Abstract
Subtle structural features in bacterial lipids such as unsaturation elements can have vast biological implications. Cyclopropane rings have been correlated with tolerance to a number of adverse conditions in bacterial phospholipids. They have been shown to play a major role in Mycobacterium tuberculosis (M. tuberculosis or Mtb) pathogenesis as they occur in mycolic acids (MAs) in the mycobacterial cell. Traditional collisional activation methods allow elucidation of basic structural features of lipids but fail to reveal the presence and position of cyclopropane rings. Here, we employ 213 nm ultraviolet photodissociation mass spectrometry (UVPD-MS) for structural characterization of cyclopropane rings in bacterial phospholipids and MAs. Upon UVPD, dual cross-ring C-C cleavages on both sides of the cyclopropane ring are observed for cyclopropyl lipids, resulting in diagnostic pairs of fragment ions spaced 14 Da apart, thus enabling cyclopropane localization. These diagnostic pairs of ions corresponding to dual cross-ring cleavage are observed in both negative and positive ion modes and afford localization of multiple cyclopropane rings within a single lipid. This method was integrated with liquid chromatography (LC) for LC/UVPD-MS analysis of cyclopropyl glycerophospholipids in Escherichia coli (E. coli) and for analysis of MAs in Mycobacterium bovis (M. bovis) and M. tuberculosis lipid extracts.
Graphical Abstract
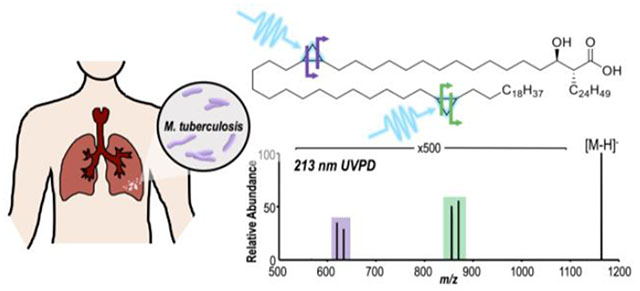
Introduction
Lipidomics, or the structural and functional analysis of the entirety of lipids that are present within a cell or organism, has become an important emerging field owing to the diversity and importance of lipids in biological systems.1,2 Mass spectrometry (MS) has proven to be an invaluable tool in the field of lipidomics, especially for the analysis of structurally complex lipids.3 MS-based lipidomics approaches used to examine ex vivo lipid profiles are comprised of two main methods, namely (1) shotgun analysis in which complex samples are introduced and analyzed directly without separation,4–6 and (2) separation-based methods.7,8 With both of these MS strategies, high resolution and high accuracy MS1 spectra provide elemental information that allow components to be matched to chemical formulas of known lipids. However, these methods alone are not able to provide structural characterization beyond elemental composition.9
Phospholipids are the main component in cell membranes, constituting approximately half of the entire bacterial membrane mass, and they are highly diverse in both their structure and size, making them suitable targets for lipidomic evaluation.10–12 Glycerophospholipids are composed of a hydrophilic headgroup containing a phosphate moiety, a glycerol backbone, and two hydrophobic fatty acid tails. The six major classes of glycerophospholipids are phosphatidylglycerol (PG), phosphatidic acid (PA), phosphatidylserine (PS), phosphatidylethanolamine (PE), phosphatidylcholine (PC), and phosphatidylinositol (PI), which all vary in their headgroup structure. In addition to headgroup variation, phospholipids can also vary in fatty acid chain lengths, sn-stereochemistry, and in their degree, stereochemistry, location and type of unsaturation – including both double bonds and cyclopropane modifications.
To address the structural analysis of complex lipids, tandem mass spectrometry (MS/MS) has been employed for investigation of phospholipids. Collision-based activation methods such as collision-induced dissociation (CID) and higher-energy collisional activation (HCD) are commonly used for structural characterization of glycerophospholipids because they typically provide fragments corresponding to lipid headgroup and acyl chain compositions.13,14 However, many of the more subtle structural features such as sn-stereochemistry and unsaturation elements including cyclopropane modifications have not been probed with these collisional-based activation methods.
Although cyclopropane modifications may seem insignificant, they have significant biological implications. Cyclopropane fatty acids (CFAs) are generated by the addition of a methylene group across the double bonds of unsaturated fatty acids (UFAs) of bacterial phospholipids.15,16 This methylene group is derived from S-adenosylmethionine, and the in situ reaction of UFA to CFA is catalyzed by Cfa synthase. During this reaction, the CFA retains the original stereochemistry of the UFA, whether cis or trans.17,18 This reaction occurs in response to adverse environmental conditions, and it is common in various gram-positive and gram-negative bacterial species including Escherichia coli (E. coli), Mycobacterium tuberculosis (M. tuberculosis or Mtb), and Streotciccys thermophilus.19,20
CFAs in bacterial phospholipids are more stable than their UFA counterparts17 and have been associated with tolerance to high osmotic pressure, high temperatures, increased organic solvent content, freeze-dry cycles, and high alcohol concentrations.15,21,22 Additionally, cyclopropane modifications in phospholipids have been correlated with acid stress resistance under low pH conditions, improved resistance to oxidative stress, and changes in membrane fluidity.15,23–25 It is thought that acid stress resistance conferred from CFAs may be due to slower transport of protons into the cell,26 while changes in membrane fluidity can result in limited permeability to antibiotics and other drugs.27,28
Mycobacteria such as M. tuberculosis, the causative agent of tuberculosis (TB), contain a unique and diverse lipidome, which includes an abundance of cyclopropyl fatty acids termed mycolic acids (MAs). MAs can be subdivided into three categories–alpha-, keto-, and methoxy-MAs–and contain one to two cyclopropane modifications depending on their subclass. These long-chain fatty acids influence lipid packing within the mycobacterial cell envelope, thereby affecting fundamental cell wall functions.29 Furthermore, antibiotic-resistant M. tuberculosis strains have been linked to distinctive remodeling of cell wall lipids.30 Previous studies have shown that these CFAs are directly correlated with M. tuberculosis persistence, indicating that they play a critical role in mycobacterial pathogenesis.27,31–33 Additionally, MAs are foreign, chemically inert compounds allowing them to be used as diagnostic markers for TB.34–36
The generation of CFAs in bacterial lipids is recognized as a crucial part of adaption to environmental stressors. These major biological ramifications make cyclopropane modifications important targets for structural characterization. Existing methods for structural characterization of cyclopropyl lipids include chemical derivatization of CFAs followed by gas-chromatography (GC) mass spectrometry and multi-stage linear ion trap MS analysis of metal-adducted lipid species.37–40 However, the derivatization methods required for GC analysis of cyclopropyl lipid analysis are time consuming and are unable to enhance sufficiently the volatilities of all classes of lipids.37,38,41 Multi-stage MS methods with collisional activation result in fragmentation at multiple locations adjacent to the cyclopropane ring, yielding convoluted spectra that can confound interpretation.39,40,42 Liquid chromatography (LC) with UV detection has previously been employed to differentiate mycobacterial species,43 and MS/MS has enabled quantitation of alpha-, keto- and methoxy-MAs in clinical samples and allowed the presence of MAs to be used as diagnostic markers for TB infection.44–46 More recently, MS/MS methods have even been used to identify MAs in archaeological material and profile the species as M. tuberculosis.47 Many of the aforementioned methods for structural characterization of cyclopropyl lipids including phospholipids and MAs are able to categorize lipids by class based on collisional dissociation and high-resolution MS1 measurements but are unable to provide information on the location and type of the unsaturation elements within these lipids.48
To overcome these types of shortcomings, alternative activation methods such as ultraviolet photodissociation (UVPD) have been employed to decipher subtle structural features such as double bonds in phospholipids, including degree and location of unsaturation, which are not accessible using traditional collision-based fragmentation methods.49,50 UVPD is able to generate unique, informative fragment ions owing to higher-energy pathways not accessed by collisional-based methods. Here, we report the use of 213 nm UVPD-MS for the characterization of CFAs in bacterial lipids, both in a shotgun approach and when integrated with chromatographic separation for the analysis of cyclopropyl lipids in complex bacterial extracts.
