Abstract
Breast cancer prevention is daunting, yet not an unsurmountable goal. Mammary stem and progenitors have been proposed as the cells‐of‐origin in breast cancer. Here, we present the concept of limiting these breast cancer precursors as a risk reduction approach in high‐risk women. A wealth of information now exists for phenotypic and functional characterization of mammary stem and progenitor cells in mouse and human. Recent work has also revealed the hormonal regulation of stem/progenitor dynamics as well as intrinsic lineage distinctions between mammary epithelial populations. Leveraging these insights, molecular marker‐guided chemoprevention is an achievable reality.
Keywords: breast cancer prevention, cell‐of‐origin, high‐risk women, mammary stem and progenitors, progesterone
Subject Categories: Cancer, Molecular Biology of Disease
Introduction
Substantial advances have been made in our understanding of breast cancer etiology, and there is avid interest in cancer prevention. However, molecular‐guided chemoprevention approaches remain limited, and primary preventive options for women at highest risk of breast cancer continue to be centered on irreversible prophylactic surgeries (Hartmann et al, 1999; Kauff et al, 2002; Eisen et al, 2005; Domchek et al, 2010; Kotsopoulos et al, 2017) that harshly impact quality of life (Guillem et al, 2006; NICE, 2013). Chemoprevention, defined by Sporn in 1976, is the use of natural, synthetic, or biological agents to reverse, suppress, or prevent either the initial phases of carcinogenesis as primary prevention or the progression of premalignant cells to invasive disease (Sporn, 1976). Two parameters intrinsic to the success of chemoprevention are that individuals requiring risk reduction measures be identifiable and that treatments to safely, effectively, and precisely target premalignant lesion be well established (Sporn, 1976). Major strides have been made toward achieving the first goal, while the second remains elusive.
Empowered with new understanding in breast biology through the identification, characterization, and regulation of mammary epithelial subpopulations, we are now poised to revolutionize chemoprevention. Normally, mammary stem cells and progenitors give rise to discrete alveolar structures that repeatedly form in the adult breast and their activity is necessary for normal mammary homeostasis. Yet, stem cells and progenitors are considered the cell‐of‐origin in many breast cancers. Therefore, pre‐emptively eliminating these cancer precursors provides the basis for targeted prevention. Here, we review the current state of chemoprevention for high‐risk patients and evidence for mammary stem and progenitors as breast cancer cells‐of‐origin. Beyond estrogen, we outline the mitogenic role of progesterone and its effectors in the adult breast, underscoring their potential in restraining unwarranted mammary cellular expansion. We highlight stem/progenitor molecular vulnerabilities uncovered through OMICs studies and propose a pipeline for discovery of targets for molecular interception. Finally, we discuss some of the challenges and open questions in the complex field of chemoprevention.
Current chemoprevention in breast cancer
Defining high‐risk women
Breast cancer continues to be the most frequent cancer in females, affecting about 1 in 8 and causing the greatest number of cancer‐related deaths in women worldwide (Bray et al, 2018). Women at high‐risk for breast cancer include those with inherited mutations in the BRCA1 or BRCA2 genes and have a ~ 72 and ~ 69% lifetime risk of developing breast cancer by the age of 80, respectively (Kuchenbaecker et al, 2017). A particularly high lifetime risk is also conferred by pathogenic mutations in PTEN (Cowden syndrome, ≥ 25–50%; Tan et al, 2012; Evans et al, 2018), TP53 (Li–Fraumeni syndrome, 49–90%; Masciari et al, 2012; Evans et al, 2018), PALB2 (33–58%; Antoniou et al, 2014), CDH1 (40–54%; Kaurah et al, 2007), and STK11 (Peutz–Jeghers syndrome, 45%; Hearle et al, 2006). Other mutations recognized to correlate with increased breast cancer risk occur in genes including ATM, BRIP1, CHEK2, MRE11A, MSH6, NBN, NF1, PMS2, RAD50, RAD51C, and SEC23B; many of which play a role in DNA damage response pathways (Walsh et al, 2006; Campeau et al, 2008; Antoniou et al, 2014; Easton et al, 2015; Kurian et al, 2017). Large genome‐wide association studies have also exposed genes positively associated with breast cancer susceptibility including FGFR2, TNRC9, MAP3K1, and LSP1, which are related to cell growth and signaling (Easton et al, 2007). Finally, genome‐wide single‐nucleotide polymorphism studies have identified > 125 loci associated with genetic breast cancer susceptibility (Easton et al, 2007; Milne et al, 2017; Evans et al, 2018).
In addition to genetic predisposition, the following criteria are also used to clinically identify women at high‐risk: (i) first‐degree relative with a breast cancer diagnosis before 50 years of age; (ii) history of atypical hyperplasia; (iii) history of lobular carcinoma in situ (LCIS); (iv) chest radiation between 10 and 30 years of age; (v) 5‐year risk of ≥ 1.7% by Gail model; and (vi) lifetime risk of ≥ 20% by International Breast Cancer Intervention Study (IBIS) model (Bevers et al, 2018). The Gail model (The Breast Cancer Risk Assessment Tool, BCRAT) is a computer‐based clinical risk assessment tool used to estimate risk in the next 5 years and up to 90 years, while IBIS, BOADICEA, and BRAPRO are examples of other risk prediction models that also incorporate the effects of genetic susceptibility. Recent NCCN guidelines recommend risk reduction strategies for women with a lifetime risk of ≥ 20% (Bevers et al, 2018). The ability to pre‐emptively identify women at increased risk for breast cancer is constantly evolving, with an expanding and better‐defined patient pool eligible for chemoprevention, whereas management and risk reduction options are lagging (Fig 1).
Figure 1. The breast: structure, risk factors and stages of cancer development.
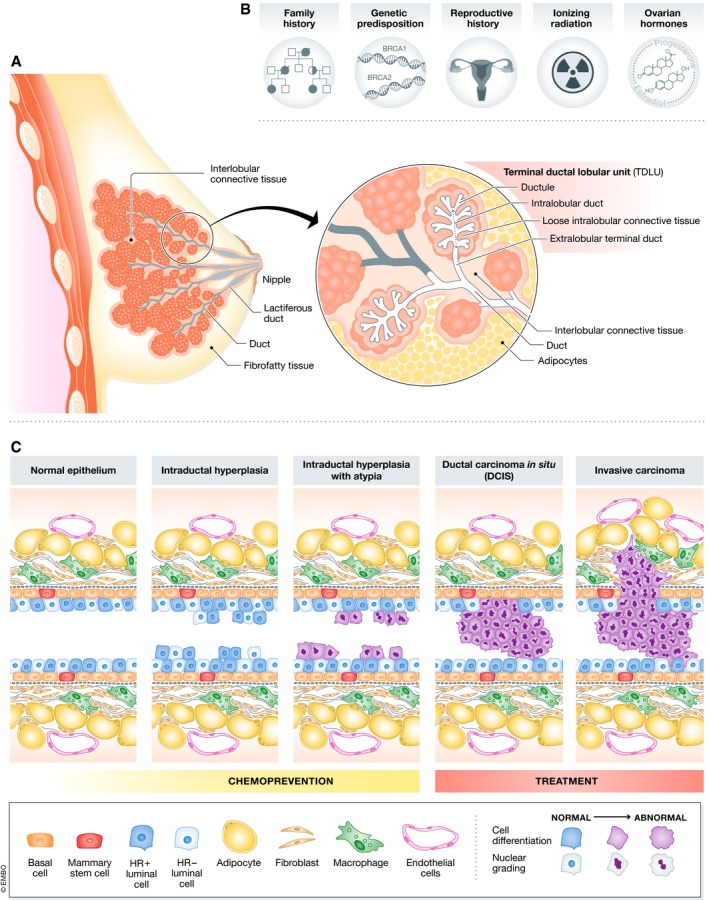
(A) Schematic of the human breast highlighting terminal ductal lobular units (TDLUs), the site of origin in a number of breast cancers. (B) Some of the major risk factors underlying high‐risk status for breast cancer. (C) Schematic of a ductal cross‐section, depicting the progression of breast cancer from normal bi‐layered epithelium to hyperplasia, to hyperplasia with atypia, to ductal carcinoma in situ, and finally to invasive disease.
Management strategies for high‐risk patients
Invasive surgery and dated hormonal therapies are the main standard of care options for intervention in women at high‐risk of developing breast cancers. Bilateral prophylactic mastectomy is generally considered for women with very strong family history and/or known gene mutations of high penetrance. It is shown to reduce breast cancer risk by at least 95% in women with BRCA1 or BRCA2 mutation and by 90% for those with a strong family history. Since BRCA1/2 carriers are also at risk of developing highly aggressive ovarian cancers with poor long‐term survival, bilateral salpingo‐oophorectomy (BSO) is also often considered. BSO, which removes the endogenous source of ovarian hormones, has been shown to reduce the risk of breast cancer by 50% in these patient groups (Rebbeck et al, 2004; Eisen et al, 2005; Domchek et al, 2010). In BRCA2 carriers, these effects are mainly observed in the first 3–5 years as they typically present with ER+ positive breast cancer and benefit from the immediate hormone deprivation (Lakhani et al, 2005; Evans et al, 2018). This efficacy of BSO was not seen in BRCA1 carriers who predominantly develop ER‐ cancers, although reduction of contralateral breast cancer with tamoxifen was observed, warranting longer follow‐up in this high‐risk group (Lakhani et al, 2005; Evans et al, 2018).
Estrogen‐centric strategies continue to dominate the breast cancer prevention field. Currently, the selective estrogen receptor modulators (SERMs) tamoxifen and raloxifene are the first‐line chemopreventive agents (King et al, 2001; Fisher et al, 2005; Vogel et al, 2006, 2010; Goss et al, 2011; Cuzick et al, 2014, 2015). Tamoxifen is recommended for 5 years to both pre‐ and post‐menopausal high‐risk women, shows a risk reduction of up to 50% and protective effects for up to 20 years after cessation of use. Women at moderate risk are also advised to consider tamoxifen for 5 years, if premenopausal (NICE, 2013). Data compiled from six studies showed tamoxifen is more effective at reducing the incidence of ER+ cancers (350 vs. 632 cases) but not the more aggressive ER− breast cancers (173 vs. 144 cases; Fisher et al, 2005; Powles et al, 2007; Veronesi et al, 2007; Goss et al, 2011; Cuzick et al, 2014, 2015; Narod, 2015). There is also concern as to whether tamoxifen use reduces mortality rates since a reduction in deaths was not observed (Narod, 2015). Moreover, tamoxifen is not recommended for patients with a past history/increased risk of thromboembolic disease or endometrial cancer and has a low compliance rate due to side‐effects (Nelson et al, 2013; Padamsee et al, 2017). Raloxifene is only recommended to post‐menopausal high‐risk individuals, and a risk reduction of 38% is anticipated. The STAR trial showed that raloxifene, although better tolerated, was inferior for longer term protection compared to tamoxifen (Vogel et al, 2010). Other SERMs such as lasofoxifene and arzoxifene have shown similar risk reduction in post‐menopausal women with osteoporosis but these agents have not been introduced into clinical practice (Suh et al, 2001; LaCroix et al, 2010; Powles et al, 2012). Beyond SERMs, aromatase inhibitors (AIs) that block the enzymatic conversion of androgen to estrogen are currently used off‐label for risk reduction but are not FDA approved for chemoprevention (Goss et al, 2011; Cuzick et al, 2014). High‐risk post‐menopausal women are offered the AI anastrozole or exemestane unless suffering from severe osteoporosis. In the IBISII trial, anastrozole reduced incidence by 50% after 3.5 years of follow‐up while a 60% reduction was observed with exemestane after 2.5 years of follow‐up in the recent MAP3 trial (Goss et al, 2011; Cuzick et al, 2014). Neither AI is recommended for premenopausal patients.
Other drugs have also been investigated as breast cancer chemopreventive agents in retrospective studies. Bisphosphonates (in osteoporosis trials) were found ineffective in decreasing the risk of invasive post‐menopausal breast cancers after 3–4 years of treatment (Hue et al, 2014); fenretinide, a retinoic acid derivative, was tested with second primary breast cancer as the endpoint in two randomized controlled trials with discordant results (Costa et al, 2006; Decensi et al, 2009). A subsequent randomized placebo‐controlled trial in 2009 (NCT01479192) was terminated early due to slow accrual, possibly due to recognized fenretinide toxicity, highlighting the importance of long‐term tolerability for prevention agents. There have also been serendipitous findings of chemopreventive agents from studying cancer‐related outcomes in patients with co‐morbidities. Diabetic patients taking metformin were found to have an overall decrease in cancer incidence compared to non‐users (Evans et al, 2005). Metformin has been reported to decrease the risk of incidence in diabetic women (Bosco et al, 2011; Chlebowski et al, 2012), whether its preventative effects would be seen in non‐diabetic women is unknown. In addition, multiple clinical studies have demonstrated that statins, the cholesterol decreasing medication, reduce the risk of breast cancer incidence (Cauley et al, 2006) and recurrence after diagnosis (Ahern et al, 2011). Other recommendations for breast cancer prevention include limiting the duration and dose of hormone replacement therapy, lowering alcohol consumption, maintaining a healthy body weight and lifestyle, and breast‐feeding. Altogether, despite the clinical introduction of breast cancer chemoprevention agents in 1998, tamoxifen remains the only FDA‐approved chemoprevention option available to premenopausal women. Given that high‐risk women develop aggressive tumors before menopause, it is necessary to develop innovative chemoprevention options. A recent review by Evans et al (2018) comprehensively delves into the management of high‐risk women.
New cellular targets in chemoprevention
The hormonal axis has been the preferred target in breast cancer prevention, but evidence pointing to epithelial subsets as the cells‐of‐origin in breast cancer is creating a paradigm shift. Specifically, mammary stem and progenitors are now purported as the cells that undergo transformation (Visvader & Stingl, 2014) and limiting these cancer precursors offers a promising approach (Casey et al, 2018). The potential of a cell‐based strategy was recognized early, beginning with pathology studies that highlighted the continuum of breast cancer progression from a premalignant cell to invasive disease (DeOme & Medina, 1969; Wellings et al, 1975; Sporn, 1976), which take years to progress. Alterations detected in normal, tumor‐adjacent epithelium that confer oncogenic potential include allelic imbalance, loss of heterozygosity, hypermethylation of promoters (e.g., p16INK4a), upregulation of epigenetic regulators (e.g., EZH2), or aberrant activation of signaling pathways (e.g., p38; Wellings et al, 1975; Sporn, 1976; Deng et al, 1996; Lakhani et al, 1999; Porter et al, 2003; Chin et al, 2004; Ellsworth et al, 2004; Larson et al, 2005; Clarke et al, 2006; Yao et al, 2006; Tripathi et al, 2008). Rational development of preventive strategies aimed at cancer precursors necessitates that we understand the cellular composition of the breast as well as the cellular subsets that underlie breast cancer subtypes.
