Abstract
Several studies suggest that upregulated expression of the long non-coding RNA HOX transcript antisense RNA (HOTAIR) is a negative predictive biomarker for numerous cancers. Herein, we performed a meta-analysis to further investigate the prognostic value of HOTAIR expression in diverse human cancers. To this end, a systematic literature review was conducted in order to select scientific studies relevant to the association between HOTAIR expression and clinical outcomes, including overall survival (OS), recurrence-free survival (RFS)/disease-free survival (DFS), and progression-free survival (PFS)/metastasis-free survival (MFS) of cancer patients. Collectively, 53 eligible studies including a total of 4873 patients were enrolled in the current meta-analysis. Pooled hazard ratios (HRs) with their corresponding 95% confidence intervals (CIs) were calculated to assess the relationship between HOTAIR and cancer patients’ survival. Elevated HOTAIR expression was found to be significantly associated with OS, RFS/DFS and PFS/MFS in diverse types of cancers. These findings were also corroborated by the results of bioinformatics analysis on overall survival. Therefore, based on our findings, HOTAIR could serve as a potential biomarker for the prediction of cancer patient survival in many different types of human cancers.
Keywords: HOTAIR, prognostic biomarker, survival, meta-analysis, cancer
1. Introduction
The long non-coding RNAs (lncRNAs) are non-protein-coding RNAs ≥ 200 bp in length, transcribed by RNA polymerase II. LncRNAs can be capped, polyadenylated and spliced, but they lack a functional open reading frame. It is estimated that approximately 27% (i.e., up to 60,000) of the annotated genes in the human genome encode lncRNAs, while the number of protein-coding genes ranges from 20,000 to 25,000 [1,2]. They are largely involved in a myriad of cellular functions, regulating gene expression at the transcriptional, post-transcriptional, and epigenetic level [1,3]. LncRNAs have emerged as critical components of cancer pathophysiology, being involved in one or more hallmarks of cancer, such as proliferation and metastasis [4,5]. They can act either as oncogenes or tumor suppressors, or indirectly through interaction with oncogenes and tumor suppressors, such as MYC proto-oncogene (MYC) and tumor protein p53 (TP53), respectively [4,5].
One of the most well-studied lncRNAs is HOX transcript antisense RNA (HOTAIR) which is located within the HOMEOBOX C (HOXC) gene cluster on chromosome 12q13.13 [6]. HOTAIR is 2158 bp long and consists of six exons. HOTAIR orthologs are restricted to eutherian mammals [7]. HOTAIR is known to bind to the Polycomb Repressive Complex 2 (PRC2) and the histone H3K4 demethylase LSD1, and serves as a scaffold to assemble these regulators at the HOXD gene cluster, where it establishes a transcriptionally repressive chromatin structure, thereby resulting in epigenetic repression of the HOXD gene locus [8]. HOTAIR has been shown to function as an oncogene since its expression is dysregulated in multiple types of cancers, including breast, lung, liver, renal, hepatocellular, gastric, nasopharyngeal, cervical, colorectal, bladder, pancreatic cancer, as well as melanoma, leukemia, etc. [9,10,11,12,13]. Furthermore, HOTAIR is suggested to promote cancer progression and contribute largely to cancer cell invasion and metastasis [14,15,16,17]. The multifunctional HOTAIR is implicated in the different aspects of cancer pathophysiology by regulating gene expression at the transcriptional, post-transcriptional, and epigenetic level [14,18,19,20]. Of note, several studies suggest that HOTAIR expression is highly predictive of cancer patient survival rates in diverse cancer types [21,22,23,24,25,26,27,28,29].
Herein, we conducted a comprehensive and updated meta-analysis to further investigate the prognostic value of HOTAIR expression for cancer patients. The potential clinical applications of our findings are also discussed towards the prognostic application of HOTAIR to multiple and different types of cancers.
2. Results
2.1. Study Selection and Charasteristics of Eligible Studies
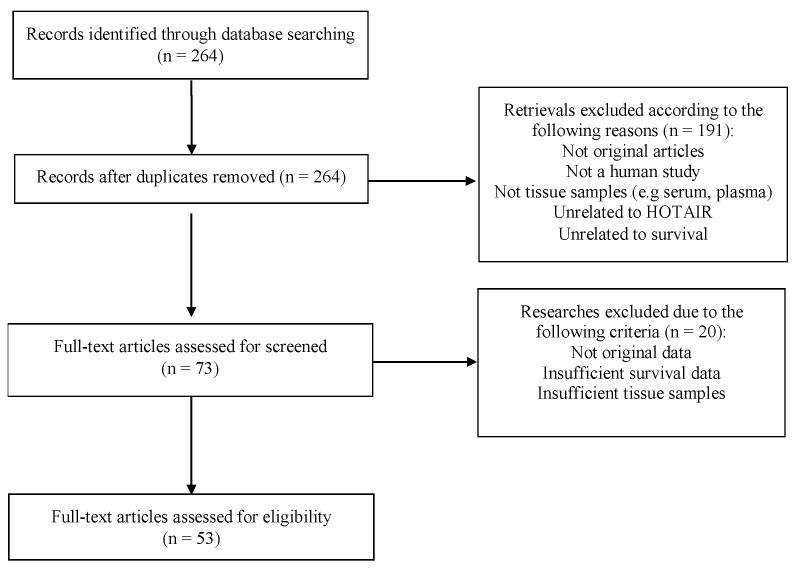
A total of 264 relevant published scientific studies were retrieved from the biomedical literature (up to 31 December 2018). According to the inclusion and exclusion criteria, 53 studies were ultimately included in this meta-analysis, as shown in Figure 1. The main characteristics of the included studies are summarized in Table 1, where the following information was recorded: first author’s surname; year of publication; country of origin; type of cancer; follow-up period (in months); total number of patients; detection assay for HOTAIR expression; HR and the corresponding 95% CI for overall survival (OS), recurrence-free survival (RFS), disease-free survival (DFS), progression-free survival (PFS), metastasis-free survival (MFS); survival data extraction method; and specimen type. Collectively, 4873 patients from 55 cohorts between 2010 and 2018 were included. The included studies reported a follow-up period ranging from 36 to 276 months. The level of HOTAIR expression was measured with quantitative reverse transcription polymerase chain reaction (qRT-PCR) in all of the included studies, except one where HOTAIR expression was estimated by microarrays (Table 1).
Figure 1.
Flow chart of the process for study selection.
Table 1.
Main characteristics of the studies included in the meta-analysis.
| Author, Year | Country | Cancer | Max. Follow-Up (Months) | Sample | Case Number | OS | DFS/RFS | MFS/PFS | Assay Method | Data Extraction Method | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| High Expression | Low Expression | Total | HR (95% CI) | p-Value | HR (95%CI) | p-Value | HR (95% CI) | p-Value | |||||||
| Gupta, 2010 [14] | USA | Breast Cancer | 240 | Tissue | 44 | 88 | 132 | 2.76 (1.45–3.3) | 0.036 | NM | NM | 3.53 (2.78–4.89) | 0.017 | qRT-PCR | K-M |
| Geng, 2011 [30] | China | HCC | 36 | Tissue | NM | NM | 50 | NM | NM | 2.24 (1.49–3.36) | 0,049 | NM | NM | qRT-PCR | K-M |
| Kogo, 2011 [31] | Japan | CRC | 60 | Tissue | 20 | 80 | 100 | 5.62 (1.52–9.57) | 0.008 | NM | NM | NM | NM | qRT-PCR | reported |
| Yang, 2011 [32] | China | HCC | 45 | Tissue | 32 | 28 | 60 | NM | NM | 3.56 (1.67–7.63) | 0.001 | NM | NM | qRT-PCR | reported |
| Lu, 2012 [33] | Italy | Breast Cancer | 108 | Tissue | NM | NM | 336 | 0.43 (0.21–0.89) | 0.022 | 0.47 (0.26–0.87) | 0.016 | NM | NM | qRT-PCR | reported |
| Niinuma, 2012 [34] | Japan | GIST | 200 | Tissue | 11 | 28 | 39 | 3.8 (0.7–21.2) | 0.123 | NM | NM | NM | NM | qRT-PCR | reported |
| Chen, 2013 [24] | China | ESCC | 60 | Tissue | 27 | 51 | 78 | 2.40 (1.35–4.28) | 0.003 | NM | NM | 2.34 (1.22–4.48) | 0.01 | qRT-PCR | reported |
| Endo, 2013 [17] | Japan | IGC | 68 | Tissue | 23 | 13 | 36 | 0.63 (0.34–1.86) | 0.137 | NM | NM | NM | NM | qRT-PCR | K-M |
| Endo, 2013 [17] | Japan | DGC | 60 | Tissue | 20 | 12 | 32 | 3.08 (1.77–5.35) | <0.01 | NM | NM | NM | NM | qRT-PCR | K-M |
| Ge, 2013 [35] | China | ESCC | 100 | Tissue | 90 | 47 | 137 | 3.16 (1.53–6.52) | 0.002 | NM | NM | 4.47 (1.99–10.06) | 0.001 | qRT-PCR | reported |
