Abstract
MADS-box family genes encode transcription factors that are involved in multiple developmental processes in plants, especially in floral organ specification, fruit development, and ripening. However, a comprehensive analysis of tomato MADS-box family genes, which is an important model plant to study flower fruit development and ripening, remains obscure. To gain insight into the MADS-box genes in tomato, 131 tomato MADS-box genes were identified. These genes could be divided into five groups (Mα, Mβ, Mγ, Mδ, and MIKC) and were found to be located on all 12 chromosomes. We further analyzed the phylogenetic relationships among Arabidopsis and tomato, as well as the protein motif structure and exon–intron organization, to better understand the tomato MADS-box gene family. Additionally, owing to the role of MADS-box genes in floral organ identification and fruit development, the constitutive expression patterns of MADS-box genes at different stages in tomato development were identified. We analyzed 15 tomato MADS-box genes involved in floral organ identification and five tomato MADS-box genes related to fruit development by qRT-PCR. Collectively, our study provides a comprehensive and systematic analysis of the tomato MADS-box genes and would be valuable for the further functional characterization of some important members of the MADS-box gene family.
Keywords: tomato, MADS-box, genome-wide analysis, floral organ, fruit development
1. Introduction
The MADS-box family genes encode transcription factors (TFs), which are widely distributed in eukaryotes and play fundamental roles in diverse biological functions [1]. The name MADS-box is derived from the initials of four transcription factors that were first discovered of this family: MINICHROMOSOME MAINTENANCE 1 (MCM1), AGAMOUS (AG), DEFICIENS (DEF), and SERUM RESPONSE FACTOR (SRF) [2]. Their N-terminal contains a highly conserved DNA-binding MADS-domain containing 56–60 amino acids [2]. Thus, a protein encoding the MADS-domain is referred to as a MADS-box protein. MADS-box genes are divided into two groups (type I and type II) throughout the eukaryotes [3]. Type I MADS-box transcription factors can be further classified into four subclasses (Mα, Mβ, Mγ, and Mδ) in view of the M domain of the encoded protein, while only a few type I genes have been characterized for their biological function [4]. Type II MADS-box transcription factors include the Myocyte Enhancer Factor 2-like (MEF2-like) group from animals and yeast and the plant-specific MIKC-type group. The name of the MIKC-type protein is derived from their four characteristic domains: MADS-box (M), intervening (I), keratin-like (K), and C-terminal (C) [5]. The MIKC type has been further subdivided into MIKCC and MIKC* types based on phylogenetic relationships and structural features [4]. The functions of the type II genes, especially the MIKCC type MADS-box genes in plants, have been reported in more detail. To date, 39 MIKCC type MADS-box genes were found in Arabidopsis thaliana [6], and 38 MIKCC type MADS-box genes were found in rice [7].
MADS-box genes are known to play important roles in different aspects of plants growth and development. The MADS-box genes AGAMOUS (AG) of Arabidopsis thaliana [8] as well as DEFICIENS (DEF) of Antirrhinum majus [2] were found to regulate floral organ formation two decades ago. The MADS-box genes were thought to be the main participants in floral organ specificity. With the analysis of the specification of floral organ identity determination, a genetic model (ABC model) was proposed in which the different steps of floral development were determined by three classes of genes (A, B, and C) [9]. Class A genes are necessary for the formation of the sepal. The class A genes, together with class B genes, determine the development of petals. The combination of class B and C genes is necessary for stamen identity, and class C genes function alone to form carpels [10]. However, there are many phenomena that this model cannot explain. For example, the constitutive co-expression of the B class genes PISTILLATA (PI) and APETALA3 (AP3) in Arabidopsis thaliana [11], does not change the identity of vegetative organs. Recent studies found that the ABC model is necessary but not sufficient to provide the floral organ identity function. Therefore, the model was advanced by subdivision into five different classes (A–E). Class D genes specify ovule development, while class E genes are necessary for the specification of petals, stamens, and carpels. In addition, these five classes of genes are mainly MADS-box genes [12]. In Arabidopsis thaliana, APETALA1 (AP1) is a typical class A gene [13], APETALA3 (AP3) and PISTILLATA (PI) belong to the class B genes [14], and AGAMOUS (AG) is a representative gene with class C function [15]. The SEPALLATA (SEP) genes are class E genes and include SEP1, SEP2, SEP3, and SEP4 in Arabidopsis thaliana [16]. The class D gene was identified by the mutant phenotype of petunia, and the sequence similarity analysis demonstrated that the corresponding gene in Arabidopsis thaliana is AGAMOUS-LIKE11 (AGL11) [17,18].
In addition, considerable evidence has revealed that MADS-box TFs share a potent effect on the regulation of fruit development and ripening [4]. There have been many studies on the function of Solanum lycopersicum MADS-box genes in fruit development and ripening. The tomato MADS-box gene RIPENING INHIBITOR (RIN) is an essential factor for fruit ripening that regulates many ripening-associated processes, including both ethylene-dependent and ethylene-independent ripening processes [19]. Moreover, the tomato MADS-box genes AGAMOUSLIKE1 (AGL1) [20,21], FRUITFULL1 (FUL1), and FRUITFULL2 (FUL2) are Arabidopsis SHATTERPROOF (SHP) and FRUITFUL (FULL) homologues respectively [22,23], and their suppression results in a phenotype that is partially similar to the ripening-defective phenotype of RIN mutant fruits. In addition to crucial roles in the regulation of plant reproductive development, MADS-box genes have also been shown to take part in plant vegetative growth processes and some stress responses in different plants such as Arabidopsis [24], rice [7,25], wheat [26], and Chinese cabbage [27]. Thus, the MADS-box protein family is an important TFs family for plant growth and development that almost affects the whole process of plant growth and development, especially plant reproductive development.
The MADS-box family of model plant species has been widely studied, including snapdragon (Antirrhinum majus) [28], rice (Oryza sativa) [7], Chinese cabbage (Brassica rapa) [27], poplar (Populus trichocarpa) [29], bread wheat (Triticum aestivumL) [30], banana (Musa acuminata) [31], petunia (Petunia hybrida) [32], apple (Malus domestica Borkh) [33], and so on. Tomato (Solanum lycopersicum) is one of the most critical model plants for studying flower and fruit development, and the MADS-box genes of tomato were among the earliest to be investigated [34]. It has been reported that 24 tomato MIKCC-type MADS-box genes had been identified in 2006 [35]. However, there has been no report to date concerning systematic information for the tomato MADS-box gene family, and confusion in the names of these genes is problematic for future research.
To obtain a genome-wide analysis of the tomato MAD-box gene family, we sorted 131 MADS-box genes from tomato that are highly homologous to MADS-box proteins known in other plant species, analyzed their phylogenetic relationships, gene structure, and conserved motifs, determined their exon–intron organization, and predicted their chromosomal localization. Furthermore, we obtained the predictions of the expression pattern of these tomato MADS-box genes in order to predict their expression pattern. In addition, the expression pattern of some MADS-box genes related to tomato flower organ identity, fruit development, and ripening were determined by qPCR analyses in different stages of tomato development. These results provide details of the tomato MADS-box family and may be useful for more comprehensive investigations of tomato MADS-box gene family members.
2. Results
2.1. Identification of MADS-Box Genes in Tomato
To extensively identify tomato MADS-box genes, a set of 131 tomato MADS-box genes that are highly homologous to the MADS-box proteins reported in other plants was recovered using BLAST searches against the NCBI and SGN databases. Redundant sequences were removed. Since 35 tomato MADS-box genes have been officially named, the newly identified 96 tomato MADS-box genes were designated as SlMADS2–SlMADS98 (Table 1). Additionally, the molecular characteristics of the 131 MADS-box genes in tomato were analyzed. The names of the tomato MADS-box genes, the locus/gene names of SGN, the length of amino acid sequence, the molecular weight, and the isoelectric point are shown in Table 1. The statistical results showed that the amino acid sequence length of the 131 predicted tomato MADS-box proteins varied from 54 (SlMBP19) to 417 (SlMADS52), the relative molecular mass ranged from 6224.26 Da (SlMBP19) to 47275.1 Da (SlMADS52), and the isoelectric point (pI) varied from 4.41 (SlMADS70) to 11.03 (SlMADS77 and SlMADS80). The homologous genes of the tomato MADS-box family genes in Arabidopsis and the references for the study of the functions in homologous genes are shown in Table S1. In addition, the functional reports of several genes that have been studied previously were listed.
Table 1.
Overview of MADS-box genes identified in tomato. List of predicted genes and related information include gene name, gene locus, molecular details, classification of two types according to the phylogenetic analysis shown in Figure S1, homologs in Arabidopsis, as well as the reference of the gene function investigation. pI, isoelectric point; MW, molecular weight.
