Abstract
High throughput sequencing has revolutionized our ability to identify aberrant RNA expression and mutations that cause or contribute to disease. These data can be used directly to design oligonucleotide-based modalities using Watson-Crick pairing to target unstructured regions in an RNA. A complementary, although more difficult, strategy to deactivate a malfunctioning RNA is to target highly structured regions with small molecules. Indeed, RNA structures are directly causative of disease. Herein, we discuss emerging strategies to design high affinity, selective, bioactive ligands targeting RNA, or small molecules interacting with RNA (SMIRNAs), and target validation and profiling methods. An experimental foundation is required for a lead identification strategy for RNA structures, constructed from a library-vs.-library screen that probes vast libraries of small molecules for binding RNA three dimensional folds. Dubbed 2-dimensional combinatorial screening (2DCS), the resulting data can be mined against transcriptomes or the composite of RNAs that are produced in an organism to define folded RNA structures that can be targeted. By applying SMIRNAs to cells and using target validation tools such as Chemical Cross-Linking and Isolation by Pull-down (Chem-CLIP) and Small Molecule Nucleic Acid Profiling by Cleavage Applied to RNA (RiboSNAP), all targets engaged in cells can be defined, along with rules for molecular recognition to affect RNA biology. This chapter will describe lessons learned in applying these approaches in vitro, in cells, and in pre-clinical animal models of disease, enabling SMIRNAs to capture opportunities in chemical biology.
1. Introduction
Affecting biology by targeting RNA has been historically focused on the bacterial ribosome. One of the first drugs to target the bacterial ribosome was streptomycin, isolated from Streptomyces bacteria in soil (Jones, Metzger, Schatz, & Waksman, 1944). Ironically, many consider Streptomycin as not “drug-like,” despite it being a transformative, first-in-class treatment for tuberculosis infections at a time when many lived out their last days quarantined in sanitariums. Indeed, much work has been completed to elucidate how antibacterials inhibit translation—by binding highly folded regions in ribosomal RNA (Poehlsgaard & Douthwaite, 2005; Tenson & Mankin, 2006). The discovery of a suite of compounds that differentially affect ribosome function has not only provided life-saving medicines but also invaluable chemical probes that enabled in-depth studies on translation, both biochemical and structural.
Another advance to target RNA was the development of antisense oligonucleotides (ASOs) that recognize sequence. Paul Zamecnik, who discovered tRNA, and coworkers used oligonucleotides complementary to mRNAs found in Rous sarcoma virus (RSV) to inhibit viral replication, transcription and translation (Stephenson & Zamecnik, 1978; Zamecnik & Stephenson, 1978). They subsequently demonstrated ASOs work against other infectious diseases, laying the foundation for treatment of human diseases. Amongst many applications, an ASO was developed to treat spinal muscular atrophy (SMA) by targeting sites in survival of motor neuron 2 (SMN2) pre-mRNA, facilitating inclusion of exon 7 and hence synthesis of a full length and functional SMN protein. This ASO is a transformative medicine for children with SMA (Morrow, 2017; Stein & Castanotto, 2017).
The above examples demonstrate that targeting RNA can improve human disease even when the therapeutic modalities are not “drug-like” (streptomycin and other antibiotics) or are of large molecular weights (ASOs). Indeed, there are exemplar cases of small molecules developed to purposefully target human RNAs despite the perception that SMIRNAs cannot be sufficiently selective or potent to affect biology or disease outcome. Herein, we describe: (i) methods to define experimentally the folded RNA structures that can be targeted by small molecules; (ii) a computational lead identification strategy for an RNA target of interest using the data defined in (i); (iii) deploying SMIRNAs to affect RNA biology and study molecular recognition; and (iv) various target validation approaches in cells to establish compound mode of action by defining the RNAs that are targeted by SMIRNAs and their binding sites within them. Collectively, these studies define principles for molecular recognition that can drive SMIRNA activity in vitro, in cells, and in pre-clinical animal models, affording chemical probes and lead medicines targeting RNA.
2. Two-dimensional combinatorial screening (2DCS), a library-vs.-library screening approach to select the RNA three dimensional folds that bind small molecules
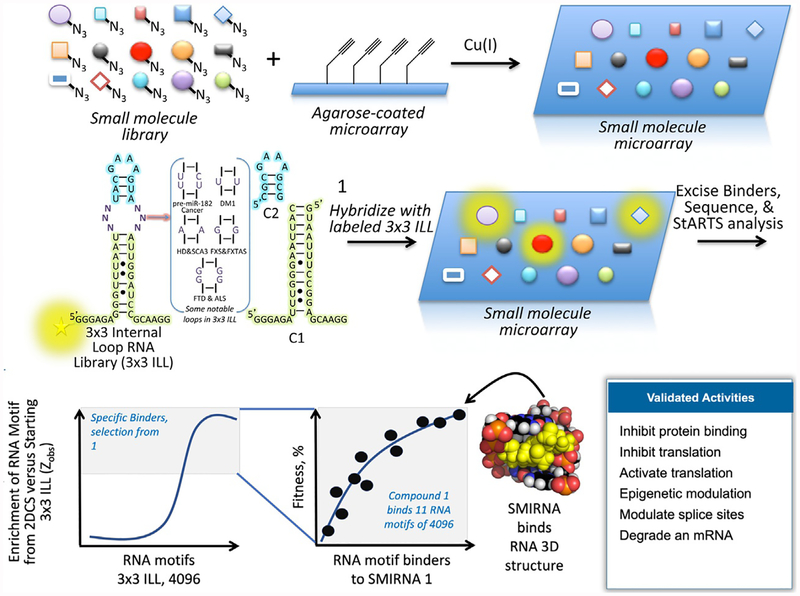
In the broadest sense, RNAs have unstructured regions and regions of highly defined structure that define their function (Rouskin, Zubradt, Washietl, Kellis, & Weissman, 2014; Spitale et al., 2015; Wan et al., 2014). This delineation has major implications for targeting RNA. For example, an ASO is tailored for the former, typically coding sequences, although regions of defined structure have been identified in some mRNAs. The latter represent ideal target sites for SMIRNAs (Angelbello et al., 2018; Disney, Dwyer, & Childs-Disney, 2018). The challenge to enable design of SMIRNAs is to define the optimal RNA three dimensional folds for a particular small molecule and chemotypes that confer avid, selective binding to RNA. As these types of data are generally lacking, a small molecule library-vs.-RNA fold library screen was developed, dubbed 2-Dimensional Combinatorial Screening (2DCS; Fig. 1). A detailed description of how to implement this approach can be found in (Aminova & Disney, 2010).
Fig. 1.