Experimental
Materials
All lipid standards including PE (16:0/17:1(c9Z)), PC (16:0/17:1(c9Z)), α-mycolic acid (C-80) and E. coli total lipid extract were purchased from Avanti Polar Lipids (Alabaster, Alabama). Structures and molecular weights of cyclopropane lipid standards are shown in Table S1. Gradient grade OmniSolv water, acetonitrile (ACN) and methanol were purchased from VWR (Radnor, PA). Chloroform and Myobacterium bovis (M. bovis) lipid extract were purchased from Sigma Aldrich (St. Louis, MO). LC grade isopropyl alcohol (IPA) and Optima LC-MS grade formic acid, ammonium formate and ammonium acetate were purchased from Thermo Fisher Scientific (Waltham, MA). Mycobacterium tuberculosis strains H37rV (NR-14837), CDC1551 (NR-14838), and HN878 (NR-14839) total lipids were obtained through BEI Resources, NIAID, NIH.
Nomenclature
Shorthand notation for phospholipid nomenclature is adapted as previously described by Liebisch et al.51 For example, PE (16:0/18:1(9Z)) indicates that the lipid contains a PE headgroup and two acyl chains with known sn-stereochemistry (as designated by the “/” as opposed to “_” when sn-stereochemistry is unknown), that the sn-1 position contains 16 carbons with zero unsaturations, and that the sn-2 position contains 18 carbons with one unsaturation element (double bond) at the 9-carbon position as counted from the acyl chain carbonyl carbon. The configuration of the double bond is indicated as cis by “9Z”, and no additional information is given in parentheses in cases where the double bond location is unknown, or “9Δ” when the stereochemistry of the double bond is unknown. Existing nomenclature was extended to describe CFAs, for example PE (16:0/17:1(c9Z)) indicates that the fatty acid at the sn-2 position contains one unsaturation, and is further specified as a cyclopropane ring at the 9-carbon position in the cis configuration. The “c” in parentheses further characterizing the nature of the unsaturation as “cyclopropane” avoids confusion with double bond unsaturation elements. If multiple different unsaturation elements exist within an acyl chain, these could be separated by commas inside the parentheses, for example PE (16:0/18:2(c9Z,11Z)) indicates the presence of a cis-cyclopropane unsaturation at the 9-carbon position and a cis-double bond unsaturation at the 11-carbon position.
Liquid Chromatography and Mass Spectrometry
For all experiments, lipid standards were diluted to 10 μM in 50:50 chloroform:methanol. All spectra were acquired on a Thermo Fisher Scientific Orbitrap Fusion Lumos Tribrid mass spectrometer equipped with a 213 nm UVPD module (San Jose, CA). For shotgun experiments, the sample (~10 μL) was loaded into a silver-coated pulled tip glass capillary of 1.2 mm outer diameter and then subsequently sprayed using a Nanospray Flex ion source (Thermo Fisher Scientific, San Jose, CA). Spectra were acquired in both positive and negative modes at a spray voltage of 1.0 kV. MS/MS spectra were collected with an isolation width of 1 m/z, an AGC target of 5e5, a resolving power of 30,000 at m/z 200, and with 25 scans averaged with 2 μscans per scan. HCD was performed at a normalized collision energy (NCE) of 25, and UVPD was performed with an activation time of 200 ms.
For all LC-MS experiments, spectra were collected with a heated electrospray ionization (H-ESI) source (Thermo Fisher Scientific, San Jose, CA) operated in negative mode at a spray voltage of of 3.8 kV. For HCD spectra, a NCE of 25 was used, along with an isolation width of 1 m/z, an AGC target of 2e5, and 2 μcans per scan. For UVPD spectra, an activation period of 100–200 ms was used, and 3 μscans per scan. All MS/MS spectra were collected using a resolving power of 15,000 at m/z 200. Alternating HCD and UVPD scans were collected in a data-dependent fashion during a total cycle time of 3–6 s between MS1 scans. Biological extracts were diluted to 50 ng/μL in 50:50 chloroform:methanol and separated using a reversed phase Acquity UPLC CSH C18 column (pore size 130 Å, 1.7 μm particle size, 2.1mm × 100 mm, Waters, Milford, MA) on a Dionex Ultimate 3000 UHPLC system (Thermo Fisher Scientific, Sunnyvale, CA). For phospholipid analysis, lipid extracts were eluted at a flow rate of 260 μL/min using a gradient of 10% B from 0 to 2 minutes, up to 45% B at 6 minutes, up to 60% B at 46 minutes, up to 95% B at 47 minutes which was held for 6 minutes, and back down to 10% B at 55 minutes for a total of 7 minutes of column re-equilibration. Mobile phase compositions were (A) 60:40 ACN:water and (B) 90:10 IPA:ACN both with 10 mM ammonium formate and 0.1% formic acid.52 For MA analysis, lipid extracts were eluted with mobile phase compositions (A) methanol with 10 mM ammonium acetate and (B) 75:25 chloroform:methanol with 10 mM ammonium acetate, similar to elution conditions described previously.34 Gradient conditions were 15% B from 0 to 5 minutes, up to 60% B from 5 to 35 minutes, held at 60% B from 35 to 41 minutes, back down to 15% B at 42 minutes, and finally 5 minutes of column re-equilibration with a constant flow rate of 400 μL/min. The injection volume for all samples was 10 μL, and the column compartment was heated to 50–65 °C during the entirety of all LC-MS runs.
