Abstract
Objective: This study aimed to investigate the role of fibroblast growth factor-5 (FGF5) in osteosarcoma (OS) and explore the potential mechanisms.
Methods: OS gene expression data was downloaded from the Gene Expression Omnibus (GEO; GSE12865) and analyzed by R software. OS tissues and cell lines were collected. The expression level of FGF5 in tumor tissues and cell lines was detected using qRT-PCR. Knockout of FGF5 was performed using CRISPR/Cas9 system. The effects of FGF5 knockout on OS cell proliferation and tumor growth were determined through cell counting kit-8 assay and xenograft nude mice, respectively. Additionally, recombinant FGF5 (rFGF5) was added into OS cell and the effects of rFGF5 on the proliferation and apoptosis of OS cell lines were assayed. Furthermore, the protein expression levels of mitogen-activated protein kinase (MAPK) signaling pathway were detected through Western blot.
Results: FGF5 was significantly upregulated in OS tissues and cells, and closely associated with poor differentiation, larger tumor size, lymph node metastasis, and advanced TNM stage. FGF5 knockout could inhibit proliferation of OS cells and tumor growth in nude mouse model. Addition of exogenous rFGF5 promoted OS cell proliferation while inhibited OS cell apoptosis. The expression levels of MAPK signaling pathway proteins in FGF5 knockout group were significantly lower than that in control when there was no rFGF5. Additionally, their expression level in rFGF5 addition group was higher than that in without rFGF5 group.
Conclusion: We demonstrated for the first time that FGF5 was overexpressed in OS cell lines and clinical tissue samples and promotes OS cell proliferation by activating MAPK signaling pathway, which indicated that FGF5 was a potential therapeutic target for OS.
Keywords: fibroblast growth factor-5, osteosarcoma, cell proliferation, MAPK signaling pathway
Introduction
Osteosarcoma (OS) is the most universal bone malignancies in children and adolescents, which is characterized by direct formation of osteoid tissue or immature bone by tumor cells.1,2 In spite of the advances in treatment strategies, including primary tumor resection, chemotherapy, or combination of surgery and chemotherapy, it is still far from satisfying the current medical requirements.3,4 Currently, genetic abnormalities have been extensively recognized to play critical roles in OS tumorigenesis, and gene therapy has been gradually reported to be available in OS treatment.5–7 However, the molecular mechanisms of OS are still elusive. Therefore, it is essential to elucidate the molecular mechanisms underlying the initiation and development of OS, which may contribute to identify reliable diagnostic and prognostic markers.
Fibroblast growth factor-5 (FGF5) is a member of the fibroblast growth factor (FGF) family.8 The FGF family constitutes 18 polypeptides which transduce signals through receptor tyrosine kinases (RTK) named FGF receptors (FGFRs) 1 to 4. Several FGFs play important roles in embryonic development, tissue maintenance, and wound healing.9 Additionally, FGFs are angiogenic and mitogenic, and overexpression or mutation of FGFs or FGFRs could contribute to malignancy.10,11 FGF5 was first identified by a screening approach for transforming oncogenes.12 Overexpression of FGF5 has been associated with prostate cancer, pancreatic cancer, breast cancer, renal cell carcinoma, and so on.13,14 However, the roles of FGF5 and its signaling pathway in OS are still unknown. Specifically, FGF5 was reported to be frequently overexpressed in embryonic tissues but scarcely expressed in adult tissues.15 Given that OS develops from embryonic cells and presents high incidence rate in adolescents, we speculated that it may be involved in the pathogenesis of OS.
Therefore, in this study, the expression level of FGF5 was detected in tumor tissues and cell lines. Further in vitro studies were performed to identify the functional role of FGF5 in proliferation and apoptosis of OS cell lines. Furthermore, the underlying molecular mechanism was also investigated.
Materials and methods
Tissue samples
A total of 15 pairs of tumor tissues and adjacent nontumor corresponding tissues obtained from 15 OS patients from 2014 and 2016 in the 107th Hospital of People’s Liberation Army and First People’s Hospital of Yancheng were used in this study. All the patients were pathologically confirmed and the tissue specimens were collected immediately after they were surgically resected, and then stored at −80°C. This study was approved by the ethics committee of 970th Hospital of People’s Liberation Army and First People’s Hospital of Yancheng and was conducted in accordance with the Declaration of Helsinki. All patients had provided their written informed consent before the study.
Cell culture
HEK293T, human normal osteoblast cells HOBC and NHOST, as well as OS cell lines U2OS, SAOS, MG63, and KHOS were purchased from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). HOBC and NHOST cells were cultured in DMEM/ham’s F-12 (Invitrogen, Carlsbad, CA, USA) containing 10% FBS(Invitrogen) and 0.3 mg/ml G418. U2OS, MG63, and SAOS cells were maintained in DMEM medium (Invitrogen) containing 10% FBS (Invitrogen), 100 U/mL penicillin and 100 g/mL streptomycin (Life Technologies, Grand Island, NY, USA). KHOS cells were cultured in RPMI 1640 medium (Invitrogen) supplemented with 10% FBS. All cells were incubated at 37°C in 5% CO2 incubator.
