Abstract
We conducted a multilaboratory assessment to determine the suitability of a new commercially available reference material with 40 cancer variants in a background of wild-type DNA at four different variant allele frequencies (VAFs): 2%, 0.50%, 0.125%, and 0%. The variants include single nucleotides, insertions, deletions, and two structural variations selected for their clinical importance and to challenge the performance of next-generation sequencing (NGS) methods. Fragmented DNA was formulated to simulate the size distribution of circulating wild-type and tumor DNA in a synthetic plasma matrix. DNA was extracted from these samples and characterized with different methods and multiple laboratories. The various extraction methods had differences in yield, perhaps because of differences in chemistry. Digital PCR assays were used to measure VAFs to compare results from different NGS methods. Comparable VAFs were observed across the different NGS methods. This multilaboratory assessment demonstrates that the new reference material is an appropriate tool to determine the analytical parameters of different measurement methods and to ensure their quality assurance.
The measurement of cell-free DNA (cfDNA) in blood plasma, a form of liquid biopsy, has great promise for early detection, treatment selection, and monitoring of cancer.1 Reference materials are needed to help develop, validate, and ensure the quality of new assays. The analysis of cfDNA is especially challenging because somatic variant alleles are typically present in low concentrations relative to germline DNA. cfDNA analyses are currently being studied for many applications, including disease detection, treatment monitoring, and assay development.2
DNA is shed into the blood from normal or abnormal cells during apoptosis and cellular metabolism and through signaling events as well as necrosis or vesicle formation.3 Cancer cells shedding circulating tumor DNA (ctDNA) into the blood often contain tumor-associated biomarkers that can be used for noninvasive detection and monitoring of cancers. However, depending on the tumor stage, anatomical site, and unknown factors, the amount of released ctDNA can be low, and variations in handling and storage conditions (preanalytical variables) can influence measurements of this class of cancer biomarkers.4, 5, 6 Furthermore, patient samples are often difficult to acquire in sufficient numbers and amounts to facilitate comparison of different measurement methods. The methods and instruments for ctDNA assays are rapidly evolving to improve sensitivity and specificity, highlighting the need for reference materials to assess and compare different assays for measuring diverse classes of cfDNA cancer biomarkers.
The National Institute of Standards and Technology (NIST) in partnership with the Early Detection Research Network, a program in the National Cancer Institute, performed a multilaboratory assessment to evaluate the suitability of a commercially available cfDNA reference material to benchmark the efficiency of multiple steps in cfDNA analysis: DNA extraction, quantitation, preparation, instrument analysis, and bioinformatics analysis of cancer biomarkers found in cfDNA samples. The technology used to make the reference material uses biosynthetic variants spiked into a background of wild-type DNA, sized to simulate patient cfDNA, and encapsulated in a lipophilic structure and formulated in a synthetic plasma matrix. The reference material used in this study contains 40 clinically relevant cancer mutations across 28 genes. The variants in the reference material include single nucleotides, insertions, deletions, and two structural variations, although in this study not all variants were measurable. Digital PCR (dPCR) measurements of the variant allele frequencies (VAFs) were compared down to 0.125% with four different varieties of next-generation sequencing (NGS), each with distinct error correction strategies.7
The individual laboratories used their own extraction, quantitation, analytics, and bioinformatics methods. The goals of the study were to demonstrate the utility of the reference material with a range of VAFs to establish the analytical parameters of different measurement methods and to monitor the long-term consistency of their ctDNA measurements to achieve measurement assurance. The reference material was formulated in a synthetic plasma reference sample designed to simulate the concentration, size, and matrix of natural plasma samples.
Methods and Materials
Reference Materials
The samples used in this study were from SeraCare Life Sciences (Milford, MA) and consist of 40 cancer DNA variants (Table 1) spiked into a background of wild-type genomic DNA (derived from cell line GM 24385, Coriell Institute for Medical Research, Camden, NJ) at VAFs of approximately 2%, 0.5%, 0.125%, and 0%. The DNA was prepared at a length of approximately 170 to 180 bp using a proprietary SeraCare process. The samples used for the NGS and the DNA extractions were the reference material samples that consisted of the Seraseq ctDNA Reference Material v2 formulated in a synthetic plasma matrix with a DNA concentration of approximately 25 ng/mL in 5 mL (total extractable DNA of approximately 125 ng, stored at 4°C). The dPCR measurements were taken using the mutation mixture that consisted of purified nucleic acids in 0.1 × TE-based buffer (10 mmol/L potassium, 1 mmol/L Tris, and 0.1 mmol/L EDTA, pH 8.0) at a concentration of 10 ng/μL (volume of 25 μL, total DNA approximately 250 ng, stored at −20°C).
Table 1.
List of Variants in the SeraCare Purified DNA and Reference Material Samples
| Gene | COSMIC ID No.∗ | Mutation type | CDS | AA | NGS method measured |
|---|---|---|---|---|---|
| AKT1 | COSM33765 | SNV | c.49 G>A | p.E17K | Archer Reveal, digital NGS |
| APC | COSM18561 | Insertion | c.4666_4667insA | p.T1556fs*3 | |
| APC | COSM13127 | SNV | c.4348C>T | p.R1450* | Digital NGS |
| ATM | COSM21924 | Deletion | c.1058_1059delGT | p.C353fs*5 | Digital NGS |
| BRAF | COSM476 | SNV | c.1799T>A | p.V600E | Archer Reveal, digital NGS, SiMSeq-Seq, DEEP-Seq |
| CTNNB1 | COSM5664 | SNV | c.121A>G | p.T41A | Archer Reveal, digital NGS |
| EGFR | COSM6225 | Deletion | c.2236_2250del15 | p.E746_A750delELREA | Archer Reveal, digital NGS, DEEP-Seq |
| EGFR | COSM12378 | Insertion | c.2310_2311insGGT | p.D770_N771insG | Digital NGS, DEEP-Seq |
| EGFR | COSM6224 | SNV | c.2573T>G | p.L858R | Archer Reveal, digital NGS, DEEP-Seq |
| EGFR | COSM6240 | SNV | c.2369C>T | p.T790M | Digital NGS, DEEP-Seq |
| ERBB2 | COSM20959 | Insertion | c.2324_2325ins12 | p.A775_G776insYVMA | Archer Reveal, digital NGS |
| FGFR3 | COSM715 | SNV | c.746C>G | p.S249C | |
| FLT3 | COSM783 | SNV | c.2503 G>T | p.D835Y | Digital NGS |
| FOXL2 | COSM33661 | SNV | c.402C>G | p.C134W | Digital NGS |
| GNA11 | COSM52969 | SNV | c.626A>T | p.Q209L | Digital NGS |
| GNAQ | COSM28758 | SNV | c.626A>C | p.Q209P | Digital NGS |
| GNAS | COSM27887 | SNV | c.601C>T | p.R201C | Digital NGS |
| IDH1 | COSM28747 | SNV | c.394C>T | p.R132C | Archer Reveal, Digital NGS |
| JAK2 | COSM12600 | SNV | c.1849 G>T | p.V617F | Digital NGS |
| KIT | COSM1314 | SNV | c.2447A>T | p.D816V | Archer Reveal, digital NGS |
| KRAS | COSM521 | SNV | c.35 G>A | p.G12D | Archer Reveal, digital NGS, SiMSeq-Seq, DEEP-Seq |
| MPL | COSM18918 | SNV | c.1544 G>T | p.W515L | Digital NGS |
| NCOA4-RET | N/A | Gene Fusion (DNA) | NCOA4{NC_000010.10}:r.1_1014+1312_ RET{NC_000010.10}:r.2327–1437_5659 |
N/A | |
| NPM1 | COSM17559 | Insertion | c.863_864insTCTG | p.W288fs*12 | |
| NRAS | COSM584 | SNV | c.182A>G | p.Q61R | Archer Reveal, digital NGS, SiMSeq-Seq, DEEP-Seq |
| PDGFRA | COSM28053 | Insertion | c.1694_1695insA | p.S566fs*6 | Archer Reveal, Digital NGS |
| PDGFRA | COSM736 | SNV | c.2525A>T | p.D842V | Archer Reveal, Digital NGS |
| PIK3CA | COSM12464 | Insertion | c.3204_3205insA | p.N1068fs*4 | Archer Reveal |
| PIK3CA | COSM763 | SNV | c.1633 G>A | p.E545K | Archer Reveal, digital NGS, DEEP-Seq |
| PIK3CA | COSM775 | SNV | c.3140A>G | p.H1047R | Archer Reveal, Digital NGS, SiMSeq-Seq, DEEP-Seq |
| PTEN | COSM5809 | Deletion | c.800delA | p.K267fs*9 | |
| PTEN | COSM4986 | Insertion | c.741_742insA | p.P248fs*5 | Digital NGS, Digital NGS |
| RET | COSM965 | SNV | c.2753T>C | p.M918T | Archer Reveal, digital NGS |
| SMAD4 | COSM14105 | Insertion | c.1394_1395insT | p.A466fs*28 | Digital NGS |
| TP53 | COSM6530 | Deletion | c.723delC | p.C242fs*5 | Archer Reveal, digital NGS |
| TP53 | COSM18610 | Deletion | c.263delC | p.S90fs*33 | Archer Reveal |
| TP53 | COSM10648 | SNV | c.524 G>A | p.R175H | Archer Reveal, digital NGS |
| TP53 | COSM10660 | SNV | c.818 G>A | p.R273H | Archer Reveal, digital NGS |
| TP53 | COSM10662 | SNV | c.743 G>A | p.R248Q | Archer Reveal, digital NGS |
| TPR-ALK | NA | Gene fusion (DNA) | TPR{NC_000001.10}:r.1_2185+246_ ALK{NC_000002.11}:r.4125-550_6265 |
NA |
AA, amino acid; CDS, coding sequence; DEEP-Seq, Deep Error Eliminating Plasma Sequencing; NA, not applicable; NGS, next-generation sequencing; SiMSeq-Seq, Simple, Multiplexed, PCR-Based Barcoding of DNA for Sensitive Mutation Detection Using Sequencing; SNV, single-nucleotide variant.
Available from the Catalog of Somatic Mutations in Cancer (COSMIC; https://cancer.sanger.ac.uk/cosmic).
Plasma Reference Material DNA Extraction Methods
The laboratories each extracted DNA from the reference material samples using different procedures (Table 2). Laboratory A used the NucleoSnap DNA Plasma Kit (Macherey-Nagel, Bethlehem, PA, catalog number 740300.50) to isolate DNA from the 5-mL reference material samples, according to the manufacturer's protocol, with a modification that the sample was heat treated at 90°C for 10 minutes after isolation, and the DNA was eluted in 5 mmol/L Tris buffer, pH 8.5, and then stored at 4°C.
Table 2.
Extraction and Quantitation Methods and DNA Yields for the Reference Material Samples
| Laboratory | Extraction kit | Elution buffer | Quantitation method | DNA yield per VAF, ng/mL |
|||
|---|---|---|---|---|---|---|---|
| 0% | 0.125% | 0.50% | 2% | ||||
| A | Nucleosnap DNA Plasma Kit | 5 mmol/L Tris, pH 8.5 | Qubit HS kit | 31.33 ± 0.12 | 23.73 ± 1.03 | 30.80 ± 2.96 | 24.07 ± 1.47 |
| B | QIAamp CNA Kit | Kit supplied buffer∗ | Qubit HS kit | 30.96 ± 4.15 | 14.60 ± 6.03 | 27.79 ± 8.21 | 21.17 ± 7.11 |
| C | Maxwell RSC LV ccfDNA Kit (automated) | 10 mmol/L Tris, pH 8.0 | Qubit HS kit | 15.24 ± 2.27 | 12.01 ± 1.72 | 16.25 ± 2.18 | 9.18 ± 5.32 |
| D | QIAsymphony DSP Circulating DNA Kit (automated) | Kit supplied buffer | Qubit HS kit | 18.20 ± 0.47 | 18.27 ± 5.85 | 17.53 ± 4.24 | 15.93 ± 0.28 |
| E | QIAamp CNA Kit | 10 mmol/L Tris, pH8.0 | Qubit HS kit | 17.51± 5.19 | 11.45 ± 1.85 | 16.64 ± 1.66 | 11.75 ± 1.27 |
| F | QIAamp CNA Kit | Kit supplied buffer∗ | Qubit BR kit | 30.71 ± 4.32 | 26.78 ± 1.75 | 30.09 ± 4.21 | 27.31 ± 5.00 |
| G | Zymo Quick-cfDNA Serum and Plasma kit | Water | Qubit HS kit | 4.34 ± 0.44 | 3.16 ±.94 | 4.32 ± 2.75 | 4.05 ± 0.34 |
Data are expressed as means ± SD. Samples were prepared in triplicate for the laboratories with the exception of laboratory G, where four samples were measured.
