Summary
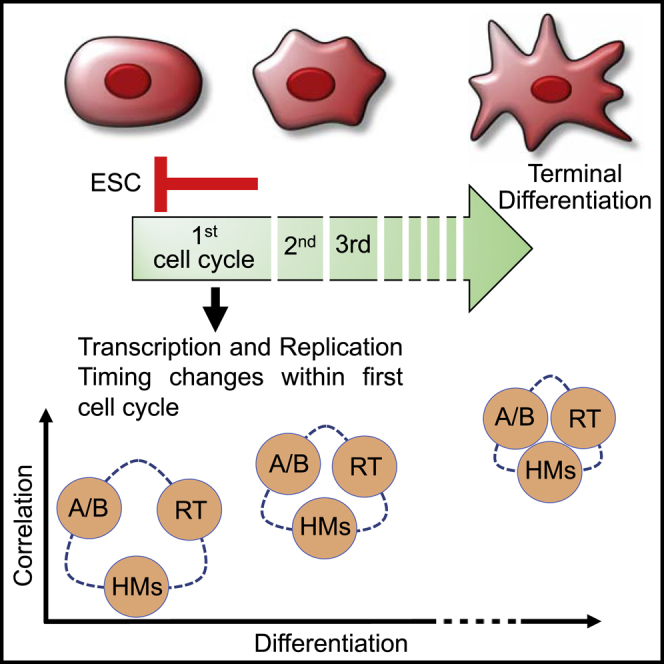
The temporal order of DNA replication is regulated during development and is highly correlated with gene expression, histone modifications and 3D genome architecture. We tracked changes in replication timing, gene expression, and chromatin conformation capture (Hi-C) A/B compartments over the first two cell cycles during differentiation of human embryonic stem cells to definitive endoderm. Remarkably, transcriptional programs were irreversibly reprogrammed within the first cell cycle and were largely but not universally coordinated with replication timing changes. Moreover, changes in A/B compartment and several histone modifications that normally correlate strongly with replication timing showed weak correlation during the early cell cycles of differentiation but showed increased alignment in later differentiation stages and in terminally differentiated cell lines. Thus, epigenetic cell fate transitions during early differentiation can occur despite dynamic and discordant changes in otherwise highly correlated genomic properties.
Keywords: replication timing, differentiation, chromatin 3D architecture, chromatin 3D organization, chromatin structure, lineage commitment, gene expression
Graphical Abstract
Highlights
-
•
Stable transcriptional reprogramming toward endoderm within one cell cycle
-
•
Replication timing can change in the first S phase of a cell fate change
-
•
Early discordance between Replication timing and Hi-C compartments
-
•
Alignment of replication timing to compartments increases in later cell cycles
The temporal order of DNA replication is regulated during development, and is highly correlated with gene expression, chromatin structure, and 3D-genome architecture. Gilbert and colleagues find that stable transcriptional changes of human embryonic stem cells to endoderm can occur within a single cell cycle, accompanied by discordance in replication timing and chromatin compartment that align in later differentiation stages.
Introduction
Replication of the eukaryotic genome occurs in a defined temporal order, regulated at the level of 400–800-kb chromosome segments that are referred to as replication domains (RDs) (Rivera-Mulia and Gilbert, 2016a, Rivera-Mulia and Gilbert, 2016b, Solovei et al., 2016, Xiang et al., 2018). This replication timing (RT) program is established during early G1 phase of each cell cycle, known as the timing decision point (TDP) (Dileep et al., 2015a, Dimitrova and Gilbert, 1999, Li et al., 2001, Lu et al., 2010, Rivera-Mulia and Gilbert, 2016a, Wu et al., 2006). During stem cell differentiation, changes in RT occur coordinately across RDs, often correlated with changes in gene activity and subnuclear position (Hiratani et al., 2004, Hiratani et al., 2008, Hiratani et al., 2010, Perry et al., 2004, Rivera-Mulia et al., 2015, Williams et al., 2006), to create cell type-specific RT programs that are highly characteristic of each cell type and are altered in human disease (Blumenfeld et al., 2017, Donley and Thayer, 2013, Fritz et al., 2013, Rivera-Mulia et al., 2017, Sasaki et al., 2017), suggesting that the RT program is associated with stable epigenetic states. Indeed, depletion and overexpression of numerous chromatin and transcription regulators has little to no effect on RT, whereas the induction of a cell fate change in stem cells induces rapid and widespread alterations in RT (Dileep et al., 2015b). Moreover, X chromosome inactivation during gastrulation in female mammals is initially reversible, but is then stabilized coincident with a switch to late replication and formation of a stable compact Barr body localized near the nuclear periphery (Barr and Bertram, 1949, Gilbert et al., 1962, Hiratani and Gilbert, 2010, Lyon, 1961, Morishima et al., 1962). Importantly, when differentiated cells are reprogrammed to the induced pluripotent stem cell (iPSC) state, clones that arrest in a stable incompletely reprogrammed state (partial-iPSCs) share a common RT profile in which domains that fail to return to early replication harbor pluripotency-specific genes that fail to reactivate (Hiratani et al., 2010), providing strong evidence that late replication is associated with a stably repressed epigenetic state that presents a barrier to reprogramming.
The longstanding correlation between early replication, transcriptional activity and active chromatin marks is nuanced with complexities (Goldman et al., 1984, Hansen et al., 1996, MacAlpine et al., 2004, Muller and Nieduszynski, 2017, Ostrow et al., 2017, Rivera-Mulia and Gilbert, 2016b, Schubeler et al., 2002, Therizols et al., 2014, White et al., 2004, Woodfine et al., 2004, Woodfine et al., 2005, Yue et al., 2014). Coordinated changes in gene expression and RT have been identified during mouse and human stem cell differentiation (Hiratani et al., 2010, Rivera-Mulia et al., 2015), but the time course of these previous differentiation protocols has precluded the ability to assess which changes first. Moreover, many genes can be either late replicating and expressed or early replicating and silent and correlations of chromatin marks and promoter content have not identified any clear differences between these sets of genes (Hiratani et al., 2010, Rivera-Mulia et al., 2015). In animals with rapid early cleavage stage embryos, the RT program is evident before the onset of transcription at the midblastula transition (Kaaij et al., 2018, Pourkarimi et al., 2016, Seller and O'Farrell, 2018, Siefert et al., 2017). Indeed, when following intermediate cell types through several lineage pathways, two-thirds of genes that experienced RT changes were expressed and late replicating in at least one intermediate stage, suggesting the existence of classes of genes that are dependent or independent of their RT (Rivera-Mulia et al., 2015).
RT is also highly correlated with 3D genome architecture. Early replicating chromatin is less densely packed and resides in the interior of nucleus while late replicating chromatin is generally more dense and localized to regions around the periphery of the nucleus and nucleoli (Dimitrova and Gilbert, 1999, Nakayasu and Berezney, 1989, O'Keefe et al., 1992). More recently, chromatin conformation capture (Hi-C) analyses have confirmed the megabase-scale folding of chromosomes into two compartments, A and B (Lieberman-Aiden et al., 2009), which show a striking correlation with RT (Dixon et al., 2012, Rivera-Mulia et al., 2018, Ryba et al., 2010, Yaffe et al., 2010). These compartments have been further divided into self-interacting units referred to as topologically associated domains (TADs), whose structural boundaries align with the functional boundaries of RDs (Pope et al., 2014). Moreover, the organization of chromatin into compartments and TADs occurs coincident with the establishment of the RT program at the TDP early in G1 (Dileep et al., 2015a) and compartment association changes coordinately with RT during differentiation, while maintaining TAD borders (Dixon et al., 2015, Takebayashi et al., 2012).
