Abstract
The dataset described in this paper provides information on the morphological features of 24 different species of the genera Bacillus, Paenibacillus, Brevibacillus, Lysinibacillus, and Rummeliibacilluswhen growing in HiCrome Bacillus agar. The species studied are common contaminants of honey. In support to the recent publication entitled “HiCrome Bacillus agar for presumptive identification of Bacillus and related species isolated from honey samples” (2), a collection of 197 bacterial isolates belonging to 24 different species of aerobic spore-forming bacteria have been screened for their colony appearance and color and any substrate color change of HiCrome Bacillus agar at 24 and 48 h of incubation. Two simple flowcharts utilizing a combination of colony and media characteristics in the chromogenic medium and a set of simple biochemical and morphological tests were developed for quick presumptive identification.
Keywords: HiCrome bacillus agar, Honey, Bacillus, Brevibacillus, Lysinibacillus, Paenibacillus, Rummeliibacillus, Aerobic spore-forming bacteria, Chromogenic media
Specifications table
| Subject area | Microbiology, Food Microbiology |
| More specific subject area | Microbiological Methods, Bioinformatics |
| Type of data | Tables, Figures, Flowcharts |
| How data was acquired | Digital camera, PCR, purification, sequencing, and Phylogenetic analysis |
| Data format | Analyzed |
| Experimental factors | Isolation of spore-forming bacteria Genomic DNA from pure bacterial cultures |
| Experimental features | Isolation and cultivation of bacteria, 16S rRNA sequencing, microbiological tests, colony morphology, and microscopy |
| Data source location | Bacteria were isolated from samples from different geographical areas The analysis was performed at CIDEFI - Facultad de Ciencias Agrarias y Forestales, Universidad Nacional de La Plata, Calles 60 y 119 S/N, 1900 La Plata, Buenos Aires, Argentina |
| Data accessibility | Data are available with this article. 16S rRNA sequences of selected bacterial strains (n = 56) isolated from honey or honeybee larvae have been deposited in GenBank (https://www.ncbi.nlm.nih.gov/genbank/) under accession numbers summarized in Table 1. |
| Related research article | HiCrome Bacillus agar for presumptive identification of Bacillus and related species isolated from honey samples by Alippi and Abrahamovich (International Journal of Food Microbiology, 2019, DOI: 10.1016/j.ijfoodmicro.2019.108245) |
Value of the data
|
1. Data
The dataset described in this paper provides information on the morphological features of 24 different species of the genera Bacillus, Paenibacillus, Brevibacillus, Lysinibacillus, and Rummeliibacillus when growing in HiCrome Bacillus agar. The species studied here have been previously reported in honey [1], [2], [3], [5], [7], [10], [12], [13], [14], [15].
A collection of 197 bacterial isolates belonging to the 24 species tested have been screened for their colony appearance and color and any substrate color change of HiCrome Bacillus agar at 24 and 48 h of incubation (Fig. 1 and Table 3). Colors of colonies and substrate observed were compared with a Pantone international chart and identified with a PMS number (http://www.cal-print.com/InkColorChart.htm).
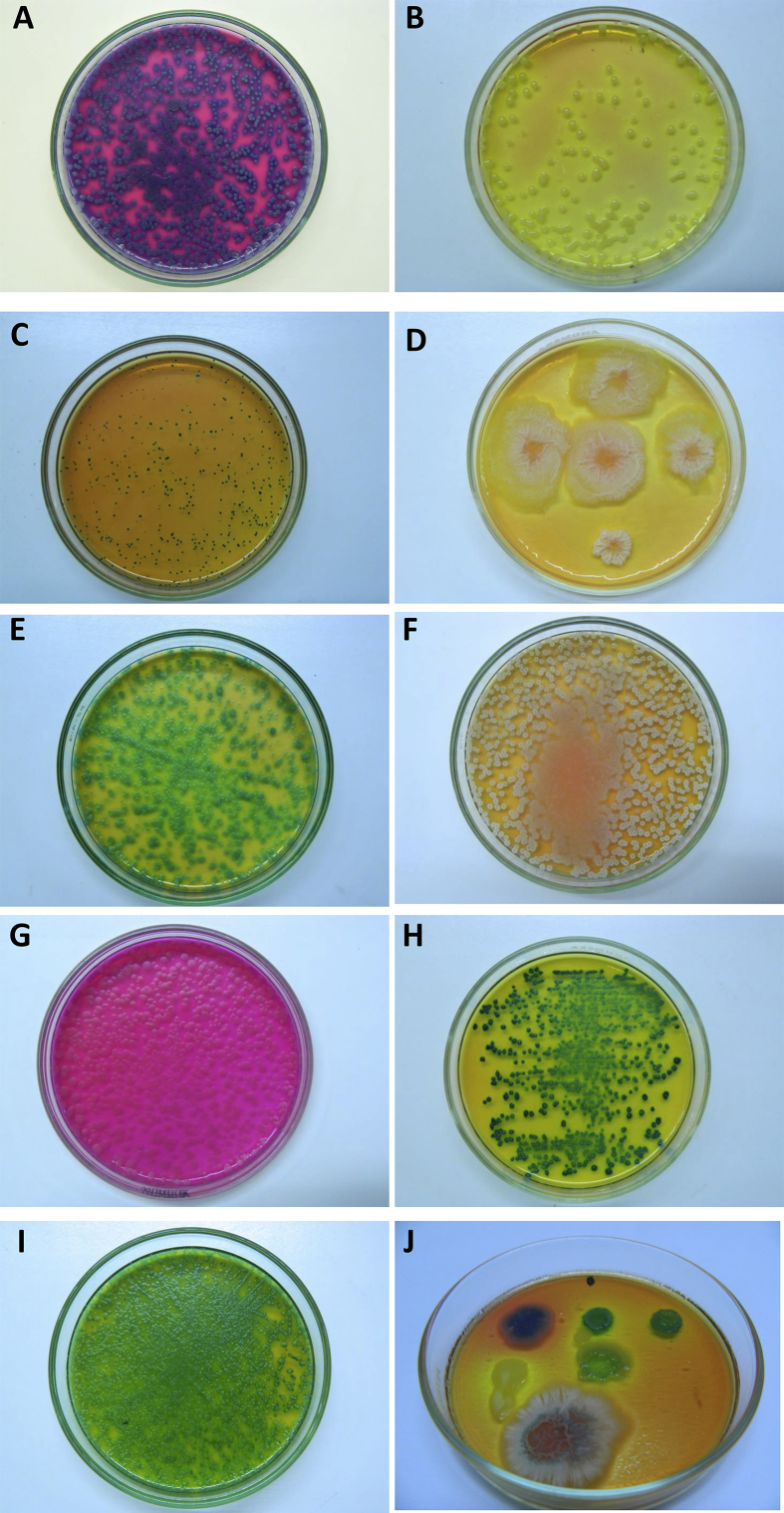
Fig. 1.