| Gene Name | Gene Locus | Protein | Type | Reference | ||
|---|---|---|---|---|---|---|
| Length (aa) | MW (Da) | pI | ||||
| SlMBP1/SlGLO1/PI/LePI-B | Solyc08g067230.2.1 | 210 | 24740.2 | 8.69 | Type II | [36] |
| SlMBP2/SlGLO2/LePI/TPI | Solyc06g059970.2.1 | 214 | 24867.4 | 9.49 | Type II | [37] |
| SlMBP3/ SlAGL11 | Solyc06g064840.3.1 | 237 | 27469.6 | 9.25 | Type II | [38] |
| SlMBP6/SlAGL6 | Solyc01g093960.2.1 | 252 | 28608.5 | 8.37 | Type II | [39,40,41] |
| SlMBP7/LeFUL2 | Solyc03g114830.2.1 | 247 | 28614.3 | 9.31 | Type II | [22,40,42,43,44] |
| SlMBP8 | Solyc12g087830.1.1 | 198 | 22643.3 | 8.97 | Type II | [45,46] |
| SlMBP10 | Solyc02g065730.1.1 | 234 | 27368.1 | 10.12 | Type II | |
| SlMBP11/AGL15-like | Solyc01g087990.2.1 | 271 | 30847 | 6.01 | Type II | [47] |
| SlMBP13 | Solyc08g080100.2.1 | 224 | 25839 | 9.76 | Type II | |
| SlMBP14 | Solyc12g056460.1.1 | 206 | 23968.4 | 6.57 | Type II | |
| SlMBP15 | Solyc12g087830.2.1 | 204 | 23667.2 | 8.35 | Type II | |
| SlMBP18/SlFYFL | Solyc03g006830.2.1 | 222 | 25369.2 | 9.46 | Type II | [48] |
| SlMBP19 | Solyc06g035570.1.1 | 54 | 6224.26 | 11.29 | Type I | |
| SlMBP20 | Solyc02g089210.2.1 | 250 | 28589.2 | 9.87 | Type II | [44] |
| SlMBP21 | Solyc12g038510.1.1 | 250 | 28442.4 | 9.26 | Type II | [49,50,51] |
| SlMBP22 | Solyc11g005120.1.1 | 238 | 27791.6 | 6.96 | Type II | |
| SlMBP23/TDR3 | Solyc10g017630.2.1 | 166 | 19196.3 | 9.49 | Type II | |
| SlMBP24 | Solyc04g076280.3.1 | 235 | 26483.6 | 7.67 | Type I | |
| SlMBP25 | Solyc05g015730.1.1 | 80 | 9311.97 | 11.01 | Type II | |
| TAG1 | Solyc02g071730.2.1 | 248 | 28723.6 | 9.97 | Type II | [52,53] |
| TAGL1 | Solyc07g055920.2.1 | 267 | 29940.5 | 9.56 | Type II | [20,21,52,54] |
| TAGL2 | Solyc05g015750.2.1 | 241 | 27579.3 | 9.07 | Type II | [55,56] |
| TAGL11 | Solyc11g028020.1.1 | 223 | 26051.7 | 9.76 | Type II | [38,56] |
| TAGL12 | Solyc11g032100.1.1 | 201 | 23104.8 | 6.94 | Type II | [56] |
| TAP3/LeAP3/LeDEF | Solyc04g081000.2.1 | 228 | 26478.2 | 9.76 | Type II | [54,57,58,59] |
| MADS-RIN | Solyc05g012020.2.1 | 242 | 27968.4 | 8.00 | Type II | [19,60,61,62,63] |
| MADS-MC | Solyc05g056620.1.1 | 244 | 28660.6 | 8.44 | Type II | [64] |
| JOINTLESS | Solyc11g010570.1.1 | 265 | 30426.3 | 7.39 | Type II | [64,65,66,67,68] |
| LeAP1 | Solyc05g012020.3.1 | 213 | 25077.3 | 5.47 | Type II | [40] |
| TM4/TDR4/LeFUL1 | Solyc06g069430.2.1 | 245 | 28290 | 9.57 | Type II | [22,42,43,59] |
| TM5/TDR5/LeSEP3 | Solyc05g015750.3.1 | 224 | 25999.3 | 9.79 | Type II | [40] |
| TM6/TDR6 | Solyc02g084630.2.1 | 225 | 26092.7 | 9.71 | Type II | [69] |
| TM8/TDR8 | Solyc03g019710.2.1 | 177 | 20703.9 | 10.44 | Type II | [70] |
| SlMADS1 | Solyc03g114840.2.1 | 246 | 28398.2 | 8.88 | Type II | [55,71] |
| SlMADS2 | Solyc01g060300.1.1 | 211 | 23122.6 | 9.36 | Type I | |
| SlMADS3 | Solyc01g060310.1.1 | 139 | 15117.5 | 8.99 | Type I | |
| SlMADS4 | Solyc01g060284.1.1 | 197 | 21565.5 | 8.63 | Type I | |
| SlMADS5 | Solyc01g066500.1.1 | 309 | 34544.4 | 4.75 | Type I | |
| SlMADS6/TM29/LeSEP1 | Solyc02g089200.2.1 | 246 | 28481.3 | 8.29 | Type II | [39,41] |
| SlMADS7 | Solyc01g103870.1.1 | 125 | 14435.6 | 9.54 | Type I | |
| SlMADS8 | Solyc06g071300.1.1 | 130 | 15009.1 | 5.14 | Type I | |
| SlMADS9 | Solyc03g007020.1.1 | 215 | 24702 | 9.21 | Type I | |
| SlMADS10 | Solyc04g064860.1.1 | 233 | 25871.3 | 7.92 | Type I | |
| SlMADS11 | Solyc04g054517.1.1 | 138 | 15663.1 | 7.75 | Type I | |
| SlMADS12 | Solyc04g056550.1.1 | 182 | 20838.7 | 5.14 | Type I | |
| SlMADS13 | Solyc04g056740.1.1 | 180 | 20521.5 | 7.44 | Type I | |
| SlMADS14 | Solyc06g054680.1.1 | 180 | 21152.8 | 7.19 | Type I | |
| SlMADS15 | Solyc06g059780.1.1 | 180 | 21057.6 | 6.37 | Type I | |
| SlMADS16 | Solyc10g050950.1.1 | 161 | 18602.2 | 6.80 | Type I | |
| SlMADS17 | Solyc10g050900.1.1 | 169 | 19796.5 | 7.83 | Type I | |
| SlMADS18 | Solyc10g050940.1.1 | 169 | 19623.3 | 7.77 | Type I | |
| SlMADS19 | Solyc01g097850.1.1 | 172 | 20073.9 | 7.35 | Type I | |
| SlMADS20 | Solyc03g034260.1.1 | 184 | 21441 | 6.29 | Type I | |
| SlMADS21 | Solyc10g018070.1.1 | 119 | 13063.9 | 10.14 | Type I | |
| SlMADS22 | Solyc10g018080.1.1 | 151 | 17218.5 | 7.04 | Type I | |
| SlMADS23 | Solyc10g018110.1.1 | 184 | 20762.4 | 6.55 | Type I | |
| SlMADS24 | Solyc04g025970.1.1 | 117 | 13333.4 | 10.16 | Type I | |
| SlMADS25 | Solyc01g010300.1.1 | 88 | 10052.7 | 10.79 | Type I | |
| SlMADS26 | Solyc02g032000.1.1 | 97 | 10944.7 | 10.32 | Type I | |
| SlMADS27 | Solyc03g119680.1.1 | 177 | 20293.2 | 7.26 | Type I | |
| SlMADS28 | Solyc09g061950.1.1 | 173 | 20138.1 | 9.33 | Type I | |
| SlMADS29 | Solyc01g106710.1.1 | 221 | 25105.7 | 10.23 | Type I | |
| SlMADS30 | Solyc04g025030.1.1 | 135 | 15411 | 10.37 | Type I | |
| SlMADS31 | Solyc04g025110.1.1 | 130 | 14313.7 | 10.26 | Type I | |
| SlMADS32 | Solyc01g106720.1.1 | 211 | 23531.2 | 10.35 | Type I | |
| SlMADS33 | Solyc01g106730.1.1 | 221 | 23888.6 | 10.14 | Type I | |
| SlMADS34 | Solyc04g047870.1.1 | 190 | 21695.2 | 10.70 | Type I | |
| SlMADS35 | Solyc10g012180.1.1 | 167 | 18824.5 | 5.38 | Type I | |
| SlMADS36 | Solyc10g012200.1.1 | 167 | 19113.1 | 8.98 | Type I | |
| SlMADS37 | Solyc11g067163.1.1 | 166 | 18919.9 | 7.67 | Type I | |
| SlMADS38 | Solyc01g066730.2.1 | 300 | 33023.4 | 5.04 | Type I | |
| SlMADS39 | Solyc01g098070.1.1 | 155 | 17200.8 | 10.09 | Type I | |
| SlMADS40 | Solyc01g098060.1.1 | 152 | 17191.5 | 9.84 | Type I | |
| SlMADS41 | Solyc03g062820.1.1 | 145 | 16350.8 | 9.23 | Type I | |
| SlMADS42 | Solyc01g098050.1.1 | 160 | 18178 | 10.45 | Type I | |
| SlMADS43 | Solyc05g013370.1.1 | 232 | 26476.7 | 5.46 | Type I | |
| SlMADS44 | Solyc05g046345.1.1 | 250 | 28489.8 | 5.47 | Type I | |
| SlMADS45 | Solyc04g025050.1.1 | 56 | 6259.52 | 11.07 | Type I | |
| SlMADS46 | Solyc07g017343.1.1 | 226 | 26532.7 | 5.2 | Type I | |
| SlMADS47 | Solyc00g179240.1.1 | 55 | 6310.11 | 11.16 | Type I | |
| SlMADS48 | Solyc07g052707.1.1 | 195 | 21878.8 | 4.86 | Type I | |
| SlMADS49 | Solyc07g052700.2.1 | 190 | 21334.2 | 7.41 | Type I | |
| SlMADS50 | Solyc11g069770.2.1 | 175 | 20171.9 | 5.45 | Type I | |
| SlMADS51 | Solyc01g106700.2.1 | 213 | 24279.5 | 7.35 | Type I | |
| SlMADS52 | Solyc03g115910.1.1 | 417 | 47275.1 | 5.82 | Type I | |
| SlMADS53 | Solyc10g012380.1.1 | 149 | 17086.7 | 5.29 | Type I | |
| SlMADS54 | Solyc10g012390.1.1 | 149 | 17086.7 | 5.29 | Type I | |
| SlMADS55 | Solyc11g069770.1.1 | 181 | 20857.8 | 5.20 | Type I | |
| SlMADS56 | Solyc12g042967.1.1 | 225 | 25552.3 | 9.05 | Type I | |
| SlMADS57 | Solyc11g020620.1.1 | 342 | 38747.9 | 5.42 | Type I | |
| SlMADS58 | Solyc11g020660.1.1 | 312 | 35558.7 | 4.61 | Type I | |
| SlMADS59 | Solyc11g020320.1.1 | 275 | 31757.7 | 6.68 | Type I | |
| SlMADS60 | Solyc01g103550.1.1 | 311 | 35839.7 | 7.87 | Type I | |
| SlMADS61 | Solyc01g060310.2.1 | 136 | 15230.6 | 6.16 | Type I | |
| SlMADS62 | Solyc12g016170.1.1 | 219 | 25087.7 | 6.57 | Type I | |
| SlMADS63 | Solyc12g016180.1.1 | 219 | 25207.8 | 6.57 | Type I | |
| SlMADS64 | Solyc12g016150.1.1 | 154 | 17645.1 | 5.38 | Type I | |
| SlMADS65 | Solyc12g017300.1.1 | 154 | 17740.1 | 5.11 | Type I | |
| SlMADS66 | Solyc12g005210.1.1 | 88 | 9985.65 | 10.41 | Type I | |
| SlMADS67 | Solyc01g102260.2.1 | 249 | 27882.8 | 9.66 | Type I | |
| SlMADS68 | Solyc05g047712.1.1 | 156 | 18199.1 | 8.89 | Type I | |
| SlMADS69 | Solyc07g043080.1.1 | 215 | 24384.8 | 6.86 | Type I | |
| SlMADS70 | Solyc06g034317.1.1 | 153 | 16949.3 | 4.41 | Type I | |
| SlMADS71 | Solyc06g048380.1.1 | 176 | 20038.6 | 4.90 | Type I | |
| SlMADS72 | Solyc06g048380.2.1 | 132 | 15027 | 4.82 | Type I | |
| SlMADS73 | Solyc06g033820.1.1 | 196 | 22657.4 | 7.25 | Type I | |
| SlMADS74 | Solyc06g033830.1.1 | 113 | 13058.8 | 8.44 | Type I | |
| SlMADS75 | Solyc07g052700.3.1 | 197 | 22133.2 | 8.44 | Type I | |
| SlMADS76 | Solyc03g115910.2.1 | 434 | 49552.6 | 5.6 | Type I | |
| SlMADS77 | Solyc12g087820.1.1 | 106 | 11969.2 | 11.03 | Type I | |
| SlMADS78 | Solyc05g051830.2.1 | 389 | 44560 | 5.91 | Type I | |
| SlMADS79 | Solyc10g017640.1.1 | 61 | 7032.22 | 11.24 | Type I | |
| SlMADS80 | Solyc04g076680.2.1 | 63 | 70492.3 | 11.03 | Type I | |
| SlMADS81 | Solyc12g088080.1.1 | 97 | 11295.2 | 10.59 | Type I | |
| SlMADS82 | Solyc01g105800.2.1 | 279 | 32104.7 | 9.11 | Type II | |
| SlMADS83 | Solyc01g106170.2.1 | 148 | 17032.2 | 10.98 | Type II | |
| SlMADS84 | Solyc10g080030.1.1 | 229 | 25791.3 | 6.37 | Type II | |
| SlMADS85 | Solyc01g105810.3.1 | 215 | 24518.2 | 9.22 | Type II | |
| SlMADS86 | Solyc04g078300.3.1 | 145 | 16547.2 | 9.9 | Type II | |
| SlMADS87 | Solyc04g078300.2.1 | 334 | 38130.7 | 5.96 | Type II | |
| SlMADS88 | Solyc03g006830.3.1 | 209 | 24310.2 | 9.43 | Type II | |
| SlMADS89 | Solyc10g044967.1.1 | 226 | 26172 | 6.14 | Type II | |
| SlMADS90 | Solyc11g032100.2.1 | 201 | 23104.8 | 6.45 | Type II | |
| SlMADS91 | Solyc05g015720.2.1 | 204 | 23433.5 | 5.36 | Type II | |
| SlMADS92 | Solyc01g093960.3.1 | 242 | 27359.1 | 8.19 | Type II | |
| SlMADS93 | Solyc02g091550.2.1 | 249 | 28342.2 | 9.07 | Type II | |
| SlMADS94 | Solyc02g091550.1.1 | 235 | 27198.1 | 9.65 | Type II | |
| SlMADS95 | Solyc04g076700.3.1 | 226 | 25933.3 | 8.59 | Type II | |
| SlMADS96 | Solyc03g019720.3.1 | 193 | 22631.1 | 9.88 | Type II | |
| SlMADS97 | Solyc12g088090.2.1 | 239 | 27695.8 | 9.84 | Type II | |
| SlMADS98/SlCMB1 | Solyc04g005320.2.1 | 238 | 27522.1 | 8.60 | Type II | [72] |
2.2. Classification and Phylogenetic Analysis of Tomato MADS-Box Genes
To study the phylogenetic relationships among MADS-box genes in tomato and Arabidopsis [6], a phylogenetic tree was drawn by the neighbor-joining (NJ) method using MEGA 5.02 (Figure S1A). Based on previous reports on Arabidopsis, the 131 tomato MADS-box genes were classified into two types: type I (81) and type II (50). Based on the phylogenetic tree, type I and type II tomato MADS-box genes were subdivided into more detailed subgroups. Then, type I could be further divided into four groups (Mα, Mβ, Mγ, and Mδ), while Type II (MIKC) could be further divided into MIKC* and MIKCC. The MIKCC-type genes included the AP3/PI, SVP, AGL15, SEPALLATA (SEP), AGL6, AP1, FLOWERING LOCUS C (FLC) SOC1, AGAMOUS (AG), TM8, and DEFICIENS (DEF)/GLOBOSA (GLO) subfamilies, similar to the MADS-box genes in other plants species. In addition, the phylogenetic tree of type I and type II MADS-box protein in tomato plants were constructed to identify the phylogenetic relationships of gene numbers in the two types of tomato MADS-box family more clearly (Figure S1B,C).