| Ishibashi, 2013 [36] | Japan | HCC | 36 | Tissue | 13 | 51 | 64 | 2.84 (1.91–4.58) | 0.041 | NM | NM | NM | NM | qRT-PCR | K-M |
| Li, 2013 [37] | China | LSCC | 60 | Tissue | 33 | 39 | 72 | 2.86 (1.15–7.07) | 0.023 | NM | NM | NM | NM | qRT-PCR | reported |
| Li, 2013 [38] | China | ESCC | 60 | Tissue | 30 | 70 | 100 | 1.91 (1.06–3.99) | 0.033 | NM | NM | NM | NM | qRT-PCR | reported |
| Liu, 2013 [39] | China | NSCLC | 60 | Tissue | 21 | 21 | 42 | 2.043 (0.91–4.58) | 0.048 | NM | NM | NM | NM | qRT-PCR | K-M |
| Lv, 2013 [40] | China | ESCC | 70 | Tissue | 49 | 44 | 93 | 1.67 (1.02–2.79) | 0.049 | NM | NM | NM | NM | qRT-PCR | K-M |
| Nakagawa, 2013 [21] | Japan | NSCLC | 50 | Tissue | 17 | 60 | 77 | NM | NM | 1.81 (1.09–3.74) | 0,047 | NM | NM | qRT-PCR | K-M |
| Nie, 2013 [41] | China | NPC | 82 | Tissue | 91 | 69 | 160 | 1.9 (1.13–3.19) | 0.012 | 1.41 (0.95–2.09) | 0.47 | 1.92 (1.11–3.31) | 0.018 | qRT-PCR | K-M |
| Sorensen, 2013 [42] | Denmark | Breast Cancer | 276 | Tissue | 79 | 85 | 164 | NM | NM | NM | NM | 1.75 (1.13–2.71) | 0.012 | Microarray | reported |
| Xu, 2013 [43] | China | Gastric cancer | 75 | Tissue | 56 | 27 | 83 | 0.47 (0.22–0.99) | 0.04 | NM | NM | NM | NM | qRT-PCR | reported |
| He, 2014 [44] | China | EC | 48 | Tissue | 62 | 83 | 145 | 3.04 (2.13–4.58) | 0.026 | NM | NM | NM | NM | qRT-PCR | K-M |
| Huang, 2014 [45] | China | Cervical cancer | 55 | Tissue | 109 | 109 | 218 | 2.86 (1.26–6.49) | 0.012 | NM | NM | NM | NM | qRT-PCR | reported |
| Lee, 2014 [46] | Korea | Gastric cancer | 48 | Tissue | 28 | 20 | 48 | NM | NM | 2.21 (0.53–9.16) | 0.141 | NM | NM | qRT-PCR | reported |
| Liu, 2014 [18] | China | Gastric cancer | 48 | Tissue | 39 | 39 | 78 | 2.7 (1.36–4.34) | 0.023 | NM | NM | NM | NM | qRT-PCR | K-M |
| Okugawa, 2014 [47] | Japan | Gastric cancer | 60 | Tissue | 77 | 73 | 150 | 1.77 (1.06–2.95) | 0.028 | NM | NM | NM | NM | qRT-PCR | reported |
| Qiu, 2014 [48] | China | EOC | 79 | Tissue | 32 | 32 | 64 | 1.87 (1.04–5.31) | 0.041 | 2.54 (1.18–5.45) | 0.034 | NM | NM | qRT-PCR | reported |
| Svoboda, 2014 [49] | Czech Republic | Colorectal cancer | 54 | Tissue | 36 | 37 | 73 | 4.46 (1.02–19.79) | 0.048 | NM | NM | NM | NM | qRT-PCR | reported |
| Wu, 2014 [50] | China | Colon Cancer | 72 | Tissue | 40 | 80 | 120 | 3.92 (1.23–12.50) | 0.021 | NM | NM | 3.88 (1.37–10.98) | 0.011 | qRT-PCR | K-M |
| Yan, 2014 [51] | China | Bladder Cancer | 60 | Tissue | 90 | 20 | 110 | 4.71 (2.89–8.71) | <0.001 | NM | NM | NM | NM | qRT-PCR | reported |
| Heubach, 2015 [52] | Germany | UHC | 200 | Tissue | 27 | 81 | 108 | 2.20 (1.23–3.93) | 0.008 | NM | NM | NM | NM | qRT-PCR | reported |
| Kim, 2015 [53] | Korea | Cervical cancer | 60 | Tissue | 89 | 22 | 111 | NM | NM | 5.28 (1.01–27.74) | 0,049 | NM | NM | qRT-PCR | reported |
| Liu, 2015 [54] | China | Gastric cancer | 40 | Tissue | 24 | 37 | 61 | NM | NM | 2.6 (1.74–3.89) | <0.001 | NM | NM | qRT-PCR | K-M |
| Ma, 2015 [55] | China | Gastric cancer | 60 | Tissue | 18 | 53 | 71 | 2.10 (1.10–4.03) | 0.022 | NM | NM | NM | NM | qRT-PCR | reported |
| Martinez-Fernandez, 2015 [56] | Spain | NMIBC | 38 | Tissue | 17 | 16 | 33 | NM | NM | NM | NM | 1.86 (0.58–5.96) | 0.296 | qRT-PCR | K-M |
| Martinez-Fernandez, 2015 [56] | Spain | NMIBC | 38 | Tissue | 30 | 33 | 63 | NM | NM | 3.78 (2.40–5.96) | <0.001 | NM | NM | qRT-PCR | K-M |
| Qiu, 2015 [57] | China | SOC | 96 | Tissue | 34 | 34 | 64 | 1.90 (1.01–3.56) | 0.046 | NM | NM | NM | NM | qRT-PCR | reported |
| Wu, 2015 [58] | China | OSCC | 60 | Tissue | 25 | 25 | 50 | 1.91 (1.33–2.74) | <0.001 | NM | NM | NM | NM | qRT-PCR | K-M |
| Wu, 2015 [59] | China | AML | 40 | Tissue | 52 | 33 | 85 | 3.37 (0.99–8.31) | 0.008 | 4.68 (2.81–7.79) | <0.001 | NM | NM | qRT-PCR | reported |
| Wu, 2015 [16] | China | OSCC | 96 | Tissue | 38 | 38 | 76 | 1.18 (0.68–2.84) | 0.03 | 1.11 (0.78–2.54) | 0.044 | NM | NM | qRT-PCR | reported |
| Xing, 2015 [60] | China | AML | 36 | Tissue | 68 | 68 | 136 | 2.03 (1.16–3.55) | 0.007 | 0.61 (0.37–1.00) | 0.034 | NM | NM | qRT-PCR | reported |
| Zhang, 2015 [61] | China | Gastric cancer | 45 | Tissue | 35 | 15 | 50 | 1.87 (1.46–2.1) | 0.028 | NM | NM | NM | NM | qRT-PCR | K-M |
| Zhao, 2015 [62] | China | Gastric cancer | 65 | Tissue | 84 | 84 | 168 | 1.47 (1.04–2.06) | 0.027 | NM | NM | NM | NM | qRT-PCR | reported |
| Luczak, 2016 [63] | Poland | EC | 96 | Tissue | 56 | 100 | 156 | 1.44 (0.81–3.19) | 0.03 | NM | NM | NM | NM | qRT-PCR | K-M |
| Luo, 2016 [64] | China | Colon cancer | 70 | Tissue | NM | NM | 80 | 1.99 (1.4–2.8) | <0.001 | NM | NM | NM | NM | qRT-PCR | K-M |
| Sun, 2016 [65] | China | Cervical cancer | 50 | Tissue | 49 | 10 | 59 | 1.31 (0.79–2.26) | 0.02 | NM | NM | NM | NM | qRT-PCR | K-M |
| Yan, 2016 [66] | China | DLBCL | 120 | Tissue | 25 | 25 | 50 | 3.13 (1.22–8.04) | 0.018 | NM | NM | NM | NM | qRT-PCR | reported |
| Zhang, 2016 [67] | China | Acute leukemia | 40 | Tissue | 19 | 77 | 96 | 2.41 (1.25–4.62) | 0.005 | NM | NM | NM | NM | qRT-PCR | K-M |
| Chen, 2017 [68] | China | Gastric cancer | 62 | Tissue | 33 | 32 | 65 | 1.99 (1.06–3.77) | 0.033 | NM | NM | NM | NM | qRT-PCR | reported |
| Hu, 2017 [69] | China | RCC | 50 | Tissue | 32 | 11 | 43 | 0.72 (0.20–2.55) | 0.62 | NM | NM | NM | NM | qRT-PCR | K-M |
| Katayama, 2017 [70] | Japan | RCC | 100 | Tissue | 21 | 43 | 64 | 1.82 (1.06–3.88) | 0.02 | NM | NM | NM | NM | qRT-PCR | K-M |
| Luan, 2017 [71] | China | MM | 60 | Tissue | 30 | 30 | 60 | 1.36 (0.79–2.83) | 0.01 | NM | NM | NM | NM | qRT-PCR | K-M |
| Xu, 2017 [72] | China | * EC | 36 | Tissue | 20 | 20 | 40 | 2.69 (1.14–6.33) | 0.032 | NM | NM | NM | NM | qRT-PCR | K-M |
| Zhang, 2017 [73] | China | Thyroid cancer | 60 | Tissue | NM | NM | 35 | 2.21 (1.38–3.54) | 0.001 | NM | NM | NM | NM | qRT-PCR | reported |
| Dong, 2018 [74] | China | Gastric cancer | 60 | Tissue | 22 | 10 | 32 | 2.26 (0.74–6.89) | 0.158 | NM | NM | NM | NM | qRT-PCR | K-M |
| Huang, 2018 [75] | China | Colorectal cancer | 110 | Tissue | 26 | 26 | 52 | 2.56 (0.91–7.35) | <0.01 | NM | NM | NM | NM | qRT-PCR | reported |
| Xiao, 2018 [76] | China | Colorectal cancer | 60 | Tissue | 52 | 52 | 104 | 1.45 (0.87–2.43) | 0.041 | NM | NM | NM | NM | qRT-PCR | K-M |
Abbreviations: OS, overall survival; RFS, recurrence-free survival; DFS, disease-free survival; MFS, metastasis-free survival; PFS, progression-free survival; HR, hazard ratio; CI, confidence interval; qRT-PCR, quantitative reverse transcription polymerase chain reaction; NM: not mentioned; K-M, Kaplan-Meier plot; AML, acute myeloid leukemia; CRC, colorectal cancer; DGC, diffuse gastric cancer; DLBCL, diffuse large B cell lymphoma; ESCC, esophageal squamous cell carcinoma; EC, endometrial carcinoma; EOC, epithelial ovarian cancer; * EC, esophageal cancer; GIST, gastrointestinal stromal tumors; HCC, hepatocellular carcinoma; IGC, intestinal gastric cancer; LSCC, laryngeal squamous cell carcinoma; MM, malignant melanoma; NSCLC, non-small cell lung cancer; NPC, nasopharyngeal carcinoma; NMIBC, non-muscle-invasive bladder cancer; OSCC, oral squamous cell carcinoma; RCC, renal cell carcinoma; SOC, serous ovarian cancer; and UHC, urothelial carcinoma.
