Abstract
Mutations in the GJB2 gene are the main cause for nonsyndromic autosomal recessive deafness 1A (DFNB1A) in many populations. GJB2 mutational spectrum and pathogenic contribution are widely varying in different populations. Significant efforts have been made worldwide to define DFNB1A molecular epidemiology, but this issue still remains open for some populations. The main aim of study is to estimate the DFNB1A prevalence and GJB2 mutational spectrum in Tuvinians—an indigenous population of the Tyva Republic (Southern Siberia, Russia). Sanger sequencing was applied to analysis of coding (exon 2) and non-coding regions of GJB2 in a cohort of Tuvinian patients with hearing impairments (n = 220) and ethnically matched controls (n = 157). Diagnosis of DFNB1A was established for 22.3% patients (28.8% of familial vs 18.6% of sporadic cases). Our results support that patients with monoallelic GJB2 mutations (8.2%) are coincidental carriers. Recessive mutations p.Trp172Cys, c.-23+1G>A, c.235delC, c.299_300delAT, p.Val37Ile and several benign variants were found in examined patients. A striking finding was a high prevalence of rare variant p.Trp172Cys (c.516G>C) in Tuvinians accounting for 62.9% of all mutant GJB2 alleles and a carrier frequency of 3.8% in controls. All obtained data provide important targeted information for genetic counseling of affected Tuvinian families and enrich current information on variability of GJB2 worldwide.
Keywords: hearing loss, nonsyndromic autosomal recessive deafness 1A (DFNB1A), GJB2, Tuvinians, Southern Siberia, Russia
1. Introduction
Hearing loss (HL) is one of the most common sensory disorders, affecting one in 500–1000 newborns, and approximately half of congenital HL cases have a genetic etiology [1]. The genetic causes for HL are extremely heterogeneous: over 400 syndromes are associated with HL [2] and over 100 different genes are causally implicated in nonsyndromic HL [3]. Despite the extraordinary genetic heterogeneity of nonsyndromic HL, homozygous or compound heterozygous mutations in the GJB2 gene (gap junction protein, β-2, OMIM 121011, 13q12.11) encoding transmembrane protein connexin 26 (Cx26) are the most common cause of nonsyndromic autosomal recessive deafness 1A (DFNB1A, OMIM 220290) in many populations [4]. Six connexin 26 molecules associate to form transmembrane hexameric hemichannels (connexons) which dock with hemichannels of adjacent cells forming gap junctions that are essential for the transport of ions and other low-molecular-weight components between cells. Cx26 plays a crucial role in potassium homeostasis in the inner ear [5]. High prevalence of the GJB2-associated HL was demonstrated in most populations [1,6,7] making GJB2 gene testing essential for the establishment of genetic diagnosis of HL.
Over 300 deafness-associated variations in GJB2 have been reported in the Human Gene Mutation Database (http://www.hgmd.cf.ac.uk) [8]. Many of them have very specific ethno-geographic prevalence patterns [7,9] being attributed to a founder effect for certain ethnic groups. The variant c.35delG (p.Gly12Valfs*2) is prevalent in deaf patients of Caucasian origin [10,11]; c.235delC (p.Leu79Cysfs*3) is common in East and Southeast Asians [12,13,14]; c.167delT (p.Leu56Argfs*26) is more specific for Ashkenazi Jews [15]; c.427C>T (p.Arg143Trp) was found with high frequency in population of Ghana (West Africa) [16]; c.71G>A (p.Trp24*) is widely distributed in Indians and European Gypsies [17,18,19]; c.109G>A (p.Val37Ile) prevails in populations of Southeast Asia [20]; splice donor variant c.-23+1G>A (originally named IVS1+1G>A) is widespread among Yakuts (Eastern Siberia, Russia) [21]; c.131G>A (p.Trp44*) was found with high frequency among descendants of the ancestral Mayan population in Guatemala [22].
Despite significant efforts made worldwide to define the molecular epidemiology of GJB2 pathogenic variants, this issue still remains open for some populations. Establishing a genetic diagnosis of HL is of great importance for clinical evaluation of deaf patients and for estimating recurrence risks for their families.
The aim of this work is to evaluate previously unknown contribution of the GJB2 mutations to HL in Tuvinians, which is the indigenous population of the Tyva Republic. The Tyva Republic (Tuva), the federal subject of the Russian Federation, is located in Southern Siberia, in the central part of the Asian continent, bordered by Mongolia in the south and the east. The Tyva Republic is divided into 17 administrative districts and includes five small towns, with the capital in the town of Kyzyl. The total population of Tuva (307,930 according to 2010 Russian Federation Census) is mainly (82%) composed of the Tuvinians (Tuvans) who descended from ancient Turkic-speaking Central Asian tribes and Mongolian-speaking groups assimilated by them [23]. Besides the Tyva Republic, relatively small groups of Tuvinians live in Mongolia and in Northwest China [24].
In our previous epidemiological study, we created a database (including information about sex, age, ethnicity, place of residence, audiological data, and age of HL onset) for approximately 1400 individuals with various hearing impairments living in the Tyva Republic [25]. The average rate of different HL cases in the Tyva Republic was estimated as 45.5 ± 1.21 per 10,000 residents or 1:220, ranging from 25.5 ± 4.44 (1:392) to 117.9 ± 12.09 (1:85) in different regions of the Tyva Republic [25]. From the total group of registered patients, based on medical history, pedigree data and after exclusion of HL cases probably due to environmental factors, we have identified a group of patients with congenital or early onset sensorineural severe-profound HL (approximately 540 individuals) likely to have genetic HL. In this study, the evaluation of the spectrum and frequency of the GJB2 gene variants was performed in a cohort of Tuvinians mainly recruited from this group of patients.
2. Materials and Methods
2.1. Patients
The cohort of patients with hearing impairment (109 females and 111 males from 1 year to 70 years old at the time of investigation) includes 220 Tuvinians from 184 unrelated families living in the Tyva Republic (Southern Siberia, Russia). In total, 76 patients were urban residents (mainly from town Kyzyl) and 144 were rural residents from numerous small settlements located in different areas of the Tyva Republic. The genomic DNA samples of patients were collected from 2010 to 2018. Thorough analysis of pedigrees and family histories of patients was performed. The examined cohort of patients included 140 single/sporadic (the only affected individual in family) and 80 familial (two or more affected family members) HL cases. Hearing status of parents was ascertained for 210 patients: 186 (88.6%) patients have both normal hearing parents (mating N x N), 9 patients (4.3%) have one deaf and another normal hearing parents (mating D x N), 15 (7.1%) patients were descendants of assortative matings (both deaf parents, mating D x D), 10 of which could be considered, according to [26], as non-complementary matings (noncomp. mating D x D) that is, the families with both deaf parents and all deaf siblings presumably having the same genetic cause of HL.
Hearing status of affected individuals was evaluated by otoscopic and pure-tone audiometry examinations that patients underwent at different times in the specialized audiological services located in town Kyzyl. Severity of hearing loss was defined as mild (25–40 dB), moderate (41–70 dB), severe (71–90 dB) or profound (above 90 dB). The majority of examined patients (210 subjects) suffered from congenital or early onset bilateral severe-to-profound HL, nine patients have moderate HL, and one patient – unilateral severe HL. Other concomitant information was collected from local unspecialized medical services and by direct interview with the patients and their relatives.
2.2. Control Sample
The ethnically matched control sample comprises 157 unrelated Tuvinians from different regions of the Tyva Republic (84 females and 73 males, aged from 10 to 73 years old). None of them were registered by the audiological services and had no complaints of hearing impairments.
2.3. Ethics Statement
Written informed consent was obtained from all individuals or their legal guardians before they participated in the study. The study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the Bioethics Commission at the Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia (Protocol No. 9, 24 April 2012).
2.4. Mutation Analysis of the GJB2 Gene
Genomic DNA was isolated from the buffy coat fraction of blood by a standard phenol-chloroform extraction method. The GJB2 coding region encompassing exon 2 and the non-coding GJB2 region (exon 1 with flanking regions) were amplified in all patients and controls. Primer pairs designed to amplify corresponding PCR products and also used for Sanger sequencing are presented in Table 1. The PCR products were purified by sorption on Agencourt Ampure XP (Beckman Coulter, Indianapolis, IN, USA) and subjected to Sanger sequencing using a BigDye Terminator V.3.1 Cycle Sequencing Kit (Applied Biosystems, Waltham, MA, USA) with subsequent unincorporated dye removal by the Sephadex G-50 gel filtration (GE Healthcare, Chicago, IL, USA). The Sanger products were analyzed on an ABI 3130XL Genetic Analyzer (Applied Biosystems) in the SB RAS Genomics Core Facility (Institute of Chemical Biology and Fundamental Medicine SB RAS, Novosibirsk, Russia). The DNA sequence variations were identified by comparison with the GJB2 gene reference sequence NG_008358.1 (https://www.ncbi.nlm.nih.gov/nuccore/NG_008358.1). Screening of common large deletion GJB6-D13S1830 (~309 kb) was performed according to [27,28] only for the group of patients with monoallelic GJB2 recessive mutations. In this group of patients, we also sequenced a region of about 0.7 kb immediately upstream of GJB2 exon 1 which includes the GJB2 basal promoter using primers PF1, PF2, and PR1 as previously described [29] (Table 1).
