Summary
Gain of 20q11.21 is one of the most common recurrent genomic aberrations in human pluripotent stem cells. Although it is known that overexpression of the antiapoptotic gene Bcl-xL confers a survival advantage to the abnormal cells, their differentiation capacity has not been fully investigated. RNA sequencing of mutant and control hESC lines, and a line transgenically overexpressing Bcl-xL, shows that overexpression of Bcl-xL is sufficient to cause most transcriptional changes induced by the gain of 20q11.21. Moreover, the differentially expressed genes in mutant and Bcl-xL overexpressing lines are enriched for genes involved in TGF-β- and SMAD-mediated signaling, and neuron differentiation. Finally, we show that this altered signaling has a dramatic negative effect on neuroectodermal differentiation, while the cells maintain their ability to differentiate to mesendoderm derivatives. These findings stress the importance of thorough genetic testing of the lines before their use in research or the clinic.
Keywords: human pluripotent stem cells, human embryonic stem cells, chromosomal abnormalities, genome instability, 20q11.21, impaired differentiation, neuroectoderm
Highlights
-
•
Bcl-xL overexpression drives the transcriptomic profile of 20q11.21 mutant lines
-
•
20q11.21 mutant lines downregulate CHCHD2, a known TGF-β pathway modulator
-
•
Mutant lines differentially express genes involved in TGF-β and SMAD signaling
-
•
Mutant lines show impaired ectoderm commitment due to TGF-β signaling deregulation
In this article Spits and colleagues show that Bcl-xL overexpression is the main driver of the distinct transcriptomic profile of human embryonic stem cells with a gain of 20q11.21. The cells have an abnormal TGF-β- and SMAD-mediated signaling, which impairs commitment to neuroectoderm while differentiation to the other lineages remains unaffected.
Introduction
Human embryonic stem cells (hESCs) can be cultured in vitro for extended periods of time without losing their ability to differentiate into all three germ layers. Their terminally differentiated derivatives have become a potent tool in disease modeling and may play important future roles in regenerative medicine, toxicology tests, and drug screening. As all these applications will require between millions and billions of cells, prolonged culture periods are almost inevitable (Avior et al., 2016, Serra et al., 2012). hESC cultures tend to acquire chromosomal abnormalities that vary in size from small structural variants to full chromosome gains and losses (Keller et al., 2018, Nguyen et al., 2013). Several aberrations are known to be recurrent, such as gains of chromosomes 1, 12, 17, and X, or parts thereof (Amps et al., 2011, Baker et al., 2007, Cowan et al., 2004, Draper et al., 2004, Herszfeld et al., 2006, Inzunza et al., 2004, Maitra et al., 2005, Mitalipova et al., 2005, Yang et al., 2010). Of special interest is the gain of 20q11.21 found in more than 20% of lines worldwide (Amps et al., 2011, Laurent et al., 2011, Lefort et al., 2008, Närvä et al., 2010, Spits et al., 2008, Wu et al., 2008). The smallest region of amplification includes the antiapoptotic gene BCL2L1 (Amps et al., 2011). We, and others, have found that the mutant cells overexpress Bcl-xL, the predominantly expressed isoform of BCL2L1. This confers a survival advantage to these mutant cells that allows them to quickly take over the culture (Avery et al., 2013, Nguyen et al., 2014).
Although it is generally assumed that chromosome abnormalities influence the functional characteristics of hESCs, particularly on differentiation, only a handful of reports on this topic have been published (Ben-David et al., 2014, Fazeli et al., 2011, Herszfeld et al., 2006, Keller et al., 2018, Werbowetski-Ogilvie et al., 2009, Yang et al., 2008). Undifferentiated chromosomally abnormal hESCs have been found to acquire characteristics such as increased cloning efficiency, decreased population doubling times (Fazeli et al., 2011), increased capacity for cell proliferation and self-renewal, antiapoptotic properties (Baker et al., 2007, Yang et al., 2008), growth factor independence, and an increase in teratoma-initiating cells (Werbowetski-Ogilvie et al., 2009). These studies also demonstrated that hESCs carrying chromosomal abnormalities show altered gene expression patterns when compared with normal cell lines, while retaining their—possibly reduced—differentiation capacity (Fazeli et al., 2011, Werbowetski-Ogilvie et al., 2009). Karyotypically abnormal hESCs tend to produce immature teratomas when injected in vivo, containing a higher proportion of poorly differentiated or undifferentiated cells when compared with normal hESCs (Herszfeld et al., 2006, Werbowetski-Ogilvie et al., 2009, Yang et al., 2008). These studies also show an upregulation of a number of oncogenes, concurrent with a downregulation of genes related to differentiation, which has been interpreted as a first step toward malignant transformation (Gopalakrishna-Pillai and Iverson, 2010, Werbowetski-Ogilvie et al., 2009, Yang et al., 2008). Furthermore, similarly to malignant cancer cells, mutant hESCs migrate faster in three-dimensional collagen gels than normal hESCs (Werbowetski-Ogilvie et al., 2009). Overall, these studies were limited in that they were carried out on a small number of lines carrying different abnormalities, or even one hESC line carrying a 20q11.21 duplication (Werbowetski-Ogilvie et al., 2009), while only one study consistently described one type of aberration using multiple carrier lines (Ben-David et al., 2014).
Our aim was to elucidate if and how the gain of 20q11.21 impacts the differentiation capacity of hESCs. This is of particular importance given that most of these gains fall below the size detection limit of conventional G-banding, which is still the most commonly used method for monitoring the karyotype of human pluripotent stem cells (hPSCs). We studied nine hESC lines, of which four carry a gain of 20q11.21 and one line transgenically overexpresses Bcl-xL. Two other genes present in the smallest common region of gain, ID1 and HM13, are not further considered in this work because, as already described, ID1 protein level does not differ in the mutant cells as compared with normal lines and the function of HM13 is limited and relates to immune function, making it a poor candidate in relation to differentiation (Nguyen et al., 2014).
Our results show that hESC lines carrying a 20q11.21 amplification have a transcriptome that is significantly different from their chromosomally normal isogenic counterparts as reflected in multiple deregulated pathways, in particular for transforming growth factor β (TGF-β)- and SMAD-mediated signaling genes. We show that Bcl-xL overexpression is the main driver of these changes. Finally, we show that this results in a strong impairment of ectodermal differentiation in the mutant cells, while their capacity for differentiation into mesendodermal derivatives remains intact.