BR, broad range; ccfDNA, circulating cell-free DNA; cfDNA, cell-free DNA; CNA, circulating nucleic acid; DSP, digital spatial profiling; HS, high sensitivity.
Carrier RNA was added in the lysis step.
Laboratories B, E, and F used the QIAamp Circulating Nucleic Acid Kit (Qiagen, Germantown, MD, catalog number 55,114) with carrier RNA (laboratories B and F) or without carrier RNA (laboratory E) added during lysis from approximately 5 mL of the reference material samples, according to the manufacturer's protocol. DNA was eluted using the kit-supplied buffer (laboratories B and F) or a 10 mmol/L Tris buffer, pH 8.0 (laboratory E), and then DNA samples were stored at 4°C or frozen at −20°C.
Laboratory C used the automated Promega Maxwell RSC LV cfDNA Custom Kit (Promega, Madison, WI, catalog number AX1115), used the heater shaker magnet protocol for preprocessing, and then completed extraction on the Maxwell RSC according to the manufacturer's protocol for each 4-mL reference material sample, and the DNA was eluted in 10 mmol/L Tris buffer, pH 8.0, and stored at 4°C.
Laboratory D used the automated QIAsymphony DSP Circulating DNA Kit (Qiagen, catalog number 937556), according to the manufacturer's circDNA_4000_DSP_V1 protocol for approximately 4.5-mL reference material samples; the DNA was eluted in kit-supplied buffer QSE1 and QSE2 and then stored at −80°C. Laboratory G used the Zymo Quick-cfDNA Serum & Plasma kit (Zymo, Irvine, CA, catalog number D4076) for the isolations according to the manufacturer's instructions. Four replicates of 3 mL of each standard plasma matrix sample were eluted in 50 μL of water (Molecular Biologicals International, Irvine, CA, catalog number NUPW-1000), dried down to completion in Eppendorf LoBind DNA tubes (Eppendorf, Hauppauge, NY, catalog number 022431021), and then resuspended in 16.5 μL of water.
DNA Concentration and Size Characterization
The extracted DNA samples were quantified by using either Qubit dsDNA High-Sensitivity Assay Kits (Thermo Fisher Scientific, Waltham, MA, catalog number Q32854) or Qubit dsDNA Broad Range Assay Kits (Thermo Fisher Scientific, catalog numbers Q32853 and Q32850) on a Qubit fluorimeter 3.0 according to the manufacturer's protocol (using 1 to 5 μL). Laboratory G quantitated the DNA before a dry-down step to concentrate the extracted DNA. Samples provided with the Qbit kit were used to determine the concentration of the samples.
Three laboratories determined the size distribution of isolated DNA. The Agilent 2100 Bioanalyzer with the Agilent DNA 1000 Kit (Agilent, Santa Clara, CA, catalog number 5067-1504) or the Agilent 4200 TapeStation (Agilent, catalog number G2991AA) with the TapeStation High Sensitivity DNA D1000 Reagent (Agilent, catalog number 5067-5585) were used to assess size distribution.
Droplet Digital PCR
The VAFs of the purified nucleic acid DNA samples were measured using a QX200 droplet digital PCR (ddPCR) system (Bio-Rad, Hercules, CA). Laboratory E measured a subset of nine variants using independently developed dPCR assays, and Laboratory F measured 39 of the 40 variants. The primers and probe sequences ddPCR assays were developed in the laboratories or from commercially available assays (PrimePCR ddPCR Mutation Assays, Bio-Rad). Laboratory E assays are given in Table 3 and laboratory F assays in Table 4 and Supplemental Table S1.
Table 3.
Primer and Probe Sequences for Digital PCR Assays Laboratory E
| Gene | COSMIC ID∗ | Forward primer sequence | Reverse primer sequence | Wild-type probe | Variant probe | Amplicon size, bp |
|---|---|---|---|---|---|---|
| AKT1 | COSM33765 | Bio-Rad developed assay (dHsaMDV2010031) | HEX | FAM | 64 | |
| BRAF | COSM476 | 5′-CCAGACAACTGTTCAAAC-3′ | 5′-ACCTCAGATATATTTCTTCATG-3′ | 5′-VIC-CTAGCTACAGTGAAATC-MGB-3′ | 5′-FAM-TAGCTACAGAGAAATC-MGB-3′ | 110 |
| EGFR | COSM6225 | 5′-CTGGATCCCAGAAGGTGAGA-3′ | 5′-CCACACAGCAAAGCAGAAAC-3′ | 5′-VIC-ATTAAGAGAAGCAACATCTCCGA-MGB-3′ | 5′-FAM-TCGCTATCAAGACATCTC-MGB-3′ | 103/118 |
| EGFR | COSM6224 | 5′-GCAGCATGTCAAGATCACAGATT-3′ | 5′-CCTCCTTCTGCATGGTATTCTTTCT-3′ | 5′-VIC-AGTTTGGCCAGCCCAA-MGB-3′ | 5′-FAM-AGTTTGGCCCGCCCAA-MGB-3′ | 78 |
| EGFR | COSM6240 | 5′-CATCTGCCTCACCTCCAC-3′ | 5′-GCCAATATTGTCTTTGTGTTCCC-3′ | 5′-HEX-T+CATC+A+C+GC/ZEN/A+GCTC-IABkFQ-3′ | 5′-FAM-T+CATC+A+T+GC/ZEN/A+GC+TC-IABkFQ-3′ | 94 |
| KIT | COSM1314 | Bio-Rad developed assay (dHsaMDV2010023) | HEX | FAM | 98 | |
| KRAS | COSM521 | 5′-AGGCCTGCTGAAAATGACTGAATAT-3′ | 5′-GCTGTATCGTCAAGGCACTCTT-3′ | 5′-VIC-TTGGAGCTGGTGGCGT-MGB-3′ | 5′-FAM-TGGAGCTGATGGCGT-MGB-3′ | 66 |
| PIK3CA | COSM12464 | 5′-GGTGGCTGGACAACAAA-3′ | 5′-TCCAGAGTGAGCTTTCATTT-3′ | 5′-VIC-CATTGAACTGAAAAGATG-MGB-3′ | 5′-FAM-CATTGAACATGAAAAGAT-MGB-3′ | 97/98 |
| PIK3CA | COSM775 | 5′-GAGCAAGAGGCTTTGGAGTA-3′ | 5′-ATGCTGTTTAATTGTGTGGAAGA-3′ | 5′-HEX-C+CATG+A+T+GT/ZEN/G+CAT-IABkFQ-3′ | 5′-FAM-C+CATG+A+C+GT/ZEN/GCAT-IABkFQ-3′ | 102 |
Available from the Catalog of Somatic Mutations in Cancer (COSMIC; https://cancer.sanger.ac.uk/cosmic).
Table 4.
Primer and Probe Sequences for Digital PCR Assays Laboratory F
| Gene | COSMIC ID∗ | Forward primer sequence | Reverse primer sequence | Wild-type probe | Variant probe | Amplicon size, bp |
|---|---|---|---|---|---|---|
| APC | COSM18561 | 5′-TCAAATGAAAACCAAGAGAAA-3′ | 5′-TCATCATCATCTGAATCATCT-3′ | HEX IDT 5′-AGGCAGAAAAAACTATTGATTC-3′ |
FAM IDT 5′-AGGCAGAAAAAAACTATTGATTC-3′ |
80,81 |
| ATM | COSM21924 | 5′-GATTTCGTAATATTGCCGTC-3′ | 5′-TTCTAAATGTGACATGACCT-3′ | HEX IDT 5′-ATGGCAGATATCTGTCACCAG-3′ |
FAM IDT 5′-ATGGCAGATATCTCACCAG-3′ |
93,91 |
| EGFR | COSM6225 | 5′-CCAGAAGGTGAGAAAGTTAAA-3′ | 5′-AAACTCACATCGAGGATTTC-3′ | HEX IDT 5′-AATTAAGAGAAGCAACATCTCCG-3′ |
FAM IDT 5′-TCGCTATCAAGACATCTCC-3′ |
95,80 |
| ERBB2 | COSM682/COSM20959 | 5′-TACCCTTGTCCCCAGG-3′ | 5′-AGAAGGCGGGAGACATA-3′ | HEX IDT 5′-AAGCATACGTGATGGCTGGTGT-3′ |
FAM IDT 5′-TGGCATACGTGATGGC-3′ |
65,78 |
| RET | N/A | 5′-CCTGACGACTCGTGCTATTT-3′ | 5′-GCCGAAATCCGAAATCTTCATC-3′ | HEX IDT 5′-TCACAGCTCGTTCATCGGGACTTG-3′ |
N/A | 105 |
| NCOA4/RET | N/A | 5′-ACACTGGGCAAGACAGTAAAT-3′ | 5′-GAGCCTCTGTTACTTCCAGAAC-3′ | N/A | FAM IDT 5′-AGTGTTCCTACTAGCACTGTCCAGGG-3′ |
111 |
| NPM1 | COSM17559 | 5′-GGTTCCTTAACCACATTTCT-3′ | 5′-GAAATAAGACGGAAAATTTTTTAAC-3′ | HEX IDT 5′-TCAAGATCTCTGGCAGTG-3′ |
FAM IDT 5′-CAAGATCTCTGTCTGGCA-3′ |
120,124 |
| PDGFRA | COSM28053 | 5′-CAGTTACCTGTCCTGGTCAT-3′ | 5′-CTGCATCGGGTCCACATAA-3′ | HEX IDT 5′CAATCAGCCC-3′ |
FAM IDT 5′-CAATACAGCCC-3′ |
110,111 |
| PIK3CA | COSM12464 | 5′-TGGTGGCTGGACAACAAA-3′ | 5′-GGAATCCAGAGTGAGCTTTCA-3′ | HEX IDT 5′-CATTGAACTGAAAAGA-3′ |
FAM IDT 5′-CATTGAACATGAAAAGA-3′ |
102,103 |
| PTEN | COSM5809 | 5′-GTGTGTGGTGATATCAAAGT-3′ | 5′-TGGATATTTCTCCCAATGAAA-3′ | HEX IDT 5′-AGAACAAGATGCTAAAAAAGGTTTG-3′ |
FAM IDT 5′-AGAACAAGATGCTAAAAAGGT-3′ |
91,90 |
| SMAD4 | COSM14105 | 5′-GGCTACTGCACAAGCTG-3′ | 5′-GCTGGAGCTATTCCACCTA-3′ | HEX IDT 5′-CCGTGGCAGG-3′ |
FAM IDT 5′-CCGTTGGCAG-3′ |
90,91 |
| TP53 | COSM6530 | 5′-TGTGATGATGGTGAGGAT-3′ | 5′-CCACCATCCACTACAACTA-3′ | HEX IDT 5′-CGCCCATGCAGGAACT-3′ |
FAM IDT 5′-CGCCCATGCAGAACT-3′ |
80,79 |
| ALK | N/A | 5′-CAACCCTTGATGGTTGTTTCAG-3′ | 5′-CAGTGGATAACAGCAGGGATAC-3′ | HEX IDT 5′-AATCCCACCGATGTCACTGTCTGC-3′ |
N/A | 96 |
| TPR/ALK | N/A | 5′-AAGGTGCATTTCAGAATCAATG-3′ | 5′-GAGCCAAAGTCAGTCATCAG-3′ | N/A | FAM IDT 5′-ACTCCCAGGAATTGGCCTGCTAAC-3′ |
97 |
Available from the Catalog of Somatic Mutations in Cancer (COSMIC; https://cancer.sanger.ac.uk/cosmic).