Table 1.
Primers for polymerase chain reaction (PCR)/Sanger sequencing.
| GJB2 | Primers | Reference |
|---|---|---|
| GJB2 coding region (exon 2) and flanking sequences | 835-F: 5’-TGCTTGCTTACCCAGACTCA-3’ 835-R: 5’-CCTCATCCCTCTCATGCTGT-3’ |
This study |
| or | ||
| F3044: 5’-AGTGCCTTTCAGCTAACGA-3’ R4242: 5’-GTGGCATCTGGAGTTTCACC-3’ | ||
| Upstream region of exon 1 (which includes the basal promoter), exon 1, donor splice site, and flanking intronic region | Ex1-F: 5’-TCTTTTCCAGAGCAAACCGC-3’ Ex1-R: 5’-CTGGGCAATGCGTTAAACTGG-3’ |
[30] |
| Ex1-792-F: 5’-GCGTTCGTTCGGATTGGT-3’ Ex1-2239-R: 5’-CGGAAACAGACCCTCGTGAAGT-3’ |
This study | |
| PF1: 5’- GGCTCAAAGGAACTAGGAGATCG-3’ PF2: 5’-CGTTCGTTCGGATTGGTGAG-3’PR1: 5’-CAGAAACGCCCGCTCCAGAA-3’ |
[29] |
F: Forward; R: Reverse.
2.5. Verification of Cis-Configuration of GJB2 Variants c.79G>A (p.Val27Ile) and c.341A>G (p.Glu114Gly)
GJB2 variants c.79G>A (p.Val27Ile, rs2274084) and c.341A>G (p.Glu114Gly, rs2274083) are located 260 bp from each other in the GJB2 coding sequence. The phase (cis or trans) of these variants was assessed in all individuals who were heterozygous for both of these variants (deaf patients, their relatives and individuals from the control sample) by either pedigree analysis or by molecular cloning and Sanger sequencing of the syntenic variants (Figure S1). Molecular cloning was performed for 27 individuals heterozygous for both of these variants by use of the CloneJET™ PCR Cloning Kit (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s protocol. The fragment of the GJB2 coding region (837 bp) including both variants c.79G>A and c.341A>G was PCR-amplified from genomic DNA using Phusion Hot Start II polymerase (Thermo Fisher Scientific) and primers 835-F and 835-R (Table 1) and then cloned into the pJET1.2/blunt vector followed by transformation in competent E.coli strain Mach-1 (Invitrogen, Carlsbad, CA, USA). Presence/absence of variants c.79G>A and c.341A>G in examined positive clones was verified by Sanger sequencing with the same primers (Figure S1).
2.6. Bioinformatics Prediction Tools
The functional effects of the c.516G>C (p.Trp172Cys) variant were predicted using PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2) [31], PROVEAN (http://provean.jcvi.org) [32], PANTHER (http://www.pantherdb.org) [33], MutationTaster (http://www.mutationtaster.org/) [34], and FATHMM (http://fathmm.biocompute.org.uk/index.html) [35].
2.7. 3D Cx26 Molecule Structure Modelling
The three-dimensional (3D) molecule structure of the wild type and the p.Trp172Cys-mutant connexin 26 (Cx26-WT and Cx26-p.Trp172Cys, respectively) was visualized by DeepView/Swiss-PdbViewer v.4.1.0 (http://www.expasy.org/spdbv/) [36]. The protein template 2ZW3.1 was found by the SWISS-MODEL (http://swissmodel.expasy.org/) [36].
2.8. Statistical Methods
Two-tailed Fisher’s exact test with significance level of p < 0.05 was applied to compare allele frequencies between patients and controls. To avoid probable bias due to the presence of related individuals in the total cohort of patients, only unrelated patients (184 out of 220 selected by pedigree analysis) were used for allele frequency calculation.
3. Results
3.1. GJB2 Genotypes Observed in Patients and Control Sample
Five pathogenic recessive GJB2 variants: c.516G>C (p.Trp172Cys), c.-23+1G>A, c.235delC (p.Leu79Cysfs*3), c.299_300delAT (p.His100Argfs*14), c.109G>A (p.Val37Ile), and also several known benign GJB2 variants were found in examined patients. The GJB2 genotypes due to the combination of GJB2 gene sequence variations observed in patients and in the control sample are presented in Table 2.
Table 2.
The GJB2 genotypes found in Tuvinian patients and control sample.
| GJB2 Genotypes * | Patients (n = 220) | Control Sample (n = 157) |
|---|---|---|
| GJB2 genotypes with biallelic recessive pathogenic variants | ||
| c.[516G>C];[516G>C] p.[Trp172Cys];[Trp172Cys] |
25 | - |
| c.[-23+1G>A];[-23+1G>A] p.[splice donor variant];[splice donor variant] |
9 | - |
| c.[-23+1G>A];[516G>C] p.[splice donor variant];[Trp172Cys] |
7 | - |
| c.[235delC];[516G>C] p.[Leu79Cysfs*3];[Trp172Cys] |
4 | - |
| c.[299_300delAT];[516G>C] p.[His100Argfs*14];[Trp172Cys] |
2 | - |
| c.[-23+1G>A];[299_300delAT] p.[splice donor variant];[His100Argfs*14] |
1 | - |
| c.[109G>A];[235delC] p.[Val37Ile];[Leu79Cysfs*3] |
1 | - |
| Total | 49 (22.3%) | - |
| GJB2 genotypes with a single recessive pathogenic variant | ||
| c.[-23+1G>A];[wt] p.[splice donor variant];[wt] |
5 | 3 |
| c.[516G>C];[wt] p.[Trp172Cys];[wt] |
9 | 6 |
| c.[-23+1G>A];[79G>A] p.[splice donor variant];[Val27Ile] |
1 | 3 |
| c.[79G>A];[516G>C] p.[Val27Ile];[Trp172Cys] |
1 | 0 |
| c.[235delC];[wt] p.[Leu79Cysfs*3];[wt] |
1 | 0 |
| c.[109G>A];[wt] p.[Val37Ile];[wt] |
1 | 2 |
| c.[79G>A];[109G>A] p.[Val27Ile];[Val37Ile] |
0 | 1 |
| Total | 18 (8.2%) | 15 (9.6%) |
| GJB2 genotypes with benign variants | ||
| c.[79G>A];[wt] p.[Val27Ile];[wt] |
27 | 31 |
| c.[79G>A;341A>G];[wt] p.[Val27Ile;Glu114Gly];[wt] |
15 | 12 |
| c.[79G>A];[79G>A] p.[Val27Ile];[Val27Ile] |
7 | 6 |
| c.[79G>A];[79G>A;341A>G] p.[Val27Ile];[Val27Ile;Glu114Gly] |
1 | 2 |
| c.[79G>A;341A>G];[571T>C] p.[Val27Ile;Glu114Gly];[Phe191Leu] |
1 | 0 |
| c.[79G>A];[571T>C] p.[Val27Ile];[Phe191Leu] |
1 | 1 |
| c.[457G>A];[wt] p.[Val153Ile];[wt] |
1 | 0 |
| c.[608T>C];[wt] p.[Ile203Thr];[wt] |
1 | 0 |
| c.[79G>A;341A>G];[457G>A] p.[Val27Ile;Glu114Gly];[Val153Ile] |
0 | 1 |
| Total | 54 (24.5%) | 53 (33.7%) |
| GJB2 genotype [wt];[wt] | 99 (45.0%) | 89 (56.7%) |
* - GJB2 variations are designated at the nucleotide level (NM_004004.6) and at the amino acid level (NP_003995.2) at the top and bottom of each line, respectively. The GJB2 sequence variant c.516G>C (p.Trp172Cys) was submitted as ‘possibly pathogenic’ to ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/) and the ClinVar accession number is SCV000852065.
Twenty-two different GJB2 genotypes found in patients can be subdivided into four groups: (1) seven genotypes with biallelic (homozygous or compound heterozygous) recessive pathogenic GJB2 variants (22.3%, 49 out of 220 patients); (2) six genotypes with single recessive pathogenic GJB2 variants in compound with a benign variant or wild type allele (8.2%, 18 out of 220 patients); (3) eight genotypes with only benign GJB2 variants (24.5%, 54 out of 220 patients); (4) wild genotype (without any changes in the GJB2 gene sequence) (45.0%, 99 out of 220 patients).
