Abstract
Phytophthora infestans causes the severe late blight disease of potato. During its infection process, P. infestans delivers hundreds of RXLR (Arg-x-Leu-Arg, x behalf of any one amino acid) effectors to manipulate processes in its hosts, creating a suitable environment for invasion and proliferation. Several effectors interact with host proteins to suppress host immunity and inhibit plant growth. However, little is known about how P. infestans regulates the host transcriptome. Here, we identified an RXLR effector, PITG_15718.2, which is upregulated and maintains a high expression level throughout the infection. Stable transgenic potato (Solanum tuberosum) lines expressing PITG_15718.2 show enhanced leaf colonization by P. infestans and reduced vegetative growth. We further investigated the transcriptional changes between three PITG_15718.2 transgenic lines and the wild type Désirée by using RNA sequencing (RNA-Seq). Compared with Désirée, 190 differentially expressed genes (DEGs) were identified, including 158 upregulated genes and 32 downregulated genes in PITG_15718.2 transgenic lines. Eight upregulated and nine downregulated DEGs were validated by real-time RT-PCR, which showed a high correlation with the expression level identified by RNA-Seq. These DEGs will help to explore the mechanism of PITG_15718.2-mediated immunity and growth inhibition in the future.
Keywords: oomycetes, late blight, effector, Solanum tuberosum, growth inhibition, RNA-Seq
1. Introduction
Plants have developed sophisticated surveillance systems to respond to pathogens and mount defenses against attack. When plants are constantly challenged by microbial parasites in the environment, they can defend themselves from most of the attacks through the innate immune system. Plants detect a multitude of potential invaders, including bacteria, fungi, and oomycetes and have evolved into a two-layered immunity system to protect themselves [1]. The pattern-triggered immunity (PTI) recognizes the microbe-associated molecular patterns (MAMPs) to trigger defense responses that can effectively defeat the vast majority of potential microbes. However, the pathogens secrete and deliver effectors into the host cells to directly manipulate the functions of immune regulators [2,3]. In turn, resistance proteins (R) recognize effectors, activating a rapid immune response known as effector-triggered immunity (ETI), which frequently results in a localized cell death known as the hypersensitive response (HR). Furthermore, effectors can evolve new functions through mutation to suppress ETI and promote disease progression [1,2].
Phytophthora infestans is a major threat to global food security, causing recurrent epidemics in the world’s fourth most important crop, potato [4]. However, the molecular interaction of this pathogen with plant hosts is poorly understood. To manipulate its hosts, P. infestans secretes effector proteins, some of which translocate into host cells [5,6,7,8]. Effectors may act outside or within plant cells to suppress immunity or modify other host processes to increase disease potential [9,10]. RXLR (Arg-x-Leu-Arg, x behalf of any one amino acid) effectors are the largest class of secreted proteins that are translocated into hosts [11,12]. Research on host targets of RXLR effectors has not only revealed essential virulence strategies of Phytophthora but also helped to identify novel components of plant immunity. The delivery of RXLR effectors inside plant cells can affect both plant PTI and ETI. Interestingly, due to the arms race between hosts and Phytophthora, RXLR effectors often act as virulence factors or avirulence factors, whether involved in PTI or ETI. For instance, the expression of PexRD2 and PITG_22798, two RXLR effectors of P. infestans, induce HR in Nicotiana benthamiana [13,14]. In addition, 11 out of 169 RXLR effectors in P. sojae can induce HR in N. benthamiana [15]. They act as elicitors to trigger the host PTI. In contrast, many RXLR effectors inside plant cells can inhibit the HR induced by MAMPs to attenuate plant PTI. Eight out of thirty-three PiRXLR effectors were identified to suppress the early Flg22-induced immune response [16]. Infestin 1 (INF1) is the best-recognized MAMP, which triggers the HR in N. benthamiana [17]. However, Avr3a can suppress the INF1-triggered HR by targeting the E3 ligase CMPG1 in the host [17,18]. PexRD54 binds the host autophagy protein ATG8CL to interfere with Joka2-mediated defense in the host [19]. Pi17316 also attenuates the INF1-triggered HR by targeting a MEK kinase (MAP3K) [20].
Alternatively, RXLR effectors play distinct roles in ETI. Currently, all known avirulence genes of potato late blight are RXLR effectors, such as Avr1, Avr2, AVR3a, Avr3b, Avr10, Avr11 and AVRblb1, which are recognized by cognate resistance proteins R1, R2/Rpi-blb3/R2-like, R3a, R3b, R10, R11 and Rpi-blb1. In addition, RXLR effectors ipiO1 and ipiO2 are recognized by RB [21,22,23,24,25,26,27]. On the other hand, some effectors can inhibit the HR triggered by the recognition of resistance proteins and the products of avirulence genes. PexRD2 directly targets and inhibits the MAPK3Kε activity and suppresses the HR mediated by the recognition of Avr4 by Cf4 [28]. Like PexRD2, Avr3a can suppress the HR triggered by the recognition of Avr4 by Cf4 [29]. Coinfiltration with Avr3b suppresses the PITG_22798 induced HR in N. benthamiana [16]. Wang et al. reported that many RXLR effectors of P. sojae inhibited the HR induced by Avh238 and Avh241 in N. benthamiana [17]. Meanwhile, successful pathogen effectors can escape the detection of resistance genes. P. infestans AVR3aK80I103 is recognized by the potato R3a protein. However, the allelic variant AVR3aE80M103 failed to be recognized by R3a [14]. Similarly, the amino acid substitution of AVRblb2 at position 69 was evolved to escape the recognization by its cognated Rpi-blb2 [15].
Moreover, to suppress the host immunity, pathogenic effectors can also alter plant development. By targeting BAK1 involved in signaling triggered by brassinosteroid (BR), multiple MAMPs and cell death, Pseudomonas syringae effectors of AvrPto and AvrPtoB suppress the plant immunity and development [30]. Overexpression of Phytoplasma effectors, SAP11, SAP54 and TENGU, lead to witches’ broom symptoms as well as enhance the susceptibility in Arabidopsis [31,32,33]. Transgenic plants carried the Ralstonia solanacearum effectors; RipAB exhibited a stunted growth in potato [34]. Phytophthora parasitica RXLR effector, penetration-specific effector 1 (PSE1), which increases pathogen susceptibility and interferes with auxin physiology [35]. Among P. infestans RXLR effectors, transgenic potato carrying Avr2 and PSR1 exhibited both enhanced susceptibility and growth inhibition, respectively [36,37]. However, the RXLR effectors exhibiting the above double functions are still limited.
In this study, we focused on an RXLR effector, PITG_15718.2, secreted by P. infestans. We found that PITG_15718.2 was upregulated during infection. Constitutive expression of PITG_15718.2 in potato promoted P. infestans infection and inhibited plant growth, causing decreased plant height. To investigate the underlying mechanisms of these effects, we employed RNA-Seq to study the transcriptome dynamics of interactions in PITG_15718.2-transformed lines and wild type. We identified 190 DEGs that were associated with the attenuated immunity and growth in PITG_15718.2-expressing potato.
2. Results
2.1. High Accumulation of the PITG_15718.2 Transcripts during P. infestans Infection
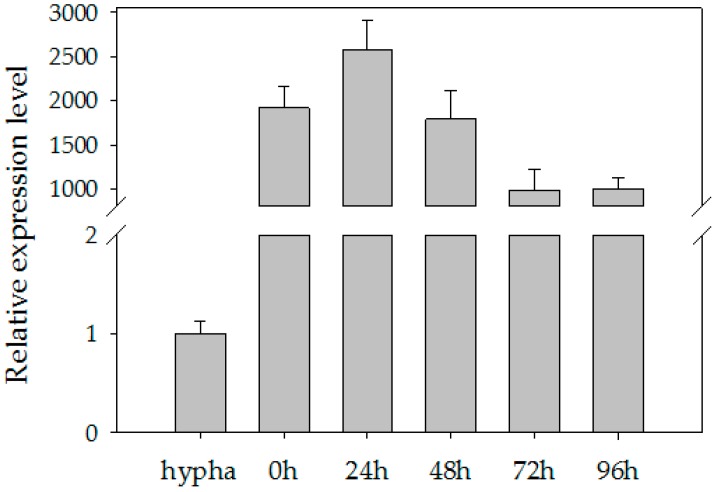
Many RXLR effectors of P. infestans are upregulated during the biotrophic phase of infection on potato plants [20]. The gene PITG_15718.2 is annotated as encoding a secreted RXLR effector protein in the P. infestans genome [11]. To investigate the expression of PITG_15718.2 in P. infestans during infection, total RNA from hyphae and potato leaves at 0, 24, 48, 72 hand 96 h post-inoculation (hpi) were purified and used for real-time RT-PCR. The expression of PITG_15718.2 was compared between lines after normalizing to the PI-O8, a repetitive element from P. infestans [38]. As shown in Figure 1, compared to hyphae (cultured nonsporulating mycelium), the expression of PITG_15718.2 was rapidly increased to over 1900-fold at 0 h, which was represented as the sporangium and zoospore attached to plant tissue. It reached the highest expression level at 24 hpi. Three repeated amplifications in independent experiments with different cDNA samples resulted in similar expression patterns, confirming that PITG_15718.2 is highly expressed during infection.