2.3. Conserved Motif and Gene Structure Analysis of Tomato MADS-Box Genes
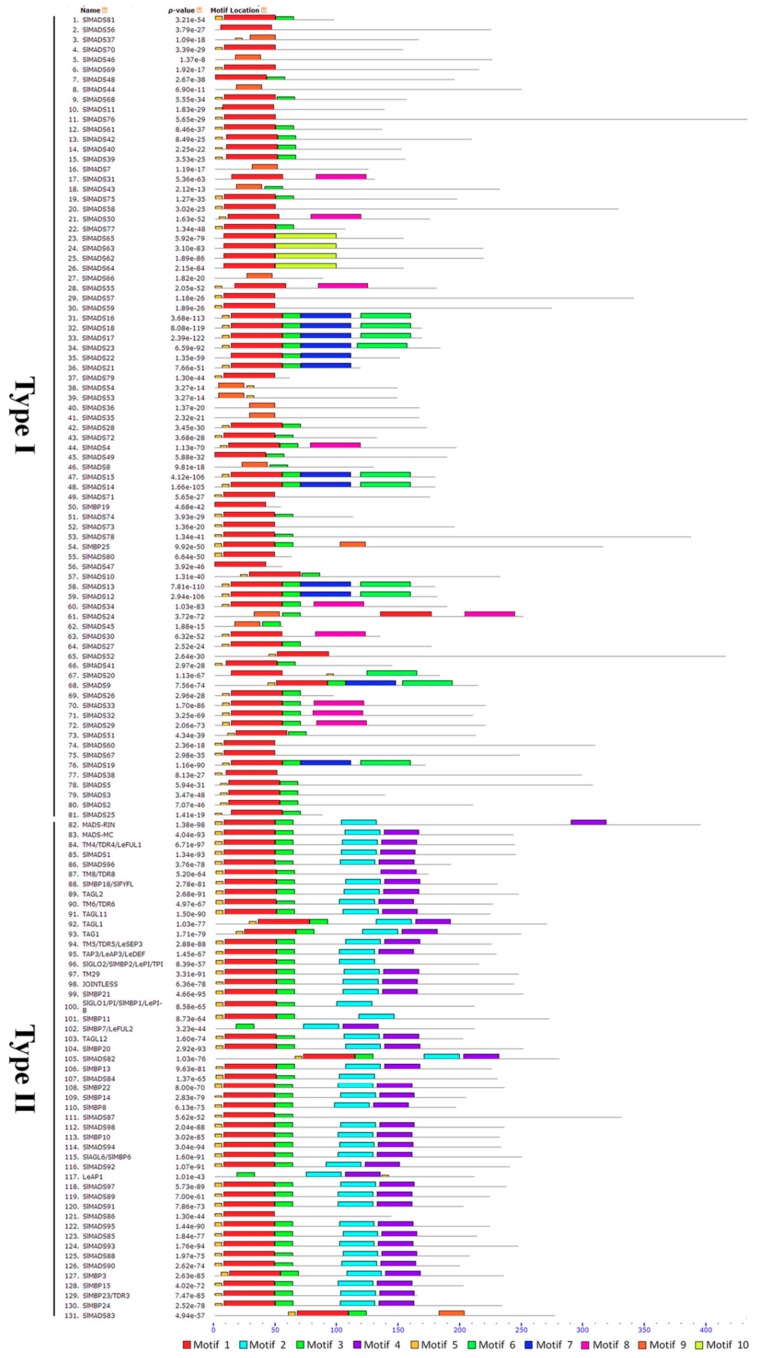
The intron–exon organization was analyzed to investigate the structural diversity and evolution of the 131 tomato MADS-box genes. As shown in Figure 1, we obtained each gene’s intron/exon arrangement by comparing their CDS with their genomic sequences using the program Gene Structure Display Server (GSDS). The number of introns in tomato MADS-box genes ranged from one to 11. Similar to Arabidopsis, the distribution of introns in tomato was different in type I and type II genes [6]. In our study, we found that the Mα, Mβ, and Mγ groups of the type I genes usually had no introns or one intron, which might be explained by the diversity of the reverse-transcribed origin or the differences in acquisition or loss introns by the ancestors of these three groups of genes [6]. Based on the genomic data, the Mδ group of type I and the type II genes contained multiple introns. Among the Mδ clade and the type II genes, 52 of 56 (92.9%) genes had more than five introns. Additionally, the gene structures of closely related genes in tomato MADS-box genes were more similar, and the differences were only in the lengths of introns and exons. However, some close gene pairs showed different intron/exon arrangements. For instance, SlMBP61 has one exon, whereas its close homologs SlMBP51 and SlMBP10 both have two, although their phylogenetic relationship displayed a high bootstrap value (Figure S1B).
Figure 1.
Gene structure analysis of MADS-box genes in tomato. The Gene Structure Display Server (GSDS) database was used to perform the exon–intron structure analyses. Lengths of exons and introns of each MADS-box gene were displayed proportionally. The blue boxes represent upstream/downstream, the yellow boxes represent exons, and the black lines represent introns.
To better analyze conserved motifs in tomato MADS-box proteins, we constructed a conserved motif figure using the Multiple EM for Motif Elicitation (MEME) program and annotated them using SMART. A total of 10 conserved motifs, named 1 to 10, were identified (Figure 2). The details of the motifs are shown in Figure S2. As expected, the same types of genes tend to possess the same motifs. Motif 1—one of the most typical MADS-box domains—comprised 42 amino acids was found in the majority of tomato MADS-box proteins. Motif 3 was also conserved across most of the tomato MADS-box proteins, including type I and type II genes. Motifs 2 and 4 represent the K domain, which plays an important role in protein–protein interactions among MADS-box proteins, and they were found only in type II MADS-box proteins. Motif 2 was identified in almost all the type II proteins except for TM8/TDR8, SlMADS86, SlMADS87, and SlMADS83. In the type II proteins, a large number of proteins had motif 4, with seven exceptions (SlGLO2, SlGLO1, SlMBP11, SlMADS84, SlMADS86, SlMADS87, and SlMADS83). Motif 9 is also a MADS-box domain that is present in a small number of tomato MADS-box proteins. However, some motifs (6, 7, 8, and 10) were shown to be weakly conserved in tomato MADS-box proteins, and they were found only in type I MADS-box proteins.
Figure 2.
Conserved motif analyses of MADS-box genes in tomato. All the conserved motifs were identified by the Multiple EM for Motif Elicitation (MEME) database using the complete amino acid sequences of tomato MADS-box genes. Ten conserved different motifs were indicated by different colors. The details of motifs refer to the Supplementary Figure S2.
2.4. Chromosomal Locations of Tomato MADS-Box Genes
According to physical genome annotation files that were obtained by using genomic sequences from the SGN and NCBI databases, 131 members of the MADS-box genes were located on all 12 tomato chromosomes, implying that the MADS-box transcription factor family may have multiple functions in tomato plants.
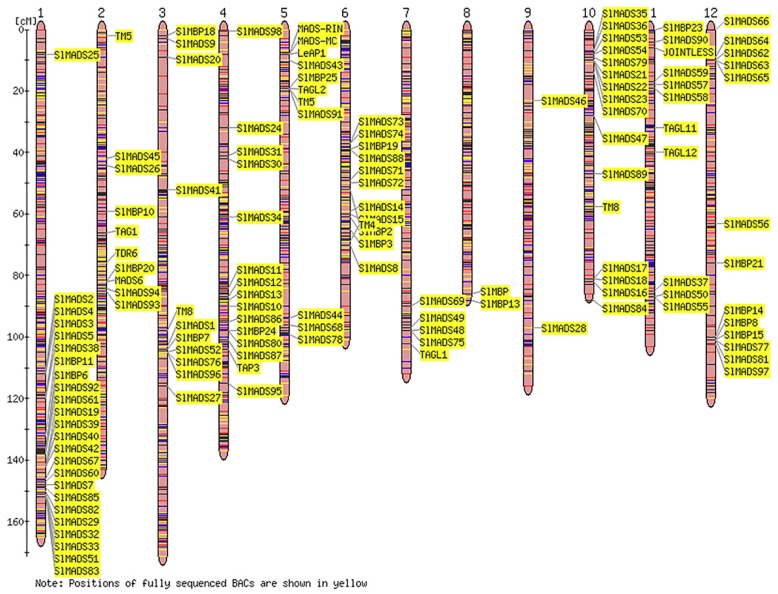
As shown in Figure 3, the tomato MADS-box genes are distributed unevenly on each chromosome. Chromosome 1 contains the most MADS-box genes (24), while chromosome 8 and 9 contain the fewest (two). Gene duplication events have a significant function in amplifying gene family numbers and genome complexity in eukaryotes [73,74]. The tandem amplification or segmental duplication of chromosomal regions can increase gene families. In this study, the results showed that chromosomes 1, 3, 4, 10, and 12 contain gene clusters or gene hotspots; in particular, chromosome 1 contains eight MADS-box genes within a short region. Additionally, we confirmed that internal chromosome duplication (tandem duplication) occurred in these genes.
Figure 3.
Chromosomal locations of tomato MADS-box genes. A total of 12 chromosomes of tomato were labeled with their names, chromosomes 1 to 12, which are indicated at the top of each bar. The position of tomato MADS-box genes on the chromosome was based on the Sol Genomics Network (SGN) and National Center for Biotechnology Information (NCBI) database and the Tomato-EXPEN 2000 was used to draw the physical map of the tomato MADS-box genes.