Determining how RT is linked with gene expression and chromatin organization will require systems that can uncouple these events. One study experimentally targeted DNA sequences to the nuclear periphery found repositioning of chromatin occurred only after mitosis, suggesting that a dismantling of the nucleus may be required to remodel subnuclear positioning (Reddy et al., 2008). However, another study showed that targeting a powerful acidic transcriptional activator was sufficient to move a peripherally localized gene off of the nuclear periphery even when the transcriptional activity of the activator was eliminated, suggesting that chromatin remodeling can reorganize 3D genome architecture (Therizols et al., 2014). One way to probe causality is to determine the order of events that occur dynamically during a cell fate transition, providing the transition is sufficiently synchronous to observe such changes within a single cell cycle. Recently, we demonstrated that differentiating human embryonic stem cells (hESCs) to definitive endoderm (DE) results in cell fate changes within one cell cycle, providing the cells are stimulated to differentiate during G1 phase (Schulz et al., 2012, Wilson et al., 2016). Here, we track the order of changes in RT, gene expression, and Hi-C A/B compartments over the first two cell cycles in this system. Remarkably, we detected transcriptional reprogramming to DE-specific transcriptional programs as early as 6 h (1/3 of a cell cycle) that could not be altered by returning to stem cell self-renewal medium conditions. This reprogramming was accompanied by changes in RT as early as 6 h after stimulation. As with other more protracted differentiation systems, transcription of some but not all genes were coincident with switches in RT. By contrast, chromatin compartment changes and RT changes were discordant during early differentiation intermediates but became more aligned in more differentiated cells and terminally differentiated cell lines. Moreover, the correlation between RT and Hi-C A/B compartments was significantly weaker in stem cells but became more aligned in more differentiated cells. A similar discordance was found for the correlations between RT changes and changes in the density of histone modifications associated with active enhancers during early differentiation cell cycles. Overall, our results reveal that epigenetic cell fate transitions during early stages of differentiation occur within the context of dynamic and discordant changes in large-scale chromatin organization and transcription that become more aligned in later differentiation stages and terminally differentiated cell types.
Results
Rapid and Stable Transcriptional Reprogramming in the First Cell Cycle of a Cell Fate Transition
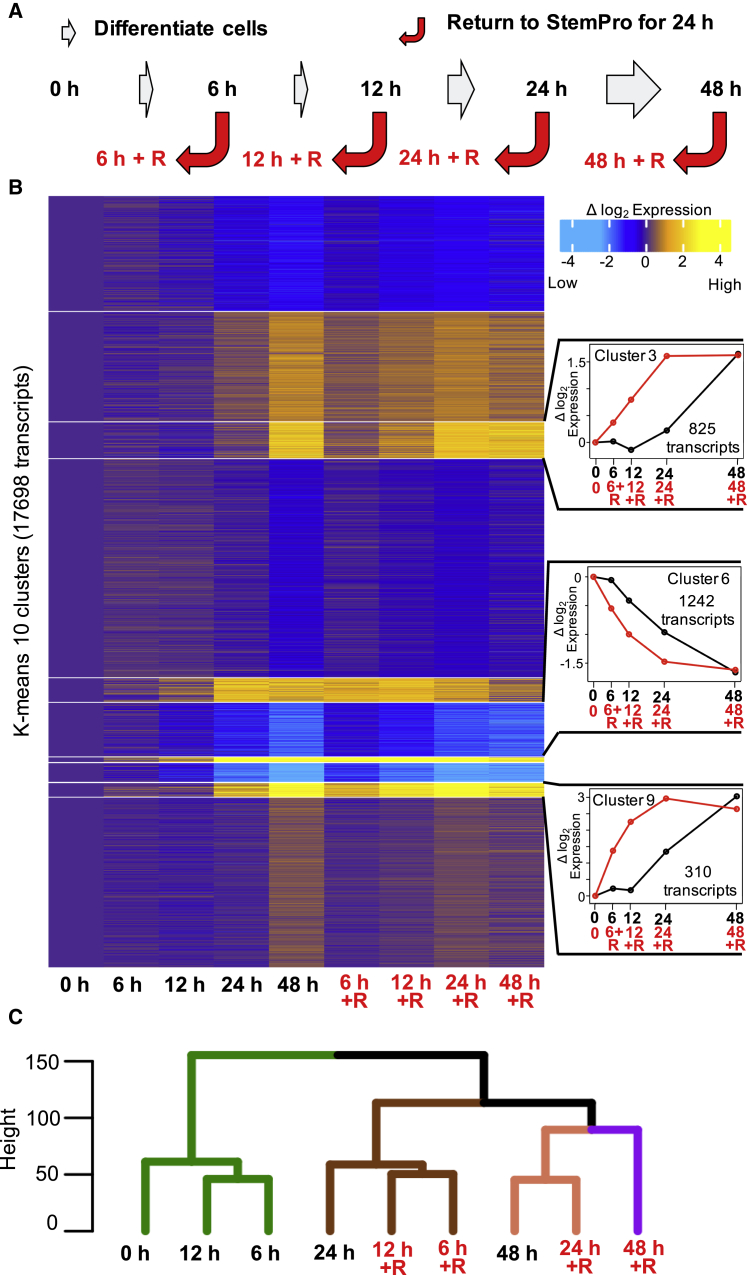
Recent reports suggest that stem cells elicit a response to differentiation factors from G1, and that once they enter S phase they do not respond until the following G1 (Pauklin and Vallier, 2013, Sela et al., 2012, Wilson et al., 2016). It has also been shown that experimental reprogramming requires passage through S phase (Tsubouchi et al., 2013), These observations suggest that transcriptional reprogramming associated with differentiation across a population of asynchronously dividing stem cells should require at least one complete cell cycle of continuous exposure to induction factors. To investigate how many cell cycles are required for stable changes in transcriptional programs, we used a robust suspension culture DE differentiation system in which >90% of cells reach DE following 48 h of induction, as indicated by the upregulation of DE-specific markers SOX17 and FOXA2 (Schulz et al., 2012). We have previously shown the hESC cell cycle is ∼18 h and does not significantly lengthen in the first few cell cycles of differentiation (Wilson et al., 2016). Cells were stimulated to differentiate for 0, 6, 12, 24, and 48 h, and half the cells were profiled for gene expression by microarray analysis, while the other half were washed with PBS, returned to normal hESC growth medium for 24 h, and then profiled (Figure 1A). Transcriptome data were filtered for genes that were upregulated or downregulated (false discovery rate [FDR] adjusted p < 0.05) during 48 h of DE differentiation, and K-means clustering was performed (Figure 1B). We reasoned cells that did not stably initiate transcriptional reprograming toward endoderm would reacquire an ESC-like transcriptome when returned to stem cell medium. Surprisingly, we found that the transcriptomes from cells stimulated for as few as 6 h proceeded to follow the same progression of transcription changes as cells maintained in differentiation medium, despite the replacement of differentiation medium with medium promoting the self-renewing ESC state (Figures 1B and S1). The transcriptome after 6 or 12 h in differentiation medium followed by 24 h of reversal in stem cell medium is strikingly similar to expression after 24 h of forward differentiation for all K-clusters (Figures 1B and S1). Further, hierarchical clustering of these datasets revealed that cells differentiated for only 6 or 12 h followed by 24 h in hESC medium clustered with cells grown under differentiation conditions for 24 h, while those differentiated for 24 h followed by 24 h in hESC medium clustered with cells grown under differentiation conditions for 48 h (Figures 1C and S1). These surprising results demonstrate that stable changes in the transcriptional program occur very rapidly and within the first cell cycle of differentiation.
Figure 1.
A Stable Transcriptional Reprogramming Occurs within One Cell Cycle
(A) Schematic of cell reversal experiment. Cells were stimulated for 0, 6, 12, 24, and 48 h. At each time point half the cells were washed and returned to stem cell medium for 24 h. At the end of each time point, gene expression was profiled by microarray analysis.
(B) Heatmap of K-means clustered average gene expression change (with respect to ESC 0 h state) during forward differentiation and reversal for all differentially regulated genes (FDR p < 0.05) during 48 h of differentiation. Exemplary plots on the right quantify change in average log2 expression with respect to 0 h for forward differentiation (black) and reversal (red) at different time points for the respective K-means cluster.
(C) Hierarchical clustering of forward differentiation and reversal time points. K-means clustering and hierarchical clustering show that 6 or 12 h of differentiation + 24 h of reversal has an expression signature more similar to 24 h of forward differentiation. Average of two independent differentiations and reversal experiments is plotted.