Comparison of colony appearance on HiCrome Bacillus agar of several Bacillus and related species commonly found in honey: A. Bacillus cereus m87, B. Bacillus megaterium m344, C. Paenibacillus alvei mv82, D. Bacillus amyloliquefaciens m39, E. Bacillus subtilis ATCC 7061, F. Bacillus subtilis m191, G. Lysinibacillus sphaericus m533, H. Bacillus licheniformis mv68, I. Bacillus pumilus mv49b, J. Complex sample of naturally contaminated honey containing (Clockwise from upper left) B. cereus, B. licheniformis, B. pumilus B. amyloliquefaciens, and B. megaterium.
Table 3.
Colony appearance and growth of selected strains tested in HiCrome Bacillus agar.
| Species | Strain/Isolate designation | Colonies in HiCrome Bacillus Agar | Ecometric Code |
||
|---|---|---|---|---|---|
| HiCrome Control | Bacillus medium | ||||
| Bacillus amyloliquefaciens | m39 | 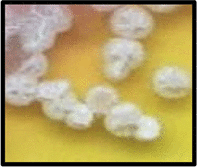 |
5 | 5 | |
| Bacillus badius | ATTC 14574 |  |
3.8 | 3.4 | |
| Bacillus cereus | ATCC 11778 | 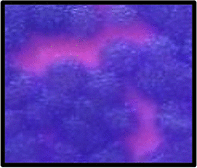 |
5 | 5 | |
| Bacillus cereus | m388 | 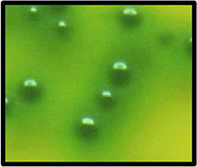 |
5 | 5 | |
| Bacillus circulans | ATCC 4515 |  |
4 | 5 | |
| Bacillus clausii | FR231 |  |
1.6 | 5 | |
| Bacillus coagulans | NRRL NRS 609 | 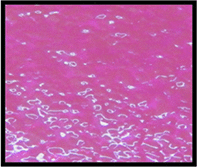 |
5 | 5 | |
| Bacillus firmus | ATCC 8247 | 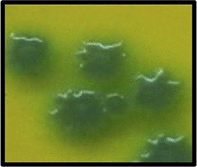 |
5 | 5 | |
| Bacillus licheniformis | NRRL B-1001 |  |
5 | 5 | |
| Bacillus megaterium | NRRL B-939 |  |
4.6 | 5 | |
| Bacillus mycoides | ATCC 10206 |  |
4 | 5 | |
| Bacillus pumilus | ATCC 7061 |  |
5 | 5 | |
| Bacillus subtilis | m191 | 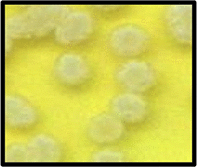 |
5 | 5 | |
| NRRL B-543 |  |
4 | 5 | ||
| Bacillus thuringiensis | ATCC 10792 | 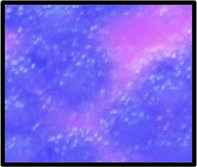 |
5 | 5 | |
| Brevibacillus borstelensis | RC | 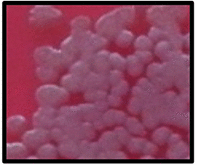 |
4.2 | 5 | |
| Brevibacillus brevis | ATCC 8246 | 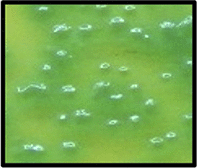 |
4.4 | 5 | |
| Brevibacillus laterosporus | CCT 0031 | 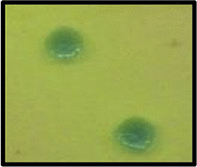 |
2.2 | 5 | |
| Lysinibacillus fusiformis | mv119 | 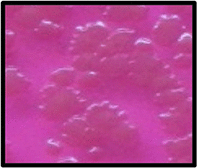 |
3.8 | 5 | |
| Lysinibacillus sphaericus | ATCC 245 | 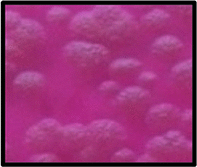 |
1.4 | 5 | |
| Paenibacillus alvei | NRRL B-383 | 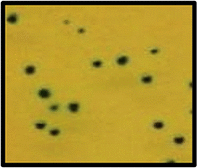 |
1 | 5 | |
| Paenibacillus amylolyticus | NRRL B-14940 | 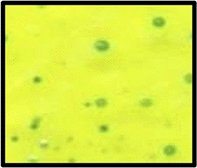 |
4 | 5 | |
| Paenibacillus apiarius | ATCC 29575 | 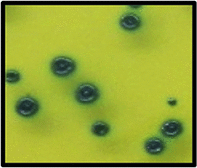 |
4 | 5 | |
| Paenibacillus larvae | ERIC I | ATCC 9545 |  |
2 | 5 |
| ERIC IV | ATCC 13537 |  |
2 | 5 | |
| ERIC II | SAG 290 |  |
3.8 | 5 | |
| Paenibacillus polymyxa | NRRL B-510 |  |
5 | 5 | |
| Rummeliibacillus stabekisii | mv111 |  |
5 | 5 | |
The Ecometric technique was used for comparative evaluation of HiCrome Bacillus agar and the control medium (Fig. 2, Fig. 3 and Table 3). E-values (Table 3) were obtained for 28 selected isolates tested by using previously published methods [2], [8].