2.5. Predictions of Expression Profiles of Tomato MADS-Box Genes in Different Organs
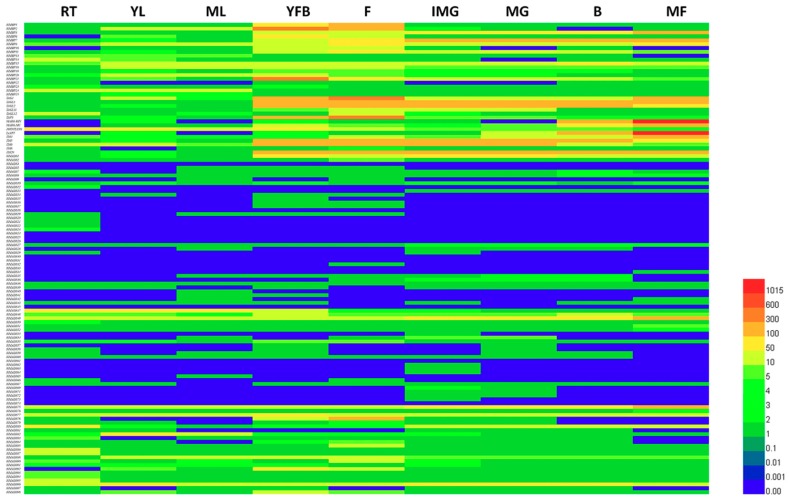
To investigate the tomato MADS-box genes expression patterns in different tissues of tomato plants, we analyzed tomato transcript expression (RNA-seq) data in nine different tomato tissues at different developmental stages. This included the expression in the whole root (RT), young leaf (YL), mature leaves (ML), young flower buds (YFB), fully open flowers (F), and at the immature green (IMG), mature green (MG), break (B), and mature (MF) stages of fruit development and ripening. These datasets were searched using the locus/gene names in SGN of 124 tomato MADS-box gene sequences, except for SlMADS4, SlMADS11, SlMADS37, SlMADS44, SlMADS46, SlMADS56, SlMADS68, SlMADS70 and SlMADS89, which were not accurately found in TFGD. Then, we constructed a hierarchical clustering heat map using these datasets (Figure 4).
Figure 4.
Heat map representation of tomato MADS-box genes in various tissues. The tissues included the whole root (Rt), young leaf (YL), mature leaves (ML), young flower buds (YFB), and fully open flowers (F), which were at the immature green (IMG), mature green (MG), break (B), and mature (MF) stages of fruit development and ripening. The bar at the bottom of the heat map represents relative gene expression values.
The expression profiles revealed that 117 genes were expressed in at least one tomato plant organ, while the other seven genes (SlMADS24, SlMADS25, SlMADS26, SlMADS33, SlMADS45, SlMADS61, and SlMADS74) were expressed at levels that were too low to be identified, or they had temporal and spatial specific expression patterns that showed no expression in the organs tested. Most tomato MADS-box genes displayed a broad expression range across all the organs and developmental stages, which is consistent with previous reports that the MADS-box genes may play multiple roles in plant growth and development [75,76]. However, some genes exhibited tissue-specific expression. For example, the expression of SlMADS12, SlMADS20, SlMADS21, SlMADS22, and SlMADS23 were restricted in whole root, and the SlMADS16, SlMADS17, and SlMADS132 transcripts were observed only during flower development. These results illustrate that these genes may be involved in the regulation of some biological process of the tomato root or in flower growth and development. Eight genes (SlMBP2, SlMBP6, SlMBP10, SlMBP21, TAP3, SlMADS78, SlMADS92, and SlMADS98) showed especially high expression in young flower buds (YFB) and fully open flowers (F), indicating that these genes may play important roles in floral organ development. We further discovered that most type II genes (SlMBP3, SlMBP7, SlMBP11, SlMBP15, MADS-RIN, MADS-MC, TAGL1, and LeAP1) were highly expressed during flower or fruit development; especially, the expression values of MADS-RIN and MADS-MC in fruits were more than 1000, suggesting that these genes may be associated with the reproductive growth of tomato. However, the expression of most type II genes showed no significant difference among tissues.
In short, these results indicate that the MADS-box genes had different expression levels in various tomato organs, and the predictions of the organ expression profiles of the tomato MADS-box gene family may provide insight for future studies on the functions of MADS-box genes in tomato plant growth and development.
2.6. The Critical Tomato MADS-Box Genes Involved in Floral Organ Development
Based on the phylogenetic analysis of the MADS-box genes that participated in floral organ development in petunia, which has been reported previously [32,77] (Figure S3), 15 tomato MADS-box genes that are possibly involved in floral organ development were screened out, including tow class A genes (MADS-MC and SlMBP20), four class B genes (TAP3, TM6, SlMBP1, and SlMBP2, two class C genes (TAG1 and TAGL1), two class D genes (SlMBP3 and SlMBP22), and five class E genes (TAGL2, TM5, SlMADS1, SlMBP21, and SlMBP6), as shown in Table S2.
According to the expression profile predictions shown in Figure 4, among the 15 MADS-box genes, nine genes (MADS-MC, TAP3, TM6, SlMBP1, SlMBP2, TAG1, SlMBP22, SlMBP21, and SlMBP6) were extremely highly expressed in flowers. However, TAGL1, SlMBP3, TAGL2, TM5, and SlMADS1 were mainly expressed during the stages of fruit development and ripening, and the expression of SlMBP20 was particularly high in leaves.
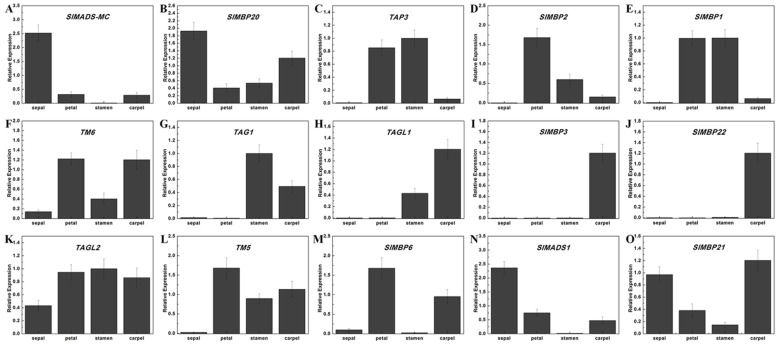
To further investigate the potential role of these MADS-box genes in floral organ development, a comparison of the expression patterns of 15 tomato MADS-box genes in four whorls of floral organs (sepal, petal, stamen, and carpel) were analyzed by qPCR. As shown in Figure 5A,B, the class A genes MADS-MC and SlMBP20 were highly expressed in sepals. The expression levels of class B genes (TAP3, SlMBP2, and SlMBP1) were notably high in the petal and stamen (Figure 5C–E), whereas the transcription of TM6 was found to be markedly higher in carpel and stamen compared with other floral organs (Figure 5F). Two class C genes, TAG1 and TAGL1, were found to be mainly expressed in stamens and carpels (Figure 5G,H). Moreover, the class D genes, SlMBP3 and SlMBP22, showed organ-specific expression patterns that were exclusively expressed in carpel (Figure 5I,J). The expression patterns of class E genes indicated that the expression of TAGL2 and TM5 were significantly higher expressed in the petal, stamen, and carpel than in the sepal (Figure 5K,L), and SlMBP6 was highly expressed in petals and carpels (Figure 5M). In comparison, the other class E genes (SlMADS1 and SlMBP21) were shown to exhibit higher expression levels in the sepal and carpel (Figure 5N,O).
Figure 5.
Expression profiles of floral organ identity genes in the four-whorl floral organs of the wild-type (WT) tomato plant for qPCR analysis. Se, sepal; Pe, petal; St, stamen; Ca, carpel. (A)–(O) Expression profiles of SlMADS-MC, SlMBP21, TAP3, SlMBP2, SlMBP1, TM6, TAG1, TAGL1, SlMBP3, SlMBP22, TAGL2, TM5, SlMBP6, SlMADS1 and SlMBP21.Each value represents the mean ± SE of three technical replicates of a single biological sample. The SlCAC gene of tomato was used as the internal standard.
In addition, the interaction networks of these 15 tomato MADS-box proteins were predicted by STRING software (Figure S4). The results showed that they established interactions with other proteins, directly or indirectly. The TAGL2, SlMBP3, TAGL1, and SlMBP22 proteins can interact directly with each other. Apart from that, the TAGL1 protein showed complex interactions with several other proteins, including the SlMBP3 and TAP3 proteins. The TM5 protein was also shown to directly interact with the TAP3 and TM6 proteins.
2.7. Differential Expression Analysis of Tomato MADS-Box Genes at Different Stages of Fruit Development and Ripening
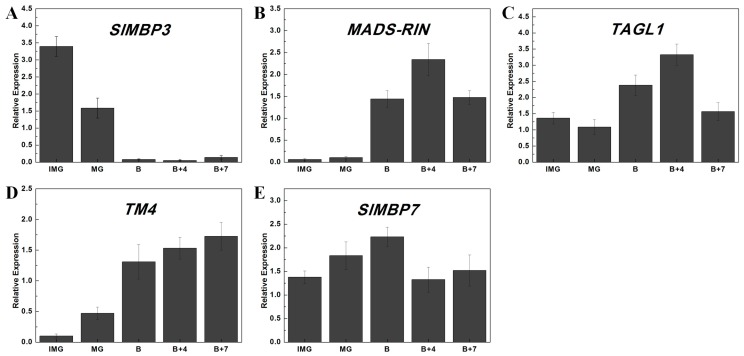
According to the expression profiles predictions, we selected five tomato MADS-box genes (SlMBP3, MADS-RIN, TAGL1, TM4, and SlMBP7) that may take part in fruit development and ripening. We analyzed their expression patterns by qPCR at five different stages of fruit development and ripening, including the immature green (IMG), mature green (MG), and break (B) stages, as well as at four days after break (B+4) and seven days after break (B+7). SlMBP3 exhibited a strikingly high expression level at the IMG stage, and showed extremely low expression at the other stages (Figure 6A). The expression levels of MADS-RIN and TAGL1 exhibited an increasing tendency from the MG to the B+4 stage, and then decreased at B+7 (Figure 6B,C). TM4 expression increased continuously during the process of fruit development and exhibited its maximum expression level at the B+7 stage (Figure 6D). Compared with the other three stages, the expression of SlMBP7 was slightly higher at B (Figure 6E). These results indicated that the qPCR data were consistent with the predictions of expression profiles and that our predictions are suitable for investigating the expression patterns of tomato MADS-box genes.
Figure 6.
Relative expression of tomato MADS-box genes in difference stages of fruit development and ripening by qPCR analysis. Expression patterns of SlMBP3 (A), MADS-RIN (B), TAGL1 (C), TM4 (D), and SlMBP7(E) in different organs, including mature green (MG), break (B), four days after break (B+4), and seven days after break (B+7). Each value represents the mean ± SE of three technical replicates of a single biological sample. The SlCAC gene of tomato was used as the internal standard.
3. Discussion
3.1. Characterization of MADS-Box Genes in Tomato
The MADS-box genes control diverse biological processes in plants, including vegetative growth and reproductive development. They mainly play key roles in the developmental processes of inflorescences, flowers, and fruits [78,79]. 24 MIKCC-type MADS-box transcription factors have been identified, and their functions and evolutions in tomatoes were thoroughly studied in 2006 [35]. However, the MIKCC-type MADS-box members are only part of the MADS-box transcription factor family, and to date, there has been no comparative report on the tomato MADS-box genes. It is well known that genome-wide analysis of gene families is a major and necessary approach to analyze the structure, evolution, and function of genes. In this study, 131 tomato MADS-box proteins were identified, and 96 new tomato MADS-box proteins with unknown functions were systemically named (Table 1). This study is the first comparative analysis of the tomato MADS-box gene family, and we believe that the resolving confusion in naming the genes will facilitate further functional analysis of the tomato MADS-box genes.