Twelve different GJB2 genotypes (including wild genotype) were found in the control sample: five genotypes included a single recessive pathogenic GJB2 variant in compound with a benign variant or wild type allele (9.6%, 15 out of 157 individuals); six genotypes—only benign GJB2 variants (33.7%, 53 out of 157 individuals); wild genotype (56.7%, 89 out of 157 individuals) (Table 2).
The contribution of the GJB2 pathogenic variants in HL of patients defined as the proportion of patients from the first group of GJB2 genotypes among all examined Tuvinian patients was estimated as 22.3%. The c.516G>C (p.Trp172Cys) variant was the most frequent (62.9%) out of all GJB2 pathogenic variants detected in patients followed by c.-23+1G>A (27.6%), c.235delC (5.2%), c.299_300delAT (2.6%), and c.109G>A (p.Val37Ile) (1.7%). Total carrier frequency of the GJB2 pathogenic variants in Tuvinian control sample was estimated as 9.6% (p.Trp172Cys – 3.8%, c.-23+1G>A – 3.8%, and p.Val37Ile – 1.9%).
3.2. GJB2 Sequence Variations in Patients and Control Sample
The c.516G>C (p.Trp172Cys) variant makes a significant contribution to deafness in Tuvinian patients since 38 (from 27 unrelated families) out of 220 examined patients are either homozygotes or compound heterozygotes for p.Trp172Cys, and also 10 patients (from seven unrelated families) have only a single allele with p.Trp172Cys (Table 2). The p.Trp172Cys variant (in compound with c.235delC) was initially found in our previous study in one deaf patient belonging to Altaians—indigenous Turkic-speaking people of the Altai Republic neighboring the Tyva Republic (Southern Siberia, Russia) [37]. Later, Tekin et al. (2010) detected p.Trp172Cys in compound with mutation c.299_300delAT in one deaf patient from Mongolia [38].
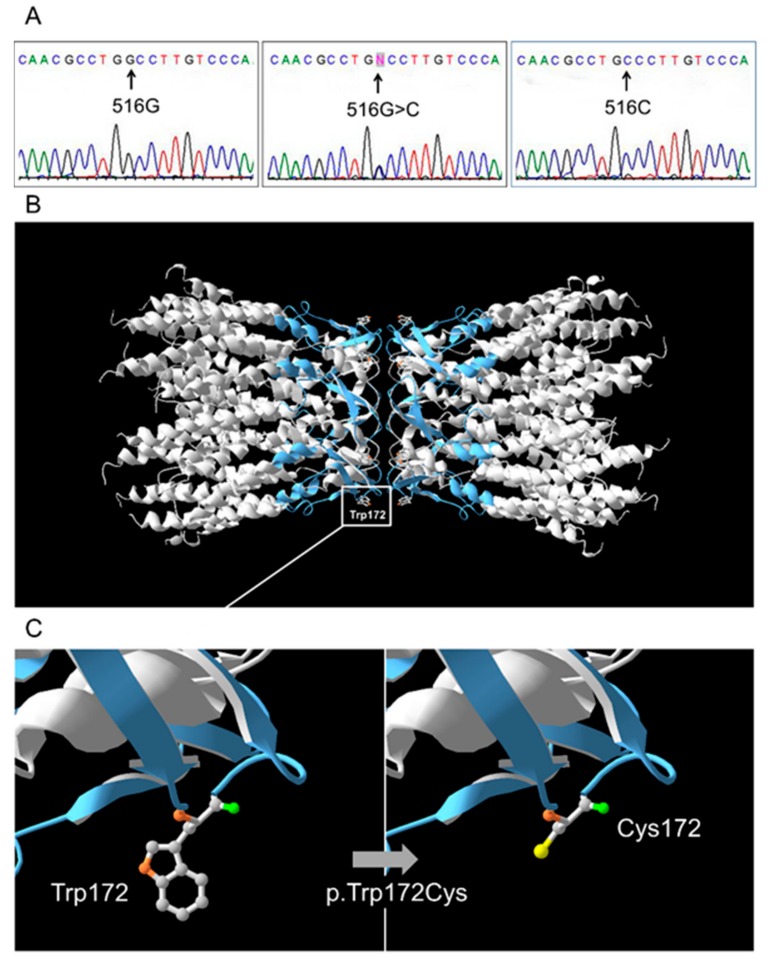
The c.516G>C substitution leads to a replacement of aromatic non-polar tryptophan by a small polar cysteine at amino acid position 172 in the second extracellular loop (E2, 155-192 amino acid residues) of the connexin 26 protein (Cx26) (Figure 1). The loop E2 is known to be essential for Cx26-hemichannel subunits connection [39]. Multiple in silico programs (scores: PolyPhen2—‘probably/possibly damaging’, PROVEAN—‘deleterious’, PANTHER—‘probably damaging’, MutationTaster—‘disease causing’, FATHMM—‘damaging’) predict a deleterious effect of p.Trp172Cys. For now, variant p.Trp172Cys is only presented in the Human Gene Mutation Database (HGMD: http://www.hgmd.cf.ac.uk/ac/) [8] with reference to our previous work [37].
Figure 1.
(A) Identification of the c.516G>C (p.Trp172Cys) mutation by Sanger sequencing. (B) The 3D structure of normal protein connexin 26 (Cx26-WT). Two adjacent Cx26-WT hemichannel subunits with designation of Trp (tryptophan) on position 172. The extracellular loop of Cx26 (E2, 155-192 amino acid residues) where variant p.Trp172Cys located is marked by blue. (C) Close-up view of Cx26-WT (Trp172) and mutant Cx26-p.Trp172Cys (Cys172) connexin 26.
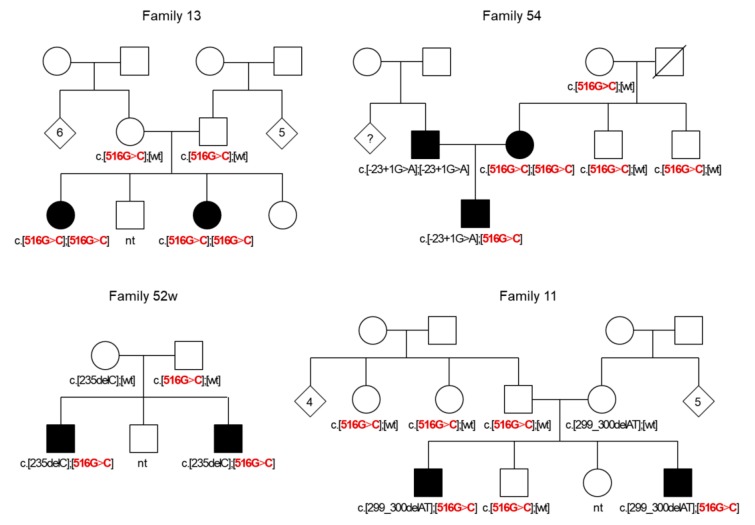
All 38 examined patients having p.Trp172Cys (biallelic or in compound with known pathogenic GJB2 variants c.-23+1G>A, c.235delC, or c.299_300delAT) were affected by nonsyndromic congenital or early onset HL. To investigate the segregation of p.Trp172Cys with HL, we have screened this variant in all available relatives of these patients and revealed a distinct segregation of homozygosity or compound heterozygosity for p.Trp172Cys with HL in affected Tuvinian families (Figure 2 and Figure S2). Moreover, the frequency of p.Trp172Cys calculated in unrelated patients (0.1386, 51/368 chromosomes) was significantly higher compared with the control sample (0.019, 6/314 chromosomes) (p < 10−8) (Table 3).
Figure 2.
The pedigrees of Tuvinian families demonstrating the segregation of variant c.516G>C (p.Trp172Cys) in a homozygous state or in compound with other recessive GJB2 mutations (c.235delC, c.-23+1G>A, and c.299_300delAT) with HL in affected family members. Deaf individuals are shown by black symbols. The variant c.516G>C (p.Trp172Cys) is shown by red.
Table 3.
Frequencies of the GJB2 gene variations in Tuvinian patients and the control sample.
| GJB2 Gene Sequence Variations | dbSNP ID | Patients (n = 184) * Number of Alleles/Frequency |
Control Sample (n = 157) Number of Alleles/Frequency |
|---|---|---|---|
| GJB2 pathogenic variants | |||
| c.516G>C (p.Trp172Cys) | not presented | 51/0.1386 | 6/0.019 |
| c.-23+1G>A | rs80338940 | 30/0.0815 | 6/0.019 |
| c.235delC | rs80338943 | 5/0.0136 | 0/0.0 |
| c.299_300delAT | rs111033204 | 2/0.0054 | 0/0.0 |
| c.109G>A (p.Val37Ile) | rs72474224 | 2/0.0054 | 3/0.0096 |
| Total | 90/0.2446 | 15/0.0478 | |
| GJB2 benign variants | |||
| c.79G>A (p.Val27Ile) | rs2274084 | 41/0.1141 | 50/0.1592 |
| c.[79G>A;341A>G] (p.[Val27Ile;Glu114Gly]) | rs2274084 + rs2274083 | 15/0.0408 | 15/0.0478 |
| c.571T>C (p.Phe191Leu) | rs397516878 | 2/0.0054 | 1/0.0032 |
| c.457G>A (p.Val153Ile) | rs111033186 | 1/0.0027 | 1/0.0032 |
| c.608T>C (p.Ile203Thr) | rs76838169 | 1/0.0027 | 0/0.0 |
* - To avoid probable bias due to the presence of related individuals in the total cohort of Tuvinian patients (n = 220), only 184 unrelated patients selected by pedigree analysis were used for this allele frequency calculation. Significant (p < 0.05) differences in frequencies between group of patients and the control group are shown in bold.