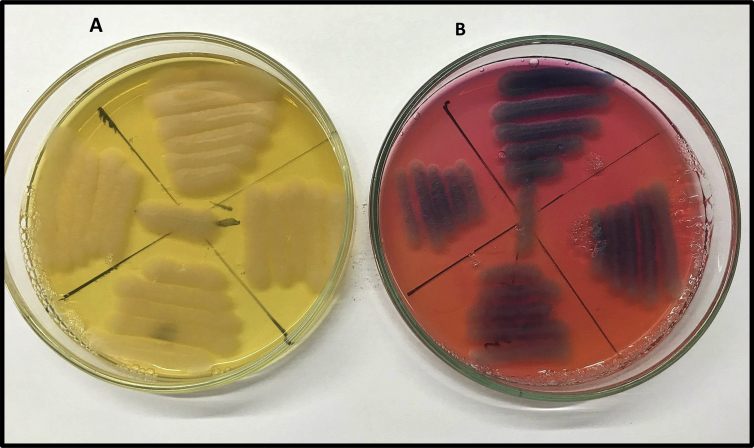
Fig. 2.
Bacillus cereus ATCC 11778 growing on A: BHIT and B. HiCrome Bacillus agar showing luxuriant growth (+++) and E value = 5.
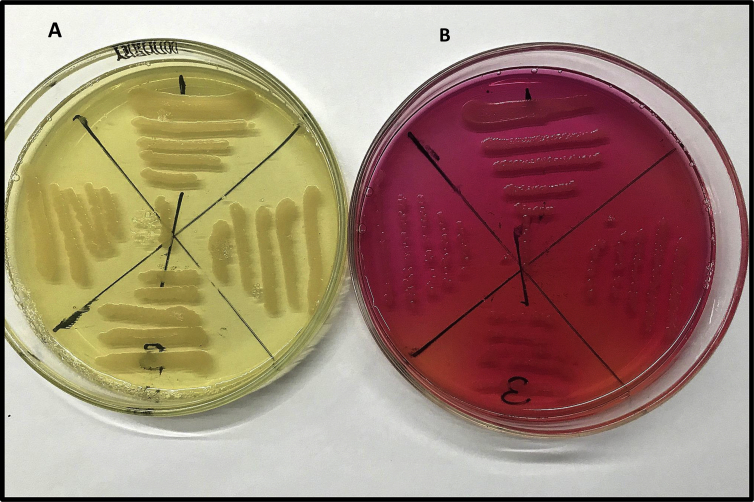
Fig. 3.
Rummeliibacillus stabekisii mv111 growing on A: MYPGP and B. HiCrome Bacillus agar showing good growth (++) and E value = 5.
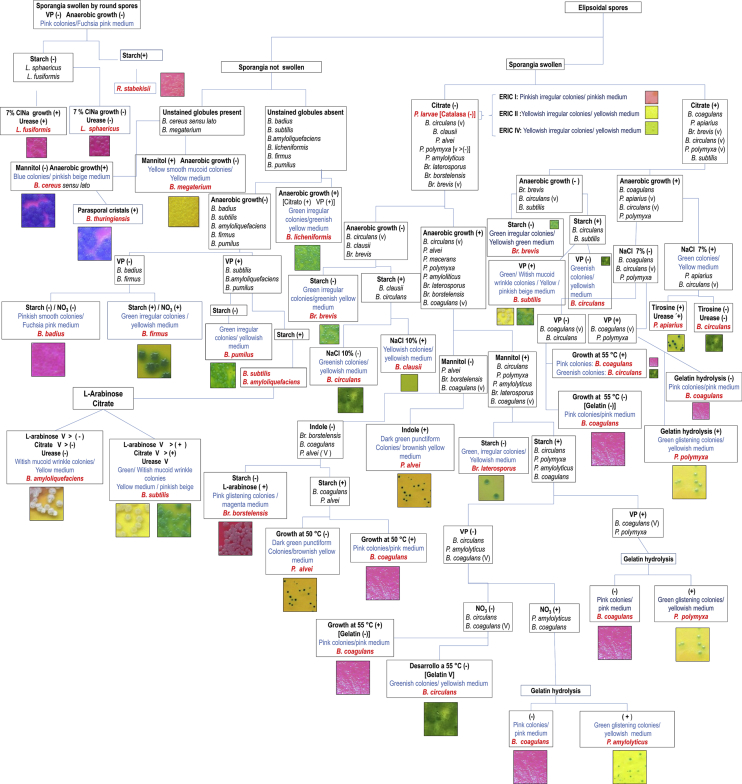
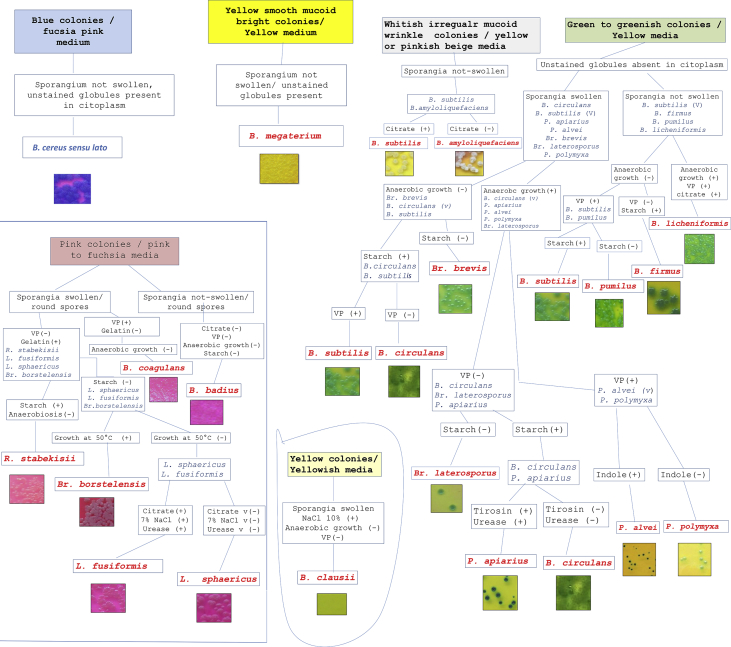
Two Flowcharts were prepared by a combination of colony and media characteristics in HiCrome Bacillus agar and a set of selected biochemical and morphological tests that are used routinely in Microbiological laboratories. The first chart (Fig. 4) permits the identification of the aerobic spore-forming bacteria reported in honey by a few simple tests. The more simplified flowchart presented in Fig. 5 allows differentiating typical strains of aerobic spore-forming species by direct isolation from honey.