All UVPD experiments were performed in the low-pressure trap of the dual linear ion trap. Default activation times were used for all HCD experiments. Mass spectral data were analyzed using XCalibur Qual Browser Version 3.1 (Thermo Fisher Scientific) and manually interpreted with the aid of ChemDraw (PerkinElmer). Mtb Lipid, MycoMass and MycoMap databases were used for secondary confirmation and comparison of proposed mycolic acid structures.53,54
Results and Discussion
Glycerophospholipids may be analyzed in the positive or negative ESI modes. The positive mode is typically used for PCs owing to the fixed positive charge on the choline headgroup, or for cation-adducted lipids. Negative ion mode, which is more commonly used for PGs, PIs, PAs and PSs, produces deprotonated species. Zwitterionic PEs are amenable to both positive and negative mode analysis. Both positive and negative modes were used in the present study owing to the variety of glycerophospholipid classes. Representative MS1 spectra are illustrated in Figure S1 for both positive mode analysis of PC (16:0/17:1(c9Z)) and negative mode analysis of PE (16:0/17:1(c9Z)). Both HCD and 213 nm UVPD were used for characterization of the glycerophospholipids to assess the unique structural information provided by each activation method. Upon HCD, singly deprotonated cyclopropane glycerophospholipid PE (16:0/17:1(c9Z)) of m/z 702 (Figure 1a) produces two dominant fragment ions of m/z 255 and m/z 267 corresponding to acyl chain product ions that are typically the most prominent products formed upon collisional activation of glycerophospholipids in the negative mode. In addition, a number of fragment ions of much lower abundance are observed that correspond to acyl chain neutral losses (m/z 434, 446, 452, 464). 213 nm UVPD of this lipid generates similar products as observed upon HCD, while additionally generating a unique pair of fragment ions of m/z 590 and m/z 604 (Figure 1b), which are absent in the corresponding HCD spectrum. This pair of fragment ions is attributed to dual C-C bond cleavages across the cyclopropane ring within the 17:1(c9Z) acyl chain, as illustrated in the fragment ion map in Figure 1c.
Figure 1.
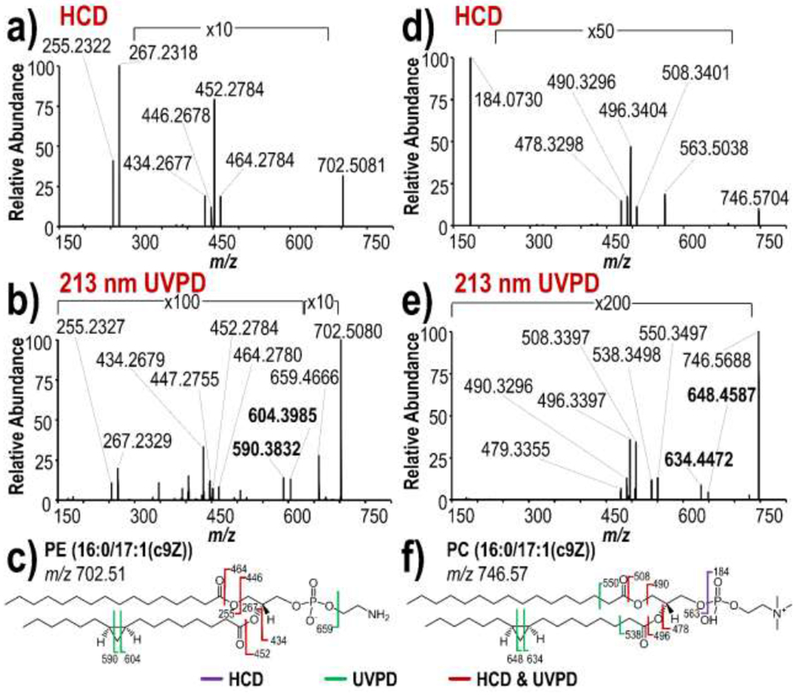
MS/MS spectra of PE (16:0/17:1(c9Z)) in negative ion mode: a) HCD, b) 213 nm UVPD and c) fragment ion map. MS/MS spectra of PC (16:0/17:1(c9Z)) in positive ion mode: d) HCD, and e) 213 nm UVPD and f) fragment ion map.
The types of products observed upon HCD and 213 nm UVPD of PC (16:0/17:1(c9Z)) of m/z 746 in the positive mode are similar to those generated by PE (16:0/17:1(c9Z)) in the negative mode. As seen in Figure 1d, HCD yields the characteristic PC headgroup ions (m/z 184, 563), while both HCD and UVPD yield acyl chain neutral loss products (m/z 478, 490, 496, 508). The diagnostic cyclopropane cleavage products separated by 14 Da (m/z 634, 648) are generated exclusively by 213 nm UVPD (Figure 1e). These results highlight the unique ability of 213 nm UVPD to localize cyclopropane rings in glycerophospholipids via a pair of fragment ions not observed upon HCD (Figure 1c,f). Importantly, this fragmentation pattern is demonstrated for both positively- and negatively-charged glycerophospholipids, making it polarity-independent.
The UVPD fragmentation efficiency is relatively low, as evidenced by the amplification factor of 100X or 200X used to enhance the diagnostic fragment ions in most of the MS/MS spectra. The low efficiency likely results from the relatively low photoabsorption cross-section of the lipids at 213 nm. Despite the low abundances of the diagnostic fragment ions, the signal-to-noise is excellent for ions detected using an Orbitrap mass analyzer, thus allowing the confident assignment of the key ions. A calibration curve for determination of the limit of detection (LOD) of cyclopropane lipids based on monitoring diagnostic fragment ions generated by UVPD is shown in Figure S2, yielding a LOD of 45 pmol (S/N 3).
