell A549 was bought from Research Facilities of PUMCH (Beijing, China). A549/G+ cell line was established from A549 by our research group via exposing A549 to the mixture of gemcitabine medium. All cells were grown in DMEM (Gibco BRL, US) containing 10–12% fetal bovine serum (FBS) (Sangon Biotech, China). Cells were incubated at 37°C and 5% in CO2 incubator (Thermo Fisher, US).
Cell counting kit-8 assay (CCK-8) and colony-forming unit assays
We used the CCK-8 assay (Dojindo, Japan) to measure the IC50 of A549 and A549/G+. We incubated cells in 96-well plates at a concentration of 3,000 cells per well. After 24 hrs of incubation in complete medium, cells were exposed to different concentrations of gemcitabine for 72 hrs. Then, we discarded the liquid in the original well, and add 100 μL 10% CCK-8 assay to each well. By using microplate reader (Bio Tek Gem5, US), we measured optical density values at 450 nm. Each drug concentration was set up in three wells and repeated at least three times.
The cells were evenly placed into a six-well plate at a density of 300 cells per well. We added different concentrations of gemcitabine to the wells, and incubate the cells for 72 hrs. Then, we switched to use the complete medium to culture the cell until they formed macroscopic colonies. After being fixed by formaldehyde, the cells were stained with 0.1% crystal violet for 10 mins.
Isolation of exosome
Beore incubation of exosomes, we rinsed the flask 4–5 times with 1× phosphate buffer saline (Gibco BRL, US) for avoiding exosomes in FBS from affecting the experiment. Each cell flask was added 5 mL DMEM only, and incubated in CO2 incubator for 72 hrs. We centrifuged the supernatant collected from the flask at 4°C, 20,000×g for 30 mins. Then, we isolated exosomes according to the procedures in the ExoQuick-TCTM kit instructions (Thermo Fisher, US).
Western blotting
The exosomal membrane is rich in transmembrane 4 superfamily, including CD63, CD81, and CD9, which is involved in vesicle trafficking. Meanwhile, there are synthesis-related proteins of multivesicular bodies such as Alix and TSG101 on the exosomal membrane. Thus, in order to confirm that the white precipitates isolated from medium suspension were exosomes, the CD9 and Alix were detected by Western blotting. The exosomes were lysed in RIPA buffer (Beyotime Biotechnology, China). The protein samples were separated by SDS-PAGE (sodium dodecyl sulfate polyacrylamide gel electrophoresis, SDS-PAGE) and transferred to a PVDF (polyvinylidene fluoride, PVDF) films. Rabbit polyclonal CD9 antibody (Abcam, Item NO.EPR2949, USA) and Alix antibody (Abcam, Item NO.EPR15314, USA) were all used at a dilution of 1:2,000, and the secondary antibody (Beyotime Biotechnology, China) at 1:1,000 dilutions. The bound antibodies were detected using ECL Western Blotting Detection system.
RNA exaction from exosomes for microarray analysis
The total RNA was extracted from the exosomes by using Trizol method, and subjected to quality inspection by NanoDrop 2000 and Agilent Bioanalyzer 2100. The qualified samples entered the stage of microarray experiment. We used Affymetrix GeneChip® miRNA 4.0 system (Thermo Fisher, US) to analysis miRNAs in exosomes, and referred to the system description to conduct the microarray experiment. The GeneChip® miRNA 4.0 arrays, containing 30,424 total mature miRNA probe sets including 2,578 mature human miRNAs, were washed and stained using the Affymetrix GeneChip Hybridization Wash and Stain Kit and were then scanned with the Affymetrix GeneChip Scanner 3000 7G (Affymetrix, Santa Clara, CA, U.S.).
Data analysis and statistical method
Transcriptome Analysis Suite Software (Thermo Fisher, US) was for miRNA 4.0 array data summarization, normalization, and quality control. We conducted the statistical analysis by using SPSS Version 20.0. Student’s t-test was used to analyze parametric continuous variables, and Mann–Whitney U test was used to analyze nonparametric continuous variables. The statistical test level was set at 0.05. The IC50 was estimated by the probit regression. DIANA-mirPath was set to use TargetScan Human 5.1 to identify possible mRNA targets. The level of significance was set at p<0.05.
Results
Establishment of A549/G+ cell line
We treated original A549 cells with continuously increasing concentration of gemcitabine from 0.1 to 3 μmol/L. Then, we began to induce cell resistance by using intermittent high-concentration dosing. The A549 cells were exposed to the concentration of 0.005 mmol/L for 72 hrs, and then the cells were cultured in complete medium. After 48–72 hrs, we continued to use concentration of 0.005 mmol/L gemcitabine to culture the cells.
