Abstract
The self‐assembly of cellular macromolecular machines such as the bacterial flagellar motor requires the spatio‐temporal synchronization of gene expression with proper protein localization and association of dozens of protein components. In Salmonella and Escherichia coli, a sequential, outward assembly mechanism has been proposed for the flagellar motor starting from the inner membrane, with the addition of each new component stabilizing the previous one. However, very little is known about flagellar disassembly. Here, using electron cryo‐tomography and sub‐tomogram averaging of intact Legionella pneumophila, Pseudomonas aeruginosa, and Shewanella oneidensis cells, we study flagellar motor disassembly and assembly in situ. We first show that motor disassembly results in stable outer membrane‐embedded sub‐complexes. These sub‐complexes consist of the periplasmic embellished P‐ and L‐rings, and bend the membrane inward while it remains apparently sealed. Additionally, we also observe various intermediates of the assembly process including an inner‐membrane sub‐complex consisting of the C‐ring, MS‐ring, and export apparatus. Finally, we show that the L‐ring is responsible for reshaping the outer membrane, a crucial step in the flagellar assembly process.
Keywords: assembly, bacterial flagellar motor, disassembly, electron cryo‐tomography, in situ imaging
Subject Categories: Membrane & Intracellular Transport; Microbiology, Virology & Host Pathogen Interaction; Structural Biology
Introduction
In order to move efficiently in their low‐Reynolds‐number environment (Purcell, 1977), bacteria have evolved a complex membrane‐embedded nanomachine known as the bacterial flagellum which exploits the flux of ions across the membrane to generate a mechanical torque that rotates a long filament (Manson et al, 1977; Hirota et al, 1981; Macnab, 1999). In general, the bacterial flagellum consists of a cell‐envelope‐embedded motor, a hook, and a filament. The motor consists of two parts: the rotor, which is composed of the basal body and the switch complex, and the stator (see (Berg, 2003; Sowa & Berry, 2008) and references therein). In the canonical flagellar systems of Salmonella enterica and Escherichia coli, the cell‐envelope‐spanning basal body consists of multiple rings: the MS (membrane/supramembrane)‐ring (formed by the protein FliF), the P (peptidoglycan)‐ring (FlgI), and the L (lipopolysaccharide)‐ring (FlgH). The P‐ and L‐rings form a bushing that allows the flagellum to rotate within the cell wall and outer membrane. These rings surround the rod structure (FliE, FlgB, FlgC, FlgF and FlgG). The C (cytoplasmic)‐ring consists of three proteins (FliG, FliM, and FliN) and forms the switch complex which is responsible for switching the direction of flagellar rotation between clockwise and counterclockwise. In E. coli and Salmonella, the stator is formed by a complex of two proteins, MotA and MotB. While MotB interacts with the peptidoglycan through a peptidoglycan‐binding domain (Koebnik, 1995; Morimoto & Minamino, 2014; Kojima et al, 2018), MotA interacts with FliG to generate the torque required for flagellar rotation (Morimoto & Minamino, 2014). The extracellular parts of the bacterial flagellum comprise the hook, which acts as a universal joint, and the filament, which serves as a helical propeller. The motor also has a type III secretion system (T3SS) export apparatus consisting of eight proteins (FliH, FliI, FliJ, FlhA, FlhB, FliP, FliQ, and FliR) located at the inner membrane. See Appendix Fig S1 for a schematic of these components.
High‐resolution structures have been solved for many components of the flagellar motor by X‐ray crystallography, NMR spectroscopy, and cryo‐EM single‐particle reconstruction, and structures of purified sub‐complexes of the Salmonella motor have been solved by electron microscopy (Francis et al, 1994; Thomas et al, 2006). Unfortunately, due to the large size of the complex and its position spanning the cell envelope, flagellar motors lose components when purified. Recently, the advent of electron cryo‐tomography (ECT) (Gan & Jensen, 2012; Oikonomou & Jensen, 2017) allowed our group and others to reveal the in situ structures of various bacterial flagellar motors within intact cells at macromolecular (~4 nm) resolution. These studies showed the diversity of flagellar motors in different species adapted to unique external environments (Chen et al, 2011; Zhao et al, 2014; Minamino & Imada, 2015; Terashima et al, 2017; Zhu et al, 2017; Kaplan et al, 2019). For instance, some species elaborate their P‐ and L‐rings with additional periplasmic components, including the T‐ring (MotX and MotY) and H‐ring (FlgO, FlgP, and FlgT) (Terashima et al, 2006, 2010; Beeby et al, 2016; Chaban et al, 2018).
The bacterial flagellum offers a prime example of the self‐assembly process of supramolecular complexes in the cell and is of interest to disciplines ranging from evolutionary biology to nanotechnology (Stock et al, 2012). Assembly requires major investments of cell energy (McCarter, 2001; Macnab, 2003). A recent study on the Salmonella motor found that only the filaments that were broken mechanically were observed to regrow, but not the filaments which were denatured using a laser pulse. This is likely because the process of flagellar assembly is so energetically demanding (Paradis et al, 2017). Our current understanding of flagellar assembly comes from studies of two enteric species of Gram‐negative bacteria, E. coli and Salmonella, which suggest an inside‐to‐outside sequential assembly process starting from the export apparatus, MS‐ring (FliF), followed by the C‐ring proteins, the rod, the P‐ and L‐rings, the hook, and finally the filament (Jones & Macnab, 1990; Kubori et al, 1992; Macnab, 2003; Li & Sourjik, 2011; Fabiani et al, 2017; Fukumura et al, 2017). The process is thought to be a cooperative one, whereby the addition of each new component stabilizes its antecedent (Li & Sourjik, 2011). The proteins forming the rod, the hook, and the filament are secreted through the T3SS export apparatus (Macnab, 2003). The P‐, L‐, T‐, and H‐ring proteins, however, are secreted to the periplasm through the conventional Sec pathway (Oliver, 1985; Homma et al, 1987; Jones et al, 1990; Macnab, 2003). Interestingly, the P‐ and L‐ring proteins in S. typhimurium were found to exist in a stable state in the periplasm in the absence of the inner membrane‐associated components (Jones et al, 1990). Despite the stability and independent export of their components, evidence suggests that P‐ and L‐rings only assemble after the assembling rod extends through the periplasm (Macnab, 2003; Cohen & Hughes, 2014). On the other hand, despite decades of work on the flagellar motor, very little is known about its disassembly. Biochemical studies on flagellar ejection in Caulobacter crescentus, which is a specific programmed developmental process, showed that the disassembly process is initiated by digestion of the C terminus of FliF, followed by ejection of the rod, the hook, and the filament as a stable sub‐complex (Jenal & Shapiro, 1996; Aldridge & Jenal, 1999; Grunenfelder et al, 2004; Kanbe, 2005). In addition, the FliG and FliM components of the C‐ring (which is connected to the export apparatus) were also shown to undergo proteolysis during the ejection process in C. crescentus (Jenal et al, 1994; Jenal & Shapiro, 1996), but the process has not been structurally verified in vivo (by imaging), and how disassembly occurs in other bacterial species also remains unknown.
Results
We recently reported the structure of intact flagellar motors in three non‐enteric Gammaproteobacteria species, Legionella pneumophila, Pseudomonas aeruginosa, and Shewanella oneidensis MR‐1, grown under different conditions (see Materials and Methods), by ECT (Kaplan et al, 2019). L. pneumophila and P. aeruginosa are human pathogens that cause serious pulmonary infections in which the flagellum is a key virulence factor (Feldman et al, 1998; Appelt & Heuner, 2017). S. oneidensis is a model system for studying extracellular respiration and is known for its production of multiheme cytochrome electron conduits and outer‐membrane appendages (Subramanian et al, 2018). All three species utilize a single, polar flagellum. We found that the flagellar motors of all three species contain elaborated P‐ and L‐rings: L. pneumophila and P. aeruginosa have an extra periplasmic ring surrounding the P‐ring, and S. oneidensis has both T‐ and H‐rings surrounding its P‐ and L‐rings, respectively (Kaplan et al, 2019). Complementing ECT imaging with bioinformatics analysis, we found a correlation between the presence of a MotY ring and the type of the stator system of the motor (Kaplan et al, 2019).