UVPD-MS localization of cyclopropane rings was further extended to MA-type lipids, found in Mycobacteria. The distinguishing structural feature of MAs is that these long-chain fatty acids contain one to two cyclopropane rings. MAs are categorized into three main classes: alpha-, keto-, and methoxy-MAs (Chart 1). Alpha-MAs contain two cyclopropane rings within their R1 chain, whereas keto- and methoxy-MAs contain one cyclopropane ring in the R1 chain in addition to a ketone or methoxy functional group, respectively.55 Cyclopropane ring locations are counted from the C1 position at which branching to R1 and R2 groups occurs. For this example, deprotonated α-MA (C-80(c16Z, c32Z)) containing two cyclopropane rings was subjected to 213 nm UVPD (Figure 2). Two pairs of diagnostic fragment ions each spaced 14.0 Da apart were observed corresponding to cross-ring cleavage at both the proximal c16Z (m/z 619, 633) and distal c32Z (m/z 855, 869) cyclopropane rings. HCD of α-MA (C-80(c16Z, c32Z)) generates a single diagnostic fragment ion of m/z 395, which corresponds to cleavage between the C2 and C3 MA carbons and reveals the R2 α-branch as C24H49 (Figure S3). UVPD-MS offers a new means to characterize cyclopropane rings in MAs, allowing localization of multiple cyclopropane rings within a single lipid. This fragmentation pathway is accessible for both phospholipids and long-chain, hydrophobic fatty acids upon UVPD. For the characterization of glycerophospholipids and MAs, combining HCD and UVPD allows the determination of both fatty acid chain composition (acyl chains and R2 α-branch) as well as cyclopropane localization based on the complementary fragmentation patterns afforded by each method.
Chart 1.
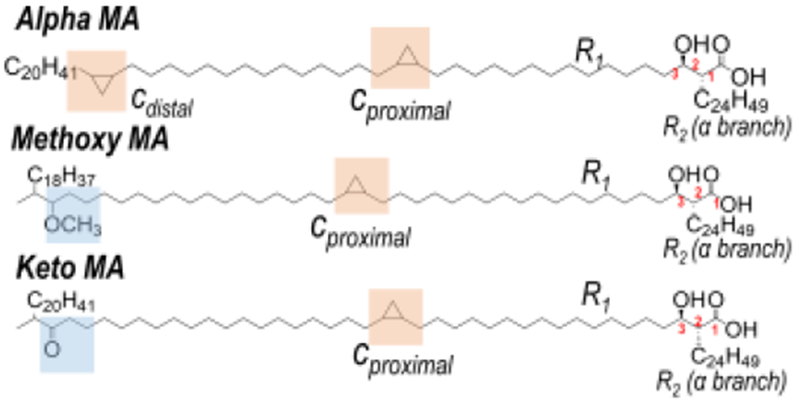
Representative structures of three major classes of mycolic acids with indicated carbon numbering scheme in red
Figure 2.
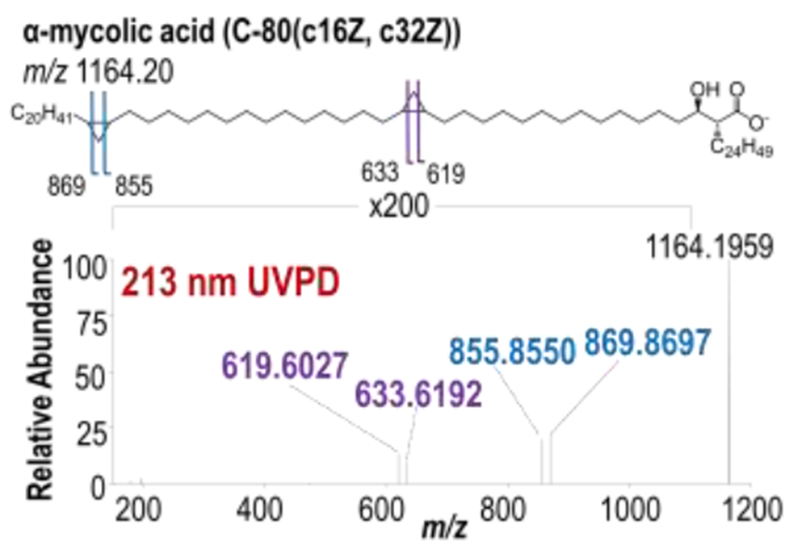
Negative mode 213 nm UVPD mass spectrum of α-mycolic acid (C-80(c16Z, c32Z)) with corresponding fragment ion map.
We speculate that cyclopropane fragmentation occurs via cross-ring cleavage on either side of the cyclopropane ring, as cross-ring fragmentation via UVPD has previously been observed in both oligosaccharides and glycans.56 Scheme S1 illustrates the alternative dual C-C bond cleavages that afford the pair of products spaced 14.0 Da apart, as shown for PE (16:0/17:1(c9Z)), corresponding to a difference of CH2. This pathway is similar to the recently proposed fragmentation route of epoxides in unsaturated lipids modified by meta-chloroperoxybenzoic acid reactions.57
For the lipids in the present study, diagnostic ions that originate from cross-ring cleavages of the cyclopropane rings may be generated upon UVPD for several reasons. The ring strain of the cyclopropyl group results in a higher ground state energy, effectively reducing the energy of the σ to σ* transition, giving the C-C bonds more pi character and increasing the photoabsorption cross-section in the UV range.58,59 The lower energy of the σ to σ* transition of a cyclopropane ring and lower bond energy of the C-C bond in cyclopropane compared to a standard C-C bond may account for the preferential cleavage upon UVPD. The maximum absorption of cyclopropane rings falls in the higher-wavelength end of the vacuum-UV range in contrast to its non-cyclic isomers, which have maximum absorption wavelengths in the lower end of the vacuum-UV range, resulting in selective absorption and fragmentation at the cyclopropane ring and not along the aliphatic carbon chain.
The diagnostic cross-ring cleavages observed upon 213 nm UVPD are also observed upon 193 nm UVPD (Figure S4). Owing to the higher power of the excimer laser used for 193 nm UVPD relative to the lower power laser used for 213 nm UVPD, the diagnostic fragment ions are more abundant upon 193 nm UVPD and are generated using a shorter activation period (20 msec) compared to the 200 ms period employed for 213 nm. The recent introduction of the 213 nm UVPD option on a commercial Orbitrap mass spectrometer platform makes it more widely available for other users.
To extend the strategy to more complex biological samples, an LC-MS workflow was implemented for the high-throughput analysis of cyclopropyl glycerophospholipids in an E. coli total lipid extract. After acquisition of survey MS1 spectra, data-dependent HCD and UVPD spectra were acquired for each precursor in alternating scans during a net 3-s cycle time. For confident identification of cyclopropyl glycerophospholipids, UVPD enabled cyclopropane localization and HCD provided structural confirmation of the lipid.