Discovering the three-dimensional RNA folds that bind small molecules using a library-vs.-library screen named 2-Dimensional Combinatorial Screening (2DCS). A small molecule library is site-selectively immobilized (shown) or absorbed onto agarose-coated microarrays. The arrays are incubated with a labeled RNA fold library that displays discrete structural elements (such as the 3 × 3 internal loop library (ILL)) in the presence of competitor oligonucleotides that mimic regions common to all library members (C1 and C2, for example). Bound RNAs are excised from the surface, and their identities are determined by RNA-seq. Statistical analysis of RNA-seq data reveals privileged RNA fold-small molecule interactions that inform design of SMIRNAs that modulate RNA biology. Along with 2DCS, we developed an approach named Structure-Activity Relationship Through Sequencing (StARTS), which allows for facile annotation of 2DCS data by scoring the relative affinity and selectivity of RNA motifs that bind to a small molecule (a metric of small molecule “fitness”).
The 2DCS approach requires encoding of both the small molecules and the RNA three dimensional folds that bind them. The small molecules are spatially arrayed and either covalently attached (Childs-Disney, Wu, Pushechnikov, Aminova, & Disney, 2007) or non-covalently absorbed (Velagapudi et al., 2018) onto microarrays and thus are spatially encoded via their position on the surface (Fig. 1). To allow for isolation of RNAs bound to the small molecules displayed on the surface, microarrays use an agarose substrate, vide infra. Arrays have been constructed by using a wide variety of compounds ranging from known nucleic acid binders to compounds with unknown RNA binding capacity to known drugs.
The arrays are hybridized with a library of three dimensionally folded RNA structural motifs that are embedded in a unimolecular hairpin that can be amplified and sequenced to decode the bound RNA folds (Fig. 1). Small molecule microarrays are hybridized with labeled RNA motif libraries in the presence of unlabeled competitor RNAs that mimic the constant regions in the RNA fold libraries; these competitor oligonucleotides are engineered such that they cannot be amplified via RT-PCR with primers that amplify the RNA fold libraries. This approach is general for many types of RNA fold libraries (internal loops, hairpins, bulges, etc.) and likely has an extended applicability beyond what has been published to date.
The members of the RNA fold library that bind to small molecules on the array are manually excised from the agarose surface, amplified, and sequenced. When sequence analyses are completed by RNA-seq with at least six-fold coverage of the selection, affinity landscapes between small molecules and the various RNA folds can be rapidly assigned (Velagapudi et al., 2017). Assigning affinity landscapes is accomplished by measuring the frequency of enrichment of a specific RNA fold selected by a small molecule to the frequency of this same fold in the starting library using a pooled population comparison. This analysis affords a Zobs score for each RNA fold, a metric of statistical significance, which correlates directly with relative affinity. A Zobs value >8 assigns a SMIRNA (RNA fold-small molecule pair) as binding selectively (Velagapudi et al., 2017). Zobs within a selection can be normalized to provide a Fitness Score for binders with Zobs >8, with a score of 100 indicating the optimal, most fit interaction. Indeed, 2DCS in conjunction with this statistical analysis has defined avid, selective RNA fold-small molecule interactions, chemotypes that confer RNA binding capacity, and the beginning of rules or framework that govern molecular recognition by SMIRNAs. Additionally, it was found that known drug classes target RNA (Velagapudi et al., 2018). It is thus provocative to think that RNA should be considered in toxicological optimization of small molecule drug candidates.
3. Inforna: A sequence-based approach to define RNAs that are druggable from the output of 2DCS
3.1. Overview of Inforna
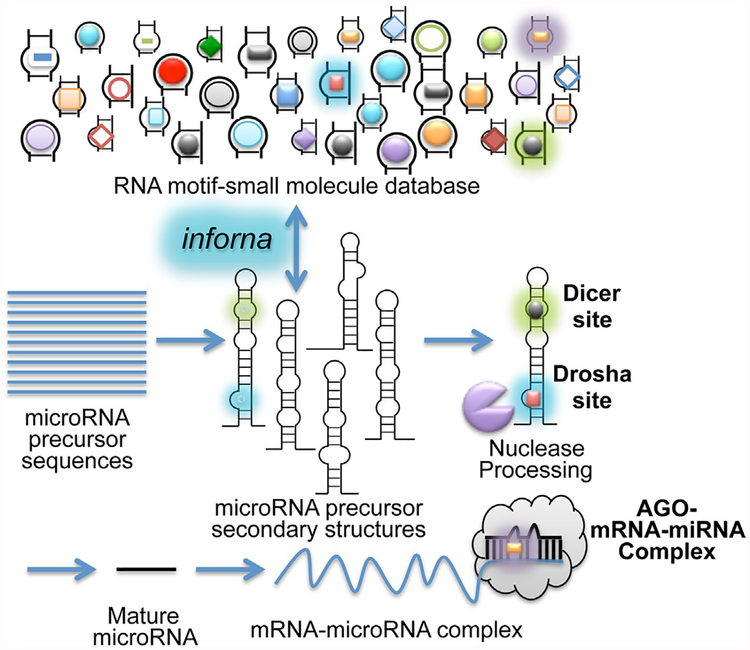
Decades of research has enabled annotation of folded structures within an RNA sequence by various methods including phylogenic comparison, free energy minimization with or without experimental constraints (Mathews et al., 2004), and a combination of conservation and free energy minimization (Mathews & Turner, 2002). Protocols to annotate RNA structure from sequence using free energy minimization are available (Mathews, 2014). By combining these advances with the RNA fold-small molecule interactions identified by 2DCS, we developed a lead identification strategy for structured RNAs named Inforna (Fig. 2) (Disney et al., 2016; Velagapudi, Gallo, & Disney, 2014).
Fig. 2.
Design of SMIRNA using Inforna, a lead identification strategy for RNA targets. Privileged RNA fold-small molecule interactions identified by 2DCS are annotated and deposited into a database. Inforna designs lead small molecules by comparing the folds in the database to folds in a cellular RNA target. Illustrated is implementation of Inforna to identify SMIRNAs for miRNA targets.
Inforna was validated by identifying folded structures in microRNA (miRNA) precursors that overlap with privileged interactions discovered by 2DCS (Velagapudi et al., 2014). MiRNAs comprise a class of non-coding (nc)RNAs that regulate gene expression and hence many biological processes including disease. They are produced as primary transcripts (pri-miRNA) that undergo two processing steps, one by the nuclear nuclease Drosha, which liberates a precursor (pre-) miRNA that is translocated to the cytoplasm, and the second by the cytoplasmic nuclease Dicer. This final mature miRNA regulates gene expression by binding to complementary 3′ untranslated regions present in a subset of mRNAs, suppressing the amount of protein that is translated (Bartel, 2004, 2009).
An analysis of the RNA structures that are presented by pri- and pre-miRNAs has provided answers to fundamental questions in RNA folding and motif redundancy (Liu et al., 2016). Among >1500 human miRNA precursors, both unique motifs, found only in a singular miRNA, and redundant motifs, found in many miRNAs, are present. A database of these and other folded structures in the human transcriptome are available in an on-line database (Liu et al., 2016).