Results
hESCs with a Gain of 20q11.21 Show an Altered Transcriptomic Profile Similar to the Bcl-xL Overexpressing Line
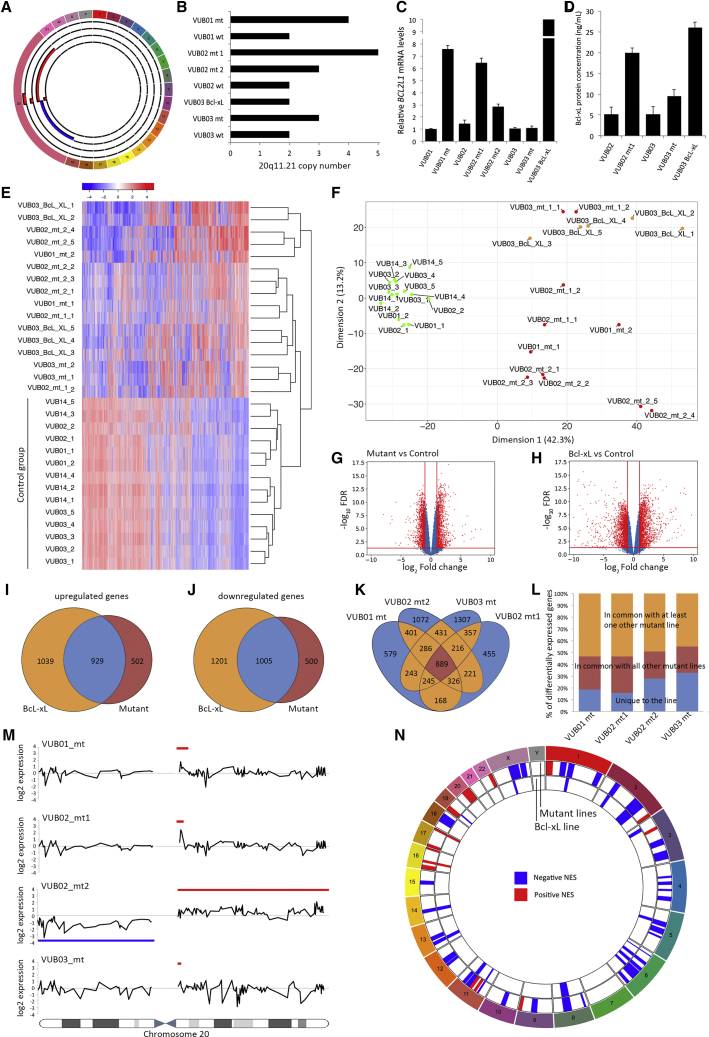
As a first step, we investigated the transcriptomic differences between lines with a gain of 20q11.21 and their genetically normal counterparts, and the role of Bcl-xL in these differences. Our study included nine hESC lines derived and kept in culture in our laboratory: four lines with gains of 20q11.21 (VUB01_mt, VUB02_mt1, VUB02_mt2, and VUB03_mt) of different sizes, their chromosomally balanced isogenic counterparts (VUB01, VUB02, and VUB03), an additional normal line (VUB14) and a line transgenically overexpressing Bcl-xL (VUB03_Bcl-xL) characterized by Nguyen et al. (2014) (Table S1). The karyotypes of all our lines were assessed by array comparative genomic hybridization (aCGH) before the experiments and the number of copies of 20q11.21 was established by real-time qPCR. Figure 1A shows a diagram of the chromosomal content of the mutant lines with a magnified chromosome 20. The size of the amplification ranged between 0.9 and 4 Mb with an identical proximal and a variable distal breakpoint. VUB02_mt2 carried an isochromosome 20 (loss of the p arm and duplication of the q arm). All the control lines had a fully balanced genetic content, and VUB03_BcL-xL also carried a gain of 1q32.1q41. The exact breakpoints of the chromosomal abnormalities, along with the passage range of each hESC line used in the study can be found in Table S1. The gain of 20q11.21 is often an amplification ranging from three to five copies rather than a simple duplication (Figure 1B). The Bcl-xL mRNA levels in the overexpressing line were nearly 70 times higher than in the control lines and were 10-fold that of VUB01_mt and VUB02_mt1 (Figure 1C), although the protein levels were similar to those of VUB02_mt1 (Figure 1D).
Figure 1.
HESCS with a Gain of 20q11.21 Show an Altered Transcriptomic Profile Similar to VUB03_BcL-xL
(A) Graphical representation of the chromosomal content of the mutant lines in this study, established by aCGH. Chromosome 20 is enlarged to show the breakpoints and region of gain. From outside to inside: VUB01_mt, VUB02_mt1, VUB02_mt2, and VUB03_mt. Regions of gain are represented in red, loss in blue.
(B) Copy number of the 20q11.21 region established by qPCR in all studied lines (n = 3).
(C and D) (C) BCL2L1 mRNA (n = 3) and (D) Bcl-xL protein levels in mutant and control hESCs and in VUB03_BcL-xL as established by sandwich ELISA. Data are shown as means ± SEM (n = 3).
(E and F) (E) Unsupervised cluster analysis and (F) multidimensional scaling plot of dimension 1 versus dimension 2 of all coding genes with a count per million greater than one in at least two samples.
(G and H) Volcano plots of the differential gene expression analysis with a cutoff value of |log2 fold change| > 1 and FDR < 0.05 for control versus mutant (G) and control versus VUB03_BcL-xL (H).
(I and J) Venn diagrams comparing the deregulated genes in VUB03_BcL-xL with those deregulated in the mutant lines. (I) Shows the upregulated genes and (J) the downregulated genes.
(K) Venn diagram comparing the deregulated genes in all mutant lines.
(L) Shows the percentage of deregulated genes that are unique or common, or common between at least two lines.
(M) Smoothened gene expression data of all genes on chromosome 20, for the four lines carrying a gain of 20q11.21. The blue lines show the region of chromosomal gain, the red line the region of loss.
(N) Gene set enrichment analysis using the MSigDB database C1, for the deregulated genes in the mutant lines and in VUB03_BcL-xL. The plot shows the chromosomal positions with significant enrichment in deregulated genes. In blue are regions with a negative normalized enrichment score (NES) and in red, with a positive NES.
We carried out RNA sequencing on two to five replicates per line collected from different dishes. We only considered coding genes with a count per million greater than one in at least two samples. Unsupervised hierarchical cluster analysis using all expressed genes shows that the mutant hESC lines and the VUB03_Bcl-xL line display an expression motif different from the control lines, resulting in the clustering of the mutant and the Bcl-xL lines separate from the control lines (Figure 1E). The same pattern appears in the multidimensional scaling plot generated considering all expressed genes, showing the clustering of the control lines separate from the mutant lines and together with VUB03_BcL-xL (Figure 1F), demonstrating that overexpression of Bcl-xL is sufficient to result in a transcriptome close to that of a mutant line. Furthermore, the clustering pattern of VUB03_Bcl-xL suggests that the impact of the overexpression of Bcl-xL on the transcriptome of the cells is stronger than that of the gain of 1q32.1q41, which the line also carries.
Differential gene expression analysis with a cutoff value of |log2 fold change| > 1 and false discovery rate q value (FDR) < 0.05 shows 1,431 upregulated and 1,505 downregulated genes in mutant versus control lines. VUB03_BcL-xL shows 1,968 upregulated and 2,206 downregulated genes compared with the normal lines (Figures 1G and 1H). Approximately 65% of the genes that are differentially expressed by the mutant lines are in common with the deregulated genes in VUB03_BcL-xL (Figures 1I and 1J). Next, we plotted the overlap in deregulated genes of the mutant lines compared with the control lines. All mutant lines have a subset of deregulated genes that are unique to the line, representing between 15% and 32% of the differential gene expression. Conversely, 22% to 30% of genes were common to all lines, and between 44% and 53% were in common with at least one other mutant line (Figures 1K and 1L). This suggests that a core of 889 genes are deregulated due to the overexpression of genes located in the common region of 20q11.21 irrespective of the line-to-line variation. Furthermore, 763 of these 889 genes (85.8%) are also deregulated in the Bcl-xL overexpressing line, strongly suggesting that Bcl-xL plays a predominant role in the transcriptomic profile of the cells with a gain of 20q11.21.
Finally, we mapped the differential gene expression to chromosomal position. First, we calculated and plotted the smoothened log2 fold-change expression of all genes on chromosome 20 for each mutant line individually, and compared this with the control lines as a group (Figure 1M, raw data in Figure S1). In VUB01_mt, VUB02_mt1, and VUB02_mt2 the overexpression of genes located in the region of gain of chromosome 20 is clearly observed. VUB03_mt carries the smallest region of gain and no differences can be observed in the expression in that region. Second, we used the gene set enrichment analysis using the MSigDB database C1 for the deregulated genes in the mutant lines and in VUB03_BcL-xL. The mutant lines have a positive enrichment score for 20q, which is absent in VUB03_BcL-xL. However, there is a similar enrichment score across the genome of the mutant lines and VUB03_BcL-xL, showing 16 of the 18 enriched regions to be in common (Figure 1N). These results illustrate a number of salient points. Expectedly, the higher copy number has an effect on the gene expression of most—but not all—genes in the amplified region, which is why gene expression data can be used to karyotype cells (Ben-David et al., 2013). However, smaller gains fall under the detection level unless they induce very strong gene expression changes. More importantly, a variant copy number of one region can strongly act in trans on the expression of genes located on other chromosomes.