Defining breast stem cells and progenitor subsets
The adult breast undergoes multiple periods of robust change marked with striking growth and structural remodeling indicative of activated stem/progenitor cell pools. The human breast develops postnatally into an organized ductal tree, and the murine mammary ductal tree is fundamentally similar. In humans, radially branching ducts end in functional pyramidal lobules termed terminal ductal lobuloalveolar units (TDLUs), and analogous lobuloalveolar structures are found in the mouse, making it an instructive model to study the mammary epithelial cell hierarchy and breast cancer. TDLUs ultimately differentiate into milk‐secreting acini during lactation but also repeatedly form and regress over the reproductive lifespan of a female. Specifically, there is a 10‐fold increase in alveoli per lobule and de novo lobular formation in pregnancy, as well as significant proliferation during each menstrual cycle, underscoring the gland's regenerative potential (Potten et al, 1988; Russo & Russo, 2004). Sustained TDLUs due to incomplete involution either age‐related or post‐gestation are known to increase breast cancer risk. Importantly, TDLUs are thought of as the site(s) of origin for the majority of human breast cancers (Wellings, 1980) and contain stem and progenitor cells (Wellings et al, 1975). Microdissected TDLUs from human breast tissue show conserved X inactivation patterns throughout, implying clonal origins, with entire TDLUs originating from the same progenitor (Tsai et al, 1996; Diallo et al, 2001). Similarly, entire ducts or lobules with identical patterns of loss of heterozygosity have been reported implicating a common progenitor (Lakhani et al, 1999). Restricting the appropriate stem/progenitor pool would block cellular expansion, diminish TDLU turnover, and intercept breast cancer establishment at its source.
At the cellular level, both ducts and TDLUs are bi‐layered with two lineages: an inner luminal epithelial layer with cells expressing cytokeratin 8 (K8), K18 as well as hormone receptors (HR, ER/PR), and an outer myoepithelial/basal layer expressing K5, K14, p63, and SMA. Importantly, these two lineages contain defined stem/progenitor‐enriched subpopulations. Cell surface markers used to segregate mammary epithelial cells include MUC1, EpCAM, CD49f, CD24, CD29, CD133, Thy1, CD10, and ALDH in humans and CD24, CD29, CD49f, EpCAM, CD49b, Sca1, Prominin‐1 (human CD133), and CD61 in the mouse (Stingl et al, 2001, 2006; Shackleton et al, 2006; Eirew et al, 2008; Shehata et al, 2012). Flow cytometry in conjunction with in vivo limiting dilution assays and in vitro colony‐forming capacity (CFC) assays has been used to enumerate stem and progenitor activity. Colonies from the human breast have been morphologically scored as basal, luminal, and mixed colonies that likely originate from basal, luminal, and bi‐potent progenitors, respectively. Commonly, EpCAM−CD49fhi is used to mark basal cells, EpCAM+CD49flo non‐clonogenic luminal cells, and EpCAM+CD49fhi for luminal progenitors, where ALDH+ is used specifically to further enrich for progenitors with an alveolar signature and this fraction expresses low levels of luminal cell differentiation (Stingl et al, 2001; Eirew et al, 2012; Shehata et al, 2012). EpCAM+MUC1− cells express high K19 and form branched structures similar to TDLUs in 3D cultures and in vivo, indicating an enrichment for a TDLU precursor (Gudjonsson et al, 2002). Other markers that further segregate luminal cells include GATA3, ErbB3, and ALDH (Asselin‐Labat et al, 2007; Ginestier et al, 2007; Shehata et al, 2012) in humans, while CD14 (Shehata et al, 2012), c‐Kit (Lim et al, 2009; Regan et al, 2012), and Elf5 (Zhou et al, 2005; Oakes et al, 2008) segregate the murine luminal compartment. Finally, Lgr5 (Van Keymeulen et al, 2011; Plaks et al, 2013), Procr (Wang et al, 2015), Tspan8 (Fu et al, 2017), Dll1 (Chakrabarti et al, 2018), and Bcl11b (Cai et al, 2017), each further enriches basal subsets for mammary stem cells, although an exclusive mammary stem cell signature remains elusive. Nuances in protocols from tissue dissociation to preferred cell surface markers and stem/progenitor readouts contribute to some discordant findings. Moreover, the potent effect of hormones on the mammary epithelium continues to be overlooked and not controlled for in most studies. However, a recent study has shown that ovarian hormones trigger a heterogeneous cell cycle response in the epithelial subsets (Shehata et al, 2018). Similarly, CD61 has been an unreliable luminal progenitor marker but the protein itself is now known to be downregulated by progesterone (Casey et al, 2018).
Knowledge of parent–progeny cell relationships and the mammary epithelial hierarchy is important to pinpoint the most effective precursor population to target in prevention. Following orthotopic transplantation in vivo, only basal cells possess the functional capacity to generate a full ductal tree, with the ability to both self‐renew and contribute to all subsequent lineages (Shackleton et al, 2006; Stingl et al, 2006; Spike et al, 2012; Wang et al, 2015). In the human breast, luminal and basal cells in the same region possess identical chromosomal alterations, implicating a shared ancestry (Deng et al, 1996) and single‐cell RNA‐Seq (scRNA‐seq) of primary human breast epithelial cells found a continuous lineage hierarchy that connected the basal lineage to two differentiated luminal branches (Nguyen et al, 2018). These studies, together with select lineage tracing reports, document the presence of a bi‐potent basal population (Rios et al, 2014; Wang et al, 2015). However, other lineage tracing studies challenge this classical view, proposing instead that the combined action of unipotent basal and luminal progenitors maintains the mammary gland (Van Keymeulen et al, 2011; Giraddi et al, 2015; Wuidart et al, 2016; Wang et al, 2017). Despite ongoing discussion in this area, a considerable body of work supports specific mammary populations as the candidate cells‐of‐origin for breast cancer subtypes. Finally, techniques such as scRNA‐seq and mass cytometry (CyTOF) are exposing heterogeneity within the basal and luminal compartments (Bach et al, 2017; Pal et al, 2017; Giraddi et al, 2018; Nguyen et al, 2018), indicating the necessity of further dissecting precursor–progeny relationships and developmental correlates of tumorigenesis.
Cells‐of‐origin in breast cancer
Cell‐of‐origin refers to a founder cell that acquires mutations leading to clonal expansion and eventual tumorigenesis (Visvader & Stingl, 2014). Stem and progenitor cells are considered cell‐of‐origin in many cancers, and their defining features of replicative potential and long cellular lifespan render them susceptible to accumulating mutations. It has been recently proposed that variations in cancer risk among tissues can be explained by the number of stem cell divisions (Tomasetti & Vogelstein, 2015; Tomasetti et al, 2017). The study's meta‐analysis of relationships between stem cell divisions and the risk of 17 different cancer types across 69 countries supports the concept that lifetime risk strongly correlates with the total number of divisions of the normal self‐renewing cells that maintain tissue's homeostasis (Tomasetti et al, 2017). This concept is especially relevant to the breast which undergoes repeated cycles of cell divisions and differentiation throughout the reproductive female lifespan (Fata et al, 2000a; Joshi et al, 2010). Two‐thirds of mutations in human cancers arise during somatic cell division due to DNA replication errors regardless of environmental factors (Tomasetti et al, 2017), and consistent with this, a large proportion of breast cancer risk genes mentioned above are involved in DNA damage repair.
Core pathways in stem cell biology have been identified as key drivers in aggressive breast cancers. Wnt is implicated in self‐renewal (Badders et al, 2009; Zeng & Nusse, 2010) and maintains adult stem cells in multiple tissues including the breast. One of the first genes whose ectopic expression was sufficient to induce mammary carcinogenesis, its overexpression in MMTV‐Wnt1 mice (Tsukamoto et al, 1988), leads to the expansion of mammary stem cell pools followed by tumorigenesis (Liu et al, 2004; Shackleton et al, 2006), and its key signaling effector β‐catenin is elevated in > 50% of breast carcinomas (Lin et al, 2000). When overexpressed, MMTV‐c‐myc results in amplification of the stem cell compartment where transformation starts in mammary ducts resulting in morphological changes that mimic characteristics of ductal carcinoma in situ (Chepko et al, 2005). Conversely, signals that decrease the active stem cell pool, such as Tgfb1, correlate with decreased tumorigenesis. Wap‐Tgfb1 mice have fewer mammary stem cells, likely due to premature stem cell aging and senescence, and reduced tumorigenesis even though the number of premalignant lesions remains comparable in hyperplastic outgrowths seen in transplantation assays (Boulanger & Smith, 2001). Finally, beyond absolute stem cell activity, decreased clonal diversity has also been postulated to contribute to age‐related cancer risk. Interestingly, aged mice with compound deletion of protease inhibitors (Timp1 and Timp3) that negatively regulate Notch activation maintain an expanded stem cell pool without increased susceptibility to carcinogen‐induced mammary tumorigenesis (Jackson et al, 2015). Hallmarks of aging such as age‐related lobular involution, increased HR+ cells, and minor hyperplasia normally detected in aged glands were also absent (Jackson et al, 2015). Stem cell loss with aging is postulated to lead to a field of premalignant cells dominated by clones with a proliferative advantage (Klein & Simons, 2011) which has important implications given that age is the primary risk factor in breast cancer, where women experience higher cancer incidence for every decade of life (Kessler, 1992).
Another line of support comes from RNA expression profiling studies where distinct stem/progenitor populations correlate with individual breast cancer subtypes. Of the five commonly accepted subtypes, HR+ luminal A and B cancers exhibit a profile similar to that of mature ER+PR+ luminal cells, although luminal B has a stronger proliferative signature (Cheang et al, 2009; Nielsen et al, 2010; Prat & Perou, 2011). A recent study that applied a large set of epithelial markers to > 15,000 normal breast cells detected 11 differentiation states for luminal cells and subsequently classified HR+ breast cancers into four new subtypes distinct from the current known categories (Santagata et al, 2014). Claudin‐low breast cancers have expression profiles similar to the mammary stem cell‐enriched ER‐PR‐ subpopulation (Lim et al, 2009; Molyneux et al, 2010). Aggressive metastatic triple‐negative (ER−PR−HER2−) breast cancers also show remarkable gene expression similarities to fetal murine mammary stem cells at embryonic days 16 and 18, stages known to have high stem cell capacity (Spike et al, 2012; Giraddi et al, 2018). Basal‐like breast cancers have expression similarities to ER−PR− luminal progenitors (Lim et al, 2009; Molyneux et al, 2010; Shehata et al, 2012). A study has shown that BRCA1 mutation carriers possess an abnormally expanded luminal progenitor pool prior to cancer onset, and mice with tissue‐specific deletion of Brca1 and p53 in the luminal lineage develop mammary cancers that resemble human BRCA1 tumors histologically (Lim et al, 2009; Molyneux et al, 2010). Luminal progenitors are of immediate interest as the cell‐of‐origin for aggressive cancers in BRCA1 mutation carriers and targeted risk reduction (Al‐Hajj et al, 2003; Lim et al, 2009; Keller et al, 2012; Visvader & Stingl, 2014). It has also been shown that luminal progenitors can give rise to basal‐like breast cancers following oncogenic insults, irrespective of BRCA1 (Koren et al, 2015; Van Keymeulen et al, 2015; Hein et al, 2016). Finally, a population responsible for giving rise to HER2+ breast cancers has yet to be pinpointed. This cancer subtype is highly heterogeneous, consisting of both ER/PR+ and ER/PR− (Konecny et al, 2003; Cancer Genome Atlas Network, 2012). Thus, resolving heterogeneity in each of the subpopulations will help to further tease out additional putative cell(s)‐of‐origin and other non‐stem cells that may serve as cancer precursors (Bu et al, 2019; Shehata et al 2019).
Targeting mitogens to limit stem cells and progenitor activity
Just as the knowledge of relevant patient pools for primary prevention is improving, so is our comprehension of breast stem/progenitor populations. Many external cues are now known as crucial regulators of these epithelial subsets during mammary gland homeostasis opening new possibilities to leverage their control over cell‐of‐origin in breast cancer. Progesterone itself is a significant culprit in breast cancer due to its effects on the mammary epithelium (Joshi et al, 2015a). However, only ~ 1/3rd of luminal cells are HR+, while basal cells, stem cells, and most progenitors lack estrogen/progesterone receptors (ER−PR−), rendering them dependent on paracrine effectors (Brisken & Duss, 2007; Shehata et al, 2012). Chimera transplant studies show that PR null mammary epithelial cells can display alveolar development if supplied with paracrine effectors and we now know that ER+PR+ cells respond to hormonal cues and in turn stimulate ER−PR− cells (Lydon et al, 1995; Humphreys et al, 1997; Brisken et al, 1998).
Progesterone, thinking beyond estrogen
Physiological, experimental, and population‐based studies all point to progesterone as a powerful mitogen in the adult breast. The Women's Health Initiative (WHI), which followed post‐menopausal women aged 50–79 years in two US multiethnic randomized clinical trials for ~ 20 years, and Million Women's studies report a significantly greater breast cancer risk associated with hormone replacement therapy formulations that also contained progestins compared to estrogens alone (Rossouw et al, 2002; Beral, 2003; Chlebowski et al, 2015). In the WHI study, the elevated risk persists long‐term post‐intervention (Chlebowski et al, 2015). Increased lifetime exposure to ovarian hormones similarly impacts risk, with a higher cumulative number of menstrual cycles correlating to greater risk (Kelsey et al, 1993). Circulating progesterone peaks during the luteal phase of the reproductive cycle, a phase when mammographic density has also been observed to be increased by some in the breast (Morrow et al, 2010). This is also reflected in mice and can be scored blindly as a function of the estrous cycle. Higher‐order epithelial branching and alveolar mammopoiesis is seen during the progesterone‐high diestrous phase by mammary whole mounts and histology which positively correlates with serum progesterone, not 17β‐estradiol (Fata et al, 2000a; Ramakrishnan et al, 2002). This highly proliferative progesterone dominant phase, accompanied by differentiation, is followed by marked regression of mammary epithelium, apoptosis, and glandular remodeling. In fact, this transient but repeated physiology generates significant gross changes in the human breast such that histology alone can infer the follicular (progesterone‐low) vs. luteal (progesterone‐high) stage of the menstrual cycle (Fata et al, 2000a; Ramakrishnan et al, 2002; Hawkins & Matzuk, 2008). A case–control study has shown high‐risk BRCA1/2 mutation carriers have 121% higher serum progesterone levels during the luteal phase compared to non‐carriers (Widschwendter et al, 2013). The same study also observed 33% higher estradiol levels and, given that estrogen is required for robust progesterone receptor expression, these increases further underscore the importance of hormone signaling in high‐risk women. Progesterone triggers mammary stem and progenitors activity during each reproductive cycle, when measured by flow cytometry and stem cell assays (Joshi et al, 2010, 2015b; Giraddi et al, 2015; Shiah et al, 2015). A 6‐fold increase in the CD24loCD49f+ basal population, a 3‐fold increase in the CD24hiCD49f− luminal population, along with a > 10‐fold increase in mammary stem cell activity occur during the progesterone‐high diestrous stage relative to the estrogen‐high estrous stage (Stingl et al, 2006; Joshi et al, 2010). Similar quantifiable cellular fluctuations have been confirmed in the premenopausal breast, with a significant increase in EpCAM+CD49fhi luminal progenitor activity enumerated by CFC assays (Joshi et al, 2015b). Given this literature, it is striking that progesterone has been largely overlooked as a target for intervention.