Fig. 4.
Flowchart - Main steps for the identification of common strains of Bacillus and related species from honey by using a combination of selected morphological and biochemical tests and HiCrome Bacillus agar.
Fig. 5.
Flowchart - Simplified steps for the identification of common strains of Bacillus and related species from honey by using isolation in HiCrome Bacillus agar and selected morphological and biochemical tests.
The bacterial identity of selected strains isolated from honey or honeybee larvae (n = 56) (Table 1) were confirmed by sequencing the 16S rDNA. Sequences were deposited in the DDBJ/EMBL/Genbank under the Accession Numbers listed in Table 1. For comparisons, 16S rRNA sequences from type cultures (n = 32 plus 1 outlier) were used and are listed in Table 2.
Table 1.
Source and accession numbers of bacterial strains and isolates used in this study.
| Species | Strain/Isolate designation | Source and geographical origin | Accesion number |
|---|---|---|---|
| Bacillus amyloliquefaciens | xx | Honeybee larvae-Argentina | KP177517.1 |
| Bacillus amyloliquefaciens | mv35 | Honey- Argentina | MG004186.1 |
| Bacillus amyloliquefaciens | m39 | Honey- Argentina | MG004187.1 |
| Bacillus amyloliquefaciens | m163b | Honey- Argentina | MG004188.1 |
| Bacillus amyloliquefaciens | m164b | Honey- Argentina | MG004193.1 |
| Bacillus amyloliquefaciens | m287b | Honey- Argentina | MG004189.1 |
| Bacillus amyloliquefaciens | m291b | Honey- Argentina | MG004190.1 |
| Bacillus badius | CCT 0196 | CCT | N/A |
| Bacillus cereus | ATCC 11778 | ATCC | AF290546.1 |
| Bacillus cereus | cm4 | Honey- Argentina | N/A |
| Bacillus cereus | m6c | Honey- Argentina | KP005456.1 |
| Bacillus cereus | cm7 | Honey- Argentina | N/A |
| Bacillus cereus | cm8 | Honey- Argentina | N/A |
| Bacillus cereus | m9a | Honey- Argentina | N/A |
| Bacillus cereus | m10a | Honey- Argentina | N/A |
| Bacillus cereus | m10b | Honey- Argentina | N/A |
| Bacillus cereus | m12 | Honey- Argentina | N/A |
| Bacillus cereus | m19 | Honey- Argentina | N/A |
| Bacillus cereus | m21 | Honey- Argentina | N/A |
| Bacillus cereus | m28 | Honey - Argentina | N/A |
| Bacillus cereus | m31 | Honey- Argentina | N/A |
| Bacillus cereus | mv33 | Honey- Argentina | KU230015.1 |
| Bacillus cereus | cm37 | Honey- Argentina | N/A |
| Bacillus cereus | mv39b | Honey- Argentina | N/A |
| Bacillus cereus | mv41x | Honey- Argentina | N/A |
| Bacillus cereus | mv54 | Honey- Argentina | N/A |
| Bacillus cereus | m54 | Honey- Argentina | N/A |
| Bacillus cereus | mv67 | Honey- Argentina | N/A |
| Bacillus cereus | m73 | Honey- Argentina | N/A |
| Bacillus cereus | mv73 | Honey- Argentina | N/A |
| Bacillus cereus | mv75 | Honey- Argentina | N/A |
| Bacillus cereus | mv76 | Honey- Argentina | N/A |
| Bacillus cereus | mv77 | Honey- Argentina | N/A |
| Bacillus cereus | mv78 | Honey- Argentina | N/A |
| Bacillus cereus | mv79 | Honey- Argentina | N/A |
| Bacillus cereus | mv80 | Honey- Argentina | N/A |
| Bacillus cereus | m84 | Honey- Argentina | N/A |
| Bacillus cereus | m85 | Honey- Argentina | N/A |
| Bacillus cereus | mv86 | Honey- Argentina | N/A |
| Bacillus cereus | mv87 | Honey- Argentina | N/A |
| Bacillus cereus | m90 | Honey- Argentina | N/A |
| Bacillus cereus | m91 | Honey- Argentina | N/A |
| Bacillus cereus | m97 | Honey- Argentina | N/A |
| Bacillus cereus | m105 | Honey- Argentina | N/A |
| Bacillus cereus | mv114 | Honey- Argentina | N/A |
| Bacillus cereus | mv117 | Honey- Argentina | N/A |
| Bacillus cereus | cm117 | Honey- Argentina | N/A |
| Bacillus cereus | cm118 | Honey- Argentina | N/A |
| Bacillus cereus | m134 | Honey- Argentina | N/A |
| Bacillus cereus | m139b | Honey- Argentina | N/A |
| Bacillus cereus | m143b | Honey- Argentina | N/A |
| Bacillus cereus | m143c | Honey- Argentina | N/A |
| Bacillus cereus | m157 | Honey- Italy | N/A |
| Bacillus cereus | m158 | Honey- Argentina | N/A |
| Bacillus cereus | m163a | Honey- Argentina | N/A |
| Bacillus cereus | m167 | Honey- Argentina | N/A |
| Bacillus cereus | m189 | Honey- Argentina | N/A |
