Abstract
In the present work, loop-mediated isothermal amplification (LAMP) and hyperbranched rolling circle amplification (HRCA) methods were developed to detect and distinguish different lethal Amanita species. Specific LAMP primers and HRCA padlock probes for species-specific identification and a set of universal LAMP primers for lethal Amanita species were designed and tested. The results indicated that the LAMP-based assay was able to discriminate introclade lethal Amanita species but was not able to discriminate intraclade species perfectly, while the HRCA-based assay could discriminate whether introclade or intraclade species. The universal LAMP primers were positive for 10 lethal species of Amanita section Phalloideae and negative for 16 species of Amanita outside section Phalloideae. The detection limits of LMAP and HRCA were 10 and 1 pg of genomic DNA per reaction, respectively. In conclusion, the two methods could be rapid, specific, sensitive and low-cost tools for the identification of lethal Amanita species.
Keywords: loop-mediated isothermal amplification, hyperbranched rolling circle amplification, ITS sequence, lethal amanitas, padlock probe
Introduction
Mushroom poisoning is the main cause of mortality in food poisoning incidents in China. According to the National Management Information System of Public Health Emergency, in China, 576 mushroom poisoning events were reported from 2004 to 2014, with 3701 poisoning cases and 786 deaths; the fatality rate of mushroom poisoning accounted for 35.57% of total food poisoning (2210) deaths (Zhou et al., 2016). More than 90% of fatal mushroom poisoning cases were caused by mistaken ingestion of lethal amanitas in Europe, North America and East Asia (Enjalbert et al., 2002; Chen et al., 2014). Lethal amanitas are a group of cyclopeptide-containing mushrooms classified in genus Amanita section Phalloideae (Fr.) Quél. (Cai et al., 2014; Tang et al., 2016). There have been approximately 50 lethal Amanita species reported worldwide (Cai et al., 2016). These lethal Amanita species have four common morphologic characteristics as bases distinguished from other taxa of Amanita, including a non-appendiculate pileus, the persistent presence of an annulus, a bulbous stipe base with a limate volva and amyloid basidiospores (Cai et al., 2014). The containing substances of various peptide toxins were another critical characteristics of lethal amanitas and the peptide toxins in Amanita can be divided into three major groups, including amatoxins, phallotoxins and virotoxins, which are bicyclic octapeptides, bicyclic heptapeptides and monocyclic heptapeptides, respectively (Wieland, 1986). The primary toxins responsible for fatal human poisoning of these lethal Amanita species are amatoxins that induce acute liver failure through binding with eukaryotic DNA-dependent RNA polymerase II and subsequently inhibiting the elongation essential to transcription (Walton, 2018). In wild, some lethal Amanita species are similar to the edible species of section Caesareae Singer, for example, A. chepangiana (edible) vs. A. exitialis (lethal) and A. hemibapha (edible) vs. A. subjunquillea (lethal), this is the main reason for mistaken collection and ingestion.
Rapid identification of poisonous mushroom species eaten by patients is very important for toxic source investigation, clinical diagnosis and proper treatment. Hence, the establishment of rapid and effective methods for detection of lethal amanitas is urgently needed. To date, the identification and detection methods for lethal Amanita species mainly depend on their morphological and anatomical evidence, toxin analysis and molecular methods, such as PCR amplification and sequencing of DNA barcoding (Enjalbert et al., 1992; Epis et al., 2010; Harper et al., 2011). However, these methods were often time consuming and complicated and dependent on expensive equipment and professionals, which were difficult to implement in primary institutions or remote areas. Thus, development of simple, rapid and low-cost detection methods would be helpful in curing mushroom poisoning at its early onset as well as investigating the toxin.
In recent years, loop-mediated isothermal amplification (LAMP) and hyperbranched rolling circle amplification (HRCA) have been widely used for molecular detection and identification of pathogenic fungi (Niessen and Vogel, 2010; Tsui et al., 2010; Dai et al., 2012; Davari et al., 2012; Duan et al., 2014; Trilles et al., 2014), in view of their rapid and sensitive detection in addition to the wide range of detection strategies available and the ability of the technique to be deployed outside conventional laboratory settings. LAMP requires a set of four primers (FIP, BIP, F3, and B3) aimed at the six different specific regions of target DNA, and the reaction happens at a constant temperature (60–65°C) catalyzed by Bst DNA polymerase (Notomi et al., 2000). A vast number of products (109–1010-fold) with a dumbbell structure, which are formed by strand displacement of the outer and inner primers, are produced by cycle amplification. The reaction time is generally about an hour, but if loop primers are added, the time consumed will be shorten by half (Nagamine et al., 2002). Unlike LAMP, the HRCA employs a linear padlock probe that hybridizes with a target DNA and is then ligated by DNA ligase to form a circular probe, which subsequently serves as the template to proceed as a turn-by-turn cascade of multiple hybridization, primer extension, and strand displacement involving two primers under isothermal conditions and finally a >109-fold amplification of products is generated from the reaction (Nilsson et al., 1994; Lizardi et al., 1998).
To date, as far as we know, only one report has been published about the LAMP-based method for rapid mushroom species identification (Vaagt et al., 2013). In this paper, the LAMP assays were used for the rapid and easy detection of the death cap mushroom Amanita phalloides from closely related edible and toxic mushroom species. Because there have been many lethal Amanita species and similarities among these species, the aims of this study are (i) to develop LAMP and HRCA methods for species-specific identification of lethal Amanita species and (ii) to design specific but universal LAMP primers for identification of all lethal Amanita species.
Materials and Methods
Mushroom Samples and Identification
A total of 26 Amanita mushroom species were used in this study, and their information is listed in Table 1, including 10 lethal species in Amanita section Phalloideae (Supplementary Figure S1) and 16 species of Amanita outside section Phalloideae. Among them, the Amanita phalloides samples were provided by Professor Li TH (Guangdong Institute of Microbiology, China), which were collected from Lazio, Rome, Italy, in October 2014; the A. bisporigera samples were collected by Zhang from Hamilton, Canada, in August 2009; and the remaining 24 tested mushroom samples were collected from China. All the mushroom materials were identified by both morphological and molecular evidence (ITS sequence) following Zhang et al. (2010) and Cai et al. (2014). The samples determined in this study were deposited in Mycological Herbarium of Hunan Normal University (MHHNU) and Mycological Herbarium of Guangdong Institute of Microbiology (GDGM).
Table 1.