An example of the base peak LC-MS/MS chromatogram from an E. coli extract is shown in Figure 3a, along with an overlay of the extracted ion chromatogram (XIC) of the highest-abundance cyclopropyl glycerophospholipid (m/z 702). The majority of glycerophospholipids elute between 17 and 35 minutes. Representative HCD (Figure 3b) and 213 nm UVPD (Figure 3c) spectra allow the confident identification of the phospholipid of m/z 702 as PE (16:0_17:1(c9Δ)) (Figure 3d). Product ions of m/z 255 and 267 in the HCD and UVPD spectra enable identification of the two acyl chains as 16:0 and 17:1, respectively, whereas the diagnostic fragment ions of m/z 590 and 604 in the UVPD spectrum confirm the location of the cyclopropane ring at the 9-carbon position within the 17:1 acyl chain. In total, the LC/UVPD-MS method allowed the identification of thirteen cyclopropyl glycerophospholipid species (Table 1), all of which were identified as either PEs or PGs. All identified cyclopropane rings were localized to either the 9- or 11-carbon position within 17:1 or 19:1 acyl chains, respectively. These results are in agreement with the previously characterized glycerophospholipid composition of E. coli,49 as 17:1 and 19:1 cyclopropane acyl chains arise from their 16:1 and 18:1 double bond acyl chain precursors (Scheme S2). Additionally, we observed that many of the identified cyclopropane phospholipids in E. coli phospholipid extract accounted for some of the most abundant phospholipids overall in the sample. All identified glycerophospholipids in E. coli total lipid extract contained at most one cyclopropane ring within each acyl chain.
Figure 3.
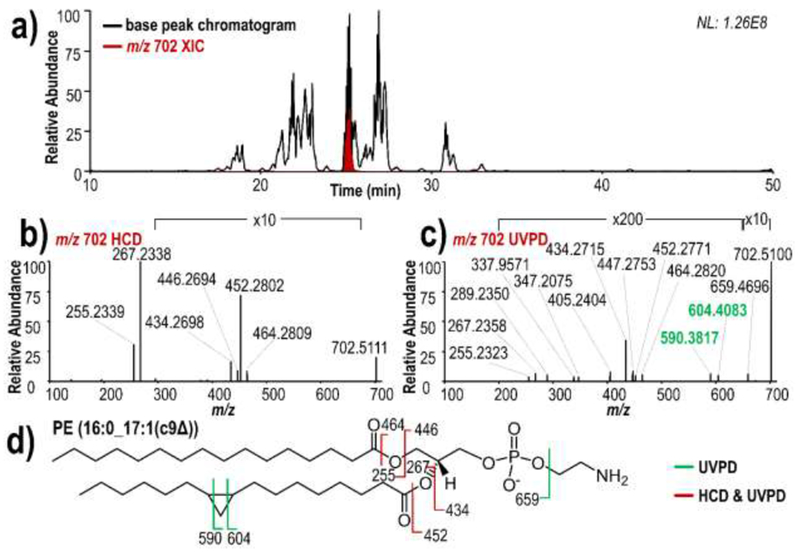
a) Negative mode LC-MS base peak chromatogram and m/z 702 extracted ion chromatogram for E. coli lipid extract, and representative b) HCD and c) 213 nm UVPD mass spectra of m/z 702, d) fragment ion map.
Table 1.
Identified cyclopropane glycerophospholipids from E. coli lipid extract by negative mode LC-MS/MS
| Headgroup | m/z [M-H]− | Fatty acid chains | m/z Diagnostic Ions | Ring Position | ||
|---|---|---|---|---|---|---|
| PG | 733.50 | 16:0 | 17.1(c) | 621.38 | 635.39 | c9Δ |
| 759.52 | 18:1 | 17:1(c) | 647.39 | 661.41 | c9Δ | |
| 761.53 | 18:0 | 17:1(c) | 649.40 | 663.43 | c9Δ | |
| 787.55 | 18:1 | 19:1(c) | 675.43 | 689.45 | c11Δ | |
| PE | 700.49 | 16:1 | 17:1(c) | 588.37 | 602.38 | c9Δ |
| 702.51 | 16:0 | 17:1(c) | 590.38 | 604.40 | c9Δ | |
| 714.50 | 17:1(c) | 17:1(c) | 602.39 | 616.40 | c9Δ | |
| 728.52 | 18:1 | 17:1(c) | 616.40 | 630.42 | c9Δ | |
| 16:1 | 19:1(c) | 616.41 | 630.41 | c11Δ | ||
| 730.54 | 16:0 | 19:1(c) | 618.41 | 632.44 | c11Δ | |
| 18:0 | 17:1(c) | 618.41 | 632.43 | c9Δ | ||
| 742.54 | 19:1(c) | 17:1(c) | 630.41 | 644.43 | c9Δ (17:1) c11Δ (19:1) |
|
| 756.65 | 18:1 | 19:1(c) | 644.42 | 658.44 | c11 | |
This LC-MS/MS approach was further applied to the analysis of MAs in complex mycobacterial lipid extracts. The base peak LC-MS/MS chromatogram of M. bovis lipid extract overlaid with the XIC of the MA of highest abundance (m/z 1136) is shown in Figure 4a with elution of MAs occurring between 10 and 18 minutes. Representative HCD (Figure 4b) and 213 nm UVPD (Figure 4c) spectra allow identification of the species of m/z 1136 as α-MA (C-78 (c14Δ, c30Δ)) (Figure 4d). The fragment ion of m/z 395 observed upon HCD enables identification of the R2 α-branch as C24H49, while two pairs of diagnostic fragment ions observed upon UVPD correspond to cross-ring cleavage at both the proximal c14Δ (m/z 591, 605) and distal c30Δ (m/z 827, 841) and cyclopropane rings. Localization of two cyclopropane rings within the R1 chain combined with high-resolution high-mass accuracy intact mass measurements permit classification of this lipid as an α-MA.
Figure 4.