First, we presented the phylogenetic relationships of 131 tomato MADS-box proteins with Arabidopsis MADS-box proteins to classify the tomato MADS-box proteins into five subfamilies (MIKC, Mα, Mβ, Mγ, and Mδ), as shown Figure S1A. Compared with Arabidopsis, a larger number of MADS-box proteins were found in tomato. In total, 81 tomato MADS-box genes were determined to be type I genes, including the Mα, Mβ, Mγ, and Mδ groups, which is more than that in Arabidopsis. We speculate that tomato type I MADS-box genes may have a higher duplication rate and/or a lower gene loss rate after duplication. Nevertheless, 50 tomato MADS-box genes were classified as type II genes, including MIKCc and MIKC*, which is comparable to that in Arabidopsis. These results indicate that the genes’ retention duplication have been different in various species, leading to different numbers of MAD-box genes among different species, with different evolutionary constraints [80]. Then, in order to investigate the phylogenetic relationships of MADS-box genes in tomato, a phylogenetic tree for two types of tomato MADS-box genes was constructed (Figure S1B,C). This showed that tomato MADS-box genes are conservative in subfamilies.
To obtain insight into the structural diversity of the tomato MADS-box genes, the intron–exon organization was analyzed (Figure 1). Previous studies have postulated that an intron-rich gene would lose multiple introns simultaneously by retrotransposition, thereby producing intron-less ancestral genes. In this study, we found that the Mα, Mβ and Mγ groups of the type I genes usually have no introns or one intron, which may experience the loss of multiple introns during MADS-box gene family diversification. In addition, the distribution of introns in tomato type I and type II genes were different, and the Mδ group of the type I and type II genes had more introns than the Mα, Mβ, and Mγ groups genes. Similar cases have also been detected in Arabidopsis and rice [7,13], suggesting the evolutionary conservation among plants. However, some close gene pairs showed different intron/exon arrangements, indicating that a more complicated gene structural evolution may exist in tomato MADS-box genes. The conserved motif analysis indicated that the same group contained most conserved motifs (Figure 2). The results suggested that these conserved motifs play important roles in group-specific functions. However, high structural divergence was found between the different groups. An analysis of the gene structures and conserved motifs could provide more clues about the evolutionary relationships of the MADS-box family in tomato.
Gene duplication (segmental duplication and tandem duplication) as well as transposition events were prevalent forces that result in the expansion of family members and genome complexity in eukaryotes [74]. The duplication of more than two genes located on one chromosome is confirmed as a tandem duplication event, whereas gene duplication that occurs on different chromosomes is identified as segmental duplication [73,81]. Both tandem and segmental duplication can play crucial roles in MADS-box gene expansion the tomato genome. In our study, a chromosomal location analysis of the tomato MADS-box genes showed that the MADS-box genes are distributed on 12 chromosomes (Figure 3). The tomato MADS-box genes had a high-density distribution on chromosome 1, which had 24 genes, suggesting that they might be caused by tandem duplications. The closely related tomato MADS-box genes formed tandem arrays on chromosomes 1, 3, 4, 10, and 12, which may help the tomato evolve distinct characterizations from other plants.
Since gene expression profiles can provide significant clues about gene function, the expression of tomato MADS-box genes in whole root (Rt), young leaf (YL), mature leaves (ML), young flower buds (YFB), fully open flowers (F), and five different fruit tissues were examined by transcription expression (RNA-seq) data. All the tested 124 tomato MAD-box genes that were expected to contain SlMADS4, SlMADS11, SlMADS37, SlMADS44, SlMADS46, SlMADS56, SlMADS68, SlMADS70, and SlMADS89 showed distinct expression patterns (Figure 4). This finding may supply insight for future studies on the functions of MADS-box genes in tomato plant growth and development. For example, we found that the SlMADS23 gene was only expressed in the root, so we speculate that the SlMADS23 gene may play a key role in root growth and development. The roots of plants determined the capacity of plants to acquire and distribute nutrients and water, as well as provide a means to suit the environmental conditions. Thus, the root architecture is extremely important for plant development and breeding. In the future, we will verify whether SlMADS23 is related to root growth by constructing a SlMADS23 overexpression vector and generating transgenic overexpression tomato plants to study the regulation of SlMADS23 gene on root growth and development. In addition, performing SlMADS23 gene mutagenesis with the CRISPR/Cas9 system transformation method may also prove to be a helpful strategy.
3.2. Prediction of MADS-Box Genes Involved in the Regulation of Flower Development and Floral Organ Identity
An investigation of the genetic and molecular basis of flower development and floral organ identity in Arabidopsis and petunia suggested that MADS-box genes play fundamental roles in floral organ identity and flower development [82]. It has been confirmed that five classes of MADS-box genes (A–E) were involved in specifying floral organ identity [83,84,85]. In Arabidopsis, the class A genes (AP1 and AP2), the class B genes (AP3 and PI), the class C gene (AG), the class D gene (AGL11), and the class E gene (SEP1, SEP2, SEP3, and SEP4) were MADS-box genes, which have been reported to be involved in the regulation of floral organ development [83,86]. In petunia, lots of MADS-box genes, including the class A genes PETUNIA FLOWERING GENE (PFG), FLORAL BINDING PROTEIN 26 (FBP26), and FBP29, the class B genes TM6, PMADS1/GP, PMADS2, and FBP1, the class C genes PMADS3, FBP6, and FBP24, the class D genes FBP11 and FBP7, as well as FBP2, FBP4, FBP5, FBP9, and the class E genes FBP23, PMADS4, and PMADS12 played important roles in flower development [17,87].
In this paper, we investigated the tomato MADS-box genes’ phylogenetic relationships with the petunia hybrid to select 15 tomato MADS-box genes that may play specific roles in flower development (Table S2). According to the expression profile predictions shown in Figure 4, the highest expression values for most of the genes (MADS-MC TAP3, TM6, SlMBP1, SlMBP2, TAG1, SlMBP22, SlMBP21, and SlMBP6) were observed in flower development stages. Furthermore, qPCR was used to study the expression patterns of four whorls of floral organs (sepal, petal, stamen, and carpel) in these 15 tomato MADS-box genes.
AP1 is an Arabidopsis A class gene, which conferred sepal identity in the first floral [88]. In petunia, the three genes PFG, FBP26, and FBP29 have been identified, which were orthologs of AP1/SQUA in Arabidopsis [17]. Our phylogenetic analysis showed that MADS-MC and SlMBP20 belonged to this clade (Figure S3) and their expression were particularly high in sepal, suggesting that they might play a similar role to AP1 (Figure 5A,B). The class B genes were involved in the identification of petal and stamen in angiosperms [89]. Regarding the class B genes, five close homologs of petunia—TOMATO MADS-BOX GENE6 (TM6), GREEN PETAL (GP)/ PETUNIA MADS BOX GENE 1 (PMADS1), PMADS2, and FBP1—were found in tomato [77]. The qPCR analysis showed that TAP3, SlMBP2, and SlMBP1 have petal and stamen specific expression, while the TM6/TDR6 transcripts were mainly detected in the petal and carpel (Figure 5C–F). These results were similar to the homologous genes of that in petunia [77]. Two tomato MADS-box genes TAG1 and TAGL1, which were involved in C functions, were from the monophyletic AGAMOUS (AG) subfamily. These two genes were mainly expressed in stamens and carpels (Figure 5G,H), which is consistent with their function in specifying stamen and carpel development [52]. SlMBP3 and SlMBP22, which are highly homologous to two petunia class D MADS-box genes, FBP11 and FBP7, were shown to be separately and exclusively expressed in carpel (Figure 5L,I). The result suggested that SlMBP3 and SlMBP22 may have similar functions to the petunia FBP7 and FBP11 genes, which are related to the establishment of real ovules or carpel-like structures [90]. Arabidopsis SEP genes, the typical class E genes, were necessary for the specification of sepal, petal, stamen, and carpel identity with interaction with the class A, B, C, and D genes [16]. In petunia, seven class E genes (FBP2, FBP4, FBP23, FBP5, FBP9, pMADS12, and pMADS4) have been determined that belong to the SEPALLATA (SEP) clade [32]. The tomato TAGL2, TM5, SlMADS1, SlMBP21, and SlMBP6 genes were homologous to these petunia class E genes (Figure S3), and some differences in expression patterns have been observed (Figure 5J–N), indicating that these five tomato class E genes may be involved in multiple floral organ identity. Thus, we believe that the expression patterns of the tomato MADS-box genes identified in our study will be an important tool for understanding the flower development mechanisms in tomato. Previous reports have found that the MADS-box proteins are able to form multiple homologous or heterodimeric complexes in plants, and the combinatorial MADS-box proteins are often deriving their regulatory specificity from other DNA binding or accessory factors. To understand how the tomato MADS-box genes can act in flower development and floral organ identity, it is necessary to identify the network of protein–protein interactions amongst them. Therefore, the predicted interaction networks of the 15 tomato MADS-box proteins, which are involved in floral organ development, were analyzed in our report (Figure S4). In many domesticated crops, it’s an important way to select inflorescence architecture with improved flower production and yield. In tomato, SlMBP21 forms protein complexes with JOINTLESS and MACROCALYX as a transcription activator for tomato flower abscission zone development [50], because SlMBP21, J, and MC have a common function in the development of the tomato flower abscission zone. In breeding, altering any of these genes will have the function on plant growth. In this study, the predicted interaction networks may help us to understand how the tomato MADS-box genes can act in flower development and floral organ identity.
3.3. Potential Functions of Tomato MADS-Box Genes during Fruit Development
To better understand the roles of tomato MADS-box genes in fruit development and ripening, we selected five tomato MADS-box genes (SlMBP3, MADS-RIN, TAGL1, TM4, and SlMBP7) that were predicted to abundantly expressed at different stages of fruit development and ripening. Then, we detected their relative expression level in fruits samples from five different stages of development by qPCR (Figure 6). As shown in Figure 6A, the expression level of SlMBP3 was found to be higher at the MG stage than at the other stages. We found that the expression levels of MADS-RIN and TAGL1 exhibited an increasing tendency during the transition from the MG stage to the B+4 stage, and then decreased at the B+7 stage (Figure 6B,C). Moreover, the expression level of TM4 was found to increase continuously in the process of fruit development (Figure 6D). The SlMBP7 gene showed a relatively high expression level at the B stage (Figure 6E). Recent reports have identified a number of MADS-box genes that are required for the regulation of fruit development and ripening. One of the most representative is the tomato MADS-RIN gene, which is one of the earliest acting ripening regulators, and plays crucial roles in fruit ripening through ethylene dependent and independent ripening regulatory pathways [19,91]. In addition, TAGL1, TM4, and SlMBP7 have been found to regulate fruit ripening in tomato [21,22]. Since our results were consistent with the functional research of these genes, the SlMBP3 gene was predicted to be particularly high in the MG stage, indicating that it might play an important function in fruit development and ripening. These results help to advance our understanding of the function of MADS-box genes in the regulation of fruit developmental and ripening processes in tomato.
4. Materials and Methods
4.1. Plant Material and Growth Condition
In this article, tomatoes (Solanum lycopersicum, ‘Ailsa Craig’ AC++) from Laboratory of molecular biology of tomato, Bioengineering College, Chongqing University, Chongqing, China, were grown in controlled greenhouse conditions of a 16-h day (25 °C)/8-h night (18 °C) cycle, 80% humidity, a 250-μmol·m−2·s−1 light intensity, and were managed routinely. The tomato flowers were tagged at anthesis and floral organs: sepals (Se), petals (Pe), stamens (St), and carpels (Ca) were collected from flowers at anthesis. The fruit color and days post-anthesis (DPA) were used to differentiate the ripening days of tomato fruits. We defined 20 DPA as the immature green (IMG), and 35 DPA as the mature green (MG), at which point the fruits are green and shiny and no obvious color change is observed. The 38-DPA tomato fruits with color change from green to yellow were characterized as breaker (B) fruits. Besides, the samples of B+4 (4 days after breaker) fruits and B+7 (7 days after breaker) fruits were also used in our study. All the samples that we used were frozen in liquid nitrogen immediately and stored at –80 °C.