2.2. Association between High HOTAIR Expression and Overall Survival in Diverse Cancers
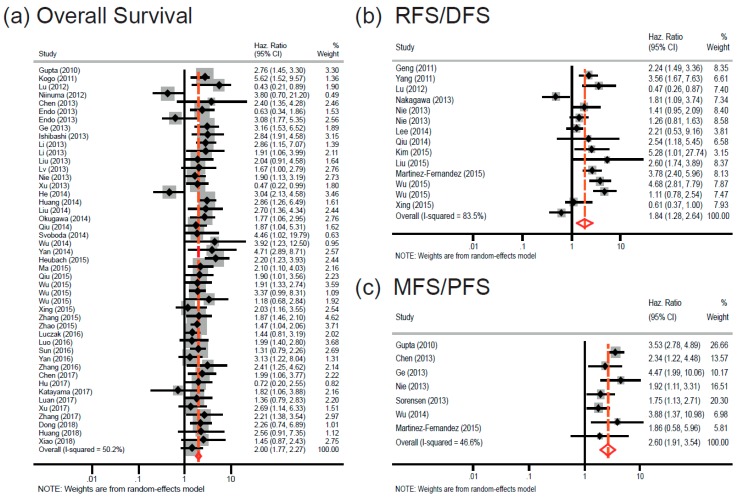
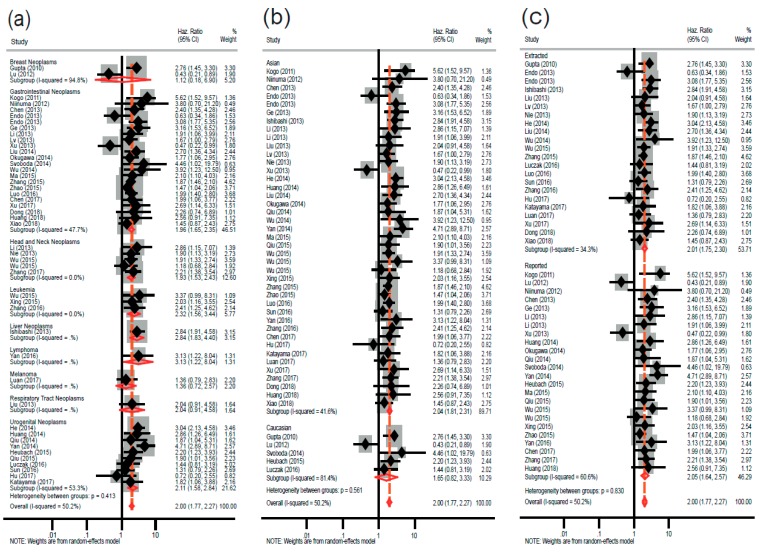
A total of 45 studies were included for overall survival (OS). We found a statistically significant relationship between elevated HOTAIR expression and poor OS (random-effects model: pooled HR = 2.00; 95% CI: 1.77–2.27; p < 0.001), with marginally moderate heterogeneity (I2 = 50.2%; Ph < 0.001) (Figure 2a). Subgroup analyses were performed based on the type of cancers, ethnic group, and data extraction method (Figure 3). When the studies were classified based on major cancer types (according to NCBI’s medical subject headings (MeSH) [77]), a significant association was found between HOTAIR overexpression and poorer OS in solid cancers, such as gastrointestinal cancers (fixed-effects model: pooled HR = 1.96; 95% CI: 1.65–2.35; p < 0.001), liver cancers (fixed-effects model: pooled HR = 2.84; 95% CI: 1.83–4.40; p < 0.001), head and neck cancers (fixed-effects model: pooled HR = 1.93; 95% CI: 1.53–2.43; p < 0.001), and urogenital cancers (random-effects model: pooled HR = 2.11; 95% CI: 1.58–2.84; p < 0.001), as well as liquid cancers, including leukemia (fixed-effects model: pooled HR = 2.32; 95% CI: 1.56–3.44; p < 0.001) and lymphoma (fixed-effects model: pooled HR = 3.13; 95% CI: 1.22–8.04; p < 0.001). Of note, the heterogeneity was reduced significantly in the individual cancer types (Figure 3a). In the subgroup analysis based on ethnicity, a statistically significant worse OS was observed for Asians (fixed-effects model: pooled HR = 2.04; 95% CI: 1.81–2.31; p < 0.001). Regarding the Caucasian subgroup, despite the relatively high HR, the relationship cannot be considered robust because the p-value is slightly higher that the cutoff value (random-effects model; pooled HR = 1.65; 95% CI: 0.82–3.33; p = 0.077) (Figure 3b). In stratified analysis, according to data extraction method, HOTAIR was found to have a significant prognostic value irrespectively of the data source. that is, the HR reported in the articles (random-effects model: pooled HR = 2.05; 95% CI: 1.64–2.57; p < 0.001) or extracted from the survival curves (fixed-effects model: pooled HR = 2.01; 95% CI: 1.75–2.30; p < 0.001) (Figure 3c).
Figure 2.
Forest plots of combined analyses on the association of survival with HOTAIR expression. (a) Forest plot of OS analysis, (b) forest plot of RFS/DFS analysis, and (c) forest plot of MFS/PFS analysis. Abbreviations: HR, Hazard ratio; OS, overall survival; RFS, recurrence-free survival; DFS, disease-free survival; MFS, metastasis-free survival; and PFS, progression-free survival.
Figure 3.
Forest plots of combined analyses for overall survival (OS) associated with HOTAIR expression in different groups. (a) Forest plot for different types of cancers, (b) forest plot for different ethnic groups, and (c) forest plot for different data extraction methods.
2.3. HOTAIR Overexpression Is Associated with Cancer Recurrence and Progression
To investigate the relationship between HOTAIR expression and cancer recurrence or relapse, the recurrence-free survival (RFS) and disease-free survival (DFS) studies were combined; collectively accounting for 14 studies. Increased HOTAIR expression was found to be strongly related to cancer recurrence (pooled HR = 1.84; 95% CI = 1.28–2.64; p = 0.001). A random-effects model was applied because of the high heterogeneity (I2 = 83.5%; Ph < 0.001) across studies (Figure 2b).
Furthermore, there are seven studies for combined metastasis-free survival (MFS) and progression-free survival (PFS). Of importance, high HOTAIR expression was predicted to be associated significantly with worse MFS/PFS (pooled HR = 2.60; 95% CI: 1.91–3.54; p < 0.001). A fixed-effects model was used because of the relatively low heterogeneity (I2 = 46.6%; Ph = 0.081) (Figure 2c).
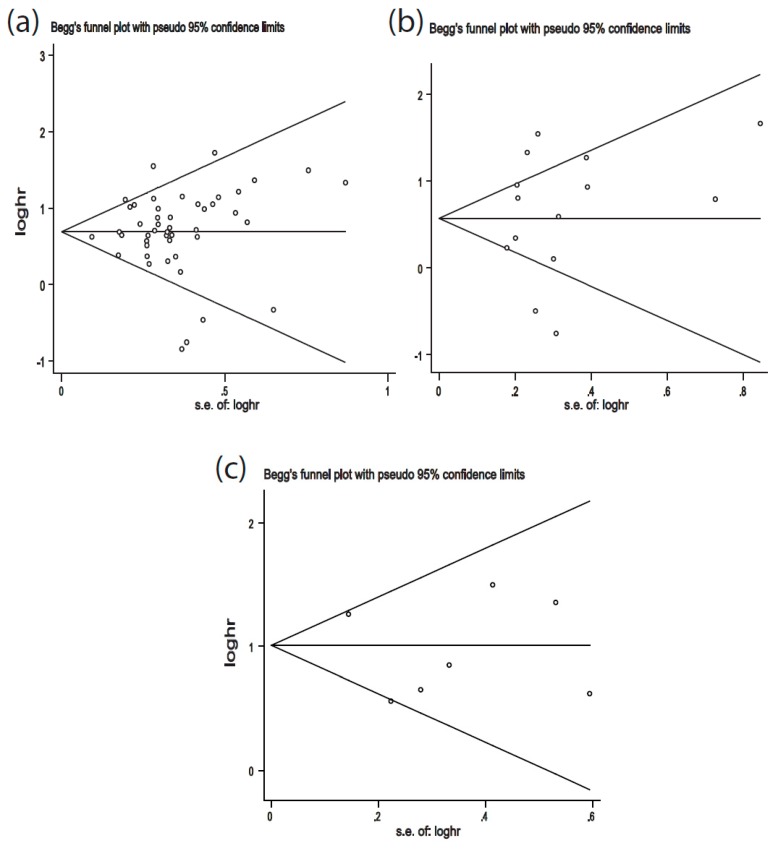
2.4. Publication Bias
Publication bias was detected by Begg’s funnel plot and Egger’s test. There was no obvious asymmetry in Begg’s funnel plots of OS, RFS/DFS, and MFS/PFS (Figure 4). Additionally, the p-values of Egger’s tests were all greater than 0.05, indicating no potential publication bias (OS: p = 0.73; RFS/DFS: p = 0.70; MFS/PFS: p = 0.64).