The PCR reaction mixture consisted of 1 × ddPCR Supermix for probes (Bio-Rad), 900 nmol/L primers, 250 nmol/L probes (final concentrations) or 1 × PrimePCR ddPCR Mutation Assay (including primers and probes), and the sample DNA (approximately 20 ng) or a non-template control in a total volume of 25 μL. Twenty microliters of the 25-μL reaction mixtures were transferred to the droplet generator DG8 cartridge. Droplet generation oil (70 μL) was added into the oil well for each channel. After droplet generation was complete, the droplets were transferred to a 96-well PCR plate and placed on a C1000 Touch thermal cycler (Bio-Rad) or an Veriti 96-well thermal cycler (Applied Biosystems). The following thermal cycling conditions were used: 95°C for 10 minutes, followed by 40 cycles of 94°C for 30 seconds and 60°C for 1 minute, then 98°C for 10 minutes, at a temperature ramp rate at 50% (3°C per second). After PCR, the 96-well PCR plate was loaded onto the QX200 droplet digital reader (Bio-Rad). Data were analyzed with QuantaSoft version 1.7.4.0917 (Bio-Rad), which calculates the concentration of the target variant and wild-type DNA sequences and their Poisson-based 95% CIs. The VAF was calculated by dividing the concentration of variant alleles by the total concentrations of the variant and wild-type alleles.
Simple, Multiplexed, PCR-Based Barcoding of DNA for Sensitive Mutation Detection Using Sequencing NGS Method
Barcoding of DNA template molecules using a short random sequence at an early stage in NGS library construction provides a way to bioinformatically identify polymerase errors that would otherwise be considered positive results. The simple, multiplexed, PCR-based barcoding of DNA for sensitive mutation detection using sequencing (SiMSen-Seq) was developed by using reduced primer concentrations, elongated PCR extension times, and hairpin-protected barcode primers to generate targeted barcoded libraries.8 SiMSen-Seq allows detection of variant alleles at <0.1%.
The SiMSen-Seq method was executed as described previously.8, 9 Briefly, barcoding of extracted 50 ng of DNA was performed using PCR in a 10-μL reaction volume that contained 1 × AccuPrime PCR Buffer II, 0.2 U AccuPrime Taq DNA Polymerase High Fidelity (Thermo Fisher Scientific), and 40 nmol/L of each primer (Integrated Device Technology, Inc., San Jose, CA). The primer sequences for PIK3CA (H1047R), KRAS (G12D), BRAF (V600E), and NRAS (Q61R) are shown in Table 5. The temperature profile was 98°C for 3 minutes, followed by three cycles of amplification (98°C for 10 seconds, 62°C for 6 minutes, and 72°C for 30 seconds), 65°C for 15 minutes, and then 95°C for 15 minutes. Twenty microliters of TE buffer, pH 8.0 (Thermo Fisher Scientific), with a final concentration of 30 ng/μL of protease (Streptomyces griseus, Sigma-Aldrich, St. Louis, MO) was added to inactivate the Taq DNA polymerase at the 65°C for a 15-minute step. A second round of PCR was performed in 40 μL using 1 × Q5 Hot Start High-Fidelity Master Mix (New England BioLabs, Ipswich, MA), with 400 nmol/L of each Illumina adaptor primer (Illumina, San Diego, CA) and 10-μL PCR products from the first round of PCR. The temperature profile was 95°C for 3 minutes followed by 18 to 30 cycles of amplification (98°C for 10 seconds, ramping from 80°C down to 72°C and up 76°C, 0.2°C per 1-second increments, 76°C for 30 seconds). Then 36-μL PCR products were purified using the Agencourt AMPure XP system (catalog number A63881, Beckman Coulter, Inc., Sykesville, MD) according to the manufacturer's instructions. The applied volume ratio between beads and PCR products ranged from 0.83 to 1.0, depending on amplicon length. The purified product was eluted in 20 μL of TE buffer, pH 8.0, and before sequencing, library products were assessed on a fragment analyzer (Agilent, Advanced Analytic Technology, Inc., Santa Clara, CA) to ensure correct sizing.
Table 5.
Primer Sequences for the Simple, Multiplexed, PCR-Based Barcoding of DNA for Sensitive Mutation Detection Using Sequencing Assays
| Assay name | Mutation | Primer sequences |
|---|---|---|
| BRAF_305 | V600E | Forward: 5′-GGACACTCTTTCCCTACACGACGCTCTTCCGATCTNNNNNNNNNNNNATGGGAAAGAGTGTCCCAAACTGATGGGACCCACTCCATCG-3′ |
| Reverse: 5′-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTGACCTCACAGTAAAAATAGGTGATTTTGGTCTAGC-3′ | ||
| KRAS_329 | G12D | Forward: 5′-GGACACTCTTTCCCTACACGACGCTCTTCCGATCTNNNNNNNNNNNNATGGGAAAGAGTGTCCGCCTGCTGAAAATGACTGAATATAAACTTG-3′ |
| Reverse: 5′-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTGCTGTATCGTCAAGGCACTCTT-3′ | ||
| NRAS_881_3 | Q61R | Forward: 5′-GGACACTCTTTCCCTACACGACGCTCTTCCGATCTNNNNNNNNNNNNATGGGAAAGAGTGTCCTTGGTCTCTCATGGCACTGTACTCTTCT-3′ |
| Reverse: 5′-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTAACCTGTTTGTTGGACATACTGGATACAGC-3′ | ||
| PIK3CA_234 | H1047R | Forward: 5′-GGACACTCTTTCCCTACACGACGCTCTTCCGATCTNNNNNNNNNNNNATGGGAAAGAGTGTCCCTGAGCAAGAGGCTTTGGAGTATTTCATG-3′ |
| Reverse: 5′-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTCCAATCCATTTTTGTTGTCCAGCCAC-3′ |
The products from the second round of PCR contained Illumina sequencing adaptor sequences and indexes and were therefore sequencer ready. To assess the amplification status of primer pairs in multiplexed SiMSen-Seq reactions, some libraries were initially sequenced at low depth using MiSeq instruments with the Nano Kit version 2 in 1 × 150 mode (Illumina). For full sequencing runs, libraries were multiplexed per lane and sequenced on MiSeq or HiSeq2500 instruments (Illumina) in single or paired 150-bp mode.
The bioinformatics workflow was implemented as previously described.8 The FASTQ files were aligned to hg19 using BWA-MEM (algorithm version 0.7.12) with output binary alignment map (BAM) files sorted by position and indexed using SAMtools version 0.1.19 (Genome Research Ltd, Cambridgeshire, UK; http://www.htslib.org). A custom pipeline was used to build consensus sequences as follows: the amplicons in each library were identified in BAM files according to library multiplexity. For example, four target amplicons were identified in four-plex experiments. Valid reads within each amplicon were identified as those that contained a barcode sequence in the correct orientation relative to the sequence of the targeting primer and hairpin stem. The remaining reads were grouped into families by amplicon and random 12mer barcode. For reads within each family, alignment information for individual reads was used to determine a consensus identity for bases (including indels) at each nucleotide position within the amplicons. This procedure is conceptually similar to that described in Schmitt et al.10 Nonreference sequences were reported in consensus sequences if they composed 100% of the reads in families with 10 to 20 reads or at least 90% of reads in families with >20 reads.
Digital NGS Method
The digital NGS method was performed with minor modifications as described previously.11 Briefly, approximately 38.4 ng of extracted DNA sample was aliquoted into a 384-well plate, with each well containing 100 pg of DNA (approximately 30 genome equivalents). This aliquoting would be expected to yield NGS reactions with VAF of approximately 3% when an aliquot contained one true mutant template, and these mutations were detected using a 1.5% VAF cutoff for calling mutations. Each of the 384 aliquots of DNA was subjected to AmpliSeq PCR amplification (the list of AmpliSeq custom panel targeted regions of the 36 variants is given in Supplemental Table S2). FuPa digestion and P1 adaptor/Xpress barcode ligation (Thermo Fisher Scientific) were performed. After library clean-up using Agencourt AMPure XP Reagent (Beckman Coulter), the libraries were eluted into low TE buffer and subsequently quantified using the Ion Quantitation Kit (Life Technologies, Carlsbad, CA). Individual ion sample libraries were equalized to 15 pM and pooled together. After introducing 20-μL pooled libraries with emulsion PCR reagents into the Ion OneTouch2 system (Life Technologies) for 5 hours, the ion sphere particles were cleaned and enriched in the Ion OneTouch ES (enrichment system; Life Technologies). The enriched ion sphere particles were loaded into a 318 chip for sequencing using an Ion Torrent Personal Genome Machine (Life Technologies). The mean sequence depth of each amplicon sequenced on the 318 chip was approximately 500 reads for each of the 384 NGS reactions (approximately 20,000 reads per target amplicon). The postsequencing raw FASTQ files were launched in NextGENe software version 2.4.2.3 (SoftGenetics, Chicago, IL) for data analysis, including alignment to the hg19 human reference genome and single-nucleotide variant SNV calling. Alignments were visually verified using the Integrative Genomics Viewer version 2.3 (Broad Institute Cambridge, MA) and the NextGENe Viewer. Variants identified by the NextGENe software were filtered to select only variants with a nucleotide score of ≥30 (reference nucleotide score + mutant nucleotide score/indel score) and no strand bias.
Archer Reveal ctDNA Targeted NGS
The NGS libraries were prepared in four testing laboratories according to the Archer Reveal ctDNA protocol for Illumina. The extracted DNA (52.5 to 25 ng) (Table 6) was used as the input into the Archer Reveal ctDNA 28 kit for Illumina (catalog number AB0021, Archer Diagnostics, Boulder, CO), and the 28-gene list can be found in Supplemental Table S3. Briefly, end repair was performed by incubation in a thermocycler at 25°C for 30 minutes with heated lid off, and the end-repaired samples were purified with AMPure XP beads (catalog number A63881, Beckman Coulter). The tailing of purified end-repaired DNA with deoxyadenosine was completed by incubation in a thermocycler at 37°C for 15 minutes followed by a clean-up with AMPure XP beads. Molecular barcode (MBC) adapter incorporation was performed by mixing the adaptors with the purified DNA, incubating at 22°C for 5 minutes, and cleaning up using Ligation Cleanup Beads included in the kit. The DNA was eluted from the beads by incubation at 75°C for 10 minutes. The entire purified DNA ligation product was mixed with 2 μL of GSP1 in the first PCR hot start reaction tubes on ice, and the first PCR was performed as follows: 95°C for 3 minutes, 15 cycles at 95°C for 30 seconds, 65°C for 5 minutes, 72°C for 3 minutes, and 4°C hold. The PCR 1 library was purified with AMPure XP beads and mixed with 2 μL of GSP2 primers into PCR 2 reagent tubes. The second PCR amplification was performed as follows: 95°C for 3 minutes, 15 cycles at 95°C for 30 seconds, 65°C for 5 minutes, 72°C for 3 minutes, and 4°C hold. The PCR 2 library was purified with AMPure XP beads and eluted in 24 μL of 10 mmol/L Tris, pH 8.0.
Table 6.
Library Preparation for the NGS Assays
| Laboratory | NGS method | NGS platform | DNA input for library, ng | PCR and other key steps | Special steps in library preparation |
|---|---|---|---|---|---|
| A | SiMSen-Seq | Illumina MiSeq | 56.8 | First round of PCR barcoding, second round of PCR adding Illumina adaptors and indexes | Elongated PCR extension times and hairpin-protected UMI primers |
| B | Digital NGS | Ion Torrent PGM | 38.4 | Each sample was aliquoted to a 384-well plate and underwent multiplexed PCR, FuPa digestion, and P1 adaptor/Xpress IonCode barcode ligation | No UMI-based target NGS |
| C | Archer Reveal ctDNA targeted NGS | Illumina NextSeq 500 | 52.5 | End repair and MBC adapter ligation, two rounds of PCR with GSP1 and GSP2 primers, respectively | Seminest-PCR based and UMI in the adapter |
| D | Illumina HiSeq 2500 | 25 | |||
| E | Illumina MiSeq | 30 | |||
| F | Illumina MiSeq | 50 | |||
| G | DEEP-Seq | Illumina MiSeq | 14.1 | Three-stage multiplexed PCR protocol: MBC-addition PCR, stage PCR, and tag PCR | UMI-based targeted deep NGS |
ctDNA, circulating tumor; DEEP-Seq, Deep Error Eliminating Plasma Sequencing; MBC, molecular barcode. NGS, next-generation sequencing; SiMSeq-Seq, Simple, Multiplexed, PCR-Based Barcoding of DNA for Sensitive Mutation Detection Using Sequencing; UMI, unique molecular identifier.