4.2. Identification of MADS-Box Genes in Tomato
The Sol Genomics Network (SGN, Available online: http://solgenomics.net/) and the National Center for Biotechnology Information (NCBI, Available online: https://www.ncbi.nlm.nih.gov/) database were used to comprehensively identify the whole MADS-box protein sequences of tomato. BLAST searches, using all the Arabidopsis and rice MADS-box protein sequences as queries, were performed to check the predicted tomato MADS-box protein sequences in the database. Subsequently, we further examined all the candidate protein sequences by the PROSITE (Available online: http://www.expasy.org/prosite/) and SMART (Available online: http://smart.embl-heidelberg.de/) databases for reliability. The tomato protein sequences, containing the typical conserved domain of the MADS-box protein family, were selected for amino acid sequence multiple alignment and phylogenetic tree analysis. Then, we obtained their DNA sequences according to their amino acid sequence from the SGN database. Additionally, the molecular weight and isoelectric points of tomato MADS-box proteins were detected by the ExPASy proteomics server.
4.3. Phylogenetic Analysis of Tomato MADS-Box Proteins
Multiple sequence alignment for the two groups of all the 131 tomato MADS-box genes (Table 1) was generated using ClustalX 1.81. The alignment results were used to conduct a phylogenetic tree by the MEGA5.02 program, and the evolutionary history was inferred using the neighbor-joining method. [92].
4.4. The Analysis of Gene Structure and Conserved Motif
The tomato MADS-box coding domain sequences (CDS) and corresponding genomic DNA sequences were collected from SGN and NCBI databases to predict gene structure. The online tool Gene Structure Display Server 2.0 (GSDS 2.0, Available online: http://gsds.cbi.pku.edu.cn/index.php), was used to construct an exon/intron map [93].
Conserved motifs of the tomato MADS-box protein sequences were identified by online software MEME Version 4.12.0 (Available online: http://meme-suite.org/tools/meme). It was performed with the following parameters: 10 different motifs, a motif width of 6–200 amino acids, and any number of repetitions. The SMART database was used to annotate the MEME motifs.
4.5. Chromosomal Locations and Identification of Interaction Network
To determine the chromosomal locations of tomato MADS-box genes, we obtained the physical genome annotation files from the SGN and NCBI database. The physical map of the tomato MADS-box genes was drawn by the Tomato-EXPEN 2000 (Available online: https://solgenomics.net/cview/map.pl).
The interaction network was conducted by STRING (functional protein association networks, Available online: https://string-db.org/) software using the search of multiple proteins sequences [94].
4.6. Digital Gene Expression Analysis of Tomato MADS-Box Genes
To obtain the expression profile of tomato MADS-box genes, the RNA-seq data based on the locus/gene names of SGN were analyzed. We downloaded the RNA-seq data from TFGD (the Tomato Functional Genomics Database), and the sequence data were obtained from various tissues in wild species LA1589 (S. pimpinellifolium). In addition, the data of the tomato lab and Tomato eFP Browser were also used to analyze the gene expression of tomato MADS-box genes. A heatmap was generated by Heml 1.0 (Heatmap illustrator, Available online: http://hemi.biocuckoo.org/) using the relative expression values or ratios of each tomato MADS-box gene [95].
4.7. Total RNA Extraction and qPCR Analysis
To study the expression patterns of the MADS-box genes involved in flower organ identity and fruit development in tomato, total RNA was extracted from the sepals (Se), petals (Pe), stamens (St), and carpels (Ca) of tomato and different developmental stages of tomato fruits at different developmental stages, including IMG (immature green), MG (mature green), B (breaker), B+4 (4 days after breaker), and B+7 (7 days after breaker) using RNAiso Plus (Takara) in accordance with the instructions. After DNase digestion (Promega, Madison, WI, USA), cDNA was synthesized with oligo(dT)20 as a primer for RNA reverse-transcription using M-MLV reverse transcriptase (Promega, Madison, WI, USA). For gene expression quantification, qPCR analysis was performed with the CFX96™ Real-Time System (Bio-Rad, Hercules, CA, USA) using the GoTaq qPCR Master Mix (Promega, Madison, WI, USA). First, 1.0 μL of mixture primers, 1.0 μL of cDNA, and 3.0 μL of ddH2O were used. NRT (no reverse transcription control) and NTC (no template control) experiments were performed to eliminate the genomic DNA and environment effects. The tomato SlCAC gene was used as an internal standard [96], and the 2−ΔΔCT method was used to perform the relative gene expression levels analysis [97]. In addition, all the experiments were performed in three biological triplicates with three technical replicates. The standard curves were run at the same time. All the primers used were designed by Primer 5.0 software and are shown in Table S3.
4.8. Data Analysis
In this study, the mean values of data are presented as mean ± standard deviation. The Origin 8.0 software (Available online: https://www.originlab.com/) was used to perform the data analysis, and mean differences were significant by t-test (* p < 0.05).
5. Conclusions
In this study, a comprehensive and systematic analysis of the tomato MADS-box transcription factor family was first conducted. A total of 131 genes encoding MADS-box transcription factors, including 81 type I and 50 type II genes, were extensively identified in the tomato genome. Then, we classified the genes according to their phylogenetic relationships between tomato and Arabidopsis. The phylogenetic relationships, gene structures, conserved motifs, chromosomal distribution, and expression patterns of the genes were characterized. The 131 tomato MADS-box genes showed differential expression levels in different organs. Since the MADS-box genes are the most powerful TFs that regulate floral organ identity and fruit development and ripening in plants, we showed that 15 tomato MADS-box genes were involved in floral organ development, and we studied the expression of five tomato MADS-box genes in different stages of fruit development and ripening. These results provide evidence of the relationship between MADS-box genes and floral organ and fruit development. Our study provides comprehensive information on the tomato MADS-box gene family, enables a better understanding of the structure–function relationships among the tomato MADS-box gene family members, and lays a solid foundation of comprehensive functional characteristics in the tomato MADS-box gene family. Furthermore, our bioinformatics and evolutionary analysis will be helpful for better understanding the underlying evolutionary relationship of the MADS-box family in higher plants.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/20/12/2961/s1.
Author Contributions
G.C. and Z.H. designed and managed the research work and improved the manuscript; Y.W. and X.G. performed the bioinformatics analysis; J.Z. performed the experiments; Y.W. wrote the manuscript; Y.W. and J.Z. prepared all figures. All the authors reviewed the manuscript.
Funding
This work was supported by National Natural Science Foundation of China (31572129), the National Natural Science Foundation of Chongqing of China (cstc2018jcyjAX0458) and Training Program of Chongqing University Bioengineering College (0221001105301).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Shore P., Sharrocks A.D. The MADS-box family of transcription factors. Eur. J. Biochem. 1995;229:1–13. doi: 10.1111/j.1432-1033.1995.tb20430.x. [DOI] [PubMed] [Google Scholar]
- 2.Schwarz-Sommer Z., Sommer H. Genetic Control of Flower Development by Homeotic Genes in Antirrhinum majus. Science. 1990;250:931–936. doi: 10.1126/science.250.4983.931. [DOI] [PubMed] [Google Scholar]
- 3.Alvarezbuylla E.R., Pelaz S., Liljegren S.J., Gold S.E., Burgeff C., Ditta G.S., Ribas d.P.L., Martínezcastilla L., Yanofsky M.F. An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. USA. 2000;97:5328–5333. doi: 10.1073/pnas.97.10.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smaczniak C., Immink R.G., Angenent G.C., Kaufmann K. Developmental and evolutionary diversity of plant MADS-domain factors: Insights from recent studies. Development. 2012;139:3081–3098. doi: 10.1242/dev.074674. [DOI] [PubMed] [Google Scholar]
- 5.Henschel K., Kofuji R., Hasebe M., Saedler H., Münster T., Theissen G. Two ancient classes of MIKC-type MADS-box genes are present in the moss Physcomitrella patens. Mol. Biol. Evol. 2002;19:801–814. doi: 10.1093/oxfordjournals.molbev.a004137. [DOI] [PubMed] [Google Scholar]
- 6.Parenicová L., De F.S., Kieffer M., Horner D.S., Favalli C., Busscher J., Cook H.E., Ingram R.M., Kater M.M., Davies B. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: New openings to the MADS world. Plant Cell. 2003;15:1538–1551. doi: 10.1105/tpc.011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arora R., Agarwal P., Ray S., Singh A.K., Singh V.P., Tyagi A.K., Kapoor S. MADS-box gene family in rice: Genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genom. 2007;8:242. doi: 10.1186/1471-2164-8-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yanofsky M.F., Ma H., Bowman J.L., Drews G.N., Feldmann K.A., Meyerowitz E.M. The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature. 1990;346:35–39. doi: 10.1038/346035a0. [DOI] [PubMed] [Google Scholar]
- 9.Coen E.S., Meyerowitz E.M. The war of the whorls: Genetic interactions controlling flower development. Nature. 1991;353:31–37. doi: 10.1038/353031a0. [DOI] [PubMed] [Google Scholar]
- 10.Causier B., Schwarz-Sommer Z., Davies B. Floral organ identity: 20 years of ABCs. Semin. Cell Dev. Biol. 2010;21:73–79. doi: 10.1016/j.semcdb.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 11.Krizek B.A., Meyerowitz E.M. The Arabidopsis homeotic genes APETALA3 and PISTILLATA are sufficient to provide the B class organ identity function. Development. 1996;122:11–22. doi: 10.1242/dev.122.1.11. [DOI] [PubMed] [Google Scholar]
- 12.Theissen G. Development of floral organ identity: Stories from the MADS house. Curr. Opin. Plant Biol. 2001;4:75–85. doi: 10.1016/S1369-5266(00)00139-4. [DOI] [PubMed] [Google Scholar]
- 13.Mandel M.A., Gustafson-Brown C., Savidge B., Yanofsky M.F. Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature. 1992;360:273–277. doi: 10.1038/360273a0. [DOI] [PubMed] [Google Scholar]