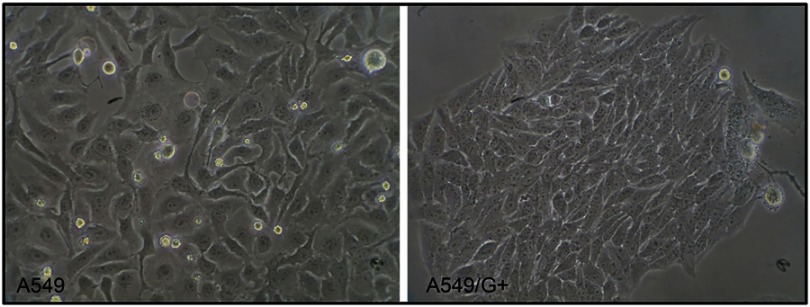
The induction of gemcitabine resistance lasted for about 39 weeks. Compared with its original cell line A549, the cell morphology of A549/G+ did not change significantly (Figure 1). As Figure 2 shown, the fold line representing the inhibition rate tended to be gentle around 0.025 mmol/L in A549, while it tended to be gentle around 0.100 mmol/L in A549/G+. Results showed that 50% inhibiting concentration (IC50) values of gemcitabine on A549 and A549/G+ were respectively 2 μmol/L (95% confidence interval: 1.0–3.0 μmol/L) and 12 μmol/L (95% confidence interval: 4.0–28.0 μmol/L). The resistance value of A549/G+ was significantly higher than that of A549 (Z=−2.366, P=0.018, α=0.05).
Figure 1.
The cell morphology of A549 and A549/G+.
Figure 2.
Inhibition rate of gemcitabine on cells.
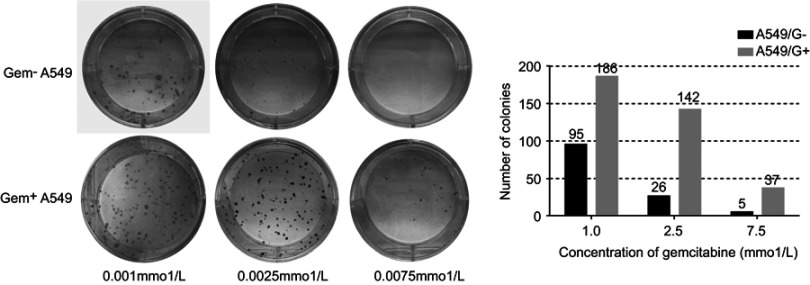
To further validate the gemcitabine resistance of A549/G+, we conducted colony-forming unit assays. Under the same concentration of gemcitabine, A549/G+ formed more cell colonies than drug-sensitive strain cells A549 (Figure 3). When the concentration of gemcitabine reached 7.5 μmol/L, A549 single cells were barely able to form colonies. Only the scattered cells could be seen in that well, and there were only five small cell colonies being found. But we could see the clear sign of colonies growth in A549/G+ which had been exposed to the concentration of 7.5 μmol/L. Both tests verified that A549/G+ has developed drug resistance to gemcitabine.
Figure 3.
The results of colony-forming unit assays.
Identification of exosomes
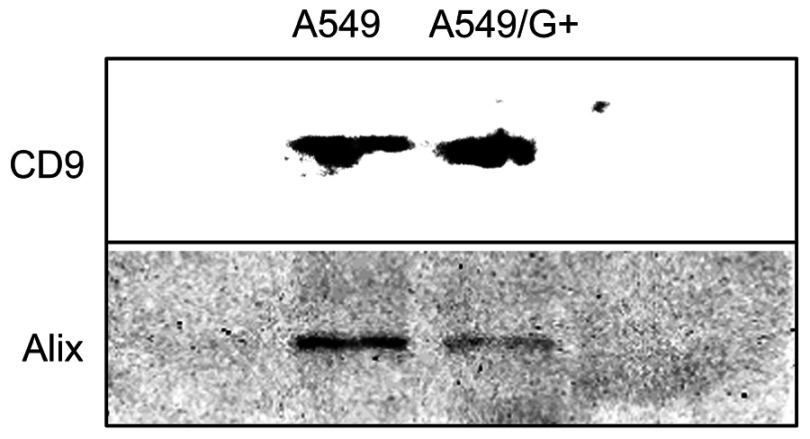
In order to confirm that the white precipitates isolated from medium suspension were exosomes, the CD9 and Alix were detected by Western Blotting. Western blotting results showed that the two marker proteins CD9 and Alix were obviously present in the exosomes extract (Figure 4). It implied that exosomes indeed existed in the extracted samples, and the samples could be used in subsequent experiments. Considering that the difference in protein expression between A549 and A549/G+ would not affect the results, we have not set any reference proteins.
