Abstract
This study describes the analytical performance of the QuantideX qPCR BCR-ABL IS Kit, the first Food and Drug Administration–cleared assay designed to monitor breakpoint cluster region–Abelson tyrosine-protein kinase 1 (BCR-ABL1) fusion transcripts isolated from peripheral blood specimens from patients with chronic myeloid leukemia. This multiplex real-time quantitative RT-PCR assay amplifies both e13a2 and e14a2 Major BCR-ABL1 transcripts and the reference target ABL1. The test results are provided in international scale (IS) values by incorporating armored RNA-based calibrators that have defined IS values tied directly to the World Health Organization BCR-ABL1 Primary Reference Materials, without the necessity of determining and maintaining conversion factors. For each batch run, the integrated interpretive software evaluates run and specimen quality control metrics (including a sufficient amount of ABL1 control transcripts to ensure a minimal limit of detection) and calculates both molecular response (MR) and %IS values for each specimen. The test has a limit of detection of MR4.7 (0.002%IS) and a linear range from MR0.3 (50%IS) to MR4.7 (0.002%IS) for both Major transcripts. Single-site and multisite precision studies demonstrated a maximum SD of 0.13 MR (30% CV within the assay range between MR0.7 and MR3.7). The performance of this BCR-ABL1 monitoring test meets all of the clinical guideline recommendations for sensitivity and IS reporting for the management of chronic myeloid leukemia patients.
There are approximately 1.8 newly diagnosed cases of chronic myeloid leukemia (CML) per 100,000 individuals per year, with the median age at diagnosis of 65 years.1 CML accounts for approximately 10% to 15% of all adult cases of leukemia. The genetic hallmark of all cases of CML is the reciprocal translocation between the long arms of chromosomes 9 and 22, termed t(9; 22) (q34.1; q11.2), generating a fusion gene breakpoint cluster region–Abelson tyrosine-protein kinase 1 (BCR-ABL1) on the derivative chromosome 22 (alias the Philadelphia chromosome).2 Most of the translocations occur between the Major breakpoint cluster region of BCR and the intron upstream of exon 2 of ABL1. The Major breakpoint cluster region occurs downstream of either exon 13 or exon 14 of BCR and results in the formation of the Major fusion transcript e13a2 or e14a2, coding for a 210-kDa protein often referred to as p210. Major fusion transcripts (ie, p210) account for >95% of CML cases, and patients can exhibit both species of Major BCR-ABL1 transcripts.3 In less than approximately 1% of CML (and two-thirds of Philadelphia-positive acute lymphoblastic leukemia), the translocation localizes to the minor breakpoint cluster region of BCR, resulting in an e1a2 transcript that produces a 190-kDa protein.3 Furthermore, a 230-kDa protein is observed in rare CML cases from translocation e19a2 and is designated as the micro breakpoint.3 Even rarer variants have been identified, including translocations at exon 3 of ABL1.4, 5 In all cases, the resulting BCR-ABL1 fusion protein is a constitutively active kinase that can drive uncontrolled proliferation in myeloid precursor cells, causing the clinical manifestation of CML.
The specific structural characteristics of the chimeric BCR-ABL1 protein allowed for the generation and subsequent Food and Drug Administration (FDA) approval of several tyrosine kinase inhibitors (TKIs) designed to inhibit the intrinsic kinase activity of the ABL1 moiety of the hybrid protein.6 The ability of these TKIs to reduce the number of leukemic cells in CML patients to levels far below previous chemotherapy- or interferon-based treatments has driven demand for a more sensitive, validated molecular monitoring method to track the dynamics of the disease. The pivotal International Randomized Study of Interferon and STI571 (IRIS) trial established quantitative RT-PCR (RT-qPCR) as the laboratory method of choice for monitoring the reduction of BCR-ABL1 transcripts in TKI-treated CML patients.7 On the basis of this and many other studies, RT-qPCR is considered the gold standard method of monitoring BCR-ABL1 transcripts, as recommended by internationally recognized guidelines for the management of CML (National Comprehensive Cancer Network, https://www.nccn.org/professionals/physician_gls/PDF/cml.pdf, last accessed February 20, 2018).8
Despite the clinical successes achieved with FDA-approved targeted therapies in CML, there remains the widespread challenge of consistently and reproducibly monitoring BCR-ABL1 transcript levels because of the variation in the design and performance characteristics of research-use-only reagents and laboratory-developed tests. To align patient monitoring to established clinical milestones and to address the portability of patient-specific data across methods and laboratories, the National Institutes of Health Consensus Group recommended the standardization of BCR-ABL1 monitoring using an international scale (IS).9 The IS is a numeric scale anchored to the standardized baseline established in the IRIS trial (100%IS) with a 3-log reduction from baseline defined as a major molecular response (MMR; MR3; 0.1%IS).7 Laboratory harmonization was established, validated, and repeatedly revalidated through sample exchange with an IS-aligned laboratory, generating a laboratory-specific conversion factor that, when applied to results from that laboratory's RT-qPCR test, aligned patient results to the IS.10 Subsequently, the National Comprehensive Cancer Network (https://www.nccn.org/professionals/physician_gls/PDF/cml.pdf) and the European LeukemiaNET8 incorporated reporting BCR-ABL1 transcripts directly on the IS into the clinical guideline recommendations for managing CML patients. To allow harmonization between methods, the World Health Organization then established the first World Health Organization International Genetics Reference Panel for quantitation of BCR-ABL translocation by RQ-PCR (National Institute for Biological Standards and Control code 09/138) in limited quantities.11
This study describes the performance characteristics of the first US FDA-cleared molecular test to monitor BCR-ABL1 transcripts in CML patients. The test reports results in both the MR scale and %IS through the use of traceable armored RNA reference materials and using automated software with integrated quality control algorithms. Over 7300 data points were generated on RNA extracted from human peripheral blood specimens to validate the test's performance characteristics, including >3600 such data points across studies for limit of detection (LOD; MR4.7), limit of quantitation (LOQ; MR4.7), linearity (from MR0.3 to MR4.7), limit of blank (LOB; undetected), and both single-site and multisite precision across four independent laboratories. Furthermore, the test is sufficiently precise to rely on singleton testing for each patient specimen using RNA isolated via typical methods from specimens up to 72 hours after venipuncture.
Materials and Methods
Specimen Preparation
Results included in this report were generated using RNA derived from CML-positive human blood specimens, nonleukemic human blood specimens, or cell line cultures. Blood specimens were obtained with patient consent under a clinical protocol under institutional review board approvals. Blood specimens were collected in EDTA anticoagulant and isolated within 72 hours of venipuncture. Leukocytes were counted by hematological analysis using a COULTER Ac·T diff Analyzer (Beckman Coulter Inc., Brea, CA) to ensure sufficient material for each study was processed into RNA. Specimens were combined with five volumes of Erythrocyte Lysis Buffer (Qiagen, Germantown, MD), incubated at room temperature for 5 minutes, and then centrifuged at 500× g at 4°C for 5 minutes to pellet remaining cells. Cell pellets were resuspended in two volumes of Erythrocyte Lysis Buffer, incubated an additional 5 minutes at room temperature, and again centrifuged for 5 minutes at 500 × g at 4°C. These leukocyte pellets were lysed in an appropriate amount of TRIzol (Thermo Fisher Scientific, Waltham, MA) to yield the equivalent of approximately 2 × 107 cells/mL, unless otherwise noted (see RNA Isolation Method). Lysates were then frozen at ≤−70°C until further processing. Unless otherwise noted, RNA was purified from blood at various scales, according to the manufacturer-recommended, isopropanol-based precipitation protocol for TRIzol, scaled appropriately for bulk isolation. RNA quality and quantity were analyzed using a NanoDrop ND-1000 (Thermo Fisher Scientific).
RT-qPCR Method
All reaction components were provided within the QuantideX qPCR BCR-ABL IS Kit (Asuragen, Inc., Austin, TX) and used in accordance with the Instructions for Use (US FDA clearance DEN160003), summarized herein. Each kit contains sufficient reagents for 60 reaction wells across a maximum of four uses. If a single run is performed, up to 49 specimens can be analyzed alongside the 11 calibrator and control wells. Briefly, the calibrators and controls included with the kit are based on Armored RNA Quant (ARQ) technology.12 Four calibrators are composed of blends of BCR-ABL1 and ABL1 RNA targets to recapitulate the CT values observed in the kit with the World Health Organization primary reference materials. RNA materials were designed to control for the relative batch run efficiency of both the RT and PCR processes. Three controls are formulated to BCR-ABL1 content that is at a high (>1%IS, MR<2), low (<0.1%IS, MR>3), or negative (ABL1 only) fusion transcript level. These materials were heat lysed and then equilibrated to room temperature.
A total of 5 μL of RT Master Mix (3.5 μL per sample of RT Buffer plus 1.5 μL per sample of RT Enzyme Mix) was distributed to reaction wells on a Fast Optical 96-well plate (Applied Biosystems by Thermo Fisher Scientific), followed by 10 μL of either Kit Calibrators (prepared in duplicate), Kit Controls (in singleton), or test specimens (1000 to 5000 ng per reaction in singleton, unless otherwise required by the specific study design). RT was performed on 7500 Fast Dx Real-Time PCR instruments (Applied Biosystems by Thermo Fisher Scientific) and run in standard mode with the following program: 25°C for 10 minutes, 42°C for 45 minutes, 93°C for 10 minutes, then hold at 25°C for up to 60 minutes. The qPCR setup was initiated during this final 60-minute hold step.
For qPCR, a 15-μL qPCR Master Mix containing 11 μL per sample of qPCR Buffer, 3.4 μL Test Primer/Probe Mix, and 0.6 μL qPCR Enzyme Mix was added to each intended-use well on a new 96-well reaction optical plate, followed by 10 μL of cDNA product from the RT. Thermal cycling was performed on 7500 Fast Dx Real-Time PCR instruments in standard mode with the following thermal cycling conditions: 95°C for 10 minutes, 40 cycles of 95°C for 15 seconds and 63°C for 1 minute, collecting data on the Cy5 (for ABL1) and FAM (for BCR-ABL1) channels, and using the ROX channel as a passive reference dye. Other instruments that contain these fluorescence channels might be used with appropriate validation. However, differences in optical system design and thermal cycling technology may yield unanticipated performance shifts.
Data Analysis
All batch run files generated by the 7500 Fast Dx system were analyzed using 21 CFR Part 11 Module 7500 Fast Real-Time PCR System Sequence Detection software version 1.4.1 (Applied Biosystems by Thermo Fisher Scientific). The background fluorescence baseline cycles and threshold levels are determined using the following parameters: Cy5 (ABL1) using a manual threshold of 0.05 and a manual baseline from cycle 5 to 13 and FAM (BCR-ABL1) using auto threshold and auto baseline. SDS files were then directly processed (without export) through QuantideX qPCR IS Kit software version 1.1 (Asuragen, Inc.) to extract the CT of each reaction, generate the standard curve, automatically assess quality control pass/fail criteria for the batch run and specimens, and calculate the result of each test specimen. Results detailed in this study are from runs that passed all quality control criteria for the calibrators, controls, and test specimens (eg, sufficient specimen ABL1 to ensure a minimal LOD for the system). The acceptance criteria for each batch run include automated review of the controls, which are also disclosed in the test system's instructions as follows: high control, MR≤2.0 (≥1%IS); low control, MR≥3.0 (≤0.1%IS); and negative control, undetected (sufficient ABL1). For cases in which any one of these three conditions was not met, the batch run was considered invalid and no data were reported for the unknown specimens. This is discussed further in Overall Performance, in Results.