Mushroom samples used in this study.
| Amanita section | Species | Specimen no. | GenBank accession no. |
|---|---|---|---|
| Sect. Phalloideae | A. bisporigera | MHHNU 7224 | KU311692 |
| A. exitialis | MHHNU 30297 | KT003192 | |
| A. fuliginea | MHHNU 30944 | KU356798 | |
| A. pallidorosea | MHHNU 8112 | KU311697 | |
| A. phalloides | GDGM 41101 | KT003193 | |
| A. rimosa | MHHNU 7954 | KU311695 | |
| A. subfuliginea | MHHNU 8812 | MH142183 | |
| A. subjunquillea | MHHNU 7751 | KR996715 | |
| A. subpallidorosea | MHHNU 8617 | KU601411 | |
| A. virosa | MHHNU 8621 | KY472227 | |
| Sect. Amanita | A. rubrovolvata | MHHNU 8591 | KU356797 |
| A. rufoferruginea | MHHNU 30943 | KU497532 | |
| A. sinensis | MHHNU 8585 | KU497533 | |
| A. sychnopyramis | MHHNU 30253 | KU497534 | |
| Sect. Caesareae | A. javanica | MHHNU 30270 | KU497535 |
| Sect. Vaginatae | A. fulva | MHHNU 8550 | KU497536 |
| A. orientifulva | MHHNU 8580 | KU497537 | |
| A. vaginata | MHHNU 30266 | KU497538 | |
| Sect. Amidella | A. neoovoidea | MHHNU 30952 | KU497539 |
| Sect. Lepidella | A. kotohiraensis | MHHNU 30259 | KU497540 |
| A. oberwinklerana | MHHNU 30819 | KT003191 | |
| A. pseudoporphyria | MHHNU 30897 | KU497541 | |
| Sect. Validae | A. citrina | MHHNU 30252 | KU497542 |
| A. orsonii | MHHNU 8562 | KU497543 | |
| A. sepiacea | MHHNU 8474 | KU497544 | |
| A. spissacea | MHHNU 8472 | KU497545 |
DNA Extraction, PCR Amplification and Sequencing
Total genomic DNA was extracted by the Fungal DNA Mini Kit (OMEGA, United States) and then diluted to 10 ng/μL as a working concentration. The primers ITS 4 and ITS 5 were used for amplification of ITS sequences. The PCR mixtures contained 1 × PCR buffer, 1.5 mM MgCl2, 0.2 mM dNTPs, 0.4 μM of each primer, 1.25 U of Taq polymerase, and 1 μL DNA template in a total volume of 25 μL. PCR was performed with an Eppendorf Mastercycler thermal cycler (Eppendorf Inc., Germany) as follows: initial denaturation at 94°C for 4 min, followed by 30 cycles of 94°C for 30 s, 54°C for 30 s, 72°C for 30 s, and a final extension at 72°C for 8 min. Amplified PCR products were detected by gel electrophoresis on a 1% agarose gel and then sent to Tsingke Biological Technology (China) for sequencing.
Phylogenetic Tree Building
Ten ITS sequences of lethal amanitas were obtained by sequencing in this study, and twenty-seven ITS sequences from GenBank were aligned by Clustal X 2.0 software (Larkin et al., 2007). Then, the alignment data of these sequences were used to construct a maximum likelihood phylogeny tree with 1000 bootstrap replicates using MEGA 6.0 software (Tamura et al., 2013).
Primers and Padlock Probes Design
ITS sequences were chosen as the candidate targets for LAMP primer and padlock probe (PLP) design. For the design, 27 ITS sequences of A. bisporigera, A. exitialis, A. fuliginea, A. pallidorosea, A. phalloides, A. rimosa, A. subfuliginea, A. subjunquillea, A. subpallidorosea and A. virosa were downloaded from NCBI GenBank and were compared and aligned using DNAMAN 7.0 software to find different target recognition regions for each species and to identify informative nucleotide polymorphic sites conserved within a single species but divergent among different species.
The ten sets of specific LAMP primers were designed by using PrimerExplorer V51. In addition, a set of universal primers for lethal amanitas were manually designed based on the multiple alignment of thirty-six published ITS sequences of fifteen lethal Amanita species. A forward inner primer (FIP) consisted of the complementary sequence of F1c and F2, a backward inner primer (BIP) consisted of B1c and B2, two outer primers (F3 and B3). Loop primers (LF or LB) were used for LAMP, and the structure of the universal primers and their complementarity to target DNA are exemplified in Figure 1.
FIGURE 1.
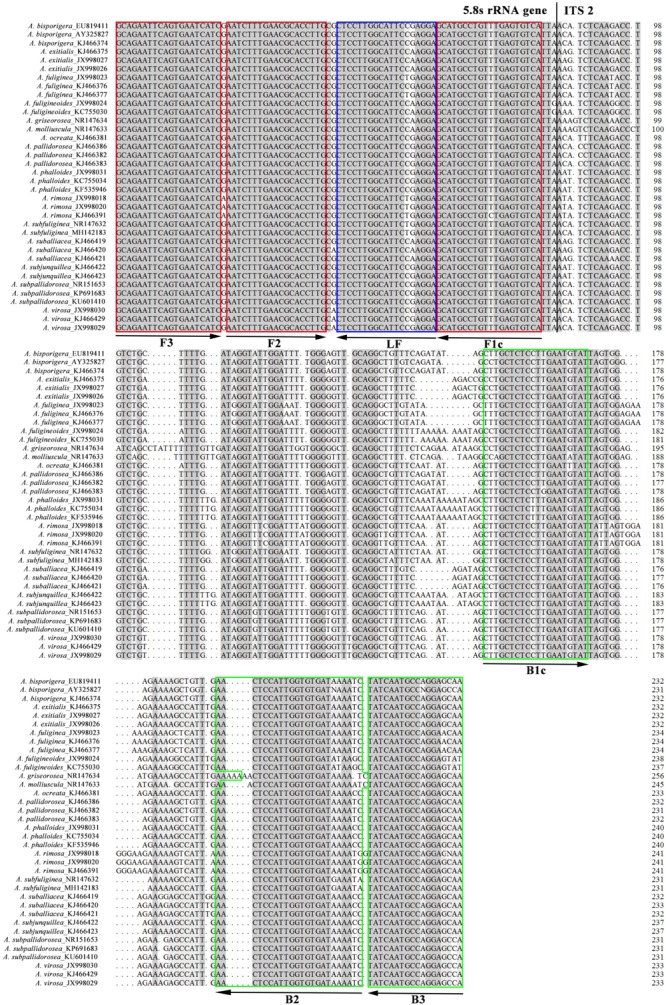
Multiple alignment of ITS sequences of fifteen lethal Amanita species. The target regions used for designing the universal primers were labeled with boxes and arrows, and the aligned sequences were partial 5.8S ribosomal RNA gene and internal transcribed spacer 2.
The ten specific PLPs were designed according to criteria as previously described by Kaocharoen et al. (2008) and Lackner et al. (2012). The PLP consists of two terminal regions complementary to a target sequence located at both ends and a linker region in the middle, which was a partial sequence of the inactive X specific transcripts (Xist) gene of Mus musculus but lacked homology for the target genes (Figure 2). To ensure the efficiency of padlock probe binding, the padlock probes were predicted with MFOLD to ensure the minimal secondary structure and were designed with the 5′-end probe binding arm Tm (62–66°C) close to or above the ligation temperature (65°C in this study, see below). To increase 3′-end binding specificity, the 3′-end probe binding arm was designed with a Tm (45–48°C) 10–15°C below ligation temperature. The 5′ terminal end of the PLP was modified by phosphorylation to allow ligation. In addition, the HRCA primers (HRCA-primer 1, 2), which are used to amplify the specific padlock probe signal during HRCA, were specifically designed to bind to the flanking linker regions of the above-designed padlock probes (Figure 2).
FIGURE 2.