Figure 4.
The Western blotting of exosomes.
Profiles of microarray and bioinformatics analysis
According to the microarray results, compared with exosomes secreted by A549 cells, 85 upregulated genes and 361 downregulated genes were detected in A549/G+ exosomes. As Table 1 shown, in the top ten differentially expressed miRNAs, let-7 family had two places. All of the top ten were down regulated in A549/G+ exosomes.
Table 1.
The top ten differentially expressed miRNAs between A549 and A549/G+
| MicroRNA | Regulation | LogFC |
|---|---|---|
| hsa-let-7i-5p | Down | −11.593143 |
| hsa-let-7d-5p | Down | −11.57318 |
| hsa-miR-107 | Down | −11.198673 |
| hsa-miR-151a-5p | Down | −11.06081 |
| hsa-miR-106a-5p | Down | −10.99788 |
| hsa-miR-26a-5p | Down | −10.66564 |
| hsa-miR-17-5p | Down | −10.54206 |
| hsa-miR-93-5p | Down | −10.23588 |
| hsa-miR-23b-3p | Down | −10.19281 |
| hsa-miR-103a-3p | Down | −10.14184 |
The target gene prediction of the top 15 differentially expressed miRNAs was performed by online prediction software for target genes. We have chosen miRWalk 3.0 which was based on three databases TargetScan, miRDB, and miRTarBase to predict the target genes. By merging the data of above three databases, we draw the following conclusions: in terms of prediction based on the 3’untranslated region of mRNA,we got 221 target genes, while 196 target genes were found when basing on the coding sequence of mRNA. Through using the String database, we plotted the network analysis of gene interactions (Figures 5 and 6).
Figure 5.
Network diagram based on genes obtained from predictions made in the coding sequence (CDS) of mRNA.
Figure 6.
Network diagram based on genes obtained from predictions made in the 3’UTR of mRNA.
Then, we employed a Gene Ontology (GO) enrichment analysis by using ENONTOLOGY database for annotation, visualization, and integrated discovery (Tables 2–4). The analysis results based on 221 target genes shown that the most GO category was related to the regulation of macromolecule biosynthetic process among the top 5 significant terms belonging to the biological process. As to other 196 target genes, results showed that the GO category of the biological process was the most relevant to negative regulation of the cellular process. Moreover, in both aspects of cellular component and molecular function, the enrichment analysis displayed that other GO categories including protein binding, intracellular part, and nuclear part were also enriched.
Table 3.
The GO category of molecular function
| No. | GO molecular function complete | Genes | FDR |
|---|---|---|---|
| Based on 3’UTR of mRNA | |||
| (GO:0005515) | Protein binding | 167 | 1.76e-05 |
| (GO:1990837) | Sequence-specific double-stranded DNA binding | 24 | 2.79e-03 |
| (GO:0001067) | Regulatory region nucleic acid binding | 26 | 2.91e-03 |
| (GO:0003690) | Double-stranded DNA binding | 26 | 2.92e-03 |
| (GO:0005488) | Binding | 190 | 3.27e-03 |
| Based on CDS of mRNA | |||
| (GO:0005515) | Protein binding | 160 | 1.84e-09 |
| (GO:0140096) | Catalytic activity, acting on a protein | 57 | 2.51e-07 |
| (GO:0005488) | Binding | 178 | 2.49e-06 |
| (GO:0000982) | Transcription factor activity, RNA polymerase II proximal promoter sequence-specific DNA binding | 19 | 3.95e-05 |
| (GO:0001067) | Regulatory region nucleic acid binding | 26 | 3.38e-04 |
Abbreviations: GO: Gene Ontology; FDR: false discovery rate; CDS: coding sequence.
Table 2.