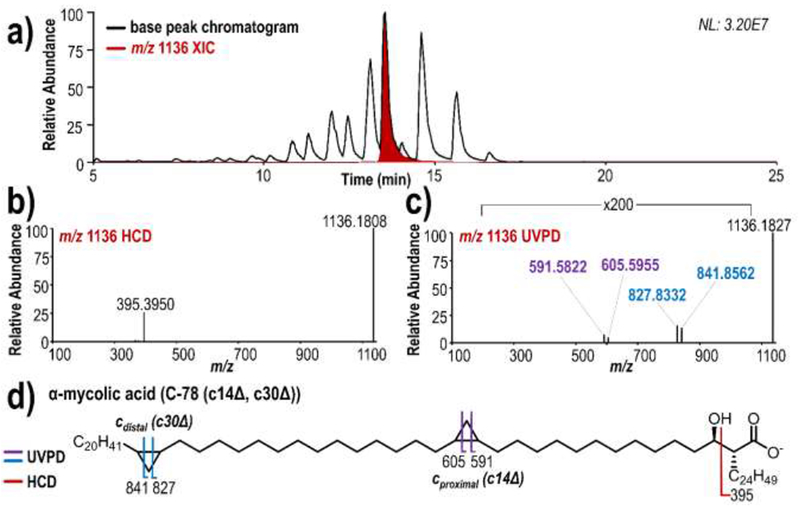
a) Negative mode LC-MS base peak chromatogram and m/z 1136 extracted ion chromatogram of m/z 1136 from M. bovis lipid extract, and representative b) HCD and c) 213 nm UVPD mass spectra of m/z 1136, d) fragment ion map.
Seven MA species were confidently assigned based on the LC-MS/MS analysis of M. bovis (Table 2). Although chromatographic separation of MAs was optimized extensively, the high hydrophobicities and minimal relative differences in hydrophobicities between MAs present considerable separation challenges. In certain cases, isomeric MAs were not baseline resolved, therefore resulting in multiple possible structural identifications for a given m/z. Therefore, chimeric MS/MS spectra. For example, activation of a methoxy-MA of m/z 1224 generated fragment ions of m/z 367 and 395 upon HCD (Figure S5a), while UVPD yielded a pair of diagnostic cyclopropane product ions of m/z 647 and 661 (Figure S5b). This compilation of fragment ions leads to two possible structural identities – a methoxy-MA with a C24H49 R2 α-branch (assigned based on the fragment ion of m/z 395) and a cyclopropane ring at the 18Δ position within the R2 chain (m/z 647, 661) as seen in Figure S5c, or a methoxy-MA with a C22H45 R2 α-branch (m/z 367) and a cyclopropane ring at the 20Δ position within the R2 chain (m/z 647, 661), as seen in Figure S5d. In the case of this type of ambiguity, all reasonable structural possibilities are included in the tables for a given precursor ion. Cyclopropane positions in M. bovis MAs as counted from the C1 position at which branching to R1 and R2 groups occurs varied from c14Δ to c20Δ for the proximal cyclopropane ring, and from c30Δ to c32Δ for the distal cyclopropane ring found exclusively in α-MAs.
Table 2.
Identified mycolic acids from M. bovis lipid extract by negative mode LC-MS/MS
| m/z [M-H]− | Chemical Formula | MA Class | m/z R2 (α branch) | Cproximal | Cdistal | ||||
|---|---|---|---|---|---|---|---|---|---|
| m/z diagnostic ions | Ring position | m/z diagnostic ions | Ring position | ||||||
| 1136.1827 | C78H156O3 | alpha | 395 | 591.5822 | 605.5955 | c14Δ | 827.8332 | 841.8562 | c30Δ |
| 1164.2144 | C80H156O3 | alpha | 395 | 619.6094 | 633.6285 | c16Δ | 855.8664 | 869.8799 | c32Δ |
| 1224.2723 | C83H164O3 | methoxy | 367, 395 | 647.6461 | 661.6635 | c18Δ (395) c20Δ (367) |
- | - | - |
| 1252.3030 | C85H168O4 | methoxy | 395 | 675.6829 | 689.6957 | c20Δ | - | - | - |
| 1264.3038 | C86H168O4 | keto | 395 | 675.6766 | 689.6908 | c20Δ | - | - | - |
| 1278.3201 | C87H170O4 | keto | 395 | 661.6508 | 675.6776 | c19Δ | - | - | - |
| 1280.3351 | C87H172O4 | methoxy | 395 | 675.6799 | 689.6937 | c20Δ | - | - | - |
In addition to M. bovis, lipid extracts from three virulent strains of M. tuberculosis, H37rV, HN878 and CDC1551, were analyzed via LC-MS/MS. Of these three strains, HN878 and CDC1551 are clinical isolates, whereas H37rV is a laboratory strain.60 HN878 is considered a “hypervirulent” Mtb strain owing to its enhanced ability to rapidly grow and eradicate infected mice in clinical studies in comparison to other clinical isolates of Mtb.61 In contrast, clinical isolate CDC1551, although not hypervirulent, has been shown to induce a more vigorous host response in vivo and in vitro.61 H37rV is a well-studied virulent laboratory Mtb strain.62
The base peak LC-MS chromatogram of Mtb H37rV lipid extract is displayed in Figure S6 showing elution of MAs between 10 and 18 minutes. Acquisition of HCD and 213 nm UVPD spectra in alternating scans and interpretation based on the key diagnostic ions described above enabled structural identification of 15 MAs in Mtb H37rV lipid extract (Table 3). Of these 15 lipids, six methoxy-, five alpha-, and four keto-MAs were identified. Cyclopropane positions varied from c12Δ to c24Δ for the proximal cyclopropane ring, and from c28Δ to c37Δ for the distal cyclopropane ring in α-MAs. M. tuberculosis showed a broader level of diversity of MA cyclopropane locations than M. bovis. In particular, many M. tuberculosis MA isomers were found not only arising from variations in R1, R2 chain lengths but also based on cyclopropane location. For example, UVPD of the methoxy-MA of m/z 1308 produced fragments of m/z 703, 717 and 731, while HCD yielded predominantly one product ion of m/z 395 ion corresponding to the R2 α-branch. This suite of ions indicates that the cyclopropane within the R1 chain could be located at either the 22Δ or 23Δ position, and that both isomers are present and were not chromatographically resolved. The tabulated identities in Table 3 include all possible combinations of R1 α-branches and R2 cyclopropane positions based on the combined HCD and UVPD spectra. LC-MS base peak chromatograms and tables of identified MAs for Mtb HN878 and CDC1551 are provided in Figure S7 (a–d). Variations in MA patterns have previously been observed between different Mtb complex strains and lineages,63 indicating there may also be biologically relevant patterns of cyclopropanation in MAs based on strain/lineage which can be connected to relevant in vivo functions. An intriguing yet unrealized opportunity is the discovery and profiling of MAs for differentiation of strains and lineages of Mtb using UVPD-MS. In order to achieve this goal, the current workflow requires additional refinement of the chromatographic method to enable successful profiling of a greater array of isomeric MAs.
Table 3.
