Abstract
Cell division control 37 (CDC37) increases the stability of heat shock protein 90 (HSP90) client proteins and is thus essential for numerous intracellular oncogenic signaling pathways, playing a key role in prostate oncogenesis. Notably, elevated expression of CDC37 was found in prostate cancer cells, although the regulatory mechanisms through which CDC37 expression becomes increased are unknown. Here we show both positive and negative regulation of CDC37 gene transcription by two members of the SREZBP-CTfin51-AW1-Number 18 cDNA (SCAN) transcription factor family—MZF1 and SCAND1, respectively. Consensus DNA-binding motifs for myeloid zinc finger 1 (MZF1/ZSCAN6) were abundant in the CDC37 promoter region. MZF1 became bound to these regulatory sites and trans-activated the CDC37 gene whereas MZF1 depletion decreased CDC37 transcription and reduced the tumorigenesis of prostate cancer cells. On the other hand, SCAND1, a zinc fingerless SCAN box protein that potentially inhibits MZF1, accumulated at MZF1-binding sites in the CDC37 gene, negatively regulated the CDC37 gene and inhibited tumorigenesis. SCAND1 was abundantly expressed in normal prostate cells but was reduced in prostate cancer cells, suggesting a potential tumor suppressor role of SCAND1 in prostate cancer. These findings indicate that CDC37, a crucial protein in prostate cancer progression, is regulated reciprocally by MZF1 and SCAND1.
Keywords: SCAN zinc finger, SCAND1, CDC37, MZF1, prostate cancer
1. Introduction
The cell division control 37 (CDC37) protein plays a fundamental role in chaperoning the protein kinase family and participates in cancer by maintaining the activity of protein kinases involved in cell proliferation and transformation. These include tyrosine kinases such as Src [1], serine/threonine kinases in the Raf-ERK pathway [2], Akt [3], the inhibitor of NF-κB kinase (IKK) [4], and cyclin-dependent kinase 4 (CDK4) [5,6,7]. CDC37 functions primarily in a complex with heat shock protein 90 (HSP90) to mediate the three-dimensional folding and structural integrity of client protein kinases [1,8,9]. CDC37 is particularly significant in prostate cancer as its overexpression leads to prostate carcinogenesis in transgenic mice [10,11,12]. It has been suggested that the high levels of oncogenic proteins present in most cancers make them dependent on molecular chaperones, a state referred to as “chaperone addiction” [13,14]. Thus, because of their large protein clienteles, the CDC37–HSP90 axis offers a critical target for inactivating multiple oncogenic pathways [13,14]. Consequently, the inhibition of HSP90 in cancer is currently a major area of research [14,15]. However, less is known regarding the regulation of CDC37 expression in cancer. Thus, we have addressed this deficiency in this study.
Examination of the CDC37 5′ upstream region and introns indicated multiple consensus sequences that are potentially bound by the transcription factor myeloid zinc finger 1 (MZF1, also known as ZSCAN6, ZNF42, ZFP98), consistent with a previous study [16]. We recently reported that the frequent amplification of MZF1 was observed in human cancers, further suggesting an oncogenic role for this factor [17]. Early studies showed that MZF1 might play roles in stemness and in the differentiation of hematopoietic stem cells and indicated its involvement in myeloma [18]. MZF1 was shown to participate in the malignant properties of several major solid tumors, including breast [19,20], lung [21], liver [22], head and neck [23], skin [24], endometrium [25], colorectal, and cervical cancers [26]. MZF1 protein structure is composed of an N-terminal SREZBP-CTfin51-AW1-Number 18 cDNA (SCAN) domain, a linker region, and a C-terminal DNA binding domain that contains 13 zinc finger (ZF) motifs and is a member of the SCAN zinc finger (SCAN-ZF) family [17,27]. The SCAN domain is a leucine-rich oligomerization domain and is highly conserved in the SCAN-ZF family, which is composed of more than 50 family members [28,29]. Zinc fingerless SCAN domain-only proteins also exist [27]. SCAND1 is a SCAN domain-only protein that has been shown to bind MZF1 and other SCAN-ZF family members. SCAN domain-only proteins have been suspected to be suppressors of intact SCAN-ZF transcriptional activity [27,28,30]. This possibility, however, has remained experimentally untested. Furthermore, we have tested the hypothesis that the relationship between MZF1 and SCAND1 regulates CDC37 expression and thereby cancer growth.
In this report we characterized the CDC37 gene promoter and determined that MZF1 indeed increases CDC37 expression, while SCAND1 represses CDC37 expression. Our findings provide insight into how MZF1-driven CDC37 expression promotes cancer progression and how SCAND1 functions as a potential tumor suppressor by repressing CDC37.
2. Results
2.1. Elevated Expression of CDC37 Is Caused by MZF1 in Prostate Cancer
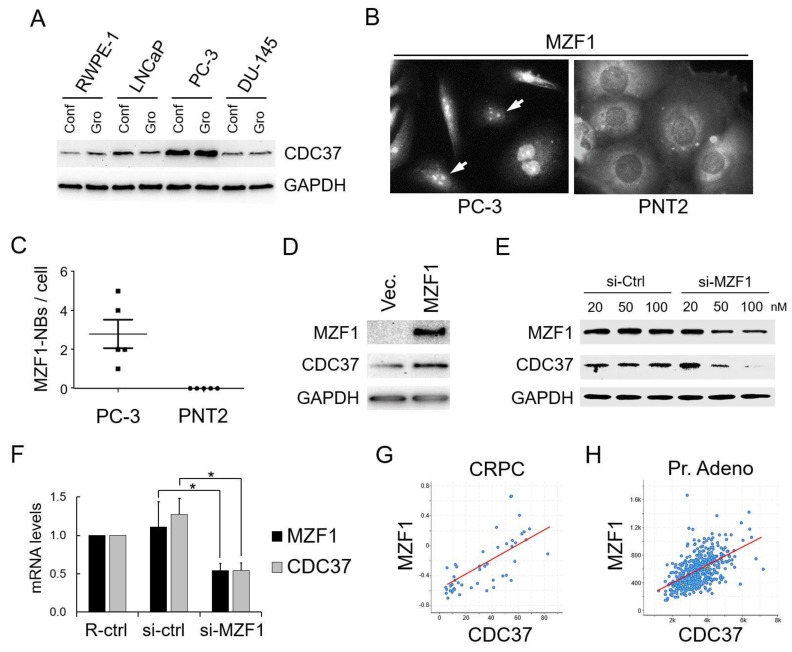
We first compared the expression levels of CDC37 between prostate cancer cell lines DU-145 and LNCaP, a castration-resistant prostate cancer (CRPC, also known as neuroendocrine prostate cancer (NEPC) cell line PC-3), and a normal prostate cell line (RWPE-1). CDC37 levels in PC-3 cells were higher than those of the other prostate cancer cells and normal prostate cells in both confluent and growing conditions (Figure 1A). CDC37 levels in the prostate cancer cells (PC-3, LNCaP, and DU-145) were higher than those of the normal cell line in confluent conditions (Figure 1A).
Figure 1.
Elevated expression of cell division control 37 (CDC37) is caused by MZF1 in prostate cancer. (A) Western blot showing CDC37 in prostate cancer cells. Cell lysates were prepared from confluent (Conf) and growing (Gro) RWPE-1, LNCaP, PC-3, and DU-145 cells. (B) Immunocytochemistry showing MZF1 expression and localization in PC-3 and PNT2 cells. Arrows indicate nuclear bodies of MZF1 (MZF1-NBs) seen in PC-3 but not in PNT2. (C) The number of MZF1-NBs per cells. (D) Western blot showing CDC37 altered by overexpressed MZF1 in DU-145. (E) Western blot showing CDC37 altered by the depletion of MZF1 in DU-145 cells. (F) mRNA levels of CDC37 and MZF1 altered by the depletion of MZF1. The MZF1-targeting siRNA mixture (A + B + C, 10 nM each) or a non-silencing control (si-ctrl) was transfected into PC-3 cells. RPL32, internal control. R-ctrl, reagent only control. * p < 0.05, n = 3. Similar results were obtained from three independent experiments. (G) Co-expression of MZF1 and CDC37 in patient samples of castration-resistant prostate cancer (CRPC). N = 114, Spearman correlation score 0.78, p = 2.60 × 10−11. (H) Co-expression of MZF1 and CDC37 in patient samples of prostate adenocarcinoma (Pr. Adeno). N = 494, Spearman correlation score 0.57, p = 7.74 × 10−44.