Progesterone proves pro‐tumorigenic in both carcinogen‐induced or Brca1 loss‐driven breast cancer pre‐clinical models (Lydon et al, 1999; Gonzalez‐Suarez et al, 2010; Schramek et al, 2010; Tanos et al, 2013; Lee et al, 2016). Administration of synthetic progestin (medroxyprogesterone acetate, MPA), used to mimic hormone replacement therapy, results in mammary tumors whereas treatment with the PR antagonists mifepristone or telapristone acetate suppresses tumorigenesis (Poole et al, 2006; Lanari et al, 2009; Lee et al, 2016). PR knockout mice similarly show a lower breast cancer incidence (Lydon et al, 1999). PR knockout mice also lack normal side‐branching and lobular alveolar development in the adult gland (Lydon et al, 1995; Ismail et al, 2002). In breast tissue with premalignant atypia, PR isoform (PRA:PRB) ratios are perturbed, with a relative loss of PRB (Mote et al, 2002). Tumor‐adjacent normal breast tissue from BRCA1 mutation carriers also has higher PR positivity (Lydon et al, 1999; King et al, 2004). Clinically, the anti‐progestins mifepristone and onapristone have shown anti‐tumor activity in women with advanced breast cancer although subsequent development was halted due to concerns around liver toxicity. On the other hand, selective progesterone receptor modulators (SPRMs), such as ulipristal acetate, are already in use for emergency contraception, uterine fibroids, or various other gynecological disorders/reproductive problems. These SPRMs are better tolerated, making repurposing for long‐term prevention studies possible which is one of the biggest hurdles in chemoprevention. Ulipristal acetate is currently being tested in a pilot study in premenopausal women at > 17% lifetime breast cancer risk to determine effects on normal breast proliferation and the luminal progenitor population (NCT02408770). Overall, leveraging the potent influence of progesterone and its effects on mammary stem/progenitors, an anti‐progestin approach for prevention offers promise (Fig 2A).
Figure 2. Progesterone‐driven cellular and molecular changes in the mammary gland.
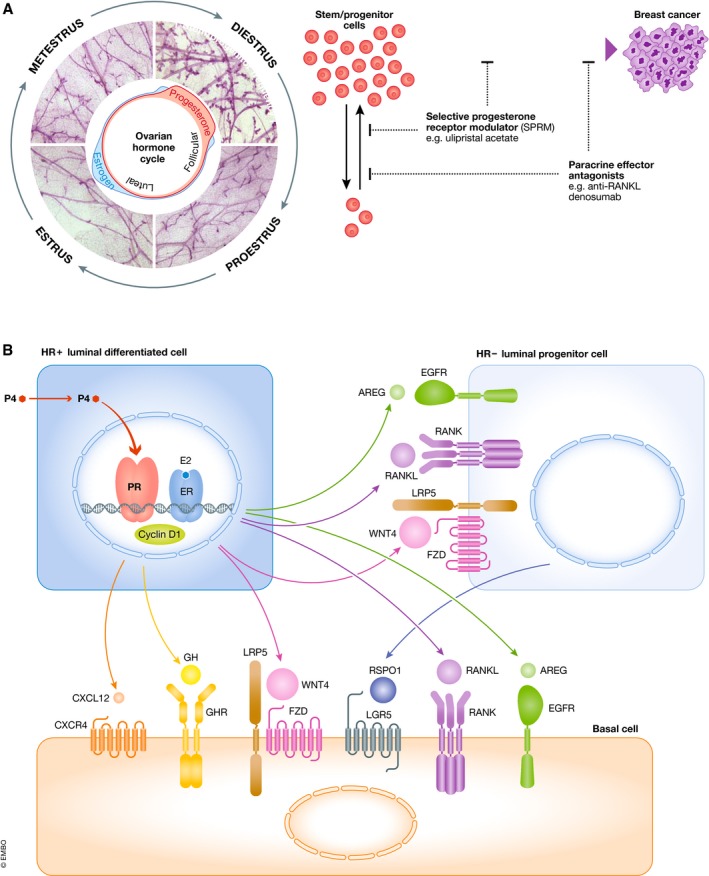
(A) The murine estrous cycle is shown with mammary whole mounts depicting the gross morphological changes elicited by fluctuations in the ovarian hormones, estrogen, and progesterone. The corresponding expansion in mammary stem and progenitors is also highlighted. Schematic illustrates the strategy of utilizing factors that limit stem/progenitor expansion as chemopreventive agents in breast cancer. Ulipristal acetate is a selective progesterone receptor modulator, and denosumab is an anti‐RANKL agent. (B) Schematic of key mitogenic paracrine effectors downstream of progesterone in the breast, some of which may prove effective as targets in future breast cancer chemoprevention strategies.
Progesterone's paracrine effectors
We now know that progesterone acts upon the ER+PR+ cells, which orchestrates a surge of potent paracrine effectors that in turn stimulate proliferation of the ER−PR− progenitor populations (Lydon et al, 1995; Humphreys et al, 1997; Brisken et al, 1998; Joshi et al, 2012; summarized in Fig 2B). Though it may have once been thought that these diverse ligands act independently of each other, work to date suggests that they are quite collaborative and converge onto this progesterone‐signaling axis responsible for side‐branching seen during the reproductive cycle and pregnancy. This is also the same axis that has been targeted to inhibit/prevent basal‐like BRCA1 breast cancer. Below we discuss the classical progesterone effectors that can potentially serve as chemoprevention targets such as RANKL, WNT, and amphiregulin (AREG).
RANKL/RANK (receptor activator of NF‐kB ligand/receptor) signaling is a core pathway in the adult mammary gland and breast cancer. It is also a key downstream paracrine effector of progesterone (Gonzalez‐Suarez et al, 2010; Schramek et al, 2010; Tanos et al, 2013; Nolan et al, 2016; Sigl et al, 2016; Kiechl et al, 2017). RANKL‐ and RANK‐deficient mice do not lactate due to defective alveolar development (Fata et al, 2000b; Gonzalez‐Suarez et al, 2010). Genetic or pharmacological inhibition of RANKL abrogates progesterone‐triggered expansion of mammary stem/progenitors essential for alveologenesis (Fata et al, 2000b; Joshi et al, 2015b). Blocking RANKL signaling also negates the normal mammary Wnt response which includes the induction of Rspondin in HR− luminal cells (Joshi et al, 2015b). Gain‐of‐function studies also show that RANK signaling promotes mammary epithelial cell proliferation and anchorage‐independent growth (Beristain et al, 2012). Loss of RANK signaling in breast cancer models drastically limits MPA‐induced and Brca1‐mediated tumorigenesis (Schramek et al, 2010; Sigl et al, 2016). In otherwise histologically normal tissue of BRCA1 mutation carriers, luminal progenitors positive for RANK expression were shown to be highly proliferative, aberrant in DNA repair, and possessed a basal‐like breast cancer molecular signature (Nolan et al, 2016). High circulating RANKL levels correlate with increased breast cancer risk even in post‐menopausal women without a genetic predisposition (Kiechl et al, 2017). The RANKL axis is a druggable pathway, effectively inhibited in humans with the monoclonal antibody denosumab. Treatment of breast organoids from pre‐neoplastic BRCA1 heterozygous tissue also resulted in attenuation of progesterone‐induced proliferation (Nolan et al, 2016). Building on these pre‐clinical data, a pilot study of denosumab has shown a reduction in proliferation of normal breast cells from women heterozygous for BRCA1 (Nolan et al, 2017). It is exciting to note that a breast cancer prevention trial is currently underway to treat BRCA1 mutation carriers with denosumab (BRCA‐P).
STAT5a‐deficient mice phenocopy PR null mice (Miyoshi et al, 2001; Cui et al, 2004). In vitro experiments have shown that progesterone treatment leads to nuclear localization of STAT5a and PR to RANKL enhancer regions. STAT5a null mammary epithelial cells fail to upregulate classical progesterone effectors such as RANKL, WNT4, and AREG in response to the PR agonist R5020 (Obr et al, 2013). Work on the inhibition of STAT5 indicates its potential in chemoprevention; reports investigating the progression of early lesions to full‐blown cancers have demonstrated a dependency on STAT5 activation in order to evade apoptosis and pharmacological inhibition of JAK (AG490, ruxolitinib) or STAT5 (C188‐9) led to regression of early lesions (Haricharan et al, 2013; Johnston et al, 2018). Four weekly injections of C188‐9 also reduced tumor incidence (Haricharan et al, 2013), ultimately leading to a prevention clinical trial on the effects of ruxolitinib on premalignant breast cancer (NCT02928978).
The CXCL12‐CXCR4 axis was identified as a ligand–receptor pair in progesterone‐driven transcriptomes of mammary epithelial subsets (Shiah et al, 2015). Inhibition of CXCR4 diminished progenitor activity and mammary stem cell frequency (Shiah et al, 2015). CXCR4 plays an important role in cancer cell survival in a variety of tissues and is implicated in metastasis. Small molecule inhibitors of CXCR4 are currently being tested in clinical trials with AMD3100 and plerixafor for hematological malignancies (Devine et al, 2004; DiPersio et al, 2009). Similar to targeting RANK with denosumab, targeting other mitogens involved in stem/progenitor cell maintenance and regulation hold therapeutic promise. Amphiregulin (AREG) is also induced by progesterone, and inhibition of its receptor EGFR (Iressa) abolishes progesterone‐induced terminal end bud formation and proliferation (Fernandez‐Valdivia et al, 2008; Aupperlee et al, 2013). The metalloprotease ADAM17 is required for AREG shedding and Adam17−/− glands phenocopy those of Egfr null mice (Sternlicht et al, 2005). Therefore, in addition to EGFR inhibitors, ADAM17 may be a viable target. Other progesterone targets that may similarly prove therapeutically useful include ID4 (Dong et al, 2011) and Cyclin D1 (Said et al, 1997).
As mentioned previously, Wnt/B‐catenin signaling is another pathway implicated both in mammary stem/progenitor cell function and mammary tumorigenesis (Li et al, 2003; Henry et al, 2004; Liu et al, 2004). Among Wnt ligands, Wnt4 is a recognized progesterone‐induced factor and Wnt4 knockout glands show decreased side‐branching during early pregnancy (Brisken et al, 2000). In transplantation assays, Wnt4−/− epithelium is impaired and reconstitutes only 50% of the fat pad in the first transplant which is lowered to 10% by the third round of transplant, underscoring its requirement in mammary stem cell maintenance (Rajaram et al, 2015). It has since been shown that Wnt4 is released from HR+ luminal cells and binds its cognate receptor Frizzled on ER−PR− progenitors. Targeting Wnt signaling may become a viable option for breast cancer prevention in the future as current phase I and II clinical trials determine the efficacy of Wnt pathway inhibitors such as β‐catenin antagonists (PRI‐724) and anti‐Frizzled agents (vantictumab, OMP‐54F28) in multiple cancer types, as reviewed elsewhere (Takebe et al, 2015). Rspondin1 is a Wnt signaling co‐factor shown to mediate mammary stem cell renewal in response to ovarian hormones and in pregnancy. It binds Lgr family proteins that in turn inhibit a negative regulator of Wnt signaling (RNF43/ZNFR3) to sustain Wnt activation. HR‐ luminal progenitors secrete Rspondin1 that acts on basal cells to amplify Wnt signaling during progesterone‐mediated expansion (Cai et al, 2014; Joshi et al, 2015b). Genetic modulation of Rspondin1 leads to reduced side‐branching, gestational alveologenesis, and mammary stem cell repopulating frequency (Chadi et al, 2009; Cai et al, 2014). Conversely, administration of recombinant Rspondin rescues Rankl−/− glands. Rspondin3 is expressed in normal basal and stromal cells. Its overexpression in breast cancers corresponds to high levels of a mesenchymal marker, whereas knockdown compromises lactogenic differentiation, xenograft growth, and lung metastasis (Tocci et al, 2018). There is currently active interest in developing neutralizing antibodies to various Rspondins (Chartier et al, 2016).
Exploiting mammary lineage vulnerabilities
It is known that tumorigenesis co‐opts normal inherent regulatory networks (Polak et al, 2015; Mayers et al, 2016; Hoadley et al, 2018) and cancers retain characteristics of normal tissues despite acquiring countless mutations (Locke et al, 2005). Thus, the molecular identity of mammary cells as dictated by chromatin confirmation, epigenomes, and proteomes will inform the development of rationalized drugs against a mammary lineage or breast cancer subtype. Profiling across multiple genomic platforms (Table 1) is yielding deeper insights into intrinsic differences between the mammary epithelial lineages and has generated powerful reference datasets. These initiatives have uncovered lineage‐specific features of basal and luminal cells, as well as the mammary stem cells and progenitors encompassed within these compartments, for subsequent targeting of cancer precursors.
Table 1.