Figure 4.
Begg’s funnel plots of publication bias. (a) Begg’s funnel plot of publication bias for OS; (b) Begg’s funnel plot of publication bias for RFS/DFS; (c) Begg’s funnel plot of publication bias for MFS/PFS. Each circle represents a separate study.
2.5. Sensitivity Analysis
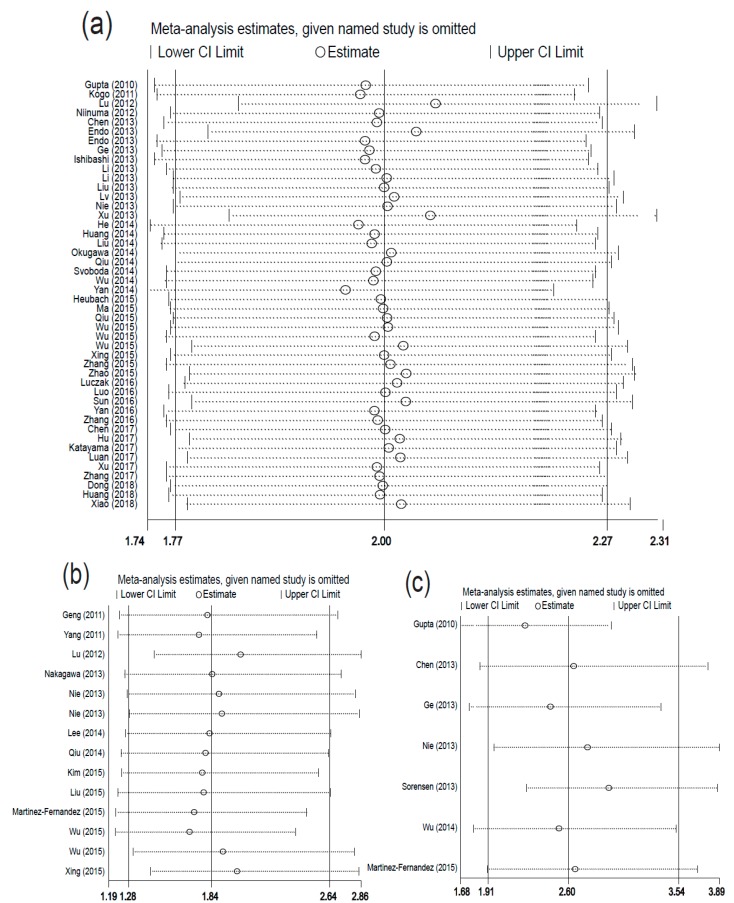
Sensitivity analyses did not indicate alterations in the results due to the inclusion of any individual study (Figure 5), that is, no single study affected the pooled HR or 95% CI.
Figure 5.
Sensitivity analysis of each eligible study. (a) OS individual studies, (b) RFS/DFS individual studies and (c) MFS/PFS individual studies.
2.6. TCGA-Derived Survival Curves
To further the clinical relevance of our work and HOTAIR importance, we explored the possibility for any association of the HOTAIR expression to overall cancer survival. It was found that HOTAIR overexpression was significantly associated with worse OS in adrenocortical carcinoma (ACC), mesothelioma (MESO), and glioblastoma multiforme (GBM) (Figure S1).
3. Discussion
HOTAIR exhibits pro-oncogenic activity since it has been shown to be overexpressed in numerous cancers and be implicated in several hallmarks of cancer, such as cellular proliferation, inhibition of apoptosis, genomic instability, angiogenesis, invasion, and metastasis [19,20].
In the current study, an updated, comprehensive meta-analysis on the prognostic value of HOTAIR in various human cancers was presented. By applying stringent inclusion and exclusion criteria, we included 53 eligible studies, a relatively large number necessary for a meta-analysis to be considered robust. Previous meta-analyses on the association of HOTAIR with clinical outcome have included a rather limited number of studies with inconclusive and inconsistent findings [28,29]. Other related studies have focused on certain types of cancers, such as head and neck squamous cell carcinoma [22] or digestive system cancers [55,78,79].
In the present study, we showed that there is a statistically significant relationship between elevated HOTAIR expression and poor OS. In the subgroup analysis, based on cancer type, HOTAIR was shown to be a significant predictor for worse prognosis for a variety of cancers, including solid cancers, such as urological cancers, head and neck neoplasms, cancers of the digestive system, and several female cancers (e.g., cervical, ovarian, and endometrial cancers), as well as the blood cancers, lymphoma and leukemia. Moreover, we complemented the findings from meta-analysis and further strengthened our hypotheses with survival information from other types of cancers, for which there were not any available eligible studies, retrieved from TCGA. It was found that there is, also, a strong relationship between HOTAIR overexpression and poor OS in neoplasms of the adrenal cortex, mesothelial neoplasms, and neuroepithelial tumors.
Taken together, the above findings lead to the suggestion that similar HOTAIR-mediated pathways might be implicated both in solid and liquid cancers [13]. In particular, in several solid tumors, HOTAIR has been shown to exert its oncogenic and metastatic potential by mediating a repressive chromatin structure through the recruitment of histone-modifying or chromatin-remodeling complexes, such as PRC2 [14,16,31]. For example, HOTAIR can promote pancreatic cancer cell proliferation by suppressing the expression of miR-663b via remodeling the chromatin structure within the miR-663b promoter [80]. In a recent study, HOTAIR was also found to recruit PRC2 to catalyze H3K27 trimethylation to transcriptionally repress E-cadherin and promote EMT in gastric cancer [81]. Similarly, high expression levels of HOTAIR and PRC2 proteins (H3K27 methylase EZH2, SUZ12, and EED) were found to be positively correlated with lymphomagenesis [82]. In addition, HOTAIR, through miRNA sponging, contributes to carcinogenesis both in blood [60] and solid tumors [83,84]. However, there is a rather limited number of studies available on major cancers, such as breast neoplasms and respiratory tract cancers. Thus, more clinical trials on these cancers would enable us to better assess the relationship between HOTAIR expression and cancer patients’ survival.
A positive correlation between HOTAIR and CDKN1A (p21) expression levels was also found (Figure S2), suggesting a possible functional and/or physical association between HOTAIR and CDKN1A (p21) in cancer pathophysiology. From a clinical perspective, there is an emerging role of CDKNIA (p21), especially in cases where p53 is mutated like in many different solid tumors. The role of p21 has been extensively viewed as an indicator of wildtype p53 activity [85]. However, recent evidence suggests that upregulated p21 can also act as an oncogenic factor in a p53-deficient environment, thereby driving a subset of atypical cancerous cells to more chemoresistant and aggressive phenotypes [86]. Therefore, we cannot exclude a possible mechanistic association between HOTAIR and p21 towards the negative regulation of target genes and a potential role in OS. Interestingly, recent studies have shown that HOTAIR expression was significantly higher in non-small-cell lung cancer (NSCLC) tissues compared to the adjacent normal tissues, and HOTAIR was negatively associated with p53 functionality rather than p53 expression [87]. In addition, HOTAIR, p21, and p53 mRNA expression in doxorubicin- or γ rays-treated oral squamous cell carcinoma (OSCC) cells was up-regulated, indicating that the DNA damage response includes HOTAIR upregulation and may be closely connected to p53 and p21 expression and/or functionality [88].
To investigate any possible effect of the genetic background and environment on the overall HRs, analyses were conducted based on the ethnic background of the participants. HOTAIR was found to be a powerful negative prediction biomarker for Asians. In the case of Caucasians, there was a link between HOTAIR overexpression and poor OS, albeit with moderate statistical significance; this is probably due to the relatively low number of available studies on patients of Caucasian origin. There were not, also, any available studies for other major ethnic groups, such as Africans or Indians, which would have further allowed us to estimate the influence of the genetic make-up on the association between HOTAIR and clinical outcome. The overall effect was similar in the stratified analysis according to data source, that is, the estimated HR reported in the articles or extrapolated from survival curves.
Therefore, high HOTAIR expression can predict an unfavorable clinical outcome in different types of cancers and possibly ethnic groups using different extraction methods. Notably, elevated expression of HOTAIR and prognosis in cancer patients is not particularly affected either by cancer type or even the patients’ genetic background.