| Bacillus cereus | m193 | Honey- Argentina | N/A |
| Bacillus cereus | m225a | Honey- Argentina | N/A |
| Bacillus cereus | m228 | Honey- Argentina | N/A |
| Bacillus cereus | m243 | Honey- Argentina | N/A |
| Bacillus cereus | m244 | Honey- Argentina | N/A |
| Bacillus cereus | m248 | Honey- Argentina | N/A |
| Bacillus cereus | m262 | Honey- Argentina | N/A |
| Bacillus cereus | m267 | Honey- Argentina | N/A |
| Bacillus cereus | cm281 | Honey- Argentina | N/A |
| Bacillus cereus | m282a | Honey- Argentina | N/A |
| Bacillus cereus | m287a | Honey- Argentina | N/A |
| Bacillus cereus | m292 | Honey- Argentina | N/A |
| Bacillus cereus | m296 | Honey- Argentina | N/A |
| Bacillus cereus | m298 | Honey- Argentina | N/A |
| Bacillus cereus | m305 | Honey- Argentina | N/A |
| Bacillus cereus | m308 | Honey- Argentina | N/A |
| Bacillus cereus | m309 | Honey- Argentina | N/A |
| Bacillus cereus | m316 | Honey- Argentina | N/A |
| Bacillus cereus | m365 | Honey- Argentina | N/A |
| Bacillus cereus | m370 | Honey- Argentina | N/A |
| Bacillus cereus | m383 | Honey- Argentina | N/A |
| Bacillus cereus | m385 | Honey- Argentina | N/A |
| Bacillus cereus | m387 | Honey- Argentina | KP005455.1 |
| Bacillus cereus | m388 | Honey- Argentina | N/A |
| Bacillus cereus | m434 | Honey- Argentina | KU230027.1 |
| Bacillus cereus | m436 | Honey- Argentina | N/A |
| Bacillus cereus | m437b | Honey- Argentina | N/A |
| Bacillus cereus | m438 | Honey- Argentina | N/A |
| Bacillus cereus | m439 | Honey- Argentina | N/A |
| Bacillus cereus | m444 | Honey- Argentina | N/A |
| Bacillus cereus | m445b | Honey - Argentina | N/A |
| Bacillus cereus | LPcer1 | Honeybee larvae- Argentina | KX431225.1 |
| Bacillus cereus | MexB | Honey- Mexico | KU230012.1 |
| Bacillus cereus | MexC | Honey- Mexico | KU230013.1 |
| Bacillus circulans | ATCC 4515 | ATCC | N/A |
| Bacillus clausii | Fr231 | Honey- France | KU230014.1 |
| Bacillus clausii | m448b | Honey- Brazil | KX685159.1 |
| Bacillus coagulans | NRRL NRS 609 | NRRL | N/A |
| Bacillus firmus | ATCC 8247 | ATCC | N/A |
| Bacillus licheniformis | mv55 | Honey-Argentina | KU230018.1 |
| Bacillus licheniformis | mv68 | Honey-Argentina | MF187633.1 |
| Bacillus licheniformis | mv72 | Honey- Argentina | N/A |
| Bacillus licheniformis | m112 | Honey- Argentina | N/A |
| Bacillus licheniformis | NRRL B-1001 | NRRL | N/A |
| Bacillus megaterium | m280 | Honey- Argentina | N/A |
| Bacillus megaterium | m327 | Honey- Argentina | MF187637.1 |
| Bacillus megaterium | m344 | Honey- Argentina | N/A |
| Bacillus megaterium | m373 | Honey- Argentina | N/A |
| Bacillus megaterium | m435 | Honey- Mexico | KU230028.1 |
| Bacillus megaterium | m441 | Honey- Argentina | N/A |
| Bacillus megaterium | m458 | Honey- Brazil | N/A |
| Bacillus megaterium | NRRL B-939 | NRRL | N/A |
| Bacillus mycoides | m336 | Honey- Argentina | MF187638.1 |
| Bacillus mycoides | m425 | Honey- Argentina | N/A |
| Bacillus mycoides | ATCC 10206 | ATCC | N/A |
| Bacillus pumilus | mv41aA | Honey- Argentina | MG366818.1 |
| Bacillus pumilus | mv49b | Honey- Argentina | KU230016.1 |
| Bacillus pumilus | mv74 | Honey- Argentina | MF972935.1 |
| Bacillus pumilus | mv81 | Honey- Argentina | KU230019.1 |
| Bacillus pumilus | m108 | Honey- Argentina | N/A |
| Bacillus pumilus | m116 | Honey- Argentina | KU230020.1 |
| Bacillus pumilus | m157 | Honey- Italy | N/A |
| Bacillus pumilus | m187 | Honey- Argentina | N/A |
| Bacillus pumilus | m225b | Honey- Argentina | N/A |
| Bacillus pumilus | m288 | Honey- Argentina | MF187635.1 |
| Bacillus pumilus | m330 | Honey- Argentina | MF187646.1 |
| Bacillus pumilus | m335 | Honey- Argentina | N/A |
| Bacillus pumilus | m339 | Honey- Argentina | MG366884.1 |
| Bacillus pumilus | m350 | Honey- Argentina | KU230023.1 |
| Bacillus pumilus | m354 | Honey- Argentina | N/A |
| Bacillus pumilus | m357 | Honey- Argentina | MF187634.1 |
| Bacillus pumilus | m358 | Honey- Argentina | MG345110.1 |
| Bacillus pumilus | m360 | Honey- Argentina | MF187636.1 |
| Bacillus pumilus | m363 | Honey- Argentina | KU230024.1 |
| Bacillus pumilus | m414 | Honey- Argentina | KU230026.1 |