The splice donor site variant c.-23+1G>A (rs80338940) located in the GJB2 non-coding region (intron 1) is the second common pathogenic GJB2 mutation (after p.Trp172Cys) in Tuvinian patients and controls. The c.-23+1G>A is apparently not a rare GJB2 mutation since it was found among deaf patients of different origins around the world [9,21,30,38,40,41,42,43,44] although not all laboratories are screening for this particular mutation since it is located outside the GJB2 coding region. The extensive accumulation of c.-23+1G>A was reported in Yakuts, indigenous Turkic-speaking people living in the subarctic region of Russia (the Sakha Republic/Yakutia). Extremely high prevalence of c.-23+1G>A could be explained by a founder effect and a probable selective advantage for the c.-23+1G>A heterozygotes in severe subarctic climate [21,43]. The c.-23+1G>A was reported as the most common GJB2 mutation in deaf Mongolian patients from Mongolia [38,44].
The GJB2 frameshift mutations c.235delC (rs80338943) and c.299_300delAT (rs111033204) found in Tuvinian patients are well known deleterious GJB2 variants associated with deafness. In many extensive studies, c.235delC and c.299_300delAT have been confirmed to be the most common GJB2 mutations in the East Asian (Chinese, Taiwanese, Japanese, Korean) populations [7,9,12,13,14,45,46] as well as in Mongolian patients living in Mongolia and in the Inner Mongolia Autonomous Region of China [38,44,47].
Mutation c.109G>A (p.Val37Ile, rs72474224) which is predominant in Southeast Asian populations [7,9,20] was rare in Tuvinian patients—only two of them had compound heterozygous and heterozygous p.Val37Ile (Table 2).
The GJB2 variants c.79G>A (p.Val27Ile, rs2274084) and c.341A>G (p.Glu114Gly, rs2274083) found in Tuvinian patients and control individuals (Table 2) are known to be common in many Asian populations. The p.Val27Ile can be detected as a single variation as well as together with p.Glu114Gly, while p.Glu114Gly is very rarely found alone. These variants are mainly considered as benign polymorphisms due to their presence in deaf patients and controls; however, some studies have not excluded a possible association of these variants combination (in particular, related to their configuration - cis or trans) with HL [38,48,49,50,51]. In vitro functional analyses were performed to clarify the possible role of p.Val27Ile and p.Glu114Gly in HL [38,49,52]. Choung et al. (2002) investigated HeLa cells expressing single variant p.Glu114Gly and revealed their gap junction functioning similar to wild type cells [52]. Tekin et al. (2010) showed that p.Val27Ile + p.Glu114Gly can significantly impair the function of the gap junctions but there were no differences in the p.Val27Ile + p.Glu114Gly haplotype frequencies between patients and control group [38]. Choi et al. (2011) showed reduced activity of the p.Glu114Gly and the p.Val27Ile + p.Glu114Gly channels and suggested that only the p.Glu114Gly homozygote or compound heterozygote for p.Glu114Gly and other GJB2 pathogenic variant can cause HL [49]. Thus, the pathogenicity of these variants remains controversial and is widely discussed in literature.
In our study, 17 Tuvinian patients and 15 control individuals were heterozygous for both p.Val27Ile and p.Glu114Gly and none of the examined individuals had p.Glu114Gly separately from p.Val27Ile (Table 2). We investigated the phase (cis or trans) of p.Val27Ile and p.Glu114Gly in all individuals heterozygous for these variants by analysis of pedigrees of patients (if relevant data were available) or by sequencing of cloned fragments spanning the variants (Figure S1). In this way, we proved their cis-configuration in all cases where p.Glu114Gly was found. Thus, we postulated the presence of allele p.[Val27Ile;Glu114Gly] in our examined samples and estimated its frequency in patients and controls as 0.0408 and 0.0478, respectively. Since the frequency of p.[Val27Ile;Glu114Gly] was slightly higher in controls than in patients (although the difference was not statistically significant, p = 0.3969), we believe that there is no association of allele p.[Val27Ile;Glu114Gly] with HL, at least in our sample of patients.
Other GJB2 benign variants found in Tuvinians were c.571T>C (p.Phe191Leu, rs397516878, c.457G>A (p.Val153Ile, rs111033186), and c.608T>C (p.Ile203Thr, rs76838169). They are listed as rare polymorphisms preferentially detected in South Asian populations (dbSNP: https://www.ncbi.nlm.nih.gov/snp/).
3.3. Group of Patients with a Single GJB2 Pathogenic Variant
We found that 18 among all 220 examined patients (8.2%) are the carriers of a single recessive GJB2 pathogenic variant: 10 patients have single c.516G>C (p.Trp172Cys), 6 patients - c.-23+1G>A, 1 patient - c.235delC, and 1 patient - c.109G>A (p.Val37Ile) (Table 2). We analyzed the pedigree and family data of patients with GJB2 monoallelic mutations. These patients represented eleven sporadic and seven familial HL cases. Among familial cases, four patients were descendants of both deaf parents (mating D x D), two patients had normal hearing parents (mating N x N), and one patient was from mating D x N (Table S1 and Figure S3).
A relatively large proportion of deaf individuals carrying only one pathogenic recessive GJB2 variant has been reported in many previous studies [6,7,41,53,54,55,56,57,58,59,60,61]. Two main assumptions have been made to resolve this issue: (1) HL of such patients could be caused by an uncertain impact of the GJB2 gene (the presence of yet undetected other pathogenic GJB2 variants or variable penetrance of pathogenic GJB2 variant due to any modulating factors); (2) these patients are only the coincidental carriers of one pathogenic GJB2 allele and other factors (other genes or environmental impacts) cause their HL.
The coding region of the GJB2 gene and the region where splice donor variant c.-23+1G>A is located were routinely analyzed in majority of studies while the involvement of other GJB2 noncoding regions in HL has not been extensively investigated. The GJB2 gene has a 128 bp basal promoter including a TATA-box (TTAAAA) at -19 to -24 and 2 GC-boxes (CCGCCC) at -76 to -81 and -88 to -93 proximal to the transcriptional start site [62].
Several studies have reported that some noncoding GJB2 variants found in upstream and basal promoter regions, exon 1, and in splice acceptor site may contribute to HL [29,63,64,65,66]. In addition, large deletions which either eliminate a hypothesized cis-acting regulatory element located far upstream of GJB2 or encompass the complete GJB2 and GJB6 genes have been reported [27,67,68,69,70,71].
We tried to clarify the cause of HL in Tuvinian patients with monoallelic GJB2 mutations. To search additional (probably missed in routine GJB2 analysis) alterations in the potentially regulatory region of GJB2, for 18 DNA samples of these patients we sequenced the 1009 bp region immediately upstream of GJB2 exon 1, which includes the basal promoter according to [29]. We also reanalyzed the GJB2 region where the acceptor splice site of the only GJB2 intron is located [66].
All variations found in these GJB2 regions are presented in Table S1. No known pathogenic variants, including c.-684-675del (previously named as −493del10 [63]), were found in patients with monoallelic GJB2 mutations. We had also screened the most common deletion GJB6-D13S1830 [27] in GJB2 heterozygous patients. None of these patients had this deletion.
Moreover, at the time of this study, our group began to test the SLC26A4 gene (OMIM 605646, 7q22.3) as a potential candidate gene for HL in GJB2-negative patients including 18 GJB2 heterozygous patients. Our preliminary results of SLC26A4 testing suggest that HL of at least 4 out of 18 (22%) patients with monoallelic GJB2 mutations was caused by homozygosity or compound heterozygosity for SLC26A4 pathogenic variants (Table S1), and consequently these patients are definitely the coincidental carriers of one pathogenic GJB2 allele.