Figure 1.
PITG_15718.2 is upregulated in sporangia and during the early stages of infection. The hyphae were scraped with a glass rod from the plants on which P. infestans grew at 18 °C for two weeks. The sporangia were collected from the suspension, then incubated on ice and stored at 4 °C for 3 h to release the zoospores. The PITG_15718 expression level was assessed by qRT-PCR at 0, 12, 24, 48, 72 and 96 h after inoculation of P. infestans. Error bars represent SD of the mean values of three biological replicates. The experiment was repeated three times with similar results.
2.2. The Constitutive Expression of PITG_15718.2 in S. tuberosum
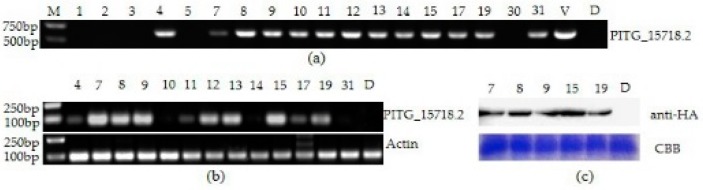
RXLR effectors often promote P. infestans virulence with specifically upregulated expression during the biotrophic phase of infection [39,40]. Désirée is a susceptible variety that is used for studying late blight disease resistance. To investigate the virulence function of PITG_15718.2 during P. infestans infection, we transformed it into potato cultivar Désirée using an Agrobacterium tumefaciens-mediated system. In brief, a construct carrying the PITG_15718.2 was cloned, with both a HA-tag and a CFP-tag fused to its N-terminus. A total of 18 transgenic plants were generated, and their DNA was extracted for positive selection. After PCR with specific primers, thirteen lines, numbered 4, 7, 8, 9, 10, 11, 12, 13, 14, 15, 17, 19 and 31, were identified to carry PITG_15718.2 (Figure 2a). To investigate the expression level of PITG_15718.2 in transgenic lines, the above thirteen positive lines were sampled to amplify the PITG_15718.2 with semiquantitative RT-PCR. The results showed that the expression of PITG_15718.2 was significantly higher in lines 7, 8, 9, 12, 13, 15 and 19 than the other lines (Figure 2b). Furthermore, lines 7, 8, 9, 15 and 19 highly expressed the PITG_15718.2 protein by western blot (Figure 2c). Thus, three transgenic lines, line 7, line 9 and line 15, were selected for further investigations.
Figure 2.
Validation of positive transgenic lines of PITG_15718.2 potato (Désirée). (a) PCR detected 18 transgenic individuals using PITG_15718.2-specific primers with agarose gel electrophoresis. A total of 13 positive lines out of 18 lines were obtained; (b) detection of the expression level of PITG_15718.2 using semiquantitative RT-PCR for 13 positive lines. The StEF gene was the endogenous control. PITG_15718.2 was highly expressed in lines 7, 8, 9 and 15; (c) western blot showing that PITG_15718.2 was highly accumulated in lines 7, 8, 9 and 15. Total protein extracts from S. tuberosum (Désirée and lines 7, 8, 9 and 15). HA-tagged fusion proteins were detected by probing western blots with HA antibodies.
2.3. PITG_15718.2 Promotes P. infestans Virulence in Potato
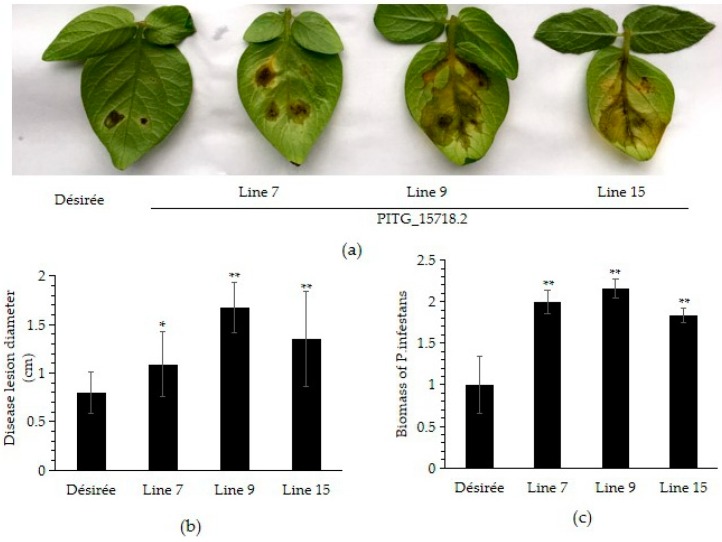
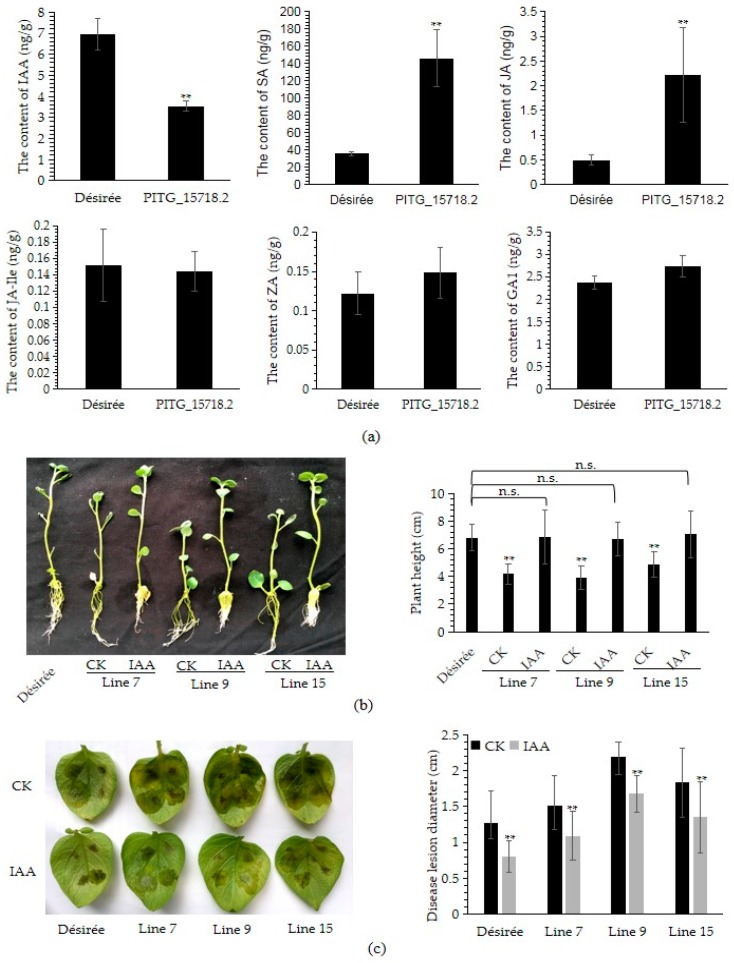
To evaluate the virulence of PITG_15718.2, detached leaves of transgenic lines 7, 9 and 15 were inoculated with P. infestans HLJ at the 8-leaf stage. As shown in Figure 3a, images were photographed at 4 dpi. Compared to Désirée, the transgenic lines were more susceptible to P. infestans HLJ. Consistent with the greater susceptibility, the average leaf disease lesion diameter was longer on all three transgenic lines than Désirée (Figure 3b). Furthermore, using the mixed DNA from potato leaves and propagated P. infestans hyphae as template, we quantified the PI-O8 element with relative quantitative PCR, which represented the relative biomass of P. infestans. All three transgenic lines showed significantly increased relative biomass of P. infestans over Désirée (Figure 3c). These results demonstrated that the transgenic lines had enhanced P. infestans colonization and propagation compared with the wild type. This suggests that PITG_15718.2 promotes P. infestans virulence in potato.