In summary, mutant lines show a different transcriptome from their normal counterparts, which can largely be attributed to the overexpression of Bcl-xL. The overexpression of other genes located within the mutation has a minimal, if any, effect.
The 20q11.21 Mutation Affects TGF-β- and SMAD-Mediated Signaling through Bcl-xL Overexpression
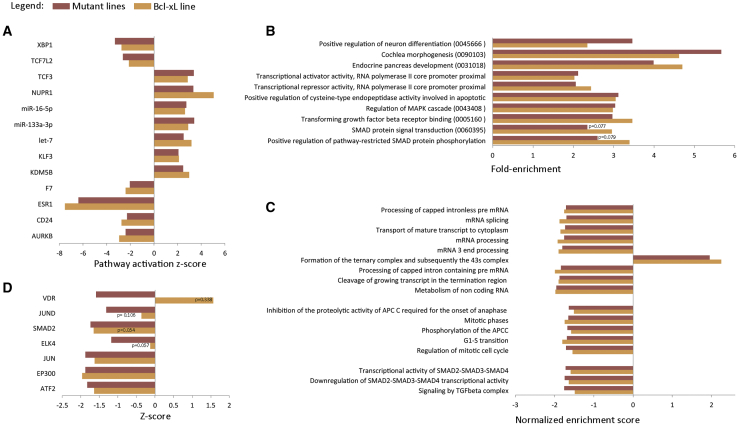
In a next step, we carried out pathway analysis using multiple bioinformatic tools. First, we carried out ingenuity pathway analysis (IPA) using all genes with a |log2 fold change| > 1 and FDR < 0.05, for both the mutant versus control groups, and the VUB03_BcL-xL line versus a control group. This analysis predicts the state of upstream regulators based on the expression of their downstream targets. IPA considers a pathway to be activated with a Z score >2, and inhibited with a score less than −2. When the p value is significant and the activation Z score is between −2 and 2, it is considered to be significantly affected, without being able to establish whether it is activated or inhibited. In our dataset, the activation score in VUB03_BcL-xL follows that of the mutant lines in all cases, and, in the case of pathways not reaching the Z score >2 or less than −2, the p value is still always significant (Figure 2A).
Figure 2.
Pathway Analysis in hESCs with a Gain of 20q11.21 and in VUB03_BcL-xL
(A) Ingenuity Pathway Analysis of the upstream regulators of pathways with an activation score between −2 and 2 and a p value < 0.05 in the wild-type (WT) versus mutant (MT) lines and their behavior in VUB03_BcL-xL.
(B) DAVID Enrichment Analysis of the 1,000 top deregulated genes in the mutant group and VUB03_BcL-xL. In parenthesis are the gene-ontology term numbers.
(C and D) Gene set enrichment analysis using the MSigDB C2 library (C) and Enrichr-based prediction of protein-protein interactions for transcription factors (D). All results are statistically significant with a p value < 0.05 unless the value is given. All the analyses were performed using the differentially expressed genes between WT versus mutant group and WT versus VUB03_BcL-xL with a cutoff value of |log2 fold change| > 1 and FDR < 0.05.
We then subjected the 1,000 genes with the highest log fold change in absolute value (563 up- and 437 downregulated for the 20q11.21 lines, 485 up- and 515 downregulated for VUB03_BcL-xL) to DAVID functional annotation enrichment analysis. The analysis retrieved a total of 75 annotations, of which we selected those gene ontology terms referring to cellular processes and that had a fold enrichment >2, and a p value < 0.05 in at least one of the groups (Figure 2B). Interestingly, the deregulated genes in both mutant lines and VUB03_BcL-xL are enriched for factors in the TGF-β and SMAD signaling, as well as in genes involved in neuron differentiation and cochlea morphogenesis, which are full or partial ectodermal derivatives, respectively. Next, we carried out gene set enrichment analysis using the C2 library (full list in Table S2). The results did not yield any significantly deregulated pathways when using the Kyoto Encyclopedia of Genes and Genomes, but revealed 17 significantly enriched processes as annotated in the Reactome library. These can broadly be categorized into related to mRNA processing, cell cycle, and TGF-β and SMAD signaling (Figure 2C).
Subsequently, we analyzed the transcription factor enrichment and protein-protein interactions in the differentially expressed genes in the mutant lines and in VUB03_BcL-xL with a |log2 fold change| > 1 and FDR < 0.05, using Enrichr (Chen et al., 2013, Kuleshov et al., 2016). Our datasets show a negative Z score for protein interaction with SMAD2 (Figures 2D and S2).
Overall, these results suggest that hESCs with a gain of 20q11.21 appear to have a perturbed TGF-β and SMAD signaling and that Bcl-xL overexpression is responsible for most of these changes. Given the pivotal role of TGF-β and SMAD signaling in gastrulation and early lineage commitment, we hypothesized that the gain of 20q11.21 may have an impact on the differentiation capacity of the cells.
Gain of 20q11.21 Results in Impaired Neuroectoderm Differentiation
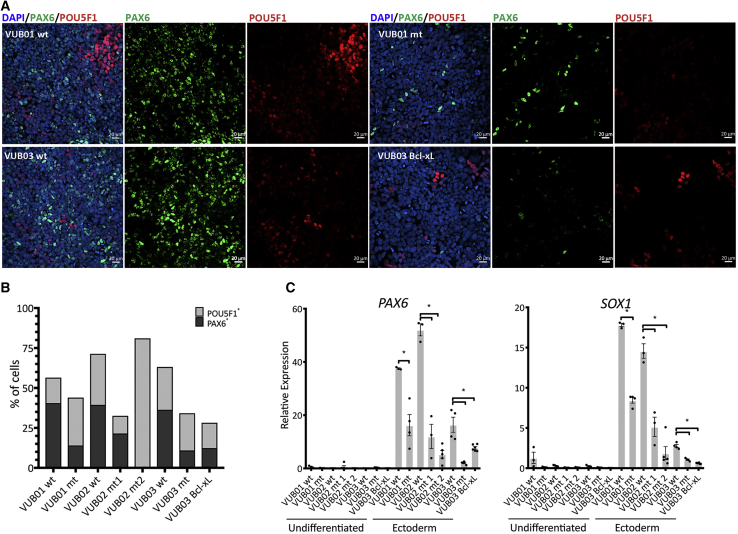
We first assessed the neuroectodermal differentiation efficiency of all our control and mutant lines and that of VUB03_BcL-xL. We performed a 4-day induction protocol recapitulating neuroectoderm commitment using Noggin and SB431542, both of which are TGF-β superfamily antagonists (Chambers et al., 2009, Chetty et al., 2013). We differentiated all our lines in three to six independent differentiation experiments to ensure the reproducibility and consistency of the experiments. At the fourth day of neuroectoderm induction, we analyzed the mRNA of markers for each lineage (endo-, neuroecto-, and mesoderm), and the stem cell markers POU5F1 and NANOG. We stained for PAX6 as neuroectodermal marker and POU5F1 as pluripotency marker and quantified the number of positive cells (Figure 3A). The data show that all samples derived from mutant lines and VUB03_BcL-xL display statistically significantly lower levels of PAX6 and SOX1 mRNA (Figure 3C), and between 1.8 and 3.3 times less PAX6-positive cells (Figures 3B and S3). Next to the neuroectoderm and stem cell markers, we also tested for the expression of mesendoderm markers (Figure S3). The results show that, while the mutant cells do exit the pluripotent state, they do not differentiate into any early, specific germ line detectable with the markers we used. The fact that VUB03_BcL-xL follows the same pattern as the mutant cells, showing practically no expression of neuroectodermal markers after the 4-day induction, supports the hypothesis that the overexpression of Bcl-xL alone is sufficient to cause this abnormal differentiation. Finally, we also differentiated control and mutant hESCs to neuroectoderm in a 12-day protocol, confirming the difference between the control and mutant hESCs after extended differentiation (Figure S3).