| Bacillus pumilus | ATCC 7061T | ATCC | AY876289.1 |
| Bacillus subtilis | m11 | Honey- Argentina | N/A |
| Bacillus subtilis | m13 | Honey - Argentina | MF187645.1 |
| Bacillus subtilis | m45 | Honey- Argentina | N/A |
| Bacillus subtilis | cm45 | Honey- Argentina | MF187639.1 |
| Bacillus subtilis | mv49a | Honey- Argentina | N/A |
| Bacillus subtilis | mv51 | Honey- Argentina | N/A |
| Bacillus subtilis | mv53b | Honey- Argentina | N/A |
| Bacillus subtilis | mv63 | Honey- Argentina | N/A |
| Bacillus subtilis | mv64 | Honey- Argentina | N/A |
| Bacillus subtilis | mv65 | Honey- Argentina | N/A |
| Bacillus subtilis | mv66 | Honey- Argentina | N/A |
| Bacillus subtilis | mv70 | Honey- Argentina | N/A |
| Bacillus subtilis | mv71 | Honey- Argentina | N/A |
| Bacillus subtilis | m107 | Honey- Argentina | N/A |
| Bacillus subtilis | m117 | Honey- Argentina | N/A |
| Bacillus subtilis | m119 | Honey- Argentina | N/A |
| Bacillus subtilis | m191 | Honey- Argentina | MF187644.1 |
| Bacillus subtilis | m192 | Honey- Argentina | N/A |
| Bacillus subtilis | m197 | Honey- Argentina | N/A |
| Bacillus subtilis | m291b | Honey- Argentina | N/A |
| Bacillus subtilis | m329 | Honey- Argentina | KU230021.1 |
| Bacillus subtilis | m334 | Honey- Argentina | KU230022.1 |
| Bacillus subtilis | m347 | Honey- Argentina | KP177515.1 |
| Bacillus subtilis | m351 | Honey- Argentina | KP177516.1 |
| Bacillus subtilis | m384 | Honey- Argentina | N/A |
| Bacillus subtilis | m386 | Honey- Argentina | N/A |
| Bacillus subtilis | m389 | Honey- Argentina | N/A |
| Bacillus subtilis | m392 | Honey- Argentina | MF187640.1 |
| Bacillus subtilis | m412 | Honey- Argentina | N/A |
| Bacillus subtilis | NRRL B-543 | NRRL | N/A |
| Bacillus thuringiensis | ATCC 10792T | ATCC | D16281.1 |
| Bacillus thuringiensis | m5 | Honey - Argentina | N/A |
| Bacillus thuringiensis | mv50b | Honey - Argentina | KU230017.1 |
| Bacillus thuringiensis | m391 | Honey - Argentina | N/A |
| Bacillus thuringiensis | m395 | Honey- Argentina | KU230025.1 |
| Bacillus thuringiensis | m401 | Honey- Argentina | N/A |
| Brevibacillus borstelensis | m348 | Honey- Argentina | MF187641.1 |
| Brevibacillus borstelensis | RC | Honey- Argentina | KP177514.1 |
| Brevibacillus brevis | ATCC 8246 | ATCC | N/A |
| Brevibacillus laterosporus | CCT 0031 | CCT | N/A |
| Brevibacillus laterosporus | BLAT169 | Honeybee larvae - Argentina | KX102627.1 |
| Brevibacillus laterosporus | BLAT170 | Honeybee larvae - Argentina | KX431223.1 |
| Brevibacillus laterosporus | BLAT171 | Honeybee larvae - Argentina | KX431224.1 |
| Lysinibacillus fusiformis | mv119 | Honey- Argentina | MG004185.1 |
| Lysinibacillus sphaericus | ATCC 245 | ATCC | N/A |
| Lysinibacillus sphaericus | m533 | Honey- Argentina | MG001492.1 |
| Lysinibacillus sphaericus | LMDZA | Honeybee larvae - Argentina | MG004191.1 |
| Paenibacillus alvei | NRRL B-383 | NRRL | N/A |
| Paenibacillus alvei | mv82 | Honey- Argentina | MF187643.1 |
| Paenibacillus alvei | m291a | Honey- Argentina | MF187632.1 |
| Paenibacillus alvei | m420 | Honey- Argentina | MF187642.1 |
| Paenibacillus amylolyticus | NRRL B-14940 | NRRL | N/A |
| Paenibacillus apiarius | ATCC 29575 | ATCC | N/A |
| Paenibacillus larvae ERIC I | ATCC 9545T | ATCC | NR_118956.1 |
| Paenibacillus larvae ERIC IV | ATCC 13537T | ATCC | KT363749.1 |
| Paenibacillus larvae ERIC I | PL38 | Honeybee larvae-Argentina | N/A |
| Paenibacillus larvae ERIC I | PL45 | Honeybee larvae- France | N/A |
| Paenibacillus larvae ERIC I | PL58 | Honeybee larvae- Sweden | N/A |
| Paenibacillus larvae ERIC II | SAG 290 | Honey - Unknown | N/A |
| Paenibacillus larvae ERIC II | SAG 10367 | Honey- Unknown | CP020557 |
| Paenibacillus larvae ERIC II | SAG 10754 | Honey- Unknown | N/A |
| Paenibacillus polymyxa | NRRL B-510 | NRRL | N/A |
| Rummeliibacillus stabekisii | mv111 | Honey- Argentina | MF972934.1 |
ATCC: American Type Culture Collection, USA; CCT: Colleçao de Culturas Tropical, Brazil; NRRL: Northern Utilization Research and Development Division, USA; SAG: Servicio Agrícola Ganadero, Chile.