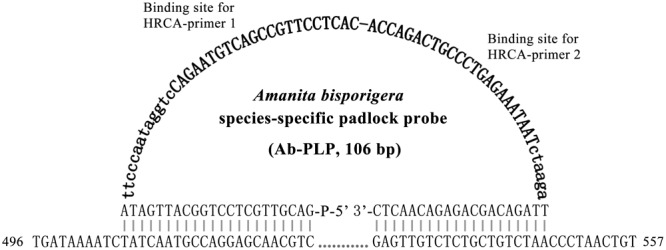
Design of a padlock probe exemplified by the Amanita bisporigera probe (Ab-PLP). Linker regions are in bold letters, and HRCA primer binding sites are in bold capital letters. Species-specific sites of the HRCA padlock probe were plain and capitalized. The line of dots indicates connection of adjacent bases, and the numbers 496 and 557 indicate the positions in the ITS region of A. bisporigera.
The primers (PAGE) and PLPs (HPLC) were synthesized by Tsingke Biological Technology (China), and their detailed sequences and lengths are shown in Table 2.
Table 2.
The information of primers and padlocks used for amplification.
| Name | Type | Sequence (5′→3′) | Length (bp) |
|---|---|---|---|
| Ab-primers | F3 | GAGGAGCATGCCTGTTTG | 18 |
| B3 | GGTCAGACAGTTAGGGTTAG | 20 | |
| FIP | CCTGCAACTCCCAAAATCCAtaatGTCATTAACATCTCAAGACCTG | 46 | |
| BIP | TTAGTGGAGAAAAGCTGTTGAACTCtataACAGCAGAGACAACTCGACG | 49 | |
| LB | AAATCTATCAATGCCAGGAGCAA | 23 | |
| Ae-primers | F3 | AATCTTTGAACGCACCTTG | 19 |
| B3 | GACAGTTAGACAGCAGAGA | 19 | |
| FIP | ACCCCCAAAATCCAATACCTATCAAtaatGAGCATGCCTGTTTGAGT | 47 | |
| BIP | GTGGAGAAAAAGCCATTTGAACTCCtataAACTAGCATTGCTCCTGG | 47 | |
| LF | TCAGACAGGTCTTGAGACTTTAATG | 25 | |
| Af-primers | F3 | TGCCTGTTTGAGTGTCATT | 19 |
| B3 | GGACATGGATTAGACAGCA | 19 | |
| FIP | GCTATACAAGCCCTGCAACCtaatAACATCTCAATACCTGTCTGC | 45 | |
| BIP | AGAAAGCTCATTGAACTCCATTGGtataAACTTGGACATTGTTCCTGG | 48 | |
| LF | CCCAATTTCCAATACCCATCAA | 22 | |
| Apa-primers | F3 | CATGCCTGTTTGAGTGTC | 18 |
| B3 | GTCAAGTGGGTCAGACAG | 18 | |
| FIP | GAAACAGCCTGCAACTCCCAtaatATTAACACCTCAAGACCTGTC | 45 | |
| BIP | TTAGTGGAGAAAAGCTGTTGAACTCtataGCAGAGACCACTTGATGTT | 48 | |
| LB | GATAAAATCTATCAATGCCAGGAGC | 25 | |
| Aph-primers | F3 | GCCTTGCTCTCTTTGAATGT | 20 |
| B3 | GATATGCTTAAGTTCAGCGG | 20 | |
| FIP | AGTGATATTGCTCCTGGCATTGtaatGAGAAAAGCCATTGAACTCC | 46 | |
| BIP | TGCTGTCTAACTGTGACTGTCTtataTAGTCCTACCTGATTTGAGGT | 47 | |
| LB | TGGATGGGGACAACTTGACC | 20 | |
| Ar-primers | F3 | TGCCTGTTTGAGTGTCATT | 19 |
| B3 | AGTTGGTCAAGTTGTCCAT | 19 | |
| FIP | CATTCAAGGAGAGCAAGCTATTTGAtaatGTCTGCTTTTGATAGGTTTCG | 50 | |
| BIP | GGTGTGATAAAATGGTATCAATGCCtataCTACAGACAGTTAGCTGAGA | 49 | |
| LF | CAACCTGCAACCCCCATAAATC | 22 | |
| Asubf-primers | F3 | TGCCTGTTTGAGTGTCATT | 19 |
| B3 | GGGTTAGACAGCAGAGAGA | 19 | |
| FIP | GCAAGCCATTTAGAAATAGCCTGCtaatGACCTGTCTGCTTTTGGA | 46 | |
| BIP | TTAGTGGAAAAAGCCATTGAACTCCtataGCTGATCATTGCTCCTGG | 47 | |
| LF | ACCCCCAAATTCCAATACCCA | 21 | |
| Asubj-primers | F3 | GCCTTGCTCTCCTTGAATGT | 20 |
| B3 | GATATGCTTAAGTTCAGCGG | 20 | |
| FIP | GTAGTGATATTGCTCCTGGCATtaatGAAAAGCCATTGAACTCCAT | 46 | |
| BIP | TGCTGTCTAACTGTGACTGTCTtataTAGTCCTACCTGATTTGAGGT | 47 | |
| LB | GGATGGGGACAACTTGACCAAC | 22 | |
| Asubp-primers | F3 | GATTTTTGGGTGTTGCA | 17 |
| B3 | AGGTCAAGTTGGTCAAGTT | 19 | |
| FIP | CCAATGGAGTTCAATGGCTCTTCtaatGCTTTTTCAGATAGCTTGCT | 47 | |
| BIP | TCTATCAATGCCAGGAGCCtataTTACAGACAACTGTGAGA | 41 | |
| LB | ATGTTAGTTCTCTCTGCTGTC | 21 | |
| Av-primers | F3 | CATCTCAAGACCTGTCTGTT | 20 |
| B3 | AGTTGGTCAAGTTGTCCAT | 19 | |
| FIP | GAGTTCAATGGCTCTTTCTCCACTAtaatGGATTTTTGGGGGTTTGC | 47 | |
| BIP | TCTATCAATGCCCAGGAGCCtataAGACAACTGTTAGCGGTTAG | 44 | |
| LF | CATTCAAGGAGAGCAAGCTATCTG | 24 | |
| Universal primers | F3 | GCAGAATTCAGTGAATCATC | 20 |
| B3 | TTGCTCCTGGCATTGATA | 18 | |
| FIP | TGACACTCAAACAGGCATGCtaatAATCTTTGAACGCACCTTG | 43 | |
| BIP | CTTGCTCTCCTTGAATGTATTAGTtataGATTTTATCACACCAATGGAGTT | 51 | |
| LF | TCCTCGGAATGCCAAGGAG | 19 | |
| Ab-PLP | P-GACGTTGCTCCTGGCATTGATATTCCCAATAGGTCCAGAATGTCAGCCGTTCCT CACACCAGACTGCCCTGAGAAATAATCTAAGATTAGACAGCAGAGACAACTC | 106 | |
| Ae-PLP | P-CATTGCTCCTGGCATTGATAGATTTTCCCAATAGGTCCAGAATGTCAGCCGTTCCTC ACACCAGACTGCCCTGAGAAATAATCTAAGAGTTAGACAGCAGAGATAACTAG | 110 | |
| Af-PLP | P-CCTGCAACCCCCAATTTCCATTCCCAATAGGTCCAGAATGTCAGCCGTTCC TCACACCAGACTGCCCTGAGAAATAATC TAAGAAGAGCAAAGCTATACAAGC | 103 | |
| Apa-PLP | P-ACTTGATGTTGCTCCTGGCATTGATTTCCCAATAGGTCCAGAATGTCAGCCGTTCCT CACACCAGACTGCCCTGAGAAATAATCTAAGAGGTTAGACAGCAGAGACC | 107 | |
| Aph-PLP | P-TTATTTGAAACAGCCTGCAACCCTTCCCAATAGGTCCAGAATGTCA GCCGTTCCTCACACCAGACTGCCCTGAGAAATAATCTAAGAAAGAGAGCAAGGCTATTT | 105 | |
| Ar-PLP | P-TTGTTCATTGCTCCTGGCATTGATATTCCCAATAGGTCCAGAATGTCAGCCGTTCCT CACACCAGACTGCCCTGAGAAATAATCTAAGAACAGTTAGCTGAGAGAACTG | 109 | |