The QuantideX IS software reports both MR values (MR in logs of reduction from the international baseline of 100%IS, or MR0) and %IS by linear regression to an IS-aligned, four-point, four-log standard curve of ΔCT(BCR-ABL1)-(ABL1) versus MR. Although most publications originally assigned MR values in bins (eg, MR4, MR4.5, or MR5 as a scoring system in Cross et al13), the concept of a continuous MR scale was introduced early in the kit's development. As such, the primary output of the standard curve is MR, using lot-specific calibrator values traced to the World Health Organization primary reference set, and requires no further correction or conversion factor for alignment to the IS. Each %IS was calculated automatically by the software as the antilog of MR (ie, %IS = 102−MR). All statistical analyses were performed on the primary MR output and, therefore, all statistical results (eg, SDs) are in the same log10 scale. The %IS measurement is a historical convention well understood in the field. However, data are not normally distributed when reported on this scale, and a long tail is often observed. Where reported, analysis of %IS was performed after a log10 transformation of the data because it is normally distributed after such transformation and the assumptions of general statistical methods therefore apply. Results are converted back to the arithmetic %IS scale using Equations 1 and 2, which are required to properly calculate the mean and SD when using data transformed from normal to nonnormal distributions (ie, SD of MR to SD of %IS). The random variable X = %IS and Y = log10(X). The mean of %IS (μx) is calculated using Equation 1 from the mean and variance of the log10 (%IS) values (μy and ). The equations for mean (Equation 1) and variance (Equation 2) are from a report by Quan and Zhang.14 Base 10 was substituted for each instance of natural base (e) in conformance with the scale of MR value.14
| (1) |
| (2) |
Test Specimens
With the exceptions of analytical specificity (exclusivity) and RNA isolation method (see below for each), all test specimens were either human CML-positive clinical specimens (whole blood stability), human CML-negative specimens (LOB), or human diagnostic-level (ie, MR<1.0) CML-positive clinical specimens diluted into human CML-negative specimens (all other studies).
Single-Site and Multisite Precision
A single RNA panel was used for both single-site precision studies (Asuragen, Inc.) and multisite precision studies (Oregon Health and Science University, Portland OR; Hospital of the University of Pennsylvania, Philadelphia, PA; Laboratory Corporation of America, Research Triangle Park, NC; and Asuragen, Inc.). Specimens were composed of five RNA isolates from residual clinical CML-positive whole blood specimens, each serially diluted into RNA from CML-negative whole blood. The CML-negative RNA samples were used either individually or mixed to achieve five distinct backgrounds of sufficient quantity and biological variability. Therefore, the panel consisted of five dilution series—two series of e13a2, two of e14a2, and one mixed e13a2/e14a2—with five target MR values per series (MR1, MR2, MR3, MR3.5, MR4; or 10%IS, 1%IS, 0.1%IS, 0.032%IS, and 0.01%IS, respectively), for a total of 25 unique specimens. Actual values were determined from data generated across 20 total runs by three operators using three kit lots and three qPCR instruments and testing five samples in duplicate per run (single-site precision) or with two operators at each of three sites across 5 days testing 25 samples in duplicate per run to investigate site- and lot-specific variability (primary arm of multisite precision). A second arm of the multisite precision study was performed at a fourth site to investigate operator- and day-specific variability. RNA concentration for each panel member was normalized to 3000 ng/RT, the middle of the kit's required input range. In total, 200 measurements were generated for the single-site precision study (n = 20 runs × 5 samples × 2 replicates = 200) and 1200 measurements were generated for the multisite precision study (arm 1: n = 3 sites × 5 days × 25 samples × 2 replicates = 750; arm 2: n = 3 operators × 3 runs × 25 samples × 2 replicates = 450). Data were analyzed using a random effect analysis of variance using the lmer function (https://www.rdocumentation.org/packages/lme4/versions/1.1-19/topics/lmer) in R software version 3.2.2 (The R Project for Statistical Computing, https://www.r-project.org). The means and SDs were calculated using unmodified mean and SD functions in R. All samples targeted to the same MR value were grouped as replicates, giving n = 40 at each of the five levels. The SD values from individual potential sources of variation are reported in Supplemental Tables S1 and S2.
The precision study's acceptance criteria were derived by a requirement of the test to distinguish specimens at the clinically relevant cut point of MR3 from those at one log lower analyte level at MR4. This supports assessment of the relapse definition of a 1-log increase in BCR-ABL1 with concomitant loss of MMR (MR3.0) (https://www.nccn.org/professionals/physician_gls/PDF/cml.pdf). For example, assuming independent error for the two samples, and SDs of 0.21 and 0.29 for samples at MR3.0 and MR4.0, respectively, the SD of the difference in measured MR values would be , implying that a 95% CI for the difference in MR between the two samples would exclude 0 (ie, MR3.0 ± 0.36 and MR4.0 ± 0.36 do not overlap, and the samples can therefore be distinguished). Although this criterion defined acceptable imprecision at this low level of analyte that approaches the anticipated LOD, greater precision is desired at higher %IS values and attainable with this technology.10 Therefore, the three bins of acceptable performance were developed in Table 1, wherein more stringent criteria are set for higher analyte levels. The MR3 and MR4 levels discussed above fall within the lower two bins. The imprecision of the single MR value is compounded from two separate measurements, each with its own level of error: BCR-ABL1 and ABL1. Consistent with data performance representation standards from the US FDA (FDA Office of Regulatory Affairs, Office of Regulatory Science, https://www.scribd.com/document/329743903/ora-laboratory-manual, last accessed September 21, 2018), the ratio of these two measurements is only as precise as the least specific quantification included in its calculation. Therefore, the accumulated variability embedded in an MR value will be equivalent to or higher than that from a single measure with a comparable level of precision.
Table 1.
Study Criteria for Precision
| Log scale | Linear scale | ||
|---|---|---|---|
| MR value | SD criteria | %IS value∗ | % CV |
| <3.5 | ≤0.21 | >0.0316 | ≤50 |
| 3.5–4.25 | ≤0.29 | 0.0316–0.0056 | ≤75 |
| >4.25 | ≤0.36 | <0.0056 | ≤100 |
Data expressed as %IS are not normally distributed. However, MR values are normally distributed, facilitating statistical analyses. The %IS values above are shown to bridge to historical perspectives only. The estimated % CV for the %IS values were calculated to be equivalent to the SD criteria set for the MR space.
IS, international scale; MR, molecular response.
Shown for reference; SD values of MR measurements formed the definitive acceptance criteria.
Limit of Blank
Specimens were composed of RNA from nonleukemic human whole blood specimens presumed to be negative or undetectable for the Major BCR-ABL1 breakpoints detected by the test. RNA isolation followed the same conditions used for patient specimens (see RNA Isolation Method). The study was designed to test the LOB of the test system, which begins with RNA extracted via a laboratory-validated method. The RT and qPCR steps for all batch runs included the standard set of calibrators and controls. Thirty specimens from unique donors, ranging from 1000 to 5000 ng/RT, were tested across multiple kit lots, operators, calendar days, and qPCR instruments, yielding 265 valid test results. Each of the nine batch runs contained a singleton of each of the 30 specimens. Any specimen that had sufficient ABL1 without detectable BCR-ABL1 (ie, no CT value within the 40 qPCR cycles performed) was considered undetected for the transcripts of the Major BCR-ABL1 translocations. Analysis followed EP17-A2 as a guide.15 The section “Probit Approach,” whereby LOB was assumed to be 0 (ie, for a qPCR assay, a sample putatively negative for the target of interest does not cross threshold within the number of cycles performed), was followed and verified through testing.
Limit of Detection
Specimens were composed of RNA from residual clinical CML-positive whole blood specimens serially diluted into RNA from nonleukemic whole blood. Four panels were generated from separate CML-positive specimens, two positive for e13a2 and two positive for e14a2, with seven members per panel ranging from approximately MR4.5 to MR6.0. Actual MR results were determined from data generated across 40 runs using four operators, using four instruments, and spanning 10 calendar days. Reaction input was normalized to 1000 ng/RT to ensure that the system's LOD was calculated using the lowest and, therefore, most challenging recommended input. In total, 60 measurements were generated for each of the 28 panel members using two kit lots, yielding 1680 possible unique measurements. Measurements that generated a CT value for BCR-ABL1 were considered positive, and a result of no CT was considered undetected. This study design and subsequent analysis used EP17-A2 as a guide.15 From 1678 valid measurements, LOD was assessed by both Probit regression and nonparametric (the “Classical Method” described in EP17-A2) analyses. Probit models were fit by R version 3.0.1 using a generalized linear model function and plotted with the ggplot2 software package version 2.2.1 (https://ggplot2.tidyverse.org). For nonparametric analysis, data for all low-level specimens targeted to the expected LOD of 4.7 (based on rangefinding via Probit) were combined into a single distribution after controlling for those exceeding a type II error rate of 5% (ie, a ≥95% true detection rate). The median of the distribution was calculated as the LOD.
Limit of Quantitation
Specimens were composed of RNA from residual clinical CML-positive whole blood specimens serially diluted into RNA from CML-negative whole blood. Six unique specimens, two positive for e13a2 and four positive for e14a2, were generated with a target value of MR4.7 at an RNA input of 1000 ng/RT. Actual MR results were determined from data generated across four runs with five replicates per specimen per run across two kit lots, generating a total of 120 possible measurements (of which 119 were valid). The SD of each specimen was compared with the allowable precision of the assay (see Precision section). This study used EP17-A2 as a guide.15
Linearity
Specimens were composed of RNA from residual clinical CML-positive whole blood specimens serially diluted into RNA from CML-negative whole blood. The test panel consisted of 18 specimens (nine levels for each breakpoint e13a2 and e14a2) ranging from targets of MR0.3 to MR4.8. Actual MR results were determined from data generated across six runs, with two runs for each of the three reagent lots. Samples targeted at MR4.3, 4.6, and 4.9 were assayed in quadruplicate. All others were assayed in duplicate. RNA input for each panel member was normalized to 3000 ng/RT, the middle of the required input range. In total, 144 measurements were generated per breakpoint (total n = 288). Per EP06-A,16 first-, second-, and third-order polynomial regression equations were calculated using the lm function in R version 3.2.2, and under the assumption of nonlinearity, the SEMs of the second- and third-order variables were checked for both significance (P ≤ 0.05) and maximum allowable deviation from linearity (selected as within ±0.5 log10).
RNA Mass Input Range
Specimens were composed of RNA from residual clinical CML-positive whole blood specimens diluted into RNA from CML-negative whole blood to target MR values of 1.0, 3.0, 4.3, or 4.7 at 600 ng/μL and then serially diluted into nuclease-free water to 500, 300, 100, 75, and 25 ng/μL. A constant 10 μL of RNA was loaded per RT reaction. Each of the six RNA input levels at each of the four MR values were tested with nine replicates, for a total of 216 valid measurements.