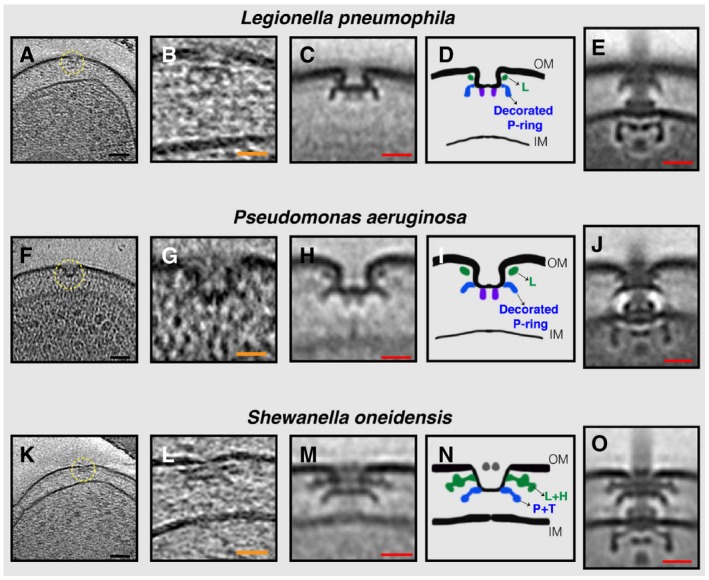
Here, we report that in addition to fully assembled flagella, in those same tomograms of all three species we observed isolated outer‐membrane complexes similar to the periplasmic P‐ and L‐rings and their associated elaborations (henceforth referred to as PL sub‐complexes) (Fig 1A–O). By performing sub‐tomogram averaging of these sub‐complexes to enhance the signal‐to‐noise ratio, we confirmed that these were PL sub‐complexes and that they lacked other flagellar components (Fig 1C, D, H, I, M and N; compare to averages of fully assembled motors in 1 E, J, and O). PL sub‐complexes had two noticeable features. First, they curved the outer membrane inward into an inverted omega shape. The membrane appeared continuous, however, and no pore was visible. Second, in L. pneumophila and P. aeruginosa two protein densities were seen extending downwards from the center of the sub‐complexes (Fig 1C, D, H and I, purple densities). These densities were less clear in the case of S. oneidensis (Fig 1M).
Figure 1. Stable PL sub‐complexes in three bacterial species imaged by ECT.
-
A–O(A, F, K) slices through electron cryo‐tomograms of L. pneumophila, P. aeruginosa, and S. oneidensis cells, respectively, highlighting a PL sub‐complex in the outer membrane (dashed yellow circle). (B, G, L) Enlarged views of the sub‐complexes. (C, H, M) Sub‐tomogram averages of PL sub‐complexes from each species. (D, I, N) Schematic representations of the sub‐tomogram averages with different rings colored and labeled. (E, J, O) Sub‐tomogram averages of fully assembled flagella from each species for comparison. Scale bars: (black) 50 nm, (orange) 25 nm, and (red) 20 nm.
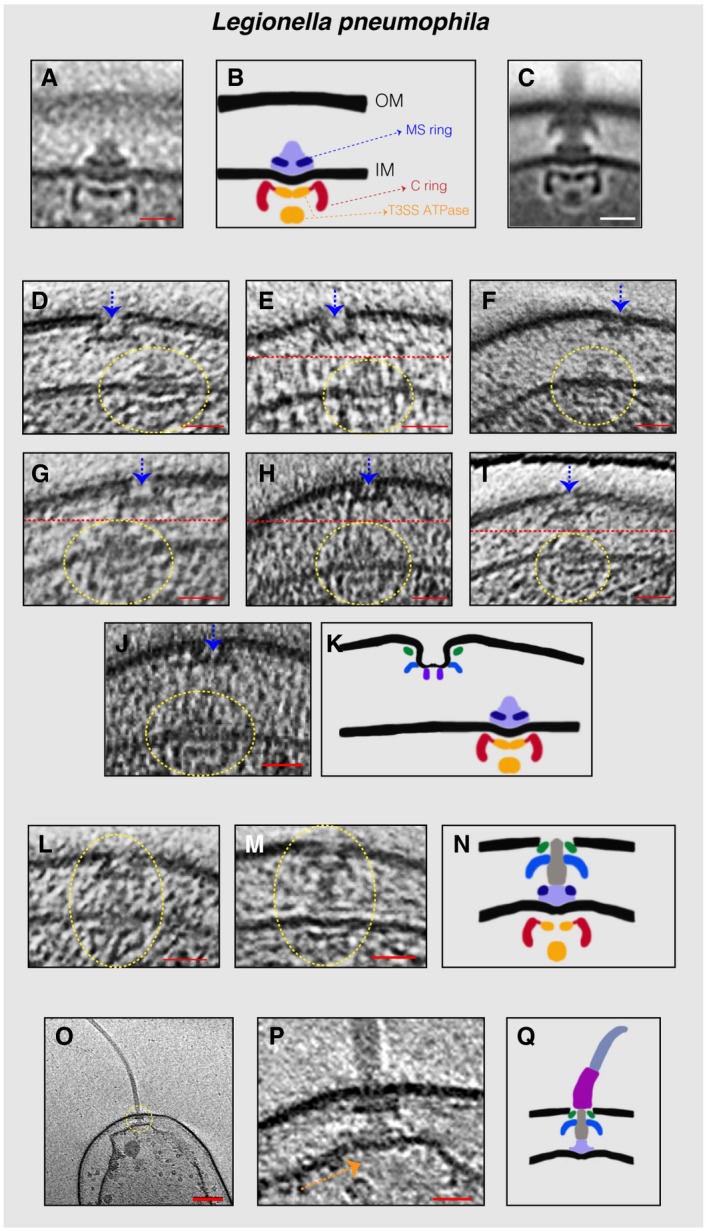
In many L. pneumophila cells, we also observed an inner‐membrane complex constituting the C‐ and MS‐rings and the T3SS export apparatus (referred to henceforth as the inner‐membrane (IM) sub‐complex) in the vicinity of the PL sub‐complexes (Fig 2A–K and Movies EV1 and EV2). In these cases, the lateral distance (in the direction of the plane parallel to the membranes) between the PL sub‐complex and the IM sub‐complex ranged from 60 nm to 5 nm. In 11 L. pneumophila cells, we found fully assembled motors lacking the hook and filament (Fig 2L–N), but no motors with only the hook (and not the filament) were observed. In one tomogram of a lysed L. pneumophila cell, we found an example of a flagellar sub‐complex missing the C‐ring, MS‐ring, and export apparatus (Fig 2O–Q). Since secretion of the rod, hook, and filament proteins into the periplasm requires the T3SS export apparatus, this must represent a disassembly product which is in agreement with what was previously described for C. crescentus (Jenal et al, 1994; Jenal & Shapiro, 1996; Aldridge & Jenal, 1999; Grunenfelder et al, 2004; Kanbe, 2005).
Figure 2. Flagellar sub‐complexes in L. pneumophila .
-
ASub‐tomogram average of the IM sub‐complex constituting the C‐ ring, MS‐ring, and export apparatus.
-
BSchematic representation of the sub‐tomogram average shown in (A) highlighting the different parts of the complex.
-
CSub‐tomogram average of the motors of fully assembled flagella.
-
D–JSlices through electron cryo‐tomograms showing neighboring PL and IM sub‐complexes. Dashed yellow circles highlight the IM sub‐complex, while dashed blue arrows highlight the PL sub‐complex. Dashed red lines mark the border between two images used to make a composite image when the PL and IM sub‐complexes were found at different levels in the tomogram.
-
KSchematic representation of an IM sub‐complex in the vicinity of a PL sub‐complex.
-
L, MSlices through electron cryo‐tomograms of L. pneumophila highlighting the presence of a fully assembled basal body lacking the hook and the filament.
-
NSchematic representation of the basal bodies in (L and M).
-
OCentral slice through an electron cryo‐tomogram of a lysed cell. The dashed yellow circle highlights the flagellar motor.
-
PEnlarged view of the same slice shown in (O). The absence of the C‐ring and the export apparatus is highlighted by the dashed orange arrow.
-
QSchematic representation of the complex found in (P).