HOTAIR was found to be a poor predictor for both cancer recurrence and progression. The similar outcomes suggest that there are similar HOTAIR-dependent mechanisms underlying these two phenomena. In particular, HOTAIR was shown to mediate recurrence and progression in bladder cancer via the histone methyltransferase EZH2 [56]. Similarly, enhanced HOTAIR expression was found to be associated both with progression and tumor recurrence in hepatocellular carcinoma by regulating the Wnt/β-catenin signal transduction pathway [89].
HOTAIR has been demonstrated to promote tumor cell invasion and metastasis by modulating epithelial-to-mesenchymal transition (EMT) [16,46,90]. Enhanced HOTAIR expression has also been shown to promote metastasis and invasion through different mechanisms including genome-wide re-targeting of PRC2 and subsequent epigenetic silencing of multiple anti-metastatic genes [14], inhibition of the expression of the metastasis suppressor gene E-cadherin by recruiting the histone methyltransferase of PRC2, EZH2 [16,90], targeting of Notch/Wnt signaling pathway-associated genes [91], and upregulating chondroitin sulfotransferase CHST15 [92], etc. HOTAIR also promotes invasion and migration by acting as a ‘miRNA sponge’, through targeting the corresponding miRNAs in the miR-1/CCND2 [93], miR-148a/SNAIL2 [72], and miR-23b/MAPK1 [94] axes.
Heterogeneity was observed within the forest plots of OS and RFS/DFS, suggesting that HRs vary across studies. For this reason, the random-effects model was applied, where the overall HR was estimated based on the weighted average of the HRs of the individual studies. Given that the overall effect for OS and RFS/DFS was not affected by any single study, according to sensitivity analyses, we could suggest that, despite heterogeneity, the pooled HR can be considered quite reliable and representative.
Moreover, potential publication bias was not detected in the present meta-analysis, probably due to the sufficient representation of eligible studies in this meta-analysis.
4. Materials and Methods
4.1. Search Strategy and Study Eligibility Criteria
This systematic review and meta-analysis was conducted by following strictly the PRISMA (preferred reporting items for systematic reviews and meta-analyses) guidelines [95].
The bibliographic database PubMed/MEDLINE [96] was manually searched for published scientific studies on the associations between HOTAIR expression and prognosis in different types of cancers by using combinations of the relevant keywords: (“HOTAIR” OR “HOX transcript antisense RNA” or “HOXC cluster antisense RNA 4” or “HOXC-AS4” OR “HOXC11-AS1”) and (“cancer” or “carcinoma” or “tumor” or “neoplasm” or “malignancy”) and (“prognosis” or “survival” or “outcome” or “mortality” or “death”). The studies had to fulfill the following inclusion criteria so as to be considered eligible: (1) studies of human clinical trials, (2) studies including more than 30 patients in total, (3) the correlation between HOTAIR expression and cancer patients’ survival was estimated, (4) availability of HR and 95% confidence interval (CI) or survival curves or sufficient data to calculate HR and 95% CI, (5) quantitative measurement (e.g., qPCR) of HOTAIR expression in cancers was included, and (6) studies published in English. Accordingly, the studies were excluded on the basis of the following exclusion criteria: (1) laboratory studies on animal models or cell lines; (2) reviews, meta-analyses, editorials, case reports, commentaries, unpublished data; (3) lack of sufficient data to estimate HR and 95% CI; and (4) samples other than tissue (e.g., blood, serum).
4.2. Study Selection, Data Extraction, and Quality Assessment
All potential studies were independently retrieved from the literature by two of the authors (H.I.T. and D.O.). Quality assessment of the studies was performed by H.I.T. and D.O. independently. Any disagreement was resolved by a third investigator (A.P.). Relevant data were extracted from the included studies and recorded into an ad hoc Excel worksheet. In the case that the HR was not reported in the corresponding article, the data were extracted from the graphical survival plots (i.e., Kaplan-Meier curves) by using the Engauge Digitizer v10.11 software, as previously described [97].
4.3. Statistical Analyses
All statistical analyses were performed with STATA statistical software version 13.0 (Stata Corporation, College Station, TX, USA) and Microsoft Excel. The heterogeneity among the included studies was estimated by Higgins I-squared (I2) statistic as follows: I2 < 25%; no heterogeneity; 25% < I2 < 50%: low heterogeneity; 50% < I2 < 75%: moderate heterogeneity; I2 >75% high heterogeneity [98,99]. In the case of statistically significant heterogeneity (I2 > 50% and Ph < 0.05), a random-effect model was applied, otherwise a fixed-effect model [100,101] was used. Sensitivity analysis was performed by consecutive omission of individual studies to verify the consistency of outcomes. Potential publication bias was detected by Begg’s funnel plot [102] and Egger’s test [103]; a p-value less than 0.05 was indicative of statistically significant publication bias.
4.4. Bioinformatics Analysis
4.4.1. Survival Analysis
Overall survival curves for different types of cancers were retrieved through the online tool GEPIA (Gene Expression Profiling Interactive Analysis) [104], which provides survival analysis based on datasets obtained from The Cancer Genome Atlas (TCGA) (https://tcga-data.nci.nih.gov).
4.4.2. Correlation Analysis
Correlation analysis between gene expression levels was performed through the web-based tool GEPIA [104] which analyzes gene expression based on RNA sequencing (RNA-Seq) data from TCGA.
5. Conclusions
In this study, we have performed a meta-analysis complemented with bioinformatics analyses towards investigating the prognostic potential of the prominent lncRNA HOTAIR in cancer. On the basis of our findings, HOTAIR represents a potential powerful predictor of prognosis of overall survival, cancer recurrence, progression, and metastasis in multiple and diverse types of cancers. Therefore, HOTAIR could be applied in the clinical setting as a universal biomarker for monitoring cancer patient survival.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6694/11/6/778/s1, Figure S1: Kaplan-Meier plots depicting the prognostic potential of HOTAIR for OS in various types of cancers. (A) ACC; (B) MESO and (C) GBM. The corresponding HRs and p-values are indicated. The CIs are denoted by dashed lines, Figure S2: Correlation between HOTAIR and CDKN1A expression.
Author Contributions
Conceptualization, A.P.; methodology, H.I.T., D.O and A.P.; software, H.I.T, D.O. and P.I.K.; validation, H.I.T, D.O. and A.P.; formal analysis, H.I.T, D.O. and A.P.; investigation, H.I.T, D.O. and A.P.; data curation, H.I.T, D.O., P.I.K. and A.P.; writing—original draft preparation, H.I.T, D.O., P.I.K., A.G.G., and A.P.; writing—review and editing, H.I.T, D.O., P.I.K., A.G.G. and A.P.; supervision, A.P.; project administration, A.P.