In this first iteration of Inforna, a target agnostic approach was taken. That is, each human miRNA precursor, with a three dimensionally folded structure, was analyzed by Inforna for overlap with privileged RNA fold-small molecule interactions from 2DCS studies (Velagapudi et al., 2014). To refine the lead small molecules designed by Inforna, only those motifs present in the Drosha or Dicer processing sites in disease-associated miRNAs were further investigated. Over two dozen disease-associated miRNA hairpin precursors were found to be targetable with small molecules, and 44% of the lead small molecules reduced levels of its cognate miRNA in cells without lead optimization (Velagapudi et al., 2014). Importantly, Inforna provides insight into potential off-targets. That is, Inforna reports if: (i) a motif is present in multiple targets (and their location within the target); and (ii) a less optimal motif for the small molecule (binds with lower affinity) is present in another target.
Among the lead interactions designed by Inforna in this inaugural study, an optimal interaction with a Fitness Score of 100% (highest affinity interaction amongst 4096 RNA folds in the selection) was identified between a benzimidazole and the Drosha site in pri-miR-96 (Velagapudi et al., 2014). In vitro, the small molecule bound to the target with low micromolar affinity, bound to the predicted site as determined by nuclease protection assays, and inhibited Drosha processing of a model pri-miR-96 RNA (Velagapudi et al., 2014). In breast cancer cells, the compound increased the levels of the pri-miR-96 and diminished levels of pre-miR-96 and miR-96 at micro-molar concentration, bolstering compound mode of action as inhibition of Drosha processing as designed (Velagapudi et al., 2014). The pro-apoptotic transcription factor Forkhead Box O1 (FOXO1) is silenced by miR-96 in many cancers. This protein functions to transactivate Bcl-2-like protein 11 (Bim), a member of the B-cell lymphoma 2 (Bcl-2) protein family that promotes apoptosis. Application of the small molecule to breast cancer cells increased FOXO1 levels and triggered apoptosis. To dissect the mechanisms by which the small molecule triggered apoptosis, an siRNA against FOXO1 was delivered; reduction of FOXO1 ablated the activity of the pri-miR-96-targeting small molecule. Notably, a control siRNA had no effect on compound activity. Full miRNA profiling in cells showed that the compound is selective as only miR-96 levels were reduced to a statistically significant extent while an antagomir directed at miR-96 reduced levels of multiple miRNAs (Velagapudi et al., 2014).
To increase the potency of our lead pri-miR-96 targeting compound such that it could be applied in vivo, Inforna was used to define a fragment that could bind to a second motif within pri-miR-96 (Velagapudi et al., 2016). Thus, the lead small molecule could be tethered to the new fragment to target two sites simultaneously thereby enhancing affinity. The compound, named TargaprimiR-96 (“Targ” indicates targets and primiR-96 indicates the RNA target) has a low nanomolar affinity, selectively triggered apoptosis in triple negative breast cancer but not healthy breast (do not express miR-96) cells, and, importantly, decreased tumor load in a mouse model (Velagapudi et al., 2016). Thus, rational design can provide lead compounds that can be optimized and reduce tumor load in a preclinical mouse model.
Using Inforna, we have successfully designed a small molecule (TargapremiR-210) that inhibits processing of another cancer-associated miRNA, miR-210 (Costales et al., 2017; Velagapudi et al., 2014). In hypoxic cancers, miR-210 targets glycerol-3-phosphate dehydrogenase 1-like (GPD1L) mRNA to repress its translation. In a normoxic environment, GPD1L protein binds prolyl hydroxylase (PHD) to promote hyper-hydroxylation of hypoxia inducible factor 1-alpha (HIF-1α) to mediate its polyubiquitination and subsequent degradation by the proteasome (Huang et al., 2009; Mathew & Simon, 2009). At low oxygen concentrations, such as in solid breast cancer tumors, miR-210 represses GPD1L mRNA. This, in turn, decreases PHD activity, stabilizing cytoplasmic HIF-1α levels, all-owing for its dimerization with hypoxia inducible factor 1-beta (HIF-1β) in the nucleus, to form the active HIF-1 transcription factor and turn on hypoxia-associated genes. As miR-210 is among the genes upregulated by HIF-1, overexpressed miR-210 triggers a positive feedback loop to drive further miR-210 expression (Kelly, Souza, Clish, & Puigserver, 2011).
Application of TargapremiR-210 to hypoxic, but not normoxic, breast cancer cell lines silenced miR-210 specifically and short-circuited the miR-210-HIF-1α pathway. In particular, it increased expression of GPD1L, decreased expression of HIF-1α, and triggered apoptosis only in hypoxic breast cancer (no effect on normoxic breast cancer cells). Further, TargapremiR-210’s activity was reduced in hypoxic cells upon forced overexpression of miR-210. Notably, TargapremiR-210 inhibited hypoxic tumor growth in a mouse model (Costales et al., 2017).
Altogether, these studies show that organic ligands can be designed to tar-get RNA using only its sequence; the only other manner in which sequence-based design had been previously possible is via oligonucleotide-based approaches. These designed compounds can provide lead Targeted Therapeutics against previously thought undruggable RNA targets both in cells and in animal models. Notably, Inforna-designed compounds can be lead optimized using chemical similarity searching (Kumar et al., 2012; Parkesh et al., 2012), traditional medicinal chemistry approaches including defining structure-activity relationships (SAR), and targeting multiple structures within the same RNA with a single small molecule, or multivalency (Rzuczek et al., 2017, 2013; Velagapudi et al., 2016; Yang, Gao, Southern, Sarkar, & Disney, 2016).
3.2. Materials
Sequence of the desired RNA target or its .CT file.
Software to generate a .CT file of the RNA target’s secondary structure, which can be generated online by mFold (http://mfold.rna.albany.edu), RNAstructure (https://rna.urmc.rochester.edu/RNAstructureWeb/) or ViennaFold (http://rna.tbi.univie.ac.at/). RNAstructure can also be downloaded and run locally (https://rna.urmc.rochester.edu/register.html).
To use Inforna, visit https://disney.florida.scripps.edu/software/ and submit a software license agreement. Upon submission, log-in credentials will be provided.
3.3. Equipment
A computer with internet access and a web browser
3.4. Procedure
A detailed procedure for using Inforna to generate lead small molecules for an RNA target of interest is available in (Disney et al., 2016) in the Supporting Information. In brief:
Generate a .CT file, a connectivity table that describes the RNA’s secondary structure. Ensure that the sequence does not contain Ts (Us only).
Log in to Inforna using the credentials provided after submission of a software license agreement.
Upload the .CT file.
Choose search options, whether to force loop nuc ost stringent) or loop nucleotides only (least stringent).
Search the database and export the results as a.csv file. The results will include the motif in the RNA target, motif in the database, simplified molecular-input line-entry system (SMILES) notations for the lead small molecules, and Fitness Scores and related meta data.