Changes in RT Can Also Be Detected within the First Cell Cycle
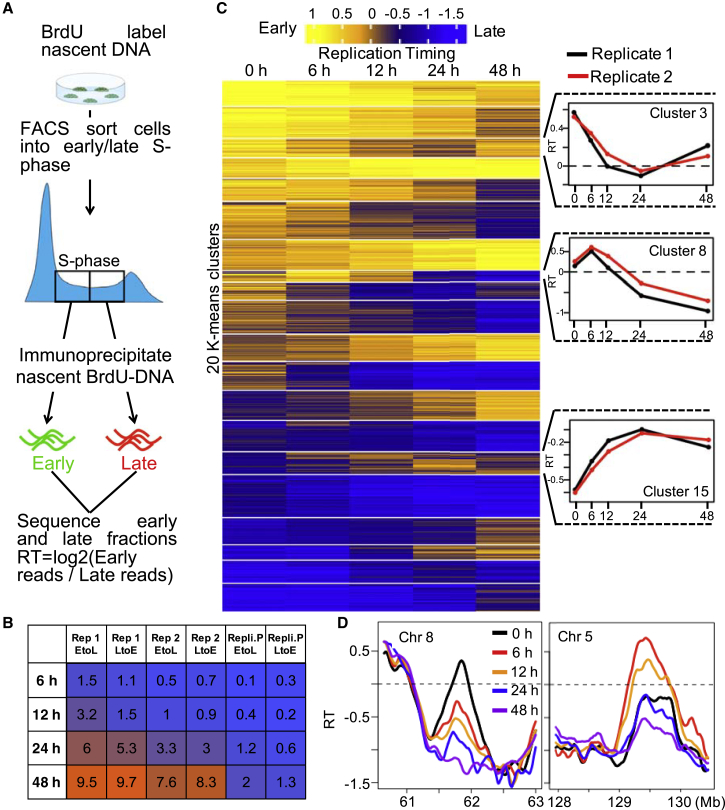
Changes in RT during differentiation are associated with changes in epigenetic state and many of the genes that are difficult to reprogram to iPSCs reside within late replicating domains (Hiratani et al., 2010), suggesting that late replication is a barrier to reprogramming. The surprising finding that stable transcriptional reprogramming occurs within less than one cell cycle prompted us to investigate whether changes in RT could occur within this short time frame. To identify regions that reproducibly change RT during differentiation, we performed two independent differentiations toward the endoderm lineage, for which cells were differentiated for 0, 6, 12, 24, and 48 h, labeled for 2 h with bromodeoxyuridine (BrdU), and then collected for analysis of RT by Repli-seq. These experiments were performed in a simpler monolayer differentiation system. Monolayer differentiation is not as efficient as the suspension system, but most cells induce SOX17 in less than two cell cycles, while the remainder either remain pluripotent or die (Davenport et al., 2016, Wilson et al., 2016). Fluorescent-activated cell sorting (FACS) analysis of the endoderm differentiation makers FOXA2 and SOX17 detected coexpression in ∼44% of cells and expression of either marker in >60% of cells at the 48 h time point (Figure S2A). Since cells need to be fixed to detect transcription factors, we could not assess the fraction of negative cells that may be nonviable. However, the kinetics of genome-wide transcriptional changes and key endoderm marker genes, measured by RNA sequencing (RNA-seq) (Figures S2B and S2C) were highly comparable with the expression microarray in Figure 1, indicating robust differentiation. To measure RT, early and late replicating cells were FACS sorted based on DNA content and nascent, BrdU incorporated, DNA was immunoprecipitated with an anti-BrdU antibody and sequenced. Sequenced DNA was then mapped back to the genome and results displayed as a Loess-smoothed log2 ratio of reads in the early versus late fractions of S phase (Figure 2A). Replicate 1 and 2 showed genome-wide changes (dRT > 0.5) (Hiratani et al., 2010), affecting 19% and 16% of the genome, respectively, by 48 h of differentiation (Figures 2B and S2D). We observed regions of genome that complete RT reprograming and remain stable thereafter at every time point, consistent with population-wide changes in RT among the viable, BrdU-incorporating cells (Figure S2E). There were also examples of domains that completed an RT transition in one direction as soon as 6 h and then transitioned in the reverse direction in later time points (Figures 2C, cluster 8 and 2D, right panel).
Figure 2.
Changes in RT Are Detectable within the First Cell Cycle
(A) Method to map genome-wide RT.
(B) Percentage of 50-kb windows that change RT (change in RT > 0.5) at each time point for independent replicate 1 and 2, and reproducible by RepliPrint (Experimental Procedures).
(C) Heatmap of RT in 50-kb windows clustered by K-means at 0, 6, 12, 24, and 48 h. Only replication domains that were reproducible between the two replicates (independent differentiations) were chosen (Experimental Procedures). Line graphs on the right shows average RT values within three exemplary K-means clusters for both replicates.
(D) Two exemplary replication domains demonstrating a change in RT between 0 and 6 h of differentiation.
To identify regions of statistically significant RT changes that occur reproducibly in both replicates, we divided the genome into 50-kb segments and then subjected the average RT for all reads within each segment from each time point to a statistical package that ranks the top regions of difference between any two groups of datasets (Repliprint [Ryba et al., 2011]). This algorithm chooses only regions that show low variance between the datasets within each group (e.g., two replicates after 6 h of differentiation) and high variance between the two comparator groups (e.g., 6 versus 0 h differentiation, 12 versus 0 h, etc.). This produced a set of high confidence 50-kb windows that reproducibly switch RT during differentiation (Experimental Procedures). To group all adjacent 50-kb windows into their corresponding domains (RDs), we consolidated adjacent windows occurring within 200 kb of each other to produce a set of high confidence switching RDs. This set of switching RDs is certainly an underestimate, as the kinetics of each differentiation can vary slightly, so domain switches that were out of synchrony between replicates would be lost (Figure 2B).
Next, to examine the kinetics of RT changes during differentiation we performed a K-means clustering analysis (20 clusters) on RT values of these high confidence switching RDs at 0, 6, 12, 24, and 48 h of differentiation (Figure 2C), which revealed distinct patterns of early to late (EtoL), or late to early (LtoE) regulation. Of the 20 clusters, 9 were progressing from LtoE (45%) and 11 were switching EtoL (55%). Since the kinetics of differentiation were slightly different for each replicate set of time points, clustering was done with one replicate, and compared with the other replicate by plotting the average RT for domains from each cluster (Figures 2C and S2F), verifying the reproducibility of these changes. Surprisingly, we detected significant RT changes within 6 h of differentiation (Figures 2C and 2D). Thus, while RT is an extremely stable cell type-specific epigenetic property that is resistant to the depletion of many chromatin regulators and architectural proteins within a given cell type (Dileep et al., 2015b), RT can nonetheless be regulated very rapidly after stimulation of differentiation.
Average Changes in Transcription Are Coordinated with RT Changes but Can Be Anticorrelated for Individual Genes
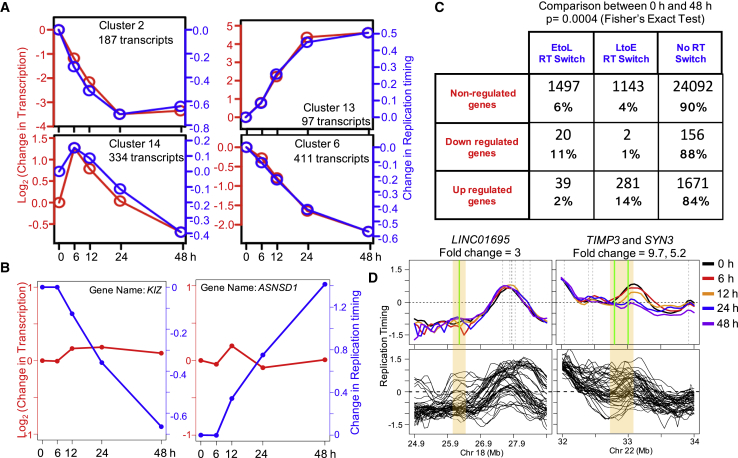
We have previously shown that many RT and transcription changes are uncoupled during differentiation to various lineages over the course of several days (Hiratani et al., 2010, Rivera-Mulia et al., 2015). To investigate how RT and transcription are coordinated during the first two cell cycles of DE lineage differentiation, we performed genome-wide RT profiling and poly(A)+ RNA-seq with the same populations of differentiating cells. First, we identified genes that were differentially regulated (FDR adjusted p < 0.05) and/or changed RT (dRT > 0.5, RT measured at transcription start site of the genes) during the course of differentiation. Next, we performed K-means clustering analysis using the expression profile of these genes at 0, 6, 12, 24, and 48 h. Then, for each K-means cluster we compared the average RT with the average transcription across all the five time points (Figures 3A, S3A, and S3B). Consistent with our previous studies performed over longer time periods (Hiratani et al., 2010, Rivera-Mulia et al., 2015), mean RT changes were remarkably coincident with mean transcriptional changes (Figures 3A, S3A, and S3B) while transcription of many individual genes did not follow RT kinetics. Figure 3B shows two individual genes wherein RT is regulated in the absence of transcriptional regulation. We also tested if the fold change and/or starting level of gene expression predicts the correlation between RT and transcriptional changes. Changes in transcription with larger magnitude tend to result in a stronger correlation between RT and transcription changes (Figure S3C). Also, upregulated genes with lower starting gene expression levels tend to result in a stronger correlation between RT and transcription changes (Figure S3D), although the inverse trend was not reproducibly observed for downregulated genes (Figure S3E). However, there were also individual genes with high fold change or low starting expression in which transcription and RT kinetics were anticorrelated (Figures S3C and S3D).