Identified mycolic acids from negative mode LC-MS/MS of Mtb H37rV lipid extract
| m/z [M-H]− | Chemical Formula | MA Class | m/z R2(α branch) | Cproximal | Cdistal | ||||
|---|---|---|---|---|---|---|---|---|---|
| m/z diagnostic ions | Ring position | m/z diagnostic ions | Ring position | ||||||
| 1108.1517 | C76H148O3 | alpha | 367, 395 | 563.5489 | 577.5628 | c12Δ (395), c14Δ (367) | 799.8055 | 813.8219 | c28Δ (395), c30Δ (367) |
| 813.8219 | 827.8329 | c29Δ (395), c31Δ (367) | |||||||
| 827.8329 | 841.8343 | c30Δ (395), c32Δ (367) | |||||||
| 1136.1832 | C78H152O3 | alpha | 367, 395 | 591.5841 | 605.6042 | c14Δ (395), c16Δ (367) | 827.8365 | 841.8579 | c30Δ (395), c32Δ (367) |
| 841.8579 | 855.8885 | c31Δ (395), c33Δ (367) | |||||||
| 1164.2152 | C80H156O3 | alpha | 367, 395 | 605.5902 | 619.6191 | c15Δ (395), c17& (367) | 855.8632 | 869.8831 | c30Δ (395), c32Δ (367) |
| 619.6191 | 633.6240 | c16Δ (395), c8Δ (367) | |||||||
| 1192.2462 | C82H160O3 | alpha | 367, 395 | 647.6431 | 661.6550 | c18Δ (395), c20Δ (367) | 883.9001 897.9147 |
897.9147 911.9293 |
c34Δ (395), c36Δ (367) c35Δ (395), c37Δ (367) |
| 1196.2405 1220.2776 |
C81H160O4 C84H164O3 |
methoxy alpha |
339, 367, 395 395 |
619.6137 675.6735 |
633.6265 689.6898 |
c16Δ (395), c18Δ (367), c20Δ (339) c20Δ |
- 911.9219 |
- 925.9453 |
- c36Δ |
| 1224.2726 | C83H164O4 | methoxy | 367, 395 | 619.6279 | 633.6438 | cl6Δ (395), c18Δ (367) | - | - | - |
| 633.6438 | 647.6468 | c17Δ (395), c19Δ (367) | - | - | - | ||||
| 647.6468 | 661.6632 | c18Δ (395), c20Δ (367) | - | - | - | ||||
| 1236.2728 | C84H164O4 | keto | 367, 395 | 647.6412 | 661.6640 | c18Δ (395), c20Δ (367) | - | - | - |
| 661.6656 | 675.6862 | c19Δ (395), c21Δ (367) | - | - | - | ||||
| 675.6862 | 689.6714 | c20Δ (395), c22Δ (367) | - | - | - | ||||
| 1250.2877 | C85H166O4 | keto | 367, 395 | 633.6160 | 647.6550 | c17Δ (395), c19Δ (367) | - | - | - |
| 1252.3036 1264.3048 |
C85H168O4 C86H168O4 |
methoxy keto |
367, 395 395 |
647.6450 | 661.6660 | c18Δ (395), c20Δ (367) | - | - | - |
| 661.6660 | 675.6946 | c19Δ (395), c21Δ (367) | - | - | - | ||||
| 675.6946 675.6756 |
689.7070 689.6970 |
c20Δ (395), c22Δ (367) c20Δ |
- - |
- - |
- - |
||||
| 1278.3203 | C87H170O4 | keto | 395 | 661.6488 | 675.6763 | c19Δ | - | - | - |
| 1280.3353 | C87H172O4 | methoxy | 395 | 675.6829 | 689.6929 | c20Δ | - | - | - |
| 689.6929 | 703.7004 | c21Δ | - | - | - | ||||
| 703.7004 | 717.7073 | c22Δ | - | - | - | ||||
| 1308.3666 | C89H176O4 | methoxy | 395 | 703.7078 717.7203 |
717.7203 731.7619 |
c22Δ c23Δ |
- - |
- - |
- - |
| 1336.3982 | c91H180O4 | methoxy | 395 | 731.7510 | 745.7446 | c24Δ | - | - | - |
Conclusions
Although traditional collisional activation methods enable characterization of basic structural lipid features, they fail to elucidate more subtle features such as cyclopropane rings. The work presented herein showcases the utility of 213 nm UVPD-MS for localization of cyclopropane rings in bacterial lipids, as UVPD results in production of a diagnostic pair of fragment ions spaced 14 Da apart corresponding to dual cross-ring C-C cleavage of each cyclopropane ring. UVPD generates the key cross-ring cleavage products for both positively and negatively charged lipids across lipid classes (glycerophospholipids and mycolic acids). For MAs, UVPD generates multiple sets of diagnostic ion pairs enabling localization of more than one cyclopropane ring within a single lipid. Additionally, UVPD-MS has also been implemented in an LC workflow for analysis of complex lipid extracts including ones from E. coli, M. bovis and M. tuberculosis. Overall, UVPD-MS presents a robust method of analysis for the localization of cyclopropane rings in bacterial lipids, enabling detailed structural characterization of subtle lipid features that are known to play major roles in cellular tolerance to adverse conditions and bacterial pathogenesis.
Supplementary Material
Acknowledgements
Funding from the NIH (RO1 GM103655), the Welch Foundation (F-1155), and the UT System for support of the UT System Proteomics Core Facility Network is gratefully acknowledged. Brendon Naicker, Duncan Cromarty and Jan A. Verschoor at the University of Pretoria are gratefully acknowledged for helpful discussions regarding mycolic acid liquid chromatography.
Footnotes
The authors declare no competing financial interest.
Associated Content
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website.
Scheme S1, possible fragmentation pathway for UVPD of cyclopropane rings in bacterial lipids as shown for PE (16:0/17:1(c9Z)); Scheme S2, in vivo generation of bacterial cyclopropane glycerophospholipid PE (16:0/17:1(c9Z)) from double bond precursor PE (16:0/16:1(9Z)); Table S1, structures of all cyclopropane lipid standards; Figure S1, MS1 spectra of PE (16:0/17:1(c9Z)) and PC (16:0/17:1(c9Z)) with corresponding structures; Figure S2, Calibration curve for PE (16:0/17:1(c9Z)) cyclopropane diagnostic ion LC/UVPD-MS peak areas as a function of injection amount ; Figure S3, HCD spectrum of α-MA (C-80(c16Z, c32Z)) with fragment ion map; Figure S4, Comparison of 193 and 213 nm UVPD for cyclopropane localization in PE (16:0/17:1(c9Z)); Figure S5, HCD and UVPD spectra of methoxy-MA of m/z 1224 with corresponding structural identifications and fragment maps; Figure S6, LC-MS base peak chromatogram of Mtb H37rV lipid extract; Figure S7, LC-MS base peak chromatograms of Mtb HN878 and CDC1551 lipid extracts with corresponding lists of identified MAs.