N/A: Not applicable.
Table 2.
Accession numbers of 16 S rRNA sequences from Type cultures used for sequence analysis.
| Species | Strain | Accession number |
|---|---|---|
| Bacillus amyloliquefaciens | NBRC 15535 | NR_112685.1 |
| Bacillus badius | ATCC 14574 | X77790.1 |
| Bacillus cereus | ATCC 11778 | NR_074540.1 |
| Bacillus circulans | ATCC 4513 | AY724690.1 |
| Bacillus clausii | DSM 8716 | X76440.1 |
| Bacillus coagulans | ATCC 7050 | DQ297928.1 |
| Bacillus firmus | NBRC 15306 | NR_112635.1 |
| Bacillus flexus | IFO15715 | NR_024691.1 |
| Bacillus licheniformis | ATCC 14580 | NR_074923.1 |
| Bacillus megaterium | IAM 13418 | D16273.1 |
| Bacillus mycoides | ATCC 6462 | NR_115993.1 |
| Bacillus niabensis | 4T19 | AY998119.2 |
| Bacillus pumilus | ATCC 7061 | AY876289.1 |
| Bacillus simplex | DSM 1321 | AJ439078 |
| Bacillus subtilis | DSM 10 | JQ424889.1 |
| Bacillus thuringiensis | IAM 12077 | D16281.1 |
| Bacillus xiamenensis | MCCC 1A00008 | NR_148244.1 |
| Brevibacillus borstelensis | DSM 6347 | AB112721 |
| Brevibacillus brevis | NBRC 15304 | NR_041524.1 |
| Brevibacillus centrosporus | NRRL NRS-664 | NR_043414.1 |
| Brevibacillus formosus | DSM 9885 | AB112712.1 |
| Brevibacillus laterosporus | IAM 12465 | D16271 |
| Lysinibacillus fusiformis | DSM 2898 | AJ310083.1 |
| Lysinibacillus sphaericus | ATCC 14577 | NR_115724.1 |
| Paenibacillus alvei | DSM 29 | AJ320491 |
| Paenibacillus amylolyticus | NRRL NRS-290 | D85396.2 |
| Paenibacillus apiarius | NRRL NRS-1438 | NR_118834.1 |
| Paenibacillus larvae subsp. larvae | ATCC 9545 | NR_118956.1 |
| Paenibacillus larvae subsp. pulvifaciens | ATCC 13537 | KT363749.1 |
| Paenibacillus macerans | IAM 12467 | NR_040886 |
| Paenibacillus polymyxa | DSM 36 | AJ320493.1 |
| Rummeliibacillus stabekisii | NBRC 104870 | NR_114270 |
| Micrococcus luteus (outlier) | DSM 20030 | AJ536198.1 |
2. Experimental design, materials, and methods
A collection of 197 bacterial isolates of Bacillus, Brevibacillus, Lysinibacillus, Paenibacillus, and Rummeliibacillus belonging to different species that have been reported in honey [1], [2], [3], [5], [7], [10], [12], [13], [14], [15] were screened for their abilities to grow and colony appearance and color, and any substrate color change by using HiCrome Bacillus agar. The collection includes 167 isolates from honey samples from different geographical areas including Argentina, Brazil, France, Italy, and Mexico; 9 isolates from honeybee larvae from different geographical areas including Argentina, France and Sweden; and 21 strains from Culture Collections used for comparison and quality control. Bacteria were maintained as stock cultures at −80 °C in the correspondent broth medium, Müller-Hinton Broth, yeast extract, potassium phosphate, glucose, and pyruvate (MYPGP) [4] or Brain heart infusion (BHI) plus 20% glycerol (v/v). For short-term storage, the strains were kept at 4 °C in screw-capped vials containing MYPGP or BHI semi-solid (0.4% agar).
Bacterial smears stained by Schaeffer-Fulton technique were examined for the presence and location of spores within cells, as well as for the size and shape of vegetative cells [9], [11]. Also, the presence of unstained globules in the cytoplasm [6], [9], [11] was examined by phase contrast microscopy (Leica, ICC50).
Bacterial cultures were also tested by catalase reaction, anaerobic growth, nitrate reduction, Voges-Proskauer reaction (VP), pH in VP broth, indol and urease production, mannitol, L-arabinose, and citrate utilization, starch and gelatin hydrolysis, decomposition of tyrosine, growth in 7% and 10% of NaCl and at different temperatures (30-37-50 and 55 °C) according to standard protocols [6], [9], [11].
The Ecometric technique was used for comparative evaluation of HiCrome Bacillus agar and the correspondent control media (BHI or MYPGP). Overnight cultures were adjusted to 0.5 Mc Farland in sterile distilled water. One loop of 10 μl of each suspension was sequentially diluted from streak to streak onto each medium by inoculating 21 streaks (5 per quadrant and 1 in the center). Growth on the plates was recorded as a score. Readings were presented as absolute growth indices with possible values of 0–5, where 0 is an absence of growth in any streak and 5 was the maximum score obtained when all of the streaks in the four quadrants and also the last streak showed visible bacterial growth [2], [8]. Twenty-eight bacterial strains with different colony types (Table 2) were used for the evaluation. Plates were inoculated and incubated in duplicate for 24–48 h at 37 °C. Scores for HiCrome and control plates were compared to estimate the degree of inhibition due to the chromogenic mixture (Table 3, Fig. 2, Fig. 3).