Figure 3.
Constitutive expression of PITG_15718.2 enhanced susceptibility to P. infestans in potato. (a) Three PITG_15718.2-expressing lines were challenged with P. infestans HLJ. Images were photographed at four days post-inoculation (dpi). Désirée was used as control; (b) the lesion size was significantly enhanced on both transgenic potato lines compared to the control. The disease lesion diameter was calculated from at least 15 leaves using the average of the longest and shortest diameters on each leaf. * indicates significant (t test, p < 0.05) differences, ** indicates extremely significant (t test, p < 0.01) differences; (c) determination of P. infestans biomass by quantitative real-time PCR. P. infestans–specific primers referred the PI-O8 element were used for PCR with the mixed-DNA template that was isolated from infected leaves (containing the DNA of P. infestans and S. tuberosum). The host gene (StEF) was used to normalize the results. ** indicates extremely significant (t test, p < 0.01) differences. The experiment was repeated three times with similar results.
2.4. PITG_15718.2 Inhibits Potato Plant Growth
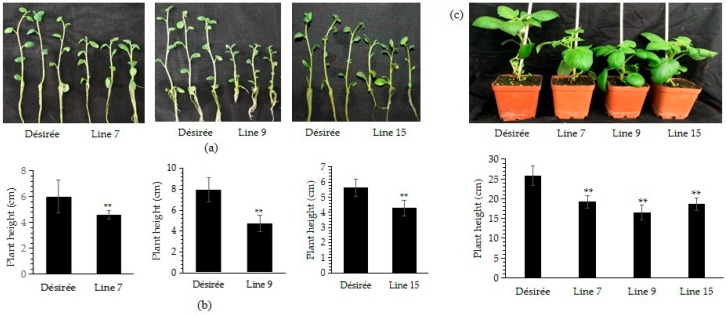
During the propagation of the tissue culture plantlets, we found that all transgenic plantlets were smaller than Désirée. Therefore, we cultured the plantlets of transgenic lines and wild type side by side in the same tissue-cultural container. After two weeks, plantlets from all three transgenic lines were smaller than Désirée (Figure 4a). The plant height was significantly shorter in the transgenic lines than in Désirée as well as the root length after measuring over 30 individuals from 10 bottles (Figure 4b and Figure S1). Furthermore, transgenic plants were exhibited the stunted phenotype three weeks after planting into soil (Figure 4c). So we concluded that the expression of PITG_15718.2 inhibits the plant growth of Désirée.
Figure 4.
Stable expression of PITG_15718.2 inhibits the vegetative growth of S. tuberosum. (a) Images of transgenic lines and controls. Each transgenic line was planted in one tissue-cultural container with Désirée. Three seedlings on each side, forming a confrontation. Images were photographed two weeks after planting; (b) the plant height was significantly dwarfed for each transgenic potato line compared to Désirée. The plant height was measured in at least 30 plantlets. ** indicates extremely significant differences determined using the Student’s t-test (p < 0.01); (c) the transgenic lines show stunted phenotype after planting in soil. Image was photographed three weeks after planting into soil. The plant height was measured in at least 20 plantlets. ** indicates extremely significant differences determined using the Student’s t-test (p < 0.01).
2.5. Transcriptional Profiling Analysis of the Transgenic Lines Constitutively Expressing PITG_15718.2
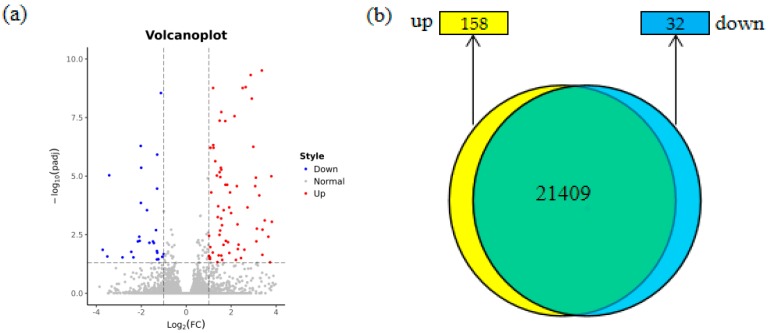
To explore the mechanism by which PITG_15718.2 suppressed potato immunity and vegetative growth, we compared the transcriptome profile of the three transgenic lines with their respective wild type controls by RNA sequencing. Six libraries were constructed and sequenced by an Illumina PE150. An average of 7 G Cleandata were obtained from each library, with over 97% of reads longer than 20 base pairs (bp) and approximately 43% G+C content (Table S1). The transcriptome sequencing reads were aligned to the potato DM genome [41] using the Hierarchical Indexing for Spliced Alignment of Transcripts (HISAT2) software. In summary, the number of RNA-Seq reads per library ranged from 49.0 to 63.9 million, in which 76.4% to 77.7% of reads mapped to the potato genome (Table S2). Then, we compared and analyzed the RNA-Seq data for the three transgenic lines and their wild type controls. We use HTSeq to calculate Reads Count and DEseq2 to compute differential expression. By defining a log2(fold change) ≥1 and adjusted p-value ≤0.05 as the standards of a differentially expressed gene (DEG), we identified 190 genes (Figure 5a), including 158 upregulated genes and 32 downregulated genes (Figure 5b), common to all three transgenic lines compared to Dall.
Figure 5.
Differentially expressed genes in transgenic lines expressing PITG_15718.2 compared with the wild type. (a) Volcano plot showing fold change and adjusted p-value of normalized read counts of the RNA-Seq data. The criteria of log2|(fold change)| ≥ 1 and padj ≤ 0.05 were used to identify the differentially expressed genes (DEGs). Blue dots indicate the downregulated DEGs (32 genes), and red dots indicate the upregulated DEGs (158 genes); (b) Venn diagram showing the overlap of the DEGs between the transgenic and wild type groups. The sum of all the numbers in the figure represents the total number of genes in the comparison. The overlapping region indicates the number of genes that were not significantly different between the groups. The yellow label indicates the number of upregulated DEGs and the cyan label indicates the number of downregulated DEGs according the set thresholds.
2.6. Functional Enrichment of the Differentially Expressed Genes
The functions of DEGs were associated with the phenotypes of immunity and growth inhibition in PITG_15718.2-expressing potato. To understand the biological functions of the DEGs, we carried out gene ontology (GO) analysis with the Hypergeometric distribution algorithm of Cluster Profiler [8] for the above 190 identified DEGs a p value less than 0.05. Among the 158-upregulated genes, their encoding proteins were classified into 22 GO enrichment summarized to three main categories including biological process, molecular function and cellular component (Table 1). For the proteins encoded by the 32-downregulated genes, 17 GO enrichments were classified and integrated into the three categories too (Table 2). Among the total of 39 GO enrichments, 35 enrichments were concentrated in biological process and molecular function. Among the upregulated DEGs, the GO term of the oxidation-reduction process, belonging to the enrichment biological process, largely overlapped with catalytic activity, belonging to molecular function. Among the downregulated DEGs, two GO terms of the biological process, response to external stimulus and symbiosis, encompassing mutualism through parasitism, were shared with most genes belonging to catalytic activity of molecular function. Whether upregulated or downregulated, the GO term of catalytic activity contained the highest number of proteins among all enrichments, indicating that they may not directly determine the PITG_15718.2 function (Table 1 and Table 2). Alternatively, we noticed that the functions were completely different under the biological process and molecular function between upregulated and downregulated proteins. The enrichment of the molecular function was concentrated on binding activity and lyase activity among upregulated and downregulated DEGs, respectively. Under biological process, the upregulated DEGs were mainly distributed into porphyrin-containing compound biosynthetic process (GO:0006779), cell differentiation (GO:0030154), cofactor biosynthetic process (GO:0051188), multiorganism process (GO:0051704), pigment biosynthetic process (GO:0046148) and oxidation-reduction process (GO:0055114). However, the biological processes of downregulated DEGs contained the enrichments of response to external stimulus (GO:0009605), response to other organism (GO:0051707), response to external biotic stimulus (GO:0043207), symbiosis, encompassing mutualism through parasitism (GO:0044403), interspecies interaction between organisms (GO:0044419) and response to biotic stimulus (GO:0009607). Those functions are clearly related to plant immunity, which may help to explain the virulence function of PITG_15718.2.