Although there are several possible mechanisms for the elevation in CDC37 expression in prostate cancer, in the present study we focused on transcriptional regulation as the most immediate level of control. To determine potential cis-acting regulatory elements in the gene, we analyzed the DNA sequence between −3.6 kbp and +2 kbp from the CDC37 transcription start site (TSS) and found numerous consensus MZF1 binding sequences (Supplementary Materials Figure S1).
To query the increased CDC37 level observed in PC-3 cells shown in Figure 1A, we next examined MZF1 expression and localization. MZF1 localized in nuclei in PC-3 but in the cytoplasm in the normal prostate cell line PNT2 (Figure 1B), suggesting that both the expression and nuclear localization of MZF1 could be involved in the increased CDC37 level in PC-3 cells. Additionally, nuclear bodies of MZF1 (designated as MZF1-NBs inasmuch as MZF1 oligomers could form nuclear bodies) were seen in PC-3 whereas cytoplasmic retention of MZF1 was seen in prostatic normal PNT2 cells, although the significance of the MZF1-NBs is currently not known (Figure 1B,C). Moreover, MZF1 overexpression led to increases in CDC37 levels in PC-3 cells (Figure 1D), while siRNA-mediated knockdown of MZF1 lowered CDC37 mRNA and protein levels (Figure 1E,F). A role for MZF1 in CDC37 transcription was thus suspected.
To investigate the potential clinical significance of MZF1 regulation of CDC37, we next examined the co-expression correlation of MZF1 and CDC37 in prostate cancer patient-derived tumor samples. Co-expression correlation was found between MZF1 and CDC37 in CRPC patient samples (Spearman’s rank correlation score: 0.78) and in prostate adenocarcinoma patient samples (Spearman’s correlation: 0.41) (Figure 1G,H). We next examined MZF1 and CDC37 expression correlation with the prognosis of prostate cancer patients. High expression levels of MZF1 (p = 0.0287) and CDC37 (p = 0.0182) were correlated with poor prognosis of patients suffering from prostate cancer and showed a higher correlation than that of PSA (p = 0.154)—named after prostate-specific antigen—which is currently used for prostate cancer screening (Table 1).
Table 1.
Correlation between protein expression and prognosis of prostate cancer patients.
| Protein Name | p Value | N (Total) | N (High) | N (Low) |
|---|---|---|---|---|
| MZF1 | 0.0287 | 494 | 99 | 395 |
| CDC37 | 0.0182 | 494 | 198 | 296 |
| PSA | 0.154 | 494 | 389 | 105 |
These data indicated MZF1 to be a potential causal transcription factor for the elevated expression of CDC37 in prostate cancer.
2.2. SCAN Zinc Finger Protein MZF1 Directly Trans-Activates the CDC37 Gene
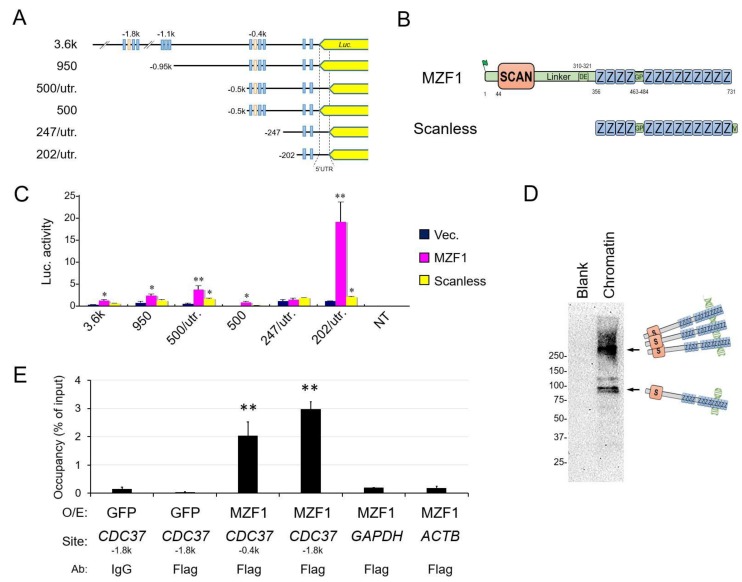
We next queried whether the SCAN zinc finger MZF1 could directly trans-regulate the CDC37 gene through direct binding to cis-elements (MZF1 binding sequences) abundantly found in the CDC37 5′ upstream region (Supplementary Materials Figure S1). We examined the activities of CDC37 promoter-driven luciferase reporter constructs containing different deletion mutants of the 5′ region and examined potential trans-regulation by MZF1 and its SCANless truncation constructs of the factor (Figure 2A,B). The full-length native MZF1 markedly increased CDC37 promoter activities of the 3.6k, 950, 500/utr, 500, and 202/utr constructs (Figure 2C). The SCANless mutant slightly increased CDC37 promoter activities (of 500/utr, 202/utr) but the transcriptional activity was greatly reduced compared with SCAN-containing native MZF1. These data indicate that the SCAN oligomerization domain is essential for the transcriptional activity of MZF1. It is notable that the 202/utr reporter was more potent in trans-activation compared to other constructs, suggesting the presence of potential inhibitory motifs upstream of the 202 sequence. We next examined whether the overexpressed MZF1 (with Flag-tag) could directly bind to cis-elements in the genomic CDC37 promoter region. MZF1 was detected by immunoblot in the crosslinked chromatin fraction and potential post-translational modification forms (upshift) and potential oligomers were detected in the chromatin (Figure 2D). The overexpressed MZF1 occupied genomic CDC37 promoter regions (−0.4k and −1.8k regions that contain MZF1-binding sites), further indicating that the CDC37 gene is regulated by MZF1 binding to these MZF1 binding sites in prostate cancer, as suggested by chromatin immunoprecipitation (ChIP) assay (Figure 2E).
Figure 2.
SCAN zinc finger MZF1 directly trans-activates the CDC37 gene. (A) Schemes of the CDC37 promoter–reporter constructs. Truncated mutants of CDC37 promoter were connected with the luciferase (Luc) gene. Blue box, MZF1 binding site. Orange box, heat shock element (HSE). 5′UTR, 5′ untranslated region. (B) The secondary structures of native MZF1 and the SCANless construct (zinc finger domain alone). S, SCAN box. Z, zinc finger motif. DE, aspartic acid- and glutamic acid-rich region. GP, glycine- and proline-rich region. MZF1 was overexpressed with an N-terminal Flag-tag. The SCANless construct was overexpressed with a C-terminal V5-tag. (C) Luciferase activities from the truncated CDC37 promoters controlled by MZF1 and the SCANless construct. Plasmid constructs shown in panels A and B were co-transfected into DU-145 cells. Vec., empty vector control. n = 3, * p < 0.05, ** p < 0.01 (vs. Vec.). (D) Chromatin Western blotting of MZF1 overexpressed in DU-145. Crosslinked chromatin was prepared from DU-145 overexpressed with MZF1. (E) ChIP-qPCR analysis showing MZF1 occupancy of CDC37. Chromatin was prepared from DU-145 overexpressed with Flag-MZF1 or GFP (control) and immunoprecipitated using anti-Flag antibody beads or the control, IgG. Co-immunoprecipitated DNA was analyzed by qPCR for CDC37 (−1.8k or −0.4k regions) or control regions in GAPDH or ACTB. N = 3, ** p < 0.01 (vs. IgG control). Similar results were obtained from three independent experiments.
These data indicate that MZF1 directly trans-activates the CDC37 gene through direct binding to the CDC37 promoter region and that SCAN domain-mediated oligomerization is essential for the full transcriptional activity of MZF1.