The GO category of biological process
| No. | GO biological process complete | Genes | FDR |
|---|---|---|---|
| Based on 3’UTR of mRNA | |||
| (GO:0010556) | Regulation of macromolecule biosynthetic process | 87 | 1.07e-06 |
| (GO:0010468) | Regulation of gene expression | 93 | 1.10e-06 |
| (GO:0031323) | Regulation of cellular metabolic process | 113 | 1.26e-06 |
| (GO:0009889) | Regulation of biosynthetic process | 92 | 1.30e-06 |
| (GO:0048523) | Negative regulation of cellular process | 90 | 1.32e-06 |
| Based on CDS of mRNA | |||
| (GO:0048523) | Negative regulation of cellular process | 94 | 5.82e-11 |
| (GO:0031323) | Regulation of cellular metabolic process | 112 | 1.95e-10 |
| (GO:0048519) | Negative regulation of biological process | 98 | 2.04e-10 |
| (GO:0036211) | Protein modification process | 73 | 4.52e-10 |
| (GO:0044260) | Cellular macromolecule metabolic process | 114 | 5.50e-10 |
Abbreviations: GO: Gene Ontology; FDR: false discovery rate; CDS: coding sequence.
Table 4.
The GO category of cellular component complete
| No. | GO cellular component complete | Genes | FDR |
|---|---|---|---|
| Based on 3’UTR of mRNA | |||
| (GO:0044424) | Intracellular part | 195 | 3.85e-07 |
| (GO:0005622) | Intracellular | 196 | 7.80e-07 |
| (GO:0043229) | Intracellular organelle | 178 | 7.91e-07 |
| (GO:0031981) | Nuclear lumen | 82 | 1.19e-06 |
| (GO:0043227) | Membrane-bounded organelle | 174 | 1.47e-06 |
| Based on CDS of mRNA | |||
| (GO:0044428) | Nuclear part | 86 | 1.14e-08 |
| (GO:0031981) | Nuclear lumen | 78 | 1.06e-07 |
| (GO:0044424) | Intracellular part | 176 | 1.29e-07 |
| (GO:0005622) | Intracellular | 178 | 1.91e-07 |
| (GO:0044428) | Nuclear part | 67 | 1.77e-06 |
Abbreviations: GO: Gene Ontology; FDR: false discovery rate; CDS: coding sequence.
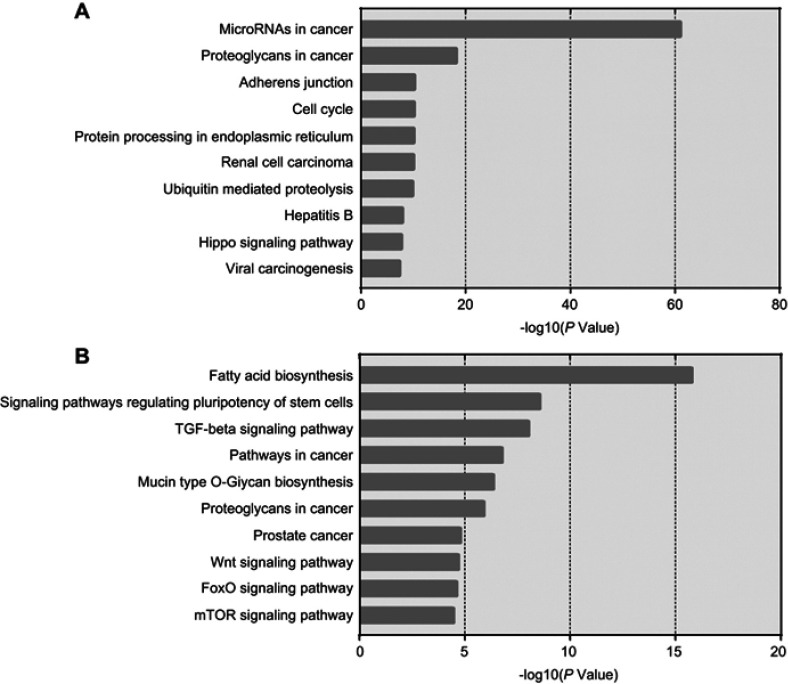
In order to characterize the predominant pathways, Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis was conducted by DIANA TOOLS. The top 10 signal pathways are shown in Figure 7. According to DIANA-Tarbase and DIANA-microT there two databases, microRNAs in cancer pathway and fatty acid biosynthesis pathway may involve in gemcitabine resistance. Our data indicated that the drug resistance of lung cancer cells was partly related to these differentially expressed miRNA.
Figure 7.
The top 10 most likely Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways.