The concentration of the libraries was quantified using quantitative PCR (Kapa Illumina Library Quantification Kit, catalog number KK4824, Kapa Biosystems, Wilmington, MA) using the provided solutions and the libraries diluted to 1:10−4, 1:10−5, 1:10−6, and 1:10−7.
Laboratory E used the MiSeq instrument. Briefly, the individual library was diluted to 4 nmol/L DNA concentration based on Kapa library real-time quantitative PCR (qPCR) quantification, and four libraries prepared from four different VAF samples were pooled into one library pool. 10% PhiX (catalog number FC-110-3001, Illumina) was added to the library pool, and 12 pM final concentration of library pool was loaded on a MiSeq using a MiSeq Reagent Kits version 3 (600-cycle, catalog number MS-102-3003, Illumina). The sample sheet with the Archer-recommended read-level depth and adaptors was used for the MiSeq run. FASTQ files were uploaded to a preview demo server for analysis using the Archer ctDNA analysis pipeline.
Laboratory F also used the MiSeq instrument. Briefly, the individual library was diluted to 4 nmol/L DNA concentration based on Kapa library qPCR quantification, and two to four prepared libraries were pooled into one library pool. Ten percent PhiX (catalog number FC-110-3001, Illumina) was added to the library pool. The library pool was then loaded on a MiSeq using a 300-cycle MiSeq Reagent Kit version 2 (catalog number MS-102-2002, Illumina). Archer software version 5.1 (Archer Diagnostics) was used for analysis.
Laboratory C used the NextSeq system, and 1.4 pM final concentration of library pool was loaded on NextSeq using a NextSeq 500/550 Mid Output version 2 Kit (catalog number FC-404-2003, Illumina) with 30% PhiX 20 pM. Archer software version 6.0 was used for analysis, with a cohort of 19 healthy control plasmas used for additional noise reduction. Custom scripts were used to evaluate results for additional primers that had been spiked into the GSP1 and GSP2 primer pools. Critical quality metrics were evaluated, results for each expected variant were tabulated, all data were analyzed by a pathologist, and a spreadsheet of key data and summary results was submitted to NIST.
Laboratory D used the HiSeq 2500 system. Briefly, individual libraries were diluted to a 2 nmol/L concentration, based on qPCR quantification, and pooled into one of the two library pools. The quantified library pool was loaded onto a HiSeq 2500 (Illumina) in Rapid Run mode (225 bp paired-end sequencing, on-board clustering). Another run was previously created using healthy donor blood from eight donors to create a healthy donor cohort for the Reveal ctDNA 28 panel that was sequenced on HiSeq 2500 in the same manner as the SeraCare samples. Resultant bcl files were converted to FASTQs and uploaded to a preview demo server for analysis using the Archer ctDNA analysis pipeline.
Data from the healthy donor cohort samples were used for background correction at each base analyzed to improve confidence in low-frequency calls. SeraCare sample results from two laboratories were derived using the healthy donor cohort and reported using the following filters from the Archer Analysis server: alternate allele observations, ≥5; unique alternate allele observations, ≥3; deep molecular bin alternate allele observations, ≥2; and background correction allele fraction outlier P value, ≤0.01; variant call was checked by JBrowse. The deep-bin allele faction (DAF; this variant call is from deep molecular bins, ie, error correctable) was used to report allele factions for the results from the Archer Reveal ctDNA Targeted assays.
Cohort allele fraction outlier P values and cohort DAF outlier P values were used to identify false-positive results because they represent the probability that a mutation was due to background noise compared with other samples such as a normal cohort. A low P value indicates that the VAF of the called variant lies significantly outside the background noise and thus provides confidence that the mutation is real. A healthy donor cohort was used for background correction at each base analyzed to improve confidence in variant calls. The NGS results from laboratories C, D, and E were derived using a healthy donor cohort (laboratories D and E used the same cohort, and laboratory C used its own cohort), and false-positive results were eliminated using the filter cohort allele fraction outlier P ≤0.01.
Deep Error Eliminating Plasma Sequencing NGS Method
The deep error eliminating plasma sequencing (DEEP-Seq) method uniquely tags each DNA input molecule with a random MBC and amplifies the corresponding product in an efficient targeted amplicon enrichment protocol similar to Peng et al.12 A 25-amplicon panel was developed to cover >500 somatic mutation hotspots across seven genes (Supplemental Table S4). Libraries were prepared from extracted DNA (approximately 14 ng) by a three-stage multiplexed PCR protocol: MBC-addition PCR, stage PCR, and tag PCR. All PCR steps used Q5 Hot Start High-Fidelity DNA Polymerase and bead-based purification between steps. Tagged PCR products were pooled and bead purified for sequencing. Pooled libraries were quantified by qPCR and sequenced on an Illumina MiSeq with version 3 reagents and eight libraries multiplexed per run.
Libraries were analyzed with an optimized bioinformatics pipeline that accounts for background noise and quantifies founder template molecules. Reads were first inspected for index-hopping errors,13 and reads with indices differing from expectation (based on library majority) were removed. The resulting FASTQ files were aligned to the hg19 reference genome using BWA-MEM version 0.7.10, and output BAM files were sorted and indexed with SAMtools version 0.1.19. Amplicons were identified by their chromosomal coordinates, and base-call quality scores were recalibrated using the Q-score recalibration module of the Genome Analysis Toolkit version 1.3-21 (Broad Institute, Cambridge, MA). Local realignment was performed using Genome Analysis Toolkit to ensure all indel reads aligned. The Q-scores for all sites were then adjusted with a Bayesian model similar to Edgar and Flybbjerg14 to account for mate-pair agreement and discordance. This model lowers the Q-scores of mismatched pairs and increases those for matched reads.
Reads were grouped by their MBC sequence and amplicon. To account for artifacts in the MBC sequence itself, read groups representing the same amplicon with barcodes separated by a Levenshtein distance of ≤1 were merged. A consensus likelihood ratio Q-score was determined to account for the quantity and quality of each read with framework and used to deduce the most probable template sequence for each read group with a method similar to Hiatt et al.15 The uncertainty of each observed nucleotide was calculated, given the quality scores of its contributing read group members. The most data-supported nucleotide was selected as the correct template nucleotide, and a corresponding Q-score was calculated for the given position. Consensus calls and their associated posterior Q-score were subsequently used for variant calling.
SNVs were identified with a site-specific model, which eliminates residual nonbiological aberrations. The DEEP-Seq model was trained on >75 diseased and healthy donor plasma and >50 commercially available cfDNA reference standard samples. Briefly, the SNV model incorporates multiple site-specific features for each observed nonreference call and estimates the posterior probability that a given event is a true variant. The DEEP-Seq indel classification model is a heuristic model based on the same training data cohort. The models were applied to the 16 libraries, and true variant status was established for all sites covered by the corresponding panel.
Results
DNA Isolation, Quantification, and Size Determination
The extraction and quantitation methods used by the different laboratories are given in Table 2. Five laboratories used manual extraction and two laboratories used automated instruments. Three of the laboratories using manual extraction used the QIAamp kit: laboratories B and F used carrier RNA during the lysis step and kit-supplied elution buffer in the elution step, whereas laboratory E extracted DNA without carrier RNA and used a Tris buffer for elution.
A one-way analysis of variance analysis was run on the compiled extraction data given in Table 2. Significant differences were noted at confidence levels of P ≤ 0.05. For the 0% (100% wild type) samples, the data from laboratories C (automated) and D (automated) were statistically different compared with the data from the manual process laboratories A, B, F, and G. For the 0.125% samples, the data from laboratory C (automated) were statistically different compared with the data from the manual process laboratories A and F; data from laboratory D (automated) were statistically different compared with the data from the manual process laboratory G. For the 0.5% samples, the data from laboratories B (automated) and C (automated) were statistically different compared with the data from the manual process laboratories A, F, and G. For the 2% samples, the data from laboratory C (automated) were statistically different compared with the data from the manual process laboratories A, B, and F; the data from laboratory D (automated) were statistically different compared to the manual process laboratories F and G.
These data indicate that the two laboratories using the QIAamp with the carrier RNA (laboratories B and F) and the laboratory using the Nucleosnap DNA Plasma kit (laboratory A), tended to have higher DNA yields compared with the laboratories using the automated instruments (laboratories C and D) and laboratory E (manual QIAamp kit with only buffer for elution) (Figure 1 and Table 2). Laboratory G had lower recoveries, possibly because of a DNA dry-down concentration step before the DNA recovery was measured (Figure 1).
Figure 1.
DNA yields for reference material samples across seven laboratories. The dotted line indicates 25 ng/mL, which is the expected DNA concentration as indicated in the manufacturer's data sheet for the 5-mL samples. Data are expressed as means ± 1 SD. n = 3 measurements (laboratories A–F); n = 4 measurements (laboratory G).
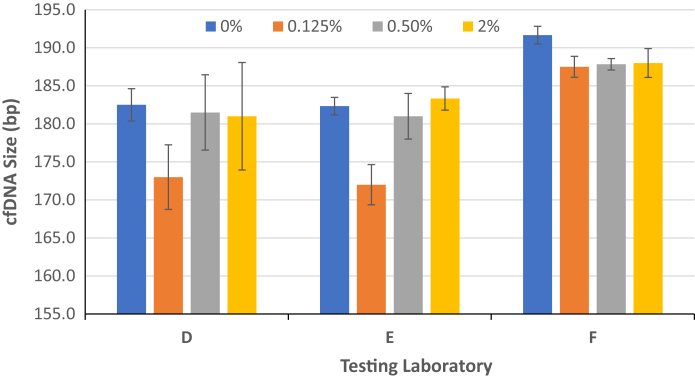
The mean calculated sizes of the DNA recovered for the different samples were similar (Figure 2). The mean size of the purified DNA sample in buffer (sample was approximately 10 ng/μL) was measured with the use of the bioanalyzer by laboratory F at approximately 188 bp. The mean size of the extracted DNA from the reference material samples after extraction (sample at concentration of approximately 1 to 2 ng/μL by laboratories D and E) was slightly lower when analyzed by TapeStation (laboratory D) and bioanalyzer (laboratory E) (Figure 3).
Figure 2.
Mean size of DNA extracted from the reference material samples. The mean DNA fragment sizes were determined by Agilent 2100 Bioanalyzer Expert Software by the peak in the DNA size range of 100 to 250 bp. The size of extracted cell-free DNA (cfDNA; at a concentration of approximately 1 to 2 ng/μL) was determined by TapeStation (laboratory D) or Agilent Bioanalyzer (laboratory E). The size of concentrated cfDNA (approximately 10 ng/μL) was determined by Agilent Bioanalyzer (laboratory F). Data are expressed as means ± 1 SD. n = 3 measurements.
Figure 3.
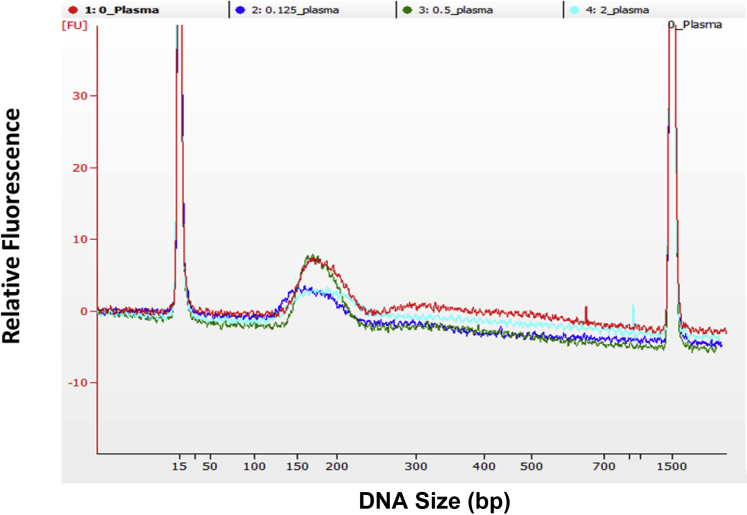
Size profiles of reference material samples. The electropherograms of extracted DNA at a concentration of approximately 2 ng/μL from reference samples were run on the bioanalyzer chips. The identity of the traces is indicated at the top section of the graph.
dPCR Measurements
Two laboratories used different dPCR assays to quantify the VAF for some of the variants. Site F performed dPCR measurements for 39 of 40 variants, whereas site E performed dPCR measurements on a subset of nine variants. Figure 4 shows the measurements of the VAF and SDs for the results of the nine variants tested by both laboratories. The dPCR measurements of the VAFs for 39 of the 40 variants performed by laboratory F are shown in Figure 5.