2.3. The Zinc Fingerless SCAND1 Factor Suppresses CDC37 Gene and Tumorigenesis of Prostate Cancer
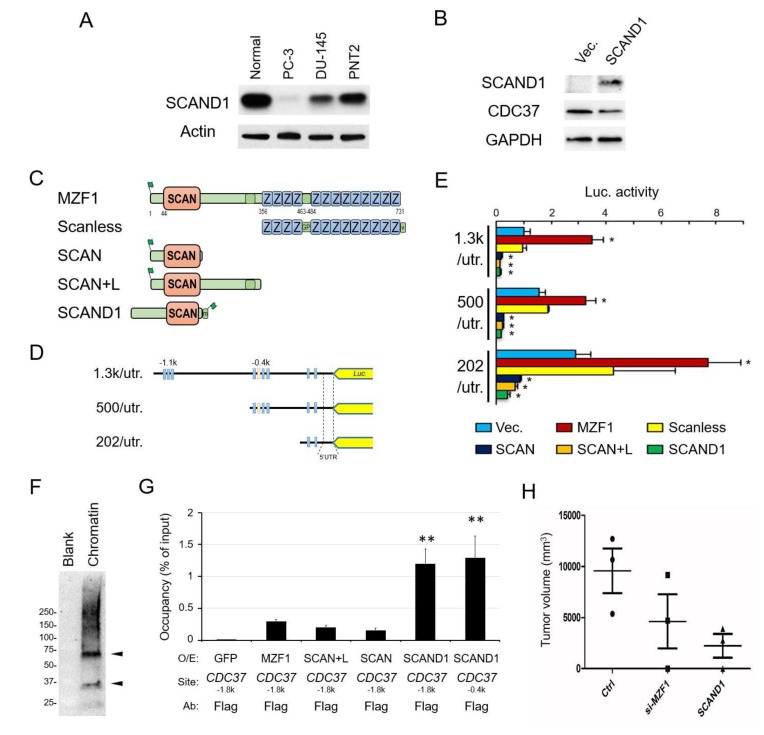
The SCAN-ZF family consists of more than 50 members and contains a few potentially inhibitory zinc fingerless members such as SCAND1. We hypothesized that SCAND1 could play a key role in limiting functions of numerous SCAN-ZF proteins such as MZF1 through hetero-oligomerization. Notably, SCAND1 is highly expressed in normal prostate tissue compared to other tissues (http://biogps.org/#goto=genereport&id=51282). We found SCAND1 to be expressed in normal prostate cells while its levels were reduced in prostate cancer PC-3 and DU-145 cells (Figure 3A), suggesting that SCAND1 expression declines along with prostate oncogenesis. Overexpression of SCAND1 led to lowered CDC37 levels in PC-3 cells (Figure 3B).
Figure 3.
Zinc fingerless SCAND1 suppresses the CDC37 gene and tumorigenesis of prostate cancer. (A) Western blot showing SCAND1 levels in normal prostate cells, PC-3, DU-145, and PNT2. (B) Western blot showing CDC37 lowered by SCAND1 overexpression. (C) Schemes of overexpression constructs. (D) Schemes of truncated CDC37 promoters fused with the luciferase reporter gene. (E) Luciferase activities from the truncated CDC37 promoters controlled by SCAND1 and truncated mutants of MZF1. Plasmid constructs shown in panels C and D were co-transfected into PC-3 cells. Vec., empty vector control. n = 3, * p < 0.05 (vs. Vec.). (F) Chromatin Western blot showing SCAND1 overexpressed in DU-145. Crosslinked chromatin was prepared from DU-145 overexpressed with SCAND1. (G) ChIP-qPCR analysis showing SCAND1 occupancy of CDC37. Chromatin was prepared from DU-145 overexpressed with SCAND1, MZF1, SCAN, SCAN + L, or GFP (control) and immunoprecipitated using anti-Flag antibody beads. Co-immunoprecipitated DNA was analyzed by qPCR for CDC37 (−1.8k or −0.4k regions). N = 3, ** p < 0.01 (vs. IgG control). For Figure 3A–G, similar data were obtained from three independent experiments. (H) Tumorigenicity of PC-3 cells lowered by SCAND1 and the depletion of MZF1. The SCAND1-overexpression plasmid, MZF1-targeted siRNA, and control GFP plasmid were transfected into PC-3 cells, which were then xenografted to immunocompromised mice subcutaneously.
To determine whether SCAND1 and a SCAN-only construct derived from MZF1 could regulate CDC37 promoter constructs, we next carried out co-transfection and reporter assays. SCAND1 and SCAN-only constructs (SCAN and SCAN + linker) derived from the truncation of MZF1 significantly repressed CDC37 promoter activities (1.3k/utr, 500/utr, 202/utr) in PC-3 cells (Figure 3C–E). We also confirmed that the native MZF1 protein activated the CDC37 promoter whereas the SCANless mutant had little effect.
We next hypothesized that SCAND1, although lacking a discernible DNA-binding domain, might be able to associate with genomic CDC37 promoter regions. Overexpressed SCAND1 was detected on chromatin in PC-3 as a monomer of the expected molecular weight. We also observed potential dimer and higher molecular weight forms in the crosslinked chromatin, suggesting the higher oligomerization of SCAND1 on chromatin, which might be consistent with a powerful repressor role (Figure 3F). The overexpressed SCAND1 (with Flag-tag) occupied the CDC37 promoter region (−1.8k and −0.4k regions, which we showed to contain MZF1-binding sites) (Figure 3G). The SCAND1 occupancy of the CDC37 regulatory site (−1.8k) was higher in magnitude than that observed with SCAN-only constructs of MZF1, as suggested by the increased enrichment in the ChIP assay. We consistently observed that overexpressed SCAND1, as well as MZF1, was found in the insoluble chromatin-containing fraction, whereas overexpressed SCAN domain (SCAN, SCAN + L) constructs tended to be found in the soluble fraction of the cell lysate (Supplementary Materials Figure S2).
These data indicate that SCAND1 could repress the CDC37 gene powerfully through association with DNA sequences that we showed to bind MZF1. Therefore, our data indicate that while MZF1 positively regulates CDC37, SCAND1 negatively regulates transcription, a finding which could be crucial in mechanisms of prostate cancer progression.
We therefore hypothesized that, in this context, MZF1 has an oncogenic influence while SCAND1 may suppress tumorigenicity. To elucidate this possibility, we asked whether tumor initiation by prostate cancer PC-3 cells could be suppressed by SCAND1 expression and by the depletion of MZF1. Indeed, we observed that SCAND1 overexpression suppressed tumorigenicity, while the depletion of MZF1 by siRNA lowered tumorigenicity in PC-3 cells (Figure 3H).