Funding
P.I.K. acknowledges support of this work by the project “ELIXIR-GR: The Greek Research Infrastructure for Data Management and Analysis in Life Sciences”, Grant Number (MIS) 5002780.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Nagano T., Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145:178–181. doi: 10.1016/j.cell.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 2.Rinn J.L., Chang H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Geisler S., Coller J. RNA in unexpected places: Long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Biol. 2013;14:699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prensner J.R., Chinnaiyan A.M. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmitt A.M., Chang H.Y. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rinn J.L., Kertesz M., Wang J.K., Squazzo S.L., Xu X., Brugmann S.A., Goodnough L.H., Helms J.A., Farnham P.J., Segal E., et al. Functional demarcation of active and silent chromatin domains in human hox loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cunningham F., Achuthan P., Akanni W., Allen J., Amode M.R., Armean I.M., Bennett R., Bhai J., Billis K., Boddu S., et al. Ensembl 2019. Nucleic Acids Res. 2019;47:D745–D751. doi: 10.1093/nar/gky1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsai M.C., Manor O., Wan Y., Mosammaparast N., Wang J.K., Lan F., Shi Y., Segal E., Chang H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai B., Song X.Q., Cai J.P., Zhang S. Hotair: A cancer-related long non-coding RNA. Neoplasma. 2014;61:379–391. doi: 10.4149/neo_2014_075. [DOI] [PubMed] [Google Scholar]
- 10.Woo C.J., Kingston R.E. Hotair lifts noncoding RNAs to new levels. Cell. 2007;129:1257–1259. doi: 10.1016/j.cell.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 11.Hao S., Shao Z. Hotair is upregulated in acute myeloid leukemia and that indicates a poor prognosis. Int. J. Clin. Exp. Pathol. 2015;8:7223–7228. [PMC free article] [PubMed] [Google Scholar]
- 12.Qu X., Alsager S., Zhuo Y., Shan B. Hox transcript antisense RNA (hotair) in cancer. Cancer Lett. 2019;454:90–97. doi: 10.1016/j.canlet.2019.04.016. [DOI] [PubMed] [Google Scholar]
- 13.Bhan A., Mandal S.S. LncRNA hotair: A master regulator of chromatin dynamics and cancer. Biochim. Biophys. Acta. 2015;1856:151–164. doi: 10.1016/j.bbcan.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta R.A., Shah N., Wang K.C., Kim J., Horlings H.M., Wong D.J., Tsai M.C., Hung T., Argani P., Rinn J.L., et al. Long non-coding RNA hotair reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng P., Yin Z., Wu Y., Xu Y., Luo Y., Zhang T.C. LncRNA hotair promotes cell migration and invasion by regulating mkl1 via inhibition mir206 expression in hela cells. Cell Commun. Signal. 2018;16:5. doi: 10.1186/s12964-018-0216-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Y., Zhang L., Zhang L., Wang Y., Li H., Ren X., Wei F., Yu W., Liu T., Wang X., et al. Long non-coding RNA hotair promotes tumor cell invasion and metastasis by recruiting ezh2 and repressing e-cadherin in oral squamous cell carcinoma. Int. J. Oncol. 2015;46:2586–2594. doi: 10.3892/ijo.2015.2976. [DOI] [PubMed] [Google Scholar]
- 17.Endo H., Shiroki T., Nakagawa T., Yokoyama M., Tamai K., Yamanami H., Fujiya T., Sato I., Yamaguchi K., Tanaka N., et al. Enhanced expression of long non-coding RNA hotair is associated with the development of gastric cancer. PLoS ONE. 2013;8:e77070. doi: 10.1371/journal.pone.0077070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu X.H., Sun M., Nie F.Q., Ge Y.B., Zhang E.B., Yin D.D., Kong R., Xia R., Lu K.H., Li J.H., et al. Lnc RNA hotair functions as a competing endogenous RNA to regulate her2 expression by sponging mir-331-3p in gastric cancer. Mol. Cancer. 2014;13:92. doi: 10.1186/1476-4598-13-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hajjari M., Salavaty A. HOTAIR: An oncogenic long non-coding RNA in different cancers. Cancer Biol. Med. 2015;12:1–9. doi: 10.7497/j.issn.2095-3941.2015.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang Q., Hann S.S. Hotair: An oncogenic long non-coding RNA in human cancer. Cell. Physiol. Biochem. 2018;47:893–913. doi: 10.1159/000490131. [DOI] [PubMed] [Google Scholar]
- 21.Nakagawa T., Endo H., Yokoyama M., Abe J., Tamai K., Tanaka N., Sato I., Takahashi S., Kondo T., Satoh K. Large noncoding RNA hotair enhances aggressive biological behavior and is associated with short disease-free survival in human non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2013;436:319–324. doi: 10.1016/j.bbrc.2013.05.101. [DOI] [PubMed] [Google Scholar]
- 22.Troiano G., Caponio V.C.A., Boldrup L., Gu X., Muzio L.L., Sgaramella N., Wang L., Nylander K. Expression of the long non-coding RNA hotair as a prognostic factor in squamous cell carcinoma of the head and neck: A systematic review and meta-analysis. Oncotarget. 2017;8:73029–73036. doi: 10.18632/oncotarget.20373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y., Zhou Y., Xu T., Tian W., Yang C., Wang X., Zhong S., Ran Q., Yang H., Zhu S. Clinical value of long noncoding RNA hotair as a novel biomarker in digestive cancers: A meta-analysis. Technol. Cancer Res. Treat. 2018;17 doi: 10.1177/1533034618756783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen F.J., Sun M., Li S.Q., Wu Q.Q., Ji L., Liu Z.L., Zhou G.Z., Cao G., Jin L., Xie H.W., et al. Upregulation of the long non-coding RNA hotair promotes esophageal squamous cell carcinoma metastasis and poor prognosis. Mol. Carcinog. 2013;52:908–915. doi: 10.1002/mc.21944. [DOI] [PubMed] [Google Scholar]
- 25.Li J., Wen W., Zhao S., Wang J., Chen J., Wang Y., Zhang Q. Prognostic role of hotair in four estrogen-dependent malignant tumors: A meta-analysis. Oncotargets Ther. 2015;8:1471–1482. doi: 10.2147/OTT.S84687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan S.K., Pastori C., Penas C., Komotar R.J., Ivan M.E., Wahlestedt C., Ayad N.G. Serum long noncoding RNA hotair as a novel diagnostic and prognostic biomarker in glioblastoma multiforme. Mol. Cancer. 2018;17:74. doi: 10.1186/s12943-018-0822-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiong Y., Wang T., Wang M., Zhao J., Li X., Zhang Z., Zhou Y., Liu J., Jia L., Han Y. Long non-coding RNAs function as novel predictors and targets of non-small cell lung cancer: A systematic review and meta-analysis. Oncotarget. 2018;9:11377–11386. doi: 10.18632/oncotarget.23994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deng Q., Sun H., He B., Pan Y., Gao T., Chen J., Ying H., Liu X., Wang F., Xu Y., et al. Prognostic value of long non-coding RNA hotair in various cancers. PLoS ONE. 2014;9:e110059. doi: 10.1371/journal.pone.0110059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang S., Chen S., Yang G., Gu F., Li M., Zhong B., Hu J., Hoffman A., Chen M. Long noncoding RNA hotair as an independent prognostic marker in cancer: A meta-analysis. PLoS ONE. 2014;9:e105538. doi: 10.1371/journal.pone.0105538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geng Y.J., Xie S.L., Li Q., Ma J., Wang G.Y. Large intervening non-coding RNA hotair is associated with hepatocellular carcinoma progression. J. Int. Med. Res. 2011;39:2119–2128. doi: 10.1177/147323001103900608. [DOI] [PubMed] [Google Scholar]
- 31.Kogo R., Shimamura T., Mimori K., Kawahara K., Imoto S., Sudo T., Tanaka F., Shibata K., Suzuki A., Komune S., et al. Long noncoding RNA hotair regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 32.Yang Z., Zhou L., Wu L.M., Lai M.C., Xie H.Y., Zhang F., Zheng S.S. Overexpression of long non-coding RNA hotair predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann. Surg. Oncol. 2011;18:1243–1250. doi: 10.1245/s10434-011-1581-y. [DOI] [PubMed] [Google Scholar]
- 33.Lu L., Zhu G., Zhang C., Deng Q., Katsaros D., Mayne S.T., Risch H.A., Mu L., Canuto E.M., Gregori G., et al. Association of large noncoding RNA hotair expression and its downstream intergenic cpg island methylation with survival in breast cancer. Breast Cancer Res. Treat. 2012;136:875–883. doi: 10.1007/s10549-012-2314-z. [DOI] [PubMed] [Google Scholar]
- 34.Niinuma T., Suzuki H., Nojima M., Nosho K., Yamamoto H., Takamaru H., Yamamoto E., Maruyama R., Nobuoka T., Miyazaki Y., et al. Upregulation of mir-196a and hotair drive malignant character in gastrointestinal stromal tumors. Cancer Res. 2012;72:1126–1136. doi: 10.1158/0008-5472.CAN-11-1803. [DOI] [PubMed] [Google Scholar]
- 35.Ge X.S., Ma H.J., Zheng X.H., Ruan H.L., Liao X.Y., Xue W.Q., Chen Y.B., Zhang Y., Jia W.H. Hotair, a prognostic factor in esophageal squamous cell carcinoma, inhibits wif-1 expression and activates wnt pathway. Cancer Sci. 2013;104:1675–1682. doi: 10.1111/cas.12296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishibashi M., Kogo R., Shibata K., Sawada G., Takahashi Y., Kurashige J., Akiyoshi S., Sasaki S., Iwaya T., Sudo T., et al. Clinical significance of the expression of long non-coding RNA hotair in primary hepatocellular carcinoma. Oncol. Rep. 2013;29:946–950. doi: 10.3892/or.2012.2219. [DOI] [PubMed] [Google Scholar]