Figure 4.
Digital PCR results. Digital PCR results from the same nine variants were compared in laboratories E and F. Laboratory E had three replicates. Laboratory F had at least eight replicates for allele frequency at 0.50% and 2% and 14 replicates for allele frequency at 0.125%. Data are expressed as means ± 1 SD.
Figure 5.
Digital PCR results from 39 variants by laboratory F. Digital PCR results from laboratory F have at least eight replicates for allele frequency at 0.50% and 2% and 14 replicates for allele frequency at 0.125%. Data are expressed as means ± 1 SD.
Summary of NGS Assays
Four laboratories used the Archer Reveal ctDNA targeted NGS method, one laboratory used the SiMSen-Seq method, one laboratory used the digital NGS method to measure the VAF of the reference material samples, and one laboratory used the DEEP-Seq NGS method. Summaries of the NGS method used, DNA input, library preparation steps, mean reads per sample, and sequencing depth are given in Tables 6 and 7 for the different methods and laboratories. The mean input DNA for library preparation ranged from 14.1 to 57 ng. The NGS assays generated reads from 1.8 million to 10 million per sample. The mean raw sequencing depth is approximately 5,000 times for all Archer Reveal ctDNA targeted amplicon NGS, 500 times for each of 384 digital NGS reactions (approximately 20,000 reads overall for each amplicon), approximately 11,000 times for DEEP-Seq NGS, and almost 13,000 times for the SiMSen-Seq NGS assay.
Table 7.
Details and Read Metrics from All NGS Assays
| Laboratory | NGS method | NGS platform | NGS replicates | Mean number of reads per sample (in millions) | Mean sequencing depth, unique reads | No. of variants detected in reference material |
|---|---|---|---|---|---|---|
| A | SiMSen-Seq | Illumina MiSeq | 3 | NA | 12,922 (Cons20)∗ | 4 (customized) |
| B | Digital NGS | Ion Torrent PGM | 3 | 6.0 | 500 | 32 |
| C | Archer Reveal ctDNA-targeted NGS | Illumina NextSeq 500 | 3 | 2.8 | 5048 | 23 |
| D | Illumina HiSeq 2500 | 4 | 10.0 | 5269 | ||
| E | Illumina MiSeq | 3 | 7.1 | 5413 | ||
| F | Illumina MiSeq | 4 | 6.9 | 5924 | ||
| G | DEEP-Seq | Illumina MiSeq | 4 | 1.8 | 10,948 | 9 |
ctDNA, circulating tumor; DEEP-Seq, Deep Error Eliminating Plasma Sequencing; NA, not applicable; NGS, next-generation sequencing; PGM, personal genome machine; SiMSeq-Seq, Simple, Multiplexed, PCR-Based Barcoding of DNA for Sensitive Mutation Detection Using Sequencing.
Cons20 is the read depth of 20 per barcode, referred to as consensus 20.
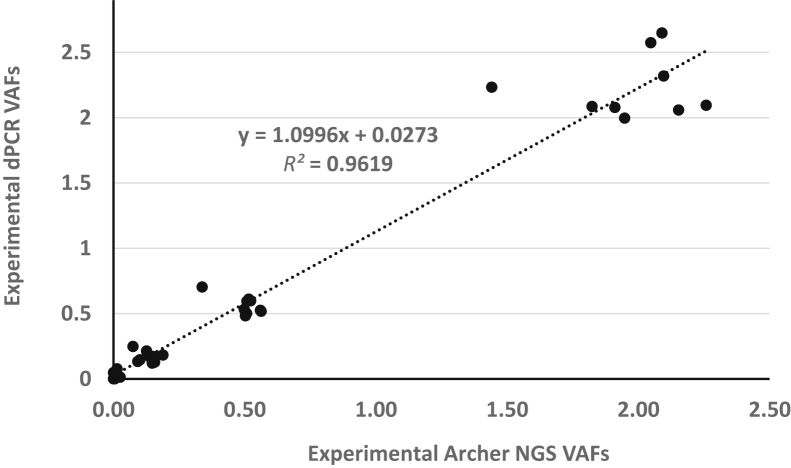
The comparison of the VAF using NGS measurements for the reference material samples is shown in Figure 6. Overall, the data shown in Figure 6 (box and whisker plot) indicate a good correlation among the different NGS methods, although this is limited by the different number of targets measured (range, 4 to 32) (Table 1) and the number of laboratories. The Archer Reveal NGS assay was performed by four laboratories and thus has data for interlaboratory comparison of the same NGS assay platform. Figure 7 shows the correlation of the Archer NGS assay results (from four laboratories) with the mean dPCR results (from two laboratories). The slope of the correlation line was approximately 1.1, indicating that the VAF from the dPCR assays were approximately 10% higher than the NGS assay results.
Figure 6.
Comparison of next-generation sequencing (NGS) results. Orange indicates laboratory A (Simple, Multiplexed, PCR-Based Barcoding of DNA for Sensitive Mutation Detection Using Sequencing NGS, four variants); dark green, laboratory B (digital NGS, 32 variants); blue, laboratory C (Archer NGS, 23 variants); purple, laboratory D (Archer NGS, 23 variants); green, laboratory E (Archer NGS, 23 variants); black, laboratory F (Archer NGS, 23 variants); and red, laboratory G (Deep Error Eliminating Plasma Sequencing NGS, nine variants). The ends of the box are the upper and lower quartiles, so the box spans the interquartile range. The median is marked by a vertical line inside the box, and the whiskers are the two lines outside the box that extend to the highest and lowest observations. n = 3 replicates (laboratories A–F); n = 4 (laboratory G).
Figure 7.
Correlation of digital PCR (dPCR) and Archer next-generation sequencing (NGS) for nine variants. The mean variant allele frequencies (VAFs) of nine variants from the means of two dPCR assays from laboratories E and Lab F were compared with the mean VAF of these variants from four laboratories using Archer Reveal circulating tumor DNA NGS assays. The dotted line is the linear regression of the data points and the line equation is on the graph.
SiMSen-Seq NGS
The SiMSen-Seq NGS method was used to measure the PIK3CA (H1047R), KRAS (G12D), BRAF (V600E), and NRAS (Q61R) mutations. The results are provided in Supplemental Figure S1.
Digital NGS
The AmpliSeq custom NGS panel was designed to identify 36 variants (Supplemental Table S2). The results yielded using digital NGS assays are shown in the Supplemental Figure S2. Four of the 36 variants (primarily mononucleotide repeats) yielded false-positive reads in the reference material VAF 0% sample and in-house healthy cohort samples consistent with the known limitations of Ion Torrent sequencing of mononucleotide repeats. Digital NGS was considered not suitable for the detection of these four variants. This is an example of the application of reference materials to help in the validation of NGS assays and benchmarking different assay platforms.
Archer Reveal ctDNA Targeted NGS
NGS assays using the Archer Reveal 28 panel kit were theoretically able to detect 23 of the 40 variants (Supplemental Table S3). Comparable sequencing depths and VAF results were obtained by the four laboratories despite differences in the Illumina platform and DNA inputs used (Tables 6 and 7, Figure 8). The DAF, representing variant calls from deep molecular bins, which is error- orrected using MBCs, was used in place of VAF. This change did not make much difference for the 0.5% and 2% reference materials, but it improved the detection limit of the NGS assays for the lowest mutant abundance 0.125% sample. When the apparent variant was ≥0.1% in VAF value, the DAF has only a few values at 0.05 or 0.06% across all of the variants in DAF values (Supplemental Table S5). The Archer NGS data for four laboratories are shown in Figure 8 and Supplemental Table S6. The VAFs correlate very well for the 0% and 0.125% samples. However, for most of the 0.125% cfDNA samples, approximately half or more of the variants were filtered out when applying background correction (VAF outlier P ≤ 0.01); background correction using a DAF outlier P value may improve confidence and sensitivity in variant calls (Supplemental Table S5). The results for the VAF 0.5% and 2% samples were similar among all seven NGS assays (Figures 6 and 7).
Figure 8.
Comparison of four Archer Reveal circulating tumor (ctDNA) next-generation sequencing (NGS) results. The data from Archer Reveal ctDNA NGS assay was run by four individual laboratories. Blue indicates laboratory C; orange, laboratory D; gray, laboratory E; and yellow, laboratory F. Data are expressed as means ± 1 SD. n = 3 measurements.
DEEP-Seq NGS Method
Isolations upstream of the DEEP-Seq method had the lowest DNA yields for three of the four reference materials (Table 2). This finding may be attributable to differences in extraction conditions relative to other laboratories (eg, 4-mL plasma inputs, isolation kit, elution buffer, and dry-down of the eluate). Consequently, DNA library input was far less than other methods (approximately 14 vs ≥25 ng). Aligned reads per sample were also lower (1.8 million vs ≥2.8 million) (Table 7). Despite lower DNA input used for library construction, the mean sequencing depth per site was high (Table 7). This finding is likely attributable to the DEEP-Seq pipeline's economical use of sequencing data, which effectively down weights the contribution of small read groups (eg, singletons) rather than discard them outright.
The DEEP-Seq panel targets >500 Catalogue of Somatic Mutations in Cancer; (COSMIC) variants (Supplemental Table S4), including nine of those included in the SeraCare reference materials. For all variants, the observed DEEP-Seq VAF closely corresponds with the expected fractions (Figures 6 and 9). There were no complete dropouts of any expected variant with this approach. However, similar to the Archer Reveal results, many of the expected 0.125% variants would be filtered out based on their model-estimated posterior probabilities (Supplemental Table S7).
Figure 9.
Deep error eliminating plasma sequencing observed variant allele frequency (VAF). Mean observed VAF for nine variants from laboratory G across the four sample types. Data are expressed as means ± 1 SD. n = 4.
Across all sixteen samples examined, the site-specific error rate for the other nonsynonymous COSMIC SNVs covered by this panel remained <0.125% (median observed frequency of 0.0067%). In the wild-type samples, no sites of interest had a median allele frequency greater than this background (adjusted U-test P > 0.07) (Supplemental Table S7). That is, the site-specific baseline for other potentially relevant sites on the DEEP-Seq panel is comparable to the sites altered in the reference materials.
All the NGS methods detected their respective target variants in the 0.5% and 2% VAF samples. The 0.125% and 0% VAF samples are more of a challenge for the NGS measurement methods. The SiMSen-Seq detected the four target variants at the 0.125% level (Supplemental Figure S1). The Digital NGS assay was able to detect 28 of 32 target variants in the 0.125% samples. The Archer Reveal assays were able to detect the 23 target variants in the 0.125% samples, as described above with adjustments to the bioinformatic detection algorithm. The DEEP-Seq detected all seven target variants in the 0.125% samples as described above using the bioinformatic processing steps.
Discussion
The development of reference materials for cfDNA is complicated by the degraded nature of the DNA strands in blood.16 The biological origin of cfDNA is not fully investigated, but the cfDNA size distribution suggests that the DNA molecules are protected by the binding of proteins (in form of nucleosomes) from digestion by nucleases in the cell or blood, producing a degradation pattern similar to the DNA degradation that occurs during apoptosis. The presence and amount of tumor derived cfDNA in patients vary with the stage and type of cancer.17 Comparisons of the size of cfDNA fragments derived from wild-type and tumor cells have shown that the tumor-derived fragments were shorter.18, 19 The sizes of the cfDNA fragments and nucleosomal positioning are different between the DNA derived from tumors and somatic cells, determined from measurements of patient samples, which complicates comparisons.20 In some patient samples, fragments corresponding to dinucleosomal and trinucleosomal sizes are measured.21 The presence of higher-molecular-weight DNA fractions in cfDNA preparations has been reported to be reduced by additional centrifugation steps before extraction, which would remove contaminating white cells in plasma.19 Alternatively, measurement uncertainty can be associated with cfDNA isolation methods.