Figure 3.
Bcl-xL Overexpression Due to a Gain of 20q11.21 Results in Impaired Neuroectoderm Differentiation
(A) Examples of immunostaining for PAX6 (green) and POU5F1 (red) in mutant and control lines and in VUB03_BcL-xL, after 4 days of neuroectoderm differentiation.
(B) Percentage of PAX6- and POU5F1-positive cells. The mutant lines and VUB03_BcL-xL yield less PAX6-positive cells than the control lines.
(C) Relative mRNA expression as measured by qPCR for ectoderm markers PAX6 and SOX1 (n = 3–6). Data are shown as means ± SEM, each dot represents an independent differentiation experiment and the horizontal bars with asterisks represent statistical significance between samples (p < 0.05, t test).
Mutant and Control Lines Equally Differentiate toward Mesendoderm Derivates
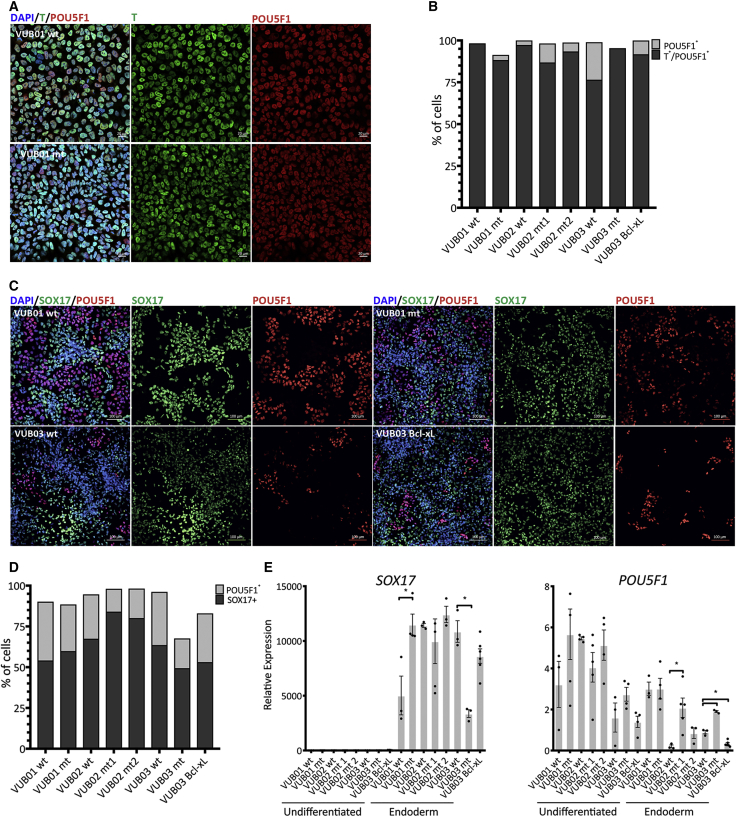
Finally, we assessed the impact of the mutation and Bcl-xL overexpression on mesendodermal lineage commitment using a 24-h protocol that drives the differentiation through the first events of the primitive streak formation by TGF-β and WNT activation. We measured the protein levels of Brachyury (T) in our lines and found that they all underwent mesendoderm induction with an equally good efficiency (Figures 4A and 4B). To investigate the ability of the lines to further commit to derivatives of this germ layer, we directed the cells toward definitive endoderm. We differentiated all eight lines (with three to six replicates per line) and evaluated the gene expression at the mRNA and protein levels. Figure 4E shows the mRNA expression results, which are in line with the immunostaining results for SOX17 and POUF51 (Figures 4C and 4D). All the lines increased their expression of endoderm markers, and no consistent expression of markers for other lineages was observed (Figure S5). NANOG and POU5F1 were overall slightly but not completely downregulated, as expected given their important roles in early endoderm differentiation (Teo et al., 2011, Ying et al., 2015). Overall, in all cases, control and mutant lines as well as VUB03_BcL-xL readily and similarly differentiated to definitive endoderm.
Figure 4.
Control and Mutant Lines and VUB03_BcL-xL Equally Differentiate to Mesendoderm Derivatives
(A) Representative examples of immunostaining for T (green) and POU5F1 (red) in control (VUB01_wt) and mutant (VUB01_mt) lines after mesendoderm induction.
(B) Percentage of T/POU5F1 double-positive (mesendoderm cells) and POU5F1-only positive cells.
(C) Examples of immunostaining for SOX17 (green) and POU5F1 (red) in control (VUB01_wt and VUB03_wt) and mutant (VUB01_wt and VUB03_mt) lines, and VUB03_BcL-xL after definitive endoderm differentiation.
(D) Percentage of SOX17- and POU5F1-positive cells.
(E) Relative mRNA expression as measured by qPCR (n = 3 to 6). Data are shown as means ± SEM, each dot represents an independent differentiation experiment and the horizontal bars with asterisks represent statistical significance between samples (p < 0.05, t test).
Downregulation of CHCHD2 Is a Transcriptomic Marker for the Gain of 20q11.21
The deregulation of the TGF-β and the SMAD signaling suggested by the DAVID analysis and the well-established relationship of this pathway to pluripotency and ectoderm differentiation prompted us to further investigate the differentially expressed genes to understand the driving mechanism for this deregulation. For this, we annotated the function of the top 100 differentially expressed genes (Table S3) in both mutant and Bcl-xL overexpressing cells, and searched the literature for their potential relationship to Bcl-xL and the TGF-β/SMAD signaling. The most interesting candidate in this list is CHCHD2 because the protein binds Bcl-xL, resulting in a decreased oligomerization of apoptotic activator Bax (Liu et al., 2015). Furthermore, low levels of CHCHD2 have been mechanistically linked to disrupted TGF-β/SMAD signaling and decreased neuroectodermal commitment in hPSCs (Zhu et al., 2016). Our RNA sequencing data showed that CHCHD2 is strongly downregulated in the lines with a gain of 20q11.21 and in VUB03_BcL-xL. Hence, we decided to investigate if CHCHD2 was effectively the mediator of the decreased neuroectoderm commitment of the mutant cells.
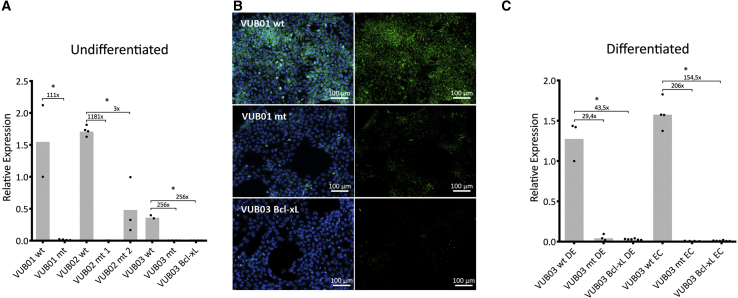
We validated the CHCHD2 RNA sequencing findings by measuring the mRNA and protein levels in normal and mutant lines as well as VUB03_BcL-xL. All mutant lines and VUB03_BcL-xL showed statistically significantly lower levels of CHCHD2 at the mRNA level, while the endogenous expression levels varied up to a 4-fold change among the control lines (Figure 5A). Immunocytochemistry of normal and mutant lines and VUB03_BcL-xL showed absence of CHCHD2 protein in the mutant and VUB03_BcL-xL cells (Figure 5B). To assess if this downregulation affected solely the pluripotent state, we differentiated the VUB03_wt, VUB03_mt, and VUB03_BcL-xL to neuroectoderm and definitive endoderm. Figure 5C shows the real-time qPCR results showing that CHCHD2 remains stably downregulated after differentiation in both VUB03_mt and VUB03_BcL-xL.
Figure 5.