CRISPR/Cas9-mediated FGF5 knockout
The gRNA targeting FGF5 gene was designed using E-CRISP database (5ʹ-3ʹ GAAAATGGGTAGAGATATGC). pX330 vector in vitro transcription of Cas9 mRNA and gRNA was prepared as previously described.16 Briefly, paired synthesized oligonucleotides were annealed and subcloned into the linearized pX330 vector. The constructed recombinant vector was used as the template. The production of gRNA by in vitro transcription was performed using the MEGAshortscript kit (Ambion, Austin, TX, USA) and then purified using the MEGAclearTM transcription clean-up kit (Ambion). Based on the Cas9 mRNA in vitro transcription vector as templates, Cas9 mRNA was produced and purified using the MEGAclearTM transcription clean-up kit (Ambion). To produce lentivirus, HEK 293T cells were transfected with pX330 vector. The lentivirus was then transfected into the MG63 and U20S cell lines. After 24 hours of transfection, puromycin was added for the selection of transfected cells. The puromycin selected cells were then expanded in regular culture medium. FGF5-knockout cell lines were determined by Western blot.
Mice
Female athymic nu/nu nude mice (AntiCancer Inc., San Diego, CA, USA), 4–6 weeks old, were purchased from Shanghai Institute of Zoology, China. They were maintained in pathogen-free conditions and were fed with autoclaved laboratory rodent diet. All animal experiments were conducted following the guidelines of Institutional Animal Care and Use Committee with ethical approval of 970th Hospital of People’s Liberation Army and First People’s Hospital of Yancheng.
Nude mouse orthotopic metastatic model
The mice were divided into four groups (N=10): MG63 transplatation (FGF5-knockout), MG63 transplatation (FGF5-control), U2OS transplantation (FGF5-knockout), and U2OS transplantation (FGF5-control). All mice were anesthetized by subcutaneous injection of 0.02 mL ketamine mixture (15.2 mg/kg xylazine, 0.48 mg/kg acepromazine, and maleate 20 mg/kg ketamine). U2OS and MG63 cells (FGF5 knockout and control) (5×105) were respectively transplanted into the tibia of the nude mice as previously described.17 The tumor size was measured once a week (six times in total). The mean tumor volume was calculated by the formula of width2× length. Mice were euthanized when tumor volume became approximately 2.0 cm3.
Expression profiling data collection and differential expression analysis
The expression profiling data (accession number: GSE12865) deposited in Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/) were extracted for analysis. The dataset included two replications of normal human osteoblasts and 12 osteosarcoma tissues. The differential expression analysis was performed using limma and pheatmap of R software18 in R3.4.1. The P-values and logFC values were adjusted into false discovery rate (FDR) using multitest package (version 2.32.0).19 FDR <0.05 and |logFC| >1 were chosen as cut-off criteria. The expression values of differentially expressed genes were extracted and performed bidirectional hierarchical clustering20 based on Euclidean distance21 using pheatmap package (version 1.0.8)22 in R3.4.1. The results were displayed using heatmaps.
Cell counting kit-8 assay
Cell proliferation was determined using cell counting kit-8 (CCK-8; Dojindo Molecular Technologies, Japan). Briefly, the cells were seeded in 96-well plates with complete growth medium (with or without 10 ng/mL recombinant FGF5 (rFGF5) protein) at a concentration of 3,000 cells per well. After 1–5 days of incubation, the growth medium in each well was removed, and then filled with 100 ml of fresh medium containing 10 ml CCK-8 solutions. After incubated for 2 h at 37 °C, cell proliferation was determined by optical density measurement at 450 nm with a microplate reader.
Colony formation assay
After transfection, cells were seeded in six-well plates at a concentration of 500 cells per well, and cultured for approximately 10 days. Then cells were fixed with 4% paraformaldehyde. Following washing, the plates were air dried, and the colonies with more than 50 cells were counted using a microscope (IX83, Olympus, Japan).
Apoptosis assay
Cell apoptosis was determined by propidium iodide (PI) and fluorescein isothiocynate (FITC)-conjugated annexin V staining. Briefly, after treatment with the rFGF5 or isometric serum-free medium for 72 hours, cells were washed with PBSand then fixed in 70% ethanol. After washing, the fixed cells were stained in PI/FITC-annexin V with 50 μg/mL RNase A and then incubated at room temperature for 1 hour in the dark. Flow cytometry analysis was performed by using a FACScan flow cytometer (Becton-Dickinson, Mountain View, CA, USA). The obtained data were analyzed using the Multicycle Cell Cycle Analysis software (Phoenix Flow System, San Diego, CA, USA).