Table 1.
Gene ontology (GO) enrichments of upregulated genes.
| GO_Accession | Description | DEG Number | Term Type |
|---|---|---|---|
| GO:0055114 | oxidation-reduction process | 12 | biological process |
| GO:0051704 | multi-organism process | 7 | |
| GO:0051188 | cofactor biosynthetic process | 3 | |
| GO:0006779 | porphyrin-containing compound biosynthetic process | 2 | |
| GO:0030154 | cell differentiation | 2 | |
| GO:0046148 | pigment biosynthetic process | 2 | |
| GO:0044427 | chromosomal part | 3 | cellular component |
| GO:0032040 | small-subunit processome | 2 | |
| GO:0030684 | Preribosome | 2 | |
| GO:0003824 | catalytic activity | 42 | molecular function |
| GO:1901363 | heterocyclic compound binding | 36 | |
| GO:0097159 | organic cyclic compound binding | 36 | |
| GO:0043168 | anion binding | 17 | |
| GO:0036094 | small molecule binding | 17 | |
| GO:0000166 | nucleotide binding | 16 | |
| GO:1901265 | nucleoside phosphate binding | 16 | |
| GO:0003677 | DNA binding | 13 | |
| GO:0020037 | heme binding | 6 | |
| GO:0046906 | tetrapyrrole binding | 6 | |
| GO:0016705 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen | 5 | |
| GO:0004674 | protein serine/threonine kinase activity | 3 | |
| GO:0045735 | nutrient reservoir activity | 2 |
Table 2.
GO enrichments of downregulated genes.
| GO_Accession | Description | DEG Number | Term Type |
|---|---|---|---|
| GO:0009058 | biosynthetic process | 8 | biological process |
| GO:0009605 | response to external stimulus | 3 | |
| GO:0044403 | symbiosis, encompassing mutualism through parasitism | 3 | |
| GO:0044419 | interspecies interaction between organisms | 3 | |
| GO:0051704 | multi-organism process | 3 | |
| GO:0006814 | sodium ion transport | 2 | |
| GO:0051707 | response to other organism | 2 | |
| GO:0043207 | response to external biotic stimulus | 2 | |
| GO:0016051 | carbohydrate biosynthetic process | 2 | |
| GO:0009607 | response to biotic stimulus | 2 | |
| GO:0015672 | monovalent inorganic cation transport | 2 | |
| GO:0044723 | single-organism carbohydrate metabolic process | 2 | |
| GO:0005886 | plasma membrane | 2 | cellular component |
| GO:0003824 | catalytic activity | 14 | molecular function |
| GO:0016829 | lyase activity | 3 | |
| GO:0016831 | carboxy-lyase activity | 2 | |
| GO:0016830 | carbon-carbon lyase activity | 2 |
2.7. Validation of DEGs by Real-Time RT-PCR
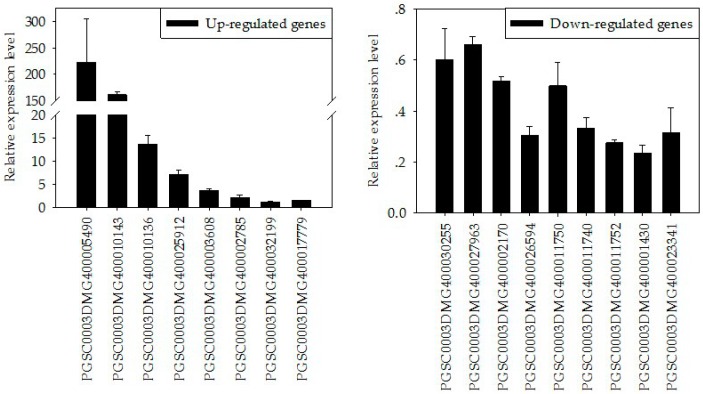
To validate the RNA-Seq results, we selected several DEGs to perform the real-time RT-PCR detection. Based on their putative functions in plant immunity or growth regulation, eight of the upregulated and nine of the downregulated genes were selected. As shown in Figure 6a,b, all seventeen genes showed differential expression in transgenic lines. Compared with the RNA-Seq fold-change numbers, except PGSC0003DMG400032199, the other 16 DEGs showed extremely tight correlations with the qRT-PCR results (Figure S2).
Figure 6.
Validating the DEGs by qRT-PCR. (a) Eight of the upregulated DEGs; (b) nine of the downregulated DEGs. Error bars represent SD of the mean of three biological replicates. The experiment was repeated three times with similar results.
2.8. Expression of PITG_15718.2 Decrease the Content of Indole-3-Acetic Acid
The expression of PITG_15718.2 increases the susceptibility to P. infestans and reduces the plant growth, indicating that phytohormones may be involved in the signaling pathway. Therefore, we quantify the content of salicylic acid (SA), jasmonic acid (JA), Ile-JA, Zeatin (ZA), indole-3-acetic acid (IAA) and gibberellic acid 1 (GA1) by liquid chromatograph-mass spectrometer (LC-MS) for both Désirée and mixed samples of three transgenic lines. As shown in Figure 7a, compared to Désirée, the content of IAA was dramatically decreased in transgenic lines while the content of SA and JA were increased. There were no significant changes for the content of Ile-JA, ZA and GA1 between Désirée and mixed transgenic samples. The high content of SA and JA were associated to enhance the potato resistance to P. infestans, and IAA was associated with promoting plant growth. So we select IAA for the growth restoration assays with PITG_15718.2 transgenic lines. As shown in Figure 7b, supplementing with 1 mg/L of IAA, all three transgenic lines can restore the plant height as well as wild type Désirée. In addition, we found that the IAA-treated plantlets show more numbers but shorter adventitious roots than untreated controls. Furthermore, we found that pre-sprayed IAA could enhance the resistance to P. infestans for both Désirée and transgenic plants (Figure 7c). Overall, the results were implied that PITG_15718.2 may suppress the plant immunity and reduce growth by decreasing the content of IAA.
Figure 7.
Decreasing of IAA is involved in PITG_15718.2-mediated inhibition of plant immunity and growth. (a) The content of salicylic acid (SA), jasmonic acid (JA), Ile-JA, Zeatin (ZA), indole-3-acetic acid (IAA) and gibberellic acid 1 (GA1). The sample was collected from leaves of 3-weeks of potato planting in soil. The sample of PITG_15718.2 was mixed with equal amount of three transgenic lines 7, 9 and 15. Four replicates for each sample were measured by LC-MS. Asterisks indicate extremely significant differences determined using the Student’s t-test (p < 0.01); (b) the plant height of transgenic lines is restored by supplementing with IAA. Images were photographed two weeks after planting. The plant height was measured with at least 20 plantlets. The concentration of IAA is 1 mg/L; (c) pre-spraying IAA enhances resistance to P. infestans. Leaves of 3-weeks of potato planting in soil was sprayed with 1 mg/L of IAA and inoculated with P. infestans 24 h later. Images were photographed four days post inoculation and disease lesion diameter were measured with at least 30 leaves. Asterisks indicate extremely significant differences determined using the Student’s t-test (p < 0.01). The above experiments (b,c) were repeated three times with the similar results.