Lines with a Gain of 20q11.21 Stably Downregulate CHCHD2 Due to Bcl-xL Overexpression
(A and B) (A) CHCHD2 expression as measured by qPCR in undifferentiated cells. All mutant lines show a significantly decreased expression compared with their normal counterparts (n = 4–9). (B) CHCHD2 immunostaining (in green) in VUB01 showing the presence of the protein in the control subline and its absence in the mutant subline.
(C) CHCHD2 expression as measured by qPCR in the three sublines of VUB03 (control, mutant, and VUB03_BcL-xL) in the differentiated state. DE, definite endoderm; EC, neuroectoderm (n = 7–10).
(A and C) Data are shown as means ± SEM, each dot represents an independent differentiation experiment and the horizontal bars with asterisks represent statistical significance between samples (p < 0.05, t test).
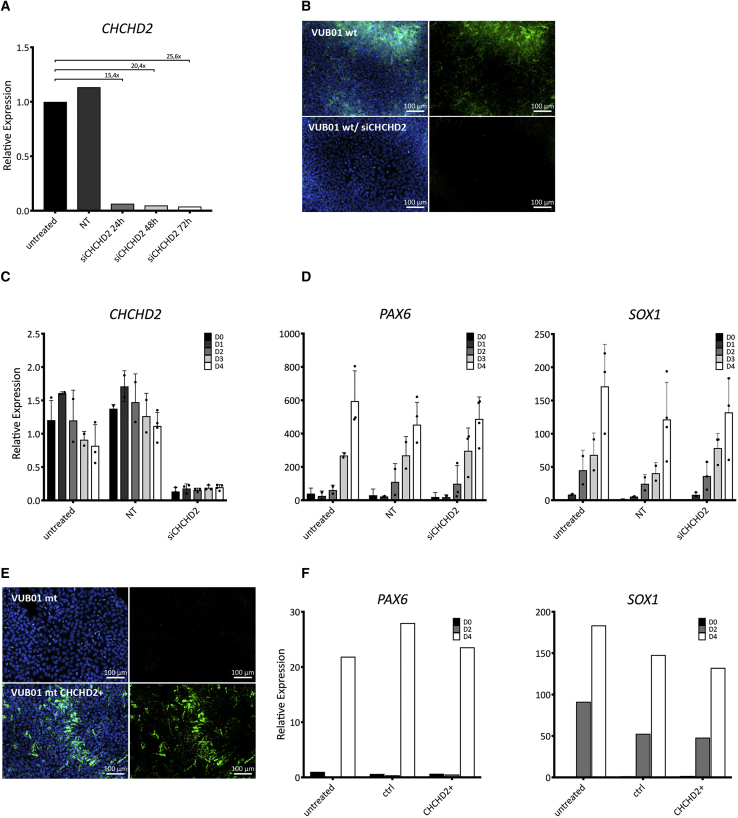
To functionally address the potential link between CHCHD2 downregulation and decreased neuroectoderm commitment, we used small interfering RNA (siRNA) to knockdown CHCHD2 in control hESCs, and mRNA transfection to exogenously overexpress CHCHD2 in mutant cells. Figure 6A shows the mRNA levels of CHCHD2 in undifferentiated VUB01_wt cells 24, 48, and 72 h after transfection, and Figure 6B shows the immunostaining for the protein 72 h posttransfection. The results show downregulation both at the mRNA and protein levels. We induced neuroectoderm commitment in VUB01_wt treated with siRNA against CHCHD2 as well as untreated cells and cells treated with a nontargeting siRNA. We measured CHCHD2, PAX6, and SOX1 expression each day of a 4-day differentiation. This experiment was repeated four times; the results are shown in Figures 6C and 6D. We found that the knockdown of CHCHD2 did not change the ability of the cells to differentiate to neuroectoderm in any of the replicates. We also exogenously overexpressed CHCHD2 in VUB01_mt (Figure 6E) and measured the PAX6 and SOX1 expression on day 0, 2, and 4 of neuroectoderm induction (Figure 6F). In line with the siRNA data, the results show that exogenous CHCHD2 expression does not rescue the effect of the gain of 20q11.21.
Figure 6.
Knockdown and Overexpression of CHCHD2 Do Not Affect Neuroectoderm Differentiation
(A) CHCHD2 expression as measured by qPCR in VUB01_wt 24, 48, and 72 h after transfection with siRNA against CHCHD2 (n = 1).
(B) Representative immunostaining of untreated and transfected cells 72 h posttransfection (the CHCHD2 protein can be seen in green).
(C and D) CHCHD2 (C) and PAX6 and SOX1 (D) expression as measured by qPCR in VUB01 WT during a 4-day induction to neuroectoderm. NT, nontargeting siRNA (n = 1).
(E) Immunostaining of untreated cells and after 72 h of exogenous CHCHD2 expression (in green).
(F) PAX6 and SOX1 expression as measured by qPCR in VUB01_mt during a 4-day induction to neuroectoderm in untreated, control (GFP) and CHCHD2-overexpressing cells (n = 1).
In summary, while CHCHD2 is consistently downregulated in mutant hESCs both before and after differentiation, our results strongly suggest that it is not the link between the Bcl-xL upregulation in the mutant cells and their decreased neuroectodermal commitment.
Discussion
The aim of our study was to investigate the impact of one of the most commonly found chromosomal abnormalities in hPSCs, a gain of 20q11.21, on their differentiation capacity (Amps et al., 2011). The awareness of genome instability in hPSCs has increased in the past years, along with the realization that we lack answers on what the functional consequences of these mutations are on differentiating cells, especially when considering clinical applications (Andrews et al., 2017). Our work was prompted by the clear need for systematic studies of the impact of recurrent chromosomal abnormalities on the differentiation capacity and malignant potential of hPSCs.
To reach our aim, we included four mutant lines and their isogenic normal counterparts, and one Bcl-xL overexpressing line. Remarkably, transcriptomic analysis showed that all mutant lines have a significantly different transcriptome from their chromosomally normal isogenic counterparts, and which is very similar to that of cells transgenically overexpressing Bcl-xL. This is despite the fact that the mutant lines carried different copy numbers, leading to a difference in overexpression of genes within the region. This strongly suggests that overexpression of Bcl-xL alone, which is also the driver for the selective advantage of the cells in their undifferentiated state (Avery et al., 2013, Nguyen et al., 2014), is sufficient to explain the majority of transcriptomic changes. Interestingly, an important part of the differential expression was of genes related to the TGF-β- and SMAD-mediated signaling, which led us to investigate the behavior of the mutant hESCs during the lineage commitment to neuroectoderm and mesendoderm. Our differentiation experiments show that the lines with a gain of 20q11.21 and those overexpressing Bcl-xL have an impaired neuroectodermal lineage commitment, with no differences in their capacity for differentiation into mesendodermal derivatives.
Next, CHCHD2 was identified as a prime candidate linking Bcl-xL to the transcriptomic changes, because it is consistently downregulated in mutant cells and the protein binds Bcl-xL, resulting in a decreased oligomerization of Bax (Liu et al., 2015). This downregulation may be through a self-regulatory feedback loop, as CHCHD2 is able to translocate to the nucleus to transactivate itself as well as other genes that contain oxygen-responsive element sequences in their promoter region (Aras et al., 2013, Grossman et al., 2017). Another interesting function of CHCHD2 is its capacity to interact with the SMADs (2/3/4) and thus modulate the transcription of TGF-β-mediated target genes (Zhu et al., 2016). Zhu et al. also showed that endogenous low levels of CHCHD2 in hPSCs result in a decreased capacity for ectodermal differentiation. The findings in our transcriptome analysis showing deregulation of both TGF-β/SMAD signaling and genes involved in neural differentiation in hESCs with a 20q11.21 mutation and Bcl-xL overexpressing cells are consistent with the known functions of CHCHD2. Finally, the TGF-β/SMAD signaling is known to have a pivotal role during the first-lineage commitment (Sumi et al., 2008). We therefore carried out functional tests, assessing the impact on neuroectoderm differentiation of siRNA knockdown of CHCHD2 in control cells and CHCHD2 mRNA transfection in mutant cells. Unexpectedly, we found that CHCHD2 does not mediate the effect on differentiation under our experimental conditions. We have no immediate explanation for these differences, although it is worth noting that Zhu et al. did not assess ectodermal differentiation in the same manner as we did, i.e., using a TGF-β inhibitor. In their work, the differentiation bias was assessed using spontaneous differentiation without modulating the TGF-β signaling, which may explain some of the differences we see. Another point of interest is that, in the last part of their work, they show that the use of the TGF-β inhibitor SB431542 during differentiation cancels the effect of downregulating CHCHD2 using siRNA. This conclusion is based on qPCR results for PAX6 between control and siRNA-treated cells. Although the authors found a statistically significant upregulation of PAX6, the relative fold change was less than 0.3 which may be not biologically relevant. This is in line with our results for siRNA CHCHD2 knockdown. Taken together, this suggests that, although Zhu et al. clearly prove that CHCHD2 interacts with SMAD4, the relationship between CHCHD2 and neuroectoderm commitment is less direct than hypothesized. Nevertheless, this gene appears as an excellent transcriptomic marker for the gain of 20q11.21.