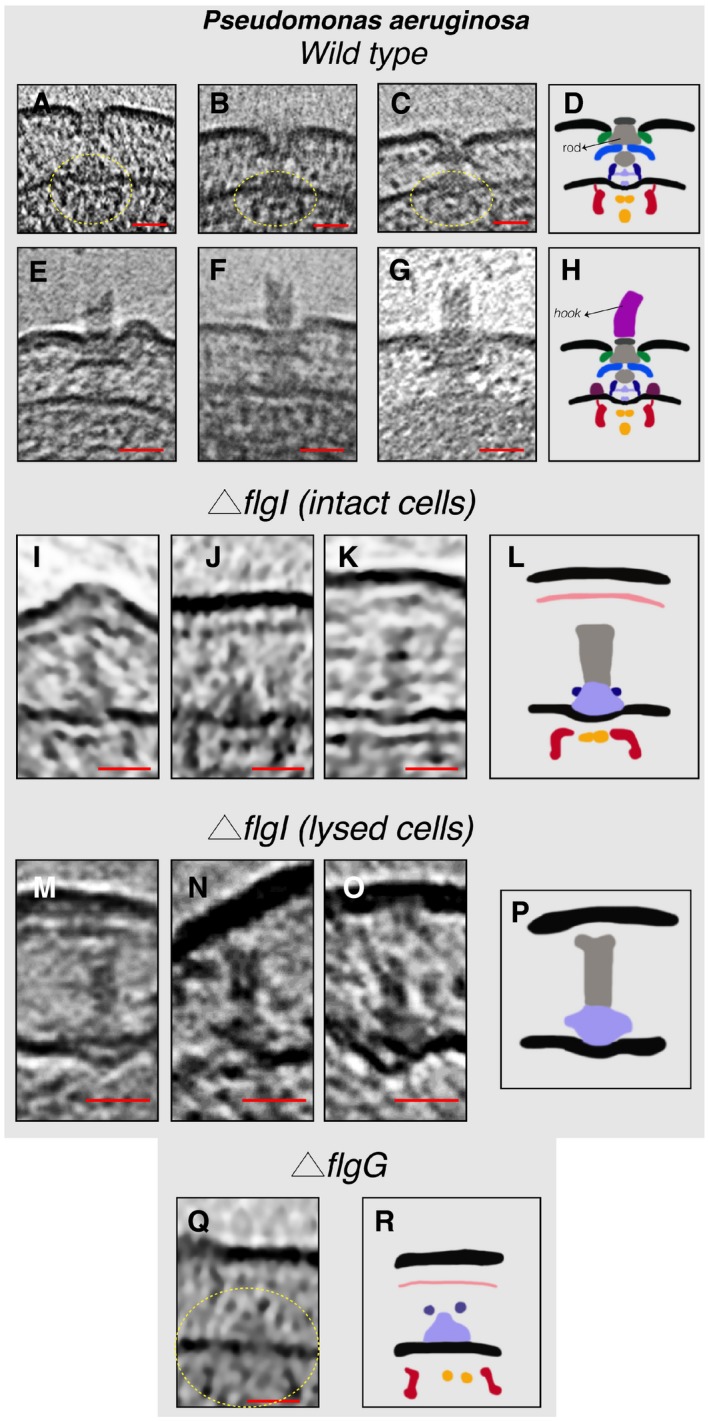
In tomograms of P. aeruginosa cells, we found (next to fully assembled flagella and PL sub‐complexes) examples of fully assembled motors both without (five cases) and with (three cases) the hook (Fig 3A–H). The low number of particles in this state suggests a fast transition from the fully assembled motor to the fully assembled flagellum. Interestingly, the upper end of the hook in two cases was not flat (Fig 3E and F) which could be either due to a breakage event or that the addition of the hook protein (FlgE) might not always proceed in a symmetrical pattern during the assembly process. To further investigate the PL sub‐complexes in P. aeruginosa and their relation to the fully assembled motor, we imaged different mutant strains of P. aeruginosa, these being: a strain lacking the P‐ring protein (FlgI, referred to henceforth as ΔflgI), a strain lacking the T3SS export apparatus protein FlhA (referred to henceforth as ΔflhA), and a strain lacking the rod protein FlgG (referred to henceforth as ΔflgG). In ΔflgI cells, no flagella or PL sub‐complexes could be detected; nevertheless, a structure comprising the inner‐membrane sub‐complex and the rod was observed (three cases) (Fig 3I–L). Interestingly, in one example this complex was pushing the outer membrane outward (Fig 3I). In a few cases of lysed ΔflgI cells, a complex consisting only of the MS‐ring and the rod was seen (three examples) (Fig 3M–P). As the export of the rod proteins to the periplasm requires the T3SS export apparatus, this complex ought to be a disassembly product. In the ΔflgG strain, the inner‐membrane sub‐complex was observed in two examples (Fig 3Q and R) and no (sub‐)complexes related to the flagellar motor could be detected in cells lacking FlhA which is believed to be one of the first proteins to be assembled (Li & Sourjik, 2011; Fabiani et al, 2017; Fukumura et al, 2017).
Figure 3. Flagellar sub‐complexes in P. aeruginosa .
-
A–CSlices through electron cryo‐tomograms showing fully assembled motors without the hook and filament. The dashed yellow circles indicate the IM sub‐complex.
-
DSchematic representation of the P. aeruginosa motors lacking the hook and filament shown in (A–C).
-
E–GSlices through electron cryo‐tomograms showing fully assembled motors with the hook and lacking the filament.
-
HSchematic representation of the motors with the hook shown in (E–G).
-
I–KSlices through electron cryo‐tomograms of intact P. aeruginosa ΔflgI cells showing the presence of the inner‐membrane sub‐complex with the rod.
-
LSchematic representation of the inner‐membrane sub‐complex with the rod shown in (I‐K).
-
M–OSlices through electron cryo‐tomograms of lysed P. aeruginosa ΔflgI cells showing the presence of the sub‐complex constituting the MS‐ring and the rod.
-
PSchematic representation of the sub‐complex described in (M–O).
-
QA slice through an electron cryo‐tomogram of a P. aeruginosa ΔflgG cell highlighting the presence of the inner‐membrane sub‐complex.
-
RSchematic representation of the inner‐membrane sub‐complex shown in (Q).
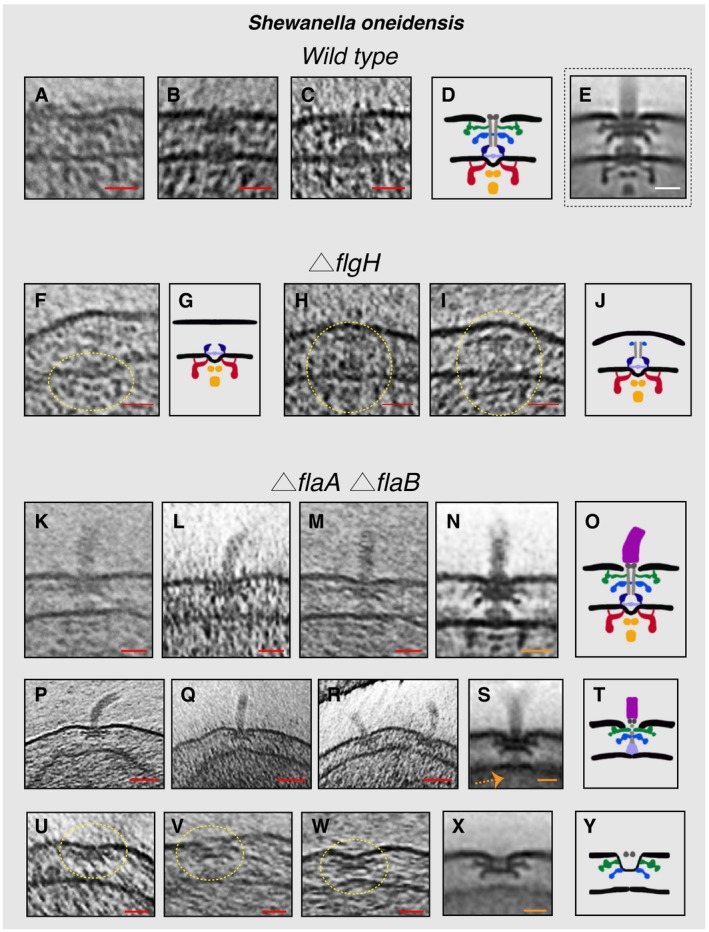
In S. oneidensis cells, we saw (four cases) fully assembled motors lacking the hook, and PL sub‐complexes which were in some cases present next to a fully assembled flagellum (Figs 4A–E and Appendix Fig S2). Additionally, we generated and imaged a S. oneidensis strain lacking the L‐ring protein FlgH. As expected, no flagella or PL sub‐complexes were seen in tomograms of ΔflgH cells. In a few cases, however, we did observe IM sub‐complexes (three examples) or the IM sub‐complex with a rod and P‐ring (five examples) (Fig 4F–J). This indicates that the non‐elaborated P‐ring can form in the absence of the L‐ring, but that FlgH is required for the full PL sub‐complex to form, the outer membrane to be reshaped, and the flagellum to proceed outside the cell. These results are consistent with studies in E. coli showing P‐ring assembly in the absence of the L‐ring (Jones et al, 1987; Ohnishi et al, 1987). However, we noticed that in the absence of FlgH the P‐ring is located much higher in the periplasm (~30 nm from the inner membrane) compared to the wild‐type motor where the P‐ring is only ~18 nm from the inner membrane. Intriguingly, the inner‐membrane sub‐complex with a rod and P‐ring appeared to push the outer membrane outward (Fig 4H and I) in a pattern reminiscent to what was seen in P. aeruginosa ΔflgI (Fig 3I). Next, we investigated a strain lacking the flagellar filament. Previous studies showed that S. oneidensis cells lacking the flagellin proteins, FlaA and FlaB, are completely non‐motile (Wu et al, 2011). In a ΔflaA ΔflaB strain, no flagellar filaments were seen, as expected. In a few cells, we observed fully assembled motors with a hook (Fig 4K–O). We also frequently observed a complex comprising the PL sub‐complex together with the rod (and possibly part of the MS‐ring) and the hook, but no IM sub‐complex or export apparatus (examples in Fig 4P–T). Again, we reasoned that this must be a disassembly intermediate, as the hook and rod proteins are secreted to the periplasm by the T3SS export apparatus. In many cases, multiple copies of this disassembly product were present at the cell pole (Fig 4R and Movie EV3), suggesting a repetitive process of attempted flagellar assembly and disassembly. Furthermore, PL sub‐complexes akin to those seen in wild‐type cells were present (Fig 4U–Y).