3. Discussion
Our data suggest a novel mechanism for prostate cancer regulation by MZF1 (Figure 1). CDC37 is a crucial molecule in prostate cancer growth through its fostering of oncogenic kinases and our data strongly indicate that the chaperone is upregulated by MZF1. We showed that MZF1 binds to sites in the CDC37 promoter and strongly activates transcription; moreover, MZF1 expression is tightly correlated with CDC37 levels in clinical prostate cancer (Figure 1 and Figure 2, Table 1). The ability of an endogenous inhibitor, the zinc fingerless SCAND1 factor, to bind the promoter and repress the transcription of MZF1 suggests a potential mechanism for prostate tumor suppression (Figure 3). These data suggest mechanisms whereby members of the SCAN transcription factor family can fine-tune prostate cancer growth by up or downregulating CDC37 transcription and thus decide the outcome of prostate tumorigenesis. It is likely that other MZF1 targets important in tumorigenesis may be regulated in a similar way [17], including oncogenes and pro-tumorigenic genes such as MYC [21], NCAD [31], YAP1 [32], TGF-beta [20], CTSB/L [33], MMP-14 [34], FOXM1 [35], CK17 [36], PAX2 [25], PRAME [24], DR5 [37], AXL [26,38], PKC-alpha [39,40], CD34, and c-Myb [18]. Besides, MZF1 might play key roles also in tumor microenvironment such as mesenchymal stem cell differentiation into cancer-associated fibroblasts [20]. However, MZF1 has also been shown to activate tumor suppressor genes such as FPN [41], NFKBIA [42], and SMAD4 [43]. In a more complex manner, MZF1 also plays a transcriptional repressor role for pro-tumorigenic genes such as MMP-2 [44] and IGF-IR [45]. Nevertheless, our data do not exactly define the mechanism through which SCAND1 represses MZF1 activity, although direct co-occupation of the CDC37 promoter appears to be involved (Figure 3). MZF1 was shown to bind DNA as a homodimer or heterodimer with other SCAN domain proteins and recruit the chromatin remodeling protein mDomino [46]. Our data pinpoint the importance of the SCAN domain, as the SCANless construct of MZF1 showed much reduced transcriptional activity compared to the powerful activity of the native MZF1 (Figure 3E). The SCAN domain is leucine-rich and mediates oligomer formation [17]. We found that MZF1 could form a higher order structure, potentially an oligomer on chromatin (Figure 2D). Therefore, it was indicated that the SCAN domain of MZF1 is required for its oligomerization and thus for the full activity for trans-activation. We also observed up-shift bands of MZF1 as potential post-translational modifications required for MZF1 activity (Figure 2D). It has been reported that MZF1 could be phosphorylated by PAK4 in a SUMO-directed manner [47] and by CK2 [31] and ERK-1/2 [36]. It was also reported that the loss of the nuclear pool of ubiquitin ligase CHIP/STUB1 unleashes the MZF1-cathepsin pro-oncogenic program [33]. Thus, upstream regulatory signals that modify specific amino acid residues of MZF1 have been recently clarified. However, little is currently known regarding the mechanisms whereby SCAND1 could repress transcription, although our data indicate that this may be associated with the CDC37 promoter in the oligomeric form at the site of MZF1 binding (Figure 3).
Our studies on CDC37 are ultimately aimed at targeting this factor in cancer as an alternative to HSP90. Numerous HSP90 inhibitors have been developed to target the abundant HSP90, which plays versatile oncogenic roles in many types of cancer [48]. Moreover, extracellular HSP90 and extracellular vesicle-associated HSP90 have been recently shown to play pro-tumorigenic roles [49,50,51,52,53]. However, it has been difficult so far to employ HSP90 inhibitors at effective doses due to normal tissue toxicity. Depletion of CDC37 reduced prostate cancer cell growth and attenuated the MEK-ERK signaling pathway and the PI3K-Akt signaling pathway [11]. Moreover, overexpression of CDC37 triggers prostatic tumorigenesis [10]. Our studies therefore suggest that further study of MZF1 and SCAND1 activities in regulating CDC37 transcription may identify a new therapeutic angle to target the cohort of oncogenic signaling processes dependent upon the HSP90–CDC37 complex.
4. Materials and Methods
4.1. Cell Culture
PC-3, DU-145, LNCaP, and RWPE-1 cells were obtained from ATCC. PC-3 was cultured in F-12K or RPMI 1640 medium containing 10% fetal bovine serum (FBS). DU-145 was cultured in DMEM containing 10% FBS. LNCaP was cultured in RPMI 1640 medium containing 10% FBS. RWPE-1 was cultured in Keratinocyte Serum-Free Medium (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with bovine pituitary extract (BPE) and human recombinant epidermal growth factor (hEGF). PNT2 was obtained from Sigma and cultured in RPMI 1640 medium supplemented with 2 mM glutamine and 10% FBS. Normal human prostate epithelial cells were purchased from Lonza (Basel, Switzerland) and cultured in Prostate Epithelial Cell Basal Medium (Lonza) supplemented with BPE, hydrocortisone, hEGF, epinephrine, transferrin, insulin, retinoic acid, triiodothyronine, and GA-1000.
4.2. Molecular Cloning
Genomic DNA was isolated from a buccal cell swab of a deidentified white male. The 3.6 kbp human CDC37 promoter region was PCR-amplified and subcloned into a pGL3-luciferase vector (Promega, Madison, WI, USA) using KpnI and BamHI. Once the pGL3-3.6 kbp CDC37promo vector was made, the 1.3 kbp, 950 kbp, 500 bp, 247 bp, and 202 bp fragments with and without the 116 bp 5′UTR were similarly subcloned into pGL3 using the same restriction sites as above. Each entire promoter fragment clone was Sanger sequenced. The complete 3.6 kbp CDC37 promoter sequence was aligned with the human reference genome GRCh37.
Human MZF1 cDNA was subcloned from pOTB-MZF1 into pcDNA3-Flag vector via TOPO directional cloning (Thermo Fisher Scientific) and designated as pcDNA3/Flag-MZF1. For SCAN-only constructs, UGA stop codons were generated in the pcDNA/Flag-MZF1 via Quickchange mutagenesis and designated as pcDNA3/Flag-C125/SCAN and pcDNA3/Flag-C252/SCAN + L. For SCANless constructs, the cDNA of the MZF1 zinc finger domain was amplified via PCR and subcloned into pcDNA/V5 vector using TOPO directional cloning and designated as pcDNA/MZF1-V5/N252 (SCANless). Human SCAND1 (NM_033630) open reading frame clone with Myc-DDK C-terminal tag (RC200079) was purchased (Origene, Rockville, MD, USA) and designated as pCMV6-ScanD1-myc-Flag. pCMV-EGFP (NEPA Gene) was used as an overexpression control.
4.3. In Silico Analysis of Promoters and Gene Bodies
Sequences of promoter regions and gene bodies of human CDC37, HSP90AA1, and HSP90AB1 were obtained from the Eukaryotic Promoter Database [54]. Binding sites for MZF1 were predicted using PROMO [55,56].
4.4. Luciferase Assay
Transient transfection and luciferase assays were performed as previously described [57]. Cells were cultured in 96-well plates. A plasmid (25 ng reporter, 100 ng effector) was transfected with 0.4 µL FuGENE HD (Roche, Basel, Switzerland) per well at a cell confluence level of 50–70%. The medium was changed at 16–20 h after transfection. At 40–48 h after transfection, 70 µL of the medium was aspirated, then 30 µL of Bright-Glo reagent (Promega) was added and mixed by pipetting. Cells were incubated for 5 min at 37 °C. The lysate (40 µL) was transferred to a 96-well white plate for the measurement of luminescence.
4.5. siRNA
The siRNA was designed based on the siRNA design method of JBioS (Japan Bio Services, Saitama, Japan). An RNA duplex of 19 bp plus TT-3′ overhangs in each strand was synthesized by Nippon Gene (Tokyo, Japan). For electroporation-transfection, a total of 40 pmol siRNA was transfected to 5 × 105 cells. For reagent-transfection, cells were transfected with siRNA at final concentrations of 20–100 nM. Non-targeting siRNA was purchased from Nippon Gene (Tokyo, Japan). The designed sequences of siRNA are listed in Supplementary Materials Table S1.
4.6. Transfection
For ChIP assay and tumorigenesis assay, electroporation-mediated transfection was performed as described previously [50,58]. To optimize electroporation for each cell type, cells (1 × 105 to 1 × 106 cells), plasmid DNA (2 to 10 µg total) or siRNA (40 pmol total), and serum-free medium were mixed to 100 µL total in a green cuvette with a 1 mm gap (NepaGene, Ichikawa, Tokyo) and placed in an NEPA21 Super Electroporator (NepaGene). The poring pulse was optimized between 100 V and 300 V for 2.5 or 5.0 ms pulse length twice with 50 ms intervals between the pulses and 10% decay rate with + polarity. The transfer pulse condition was five pulses at 20 V for 50 ms pulse length with 50 ms intervals between the pulses and 40% decay rate with +/− polarity. After electroporation, cells were recovered in serum-containing media. PC-3 cells (5 × 105 cells) were electroporation-transfected with 10 µg plasmid DNA or 40 pmol siRNA with a poring pulse at 175 V for 2.5 ms pulse length twice.
For the overexpression ChIP assay, DU-145 cells (5 × 106 cells) were transfected with 15 µg of pcDNA/Flag-MZF1 or pCMV-GFP with a poring pulse at 175 V for 5 ms pulse length twice, cultured for 4 days in a 15 cm dish, and then used for ChIP assay.