4. Validating the targets of SMIRNAs in cells
4.1. Overview of target validation
Target validation is critical to define SMIRNA mode of action and to define occupied off-targets. Target validation methods for RNA, however, have generally not been developed (Childs-Disney & Disney, 2016) and have not been considered broadly when developing SMIRNAs. In the protein world, activity-based protein profiling (Cravatt, Wright, & Kozarich, 2008) and other cross-linking methods (Parker et al., 2017) have been used to validate targets. The most common method to identify RNA-small molecule binding sites is to monitor sites of protection from nuclease cleavage or chemical modification reagents. For example, in vitro mapping studies identified sites in the ribosome that bound antibiotics (Moazed & Noller, 1987; Stern, Moazed, & Noller, 1988). Some binding sites, however, can go undetected: (i) if nucleotides within a binding site do not react with a chemical modifying reagent; (ii) the irreversible nature of the reaction with the probe, which would require long SMIRNA residence time. These are not trivial issues and significantly complicate analysis and detection.
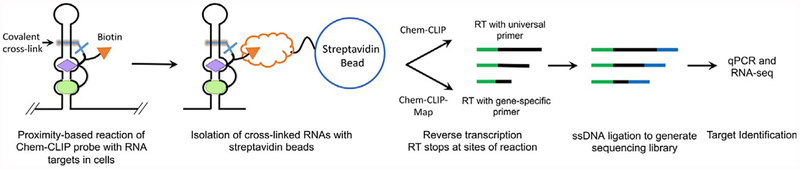
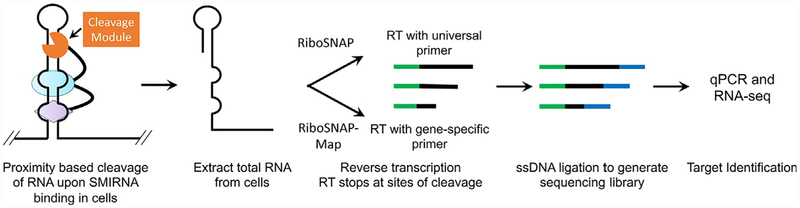
In this section, we describe the development and implementation of two target validation and profiling approaches for SMIRNAs that rely on cross-linking and cleavage, named Chemical Cross-Linking and Isolation by Pull-down (Chem-CLIP; Fig. 3) (Guan & Disney, 2013a; Yang, Wilson, Velagapudi, & Disney, 2015) and Small Molecule Nucleic Acid Profiling by Cleavage Applied to RNA (RiboSNAP; Fig. 4) (Li & Disney, 2018; Rzuczek et al., 2017), respectively. Indeed, both approaches have been further developed to map the binding sites of small molecules in cells. Because Chem-CLIP and RiboSNAP probes undergo a proximity-based reaction with the RNA target in a live cell, it mitigates complications observed with competitive chemical modification or nuclease protection discussed above.
Fig. 3.
Overview of the target validation and profiling method Chemical Cross-Linking and Isolation by Pull-down (Chem-CLIP). A Chem-CLIP probe is an SMIRNA appended with a cross-linking module (typically the nitrogen mustard chlorambucil) and a biotin purification module. Thus, RNA targets that bind the SMIRNA are brought into close proximity with the cross-linking module, forming a covalent adduct upon reaction. In brief, an appropriate cell line is incubated with the Chem-CLIP probe, followed by isolation of total RNA. RNAs that reacted with the Chem-CLIP probe are isolated and purified via its biotin tag and streptavidin beads. After elution of the adducts, the RNAs are reverse transcribed with either a universal primer (Chem-CLIP to identify all targets) or a gene-specific primer (Chem-CLIP-Map to identify the SMIRNA binding site). After RT, a single stranded DNA adaptor is ligated to enable generation of a sequencing library; qPCR and sequencing (RNA-seq) are then completed.
Fig. 4.
Overview of the target validation and profiling method Small Molecule Nucleic Acid Profiling by Cleavage Applied to RNA (RiboSNAP). A RiboSNAP probe is an SMIRNA appended with a cleavage module, which can be activated by light (such as N-HPT), recruit a nuclease to effect cleavage at the target site (RIBOTACs), or cleave the target directly (bleomycin). RNA targets that bind the SMIRNA bring the cleavage module into close proximity such that the RNA is cleaved. Briefly, cells are treated with a RiboSNAP probe and total RNA is isolated. In contrast to Chem-CLIP studies, total RNA can be analyzed directly, avoiding the pull-down step. Reverse transcription is either completed with a universal primer to identify all targets (RiboSNAP) or with a gene-specific primer to localize the SMIRNA binding site (RiboSNAP-Map). A sequencing library is then generated by ligation of a single stranded DNA adaptor and subsequent qPCR and RNA-seq analysis.
4.2. Chemical cross-linking and isolation by pull-down (Chem-CLIP)
The Chem-CLIP approach is a chemical method of cross-linking that is operationally similar to the technique described by Darnell and co-workers to define protein-RNA binding sites via Cross-linking and immunoprecipitation (CLIP) (Licatalosi et al., 2008; Ule et al., 2003). In Chem-CLIP, two functionalities are installed onto a SMIRNA; (i) a proximity-based chemical crosslinking unit; and (ii) a tag that allows for purification of cross-linked material (Fig. 3) (Guan & Disney, 2013a; Yang et al., 2015). These units must be installed in positions that do not significantly affect molecular recognition. The position used for covalent attachment to array surfaces in 2DCS studies or the scaffold used to link the fragments (i.e., a peptidomimetic such as a peptoid). After incubating cells with a Chem-CLIP probe, biotin is used to isolate and purify RNA targets that reacted with the cross-linker (pull-down) (Fig. 3). The targets are identified by RT-qPCR or RNA-seq.
The molecular recognition of TargapremiR-210 was studied in cells by using Chem-CLIP. In these studies, the interaction the SMIRNA with hundreds of cellular RNAs of different types was investigated (Costales et al., 2017). Indeed, the TargapremiR-210 Chem-CLIP probe bound the desired target, pre-miR-210, in cells to the greatest extent, as determined by measuring its enrichment in the pulled down fractions vs. lysate. Important control experiments included Competitive Chem-CLIP (C-Chem-CLIP) in which cells are co-treated with the Chem-CLIP probe and parent SMIRNA; the SMIRNA competes for binding to the RNA target and prevents reaction with the Chem-CLIP probe. C-Chem-CLIP studies with TargapremiR-210 showed that the parent SMIRNA bound pre-miR-210 in breast cancer cells. Furthermore, C-Chem-CLIP allows the relative occupancy of a series of SMIRNAs to a target (Costales et al., 2017; Disney et al., 2012; Yang et al., 2015).
By analysis of the pulled down material from a Chem-CLIP study, differential selectivity of a SMIRNA in cells can be assessed. As mentioned above, TargapremiR-210 engaged the desired target to the greatest extent, demonstrating specific binding. These studies revealed that other RNA targets were occupied, albeit to lesser extent, yet their biological function was unaffected. For example, the motif in pre-miR-210’s Dicer site is also present in pre-miR-497, and both are occupied by the SMIRNA. How-ever, only miR-210 abundance is affected because the binding site is located in a functional, Dicer processing site.