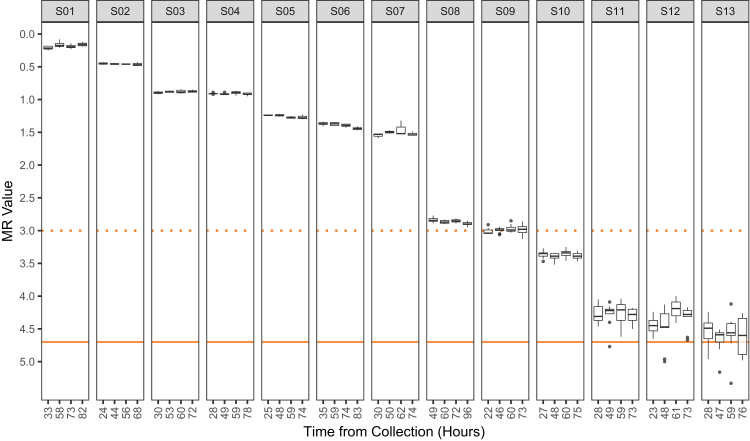
Whole Blood Specimen Stability
Thirteen separate (not mixed or otherwise preprocessed), CML-positive whole blood specimens were received from collection sites in 10-mL EDTA vacutainers (two to three vacutainers per specimen) shipped on cold packs pulled from storage at 2°C to 8°C. On receipt, each specimen was time stamped, consolidated into a 50-mL centrifuge tube, analyzed for white blood cell count, portioned into four separate EDTA vacutainers, and then stored at 2°C to 8°C. Processing into cell lysate for each aliquot in the specimen set occurred either immediately (ie, as soon as possible after receipt, usually approximately 24 hours after venipuncture) or at approximately 48, 60, or 72 hours after venipuncture. Lysates were stored at ≤−70°C until RNA isolation and subsequent RT-qPCR. Each specimen was tested in three to nine replicates per time point (1000 ng/RT), dependent on the total mass of RNA recovered from each vacutainer.
Because of geographical distance and, therefore, the necessity of overnight shipment from the collection site to the processing facility, CML-positive specimens could not be processed within 24 hours after venipuncture. To establish performance of the time course with a baseline closer to blood draw, the stability of RNA was assessed before 24 hours in a supplemental study using three nonleukemic human blood specimens, drawn at the processing facility, and then processed in parallel at 4-, 24-, 48-, 72-, and 96-hour time points. Four hours was the minimum amount of time needed for samples to be drawn, submitted, transferred, accessioned, and processed within the facility at Asuragen, Inc. After RT-qPCR, the ABL1 CT values were compared across time as the only measure that could be obtained for these specimens (as BCR-ABL1 was undetected in these nonleukemic specimens). For each specimen, the lysates were processed into RNA simultaneously to minimize uncontrolled variables.
Traceability to the International Scale
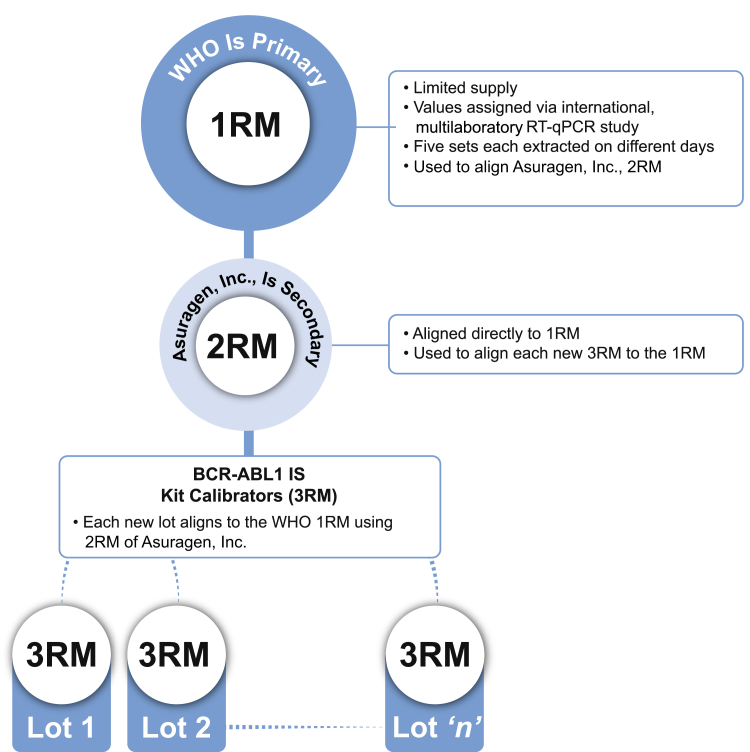
The World Health Organization generated a limited set of primary reference materials (1RM) from lyophilized cell-line mixtures with BCR-ABL1/control gene values reported on the IS11 and available to selected manufacturers. ARQ technology was used to generate stable, synthetic RNA molecules encapsidated in a bacteriophage protein coat to resist degradation.17, 18, 19 A master lot of ARQ secondary reference materials (2RM) was manufactured and assigned values after alignment to the 1RM (data not shown), and subsequently used to align each new lot of kit calibrators (tertiary reference materials, or 3RM) to the 1RM (Figure 1). A unique set of 3RM calibrators is generated for each batch lot of test with unique values assigned through traceable testing. The traceability of the 3RM to the 1RM was validated by measuring the MR values of each of the four 1RM panel members in duplicate using three different lots of kit (each of which includes a separate lot of 3RM) across three runs (one run per lot).
Figure 1.
Schematic of traceability to the primary World Health Organization (WHO) standard reference materials (1RM). The schematic depicts the manufactured lots of kit calibrators as tertiary reference materials (3RM) aligned to the World Health Organization primary reference materials (1RM) through a master lot of secondary standards (2RM) built using Armored RNA Quant technology. RT-qPCR, real-time quantitative RT-PCR.
RNA Isolation Method
A panel of white blood cells enriched via erythrocyte lysis from diagnostic-level human CML-positive whole blood was serially diluted across four logs into human CML-negative whole blood. This white blood cell–into–whole blood scheme was used to avoid coagulation due to histoincompatibility while still providing an RNA matrix derived entirely from human whole blood. However, as the initial MR value of the freshly drawn CML-positive source sample was unknown a priori, targeting highly accurate MR values was not feasible. Each specimen was divided and subjected to RNA extraction using three methods: i) TRIzol guanidinium thiocyanate-phenol-chloroform extraction with isopropanol precipitation (Thermo Fisher Scientific), ii) column-based RNeasy Mini Kit (Qiagen), and iii) an automated, customized magnetic bead–based isolation [Kingfisher Flex (Thermo Fisher Scientific); RNAClean XL (Beckman Coulter)]. RNA samples were further divided and assayed across two kit lots. All three isolation methods were evaluated for equivalency by assessing the variability of all MR values per specimen against the acceptable error of the method (see Single-Site and Multisite Precision).
Analytical Specificity
Eleven off-target fusion transcripts were evaluated to assess analytical specificity of the assay for detection of the Major p210 fusion transcripts of BCR-ABL1 (e13a2, e14a2). Testing included cell line–derived RNA (because of difficulty in collecting certain primary human materials) for the panel: four acute myeloid leukemia fusions (RUNX1-RUNX1T1, PML-RARA, CBFB-MYH11, and MLLT3-KMT2A), four acute lymphoblastic leukemia fusions (ETV6-RUNX1, TCF3-PBX1, KMT2A-AFF1 e9e5, and KMT2A-AFF1 e10e4), and one CML fusion (BCR-ABL1 e1a2 minor breakpoint cluster region). Two in vitro transcripts were also generated because of difficulty in obtaining certain cell lines. These in vitro transcripts were then blended with nonleukemic cellular RNA: one acute myeloid leukemia fusion (RBM15-MKL1) and one CML fusion (BCR-ABL1 e19a2 micro breakpoint). The in vitro transcript for this breakpoint contained only the exons BCR e19 and ABL1 a2, which is a limitation. Because native e19a2 mRNA encompasses the BCR exon that contains one of the system's primer binding regions, it may inefficiently amplify a large amplicon and, therefore, generate a low-level, false-positive signal. Each panel specimen was tested in triplicate across three lots to validate exclusivity. Inclusivity was inherently supported by specific positive detection across all BCR-ABL1–positive specimens in this study.
Results
Single-Site and Multisite Precision
The specimens within the challenge panel were measured from MR0.7 to MR3.7. The small shift from the target levels (MR1.0 to MR4.0, respectively) was attributed to ABL1 contribution from the diagnostic-level CML-positive source specimens (compared with the ABL1 in the CML-negative background diluent at the same RNA concentration), which affected the initial dilution to MR1.0 (data not shown) and was propagated to subsequent dilutions.
The observed SDs for all specimens satisfied all predetermined precision criteria (Table 1 and Table 2). Because each of the five dilution series was a unique formulation event, the components of variability were initially characterized separately for each of the 25 samples (Supplemental Tables S1 and S2). Further analysis was performed by combining the data for all five samples within each of the five target MR levels (Table 2). The maximum SD in this analysis was 0.13 MR units (18.9% CV for %IS) observed during the single-site precision study and 0.13 MR units (29.9% CV for %IS) observed during the multisite precision study (Table 2). Combining the data by each target MR value in this precision analysis includes variability of the test system as well as the five independent sample dilutions. Each of the five samples targeted to each MR level were expected to give slightly different MR values. Indeed, mean MR values were observed at the lowest analyte level in the single-site study of 3.54, 3.60, 3.67, 3.88, and 3.68 for dilution series 1, 2, 3, 4, and 5, respectively. However, the precision measured across independent dilution series was comparable to the individual sample precision in most cases.
Table 2.
Single-Site and Multisite Precision
| Target MR | Mean MR | SD | Target %IS | Mean %IS | % CV | N |
|---|---|---|---|---|---|---|
| Single-site precision | ||||||
| 1 | 0.70 | 0.08 | 10 | 20.3613 | 16.6 | 40 |
| 2 | 1.63 | 0.08 | 1 | 2.3423 | 11.7 | 40 |
| 3 | 2.66 | 0.08 | 0.1 | 0.2211 | 11.0 | 40 |
| 3.5 | 3.18 | 0.10 | 0.032 | 0.0663 | 16.8 | 40 |
| 4 | 3.68 | 0.13 | 0.01 | 0.0215 | 18.9 | 40 |
| Multisite precision | ||||||
| 1 | 0.74 | 0.06 | 10 | 18.1906 | 13.7 | 240 |
| 2 | 1.69 | 0.07 | 1 | 2.0787 | 15.8 | 240 |
| 3 | 2.70 | 0.09 | 0.1 | 0.2031 | 20.9 | 240 |
| 3.5 | 3.22 | 0.13 | 0.032 | 0.0628 | 29.4 | 240 |
| 4 | 3.72 | 0.13 | 0.01 | 0.0197 | 29.9 | 240 |
The precision of the assay was evaluated with both single-site and multisite studies by testing five separate dilution series across five target MR values. Data herein were combined per target MR level. SD values of MR measurements formed the definitive acceptance criteria. %IS values are shown for historical reference. Supplemental Tables S1 and S2 detail each specimen and source of variation separately.
IS, international scale; MR, molecular response.
Limit of Blank
The assumption of LOB = 0 (or undetected in the kit) was confirmed by singleton testing of 30 distinct nonleukemic human RNA specimens across nine replicate plates, with a maximum false-positive rate of 5% allowed for confirmation. Of 265 valid measurements, only two positive results were observed (one positive result each from two specimens), at MR5.71 (0.0002%IS) and MR5.35 (0.0004%IS), indicating a true negative rate of >99%. As the 95th percentile (95% CI, 98.6%–100%) of results was, therefore, 0, an LOB of 0 (or undetected) was confirmed. A related study was performed to assess the inherent propensity of the test system to generate cross-contaminating signal. No carryover contamination was observed when 25 wells of a high-positive (MR0.8 or 16%IS) specimen were contiguously alternated in checkerboard manner with 25 wells of a CML-negative specimen across a 96-well plate. All CML-negative specimens were reported as undetected (sufficient ABL1) (data not shown).