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted using TRIzol reagent (Life Technologies Corporation, Carlsbad, CA, USA) and was reverse-transcribed into cDNA by Reverse Transcription kit (TaKaRa, Tokyo, Japan). The One Step SYBR® PrimeScript® PLUS RT-RNA PCR Kit (TaKaRa Biotechnology, Dalian, China) was to detect the expression level of FGF5 by real-time PCR analysis with GAPDH as internal reference. The relative expression levels were calculated by relative quantification (2−ΔΔCt) method.
Western blot
Protein was extracted using RIPA lysis buffer (Beyotime Biotechnology, Shanghai, China) containing protease inhibitors (Roche, Guangzhou, China) and then quantified with BCA™ Protein Assay Kit (Pierce, Appleton, WI, USA). The Western blot system was constructed using a Bio-Rad Bis-Tris Gel system according to the manufacturer’s instructions. Primary antibodies (anti-FGF5, FGFR1, PARP, p-MEK2, p-ERK, p-Elk-1, and p-MNK1/2) were prepared in 5% blocking buffer at a dilution of 1:1000, and then incubated with the membrane at 4°C overnight. After washing, the membrane was incubated with secondary antibody marked by horseradish peroxidase for 1 hour at room temperature. The polyvinylidene difluoride membrane carrying blots and antibodies was transferred into the Bio-Rad ChemiDoc™ XRS system, with coverage of 200 μL Immobilon Western Chemiluminescent HRP Substrate (Merck Millipore, Billerica, MA, USA). The signals were captured and the intensity of the bands was quantified using ImageJ software.
Statistical analysis
All experiments were repeated three times and the data of multiple experiments are presented as mean ± SD. Statistical analyses were performed using SPSS 16.0 and Graphpad 6.0. The P-values were calculated using a one-way ANOVA. A P-value of <0.05 was considered as statistically significant.
Results
FGF5 was upregulated in OS
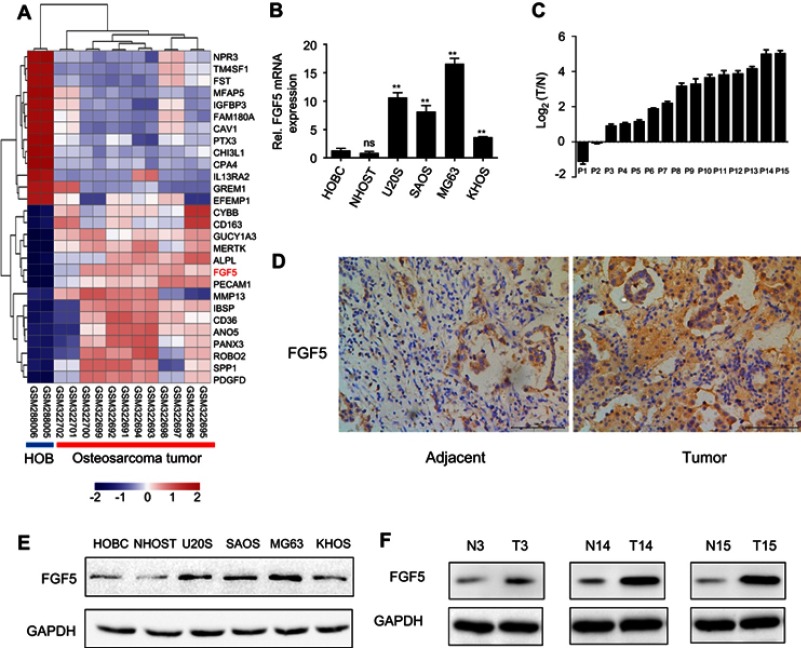
After differential expression analysis of the dataset in GSE12865, FGF5 was found to be significantly upregulated between tumor and control groups, as shown in Figure 1A. To validate the analysis results above, the expression levels of FGF5 in tumor cells and tissues of all patients were determined using qRT-PCR and Western blot. As shown in Figure 1B, FGF5 was significantly upregulated in five OS cell lines compared with that in normal osteoblast cells HOBC and NHOST (P<0.01). Additionally, FGF5 was found to be upregulated in OS tissues of 13 patients compared with adjacent nontumor tissues collected by us (Figure 1C). Moreover, the results of IHC staining and Western blot also showed that FGF5 was overexpressed in OS cells and tissues (Figure 1D–F). Importantly, high FGF5 expression was closely related to poor differentiation (P=0.035), larger tumor size (P=0.047), lymph node metastasis (P=0.011), and advanced TNM stage (P=0.044) (Table 1).
Figure 1.