Chem-CLIP was also applied to fragment assembled compounds that target pri-miR-96 (Velagapudi et al., 2016) and to RNA repeat expansions including the r(CUG) repeat that causes myotonic dystrophy type 1 (Rzuczek et al., 2017), the r(CGG) repeat that causes fragile X-associated tremor ataxia syndrome (Yang et al., 2015, 2016), and the r(G4C2) repeat that causes C9orf72 amyotrophic lateral sclerosis and frontotemporal dementia (c9ALS/FTD) (Su et al., 2014; Wang et al., 2018). Interestingly, these studies showed that SMIRNAs selectively recognize mutant alleles containing the RNA repeat expansion. For the case of r(CUG)-targeting ligands, the SMIRNA targets disease driving repeats over other transcripts with shorter, non-toxic repeats of r(CUG), including the wild type allele (Rzuczek et al., 2017). Chem-CLIP has also been used to show that known drugs target microRNA precursors in cancer cells, supporting the notion that anti-cancer drugs target RNA and affect their biology (Velagapudi et al., 2018). Perhaps other drugs target RNA and affect its biology.
As mentioned above, Chem-CLIP has been further developed to map the binding sites of SMIRNAs in cells, or Chem-CLIP-Map (Fig. 3). In the first iteration, binding sites are revealed by completing a traditional Chem-CLIP experiment, digesting the captured RNAs with antisense oligonucle-otides and RNase H, and analyzing the fragments by RT-qPCR. This technique was developed for RNA repeat expansions as it is difficult for reverse transcriptase (RT) to read through the repeats and to PCR amplify. In multiple examples, this method localized the binding of an SMIRNA to its cognate repeat expansion (Disney et al., 2012; Rzuczek et al., 2017; Yang et al., 2015; Yang, He, et al., 2016). These binding sites gratifyingly are in accordance with Inforna-based design.
For other RNA targets, RT-PCR, including RNA-seq analysis, can be used to define cellular binding sites. In these experiments, the site that the probe reacts with in an RNA target can be detected as an “RT stop” (Sexton, Wang, Rutenberg-Schoenberg, & Simon, 2017; Velagapudi, Li, & Disney, 2019). Thus, pulled down RNA can be reverse transcribed with a gene-specific RT primer, followed by addition of adaptors and amplification via RT-PCR. Chem-CLIP-Map-Seq was validated with Targaprimir-96, demonstrating that the reaction site for the Chem-CLIP probe is proximal to the SMIRNA binding site and in accordance with Inforna design and modeling (Velagapudi et al., 2019).
4.2.1. Materials
Chem-CLIP probe
Disease-relevant cell line and corresponding growth medium
TRIzol reagent (Ambion)
miRNeasy Kit (Qiagen)
Quick-RNA Miniprep Kit (Zymo Research)
Dynabeads™ MyOne™ Streptavidin C1 (Thermo Fisher)
Ethanol and isopropanol
1 × Wash Buffer: 10mM Tris-HCl, pH 7.0, 1mM EDTA, 4M NaCl, 0.2% (v/v) Tween-20
1 × Elution Buffer: 10mM Tris-HCl pH 8.0, 5mM EDTA, 1mM bio-tin, and 1μL SUPERase In RNase Inhibitor (20units/μL; Thermo Fisher), 2.5μL Proteinase K
Gene specific primers
qScript cDNA Synthesis Kit (Quantabio)
miScript II RT kit (Qiagen)
Power SYBR Green Master Mix (Thermo Fisher Scientific)
Superscript™ III Reverse Transcription Kit (Thermo Fisher Scientific)
RNase H (2units/μL; New England Biolabs)
RNase A (10mg/mL; ThermoFisher Scientific)
Agencourt RNAClean XP (Beckman Coulter)
ssDNA adaptor: 5′ phosphate-NNNAGATCGGAAGAGCG-TCGTGTAG-3C spacer
T4 RNA Ligase I (NEB)
1mM ATP
50% PEG-8000
Phusion polymerase (NEB)
AMPure XP beads (Beckman Coulter)
Agencourt XP beads (Beckman Coulter)
PCR Cloning Kit (such as NEB® PCR Cloning Kit, catalog #: E1202)
4.2.2. Equipment
Clinical centrifuge
Microcentrifuge
qPCR thermocycler (such as an Applied Biosystem’s 7900HT Fast Real Time PCR System)
PCR thermocycler
Heat block, water bath, incubator
Nanodrop UV spectrophotometer
Magnetic rack
Gel electrophoresis equipment
Phosphorimager (such as Molecular Dynamics Typhoon; GE Healthcare Life Sciences)
4.2.3. Protocol (Fig. 3)
Culture the appropriate cell line in complete growth medium (100mm dishes) to about 70–80% confluency. Treat cells with appropriate Chem-CLIP probe in growth medium for 18–24h. Although the concentration of Chem-CLIP probe is likely unique for each target, we typically use the concentration that reduces levels of the mature miRNA by 50% in an appropriate cell line, as determined by RT-qPCR. In the case of miR-96, the concentration of the Chem-CLIP probe used was 1μM, where levels of mature miR-96 were reduced by 50% in MDA-MB-231 cells.
Extract total RNA using either TRIzol reagent, Quick-RNA Miniprep Kit, or miRNeasy Kit as appropriate per manufacturer’s protocol. Quantify the amount of RNA isolated using a Nanodrop spectrophotometer.
Using approximately 30μg of total RNA, pull down the RNAs that reacted with the Chem-CLIP probe by incubating with 50μL of Dynabeads MyOne Streptavidin C1 beads in 1 PBS and gently shaking for 30min at room temperature.
Separate the beads from the supernatant containing unbound RNAs by using a magnetic rack. Remove the supernatant.
Wash the beads containing RNA-compound adducts three times with 1 × Wash Buffer and then twice with nuclease free water.
Elute bound RNA in 200μL of 1 × Elution Buffer by heating at 37°C for 30min. Repeat this step, combining the fractions.
Purify the eluted RNA using Zymo’s Quick-RNA Miniprep Kit per the manufacturer’s protocol.
- Amplification
- Chem-CLIP:
- Reverse transcribe approximately 200–1000ng of total RNA, which can be completed using a miScript II RT Kit (for miRNAs) or a qScript cDNA Synthesis Kit (for longer RNAs) per the manufacturer’s protocol.
- Complete qPCR using Power SYBR Green Master Mix per the manufacturer’s recommended protocol and a Real Time PCR amplification thermocycler such as an Applied Biosystem’s 7900HT Fast Real Time PCR System. Normalize expression levels to a housekeeping gene or to 18S rRNA.