Discussion
Until the late 1990s, regardless of the histological subtype of advanced lung cancer, the chemotherapy regimen was to simply follow using platinum-based combination therapy. With the use of “third-generation” cytotoxic drugs such as gemcitabine, paclitaxel, and docetaxel, the treatment regimen of NSCLC has been optimized, and the overall survival of patients has increased to 8 months in clinical practice.13 However, acquired drug resistance greatly limited the clinical efficacy of gemcitabine therapy. Thus, it has become particularly important to find one or more biomarkers that can be used to evaluate the sensitivity of individual gemcitabine treatment. This will not only benefit patients from the gemcitabine treatment regimen, but also improve the clinical effect, and the precise and individualized treatment plan will also avoid the patient getting astray in treatment.
At present, gemcitabine has rarely been reported in the NSCLC precision treatment. However, the application and research of gemcitabine in the pancreatic cancer precise treatment provide research route for NSCLC. In a 4-year tracing test of 538 patients with pancreatic cancer, researchers found that hENT1 (human nucleotide balance transporter 1, hENT1) expression was significantly associated with overall survival and disease-free survival in patients who have been receiving gemcitabine.14 Therefore, the researchers believed that hENT1 immunohistochemistry has clinical value in evaluating the efficacy of gemcitabine chemotherapy in patients with pancreatic cancer. Subsequent similar reports15–18 have also demonstrated that precision treatment can improve the efficacy of gemcitabine in pancreatic cancer.
MiRNAs are a type of non-coding small RNAs that alter the genetic information flow by degrading mRNA, or directly inhibiting mRNA translation.19 A large amount of evidence demonstrated that miRNAs have been involved in the process of NSCLC resistance to chemotherapeutic drugs. Zhao et al, proved that high expression of cyclin-dependent knase inhibitor 1A and RAD21 cohesin complex component (Rad21) gene occurred by low expression of miR-17 and miR-92 family could not only prevent cisplatin (DDP) from inducing apoptosis of NSCLC cells, but also promote the DNA of cancer cells repair, and this would maintain the drug resistance of NSCLC cells to DDP. Another study found that increased expression of miRNA-135a accompanied by a decreased expression of the APC gene was observed in nude mice which were inoculated subcutaneously with a NSCLC cell line and induced to paclitaxel resistance.20 In addition, the study still proved that paclitaxel-sensitive cell line would develop into resistance to paclitaxel if the expression of miRNA-135a decreased in vitro. Therefore, our study tempted to develop a gemcitabine-resistant NSCLC cell line A549/G+ by treating gemcitabine for a prolonged period, and analyzed the differences in expression of miRNAs between A549 and A549/G+.
Through using the miRNA microarray technique, compared with A549, the expressions of let-7d-5p, let-7i-5p, miR-17-5p, miR-23b-3p, miR-26a-5p, miR-93-5p ten miRNAs, miR-103a-3p, miR-106a-5p, miR-107, and miR-151a-5p were most significantly decreased in A549/G+. Among these ten miRNAs, miR-17, and miR-106 family have been proposed to be closely related to the drug resistance of NSCLC in previous reports. Zhao et al, reported that low expression of miR-17 could maintain DDP resistance of NSCLC.21 Moreover, Yu et al, found that upregulation of miR-106b-5p would increase the expression of polycystic kidney disease type 2 gene, and it could result in increasing sensitivity to DDP in the DPP resistance cell line A549/DDP.22 Among the top 10 differentially expressed miRNAs, miRNA let-7 family took two places. It reported that let-7 family was closely related to the lung cancer. The prognosis of lung cancer patients with low expression of let-7 was far from ideal. And it showed the increased expression of let-7 had a greater inhibitory effect on tumor growth. Johnson et al, identified K-ras as a target gene for let-7.23 Additionally, their works also showed that upregulation of let-7 expression could inhibit NSCLC cells division, and lead to cell cycle arrest. It would not be difficult to understand why A549/G+ maintained a relatively strong growth in the cell cycle inhibitory drug gemcitabine. A549/G+ with low-level expression of let-7 may offset the effects of gemcitabine via a series of pathways. The rest of the top 10 miRNAs including miR-23b-3p, miR-26a-5p, miR-93-5p, miR-103a-3p, miR-107, and miR-151a-5p were first discovered to be associated with NSCLC resistance in our study. Particularly, miRNA-26a had been reported to be associated with doxorubicin (Dox) resistance in liver cancer.24 The study suggested that miR-26a may target UNC-51-like kinas (ULK1) gene which was an essential promoter of autophagy. Increasing expression of ULK1 was able to inhibit autophagy and promote cell apoptosis that it could increase the Dox sensitivity of hepatocellular carcinoma cells.