For Western blot analysis with overexpression of MZF1 and SCAND1, PC-3 cells were transfected with pcDNA3.1(–), pcDNA/Flag-MZF1, and pCMV6-SCAND1-myc-Flag using FuGENE6 (Roche).
For qRT-PCR after MZF1 depletion, PC-3 cells were transfected with a mixture of MZF1-siRNA A, B, and C (SR305183, OriGene, Rockville, MD) or non-silencing siRNA (SR30004, OriGene) using Lipofectamine RNAi max (Thermo Fisher Scientific).
For the depletion of MZF1 and Western blotting, DU-145 cells cultured in a 6-well plate were transfected with siRNA hMZF1-all-NM_003422-53 using Lipofectamine RNAi Max. The medium was replaced with a fresh one at 24 h after transfection. Cells were lysed at 48 h after transfection.
4.7. Chromatin
For protein overexpression and ChIP assay, DU-145 cells (5 × 106 cells) were electroporation-transfected with pCMV6-ScanD1-myc-Flag, pcDNA3/Flag-MZF1, pcDNA3/Flag-SCAN, pcDNA/Flag-SCAN + L, and pCMV-GFP and then cultured for 3 days in 150 mm dishes. ChIP was conducted using a SimpleChIP enzymatic chromatin IP kit with magnetic beads (Cell Signaling Technology). Briefly, proteins/DNA were crosslinked with formaldehyde. For optimization, nuclear fractions were treated with MNase (0, 0.25, 0.5, 0.75, and 1 µL per reaction) and subsequent ultra-sonication at high and low power using an Ultrasonic Homogenizer Smurt NR-50M (Microtec Nition), and then analyzed within 2% agarose gel electrophoresis. After the optimization, the nuclear fractions were sheared with MNase (0.75 µL per reaction) and ultra-sonication at high power and then centrifuged at 10,000× g for 10 min at 4 °C. The supernatants were used as crosslinked chromatin fractions. Overexpressed proteins in the chromatin fraction were analyzed by Western blotting using anti-MZF1 antibody (Assay Biotechnology, Fremont, CA, USA) and anti-SCAND1 antibody (ab64828, Abcam, Cambridge, UK). Anti-Flag M2 magnetic beads (Sigma, St. Louis, MO, USA) were used for ChIP. DNA was purified from the eluate and from 1% and 10% input chromatin using a QIA-Quick DNA purification kit (Qiagen, Hilden, Germany) and eluted within 50 μL of ddH2O. The purified DNA (3–5 μL) was used for ChIP-qPCR. Amplification specificity and efficiency were confirmed with melting curve analysis, slope analysis, and agarose gel electrophoresis. Primers are listed in Supplementary Materials Table S2.
4.8. Tumorigenesis
The efforts of all animal experiments were made to minimize suffering. The studies were carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the Japanese Pharmacological Society. The protocol was approved by the Animal Care and Use Committee, Okayama University (Permit Number: OKU-2016219). All animals were held under specific pathogen-free conditions. PC-3 cells (5 × 105) were transfected with 40 pmol MZF1-targeted siRNA, 10 µg pCMV6-ScanD1-myc-Flag, or 10 µg pCMV-EGFP via electroporation, cultured for 5 days, and then 1 × 106 cells were subcutaneously injected into the back of a 6–7 year old CB17/IcrJcl-Pkrdcscid (SCID) mouse (CLEA, Japan, Tokyo). Tumor volumes were measured at day 41 post-injection. The major axis (a) and minor axis (b) of tumors were measured with a caliper. The tumors were deemed to be ellipsoid and the volumes were calculated as follows: tumor volume (V) ≒ 4πab2/3.
4.9. Western Blot
To compare CDC37 levels among several types of cells, cells were cultured to reach confluence in a 6 cm dish or sparsely grown in a 10 cm dish. For endogenous SCAND1, protein samples were collected when cells reached sub-confluence. At day 2 after medium replacement, cells were washed with phosphate buffered saline (PBS) and lysed within CelLytic M (Sigma) for 15 min with gentle shaking. Lysates were collected and centrifuged at 14,000× g for 15 min. Protein concentration was measured using BCA protein assay (Thermo Fisher Scientific). Protein samples (10–40 µg: equal amount) were loaded to SDS-PAGE. Proteins were transferred to a polyviniylidine difluoride (PVDF) membrane with a semi-dry method. The membranes were blocked within 5% skimmed milk in Tris buffered saline with 0.1% Tween 20 (TBST) for 1 h. The membranes were incubated with primary antibodies overnight at 4 °C and with secondary antibodies for 1 h at room temperature (RT). The membranes were washed three times with TBST for 15 min after each antibody reaction. Chemiluminescence was detected with a ChemiDoc MP Imaging System (Bio-Rad, Hercules, CA, USA).
For crosslinked chromatin Western blotting, see the Section 4.7. For regular Western blotting after the overexpression of MZF1 and SCAND1, cells were transfected with pcDNA3/Flag-MZF1 or pcDNA3(-) using FuGENE6 (Roche) and cultured for 24 h. Cells were washed with ice-cold PBS twice and lysed within RIPA buffer supplemented with a protease phosphatase inhibitor cocktail (Thermo Fisher Scientific) at 4 °C for 15 min, and then collected. The lysed cells were homogenized 10 times using a 25 gauge needle attached to a 1 mL syringe and then incubated on ice for 30 min. The lysate was centrifuged at 12,000× g for 20 min at 4 °C to remove debris. The protein sample amounts (10–50 µg) were used for SDS-PAGE and semi-dry transfer.
For the depletion of MZF1, DU-145 cells cultured in a 6-well plate were transfected with siRNA hMZF1-all-NM_003422-53 using Lipofectamine RNAi Max following the manufacturer’s instruction. The medium was replaced with a fresh amount at 24 h after transfection. Cells were collected using trypsin and counted at 48 h after transfection. The cell lysate was prepared using RIPA buffer as described above. The equal amount of lysate (25 µg) was loaded to each lane for SDS-PAGE (4–20% TGX gel, Bio-Rad). The proteins were transferred to a PVDF membrane with the wet-transfer method at 40 V for 16 h on ice with a transfer buffer (25 mM Tris, 192 mM glycine, 10% methanol, and 0.05% SDS). The membrane was washed three times with TBST for 15 min and then blocked for 1 h within 5% skimmed milk. MZF1 and CDC37 were detected by Western blotting.
The antibodies used were anti-MZF1 (C10502, Assay Biotechnology, 1:500), anti-CDC37 (D11A3, Cell Signaling Technology, 1:1000), anti-FLAG (Clone M2, Sigma), anti-SCAND1 (ab64828, Abcam, Cambridge, UK), and HRP-conjugated anti-GAPDH antibody (Clone 5A12, Fujifilm Wako, 1:5000).
4.10. Immunocytochemistry
Immunocytochemistry was performed as described [59]. Cells cultured in 4-well chamber slides were fixed with 4% (w/v) paraformaldehyde in PBS for 15 min. Cells were permeabilized with 0.2% Triton X-100 in PBS for 15 min. Cells were blocked in IHC/ICC blocking buffer high protein (eBioscience, San Diego, CA, USA) for 10 min and then reacted with anti-MZF1 antibody (1:50, C10502; Assay Biotechnology) and AlexaFluor488 secondary antibody (Thermo Fisher Scientific, 1:1000) in the blocking buffer. Cells were washed with PBS for 5 min twice between the steps. Cells were mounted with ProLong Gold AntiFade Reagent (Thermo Fisher Scientific). Fluorescence images were captured using an Axio Vision microscope equipped with an AxioCam MR3 (Zeiss, Oberkochen, Germany).
4.11. RT-qPCR
RT-qPCR was performed as previously described [59,60]. Total RNA was prepared using an RNeasy RNA purification system (Qiagen, Hilden, Germany) with DNase I treatment. cDNA was synthesized using an RT kit for RT-qPCR (Qiagen) with a mixture of oligo dT and random primers. The cDNA pool was diluted 5- to 20-fold. The cDNA standard with permissible slopes of PCR efficiencies was prepared by step dilution of the cDNA pool for the relative quantification of mRNA levels. In 20 µL of qPCR mix, 0.25 µM of each primer, 4–10 µL of diluted cDNA, and 10 µL SYBR green 2× Master Mix (Applied Biosystems, Waltham, MA) were added and reacted at 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min. Dissociation curves with specific single-peaked PCR and proper amplification slopes for PCR efficiency were confirmed. LinRegPCR software was used for baseline fluorescence correction, and for the calculation of Cq values and PCR efficiency of amplicons [61]. Primer sequences are listed in Supplementary Materials Table S3.