| Asubf-PLP | P-TCAAGAGACCAGTCAAAAGTCTCTCATTTCCCAATAGGTCCAGA ATGTCAGCCGTTCCTCACACCAGACTGCCCTGAGAAATA ATCTAAGAGATTCCAATTCAAATCAAT | 110 | |
| Asubj-PLP | P-AGTGATATTGCTCCTGGCATTGATATTCCCAATAGGTCCAGAATGTCAGCCGTTCCTCACA CCAGACTGCCCTGAGAAATAATCTAAGAGTTAGACAGCAGAGAGAAGT | 109 | |
| Asubp-PLP | P-GTTAGACAGCAGAGAGAACTAACATGGCTTCCCAATAGGTCCAG AATGTCAGCCGTTCCTCACACCAGACTGCCCTGAGAAATA ATCTAAGATTTTACAGACAACTGTGAGA | 112 | |
| Av-PLP | P-GTTAGACAGCAGAGAGAACTAACATGGCTTCCCAATAGGTCCAGAATGTCAGCCGTT CCTCACACCAG ACTGCCCTGAGAAATAATCTAAGATTACAGACAACTGTTAGCG | 111 | |
| HRCA-primer 1 | GTGAGGAACGGCTGACATTCTG | 22 | |
| HRCA-primer 2 | ACCAGACTGCCCTGAGAAATAAT | 23 |
HRCA, hyperbranched rolling circle amplification; PLP, padlock probe; Ab, A. bisporigera; Ae, A. exitialis; Af, A. fuliginea; Apa, A. pallidorosea; Aph, A. phalloides; Ar, A. rimosa; Asubf, A. subfuliginea; Asubj, A. subjunquillea; Ab, A. subpallidorosea; Av, A. virosa. taat, tata were the spacers between F1c and F2, B1c and B2, respectively. P represents phosphorylation and bold represents the 5′ and 3′ arms of the PLP matched to the target region.
LAMP Reaction and Product Detection
The LAMP reaction was carried out in 10 μL reaction mixtures: 1 × ThermoPol buffer (20 mM Tris–HCl, 10 mM (NH4)2SO4, 10 mM KCl, 2 mM MgSO4, 0.1% Triton X-100, PH 8.8), 4 mM MgSO4, 1.4 mM dNTP mix, 1.28 μM FIP, 1.28 μM BIP, 0.12 μM F3, 0.12 μM B3, 3.2 U Bst DNA polymerase (NEB, United States), 1 μL DNA template (10 ng), 150 μM HNB, and ddH2O to 10 μL. The reaction was performed in a 0.2 mL tube with a water bath incubated at 62°C for 60 min and finally 80°C for 10 min to termination.
Two approaches were used to analyze DNA amplification, including direct visual inspection of the color of the LAMP mixture with HNB dye and 2% agarose gel electrophoresis.
HRCA Reaction and Product Detection
The ligation was carried out in a 10 μL mixture containing: 1 × Taq DNA ligase buffer (20 mM Tris–HCl, 25 mM KAc, 10 mM Mg(Ac)2, 10 mM DTT, 1 mM NAD, 0.1% Triton X-100), 10 pM linear padlock probe, 12 U of Taq DNA ligase (NEB, United States) and 1 μL of DNA template (10 ng). The ligation mixture was incubated at 65°C for 1 h.
After ligation, 1 μL of ligation product was added into an HRCA reaction mixture containing 1 × ThermoPol buffer (20 mM Tris–HCl, 10 mM (NH4)2SO4, 10 mM KCl, 2 mM MgSO4, 0.1% Triton X-100), 0.4 mM dNTP mix, 0.5 μM of each HRCA primers and 1.6 U Bst DNA polymerase (NEB, United States) with a total 10 μL volume. The reaction was performed in a 0.2 mL tube in a water bath incubated at 62°C for 60 min.
The results were judged by the appearance of color after adding 1 μl of 1000 × SYBR Green I dye to the system after the reaction or 2% agarose gel electrophoresis of the HRCA product.
Specificity of LAMP Primers and HRCA PLPs
To test the specificity of the ten sets of specific LAMP primers and HRCA PLPs designed above, genomic DNA extracted from A. bisporigera, A. exitialis, A. fuliginea, A. pallidorosea, A. phalloides, A. rimosa, A. subfuliginea, A. subjunquillea, A. subpallidorosea, and A. virosa was used for cross reaction testing.
For the specificity of the universal primers, genomic DNA from twenty-six Amanita species listed in Table 1 was tested by the LAMP method.
Sensitivity of LAMP and HRCA
To determine the detection limit, the LAMP and HRCA assays were performed using a 10-fold dilution series of genomic DNA from A. fuliginea ranging from 10 ng to 10 fg.
Results
Specificity of LAMP and HRCA
Genomic DNA from ten lethal Amanita mushrooms was used to test the specificity of the corresponding sets of specific LAMP primers and specific HRCA PLPs.
As shown in Figure 3A, the LAMP reactions were analyzed by HNB dye staining and agarose gel electrophoresis. The results of the two detection methods were consistent. Positive reactions were observed with a sky-blue mixture and typical ladder-like banding, whereas for the negative reactions, the color of the tubes remained violet, and no bands were detected after electrophoresis. The six primer sets, Ae-primers, Af-primers Ar-primers, Asubf-primers, Asubp-primers and Av-primers, could clearly recognize and distinguish the expected Amanita species. However, cross reaction occurred between the Ab-primers and Apa-primers and the Aph-primers and Asubj-primers.
FIGURE 3.