There are hundreds of miRNAs and their subtypes existing in cells. Simply studying miRNAs originated from the cells may lead to extremely high workload and research costs. A large number of research reports have confirmed that exosomes secreted by drug-resistant cancer cells could carry effective miRNAs to transfer information between cells, which can turn drug-sensitive host cells into drug-resistant cells.25–27 It is inferred that exosomes secreted by gemcitabine-resistant NSCLC cells must contain miRNAs which closely related to drug resistance. Meanwhile, in order to eliminate those miRNAs that would influence the experimental results, we selected exosomes as entry points for studying gemcitabine resistance in NSCLC. In addition, studies have shown that exosomes can increase the sensitivity of miRNA detection and reduce the incidence of false negative results.28 That is why we need to study miRNA through exosomes.
However, a frustrating aspect of miRNA research is that individual miRNAs have opposite functions in different systems. For instance, miRNA-125 was downregulated in hepatocellular carcinoma, breast cancer, and lung cancer, but overexpressed in colorectal cancer, pancreatic cancer, and gastric cancer.29 This phenomenon indicated that miR-125b may act as both oncogene and anti-oncogene. Moreover, in the same type of cancer, the role of miRNAs was reported to be different. The results of Yan et al, were contrary to Zhao21 cited above. Yan et al, found that inhibition of mi-92 expression can make NSCLC cells sensitive to DDP. Besides, Ma et al, indicated that A549 would be resistant to DDP after transfection with miR-106a.30 This result was in stark contrast to our array results that downregulation of miR-106a may be one of the reasons leading to NSCLC cell resistance to gemcitabine.
Therefore, some researchers have suggested that the regulation of targeting mRNA by miRNA was accomplished by the coordinated action of two or several miRNAs, and was not a “credit“ for one single miRNA.31 The study by Mitra et al, demonstrated that miR-9-5p and miR-130b-3p can directly bind to the 3’ non-coding region of the transforming growth factor beta receptor 2 (TGFBR2) gene at the same time.32 Increased expression of both miRNAs could induce the expression of TGFBR2 in NSCLC. In our study, DIANA TOOLS pathway prediction software was used to analyze differentially expressed miRNAs to predict possible biological signal pathways related to NSCLC gemcitabine resistance. And the KEGG analysis was based on the two databases DIANA-TarBase and DIANA-microT in the software. The former is a comprehensive collection of miRNA target databases that have been verified by experiments, while the latter can predict and analyze target gene based on the 3’UTR and coding sequence (CDS) regions of mRNA.33,34 We found that let-7, miR-107, and miR-16 were involved in the development of NSCLC in the “MicroRNAs in cancer” pathway. They, respectively, regulate cdc25, cdk6, and cyclins genes which were closely related to the cell cycle. Particularly, we also found that miR103, miR107, miR222, and Let7 are also involved in the process of breast cancer drug tolerance in this pathway. This provides a reference for the next research. Meanwhile, we found fatty acid synthase (FASN) in “Fatty acid biosynthesis” pathway was reported to be overexpressed in both NSCLC cisplatin-resistance cell line and ovarian cancer cisplatin-resistance cell line.35 It is suggested that FASN plays an important role in the process of cancer cell resistance. Moreover, for fully screening target genes, prediction of 3’-UTR and CDS miRNA-binding sites was conducted. We found that 3’-UTR and CDS miRNA-binding sites of genes had similar functions in terms of GO enrichment analysis. For instance, both of them were enriched in negative regulation of cellular process and protein binding. The results suggested we are most likely to find targeted genes in these enrichments. Although bioinformatics analysis provided a reference for the next practical research, the results still need to be verified by other tests such as real-time PCR for high positive rate brought by molecular hybridization.
Conclusion
Based on bioinformatics analysis, we speculated that the gemcitabine-resistance to NSCLC may possibly be the result of the interaction of several above-mentioned miRNAs including let-7d-5p, let-7i-5p, miR-17-5p, and miR-23b-3p. These miRNAs may be used as potential biomarkers to provide references for the diagnosis of gemcitabine tolerance in NSCLC patients. And they may also provide evidence for the development of precise treatment options. We believed that following further research may find an entire biological information chain to explain the complete process of NSCLC resistance to gemcitabine.
Acknowledgment
This work was supported by the National Natural Science Foundation of China (No. 81071853).