4.12. Gene and Protein Expression in Clinical Samples
Co-expression of MZF1 and CDC37 was analyzed in TCGA in cBioPortal. Data sets of NEPC/CRPC (Trento/Cornell/Broad 2016, 114 samples) and prostate adenocarcinomas (TCGA, PanCancer Atlas; 494 patients/samples) were analyzed with Spearman’s rank correlation coefficient of co-expression.
For the correlation of protein expression with patient prognosis, Kaplan–Meier survival analysis was performed using 494 prostate cancer patient samples in the Human Protein Atlas Database.
4.13. Statistics
Data were expressed as the means ± SD unless otherwise specified. Comparisons of two were performed with an unpaired Student’s t-test.
5. Conclusions
In conclusion, it was demonstrated that pro-oncogenic MZF1 and its potential antagonist SCAND1 coordinately regulate the expression of CDC37, a factor that is essential for prostate cancer tumorigenesis. The SCAN domain is required for the full transcriptional activity of MZF1, whereas the SCAN-only factor SCAND1 is powerfully associated with chromatin and represses the tumorigenesis of prostate cancer cells.
Acknowledgments
This paper is dedicated to the memory of one of our mentors, Ken-ichi Kozaki, who passed away on May 29, 2016. The authors thank Barbara Wegiel, Eva Csizmadia, Ayesha Murshid, Yuka Okusha, Kuniaki Okamoto, and Yasutomo Nasu for valuable, illuminating discussion and encouragement. The authors thank Nusrat Rahman and Sachin Doshi for technical assistance.
Abbreviations
| CDC37 | Cell division control 37 |
| CRPC | Castration-resistant prostate cancer |
| HSP | Heat shock protein |
| MZF1 | Myeloid zinc finger 1 |
| NEPC | Neuroendocrine prostate cancer |
| SCAN | SREZBP-CTfin51-AW1-Number 18 cDNA |
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6694/11/6/792/s1, Figure S1: Binding sites for MZF1 and HSF1 in the CDC37 promoter, Figure S2: Western blotting showing overexpressed MZF1, its truncated mutants, and SCAND1, Table S1: List of siRNA sequences, Table S2: List of primers for ChIP-qPCR. Table S3: List of primers for RT-qPCR.
Author Contributions
T.E., T.L.P. and S.K.C. conceptualized and designed the study. T.E., S.K.C. and T.L.P. acquired funding. T.L.P. and T.E. constructed materials. T.E., M.T.T., T.L.P., C.S. and B.J.L. devised methodologies and performed the experimentation. T.E. wrote the original manuscript. S.K.C., T.L.P., B.J.L., and T.E. edited the manuscript. All authors reviewed the manuscript.
Funding
This work was supported by JSPS KAKENHI grants JP17K11642, JP17K11669-KOh, and JP17K11643 (T.E.), by a JCRT foundation grant at HMS (T.L.P., S.K.C.), and by NIH research grants RO1CA119045, RO1CA47407, and RO1CA176326 (S.K.C.).
Conflicts of Interest
The authors declare no conflict of interest with the content of this study.
References
- 1.Kimura Y., Rutherford S.L., Miyata Y., Yahara I., Freeman B.C., Yue L., Morimoto R.I., Lindquist S. Cdc37 is a molecular chaperone with specific functions in signal transduction. Genes Dev. 1997;11:1775–1785. doi: 10.1101/gad.11.14.1775. [DOI] [PubMed] [Google Scholar]
- 2.Silverstein A.M., Grammatikakis N., Cochran B.H., Chinkers M., Pratt W.B. p50 cdc37 binds directly to the catalytic domain of Raf as well as to a site on hsp90 that is topologically adjacent to the tetratricopeptide repeat binding site. J. Biol. Chem. 1998;273:20090–20095. doi: 10.1074/jbc.273.32.20090. [DOI] [PubMed] [Google Scholar]
- 3.Basso A.D., Solit D.B., Chiosis G., Giri B., Tsichlis P., Rosen N. Akt forms an intracellular complex with heat shock protein 90 (Hsp90) and Cdc37 and is destabilized by inhibitors of Hsp90 function. J. Biol. Chem. 2002;277:39858–39866. doi: 10.1074/jbc.M206322200. [DOI] [PubMed] [Google Scholar]
- 4.Chen G.Y., Cao P., Goeddel D.V. TNF-induced recruitment and activation of the IKK complex require Cdc37 and Hsp90. Mol. Cell. 2002;9:401–410. doi: 10.1016/S1097-2765(02)00450-1. [DOI] [PubMed] [Google Scholar]
- 5.Dai K., Kobayashi R., Beach D. Physical interactionof mammalian CDC37 with CDK4. J. Biol. Chem. 1996;271:22030–22034. doi: 10.1074/jbc.271.36.22030. [DOI] [PubMed] [Google Scholar]
- 6.Stepanova L., Leng X., Parker S.B., Harper J.W. Mammalian p50 Cdc37 is aprotein kinase-targeting subunit of Hsp90 that binds and stabilize Cdk4. Genes Dev. 1996;10:1491–1502. doi: 10.1101/gad.10.12.1491. [DOI] [PubMed] [Google Scholar]
- 7.Vaughan C.K., Gohlke U., Sobott F., Good V.M., Ali M.M., Prodromou C., Robinson C.V., Saibil H.R., Pearl L.H. Structure of an Hsp90-Cdc37-Cdk4 complex. Mol. Cell. 2006;23:697–707. doi: 10.1016/j.molcel.2006.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siligardi G., Panaretou B., Meyer P., Singh S., Woolfson D.N., Piper P.W., Pearl L.H., Prodromou C. Regulation of Hsp90 ATPase activity by the co-chaperone Cdc37p/p50cdc37. J. Biol. Chem. 2002;277:20151–20159. doi: 10.1074/jbc.M201287200. [DOI] [PubMed] [Google Scholar]
- 9.Roe S.M., Ali M.M.U., Meyer P., Vaughan C.K., Panaretou B., Piper P.W., Prodromou C., Pearl L.H. The mechanism of Hsp90 regulation by the protein kinase-specific cochaperone p50 cdc37. Cell. 2004;116:87–98. doi: 10.1016/S0092-8674(03)01027-4. [DOI] [PubMed] [Google Scholar]
- 10.Stepanova L., Yang G., DeMayo F., Wheeler T.M., Finegold M., Thompson T.C., Harper J.W. Induction of human CDC37 in prostate cancer correlates with the ability of targeted CDC37 expression to promote prostatic hyperplasia. Oncogene. 2000;19:2186–2193. doi: 10.1038/sj.onc.1203561. [DOI] [PubMed] [Google Scholar]
- 11.Gray P.J., Jr., Stevenson M.A., Calderwood S.K. Targeting Cdc37 inhibits multiple signaling pathways and induces growth arrest in prostate cancer cells. Cancer Res. 2007;67:11942–11950. doi: 10.1158/0008-5472.CAN-07-3162. [DOI] [PubMed] [Google Scholar]
- 12.Gray P.J., Prince T., Cheng J., Stevenson M.A., Calderwood S.K. Targeting the oncogene and kinome chaperone CDC37. Nat. Rev. Cancer. 2008;8:491–495. doi: 10.1038/nrc2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calderwood S.K. Cdc37 as a co-chaperone to Hsp90. Subcell. Biochem. 2015;78:103–112. doi: 10.1007/978-3-319-11731-7_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith J.R., de Billy E., Hobbs S., Powers M., Prodromou C., Pearl L., Clarke P.A., Workman P. Restricting direct interaction of CDC37 with HSP90 does not compromise chaperoning of client proteins. Oncogene. 2015;34:15–26. doi: 10.1038/onc.2013.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neckers L., Workman P. Hsp90 molecular chaperone inhibitors: Are we there yet? Clin. Cancer Res. 2012;18:64–76. doi: 10.1158/1078-0432.CCR-11-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang L., Grammatikakis N., Toole B.P. Organization of the Chick CDC37 Gene. J. Biol. Chem. 1998;273:3598–3603. doi: 10.1074/jbc.273.6.3598. [DOI] [PubMed] [Google Scholar]