Figure 4. Flagellar sub‐complexes in S. oneidensis wild‐type and mutant cells.
-
A–CSlices through electron cryo‐tomograms of wild‐type cells showing fully assembled motors without the hook and filament.
-
DSchematic representation of the motors lacking the hook and filament shown in (A–C).
-
ESub‐tomogram average of the motors of fully assembled flagella.
-
FSlice through an electron cryo‐tomogram of a ΔflgH cell showing an IM sub‐complex, indicated by the dashed yellow circle.
-
GSchematic representation of the IM sub‐complex shown in (F).
-
H, ISlices through electron cryo‐tomograms of ΔflgH cells showing the IM sub‐complex with the rod and the P‐ring, indicated by the dashed yellow circles.
-
JSchematic representation of the structures shown in (H and I).
-
K–MSlices through electron cryo‐tomograms of ΔflaA/B cells highlighting the flagellar motor and the hook (without the filament).
-
NSub‐tomogram average of the flagellar motor and the hook structure found in ΔflaA/B cells.
-
OSchematic representation of the sub‐tomogram average shown in (N).
-
P–RSlices through electron cryo‐tomograms of ΔflaA/B cells illustrating a disassembly product constituting the PL sub‐complex, the rod, and the hook.
-
SSub‐tomogram average of the disassembly complex shown in (P–R). The dashed orange arrow indicates the absence of the IM sub‐complex in this structure.
-
TSchematic representation of the disassembly product shown in (P–S).
-
U–WSlices through electron cryo‐tomograms of ΔflaA/B cells highlighting PL sub‐complexes (dashed yellow circles).
-
XSub‐tomogram average of the PL sub‐complexes in ΔflaA/B cells.
-
YSchematic representation of the sub‐tomogram average shown in (X).
To confirm whether the observed PL sub‐complexes are assembly or disassembly products, we imaged a Pseudomonas aeruginosa strain lacking the MS protein FliF and generated and imaged the same mutant in S. oneidensis (referred to henceforth as ΔfliF). In the classical assembly pathway, FliF is one of the first proteins that is required to assemble and recruit other flagellar proteins during the process. If the PL sub‐complexes are assembled independently of other flagellar proteins, they should still be present in this mutant strain. In 150 tomograms of the P. aeruginosa ΔfliF strain, no flagella or outer‐membrane sub‐complexes were observed, indicating that PL sub‐complexes do not assemble in the absence of the motor and we therefore conclude they are disassembly products. Corroborating this notion is the absence of PL sub‐complexes in 45 tomograms of S. oneidensis ΔfliF cells. Interestingly, we observed the presence of chemoreceptor arrays in the S. oneidensis ΔfliF strain (Appendix Fig S3). It is known that the flagellar motor and chemotaxis genes are clustered into three different groups that are expressed in a hierarchical manner (see Chilcott and Hughes (2000) for a discussion about E. coli and Salmonella systems and Wu et al (2011) for a discussion about the S. oneidensis system); therefore, the presence of chemoreceptor arrays indicates that (at least some of the) flagellar/chemotaxis genes are being expressed in this mutant.
As many of our tomograms were from cells grown in overnight cultures to high OD600, we set out to explore the relationship between the flagellar disassembly process and the cell density (OD600). To this end, we prepared two S. oneidensis samples with different OD600 (one grown overnight to high OD600 and the other grown for a few hours to low OD600, see Materials and Methods). We visualized them by negative‐stain electron microscopy and counted the number of unambiguously cell‐attached flagella in 100 randomly selected cells in each sample. We found 20 cell‐attached flagella in the high OD600 sample and 48 in the sample with low OD600. Moreover, we repeated the same experiment for S. oneidensis cells grown either in fresh LB medium, LB medium of an overnight culture (spent LB medium), minimal medium, or minimal medium supplemented with fresh LB for 3 h (see Materials and Methods). While 44 cell‐attached flagella were counted in 60 randomly selected cells from the sample grown in fresh LB medium, only 9 were found in the cells grown in the spent LB medium. In accordance, 8 cell‐attached flagella were observed in 60 cells grown in minimal medium compared to 19 from the cells grown in minimal medium supplemented with LB. These results, together with the fact that we observed the PL sub‐complexes in tomograms of cells grown to high OD600, indicate that the cells disassemble their flagella when they enter the stationary phase at high OD600 with decreased level of nutrients in the medium; therefore, we believe that this process is of a different nature than the programmed flagellar ejection process that happens during the lifecycle of C. crescentus.
As mentioned earlier, while flagellar disassembly leaves stable PL sub‐complexes in the outer membrane, the membrane remains sealed with no apparent hole. This membrane‐sealing process might be due to a conformational change that happens in the PL sub‐complex upon flagellar disassembly, or might involve a novel protein(s). To investigate possible novel candidates involved in this process (other than the PL sub‐complex proteins), we looked at protein fold‐changes in isolated membrane fractions from S. oneidensis grown to high OD600 (~5, referred to henceforth as HOD) and low OD600 (~0.6, referred to henceforth as LOD) using mass spectrometry (raw MS data have been deposited in the PRIDE archive, see Materials and Methods). While flagellar proteins associated with the inner‐membrane sub‐complex (like FliF and FliG) did not show a marked fold‐change between the two samples (Appendix Table S1), proteins of the T‐ and H‐rings (MotX, MotY, FlgP, FlgO, and FlgT, which are part of the PL sub‐complex) had a fold‐change of 2‐3 (HOD/LOD, see Appendix Table S2). This fold‐change reflects the fact that more PL sub‐complexes are present at higher OD600 which is in agreement with the difference in the number of unambiguously cell‐attached flagella between the two samples as observed by negative‐stain EM. Moreover, the fact that FliF and FliG do not show a fold‐change at different OD600 values is in accordance with ECT data where more PL sub‐complexes were seen compared to inner‐membrane sub‐complexes in the three imaged species, implying that the IM sub‐complex is less stable than the OM‐associated one. Based on these observations, we looked for candidate proteins(s) that might be involved in sealing the membrane: (i) We reasoned that this protein(s) has to be periplasmic; therefore, we made a list of all proteins with a signal peptide in S. oneidensis (520 proteins, see Dataset EV1). (ii) As PL sub‐complexes were observed in all three species investigated here, we expected the protein(s) involved in the membrane‐sealing process also to have homologues in all three species (50 proteins, see Dataset EV2). (iii) As the T‐ and H‐rings proteins showed a fold‐change (HOD/LOD) between 1.6 and 2.8, we searched for proteins in the second list (Dataset EV2) with a similar fold‐change between 1 and 10 (11 proteins, see Dataset EV3). It will be interesting in future work to see whether any of these 11 candidates is involved in the outer membrane‐sealing mechanism.
Discussion
Despite being one of the most studied nanomachines, the process of flagellar motor assembly has previously only been imaged directly in situ in the periplasmic flagellum of the spirochete Borrelia burgdorferi (Zhao et al, 2013). In this work, we used ECT to visualize flagellar motor assembly and disassembly in situ in three additional species. Our observations from L. pneumophila, P. aeruginosa, and S. oneidensis are summarized in Fig 5 (see also Appendix Fig S4 for the color code). Specifically, we found that in all three species the embellished P‐ and L‐rings formed an independent stable disassembly complex embedded in the outer membrane. These sub‐complexes bent the outer membrane but there was no apparent central pore, which raises the question of how the outer‐membrane hole necessary for flagellum assembly is sealed after the flagellum disassembles. It also remains unclear whether the two densities at the base of these complexes are present in the full motor or are specific to the isolated PL sub‐complexes, for instance because they are part of the membrane resealing mechanism. The fact that these PL sub‐complexes remain stable in the outer membrane may be either because they have a specific function or because they are difficult to degrade [for example, because proteolytic degradation might be less active or more difficult to regulate in the periplasm compared to the cytoplasm (Talmadge & Gilbert, 1982)]. Whether PL sub‐complexes persist after flagellar motor disassembly in other species remains unknown. Previous ECT studies of the flagellar motor in other species may not have observed PL sub‐complexes because they lack the ornamentation of the T‐ and/or H‐rings, or because they do not reshape the outer membrane significantly, which enhances visibility.
Figure 5. Summary of observations in the three bacterial species investigated in this study.