Abbreviations list
A549/G+, gemcitabine-resistant lung adenocarcinoma cell line A549; NSCLC, Non-small cell lung cancer; NCCN, National Comprehensive Cancer Network; NICE, National Institute for Health and Care Excellence; ORR, objective response rate; RRM1, ribonucleotide reductase subunit 1; ABCC5, ATP-binding cassette C5; CCK-8, cell counting kit-8 assay; TM4SF, transmembrane 4 superfamily; IC50, 50% inhibiting concentration; DDP, cisplatin.
Disclosure
All authors report no conflicts of interest in this work.
References
- 1.She J, Yang P, Hong Q, Bai C. Lung cancer in China: challenges and interventions. Chest. 2013;143(4):1117–1126. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. [DOI] [PubMed] [Google Scholar]
- 3.Tokunaga Y, Liu D, Nakano J, et al. Potent effect of adenoviral vector expressing short hairpin RNA targeting ribonucleotide reductase large subunit M1 on cell viability and chemotherapeutic sensitivity to gemcitabine in non-small cell lung cancer cells. Eur J Cancer. 2015;51(16):2480–2489. [DOI] [PubMed] [Google Scholar]
- 4.Ricci S, Antonuzzo A, Galli L, et al. Gemcitabine monotherapy in elderly patients with advanced non-small cell lung cancer: a multicenter phase II study. Lung Cancer. 2000;27(2):75. [DOI] [PubMed] [Google Scholar]
- 5.Gridelli C, Cigolari S, Gallo C. Activity and toxicity of gemcitabine and gemcitabine+vinorelbine in advanced non-small-cell lung cancer elderly patients. Lung Cancer. 2001;31(2–3):277–284. [DOI] [PubMed] [Google Scholar]
- 6.Davidson JD, Ma L, Flagella M, Geeganage S, Gelbert LM, Slapak CA. An increase in the expression of ribonucleotide reductase large subunit 1 is associated with gemcitabine resistance in non-small cell lung cancer cell lines. Cancer Res. 2004;64(11):3761–3766. [DOI] [PubMed] [Google Scholar]
- 7.Oguri T, Achiwa H, Sato S, et al. The determinants of sensitivity and acquired resistance to gemcitabine differ in non-small cell lung cancer: a role of ABCC5 in gemcitabine sensitivity. Mol Cancer Ther. 2006;5(7):1800–1806. [DOI] [PubMed] [Google Scholar]
- 8.Qu Y, Yang Y, Liu B, Xiao W. Comparative proteomic profiling identified sorcin being associated with gemcitabine resistance in non-small cell lung cancer. Med Oncol. 2010;27(4):1303–1308. [DOI] [PubMed] [Google Scholar]
- 9.Azmi AS, Bao B, Sarkar FH. Exosomes in cancer development, metastasis, and drug resistance: a comprehensive review. Cancer Metastasis Rev. 2013;32(3–4):623–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu CY, Du SL, Zhang J, Liang AL, Liu YJ. Exosomes and breast cancer: a comprehensive review of novel therapeutic strategies from diagnosis to treatment. Cancer Gene Ther. 2016;24(1).6. [DOI] [PubMed] [Google Scholar]
- 11.Yu DD, Wu Y, Zhang XH, et al. Exosomes from adriamycin-resistant breast cancer cells transmit drug resistance partly by delivering miR-222. Tumor Biol. 2016;37(3):3227–3235. [DOI] [PubMed] [Google Scholar]
- 12.Chen WX, Cai YQ, Lv MM, et al. Exosomes from docetaxel-resistant breast cancer cells alter chemosensitivity by delivering microRNAs. Tumour Biol. 2014;35(10):9649–9659. [DOI] [PubMed] [Google Scholar]
- 13.Schiller JH, Harrington D, Belani CP, et al. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002;346(2):92–98. [DOI] [PubMed] [Google Scholar]
- 14.Farrell JJ, Elsaleh H, Garcia M, et al. Human equilibrative nucleoside transporter 1 levels predict response to gemcitabine in patients with pancreatic cancer. Gastroenterology. 2009;136(1):187–195. [DOI] [PubMed] [Google Scholar]
- 15.Spratlin J, Sangha R, Glubrecht D, et al. The absence of human equilibrative nucleoside transporter 1 is associated with reduced survival in patients with gemcitabine-treated pancreas adenocarcinoma. Clin Cancer Res. 2004;10(20):6956–6961. [DOI] [PubMed] [Google Scholar]
- 16.Marechal R, Bachet JB, Mackey JR, et al. Levels of gemcitabine transport and metabolism proteins predict survival times of patients treated with gemcitabine for pancreatic adenocarcinoma. Gastroenterology. 2012;143(3):664–674. [DOI] [PubMed] [Google Scholar]