In summary, our work shows the significant impact genetic changes can have on hPSCs, and provides, to the best of our knowledge, one of the few studies pinpointing a specific effect of a recurrent aberration and identifies the mechanisms behind it. The results of our study are of particular importance when taking into account that the 20q11.21 amplification occurs in more than 20% of hPSC lines worldwide and that G-banding as the most widely used method for karyotyping fails to detect it. The mutation may bias experimental data in a research setting if unnoticed, although it is unlikely that it would be undetected in a clinical setting, because in clinical trials the genetic screening protocols are more stringent. Nevertheless, if the mutation is in the process of culture take-over, it could remain undetected and result in poor differentiation in the clinically relevant cell types.
Experimental Procedures
hESC Lines and Culture
hESC were derived and characterized as described previously (Mateizel et al., 2006, Mateizel et al., 2009). All lines are registered with the EU hPSC registry (https://hpscreg.eu/). hESCs were cultured on dishes coated with 10 μg/mL laminin-521 (Biolamina) in NutriStem hESC XF medium (NS medium; Biological Industries) with 100 U/mL penicillin/streptomycin (Thermo Fisher Scientific), and passaged as single cells in a 1:10 to 1:100 ration using TrypLE Express (Thermo Fisher Scientific) when 70%–80% confluent. The cells were grown at 37°C in 5% CO2. VUB03_BcL-xL was generated by lentiviral transduction, as described by Nguyen et al. (2014) and in the Supplemental Information.
A large bulk of frozen cells was generated for each line. After thawing, the lines were used at the lowest passage possible (Table S1), and were checked for the 20q11.21 amplification by copy-number assay 3 to 4 passages after thawing.
aCGH
Oligonucleotide aCGH was carried out based on the protocol provided by the manufacturer (Agilent Technologies). A total of 400 ng of DNA was labeled with Cy3, while the reference DNA (Promega) was labeled with Cy5. The samples are hybridized on the microarray slide (4 × 44K Human Genome CGH Microarray, Agilent Technologies). The slides were scanned using an Agilent dual laser DNA microarray scanner G2566AA. Only arrays with an SD ≤ 0.20, signal intensity > 50, background noise < 5, and a derivative log-ratio < 0.2 were taken into account. Cutoff values were set at three consecutive probes with an average log2 ratio over 0.3 for gains and of −0.45 for loss.
Gene Copy Number Using Real-Time qPCR
Copy-number quantification was performed on the ViiA 7 thermocycler (Thermo Fisher Scientific), ViiA 7 software v.1.2 (Thermo Fisher Scientific), and Applied Biosystems Copy Caller v.2.1. We used the copy-number assays for RNAseP (4403326) as a reference and POFUT1 (Hs02487189_cn) to assess the number of copies of the 20q11.21 region.
RNA Sequencing
RNA (150 ng) was used to perform an Illumina sequencing library preparation using the QuantSeq 3′ mRNA-Seq Library Prep Kits (Lexogen) according to manufacturer's protocol. During library preparation 17 PCR cycles were used. Libraries were quantified by qPCR, according to Illumina's protocol “Sequencing Library qPCR Quantification protocol guide,” version February 2011. A high-sensitivity DNA chip (Agilent Technologies) was used to control the library's size distribution and quality. Sequencing was performed on a high-throughput Illumina NextSeq 500 flow cell generating 75 bp single reads. Details on the bioinformatics processing can be found in the Supplemental Information.
hESC Differentiation
The protocol for neuroectoderm was adapted from Chetty et al. (2013) and Chambers et al. (2009). hESCs were passaged on laminin-521, as described above, 1–2 days before EC differentiation in a ratio of 50,000 cells per cm2 so that they were 90% confluent on the starting day. The neuroectoderm differentiation medium was refreshed daily, and consisted of KnockOut DMEM (Thermo Fisher Scientific) with 10% KnockOut Serum Replacement (Thermo Fisher Scientific) and supplemented with 500 ng/mL Recombinant Human Noggin Protein (R&D Systems) and 10 μM SB431542 (Tocris). Endoderm differentiation was carried out using a protocol based on Sui et al. (2012). hESCs were passaged on laminin-521, as described above, 1–2 days before differentiation in a ratio of 30,000 cells per cm2 so that they were 80%–90% confluent on the starting day. Definitive endoderm differentiation medium contains RPMI 1640 supplemented by GlutaMAX (Thermo Fisher Scientific), 0.5% B27 supplement (Thermo Fisher Scientific), 100 ng recombinant human/mouse/rat activin A (R&D Systems) and 3 μM CHIR99021 (Stemgent). One day later the medium was changed to differentiation medium without CHIR99021 and the cells were cultured for 2 more days. For mesendoderm induction, the same protocol as for endoderm was used, for 1 day.
Protein and mRNA Analysis
Real-time qPCR was carried out on a ViiA 7 thermocycler (Thermo Fisher Scientific) and using standard protocol as provided by the manufacturer. Details on the probes, assays and primers are listed in the Supplemental Information. Sandwich ELISA was conducted using the human total Bcl-xL ELISA kit for Bcl-xL (DuoSet IC, R&D Systems) following the manufacturer's protocol. All samples were analyzed in triplicate. Immunostaining was carried out on cells fixed and permeabilized with 4% paraformaldehyde and 100% methanol (Sigma-Aldrich), and blocked with fetal bovine serum (Thermo Fischer Scientific). The list with antibodies can be found in the Supplemental Information. Imaging was performed on a LSM800 confocal microscope (Carl Zeiss), and cell counts were done using the Zen 2 (blue edition) imaging software.
Statistics
Data are presented as means. Unpaired two-tailed t test (GraphPad Prism 5.0b) was used to determine significance between groups. For all statistical tests, the 0.05 level of confidence was accepted for statistical significance.
siRNA Knockdown
The siRNAs used in this study were purchased from Healthcare/Dharmacon: ON-TARGETplus Human CHCHD2 siRNA SMARTpool (cat. no L-019120-100005) and ON-TARGETplus no-targeting siRNA as a negative control (cat. no. D-001810-01-05). The siRNA was dissolved in 1× siRNA buffer and a final concentration of 50 nM was transfected with lipofectamine RNAiMAX (13778150) in Opti-MEM (11058021) following the manufacturer's protocols (Thermo Fischer Scientific).
Plasmid Preparation and In Vitro Transcription of Capped mRNA
The synthetic gene encoding for CHCHD2 was purchased from Integrated DNA Technologies and cloned as NcoI-XhoI fragment in the vector pEtheRNA-v2 as previously described (Van Lint et al., 2016). In vitro mRNA transcription and quality controls were performed as described in Bonehill et al. (2004). In brief, pEtheRNA-v2 plasmid was linearized with BfuAI (Fermentas) and in vitro mRNA transcription was performed with T7 polymerase according to the manufacturer instructions. mRNA integrity, concentration, and purity were tested on Agilent 2100 Bioanalyzer (Agilent Technologies).