- 37.Li D., Feng J., Wu T., Wang Y., Sun Y., Ren J., Liu M. Long intergenic noncoding RNA hotair is overexpressed and regulates pten methylation in laryngeal squamous cell carcinoma. Am. J. Pathol. 2013;182:64–70. doi: 10.1016/j.ajpath.2012.08.042. [DOI] [PubMed] [Google Scholar]
- 38.Li X., Wu Z., Mei Q., Li X., Guo M., Fu X., Han W. Long non-coding RNA hotair, a driver of malignancy, predicts negative prognosis and exhibits oncogenic activity in oesophageal squamous cell carcinoma. Br. J. Cancer. 2013;109:2266–2278. doi: 10.1038/bjc.2013.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu X.H., Liu Z.L., Sun M., Liu J., Wang Z.X., De W. The long non-coding RNA hotair indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer. 2013;13:464. doi: 10.1186/1471-2407-13-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lv X.B., Lian G.Y., Wang H.R., Song E., Yao H., Wang M.H. Long noncoding RNA hotair is a prognostic marker for esophageal squamous cell carcinoma progression and survival. PLoS ONE. 2013;8:e63516. doi: 10.1371/journal.pone.0063516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nie Y., Liu X., Qu S., Song E., Zou H., Gong C. Long non-coding RNA hotair is an independent prognostic marker for nasopharyngeal carcinoma progression and survival. Cancer Sci. 2013;104:458–464. doi: 10.1111/cas.12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sorensen K.P., Thomassen M., Tan Q., Bak M., Cold S., Burton M., Larsen M.J., Kruse T.A. Long non-coding RNA hotair is an independent prognostic marker of metastasis in estrogen receptor-positive primary breast cancer. Breast Cancer Res. Treat. 2013;142:529–536. doi: 10.1007/s10549-013-2776-7. [DOI] [PubMed] [Google Scholar]
- 43.Xu Z.Y., Yu Q.M., Du Y.A., Yang L.T., Dong R.Z., Huang L., Yu P.F., Cheng X.D. Knockdown of long non-coding RNA hotair suppresses tumor invasion and reverses epithelial-mesenchymal transition in gastric cancer. Int. J. Biol. Sci. 2013;9:587–597. doi: 10.7150/ijbs.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.He X., Bao W., Li X., Chen Z., Che Q., Wang H., Wan X.P. The long non-coding RNA hotair is upregulated in endometrial carcinoma and correlates with poor prognosis. Int. J. Mol. Med. 2014;33:325–332. doi: 10.3892/ijmm.2013.1570. [DOI] [PubMed] [Google Scholar]
- 45.Huang L., Liao L.M., Liu A.W., Wu J.B., Cheng X.L., Lin J.X., Zheng M. Overexpression of long noncoding RNA hotair predicts a poor prognosis in patients with cervical cancer. Arch. Gynecol. Obstet. 2014;290:717–723. doi: 10.1007/s00404-014-3236-2. [DOI] [PubMed] [Google Scholar]
- 46.Lee N.K., Lee J.H., Park C.H., Yu D., Lee Y.C., Cheong J.H., Noh S.H., Lee S.K. Long non-coding RNA hotair promotes carcinogenesis and invasion of gastric adenocarcinoma. Biochem. Biophys. Res. Commun. 2014;451:171–178. doi: 10.1016/j.bbrc.2014.07.067. [DOI] [PubMed] [Google Scholar]
- 47.Okugawa Y., Toiyama Y., Hur K., Toden S., Saigusa S., Tanaka K., Inoue Y., Mohri Y., Kusunoki M., Boland C.R., et al. Metastasis-associated long non-coding RNA drives gastric cancer development and promotes peritoneal metastasis. Carcinogenesis. 2014;35:2731–2739. doi: 10.1093/carcin/bgu200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qiu J.J., Lin Y.Y., Ye L.C., Ding J.X., Feng W.W., Jin H.Y., Zhang Y., Li Q., Hua K.Q. Overexpression of long non-coding RNA hotair predicts poor patient prognosis and promotes tumor metastasis in epithelial ovarian cancer. Gynecol. Oncol. 2014;134:121–128. doi: 10.1016/j.ygyno.2014.03.556. [DOI] [PubMed] [Google Scholar]
- 49.Svoboda M., Slyskova J., Schneiderova M., Makovicky P., Bielik L., Levy M., Lipska L., Hemmelova B., Kala Z., Protivankova M., et al. Hotair long non-coding RNA is a negative prognostic factor not only in primary tumors, but also in the blood of colorectal cancer patients. Carcinogenesis. 2014;35:1510–1515. doi: 10.1093/carcin/bgu055. [DOI] [PubMed] [Google Scholar]
- 50.Wu Z.H., Wang X.L., Tang H.M., Jiang T., Chen J., Lu S., Qiu G.Q., Peng Z.H., Yan D.W. Long non-coding RNA hotair is a powerful predictor of metastasis and poor prognosis and is associated with epithelial-mesenchymal transition in colon cancer. Oncol. Rep. 2014;32:395–402. doi: 10.3892/or.2014.3186. [DOI] [PubMed] [Google Scholar]
- 51.Yan T.H., Lu S.W., Huang Y.Q., Que G.B., Chen J.H., Chen Y.P., Zhang H.B., Liang X.L., Jiang J.H. Upregulation of the long noncoding RNA hotair predicts recurrence in stage ta/t1 bladder cancer. Tumour Biol. 2014;35:10249–10257. doi: 10.1007/s13277-014-2344-8. [DOI] [PubMed] [Google Scholar]
- 52.Heubach J., Monsior J., Deenen R., Niegisch G., Szarvas T., Niedworok C., Schulz W.A., Hoffmann M.J. The long noncoding RNA hotair has tissue and cell type-dependent effects on hox gene expression and phenotype of urothelial cancer cells. Mol. Cancer. 2015;14:108. doi: 10.1186/s12943-015-0371-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim H.J., Lee D.W., Yim G.W., Nam E.J., Kim S., Kim S.W., Kim Y.T. Long non-coding RNA hotair is associated with human cervical cancer progression. Int. J. Oncol. 2015;46:521–530. doi: 10.3892/ijo.2014.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Y.W., Sun M., Xia R., Zhang E.B., Liu X.H., Zhang Z.H., Xu T.P., De W., Liu B.R., Wang Z.X. Linchotair epigenetically silences mir34a by binding to prc2 to promote the epithelial-to-mesenchymal transition in human gastric cancer. Cell Death Dis. 2015;6:e1802. doi: 10.1038/cddis.2015.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ma G., Wang Q., Lv C., Qiang F., Hua Q., Chu H., Du M., Tong N., Jiang Y., Wang M., et al. The prognostic significance of hotair for predicting clinical outcome in patients with digestive system tumors. J. Cancer Res. Clin. Oncol. 2015;141:2139–2145. doi: 10.1007/s00432-015-1980-8. [DOI] [PubMed] [Google Scholar]
- 56.Martinez-FeRNAndez M., Feber A., Duenas M., Segovia C., Rubio C., FeRNAndez M., Villacampa F., Duarte J., Lopez-Calderon F.F., Gomez-Rodriguez M.J., et al. Analysis of the polycomb-related lncRNAs hotair and anril in bladder cancer. Clin. Epigenet. 2015;7:109. doi: 10.1186/s13148-015-0141-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qiu J.J., Wang Y., Ding J.X., Jin H.Y., Yang G., Hua K.Q. The long non-coding RNA hotair promotes the proliferation of serous ovarian cancer cells through the regulation of cell cycle arrest and apoptosis. Exp. Cell Res. 2015;333:238–248. doi: 10.1016/j.yexcr.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 58.Wu J., Xie H. Expression of long noncoding RNA-hox transcript antisense intergenic RNA in oral squamous cell carcinoma and effect on cell growth. Tumour Biol. 2015;36:8573–8578. doi: 10.1007/s13277-015-3598-5. [DOI] [PubMed] [Google Scholar]
- 59.Wu S., Zheng C., Chen S., Cai X., Shi Y., Lin B., Chen Y. Overexpression of long non-coding RNA hotair predicts a poor prognosis in patients with acute myeloid leukemia. Oncol. Lett. 2015;10:2410–2414. doi: 10.3892/ol.2015.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xing C.Y., Hu X.Q., Xie F.Y., Yu Z.J., Li H.Y., Bin Z., Wu J.B., Tang L.Y., Gao S.M. Long non-coding RNA hotair modulates c-kit expression through sponging mir-193a in acute myeloid leukemia. FEBS Lett. 2015;589:1981–1987. doi: 10.1016/j.febslet.2015.04.061. [DOI] [PubMed] [Google Scholar]
- 61.Zhang Z.Z., Shen Z.Y., Shen Y.Y., Zhao E.H., Wang M., Wang C.J., Cao H., Xu J. Hotair long noncoding RNA promotes gastric cancer metastasis through suppression of poly r(c)-binding protein (pcbp) 1. Mol. Cancer Ther. 2015;14:1162–1170. doi: 10.1158/1535-7163.MCT-14-0695. [DOI] [PubMed] [Google Scholar]
- 62.Zhao W., Dong S., Duan B., Chen P., Shi L., Gao H., Qi H. Hotair is a predictive and prognostic biomarker for patients with advanced gastric adenocarcinoma receiving fluorouracil and platinum combination chemotherapy. Am. J. Transl. Res. 2015;7:1295–1302. [PMC free article] [PubMed] [Google Scholar]
- 63.Luczak A., SupeRNAt A., Lapinska-Szumczyk S., Jachimowicz D., Majewska H., Gulczynski J., Zaczek A.J. Hotair in relation to epithelial-mesenchymal transition and cancer stem cells in molecular subtypes of endometrial cancer. Int. J. Biol. Markers. 2016;31:e245–e251. doi: 10.5301/jbm.5000187. [DOI] [PubMed] [Google Scholar]
- 64.Luo Z.F., Zhao D., Li X.Q., Cui Y.X., Ma N., Lu C.X., Liu M.Y., Zhou Y. Clinical significance of hotair expression in colon cancer. World J. Gastroenterol. 2016;22:5254–5259. doi: 10.3748/wjg.v22.i22.5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun J., Chu H., Ji J., Huo G., Song Q., Zhang X. Long non-coding RNA hotair modulates hla-g expression by absorbing mir-148a in human cervical cancer. Int. J. Oncol. 2016;49:943–952. doi: 10.3892/ijo.2016.3589. [DOI] [PubMed] [Google Scholar]