3. Discussion
P. infestans is a particularly destructive pathogen causing late blight that leads to significant potato crop losses due to early defoliation and tuber infection. The pathogen spreads rapidly, causing complete crop loss within days without successful management [42]. RXLR effectors are secreted by P. infestans to manipulate the host’s physiology to promote infection, including debilitation of the immune system. The genome sequence shows that there are more than 500 RXLR effectors in P. infestans. The RXLR effectors promoting the infection of P. infestans are upregulated during the infection stage [39,40]. Assays of upregulated RXLRs have indicated that 45 out of 52 RXLRs promote pathogen leaf colonization when expressed in N. benthamiana [43]. Nevertheless, for the majority of RXLR effectors, their biological functions and potential host targets are unknown. In this study, we identified that the RXLR effector encoded by PITG_15718.2 is upregulated during the infection stage of P. infestans and maintains a high expression throughout the infection phase. Consistent with previous reports, constitutive expression of PITG_15718.2 increased the susceptibility of potato to P. infestans in detached leaves. We conclude that PITG_15718.2 acts as a virulence factor in the interaction of P. infestans and S. tuberosum, providing another example of an RXLR effector as a virulence factor.
Most identified RXLR effectors of P. infestans are involved in regulating plant immunity, whether directly as virulence factors as discussed above, or indirectly as avirulence factors recognized by their cognate R proteins. As virulence factors, knowledge of their functions in regulating plant growth are still limited. Phytohormones, such as SA, JA and IAA, broadly involved in regulating both plant immunity and growth. SA antagonize JA and repress auxin accumulation in plant defense. However, the crosstalk of phytohormones mediated by the RXLR effector is poorly understood. Constitutively expressed Avr2, an RXLR effector, interferes with plant growth through overactive brassinosteroid (BR) hormone signaling [36]. Overexpression of StPSR1 could inhibit the growth of Arabidopsis, and the root hairs phenotype could significantly decrease about 80% after IAA treatment [37]. Here, we showed that expressing PITG_15718.2 also significantly decreased plant height (Figure 4a) and the content of IAA (Figure 7a). IAA treatment could restore the plantlets height and increase the disease resistance to P. infestans (Figure 7b,c). It was demonstrated that both the PITG_15718.2-mediated susceptibility and growth inhibition were partially caused by decreasing of the IAA content. It is similar as the Phytoplasma effector TENGU that promoted the infection and led to dwarfism by downregulating the auxin-related genes [31]. Meanwhile, we also have noticed the accumulation of higher content of SA in transgenic plants than in Désirée (Figure 7a). High accumulation of SA would reduce the IAA accumulation and cause the inhibition of plant growth. Additionally treatment with SA has been previously reported to increase the resistance or susceptibility to P. infestans on susceptible potato [44]. Whether and how the PITG_15718.2 regulates the SA and IAA in Désirée needs to be further studied.
Next-generation sequencing (NGS) technologies are fast evolving and transforming biology research [45]. RNA-Seq is an approach to study the transcriptome [46,47]. RNA-Seq is a valuable method for transcriptome dynamics analysis in tetraploid potato [48,49,50]. This suggests that RNA-Seq could be used to study potato immunity and growth. However, to understand the function of RXLR effectors, previous studies mainly used the protein–proteininteraction system to identify their host targets, as well as to determine the biological functions in regulating host immunity. For instance, Avr3a modifies the E3 ubiquitin ligase CMPG1, resulting in accumulation of CMPG1 and making plants more susceptible [14]. Quite coincidentally, to weaken the plant immunity, the Pi02860 target host NRL1, a BTB-domain protein that forms the substrate adaptor component of a CULLIN3 ubiquitin E3 ligase, degrades SWAP70, a positive regulator of immunity [51]. PexRD2 targets MAPKKKε to suppress host immunity [28]. Avrblb2 interacts with the cysteine caspase C14 to prevent C14 secretion and destroy host immunity [52]. PexRD54 is able to combine with ATG8 to interfere with the host selective autophagy [19]. However, after these protein–protein interactions, the downstream signaling transduction pathways are still unknown. Recently, by analyzing BR-treatment microarray data, a BR-inducible bHLH transcription factor, StCHL1, was identified to be upregulated by AVR2 and to be required for its suppression of INF1-mediated HR [36]. To explore the mechanism by which PITG_15718.2 affects plant immunity and growth, we performed the RNA-Seq analysis of transgenic plants. We conducted the RNA-Seq on three independent transgenic lines in parallel to eliminate the effects caused by different T-DNA insertions. Only 190 DEGs, including 158 upregulated and 32 downregulated, were identified.
Interestingly, among upregulated DEGs, PGSC0003DMG400003608, encoding an abscisic acid 8’-hydroxylase, is reminiscent of a previous finding of negatively regulated plant growth because overexpression of the homologue of GhABAH was found to inhibit the growth in transgenic tomatoes and cotton [53]. Overexpression of ShMKS1 in Arabidopsis thaliana and Nicotiana tabacum also caused serious growth defects [54]. The upregulated DEG PGSC0003DMG400025912 is a homologue of ShMKS, which encodes a salicylic acid-binding protein and may negatively regulate plant growth. PGSC0003DMG400010143 encodes a cysteine protease inhibitor gene, which was upregulated during P. infestans infection of tomato, suggesting a role during P. infestans-host interactions [55]. These data indicate that the PITG_15718.2-induced DEGs may negatively regulate the plant immunity or vegetative growth. In contrast to upregulated DEGs, among downregulated genes, cytochrome P450 (PGSC0003DMG400026594 and PGSC0003DMG400011750) are essential for lignification and defense against predators and pathogens [56]. The transgenic potato carrying the antisense of Gibberellin 20 oxidase (PGSC0003DMG400027963) cDNA had a shorter stem, a decreased length of the internodes and tuberized earlier than control plants, showing increased tuber yields [57]. Mutants at the PROCUSTE1 (PRC1) or RSW1 (CesA1) loci (cellulose synthase, PGSC0003DMG400011752) showed decreased cell elongation, specifically in roots and dark-grown hypocotyls [58]. PGSC0003DMG400030255 encodes a member of YUCCA gene family, which is an important regulator of IAA biosynthesis. Consistent with its downregulated expression, the content of IAA is dramatically reduced in transgenic lines than in Désirée (Figure 7a). The above cases suggest that the downregulated DEGs in PITG_15718.2 transgenic potato may positively regulate immunity and plant growth. They are likely to be key components downstream of the interaction between PITG_15718.2 and host targets that need to be investigated in the future.
4. Materials and Methods
4.1. Microorganism and Plant Materials
Escherichia coli DH5a and Agrobacterium tumefaciens GV3101 and AGL1were routinely grown in Luria–Bertani (LB: 10 g of bacteriological peptone, 10 g NaCl, and 5 g yeast extract in 1 L of distilled water) medium plus with appropriate antibiotics at 37 and 28 °C, respectively. P. infestans strain HLJ, originally isolated from the province of Heilongjiang, north of China, was grown in the dark at 18 °C using rye A agar. Unless noted, S. tuberosum and N. benthamiana plants were grown at 22 and 25 °C, respectively, under 70% humidity with 16 h of light and 8 h of dark.
4.2. Construction of Recombinant Binary Vector
Since none of the tested Avr genes contained predicted introns, we amplified each of the genes directly from genomic DNA from the P. infestans HLJ, using the Cobuddy DNA Polymerase (CWBIO, Beijing, China) and specific primers. The PCR products were digested with the appropriate enzymes and cloned into the recombinant binary vector pEarleyGate 102 (C-CFP-HA) [59] and PVX vector pGR106. The constructs were validated by DNA sequencing then introduced into Agrobacterium tumefaciens GV3101 or AGL1.
4.3. Transformation of Potato
Agrobacterium tumefaciens AGL1 containing pEarleyGate 102-PITG_15718.2 was used for transformation with the stem of the potato cv. Désirée as previously reported [60]. Positive lines were first screened on differential medium (MS + 0.2 mg·L−1 NAA + 0.02 mg·L−1 GA3 + 2.5 mg·L−1 zeatin riboside + 500 mg·L−1 cefotaxime) and then transferred to root generation medium (MS + 0.02 mg·L−1 NAA + 0.02 mg·L−1 GA3 + 2 mg·L−1 zeatin riboside + 500 mg·L−1 cefotaxime + 50 mg/L glufosinate). The positive selection and the expression level of PITG_15718.2 was investigated with PCR, semiquantitative RT-PCR and western blot.