(A) Heatmap of differentially expressed genes between osteosarcoma tissues and tissues adjacent to cancer. (B and C) Expression levels of FGF5 in osteosarcoma cell lines and tissues detected by qRT-PCR, the results showed that FGF5 mRNA expression was significantly increased in osteosarcoma cell lines and tissues. (D) ICH staining for FGF5 in osteosarcoma and matched normal tissues. (E and F) Protein expression levels of FGF5 in osteosarcoma cell lines and tissues detected by Western blot, the results showed that FGF5 protein expression was upregulated in osteosarcoma cell lines and tissues. Error bars indicated mean ± SD (*P<0.01).
Abbreviations: HOB, human normal osteoblast; FGF5, fibroblast growth factor-5; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Table 1.
Correlation between FGF5 expression and clinical-pathological characteristic in 15 osteosarcoma patients
| FGF5 expression | ||||
|---|---|---|---|---|
| Characteristics | Case number | Low | High | P-value |
| Gender | ||||
| Male | 9 | 4 | 6 | 0.706 |
| Female | 6 | 2 | 3 | |
| Age | ||||
| ≤25 | 12 | 6 | 6 | 0.185 |
| 25 | 3 | 0 | 3 | |
| Tumor location | ||||
| Tibia/femur | 10 | 4 | 6 | 0.706 |
| Elsewhere | 5 | 2 | 3 | |
| Histology types | ||||
| Osteoblast type | 8 | 5 | 3 | |
| Chondroblast type | 4 | 2 | 2 | 0.467 |
| Other type | 3 | 1 | 2 | |
| X-ray types | ||||
| Osteogenesis type | 5 | 3 | 2 | |
| Osteolysis type | 7 | 3 | 4 | 0.35 |
| Mixed type | 3 | 1 | 2 | |
| Differentiated degree | ||||
| High | 9 | 2 | 7 | 0.035 |
| Low | 6 | 5 | 1 | |
| Tumor size | ||||
| 8 cm | 5 | 4 | 1 | 0.047 |
| 8 cm | 10 | 2 | 8 | |
| Lymph node metastasis | ||||
| Yes | 11 | 2 | 9 | 0.011 |
| No | 4 | 4 | 0 | |
| Tumor stage | ||||
| IIA | 26 | 3 | 0 | 0.044 |
| IIB/III | 34 | 3 | 9 | |
Downregulation of FGF5 decreased OS cell proliferation and tumor growth
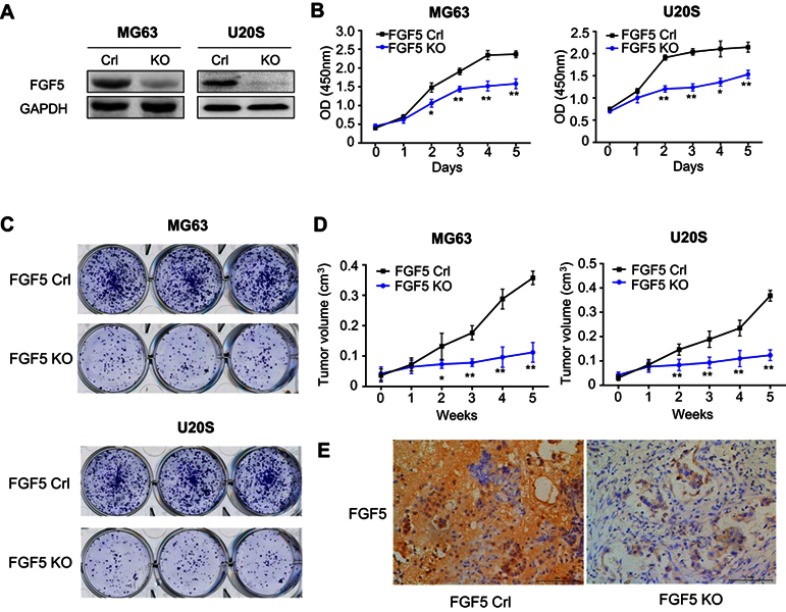
To explore the role of FGF5 in OS progression, we examined the effects of FGF5 suppression on OS cell proliferation, colony formation, and tumor growth. Because FGF5 had higher expression level in MG63 and U20S cell lines, the two cell lines were selected for this experiment. FGF5 in MG63 and U20S cell lines was knocked out by CRISPR/Cas9, which was confirmed by Western blot (Figure 2A). According to the OD450 value determined by CCK-8 assay, the OS cells proliferation in FGF5 suppression group was significantly inhibited compared with control group from the second day (P<0.05, Figure 2B). Similar results were found in colony formation assay. As shown in Figure 2C, colony formation experiment revealed that FGF5 knockout obviously inhibited colony formation. Additionally, by establishment of nude mouse orthotopic metastatic model, we found that mice with FGF5 knockout exhibited low FGF5 expression and the tumor value in mice significantly reduced in FGF5 knockout group from the second week (P<0.05, Figure 2D–E).