- 66.Yan Y., Han J., Li Z., Yang H., Sui Y., Wang M. Elevated RNA expression of long noncoding hotair promotes cell proliferation and predicts a poor prognosis in patients with diffuse large b cell lymphoma. Mol. Med. Rep. 2016;13:5125–5131. doi: 10.3892/mmr.2016.5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang Y.Y., Huang S.H., Zhou H.R., Chen C.J., Tian L.H., Shen J.Z. Role of hotair in the diagnosis and prognosis of acute leukemia. Oncol. Rep. 2016;36:3113–3122. doi: 10.3892/or.2016.5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen W.M., Chen W.D., Jiang X.M., Jia X.F., Wang H.M., Zhang Q.J., Shu Y.Q., Zhao H.B. Hox transcript antisense intergenic RNA represses e-cadherin expression by binding to ezh2 in gastric cancer. World J. Gastroenterol. 2017;23:6100–6110. doi: 10.3748/wjg.v23.i33.6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hu G., Dong B., Zhang J., Zhai W., Xie T., Huang B., Huang C., Yao X., Zheng J., Che J., et al. The long noncoding RNA hotair activates the hippo pathway by directly binding to sav1 in renal cell carcinoma. Oncotarget. 2017;8:58654–58667. doi: 10.18632/oncotarget.17414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Katayama H., Tamai K., Shibuya R., Nakamura M., Mochizuki M., Yamaguchi K., Kawamura S., Tochigi T., Sato I., Okanishi T., et al. Long non-coding RNA hotair promotes cell migration by upregulating insulin growth factor-binding protein 2 in renal cell carcinoma. Sci. Rep. 2017;7:12016. doi: 10.1038/s41598-017-12191-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Luan W., Li R., Liu L., Ni X., Shi Y., Xia Y., Wang J., Lu F., Xu B. Long non-coding RNA hotair acts as a competing endogenous RNA to promote malignant melanoma progression by sponging mir-152-3p. Oncotarget. 2017;8:85401–85414. doi: 10.18632/oncotarget.19910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu F., Zhang J. Long non-coding RNA hotair functions as miRNA sponge to promote the epithelial to mesenchymal transition in esophageal cancer. Biomed. Pharmacother. 2017;90:888–896. doi: 10.1016/j.biopha.2017.03.103. [DOI] [PubMed] [Google Scholar]
- 73.Zhang Y., Yu S., Jiang L., Wang X., Song X. Hotair is a promising novel biomarker in patients with thyroid cancer. Exp. Ther. Med. 2017;13:2274–2278. doi: 10.3892/etm.2017.4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dong X., He X., Guan A., Huang W., Jia H., Huang Y., Chen S., Zhang Z., Gao J., Wang H. Long non-coding RNA hotair promotes gastric cancer progression via mir-217-gpc5 axis. Life Sci. 2019;217:271–282. doi: 10.1016/j.lfs.2018.12.024. [DOI] [PubMed] [Google Scholar]
- 75.Huang K.B., Zhang S.P., Zhu Y.J., Guo C.H., Yang M., Liu J., Xia L.G., Zhang J.F. Hotair mediates tumorigenesis through recruiting ezh2 in colorectal cancer. J. Cell. Biochem. 2019;120:6071–6077. doi: 10.1002/jcb.27893. [DOI] [PubMed] [Google Scholar]
- 76.Xiao Z., Qu Z., Chen Z., Fang Z., Zhou K., Huang Z., Guo X., Zhang Y. LncRNA hotair is a prognostic biomarker for the proliferation and chemoresistance of colorectal cancer via mir-203a-3p-mediated wnt/ss-catenin signaling pathway. Cell. Physiol. Biochem. 2018;46:1275–1285. doi: 10.1159/000489110. [DOI] [PubMed] [Google Scholar]
- 77.Coordinators N.R. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2018;46:D8–D13. doi: 10.1093/nar/gkx1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang Y., Wang L.J., Li W.F., Zhang X., Yang X.J. The prognostic value of hotair for predicting long-term prognosis of patients with gastrointestinal cancers. Medicine. 2018;97:e11139. doi: 10.1097/MD.0000000000011139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Abdeahad H., Avan A., Pashirzad M., Khazaei M., Soleimanpour S., Ferns G.A., Fiuji H., Ryzhikov M., Bahrami A., Hassanian S.M. The prognostic potential of long noncoding RNA hotair expression in human digestive system carcinomas: A meta-analysis. J. Cell. Physiol. 2019;234:10926–10933. doi: 10.1002/jcp.27918. [DOI] [PubMed] [Google Scholar]
- 80.Cai H., An Y., Chen X., Sun D., Chen T., Peng Y., Zhu F., Jiang Y., He X. Epigenetic inhibition of mir-663b by long non-coding RNA hotair promotes pancreatic cancer cell proliferation via up-regulation of insulin-like growth factor 2. Oncotarget. 2016;7:86857–86870. doi: 10.18632/oncotarget.13490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Song Y., Wang R., Li L.W., Liu X., Wang Y.F., Wang Q.X., Zhang Q. Long non-coding RNA hotair mediates the switching of histone h3 lysine 27 acetylation to methylation to promote epithelial-to-mesenchymal transition in gastric cancer. Int. J. Oncol. 2019;54:77–86. doi: 10.3892/ijo.2018.4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Oh E.J., Kim S.H., Yang W.I., Ko Y.H., Yoon S.O. Long non-coding RNA hotair expression in diffuse large b-cell lymphoma: In relation to polycomb repressive complex pathway proteins and h3k27 trimethylation. J. Pathol. Transl. Med. 2016;50:369–376. doi: 10.4132/jptm.2016.06.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jiang C., Yang Y., Yang Y., Guo L., Huang J., Liu X., Wu C., Zou J. Long noncoding RNA (lncRNA) hotair affects tumorigenesis and metastasis of non-small cell lung cancer by upregulating mir-613. Oncol. Res. 2018;26:725–734. doi: 10.3727/096504017X15119467381615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang Z., Cheng J., Wu Y., Qiu J., Sun Y., Tong X. LncRNA hotair controls the expression of rab22a by sponging mir-373 in ovarian cancer. Mol. Med. Rep. 2016;14:2465–2472. doi: 10.3892/mmr.2016.5572. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 85.Georgakilas A.G., Martin O.A., Bonner W.M. P21: A two-faced genome guardian. Trends Mol. Med. 2017;23:310–319. doi: 10.1016/j.molmed.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 86.Galanos P., Vougas K., Walter D., Polyzos A., Maya-Mendoza A., Haagensen E.J., Kokkalis A., Roumelioti F.M., Gagos S., Tzetis M., et al. Chronic p53-independent p21 expression causes genomic instability by deregulating replication licensing. Nat. Cell Biol. 2016;18:777–789. doi: 10.1038/ncb3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhai N., Xia Y., Yin R., Liu J., Gao F. A negative regulation loop of long noncoding RNA hotair and p53 in non-small-cell lung cancer. Oncotargets Ther. 2016;9:5713–5720. doi: 10.2147/OTT.S110219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu H., Li Z., Wang C., Feng L., Huang H., Liu C., Li F. Expression of long non-coding RNA-hotair in oral squamous cell carcinoma tca8113 cells and its associated biological behavior. Am. J. Transl. Res. 2016;8:4726–4734. [PMC free article] [PubMed] [Google Scholar]
- 89.Gao J.Z., Li J., Du J.L., Li X.L. Long non-coding RNA hotair is a marker for hepatocellular carcinoma progression and tumor recurrence. Oncol. Lett. 2016;11:1791–1798. doi: 10.3892/ol.2016.4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zheng J., Xiao X., Wu C., Huang J., Zhang Y., Xie M., Zhang M., Zhou L. The role of long non-coding RNA hotair in the progression and development of laryngeal squamous cell carcinoma interacting with ezh2. Acta Oto-Laryngol. 2017;137:90–98. doi: 10.1080/00016489.2016.1214982. [DOI] [PubMed] [Google Scholar]
- 91.Lee M., Kim H.J., Kim S.W., Park S.A., Chun K.H., Cho N.H., Song Y.S., Kim Y.T. The long non-coding RNA hotair increases tumour growth and invasion in cervical cancer by targeting the notch pathway. Oncotarget. 2016;7:44558–44571. doi: 10.18632/oncotarget.10065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liu L.C., Wang Y.L., Lin P.L., Zhang X., Cheng W.C., Liu S.H., Chen C.J., Hung Y., Jan C.I., Chang L.C., et al. Long noncoding RNA hotair promotes invasion of breast cancer cells through chondroitin sulfotransferase chst15. Int. J. Cancer. 2019 doi: 10.1002/ijc.32319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Di W., Li Q., Shen W., Guo H., Zhao S. The long non-coding RNA hotair promotes thyroid cancer cell growth, invasion and migration through the mir-1-ccnd2 axis. Am. J. Cancer Res. 2017;7:1298–1309. [PMC free article] [PubMed] [Google Scholar]
- 94.Li Q., Feng Y., Chao X., Shi S., Liang M., Qiao Y., Wang B., Wang P., Zhu Z. Hotair contributes to cell proliferation and metastasis of cervical cancer via targetting mir-23b/mapk1 axis. Biosci. Rep. 2018;38 doi: 10.1042/BSR20171563. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 95.Liberati A., Altman D.G., Tetzlaff J., Mulrow C., Gotzsche P.C., Ioannidis J.P., Clarke M., Devereaux P.J., Kleijnen J., Moher D. The prisma statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: Explanation and elaboration. J. Clin. Epidemiol. 2009;62:e1–e34. doi: 10.1016/j.jclinepi.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 96.Fiorini N., Lipman D.J., Lu Z. Towards pubmed 2.0. eLife. 2017;6:e28801. doi: 10.7554/eLife.28801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tierney J.F., Stewart L.A., Ghersi D., Burdett S., Sydes M.R. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Higgins J.P., Thompson S.G., Deeks J.J., Altman D.G. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Higgins J.P., Thompson S.G. Quantifying heterogeneity in a meta-analysis. Stat. Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 100.DerSimonian R., Laird N. Meta-analysis in clinical trials. Control. Clin. Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 101.DerSimonian R., Laird N. Meta-analysis in clinical trials revisited. Contemp. Clin. Trials. 2015;45:139–145. doi: 10.1016/j.cct.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Begg C.B., Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. doi: 10.2307/2533446. [DOI] [PubMed] [Google Scholar]
- 103.Egger M., Davey Smith G., Schneider M., Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z. Gepia: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.