Limit of Detection
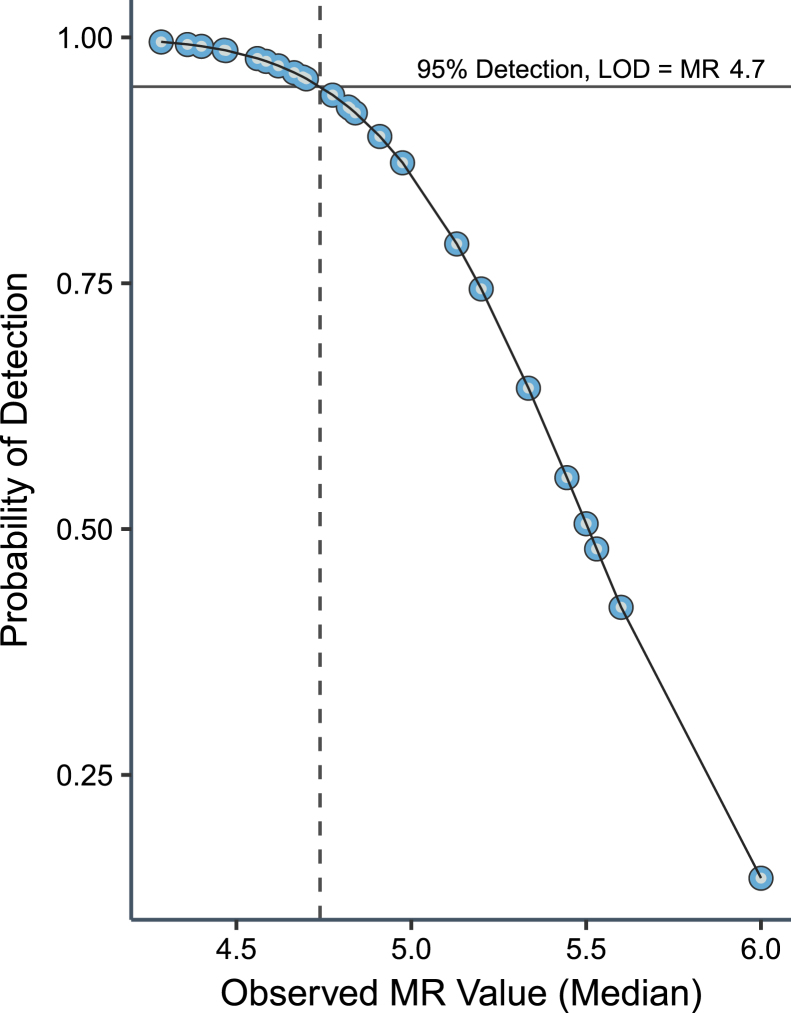
Probit analysis of the full data set, including both breakpoints and both reagent lots, yielded a 95% detection estimate of MR4.74 (0.0018%IS) with an SEM of ±0.04 (Figure 2). The goodness of fit of the Probit model was validated by the Pearson χ2 test. Probit analysis split by either lot or BCR-ABL1 breakpoint supported an LOD within the margin of error (data not shown). Other probabilities determined from the Probit model included 85% at MR5.02 (0.0010%IS), 75% at MR5.19 (0.0006%IS), 50% at MR5.51 (0.0003%IS), and 25% at MR5.82 (0.0002%IS). In other words, a specimen at MR5.5, for example, will be detected approximately half of the times that it is assayed. This detection limit was validated by nonparametric analysis, which yielded 96.1% detection at MR4.70 for the e13a2 breakpoint and 95.2% detection at MR4.70 for the e14a2 breakpoint (Table 3).
Figure 2.
Determination of analytical sensitivity. The limit of detection (LOD) of the assay was established at molecular response (MR) 4.7 (0.002% international scale) from 1678 valid data points generated from human clinical RNA specimens across 40 test runs by four operators and using two lots of reagents. Data were used to assess the limit of detection by Probit analysis, where the x axis represents median MR value of 60 replicates at each of the 28 dilution points and the y axis represents the probability of a positive result. This yielded an estimate at the 95% probability level (positivity fraction; solid horizontal line) of MR4.74, which is shown as a vertical dotted line. Other probabilities were determined from the model as 85% at MR5.02, 75% at MR5.19, 50% at MR5.51, and 25% at MR5.82.
Table 3.
LOD Data
| Transcript identity | Unique specimens | Valid measurements | Positive results | Detected, % | LOD (MR) | LOD (%IS) |
|---|---|---|---|---|---|---|
| e13a2 | 3 | 179 | 172 | 96.1 | 4.70 | 0.002 |
| e14a2 | 7 | 420 | 400 | 95.2 | 4.70 | 0.002 |
The LOD of the test was estimated using Probit analysis (Figure 2) and validated using the nonparametric method, wherein >95% of all low-level specimens targeted near the expected LOD of MR4.7 (≥0.002%IS) were detected. MR measurements formed the definitive acceptance criteria. %IS values are shown for historical reference.
IS, international scale; LOD, limit of detection; MR, molecular response.
Limit of Quantitation
LOQ was determined by identifying the challenge panel's specimen with the least analyte (ie, highest MR) whose measured variability was within the preestablished allowable variability of the assay (set at SD ≤ 0.36 per the precision studies). When the SD is calculated from all data generated within a specimen across lots, all six specimens met these acceptance criteria (Table 4). The highest measured value was a mean of MR4.87. Results were similar when separated by lot (data not shown). Because MR4.87 is a lower level of analyte than the validated LOD, the LOQ is constrained by LOD at MR4.7. Such an observation can occur as LOD is based on positivity, whereas LOQ is based on imprecision.
Table 4.
LOQ Data
| Specimen no. | Transcript identity | Valid measurements | MR (mean) | SD | Result |
|---|---|---|---|---|---|
| 1 | e13a2 | 20 | 4.79 | 0.27 | Pass |
| 2 | e14a2 | 20 | 4.67 | 0.23 | Pass |
| 3 | e14a2 | 19 | 4.80 | 0.34 | Pass |
| 4 | e14a2 | 20 | 4.60 | 0.24 | Pass |
| 5 | e14a2 | 20 | 4.87 | 0.25 | Pass |
| 6 | e13a2 | 20 | 4.82 | 0.25 | Pass |
The LOQ of the assay was validated as MR4.7 (0.002% international scale) by measuring six low-level specimens across 119 valid measurements using two lots of the kit across 3 testing days. All specimens satisfied the error criteria (SDMR ≤ 0.36), supporting a limit beyond (and, therefore, constrained by) the limit of detection of MR4.70.
LOQ, limit of quantitation; MR, molecular response.
Linearity
Linearity was evaluated by three assessments (per EP06-A).16 First, the SD of test specimens must be within the allowable precision of the assay (see Single-Site and Multisite Precision). Second, the second- and third-order regression statistics for the second- and third-order polynomials should be nonsignificant (t-test, P < 0.05). Third, for any results found to be significant, the absolute deviation from linearity must be reported.
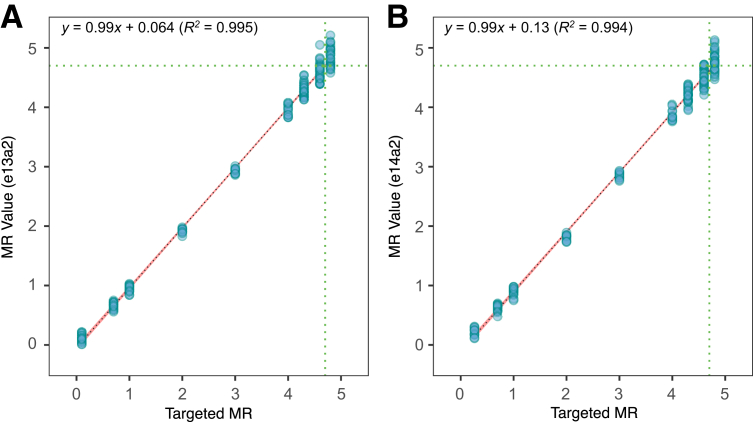
Combining data from two lots of the kit, linear regression equations were calculated with slopes of 1.01 and 1.01 and intercepts of −0.11 and −0.05 for e13a2 and e14a2, respectively (Figure 3). Breakpoint e13a2 measured MR0.12 to MR4.84, with a maximum SD of 0.17 at the highest MR value tested. Breakpoint e14a2 measured MR 0.22 to MR 4.78, also with a maximum SD of 0.17 at the highest MR value tested.
Figure 3.
Determination of the linear range of the test. Linearity of the assay was established by measuring a range spanning nearly five orders of magnitude across 288 valid measurements across two lots. The e13a2 and e14a2 breakpoints were evaluated and charted separately (A and B, respectively), with the x axis representing the targeted molecular response (MR) value of the specimen and the y axis representing the measured MR value. The symbols are depicted with 50% transparency to clarify overplotting. The 95% CI is shown in red around the largely overlapping, dotted linear regression line. The green dotted lines are drawn at the limit of detection of MR4.7. The linear regression spans MR0.1 to MR4.8, with the equations indicated for e13a2 and e14a2.
For both breakpoints, the second-order polynomial regression had small, but statistically significant, second-order coefficients, suggesting a small degree of nonlinearity: 0.02 (P = 2 × 10−4) for e13a2 and 0.03 (P = 1 × 10−5) for e14a2. However, the deviation from linearity for the second-order polynomial was trivial (ie, less than a tenth of a log at ≤0.08 absolute MR units across the entire range). Taken together, the linear range spans from at least MR0.3 (50%IS) to MR4.7 (0.002%IS).
RNA Input Range
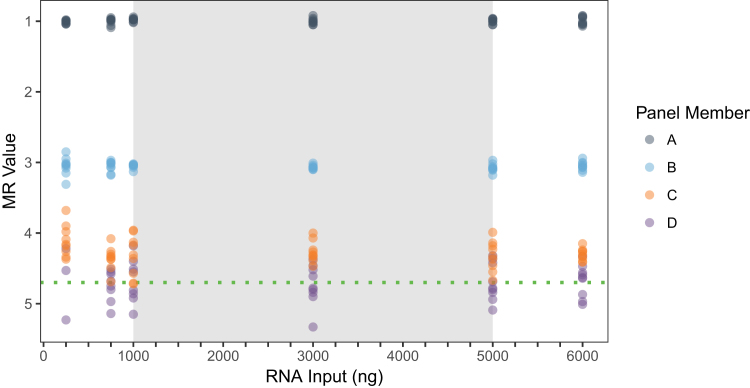
The ability of the system to maintain a similar measurement (MR value) across the range of inputs recommended by the instructions (1000 to 5000 ng per RT reaction) was evaluated. A panel of samples challenged this performance characteristic from 250 to 6000 ng RNA per RT as well as spanned a broad range of MR values (MR1.0 to MR4.7). Results are depicted in Figure 4, which suggests that at the lowest input of 250 ng, a shift may be present in the two samples of lowest analyte concentration (MR4.3 and MR4.7).
Figure 4.
RNA input range. The RNA input range of the assay was confirmed by measuring specimens at four distinct molecular response (MR) values with six total RNA input amounts, including three input amounts outside of the recommended input range: 250, 500, and 6000 ng. Dilution series labeled A, B, C, and D were targeted to MR1.0, MR3.0, MR4.3, and MR4.5, respectively. Actual values were determined on testing. The x axis represents total RNA amount included in the RT, and the y axis represents measured MR value (with highest BCR-ABL1 analyte level at the top); symbol colors indicate different specimens (depicted with 50% transparency to clarify overplotting); the gray region is the recommended input range (1000 to 5000 ng); and the green dotted line is the test's limit of detection of MR4.7.