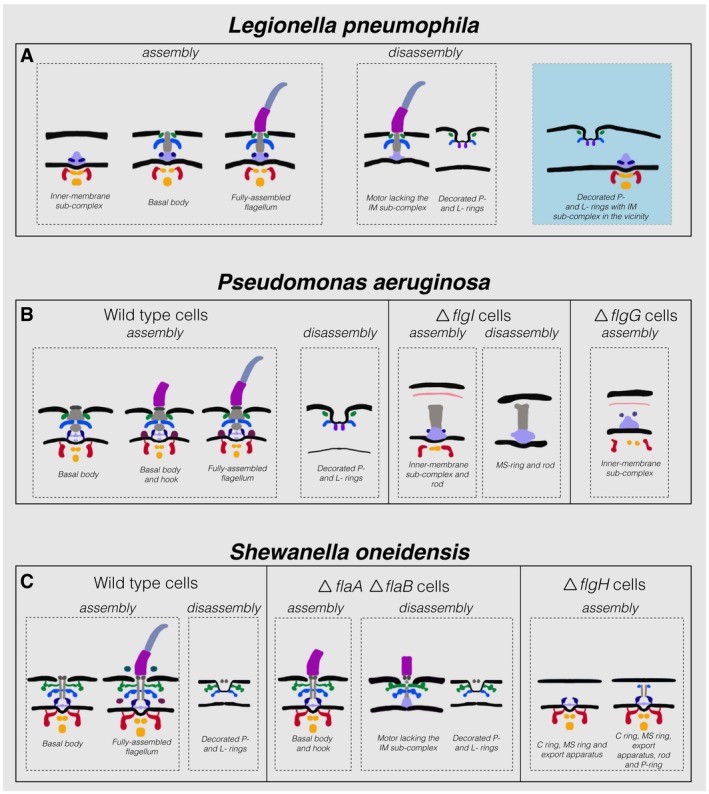
-
A–CSchematic representations of the various assembly and disassembly sub‐complexes observed in this study in L. pneumophila, P. aeruginosa, and S. oneidensis (wild‐type and mutant strains), respectively. The blue shaded area in (A) represents an intact IM sub‐complex in the vicinity of a PL sub‐complex. Note that the boundaries between the different parts in these schematics are tentative.
In addition to the PL sub‐complex in the outer membrane, in L. pneumophila, a ΔflgH mutant of S. oneidensis and a ΔflgG mutant of P. aeruginosa, we found an independent IM sub‐complex (C‐ and MS‐rings with the export apparatus) embedded in the inner membrane. The fact that these IM sub‐complexes have an intact MS‐ring (Fig 2A and B) strongly suggests that they are products of an assembly process, since the disassembly process begins with the digestion of the MS protein FliF (Jenal et al, 1994; Jenal & Shapiro, 1996; Aldridge & Jenal, 1999; Grunenfelder et al, 2004; Kanbe, 2005). This is also corroborated by our findings of disassembly products in L. pneumophila, S. oneidensis ΔflaA ΔflaB, and P. aeruginosa ΔflgI that lacked intact IM sub‐complexes. The disassembly intermediate in P. aeruginosa ΔflgI is reminiscent of the structure (extended rivet) described in reference (Kubori et al, 1992) for the same mutant in Salmonella. However, in L. pneumophila, these IM sub‐complexes (with intact MS‐ring; Fig 2A) were found in the vicinity of the PL sub‐complexes. In some cases (for example, Fig 2D, F, and J), IM sub‐complexes were close enough to the PL sub‐complexes that assembling new embellished PL rings would result in steric clashes with the already existing PL sub‐complexes. One plausible explanation for this observation could be due to a breakage event in which the cell membranes are pulled apart. In addition to PL sub‐complexes, we detected other flagellar disassembly products in different flagellar mutants in the three species. These disassembly products suggest that the disassembly process also starts at the inner‐membrane sub‐complex in a pattern reminiscent of what has previously been reported for C. crescentus which starts by digesting the MS‐ring protein, FliF (Jenal & Shapiro, 1996; Aldridge & Jenal, 1999; Grunenfelder et al, 2004; Kanbe, 2005).
Moreover, we explored the physiological context of the flagellar disassembly process reported here. Unlike the programmed flagellar ejection process described previously in C. crescentus, our results indicate that the process in the three species scrutinized here is related to the cell density and growth phase (OD600). Our experiments (mass spectrometry of isolated membrane fractions and negative‐stain electron microscopy) revealed that the flagellar disassembly process happens more frequently at high OD600 values where the cells enter the stationary phase due to low nutrient levels in the environment. While this manuscript was under review, an independent study appeared describing a similar observation in other Gammaproteobacteria species (Ferreira et al, 2019), suggesting that this phenomenon is ubiquitous in this class of bacteria and raising the question whether a similar behavior is present in other bacterial classes. We also explored the process by which the outer membrane becomes sealed after flagellar disassembly and provided a list of a handful of candidate proteins that might be involved in this process. However, we cannot exclude that proteins already present in the motor before its disassembly (like PL sub‐complex proteins) are responsible for membrane sealing. Further work has to be conducted in the future to decipher this important process at the molecular level.
Regarding the motor's assembly process, the various intermediate states of flagellar assembly we imaged are consistent with the classical model described for Salmonella and E. coli (Jones & Macnab, 1990; Kubori et al, 1992; Macnab, 2003; Li & Sourjik, 2011). However, the intermediates described in reference (Kubori et al, 1992) lacked the switch complex which is present in our in situ imaged intermediates. This can be explained by the fact that early flagellar motor isolation protocols did not retain the switch complex (Francis et al, 1994). Moreover, our results reveal the important role that the L‐ring plays in the assembly process. In the absence of the L‐ring, we did not observe the reshaping of the outer membrane that is necessary for the assembly of the flagellum. Furthermore, in the absence of the L‐ring, the P‐ring appeared much higher in the periplasm, suggesting that the reshaping of the outer membrane by the L‐ring might be responsible for correctly locating the P‐ring in the motor.
Materials and Methods
Strains and growth conditions
Shewanella oneidensis ΔflaA ΔflaB cells were grown using the batch culture method, and S. oneidensis MR‐1 wild‐type cells were grown using either the chemostat or the batch culture method. Detailed description of both methods can be found in Subramanian et al (2018). Briefly, in the chemostat method, 5 ml of a stationary‐phase overnight LB culture was injected into a continuous‐flow bioreactor containing an operating liquid volume of 1 l of a defined medium (Pirbadian et al, 2014), while dissolved oxygen tension (DOT) was maintained at 20%. After 20 h, and as the culture reached stationary phase, continuous‐flow of the defined medium (Pirbadian et al, 2014) was started with a dilution rate of 0.05/h while DOT was still maintained at 20%. After 48 h of aerobic growth under continuous flow conditions, the DOT was manually reduced to 0%. O2 served as the sole terminal electron acceptor throughout the experiment. pH was maintained at 7.0, temperature at 30°C, and agitation at 200 rpm. Either 24 or 40 h after DOT reached 0%, samples were taken from the chemostat for ECT imaging.
In the batch culture method, 200 μl of an overnight LB culture of S. oneidensis cells was added to each of two sealed and autoclaved serum bottles containing 60 ml of a defined medium (Pirbadian et al, 2014). One of the two bottles acted as a control and was not used for imaging. To this control bottle, 5 μM resazurin was added to indicate the O2 levels in the medium. The bottles were then placed in an incubator at 30°C, with shaking at 150 rpm until the color due to resazurin in the control bottle completely faded, indicating anaerobic conditions. At this point, samples were taken for ECT imaging from the bottle that did not contain resazurin.
The ΔflgH mutant was constructed by a markerless in‐frame deletion in the S. oneidensis MR‐1 background made by homologous recombination using the pSMV3 suicide vector (Saltikov & Newman, 2003) containing up‐ and downstream regions cloned using BamHI and SacI. The deletion was confirmed by PCR and swim‐plate assay (lack of swimming on 0.3% LB agar, and complementation by plasmid‐expressed FlgH) and verified by Sanger sequencing with flanking primers. Primers for the deletion construct and flanking region are as follows:
HdelUpF, ACGGGATCCCGGCAACGCACAAATGATGCG;
HdelUpR, CCAGTCGCTCATAAAGAACTGGCTGAGCGCAGCGGCCAATAGTAA;
HdelDnF, TTACTATTGGCCGCTGCGCTCAGCCAGTTCTTTATGAGCGACTGG;
HdelDnR, ACGGAGCTCGGCGCTGCACCCACTAAGTTT;
HdelFlankF, GGAAGTCGTCGAAGAGGTTGGAC; and
HdelFlankR, CCATGCAAAGCTCCTGCCACTT.
The ΔfliF mutant was constructed by markerless in‐frame deletion in the wild‐type S. oneidensis MR‐1 background. Briefly, 1.5 kb homology arms were amplified and joined by overlap PCR, then digested with SacI and SpeI, and inserted into the suicide vector pSMV3 (Saltikov & Newman, 2003). WT MR‐1 cells were transformed by electroporation (Corts et al, 2019), and mutants were obtained by homologous recombination with counterselection on sucrose. fliF deletion was confirmed by PCR and swim‐plate assay (lack of swimming on 0.3% LB agar), and the flanking sequence was verified by Sanger sequencing. Primers for the deletion construct and outside flanking region are as follows:
| fliF_upF | ACGACTAGTGAGCCTCTCGTTATTGTCCTTGGC |
| fliF_upR | CTCAGCCATTGTCTTGTAACCAGTTCGATCCGACAATCATTTCTGTGCTCAC |
| fliF_dnF | GTGAGCACAGAAATGATTGTCGGATCGAACTGGTTACAAGACAATGGCTGAG |
| fliF_dnR | ACGGAGCTCAAGGTATGCTCAATATCGCCATCAAG |
| fliF_flankF | CCCTGAGGTGTTATTAAACCAAGTATCC |
| fliF_flankR | GATTGACAGTTTGTTCCTTAATCGGGAGAG |
Shewanella oneidensis ΔflgH and ΔfliF cells were grown aerobically in LB culture at 30°C with shaking at 175 rpm to an OD600 of 2.4–2.8.