Digital NGS and dPCR methods are the measurement tools used to determine the concentrations of target sequences (variant or wild type). The dPCR assays are both dependent on the following steps working efficiently: primers binding specifically to the DNA, efficient copying of both DNA strands by the polymerase, and probe binding the target sequence to release the fluorescent label that allows detection. The different NGS assays have their own sources of bias, depending on the steps used for library preparation, detection, and data processing. Some of these steps include different manipulations of the cfDNA fragments (library construction), including end repair, ligation of oligonucleotides, in some cases hybrid selection, PCR amplification, fluorescence or semiconductor detection, and bioinformatics calling of sequences. Different forms of hybrid selection can target the tumor-derived nucleosomal DNA in preference to the larger genomic DNA derived from somatic sources.22
The sites of nuclease cleavage in cfDNA and ctDNA are not random19 because of differences in nucleosome positioning determined by the biology of the tumor and normal cells. Because dPCR and NGS assays have different processing steps and target analyte requirements, it is expected that they will have different biases that will influence the values of the respective measurements. The VAF measurements require accurate measurements of both variant and wild-type observations, and method-specific biases in either of the targets will result in inconsistent measurements. This finding is also true for reference materials used for assignment of VAF measurements; reference materials should mimic the target analytes found in patient samples as closely as possible to achieve commutability.23
The preanalytical steps in the measurements of cfDNA in patient samples are important to obtain reliable results. A recent review of the clinical applications and preanalytical processing of cfDNA from patient samples provides the current evidence on the effects of sample collection, processing, analytical validity, interpretation and reporting, and clinical validity and utility.24
Previous studies have found that different extraction methods can result in different yields of cfDNA from patient plasma.21, 25, 26, 27 Several recent studies focusing on the recovery of cfDNA from blood using commercial kits have confirmed the differences in the total yield and different recovery, depending on the size of the DNA.28, 29
The efficient isolation of circulating cfDNA from a limited amount of plasma is important in the analysis of ctDNA that can comprise well <1% of cfDNA. Studies have reported that different cfDNA extraction methods appear to have different efficiencies.26, 28, 30 These data are consistent with those studies in which the manual QIAGEN QIAamp Circulating Nucleic Acid kit resulted in some of the highest reported yields from the reference materials, and this kit was also used by the manufacturer to develop the reference materials. Differences between the extraction methods used here include the mechanism of DNA binding to a solid phase (eg, glass or anion exchange), (if used) the chaotropic salt and any organic solvents that promote the binding of nucleic acids, and additional additives that promote the recovery of circulating cfDNA. In addition, the reference material used in this study was a combination of synthetic plasma and DNA stabilized by encapsulation within lipid bilayers. Similarly, a portion of circulating cfDNA is likely still associated with cellular debris and plasma components. Thus, any extraction method that is less efficient at recovering such DNA would be expected to have a lower yield.
Consistent with these studies, these limited results by the laboratories that used different methods, modifications, and personnel indicated that reference material DNA yields were influenced by the extraction method and automated methods tended to have lower yields compared with some of the manual procedures, although the data were limited in this study and more data are needed. The addition of carrier RNA in the manual method lysis buffer appears to help improve the recovery of the DNA.
A recent study used ddPCR assays for seven different gene targets to quantify cfDNA in patient samples.25 Some of the target gene measurements varied by more than twofold compared with the other targets, and they recommended a multireference gene approach for quantitation. A multiplex dPCR assay targeting nine different genomic regions with short (mean, 71 bp) and long (mean, 471 bp) amplicons was used to investigate size and recovery of patient cfDNA with extraction method and blood collection protocols.27 Even with pristine genomic DNA extracted from cells, in our experience, the quantitation of genomic DNA can be a challenge.31 Absorbance measurements are straightforward but require pure DNA of sufficient concentration to produce good results. Fluorescence dye binding methods widely used are sensitive but can be biased by contaminants or degraded DNA.32, 33 An appropriate reference material for cfDNA quantitation would be useful for assay calibration and validation.
The dPCR assays used to characterize the reference material are reliable, are economical, and have a direct sample preparation, and the data can be used to assign values that are useful with the limitations of this measurement method. The dPCR assays and the NGS measurements have their own biases, as have been discussed. The data from this evaluation study indicate a reasonable correlation between the dPCR assays (two laboratories) and the NGS assays (seven laboratories using four different methods) (Figures 6 and 7). The direct comparisons of the NGS methods are not straight forward because they are measuring different numbers of variants and different targets. In this study, the targets for the NGS methods ranged from four to 32, including SNVs and indels. The read depth of the variants from the NGS methods will depend on not only the amount of DNA used for the library preparation but also the steps in the library preparation, the amount of multiplexing targets, the amount of library loaded on the specific type of sequencer, and the bioinformatic methods used.
The detection of variants in the 0% VAF sample can be attributable to the presence of variants in the DNA used as the background host DNA (true-positive results) or errors (false-positive results) that could be attributable to problems in the processing steps in the preparation of the reference material, the detection method, or the bioinformatic processing of the data. The presence of true-positive results in the background host DNA (albeit at low VAF) has implications in developing even more sensitive assays, although the clinical significance of the presence of variants at low frequencies needs to be investigated. The availability of well-characterized reference materials will facilitate the implementation of sensitive reliable measurement methods for patient samples to help determine the significance of very low concentrations of mutations.
Using an orthogonal measurement method can help to distinguish between false- and true-positive results. In several cases, the dPCR assays from two laboratories detected variants in the wild-type sample [for instance AKT1 (E17K) and PIK3CA (H1047R)] (Figure 4), and three of the laboratories using the Archer Reveal ctDNA NGS assay also detected the variants in the wild-type sample (Figure 8). We plan on investigating the source of the positive results in the background host DNA samples.
Gene fusions and copy number alterations are important variants for cancer diagnostics and treatment targets. This reference material contains two gene fusions (Table 1) that were not targeted in this study. Gene fusion target assays are complicated by the variable length of DNA territory (introns) that must be screened to detect the different junction points in patient samples. Gene copy number variants are challenging to measure in the ctDNA environment given the high background of gene targets derived from the normal cells and tissues present in the blood. NIST has developed gene copy number reference materials based on cell line genomic DNA for ERBB2 (NIST SRM 2373) and EGFR and MET (NIST RM 8366). We are working on developing reference materials for gene fusions and copy number alterations in a form that is a suitable simulant for ctDNA measurement methods.
It is a challenge to reliably measure low VAF patient samples and reference materials will assist in these measurements. It is important to standardize the preanalytical steps, including sample collection, storage, and processing, in addition to optimizing the sample preparation, library preparation, instrument, and bioinformatic processing steps to detect the low VAF samples.
Newman et al34 used an integrated digital error suppression software approach to eliminate background artifacts and identify cfDNA using barcoding (Cancer Personalized Profiling by Deep Sequencing). The SiMSen-Seq was introduced by Ståhlberg et al8, 9 by using reduced primer concentrations, elongated PCR extension times, and hairpin-protected barcode primers to generate targeted barcoded libraries. SiMSen-Seq is able to achieve detection of variant alleles at <0.1% VAF.
The Archer Reveal ctDNA targeted kit uses a barcoding and seminested PCR–based library preparation method and achieves low VAF detection through barcoding correction and noise reduction through cohort comparison that surveys background noise at each nucleotide position. The digital NGS method was developed to improve reliable detection of lower VAF without the need of barcoding.11, 35
In the current study, some variants were not detected at the 0.125% concentration. The mean read depth and mean reads per sample may correlate with the ability to discriminate lower VAFs from the wild-type control background. Using the cohort outlier P values was helpful to distinguish background noise and increase detection of true variants.
Sequence variants identified using NGS-based tests need to be present at sufficient concentrations to be measured as true mutations rather than background sequencing errors.36 NGS is being evaluated as a test to detect low-abundance somatic mutations, but the rate of sequencing errors generated by NGS assays poses challenges to the detection of low-abundance mutations. In principle, the ability of NGS to accurately detect low-abundance mutations could be improved by using digital strategies, analogous to dPCR. A digital NGS method for this purpose was applied by performing discrete NGS analyses on many (n = 384) individual aliquots of DNA from a single biological sample in which each aliquot contains only a few genome equivalents of DNA. Each aliquot can then be expected to have either zero or one mutation-containing DNA template at each nucleotide of interest in addition to a small number of wild-type templates. True somatic mutations should be detectable in more than one aliquot.
Tables 6 and 7 list the major differences in the NGS assays used in this interlaboratory evaluation. There were comparable results between the NGS and PCR methods compared in this study. The reference material evaluated in this study provides a useful tool to help evaluate some of the processing steps in ctDNA measurements, including extraction efficiency, and it provides a uniform sample for the 40 variants that can be used to establish a baseline, evaluate measurement consistency over time, and assess interlaboratory results for the analytical and bioinformatics steps. This study focused on the utility of cfDNA reference materials as important tools to monitor the consistency of measurements using dPCR and NGS assays for variant measurements over time, with different personnel, processes, and reagent changes.
The dPCR assays used an input of 20 ng of DNA, using the mass of a normal human haploid genome of 3.3 pg,31 which is equivalent to approximately 6060 genomes. The 0.125% VAF sample would be expected to have approximately seven variant copies. This is a good test of the detection limit of for a dPCR assay. The wild-type DNA sample in this case is derived from a cell line (GM 24385). Cell lines can accumulate somatic variants during growth in culture. The Genome in a Bottle reference materials (NIST RM 8391, derived from GM 24385) is a well annotated genome that will be a useful material to benchmark the performance of NGS panels.37 The Global Alliance for Genomics and Health have established best practices for benchmarking germline small variant calls in human genomes.38 It is useful to have reference materials with high confidence allele calls for these important variant locations to help determine whether the variants that occur at low allele frequencies are attributable to false-positive results because of the measurement method or present in the materials at low frequency. We are planning additional measurements to determine the source of the low-frequency variants in the wild-type DNA.
The establishment of certified values for the variants in reference material will require careful characterization of the materials and investigation into the sources of uncertainty in the measurements. Determination of the biases in the methods variant measurement and the quantitation of the extracted DNA, different assays, and bioinformatics analysis of the data are essential.
Footnotes
Supported in part by an interagency agreement between the National Cancer Institute and the National Institute of Standards and Technology; NIH Small Business Innovative Research grant 12475715 (J. L. L., L.R., and B.C.H.); NIH grant CA210170 (M.G.); Sahlgrenska Academy (ALF) at University of Gothenburg grant 716321; Knut and Alice Wallenberg Foundation, Wallenberg Center for Molecular and Translational Medicine, University of Gothenburg; Swedish Cancer Society grant 2016-438; Swedish Research Council grant 2017-01392; Swedish Childhood Cancer Foundation grant 2017-0043; the University of North Carolina (UNC) Department of Pathology and Laboratory Medicine; the University Cancer Research Fund; and the UNC Integrated Translational Science Center award NCI 5-U10CA181009-05.
Disclosures: Certain commercial equipment, instruments, and materials are identified to specify the experimental procedure. In no case does such identification imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the materials or equipment are necessarily the best available for the purpose. Y.K., F.L.T., and R.G. are employed by SeraCare Life Sciences. J.L.L., L.R., and B.C.H. are employed by Asuragen, Inc. A.G. is employed by and owns stock in Archer Diagnostics, Inc. T.E.G. and A.S. are co-inventors of SiMSen-Seq that is patent pending (US serial number 15/552,618). The patent applicant was the Trustees of Boston University, and the patent is specifically covering the use of molecular hairpins to protect molecular barcodes hairpins (SiMSen-Seq).
Portions of this were work presented at the Association for Cancer Research Annual Meeting; April 14–18, 2018; Chicago, IL.
Current address of E.V.S., QIAGEN, Enterprise Genomic Services, QIAGEN Sciences Inc., Frederick, Maryland.
Supplemental material for this article can be found at https://doi.org/10.1016/j.jmoldx.2019.03.006.
Supplemental Data
SiMSen-Seq next-generation sequencing (NGS) data for reference materials for four variants: PIK3CA (p.H1047R), KRAS (p.G12D), BRAF (p.V600E), and NRAS (p.Q61R). Data are expressed as means ± 1 SD. n = 3 measurements.