4.4. Gene Expression Analysis by qRT-PCR
Total RNA was isolated with 100 mg fresh tissue using the TRI Reagent (Merck, Darmstadt, Germany). The residual DNA removal and first-strand cDNA synthesis were conducted with ReverTra Ace qPCR RT Master Mix with gDNA Remover kit (TOYOBO, Osaka, Japan). The real-time RT-PCR was done with KOD SYBR qPCR Mix (TOYOBO, Osaka, Japan). For PCR, samples were preheated at 95 °C for 5 min. Then, 40 amplification cycles were run: 15 s at 95 °C, 30 s at 60 °C. Fluorescence (520 nm) was detected at the end of the elongation phase for each cycle. After the final amplification cycle, a melting curve was done by heating to 95 °C, cooling to 65 °C and slowly heating to 95 °C at 0.1 °C·s−1 with continuous measurement of fluorescence at 520 nm.
4.5. Detached Leaf Assays
P. infestans HLJ was grown in petri dishes in Rye A for two weeks at 18 °C. Plates were flooded with 5 mL H2O and scraped with a glass rod to release sporangia. The suspension was poured into a clean Petri dish, placed on ice and stored in the 4 °C for 3 h to release zoospores. The sporangia were counted using a hemocytometer and adjusted to 30,000 sporangia per milliliter. The leaves were placed on moistened tissue paper in a 25-by-25-cm plate, then inoculated with 20 μL droplets of a zoospores and sporangia suspension. Inoculated leaves were incubated at 18 °C (16 h of light and 8 h of dark). The lesions were measured at 5 dpi. For IAA treatment, detached leaves were sprayed with the 1 mg/L of IAA and incubated for 24 h, then inoculated with P. infestans.
4.6. Determination of Pathogen Biomass by Quantitative Real-Time PCR
The biomass of P. infestans was determined using the modified method of Kobayashi [61]. To determine the growth of P. infestans, inoculated potato leaves were used for quantitative real-time PCR. Tissue was broken in liquid nitrogen. The DNA was isolated with the CTAB method. To amplify and detect P. infestans and plant specific DNA sequences [38], the following primers were used: O8-3 (5′-GAAAGGCATAGAAGGTAGA-3′) and O8-4 (5′-TAACCGACCAAGTAGTAAA-3′) for PI-O8 element of P. infestans; and StEF-1a -F(5′-GGTCTACCAACCTCGACTGGTAC-3′) and StEF-1a -R(5′-GGGTTTGTCTGATGGCCTCTTGG-3′) for potato StEF (AB061263) gene. The PCR system was as described above.
4.7. RNA-Seq
The plantlets of three PITG_15718.2 transgenic lines and the wild type were planted side by side in the same tissue culture bottle for 20 days at 18 °C. Each bottle contained three plantlets for transgenic and three for Dhree f. There were at least five bottles for each experiment. For preparing each pooled RNA sample, a total of 15 seedling plants from five bottles were collected and submitted to total RNA extraction as described above. The quantity and quality of RNA samples were assessed using a NanoDrop One (Thermo Scientific, Waltham, MA, USA), Qubit 3.0 (Thermo Scientific) and 4200 TapeStation (Agilent, Palo Alto, CA, USA). High-quality total RNA (5 µg, 100 ng/μL) samples were sent to the HaploX Genomics Center (www.haplox.cn, shangrao, China) for RNA-Seq with an Illumina PE150.
4.8. Determination of the Phytohormones Concentration
The concentration of SA, IAA, JA, Ile-JA, ZA and GA1 was determined using the Shimadzu LCMS 8040 system (Shimadzu, Tokyo, Japan) as previously described [62]. Approximately 200 mg of leaf tissue was collected from the upper-full-expand leaves of 3-week-old wild type and ground in liquid nitrogen. The hormones were extracted with 1 mL of ethyl acetate spiked with each internal standard and quantified with LC-MS. Four replicates were conducted for each sample.
4.9. Deposition of Sequences
The data sets supporting the results of this article are included within the article and its additional files. The nucleic acid sequence of PITG_15718.2 is available in NCBI’s database with accession number XM_002996840.1 (http://www.ncbi.nlm.nih.gov/nucleotide). The RNA-Seq data supporting the results of this article are available in the NCBI’s SRA with the accession number SRR8392524 (http://www.ncbi.nlm.nih.gov/sra/).
5. Conclusions
In this study, we identified a novel RXLR effector of P. infestans, PITG_15718.2, a virulence factor that promoted the colonization of P. infestans and inhibited the vegetative growth of potato. We also identified 190 DEGs and the content of IAA that were associated with the attenuated immunity and growth in PITG_15718.2-expressing potato.
Abbreviations
| DEG | differentially expressed gene |
| ETI | effector-triggered immunity |
| HR | hypersensitive response |
| LD | linear dichroism |
| MAMPs | microbe-associated molecular patterns |
| PTI | pattern-triggered immunity |
| R | resistance protein |
| RNA-Seq | RNA sequencing |
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/20/12/3031/s1.
Author Contributions
Z.C. designed and supervised the experiments; J.W., C.G., R.D. and L.L. performed the transformation and disease assay; J.W. and C.G. performed the RNA-Seq and data analysis; J.W. and W.C. performed the qRT-PCR and data analysis; X.D. and C.Z. provided project administration and discussion; J.W. and Z.C. wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31801722) and the Funds of Shandong “Double Tops” Program (2017). X.D. was funded by the Taishan Scholar Project (20150621).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Jones J.D.G., Dangl J.L. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 2.Dou D., Zhou J.M. Phytopathogen effectors subverting host immunity: Different foes, similar battleground. Cell Host Microbe. 2012;12:484–495. doi: 10.1016/j.chom.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 3.Dangl J.L., Horvath D.M., Staskawicz B.J. Pivoting the plant immune system from dissection to deployment. Science. 2013;341:746–751. doi: 10.1126/science.1236011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.David E.L.C., Liliana M.C., Sylvain R., Ruairidh A.B., Louise R.C., Graham J.E., Kenneth L.D., Rhys A.F., Eleanor M.G., Erica M.G., et al. Genome analyses of an aggressive and invasive lineage of the Irish potato famine pathogen. PLoS Pathog. 2012;8:e1002940. doi: 10.1371/journal.ppat.1002940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birch P.R., Rehmany A.P., Pritchard L., Kamoun S., Beynon L. Trafficking arms: Oomycete effectors enter host plant cells. Trends Microbiol. 2006;14:8–11. doi: 10.1016/j.tim.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 6.Kamoun S.A. Catalogue of the effector secretome of plant pathogenic oomycetes. Annu. Rev. Phytopathol. 2006;44:41–60. doi: 10.1146/annurev.phyto.44.070505.143436. [DOI] [PubMed] [Google Scholar]
- 7.Whisson S.C., Boevink P.C., Moleleki L., Avrova A.O., Morales J.G., Gilroy E.M., Armstrong M.R., Grouffaud S., van West P., Chapman S., et al. A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature. 2007;450:115–118. doi: 10.1038/nature06203. [DOI] [PubMed] [Google Scholar]
- 8.Schornack S., van Damme M., Bozkurt T.O., Cano L.M., Smoker M., Thines M., Gaulin E., Kamoun S., Huitema E. Ancient class of translocated oomycete effectors targets the host nucleus. Proc. Natl. Acad. Sci. USA. 2010;107:17421–17426. doi: 10.1073/pnas.1008491107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boller T., He S.Y. Innate immunity in plants: An arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science. 2009;324:742–744. doi: 10.1126/science.1171647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chisholm S.T., Coaker G., Day B., Staskawicz B.J. Host-microbe interactions: Shaping the evolution of the plant immune response. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 11.Haas B.J., Kamoun S., Zody M.C., Jiang R.H., Handsaker R.E., Cano L.M., Grabherr M., Kodira C.D., Raffaele S., Torto-Alalibo T., et al. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature. 2009;461:393–398. doi: 10.1038/nature08358. [DOI] [PubMed] [Google Scholar]