- 17.Morinaga S, Nakamura Y, Watanabe T, et al. Immunohistochemical analysis of human equilibrative nucleoside transporter-1 (hENT1) predicts survival in resected pancreatic cancer patients treated with adjuvant gemcitabine monotherapy. Ann Surg Oncol. 2012;19(Suppl 3):S558–564. [DOI] [PubMed] [Google Scholar]
- 18.Greenhalf W, Ghaneh P, Neoptolemos JP, et al. Pancreatic cancer hENT1 expression and survival from gemcitabine in patients from the ESPAC-3 trial. J Natl Cancer Inst. 2014;106(1):djt347. [DOI] [PubMed] [Google Scholar]
- 19.Zhao Y, Srivastava D. A developmental view of microRNA function. Trends Biochem Sci. 2007;32(4):189–197. [DOI] [PubMed] [Google Scholar]
- 20.Holleman A, Chung I, Olsen RR, et al. miR-135a contributes to paclitaxel resistance in tumor cells both in vitro and in vivo. Oncogene. 2011;30(43):4386–4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao J, Fu W, Liao H, et al. The regulatory and predictive functions of miR-17 and miR-92 families on cisplatin resistance of non-small cell lung cancer. BMC Cancer. 2015;15:731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu S, Qin X, Chen T, Zhou L, Xu X, Feng J. MicroRNA-106b-5p regulates cisplatin chemosensitivity by targeting polycystic kidney disease-2 in non-small-cell lung cancer. Anticancer Drugs. 2017;28(8):852–860. [DOI] [PubMed] [Google Scholar]
- 23.Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120(5):635–647. [DOI] [PubMed] [Google Scholar]
- 24.Jin F, Wang Y, Li M, et al. MiR-26 enhances chemosensitivity and promotes apoptosis of hepatocellular carcinoma cells through inhibiting autophagy. Cell Death Dis. 2017;8(1):e2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Challagundla KB, Wise PM, Neviani P, et al. Exosome-mediated transfer of microRNAs within the tumor microenvironment and neuroblastoma resistance to chemotherapy. J Natl Cancer Inst. 2015;107(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lv MM, Zhu XY, Chen WX, et al. Exosomes mediate drug resistance transfer in MCF-7 breast cancer cells and a probable mechanism is delivery of P-glycoprotein. Tumour Biol. 2014;35(11):10773–10779. [DOI] [PubMed] [Google Scholar]
- 27.Wei F, Ma C, Zhou T, et al. Exosomes derived from gemcitabine-resistant cells transfer malignant phenotypic traits via delivery of miRNA-222-3p. Mol Cancer. 2017;16(1):132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gallo A, Tandon M, Alevizos I, Illei GG. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS One. 2012;7(3):e30679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Banzhaf-Strathmann J, Edbauer D. Good guy or bad guy: the opposing roles of microRNA 125b in cancer. Cell Commun Signal. 2014;12:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma Y, Li X, Cheng S, Wei W, Li Y. MicroRNA-106a confers cisplatin resistance in non-small cell lung cancer A549 cells by targeting adenosine triphosphatase-binding cassette A1. Mol Med Rep. 2015;11(1):625–632. [DOI] [PubMed] [Google Scholar]
- 31.Mohr AM, Mott JL. Overview of microRNA biology. Semin Liver Dis. 2015;35(1):3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitra R, Edmonds MD, Sun J, et al. Reproducible combinatorial regulatory networks elucidate novel oncogenic microRNAs in non-small cell lung cancer. RNA. 2014;20(9):1356–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vlachos IS, Paraskevopoulou MD, Karagkouni D, et al. DIANA-TarBase v7.0: indexing more than half a million experimentally supported miRNA: mRNAinteractions. Nucleic Acids Res. 2015;43(Database issue):D153–D159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paraskevopoulou MD, Georgakilas G, Kostoulas N, et al. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013;41(Web Server issue):W169–W173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang L, Zhang F, Wang X, et al. A FASN-TGF-β1-FASN regulatory loop contributes to high EMT/metastatic potential of cisplatin-resistant non-small cell lung cancer. Oncotarget. 2016;7(34):55543–55554. [DOI] [PMC free article] [PubMed] [Google Scholar]