Global profiling datasets of mammary epithelial cells
| Author | Technique | Populations (time points) | Species | Hormones |
|---|---|---|---|---|
| Giraddi et al (2018) | scRNA‐seq | Fetal (E16, 18), Adult MaSC (10–16 weeks) | Mouse | – |
| Nguyen et al (2018) | scRNA‐seq | Total luminal and basal | Human | – |
| Pal et al (2017) | scRNA‐seq | Total Mammary Gland (2, 5, 10 weeks) | Mouse | EstrusDiestrus |
| Bach et al (2017) | scRNA‐seq | Total EpCAM population | Mouse | Nulliparous (8 weeks)Gestation (14.5 D)Lactation (6 D)Involution (Post 11 D) |
| Knapp et al (2017) | CyTOF | Total epithelium | Human | – |
| Pal et al (2013) | ChIP‐seq (H3K4me3, H3K27me3, H3K9me2) | Adult LP, LM, B (8 weeks) | Mouse | – |
| Pellacani et al (2016) | ChIP‐seq (H3K4me3, H3K4me1, H3K27ac, H3K27me3, H3K9me3, and H3K36me3)WGBS (DNA Methylation)RNA‐seq | LP, LM, B | Human | – |
| Maruyama et al (2011) | ChIP‐seq (H3K4me3, H3K27me3)SAGE‐seq (gene expression)MSDK‐seq (DNA Methylation) | CD24+ and CD44+ | Human | – |
| Dos Santos et al (2015) | WGBS (DNA Methylation) | LP, LM, B | Mouse | Post‐pubertal (nulliparous, 8–15 weeks)Post‐pregnancy (parous, > 12 weeks) |
| Casey et al (2018) | ATAC‐seq (Open chromatin)RRBS (DNA Methylation)UPLC‐MS (Proteomics) | Adult LP, LM, B (8–12 weeks) | Mouse | Hormone pellets |
| Dravis et al (2018) | ATAC‐seq (Open chromatin)RNA‐seqChIP‐seq (H3K27ac) |
Fetal MaSC (E18) Adult LM, LP and B (6–10 weeks) |
Mouse | – |
| Gascard et al (2015) | RNA‐seqmiRNA‐seqChIP‐seq (H3K36me3)MeDIP‐seq, MRE‐seq, WGBS (DNA methylation) | Myoepithelial, luminal, stem‐like | Human | – |
| Shiah et al (2015) | Microarray | Adult total luminal and basal (8–12 weeks) | Mouse | Hormone pellets |
OMICs‐based lineage distinctions
Microarrays of FACS‐purified mouse and human mammary subsets show that the basal and luminal lineages are separate entities (Kendrick et al, 2008; Lim et al, 2010; Pardo et al, 2014; Shiah et al, 2015). Kendrick et al (2008) found differentially expressed genes in basal (861), HR+ (326), and HR− (488) luminal populations, and ovarian hormone‐induced transcriptomes have also been reported (Casey et al, 2018). In normal breast epithelium, 255 genes (~ 1.4% of the transcriptome) are differentially expressed between the two phases of menstrual cycle, with ~ 87% of genes being higher in the luteal phase (Pardo et al, 2014). Distinct molecular programs are reflected in lineage‐specific expression of key transcriptional factors and signaling components, including the Notch (Raouf et al, 2008), WNT (Zeng & Nusse, 2010; van Amerongen et al, 2012; Gu et al, 2013; Arendt et al, 2014), and Hippo pathways (Chen et al, 2014; Skibinski et al, 2014; Britschgi et al, 2017). Proteomic landscapes of murine mammary subpopulations similarly point to lineage‐restricted and progesterone‐driven protein changes (Fig 3; Casey et al, 2018). Parallel profiling of open chromatin regions (ATAC‐Seq) and methylomes (RRBS‐Seq) on the same basal and luminal samples permitted quantification of system‐level relationships between chromatin–DNA–RNA–protein states. These data also revealed differential DNA methylation patterns at transcription factor binding sites, identifying motifs hypomethylated and/or enriched in open chromatin regions in basal vs. luminal cells; motifs for key transcription factors included FOXA1, ELF5, GATA3, TP63, known essential regulators of mammary morphogenesis, cell fate, differentiation, and lineage identity. Novel lineage associations were noted for TP53 and EGR1 motifs in basal cells and for FOXA2, SPI1, and FOXP1 motifs in luminal cells (Casey et al, 2018). Integration of human breast cell epigenomes and transcriptomes shows that luminal cell genomes harbor more than twice the number of hypomethylated enhancer elements than basal cells (Gascard et al, 2015). Unique methylation patterns of breast subsets also illustrate extensive lineage specificity as well as distinct patterns across breast cancer subtypes (Bediaga et al, 2010; Holm et al, 2010; Gascard et al, 2015; Pellacani et al, 2016; Casey et al, 2018). For instance, in a meta‐analysis of 40 studies, BRCA1 promoter methylation was statistically significantly higher in breast cancers negative for both ER, PR and with a triple‐negative phenotype (Zhang & Long, 2015).
Figure 3. Lineage‐specific molecular programs and epigenetic regulators.
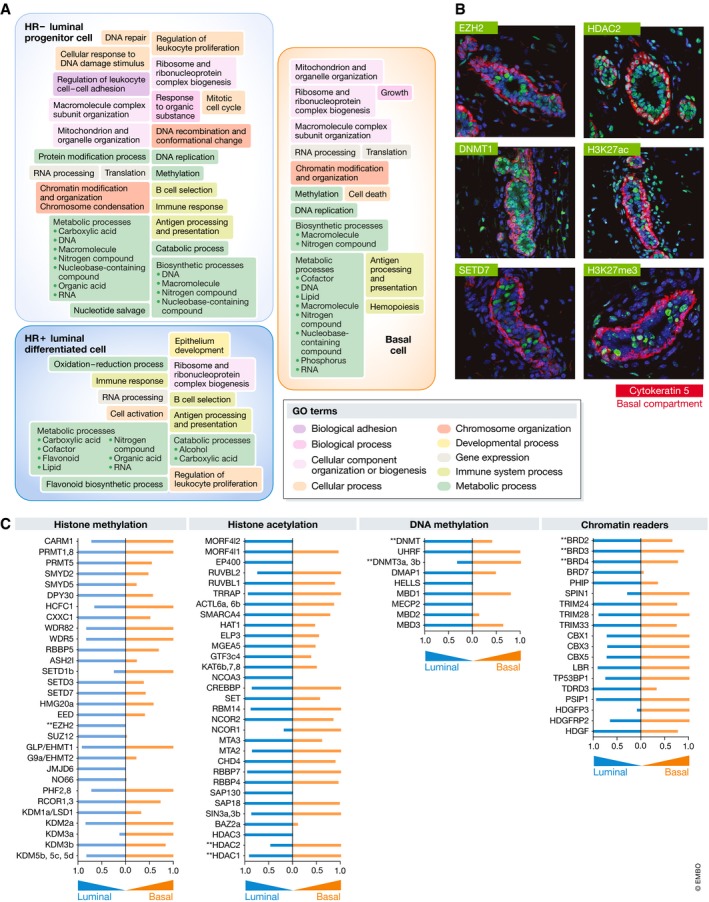
(A) Schematic illustrating the main GO terms biological processes (≥ 3‐fold upregulated, FDR ≤ 0.01) enriched in the three distinct mammary cell subpopulations in response to progesterone, based on proteomic analysis from Casey et al. (B) Visualization of the indicated epigenetic proteins or histone marks (green) in situ. Lineage specificity is observed for select proteins (EZH2, DNMT1, and SETD7). Cytokeratin 5 (red) marks the basal compartment. (C) Comparative abundance of epigenetic regulatory proteins detected in luminal vs. basal compartment.
Given the current momentum in utilizing drugs tailored to epigenetic machinery in oncology, understanding the epigenomes of mammary lineages has become an active area (Maruyama et al, 2011; Pellacani et al, 2016; Dravis et al, 2018). Mammary cell proteomes have exposed the enrichment of key epigenetic regulators in the luminal lineage (Casey et al, 2018). The study short‐listed 13 rationalized inhibitors corresponding to histone or chromatin modifiers and evaluated their capacity to abrogate luminal and basal CFCs. Specifically, TSA/SAHA (HDAC inhibitors), decitabine (DAC, DNMT1, 3a, 3b inhibitor), and JQ1 (BRD2, 3, 4 & T) stably reduced mammary stem/progenitor function in vivo, where DAC also significantly delayed tumor latency and reduced tumor incidence of p53‐driven mammary cancer. Notably, select drugs effectively inhibited the clonogenicity of breast cells from women with BRCA1/2 mutations. Altogether, inhibitors depleted mammary stem/progenitors, delayed aggressive breast cancer tumorigenesis, and demonstrated cytostatic effects underscoring their potential as intervention agents. The histone methyltransferase EZH2 is another candidate for intervention as it is overexpressed in BRCA1 normal breast tissue and implicated in mammary stem cell expansion as well as breast cancer (Ding et al, 2006). Trials addressing the clinic utility of epigenetic therapies in breast cancers are already underway. Combinations of HDAC inhibitors with DNMT inhibitors show superior ER re‐expression in breast cancer cell lines, and testing of the HDAC inhibitor entinostat + exemestane is ongoing in HR+ breast cancer patients (NCT02115282; Yang et al, 2000; Connolly & Stearns, 2012).
Known intrinsic capacities
In addition to differences in DNA–RNA–protein composition across the two mammary lineages, divergent stress responses and cellular features are observed. For instance, purified human luminal progenitors, which have higher levels of reactive oxygen species (ROS) compared to basal cells, have efficient antioxidant mechanisms and possess higher levels of several glutathione peroxidases (Kannan et al, 2014) ROS arise during normal cellular activity but high levels can damage DNA, proteins, and lipids (Gorrini et al, 2013), rendering basal cells more vulnerable (Kannan et al, 2014). Along the same lines, molecular profiling indicates that luminal progenitors are cells with active transcription. A higher accumulation of R‐loops, which naturally occur as by‐products of transcription, was noted in luminal progenitors compared to basal cells (Zhang et al, 2017). This is relevant since luminal progenitors are the likely cell‐of‐origin for BRCA1‐mutated breast cancer, and the dysregulated levels of R‐loops seen in cancers lead to genomic instability (Aguilera & García‐Muse, 2012). Unusually, short telomeres (< 3 kb) were observed in luminal progenitors irrespective of donor age, while contrastingly, basal cells had telomere lengths of ~ 6–8 kb. Only luminal progenitor subsets express human telomerase hTERT, the enzyme responsible for elongating telomere ends, and also express higher levels of several telomere‐associated genes, some crucial for DNA damage repair (MRE11, RAD50, ATM, ATR and BLM; Kannan et al, 2013). Oncogene‐induced DNA damage response differs across human luminal and basal populations; luminal subset exhibits copious DNA damage and repair activation (more γH2AX & 53BP1 foci), whereas basal cells show little response (Morel et al, 2017). Thus, luminal progenitors appear to be better equipped with DNA damage machinery, ultimately impacting their ability for transformation. Other findings suggest that select basal cells are the most likely cell‐of‐origin for a subclass of triple‐negative breast cancers with low chromosomal instability and mutational load (Morel et al, 2017). PARP inhibitors specifically leverage DNA damage responses to induce synthetic lethality; therefore, lineage‐specific repair capacities hold clinical value. Overall, recognizing lineage‐imposed differences will guide the generation of appropriate intervention strategies against specific cells‐of‐origin.
Molecular‐guided prevention pipeline
Table 1 summarizes the current data resources available to the breast research community that span breast mammopoiesis. From fetal mammary stem cells to the adult gland under defined hormone settings, these bodies of work provide a foundation for the discovery of novel chemopreventive agents using a systems approach to mammary cell biology (Fig 4). Mining these datasets (genomes, epigenomes, and proteomes) aids to unravel the molecular nature of distinct cells‐of‐origin within the breast. Specifically, these data help expose novel and/or unique biological pathways and highlight putative vulnerabilities intrinsic to specific mammary subpopulations. The corresponding pharmacological agents that are subsequently identified require successive evaluation in rigorous experiments for functional and therapeutic validation pertaining to distinct aspects of breast cancer development.
Figure 4. A path to OMICS‐guided chemoprevention.
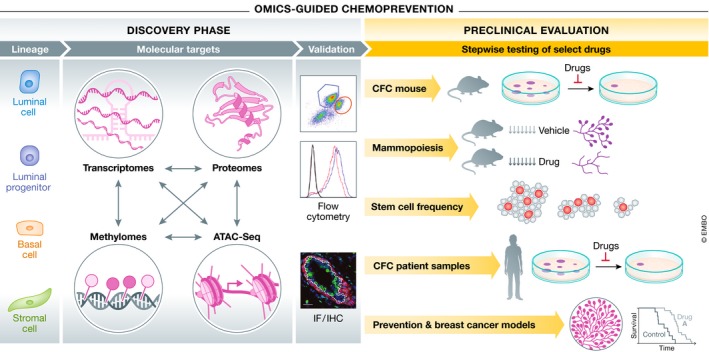
A workflow modeling a discovery‐to‐intervention pipeline for OMICs‐guided chemoprevention. FACS‐purified mammary cell populations are the input for integrative molecular profiling, target validation, rationalized drug identification, and evaluation in a series of biological, pre‐clinical assays.
Utilizing stem cell function (clonogenicity, in vivo mammopoiesis, and limiting dilution) and tumor onset, vital pre‐clinical data can be generated. For instance, mouse or human breast epithelial cell 2D colony‐forming capacity assays are a simple, cost‐effective method to screen for drugs that decrease clonogenicity. Excitingly, 3D organoids have also been reported that form bi‐layered morphological structures mimicking the complexity of breast terminal ductal lobular units. Although these culture systems are still being optimized, these structures have been reported as exclusively arising from bi‐potent stem/progenitor cells in the basal compartment and may prove useful as screening tools similar to 2D colony assays (Linnemann et al, 2015; Sokol et al, 2016). Casey et al showed how in vivo mammopoiesis assays that measure effects on progesterone‐driven side‐branching and lobuloalveolar development (the putative sites of tumorigenesis) can also be successfully used to further short‐list agents with potential in chemoprevention and ultimately limit cancer incidence in breast cancer models. Limiting dilution assays that enumerate mammary repopulating potential can similarly pinpoint agents effective against mammary stem cells, the putative cell‐of‐origin in select breast cancers. Finally, a number of informative breast cancer models exist, yet are unsuitable for the study of chemoprevention due to their overtly aggressive nature. Genetically engineered mouse models deficient in genes such as Brca1 and p53 exhibit pre‐neoplastic events such as increased numbers of mammary stem/progenitors and hyperplasia followed by multiple mammary tumors (Brodie et al, 2001; Evers & Jonkers, 2006). Evaluating drug efficacy in limiting these pre‐neoplastic events provides essential evidence necessary to accelerate translation into clinical trials, as previously demonstrated (Nolan et al, 2016; Sigl et al, 2016). This overarching workflow is depicted in Fig 4.
Open questions
In the upcoming years, new approaches to breast cancer prevention are bound to flourish. The World Health Organization Global Action Plan for the Prevention and Control of Noncommunicable Diseases hopes for a 25% reduction in cancer mortality rates by 2025, and the United Nations Sustainable Development Goals program strives for a 33% reduction by 2030, although < 10% of current research funding is dedicated to prevention research (Song et al, 2018). Outlining the contributions of lineage and hormones, this review highlights how relevant mammary biology empowers identification of new therapeutic targets to limit unwarranted stem cells and progenitors, the seeds of transformation. However, barriers still hinder wide clinical implementation of chemoprevention, from the patient, clinical, and research standpoint.
Ensuring minimal disruption to reproductive health and drug cytotoxicity is essential in breast cancer chemoprevention. How long should a patient receive a drug and how often? High‐risk groups have an increased lifetime risk so when should treatment begin? Challenges of balancing long‐term compliance against lifetime risk call for the development of new intermittent drug regimens. Stalling expansion of stem and progenitor cell populations in a reversible manner may allow normal activity such as lactation. Anti‐progestins may address these needs as their use can be restricted to a specific reproductive phase. Further, repurposing drugs with known safety profiles, such as ulipristal acetate, denosumab, and metformin, allow immediate translation.
From a clinical standpoint, important questions center on how to design prevention trials. What would be considered a benchmark of effective prevention? Some interesting questions remain if it is sufficient to stabilize breast cancer precursor populations vs. eradicating them? Careful consideration of how to define appropriate clinical endpoints of therapeutic response in prevention will require further investigation. Are there surrogate indicators that can be used to monitor prevention? There is an unmet need for better surrogate indicators, especially for trials of agents with poorly established potential for delayed/long‐term toxicities. Innovation in non‐invasive molecular imaging and liquid biopsy‐based monitoring may resolve some of these challenges.