- 12.Raffaele S., Farrer R.A., Cano L.M., Studholme D.J., MacLean D., Thines M., Jiang R.H., Zody M.C., Kunjeti S.G., Donofrio N.M., et al. Genome evolution following host jumps in the Irish potato famine pathogen lineage. Science. 2010;330:1540–1543. doi: 10.1126/science.1193070. [DOI] [PubMed] [Google Scholar]
- 13.Oh S.K., Young C., Lee M., Oliva R., Bozkurt T.O., Cano L.M., Win J., Bos J.I., Liu H.Y., van Damme M., et al. In planta expression screens of Phytophthora infestans RXLR effectors reveal diverse phenotypes, including activation of the Solanum bulbocastanum disease resistance protein Rpi-blb2. Plant Cell. 2009;21:2928–2947. doi: 10.1105/tpc.109.068247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H., Ren Y., Zhou J., Du J., Hou J., Jiang R., Wang H., Tian Z., Xie C. The cell death triggered by the nuclear localized RXLR effector PITG_22798 from Phytophthora infestans is suppressed by the effector Avr3b. Int. J. Mol. Sci. 2017;18:409. doi: 10.3390/ijms18020409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Q., Han C., Ferreira A.O., Yu X., Ye W., Tripathy S., Kale S.D., Gu B., Sheng Y., Sui Y., et al. Transcriptional programming and functional interactions within the Phytophthora sojae RXLR effector repertoire. Plant Cell. 2011;23:2064–2086. doi: 10.1105/tpc.111.086082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng X., McLellan H., Fraiture M., Liu X., Boevink C.P., Gilroy M.E., Chen Y., Kandel K., Sessa G., Birch R.J.P., et al. Functionally redundant RXLR effectors from Phytophthora infestans act at different steps to suppress early flg22-triggered immunity. PLoS Pathog. 2014;10:506–513. doi: 10.1371/journal.ppat.1004057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindsay J.W.L., Lindsay D. Resistance of Nicotiana to Phytophthora infestans is mediated by the recognition of elicitor protein INF1. Plant Cell. 1998;10:1413–1425. doi: 10.2307/3870607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bos J.I., Kanneganti T.D., Young C., Cakir C., Huitema E., Win J., Armstrong M.R., Birch P.R.J., Kamoun S. The C-terminal half of Phytophthora infestans RXLR effector AVR3a is sufficient to trigger R3a-mediated hypersensitivity and suppress INF1-induced cell death in Nicotiana benthamiana. Plant J. 2006;48:165–176. doi: 10.1111/j.1365-313X.2006.02866.x. [DOI] [PubMed] [Google Scholar]
- 19.Abbas M., Hughes R.K., Dagdas Y.F., Nicholas T., Erin Z., Khaoula B. Structural basis of host autophagy-related protein 8 (ATG8) binding by the Irish potato famine pathogen effector protein PexRD54. J. Biol. Chem. 2016;291:20270–20282. doi: 10.1074/jbc.M116.744995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murphy F., He Q., Armstrong M., Giuliani L.M., Boevink P.C., Zhang W., Tian Z., Birch P.R.J., Gilroy E.M. Potato MAP3K StVIK is required for Phytophthora infestans RXLR Effector Pi17316 to promote disease. Plant Physiol. 2018 doi: 10.1104/pp.18.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shan W., Cao M., Leung D., Tyler B.M. The Avr1b locus of Phytophthora sojae encodes an elicitor and a regulator required for avirulence on soybean plants carrying resistance gene Rps1b. Mol. Plant Microbe Interact. 2004;17:394–403. doi: 10.1094/MPMI.2004.17.4.394. [DOI] [PubMed] [Google Scholar]
- 22.Vleeshouwers V.G., Rietman H., Krenek P., Champouret N., Young C., Oh S.K., Wang M., Bouwmeester K., Vosman B., Visser R.G., et al. Effector genomics accelerates discovery and functional profiling of potato disease resistance and Phytophthora infestans avirulence genes. PLoS ONE. 2008;3:e2875. doi: 10.1371/journal.pone.0002875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lokossou A.A., Park T.H., van Arkel G., Arens M., Ruyter-Spira C., Morales J., Whisson S.C., Birch P.R., Visser R.G., Jacobsen E., et al. Exploiting knowledge of R/Avr genes to rapidly clone a new LZ-NBS-LRR family of late blight resistance genes from potato linkage group IV. Mol. Plant Microbe Interact. 2009;22:630–641. doi: 10.1094/MPMI-22-6-0630. [DOI] [PubMed] [Google Scholar]
- 24.Van W.P., de Jong A.J., Judelson H.S., Emons A.M., Govers F. The ipiO gene of Phytophthora infestans is highly expressed in invading hyphae during infection. Fungal Genet. 1998;23:126–138. doi: 10.1006/fgbi.1998.1036. [DOI] [PubMed] [Google Scholar]
- 25.Champouret N., Bouwmeester K., Rietman H., van der Lee T., Maliepaard C., Heupink A., van de Vondervoort P.J., Jacobsen E., Visser R.G., van der Vossen E.A., et al. Phytophthora infestans isolates lacking class I ipiO variants are virulent on Rpi-blb1 potato. Mol. Plant Microbe Interact. 2009;22:1535–1545. doi: 10.1094/MPMI-22-12-1535. [DOI] [PubMed] [Google Scholar]
- 26.Jiang R.H., Weide R., Peter J.I., van de Vondervoort P.J., Francine G. Amplification generates modular diversity at an avirulence locus in the pathogen Phytophthora. Genome Res. 2006;16:827–840. doi: 10.1101/gr.5193806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ballvora A., Ercolano M.R., Weiss J., Meksem K., Bormann C.A., Oberhagemann P., Salamini F., Gebhardt C. The R1 gene for potato resistance to late blight (Phytophthora infestans) belongs to the leucine zipper/NBS/LRR class of plant resistance genes. Plant J. 2002;30:361–371. doi: 10.1046/j.1365-313X.2001.01292.x. [DOI] [PubMed] [Google Scholar]
- 28.King S.R., McLellan H., Boevink P.C., Armstrong M.R., Bukharova T., Sukarta O., Win J., Kamoun S., Birch P.R., Banfield M.J. Phytophthora infestans RXLR Effector PexRD2 Interacts with Host MAPKKK ε to suppress plant immune signaling. Plant Cell. 2014;26:1345–1359. doi: 10.1105/tpc.113.120055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang L., Mclellan H., Naqvi S., He Q., Boevink P.C., Armstrong M., Giuliani L.M., Zhang W., Tian Z., Zhan J., et al. Potato NPH3/RPT2-like protein StNRL1, targeted by a Phytophthora infestans RXLR effector, is a susceptibility factor. Plant Physiol. 2016;171:645–657. doi: 10.1104/pp.16.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu D., He P., Shan L. Bacterial effectors target BAK1-associated receptor complexes: One stone two birds. Commun. Integr. Biol. 2010;3:80–83. doi: 10.4161/cib.3.2.10301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoshi A., Oshima K., Kakizawa S., Ishii Y., Ozeki J., Hashimoto M., Komatsu K., Kagiwada S., Yamaji Y., Namna S. A unique virulence factor for proliferation and dwarfism in plants identified from a phytopathogenic bacterium. Proc. Natl. Acad. Sci. USA. 2009;106:6416–6421. doi: 10.1073/pnas.0813038106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sugio A., Kingdom H.N., MacLean A.M., Grieve V.M., Hogenhout S.A. Phytoplasma protein effector SAP11 enhances insect vector reproduction by manipulating plant development and defense hormone biosynthesis. Proc. Natl. Acad. Sci. USA. 2011;108:1254–1263. doi: 10.1073/pnas.1105664108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacLean A.M., Orlovskis Z., Kowitwanich K., Zdziarska A.M., Angenent G.C., Immink R.G., Hogenhout S.A. Phytoplasma effector SAP54 hijacks plant reproduction by degrading MADS-box proteins and promotes insect colonization in a RAD23-dependent manner. PLoS Biol. 2014;12:e1001835. doi: 10.1371/journal.pbio.1001835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng X., Li X., Wang B., Cheng D., Li Y., Li W., Huang M., Tan X., Zhao G., Song B., et al. A systematic screen of conserved Ralstonia solanacearum effectors reveals the role of RipAB, a nuclear-localized effector that suppresses immune responses in potato. Mol. Plant Pathol. 2019;20:547–561. doi: 10.1111/mpp.12774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Evangelisti E., Govetto B., Minet-Kebdani N., Kuhn M.L., Attard A., Ponchet M., Panabières F., Gourgues M. The Phytophthora parasitica RXLR effector penetration-specific effector 1 favours Arabidopsis thaliana infection by interfering with auxin physiology. New Phytol. 2013;199:476–489. doi: 10.1111/nph.12270. [DOI] [PubMed] [Google Scholar]