Assuming that HL in patients with monoallelic GJB2 mutations is not associated with the GJB2 gene, the frequency of GJB2 mutations in a sample of non-DFNB1A patients (that is, patients that do not have biallelic GJB2 mutations) should be the same as the frequency in control samples. To test this hypothesis, we compared the frequencies of GJB2 pathogenic variants among non-DFNB1A patients and controls. To avoid probable bias due to the presence of related individuals in the cohort of Tuvinian patients, only unrelated patients selected by pedigree analysis (in total, 146 patients including 14 GJB2 heterozygous patients and 132 patients without the GJB2 mutations) were used for this calculation. Overall frequency of monoallelic GJB2 mutations among this group of patients was calculated to be 0.0479. Among control samples, 15 individuals were heterozygous for one of any GJB2 mutations (Table 2), and thus the total frequency of the GJB2 mutations was estimated as 0.0478 (Table 3). As a result, there were no statistically significant differences in the frequency of monoallelic GJB2 mutations among patients compared to the control sample.
4. Discussion
A high variability in the GJB2 mutation spectrum and frequencies has been reported among different worldwide populations. Nevertheless, the contribution of the GJB2 gene to HL (DFNB1A) remains unknown for certain ethnic groups in different regions of the world.
In this study, we present for the first time data on the spectrum and frequency of the GJB2 gene variants in a large cohort of Tuvinian patients with HL (n = 220) and also in unrelated normal hearing Tuvinians (n = 157) living in the Tyva Republic (Southern Siberia, Russia). As the result of sequencing the GJB2 coding (exon 2) and non-coding (exon 1) regions with flanking sequences, a genetic diagnosis “Nonsyndromic autosomal recessive deafness 1A (DFNB1A)” due to the presence of biallelic GJB2 pathogenic variants was established for 22.3% (49 out of 220) patients. In total, 8.2% (18 out of 220) of patients were the carriers of a single recessive GJB2 pathogenic variant, and 69.5% (153 out of 220) of patients were GJB2-negative (without GJB2 pathogenic variants).
In total, 26 out of 49 DFNB1A-patients represent sporadic HL cases and 23–familial cases. Thus, the DFNB1A prevalence among all sporadic cases (n = 140) could be estimated as 18.6%, whereas the proportion of DFNB1A among familial cases (n = 80)—as 28.8%. A lower frequency of DFNB1A in sporadic cases compared with familial cases may partly be explained by the inclusion of potential nongenetic HL cases. Based on the pedigree data, we found that 20 out of 23 familial DFNB1A-patients were from 11 unrelated families with two and more deaf siblings representing typical autosomal recessive type of HL inheritance, and although not all affected individuals in these families were examined by us, untested siblings in these Tuvinian families obviously have the same type of HL (DFNB1A). Two familial DFNB1A-cases were represented by patients who were descendants from non-complementary assortative matings (noncomp. mating D x D) [26]; therefore, HL of their untested deaf parents and siblings is also highly likely due to the GJB2 mutations. The last familial DFNB1A-patient was from mating D x N, representing a pseudo-dominant type of inheritance. Thus, based on pedigree data, we suggest that contribution of the GJB2 mutations to HL in deaf Tuvinians may be more than 22.3% due to probable DFNB1A-cases in untested affected relatives of examined patients, making our results applicable in genetic testing and counseling for a larger number of deaf patients in the Tyva Republic.
A large number of deaf individuals with a single recessive GJB2 pathogenic allele was detected in different studies, which complicates the molecular diagnosis for their HL [6,7,41,53,54,55,56,57,58,59,60,61,72]. These findings may partly be explained by the fact that only the GJB2 coding region (exon 2) was tested in some older studies; therefore, potential pathogenic variations in the GJB2 non-coding or regulatory regions could be missed.
We thoroughly analyzed a group of 18 Tuvinian patients with a single recessive GJB2 pathogenic variant (8.2% out of all examined patients) and concluded that they are most likely the coincidental carriers of one pathogenic GJB2 allele and the cause of their HL could be attributed to mutations in other genes associated with HL or environmental impacts. The absence of differences in the frequency of monoallelic GJB2 mutations in patients and in the control sample may be additional evidence for this conclusion.
Ten GJB2 sequence variations including five pathogenic (p.Trp172Cys, c.-23+1G>A, c.235delC, c.299_300delAT, and p.Val37Ile) and five benign (p.Val27Ile, p.Glu114Gly, p.Phe191Leu, p.Val153Ile, and p.Ile203Thr) variants were identified among 220 examined Tuvinian patients.
A striking finding was the discovery of common GJB2 variant c.516G>C (p.Trp172Cys) found with frequency 62.9% among all GJB2 mutated alleles detected in deaf Tuvinian patients. The p.Trp172Cys carrier frequency in Tuvinian control group was estimated as 3.8%.
The c.516G>C substitution leads to a replacement of aromatic non-polar tryptophan by a small polar cysteine at conservative amino acid position 172 (p.Trp172Cys) in the second extracellular loop (E2, 155-192 amino acid residues) of protein Cx26 (Figure 1). Both extracellular loops (E1 and E2) of Cx26 molecules contain six conserved cysteines (C53, C60, C64 in E1 and C169, C174, C180 in E2) which form intramolecular disulfide bonds, playing an essential role in Cx26 hemichannel subunits docking [39]. To our knowledge, the only other substitution of tryptophan at position 172 to arginine (p.Trp172Arg) caused by substitution c.514T>A has been described in the study by Mani et al. (2009) where two deaf patients from India were found to be homozygous for p.Trp172Arg [64]. Mani et al. showed that Trp172Arg-mutated Cx26 exhibited normal plasma membrane localization but defective gap junction channel activity and the authors suggested that replacement of tryptophan 172, located in close proximity to at least two of three cysteine residues in E2, could result in defective docking of two opposing connexon hemichannels [64].
Other lines of evidences also support pathogenicity of c.516G>C (p.Trp172Cys). A distinct segregation of homozygosity or compound heterozygosity for p.Trp172Cys with HL was revealed in affected Tuvinian families, and the p.Trp172Cys frequency among Tuvinian patients (0.1386) was significantly higher than in Tuvinian controls (0.019) (p < 10−8). Multiple in silico tools (PolyPhen2, PROVEAN, PANTHER, MutationTaster, FATHMM) predict a deleterious effect of p.Trp172Cys.
The p.Trp172Cys variant is yet unreported in the world human genome databases (dbSNP, Exome Variant Server, Exome Aggregation Consortium (ExAC), 1000 Genomes Project). Recently, we submitted c.516G>C (p.Trp172Cys) to ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/) as a ‘likely pathogenic’ variant (ClinVar accession number SCV000852065) because p.Trp172Cys meets main genetic criteria [73] for such classification: (i) strong segregation of homozygosity for p.Trp172Cys with recessive HL in multiple affected subjects from unrelated families; (ii) significantly higher frequency of p.Trp172Cys in patients compared with ethnically matched controls; (iii) multiple computational predictions of deleterious effect of p.Trp172Cys; (iiii) extreme rarity of p.Trp172Cys.
Chronologically, the p.Trp172Cys variant was initially found in our previous study in one patient belonging to Altaians - indigenous Turkic-speaking people of the Altai Republic (Southern Siberia, Russia) neighboring the Tyva Republic [37] and later Tekin et al. (2010) detected p.Trp172Cys in one Mongolian deaf patient from Mongolia [38]. Our recent data obtained on extended sample of Altaian deaf patients [74] revealed slightly elevated prevalence of p.Trp172Cys in Altaians whereas recent studies did not detect p.Trp172Cys among deaf Mongolian patients from Mongolia [44] as well as in Mongolian patients living in China [47,75]. Therefore, we suggest that p.Trp172Cys is the GJB2 mutation endemic to Tuvinians, indigenous Turkic-speaking people of the Tyva Republic, since apart from them p.Trp172Cys has only minor incidence in indigenous peoples living in the neighboring Altai Republic and Mongolia.
5. Conclusions
The data on spectrum and frequency of GJB2 gene variants were obtained for the first time for a large cohort of 220 deaf Tuvinian patients from the Tyva Republic (Southern Siberia, Russia) and ethnically matched cohort of 157 subjects with normal hearing. The GJB2 biallelic pathogenic variants were found in 22.3% patients. We suggest that patients with monoallelic GJB2 mutations (8.2%) are coincidental carriers. High prevalence of rare pathogenic variant p.Trp172Cys was found in Tuvinians while other GJB2 mutations are shared by nearby populations. All obtained data provide important targeted information for genetic counseling of affected Tuvinian families and enrich current information on variability of GJB2 worldwide.
Acknowledgments
The authors are sincerely grateful to all participants of the study.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/10/6/429/s1, Figure S1: Verification of cis-configuration of variants c.79G>A (p.Val27Ile) and c.341A>G (p.Glu114Gly), Figure S2: Additional pedigrees of Tuvinian families demonstrating the segregation of variant c.516G>C (p.Trp172Cys) in homozygous or in compound heterozygous state with HL in affected family members, Figure S3: The familial GJB2 monoallelic cases, Table S1: The detailed data on the patients with a single GJB2 pathogenic variant.