- 17.Eguchi T., Prince T., Wegiel B., Calderwood S.K. Role and Regulation of Myeloid Zinc Finger Protein 1 in Cancer. J. Cell. Biochem. 2015;116:2146–2154. doi: 10.1002/jcb.25203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perrotti D., Melotti P., Skorski T., Casella I., Peschle C., Calabretta B. Overexpression of the zinc finger protein MZF1 inhibits hematopoietic development from embryonic stem cells: Correlation with negative regulation of CD34 and c-myb promoter activity. Mol. Cell. Biol. 1995;15:6075–6087. doi: 10.1128/MCB.15.11.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rafn B., Nielsen C.F., Andersen S.H., Szyniarowski P., Corcelle-Termeau E., Valo E., Fehrenbacher N., Olsen C.J., Daugaard M., Egebjerg C., et al. ErbB2-driven breast cancer cell invasion depends on a complex signaling network activating myeloid zinc finger-1-dependent cathepsin B expression. Mol. Cell. 2012;45:764–776. doi: 10.1016/j.molcel.2012.01.029. [DOI] [PubMed] [Google Scholar]
- 20.Weber C.E., Kothari A.N., Wai P.Y., Li N.Y., Driver J., Zapf M.A., Franzen C.A., Gupta G.N., Osipo C., Zlobin A., et al. Osteopontin mediates an MZF1-TGF-beta1-dependent transformation of mesenchymal stem cells into cancer-associated fibroblasts in breast cancer. Oncogene. 2015;34:4821–4833. doi: 10.1038/onc.2014.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsai L.H., Wu J.Y., Cheng Y.W., Chen C.Y., Sheu G.T., Wu T.C., Lee H. The MZF1/c-MYC axis mediates lung adenocarcinoma progression caused by wild-type lkb1 loss. Oncogene. 2015;34:1641–1649. doi: 10.1038/onc.2014.118. [DOI] [PubMed] [Google Scholar]
- 22.Hsieh Y.H., Wu T.T., Huang C.Y., Hsieh Y.S., Liu J.Y. Suppression of tumorigenicity of human hepatocellular carcinoma cells by antisense oligonucleotide MZF-1. Chin. J. Physiol. 2007;50:9–15. [PubMed] [Google Scholar]
- 23.Ko C.P., Yang L.C., Chen C.J., Yeh K.T., Lin S.H., Yang S.F., Chen M.K., Lin C.W. Expression of myeloid zinc finger 1 and the correlation to clinical aspects of oral squamous cell carcinoma. Tumour Biol. 2015;36:7099–7105. doi: 10.1007/s13277-015-3419-x. [DOI] [PubMed] [Google Scholar]
- 24.Lee Y.K., Park U.H., Kim E.J., Hwang J.T., Jeong J.C., Um S.J. Tumor antigen PRAME is up-regulated by MZF1 in cooperation with DNA hypomethylation in melanoma cells. Cancer Lett. 2017;403:144–151. doi: 10.1016/j.canlet.2017.06.015. [DOI] [PubMed] [Google Scholar]
- 25.Jia N., Wang J., Li Q., Tao X., Chang K., Hua K., Yu Y., Wong K.K., Feng W. DNA methylation promotes paired box 2 expression via myeloid zinc finger 1 in endometrial cancer. Oncotarget. 2016;7:84785–84797. doi: 10.18632/oncotarget.12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mudduluru G., Vajkoczy P., Allgayer H. Myeloid zinc finger 1 induces migration, invasion, and in vivo metastasis through Axl gene expression in solid cancer. Mol. Cancer Res. 2010;8:159–169. doi: 10.1158/1541-7786.MCR-09-0326. [DOI] [PubMed] [Google Scholar]
- 27.Edelstein L.C., Collins T. The SCAN domain family of zinc finger transcription factors. Gene. 2005;359:1–17. doi: 10.1016/j.gene.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 28.Williams A.J., Blacklow S.C., Collins T. The zinc finger-associated SCAN box is a conserved oligomerization domain. Mol. Cell. Biol. 1999;19:8526–8535. doi: 10.1128/MCB.19.12.8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schumacher C., Wang H., Honer C., Ding W., Koehn J., Lawrence Q., Coulis C.M., Wang L.L., Ballinger D., Bowen B.R., et al. The SCAN domain mediates selective oligomerization. J. Biol. Chem. 2000;275:17173–17179. doi: 10.1074/jbc.M000119200. [DOI] [PubMed] [Google Scholar]
- 30.Sander T.L., Haas A.L., Peterson M.J., Morris J.F. Identification of a novel SCAN box-related protein that interacts with MZF1B. The leucine-rich SCAN box mediates hetero- and homoprotein associations. J. Biol. Chem. 2000;275:12857–12867. doi: 10.1074/jbc.275.17.12857. [DOI] [PubMed] [Google Scholar]
- 31.Ko H., Kim S., Yang K., Kim K. Phosphorylation-dependent stabilization of MZF1 upregulates N-cadherin expression during protein kinase CK2-mediated epithelial-mesenchymal transition. Oncogenesis. 2018;7:27. doi: 10.1038/s41389-018-0035-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verma N.K., Gadi A., Maurizi G., Roy U.B., Mansukhani A., Basilico C. Myeloid Zinc Finger 1 and GA Binding Protein Co-Operate with Sox2 in Regulating the Expression of Yes-Associated Protein 1 in Cancer Cells. Stem Cells. 2017;35:2340–2350. doi: 10.1002/stem.2705. [DOI] [PubMed] [Google Scholar]
- 33.Luan H., Mohapatra B., Bielecki T.A., Mushtaq I., Mirza S., Jennings T.A., Clubb R.J., An W., Ahmed D., El-Ansari R., et al. Loss of the Nuclear Pool of Ubiquitin Ligase CHIP/STUB1 in Breast Cancer Unleashes the MZF1-Cathepsin Pro-oncogenic Program. Cancer Res. 2018;78:2524–2535. doi: 10.1158/0008-5472.CAN-16-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng L., Jiao W., Mei H., Song H., Li D., Xiang X., Chen Y., Yang F., Li H., Huang K., et al. miRNA-337-3p inhibits gastric cancer progression through repressing myeloid zinc finger 1-facilitated expression of matrix metalloproteinase 14. Oncotarget. 2016;7:40314–40328. doi: 10.18632/oncotarget.9739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen P.M., Cheng Y.W., Wang Y.C., Wu T.C., Chen C.Y., Lee H. Up-regulation of FOXM1 by E6 oncoprotein through the MZF1/NKX2-1 axis is required for human papillomavirus-associated tumorigenesis. Neoplasia. 2014;16:961–971. doi: 10.1016/j.neo.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu L., Han L., Zhou C., Wei W., Chen X., Yi H., Wu X., Bai X., Guo S., Yu Y., et al. TGF-beta1-induced CK17 enhances cancer stem cell-like properties rather than EMT in promoting cervical cancer metastasis via the ERK1/2-MZF1 signaling pathway. FEBS J. 2017;284:3000–3017. doi: 10.1111/febs.14162. [DOI] [PubMed] [Google Scholar]