- 14.Jack T., Brockman L.L., Meyerowitz E.M. The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell. 1992;68:683–697. doi: 10.1016/0092-8674(92)90144-2. [DOI] [PubMed] [Google Scholar]
- 15.Mizukami Y., Ma H. Ectopic expression of the floral homeotic gene AGAMOUS in transgenic Arabidopsis plants alters floral organ identity. Cell. 1992;71:119–131. doi: 10.1016/0092-8674(92)90271-D. [DOI] [PubMed] [Google Scholar]
- 16.Pelaz S., Ditta G.S., Baumann E., Wisman E., Yanofsky M.F. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature. 2000;405:200–203. doi: 10.1038/35012103. [DOI] [PubMed] [Google Scholar]
- 17.Angenent G.C., Franken J., Busscher M., Van D.A., van Went J.L., Dons H.J., van Tunen A.J. A novel class of MADS box genes is involved in ovule development in petunia. Plant Cell. 1995;7:1569–1582. doi: 10.1105/tpc.7.10.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colombo L., Franken J., Koetje E., Went J.V., Dons H.J., Angenent G.C., Tunen A.J.V. The petunia MADS box gene FBP11 determines ovule identity. Plant Cell. 1995;7:1859–1868. doi: 10.1105/tpc.7.11.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vrebalov J., Ruezinsky D., Padmanabhan V., White R., Medrano D., Drake R., Schuch W., Giovannoni J. A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science. 2002;296:343–346. doi: 10.1126/science.1068181. [DOI] [PubMed] [Google Scholar]
- 20.Itkin M., Seybold H., Breitel D., Rogachev I., Meir S., Aharoni A. TOMATO AGAMOUS-LIKE 1 is a component of the fruit ripening regulatory network. Plant J. Cell Mol. Biol. 2009;60:1081–1095. doi: 10.1111/j.1365-313X.2009.04064.x. [DOI] [PubMed] [Google Scholar]
- 21.Arroyo A.J.M., Mcquinn R., Poole M., Seymour G., Giovannoni J., Irish V.F., Vrebalov J., Pan I.L., Chung M., Rose J. Fleshy fruit expansion and ripening are regulated by the Tomato SHATTERPROOF gene TAGL1. Plant Cell. 2009;21:3041–3062. doi: 10.1105/tpc.109.066936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bemer M., Karlova R., Ballester A.R., Tikunov Y.M., Bovy A.G., Wolters-Arts M., Rossetto P.B., Angenent G.C., de Maagd R.A. The Tomato FRUITFULL Homologs TDR4/FUL1 and MBP7/FUL2 Regulate Ethylene-Independent Aspects of Fruit Ripening. Plant Cell. 2012;24:4437–4451. doi: 10.1105/tpc.112.103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shima Y., Kitagawa M., Fujisawa M., Nakano T., Kato H., Kimbara J., Kasumi T., Ito Y. Tomato FRUITFULL homologues act in fruit ripening via forming MADS-box transcription factor complexes with RIN. Plant Mol. Biol. 2013;82:427–438. doi: 10.1007/s11103-013-0071-y. [DOI] [PubMed] [Google Scholar]
- 24.Byeong-ha Lee D.A.H., Zhu J.-K. The Arabidopsis Cold-Responsive Transcriptome and Its Regulation by ICE1. Plant Cell. 2005;17:3155–3175. doi: 10.1105/tpc.105.035568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee I., Ronald P.C. Genetic dissection of the biotic stress response using a genome-scale gene network for rice. Proc. Natl. Acad. Sci. USA. 2011;108:18548–18553. doi: 10.1073/pnas.1110384108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tardif G., Kane N.A., Adam H., Labrie L., Major G., Gulick P., Sarhan F., Laliberté J.F. Interaction network of proteins associated with abiotic stress response and development in wheat. Plant Mol. Biol. 2007;63:703–718. doi: 10.1007/s11103-006-9119-6. [DOI] [PubMed] [Google Scholar]
- 27.Duan W., Song X., Liu T., Huang Z., Ren J., Hou X., Li Y. Genome-wide analysis of the MADS-box gene family in Brassica rapa (Chinese cabbage) Mol. Genet. Genom. 2015;290:239–255. doi: 10.1007/s00438-014-0912-7. [DOI] [PubMed] [Google Scholar]
- 28.Sommer H., Beltrán J.P., Huijser P., Pape H., Lönnig W.E., Saedler H., Schwarz-Sommer Z. Deficiens, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus: The protein shows homology to transcription factors. Embo J. 1990;9:605–613. doi: 10.1002/j.1460-2075.1990.tb08152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leseberg C.H., Li A., Kang H., Duvall M., Mao L. Genome-wide analysis of the MADS-box gene family in Populus trichocarpa. Gene. 2006;378:84–94. doi: 10.1016/j.gene.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 30.Jian M., Yang Y., Wei L., Yang C., Ding P., Liu Y., Qiao L., Chang Z., Geng H., Wang P. Genome-wide identification and analysis of the MADS-box gene family in bread wheat (Triticum aestivum L.) PLoS ONE. 2017;12:e0181443. doi: 10.1371/journal.pone.0181443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu J., Zhang J., Zhang J., Miao H., Wang J., Gao P., Hu W., Jia C., Wang Z., Xu B. Genome-wide analysis of banana MADS-box family closely related to fruit development and ripening. Sci. Rep. 2017;7:3467. doi: 10.1038/s41598-017-03897-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Busscherlange J. Analysis of the petunia MADS-box transcription factor family. Mol. Genet. Genom. 2003;268:598–606. doi: 10.1007/s00438-002-0781-3. [DOI] [PubMed] [Google Scholar]
- 33.Velasco R., Zharkikh A., Affourtit J., Dhingra A., Cestaro A., Kalyanaraman A., Fontana P., Bhatnagar S.K., Troggio M., Pruss D., et al. The genome of the domesticated apple (Malus x domestica Borkh.) Nat. Genet. 2010;42:833–839. doi: 10.1038/ng.654. [DOI] [PubMed] [Google Scholar]
- 34.Leseberg C.H., Eissler C.L., Wang X., Johns M.A., Duvall M.R., Mao L. Interaction study of MADS-domain proteins in tomato. J. Exp. Bot. 2008;59:2253–2265. doi: 10.1093/jxb/ern094. [DOI] [PubMed] [Google Scholar]
- 35.Hileman L.C., Sundstrom J.F., Litt A., Chen M., Shumba T., Irish V.F. Molecular and phylogenetic analyses of the MADS-box gene family in tomato. Mol. Biol. Evol. 2006;23:2245–2258. doi: 10.1093/molbev/msl095. [DOI] [PubMed] [Google Scholar]
- 36.Guo X., Hu Z., Yin W., Yu X., Zhu Z., Zhang J., Chen G. The tomato floral homeotic protein FBP1-like gene, SlGLO1, plays key roles in petal and stamen development. Sci. Rep. 2016;6:20454. doi: 10.1038/srep20454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martino G.D., Pan I., Emmanuel E., Levy A., Irish V.F. Functional Analyses of Two Tomato APETALA3 Genes Demonstrate Diversification in Their Roles in Regulating Floral Development. Plant Cell. 2006;18:1833–1845. doi: 10.1105/tpc.106.042978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang B., Routaboul J.M., Liu M., Deng W., Maza E., Mila I., Hu G., Zouine M., Frasse P., Vrebalov J.T. Overexpression of the class D MADS-box gene Sl-AGL11 impacts fleshy tissue differentiation and structure in tomato fruits. J. Exp. Bot. 2017;68:4869–4884. doi: 10.1093/jxb/erx303. [DOI] [PubMed] [Google Scholar]
- 39.Ampomahdwamena C., Morris B.A., Sutherland P., Veit B., Yao J.L. Down-regulation of TM29, a tomato SEPALLATA homolog, causes parthenocarpic fruit development and floral reversion. Plant Physiol. 2002;130:605–617. doi: 10.1104/pp.005223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Litt A., Irish V.F. Duplication and Diversification in the APETALA1/FRUITFULL Floral Homeotic Gene Lineage: Implications for the Evolution of Floral Development. Genetics. 2003;165:821–833. doi: 10.1093/genetics/165.2.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yao J.L., Ampomah-Dwamena C. The tomato SEPALLATA homolog TM29 has diverse functions in regulating flower and fruit development. Plant Physiol. 2003;35:28–33. [Google Scholar]
- 42.Wang S., Gang L., Zheng H., Luo Z., Wang T., Li H., Zhang J., Ye Z. Members of the tomato FRUITFULL MADS-box family regulate style abscission and fruit ripening. J. Exp. Bot. 2014;65:3005–3014. doi: 10.1093/jxb/eru137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shima Y., Fujisawa M., Kitagawa M., Nakano T., Kimbara J., Nakamura N., Shiina T., Sugiyama J., Nakamura T., Kasumi T. Tomato FRUITFULL homologs regulate fruit ripening via ethylene biosynthesis. J. Agric. Chem. Soc. Jpn. 2014;78:231–237. doi: 10.1080/09168451.2014.878221. [DOI] [PubMed] [Google Scholar]
- 44.Burko Y., Shleizer-Burko S., Yanai O., Shwartz I., Zelnik I.D., Jacob-Hirsch J., Kela I., Eshed-Williams L., Ori N. A role for APETALA1/fruitfull transcription factors in tomato leaf development. Plant Cell. 2013;25:2070–2083. doi: 10.1105/tpc.113.113035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yin W., Hu Z., Hu J., Zhu Z., Yu X., Cui B., Chen G. Tomato (Solanum lycopersicum) MADS-box transcription factor SlMBP8 regulates drought, salt tolerance and stress-related genes. Plant Growth Regul. 2017;83:55–68. doi: 10.1007/s10725-017-0283-2. [DOI] [Google Scholar]
- 46.Yin W., Hu Z., Cui B., Guo X., Hu J., Zhu Z., Chen G. Suppression of the MADS-box gene SlMBP8 accelerates fruit ripening of tomato (Solanum lycopersicum) Plant Physiol. Biochem. 2017;118:235–244. doi: 10.1016/j.plaphy.2017.06.019. [DOI] [PubMed] [Google Scholar]
- 47.Guo X., Chen G., Cui B., Gao Q., Guo J.E., Li A., Zhang L., Hu Z. Solanum lycopersicum agamous-like MADS-box protein AGL15-like gene, SlMBP11, confers salt stress tolerance. Mol. Breed. 2016;36:125. doi: 10.1007/s11032-016-0544-1. [DOI] [Google Scholar]
- 48.Xie Q., Hu Z., Zhu Z., Dong T., Zhao Z., Cui B., Chen G. Overexpression of a novel MADS-box gene SlFYFL delays senescence, fruit ripening and abscission in tomato. Sci. Rep. 2014;4:4367. doi: 10.1038/srep04367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li N., Huang B., Tang N., Jian W., Zou J., Chen J., Cao H., Habib S., Dong X., Wei W. The MADS-box Gene SlMBP21 Regulates Sepal Size Mediated by Ethylene and Auxin in Tomato. Plant Cell Physiol. 2017;58:2241–2256. doi: 10.1093/pcp/pcx158. [DOI] [PubMed] [Google Scholar]
- 50.Liu D., Wang D., Qin Z., Zhang D., Yin L., Wu L., Colasanti J., Li A., Mao L. The SEPALLATA MADS-box protein SLMBP21 forms protein complexes with JOINTLESS and MACROCALYX as a transcription activator for development of the tomato flower abscission zone. Plant J. 2014;77:284–296. doi: 10.1111/tpj.12387. [DOI] [PubMed] [Google Scholar]
- 51.Roldan M.V.G., Périlleux C., Morin H., Huerga-Fernandez S., Latrasse D., Benhamed M., Bendahmane A. Natural and induced loss of function mutations inSlMBP21MADS-box gene led tojointless-2phenotype in tomato. Sci. Rep. 2017;7:4402. doi: 10.1038/s41598-017-04556-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gimenez E., Castañeda L., Pineda B., Pan I.L., Moreno V., Angosto T., Lozano R. TOMATO AGAMOUS1 and ARLEQUIN/TOMATO AGAMOUS—LIKE1 MADS-box genes have redundant and divergent functions required for tomato reproductive development. Plant Mol. Biol. 2016;91:513–531. doi: 10.1007/s11103-016-0485-4. [DOI] [PubMed] [Google Scholar]