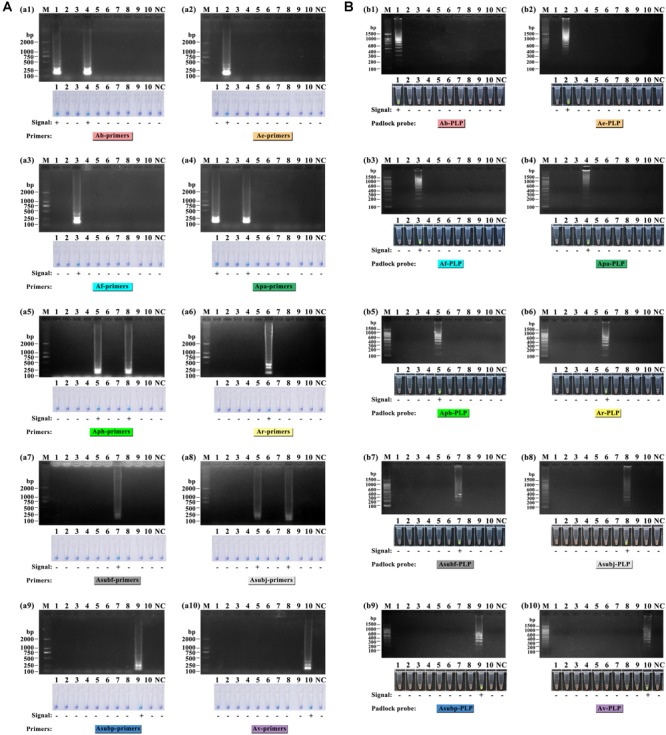
Specificity test of the ten sets of LAMP primers (A) and ten HRCA padlock probes (B) for lethal amanitas. M: DL2000 or 100 bp ladder, 1: A. bisporigera, 2: A. exitialis, 3: A. fuliginea, 4: A. pallidorosea, 5: A. phalloides, 6: A. rimosa, 7: A. subfuliginea, 8: A. subjunquillea, 9: A. subpallidorosea, 10: A. virosa, NC: negative control.
For the HRCA, amplification products were detected by SYBR Green I dye staining and agarose gel electrophoresis. Positive HRCA results generated a typical ladder-like pattern of fragments increasing in size, comprising the monomer and multimer repeats of the amplified product formed by single and multiple copies of the circularized padlock probe, while negative reactions had a clean background. The HRCA signal was also determined by adding SYBR Green I dye after the reactions; positive reactions turned green while negative reactions remained orange. From Figure 3B, it could be seen that the probes could specifically detect their corresponding targets, and no false-positive reaction was observed. The results from analysis with SYBR Green I dye were compatible with those obtained with electrophoresis.
In addition, the phylogenetic relationship of the lethal Amanita species based on ITS sequences was analyzed, and the resulting tree (Figure 4) strongly resolved the examined taxa into seven clades comprising ten phylogenetic species. These results are consistent with the previous results of Cai et al. (2014). A. exitialis, A. fuliginea, A. rimosa, and A. subfuliginea formed a clade alone with 99 or 100% bootstrap percentages, while A. bisporigera and A. pallidorosea, A. phalloides and A. subjunquillea, and A. subpallidorosea and A. virosa formed a clade but were classified into two branches with 99, 98, and 96% bootstrap, respectively. By combining the tree and the amplification signals above, it could be intuitively found that LAMP was capable of discriminating interclade lethal Amanita species but could not perfectly discriminate the intraclade species (Clade 1 and 5 failed, Clade 3 succeeded); however, HRCA could discriminate intraclade species well. Hence, it could be concluded that the specificity of HRCA was clearly higher than LAMP.
FIGURE 4.
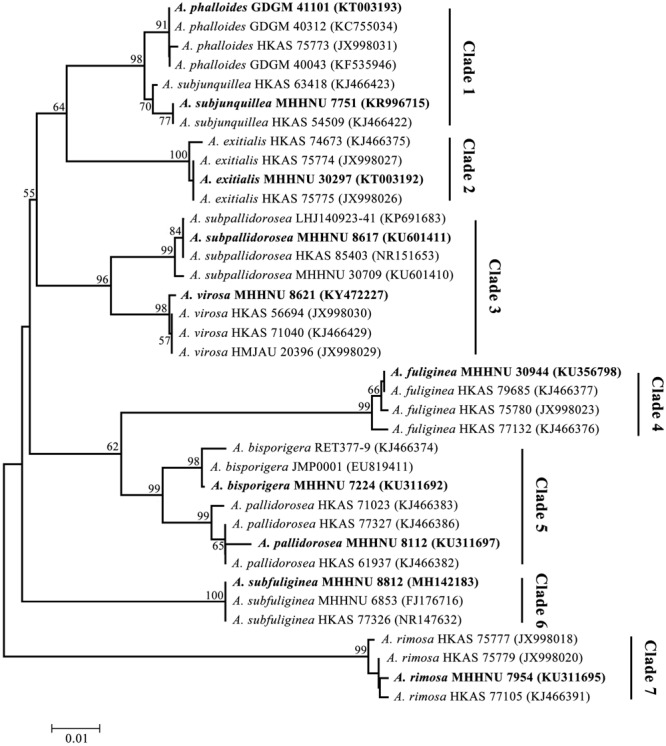
Phylogenetic tree generated from maximum likelihood analysis based on ITS sequences. Bootstrap percentages (>50%) based on 1000 replications are shown at nodes. Bar, a substitution per 100 nucleotides. Sequences in bold were obtained in this study, and the others were from NCBI GenBank.
Evaluation of Universal LAMP Primers
To verify the specificity and universality of the universal primers for lethal amanitas, the LAMP reactions were carried out with genomic DNA extracted from 10 lethal species from Amanita section Phalloideae and 16 species of Amanita outside section Phalloideae. As shown in Figure 5, the result showed that positive LAMP reaction occurred only in lethal Amanita species, while the other species were negative.
FIGURE 5.
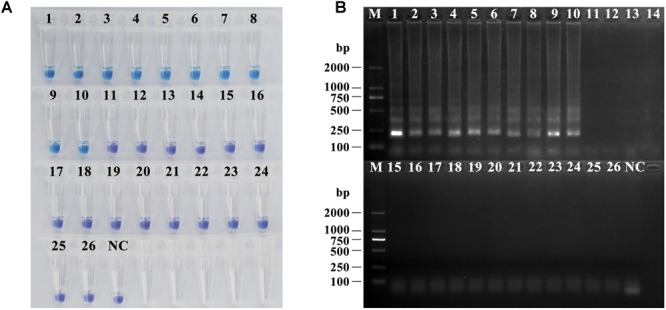
Specificity and universality tests of the universal primers for lethal amanitas. (A) Coloration of LAMP by adding HNB dye; (B) Electrophoresis analysis of LAMP-amplified products. M: DL2000, 1: A. bisporigera, 2: A. exitialis, 3: A. fuliginea, 4: A. pallidorosea, 5: A. phalloides, 6: A. rimosa, 7: A. subfuliginea, 8: A. subjunquillea, 9: A. subpallidorosea, 10: A. virosa, 11: Amanita rubrovolvata, 12: A. rufoferruginea, 13: A. sinensis, 14: A. sychnopyramis, 15: A. javanica, 16: A. fulva, 17: A. orientifulva, 18: A. vaginata, 19: A. neoovoidea, 20: A. kotohiraensis, 21: A. oberwinklerana, 22: A. pseudoporphyria, 23: A. citrina 24: A. orsonii, 25: A. sepiacea, 26: A. spissacea, NC: negative control.
Sensitivity of LAMP and HRCA
To determine the detection limit, the LAMP reactions were performed using a serial 10-fold dilution ranging from 10 ng to 10 fg of DNA template of A. fuliginea. The detection limit of LAMP and HRCA were 10 pg and 1 pg per reaction, respectively (Figure 6). These results suggested that the detection sensitivity of HRCA was ten times higher than that of LAMP.