Figure 3.
Average Changes in Transcription Are Coincident with RT Changes
(A) Line graphs of average transcriptional change (red line, log2 scale) compared with average RT change at the transcription start site (TSS) (blue line) of all genes within exemplary K-means clusters. K-means clusters were defined with transcripts that have either an expression change (FDR p < 0.05 using two independent differentiations) and/or an RT change (dRT > 0.5). The replicate with matched RT data is plotted. The changes were calculated with respect to the ESC 0 h time point.
(B) Two exemplary genes illustrating that changes in RT do not cause a corresponding change in transcription. The expression level of the gene is indicated by the red line and the RT of the gene at its TSS is indicated by the blue line.
(C) Table quantifying the number of transcripts within categories defined by transcriptional regulation and RT regulation between 0 and 48 h time point. The threshold for transcriptional regulation and replication regulation were FDR p < 0.05 and 0.5, respectively. The values are also expressed as percentages within each row category.
(D) Top: exemplary regions (highlighted) harboring a gene that is upregulated during 48 h of DE differentiation (FDR p < 0.05, fold change > 2), but does not exhibit a corresponding change in RT. Green vertical lines mark upregulated genes and gray lines indicate nonregulated genes. Bottom: RT in 48 different cell lines/cell types/differentiation states demonstrate that the RT at this location can be regulated.
To quantify this variability, we grouped genes as up, downregulated (FDR p < 0.05) or nonregulated based on the expression at 48 and 0 h. Then we quantified how many of them exhibited an RT change in the matched RT dataset (Figure 3C). Consistent with Figure 3A, when there is a change in RT and transcription, they were generally correlated, but individual gene expression patterns did not always track with RT changes.
Further stratification of RT changes into early (E), middle early (ME), middle late (ML), and late (L) showed that a large number of RT transitions were confined to E, ME, and ML with very few genes showing a drastic EtoL or LtoE (Figures S3F and S3G). This is consistent with the general observation that there are fewer genes in the very late replicating portion of the genome (Hiratani et al., 2010, Rivera-Mulia et al., 2015). In addition, we found domains that harbor genes induced more than 2-fold (FDR p < 0.05), reside in domains that have the potential to switch RT (i.e., replicate early in other cell types), yet fail to undergo an LtoE change in RT in response to transcriptional induction (Figure 3D). These results demonstrate that, even within the time resolution of the first two cell cycles of a cell fate transition, RT changes are insufficient to alter transcription and transcription changes are insufficient to alter RT.
Changes in RT and Subnuclear A/B Compartment Are Uncoupled Early during Differentiation
Since we have previously shown that RT and the 3D compartmentalization of chromatin are highly correlated and are established within the same short interval of early G1 phase at the TDP (Dileep et al., 2015a), we expected RT changes to be closely coordinated with changes in A/B compartment, reflecting changes in subnuclear position. Our results described above demonstrate that RT changes can occur rapidly after induction of differentiation and RT is progressively remodeled with continued differentiation. To determine whether chromatin compartments change coordinately with RT, we performed Hi-C in cells differentiated for 24 h, representing slightly more than one cell cycle, and 48 h representing more than two cell cycles (Figure S4; Table S1).
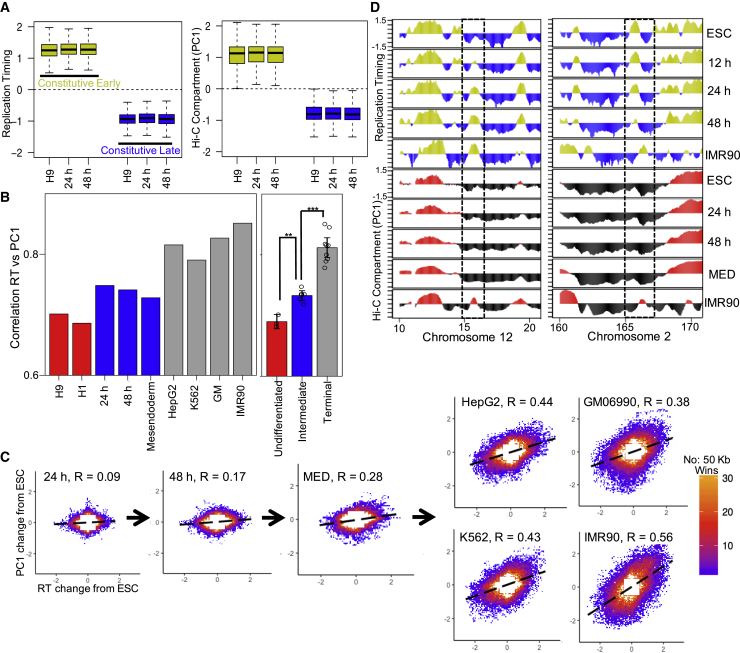
The first principal component (PC1) derived from Hi-C data reflects the active and repressed A/B compartments of the nucleus (Lieberman-Aiden et al., 2009), which are highly correlated with RT (Ryba et al., 2010), a correlation that is particularly strong for the segments of the genome that do not change RT across different cell types (constitutively early and late regions) (Dileep et al., 2015a). Consistently, the constitutively early and late replicating regions of the genome in hESCs and their differentiating derivatives showed a very distinct separation in their nuclear compartments (Figure 4A) However, we found a weaker global correlation of RT to PC1/eigenvector-defined A/B compartments than previously reported using established cell lines (Ryba et al., 2010). This raised the possibility that RT and A/B compartments become more aligned in later stages of differentiation. To address this hypothesis, we compared the Hi-C PC1 with RT datasets of our time course differentiation to several different previously published Hi-C datasets from cell lines and a previously published Hi-C dataset of hESC line H1 before and after differentiation to mesendoderm (Table S1). Figure 4B shows the Pearson correlation between RT and PC1 for each cell line. We found significantly higher correlation in early differentiation intermediats vs. stem cells and again in cell lines versus early differentiation intermediates (Figures 4B and S5A–S5C).
Figure 4.
Increased Correlation between Chromatin Compartments and RT in Terminally Differentiated Cells Compared to Early Stages of Differentiation
(A) Left: boxplot of RT for 50-kb windows that are constitutively early replicating (yellow) or late replicating (blue) in all cell types (Experimental Procedures). Right: boxplot of compartment (PC1) values for the 50-kb bins in top panel.
(B) Correlation between RT and compartments (PC1) in undifferentiated ESCs (red), early differentiation intermediates (blue), and terminally differentiated cells (gray). Left panel shows data from individual differentiation states or cell lines (average of replicates). Right panel shows all replicates grouped into undifferentiated, intermediate, or terminal differentiation. Differences were significant with ∗∗p = 0.004, ∗∗∗p = 2.595 × 10−6 (t test). Error bars indicate SEM.
(C) Scatterplot showing correlation between changes in RT and changes in PC1 for each differentiation stage (compared with undifferentiated state). R is Pearson’s correlation. MED is mesendoderm.
(D) Exemplary plots show discordance between RT and compartments in ESC and early differentiation stages, but becomes concordant in IMR90 (terminally differentiated cell line). For all panels except the individual tracks in (D), plots of samples 24 and 48 h, IMR90, K562, H1ESC, and mesendoderm are average of two independent differentiations or cell collections, IMR90 is average of four independent cell collections.