Figure 2.
(A) The expression of FGF5 in two osteosarcoma cell lines after FGF5 knockout by Crispr/Cas9, the results showed that FGF5 was dramatically decreased in FGF5-KO MG63 an U20S cells. (B and C) Effects of FGF5 on cell proliferation and colony formation assessed by CCK-8 and colony formation assays. The results showed that knockout of FGF5 inhibited cell proliferation. (D) Effects of FGF5 on tumor growth determined by nude mouse orthotopic metastatic model. (E) ICH staining for FGF5 in nude mice. Error bars indicated mean ± SD (*P<0.05, **P<0.01).
Abbreviations: Crl, control; KO, knockout; FGF5, fibroblast growth factor-5; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
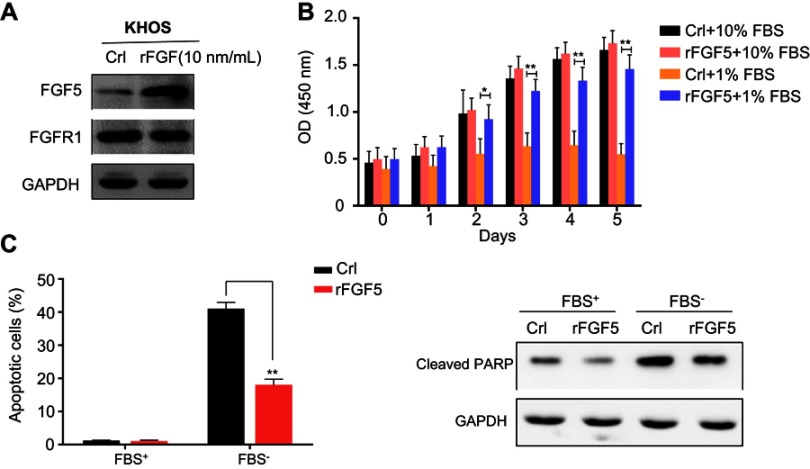
Exogenous rFGF5 promoted OS cell proliferation while inhibited OS cell apoptosis
To further investigate the effects of FGF5 on OS cell proliferation and apoptosis, we added 10 ng/mL exogenous rFGF5 into KHOS cell line. KHOS cell line was selected because FGF5 had a lower expression level in KHOS cell line than in the other three PS cell lines. The protein expression of FGF5 and its receptor FGFR1 was detected using Western blot. As shown in Figure 3A, FGF5 expression significantly increased after the addition of rFGF5, while the FGFR1 expression showed no obvious change. CCK-8 assay revealed that under 10% FBS, the proliferation promotion effect of rFGF5 addition on OS cells was not obvious. Whereas, under 1% FBS, addition of rFGF5 significantly increased cell proliferation from the third day (P<0.05, Figure 3B). Furthermore, flow cytometry analysis revealed that rFGF5 addition significantly inhibited apoptosis of OS cells under 1% FBS. The protein expression of cleaved PARP significantly decreased in rFGF5 group compared with control under 1% FBS (Figure 3C).
Figure 3.
(A) The expression of FGF5 and FGFR1 in osteosarcoma cell after addition of exogenous rFGF5 into KHOS cell line. (B) The effect of rFGF5 on cell proliferation detected by CCK-8 assay. The results showed that exogenous rFGF5 enhanced cell proliferation. (C) The effect of rFGF5 on apoptosis detected by flow cytometry analysis and the protein expression of cleaved PARP. The results showed that exogenous rFGF5 inhibited cell apoptosis. Error bars indicated mean ±SD (*P<0.05, **P<0.01).
Abbreviations: Crl, control; FGF5, fibroblast growth factor-5; FGFR1, FGF receptor-1; rFGF5, recombinant FGF5; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; CCK-8, cell counting kit-8; PARP, poly (adenosine diphosphate) ribose polymerase.