To assess similarity of measurement from a quantitative standpoint, the SD criteria used for precision were used herein as an estimate of total analytical error (ie, the combined effects of random variability plus any bias due to the amount of RNA introduced into the system). The SD calculated from measurements across all RNA inputs should not exceed the predefined necessary precision of the assay. When data within a comparable targeted MR value are combined across input amounts, the total analytical error of samples at each MR was within the acceptable criteria (Supplemental Table S3), indicating minimal bias due to RNA input. Specifically, the SDs calculated within each of the four panel members were 0.04, 0.07, 0.21, and 0.27 at MR1.0, MR3.1, MR4.3, and MR4.7, respectively. The SD increased and hit rate (percentage of positive replicate results) decreased for samples near LOD at the lowest level of input: MR4.7 at 33% (3/9) positive and SD = 0.52 at 250 ng/RT (Supplemental Table S3). Therefore, despite maintenance of MR value within desired system precision when data were combined throughout the range tested, using <750 ng RNA may reduce the system's ability to reliably detect and quantify the LOD of MR4.7.
Whole Blood Specimen Stability
As the analyte of interest (RNA) is subject to degradation through normal cellular processes, reviews and laboratory guidelines have recommended isolating specimens within 24 hours of venipuncture to obtain the most quantifiable, sensitive measurements of BCR-ABL1.9, 20 However, this is often difficult to achieve because many testing facilities receive CML specimens from multiple clinical practices via next-day mailing services. Therefore, we evaluated the ability of the system to maintain a similar measurement (MR value) for a given blood specimen from receipt to processing (approximately 24 and 72 hours, respectively). Thirteen CML-positive blood specimens were received and then processed at multiple time points. The samples spanned from MR0.2 to MR4.5 (Figure 5). As expected, higher variability was observed in the samples approaching the LOD of MR4.7. For each sample, the difference in MR value for each time point was calculated compared with its baseline value. The range of MR value differences for individual time points was −0.13 (specimen S13 at MR4.6 tested at the second of four points at 47 hours) to 0.26 (specimen S12 at MR4.4 tested at the third of four points at 61 hours), and the overall mean difference across all 13 specimens (n = 39 time points) was 0.005 MR units. A review of hit rate (portion of positive measurements) showed that all specimens were detected (defined as a valid CT value for BCR-ABL1), indicating no decrease in positivity through the tested time points.
Figure 5.
Whole blood specimen stability. The stability of whole blood RNA for BCR-ABL1 monitoring in the context of the kit was determined by processing aliquots of human blood specimens from chronic myeloid leukemia (CML)–positive patients over an approximate time frame of 72 hours. Each isolate of RNA was tested with the kit a minimum of three times, and in some cases up to nine times (depending on RNA yield). Molecular response (MR) values for each replicate are plotted according to the time they were processed. The median, upper, and lower quartiles are shown by the box-and-whisker plot. Following the recommendations of Tukey,21 the whiskers extend to 1.5 times the upper and lower quantiles (first and third, respectively) and gray points denote values measured outside this range. Actual time point of each specimen's processing is shown rounded to the nearest hour after venipuncture. The dotted orange line is drawn at MR3 (equivalent to 0.1% and major molecular response). The y axis represents measured MR value (with highest BCR-ABL1 analyte level at the top), and the solid orange line is drawn at MR4.7, the limit of detection of the test. Supplemental Table S4 shows estimates of imprecision of ABL1 CT value across post-venipuncture time points for three CML-negative donors.
A study was also performed with an earlier baseline and parallel processing using blood from three simultaneously drawn CML-negative donors. It was predicted that degradation of RNA under constant mass input on the same qPCR batch run would be detected as an increase in CT value for ABL1. All time points showed similar mean CT values and overlapping 95% CI ranges (Supplemental Table S4), indicating no gross decay of ABL1 mRNA for up to 96 hours after collection.
Traceability to the International Scale
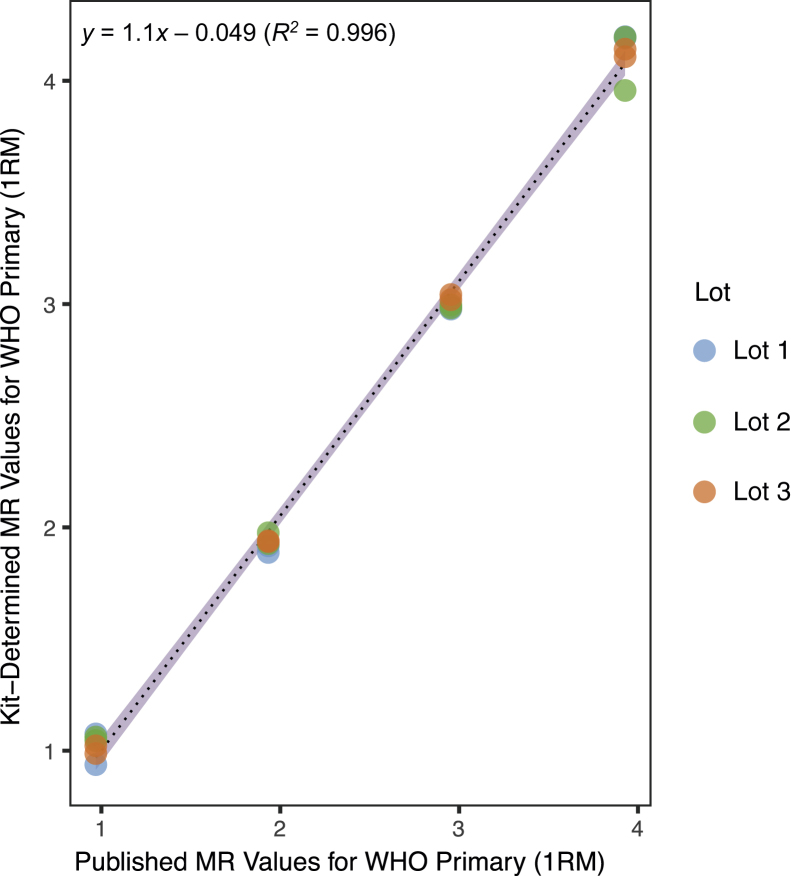
To standardize reporting of patient values on the IS, the World Health Organization generated a limited set of primary reference materials (1RM) from lyophilized cell-line mixtures with values reported on the IS.11 ARQ technology was used to generate nuclease-resistant RNA for use as calibrators in the test.17, 18, 19 The 2RM were aligned to the 1RM and were then used to align the calibrators included in each kit lot (ie, 3RM) (Figure 1). The traceability of the 3RM was validated to the 1RM by testing the commutability of values. The four 1RM panel members were assayed using three different lots of kit. The MR values of the 1RM, as measured by these three independent lots of 3RM, were plotted against the values published by the World Health Organization for the 1RM (Figure 6). The data demonstrated high correlation between the measured and published 1RM values; linear regression yielded a slope of nearly 1 at 1.1, a y-intercept of nearly 0 at −0.049, and a coefficient of determination near 1 at 0.996 across all three lots, with similar values within each lot (data not shown).
Figure 6.
Validation of traceability to the primary World Health Organization (WHO) standard reference materials (1RM). This study assessed the traceability of tertiary reference materials (kit calibrators) to the 1RM across three different manufacturing lots of the kit. Empirical molecular response (MR) values for the World Health Organization primary (1RM) materials generated using the kit (y axis) are plotted against the MR values published in the World Health Organization primary reference panel's instructions for use (x axis). The three lots are represented by blue (lot 1), green (lot 2), and orange (lot 3) data points, each depicted with 25% transparency to clarify overplotting (seen as darker areas on the chart's points). Regardless, the similarity of the data points leads to lot 1 data at MR3 and MR4 being obscured by the other two lots. The 95% CI is plotted in violet around the black dotted linear regression line. Regression analysis and CIs are based on all data in aggregate.
RNA Isolation Method
A panel of blood samples was generated and then isolated to RNA via three methods: guanidinium thiocyanate-phenol-chloroform with isopropanol precipitation (TRIzol), manual spin columns (RNeasy Mini Kit), and a custom, automated magnetic bead–based isolation (Kingfisher Flex; RNAClean XL). Maximum SD within each RNA isolation method was 0.08 MR units for all BCR-ABL1 levels tested (Table 5). It was predicted that if the measured MR values diverged between methods enough to affect interpretation, then data combined across methods would show variability that exceeds the precision limits set for the system. This was not observed. When all data generated within a specimen across all three isolation methods were combined, the SDs showed a maximum of 0.08 MR units. This variability was within the criteria established for precision.
Table 5.
RNA Isolation Method
| Specimen no. | Isolation | Mean MR | Median MR | SD | n |
|---|---|---|---|---|---|
| 1 | Kingfisher | 0.61 | 0.62 | 0.06 | 16 |
| 1 | RNeasy | 0.62 | 0.64 | 0.07 | 16 |
| 1 | TRIzol | 0.73 | 0.75 | 0.05 | 16 |
| 1 | All | 0.66 | 0.67 | 0.08 | 48 |
| 2 | Kingfisher | 1.48 | 1.48 | 0.03 | 16 |
| 2 | RNeasy | 1.47 | 1.47 | 0.02 | 16 |
| 2 | TRIzol | 1.54 | 1.55 | 0.03 | 16 |
| 2 | All | 1.50 | 1.49 | 0.04 | 48 |
| 3 | Kingfisher | 2.49 | 2.47 | 0.04 | 16 |
| 3 | RNeasy | 2.48 | 2.48 | 0.03 | 16 |
| 3 | TRIzol | 2.56 | 2.55 | 0.03 | 16 |
| 3 | All | 2.51 | 2.50 | 0.05 | 48 |
| 4 | Kingfisher | 3.52 | 3.51 | 0.07 | 16 |
| 4 | RNeasy | 3.47 | 3.46 | 0.07 | 16 |
| 4 | TRIzol | 3.56 | 3.56 | 0.08 | 16 |
| 4 | All | 3.52 | 3.51 | 0.08 | 48 |
Freshly drawn, enriched, human white blood cells from a chronic myeloid leukemia (CML)–positive donor were serially diluted across four logs into CML-negative anticoagulated whole blood, generating specimens 1, 2, 3, and 4. The resulting specimens were subjected to RNA extraction by three methods: TRIzol guanidinium thiocyanate-phenol-chloroform extraction with isopropanol precipitation, the column-based RNeasy Mini Kit (Qiagen), and an automated, customized magnetic bead–based RNA isolation (Kingfisher Flex using RNAClean XL magnetic beads). All three isolation methods were evaluated for similarity by assessing the variability of all MR values per specimen across the three methods against the acceptable precision of the method. Shown are MR values (mean and median), SD, and number of valid measurements (n). All refers to the aggregate data of all three isolation methods within a specimen.
MR, molecular response.
Analytical Specificity
The exclusivity of the system to detect the Major breakpoint cluster region was assessed against 11 other leukemic targets. Only one false-positive value was observed in 116 valid results: MR6.1 for KMT2A/AFF1 [t(4;11)(q21;q23), previously known as MLL/AF4], a result that is 1.4 logs below the validated LOD of the test. Furthermore, the other eight replicates for this sample were negative. On the basis of the data, the test is interpreted to be specific for the BCR-ABL1 e13a2 and e14a2 transcripts across its linear range. This was the predicted outcome based on in silico analyses performed early in the primer development and selection process. Assessments were also performed in silico for polymorphisms in the primer binding sites. No allelic variations were found within three nucleotides of the 3′ end of any of the assay's primers, and all identified allele frequencies upstream of this were <1% (not shown).