At the basic level, heterogeneity of adult breast tissue as a function of age, reproductive history, and ethnicity continues to pose challenges. How do other known risk factors, such as obesity, relate to breast cancer cells‐of‐origin? Given the sheer amount of non‐epithelial components in the breast, such as stroma, adipocytes, and immune cells, what is their influence on the nature of risk and their role as targets of prevention? Interestingly, a recent study demonstrates a mesenchymal adipocyte contribution to the mammary epithelium (Joshi et al, 2019). Moreover, as we continue to dissect cellular heterogeneity within normal epithelium and breast cancers, the cell‐of‐origin field will correspondingly move forward and further advance molecular‐guided chemoprevention.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgements
We thank Dr. P. Waterhouse and other members of the Khokha Lab for reviewing this manuscript. This work is supported by funding from the Canadian Breast Cancer Foundation (CBCF; grant 313420), Canadian Cancer Society Research Institute (grants 702294 and 705458), Prairie Women on Snowmobiles, and Hold'em For Life. P.T. holds a CBCF/CCSRI doctoral fellowship; M.M. received a Canadian Institutes of Health Research (CIHR) Masters Award.
The EMBO Journal (2019) 38: e100852
References
- Aguilera A, García‐Muse T (2012) R loops: from transcription byproducts to threats to genome stability. Mol Cell 46: 115–124 [DOI] [PubMed] [Google Scholar]
- Ahern TP, Pedersen L, Tarp M, Cronin‐Fenton DP, Garne JP, Silliman RA, Sørensen HT, Lash TL (2011) Statin prescriptions and breast cancer recurrence risk: a Danish nationwide prospective cohort study. J Natl Cancer Inst 103: 1461–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al‐Hajj M, Wicha MS, Benito‐Hernandez A, Morrison SJ, Clarke MF (2003) Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA 100: 3983–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Amerongen R, Fuerer C, Mizutani M, Nusse R (2012) Wnt5a can both activate and repress Wnt/beta‐catenin signaling during mouse embryonic development. Dev Biol 369: 101–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniou AC, Casadei S, Heikkinen T, Barrowdale D, Pylkäs K, Roberts J, Lee A, Subramanian D, De Leeneer K, Fostira F et al (2014) Breast‐cancer risk in families with mutations in PALB2. N Engl J Med 371: 497–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendt LM, St Laurent J, Wronski A, Caballero S, Lyle SR, Naber SP, Kuperwasser C (2014) Human breast progenitor cell numbers are regulated by WNT and TBX3. PLoS One 9: e111442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselin‐Labat ML, Sutherland KD, Barker H, Thomas R, Shackleton M, Forrest NC, Hartley L, Robb L, Grosveld FG, van der Wees J et al (2007) Gata‐3 is an essential regulator of mammary‐gland morphogenesis and luminal‐cell differentiation. Nat Cell Biol 9: 201–209 [DOI] [PubMed] [Google Scholar]
- Aupperlee MD, Leipprandt JR, Bennett JM, Schwartz RC, Haslam SZ (2013) Amphiregulin mediates progesterone‐induced mammary ductal development during puberty. Breast Cancer Res 15: R44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach K, Pensa S, Grzelak M, Hadfield J, Adams DJ, Marioni JC, Khaled WT (2017) Differentiation dynamics of mammary epithelial cells revealed by single‐cell RNA sequencing. Nat Commun 8: 2128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badders NM, Goel S, Clark RJ, Klos KS, Kim S, Bafico A, Lindvall C, Williams BO, Alexander CM (2009) The Wnt receptor, Lrp5, is expressed by mouse mammary stem cells and is required to maintain the basal lineage. PLoS One 4: e6594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bediaga NG, Acha‐Sagredo A, Guerra I, Viguri A, Albaina C, Ruiz Diaz I, Rezola R, Alberdi MJ, Dopazo J, Montaner D et al (2010) DNA methylation epigenotypes in breast cancer molecular subtypes. Breast Cancer Res 12: R77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beral V, Million Women Study Collaborators (2003) Breast cancer and hormone‐replacement therapy in the Million Women Study. Lancet 362: 419–427 [DOI] [PubMed] [Google Scholar]
- Beristain AG, Narala SR, Di Grappa MA, Khokha R (2012) Homotypic RANK signaling differentially regulates proliferation, motility and cell survival in osteosarcoma and mammary epithelial cells. J Cell Sci 125: 943–955 [DOI] [PubMed] [Google Scholar]
- Bevers TB, Helvie M, Bonaccio E, Calhoun KE, Daly MB, Farrar WB, Garber JE, Gray R, Greenberg CC, Greenup R et al (2018) Breast cancer screening and diagnosis, version 3.2018, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw 16: 1362–1389 [DOI] [PubMed] [Google Scholar]
- Bosco JLF, Antonsen S, Sørensen HT, Pedersen L, Lash TL (2011) Metformin and incident breast cancer among diabetic women: a population‐based case‐control study in Denmark. Cancer Epidemiol Biomarkers Prev 20: 101–111 [DOI] [PubMed] [Google Scholar]
- Boulanger CA, Smith GH (2001) Reducing mammary cancer risk through premature stem cell senescence. Oncogene 20: 2264 [DOI] [PubMed] [Google Scholar]
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68: 394–424 [DOI] [PubMed] [Google Scholar]
- Brisken C, Park S, Vass T, Lydon JP, O'Malley BW, Weinberg RA (1998) A paracrine role for the epithelial progesterone receptor in mammary gland development. Proc Natl Acad Sci USA 95: 5076–5081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisken C, Heineman A, Chavarria T, Elenbaas B, Tan J, Dey SK, McMahon JA, McMahon AP, Weinberg RA (2000) Essential function of Wnt‐4 in mammary gland development downstream of progesterone signaling. Genes Dev 14: 650–654 [PMC free article] [PubMed] [Google Scholar]
- Brisken C, Duss S (2007) Stem cells and the stem cell niche in the breast: an integrated hormonal and developmental perspective. Stem Cell Rev 3: 147–156 [DOI] [PubMed] [Google Scholar]
- Britschgi A, Duss S, Kim S, Couto JP, Brinkhaus H, Koren S, De Silva D, Mertz KD, Kaup D, Varga Z et al (2017) The Hippo kinases LATS1 and 2 control human breast cell fate via crosstalk with ERα. Nature 541: 541–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodie SG, Xu X, Qiao W, Li WM, Cao L, Deng CX (2001) Multiple genetic changes are associated with mammary tumorigenesis in Brca1 conditional knockout mice. Oncogene 20: 7514–7523 [DOI] [PubMed] [Google Scholar]
- Bu W, Liu Z, Jiang W, Nagi C, Huang S, Edwards DP, Jo E, Mo Q, Creighton CJ, Hilsenbeck SG et al (2019) Mammary precancerous stem and non‐stem cells evolve into cancers of distinct subtypes. Cancer Res 79: 61–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai C, Yu QC, Jiang W, Liu W, Song W, Yu H, Zhang L, Yang Y, Zeng YA (2014) R‐spondin1 is a novel hormone mediator for mammary stem cell self‐renewal. Genes Dev 28: 2205–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S, Kalisky T, Sahoo D, Dalerba P, Feng W, Lin Y, Qian D, Kong A, Yu J, Wang F et al (2017) A quiescent Bcl11b high stem cell population is required for maintenance of the mammary gland. Cell Stem Cell 20: 247–260.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campeau PM, Foulkes WD, Tischkowitz MD (2008) Hereditary breast cancer: new genetic developments, new therapeutic avenues. Hum Genet 124: 31–42 [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network (2012) Comprehensive molecular portraits of human breast tumours. Nature 490: 61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey AE, Sinha A, Singhania R, Livingstone J, Waterhouse P, Tharmapalan P, Cruickshank J, Shehata M, Drysdale E, Fang H et al (2018) Mammary molecular portraits reveal lineage‐specific features and progenitor cell vulnerabilities. J Cell Biol 217: 2951–2974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauley JA, McTiernan A, Rodabough RJ, LaCroix A, Bauer DC, Margolis KL, Paskett ED, Vitolins MZ, Furberg CD, Chlebowski RT et al (2006) Statin use and breast cancer: prospective results from the Women's Health Initiative. J Natl Cancer Inst 98: 700–707 [DOI] [PubMed] [Google Scholar]
- Chadi S, Buscara L, Pechoux C, Costa J, Laubier J, Chaboissier M‐C, Pailhoux E, Vilotte J‐L, Chanat E, Le Provost F (2009) R‐spondin1 is required for normal epithelial morphogenesis during mammary gland development. Biochem Biophys Res Commun 390: 1040–1043 [DOI] [PubMed] [Google Scholar]
- Chakrabarti R, Celià‐Terrassa T, Kumar S, Hang X, Wei Y, Choudhury A, Hwang J, Peng J, Nixon B, Grady JJ et al (2018) Notch ligand Dll1 mediates cross‐talk between mammary stem cells and the macrophageal niche. Science 360: eaan4153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartier C, Raval J, Axelrod F, Bond C, Cain J, Dee‐Hoskins C, Ma S, Fischer MM, Shah J, Wei J et al (2016) Therapeutic targeting of tumor‐derived R‐spondin attenuates β‐catenin signaling and tumorigenesis in multiple cancer types. Cancer Res 76: 713–723 [DOI] [PubMed] [Google Scholar]
- Cheang MCU, Chia SK, Voduc D, Gao D, Leung S, Snider J, Watson M, Davies S, Bernard PS, Parker JS et al (2009) Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst 101: 736–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Zhang N, Gray RS, Li H, Ewald AJ, Zahnow CA, Pan D (2014) A temporal requirement for Hippo signaling in mammary gland differentiation, growth, and tumorigenesis. Genes Dev 28: 432–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chepko G, Slack R, Carbott D, Khan S, Steadman L, Dickson RB (2005) Differential alteration of stem and other cell populations in ducts and lobules of TGFalpha and c‐Myc transgenic mouse mammary epithelium. Tissue Cell 37: 393–412 [DOI] [PubMed] [Google Scholar]
- Chin K, de Solorzano CO, Knowles D, Jones A, Chou W, Rodriguez EG, Kuo W‐L, Ljung B‐M, Chew K, Myambo K et al (2004) In situ analyses of genome instability in breast cancer. Nat Genet 36: 984 [DOI] [PubMed] [Google Scholar]
- Chlebowski RT, McTiernan A, Wactawski‐Wende J, Manson JE, Aragaki AK, Rohan T, Ipp E, Kaklamani VG, Vitolins M, Wallace R et al (2012) Diabetes, metformin, and breast cancer in postmenopausal women. J Clin Oncol 30: 2844–2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chlebowski RT, Rohan TE, Manson JE, Aragaki AK, Kaunitz A, Stefanick ML, Simon MS, Johnson KC, Wactawski‐Wende J, O'Sullivan MJ et al (2015) Breast cancer after use of estrogen plus progestin and estrogen alone: analyses of data from 2 women's health initiative randomized clinical trials. JAMA Oncol 1: 296–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke CL, Sandle J, Jones AA, Sofronis A, Patani NR, Lakhani SR (2006) Mapping loss of heterozygosity in normal human breast cells from BRCA1/2 carriers. Br J Cancer 95: 515–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly R, Stearns V (2012) Epigenetics as a therapeutic target in breast cancer. J Mammary Gland Biol Neoplasia 17: 191–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A, Decensi A, Marubini E, Formelli F, De Palo G, Mariani L, Sporn MB, Di Mauro MG, Miceli R, Camerini T et al (2006) Fifteen‐year results of a randomized phase III trial of fenretinide to prevent second breast cancer. Ann Oncol 17: 1065–1071 [DOI] [PubMed] [Google Scholar]
- Cui Y, Riedlinger G, Miyoshi K, Tang W, Li C, Deng C‐X, Robinson GW, Hennighausen L (2004) Inactivation of Stat5 in mouse mammary epithelium during pregnancy reveals distinct functions in cell proliferation, survival, and differentiation. Mol Cell Biol 24: 8037–8047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuzick J, Sestak I, Forbes JF, Dowsett M, Knox J, Cawthorn S, Saunders C, Roche N, Mansel RE, von Minckwitz G et al (2014) Anastrozole for prevention of breast cancer in high‐risk postmenopausal women (IBIS‐II): an international, double‐blind, randomised placebo‐controlled trial. Lancet 383: 1041–1048 [DOI] [PubMed] [Google Scholar]
- Cuzick J, Sestak I, Cawthorn S, Hamed H, Holli K, Howell A, Forbes JF, IBIS‐I Investigators (2015) Tamoxifen for prevention of breast cancer: extended long‐term follow‐up of the IBIS‐I breast cancer prevention trial. Lancet Oncol 16: 67–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decensi A, Robertson C, Guerrieri‐Gonzaga A, Serrano D, Cazzaniga M, Mora S, Gulisano M, Johansson H, Galimberti V, Cassano E et al (2009) Randomized double‐blind 2 × 2 trial of low‐dose tamoxifen and fenretinide for breast cancer prevention in high‐risk premenopausal women. J Clin Oncol 27: 3749–3756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng G, Lu Y, Zlotnikov G, Thor AD, Smith HS (1996) Loss of heterozygosity in normal tissue adjacent to breast carcinomas. Science 274: 2057–2059 [DOI] [PubMed] [Google Scholar]
- DeOme KB, Medina D (1969) A new approach to mammary tumorigenesis in rodents. Cancer 24: 1255–1258 [DOI] [PubMed] [Google Scholar]
- Devine SM, Flomenberg N, Vesole DH, Liesveld J, Weisdorf D, Badel K, Calandra G, DiPersio JF (2004) Rapid mobilization of CD34+ cells following administration of the CXCR49 antagonist AMD3100 to patients with multiple myeloma and non‐Hodgkin's lymphoma. J Clin Oncol 22: 1095–1102 [DOI] [PubMed] [Google Scholar]
- Diallo R, Schaefer K‐L, Poremba C, Shivazi N, Willmann V, Buerger H, Dockhorn‐Dworniczak B, Boecker W (2001) Monoclonality in normal epithelium and in hyperplastic and neoplastic lesions of the breast. J Pathol 193: 27–32 [DOI] [PubMed] [Google Scholar]
- Ding L, Erdmann C, Chinnaiyan AM, Merajver SD, Kleer CG (2006) Identification of EZH2 as a molecular marker for a precancerous state in morphologically normal breast tissues. Cancer Res 66: 4095–4099 [DOI] [PubMed] [Google Scholar]
- DiPersio JF, Micallef IN, Stiff PJ, Bolwell BJ, Maziarz RT, Jacobsen E, Nademanee A, McCarty J, Bridger G, Calandra G et al (2009) Phase III prospective randomized double‐blind placebo‐controlled trial of plerixafor plus granulocyte colony‐stimulating factor compared with placebo plus granulocyte colony‐stimulating factor for autologous stem‐cell mobilization and transplantation for. J Clin Oncol 27: 4767–4773 [DOI] [PubMed] [Google Scholar]