Next, we analyzed the coordination between changes in RT and changes in chromatin compartments. There was a weak correlation between the changes in A/B compartments and changes in RT at 24 h and this correlation increased at 48 h while terminally differentiated cells showed a strikingly higher correlation between their differences with hESCs in A/B compartment versus RT (Figure 4C). Figure 4D shows exemplary regions where there is a discordance between RT and PC1, which is strongly correlated in a terminally differentiated cell line (IMR90). To determine whether there is an increase in the correlation between RT and A/B compartment when cells are differentiated beyond the DE stage, we performed a more protracted endoderm differentiation, collecting downstream intermediates in the pancreatic lineage, including DE, pancreatic progenitor, poly-hormonal cells and finally pancreatic beta-like cells (Figure S5D). We measured RT and Hi-C using a 4-bp cutter restriction enzyme to obtain improved A/B compartment resolution (Experimental Procedures). Consistent with the results shown in Figures 4B and S5A–S5C, we detected an increase in the global correlation between RT and Hi-C when hESCs commit to DE (Figure S5C). This global correlation did not further increase as differentiation progressed past DE but, terminally differentiated cell lines still showed a higher correlation than either stem cells or any of the differentiation intermediates (Figure S5C). However, when the coordination between significant changes in RT versus A/B compartment was compared, there was no difference between terminally differentiated cell lines and all differentiation intermediates beyond DE (Figure S5E). Taken together, we find evidence for discordance between changes in RT and chromatin compartments during early stages of differentiation that becomes resolved by the DE stage. At the same time, there is a global increase in correlation between RT and chromatin compartments 24 hours after stem cells are stimulated to differentiate that does not increase at least through to the insulin expressing pancreatic beta like cell stage, likely owing to small increases in alignment occuring globally across the genome. This correlation increases further in cell lines, which could either be related to terminal differentiation or to changes occuring during establishment of cell lines.
Changes in Chromatin Marks and RT Correlate More Strongly when Stem Cells Are Compared with More Differentiated Cell Types
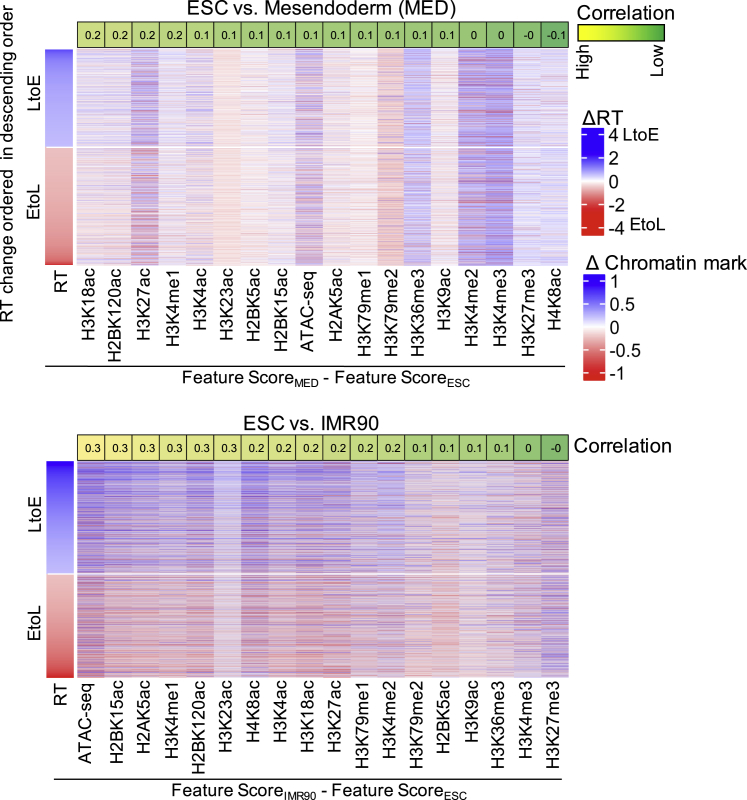
RT has been correlated with myriad epigenetic marks in many systems and cell types (Eaton et al., 2011, Lande-Diner et al., 2009, Schwaiger et al., 2009, Suzuki et al., 2011, Zhang et al., 2002). We previously correlated differences in RT to differences in many histone marks analyzed by the ENCODE consortium (Yue et al., 2014) and identified strong correlations between early replication and increased density of enhancer marks such as H3K4me1 and H3K27ac, but no correlation with changes in repressive marks such as H3K9me3 and H3K27me3. To determine which of these changes occurs first during differentiation, we analyzed public chromatin immunoprecipitation (ChIP) data for H1 and H9 cells differentiated to mesendoderm (Roadmap Epigenetics) and compared the coordination between differences in histone modifications and RT changes during early development (ESC versus mesendoderm) and terminal differentiation (ESC versus IMR90). Similar to the relationship between RT and A/B compartment, there was lower correlation between histone modification changes and RT changes during mesendoderm differentiation, whereas the previously reported correlations were easily detectable in IMR90 (Figure 5). These results demonstrate that the density of chromatin marks is less coordinated with RT changes during the early stages of differentiation and suggest that they may also become resolved at later stages.
Figure 5.
Changes in Chromatin Marks and RT Correlate More Strongly when Stem Cells Are Compared with More Differentiated Cell Types
Heatmap of changes in chromatin modification and chromatin accessibility (from roadmap epigenomics, independent replicates) at regions that change RT (absolute change in RT > 0.5) for ESC versus mesendoderm and ESC versus IMR90. The rows are ordered according to descending order of change in RT. The values on the top of the columns indicate Pearson correlation (rounded) between changes in chromatin feature versus changes in RT. Feature score is the RPKM values binned to 1 kb, normalized with input and binned into 50 kb bins to match the resolution of the replication timing data (see Experimental Procedures).
Discussion
Here we find that transcriptional programs become irreversibly altered within the first hours after cells are stimulated to differentiate. RT changes generally correlate with transcriptional changes but are clearly uncoupled at many loci. Further, we find a discordance between RT, chromatin structure and chromatin compartments during the earliest stages of differentiation. These results demonstrate that changes in these properties are not sufficient to dictate changes in the alternate properties, but rather that they can be independently regulated and become aligned as differentiation proceeds.
RT and Transcription Changes Are Globally Correlated but Can Be Locally Independent
Previous attempts to elucidate the order of RT and transcription changes during differentiation found that approximately one-third of genes change both RT and transcription during differentiation of mouse or human ESCs (Hiratani et al., 2010, Rivera-Mulia et al., 2015). Two-thirds of those genes were expressed and late replicating in at least one intermediate stage clearly demonstrating that transcription is not sufficient for early replication. But, for many genes, the protracted time course of differentiation precluded the ability to determine which changed first (Rivera-Mulia et al., 2015). Here, we used a differentiation system that is sufficiently rapid as to permit us to track these events within a single cell cycle. Our results demonstrate that RT and transcription changes can occur simultaneously within the first cell cycle following stimulation. The short time periods of differentiation used here permitted us to conclude that RT and transcription can change in either order or can change simultaneously. We also found genes that undergo significant changes in transcription without a corresponding change in RT and vice versa. No consistent pattern emerged, except that, when the RT and transcription for many genes together were averaged, a very strong coordination of the two emerged. Collectively, these data show that while RT is highly correlated with transcription, transcription itself is neither necessary nor sufficient to drive changes in RT.
RT and Chromatin Compartments Are Regulated Independently
RT is most strongly correlated with Hi-C A/B chromatin interaction compartments that consolidate coordinately over the course of protracted (several days) differentiation schemes (Dixon et al., 2012, Dixon et al., 2015, Rivera-Mulia et al., 2018, Ryba et al., 2010, Takebayashi et al., 2012). Moreover, RT is regulated in chromosome units that align with TADs, and both the acquisition of TAD boundaries and the appearance of A/B compartments occur simultaneous with the establishment of an RT program early during G1 phase (Dileep et al., 2015a, Pope et al., 2014). Altogether, these observations give rise to the expectation that a change in one of these properties would be a proxy for a change in the other. On the other hand, exceptional domains that replicate at a time not consistent with their compartment do exist and we have also shown that, during G2 phase, properties of chromatin that dictate the RT program are lost even though TADs and A/B compartments can still be detected (Dileep et al., 2015a), demonstrating that the two can be uncoupled in certain conditions. Here we show that, during the first cell cycles of differentiation, changes in RT and A/B compartments occur independently in different domains, providing a clear example of the uncoupling of regulatory mechanisms controlling these properties. However, these discordances become increasingly aligned as lineage commitment proceeds. Consistently, a recent report found that compartments become weaker during reprogramming of terminally differentiated cells back to induced pluripotent cells (Stadhouders et al., 2018). It is possible that this discordance is representative of the uncommitted mesendoderm state seen in the mammalian primitive streak (Wang and Chen, 2016). It will be interesting to perform similar experiments to those described here with other differentiation systems.