- Chem-CLIP-Map-Seq:
- Reverse transcription: Reverse transcribe ~500ng of pulled down RNA with a gene-specific primer with an adapter sequence at the 3′ end (GATCTCGCAGCTGCGGGTCCT) using Superscript III (SSIII) per the manufacturer’s protocol. Next, add 0.5μL of 2units/μL RNase H and 0.5μL of 10mg/mL RNase A and incubate at 37°C for 30min to degrade RNA. Clean up the RT reaction using an Agencourt RNAClean XP Kit per manufacturer’s protocol for small RNAs by adding 1.8 volume Agencourt RNAClean XP beads, 3 volumes 2.5M NaCl, 20% PEG-8000, and 1 volume isopropanol.
- ssDNA ligation: Ligate the purified RT reaction to a ssDNA adaptor (5′ phosphate-NNNAGATCGGAAGAGCGTCGTGTAG-3C spacer) using T4 RNA ligase I. In a 20μL total volume, add together 2μL 10 T4 RNA ligase buffer, 1μL 1Mm ATP, 10μL 50% PEG-8000, 5μL cDNA (from step i), 1μL of 20μM ssDNA adaptor, and 1μL of T4 RNA ligase and incubate overnight at room temperature. After overnight incubation, clean up using RNA Clean XP as described above in step i.
- Amplification: PCR amplify the cDNA from step ii using Phusion polymerase using the following primers: 5′-CTACACGACGCTCTTCCGATCT-3′ and 5′-CAGA-CGTGCTCTTCCGAT-3′. Clean up the PCR reaction using Ampure XP beads by adding 1.8 volume Agencourt Ampure XP beads, 3 volumes of 2.5 M NaCl, 20% PEG-8000, and 1 volume isopropanol.
- Generation of sequencing library: Generate a sequencing library using NEBNext® Multiplex Oligos for Illumina® (NEB) per the manufacturer’s recommended protocol.
- Sequence the library: Either clone the library generated in (iv) into a vector for sequencing or sequence directly with an Illumina® high throughput sequencer.
- Analyze the sequencing data by comparing probe-treated and vehicle-treated samples. In particular, calculate the percentage of sequencing reads that terminate at a given nucleotide in both data sets and then determined statistical significance using a two-tailed Student t-test or the like. Statistically significant differences identify the cross-linked region and hence the binding site.
4.3. Small molecule nucleic acid profiling by cleavage applied to RNA (Ribo-SNAP)
In the Chem-CLIP studies, target profiling is accomplished by pulling down cross-linked RNA targets. Pull-down and isolation of the targets can be laborious. Thus, we sought to develop an approach to profile the binding of SMIRNAs directly from total RNA harvested from cells by using small molecules that cleave an RNA target. This RiboSNAP approach can be implemented in three different ways, each of which has a subset of ideal targets (Fig. 4). Importantly, this approach has been used to demonstrate exquisite selectively for SMIRNAs in animal models. Compounds have cleaved a designed target and have not cleavage off-targets as assessed via RNA-seq. from mouse tissues (Angelbello et al., 2019).
In the first RiboSNAP method reported, a SMIRNA was appended with a cleavage module that is activated by light (Guan & Disney, 2013b). Briefly, 1-hydroxy-6-thioxo-1,6-dihydropyridine-2-carboxylic acid (N-HPT) was appended to an RNA binder. Upon irradiation with light, N-HPT produced hydroxyl radicals that cleaved the desired RNA (Guan & Disney, 2013b). As light is required for targeted cleavage, the tissues to which this approach can be applied are limited.
The second approach uses a small molecule to recruit an endogenous nuclease to the desired target to induce its cleavage. As the cleavage-inducing probe is comprised of RNA-binding and nuclease-recruiting modules, they were dubbed ribonuclease targeting chimeras, or RIBOTACs (Costales, Matsumoto, Velagapudi, & Disney, 2018). Because cleavage is affected by a recruited nuclease and not the small molecule itself, the sites of cleavage can be complex and distal to the SMIRNA binding site.
To alleviate issues encountered with light-activated cleavage modules (N-HPT) and complex cleavage patterns (RIBOTACs), we employed bleomycin A5 as cleaving module for RiboSNAP probes (Fig. 4). Bleomycin A5, a derivative of the well-known DNA cleavage anti-tumor agent bleomycin, has a free amine that drives DNA-binding affinity and cleavage. Fortuitously, this amine provides a conjugation site to a SMIRNA while reducing DNA affinity and cleavage. Indeed, conjugation of the cleaving module at this site allows for RNA specific cleavage in vitro and in cells (Li & Disney, 2018; Rzuczek et al., 2017).
We have validated using bleomycin A5 as the cleavage module, showing in both cases that conjugation to an RNA binder allows for selective cleavage. That is, it appears that conjugation of any RNA-binder to bleomycin A5’s free amine direct cleavage away from DNA and to the RNA target to which the SMIRNA binds. For example, when bleomycin A5 was appended to a dimeric SMIRNA that selectively recognizes r(CUG) repeat expansion, cleavage was not only selective for the mutant allele over wild type but also over transcripts with short, non-disease-causing r(CUG) repeats, mirroring the results obtained from the corresponding Chem-CLIP probe (Rzuczek et al., 2017). Likewise, conjugation of bleomycin A5 to Targaprimir-96 led to selective cleavage of pri-miR-96 in vitro and in cells. Further, profiling of all miRNAs expressed in triple negative breast cancer cells showed that only miR-96 was significantly affected (Li & Disney, 2018).
The exact binding sites of bleomycin RiboSNAP probes, or RiboSNAP-Map, can be determined similarly as described for Chem-CLIP-Map (Fig. 4). After incubation with the RiboSNAP probe, total RNA is harvested and subjected to gene-specific RT for the RNA target that was depleted in profiling experiments (e.g., pri-miR-96), followed by ligation of DNA adaptors to which PCR products are complementary. After PCR amplification, the products are sequenced, and binding sites are revealed where amplification stops. Akin to the results observed for Chem-CLIP-Map experiments, the binding site of Targaprimir-96 was proximal to the ligand binding site, in corroboration with Inforna design and three-dimensional modeling (Li & Disney, 2018). Below, we describe implementation of RiboSNAP using bleomycin A5 as the cleavage module to map SMIRNA binding sites.
4.3.1. Materials
Please see Section 4.2.1, except item #1 is a RiboSNAP probe
4.3.2. Equipment
Please see Section 4.2.2
4.3.3. Protocol (Fig. 4)
Culture the appropriate cell line in complete growth medium in 100mm dishes to ~70% confluency. Treat with a bleomycin RiboSNAP probe, prepared in growth medium, for 6h (optimized to obtain specific cleavage sites without destroying the target RNA and is likely specific to each target). Akin to Chem-CLIP studies described above, the concentration of RiboSNAP probe typically used reduces the cellular levels of the RNA of interest by ~50%, as determined by RT-qPCR. In the case of miR-96, the concentration of bleomycin RiboSNAP probe was chosen based on the above criterion, or 500nM in MDA-MB-231 cells.