Overall Performance
A high-batch run pass rate was observed throughout the studies, with assignable causes for failing specimens and runs. In total, 252 batch runs were performed. Nine batch runs (9/252, 3.6%) failed, all with assignable causes. Two (2/252, 0.8%) were attributed to instrument error, both of which were properly identified by the kit's interpretive software: one failure of the instrument's software to call a correct baseline and one failure of its optical system. Seven failures (7/252, 2.8%) were attributed to operator error: not starting the qPCR run before leaving the instrument; prematurely ending the qPCR run; poor plate sealing, resulting in evaporation; pipetting errors on the negative control; one low calibrator R2 value; and two instances of a false-negative calibrator. All of these errors were identified by the interpretive software. The 243 valid batch runs (243/252, 96.4%) contained 7662 reaction wells. Thirteen (13/7662, 0.17%) wells failed, all with assignable causes: 12 occurrences (12/7662, 0.16%) of failure of the ABI 7500 Fast Dx instrument's software to call a correct baseline and 1 occurrence (1/7662, 0.01%) of no ABL1 signal due to operator error in pipetting.
Discussion
The robust patient responses to the approved, first-generation BCR-ABL1 TKI imatinib were shown to reduce the leukemic burden far below that of conventional cytotoxic and interferon-based therapies, which drove the development and incorporation of highly sensitive qPCR technologies to monitor BCR-ABL1 transcripts for improved CML patient management. Two clinically relevant BCR-ABL1 measurements were generated from the pivotal IRIS trial: a standardized study baseline that was calculated as the median pretreatment BCR-ABL1 levels of 30 chronic phase CML patients (now 100%IS or MR0) and MMR at a 3-log reduction of normalized BCR-ABL1 levels (now 0.1%IS or MR3).7 This and subsequent studies identified a critical need for standardization in an attempt to define clinical outcomes, made clear by the difficulties in aligning measurements across different laboratories using different monitoring procedures. In 2005, the National Institute of Health Consensus Group proposed the use of the IS to monitor BCR-ABL1 transcripts by qPCR methods in CML patients,9 which established a worldwide standard for clinical practice and reporting guidelines that remain in effect today (https://www.nccn.org/professionals/physician_gls/PDF/cml.pdf).8
Standardization efforts initially required a sample exchange program be performed with a recognized laboratory to establish, validate, and periodically revalidate laboratory-specific conversion factors for proper alignment to the IS.10 In 2010, the World Health Organization generated a limited set of primary reference materials with values reported on the IS to bring improved comparability of test results between sites.11 This material was made available to certain manufacturers to produce secondary materials aligned to the World Health Organization primary standards. Calibrators aligned to the primary reference standard set in this manner have been shown to be effective materials for laboratories to generate their own conversion factor and report directly on the IS without sample exchange.17, 18, 19 Because the analyte for monitoring CML is RNA, ARQ technology is particularly well suited for the generation of secondary BCR-ABL1 standards as it accounts for the relative batch run efficiency of the RT step.11 In our experience, RT is a large source of variation in gene expression assays that is not adequately addressed by using plasmids or other DNA-based controls and calibrators.22
A master lot of 2RM was generated using ARQ technology and aligned to the World Health Organization primary reference materials (1RM) (Figure 1). The 2RM set is then used to align manufactured, tertiary calibrators (3RM) in each batch of kit to the World Health Organization standards, maintaining traceability of the %IS and MR values of every specimen without the need for laboratory-specific conversion factors. This assay was, thus, the first FDA-cleared BCR-ABL1 test to report specimen values directly on the IS.
ABL1 was chosen as the endogenous control gene because the Europe Against Cancer Program study determined that ABL1 is one of the few suitable control genes to normalize BCR-ABL1 transcript levels in myeloid cells, and ABL1 is the most commonly used endogenous control gene in tests.23, 24 A limitation of the use of this endogenous control gene is that the analyte of interest, BCR-ABL1 transcript, contains the ABL1 sequence; therefore, BCR-ABL1 signal may contribute to the total signal observed for the endogenous control gene. Although the effect is predicted to be negligible at low analyte levels, at extremely high levels (eg, shortly after diagnosis), the overall %IS is predicted to read lower than expected. The linearity study demonstrated a linear range of MR0.3 (50%IS) to MR4.7 (0.002%IS), indicating that any bias in measurement at high BCR-ABL1 content is trivial at levels relevant for disease monitoring. This performance characteristic is especially important as interest increases in studying early molecular response against a patient-specific baseline.25
Indefinite treatment of CML patients with TKI therapy was long considered the standard management practice for CML.26 Although the TKIs for CML are generally well tolerated, toxicities are well documented and sometimes require adjustments in treatment.27 Furthermore, the economic burden on patients and the health care system from the indefinite use of TKI therapy is a growing health care industry concern.28, 29 Ongoing clinical trials have shown that treatment-free remission (TFR) for patients who have experienced prolonged deep molecular response offers a possible solution to both problems.30, 31 Therefore, the National Comprehensive Cancer Network released new guidelines for BCR-ABL1 qPCR monitoring tests that are sensitive enough to detect at least a 4.5-log reduction from the IS baseline (MR4.5, 0.0032%IS) to identify candidates for TFR (https://www.nccn.org/professionals/physician_gls/PDF/cml.pdf). Presented herein are the validated performance characteristics of an assay with LOD and quantitation both at MR4.7 (Figure 2 and Tables 1 and 2), surpassing the recent 2018 National Comprehensive Cancer Network guidelines (https://www.nccn.org/professionals/physician_gls/PDF/cml.pdf).
Although the lower limits of detection and quantitation are among the most critical parameters for BCR-ABL1 monitoring, National Comprehensive Cancer Network guidelines specify several clinically relevant milestones spanning a large dynamic range up to 10%IS (MR1.0). Thus, it is important that such a test maintain linearity across a large range of values, ideally exceeding four orders of magnitude. Herein, a validated linear range was established between MR0.3 and MR4.7 (50%IS to 0.002%IS, respectively) (Figure 3). In addition, the single-site and multisite precision studies validated that the repeatability, within-site precision, and multisite precision of the test were consistently high across the linear range of the test, demonstrating that the test produces reliable measurements at all clinically relevant levels (Table 2 and Supplemental Tables S1 and S2). Targeting MR4 allowed us to validate precision to a level of analyte a full log below the medical decision point of MR3 (MMR) that was also used in our clinical trials as well as in the pivotal IRIS trial.7 This provided analyte targeted at high (MR1), low (MR4), and multiple levels in between. The low (MR4) level was assessed in response to an indication of precision at deeper response levels that were emerging in anticipation of investigation of TFR. Because deep molecular response at MR4.5 is now used to identify patients who are eligible to attempt TFR, the precision of an assay at the level of analyte has become a critical topic. The LOD study's data were reviewed to obtain an estimate at MR4.5. Two specimens measured a mean of MR4.5 with 100% positive detection: one with an SD of 0.22 (n = 60) and another with an SD of 0.26 (n = 60), both of which were detected in 100% of the replicates.
In quantitative tests, the amount of replication required is informed by analytical sensitivity and precision. Using fewer replicates requires less reagent volume, maximizes qPCR plate and instrument use, and requires less specimen RNA. Most laboratory-developed tests recommend replicate testing because of concerns that imprecision will lead to error in distinguishing clinical cut points. For qPCR-based assays designed to monitor BCR-ABL1 transcripts at deep MR levels (ie, MR ≥ 4.5), specific preanalytical recommendations have been published to achieve amounts of RNA from patient whole blood samples suitable for such test sensitivity.8, 20 Namely, EDTA whole-blood samples should be processed quickly after collection (up to 72 hours), with a target of 2 × 107 nucleated cells per RNA isolation. In the present studies, the precision of the system was validated in the context of singleton testing. This approach lessens the burden of RNA isolation from clinical samples in comparison to other methods that rely on replicate testing. Moreover, despite the challenges in obtaining sufficient human materials to generate such large-scale challenge panels, the overwhelming majority of the validation data herein were derived from human peripheral blood specimens rather than cell line–derived RNA or other contrived materials (eg, in vitro transcripts or plasmids). Human peripheral blood specimens were used extensively during assessment of the test's analytical performance to ensure maximum commutability with clinical blood specimens, eliminate the concern of noncomparability with commonly used cell line RNA, and ensure that users of the assay can expect similar performance when monitoring clinical CML patients.
There are certain limitations of the system validated in this study. For example, the use of RNA extracted from bone marrow aspirates or monitoring of Philadelphia-positive precursor acute lymphoblastic leukemia was not validated. Additional studies would be required to extend monitoring for these applications. Furthermore, the test is only designed to detect, but not distinguish between, the BCR-ABL1 fusion transcripts e13a2 (b2a2) and e14a2 (eg, sb3a2). The ability to detect other fusion transcripts has not been evaluated beyond those described in this report. Hence, it does not detect minor (e1a2), micro (e19a2), or rare (e13a3) breakpoints, microdeletions, or mutations. Therefore, the test does not cover the <1% of CML cases that are defined by the minor breakpoint. The validation studies were performed on only one instrument model (7500 Fast Dx Instrument). Additional studies have been performed with comparable performance on the cobas z 480 instrument (Roche, Basel, Switzerland) (data not shown), but this application has not been cleared by the FDA.
Another consideration in the achievement of the level of analytical sensitivity disclosed in this report, each RT requires at least 1000 ng RNA in ≤10 μL. Although this is generally possible with traditional methods (TRIzol and alcohol precipitation), it can be difficult to achieve with more recent methods—especially automated methods that are locked into high-volume (and, therefore, low-concentration) elutions. Furthermore, RNA quality and quantity can affect the results; for example, samples of low OD 260/OD 230 ratios (below approximately 1.2) have been observed to interfere with detection of passive reference dye, which may lead to software errors and/or misquantification (data not shown). And, as with any quantitative system, patients with low levels of BCR-ABL1 transcript (MR >4.7 or %IS <0.002%) may be reported as undetected (sufficient ABL). Hence, an undetected result does not preclude the presence of low levels of leukemic cells in the patient.
From a regulatory perspective, the kit validated in these studies is compliant with the special controls issued recently by the FDA for BCR-ABL1 quantitation tests. Specifically, 21 CFR 866.6060(b)(3) lays out multiple requirements, including those critical for analytical consideration. The system must incorporate an RT-qPCR test with results on the IS for monitoring CML. Analytical validation must include sensitivity (as LOB, LOD, and LOQ), specificity (including interference and cross-contamination), kit stability, multisite precision, linearity, and reportable range. All of these were determined for system and reported herein—with the exception of interference and stability, both of which passed predetermined specifications (data not shown). Of importance, the regulation states that the “device output must include results on the International Scale (IS) and your assay must include multipoint calibration controls traceable to a relevant international reference panel (eg, the World Health Organization International Genetic Reference Panel for quantitation of BCR-ABL mRNA).” The calibrators in the kit are across four points designed to recapitulate the approximate CT values seen with the kit when using the World Health Organization primary reference materials.
In the future, additional clinical and analytical studies may be of interest for the system. For example, predicting successful TFR is of increased interest because recent studies show 40% to 60% of patients relapsing within the first 6 months of TKI cessation.31 Does a test with this level of analytical sensitivity make higher-confidence predictions of TFR and more sensitive monitoring for early post-cessation relapse? In addition, a validation of diagnostic use may be helpful to reduce the cost and burden of the diagnostic workup on a new CML patient. Extension of the test to diagnose and monitor Philadelphia-positive acute lymphoblastic leukemia may also provide cost, workflow, and clinical benefits. In conclusion, analytical validation demonstrated that the test performs with high precision, accuracy, reportable range, specimen stability, robustness, and analytical sensitivity.