- Domchek SM, Friebel TM, Singer CF, Evans DG, Lynch HT, Isaacs C, Garber JE, Neuhausen SL, Matloff E, Eeles R et al (2010) Association of risk‐reducing surgery in BRCA1 or BRCA2 mutation carriers with cancer risk and mortality. JAMA 304: 967–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Huang S, Caikovski M, Ji S, McGrath A, Custorio MG, Creighton CJ, Maliakkal P, Bogoslovskaia E, Du Z et al (2011) ID4 regulates mammary gland development by suppressing p38MAPK activity. Development 138: 5247–5256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Santos CO, Dolzhenko E, Hodges E, Smith AD, Hannon GJ (2015) An Epigenetic Memory of Pregnancy in the Mouse Mammary Gland. Cell Rep 11: 1102–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dravis C, Chung C‐Y, Lytle NK, Herrera‐Valdez J, Luna G, Trejo CL, Reya T, Wahl GM (2018) Epigenetic and transcriptomic profiling of mammary gland development and tumor models disclose regulators of cell state plasticity. Cancer Cell 34: 466–482.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton DF, Pooley KA, Dunning AM, Pharoah PDP, Thompson D, Ballinger DG, Struewing JP, Morrison J, Field H, Luben R et al (2007) Genome‐wide association study identifies novel breast cancer susceptibility loci. Nature 447: 1087–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton DF, Pharoah PDP, Antoniou AC, Tischkowitz M, Tavtigian SV, Nathanson KL, Devilee P, Meindl A, Couch FJ, Southey M et al (2015) Gene‐panel sequencing and the prediction of breast‐cancer risk. N Engl J Med 372: 2243–2257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eirew P, Stingl J, Raouf A, Turashvili G, Aparicio S, Emerman JT, Eaves CJ (2008) A method for quantifying normal human mammary epithelial stem cells with in vivo regenerative ability. Nat Med 14: 1384–1389 [DOI] [PubMed] [Google Scholar]
- Eirew P, Kannan N, Knapp DJHF, Vaillant F, Emerman JT, Lindeman GJ, Visvader JE, Eaves CJ (2012) Aldehyde dehydrogenase activity is a biomarker of primitive normal human mammary luminal cells. Stem Cells 30: 344–348 [DOI] [PubMed] [Google Scholar]
- Eisen A, Lubinski J, Klijn J, Moller P, Lynch HT, Offit K, Weber B, Rebbeck T, Neuhausen SL, Ghadirian P et al (2005) Breast cancer risk following bilateral oophorectomy in BRCA1 and BRCA2 mutation carriers: an International Case‐Control Study. J Clin Oncol 23: 7491–7496 [DOI] [PubMed] [Google Scholar]
- Ellsworth DL, Ellsworth RE, Love B, Deyarmin B, Lubert SM, Mittal V, Shriver CD (2004) Genomic patterns of allelic imbalance in disease free tissue adjacent to primary breast carcinomas. Breast Cancer Res Treat 88: 131–139 [DOI] [PubMed] [Google Scholar]
- Evans JMM, Donnelly LA, Emslie‐Smith AM, Alessi DR, Morris AD (2005) Metformin and reduced risk of cancer in diabetic patients. BMJ 330: 1304–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DG, Howell SJ, Howell A (2018) Personalized prevention in high risk individuals: managing hormones and beyond. Breast 39: 139–147 [DOI] [PubMed] [Google Scholar]
- Evers B, Jonkers J (2006) Mouse models of BRCA1 and BRCA2 deficiency: past lessons, current understanding and future prospects. Oncogene 25: 5885–5897 [DOI] [PubMed] [Google Scholar]
- Fata JE, Ho AT, Leco KJ, Moorehead RA, Khokha R (2000a) Cellular turnover and extracellular matrix remodeling in female reproductive tissues: functions of metalloproteinases and their inhibitors. Cell Mol Life Sci 57: 77–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fata JE, Kong Y‐Y, Li J, Sasaki T, Irie‐Sasaki J, Moorehead RA, Elliott R, Scully S, Voura EB, Lacey DL et al (2000b) The osteoclast differentiation factor osteoprotegerin‐ligand is essential for mammary gland development. Cell 103: 41–50 [DOI] [PubMed] [Google Scholar]
- Fernandez‐Valdivia R, Mukherjee A, Creighton CJ, Buser AC, DeMayo FJ, Edwards DP, Lydon JP (2008) Transcriptional response of the murine mammary gland to acute progesterone exposure. Endocrinology 149: 6236–6250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher B, Costantino JP, Wickerham DL, Cecchini RS, Cronin WM, Robidoux A, Bevers TB, Kavanah MT, Atkins JN, Margolese RG et al (2005) Tamoxifen for the prevention of breast cancer: current status of the National Surgical Adjuvant Breast and Bowel Project P‐1 study. J Natl Cancer Inst 97: 1652–1662 [DOI] [PubMed] [Google Scholar]
- Fu NY, Rios AC, Pal B, Law CW, Jamieson P, Liu R, Vaillant F, Jackling F, Liu KH, Smyth GK et al (2017) Identification of quiescent and spatially restricted mammary stem cells that are hormone responsive. Nat Cell Biol 19: 164 [DOI] [PubMed] [Google Scholar]
- Gascard P, Bilenky M, Sigaroudinia M, Zhao J, Li L, Carles A, Delaney A, Tam A, Kamoh B, Cho S et al (2015) Epigenetic and transcriptional determinants of the human breast. Nat Commun 6: 6351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginestier C, Hur MH, Charafe‐Jauffret E, Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG, Liu S et al (2007) ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell 1: 555–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraddi RR, Shehata M, Gallardo M, Blasco MA, Simons BD, Stingl J (2015) Stem and progenitor cell division kinetics during postnatal mouse mammary gland development. Nat Commun 6: 8487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraddi RR, Chung C‐Y, Heinz RE, Balcioglu O, Novotny M, Trejo CL, Dravis C, Hagos BM, Mehrabad EM, Rodewald LW et al (2018) Single‐cell transcriptomes distinguish stem cell state changes and lineage specification programs in early mammary gland development. Cell Rep 24: 1653–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez‐Suarez E, Jacob AP, Jones J, Miller R, Roudier‐Meyer MP, Erwert R, Pinkas J, Branstetter D, Dougall WC (2010) RANK ligand mediates progestin‐induced mammary epithelial proliferation and carcinogenesis. Nature 468: 103–107 [DOI] [PubMed] [Google Scholar]
- Gorrini C, Harris IS, Mak TW (2013) Modulation of oxidative stress as an anticancer strategy. Nat Rev Drug Discov 12: 931–947 [DOI] [PubMed] [Google Scholar]
- Goss PE, Ingle JN, Alés‐Martínez JE, Cheung AM, Chlebowski RT, Wactawski‐Wende J, McTiernan A, Robbins J, Johnson KC, Martin LW et al (2011) Exemestane for breast‐cancer prevention in postmenopausal women. N Engl J Med 364: 2381–2391 [DOI] [PubMed] [Google Scholar]
- Gu B, Watanabe K, Sun P, Fallahi M, Dai X (2013) Chromatin effector Pygo2 mediates Wnt‐notch crosstalk to suppress luminal/alveolar potential of mammary stem and basal cells. Cell Stem Cell 13: 48–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudjonsson T, Villadsen R, Nielsen HL, Ronnov‐Jessen L, Bissell MJ, Petersen OW (2002) Isolation, immortalization, and characterization of a human breast epithelial cell line with stem cell properties. Genes Dev 16: 693–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillem JG, Wood WC, Moley JF, Berchuck A, Karlan BY, Mutch DG, Gagel RF, Weitzel J, Morrow M, Weber BL et al (2006) ASCO/SSO review of current role of risk‐reducing surgery in common hereditary cancer syndromes. J Clin Oncol 24: 4642–4660 [DOI] [PubMed] [Google Scholar]
- Haricharan S, Dong J, Hein S, Reddy JP, Du Z, Toneff M, Holloway K, Hilsenbeck SG, Huang S, Atkinson R et al (2013) Mechanism and preclinical prevention of increased breast cancer risk caused by pregnancy. Elife 2: e00996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann LC, Schaid DJ, Woods JE, Crotty TP, Myers JL, Arnold PG, Petty PM, Sellers TA, Johnson JL, McDonnell SK et al (1999) Efficacy of bilateral prophylactic mastectomy in women with a family history of breast cancer. N Engl J Med 340: 77–84 [DOI] [PubMed] [Google Scholar]
- Hawkins SM, Matzuk MM (2008) The menstrual cycle: basic biology. Ann N Y Acad Sci 1135: 10–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearle N, Schumacher V, Menko FH, Olschwang S, Boardman LA, Gille JJP, Keller JJ, Westerman AM, Scott RJ, Lim W et al (2006) Frequency and spectrum of cancers in the Peutz‐Jeghers syndrome. Clin Cancer Res 12: 3209–3215 [DOI] [PubMed] [Google Scholar]
- Hein SM, Haricharan S, Johnston AN, Toneff MJ, Reddy JP, Dong J, Bu W, Li Y (2016) Luminal epithelial cells within the mammary gland can produce basal cells upon oncogenic stress. Oncogene 35: 1461–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry MD, Triplett AA, Oh KB, Smith GH, Wagner K‐U (2004) Parity‐induced mammary epithelial cells facilitate tumorigenesis in MMTV‐neu transgenic mice. Oncogene 23: 6980–6985 [DOI] [PubMed] [Google Scholar]
- Hoadley KA, Yau C, Hinoue T, Wolf DM, Lazar AJ, Drill E, Shen R, Taylor AM, Cherniack AD, Thorsson V et al (2018) Cell‐of‐origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell 173: 291–304.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm K, Hegardt C, Staaf J, Vallon‐Christersson J, Jönsson G, Olsson H, Borg A, Ringnér M (2010) Molecular subtypes of breast cancer are associated with characteristic DNA methylation patterns. Breast Cancer Res 12: R36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hue TF, Cummings SR, Cauley JA, Bauer DC, Ensrud KE, Barrett‐Connor E, Black DM (2014) Effect of bisphosphonate use on risk of postmenopausal breast cancer: results from the randomized clinical trials of alendronate and zoledronic acid bisphosphonates and postmenopausal breast cancer bisphosphonates and postmenopausal breast cancer. JAMA Intern Med 174: 1550–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys RC, Lydon JP, O'Malley BW, Rosen JM (1997) Use of PRKO mice to study the role of progesterone in mammary gland development. J Mammary Gland Biol Neoplasia 2: 343–354 [DOI] [PubMed] [Google Scholar]
- Ismail PM, Li J, DeMayo FJ, O'Malley BW, Lydon JP (2002) A novel LacZ reporter mouse reveals complex regulation of the progesterone receptor promoter during mammary gland development. Mol Endocrinol 16: 2475–2489 [DOI] [PubMed] [Google Scholar]
- Jackson HW, Waterhouse P, Sinha A, Kislinger T, Berman HK, Khokha R (2015) Expansion of stem cells counteracts age‐related mammary regression in compound Timp1/Timp3 null mice. Nat Cell Biol 17: 217–227 [DOI] [PubMed] [Google Scholar]
- Johnston AN, Bu W, Hein S, Garcia S, Camacho L, Xue L, Qin L, Nagi C, Hilsenbeck SG, Kapali J et al (2018) Hyperprolactinemia‐inducing antipsychotics increase breast cancer risk by activating JAK‐STAT5 in precancerous lesions. Breast Cancer Res 20: 42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi PA, Jackson HW, Beristain AG, Di Grappa MA, Mote PA, Clarke CL, Stingl J, Waterhouse PD, Khokha R (2010) Progesterone induces adult mammary stem cell expansion. Nature 465: 803–807 [DOI] [PubMed] [Google Scholar]
- Joshi PA, Di Grappa MA, Khokha R (2012) Active allies: hormones, stem cells and the niche in adult mammopoiesis. Trends Endocrinol Metab 23: 299–309 [DOI] [PubMed] [Google Scholar]
- Joshi PA, Goodwin PJ, Khokha R (2015a) Progesterone exposure and breast cancer risk understanding the biological roots. JAMA Oncol 1: 283–285 [DOI] [PubMed] [Google Scholar]
- Joshi PA, Waterhouse PD, Kannan N, Narala S, Fang H, Di Grappa MA, Jackson HW, Penninger JM, Eaves C, Khokha R (2015b) RANK signaling amplifies WNT‐responsive mammary progenitors through R‐SPONDIN1. Stem Cell Rep 5: 31–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi PA, Waterhouse PD, Kasaian K, Fang H, Gulyaeva O, Sul HS, Boutros PC, Khokha R (2019) PDGFRα+ stromal adipocyte progenitors transition into epithelial cells during lobulo‐alveologenesis in the murine mammary gland. Nat Commun 10: 1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan N, Huda N, Tu L, Droumeva R, Aubert G, Chavez E, Brinkman RR, Lansdorp P, Emerman J, Abe S et al (2013) The luminal progenitor compartment of the normal human mammary gland constitutes a unique site of telomere dysfunction. Stem Cell Rep 1: 28–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan N, Nguyen LV, Makarem M, Dong Y, Shih K, Eirew P, Raouf A, Emerman JT, Eaves CJ (2014) Glutathione‐dependent and ‐independent oxidative stress‐control mechanisms distinguish normal human mammary epithelial cell subsets. Proc Natl Acad Sci USA 111: 7789–7794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauff ND, Satagopan JM, Robson ME, Scheuer L, Hensley M, Hudis CA, Ellis NA, Boyd J, Borgen PI, Barakat RR et al (2002) Risk‐reducing salpingo‐oophorectomy in women with a BRCA1 or BRCA2 mutation. N Engl J Med 346: 1609–1615 [DOI] [PubMed] [Google Scholar]
- Kaurah P, MacMillan A, Boyd N, Senz J, De Luca A, Chun N, Suriano G, Zaor S, Van Manen L, Gilpin C et al (2007) Founder and recurrent CDH1 mutations in families with hereditary diffuse gastric cancer. JAMA 297: 2360–2372 [DOI] [PubMed] [Google Scholar]
- Keller PJ, Arendt LM, Skibinski A, Logvinenko T, Klebba I, Dong S, Smith AE, Prat A, Perou CM, Gilmore H et al (2012) Defining the cellular precursors to human breast cancer. Proc Natl Acad Sci USA 109: 2772–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsey JL, Gammon MD, John EM (1993) Reproductive factors and breast cancer. Epidemiol Rev 15: 36–47 [DOI] [PubMed] [Google Scholar]
- Kendrick H, Regan JL, Magnay F‐A, Grigoriadis A, Mitsopoulos C, Zvelebil M, Smalley MJ (2008) Transcriptome analysis of mammary epithelial subpopulations identifies novel determinants of lineage commitment and cell fate. BMC Genom 9: 591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler LG (1992) The relationship between age and incidence of breast cancer. Population and screening program data. Cancer 69: 1896–1903 [DOI] [PubMed] [Google Scholar]