Legionella pneumophila Lp02 strain (thyA hsdR rpsL) is a derivative of the clinical isolate L. pneumophila Philadelphia‐1. Legionella pneumophila cells were grown on ACES [N‐(2‐acetamido)‐2‐aminoethanesulfonic acid]‐buffered charcoal yeast extract agar (CYE) or in ACES‐buffered yeast extract broth (AYE). The culture media (CYE and AYE) were supplemented with ferric nitrate and cysteine hydrochloride. Legionella pneumophila Lp02 strain is a thymidine auxotroph, so cells were grown in the presence of thymidine (100 μg/ml). Cells were grown to early stationary phase (OD600 ~ 2.5) and subsequently harvested for ECT sample preparation.
Pseudomonas aeruginosa PAO1 cells were grown on LB plates overnight at 37°C. After that, cells were inoculated into 5 ml MOPS [(3‐(N‐morpholino)propanesulfonic acid)] Minimal Media Limited Nitrogen and grown for ~ 24 h at 30°C. Pseudomonas aeruginosa PA14 ΔfliF, ΔflgI, ΔflgG, and ΔflhA cells were first grown on agar plates at 37°C overnight from a −80°C stock with 100 μg/ml gentamycin. Subsequently, one colony was inoculated into 2 ml of LB with 100 μg/ml gentamycin and grown at 37°C with shaking overnight. All P. aeruginosa mutants were kindly supplied by Dianne Newman's laboratory at Caltech from PA14 Transposon Insertion Mutant Library.
Many of the tomograms analyzed here were recorded for more than one purpose. The S. oneidensis MR‐1, for instance, were grown under different growth conditions for the purpose of studying the nanowires formed by these cells (Subramanian et al, 2018). Legionella pneumophila tomograms were also used to study the bacterial type IV secretion system (Ghosal et al, 2017, 2019a) and the bacterial type II secretion system (preprint: Ghosal et al, 2019b). For further details, please see the Materials and Methods section of reference (Kaplan et al, 2019).
Preparing Shewanella oneidensis cells for negative‐staining experiments
Shewanella oneidensis MR‐1 cells were grown in 2 ml of LB at 30°C (shaking 175 rpm) overnight. Next day, 2 μl of this overnight culture was inoculated in 2 ml of fresh LB medium and both samples were further incubated at 30°C (shaking 175 rpm) for 6 more hours. After that, 2 μl of each sample was added to a glow‐discharged Formvar grid and negatively stained using 2% uranyl acetate. Subsequently, the number of unambiguously cell‐attached flagella was counted in 100 randomly selected cells in each sample using a FEI T12 TEM equipped with LaB6 electron source.
For the second negative‐staining experiment, S. oneidensis cells were grown in fresh LB medium for 6 h at 30°C (shaking 175 rpm). Thereafter, the cells were spun down at 3,000 g for 10 min at room temperature. Subsequently, the pellet was resuspended in 100 μl of the supernatant and 25 μl was added to 1 ml of each of the following:
Minimal medium.
A medium consisting of half ml of minimal medium and half ml of fresh LB.
Fresh LB.
Spent LB medium obtained from an overnight culture of S. oneidensis (grown at 30°C with shaking at 175 rpm). The overnight culture was spun down, and 1 ml of the supernatant was used as the spent LB medium.
After that, the four cultures were incubated at 30°C with shaking at 175 rpm for another 3 h. Sample preparation for negative‐staining experiments was done as described above in this section for the first experiment.
Sample preparation for mass spectrometry
Shewanella oneidensis MR‐1 cells were grown in 250 ml of LB medium overnight at 30°C with shaking (175 rpm). This is the sample referred to as high OD600 (HOD). The next morning, 50 μl of the overnight culture was inoculated in a fresh 250 ml of LB [this sample is referred to as low OD600 (LOD)] and both samples were incubated under the above‐mentioned growth conditions for 5 more hours. Both samples were spun at 3,000 × g for 20 min, and pelleted cells were washed once with ice‐cold PBS. The cells were spun down again and resuspended in lysis buffer (50 mM Tri–HCl and 200 mM NaCl pH 8). Subsequently, the cells were lysed using a microfluidizer in the presence of protease inhibitors and DNase. Thereafter, both samples were spun down at low speed (3,000 × g for 15 min at 4°C) to remove cell debris. To isolate membrane fractions, samples were spun down at 150,000 × g for 11 min at 4°C. The pellets were resuspended in 10 mM HEPES pH 7 buffer and used for mass spectrometry experiments. This protocol is adapted from references (Kaplan et al, 2015; Baker et al, 2015).
Mass spectrometry
Isolated membrane fractions were prepared for mass spectrometry as described in reference (Kaplan et al, 2016). Then, digested samples were subjected to LC‐MS/MS analysis on a nanoflow LC system, EASY‐nLC 1000 (Thermo Fisher Scientific, Bremen, Germany) coupled to a LTQ Orbitrap Elite mass spectrometer (Thermo Fisher Scientific).
For the EASY‐nLC 1000 system, solvent A consisted of 97.8% H2O, 2% ACN, and 0.2% formic acid and solvent B consisted of 19.8% H2O, 80% ACN, and 0.2% formic acid. Samples were directly loaded onto a 25‐cm analytical HPLC column (50 μm ID) packed in‐house with ReproSil‐Pur C18AQ 3 μm resin (120A° pore size; Dr. Maisch, Ammerbuch, Germany). The column was heated to 55°C. The peptides were separated with a 96‐min gradient at a flow rate of 220 nl/min. The gradient was as follows: 2–30% solvent B (85 min), 30–100% B (1 min), and 100% B (9 min). Eluted peptides were then ionized using a Nanospray Flex ion source (Thermo Fisher Scientific) and introduced into the mass spectrometer. The LTQ Orbitrap Elite was operated in a data‐dependent mode, automatically alternating between a full scan (m/z 400–1,600, 120K resolution) in the Orbitrap and subsequent MS/MS scans of the 20 most abundant peaks in the linear ion trap (Top20 method). Data acquisition was controlled by Xcalibur 2.2 and LTQ Tune Plus 2.7 software (Thermo Fisher Scientific).
Analysis of MS data and bioinformatics
Raw files were processed using MaxQuant (v. 1.6.3.3; Cox et al, 2011; Cox & Mann, 2008). Mass spectra were searched against the UniProt S. oneidensis (strain MR‐1) database (4,068 sequences) as well as a common protein contaminant database (246 sequences). Trypsin was specified as the digestion enzyme with up to two missed cleavages allowed. Precursor mass tolerance was 4.5 ppm after mass recalibration, and fragment ion tolerance was 0.5 Da. Carbamidomethylation of cysteine was specified as a fixed modification, and oxidation of methionine and protein N‐terminal acetylation were specified as variable modifications. “Match between runs” was enabled as well as “Label‐free quantitation” (Cox et al, 2014). Protein and peptide score thresholds were determined so as to achieve a 1% protein and peptide false discovery rate as estimated by a target‐decoy approach. Raw data and search results have been deposited in the PRIDE archive with accession PXD012554.
Protein fold‐changes between HOD and LOD samples were calculated by dividing the HOD LFQ intensity by the LOD LFQ intensity.
All protein sequences were obtained from the MiST3 database (Ulrich & Zhulin, 2010). Information about signal peptide prediction in S. oneidensis was obtained from UniProt (Apweiler, 2004). Similarity searches were performed using NCBI's Protein‐Protein BLAST 2.8.1+ (Camacho et al, 2009). We downloaded the genomes of P. aeruginosa (GCF_000006765.1), L. pneumophila (GCF_000008485.1), and S. oneidensis (GCF_000146165.2) to build a BLAST database. We selected S. oneidensis genes with predicted signal peptide and perform a similarity search against the custom database. Because we wanted proteins with homologues in all three species, proteins with the lowest negative log10 of the E‐value of the best BLAST hit in each genome above 30 were reported.