FIGURE 6.
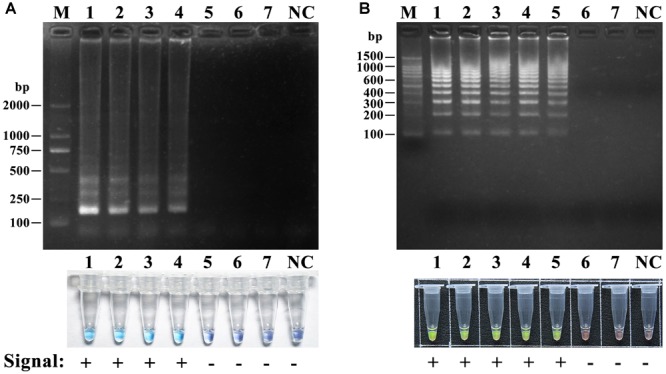
Sensitivity of the LAMP assay (A) and the HRCA (B) assay for A. fuliginea. M: DL 2000; NC, negative control. A dilution series of A. fuliginea DNA was as follows: 1, 10 ng; 2, 1 ng; 3, 100 pg, 4, 10 pg; 5, 1 pg; 6, 100 fg; 7, 10 fg.
Discussion
In the last 10 years, molecular detection based on ITS sequence has provided a promising alternative strategy for the identification of poisonous Amanita species; the ITS sequences could be used as a DNA barcode marker for lethal amanitas (Zhang et al., 2010; Cai et al., 2014). The phylogenetic analysis of the ITS data showed that lethal amanitas (Amanita section Phalloideae) were robustly supported as a monophyletic group, in which twenty-eight phylogenetic species were divided into nine major clades (Cai et al., 2014). Furthermore, these important molecular characteristics provide a great opportunity for us to design specific or universal primers for the rapid identification of lethal amanitas based on isothermal amplification methods. Vaagt et al. (2013) designed a set of LAMP primers based on the ITS sequence for the specific detection of death cap A. phalloides. The limited number of species of Amanita in the institute collection did not represent all species of Amanita; the related Amanita species tested in our study, such as A. muscaria, A. citrina, A. pantherina, and A. rubescens, are species outside section Phalloideae. In our present study, we endeavored to develop a series of species-specific LAMP primers capable of distinguishing each lethal amanitas within section Phalloideae. The results showed that the LAMP-based method could distinguish available interclade Amanita species but mostly failed to distinguish intraclade Amanita species. Some lethal Amanita species are very closely evolutionarily related based on small variations in ITS sequences, which are highly similar and identical (Zhang et al., 2010; Cai et al., 2014, 2016). For example, for A. bisporigera and A. pallidorosea, their ITS are almost the same, with a 98% identity, and they are in the same clade but classified into two branches in the phylogenetic tree (Figure 4), which indicated that LAMP has a certain limitation, and the specificity of the method is not applicable for highly identical templates. Indeed, it was reported that SNP-LAMP was developed to detect allele specific detection or single nucleotide polymorphisms (Fukuta et al., 2006; Ayukawa et al., 2017; Yongkiettrakul et al., 2017). However, it should be noted that some objective factors, such as the base composition of the target template, SNP distribution and amount, melting temperature and GC content of the primer, could affect the final result of SNP-LAMP detection. As reported, Yongkiettrakul et al. (2017) failed to distinguished between the wild-type and quadruple mutant dhfr gene of Plasmodium falciparum by SNP-LAMP. In our study, many attempts were made to design SNP-LAMP primers for intraclade Amanita species; however, only A. subpallidorosea and A. virosa (Clade3) were distinguished successfully, and the other two intraclade Amanita species, A. bisporigera and A. pallidorosea (Clade5) and A. phalloides and A. subjunquillea (Clade1) were not distinguished.
It has been reported hyperbranched rolling cycle amplification coupled with PLP was a particularly useful tool to discriminate closely related species and even subtypes of species with minimal nucleotide polymorphisms (Tong et al., 2007; Najafzadeh et al., 2013; Lin et al., 2018). Therefore, 10 specific PLPs were subsequently designed for each of the 10 lethal amanitas in our present experiment. The results suggested that the HRCA-based assay was able to determine whether each species of 10 lethal amanitas was interclade or intraclade. Notably, A. bisporigera was clearly distinguished from A. pallidorosea, and A. phalloides was also clearly distinguished from A. subpallidorosea by HRCA. Even though these two pairs failed to be distinguished by LAMP, these results indicated that HRCA had a higher specificity than LAMP. The high specificity of HRCA resulted from the single base recognition capability of the PLP, which is sensitive to mismatches between the probe and the target (Pickering et al., 2002; Szemes et al., 2005). It was confirmed that mismatches positioned at the 3′ end of PLP were strongly discriminating (Pickering et al., 2002; Szemes et al., 2005), which confers definite and informative target sites for detection. Next, increasing the hybridization temperature and shortening the 3′ arm of the PLP with melting temperature below the ligation temperature are considered to further improve specificity (Faruqi et al., 2001; van Doorn et al., 2007). According these rules, the PLPs designed in our study were preferred with more discriminating bases in the 3′ terminal and short 3′ arms, which induce extremely high specificity.
Furthermore, to distinguish the lethal Amanita species in section Phalloideae from the other Amanita species outside of section Phalloideae, a set of universal primers was designed based on the multiple alignment of thirty-six published ITS sequences of fifteen lethal Amanita species. The results showed that a positive LAMP reaction occurred only in lethal Amanita species, while the rest were negative, which indicated this LAMP method could distinguish the lethal Amanita species from the other Amanita species outside of section Phalloideae. Because these lethal Amanita species account for over 90% of all fatal mushroom poisonings worldwide, amatoxins are the common chemical property of these Amanita species, which induce acute liver failure (Walton, 2018). In the treatment of clinical poisoning, it is sometimes more important to determine the nature of the species than to determine the accurate species; in this case, this universal LAMP method could be used to rapidly determine whether the species is lethal.
Loop-mediated isothermal amplification and HRCA possess a sensitivity advantage that is 10–100 times higher than conventional PCR (Wang et al., 2012, 2015). Our results showed that the detection limits of the two methods could be at the pg level for the mushroom genomic DNA, and the sensitivity of HRCA was 10 times higher than that of LAMP, which was consistent with Wang et al. (2012). Compared to HRCA, the LAMP detection test was more rapid and simple, where white precipitate was generated by the naked eye within approximately 1 h. However, this technique had a very high risk for contamination, and the precipitate was inconveniently observational. Therefore, hydroxyl naphthol blue (HNB) was used as an indicative dye for the LAMP reaction in this study. When HNB was added before the reaction, a positive reaction will produce large amounts of magnesium pyrophosphate precipitate, thus producing Mg2+ and a pH change, and the color of the reaction solution change from violet into blue, so the result is easy to observe, and it does not cause aerosol pollution without opening the tube (Goto et al., 2009). In contrast, HRCA was relatively complicated and needed hours for completion for the extra PLP ligation and exonucleolysis steps. Nevertheless, we proved that exonucleolysis could be omitted because background signals caused by linear probes were almost invisible and insusceptible (data not shown), as was also found by Lackner et al. (2012) and Lin et al. (2018). Thus, the exclusion of an exonuclease reaction shortened the procedure by at least 2 h, and HRCA detection could be completed within 2 h (an hour for PLP cyclization and another hour for amplification) in our study. Despite more reagents and procedures, HRCA is more specific and sensitive than LAMP as described earlier. PCR amplification and sequencing of the ITS is the gold standard for mushroom species identification. However, compared with the two isothermal amplification methods above PCR-based method requires the expensive instrument for thermal cycling and extra time and cost for gel electrophoresis and sequencing and the species identification period using sequencing of ITS usually takes 1–2 working day. But for LAMP and HRCA, the identification only requires a water bath for the reaction and the detection can be completed and judged by dye staining within several hours. Therefore, LAMP and HRCA detection are rapider and require lower cost than PCR.