Extract total RNA using TRIzol reagent per the manufacturer’s recommended protocol and quantify the amount present by using a Nanodrop spectrophotometer.
- Reverse transcription and PCR amplification
- RiboSNAP
- Reverse transcribe approximately 1μg of total RNA using a miS-cript II RT Kit (for miRNAs) or a qScript cDNA Synthesis Kit (for longer RNAs) per the manufacturer’s protocol.
- Complete qPCR using Power SYBR Green Master Mix per the manufacturer’s recommended protocol and Real Time PCR amplification thermocycler. Normalize expression levels to a housekeeping gene or to 18S rRNA.
- RiboSNAP-Map
- Reverse transcribe total RNA using a gene specific primer containing a 5′ adapter and SuperScript™ III. Anneal the primer by incubating the following at 65°C for 5min followed by ice for 5min: 10μg of total RNA, 2pmol primer, and 1μL of 10mM dNTP mix in a total volume of 13μL. Then, add 4μL 5 First-Strand Buffer, 1μL of 0.1M DTT, 1μL of RNaseOUT, and 1μL of SSIII RT. Incubate the samples at at 50°C for 1h and then at 85°C for 10min to inactivate the RT.
- Add 0.5μL of 2units/μL RNase H and 0.5μL of 10mg/mL RNase A to the RT reaction and incubate at 37°C for 30min to degrade the RNA.
- Follow steps (ii)–(iv) in Section 4.2.3, Chem-CLIP-Map-Seq to construct libraries for sequencing and hence to identify binding sites.
5. Summary
Herein, we have provided an overview for the design of small molecules that target RNA structures from sequence (Figs. 1 and 2). Indeed, many RNA structures are implicated in disease, particularly neurological diseases (Bernat & Disney, 2015). As oligonucleotides are best suited for targeting unstructured regions, small molecules provide an important complementary strategy for deactivating disease-causing RNAs. Essential to the development of SMIRNAs are target validation methods to confirm compound mode of action. Although these methods have been developed and implemented in the protein-targeting field, they have generally been lacking for RNA. A detailed description of two methods, Chem-CLIP (based on covalent cross-linking upon binding of the SMIRNA to the target; Fig. 3) and RiboSNAP (based on cleavage of the RNA target upon SMIRNA binding; Fig. 4), including advantages and limitations have been provided along with protocols for their implementation.
Acknowledgment
This work was funded by the National Institutes of Health Grants R01-GM097455, DP1-NS096898, and P01-NS099114 to M.D.D.
References
- Aminova O, & Disney MD (2010). A microarray-based method to perform nucleic acid selections. Methods in Molecular Biology, 669, 209–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelbello AJ, Chen JL, Childs-Disney JL, Zhang P, Wang ZF, & Disney MD (2018). Using genome sequence to enable the design of medicines and chemical probes. Chemical Reviews, 118(4), 1599–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelbello AJ, Rzuczek SG, Mckee KK, Chen JL, Olafson H, Cameron MD, et al. (2019). Precise small-molecule cleavage of an r(CUG) repeat expansion in a myotonic dystrophy mouse model. Proceedings of the National Academy of Sciences of the United States of America, 116(16), 7799–7804. 10.1073/pnas.1901484116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell, 116(2), 281–297. [DOI] [PubMed] [Google Scholar]
- Bartel DP (2009). MicroRNAs: Target recognition and regulatory functions. Cell, 136(2), 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernat V, & Disney MD (2015). RNA structures as mediators of neurological diseases and as drug targets. Neuron, 87(1), 28–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs-Disney JL, & Disney MD (2016). Approaches to validate and manipulate RNA targets with small molecules in cells. Annual Reviews of Pharmacology and Toxicology, 56, 123–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs-Disney JL, Wu M, Pushechnikov A, Aminova O, & Disney MD (2007). A small molecule microarray platform to select RNA internal loop-ligand interactions. ACS Chemical Biology, 2(11), 745–754. [DOI] [PubMed] [Google Scholar]
- Costales MG, Haga CL, Velagapudi SP, Childs-Disney JL, Phinney DG, & Disney MD (2017). Small molecule inhibition of microRNA-210 reprograms an oncogenic hypoxic circuit. Journal of the American Chemical Society, 139(9), 3446–3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costales MG, Matsumoto Y, Velagapudi SP, & Disney MD (2018). Small molecule targeted recruitment of a nuclease to RNA. Journal of the American Chemical Society, 140(22), 6741–6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cravatt BF, Wright AT, & Kozarich JW (2008). Activity-based protein profiling: From enzyme chemistry to proteomic chemistry. Annual Reviews of Biochemistry, 77, 383–414. [DOI] [PubMed] [Google Scholar]
- Disney MD, Dwyer BG, & Childs-Disney JL (2018). Drugging the RNA world. Cold Spring Harbor Perspectives in Biology, 10(11), a034769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disney MD, Liu B, Yang WY, Sellier C, Tran T, Charlet-Berguerand N, et al. (2012). A small molecule that targets r(CGG)exp and improves defects in fragile X-associated tremor ataxia syndrome. ACS Chemical Biology, 7(10), 1711–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disney MD, Winkelsas AM, Velagapudi SP, Southern M, Fallahi M, & Childs-Disney JL (2016). Inforna 2.0: A platform for the sequence-based design of small molecules targeting structured RNAs. ACS Chemical Biology, 11(6), 1720–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan L, & Disney MD (2013a). Covalent small-molecule-RNA complex formation enables cellular profiling of small-molecule-RNA interactions. Angewandte Chemie International Edition in English, 1(10), 201301639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan L, & Disney MD (2013b). Small molecule-mediated cleavage of RNA in living cells. Angewandte Chemie International Edition in English, 52(5), 1462–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Ding L, Bennewith KL, Tong RT, Welford SM, Ang KK, et al. (2009). Hypoxia-inducible mir-210 regulates normoxic gene expression involved in tumor initiation. Molecular Cell, 35(6), 856–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D, Metzger HJ, Schatz A, & Waksman SA (1944). Control of gram-negative bacteria in experimental animals by streptomycin. Science, 100(2588), 103–105. [DOI] [PubMed] [Google Scholar]
- Kelly TJ, Souza AL, Clish CB, & Puigserver P (2011). A hypoxia-induced positive feedback loop promotes hypoxia-inducible factor 1alpha stability through miR-210 suppression of glycerol-3-phosphate dehydrogenase 1-like. Molecular and Cellular Biology, 31(13), 2696–2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Parkesh R, Sznajder LJ, Childs-Disney JL, Sobczak K, & Disney MD (2012). Chemical correction of pre-mRNA splicing defects associated with sequestration of muscleblind-like 1 protein by expanded r(CAG)-containing transcripts. ACS Chemical Biology, 7(3), 496–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, & Disney MD (2018). Precise small molecule degradation of a noncoding RNA identifies cellular binding sites and modulates an oncogenic phenotype. ACS Chemical Biology, 13, 3065–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, et al. (2008). HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature, 456(7221), 464–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Childs-Disney JL, Znosko BM, Wang D, Fallahi M, Gallo SM, et al. (2016). Analysis of secondary structural elements in human microRNA hairpin precursors. BMC Bioinformatics, 17, 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew LK, & Simon MC (2009). Mir-210: A sensor for hypoxic stress during tumorigenesis. Molecular Cell, 35(6), 737–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews DH (2014). RNA secondary structure analysis using RNAstructure. Current Protocols in Bioinformatics, 46, [12.16.11–12.16.25]. [DOI] [PubMed] [Google Scholar]
- Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, & Turner DH (2004). Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proceedings of the National Academy of Sciences of the United States of America, 101(19), 7287–7292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews DH, & Turner DH (2002). Dynalign: An algorithm for finding the secondary structure common to two RNA sequences. Journal of Molecular Biology, 317(2), 191–203. [DOI] [PubMed] [Google Scholar]
- Moazed D, & Noller HF (1987). Interaction of antibiotics with functional sites in 16S ribosomal-RNA. Nature, 327(6121), 389–394. [DOI] [PubMed] [Google Scholar]
- Morrow T (2017). New therapy for spinal muscular atrophy offers modest bang for pharamaceutical buck. Managed Care, 26(2), 36–37. [PubMed] [Google Scholar]
- Parker CG, Galmozzi A, Wang Y, Correia BE, Sasaki K, Joslyn CM, et al. (2017). Ligand and target discovery by fragment-based screening in human cells. Cell, 168(3), 527–541. [e529]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkesh R, Childs-Disney JL, Nakamori M, Kumar A, Wang E, Wang T, et al. (2012). Design of a bioactive small molecule that targets the myotonic dystrophy type 1 RNA via an RNA motif-ligand database and chemical similarity searching. Journal of the American Chemical Society, 134(10), 4731–4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poehlsgaard J, & Douthwaite S (2005). The bacterial ribosome as a target for antibiotics. Nature Reviews Microbiology, 3(11), 870–881. [DOI] [PubMed] [Google Scholar]
- Rouskin S, Zubradt M, Washietl S, Kellis M, & Weissman JS (2014). Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature, 505(7485), 701–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzuczek SG, Colgan LA, Nakai Y, Cameron MD, Furling D, Yasuda R, et al. (2017). Precise small-molecule recognition of a toxic CUG RNA repeat expansion. Nature Chemical Biology, 13(2), 188–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzuczek SG, Gao Y, Tang ZZ, Thornton CA, Kodadek T, & Disney MD (2013). Features of modularly assembled compounds that impart bioactivity against an RNA target. ACS Chemical Biology, 8(10), 2312–2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton AN, Wang PY, Rutenberg-Schoenberg M, & Simon MD (2017). Interpreting reverse transcriptase termination and mutation events for greater insight into the chemical probing of RNA. Biochemistry, 56(35), 4713–4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitale RC, Flynn RA, Zhang QC, Crisalli P, Lee B, Jung JW, et al. (2015). Structural imprints in vivo decode RNA regulatory mechanisms. Nature, 519(7544), 486–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein CA, & Castanotto D (2017). FDA-approved oligonucleotide therapies in 2017. Molecular Therapy, 25(5), 1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson ML, & Zamecnik PC (1978). Inhibition of Rous sarcoma viral RNA translation by a specific oligodeoxyribonucleotide. Proceedings of the National Academy of Sciences of the United States of America, 75(1), 285–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern S, Moazed D, & Noller HF (1988). Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods in Enzymology, 164, 481–489. [DOI] [PubMed] [Google Scholar]
- Su Z, Zhang Y, Gendron TF, Bauer PO, Chew J, Yang WY, et al. (2014). Discovery of a biomarker and lead small molecules to target r(GGGGCC)-associated defects in c9FTD/ALS. Neuron, 83(5), 1043–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenson T, & Mankin A (2006). Antibiotics and the ribosome. Molecular Microbiology, 59(6), 1664–1677. [DOI] [PubMed] [Google Scholar]
- Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, & Darnell RB (2003). CLIP identifies Nova-regulated RNA networks in the brain. Science, 302(5648), 1212–1215. [DOI] [PubMed] [Google Scholar]
- Velagapudi SP, Cameron MD, Haga CL, Rosenberg LH, Lafitte M, Duckett DR, et al. (2016). Design of a small molecule against an oncogenic noncoding RNA. Proceedings of the National Academy of Sciences of the United States of America, 113(21), 5898–5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velagapudi SP, Costales MG, Vummidi BR, Nakai Y, Angelbello AJ, Tran T, et al. (2018). Approved anti-cancer drugs target oncogenic non-coding RNAs. Cell Chemical Biology, 25(9), 1086–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velagapudi SP, Gallo SM, & Disney MD (2014). Sequence-based design of bioactive small molecules that target precursor microRNAs. Nature Chemical Biology, 10(4), 291–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velagapudi SP, Li Y, & Disney MD (2019). A cross-linking approach to map small molecule-RNA binding sites in cells. Bioorganic & Medical Chemistry Letters, 29(12), 1532–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velagapudi SP, Luo Y, Tran T, Haniff HS, Nakai Y, Fallahi M, et al. (2017). Defining RNA-small molecule affinity landscapes enables design of a small molecule inhibitor of an oncogenic noncoding RNA. ACS Central Science, 3(3), 205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Qu K, Zhang QC, Flynn RA, Manor O, Ouyang Z, et al. (2014). Landscape and variation of RNA secondary structure across the human transcriptome. Nature, 505(7485), 706–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZF, Ursu A, Childs-Disney JL, Guertler R, Yang WY, Bernat V, et al. (2018). The hairpin form of r(G4 C2)exp in c9ALS/FTD is repeat-associated non-ATG translated and a target for bioactive small molecules. Cell Chemical Biology, 26, 179–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WY, Gao R, Southern M, Sarkar PS, & Disney MD (2016). Design of a bioactive small molecule that targets r(AUUCU) repeats in spinocerebellar ataxia 10. Nature Communications, 7 11647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WY, He F, Strack RL, Oh SY, Frazer M, Jaffrey SR, et al. (2016). Small molecule recognition and tools to study modulation of r(CGG)exp in fragile X-associated tremor ataxia syndrome. ACS Chemical Biology, 11(9), 2456–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WY, Wilson HD, Velagapudi SP, & Disney MD (2015). Inhibition of non-ATG translational events in cells via covalent small molecules targeting RNA. Journal of the American Chemical Society, 137(16), 5336–5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamecnik PC, & Stephenson ML (1978). Inhibition of Rous sarcoma virus replication and cell transformation by a specific oligodeoxynucleotide. Proceeding of the National Academy of Sciences of the United States of America, 75(1), 280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]