- 36.Turnbull D., Yang L., Naqvi S., Breen S., Welsh L., Stephens J., Morris J., Boevink P.C., Hedley P.E., Zhan J., et al. RXLR effector AVR2 up-regulates a brassinosteroid responsive bHLH transcription factor to suppress immunity. Plant Physiol. 2017;174:356–369. doi: 10.1104/pp.16.01804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang P., Jia Y., Shi J., Chen C., Ye W., Wang Y., Ma W., Qiao Y. The WY domain in the Phytophthora effector PSR1 is required for infection and RNA silencing suppression activity. New Phytol. 2019 doi: 10.1111/nph.15836. [DOI] [PubMed] [Google Scholar]
- 38.Judelson H.S., Tooley P.W. Enhanced polymerase chain reaction methods for detecting and quantifying Phytophthora infestans in plants. Phytopathology. 2000;90:1112–1119. doi: 10.1094/PHYTO.2000.90.10.1112. [DOI] [PubMed] [Google Scholar]
- 39.Armstrong M.R., Whisson S.C., Pritchard L., Bos J.I., Venter E., Avrova A.O., Rehmany A.P., Böhme U., Brooks K., Cherevach I., et al. An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proc. Natl. Acad. Sci. USA. 2005;102:7766–7771. doi: 10.1073/pnas.0500113102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yin W., Dong S., Zhai L., Lin Y., Zheng X., Wang Y. The Phytophthora sojae Avr1d gene encodes an RxLR-dEER effector with presence and absence polymorphisms among pathogen strains. Mol. Plant Microbe Interact. 2013;26:958–968. doi: 10.1094/MPMI-02-13-0035-R. [DOI] [PubMed] [Google Scholar]
- 41.Xu X., Pan S., Cheng S., Zhang B., Mu D., Ni P., Zhang G., Yang S., Li R., Wang J., et al. Genome sequence and analysis of the tuber crop potato. Nature. 2011;475:189–195. doi: 10.1038/nature10158. [DOI] [PubMed] [Google Scholar]
- 42.William F. Phytophthora infestans: The plant (and R gene) destroyer. Mol. Plant Pathol. 2010;9:385–402. doi: 10.1111/j.1364-3703.2007.00465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang S., Mclellan H., Bukharova T., He Q., Murphy F., Shi J., Sun S., Weymers P., Ren Y., Thilliez G., et al. Phytophthora infestans RXLR effectors act in concert at diverse subcellular localisations to enhance host colonisation. J. Exp. Bot. 2019;70:343–356. doi: 10.1093/jxb/ery360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quintanilla P., Brishammar S. Systemic induced resistance to late blight in potato by treatment with salicylic acid andPhytophthora cryptogea. Potato Res. 1998;41:135–142. doi: 10.1007/BF02358436. [DOI] [Google Scholar]
- 45.Morozova O., Marra M.A. Applications of next-generation sequencing technologies in functional genomics. Genomics. 2008;92:255–264. doi: 10.1016/j.ygeno.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 46.Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 47.Marioni J.C., Mason C.E., Mane S.M., Stephens M., Gilad Y. RNA-seq: An assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao L.L., Tu Z.J., Millett B.P., Bradeen J.M. Insights into organ-specific pathogen defense responses in plants: RNA-seq analysis of potato tuber-Phytophthora infestans interactions. BMC Genom. 2013;14:340. doi: 10.1186/1471-2164-14-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aulakh S.S., Veilleux R.E., Dickerman A.W., Tang G.Z., Flinn B.S. Characterization and RNA-seq analysis of underperformer, an activation-tagged potato mutant. Plant. Mol. Biol. 2014;84:635–658. doi: 10.1007/s11103-013-0159-4. [DOI] [PubMed] [Google Scholar]
- 50.Zhang N., Liu B.L., Ma C.Y., Zhang G.D., Chang J., Si H.J., Wang D. Transcriptome characterization and sequencing-based identification of drought-responsive genes in potato. Mol. Biol. Rep. 2014;41:505–517. doi: 10.1007/s11033-013-2886-7. [DOI] [PubMed] [Google Scholar]
- 51.Qin H., Shaista N., Hazel M.L., Petra C.B., Nicolas C., Ingo H., Paul R.J.B. Plant pathogen effector utilizes host susceptibility factor NRL1 to degrade the immune regulator SWAP70. Proc. Natl. Acad. Sci. USA. 2018;115:7834–7843. doi: 10.1073/pnas.1808585115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bozkurt TO., Schornack S., Win J., Shindo T., Ilyas M., Oliva R. Phytophthora infestans effector Avrblb2 prevents secretion of a plant immune protease at the haustorial interface. Proc. Natl. Acad. Sci. USA. 2011;108:20832–20837. doi: 10.1073/pnas.1112708109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ye S. Master’s Thesis. Southwest University; Chongqing, China: 2014. Functional Characterization of Abscisic Acid 8’-Hydroxylases Gene from Upland Cotton (Gossypium hirsutum) [Google Scholar]
- 54.Yu G., Pichersky E. Heterologous expression of methylketone synthase1 and methylketone synthase2 leads to production of methylketones and myristic acid in transgenic plants. Plant. Physiol. 2014;164:612–622. doi: 10.1104/pp.113.228502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tian M., Win J., Song J., van der Hoorn R., van der Knaap E., Kamoun S. A Phytophthora infestans Cystatin-like protein targets a novel tomato papain-like apoplastic protease. Plant. Physiol. 2007;143:364–377. doi: 10.1104/pp.106.090050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Teutsch H.G., Hasenfratz M.P., Lesot A., Stoltz C., Garnier J.M., Jeltsch J.M., Durst F., Werck-Reichhart D. Isolation and sequence of a cDNA encoding the jerusalem artichoke cinnamate 4-hydroxylase, a major plant cytochrome P450 involved in the general phenylpropanoid pathway. Proc. Natl. Acad. Sci. USA. 1993;90:4102–4106. doi: 10.1073/pnas.90.9.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carrera E., Bou J., García-Martínez J.L., Prat S. Changes in GA 20-oxidase gene expression strongly affect stem length, tuber induction and tuber yield of potato plants. Plant. J. 2000;22:247–256. doi: 10.1046/j.1365-313x.2000.00736.x. [DOI] [PubMed] [Google Scholar]
- 58.Fagard M., Desnos T., Desprez T., Goubet F., Refregier G.M., McCann M., Rayon C., Vernhettes S., Höftea H. PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of arabidopsis. Plant. Cell. 2000;12:2409–2423. doi: 10.1105/tpc.12.12.2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu R., Malcuit I., Moffett P., Ruiz M.T., Peart J., Wu A.J., Rathjen J.P., Bendahmane A., Day L., Baulcombe D.C. High throughput virus-induced gene silencing implicates heat shock protein 90 in plant disease resistance. Eur. Mol. Biol. Organ. J. 2003;22:5690–5699. doi: 10.1093/emboj/cdg546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Y., Tang W., Chen J., Jia R., Ma L., Wang S., Wang J., Shen X., Chu Z., Zhu C., et al. Development of marker-free transgenic potato tubers enriched in caffeoyl quinic acids and flavonols. J. Agric. Food Chem. 2016;64:2932–2940. doi: 10.1021/acs.jafc.6b00270. [DOI] [PubMed] [Google Scholar]
- 61.Kobayashi M., Yoshioka M., Asai S., Nomura H., Kuchimura K., Mori H., Doke N., Yoshioka H. StCDPK5 confers resistance to late blight pathogen but increases susceptibility to early blight pathogen in potato via reactive oxygen species burst. New Phytol. 2012;196:223–237. doi: 10.1111/j.1469-8137.2012.04226.x. [DOI] [PubMed] [Google Scholar]
- 62.Li X., Han H., Chen M., Yang W., Liu L., Li N., Ding X., Chu Z. Overexpression of OsDT11, which encodes a novel cysteine-rich peptide, enhances drought tolerance and increases ABA concentration in rice. Plant. Mol. Biol. 2017;93:21–34. doi: 10.1007/s11103-016-0544-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.