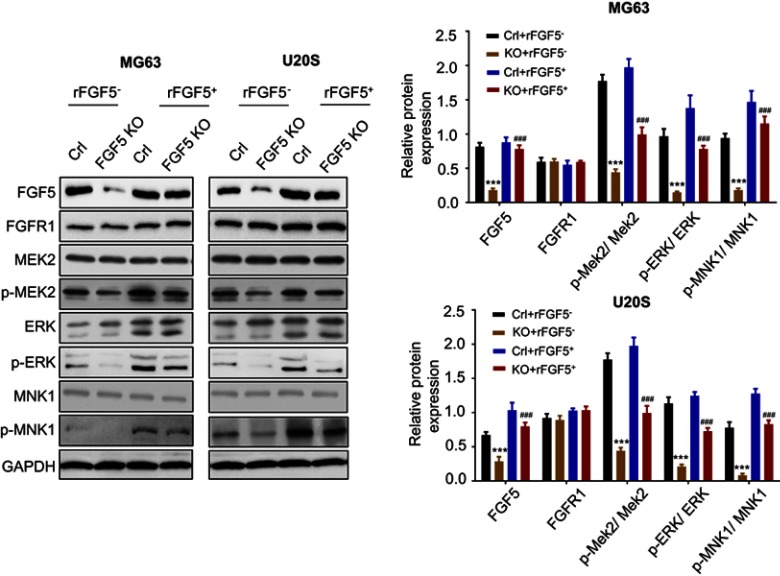
FGF5 promoted OS cell proliferation via MAPK signaling pathway
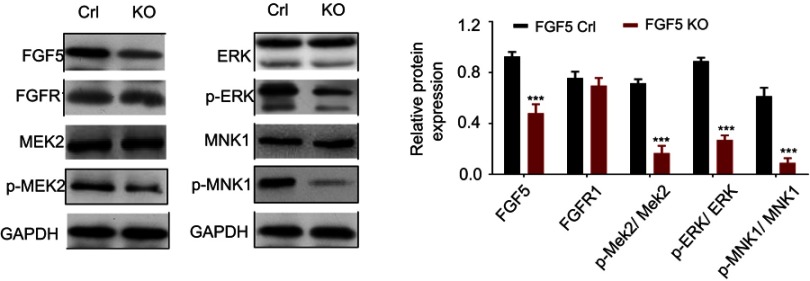
Study has reported that mitogen-activated protein kinase (MAPK) signaling plays an important role in OS cell growth.23 We speculated that FGF5 may function in OS cell proliferation through MAPK signaling pathway. Therefore, the expression level of FGF5, FGFR1 and MAPK signaling pathway associated proteins (p-MEK2, p-ERK, p-Elk-1, and p-MNK1/2) on OS cells without rFGF5 (FGF5 knockout and control) and with rFGF5 (FGF5 knockout and control) were detected through Western blot. As shown in Figure 4, the expression levels of FGF5, p-MEK2, p-ERK, p-Elk-1, and p-MNK1/2 in FGF5 knockout group were significantly lower than that in control when there was no rFGF5, however, the expression of total MEK2, ERK, and MNK1 remained unchanged. Additionally, their expression level in rFGF5 addition group was higher than that in rFGF5 without group. Moreover, the Western blot results in vivo also confirmed this finding (Figure 5).
Figure 4.
The expression level of FGF5, FGFR1 and MAPK signaling pathway associated proteins (p-MEK2, MEK2, p-ERK, ERK, p-MNK1, and MNK1) on control or FGF5 knockout OS cells without or with rFGF5 treatment and detected by Western blot. The results showed that FGF5 activiated the MAPK signaling. *** P <0.001 vs. Crl; ### P <0.001 vs rFGF5-
Abbreviations: Crl, control; KO, knockout; FGF5, fibroblast growth factor-5; FGFR1, FGF receptor-1; rFGF5, recombinant FGF5; OS, osteosarcoma; MAPK, mitogen-activated protein kinase; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Figure 5.
The expression levels of FGF5, FGFR1 and MAPK signaling pathway associated proteins (p-MEK2, MEK2, p-ERK, ERK, p-MNK1, and MNK1) in control and FGF5 knockout nude mice detected by Western blot. The results showed that FGF5 knockout dampen MAPK signaling in vivo. *** P <0.001
Abbreviations: Crl, control; KO, knockout; FGF5, fibroblast growth factor-5; FGFR1, FGF receptor-1; MAPK, mitogen-activated protein kinase; MAPK, mitogen-activated protein kinase; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Discussion
In the present study, FGF5 was found to be significantly upregulated in OS tissues and cells, and FGF5 knockout could inhibit OS cell proliferation and tumor growth in nude mouse model. Subsequent experiments revealed that addition of exogenous rFGF5 promoted OS cell proliferation while inhibited OS cell apoptosis. Finally, we for the first time presented evidence that FGF5 exerted oncogenic activities in OS by activating MAPK signaling pathway.
FGFs are small polypeptide growth factors containing signal peptides to secrete to the extracellular environment to promote embryonic development, cell growth, morphogenesis, tumor growth and invasion.9 In addition, FGF signaling has been implicated in a variety of pathological processes in angiogenesis.24 FGF5 is originally identified as a transforming proto-oncogene, but the related data on oncogenic functions of FGF5 are relatively limited. Kornmann et al25,26 have reported that overexpression of FGF5 is associated with the development of human pancreatic cancer. Hanada et al27 have also reported that FGF5 is an overexpressed antigen in multiple human adenocarcinomas. In accordance with the findings above, our study also revealed a high expression of FGF5 in OS, further indicating the oncogenic potential of FGF5 in cancers.