The identity of selected strains (n = 56) was confirmed by sequencing the 16S rDNA. Universal eubacterial primers used for 16S rDNA sequence analysis were 27f (5′-AGAGTTTGATCMTGGCTCAG - 3′) and 1492r (5′-TACGGYTACCTTGTTACGACTT- 3′).
For purification of PCR products the following enzymatic procedure was used: The mixture contained 0.5 μl Antarctic phosphatase buffer (NEB, Migliore Lacaustra, Argentina), 0.6 μl Antarctic phosphatase (NEB, Migliore Lacaustra, Argentina), 0.6 μl Exonuclease I (NEB, Migliore Lacaustra, Argentina), 4 μl unpurified PCR product and 3.3 μl double distilled sterile deionized water. The Thermal cycler protocol consisted of one step of 37 °C for 20 min and the second of 80 °C for 20 min.
The quality and quantity of PCR products were assessed by gel electrophoresis (1 μl/1.6% agarose/molecular weight marker QuantiMarker, Promega, Argentina) and DNA concentration was estimated by using a Genova Nano spectrophotometer (JenWay). The purified PCR products of approximately 1,400 bp were sequenced by the dideoxy termination method by the commercial services of Macrogen Inc. (Seoul, Korea) or Unidad de Genómica, Instituto de Biotecnología, CICVYA - INTA (Hurlingham, Argentina). Sequence assembly and contig editing were performed by using CodonCode Aligner software (Codon Code Corporation, MA, USA). The partial sequences obtained were subjected to both Blast-N (http://www.ncbi.nlm.nih.gov), and EZBiocloud (http://www.ezbiocloud.n) search to identify sequences with the highest similarity by comparison only with sequences obtained from Type Cultures [2].
Acknowledgments
This work was financially supported by the Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT) [PICT 2012/0189 and PICT 2017/2014] and the Comisión de Investigaciones Científicas de la Provincia de Buenos Aires (CICBA) [Grants 1194/14 and 274/16]. AMA is a member of the Scientific Research Career of CICBA. I thank Dr. Ivo Siegrist for his helpful advice on HiCrome Bacillus agar and Dr. Eliana Abrahamovich for helping with the artwork and photographs.
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Alippi A.M. Detection of Bacillus larvae spores in Argentinian honeys by using a semi-selective medium. Microbiología (Madrid) 1995;11:343–350. [PubMed] [Google Scholar]
- 2.Alippi A.M., Abrahamovich E. HiCrome Bacillus agar for presumptive identification of Bacillus and related species isolated from honey samples. Int. J. Food Microbiol. 2019 doi: 10.1016/j.ijfoodmicro.2019.108245. [DOI] [PubMed] [Google Scholar]
- 3.Alippi A.M., Reynaldi F.J., López A.C., De Giusti M.R., Aguilar O.M. Molecular epidemiology of Paenibacillus larvae larvae and incidence of American foulbrood in argentinean honeys from Buenos Aires Province. J. Apic. Res. 2004;43:135–143. [Google Scholar]
- 4.Dingman D.W., Stahly D.P. Medium promoting sporulation of Bacillus larvae and metabolism of medium components. Appl. Environ. Microbiol. 1983;46:860–869. doi: 10.1128/aem.46.4.860-869.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilliam M. Microbiology of pollen and bee bread: the genus Bacillus. Apidologie. 1979;10:269–274. [Google Scholar]
- 6.Gordon R.E., Haynes W.C., Pang C.H.-N. Agricultural Research Service, USDA; Washington, D.C.: 1973. The Genus Bacillus: Agriculture Handbook No. 427. [Google Scholar]
- 7.Iurlina M.O., Fritz R. Characterization of microorganisms in Argentinean honeys from different sources. Int. J. Food Microbiol. 2005;105:297–304. doi: 10.1016/j.ijfoodmicro.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 8.Kornacki J.L., Gurtler J.B., Yan Z., Cooper C.H. Evaluation of several modifications of the ecometric technique for assessment of media performance. J. Food Prot. 2003;66:1727–1732. doi: 10.4315/0362-028x-66.9.1727. [DOI] [PubMed] [Google Scholar]
- 9.Parry J.M., Turnbull P.C.B., Gibson J.R. Wolfe Medical Publications Ltd.; Ipswich, England: 1983. A Colour Atlas of Bacillus Species. [Google Scholar]
- 10.Piccini C., Antúnez K., Zunino P. An approach to the characterization of the honey bee hive bacterial flora. J. Apic. Res. 2004;43:101–104. [Google Scholar]
- 11.Priest F.G., Goodfellow M., Todd C. A numerical classification of the genus Bacillus. J. Gen. Microbiol. 1988;143:1847–1882. doi: 10.1099/00221287-134-7-1847. [DOI] [PubMed] [Google Scholar]
- 12.Silva M.S., Rabadzhiev Y., Eller M.R., Iliev I., Ivanova I., Santana W.C. Chapter 11: Microorganisms in honey. In: de Toledo V.A., editor. Honey Analysis. 2017. pp. 233–234. [Google Scholar]
- 13.Sinacori M., Francesca N., Alfonzo A., Cruciata M., Sannino C., Settanni L., Moschetti G. Cultivable microorganisms associated with honeys of different geographical and botanical origin. Food Microbiol. 2014;38:284–294. doi: 10.1016/j.fm.2013.07.013. [DOI] [PubMed] [Google Scholar]
- 14.Snowdon J.A., Cliver D.O. Microorganisms in honey. Int. J. Food Microbiol. 1996;31:1–26. doi: 10.1016/0168-1605(96)00970-1. [DOI] [PubMed] [Google Scholar]
- 15.Wen Y., Wang L., Yue J., Zhang J., Su L., Zhang X., Zhou J., Li Y. The microbial community dynamics during the vitex honey ripening process in the honeycomb. Front. Microbiol. 2017;8:1649. doi: 10.3389/fmicb.2017.01649. [DOI] [PMC free article] [PubMed] [Google Scholar]