- Kiechl S, Schramek D, Widschwendter M, Fourkala E‐O, Zaikin A, Jones A, Jaeger B, Rack B, Janni W, Scholz C et al (2017) Aberrant regulation of RANKL/OPG in women at high risk of developing breast cancer. Oncotarget 8: 3811–3825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MC, Wieand S, Hale K, Lee M, Walsh T, Owens K, Tait J, Ford L, Dunn BK, Costantino J et al (2001) Tamoxifen and breast cancer incidence among women with inherited mutations in BRCA1 and BRCA2: National Surgical Adjuvant Breast and Bowel Project (NSABP‐P1) Breast Cancer Prevention Trial. JAMA 286: 2251–2256 [DOI] [PubMed] [Google Scholar]
- King TA, Gemignani ML, Li W, Giri DD, Panageas KS, Bogomolniy F, Arroyo C, Olvera N, Robson ME, Offit K et al (2004) Increased progesterone receptor expression in benign epithelium of BRCA1‐related breast cancers. Cancer Res 64: 5051–5053 [DOI] [PubMed] [Google Scholar]
- Klein AM, Simons BD (2011) Universal patterns of stem cell fate in cycling adult tissues. Development 138: 3103–3111 [DOI] [PubMed] [Google Scholar]
- Knapp DJHF, Kannan N, Pellacani D, Eaves CJ (2017) Mass Cytometric Analysis Reveals Viable Activated Caspase‐3+Luminal Progenitors in the Normal Adult Human Mammary Gland. Cell Rep 21: 1116–1126 [DOI] [PubMed] [Google Scholar]
- Konecny G, Pauletti G, Pegram M, Untch M, Dandekar S, Aguilar Z, Wilson C, Rong H‐M, Bauerfeind I, Felber M et al (2003) Quantitative association between HER‐2/neu and steroid hormone receptors in hormone receptor‐positive primary breast cancer. J Natl Cancer Inst 95: 142–153 [DOI] [PubMed] [Google Scholar]
- Koren S, Reavie L, Couto JP, De Silva D, Stadler MB, Roloff T, Britschgi A, Eichlisberger T, Kohler H, Aina O et al (2015) PIK3CA(H1047R) induces multipotency and multi‐lineage mammary tumours. Nature 525: 114–118 [DOI] [PubMed] [Google Scholar]
- Kotsopoulos J, Huzarski T, Gronwald J, Singer CF, Moller P, Lynch HT, Armel S, Karlan B, Foulkes WD, Neuhausen SL et al (2017) Bilateral oophorectomy and breast cancer risk in BRCA1 and BRCA2 mutation carriers. J Natl Cancer Inst 109: djw177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchenbaecker KB, Hopper JL, Barnes DR, Phillips K‐A, Mooij TM, Roos‐Blom M‐J, Jervis S, van Leeuwen FE, Milne RL, Andrieu N et al (2017) Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA 317: 2402–2416 [DOI] [PubMed] [Google Scholar]
- Kurian AW, Hughes E, Handorf EA, Gutin A, Allen B, Hartman A‐R, Hall MJ (2017) Breast and ovarian cancer penetrance estimates derived from germline multiple‐gene sequencing results in women. JCO Precis Oncol 1: 1–12 [DOI] [PubMed] [Google Scholar]
- LaCroix AZ, Powles T, Osborne CK, Wolter K, Thompson JR, Thompson DD, Allred DC, Armstrong R, Cummings SR, Eastell R et al (2010) Breast cancer incidence in the randomized pearl trial of lasofoxifene in postmenopausal osteoporotic women. J Natl Cancer Inst 102: 1706–1715 [DOI] [PubMed] [Google Scholar]
- Lakhani SR, Chaggar R, Davies S, Jones C, Collins N, Odel C, Stratton MR, O'Hare MJ (1999) Genetic alterations in ‘normal’ luminal and myoepithelial cells of the breast. J Pathol 189: 496–503 [DOI] [PubMed] [Google Scholar]
- Lakhani SR, Reis‐Filho JS, Fulford L, Penault‐Llorca F, van der Vijver M, Parry S, Bishop T, Benitez J, Rivas C, Bignon Y‐J et al (2005) Prediction of BRCA1 status in patients with breast cancer using estrogen receptor and basal phenotype. Clin Cancer Res 11: 5175–5180 [DOI] [PubMed] [Google Scholar]
- Lanari C, Lamb CA, Fabris VT, Helguero LA, Soldati R, Bottino MC, Giulianelli S, Cerliani JP, Wargon V, Molinolo A (2009) The MPA mouse breast cancer model: evidence for a role of progesterone receptors in breast cancer. Endocr Relat Cancer 16: 333–350 [DOI] [PubMed] [Google Scholar]
- Larson PS, Schlechter BL, de las Morenas A, Garber JE, Cupples LA, Rosenberg CL (2005) Allele imbalance, or loss of heterozygosity, in normal breast epithelium of sporadic breast cancer cases and BRCA1 gene mutation carriers is increased compared with reduction mammoplasty tissues. J Clin Oncol 23: 8613–8619 [DOI] [PubMed] [Google Scholar]
- Lee O, Choi M‐R, Christov K, Ivancic D, Khan SA (2016) Progesterone receptor antagonism inhibits progestogen‐related carcinogenesis and suppresses tumor cell proliferation. Cancer Lett 376: 310–317 [DOI] [PubMed] [Google Scholar]
- Li Y, Welm B, Podsypanina K, Huang S, Chamorro M, Zhang X, Rowlands T, Egeblad M, Cowin P, Werb Z et al (2003) Evidence that transgenes encoding components of the Wnt signaling pathway preferentially induce mammary cancers from progenitor cells. Proc Natl Acad Sci USA 100: 15853–15858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, Asselin‐Labat ML, Gyorki DE, Ward T, Partanen A et al (2009) Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med 15: 907–913 [DOI] [PubMed] [Google Scholar]
- Lim E, Wu D, Pal B, Bouras T, Asselin‐Labat M‐L, Vaillant F, Yagita H, Lindeman GJ, Smyth GK, Visvader JE (2010) Transcriptome analyses of mouse and human mammary cell subpopulations reveal multiple conserved genes and pathways. Breast Cancer Res 12: R21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SY, Xia W, Wang JC, Kwong KY, Spohn B, Wen Y, Pestell RG, Hung MC (2000) Beta‐catenin, a novel prognostic marker for breast cancer: its roles in cyclin D1 expression and cancer progression. Proc Natl Acad Sci USA 97: 4262–4266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnemann JR, Miura H, Meixner LK, Irmler M, Kloos UJ, Hirschi B, Bartsch HS, Sass S, Beckers J, Theis FJ et al (2015) Quantification of regenerative potential in primary human mammary epithelial cells. Development 142: 3239–3251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu BY, McDermott SP, Khwaja SS, Alexander CM (2004) The transforming activity of Wnt effectors correlates with their ability to induce the accumulation of mammary progenitor cells. Proc Natl Acad Sci USA 101: 4158–4163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke M, Heywood M, Fawell S, Mackenzie IC (2005) Retention of intrinsic stem cell hierarchies in carcinoma‐derived cell lines. Cancer Res 65: 8944–8950 [DOI] [PubMed] [Google Scholar]
- Lydon JP, DeMayo FJ, Funk CR, Mani SK, Hughes AR, Montgomery CA, Shyamala G, Conneely OM, O'Malley BW (1995) Mice lacking progesterone receptor exhibit pleiotropic reproductive abnormalities. Genes Dev 9: 2266–2278 [DOI] [PubMed] [Google Scholar]
- Lydon JP, Ge G, Kittrell FS, Medina D, O'Malley BW (1999) Murine mammary gland carcinogenesis is critically dependent on progesterone receptor function. Cancer Res 59: 4276–4284 [PubMed] [Google Scholar]
- Maruyama R, Choudhury S, Kowalczyk A, Bessarabova M, Beresford‐Smith B, Conway T, Kaspi A, Wu Z, Nikolskaya T, Merino VF et al (2011) Epigenetic regulation of cell type‐specific expression patterns in the human mammary epithelium. PLoS Genet 7: e1001369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masciari S, Dillon DA, Rath M, Robson M, Weitzel JN, Balmana J, Gruber SB, Ford JM, Euhus D, Lebensohn A et al (2012) Breast cancer phenotype in women with TP53 germline mutations: a Li‐Fraumeni syndrome consortium effort. Breast Cancer Res Treat 133: 1125–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayers JR, Torrence ME, Danai LV, Papagiannakopoulos T, Davidson SM, Bauer MR, Lau AN, Ji BW, Dixit PD, Hosios AM et al (2016) Tissue of origin dictates branched‐chain amino acid metabolism in mutant Kras‐driven cancers. Science 353: 1161–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne RL, Kuchenbaecker KB, Michailidou K, Beesley J, Kar S, Lindström S, Hui S, Lemaçon A, Soucy P, Dennis J et al (2017) Identification of ten variants associated with risk of estrogen‐receptor‐negative breast cancer. Nat Genet 49: 1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi K, Shillingford JM, Smith GH, Grimm SL, Wagner KU, Oka T, Rosen JM, Robinson GW, Hennighausen L (2001) Signal transducer and activator of transcription (Stat) 5 controls the proliferation and differentiation of mammary alveolar epithelium. J Cell Biol 155: 531–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneux G, Geyer FC, Magnay FA, McCarthy A, Kendrick H, Natrajan R, MacKay A, Grigoriadis A, Tutt A, Ashworth A et al (2010) BRCA1 basal‐like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell 7: 403–417 [DOI] [PubMed] [Google Scholar]
- Morel AP, Ginestier C, Pommier RM, Cabaud O, Ruiz E, Wicinski J, Devouassoux‐Shisheboran M, Combaret V, Finetti P, Chassot C et al (2017) A stemness‐related ZEB1‐MSRB3 axis governs cellular pliancy and breast cancer genome stability. Nat Med 23: 568–578 [DOI] [PubMed] [Google Scholar]
- Morrow M, Chatterton RT, Rademaker AW, Hou N, Jordan VC, Hendrick RE, Khan SA (2010) A prospective study of variability in mammographic density during the menstrual cycle. Breast Cancer Res Treat 121: 565–574 [DOI] [PubMed] [Google Scholar]
- Mote PA, Bartow S, Tran N, Clarke CL (2002) Loss of co‐ordinate expression of progesterone receptors A and B is an early event in breast carcinogenesis. Breast Cancer Res Treat 72: 163–172 [DOI] [PubMed] [Google Scholar]
- Narod SA (2015) Tamoxifen chemoprevention—end of the road? JAMA Oncol 1: 1033–1034 [DOI] [PubMed] [Google Scholar]
- Nelson HD, Smith MEB, Griffin JC, Fu R (2013) Use of medications to reduce risk for primary breast cancer: a systematic review for the US. Preventive Services Task Force. Ann Intern Med 158: 604–614 [DOI] [PubMed] [Google Scholar]
- Nguyen QH, Pervolarakis N, Blake K, Ma D, Davis RT, James N, Phung AT, Willey E, Kumar R, Jabart E et al (2018) Profiling human breast epithelial cells using single cell RNA sequencing identifies cell diversity. Nat Commun 9: 2028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- NICE (2013) Familial breast cancer: classification, care and managing breast cancer and related risks in people with a family history of breast cancer. Cardiff, UK: National Collaborating Centre for Cancer; Natl. Institute Heal. Care Excell. 48 [Google Scholar]
- Nielsen TO, Parker JS, Leung S, Voduc D, Ebbert M, Vickery T, Davies SR, Snider J, Stijleman IJ, Reed J et al (2010) A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen‐treated estrogen receptor‐positive breast cancer. Clin Cancer Res 16: 5222–5232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan E, Vaillant F, Branstetter D, Pal B, Giner G, Whitehead L, Lok SW, Mann GB, Kathleen Cuningham Foundation Consortium for Research into Familial Breast Cancer (kConFab) , Rohrbach K et al (2016) RANK ligand as a potential target for breast cancer prevention in BRCA1‐mutation carriers. Nat Med 22: 933–939 [DOI] [PubMed] [Google Scholar]
- Nolan E, Lindeman GJ, Visvader JE (2017) Out‐RANKing BRCA1 in mutation carriers. Cancer Res 77: 595–600 [DOI] [PubMed] [Google Scholar]
- Oakes SR, Naylor MJ, Asselin‐Labat M‐L, Blazek KD, Gardiner‐Garden M, Hilton HN, Kazlauskas M, Pritchard MA, Chodosh LA, Pfeffer PL et al (2008) The Ets transcription factor Elf5 specifies mammary alveolar cell fate. Genes Dev 22: 581–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obr AE, Grimm SL, Bishop KA, Pike JW, Lydon JP, Edwards DP (2013) Progesterone receptor and Stat5 signaling cross talk through RANKL in mammary epithelial cells. Mol Endocrinol 27: 1808–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padamsee TJ, Wills CE, Yee LD, Paskett ED (2017) Decision making for breast cancer prevention among women at elevated risk. Breast Cancer Res 19: 34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal B, Bouras T, Shi W, Vaillant F, Sheridan JM, Fu N, Breslin K, Jiang K, Ritchie ME, Young M, Lindeman GJ, Smyth GK, Visvader JE (2013) Global Changes in the Mammary Epigenome Are Induced by Hormonal Cues and Coordinated by Ezh2. Cell Rep 3: 411–426 [DOI] [PubMed] [Google Scholar]
- Pal B, Chen Y, Vaillant F, Jamieson P, Gordon L, Rios AC, Wilcox S, Fu N, Liu KH, Jackling FC et al (2017) Construction of developmental lineage relationships in the mouse mammary gland by single‐cell RNA profiling. Nat Commun 8: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo I, Lillemoe HA, Blosser RJ, Choi M, Sauder CAM, Doxey DK, Mathieson T, Hancock BA, Baptiste D, Atale R et al (2014) Next‐generation transcriptome sequencing of the premenopausal breast epithelium using specimens from a normal human breast tissue bank. Breast Cancer Res 16: R26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellacani D, Bilenky M, Kannan N, Heravi‐Moussavi A, Knapp DJHF, Gakkhar S, Moksa M, Carles A, Moore R, Mungall AJ et al (2016) Analysis of normal human mammary epigenomes reveals cell‐specific active enhancer states and associated transcription factor networks. Cell Rep 17: 2060–2074 [DOI] [PubMed] [Google Scholar]
- Plaks V, Brenot A, Lawson DA, Linnemann JR, Van Kappel EC, Wong KC, de Sauvage F, Klein OD, Werb Z (2013) Lgr5‐expressing cells are sufficient and necessary for postnatal mammary gland organogenesis. Cell Rep 3: 70–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak P, Karlic R, Koren A, Thurman R, Sandstrom R, Lawrence MS, Reynolds A, Rynes E, Vlahovicek K, Stamatoyannopoulos JA et al (2015) Cell‐of‐origin chromatin organization shapes the mutational landscape of cancer. Nature 518: 360–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole AJ, Li Y, Kim Y, Lin S‐CJ, Lee W‐H, Lee EY‐HP (2006) Prevention of Brca1‐mediated mammary tumorigenesis in mice by a progesterone antagonist. Science 314: 1467–1470 [DOI] [PubMed] [Google Scholar]
- Porter D, Lahti‐Domenici J, Keshaviah A, Bae YK, Argani P, Marks J, Richardson A, Cooper A, Strausberg R, Riggins GJ et al (2003) Molecular markers in ductal carcinoma in situ of the breast. Mol Cancer Res 1: 362–375 [PubMed] [Google Scholar]