Author Contributions
Conceptualization, O.L.P.; Methodology, O.L.P., A.A.B. and I.V.M; Formal analysis, M.V.Z., V.Y.D., E.A.M., M.S.B.-K., A.A.B. and I.V.M.; Investigation, M.V.Z., V.Y.D., E.A.M., M.S.B.-K., A.A.B. and I.V.M; Resources, O.L.P. and M.S.B.-K.; Data curation, O.L.P. and M.S.B.-K.; Writing—original draft preparation, O.L.P., M.V.Z., V.Y.D., E.A.M. and M.S.B.-K.; Writing—review and editing, O.L.P., M.V.Z., V.Y.D., E.A.M., N.A.B., A.A.B., I.V.M and V.N.M.; Supervision, M.I.V.; Project administration, O.L.P. and M.I.V.; Funding acquisition, O.L.P.; M.I.V.; M.V.Z.; N.A.B.
Funding
This work was supported by Project #0324-2019-0041 of Institute of Cytology and Genetics SB RAS (to O.L.P., M.V.Z., V.Y.D., E.A.M., V.N.M., M.I.V.), and by the RFBR grants: #17-29-06016_ofi_m (to O.L.P., M.V.Z., M.S.B.-K., V.Y.D., E.A.M., N.A.B., A.A.B., I.V.M), #18-34-00166_mol-a (to M.V.Z., V.Y.D., E.A.M.), #18-013-00738_A and #18-05-600035_Arctica (to N.A.B.).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Morton C.C., Nance W.E. Newborn hearing screening—A silent revolution. N. Engl. J. Med. 2006;354:2151–2164. doi: 10.1056/NEJMra050700. [DOI] [PubMed] [Google Scholar]
- 2.Toriello H.V., Smith S.D. Hereditary Hearing Loss and Its Syndromes. 3rd ed. Oxford University Press; Oxford, UK: 2013. p. 756. [Google Scholar]
- 3.Van Camp G., Smith R.J.H. Hereditary Hearing Loss Homepage. [(accessed on 4 April 2019)]; Available online: https://hereditaryhearingloss.org.
- 4.Del Castillo F.J., del Castillo I. DFNB1 Non-syndromic hearing impairment: diversity of mutations and associated phenotypes. Front. Mol. Neurosci. 2017;10:428. doi: 10.3389/fnmol.2017.00428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruzzone R., White T.W., Paul D.L. Connections with connexins: The molecular basis of direct intercellular signaling. Eur. J. Biochem. 1996;238:1–27. doi: 10.1111/j.1432-1033.1996.0001q.x. [DOI] [PubMed] [Google Scholar]
- 6.Kenneson A., Van Naarden Braun K., Boyle C. GJB2 (connexin 26) variants and nonsyndromic sensorineural hearing loss: A HuGE review. Genet. Med. 2002;4:258–274. doi: 10.1097/00125817-200207000-00004. [DOI] [PubMed] [Google Scholar]
- 7.Chan D.K., Chang K.W. GJB2-associated hearing loss: Systematic review of worldwide prevalence, genotype, and auditory phenotype: Systematic review of Cx-26-associated hearing loss. Laryngoscope. 2014;124:E34–E53. doi: 10.1002/lary.24332. [DOI] [PubMed] [Google Scholar]
- 8.Stenson P.D., Mort M., Ball E.V., Evans K., Hayden M., Heywood S., Hussain M., Phillips A.D., Cooper D.N. The human gene mutation database: Towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum. Genet. 2017;136:665–677. doi: 10.1007/s00439-017-1779-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsukada K., Nishio S.Y., Hattori M., Usami S. Ethnic-specific spectrum of GJB2 and SLC26A4 mutations: Their origin and a literature review. Ann. Otol. Rhinol. Laryngol. 2015;124:61S–76S. doi: 10.1177/0003489415575060. [DOI] [PubMed] [Google Scholar]
- 10.Gasparini P., Rabionet R., Barbujani G., Melchionda S., Petersen M., Brøndum-Nielsen K., Metspalu A., Oitmaa E., Pisano M., Fortina P., et al. High carrier frequency of the 35delG deafness mutation in European populations. Eur. J. Hum. Genet. 2000;8:19–23. doi: 10.1038/sj.ejhg.5200406. [DOI] [PubMed] [Google Scholar]
- 11.Mahdieh N., Rabbani B. Statistical study of 35delG mutation of GJB2 gene: A meta-analysis of carrier frequency. Int. J. Audiol. 2009;48:363–370. doi: 10.1080/14992020802607449. [DOI] [PubMed] [Google Scholar]
- 12.Liu X., Xia X., Ke X., Ouyang X., Du L., Liu Y., Angeli S., Telischi F., Nance W., Balkany T., et al. The prevalence of connexin 26 (GJB2) mutations in the Chinese population. Hum. Genet. 2002;111:394–397. doi: 10.1007/s00439-002-0811-6. [DOI] [PubMed] [Google Scholar]
- 13.Ohtsuka A., Yuge I., Kimura S., Namba A., Abe S. GJB2 deafness gene shows a specific spectrum of mutations in Japan, including a frequent founder mutation. Hum. Genet. 2003;112:329–333. doi: 10.1007/s00439-002-0889-x. [DOI] [PubMed] [Google Scholar]
- 14.Yao J., Lu Y., Wei Q., Cao X., Xing G. A systematic review and meta-analysis of 235delC mutation of GJB2 gene. J. Transl. Med. 2012;10:136. doi: 10.1186/1479-5876-10-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morell R.J., Kim H.J., Hood L.J., Goforth L., Friderici K., Fisher R., van Camp G., Berlin C.I., Oddoux C., Ostrer H., et al. Mutations in the Connexin 26 Gene (GJB2) among Ashkenazi Jews with nonsyndromic recessive deafness. N. Engl. J. Med. 1998;339:1500–1505. doi: 10.1056/NEJM199811193392103. [DOI] [PubMed] [Google Scholar]
- 16.Hamelmann C., Amedofu G.K., Albrecht K., Muntau B., Gelhaus A., Brobby G.W., Horstmann R.D. Pattern of connexin 26 (GJB2) mutations causing sensorineural hearing impairment in Ghana. Hum. Mutat. 2001;18:84–85. doi: 10.1002/humu.1156. [DOI] [PubMed] [Google Scholar]
- 17.RamShankar M., Girirajan S., Dagan O., Ravi Shankar H.M., Jalvi R., Rangasayee R., Avraham K.B., Anand A. Contribution of connexin26 (GJB2) mutations and founder effect to non-syndromic hearing loss in India. J. Med. Genet. 2003;40:e68. doi: 10.1136/jmg.40.5.e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Minárik G., Ferák V., Feráková E., Ficek A., Poláková H., Kádasi Ľ. High frequency of GJB2 mutation W24X among Slovak Romany (Gypsy) patients with non-syndromic hearing loss (NSHL) Gen. Physiol. Biophys. 2003;22:549–556. [PubMed] [Google Scholar]
- 19.Álvarez A., del Castillo I., Villamar M., Aguirre L.A., González-Neira A., López-Nevot A., Moreno-Pelayo M.A., Moreno F. High prevalence of theW24X mutation in the gene encoding connexin-26 (GJB2) in Spanish Romani (gypsies) with autosomal recessive non-syndromic hearing loss. Am. J. Med. Genet. A. 2005;137A:255–258. doi: 10.1002/ajmg.a.30884. [DOI] [PubMed] [Google Scholar]
- 20.Wattanasirichaigoon D., Limwongse C., Jariengprasert C., Yenchitsomanus P., Tocharoenthanaphol C., Thongnoppakhun W., Thawil C., Charoenpipop D., Pho-iam T., Thongpradit S., et al. High prevalence of V37I genetic variant in the connexin-26 (GJB2) gene among non-syndromic hearing-impaired and control Thai individuals: High prevalence of V37I among Thai subjects. Clin. Genet. 2004;66:452–460. doi: 10.1111/j.1399-0004.2004.00325.x. [DOI] [PubMed] [Google Scholar]
- 21.Barashkov N.A., Dzhemileva L.U., Fedorova S.A., Teryutin F.M., Posukh O.L., Fedotova E.E., Lobov S.L., Khusnutdinova E.K. Autosomal recessive deafness 1A (DFNB1A) in Yakut population isolate in Eastern Siberia: Extensive accumulation of the splice site mutation IVS1+1G>A in GJB2 gene as a result of founder effect. J. Hum. Genet. 2011;56:631–639. doi: 10.1038/jhg.2011.72. [DOI] [PubMed] [Google Scholar]
- 22.Carranza C., Menendez I., Herrera M., Castellanos P., Amado C., Maldonado F., Rosales L., Escobar N., Guerra M., Alvarez D., et al. A Mayan founder mutation is a common cause of deafness in Guatemala. Clin. Genet. 2016;89:461–465. doi: 10.1111/cge.12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vainshtein S.I., Mannay-ool M.H. History of Tyva. 2nd ed. Science; Novosibirsk, Russia: 2001. (In Russian) [Google Scholar]
- 24.Mongush M.V. Tuvans of Mongolia and China. Int. J. Cent. Asian Stud. 1996;1:225–243. [Google Scholar]