Author Contributions
C.M. carried out all of the experiments unless stated otherwise and co-wrote the manuscript. E.C.D.D. did the bioinformatics analysis. M.R. performed the microscopy and cell counting. H.T.N. provided the Bcl-xL overexpressing line. F.Z. assisted with the gene expression analysis. A.K. provided the isochromosome line. D.D. provided the control samples for RNA sequencing. J.D.K. assisted with the microscopy. L.T. and F.V.N. performed the RNA sequencing and assisted in the bio-informatic analysis. L.F. supervised the mRNA production. K.S. proofread the paper. M.G. proofread the paper and supervised the experimental work. C.S. co-wrote the paper, designed and supervised the experimental work.
Acknowledgments
The authors acknowledge their colleagues from the human embryonic stem cell lab for the derivation and culture of the lines and professor Kris Thielemans from the LMCT laboratory of the VUB for providing the mRNA for overexpressing CHCHD2. This work was supported by the Fonds for Scientific Research in Flanders (Fonds Wetenschappelijk Onderzoek – Vlaanderen [FWO]) and the Methusalem Grant of the Research Council of the VUB. C.M. is a doctoral fellow supported by the Instituut voor Innovatie door Wetenschap en Technologie (IWT). A.K. and D.D. are doctoral fellows at the FWO.
Published: June 6, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2019.05.005.
Accession Numbers
All data have been deposited in the GEO repository with accession number GEO: GSE116372.
Supplemental Information
References
- Amps K., Andrews P.W., Anyfantis G., Armstrong L., Avery S., Baharvand H., Baker J., Baker D., Munoz M.B., Beil S. Screening ethnically diverse human embryonic stem cells identifies a chromosome 20 minimal amplicon conferring growth advantage. Nat. Biotechnol. 2011;29:1132–1144. doi: 10.1038/nbt.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews P.W., Ben-David U., Benvenisty N., Coffey P., Eggan K., Knowles B.B., Nagy A., Pera M., Reubinoff B., Rugg-Gunn P.J. Assessing the safety of human pluripotent stem cells and their derivatives for clinical applications. Stem Cell Reports. 2017;9:1–4. doi: 10.1016/j.stemcr.2017.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aras S., Pak O., Sommer N., Finley R., Hüttemann M., Weissmann N., Grossman L.I., Grossman L.I. Oxygen-dependent expression of cytochrome c oxidase subunit 4-2 gene expression is mediated by transcription factors RBPJ, CXXC5 and CHCHD2. Nucleic Acids Res. 2013;41:2255–2266. doi: 10.1093/nar/gks1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery S., Hirst A.J., Baker D., Lim C.Y., Alagaratnam S., Skotheim R.I., Lothe R.A., Pera M.F., Colman A., Robson P. BCL-XL mediates the strong selective advantage of a 20q11.21 amplification commonly found in human embryonic stem cell cultures. Stem Cell Reports. 2013;1:1–8. doi: 10.1016/j.stemcr.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avior Y., Sagi I., Benvenisty N. Pluripotent stem cells in disease modelling and drug discovery. Nat. Rev. Mol. Cell Biol. 2016;17:170–182. doi: 10.1038/nrm.2015.27. [DOI] [PubMed] [Google Scholar]
- Baker D.E.C., Harrison N.J., Maltby E., Smith K., Moore H.D., Shaw P.J., Heath P.R., Holden H., Andrews P.W. Adaptation to culture of human embryonic stem cells and oncogenesis in vivo. Nat. Biotechnol. 2007;25:207–215. doi: 10.1038/nbt1285. [DOI] [PubMed] [Google Scholar]
- Ben-David U., Mayshar Y., Benvenisty N. Virtual karyotyping of pluripotent stem cells on the basis of their global gene expression profiles. Nat. Protoc. 2013;8:989–997. doi: 10.1038/nprot.2013.051. [DOI] [PubMed] [Google Scholar]
- Ben-David U., Arad G., Weissbein U., Mandefro B., Maimon A., Golan-Lev T., Narwani K., Clark A.T., Andrews P.W., Benvenisty N. Aneuploidy induces profound changes in gene expression, proliferation and tumorigenicity of human pluripotent stem cells. Nat. Commun. 2014;5:4825. doi: 10.1038/ncomms5825. [DOI] [PubMed] [Google Scholar]
- Bonehill A., Heirman C., Tuyaerts S., Michiels A., Breckpot K., Brasseur F., Zhang Y., Van Der Bruggen P., Thielemans K. Messenger RNA-electroporated dendritic cells presenting MAGE-A3 simultaneously in HLA class I and class II molecules. J. Immunol. 2004;172:6649–6657. doi: 10.4049/jimmunol.172.11.6649. [DOI] [PubMed] [Google Scholar]
- Chambers S.M., Fasano C.A., Papapetrou E.P., Tomishima M., Sadelain M., Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma’ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetty S., Pagliuca F.W., Honore C., Kweudjeu A., Rezania A., Melton D.A. A simple tool to improve pluripotent stem cell differentiation. Nat. Methods. 2013;10:553–556. doi: 10.1038/nmeth.2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan C.A., Klimanskaya I., McMahon J., Atienza J., Witmyer J., Zucker J.P., Wang S., Morton C.C., McMahon A.P., Powers D. Derivation of embryonic stem-cell lines from human blastocysts. N. Engl. J. Med. 2004;350:1353–1356. doi: 10.1056/NEJMsr040330. [DOI] [PubMed] [Google Scholar]
- Draper J.S., Smith K., Gokhale P., Moore H.D., Maltby E., Johnson J., Meisner L., Zwaka T.P., Thomson J.A., Andrews P.W. Recurrent gain of chromosomes 17q and 12 in cultured human embryonic stem cells. Nat. Biotechnol. 2004;22:53–54. doi: 10.1038/nbt922. [DOI] [PubMed] [Google Scholar]
- Fazeli A., Liew C.-G., Matin M.M., Elliott S., Jeanmeure L.F.C., Wright P.C., Moore H., Andrews P.W. Altered patterns of differentiation in karyotypically abnormal human embryonic stem cells. Int. J. Dev. Biol. 2011;55:175–180. doi: 10.1387/ijdb.103177af. [DOI] [PubMed] [Google Scholar]
- Gopalakrishna-Pillai S., Iverson L.E. Astrocytes derived from trisomic human embryonic stem cells express markers of astrocytic cancer cells and premalignant stem-like progenitors. BMC Med. Genomics. 2010;3:12. doi: 10.1186/1755-8794-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman L.I., Purandare N., Arshad R., Gladyck S., Somayajulu M., Hüttemann M., Aras S. MNRR1, a biorganellar regulator of mitochondria. Oxid. Med. Cell. Longev. 2017;2017:6739236. doi: 10.1155/2017/6739236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herszfeld D., Wolvetang E., Langton-Bunker E., Chung T.-L., Filipczyk A.A., Houssami S., Jamshidi P., Koh K., Laslett A.L., Michalska A. CD30 is a survival factor and a biomarker for transformed human pluripotent stem cells. Nat. Biotechnol. 2006;24:351–357. doi: 10.1038/nbt1197. [DOI] [PubMed] [Google Scholar]
- Inzunza J., Sahlén S., Holmberg K., Strömberg A.M., Teerijoki H., Blennow E., Hovatta O., Malmgren H. Comparative genomic hybridization and karyotyping of human embryonic stem cells reveals the occurrence of an isodicentric X chromosome after long-term cultivation. Mol. Hum. Reprod. 2004;10:461–466. doi: 10.1093/molehr/gah051. [DOI] [PubMed] [Google Scholar]