ECT sample preparation and imaging
BSA‐treated 10‐nm colloidal gold solution was mixed with cells from the three species, and 4 μl of this mixture was applied to a glow‐discharged, carbon‐coated, R2/2, 200 mesh copper Quantifoil grid (Quantifoil Micro Tools) in a Vitrobot chamber (FEI). Excess liquid was blotted off, and the grid was plunge‐frozen for ECT imaging. Imaging of all ECT samples was performed on either an FEI Polara 300‐keV field emission gun electron microscope (FEI company, Hillsboro, OR, USA) equipped with a Gatan imaging filter and K2 Summit direct electron detector in counting mode (Gatan, Pleasanton, CA, USA), or an FEI Titan Krios 300‐kV field emission gun transmission electron microscope equipped with a Gatan imaging filter and a K2 Summit direct detector in counting mode (Gatan). Data were collected using the UCSF Tomography software (Zheng et al, 2007) or SerialEM, with each tilt series ranging from −60° to 60° in 1° increments and an underfocus of ~ 5–10 μm. A cumulative electron dose of ~ 130–160 e−/A2 for each individual tilt series was used for S. oneidensis and P. aeruginosa, while a cumulative dose of ˜ 100 e−/A2 was used for L. pneumophila.
Image processing and sub‐tomogram averaging
The IMOD software package was used to calculate three‐dimensional reconstructions of tilt series (Kremer et al, 1996). Alternatively, the images were aligned and contrast transfer function corrected (for L. pneumophila data) using the IMOD software package before producing SIRT reconstructions using the TOMO3D program (Agulleiro & Fernandez, 2011). Sub‐tomogram averages with twofold symmetrization along the particle y‐axis were produced using the PEET program (Nicastro, 2006). Ten PL sub‐complexes were averaged for L. pneumophila, 20 PL sub‐complexes for P. aeruginosa, and 18 PL sub‐complexes for S. oneidensis. Twofold symmetrization along the y‐axis was performed for the S. oneidensis average only. Ten IM sub‐complexes were averaged for L. pneumophila.
Author contributions
MK and GJJ designed research. MK, PS, DG, and RS‐L prepared samples. MK, PS, and DG recorded ECT data. MK performed MS experiments. MK and DRO performed bioinformatics analysis. SP, SKM, JAG, and MYE‐N supplied reagents. MK, CMO, and GJJ analyzed data. MK, CMO, and GJJ wrote the manuscript, and all authors edited it.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Dataset EV1
Dataset EV2
Dataset EV3
Movie EV1
Movie EV2
Movie EV3
Review Process File
Acknowledgements
This work was supported by the National Institutes of Health (NIH, grant R01 AI127401 to G.J.J.). M.K. is supported by a postdoctoral Rubicon fellowship from De Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO). S.P. and M.Y.E.‐N. are supported by the Air Force Office of Scientific Research Presidential Early Career Award for Scientists and Engineers (FA955014‐1‐0294, to M.Y.E.‐N.). R.S.L. and J.A.G acknowledge support from the Office of Naval Research (N00014‐18‐1‐2632 to J.A.G.). We would like to thank Dr. Spiros Garbis, Dr. Annie Moradian, Dr. Michael Sweredoski, and Dr. Brett Lomenick from the Caltech Proteome Exploration Laboratory for their help in mass spectrometry experiments and data analysis. All P. aeruginosa mutants were kindly supplied by Dianne Newman's laboratory at Caltech. ECT was performed in the Beckman Institute Resource Center for Cryo‐EM.
The EMBO Journal (2019) 38: e100957
References
- Agulleiro JI, Fernandez JJ (2011) Fast tomographic reconstruction on multicore computers. Bioinformatics 27: 582–583 [DOI] [PubMed] [Google Scholar]
- Aldridge P, Jenal U (1999) Cell cycle‐dependent degradation of a flagellar motor component requires a novel‐type response regulator. Mol Microbiol 32: 379–391 [DOI] [PubMed] [Google Scholar]
- Appelt S, Heuner K (2017) The Flagellar Regulon of Legionella—A review. Front Cell Infect Microbiol 7: 454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler R (2004) UniProt: the universal protein knowledgebase. Nucleic Acids Res 32: 115D–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker LA, Folkers GE, Sinnige T, Houben K, Kaplan M, van der Cruijsen EAW, Baldus M (2015) Magic‐angle‐spinning solid‐state NMR of membrane proteins In Methods in enzymology, Shukla AK. (ed.), pp 307–328. Elsevier; [DOI] [PubMed] [Google Scholar]
- Beeby M, Ribardo DA, Brennan CA, Ruby EG, Jensen GJ, Hendrixson DR (2016) Diverse high‐torque bacterial flagellar motors assemble wider stator rings using a conserved protein scaffold. Proc Natl Acad Sci USA 113: E1917–E1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg HC (2003) The rotary motor of bacterial flagella. Annu Rev Biochem 72: 19–54 [DOI] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2009) BLAST+: architecture and applications. BMC Bioinformatics 10: 421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaban B, Coleman I, Beeby M (2018) Evolution of higher torque in Campylobacter‐type bacterial flagellar motors. Sci Rep 8: 97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Beeby M, Murphy GE, Leadbetter JR, Hendrixson DR, Briegel A, Li Z, Shi J, Tocheva EI, Müller A et al (2011) Structural diversity of bacterial flagellar motors: structural diversity of bacterial flagellar motors. EMBO J 30: 2972–2981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilcott GS, Hughes KT (2000) Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar typhimurium and Escherichia coli . Microbiol Mol Biol Rev MMBR 64: 694–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen EJ, Hughes KT (2014) Rod‐to‐hook transition for extracellular flagellum assembly is catalyzed by the L‐ring‐dependent rod scaffold removal. J Bacteriol 196: 2387–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corts AD, Thomason LC, Gill RT, Gralnick JA (2019) A new recombineering system for precise genome‐editing in Shewanella oneidensis strain MR‐1 using single‐stranded oligonucleotides. Sci Rep 9: 39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.‐range mass accuracies and proteome‐wide protein quantification. Nat Biotechnol 26: 1367–1372 [DOI] [PubMed] [Google Scholar]
- Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M (2011) Andromeda: a peptide search engine integrated into the maxquant environment. J Proteome Res 10: 1794–1805 [DOI] [PubMed] [Google Scholar]
- Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M (2014) Accurate proteome‐wide label‐free quantification by delayed normalization and maximal peptide ratio extraction termed MaxLFQ. Mol Cell Proteomics 13: 2513–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabiani FD, Renault TT, Peters B, Dietsche T, Gálvez EJC, Guse A, Freier K, Charpentier E, Strowig T, Franz‐Wachtel M et al (2017) A flagellum‐specific chaperone facilitates assembly of the core type III export apparatus of the bacterial flagellum. PLoS Biol 15: e2002267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M, Bryan R, Rajan S, Scheffler L, Brunnert S, Tang H, Prince A (1998) Role of flagella in pathogenesis of Pseudomonas aeruginosa pulmonary infection. Infect Immun 66: 43–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira JL, Gao FZ, Rossmann FM, Nans A, Brenzinger S, Hosseini R, Wilson A, Briegel A, Thormann KM, Rosenthal PB et al (2019) γ‐proteobacteria eject their polar flagella under nutrient depletion, retaining flagellar motor relic structures. PLoS Biol 17: e3000165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis NR, Sosinsky GE, Thomas D, DeRosier DJ (1994) Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J Mol Biol 235: 1261–1270 [DOI] [PubMed] [Google Scholar]
- Fukumura T, Makino F, Dietsche T, Kinoshita M, Kato T, Wagner S, Namba K, Imada K, Minamino T (2017) Assembly and stoichiometry of the core structure of the bacterial flagellar type III export gate complex. PLoS Biol 15: e2002281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan L, Jensen GJ (2012) Electron tomography of cells. Q Rev Biophys 45: 27–56 [DOI] [PubMed] [Google Scholar]
- Ghosal D, Chang Y‐W, Jeong KC, Vogel JP, Jensen GJ (2017) In situ structure of the Legionella Dot/Icm type IV secretion system by electron cryotomography. EMBO Rep 18: 726–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosal D, Chang Y‐W, Jeong KC, Vogel JP, Jensen G (2019a) Molecular architecture, polar targeting and biogenesis of the Legionella Dot/Icm T4SS. Nat Microbiol 4: 1173–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosal D, Kim KW, Zheng H, Kaplan M, Vogel JP, Cianciotto NP, Jensen GJ (2019b) In vivo structure of the Legionella type II secretion system by electron cryotomography. bioRxiv 10.1101/525063 [PREPRINT] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunenfelder B, Tawfilis S, Gehrig S, Osteras M, Eglin D, Jenal U (2004) Identification of the protease and the turnover signal responsible for cell cycle‐dependent degradation of the caulobacter FliF motor protein. J Bacteriol 186: 4960–4971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota N, Kitada M, Imae Y (1981) Flagellar motors of alkalophilic bacillus are powered by an electrochemical potential gradient of Na+ . FEBS Lett 132: 278–280 [Google Scholar]