- 25.Bady-Khoo M.S., Posukh O.L., Zorkoltseva I.V., Skidanova O.V., Barashkov N.A., Omzar O.S., Mongush R.S., Bamba O.M., Tukar V.M., Zytsar M.V., et al. Study of hereditary hearing loss in the Republic of Tuva. I. Epidemiology of hearing impairments in the Republic of Tuva. Med. Genetika. 2014;13:17–26. (In Russian) [Google Scholar]
- 26.Nance W.E., Liu X.-Z., Pandya A. Relation between choice of partner and high frequency of connexin-26 deafness. Lancet. 2000;356:500–501. doi: 10.1016/S0140-6736(00)02565-4. [DOI] [PubMed] [Google Scholar]
- 27.Del Castillo I., Villamar M., Moreno-Pelayo M.A., del Castillo F.J., Álvarez A., Tellería D., Menéndez I., Moreno F. A deletion involving the Connexin 30 Gene in nonsyndromic hearing impairment. N. Engl. J. Med. 2002;346:243–249. doi: 10.1056/NEJMoa012052. [DOI] [PubMed] [Google Scholar]
- 28.Brown K.K., Rehm H.L. Molecular diagnosis of hearing loss. Curr. Protoc. Hum. Genet. 2012;72:9.16.1–9.16.16.. doi: 10.1002/0471142905.hg0916s72. [DOI] [PubMed] [Google Scholar]
- 29.Matos T.D., Caria H., Simoes-Teixeira H., Aasen T., Nickel R., Jagger D.J., O’Neill A., Kelsell D.P., Fialho G. A novel hearing loss-related mutation occurring in the GJB2 basal promoter. J. Med. Genet. 2007;44:721–725. doi: 10.1136/jmg.2007.050682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sirmaci A., Akcayoz-Duman D., Tekin M. The c.IVS1+1G>A mutation in the GJB2 gene is prevalent and large deletions involving the GJB6 gene are not present in the Turkish population. J. Genet. 2006;85:213–216. doi: 10.1007/BF02935334. [DOI] [PubMed] [Google Scholar]
- 31.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi Y., Chan A.P. PROVEAN web server: A tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31:2745–2747. doi: 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang H., Thomas P.D. PANTHER-PSEP: Predicting disease-causing genetic variants using position-specific evolutionary preservation. Bioinformatics. 2016;32:2230–2232. doi: 10.1093/bioinformatics/btw222. [DOI] [PubMed] [Google Scholar]
- 34.Schwarz J.M., Cooper D.N., Schuelke M., Seelow D. MutationTaster2: Mutation prediction for the deep-sequencing age. Nat. Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 35.Shihab H.A., Gough J., Cooper D.N., Stenson P.D., Barker G.L.A., Edwards K.J., Day I.N.M., Gaunt T.R. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden markov models. Hum. Mutat. 2013;34:57–65. doi: 10.1002/humu.22225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 37.Posukh O., Pallares-Ruiz N., Tadinova V., Osipova L., Claustres M., Roux A.-F. First molecular screening of deafness in the Altai Republic population. BMC Med. Genet. 2005;6:12. doi: 10.1186/1471-2350-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tekin M., Xia X.-J., Erdenetungalag R., Cengiz F.B., White T.W., Radnaabazar J., Dangaasuren B., Tastan H., Nance W.E., Pandya A. GJB2 mutations in Mongolia: Complex alleles, low frequency, and reduced fitness of the deaf. Ann. Hum. Genet. 2010;74:155–164. doi: 10.1111/j.1469-1809.2010.00564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maeda S., Nakagawa S., Suga M., Yamashita E., Oshima A., Fujiyoshi Y., Tsukihara T. Structure of the connexin 26 gap junction channel at 3.5 Å resolution. Nature. 2009;458:597–602. doi: 10.1038/nature07869. [DOI] [PubMed] [Google Scholar]
- 40.Seeman P., Sakmaryová I. High prevalence of the IVS 1 + 1 G to A/GJB2 mutation among Czech hearing impaired patients with monoallelic mutation in the coding region of GJB2: IVS1 + 1 G to A GJB2 mutation in Czech. Clin. Genet. 2006;69:410–413. doi: 10.1111/j.1399-0004.2006.00602.x. [DOI] [PubMed] [Google Scholar]
- 41.Yuan Y., Yu F., Wang G., Huang S., Yu R., Zhang X., Huang D., Han D., Dai P. Prevalence of the GJB2 IVS1+1G > A mutation in Chinese hearing loss patients with monoallelic pathogenic mutation in the coding region of GJB2. J. Transl. Med. 2010;8:127. doi: 10.1186/1479-5876-8-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bazazzadegan N., Nikzat N., Fattahi Z., Nishimura C., Meyer N., Sahraian S., Jamali P., Babanejad M., Kashef A., Yazdan H., et al. The spectrum of GJB2 mutations in the Iranian population with non-syndromic hearing loss—A twelve year study. Int. J. Pediatr. Otorhinolaryngol. 2012;76:1164–1174. doi: 10.1016/j.ijporl.2012.04.026. [DOI] [PubMed] [Google Scholar]
- 43.Barashkov N.A., Pshennikova V.G., Posukh O.L., Teryutin F.M., Solovyev A.V., Klarov L.A., Romanov G.P., Gotovtsev N.N., Kozhevnikov A.A., Kirillina E.V., et al. Spectrum and frequency of the GJB2 gene pathogenic variants in a large cohort of patients with hearing impairment living in a subarctic region of Russia (the Sakha Republic) PLoS ONE. 2016;11:e0156300. doi: 10.1371/journal.pone.0156300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Erdenechuluun J., Lin Y.-H., Ganbat K., Bataakhuu D., Makhbal Z., Tsai C.-Y., Lin Y.-H., Chan Y.-H., Hsu C.-J., Hsu W.-C., et al. Unique spectra of deafness-associated mutations in Mongolians provide insights into the genetic relationships among Eurasian populations. PLoS ONE. 2018;13:e0209797. doi: 10.1371/journal.pone.0209797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu C.-C., Chen P.-J., Chiu Y.-H., Lu Y.-C., Wu M.-C., Hsu C.-J. Prospective mutation screening of three common deafness genes in a large taiwanese cohort with idiopathic bilateral sensorineural hearing impairment reveals a difference in the results between families from hospitals and those from rehabilitation facilities. Audiol. Neurotol. 2008;13:172–181. doi: 10.1159/000112425. [DOI] [PubMed] [Google Scholar]
- 46.Dai P., Yu F., Han B., Liu X., Wang G., Li Q., Yuan Y., Liu X., Huang D., Kang D., et al. GJB2 mutation spectrum in 2063 Chinese patients with nonsyndromic hearing impairment. J. Transl. Med. 2009;7:26. doi: 10.1186/1479-5876-7-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Y., Ao L., Ding H., Zhang D. Genetic frequencies related to severe or profound sensorineural hearing loss in Inner Mongolia Autonomous Region. Genet. Mol. Biol. 2016;39:567–572. doi: 10.1590/1678-4685-gmb-2015-0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Putcha G.V., Bejjani B.A., Bleoo S., Booker J.K., Carey J.C., Carson N., Das S., Dempsey M.A., Gastier-Foster J.M., Greinwald J.H., et al. A multicenter study of the frequency and distribution of GJB2 and GJB6 mutations in a large North American cohort. Genet. Med. 2007;9:413–426. doi: 10.1097/GIM.0b013e3180a03276. [DOI] [PubMed] [Google Scholar]
- 49.Choi S.-Y., Lee K.Y., Kim H.-J., Kim H.-K., Chang Q., Park H.-J., Jeon C.-J., Lin X., Bok J., Kim U.-K. Functional evaluation of GJB2 variants in nonsyndromic hearing loss. Mol. Med. 2011;17:550–556. doi: 10.2119/molmed.2010.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao F.-F., Ji Y.-B., Wang D.-Y., Lan L., Han M.-K., Li Q., Zhao Y., Rao S., Han D., Wang Q.-J. Phenotype–genotype correlation in 295 Chinese deaf subjects with biallelic causative mutations in the GJB2 gene. Genet. Test. Mol. Biomark. 2011;15:619–625. doi: 10.1089/gtmb.2010.0192. [DOI] [PubMed] [Google Scholar]
- 51.Chen W., Huang Y., Yang X., Duan B., Lu P., Wang Y., Xu Z. The homozygote p.V27I/p.E114G variant of GJB2 is a putative indicator of nonsyndromic hearing loss in Chinese infants. Int. J. Pediatr. Otorhinolaryngol. 2016;84:48–51. doi: 10.1016/j.ijporl.2016.02.024. [DOI] [PubMed] [Google Scholar]