- 53.Pnueli L., Hareven D., Rounsley S.D., Yanofsky M.F., Lifschitz E. Isolation of the Tomato AGAMOUS Gene TAG1 and Analysis of Its Homeotic Role in Transgenic Plants. Plant Cell. 1994;6:163–173. doi: 10.1105/tpc.6.2.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Giménez E., Pineda B., Capel J., Antón M.T., Atarés A., Pérezmartín F., Garcíasogo B., Angosto T., Moreno V., Lozano R. Functional analysis of the Arlequin mutant corroborates the essential role of the ARLEQUIN/TAGL1 gene during reproductive development of tomato. PLoS ONE. 2010;5:e14427. doi: 10.1371/journal.pone.0014427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gaffe J., Lemercier C., Alcaraz J.P., Kuntz M. Identification of three tomato flower and fruit MADS-box proteins with a putative histone deacetylase binding domain. Gene. 2011;471:19–26. doi: 10.1016/j.gene.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 56.Busi M.V., Bustamante C., D’Angelo C., Hidalgocuevas M., Boggio S.B., Valle E.M., Zabaleta E. MADS-box genes expressed during tomato seed and fruit development. Plant Mol. Biol. 2003;52:801–815. doi: 10.1023/a:1025001402838. [DOI] [PubMed] [Google Scholar]
- 57.Kramer E.M., Dorit R.L., Irish V.F. Molecular evolution of genes controlling petal and stamen development: Duplication and divergence within the APETALA3 and PISTILLATA MADS-box gene lineages. Genetics. 1998;149:765–783. doi: 10.1093/genetics/149.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pracros P., Renaudin J., Eveillard S., Mouras A., Hernould M. Tomato flower abnormalities induced by stolbur phytoplasma infection are associated with changes of expression of floral development genes. Mol. Plant Microbe Interact. 2006;19:62–68. doi: 10.1094/MPMI-19-0062. [DOI] [PubMed] [Google Scholar]
- 59.Fujisawa M., Shima Y., Nakagawa H., Kitagawa M., Kimbara J., Nakano T., Kasumi T., Ito Y. Transcriptional regulation of fruit ripening by tomato FRUITFULL homologs and associated MADS box proteins. Plant Cell. 2014;26:89–101. doi: 10.1105/tpc.113.119453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ito Y. Regulation of Tomato Fruit Ripening by MADS-Box Transcription Factors. Jpn. Agric. Res. Q. 2016;50:33–38. doi: 10.6090/jarq.50.33. [DOI] [Google Scholar]
- 61.Martel C., Vrebalov J., Tafelmeyer P., Giovannoni J.J. The tomato (Solanum lycopersicum) MADS-box transcription factor RIN interacts with promoters involved in numerous ripening processes in a CNR dependent manner. Plant Physiol. 2011;157:1568–1579. doi: 10.1104/pp.111.181107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ito Y., Kitagawa M., Ihashi N., Yabe K., Kimbara J., Yasuda J., Ito H., Inakuma T., Hiroi S., Kasumi T. DNA-binding specificity, transcriptional activation potential, and the rin mutation effect for the tomato fruit-ripening regulator RIN. Plant J. 2008;55:212–223. doi: 10.1111/j.1365-313X.2008.03491.x. [DOI] [PubMed] [Google Scholar]
- 63.Li L., Zhu B., Fu D., Luo Y. RIN transcription factor plays an important role in ethylene biosynthesis of tomato fruit ripening. J. Sci. Food Agric. 2011;91:2308–2314. doi: 10.1002/jsfa.4475. [DOI] [PubMed] [Google Scholar]
- 64.Nakano T., Kimbara J., Fujisawa M., Kitagawa M., Ihashi N., Maeda H., Kasumi T., Ito Y. MACROCALYX and JOINTLESS interact in the transcriptional regulation of tomato fruit abscission zone development. Plant Physiol. 2012;158:439–450. doi: 10.1104/pp.111.183731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang X., Yin J. Functional Studies of JOINTLESS and Its Interacting MADS-domain Proteins in Tomato. Acta Hortic. Sin. 2011;38:701–708. [Google Scholar]
- 66.Nakano T., Kato H., Shima Y., Ito Y. Apple SVP family MADS-box proteins and the tomato pedicel abscission zone regulator JOINTLESS have similar molecular activities. Plant Cell Physiol. 2015;56:1097–1106. doi: 10.1093/pcp/pcv034. [DOI] [PubMed] [Google Scholar]
- 67.Szymkowiak E.J., Irish E.E. JOINTLESS suppresses sympodial identity in inflorescence meristems of tomato. Planta. 2006;223:646–658. doi: 10.1007/s00425-005-0115-x. [DOI] [PubMed] [Google Scholar]
- 68.Mao L., Begum D., Chuang H.W., Budiman M.A., Szymkowiak E.J., Irish E.E., Wing R.A. JOINTLESS is a MADS-box gene controlling tomato flower abscission zone development. Nature. 2000;406:910–913. doi: 10.1038/35022611. [DOI] [PubMed] [Google Scholar]
- 69.Pnueli L., Abuabeid M., Zamir D., Nacken W., Schwarzsommer Z., Lifschitz E. The MADS box gene family in tomato: Temporal expression during floral development, conserved secondary structures and homology with homeotic genes from Antirrhinum and Arabidopsis. Plant J. 1991;1:255–266. doi: 10.1111/j.1365-313X.1991.00255.x. [DOI] [PubMed] [Google Scholar]
- 70.Daminato M., Masiero S., Resentini F., Lovisetto A., Casadoro G. Characterization of TM8, a MADS-box gene expressed in tomato flowers. BMC Plant Biol. 2014;14:319. doi: 10.1186/s12870-014-0319-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dong T., Hu Z., Deng L., Wang Y., Zhu M., Zhang J., Chen G. A Tomato MADS-Box Transcription Factor, SlMADS1, Acts as a Negative Regulator of Fruit Ripening. Plant Physiol. 2013;163:1026–1036. doi: 10.1104/pp.113.224436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang J., Hu Z., Yao Q., Guo X., Nguyen V., Li F., Chen G. A tomato MADS-box protein, SlCMB1, regulates ethylene biosynthesis and carotenoid accumulation during fruit ripening. Sci. Rep. 2018;8:3413. doi: 10.1038/s41598-018-21672-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang R., Ming M., Li J., Shi D., Qiao X., Li L., Zhang S., Wu J. Genome-wide identification of the MADS-box transcription factor family in pear (Pyrus bretschneideri) reveals evolution and functional divergence. Peerj. 2017;5:e3776. doi: 10.7717/peerj.3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hughes A.L. The evolution of functionally novel proteins after gene duplication. Proc. Biol. Sci. 1994;256:119–124. doi: 10.1098/rspb.1994.0058. [DOI] [PubMed] [Google Scholar]
- 75.Alvarez-Buylla E.R., Liljegren S.J., Pelaz S., Gold S.E., Burgeff C., Ditta G.S., Vergara-Silva F., Yanofsky M.F. MADS-box gene evolution beyond flowers: Expression in pollen, endosperm, guard cells, roots and trichomes. Plant J. 2000;24:457–466. doi: 10.1046/j.1365-313x.2000.00891.x. [DOI] [PubMed] [Google Scholar]
- 76.Causier B., Kieffer M., Davies B. Plant biology. MADS-box genes reach maturity. Science. 2002;296:275–276. doi: 10.1126/science.1071401. [DOI] [PubMed] [Google Scholar]
- 77.Rijpkema A., Gerats T., Vandenbussche M. Genetics of Floral Development in Petunia. Adv. Bot. Res. 2006;44:237–278. [Google Scholar]
- 78.Becker A., Theißen G. The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet. Evol. 2003;29:464–489. doi: 10.1016/S1055-7903(03)00207-0. [DOI] [PubMed] [Google Scholar]
- 79.Rounsley S.D., Ditta G.S., Yanofsky M.F. Diverse roles for MADS box genes in Arabidopsis development. Plant Cell. 1995;7:1259–1269. doi: 10.1105/tpc.7.8.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Airoldi C.A., Davies B. Gene Duplication and the Evolution of Plant MADS-box Transcription Factors. J. Genet. Genom. 2012;39:157–165. doi: 10.1016/j.jgg.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 81.Liu Y., Jiang H.Y., Chen W., Qian Y., Ma Q., Cheng B., Zhu S. Genome-wide analysis of the auxin response factor (ARF) gene family in maize (Zea mays) Plant Growth Regul. 2011;63:225–234. doi: 10.1007/s10725-010-9519-0. [DOI] [Google Scholar]
- 82.Ma H. The unfolding drama of flower development: Recent results from genetic and molecular analyses. Genes Dev. 1994;8:745–756. doi: 10.1101/gad.8.7.745. [DOI] [PubMed] [Google Scholar]
- 83.Pelaz S., Tapialópez R., Alvarezbuylla E.R., Yanofsky M.F. Conversion of leaves into petals in Arabidopsis. Curr. Biol. 2001;11:182–184. doi: 10.1016/S0960-9822(01)00024-0. [DOI] [PubMed] [Google Scholar]
- 84.Davies B., Egeacortines M., De E.A.S., Saedler H., Sommer H. Multiple interactions amongst floral homeotic MADS box proteins. Embo J. 1996;15:4330–4343. doi: 10.1002/j.1460-2075.1996.tb00807.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim S., Koh J., Yoo M.J., Kong H., Hu Y., Ma H., Soltis P.S., Soltis D.E. Expression of floral MADS-box genes in basal angiosperms: Implications for the evolution of floral regulators. Plant J. Cell Mol. Biol. 2005;43:724–744. doi: 10.1111/j.1365-313X.2005.02487.x. [DOI] [PubMed] [Google Scholar]
- 86.Theissen G., Becker A., Rosa A.D., Kanno A., Kim J.T., Münster T., Winter K.U., Saedler H. A short history of MADS-box genes in plants. Plant Mol. Biol. 2000;42:115–149. doi: 10.1023/A:1006332105728. [DOI] [PubMed] [Google Scholar]
- 87.Angenent G.C., Franken J., Busscher M., Colombo L., van Tunen A.J. Petal and stamen formation in petunia is regulated by the homeotic gene fbp1. Plant J. 1993;4:101–112. doi: 10.1046/j.1365-313X.1993.04010101.x. [DOI] [PubMed] [Google Scholar]
- 88.Bowman J.L., Alvarez J., Weigel D., Meyerowitz E.M., Smyth D.R. Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development. 1993;119:721–743. [Google Scholar]
- 89.Poupin M., Federici F., Medina C., Mattis J., Timmermann T.J.P. Isolation of the three grape sub-lineages of B-class MADS-box TM6, PISTILLATA and APETALA3 genes which are differentially expressed during flower and fruit development. Gene. 2007;404:10–24. doi: 10.1016/j.gene.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 90.Cheng X.F., Wittich P.E., Kieft H., Angenent G., Xuhan X., Lammeren A.A.M.V. Temporal and Spatial Expression of MADS Box Genes, FBP7 and FBP11, During Initiation and Early Development of Ovules in Wild Type and Mutant Petunia hybrida. Plant Biol. 2000;2:693–702. doi: 10.1055/s-2000-16640. [DOI] [Google Scholar]
- 91.Fujisawa M., Nakano T., Shima Y., Ito Y. A large-scale identification of direct targets of the tomato MADS box transcription factor RIPENING INHIBITOR reveals the regulation of fruit ripening. Plant Cell. 2013;25:371–386. doi: 10.1105/tpc.112.108118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2014;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Guo A.Y., Zhu Q.H., Chen X., Luo J.C. GSDS: A gene structure display server. Hereditas. 2007;29:1023–1026. doi: 10.1360/yc-007-1023. [DOI] [PubMed] [Google Scholar]
- 94.Lu L.J., Zhang M. Protein-Protein Interaction Networks. Springer; New York, NY, USA: 2013. pp. 141–181. [Google Scholar]
- 95.Deng W., Wang Y., Liu Z., Cheng H., Xue Y. HemI: A toolkit for illustrating heatmaps. PLoS ONE. 2014;9:e111988. doi: 10.1371/journal.pone.0111988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Expósito-Rodríguez M., Borges A.A., Borges-Pérez A., Pérez J.A. Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol. 2008;8:131. doi: 10.1186/1471-2229-8-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.