Digital next-generation sequencing (NGS) results for 32 variants. Data are expressed as means ± 1 SD. n = 3 measurements.
References
- 1.Husain H., Velculescu V.E. Cancer DNA in the circulation the liquid biopsy. JAMA Insights. 2017;318:1272–1274. doi: 10.1001/jama.2017.12131. [DOI] [PMC free article] [PubMed] [Google Scholar]; Husain H, Velculescu VE: Cancer DNA in the Circulation The Liquid Biopsy. JAMA Insights 2017, 318:1272-1274. [DOI] [PMC free article] [PubMed]
- 2.Volik S., Alcaide M., Morin R.D., Collins C. Cell-free DNA (cfDNA): clinical significance and utility in cancer shaped by emerging technologies. Mol Cancer Res. 2016;14:898–908. doi: 10.1158/1541-7786.MCR-16-0044. [DOI] [PubMed] [Google Scholar]; Volik S, Alcaide M, Morin RD, Collins C: Cell-free DNA (cfDNA): Clinical Significance and Utility in Cancer Shaped By Emerging Technologies. Mol Cancer Res 2016, 14:898-908. [DOI] [PubMed]
- 3.Wan J.C.M., Massie C., Garcia-Corbacho J., Mouliere F., Brenton J.D., Caldas C., Pacey S., Baird R., Rosenfeld N. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017;17:223–238. doi: 10.1038/nrc.2017.7. [DOI] [PubMed] [Google Scholar]; Wan JCM, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD, Caldas C, Pacey S, Baird R, Rosenfeld N: Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 2017, 17:223-238. [DOI] [PubMed]
- 4.Chang Y., Tolani B., Nie X., Zhi X., Hu M., He B. Review of the clinical applications and technological advances of circulating tumor DNA in cancer monitoring. Ther Clin Risk Manag. 2017;13:1363–1374. doi: 10.2147/TCRM.S141991. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chang Y, Tolani B, Nie X, Zhi X, Hu M, He B: Review of the clinical applications and technological advances of circulating tumor DNA in cancer monitoring. Ther Clin Risk Manag 2017, 13:1363-1374. [DOI] [PMC free article] [PubMed]
- 5.Diaz L.A., Jr., Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014;32:579–586. doi: 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]; Diaz LA, Jr., Bardelli A: Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 2014, 32:579-586. [DOI] [PMC free article] [PubMed]
- 6.Aravanis A.M., Lee M., Klausner R.D. Next-generation sequencing of circulating tumor DNA for early cancer detection. Cell. 2017;168:571–574. doi: 10.1016/j.cell.2017.01.030. [DOI] [PubMed] [Google Scholar]; Aravanis AM, Lee M, Klausner RD: Next-Generation Sequencing of Circulating Tumor DNA for Early Cancer Detection. Cell 2017, 168:571-574. [DOI] [PubMed]
- 7.Salk J.J., Schmitt M.W., Loeb L.A. Enhancing the accuracy of next-generation sequencing for detecting rare and subclonal mutations. Nat Rev Genet. 2018;19:269–285. doi: 10.1038/nrg.2017.117. [DOI] [PMC free article] [PubMed] [Google Scholar]; Salk JJ, Schmitt MW, Loeb LA: Enhancing the accuracy of next-generation sequencing for detecting rare and subclonal mutations. Nat Rev Genet 2018, 19:269-285. [DOI] [PMC free article] [PubMed]
- 8.Ståhlberg A., Krzyzanowski P.M., Jackson J.B., Egyud M., Stein L., Godfrey T.E. Simple, multiplexed, PCR-based barcoding of DNA enables sensitive mutation detection in liquid biopsies using sequencing. Nucleic Acids Res. 2016;44:e105. doi: 10.1093/nar/gkw224. [DOI] [PMC free article] [PubMed] [Google Scholar]; Stahlberg A, Krzyzanowski PM, Jackson JB, Egyud M, Stein L, Godfrey TE: Simple, multiplexed, PCR-based barcoding of DNA enables sensitive mutation detection in liquid biopsies using sequencing. Nucleic Acids Res 2016, 44:e105. [DOI] [PMC free article] [PubMed]
- 9.Ståhlberg A., Krzyzanowski P.M., Egyud M., Filges S., Stein L., Godfrey T.E. Simple multiplexed PCR-based barcoding of DNA for ultrasensitive mutation detection by next-generation sequencing. Nat Protoc. 2017;12:664–682. doi: 10.1038/nprot.2017.006. [DOI] [PubMed] [Google Scholar]; Stahlberg A, Krzyzanowski PM, Egyud M, Filges S, Stein L, Godfrey TE: Simple multiplexed PCR-based barcoding of DNA for ultrasensitive mutation detection by next-generation sequencing. Nat Protoc 2017, 12:664-682. [DOI] [PubMed]
- 10.Schmitt M.W., Kennedy S.R., Salk J.J., Fox E.J., Hiatt J.B., Loeb L.A. Detection of ultra-rare mutations by next-generation sequencing. Proc Natl Acad Sci U S A. 2012;109:14508–14513. doi: 10.1073/pnas.1208715109. [DOI] [PMC free article] [PubMed] [Google Scholar]; Schmitt MW, Kennedy SR, J.J. S, Fox EJ, Hiatt JB, Loeb LA: Detection of ultra-rare mutations by next-generation sequencing. Proc Natl Acad Sci U S A 2012, 109:14508-14513. [DOI] [PMC free article] [PubMed]
- 11.Yu J., Sadakari Y., Shindo K., Suenaga M., Brant A., Almario J.A.N., Borges M., Barkley T., Fesharakizadeh S., Ford M., Hruban R.H., Shin E.J., Lennon A.M., Canto M.I. Digital next-generation sequencing identifies low-abundance mutations in pancreatic juice samples collected from the duodenum of patients with pancreatic cancer and intraductal papillary mucinous neoplasms. Gut. 2017;66:1677–1687. doi: 10.1136/gutjnl-2015-311166. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yu J, Sadakari Y, Shindo K, Suenaga M, Brant A, Almario JAN, Borges M, Barkley T, Fesharakizadeh S, Ford M, Hruban RH, Shin EJ, Lennon AM, Canto MI: Digital next-generation sequencing identifies low-abundance mutations in pancreatic juice samples collected from the duodenum of patients with pancreatic cancer and intraductal papillary mucinous neoplasms. Gut 2017, 66:1677-1687. [DOI] [PMC free article] [PubMed]
- 12.Peng Q., Vijaya S.R., Lewis M., Randad P., Wang Y. Reducing amplification artifacts in high multiplex amplicon sequencing by using molecular barcodes. BMC Genomics. 2015;16:589. doi: 10.1186/s12864-015-1806-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; Peng Q, Vijaya S, R., Lewis M, Randad P, Wang Y: Reducing amplification artifacts in high multiplex amplicon sequencing by using molecular barcodes. BMC Genomics 2015, 16:589. [DOI] [PMC free article] [PubMed]
- 13.Kircher M., Sawyer S., Meyer M. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res. 2012;40:e3. doi: 10.1093/nar/gkr771. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kircher M, Sawyer S, Meyer M: Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res 2012, 40:e3. [DOI] [PMC free article] [PubMed]
- 14.Edgar R., Flybbjerg H. Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics. 2015;31:3476–3482. doi: 10.1093/bioinformatics/btv401. [DOI] [PubMed] [Google Scholar]; Edgar R, Flybbjerg H: Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics 2015, 31:3476-3482. [DOI] [PubMed]
- 15.Hiatt J.B., Pritchard C.C., Salipante S.J., O'Roak B.J., Shendure J. Single molecule molecular inversion probes for targeted, high-accuracy detection of low-frequency variation. Genome Res. 2013;23:843–854. doi: 10.1101/gr.147686.112. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hiatt JB, Pritchard CC, Salipante SJ, O'Roak BJ, Shendure J: Single molecule molecular inversion probes for targeted, high-accuracy detection of low-frequency variation. Genome Res 2013, 23:843-854. [DOI] [PMC free article] [PubMed]
- 16.Jiang P., Lo Y.M.D. The long and short of circulating cell-free DNA and the ins and outs of molecular diagnostics. Trends Genet. 2016;32:360–371. doi: 10.1016/j.tig.2016.03.009. [DOI] [PubMed] [Google Scholar]; Jiang P, Lo YMD: The Long and Short of Circulating Cell-Free DNA and the Ins and Outs of Molecular Diagnostics. Trends Genet 2016, 32:360-371. [DOI] [PubMed]
- 17.Bettegowda C., Sausen M., Leary R.J., Kinde I., Wang Y., Agrawal N. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, Antonarakis ES, Azad NS, Bardelli A, Brem H, Cameron JL, Lee CC, Fecher LA, Gallia GL, Gibbs P, Le D, Giuntoli RL, Goggins M, Hogarty MD, Holdhoff M, Hong SM, Jiao Y, Juhl HH, Kim JJ, Siravegna G, Laheru DA, Lauricella C, Lim M, Lipson EJ, Marie SK, Netto GJ, Oliner KS, Olivi A, Olsson L, Riggins GJ, Sartore-Bianchi A, Schmidt K, Shih lM, Oba-Shinjo SM, Siena S, Theodorescu D, Tie J, Harkins TT, Veronese S, Wang TL, Weingart JD, Wolfgang CL, Wood LD, Xing D, Hruban RH, Wu J, Allen PJ, Schmidt CM, Choti MA, Velculescu VE, Kinzler KW, Vogelstein B, Papadopoulos N, Diaz LAJ: Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014, 6:224ra24. [DOI] [PMC free article] [PubMed]
- 18.Jiang P., Chan C.W., Chan K.C., Cheng S.H., Wong J., Wong V.W., Wong G.L., Chan S.L., Mok T.S., Chan H.L., Lai P.B., Chiu R.W., Lo Y.M. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc Natl Acad Sci U S A. 2015;112:E1317–E1325. doi: 10.1073/pnas.1500076112. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jiang P, Chan CW, Chan KC, Cheng SH, Wong J, Wong VW, Wong GL, Chan SL, Mok TS, Chan HL, Lai PB, Chiu RW, Lo YM: Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc Natl Acad Sci U S A 2015, 112:E1317-E1325. [DOI] [PMC free article] [PubMed]
- 19.Raymond C.K., Hernandez J., Karr R., Hill K., Li M. Collection of cell-free DNA for genomic analysis of solid tumors in a clinical laboratory setting. PLoS One. 2017;12:e0176241. doi: 10.1371/journal.pone.0176241. [DOI] [PMC free article] [PubMed] [Google Scholar]; Raymond CK, Hernandez J, Karr R, Hill K, Li M: Collection of cell-free DNA for genomic analysis of solid tumors in a clinical laboratory setting. PLoS One 2017, 12:e0176241. [DOI] [PMC free article] [PubMed]
- 20.Mouliere F., Rosenfeld N. Circulating tumor-derived DNA is shorter than somatic DNA in plasma. Proc Natl Acad Sci U S A. 2015;112:3178–3179. doi: 10.1073/pnas.1501321112. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mouliere F, Rosenfeld N: Circulating tumor-derived DNA is shorter than somatic DNA in plasma. Proc Natl Acad Sci U S A 2015, 112:3178-3179. [DOI] [PMC free article] [PubMed]
- 21.Pérez-Barrios C., Nieto-Alcolado I., Torrente M., Jiménez-Sánchez C., Calvo V., Gutierrez-Sanz L., Palka M., Donoso-Navarro E., Provencio M., Romero A. Comparison of methods for circulating cell-free DNA isolation using blood from cancer patients: impact on biomarker testing. Transl Lung Cancer Res. 2016;5:665–672. doi: 10.21037/tlcr.2016.12.03. [DOI] [PMC free article] [PubMed] [Google Scholar]; Perez-Barrios C, Nieto-Alcolado I, Torrente M, Jimenez-Sanchez C, Calvo V, Gutierrez-Sanz L, Palka M, Donoso-Navarro E, Provencio M, Romero A: Comparison of methods for circulating cell-free DNA isolation using blood from cancer patients: impact on biomarker testing. Transl Lung Cancer Res 2016, 5:665-672. [DOI] [PMC free article] [PubMed]