What Is Epigenetic Commitment and Cell Fate Determination?
Previous reports have established that G1 represents a critical window to respond to differentiation signals (Pauklin and Vallier, 2013, Singh et al., 2013, Wilson et al., 2016), and in endoderm differentiation seems to be linked to cyclin D accumulation during G1, which facilitates recruitment of transcriptional regulators to developmental genes (Pauklin et al., 2016). Experiments reported here show a rapid and stable transcriptional reprogramming as early as 6 h of differentiation. These cells continued to progress toward the endoderm lineage even after returning to stem cell medium for up to 24 h suggests that these cells have become committed. Our experimental conditions cannot rule out the possibility that some cells in the population secrete factors that drive differentiation forward even after return to stem cell medium. However, this seems unlikely, because stem cell medium contains high insulin growth factor and Heregulin beta-1, both of which activate phosphatidylinositol 3-kinase signaling, which should inhibit endoderm differentiation even in the presence of endodermal growth factors (McLean et al., 2007). It will be interesting to pursue these experiments with different plating densities, longer reversal times, and single cell measurements.
Nonetheless, before the experiments reported here, there was no expectation that early stem cell lineage specification would be so rapid and irreversible. For example, irreversibility of X inactivation occurs over the course of many days (Hansen et al., 1996, Hiratani and Gilbert, 2010). Moreover, studies of cell specification versus determination in mouse embryos have revealed a great deal of plasticity (Bedzhov et al., 2014). An important motivation for these studies is that we have shown that cells that fail to reprogram back to the ESC state (partially iPSCs) share a unique partially iPSC-specific RT profile, suggesting that some RT changes may represent an epigenetic barrier to reprogramming (Hiratani et al., 2010, Hiratani and Gilbert, 2010). However, in the study reported here, genes that are included in these “difficult to reprogram” domains such as DPPA2, DPPA4, and REX1 do not switch from EtoL replication until 24–48 h after stimulation to differentiate, suggesting that the barrier to reprogramming detected by iPSCs is downstream of whatever is driving transcriptional reprogramming detected in Figure 1. The fact that irreversible transcriptional reprograming occurred so rapidly in our hands, coincident with discordant changes in RT, Hi-C A/B compartments and chromatin modifications, leaves us uncertain as to whether alterations in these properties are linked to epigenetic commitment or whether the changes in transcriptional regulatory networks themselves are sufficient for cell fate determination.
Experimental Procedures
Experimental procedures for RNA-seq, Expression microarray and ChIP sequencing (ChiP-seq) datasets are explained in the Supplemental Experimental Procedures.
Cell Culture
H9 hESCs were cultured under feeder-free conditions on Geltrex (Thermo Fisher Scientific, A14133)-coated dishes, and maintained in StemPro (Thermo Fisher Scientific, A100701) culture medium per manufacture's specifications. Cell passaging was achieved by brief treatment (∼6–8 min) with Accutase (Thermo Fisher Scientific, A1110501). After detachment, cells were gently collected, centrifuged for 5 min at 200 × g, and replated on freshly coated Geltrex dishes.
DE Differentiation
Differentiation conditions for the reversal experiments described in Figure 1 were as described in (Schulz et al., 2012). The differentiation to DE for RT and RNA-seq were carried out after adapting this protocol to adherent culture. In brief, cultures at ∼70% confluency were washed twice with cold PBS and stimulated to differentiate by the addition of RPMI (Invitrogen), 50 ng/mL Wnt (CellGS, GFM77), 100 ng/mL Activin A (CellGS, GFH6). Twenty-four hours after stimulation, medium was replaced with RPMI containing 0.2% fetal bovine serum, and 100 ng/mL Activin A.
Differentiation toward Pancreatic Beta Cells and Intermediates
hESCs were differentiated toward DE, pancreatic progenitor, poly-hormonal cells and pancreatic beta-like cells as previously described with modifications (Pagliuca et al., 2014). Details are in the Supplemental Experimental Procedures.
Genome-wide RT Analysis
Genome-wide population RT was measured using Repli-seq protocol as described previously (Marchal et al., 2018). Details are in the Supplemental Experimental Procedures.
Hi-C
Hi-C was performed, using HindIII or DpnII restriction enzyme, as described previously (Dixon et al., 2015, Rao et al., 2014). PC1 values were calculated using the software package Homer with 50-kb bins (Homer parameters resolution = 50 kb and super-resolution = 500 kb). Sex chromosomes were removed from the analysis to enable comparisons between male and female cell lines. Also, Chr 4 and 19 were removed because the PC1 values of these chromosomes did not reflect the nuclear compartmentalization in certain datasets.
Author Contributions
K.A.W., V.D., and D.M.G. designed the study. K.W. performed most of the experiments. V.D. performed most of the computational analyses. C.M. and V.D. performed the Hi-C analysis. X.L. and V.G.C. performed the Hi-C experiments. P.A.Z. performed the pancreatic progenitor Repli-seq. B.L. and Z.Q. helped with RNA-seq analysis. A.P. and D.A.B. organized the ChIP-seq datasets. A.J.R. and T.C.S. performed the differentiation for the reversal experiment. M.J.K. and S.D. performed the pancreatic progenitor differentiation. J.C.R.-M. helped with microarray experiments and analysis. J.D. and R.P.M. generated HepG2 Hi-C data. V.D., K.A.W., and D.M.G. wrote the manuscript with input from all authors.
Acknowledgments
We thank R. Didier for assistance with flow cytometry. This work was funded by NIH grants GM085354 and GM083337 to D.M.G. and HG003143 to J.D. A.J.R. and T.C.S. are employees of Viacyte receiving salary and stock options.
Published: June 20, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2019.05.021.
Accession Numbers
All datasets generated in this study are deposited in the NCBI GEO under accession number GEO: GSE130547.