In conclusion, the LAMP and HRCA-based assays established in this study provided rapid, specific, sensitive and cost-effective tools for the detection and identification of lethal amanitas.
Data Availability
All datasets generated for this study are included in the manuscript and/or the Supplementary Files.
Author Contributions
ZC conceived and designed the experiments. ZH, YS, and SL carried out the LAMP and HRCA assay. PL carried out the analysis of ITS DNA sequences of all species and phylogenetic tree building. PZ provided some Amanita materials and identified the species. ZH and ZC wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This study was supported by the National Natural Science Foundation of China (Grant No. 31872616), Hunan Provincial Construct Program of the Key Discipline in Ecology, China (Grant No. 0713), and Hunan Provincial Cooperative Innovation Center of Engineering and New Products for Developmental Biology, China (Grant No. 20134486).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01523/full#supplementary-material
Basidiomata of nine lethal Amanita species. (A) A. bisporigera (MHHNU 7224); (B) A. exitialis (MHHNU 30297); (C) A. fuliginea (MHHNU 30944); (D) A. pallidorosea (MHHNU 8112); (E) A. rimosa (MHHNU 7954); (F) A. subfuliginea (MHHNU 8812); (G) A. subjunquillea (MHHNU 7751); (H) A. subpallidorosea (MHHNU 8617); (I) A. virosa (MHHNU 8621; photos A, D, E, F, G, and H by PZ; photos B, C, and I by ZC).
References
- Ayukawa Y., Hanyuda S., Fujita N., Komatsu K., Arie T. (2017). Novel loop-mediated isothermal amplification (LAMP) assay with a universal QProbe can detect SNPs determining races in plant pathogenic fungi. Sci. Rep. 7:4253. 10.1038/s41598-017-04084-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q., Cui Y. Y., Yang Z. L. (2016). Lethal Amanita species in China. Mycologia 108 993–1009. 10.3852/16-008 [DOI] [PubMed] [Google Scholar]
- Cai Q., Tulloss R. E., Tang L. P., Tolgor B., Zhang P., Chen Z. H., et al. (2014). Multi-locus phylogeny of lethal amanitas: implications for species diversity and historical biogeography. BMC Evol. Biol. 14:143. 10.1186/1471-2148-14-143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. H., Zhang P., Zhang Z. G. (2014). Investigation and analysis of 102 mushroom poisoning cases in Southern China from 1994 to 2012. Fungal Divers. 64 123–131. 10.1007/s13225-013-0260-7 [DOI] [Google Scholar]
- Dai T. T., Lu C. C., Lu J., Dong S., Ye W., Wang Y., et al. (2012). Development of a loop-mediated isothermal amplification assay for detection of Phytophthora sojae. FEMS Microbiol. Lett. 334 27–34. 10.1111/j.1574-6968.2012.02619.x [DOI] [PubMed] [Google Scholar]
- Davari M., Diepeningen A. D. V., Babai-Ahari A., Arzanlou M., Najafzadeh M. J., Lee T. A., et al. (2012). Rapid identification of Fusarium graminearum species complex using rolling circle amplification (RCA). J. Microbiol. Meth. 89 63–70. 10.1016/j.mimet.2012.01.017 [DOI] [PubMed] [Google Scholar]
- Duan Y. B., Ge C. Y., Zhang X. K., Wang J. X., Zhou M. G. (2014). Development and evaluation of a novel and rapid detection assay for Botrytis cinerea based on loop-mediated isothermal amplification. PLoS One 9:e111094. 10.1371/journal.pone.0111094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjalbert F., Gallion C., Jehl F., Monteil H., Faulstich H. (1992). Simultaneous assay for amatoxins and phallotoxins in Amanita phalloides Fr. by high-performance liquid chromatography. J. Chromatogr. 598 227–236. 10.1016/0021-9673(92)85052-U [DOI] [PubMed] [Google Scholar]
- Enjalbert F., Rapior S., Nouguier-Soulé J., Guillon S., Amouroux N., Cabot C. (2002). Treatment of amatoxin poisoning: 20-year retrospective analysis. J. Toxicol. clin. Toxic. 40 715–757. 10.1081/CLT-120014646 [DOI] [PubMed] [Google Scholar]
- Epis S., Matinato C., Gentili G., Varotto F., Bandi C., Sassera D. (2010). Molecular detection of poisonous mushrooms in different matrices. Mycologia 102 747–754. 10.3852/09-124 [DOI] [PubMed] [Google Scholar]
- Faruqi A. F., Hosono S., Driscoll M. D., Dean F. B., Alsmadi O., Bandaru R., et al. (2001). High-throughput genotyping of single nucleotide polymorphisms with rolling circle amplification. BMC Genomics 2:4 10.1186/1471-2164-2-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuta S., Mizukami Y., Ishida A., Kanbe M. (2006). Development of loop-mediated isothermal amplification (LAMP)-based SNP markers for shelf-life in melon (Cucumis melo L.). J. Appl. Genet. 47 303–308. 10.1007/BF03194639 [DOI] [PubMed] [Google Scholar]
- Goto M., Honda E., Oqura A., Nomoto A., Hanaki K. (2009). Clorimetric detection of loop-mediated isothermal amplification reaction by using hydroxy naphthol blue. Biotechniques 46 167–172. 10.2144/000113072 [DOI] [PubMed] [Google Scholar]
- Harper K. A., Smart C. D., Davis R. M. (2011). Development of a DNA-based macroarray for the detection and identification of Amanita species. J. Forensic Sci. 56 1003–1009. 10.1111/j.1556-4029.2011.01739.x [DOI] [PubMed] [Google Scholar]
- Kaocharoen S., Wang B., Tsui K. M., Trilles L., Kong F., Meyer W. (2008). Hyperbranched rolling circle amplification as a rapid and sensitive method for species identification within the Cryptococcus species complex. Electrophoresis 29 3183–3191. 10.1002/elps.200700903 [DOI] [PubMed] [Google Scholar]
- Lackner M., Najafzadeh M. J., Sun J., Lu Q., de Hoog G. S. (2012). Rapid identification of Pseudallescheria and Scedosporium strains by using rolling circle amplification. Appl. Environ. Microb. 78 126–133. 10.1128/AEM.05280-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M. A., Blackshields G., Brown N. P., Chenna R., Mcgettigan P. A., Mcwilliam H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23 2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Lin H., Jiang X., Yi J., Wang X., Zuo R., Jiang Z., et al. (2018). Molecular identification of Neofabraea species associated with bull’s eye rot on apple using rolling-circle amplification of partial EF-1α sequence. Can. J. Microbiol. 64 57–68. 10.1139/cjm-2017-0448 [DOI] [PubMed] [Google Scholar]