It is well known that sustaining proliferative signaling as well as resisting cell death are the hallmarks of tumors.28 To demonstrate the role of FGF5 in OS cell proliferation and apoptosis, we knocked out the FGF5 in OS cell lines through CRISPR/Cas9 system and added rFGF5 in OS cells to investigate the changes of OS cell proliferation and apoptosis. The results showed that silenced FGF5 significantly inhibited cell proliferation, while rFGF5 could promote OS cells proliferation but inhibit their apoptosis, which was in line with a previous study of Allerstorfer et al.15They have suggested that siRNA-mediated FGF5 suppression significantly decreases glioblastoma cells proliferation, while rFGF5 significantly increases this parameter.
To further elucidate the molecular mechanisms underlying the oncogenic activities of FGF5, we detected the expressions of MAPK signaling pathways. MAPK is an insulin-mitogen activated protein kinase, which is related with cell survival, growth, and migration by regulating the signals from cell surface to nucleus through phosphorylation.29,30 The MAPK pathway exerts a key role in integrating external signals from mitogens into signaling events to promote cell proliferation. Aberrant activation of MAPK signaling pathway has been demonstrated to be associated with tumor cell growth and proliferation, including in OS.31,32 The present results showed that MAPK signaling pathway was activated in OS cell without FGF5 knockout, suggesting that FGF5 may promote OS cell proliferation via activation of MAPK signaling pathway.
Conclusion
We demonstrated for the first time that FGF5 was overexpressed in OS cell lines and clinical tissue samples. High expression of FGF5 could promote OS cell proliferation. Importantly, we suggested that FGF5 may exert oncogenic function in OS cells by activating the MAPK signaling pathway. Therefore, FGF5 may serve as a potential therapeutic target for OS, and abrogation of FGF5 dependent signaling may have a role in OS therapy.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. Ca A Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 2.Meyers PA, Gorlick R. Osteosarcoma. Pediatr Clin North Am. 1997;44(4):973–989. [DOI] [PubMed] [Google Scholar]
- 3.Yuan G, Chen J, Wu D, Gao C. Neoadjuvant chemotherapy combined with limb salvage surgery in patients with limb osteosarcoma of Enneking stage II: a retrospective study. Onco Targets Ther. 2017;10:2745–2750. doi: 10.2147/OTT [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bacci, G., Longhi, A., Versari, M., et al. Prognostic factors for osteosarcoma of the extremity treated with neoadjuvant chemotherapy: 15-year experience in 789 patients treated at a single institution. Cancer. 2006;106(5):1154–1161. doi: 10.1002/cncr.21730 [DOI] [PubMed] [Google Scholar]
- 5.Simpson, S., Dunning, MD., de Brot, S., et al. Comparative review of human and canine osteosarcoma: morphology, epidemiology, prognosis, treatment and genetics. Acta Vet Scand. 2017;59(1):71. doi: 10.1186/s13028-017-0347-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the Surveillance, Epidemiology, and End Results Program. Cancer. 2009;115(7):1531–1543. doi: 10.1002/cncr.v115:7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amankwah EK, Conley AP, Reed DR. Epidemiology and therapies for metastatic sarcoma. Clin Epidemiol. 2013;5(Issue 1):147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernig DG, Gallagher JT. Fibroblast growth factors and their receptors: an information network controlling tissue growth, morphogenesis and repair. Prog Growth Factor Res. 1994;5(4):353. doi: 10.1016/0955-2235(94)00007-8 [DOI] [PubMed] [Google Scholar]
- 9.Powers CJ, Mcleskey SW, Wellstein A. Fibroblast growth factors, their receptors and signaling. Endocr Relat Cancer. 2000;7(3):165–197. [DOI] [PubMed] [Google Scholar]
- 10.Weiss J, Sos ML, Seidel D, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med. 2010;2(62):62ra93. doi: 10.1126/scitranslmed.3001451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elbauomy ES, Green AR, Lambros MB, et al. FGFR1 amplification in breast carcinomas: a chromogenic in situ hybridisation analysis. Breast Cancer Res. 2007;9(2):R23. doi: 10.1186/bcr1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhan X, Bates B, Hu XG, Goldfarb M. The human FGF-5 oncogene encodes a novel protein related to fibroblast growth factors. Mol Cel Biol. 1988;8(8):3487–3495. doi: 10.1128/MCB.8.8.3487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kornmann M, Ishiwata T, Beger HG, Korc M. Fibroblast growth factor-5 stimulates mitogenic signaling and is overexpressed in human pancreatic cancer: evidence for autocrine and paracrine actions. Oncogene. 1997;15(12):1417–1424. doi: 10.1038/sj.onc.1201307 [DOI] [PubMed] [Google Scholar]