- Homma M, Komeda Y, Iino T, Macnab RM (1987) The flaFIX gene product of Salmonella typhimurium is a flagellar basal body component with a signal peptide for export. J Bacteriol 169: 1493–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal U, White J, Shapiro L (1994) Caulobacter flagellar function, but not assembly, requires FliL, a non‐polarly localized membrane protein present in all cell types. J Mol Biol 243: 227–244 [DOI] [PubMed] [Google Scholar]
- Jenal U, Shapiro L (1996) Cell cycle‐controlled proteolysis of a flagellar motor protein that is asymmetrically distributed in the caulobacter predivisional cell. EMBO J 15: 2393–2406 [PMC free article] [PubMed] [Google Scholar]
- Jones CJ, Homma M, Macnab RM (1987) Identification of proteins of the outer (L and P) rings of the flagellar basal body of Escherichia coli . J Bacteriol 169: 1489–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CJ, Macnab RM (1990) Flagellar assembly in Salmonella typhimurium: analysis with temperature‐sensitive mutants. J Bacteriol 172: 1327–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CJ, Macnab RM, Okino H, Aizawa S‐I (1990) Stoichiometric analysis of the flagellar hook‐(basal‐body) complex of Salmonella typhimurium . J Mol Biol 212: 377–387 [DOI] [PubMed] [Google Scholar]
- Kanbe M (2005) Protease susceptibility of the Caulobacter crescentus flagellar hook‐basal body: a possible mechanism of flagellar ejection during cell differentiation. Microbiology 151: 433–438 [DOI] [PubMed] [Google Scholar]
- Kaplan M, Cukkemane A, van Zundert GCP, Narasimhan S, Daniëls M, Mance D, Waksman G, Bonvin AMJJ, Fronzes R, Folkers GE et al (2015) Probing a cell‐embedded megadalton protein complex by DNP‐supported solid‐state NMR. Nat Methods 12: 649–652 [DOI] [PubMed] [Google Scholar]
- Kaplan M, Narasimhan S, de Heus C, Mance D, van Doorn S, Houben K, Popov‐Čeleketić D, Damman R, Katrukha EA, Jain P et al (2016) EGFR dynamics change during activation in native membranes as revealed by NMR. Cell 167: 1241–1251.e11 [DOI] [PubMed] [Google Scholar]
- Kaplan M, Ghosal D, Subramanian P, Oikonomou CM, Kjaer A, Pirbadian S, Ortega DR, Briegel A, El‐Naggar MY, Jensen GJ (2019) The presence and absence of periplasmic rings in bacterial flagellar motors correlates with stator type. Elife 8: e43487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koebnik R (1995) Proposal for a peptidoglycan‐associating alpha‐helical motif in the C‐terminal regions of some bacterial cell‐surface proteins. Mol Microbiol 16: 1269–1270 [DOI] [PubMed] [Google Scholar]
- Kojima S, Takao M, Almira G, Kawahara I, Sakuma M, Homma M, Kojima C, Imada K (2018) The helix rearrangement in the periplasmic domain of the flagellar stator B subunit activates peptidoglycan binding and ion influx. Structure 26: 590–598.e5 [DOI] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR (1996) Computer visualization of three‐dimensional image data using IMOD. J Struct Biol 116: 71–76 [DOI] [PubMed] [Google Scholar]
- Kubori T, Shimamoto N, Yamaguchi S, Namba K, Aizawa S‐I (1992) Morphological pathway of flagellar assembly in Salmonella typhimurium . J Mol Biol 226: 433–446 [DOI] [PubMed] [Google Scholar]
- Li H, Sourjik V (2011) Assembly and stability of flagellar motor in Escherichia coli: flagellar motor assembly. Mol Microbiol 80: 886–899 [DOI] [PubMed] [Google Scholar]
- Macnab RM (1999) The bacterial flagellum: reversible rotary propellor and type III export apparatus. J Bacteriol 181: 7149–7153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnab RM (2003) How bacteria assemble flagella. Annu Rev Microbiol 57: 77–100 [DOI] [PubMed] [Google Scholar]
- Manson MD, Tedesco P, Berg HC, Harold FM, Van der Drift C (1977) A protonmotive force drives bacterial flagella. Proc Natl Acad Sci USA 74: 3060–3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarter LL (2001) Polar flagellar motility of the vibrionaceae. Microbiol Mol Biol Rev 65: 445–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T, Imada K (2015) The bacterial flagellar motor and its structural diversity. Trends Microbiol 23: 267–274 [DOI] [PubMed] [Google Scholar]
- Morimoto Y, Minamino T (2014) Structure and function of the bi‐directional bacterial flagellar motor. Biomolecules 4: 217–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicastro D (2006) The molecular architecture of axonemes revealed by cryoelectron tomography. Science 313: 944–948 [DOI] [PubMed] [Google Scholar]
- Ohnishi K, Homma M, Kutsukake K, Iino T (1987) Formation of flagella lacking outer rings by flaM, flaU, and flaY mutants of Escherichia coli . J Bacteriol 169: 1485–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikonomou CM, Jensen GJ (2017) A new view into prokaryotic cell biology from electron cryotomography. Nat Rev Microbiol 15: 128 [DOI] [PubMed] [Google Scholar]
- Oliver D (1985) Protein secretion in Escherichia coli . Annu Rev Microbiol 39: 615–648 [DOI] [PubMed] [Google Scholar]
- Paradis G, Chevance FFV, Liou W, Renault TT, Hughes KT, Rainville S, Erhardt M (2017) Variability in bacterial flagella re‐growth patterns after breakage. Sci Rep 7: 1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirbadian S, Barchinger SE, Leung KM, Byun HS, Jangir Y, Bouhenni RA, Reed SB, Romine MF, Saffarini DA, Shi L et al (2014) Shewanella oneidensis MR‐1 nanowires are outer membrane and periplasmic extensions of the extracellular electron transport components. Proc Natl Acad Sci USA 111: 12883–12888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell EM (1977) Life at low reynolds number. Am J Phys 45: 3–11 [Google Scholar]
- Saltikov CW, Newman DK (2003) Genetic identification of a respiratory arsenate reductase. Proc Natl Acad Sci USA 100: 10983–10988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa Y, Berry RM (2008) Bacterial flagellar motor. Q Rev Biophys 41: 103–132 [DOI] [PubMed] [Google Scholar]
- Stock D, Namba K, Lee LK (2012) Nanorotors and self‐assembling macromolecular machines: the torque ring of the bacterial flagellar motor. Curr Opin Biotechnol 23: 545–554 [DOI] [PubMed] [Google Scholar]
- Subramanian P, Pirbadian S, El‐Naggar MY, Jensen GJ (2018) Ultrastructure of Shewanella oneidensis MR‐1 nanowires revealed by electron cryotomography. Proc Natl Acad Sci USA 115: E3246–E3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talmadge K, Gilbert W (1982) Cellular location affects protein stability in Escherichia coli . Proc Natl Acad Sci USA 79: 1830–1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terashima H, Fukuoka H, Yakushi T, Kojima S, Homma M (2006) The Vibrio motor proteins, MotX and MotY, are associated with the basal body of Na + ‐driven flagella and required for stator formation. Mol Microbiol 62: 1170–1180 [DOI] [PubMed] [Google Scholar]
- Terashima H, Koike M, Kojima S, Homma M (2010) The flagellar basal body‐associated protein flgt is essential for a novel ring structure in the sodium‐driven vibrio motor. J Bacteriol 192: 5609–5615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terashima H, Kawamoto A, Morimoto YV, Imada K, Minamino T (2017) Structural differences in the bacterial flagellar motor among bacterial species. Biophys Physicobiol 14: 191–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DR, Francis NR, Xu C, DeRosier DJ (2006) The three‐dimensional structure of the flagellar rotor from a clockwise‐locked mutant of Salmonella enterica Serovar Typhimurium. J Bacteriol 188: 7039–7048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich LE, Zhulin IB (2010) The MiST2 database: a comprehensive genomics resource on microbial signal transduction. Nucleic Acids Res 38: D401–D407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Wang J, Tang P, Chen H, Gao H (2011) Genetic and molecular characterization of flagellar assembly in Shewanella oneidensis . PLoS ONE 6: e21479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Zhang K, Boquoi T, Hu B, Motaleb MA, Miller KA, James ME, Charon NW, Manson MD, Norris SJ et al (2013) Cryoelectron tomography reveals the sequential assembly of bacterial flagella in Borrelia burgdorferi . Proc Natl Acad Sci USA 110: 14390–14395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Norris SJ, Liu J (2014) Molecular architecture of the bacterial flagellar motor in cells. Biochemistry 53: 4323–4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Keszthelyi B, Branlund E, Lyle JM, Braunfeld MB, Sedat JW, Agard DA (2007) UCSF tomography: an integrated software suite for real‐time electron microscopic tomographic data collection, alignment, and reconstruction. J Struct Biol 157: 138–147 [DOI] [PubMed] [Google Scholar]
- Zhu S, Nishikino T, Hu B, Kojima S, Homma M, Liu J (2017) Molecular architecture of the sheathed polar flagellum in Vibrio alginolyticus . Proc Natl Acad Sci USA 114: 10966–10971 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Dataset EV1
Dataset EV2
Dataset EV3
Movie EV1
Movie EV2
Movie EV3
Review Process File