- 37.Horinaka M., Yoshida T., Tomosugi M., Yasuda S., Sowa Y., Sakai T. Myeloid zinc finger 1 mediates sulindac sulfide-induced upregulation of death receptor 5 of human colon cancer cells. Sci. Rep. 2014;4:6000. doi: 10.1038/srep06000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee E.H., Ji K.Y., Kim E.M., Kim S.M., Song H.W., Choi H.R., Chung B.Y., Choi H.J., Bai H.W., Kang H.S. Blockade of Axl signaling ameliorates HPV16E6-mediated tumorigenecity of cervical cancer. Sci. Rep. 2017;7:5759. doi: 10.1038/s41598-017-05977-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee C.J., Hsu L.S., Yue C.H., Lin H., Chiu Y.W., Lin Y.Y., Huang C.Y., Hung M.C., Liu J.Y. MZF-1/Elk-1 interaction domain as therapeutic target for protein kinase Calpha-based triple-negative breast cancer cells. Oncotarget. 2016;7:59845–59859. doi: 10.18632/oncotarget.11337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yue C.H., Huang C.Y., Tsai J.H., Hsu C.W., Hsieh Y.H., Lin H., Liu J.Y. MZF-1/Elk-1 Complex Binds to Protein Kinase Calpha Promoter and Is Involved in Hepatocellular Carcinoma. PLoS ONE. 2015;10:e0127420. doi: 10.1371/journal.pone.0127420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Y., Zhang Z., Yang K., Du J., Xu Y., Liu S. Myeloid zinc-finger 1 (MZF-1) suppresses prostate tumor growth through enforcing ferroportin-conducted iron egress. Oncogene. 2015;34:3839–3847. doi: 10.1038/onc.2014.310. [DOI] [PubMed] [Google Scholar]
- 42.Lin S., Wang X., Pan Y., Tian R., Lin B., Jiang G.S., Chen K., He Y.Q., Zhang L., Zhai W., et al. Transcription Factor Myeloid Zinc-Finger 1 Suppresses Human Gastric Carcinogenesis by Interacting with Metallothionein 2A. Clin. Cancer Res. 2019 doi: 10.1158/1078-0432.CCR-18-1281. [DOI] [PubMed] [Google Scholar]
- 43.Lee J.H., Kim S.S., Lee H.S., Hong S., Rajasekaran N., Wang L.H., Choi J.S., Shin Y.K. Upregulation of SMAD4 by MZF1 inhibits migration of human gastric cancer cells. Int. J. Oncol. 2017;50:272–282. doi: 10.3892/ijo.2016.3793. [DOI] [PubMed] [Google Scholar]
- 44.Tsai S.J., Hwang J.M., Hsieh S.C., Ying T.H., Hsieh Y.H. Overexpression of myeloid zinc finger 1 suppresses matrix metalloproteinase-2 expression and reduces invasiveness of SiHa human cervical cancer cells. Biochem. Biophys. Res. Commun. 2012;425:462–467. doi: 10.1016/j.bbrc.2012.07.125. [DOI] [PubMed] [Google Scholar]
- 45.Vishwamitra D., Curry C.V., Alkan S., Song Y.H., Gallick G.E., Kaseb A.O., Shi P., Amin H.M. The transcription factors Ik-1 and MZF1 downregulate IGF-IR expression in NPM-ALK(+) T-cell lymphoma. Mol. Cancer. 2015;14:53. doi: 10.1186/s12943-015-0324-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ogawa H., Ueda T., Aoyama T., Aronheim A., Nagata S., Fukunaga R. A SWI2/SNF2-type ATPase/helicase protein, mDomino, interacts with myeloid zinc finger protein 2A (MZF-2A) to regulate its transcriptional activity. Genes Cells. 2003;8:325–339. doi: 10.1046/j.1365-2443.2003.00636.x. [DOI] [PubMed] [Google Scholar]
- 47.Brix D.M., Tvingsholm S.A., Hansen M.B., Clemmensen K.B., Ohman T., Siino V., Lambrughi M., Hansen K., Puustinen P., Gromova I., et al. Release of transcriptional repression via ErbB2-induced, SUMO-directed phosphorylation of myeloid zinc finger-1 serine 27 activates lysosome redistribution and invasion. Oncogene. 2019;38:3170–3184. doi: 10.1038/s41388-018-0653-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Workman P., Burrows F., Neckers L., Rosen N. Drugging the cancer chaperone HSP90: Combinatorial therapeutic exploitation of oncogene addiction and tumor stress. Ann. N. Y. Acad. Sci. 2007;1113:202–216. doi: 10.1196/annals.1391.012. [DOI] [PubMed] [Google Scholar]
- 49.Eguchi T., Sogawa C., Okusha Y., Uchibe K., Iinuma R., Ono K., Nakano K., Murakami J., Itoh M., Arai K., et al. Organoids with Cancer Stem Cell-like Properties Secrete Exosomes and HSP90 in a 3D NanoEnvironment. PLoS ONE. 2018;13:e0191109. doi: 10.1371/journal.pone.0191109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ono K., Eguchi T., Sogawa C., Calderwood S.K., Futagawa J., Kasai T., Seno M., Okamoto K., Sasaki A., Kozaki K.I. HSP-enriched properties of extracellular vesicles involve survival of metastatic oral cancer cells. J. Cell. Biochem. 2018 doi: 10.1002/jcb.27039. [DOI] [PubMed] [Google Scholar]
- 51.Nolan K.D., Kaur J., Isaacs J.S. Secreted heat shock protein 90 promotes prostate cancer stem cell heterogeneity. Oncotarget. 2017;8:19323–19341. doi: 10.18632/oncotarget.14252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de la Mare J.A., Jurgens T., Edkins A.L. Extracellular Hsp90 and TGFbeta regulate adhesion, migration and anchorage independent growth in a paired colon cancer cell line model. BMC Cancer. 2017;17:202. doi: 10.1186/s12885-017-3190-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dong H., Zou M., Bhatia A., Jayaprakash P., Hofman F., Ying Q., Chen M., Woodley D.T., Li W. Breast Cancer MDA-MB-231 Cells Use Secreted Heat Shock Protein-90alpha (Hsp90alpha) to Survive a Hostile Hypoxic Environment. Sci. Rep. 2016;6:20605. doi: 10.1038/srep20605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dreos R., Ambrosini G., Groux R., Cavin Perier R., Bucher P. The eukaryotic promoter database in its 30th year: Focus on non-vertebrate organisms. Nucleic Acids Res. 2017;45:D51–D55. doi: 10.1093/nar/gkw1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Messeguer X., Escudero R., Farre D., Nunez O., Martinez J., Alba M.M. PROMO: Detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. 2002;18:333–334. doi: 10.1093/bioinformatics/18.2.333. [DOI] [PubMed] [Google Scholar]
- 56.Eguchi T., Kubota S., Takigawa M. Promoter Analyses of CCN Genes. Methods Mol. Biol. 2017;1489:177–185. doi: 10.1007/978-1-4939-6430-7_18. [DOI] [PubMed] [Google Scholar]
- 57.Eguchi T., Calderwood S.K., Takigawa M., Kubota S., Kozaki K.I. Intracellular MMP3 Promotes HSP Gene Expression in Collaboration With Chromobox Proteins. J. Cell. Biochem. 2017;118:43–51. doi: 10.1002/jcb.25607. [DOI] [PubMed] [Google Scholar]
- 58.Namba Y., Sogawa C., Okusha Y., Kawai H., Itagaki M., Ono K., Murakami J., Aoyama E., Ohyama K., Asaumi J.I., et al. Depletion of Lipid Efflux Pump ABCG1 Triggers the Intracellular Accumulation of Extracellular Vesicles and Reduces Aggregation and Tumorigenesis of Metastatic Cancer Cells. Front. Oncol. 2018;8:376. doi: 10.3389/fonc.2018.00376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eguchi T., Kubota S., Kawata K., Mukudai Y., Uehara J., Ohgawara T., Ibaragi S., Sasaki A., Kuboki T., Takigawa M. Novel transcription-factor-like function of human matrix metalloproteinase 3 regulating the CTGF/CCN2 gene. Mol. Cell. Biol. 2008;28:2391–2413. doi: 10.1128/MCB.01288-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eguchi T., Watanabe K., Hara E.S., Ono M., Kuboki T., Calderwood S.K. OstemiR: A novel panel of microRNA biomarkers in osteoblastic and osteocytic differentiation from mesencymal stem cells. PLoS ONE. 2013;8:e58796. doi: 10.1371/journal.pone.0058796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ruijter J.M., Ramakers C., Hoogaars W.M., Karlen Y., Bakker O., van den Hoff M.J., Moorman A.F. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009;37:e45. doi: 10.1093/nar/gkp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.