- 14.Hanada, K., Perry-Lalley, DM., Ohnmacht, GA., et al. Identification of fibroblast growth factor-5 as an overexpressed antigen in multiple human adenocarcinomas. Cancer Res. 2001;61(14):5511. [PubMed] [Google Scholar]
- 15.Allerstorfer S, Sonvilla G, Fischer H, et al. FGF5 as an oncogenic factor in human glioblastoma multiforme: autocrine and paracrine activities. Oncogene. 2008;27(30):4180. doi: 10.1038/onc.2008.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou J, Shen B, Zhang W, et al. One-step generation of different immunodeficient mice with multiple gene modifications by CRISPR/Cas9 mediated genome engineering. Int J Biochem Cell Biol. 2014;46(1):49–55. doi: 10.1016/j.biocel.2013.10.010 [DOI] [PubMed] [Google Scholar]
- 17.Tome Y, Tsuchiya H, Hayashi K, et al. In vivo gene transfer between interacting human osteosarcoma cell lines is associated with acquisition of enhanced metastatic potential. J Cell Biochem. 2010;108(2):362–367. doi: 10.1002/jcb.v108:2 [DOI] [PubMed] [Google Scholar]
- 18.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carvajalrodríguez A, Uñaalvarez JD, Rolánalvarez E. A new multitest correction (SGoF) that increases its statistical power when increasing the number of tests. BMC Bioinformatics. 2009;10(1):209. doi: 10.1186/1471-2105-10-209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szekely GJ, Rizzo ML. Hierarchical clustering via joint between-within distances: extending ward‘s minimum variance method. J Classification. 2005;22(2):151–183. doi: 10.1007/s00357-005-0012-9 [DOI] [Google Scholar]
- 21.Deza MM, Deza E. Encyclopedia of Distances‘. Ref Rev. 2009;24(6):1–583. [Google Scholar]
- 22.Wang, L., Cao, C., Ma, Q., et al. RNA-seq analyses of multiple meristems of soybean: novel and alternative transcripts, evolutionary and functional implications. BMC Plant Biol. 2014;14(1):169. doi: 10.1186/s12870-014-0243-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng D-D, Zhu B, Li S-J, Yuan T, Yang Q-C, Fan C-Y. Down-regulation of RPS9 inhibits osteosarcoma cell growth through inactivation of MAPK signaling pathway. J Cancer. 2017;8(14):2720. doi: 10.7150/jca.19130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang, X., Ibrahimi, OA., Olesen SK., et al. Receptor specificity of the fibroblast growth factor family the complete mammalian fgf family. J Biol Chem. 2006;281(23):15694–15700. doi: 10.1074/jbc.M601252200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kornmann, M., Ishiwata, T., Beger, HG., et al. Fibroblast growth factor-5 stimulates mitogenic signaling and is overexpressed in human pancreatic cancer: evidence for autocrine and paracrine actions. Oncogene. 1997;15(12):1417. doi: 10.1038/sj.onc.1201351 [DOI] [PubMed] [Google Scholar]
- 26.Kornmann M, Lopez ME, Beger HG, Korc M. Expression of the IIIc variant of FGF receptor-1 confers mitogenic responsiveness to heparin and FGF-5 in TAKA-1 pancreatic ductal cells. Int J Pancreatol. 2001;29(2):85–92. doi: 10.1385/IJGC:29:2:085 [DOI] [PubMed] [Google Scholar]
- 27.Hanada K-I, Perry-Lalley DM, Ohnmacht GA, Bettinotti MP, Yang JC. Identification of fibroblast growth factor-5 as an overexpressed antigen in multiple human adenocarcinomas. Cancer Res. 2001;61(14):5511–5516. [PubMed] [Google Scholar]
- 28.Douglas H, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 29.Avruch J. MAP kinase pathways: the first twenty years. Biochimica Et Biophysica Acta (Bba)-Mol Cell Res. 2007;1773(8):1150–1160. doi: 10.1016/j.bbamcr.2006.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang L, Karin M. Mammalian MAP kinase signalling cascades. Nature. 2001;410(6824):37. doi: 10.1038/35065000 [DOI] [PubMed] [Google Scholar]
- 31.Ghavami S, Chitayat S, Hashemi M, et al. S100A8/A9: a Janus-faced molecule in cancer therapy and tumorgenesis. Eur J Pharmacol. 2009;625(1–3):73–83. doi: 10.1016/j.ejphar.2009.08.044 [DOI] [PubMed] [Google Scholar]
- 32.Cheng S, Zhang X, Huang N, Qiu Q, Jin Y, Jiang D. Down-regulation of S100A9 inhibits osteosarcoma cell growth through inactivating MAPK and NF-κB signaling pathways. BMC Cancer. 2016;16(1):253. doi: 10.1186/s12885-016-2294-1 [DOI] [PMC free article] [PubMed] [Google Scholar]