Supplemental Information
References
- Barr M.L., Bertram E.G. A morphological distinction between neurones of the male and female, and the behaviour of the nucleolar satellite during accelerated nucleoprotein synthesis. Nature. 1949;163:676. doi: 10.1038/163676a0. [DOI] [PubMed] [Google Scholar]
- Bedzhov I., Graham S.J., Leung C.Y., Zernicka-Goetz M. Developmental plasticity, cell fate specification and morphogenesis in the early mouse embryo. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014;369 doi: 10.1098/rstb.2013.0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenfeld B., Ben-Zimra M., Simon I. Perturbations in the replication program contribute to genomic instability in cancer. Int. J. Mol. Sci. 2017;18 doi: 10.3390/ijms18061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport C., Diekmann U., Naujok O. A quick and efficient method for the purification of endoderm cells generated from human embryonic stem cells. J. Vis. Exp. 2016 doi: 10.3791/53655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dileep V., Ay F., Sima J., Vera D.L., Noble W.S., Gilbert D.M. Topologically-associating domains and their long-range contacts are established during early G1 coincident with the establishment of the replication timing program. Genome Res. 2015;25:1104–1113. doi: 10.1101/gr.183699.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dileep V., Rivera-Mulia J.C., Sima J., Gilbert D.M. Large-scale chromatin structure-function relationships during the cell cycle and development: insights from replication timing. Cold Spring Harb. Symp. Quant. Biol. 2015;80:53–63. doi: 10.1101/sqb.2015.80.027284. [DOI] [PubMed] [Google Scholar]
- Dimitrova D.S., Gilbert D.M. The spatial position and replication timing of chromosomal domains are both established in early G1-phase. Mol. Cell. 1999;4:983–993. doi: 10.1016/s1097-2765(00)80227-0. [DOI] [PubMed] [Google Scholar]
- Dixon J.R., Jung I., Selvaraj S., Shen Y., Antosiewicz-Bourget J.E., Lee A.Y., Ye Z., Kim A., Rajagopal N., Xie W. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518:331–336. doi: 10.1038/nature14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donley N., Thayer M.J. DNA replication timing, genome stability and cancer: late and/or delayed DNA replication timing is associated with increased genomic instability. Semin. Cancer Biol. 2013;23:80–89. doi: 10.1016/j.semcancer.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton M.L., Prinz J.A., MacAlpine H.K., Tretyakov G., Kharchenko P.V., MacAlpine D.M. Chromatin signatures of the Drosophila replication program. Genome Res. 2011;21:164–174. doi: 10.1101/gr.116038.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz A., Sinha S., Marella N., Berezney R. Alterations in replication timing of cancer-related genes in malignant human breast cancer cells. J. Cell. Biochem. 2013;114:1074–1083. doi: 10.1002/jcb.24447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C.W., Muldal S., Lajtha L.G., Rowley J. Time-sequence of human chromosome duplication. Nature. 1962;195:869–873. doi: 10.1038/195869a0. [DOI] [PubMed] [Google Scholar]
- Goldman M.A., Holmquist G.P., Gray M.C., Caston L.A., Nag A. Replication timing of genes and middle repetitive sequences. Science. 1984;224:686–692. doi: 10.1126/science.6719109. [DOI] [PubMed] [Google Scholar]
- Hansen R.S., Canfield T.K., Fjeld A.D., Gartler S.M. Role of late replication timing in the silencing of X-linked genes. Hum. Mol. Genet. 1996;5:1345–1353. doi: 10.1093/hmg/5.9.1345. [DOI] [PubMed] [Google Scholar]
- Hiratani I., Gilbert D.M. Autosomal lyonization of replication domains during early mammalian development. In: Meshorer E., Plath K., editors. The Cell Biology of Stem Cells. Landes and Springer; 2010. pp. 41–58. [DOI] [PubMed] [Google Scholar]
- Hiratani I., Leskovar A., Gilbert D.M. Differentiation-induced replication-timing changes are restricted to AT-rich/long interspersed nuclear element (LINE)-rich isochores. Proc. Natl. Acad. Sci. U S A. 2004;101:16861–16866. doi: 10.1073/pnas.0406687101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I., Ryba T., Itoh M., Rathjen J., Kulik M., Papp B., Fussner E., Bazett-Jones D.P., Plath K., Dalton S. Genome-wide dynamics of replication timing revealed by in vitro models of mouse embryogenesis. Genome Res. 2010;20:155–169. doi: 10.1101/gr.099796.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I., Ryba T., Itoh M., Yokochi T., Schwaiger M., Chang C.W., Lyou Y., Townes T.M., Schubeler D., Gilbert D.M. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol. 2008;6:e245. doi: 10.1371/journal.pbio.0060245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaaij L.J.T., van der Weide R.H., Ketting R.F., de Wit E. Systemic loss and gain of chromatin architecture throughout zebrafish development. Cell Rep. 2018;24:1–10.e4. doi: 10.1016/j.celrep.2018.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande-Diner L., Zhang J., Cedar H. Shifts in replication timing actively affect histone acetylation during nucleosome reassembly. Mol. Cell. 2009;34:767–774. doi: 10.1016/j.molcel.2009.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Chen J., Izumi M., Butler M.C., Keezer S.M., Gilbert D.M. The replication timing program of the Chinese hamster beta-globin locus is established coincident with its repositioning near peripheral heterochromatin in early G1 phase. J. Cell Biol. 2001;154:283–292. doi: 10.1083/jcb.200104043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E., van Berkum N.L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B.R., Sabo P.J., Dorschner M.O. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Li F., Murphy C.S., Davidson M.W., Gilbert D.M. G2 phase chromatin lacks determinants of replication timing. J. Cell Biol. 2010;189:967–980. doi: 10.1083/jcb.201002002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon M.F. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- MacAlpine D.M., Rodriguez H.K., Bell S.P. Coordination of replication and transcription along a Drosophila chromosome. Genes Dev. 2004;18:3094–3105. doi: 10.1101/gad.1246404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchal C., Sasaki T., Vera D., Wilson K., Sima J., Rivera-Mulia J.C., Trevilla-Garcia C., Nogues C., Nafie E., Gilbert D.M. Genome-wide analysis of replication timing by next-generation sequencing with E/L Repli-seq. Nat. Protoc. 2018;13:819–839. doi: 10.1038/nprot.2017.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean A.B., D'Amour K.A., Jones K.L., Krishnamoorthy M., Kulik M.J., Reynolds D.M., Sheppard A.M., Liu H., Xu Y., Baetge E.E. Activin A efficiently specifies definitive endoderm from human embryonic stem cells only when phosphatidylinositol 3-kinase signaling is suppressed. Stem Cells. 2007;25:29–38. doi: 10.1634/stemcells.2006-0219. [DOI] [PubMed] [Google Scholar]
- Morishima A., Grumbach M.M., Taylor J.H. Asynchronous duplication of human chromosomes and the origin of sex chromatin. Proc. Natl. Acad. Sci. U S A. 1962;48:756–763. doi: 10.1073/pnas.48.5.756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller C.A., Nieduszynski C.A. DNA replication timing influences gene expression level. J. Cell Biol. 2017;216:1907–1914. doi: 10.1083/jcb.201701061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayasu H., Berezney R. Mapping replicational sites in the eucaryotic cell nucleus. J. Cell Biol. 1989;108:1–11. doi: 10.1083/jcb.108.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Keefe R.T., Henderson S.C., Spector D.L. Dynamic organization of DNA replication in mammalian cell nuclei - spatially and temporally defined replication of chromosome-specific alpha-satellite DNA sequences. J. Cell Biol. 1992;116:1095–1110. doi: 10.1083/jcb.116.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrow A.Z., Kalhor R., Gan Y., Villwock S.K., Linke C., Barberis M., Chen L., Aparicio O.M. Conserved forkhead dimerization motif controls DNA replication timing and spatial organization of chromosomes in S. cerevisiae. Proc. Natl. Acad. Sci. U S A. 2017;114:E2411–E2419. doi: 10.1073/pnas.1612422114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliuca F.W., Millman J.R., Gurtler M., Segel M., Van Dervort A., Ryu J.H., Peterson Q.P., Greiner D., Melton D.A. Generation of functional human pancreatic beta cells in vitro. Cell. 2014;159:428–439. doi: 10.1016/j.cell.2014.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauklin S., Madrigal P., Bertero A., Vallier L. Initiation of stem cell differentiation involves cell cycle-dependent regulation of developmental genes by Cyclin D. Genes Dev. 2016;30:421–433. doi: 10.1101/gad.271452.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauklin S., Vallier L. The cell-cycle state of stem cells determines cell fate propensity. Cell. 2013;155:135–147. doi: 10.1016/j.cell.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry P., Sauer S., Billon N., Richardson W.D., Spivakov M., Warnes G., Livesey F.J., Merkenschlager M., Fisher A.G., Azuara V. A dynamic switch in the replication timing of key regulator genes in embryonic stem cells upon neural induction. Cell Cycle. 2004;3:1645–1650. [PubMed] [Google Scholar]