Acknowledgments
We thank John N. Milligan, Scott Shell, and Annette Schlageter for assistance with the manuscript.
J.T.B. is the guarantor of this work and, as such, had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
Supported, in part, by the NIH National Cancer Institute contract HHSN261201500009C (principal investigator: J.T.B.).
Disclosures: Asuragen, Inc., provided reagents to each site to support this study. J.T.B., I.J.B., W.L.-W., M.E.F., K.L.J., A.K.R., J.B.H., and B.F.A. are employees of Asuragen, Inc. Asuragen, Inc., employees have or may have stock in Asuragen, Inc.
Supplemental material for this article can be found at http://doi.org/10.1016/j.jmoldx.2019.03.002.
Supplemental Data
References
- 1.Noone A.M., Howlader N., Krapcho M., Miller D., Brest A., Yu M., Ruhl J., Tatalovich Z., Mariotto A., Lewis D.R., Chen H.S., Feuer E.J., Cronin K.A., editors. SEER Cancer Statistics Review, 1975-2015. National Cancer Institute; Bethesda, MD: 2018. [Google Scholar]; SEER Cancer Statistics Review, 1975-2015. Edited by Noone AM, Howlader N, Krapcho M, Miller D, Brest A, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, Chen HS, Feuer EJ, Cronin KA. National Cancer Institute. Bethesda, MD, based on November 2017 SEER data submission, posted April 2018 to the SEER web site Available at: https://seer.cancer.gov/csr/1975_2015/.
- 2.Faderl S., Talpaz M., Estrov Z., O'Brien S., Kurzrock R., Kantarjian H.M. The biology of chronic myeloid leukemia. N Engl J Med. 1999;341:164–172. doi: 10.1056/NEJM199907153410306. [DOI] [PubMed] [Google Scholar]; Faderl S, Talpaz M, Estrov Z, O'Brien S, Kurzrock R, Kantarjian HM: The biology of chronic myeloid leukemia. N Engl J Med 1999, 341:164-172. [DOI] [PubMed]
- 3.Quintas-Cardama A., Cortes J. Molecular biology of bcr-abl1-positive chronic myeloid leukemia. Blood. 2009;113:1619–1630. doi: 10.1182/blood-2008-03-144790. [DOI] [PMC free article] [PubMed] [Google Scholar]; Quintas-Cardama A, Cortes J: Molecular biology of bcr-abl1-positive chronic myeloid leukemia. Blood 2009, 113:1619-1630. [DOI] [PMC free article] [PubMed]
- 4.Arun A.K., Senthamizhselvi A., Mani S., Vinodhini K., Janet N.B., Lakshmi K.M., Abraham A., George B., Srivastava A., Srivastava V.M., Mathews V., Balasubramanian P. Frequency of rare BCR-ABL1 fusion transcripts in chronic myeloid leukemia patients. Int J Lab Hematol. 2017;39:235–242. doi: 10.1111/ijlh.12616. [DOI] [PubMed] [Google Scholar]; Arun AK, Senthamizhselvi A, Mani S, Vinodhini K, Janet NB, Lakshmi KM, Abraham A, George B, Srivastava A, Srivastava VM, Mathews V, Balasubramanian P: Frequency of rare BCR-ABL1 fusion transcripts in chronic myeloid leukemia patients. Int J Lab Hematol 2017, 39:235-242. [DOI] [PubMed]
- 5.Burmeister T., Reinhardt R. A multiplex PCR for improved detection of typical and atypical BCR-ABL fusion transcripts. Leuk Res. 2008;32:579–585. doi: 10.1016/j.leukres.2007.08.017. [DOI] [PubMed] [Google Scholar]; Burmeister T, Reinhardt R: A multiplex PCR for improved detection of typical and atypical BCR-ABL fusion transcripts. Leuk Res 2008, 32:579-585. [DOI] [PubMed]
- 6.Jabbour E.J., Cortes J.E., Kantarjian H.M. Tyrosine kinase inhibition: a therapeutic target for the management of chronic-phase chronic myeloid leukemia. Expert Rev Anticancer Ther. 2013;13:1433–1452. doi: 10.1586/14737140.2013.859074. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jabbour EJ, Cortes JE, Kantarjian HM: Tyrosine kinase inhibition: a therapeutic target for the management of chronic-phase chronic myeloid leukemia. Expert Rev Anticancer Ther 2013, 13:1433-1452. [DOI] [PMC free article] [PubMed]
- 7.Hughes T.P., Kaeda J., Branford S., Rudzki Z., Hochhaus A., Hensley M.L., Gathmann I., Bolton A.E., van Hoomissen I.C., Goldman J.M., Radich J.P. International Randomised Study of Interferon versus STISG: frequency of major molecular responses to imatinib or interferon alfa plus cytarabine in newly diagnosed chronic myeloid leukemia. N Engl J Med. 2003;349:1423–1432. doi: 10.1056/NEJMoa030513. [DOI] [PubMed] [Google Scholar]; Hughes TP, Kaeda J, Branford S, Rudzki Z, Hochhaus A, Hensley ML, Gathmann I, Bolton AE, van Hoomissen IC, Goldman JM, Radich JP, International Randomised Study of Interferon versus STISG: frequency of major molecular responses to imatinib or interferon alfa plus cytarabine in newly diagnosed chronic myeloid leukemia. N Engl J Med 2003, 349:1423-1432. [DOI] [PubMed]
- 8.Baccarani M., Deininger M.W., Rosti G., Hochhaus A., Soverini S., Apperley J.F., Cervantes F., Clark R.E., Cortes J.E., Guilhot F., Hjorth-Hansen H., Hughes T.P., Kantarjian H.M., Kim D.W., Larson R.A., Lipton J.H., Mahon F.X., Martinelli G., Mayer J., Muller M.C., Niederwieser D., Pane F., Radich J.P., Rousselot P., Saglio G., Saussele S., Schiffer C., Silver R., Simonsson B., Steegmann J.L., Goldman J.M., Hehlmann R. European LeukemiaNet recommendations for the management of chronic myeloid leukemia: 2013. Blood. 2013;122:872–884. doi: 10.1182/blood-2013-05-501569. [DOI] [PMC free article] [PubMed] [Google Scholar]; Baccarani M, Deininger MW, Rosti G, Hochhaus A, Soverini S, Apperley JF, Cervantes F, Clark RE, Cortes JE, Guilhot F, Hjorth-Hansen H, Hughes TP, Kantarjian HM, Kim DW, Larson RA, Lipton JH, Mahon FX, Martinelli G, Mayer J, Muller MC, Niederwieser D, Pane F, Radich JP, Rousselot P, Saglio G, Saussele S, Schiffer C, Silver R, Simonsson B, Steegmann JL, Goldman JM, Hehlmann R: European LeukemiaNet recommendations for the management of chronic myeloid leukemia: 2013. Blood 2013, 122:872-884. [DOI] [PMC free article] [PubMed]
- 9.Hughes T., Deininger M., Hochhaus A., Branford S., Radich J., Kaeda J., Baccarani M., Cortes J., Cross N.C., Druker B.J., Gabert J., Grimwade D., Hehlmann R., Kamel-Reid S., Lipton J.H., Longtine J., Martinelli G., Saglio G., Soverini S., Stock W., Goldman J.M. Monitoring CML patients responding to treatment with tyrosine kinase inhibitors: review and recommendations for harmonizing current methodology for detecting BCR-ABL transcripts and kinase domain mutations and for expressing results. Blood. 2006;108:28–37. doi: 10.1182/blood-2006-01-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hughes T, Deininger M, Hochhaus A, Branford S, Radich J, Kaeda J, Baccarani M, Cortes J, Cross NC, Druker BJ, Gabert J, Grimwade D, Hehlmann R, Kamel-Reid S, Lipton JH, Longtine J, Martinelli G, Saglio G, Soverini S, Stock W, Goldman JM: Monitoring CML patients responding to treatment with tyrosine kinase inhibitors: review and recommendations for harmonizing current methodology for detecting BCR-ABL transcripts and kinase domain mutations and for expressing results. Blood 2006, 108:28-37. [DOI] [PMC free article] [PubMed]
- 10.Branford S., Fletcher L., Cross N.C., Muller M.C., Hochhaus A., Kim D.W., Radich J.P., Saglio G., Pane F., Kamel-Reid S., Wang Y.L., Press R.D., Lynch K., Rudzki Z., Goldman J.M., Hughes T. Desirable performance characteristics for BCR-ABL measurement on an international reporting scale to allow consistent interpretation of individual patient response and comparison of response rates between clinical trials. Blood. 2008;112:3330–3338. doi: 10.1182/blood-2008-04-150680. [DOI] [PubMed] [Google Scholar]; Branford S, Fletcher L, Cross NC, Muller MC, Hochhaus A, Kim DW, Radich JP, Saglio G, Pane F, Kamel-Reid S, Wang YL, Press RD, Lynch K, Rudzki Z, Goldman JM, Hughes T: Desirable performance characteristics for BCR-ABL measurement on an international reporting scale to allow consistent interpretation of individual patient response and comparison of response rates between clinical trials. Blood 2008, 112:3330-3338. [DOI] [PubMed]
- 11.White H.E., Matejtschuk P., Rigsby P., Gabert J., Lin F., Lynn Wang Y., Branford S., Muller M.C., Beaufils N., Beillard E., Colomer D., Dvorakova D., Ehrencrona H., Goh H.G., El Housni H., Jones D., Kairisto V., Kamel-Reid S., Kim D.W., Langabeer S., Ma E.S., Press R.D., Romeo G., Wang L., Zoi K., Hughes T., Saglio G., Hochhaus A., Goldman J.M., Metcalfe P., Cross N.C. Establishment of the first World Health Organization International Genetic Reference Panel for quantitation of BCR-ABL mRNA. Blood. 2010;116:e111–e117. doi: 10.1182/blood-2010-06-291641. [DOI] [PubMed] [Google Scholar]; White HE, Matejtschuk P, Rigsby P, Gabert J, Lin F, Lynn Wang Y, Branford S, Muller MC, Beaufils N, Beillard E, Colomer D, Dvorakova D, Ehrencrona H, Goh HG, El Housni H, Jones D, Kairisto V, Kamel-Reid S, Kim DW, Langabeer S, Ma ES, Press RD, Romeo G, Wang L, Zoi K, Hughes T, Saglio G, Hochhaus A, Goldman JM, Metcalfe P, Cross NC: Establishment of the first World Health Organization International Genetic Reference Panel for quantitation of BCR-ABL mRNA. Blood 2010, 116:e111-e117. [DOI] [PubMed]
- 12.Stevenson J., Hymas W., Hillyard D. The use of Armored RNA as a multi-purpose internal control for RT-PCR. J Virol Methods. 2008;150:73–76. doi: 10.1016/j.jviromet.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; Stevenson J, Hymas W, Hillyard D: The use of Armored RNA as a multi-purpose internal control for RT-PCR. J Virol Methods 2008, 150:73-76. [DOI] [PMC free article] [PubMed]
- 13.Cross N.C., White H.E., Colomer D., Ehrencrona H., Foroni L., Gottardi E., Lange T., Lion T., Machova Polakova K., Dulucq S., Martinelli G., Oppliger Leibundgut E., Pallisgaard N., Barbany G., Sacha T., Talmaci R., Izzo B., Saglio G., Pane F., Muller M.C., Hochhaus A. Laboratory recommendations for scoring deep molecular responses following treatment for chronic myeloid leukemia. Leukemia. 2015;29:999–1003. doi: 10.1038/leu.2015.29. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cross NC, White HE, Colomer D, Ehrencrona H, Foroni L, Gottardi E, Lange T, Lion T, Machova Polakova K, Dulucq S, Martinelli G, Oppliger Leibundgut E, Pallisgaard N, Barbany G, Sacha T, Talmaci R, Izzo B, Saglio G, Pane F, Muller MC, Hochhaus A: Laboratory recommendations for scoring deep molecular responses following treatment for chronic myeloid leukemia. Leukemia 2015, 29:999-1003. [DOI] [PMC free article] [PubMed]
- 14.Quan H., Zhang J. Estimate of standard deviation for a log-transformed variable using arithmetic means and standard deviations. Stat Med. 2003;22:2723–2736. doi: 10.1002/sim.1525. [DOI] [PubMed] [Google Scholar]; Quan H, Zhang J: Estimate of standard deviation for a log-transformed variable using arithmetic means and standard deviations. Stat Med 2003, 22:2723-2736. [DOI] [PubMed]
- 15.CLSI . Clinical and Laboratory Standards Institute; Wayne, PA: 2012. Evaluation of Detection Capability for Clinical Laboratory Measurement Procedures; Approved Guideline-Second Edition. CLSI document. [Google Scholar]; CLSI: Evaluation of Detection Capability for Clinical Laboratory Measurement Procedures; Approved Guideline-Second Edition. CLSI document. Wayne, PA: Clinical and Laboratory Standards Institute, 2012.