- Potten CS, Watson RJ, Williams GT, Tickle S, Roberts SA, Harris M, Howell A (1988) The effect of age and menstrual cycle upon proliferative activity of the normal human breast. Br J Cancer 58: 163–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powles TJ, Ashley S, Tidy A, Smith IE, Dowsett M (2007) Twenty‐year follow‐up of the Royal Marsden randomized, double‐blinded tamoxifen breast cancer prevention trial. J Natl Cancer Inst 99: 283–290 [DOI] [PubMed] [Google Scholar]
- Powles TJ, Diem SJ, Fabian CJ, Neven P, Wickerham DL, Cox DA, Muram D, Agnusdei D, Dowsett SA, Amewou‐Atisso M et al (2012) Breast cancer incidence in postmenopausal women with osteoporosis or low bone mass using arzoxifene. Breast Cancer Res Treat 134: 299–306 [DOI] [PubMed] [Google Scholar]
- Prat A, Perou CM (2011) Deconstructing the molecular portraits of breast cancer. Mol Oncol 5: 5–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajaram RD, Buric D, Caikovski M, Ayyanan A, Rougemont J, Shan J, Vainio SJ, Yalcin‐Ozuysal O, Brisken C (2015) Progesterone and Wnt4 control mammary stem cells via myoepithelial crosstalk. EMBO J 34: 641–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan R, Khan SA, Badve S (2002) Morphological changes in breast tissue with menstrual cycle. Mod Pathol 15: 1348–1356 [DOI] [PubMed] [Google Scholar]
- Raouf A, Zhao Y, To K, Stingl J, Delaney A, Barbara M, Iscove N, Jones S, McKinney S, Emerman J et al (2008) Transcriptome analysis of the normal human mammary cell commitment and differentiation process. Cell Stem Cell 3: 109–118 [DOI] [PubMed] [Google Scholar]
- Rebbeck TR, Friebel T, Lynch HT, Neuhausen SL, van ‘t Veer L, Garber JE, Evans GR, Narod SA, Isaacs C, Matloff E et al (2004) Bilateral prophylactic mastectomy reduces breast cancer risk in BRCA1 and BRCA2 mutation carriers: the PROSE Study Group. J Clin Oncol 22: 1055–1062 [DOI] [PubMed] [Google Scholar]
- Regan JL, Kendrick H, Magnay FA, Vafaizadeh V, Groner B, Smalley MJ (2012) c‐Kit is required for growth and survival of the cells of origin of Brca1‐mutation‐associated breast cancer. Oncogene 31: 869–883 [DOI] [PubMed] [Google Scholar]
- Rios AC, Fu NY, Lindeman GJ, Visvader JE (2014) In situ identification of bipotent stem cells in the mammary gland. Nature 506: 322–327 [DOI] [PubMed] [Google Scholar]
- Rossouw JE, Anderson GL, Prentice RL, LaCroix AZ, Kooperberg C, Stefanick ML, Jackson RD, Beresford SAA, Howard BV, Johnson KC et al (2002) Risks and benefits of estrogen plus progestin in healthy postmenopausal women: principal results From the Women's Health Initiative randomized controlled trial. JAMA 288: 321–333 [DOI] [PubMed] [Google Scholar]
- Russo J, Russo IH (2004) Development of the human breast. Maturitas 49: 2–15 [DOI] [PubMed] [Google Scholar]
- Said TK, Conneely OM, Medina D, O'Malley BW, Lydon JP (1997) Progesterone, in addition to estrogen, induces cyclin D1 expression in the murine mammary epithelial cell, in vivo . Endocrinology 138: 3933–3939 [DOI] [PubMed] [Google Scholar]
- Santagata S, Thakkar A, Ergonul A, Wang B, Woo T, Hu R, Harrell JC, McNamara G, Schwede M, Culhane AC et al (2014) Taxonomy of breast cancer based on normal cell phenotype predicts outcome. J Clin Invest 124: 859–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramek D, Leibbrandt A, Sigl V, Kenner L, Pospisilik JA, Lee HJ, Hanada R, Joshi PA, Aliprantis A, Glimcher L et al (2010) Osteoclast differentiation factor RANKL controls development of progestin‐driven mammary cancer. Nature 468: 98–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin‐Labat M‐L, Wu L, Lindeman GJ, Visvader JE (2006) Generation of a functional mammary gland from a single stem cell. Nature 439: 84–88 [DOI] [PubMed] [Google Scholar]
- Shehata M, Kim H, Vellanki R, Waterhouse PD, Mahendralingam M, Casey AE, Koritzinsky M, Khokha R (2019) Identifying the murine mammary cell target of metformin exposure. Commun Biol 2: 192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehata M, Teschendorff A, Sharp G, Novcic N, Russell A, Avril S, Prater M, Eirew P, Caldas C, Watson CJ et al (2012) Phenotypic and functional characterization of the luminal cell hierarchy of the mammary gland. Breast Cancer Res 14: R134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehata M, Waterhouse PD, Casey AE, Fang H, Hazelwood L, Khokha R (2018) Proliferative heterogeneity of murine epithelial cells in the adult mammary gland. Commun Biol 1: 111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiah Y‐J, Tharmapalan P, Casey AE, Joshi PA, McKee TD, Jackson HW, Beristain AG, Chan‐Seng‐Yue MA, Bader GD, Lydon JP et al (2015) A progesterone‐CXCR178 axis controls mammary progenitor cell fate in the adult gland. Stem Cell Rep 4: 313–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigl V, Owusu‐Boaitey K, Joshi PA, Kavirayani A, Wirnsberger G, Novatchkova M, Kozieradzki I, Schramek D, Edokobi N, Hersl J et al (2016) RANKL/RANK control Brca1 mutation‐driven mammary tumors. Cell Res 26: 761–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibinski A, Breindel JL, Prat A, Galván P, Smith E, Rolfs A, Gupta PB, LaBaer J, Kuperwasser C (2014) The Hippo transducer TAZ interacts with the SWI/SNF complex to regulate breast epithelial lineage commitment. Cell Rep 6: 1059–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol ES, Miller DH, Breggia A, Spencer KC, Arendt LM, Gupta PB (2016) Growth of human breast tissues from patient cells in 3D hydrogel scaffolds. Breast Cancer Res 18: 19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M, Vogelstein B, Giovannucci EL, Willett WC, Tomasetti C (2018) Cancer prevention: molecular and epidemiologic consensus. Science 361: 1317–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spike BT, Engle DD, Lin JC, Cheung SK, La J, Wahl GM (2012) A mammary stem cell population identified and characterized in late embryogenesis reveals similarities to human breast cancer. Cell Stem Cell 10: 183–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporn MB (1976) Approaches to prevention of epithelial cancer during the preneoplastic period. Cancer Res 36: 2699–2702 [PubMed] [Google Scholar]
- Sternlicht MD, Sunnarborg SW, Kouros‐Mehr H, Yu Y, Lee DC, Werb Z (2005) Mammary ductal morphogenesis requires paracrine activation of stromal EGFR via ADAM17‐dependent shedding of epithelial amphiregulin. Development 132: 3923–3933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingl J, Eaves CJ, Zandieh I, Emerman JT (2001) Characterization of bipotent mammary epithelial progenitor cells in normal adult human breast tissue. Breast Cancer Res Treat 67: 93–109 [DOI] [PubMed] [Google Scholar]
- Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, Li HI, Eaves CJ (2006) Purification and unique properties of mammary epithelial stem cells. Nature 439: 993–997 [DOI] [PubMed] [Google Scholar]
- Suh N, Glasebrook AL, Palkowitz AD, Bryant HU, Burris LL, Starling JJ, Pearce HL, Williams C, Peer C, Wang Y et al (2001) Arzoxifene, a new selective estrogen receptor modulator for chemoprevention of experimental breast cancer. Cancer Res 61: 8412–8415 [PubMed] [Google Scholar]
- Takebe N, Miele L, Harris PJ, Jeong W, Bando H, Kahn M, Yang SX, Ivy SP (2015) Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: clinical update. Nat Rev Clin Oncol 12: 445–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M‐H, Mester JL, Ngeow J, Rybicki LA, Orloff MS, Eng C (2012) Lifetime cancer risks in individuals with germline PTEN mutations. Clin Cancer Res 18: 400–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanos T, Sflomos G, Echeverria PC, Ayyanan A, Gutierrez M, Delaloye J‐F, Raffoul W, Fiche M, Dougall W, Schneider P et al (2013) Progesterone/RANKL is a major regulatory axis in the human breast. Sci Transl Med 5: 182ra55 [DOI] [PubMed] [Google Scholar]
- Tocci JM, Felcher CM, García Solá ME, Goddio MV, Zimberlin MN, Rubinstein N, Srebrow A, Coso OA, Abba MC, Meiss RP et al (2018) R‐spondin3 is associated with basal‐progenitor behavior in normal and tumor mammary cells. Cancer Res 78: 4497–4511 [DOI] [PubMed] [Google Scholar]
- Tomasetti C, Vogelstein B (2015) Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science 347: 78–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasetti C, Li L, Vogelstein B (2017) Stem cell divisions, somatic mutations, cancer etiology, and cancer prevention. Science 355: 1330–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi A, King C, de la Morenas A, Perry VK, Burke B, Antoine GA, Hirsch EF, Kavanah M, Mendez J, Stone M et al (2008) Gene expression abnormalities in histologically normal breast epithelium of breast cancer patients. Int J Cancer 122: 1557–1566 [DOI] [PubMed] [Google Scholar]
- Tsai YC, Lu Y, Nichols PW, Zlotnikov G, Jones PA, Smith HS (1996) Contiguous patches of normal human mammary epithelium derived from a single stem cell: implications for breast carcinogenesis. Cancer Res 56: 402–404 [PubMed] [Google Scholar]
- Tsukamoto AS, Grosschedl R, Guzman RC, Parslow T, Varmus HE (1988) Expression of the int‐1 gene in transgenic mice is associated with mammary gland hyperplasia and adenocarcinomas in male and female mice. Cell 55: 619–625 [DOI] [PubMed] [Google Scholar]
- Van Keymeulen A, Rocha AS, Ousset M, Beck B, Bouvencourt G, Rock J, Sharma N, Dekoninck S, Blanpain C (2011) Distinct stem cells contribute to mammary gland development and maintenance. Nature 479: 189–193 [DOI] [PubMed] [Google Scholar]
- Van Keymeulen A, Lee MY, Ousset M, Brohée S, Rorive S, Giraddi RR, Wuidart A, Bouvencourt G, Dubois C, Salmon I et al (2015) Reactivation of multipotency by oncogenic PIK3CA induces breast tumour heterogeneity. Nature 525: 119–123 [DOI] [PubMed] [Google Scholar]
- Veronesi U, Maisonneuve P, Rotmensz N, Bonanni B, Boyle P, Viale G, Costa A, Sacchini V, Travaglini R, D'Aiuto G et al (2007) Tamoxifen for the prevention of breast cancer: late results of the Italian Randomized Tamoxifen prevention trial among women with hysterectomy. J Natl Cancer Inst 99: 727–737 [DOI] [PubMed] [Google Scholar]
- Visvader JE, Stingl J (2014) Mammary stem cells and the differentiation hierarchy: current status and perspectives. Genes Dev 28: 1143–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel VG, Costantino JP, Wickerham DL, Cronin WM, Cecchini RS, Atkins JN, Bevers TB, Fehrenbacher L, Pajon ER, Wade JL et al (2006) Effects of tamoxifen vs raloxifene on the risk of developing invasive breast cancer and other disease outcomes: the NSABP Study of Tamoxifen and Raloxifene (STAR) P‐2 trial. JAMA 295: 2727–2741 [DOI] [PubMed] [Google Scholar]
- Vogel VG, Costantino JP, Wickerham DL, Cronin WM, Cecchini RS, Atkins JN, Bevers TB, Fehrenbacher L, Pajon ER, Wade JL et al (2010) Update of the National Surgical Adjuvant Breast and Bowel Project Study of Tamoxifen and Raloxifene (STAR) P‐2 Trial: preventing breast cancer. Cancer Prev Res (Phila) 3: 696–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh T, Casadei S, Coats KH, Swisher E, Stray SM, Higgins J, Roach KC, Mandell J, Lee MK, Ciernikova S et al (2006) Spectrum of mutations in BRCA1, BRCA2, CHEK2, and TP53 in families at high risk of breast cancer. JAMA 295: 1379–1388 [DOI] [PubMed] [Google Scholar]
- Wang D, Cai C, Dong X, Yu QC, Zhang X‐O, Yang L, Zeng YA (2015) Identification of multipotent mammary stem cells by protein C receptor expression. Nature 517: 81–84 [DOI] [PubMed] [Google Scholar]
- Wang C, Christin JR, Oktay MH, Guo W (2017) Lineage‐biased stem cells maintain estrogen‐receptor‐positive and ‐negative mouse mammary luminal lineages. Cell Rep 18: 2825–2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellings SR, Jensen HM, Marcum RG (1975) An atlas of subgross pathology of the human breast with special reference to possible precancerous lesions. J Natl Cancer Inst 55: 231–273 [PubMed] [Google Scholar]
- Wellings SR (1980) A hypothesis of the origin of human breast cancer from the terminal ductal lobular unit. Pathol Res Pract 166: 515–535 [DOI] [PubMed] [Google Scholar]
- Widschwendter M, Rosenthal AN, Philpott S, Rizzuto I, Fraser L, Hayward J, Intermaggio MP, Edlund CK, Ramus SJ, Gayther SA et al (2013) The sex hormone system in carriers of BRCA1/2 mutations: a case‐control study. Lancet Oncol 14: 1226–1232 [DOI] [PubMed] [Google Scholar]
- Wuidart A, Ousset M, Rulands S, Simons BD, Van Keymeulen A, Blanpain C (2016) Quantitative lineage tracing strategies to resolve multipotency in tissue‐specific stem cells. Genes Dev 30: 1261–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Ferguson AT, Nass SJ, Phillips DL, Butash KA, Wang SM, Herman JG, Davidson NE (2000) Transcriptional activation of estrogen receptor alpha in human breast cancer cells by histone deacetylase inhibition. Cancer Res 60: 6890–6894 [PubMed] [Google Scholar]
- Yao J, Weremowicz S, Feng B, Gentleman RC, Marks JR, Gelman R, Brennan C, Polyak K (2006) Combined cDNA array comparative genomic hybridization and serial analysis of gene expression analysis of breast tumor progression. Cancer Res 66: 4065–4078 [DOI] [PubMed] [Google Scholar]
- Zeng YA, Nusse R (2010) Wnt proteins are self‐renewal factors for mammary stem cells and promote their long‐term expansion in culture. Cell Stem Cell 6: 568–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Long X (2015) Association of BRCA1 promoter methylation with sporadic breast cancers: evidence from 40 studies. Sci Rep 5: 17869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Chiang H‐C, Wang Y, Zhang C, Smith S, Zhao X, Nair SJ, Michalek J, Jatoi I, Lautner M et al (2017) Attenuation of RNA polymerase II pausing mitigates BRCA1‐associated R‐loop accumulation and tumorigenesis. Nat Commun 8: 15908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Chehab R, Tkalcevic J, Naylor MJ, Harris J, Wilson TJ, Tsao S, Tellis I, Zavarsek S, Xu D et al (2005) Elf5 is essential for early embryogenesis and mammary gland development during pregnancy and lactation. EMBO J 24: 635–644 [DOI] [PMC free article] [PubMed] [Google Scholar]