- Keller A., Dziedzicka D., Zambelli F., Markouli C., Sermon K., Spits C., Geens M. Genetic and epigenetic factors which modulate differentiation propensity in human pluripotent stem cells. Hum. Reprod. Update. 2018;24:162–175. doi: 10.1093/humupd/dmx042. [DOI] [PubMed] [Google Scholar]
- Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent L.C., Ulitsky I., Slavin I., Tran H., Schork A., Morey R., Lynch C., Harness J.V., Lee S., Barrero M.J. Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell. 2011;8:106–118. doi: 10.1016/j.stem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefort N., Feyeux M., Bas C., Féraud O., Bennaceur-Griscelli A., Tachdjian G., Peschanski M., Perrier A.L. Human embryonic stem cells reveal recurrent genomic instability at 20q11.21. Nat. Biotechnol. 2008;26:1364–1366. doi: 10.1038/nbt.1509. [DOI] [PubMed] [Google Scholar]
- Van Lint S., Renmans D., Broos K., Goethals L., Maenhout S., Benteyn D., Goyvaerts C., Du Four S., Van der Jeught K., Bialkowski L. Intratumoral delivery of TriMix mRNA results in T-cell activation by cross-presenting dendritic cells. Cancer Immunol. Res. 2016;4:146–156. doi: 10.1158/2326-6066.CIR-15-0163. [DOI] [PubMed] [Google Scholar]
- Liu Y., Clegg H.V., Leslie P.L., Di J., Tollini L.A., He Y., Kim T.-H., Jin A., Graves L.M., Zheng J. CHCHD2 inhibits apoptosis by interacting with Bcl-x L to regulate Bax activation. Cell Death Differ. 2015;22:1035–1046. doi: 10.1038/cdd.2014.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitra A., Arking D.E., Shivapurkar N., Ikeda M., Stastny V., Kassauei K., Sui G., Cutler D.J., Liu Y., Brimble S.N. Genomic alterations in cultured human embryonic stem cells. Nat. Genet. 2005;37:1099–1103. doi: 10.1038/ng1631. [DOI] [PubMed] [Google Scholar]
- Mateizel I., De Temmerman N., Ullmann U., Cauffman G., Sermon K., Van de Velde H., De Rycke M., Degreef E., Devroey P., Liebaers I. Derivation of human embryonic stem cell lines from embryos obtained after IVF and after PGD for monogenic disorders. Hum. Reprod. 2006;21:503–511. doi: 10.1093/humrep/dei345. [DOI] [PubMed] [Google Scholar]
- Mateizel I., Spits C., Verloes A., Mertzanidou A., Liebaers I., Sermon K. Characterization of CD30 expression in human embryonic stem cell lines cultured in serum-free media and passaged mechanically. Hum. Reprod. 2009;24:2477–2489. doi: 10.1093/humrep/dep234. [DOI] [PubMed] [Google Scholar]
- Mitalipova M.M., Rao R.R., Hoyer D.M., Johnson J.A., Meisner L.F., Jones K.L., Dalton S., Stice S.L. Preserving the genetic integrity of human embryonic stem cells. Nat. Biotechnol. 2005;23:19–20. doi: 10.1038/nbt0105-19. [DOI] [PubMed] [Google Scholar]
- Närvä E., Autio R., Rahkonen N., Kong L., Harrison N., Kitsberg D., Borghese L., Itskovitz-Eldor J., Rasool O., Dvorak P. High-resolution DNA analysis of human embryonic stem cell lines reveals culture-induced copy number changes and loss of heterozygosity. Nat. Biotechnol. 2010;28:371–377. doi: 10.1038/nbt.1615. [DOI] [PubMed] [Google Scholar]
- Nguyen H.T., Geens M., Spits C. Genetic and epigenetic instability in human pluripotent stem cells. Hum. Reprod. Update. 2013;19:187–205. doi: 10.1093/humupd/dms048. [DOI] [PubMed] [Google Scholar]
- Nguyen H.T., Geens M., Mertzanidou A., Jacobs K., Heirman C., Breckpot K., Spits C. Gain of 20q11.21 in human embryonic stem cells improves cell survival by increased expression of Bcl-xL. Mol. Hum. Reprod. 2014;20:168–177. doi: 10.1093/molehr/gat077. [DOI] [PubMed] [Google Scholar]
- Serra M., Brito C., Correia C., Alves P.M. Process engineering of human pluripotent stem cells for clinical application. Trends Biotechnol. 2012;30:350–359. doi: 10.1016/j.tibtech.2012.03.003. [DOI] [PubMed] [Google Scholar]
- Spits C., Mateizel I., Geens M., Mertzanidou A., Staessen C., Vandeskelde Y., Van der Elst J., Liebaers I., Sermon K. Recurrent chromosomal abnormalities in human embryonic stem cells. Nat. Biotechnol. 2008;26:1361–1363. doi: 10.1038/nbt.1510. [DOI] [PubMed] [Google Scholar]
- Sui L., Mfopou J.K., Geens M., Sermon K., Bouwens L. FGF signaling via MAPK is required early and improves Activin A-induced definitive endoderm formation from human embryonic stem cells. Biochem. Biophys. Res. Commun. 2012;426:380–385. doi: 10.1016/j.bbrc.2012.08.098. [DOI] [PubMed] [Google Scholar]
- Sumi T., Tsuneyoshi N., Nakatsuji N., Suemori H. Defining early lineage specification of human embryonic stem cells by the orchestrated balance of canonical Wnt/beta-catenin, Activin/Nodal and BMP signaling. Development. 2008;135:2969–2979. doi: 10.1242/dev.021121. [DOI] [PubMed] [Google Scholar]
- Teo A.K.K., Arnold S.J., Trotter M.W.B., Brown S., Ang L.T., Chng Z., Robertson E.J., Dunn N.R., Vallier L. Pluripotency factors regulate definitive endoderm specification through eomesodermin. Genes Dev. 2011;25:238–250. doi: 10.1101/gad.607311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werbowetski-Ogilvie T.E., Bossé M., Stewart M., Schnerch A., Ramos-Mejia V., Rouleau A., Wynder T., Smith M.-J., Dingwall S., Carter T. Characterization of human embryonic stem cells with features of neoplastic progression. Nat. Biotechnol. 2009;27:91–97. doi: 10.1038/nbt.1516. [DOI] [PubMed] [Google Scholar]
- Wu H., Kim K.J., Mehta K., Paxia S., Sundstrom A., Anantharaman T., Kuraishy A.I., Doan T., Ghosh J., Pyle A.D. Copy number variant analysis of human embryonic stem cells. Stem Cells. 2008;26:1484–1489. doi: 10.1634/stemcells.2007-0993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Lin G., Tan Y., Zhou D., Deng L., Cheng D., Luo S., Liu T., Zhou X., Sun Z. Tumor progression of culture-adapted human embryonic stem cells during long-term culture. genes. Chromosomes Cancer. 2008;47:665–679. doi: 10.1002/gcc.20574. [DOI] [PubMed] [Google Scholar]
- Yang S., Lin G., Tan Y.-Q., Deng L.-Y., Yuan D., Lu G.-X. Differences between karyotypically normal and abnormal human embryonic stem cells. Cell Prolif. 2010;43:195–206. doi: 10.1111/j.1365-2184.2010.00669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying L., Mills J.A., French D.L., Gadue P. OCT4 coordinates with WNT signaling to pre-pattern chromatin at the SOX17 locus during human ES cell differentiation into definitive endoderm. Stem Cell Reports. 2015;5:490–498. doi: 10.1016/j.stemcr.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Gomez-Duran A., Saretzki G., Jin S., Tilgner K., Melguizo-Sanchis D., Anyfantis G., Al-Aama J., Vallier L., Chinnery P. The mitochondrial protein CHCHD2 primes the differentiation potential of human induced pluripotent stem cells to neuroectodermal lineages. J. Cell Biol. 2016;215:187–202. doi: 10.1083/jcb.201601061. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.