- 16.CLSI . Clinical and Laboratory Standards Institute; Wayne, PA: 2003. Evaluation of the Linearity of Quantitative Measurement Procedures: A Statistical Approach; Approved Guidelines. [Google Scholar]; CLSI: Evaluation of the Linearity of Quantitative Measurement Procedures: A Statistical Approach; Approved Guidelines. Wayne, PA: Clinical and Laboratory Standards Institute, 2003.
- 17.Brown J.T., Laosinchai-Wolf W., Hedges J.B., Watt C.D., Van Deerlin V.M., Fletcher L., Branford S., Labourier E. Establishment of a standardized multiplex assay with the analytical performance required for quantitative measurement of BCR-ABL1 on the international reporting scale. Blood Cancer J. 2011;1:e13. doi: 10.1038/bcj.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]; Brown JT, Laosinchai-Wolf W, Hedges JB, Watt CD, Van Deerlin VM, Fletcher L, Branford S, Labourier E: Establishment of a standardized multiplex assay with the analytical performance required for quantitative measurement of BCR-ABL1 on the international reporting scale. Blood Cancer J 2011, 1:e13. [DOI] [PMC free article] [PubMed]
- 18.Jennings L.J., George D., Czech J., Yu M., Joseph L. Detection and quantification of BCR-ABL1 fusion transcripts by droplet digital PCR. J Mol Diagn. 2014;16:174–179. doi: 10.1016/j.jmoldx.2013.10.007. [DOI] [PubMed] [Google Scholar]; Jennings LJ, George D, Czech J, Yu M, Joseph L: Detection and quantification of BCR-ABL1 fusion transcripts by droplet digital PCR. J Mol Diagn 2014, 16:174-179. [DOI] [PubMed]
- 19.White H.E., Hedges J., Bendit I., Branford S., Colomer D., Hochhaus A., Hughes T., Kamel-Reid S., Kim D.W., Modur V., Muller M.C., Pagnano K.B., Pane F., Radich J., Cross N.C., Labourier E. Establishment and validation of analytical reference panels for the standardization of quantitative BCR-ABL1 measurements on the international scale. Clin Chem. 2013;59:938–948. doi: 10.1373/clinchem.2012.196477. [DOI] [PubMed] [Google Scholar]; White HE, Hedges J, Bendit I, Branford S, Colomer D, Hochhaus A, Hughes T, Kamel-Reid S, Kim DW, Modur V, Muller MC, Pagnano KB, Pane F, Radich J, Cross NC, Labourier E: Establishment and validation of analytical reference panels for the standardization of quantitative BCR-ABL1 measurements on the international scale. Clin Chem 2013, 59:938-948. [DOI] [PubMed]
- 20.Soverini S., De Benedittis C., Mancini M., Martinelli G. Best practices in chronic myeloid leukemia monitoring and management. Oncologist. 2016;21:626–633. doi: 10.1634/theoncologist.2015-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]; Soverini S, De Benedittis C, Mancini M, Martinelli G: Best practices in chronic myeloid leukemia monitoring and management. Oncologist 2016, 21:626-633. [DOI] [PMC free article] [PubMed]
- 21.Frigge M., Hoaglin D.C., Iglewicz B. Some implementations of the boxplot. Am Stat. 1989;43:50–54. [Google Scholar]; Frigge M, Hoaglin DC, Iglewicz B: Some implementations of the boxplot. Am Stat 1989, 43:50-54.
- 22.Stahlberg A., Hakansson J., Xian X., Semb H., Kubista M. Properties of the reverse transcription reaction in mRNA quantification. Clin Chem. 2004;50:509–515. doi: 10.1373/clinchem.2003.026161. [DOI] [PubMed] [Google Scholar]; Stahlberg A, Hakansson J, Xian X, Semb H, Kubista M: Properties of the reverse transcription reaction in mRNA quantification. Clin Chem 2004, 50:509-515. [DOI] [PubMed]
- 23.Beillard E., Pallisgaard N., van der Velden V.H., Bi W., Dee R., van der Schoot E., Delabesse E., Macintyre E., Gottardi E., Saglio G., Watzinger F., Lion T., van Dongen J.J., Hokland P., Gabert J. Evaluation of candidate control genes for diagnosis and residual disease detection in leukemic patients using “real-time” quantitative reverse-transcriptase polymerase chain reaction (RQ-PCR): a Europe against cancer program. Leukemia. 2003;17:2474–2486. doi: 10.1038/sj.leu.2403136. [DOI] [PubMed] [Google Scholar]; Beillard E, Pallisgaard N, van der Velden VH, Bi W, Dee R, van der Schoot E, Delabesse E, Macintyre E, Gottardi E, Saglio G, Watzinger F, Lion T, van Dongen JJ, Hokland P, Gabert J: Evaluation of candidate control genes for diagnosis and residual disease detection in leukemic patients using “real-time” quantitative reverse-transcriptase polymerase chain reaction (RQ-PCR): a Europe against cancer program. Leukemia 2003, 17:2474-2486. [DOI] [PubMed]
- 24.Wang Y.L., Lee J.W., Cesarman E., Jin D.K., Csernus B. Molecular monitoring of chronic myelogenous leukemia: identification of the most suitable internal control gene for real-time quantification of BCR-ABL transcripts. J Mol Diagn. 2006;8:231–239. doi: 10.2353/jmoldx.2006.040404. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wang YL, Lee JW, Cesarman E, Jin DK, Csernus B: Molecular monitoring of chronic myelogenous leukemia: identification of the most suitable internal control gene for real-time quantification of BCR-ABL transcripts. J Mol Diagn 2006, 8:231-239. [DOI] [PMC free article] [PubMed]
- 25.Harrington P., Kizilors A., de Lavallade H. The role of early molecular response in the management of chronic phase CML. Curr Hematol Malig Rep. 2017;12:79–84. doi: 10.1007/s11899-017-0375-0. [DOI] [PMC free article] [PubMed] [Google Scholar]; Harrington P, Kizilors A, de Lavallade H: The role of early molecular response in the management of chronic phase CML. Curr Hematol Malig Rep 2017, 12:79-84. [DOI] [PMC free article] [PubMed]
- 26.Jain P., Kantarjian H., Alattar M.L., Jabbour E., Sasaki K., Nogueras Gonzalez G., Dellasala S., Pierce S., Verstovsek S., Wierda W., Borthakur G., Ravandi F., O'Brien S., Cortes J. Long-term molecular and cytogenetic response and survival outcomes with imatinib 400 mg, imatinib 800 mg, dasatinib, and nilotinib in patients with chronic-phase chronic myeloid leukaemia: retrospective analysis of patient data from five clinical trials. Lancet Haematol. 2015;2:e118–e128. doi: 10.1016/S2352-3026(15)00021-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jain P, Kantarjian H, Alattar ML, Jabbour E, Sasaki K, Nogueras Gonzalez G, Dellasala S, Pierce S, Verstovsek S, Wierda W, Borthakur G, Ravandi F, O'Brien S, Cortes J: Long-term molecular and cytogenetic response and survival outcomes with imatinib 400 mg, imatinib 800 mg, dasatinib, and nilotinib in patients with chronic-phase chronic myeloid leukaemia: retrospective analysis of patient data from five clinical trials. Lancet Haematol 2015, 2:e118-e128. [DOI] [PMC free article] [PubMed]
- 27.Caldemeyer L., Dugan M., Edwards J., Akard L. Long-term side effects of tyrosine kinase inhibitors in chronic myeloid leukemia. Curr Hematol Malig Rep. 2016;11:71–79. doi: 10.1007/s11899-016-0309-2. [DOI] [PubMed] [Google Scholar]; Caldemeyer L, Dugan M, Edwards J, Akard L: Long-term side effects of tyrosine kinase inhibitors in chronic myeloid leukemia. Curr Hematol Malig Rep 2016, 11:71-79. [DOI] [PubMed]
- 28.Knopf K.B., Divino V., McGarry L., Chen Y.J., Pokras S., Munakata J., Taylor C., Ng D., Nieset C., Huang H. Economic burden of tyrosine kinase inhibitor treatment failure in chronic myeloid leukemia. Clin Lymphoma Myeloma Leuk. 2015;15:e163–e171. doi: 10.1016/j.clml.2015.07.647. [DOI] [PubMed] [Google Scholar]; Knopf KB, Divino V, McGarry L, Chen YJ, Pokras S, Munakata J, Taylor C, Ng D, Nieset C, Huang H: Economic burden of tyrosine kinase inhibitor treatment failure in chronic myeloid leukemia. Clin Lymphoma Myeloma Leuk 2015, 15:e163-e171. [DOI] [PubMed]
- 29.Latremouille-Viau D., Guerin A., Gagnon-Sanschagrin P., Dea K., Cohen B.G., Joseph G.J. Health care resource utilization and costs in patients with chronic myeloid leukemia with better adherence to tyrosine kinase inhibitors and increased molecular monitoring frequency. J Manag Care Spec Pharm. 2017;23:214–224. doi: 10.18553/jmcp.2017.23.2.214. [DOI] [PMC free article] [PubMed] [Google Scholar]; Latremouille-Viau D, Guerin A, Gagnon-Sanschagrin P, Dea K, Cohen BG, Joseph GJ: Health care resource utilization and costs in patients with chronic myeloid leukemia with better adherence to tyrosine kinase inhibitors and increased molecular monitoring frequency. J Manag Care Spec Pharm 2017, 23:214-224. [DOI] [PMC free article] [PubMed]
- 30.Hughes T.P., Ross D.M. Moving treatment-free remission into mainstream clinical practice in CML. Blood. 2016;128:17–23. doi: 10.1182/blood-2016-01-694265. [DOI] [PubMed] [Google Scholar]; Hughes TP, Ross DM: Moving treatment-free remission into mainstream clinical practice in CML. Blood 2016, 128:17-23. [DOI] [PubMed]
- 31.Saussele S., Richter J., Hochhaus A., Mahon F.X. The concept of treatment-free remission in chronic myeloid leukemia. Leukemia. 2016;30:1638–1647. doi: 10.1038/leu.2016.115. [DOI] [PMC free article] [PubMed] [Google Scholar]; Saussele S, Richter J, Hochhaus A, Mahon FX: The concept of treatment-free remission in chronic myeloid leukemia. Leukemia 2016, 30:1638-1647. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.