- 52.Choung Y.H., Moon S.-K., Park H.-J. Functional study of GJB2 in hereditary hearing loss. Laryngoscope. 2002;112:1667–1671. doi: 10.1097/00005537-200209000-00026. [DOI] [PubMed] [Google Scholar]
- 53.Wilcox S.A., Saunders K., Osborn A.H., Arnold A., Wunderlich J., Kelly T., Collins V., Wilcox L.J., McKinlay Gardner R., Kamarinos M., et al. High frequency hearing loss correlated with mutations in the GJB2 gene. Hum. Genet. 2000;106:399–405. doi: 10.1007/s004390000273. [DOI] [PubMed] [Google Scholar]
- 54.Del Castillo I., Moreno-Pelayo M.A., del Castillo F.J., Brownstein Z., Marlin S., Adina Q., Cockburn D.J., Pandya A., Siemering K.R., Chamberlin G.P., et al. Prevalence and evolutionary origins of the del(GJB6-D13S1830) mutation in the DFNB1 locus in hearing-impaired subjects: A multicenter study. Am. J. Hum. Genet. 2003;73:1452–1458. doi: 10.1086/380205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Azaiez H., Chamberlin G.P., Fischer S.M., Welp C.L., Prasad S.D., Taggart R.T., Castillo I., del Camp G.V., Smith R.J.H. GJB2: The spectrum of deafness-causing allele variants and their phenotype. Hum. Mutat. 2004;24:305–311. doi: 10.1002/humu.20084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Angeli S.I. Phenotype/genotype correlations in a DFNB1 cohort with ethnical diversity. Laryngoscope. 2008;118:2014–2023. doi: 10.1097/MLG.0b013e31817fb7ad. [DOI] [PubMed] [Google Scholar]
- 57.Pollak A., Mueller-Malesinska M., Skorka A., Kostrzewa G., Oldak M., Korniszewski L., Skarzynski H., Ploski R. GJB2 and hearing impairment: Promoter defects do not explain the excess of monoallelic mutations. J. Med. Genet. 2008;45:607–608. doi: 10.1136/jmg.2008.059873. [DOI] [PubMed] [Google Scholar]
- 58.Kashef A., Nikzat N., Bazzazadegan N., Fattahi Z., Sabbagh-Kermani F., Taghdiri M., Azadeh B., Mojahedi F., Khoshaeen A., Habibi H., et al. Finding mutation within non-coding region of GJB2 reveals its importance in genetic testing of Hearing Loss in Iranian population. Int. J. Pediatr. Otorhinolaryngol. 2015;79:136–138. doi: 10.1016/j.ijporl.2014.11.024. [DOI] [PubMed] [Google Scholar]
- 59.Burke W.F., Warnecke A., Schöner-Heinisch A., Lesinski-Schiedat A., Maier H., Lenarz T. Prevalence and audiological profiles of GJB2 mutations in a large collective of hearing impaired patients. Hear. Res. 2016;333:77–86. doi: 10.1016/j.heares.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 60.Kim S.Y., Kim A.R., Kim N.K.D., Lee C., Kim M.Y., Jeon E.-H., Park W.-Y., Choi B.Y. Unraveling of enigmatic hearing-impaired GJB2 single heterozygotes by massive parallel sequencing: DFNB1 or not? Medicine (Baltimore) 2016;95:e3029. doi: 10.1097/MD.0000000000003029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parzefall T., Frohne A., Koenighofer M., Kirchnawy A., Streubel B., Schoefer C., Frei K., Lucas T. Whole-exome sequencing to identify the cause of congenital sensorineural hearing loss in carriers of a heterozygous GJB2 mutation. Eur. Arch. Otorhinolaryngol. 2017;274:3619–3625. doi: 10.1007/s00405-017-4699-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tu Z.J., Kiang D.T. Mapping and characterization of the basal promoter of the human connexin26 gene. Biochim. Biophys. Acta—Gene Struct. Expr. 1998;1443:169–181. doi: 10.1016/S0167-4781(98)00212-7. [DOI] [PubMed] [Google Scholar]
- 63.Zoll B., Petersen L., Lange K., Gabriel P., Kiese-Himmel C., Rausch P., Berger J., Pasche B., Meins M., Gross M., et al. Evaluation of Cx26/GJB2 in German hearing impaired persons: Mutation spectrum and detection of disequilibrium between M34T (c.101T>C) and −493del10. Hum. Mutat. 2003;21:98. doi: 10.1002/humu.9098. [DOI] [PubMed] [Google Scholar]
- 64.Mani R.S., Ganapathy A., Jalvi R., Srikumari Srisailapathy C.R., Malhotra V., Chadha S., Agarwal A., Ramesh A., Rangasayee R.R., Anand A. Functional consequences of novel connexin 26 mutations associated with hereditary hearing loss. Eur. J. Hum. Genet. 2009;17:502–509. doi: 10.1038/ejhg.2008.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Matos T.D., Simões-Teixeira H., Caria H., Cascão R., Rosa H., O’Neill A., Dias Ó., Andrea M.E., Kelsell D.P., Fialho G. Assessing noncoding sequence variants of GJB2 for hearing loss association. Genet. Res. Int. 2011;2011:1–8. doi: 10.4061/2011/827469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gandía M., del Castillo F.J., Rodríguez-Álvarez F.J., Garrido G., Villamar M., Calderón M., Moreno-Pelayo M.A., Moreno F., del Castillo I. A novel splice-site mutation in the GJB2 gene causing mild postlingual hearing impairment. PLoS ONE. 2013;8:e73566. doi: 10.1371/journal.pone.0073566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.del Castillo F.J. A novel deletion involving the connexin-30 gene, del(GJB6-d13s1854), found in trans with mutations in the GJB2 gene (connexin-26) in subjects with DFNB1 non-syndromic hearing impairment. J. Med. Genet. 2005;42:588–594. doi: 10.1136/jmg.2004.028324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Feldmann D., Le Maréchal C., Jonard L., Thierry P., Czajka C., Couderc R., Ferec C., Denoyelle F., Marlin S., Fellmann F. A new large deletion in the DFNB1 locus causes nonsyndromic hearing loss. Eur. J. Med. Genet. 2009;52:195–200. doi: 10.1016/j.ejmg.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 69.Wilch E., Azaiez H., Fisher R., Elfenbein J., Murgia A., Birkenhäger R., Bolz H., Da Silva-Costa S., del Castillo I., Haaf T., et al. A novel DFNB1 deletion allele supports the existence of a distant cis-regulatory region that controls GJB2 and GJB6 expression. Clin. Genet. 2010;78:267–274. doi: 10.1111/j.1399-0004.2010.01387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bliznetz E.A., Makienko O.N., Okuneva E.G., Markova T.G., Polyakov A.V. New recurrent large deletion, encompassing both GJB2 and GJB6 genes, results in isolated sensorineural hearing impairment with autosomal recessive mode of inheritance. Russ. J. Genet. 2014;50:415–420. doi: 10.1134/S1022795414020045. [DOI] [PubMed] [Google Scholar]
- 71.Tayoun A.N.A., Mason-Suares H., Frisella A.L., Bowser M., Duffy E., Mahanta L., Funke B., Rehm H.L., Amr S.S. Targeted droplet-digital PCR as a tool for novel deletion discovery at the DFNB1 locus. Hum. Mutat. 2016;37:119–126. doi: 10.1002/humu.22912. [DOI] [PubMed] [Google Scholar]
- 72.Pshennikova V.G., Barashkov N.A., Solovyev A.V., Romanov G.P., Diakonov E.E., Sazonov N.N., Morozov I.V., Bondar A.A., Posukh O.L., Dzhemileva L.U., et al. Analysis of GJB6 (Cx30) and GJB3 (Cx31) genes in deaf patients with monoallelic mutations in GJB2 (Cx26) gene in the Sakha Republic (Yakutia) Russ. J. Genet. 2017;53:688–697. doi: 10.1134/S1022795417030103. [DOI] [Google Scholar]
- 73.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Posukh O.L. (Institute of Cytology and Genetics, Novosibirsk, Russia). Personal Communication. 2019
- 75.Yang X.-L., Xu B.-C., Chen X.-J., Bian P.-P., Ma J.-L., Liu X.W., Zhang Z.-W., Wan D., Zhu Y.-M., Guo Y.-F. Common molecular etiology of patients with nonsyndromic hearing loss in Tibetan, Tu nationality, and Mongolian patients in the northwest of China. Acta Otolaryngol. (Stockh.) 2013;133:930–934. doi: 10.3109/00016489.2013.795288. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.