- 22.Paweletz C.P., Sacher A.G., Raymond C.K., Alden R.S., O'Connell A., Mach S.L., Kuang Y., Gandhi L., Kirschmeier P., English J.M., Lim L.P., Jänne P.A., Oxnard G.R. Bias-corrected targeted next-generation sequencing for rapid, multiplexed detection of actionable alterations in cell-free DNA from advanced lung cancer patients. Clin Cancer Res. 2016;22:915–922. doi: 10.1158/1078-0432.CCR-15-1627-T. [DOI] [PMC free article] [PubMed] [Google Scholar]; Paweletz CP, Sacher AG, Raymond CK, Alden RS, O'Connell A, Mach SL, Kuang Y, Gandhi L, Kirschmeier P, English JM, Lim LP, Janne PA, Oxnard GR: Bias-Corrected Targeted Next-Generation Sequencing for Rapid, Multiplexed Detection of Actionable Alterations in Cell-Free DNA from Advanced Lung Cancer Patients. Clin Cancer Res 2016, 22:915-922. [DOI] [PMC free article] [PubMed]
- 23.Vesper H.W., Miller W.G., Myers G.L. Reference materials and commutability. Clin Biochem Rev. 2007;28:139–147. [PMC free article] [PubMed] [Google Scholar]; Vesper HW, Miller WG, Myers GL: Reference materials and commutability. Clin Biochem Rev 2007, 28:139-147. [PMC free article] [PubMed]
- 24.Merker J.D., Oxnard G.R., Compton C., Diehn M., Hurley P., Lazar A.J., Lindeman N., Lockwood C.M., Rai A.J., Schilsky R.L., Tsimberidou A.M., Vasalos P., Billman B.L., Oliver T.K., Bruinooge S.S., Hayes D.F., Turner N.C. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists joint review. J Clin Oncol. 2018;36:1631–1641. doi: 10.1200/JCO.2017.76.8671. [DOI] [PubMed] [Google Scholar]; Merker JD, Oxnard GR, Compton C, Diehn M, Hurley P, Lazar AJ, Lindeman N, Lockwood CM, Rai AJ, Schilsky RL, Tsimberidou AM, Vasalos P, Billman BL, Oliver TK, Bruinooge SS, Hayes DF, Turner NC: Circulating Tumor DNA Analysis in Patients With Cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J Clin Oncol 2018, 36:1631-1641. [DOI] [PubMed]
- 25.Devonshire A.S., Whale A.S., Gutteridge A., Jones G., Cowen S., Foy C.A., Huggett J.F. Towards standardisation of cell-free DNA measurement in plasma: controls for extraction efficiency, fragment size bias and quantification. Anal Bioanal Chem. 2014;406:6499–6512. doi: 10.1007/s00216-014-7835-3. [DOI] [PMC free article] [PubMed] [Google Scholar]; Devonshire AS, Whale AS, Gutteridge A, Jones G, Cowen S, Foy CA, Huggett JF: Towards standardisation of cell-free DNA measurement in plasma: controls for extraction efficiency, fragment size bias and quantification. Anal Bioanal Chem 2014, 406:6499-6512. [DOI] [PMC free article] [PubMed]
- 26.Sorber L., Zwaenepoel K., Deschoolmeester V., Roeyen G., Lardon F., Rolfo C., Pauwels P. A comparison of cell-free DNA isolation kits: isolation and quantification of cell-free DNA in plasma. J Mol Diagn. 2017;19:162–168. doi: 10.1016/j.jmoldx.2016.09.009. [DOI] [PubMed] [Google Scholar]; Sorber L, Zwaenepoel K, Deschoolmeester V, Roeyen G, Lardon F, Rolfo C, Pauwels P: A Comparison of Cell-Free DNA Isolation Kits: Isolation and Quantification of Cell-Free DNA in Plasma. J Mol Diagn 2017, 19:162-168. [DOI] [PubMed]
- 27.Markus H., Contente-Cuomo T., Farooq M., Liang W.S., Borad M.J., Sivakumar S., Gollins S., Tran N.L., Dhruv H.D., Berens M.E., Bryce A., Sekulic A., Ribas A., Trent J.M., LoRusso P.M., Murtaza M. Evaluation of pre-analytical factors affecting plasma DNA analysis. Sci Rep. 2018;8:7375. doi: 10.1038/s41598-018-25810-0. [DOI] [PMC free article] [PubMed] [Google Scholar]; Markus H, Contente-Cuomo T, Farooq M, Liang WS, Borad MJ, Sivakumar S, Gollins S, Tran NL, Dhruv HD, Berens ME, Bryce A, Sekulic A, Ribas A, Trent JM, LoRusso PM, Murtaza M: Evaluation of pre-analytical factors affecting plasma DNA analysis. Sci Rep 2018, 8:7375. [DOI] [PMC free article] [PubMed]
- 28.Diefenbach R.J., Lee J.H., Kefford R.F., Rizos H. Evaluation of commercial kits for purification of circulating free DNA. Cancer Genet. 2018;228-229:21–27. doi: 10.1016/j.cancergen.2018.08.005. [DOI] [PubMed] [Google Scholar]; Diefenbach RJ, Lee JH, Kefford RF, Rizos H: Evaluation of commercial kits for puriþcation of circulating free DNA. Cancer Genet 2018, in press. [DOI] [PubMed]
- 29.Warton K., Samimi G. Methylation of cell-free circulating DNA in the diagnosis of cancer. Front Mol Biosci. 2015;F2:13. doi: 10.3389/fmolb.2015.00013. eCollection 2015:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]; Warton K, Samimi G: Methylation of cell-free circulating DNA in the diagnosis of cancer. Front Mol Biosci 2015, F2:13. eCollection 2015:1-10. [DOI] [PMC free article] [PubMed]
- 30.van Dessel L.F., Vitale S.R., Helmijr J.C.A., Wilting S.M., van der Vlugt-Daane M., Oomen-de Hoop E., Sleijfer S., Martens J.W.M., Jansen M.P.H.M., Lolkema M.P. High-throughput isolation of circulating tumor DNA: a comparison of automated platforms. Mol Oncol. 2019;13:392–402. doi: 10.1002/1878-0261.12415. [DOI] [PMC free article] [PubMed] [Google Scholar]; van Dessel LF, Vitale SR, Helmijr JCA, Wilting SM, van der Vlugt-Daane M, Oomen-de Hoop E, Sleijfer S, Martens JWM, Jansen MPHM, Lolkema MP: High-throughput isolation of circulating tumor DNA: a comparison of automated platforms. Mol Oncol 2019, 13:392-402. [DOI] [PMC free article] [PubMed]
- 31.He H.J., Stein E.V., DeRose P., Cole K.D. Limitations of methods for measuring the concentration of human genomic DNA and oligonucleotide samples. Biotechniques. 2018;64:59–68. doi: 10.2144/btn-2017-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]; He HJ, Stein EV, DeRose P, Cole KD: Limitations of methods for measuring the concentration of human genomic DNA and oligonucleotide samples. Biotechniques 2018, 64:59-68. [DOI] [PMC free article] [PubMed]
- 32.Georgiou C.D., Papapostolou I. Assay for the quantification of intact/fragmented genomic DNA. Anal Biochem. 2006;358:247–256. doi: 10.1016/j.ab.2006.07.035. [DOI] [PubMed] [Google Scholar]; Georgiou CD, Papapostolou I: Assay for the quantification of intact/fragmented genomic DNA. Anal Biochem 2006, 358:247-256. [DOI] [PubMed]
- 33.Li X., Wu Y., Zhang L., Cao Y., Li Y., Li J., Zhu L., Wu G. Comparison of three common DNA concentration measurement methods. Anal Biochem. 2014;451:18–24. doi: 10.1016/j.ab.2014.01.016. [DOI] [PubMed] [Google Scholar]; Li X, Wu Y, Zhang L, Cao Y, Li Y, Li J, Zhu L, Wu G: Comparison of three common DNA concentration measurement methods. Anal Biochem 2014, 451:18-24. [DOI] [PubMed]
- 34.Newman A.M., Lovejoy A.F., Klass D.M., Kurtz D.M., Chabon J.J., Scherer F., Stehr H., Liu C.L., Bratman S.V., Say C., Zhou L., Carter J.N., West R.B., Sledge G.W., Shrager J.B., Loo B.W., Neal J.W., Wakelee H.A., Diehn M., Alizadeh A.A. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol. 2016;34:547–555. doi: 10.1038/nbt.3520. [DOI] [PMC free article] [PubMed] [Google Scholar]; Newman AM, Lovejoy AF, Klass DM, Kurtz DM, Chabon JJ, Scherer F, Stehr H, Liu CL, Bratman SV, Say C, Zhou L, Carter JN, West RB, Sledge GW, Shrager JB, Loo BW, Neal JW, Wakelee HA, Diehn M, Alizadeh AA: Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol 2016, 34:547-555. [DOI] [PMC free article] [PubMed]
- 35.Suenaga M., Yu J., Shindo K., Tamura K., Almario J.A.N., Zaykoski C.M., Witmer P.D., Fesharakizadeh S., Borges M., Lennon A.M., Shin E.J., Canto M.I., Goggins M.G. Pancreatic juice mutation concentrations can help predict the grade of dysplasia in patients undergoing pancreatic surveillance. Clin Cancer Res. 2018;24:2963–2974. doi: 10.1158/1078-0432.CCR-17-2463. [DOI] [PMC free article] [PubMed] [Google Scholar]; Suenaga M, Yu J, Shindo K, Tamura K, Almario JAN, Zaykoski CM, Witmer PD, Fesharakizadeh S, Borges M, Lennon AM, Shin EJ, Canto MI, Goggins MG: Pancreatic juice mutation concentrations can help predict the grade of dysplasia in patients undergoing pancreatic surveillance. Clinical cancer research : an official journal of the American Association for Cancer Research 2018. [DOI] [PMC free article] [PubMed]
- 36.Quail M.A., Smith M., Coupland P., Otto T.D., Harris S.R., Connor T.R., Bertoni A., Swerdlow H.P., Gu Y. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics. 2012;13:341. doi: 10.1186/1471-2164-13-341. [DOI] [PMC free article] [PubMed] [Google Scholar]; Quail MA, Smith M, Coupland P, Otto TD, Harris SR, Connor TR, Bertoni A, Swerdlow HP, Gu Y: A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics 2012, 13:341. [DOI] [PMC free article] [PubMed]
- 37.Cleveland M.H., Zook J.M., Salit M., Vallone P.M. Determining performance metrics for targeted next-generation sequencing panels using reference materials. J Mol Diagn. 2018;20:583–590. doi: 10.1016/j.jmoldx.2018.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cleveland MH, Zook JM, Salit M, Vallone PM: Determining Performance Metrics for Targeted Next-Generation Sequencing Panels Using Reference Materials. J Mol Diagn 2018, 20:583-590. [DOI] [PMC free article] [PubMed]
- 38.Krusche P., Trigg L., Boutros P.C., Mason C.E., De La Vega F.M., Moore B.L., Gonzalez-Porta M., Eberle M.A., Tezak Z., Labadibi S., Truty R., Asimenos G., Funke B., Fleharty M., Salit M., Zook J.M. Best practices for benchmarking germline small variant calls in human genomes. Nat Biotechnol. 2019;37:555–560. doi: 10.1038/s41587-019-0054-x. [DOI] [PMC free article] [PubMed] [Google Scholar]; Krusche P, Trigg L, Boutros PC, Mason CE, De La Vega FM, Moore BL, Gonzalez-Porta M, Eberle MA, Tezak Z, Labadibi S, Truty R, Asimenos G, Funke B, Fleharty M, Salit M, Zook JM: Best Practices for Benchmarking Germline Small Variant Calls in Human Genomes BioRxiv 2018, doi: https://doi.org/10.1101/270157
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SiMSen-Seq next-generation sequencing (NGS) data for reference materials for four variants: PIK3CA (p.H1047R), KRAS (p.G12D), BRAF (p.V600E), and NRAS (p.Q61R). Data are expressed as means ± 1 SD. n = 3 measurements.
Digital next-generation sequencing (NGS) results for 32 variants. Data are expressed as means ± 1 SD. n = 3 measurements.