- Pope B.D., Ryba T., Dileep V., Yue F., Wu W., Denas O., Vera D.L., Wang Y., Hansen R.S., Canfield T.K. Topologically associating domains are stable units of replication-timing regulation. Nature. 2014;515:402–405. doi: 10.1038/nature13986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourkarimi E., Bellush J.M., Whitehouse I. Spatiotemporal coupling and decoupling of gene transcription with DNA replication origins during embryogenesis in C. elegans. Elife. 2016;5 doi: 10.7554/eLife.21728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S.S., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy K.L., Zullo J.M., Bertolino E., Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–247. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- Rivera-Mulia J.C., Buckley Q., Sasaki T., Zimmerman J., Didier R.A., Nazor K., Loring J.F., Lian Z., Weissman S.M., Robins A.J. Dynamic changes in replication timing and gene expression during lineage specification of human pluripotent stem cells. Genome Res. 2015;25:1091–1103. doi: 10.1101/gr.187989.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Mulia J.C., Desprat R., Trevilla-Garcia C., Cornacchia D., Schwerer H., Sasaki T., Sima J., Fells T., Studer L., Lemaitre J.M. DNA replication timing alterations identify common markers between distinct progeroid diseases. Proc. Natl. Acad. Sci. U S A. 2017;114:E10972–E10980. doi: 10.1073/pnas.1711613114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Mulia J.C., Dimond A., Vera D., Trevilla-Garcia C., Sasaki T., Zimmerman J., Dupont C., Gribnau J., Fraser P., Gilbert D.M. Allele-specific control of replication timing and genome organization during development. Genome Res. 2018;28:800–811. doi: 10.1101/gr.232561.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Mulia J.C., Gilbert D.M. Replicating large genomes: divide and conquer. Mol. Cell. 2016;62:756–765. doi: 10.1016/j.molcel.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Mulia J.C., Gilbert D.M. Replication timing and transcriptional control: beyond cause and effect - part III. Curr. Opin. Cell Biol. 2016;40:168–178. doi: 10.1016/j.ceb.2016.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba T., Hiratani I., Lu J., Itoh M., Kulik M., Zhang J., Schulz T.C., Robins A.J., Dalton S., Gilbert D.M. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res. 2010;20:761–770. doi: 10.1101/gr.099655.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba T., Hiratani I., Sasaki T., Battaglia D., Kulik M., Zhang J., Dalton S., Gilbert D.M. Replication timing: a fingerprint for cell identity and pluripotency. PLoS Comput. Biol. 2011;7:e1002225. doi: 10.1371/journal.pcbi.1002225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T., Rivera-Mulia J.C., Vera D., Zimmerman J., Das S., Padget M., Nakamichi N., Chang B.H., Tyner J., Druker B.J. Stability of patient-specific features of altered DNA replication timing in xenografts of primary human acute lymphoblastic leukemia. Exp. Hematol. 2017;51:71–82.e3. doi: 10.1016/j.exphem.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubeler D., Scalzo D., Kooperberg C., Van Steensel B., Delrow J., Groudine M. Genome-wide DNA replication profile for Drosophila melanogaster: a link between transcription and replication timing. Nat. Genet. 2002;32:438–442. doi: 10.1038/ng1005. [DOI] [PubMed] [Google Scholar]
- Schulz T.C., Young H.Y., Agulnick A.D., Babin M.J., Baetge E.E., Bang A.G., Bhoumik A., Cepa I., Cesario R.M., Haakmeester C. A scalable system for production of functional pancreatic progenitors from human embryonic stem cells. PLoS One. 2012;7:e37004. doi: 10.1371/journal.pone.0037004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaiger M., Stadler M.B., Bell O., Kohler H., Oakeley E.J., Schubeler D. Chromatin state marks cell-type- and gender-specific replication of the Drosophila genome. Genes Dev. 2009;23:589–601. doi: 10.1101/gad.511809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela Y., Molotski N., Golan S., Itskovitz-Eldor J., Soen Y. Human embryonic stem cells exhibit increased propensity to differentiate during the G1 phase prior to phosphorylation of retinoblastoma protein. Stem Cells. 2012;30:1097–1108. doi: 10.1002/stem.1078. [DOI] [PubMed] [Google Scholar]
- Seller C.A., O'Farrell P.H. Rif1 prolongs the embryonic S phase at the Drosophila mid-blastula transition. PLoS Biol. 2018;16:e2005687. doi: 10.1371/journal.pbio.2005687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siefert J.C., Georgescu C., Wren J.D., Koren A., Sansam C.L. DNA replication timing during development anticipates transcriptional programs and parallels enhancer activation. Genome Res. 2017;27:1406–1416. doi: 10.1101/gr.218602.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A.M., Chappell J., Trost R., Lin L., Wang T., Tang J., Matlock B.K., Weller K.P., Wu H., Zhao S. Cell-cycle control of developmentally regulated transcription factors accounts for heterogeneity in human pluripotent cells. Stem Cell Reports. 2013;1:532–544. doi: 10.1016/j.stemcr.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovei I., Thanisch K., Feodorova Y. How to rule the nucleus: divide et impera. Curr. Opin. Cell Biol. 2016;40:47–59. doi: 10.1016/j.ceb.2016.02.014. [DOI] [PubMed] [Google Scholar]
- Stadhouders R., Vidal E., Serra F., Di Stefano B., Le Dily F., Quilez J., Gomez A., Collombet S., Berenguer C., Cuartero Y. Transcription factors orchestrate dynamic interplay between genome topology and gene regulation during cell reprogramming. Nat. Genet. 2018;50:238–249. doi: 10.1038/s41588-017-0030-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M., Oda M., Ramos M.P., Pascual M., Lau K., Stasiek E., Agyiri F., Thompson R.F., Glass J.L., Jing Q. Late-replicating heterochromatin is characterized by decreased cytosine methylation in the human genome. Genome Res. 2011;21:1833–1840. doi: 10.1101/gr.116509.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebayashi S., Dileep V., Ryba T., Dennis J.H., Gilbert D.M. Chromatin-interaction compartment switch at developmentally regulated chromosomal domains reveals an unusual principle of chromatin folding. Proc. Natl. Acad. Sci. U S A. 2012;109:12574–12579. doi: 10.1073/pnas.1207185109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therizols P., Illingworth R.S., Courilleau C., Boyle S., Wood A.J., Bickmore W.A. Chromatin decondensation is sufficient to alter nuclear organization in embryonic stem cells. Science. 2014;346:1238–1242. doi: 10.1126/science.1259587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi T., Soza-Ried J., Brown K., Piccolo F.M., Cantone I., Landeira D., Bagci H., Hochegger H., Merkenschlager M., Fisher A.G. DNA synthesis is required for reprogramming mediated by stem cell fusion. Cell. 2013;152:873–883. doi: 10.1016/j.cell.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Chen Y.G. Signaling control of differentiation of embryonic stem cells toward mesendoderm. J. Mol. Biol. 2016;428:1409–1422. doi: 10.1016/j.jmb.2015.06.013. [DOI] [PubMed] [Google Scholar]
- White E.J., Emanuelsson O., Scalzo D., Royce T., Kosak S., Oakeley E.J., Weissman S., Gerstein M., Groudine M., Snyder M. DNA replication-timing analysis of human chromosome 22 at high resolution and different developmental states. Proc. Natl. Acad. Sci. U S A. 2004;101:17771–17776. doi: 10.1073/pnas.0408170101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R.R., Azuara V., Perry P., Sauer S., Dvorkina M., Jorgensen H., Roix J., McQueen P., Misteli T., Merkenschlager M. Neural induction promotes large-scale chromatin reorganisation of the Mash1 locus. J. Cell Sci. 2006;119:132–140. doi: 10.1242/jcs.02727. [DOI] [PubMed] [Google Scholar]
- Wilson K.A., Elefanty A.G., Stanley E.G., Gilbert D.M. Spatio-temporal re-organization of replication foci accompanies replication domain consolidation during human pluripotent stem cell lineage specification. Cell Cycle. 2016;15:2464–2475. doi: 10.1080/15384101.2016.1203492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodfine K., Beare D.M., Ichimura K., Debernardi S., Mungall A.J., Fiegler H., Collins V.P., Carter N.P. Replication timing of human chromosome 6. Cell Cycle. 2005;4:172–176. doi: 10.4161/cc.4.1.1350. [DOI] [PubMed] [Google Scholar]
- Woodfine K., Fiegler H., Beare D.M., Collins J.E., McCann O.T., Young B.D., Debernardi S., Mott R., Dunham I., Carter N.P. Replication timing of the human genome. Hum. Mol. Genet. 2004;13:191–202. doi: 10.1093/hmg/ddh016. [DOI] [PubMed] [Google Scholar]
- Wu R., Singh P.B., Gilbert D.M. Uncoupling global and fine-tuning replication timing determinants for mouse pericentric heterochromatin. J. Cell Biol. 2006;174:185–194. doi: 10.1083/jcb.200601113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang W., Roberti M.J., Heriche J.K., Huet S., Alexander S., Ellenberg J. Correlative live and super-resolution imaging reveals the dynamic structure of replication domains. J. Cell Biol. 2018;217:1973–1984. doi: 10.1083/jcb.201709074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe E., Farkash-Amar S., Polten A., Yakhini Z., Tanay A., Simon I. Comparative analysis of DNA replication timing reveals conserved large-scale chromosomal architecture. PLoS Genet. 2010;6:e1001011. doi: 10.1371/journal.pgen.1001011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue F., Cheng Y., Breschi A., Vierstra J., Wu W., Ryba T., Sandstrom R., Ma Z., Davis C., Pope B.D. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515:355–364. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Xu F., Hashimshony T., Keshet I., Cedar H. Establishment of transcriptional competence in early and late S phase. Nature. 2002;420:198–202. doi: 10.1038/nature01150. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.