- Lizardi P. M., Huang X., Zhu Z., Bray-Ward P., Thomas D. C., Ward D. C. (1998). Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nat. Genet. 19 225–232. 10.1038/898 [DOI] [PubMed] [Google Scholar]
- Nagamine K., Hase T., Notomi T. (2002). Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probe. 16 223–229. 10.1006/mcpr.2002.0415 [DOI] [PubMed] [Google Scholar]
- Najafzadeh M. J., Dolatabadi S., Keisari M. S., Naseri A., Feng P., de Hoog G. S. (2013). Detection and identification of opportunistic Exophiala species using the rolling circle amplification of ribosomal internal transcribed spacers. J. Microbiol. Meth. 94 338–342. 10.1016/j.mimet.2013.06.026 [DOI] [PubMed] [Google Scholar]
- Niessen L., Vogel R. F. (2010). Detection of Fusarium graminearum DNA using a loop-mediated isothermal amplification (LAMP) assay. Int. J. Food Microbiol. 140 183–191. 10.1016/j.ijfoodmicro.2010.03.036 [DOI] [PubMed] [Google Scholar]
- Nilsson M., Malmgren H., Samiotaki M., Kwiatkowski M., Chowdhary B. P., Landegren U. (1994). Padlock probes: circularizing oligonucleotides for localized DNA detection. Science 265 2085–2088. 10.1126/science.7522346 [DOI] [PubMed] [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., et al. (2000). Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:E63. 10.1097/RLU.0b013e3181f49ac7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering J., Bamford A., Godbole V., Briggs J., Scozzafava G., Roe P., et al. (2002). Integration of DNA ligation and rolling circle amplification for the homogeneous, end-point detection of single nucleotide polymorphisms. Nucleic Acids Res. 30:e60. 10.1093/nar/gnf060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szemes M., Bonants P., Weerdt M., Baner J., Landegren U., Schoen C. D. (2005). Diagnostic application of padlock probes-multiplex detection of plant pathogens using universal microarray. Nucleic Acids Res. 33:e70. 10.1093/nar/gni069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis Version 6.0. Mol. Biol. Evol. 30 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S. S., Zhou Q., He Z. M., Luo T., Zhang P., Cai Q., et al. (2016). Cyclopeptide toxins of lethal amanitas: compositions, distribution and phylogenetic implication. Toxicon 120 78–88. 10.1016/j.toxicon.2016.07.018 [DOI] [PubMed] [Google Scholar]
- Tong Z., Kong F., Wang B., Zeng X., Gilbert G. L. (2007). A practical method for subtyping of Streptococcus agalactiae serotype III, of human origin, using rolling circle amplification. J. Microbiol. Meth. 70 39–44. 10.1016/j.mimet.2007.03.010 [DOI] [PubMed] [Google Scholar]
- Trilles L., Wang B., Firacative C., Lazéra M. S., Wanke B., Meyer W. (2014). Identification of the major molecular types of Cryptococcus neoformans and C. gattii by Hyperbranched rolling circle amplification. PLoS One 9:e94648. 10.1371/journal.pone.0094648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui C. K., Wang B., Khadempour L., Alamouti S. M., Bohlmann J., Murray B. W., et al. (2010). Rapid identification and detection of pine pathogenic fungi associated with mountain pine beetles by padlock probes. J. Microbiol. Meth. 83 26–33. 10.1016/j.mimet.2010.07.016 [DOI] [PubMed] [Google Scholar]
- Vaagt F., Haase I., Fischer M. (2013). Loop-mediated isothermal amplification LAMP)-based method for rapid mushroom species identification. J. Agr. Food Chem. 61 1833–1840. 10.1021/jf304824b [DOI] [PubMed] [Google Scholar]
- van Doorn R., Szemes M., Bonants P., Kowalchuk G. A., Salles J. F., Ortenberg E., et al. (2007). Quantitative multiplex detection of plant pathogens using a novel ligation probe-based system coupled with universal, high-throughput real-time PCR on OpenArraysTM. BMC Genomics 8:276. 10.1186/1471-2164-8-276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton J. (2018). “Biological activities of the Amanita peptide toxins,” in The Cyclic Peptide Toxins of Amanita and Other Poisonous Mushrooms ed. Walton J. (Cham: Springer-Verlag Press; ) 131–142. [Google Scholar]
- Wang X., Teng D., Tian F., Guan Q., Wang J. (2012). Comparison of three DNA extraction methods for feed products and four amplification methods for the 5’-junction fragment of Roundup Ready soybean. J. Agr. Food Chem. 60 4586–4595. 10.1021/jf300827q [DOI] [PubMed] [Google Scholar]
- Wang X. R., Wu L. F., Wang Y., Ma Y. Y., Chen F. H., Ou H. L. (2015). Rapid detection of Staphylococcus Aureus by loop-mediated isothermal amplification. Appl. Biochem. Biotech. 175 882–891. 10.1007/s12010-014-1328-x [DOI] [PubMed] [Google Scholar]
- Wieland T. (1986). Peptides of Poisonous Amanita Mushrooms. New York, NY: Springer-Velag Press. [Google Scholar]
- Yongkiettrakul S., Kampeera J., Chareanchim W., Rattanajak R., Pornthanakasem W., Kiatpathomchai W., et al. (2017). Simple detection of single nucleotide polymorphism in Plasmodium falciparum by SNP-LAMP assay combined with lateral flow dipstick. Parasitol. Int. 66 964–971. 10.1016/j.parint.2016.10.024 [DOI] [PubMed] [Google Scholar]
- Zhang P., Chen Z. H., Xiao B., Tolgor B., Bao H. Y., Yang Z. L. (2010). Lethal amanitas of East Asia characterized by morphological and molecular data. Fungal Divers. 42 119–133. 10.1007/s13225-010-0018-4 [DOI] [Google Scholar]
- Zhou J., Yuan Y., Lang N., Yin Y., Sun C. Y. (2016). Analysis of hazard in mushroom poisoning incidents in China mainland. Chin. J. Emerg. Med. 25 724–728. 10.3760/cma.j.issn.1671-0282.2016.06.008 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Basidiomata of nine lethal Amanita species. (A) A. bisporigera (MHHNU 7224); (B) A. exitialis (MHHNU 30297); (C) A. fuliginea (MHHNU 30944); (D) A. pallidorosea (MHHNU 8112); (E) A. rimosa (MHHNU 7954); (F) A. subfuliginea (MHHNU 8812); (G) A. subjunquillea (MHHNU 7751); (H) A. subpallidorosea (MHHNU 8617); (I) A. virosa (MHHNU 8621; photos A, D, E, F, G, and H by PZ; photos B, C, and I by ZC).
Data Availability Statement
All datasets generated for this study are included in the manuscript and/or the Supplementary Files.