References
- (1).Rolim AE; Henrique-Araújo R; Ferraz EG; De FAAD; Fernandez LG Lipidomics in the Study of Lipid Metabolism: Current Perspectives in the Omic Sciences. Gene 2015, 554, 131–139. [DOI] [PubMed] [Google Scholar]
- (2).Han X Lipidomics for Studying Metabolism. Nat. Rev. Endocrinol. 2016, 12, 668–679. [DOI] [PubMed] [Google Scholar]
- (3).Rustam YH; Reid GE Analytical Challenges and Recent Advances in Mass Spectrometry Based Lipidomics. Anal. Chem. 2018, 90, 374–397. [DOI] [PubMed] [Google Scholar]
- (4).Han X; Gross RW Shotgun Lipidomics: Electrospray Ionization Mass Spectrometric Analysis and Quantitation of Cellular Lipidomes Directly from Crude Extracts of Biological Samples. Mass Spectrom. Rev. 2005, 24, 367–412. [DOI] [PubMed] [Google Scholar]
- (5).Han X; Gross RW Shotgun Lipidomics: Multidimensional MS Analysis of Cellular Lipidomes. Expert Rev. Proteomics 2005, 2, 253–264. [DOI] [PubMed] [Google Scholar]
- (6).Yang K; Zhao Z; Gross RW; Han X Identification and Quantitation of Unsaturated Fatty Acid Isomers by Electrospray Ionization Tandem Mass Spectrometry: A Shotgun Lipidomics Approach. Anal. Chem. 2011, 83, 4243–4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Cajka T; Fiehn O Comprehensive Analysis of Lipids in Biological Systems by Liquid Chromatography-Mass Spectrometry. TrAC Trends in Analytical Chemistry 2014, 61, 192–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Retra K; Bleijerveld OB; van Gestel RA; Tielens AGM; van Hellemond JJ; Brouwers JF A Simple and Universal Method for the Separation and Identification of Phospholipid Molecular Species. Rapid Commun. Mass Spectrom. 2008, 22, 1853–1862. [DOI] [PubMed] [Google Scholar]
- (9).Pulfer M; Murphy RC Electrospray Mass Spectrometry of Phospholipids. Mass Spectrom. Rev. 2003, 22, 332–364. [DOI] [PubMed] [Google Scholar]
- (10).Brügger B Lipidomics: Analysis of the Lipid Composition of Cells and Subcellular Organelles by Electrospray Ionization Mass Spectrometry. Annual Review of Biochemistry 2014, 83, 79–98. [DOI] [PubMed] [Google Scholar]
- (11).Shevchenko A; Simons K Lipidomics: Coming to Grips with Lipid Diversity. Nat. Rev. Mol. Cell Biol. 2010, 11, 593–598. [DOI] [PubMed] [Google Scholar]
- (12).Brown SHJ; Mitchell TW; Oakley AJ; Pham HT; Blanksby SJ Time to Face the Fats: What Can Mass Spectrometry Reveal about the Structure of Lipids and Their Interactions with Proteins? J. Am. Soc. Mass Spectrom. 2012, 23, 1441–1449. [DOI] [PubMed] [Google Scholar]
- (13).Hsu F-F; Turk J Electrospray Ionization with Low-Energy Collisionally Activated Dissociation Tandem Mass Spectrometry of Glycerophospholipids: Mechanisms of Fragmentation and Structural Characterization. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci 2009, 877, 2673–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Hsu F-F; Turk J Electrospray Ionization/Tandem Quadrupole Mass Spectrometric Studies on Phosphatidylcholines: The Fragmentation Processes. J. Am. Soc. Mass Spectrom. 2003, 14, 352–363. [DOI] [PubMed] [Google Scholar]
- (15).Poger D; Mark AE A Ring to Rule Them All: The Effect of Cyclopropane Fatty Acids on the Fluidity of Lipid Bilayers. J. Phys. Chem. B 2015, 119, 5487–5495. [DOI] [PubMed] [Google Scholar]
- (16).Wehrli PM; Angerer TB; Farewell A; Fletcher JS; Gottfries J Investigating the Role of the Stringent Response in Lipid Modifications during the Stationary Phase in E. Coli by Direct Analysis with Time-of-Flight-Secondary Ion Mass Spectrometry. Anal. Chem. 2016, 88, 8680–8688. [DOI] [PubMed] [Google Scholar]
- (17).Zhang Y-M; Rock CO Membrane Lipid Homeostasis in Bacteria. Nat. Rev. Microbiol. 2008, 6, 222–233. [DOI] [PubMed] [Google Scholar]
- (18).Cronan JE Phospholipid Modifications in Bacteria. Curr. Opin. Microbiol. 2002, 5, 202–205. [DOI] [PubMed] [Google Scholar]
- (19).To TMH; Grandvalet C; Tourdot-Maréchal R Cyclopropanation of Membrane Unsaturated Fatty Acids Is Not Essential to the Acid Stress Response of Lactococcus Lactis Subsp. Cremoris. Appl. Environ. Microbiol. 2011, 77, 3327–3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Beal C; Fonseca F; Corrieu G Resistance to Freezing and Frozen Storage of Streptococcus Thermophilus Is Related to Membrane Fatty Acid Composition. J. Dairy Sci. 2001, 84, 2347–2356. [DOI] [PubMed] [Google Scholar]
- (21).Pini C; Bernal P; Godoy P; Ramos J; Segura A Cyclopropane Fatty Acids Are Involved in Organic Solvent Tolerance but Not in Acid Stress Resistance in Pseudomonas Putida DOT-T1E. Microb. Biotechnol. 2009, 2, 253–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Muñoz-Rojas J; Bernal P; Duque E; Godoy P; Segura A; Ramos J-L Involvement of Cyclopropane Fatty Acids in the Response of Pseudomonas Putida KT2440 to Freeze-Drying. Appl. Environ. Microbiol. 2006, 72, 472–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Shabala L; Ross T Cyclopropane Fatty Acids Improve Escherichia Coli Survival in Acidified Minimal Media by Reducing Membrane Permeability to H+ and Enhanced Ability to Extrude H+. Res. Microbiol. 2008, 159, 458–461. [DOI] [PubMed] [Google Scholar]
- (24).Grogan DW; Cronan JE Cyclopropane Ring Formation in Membrane Lipids of Bacteria. Microbiol. Mol. Biol. Rev. 1997, 61, 429–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Chen YY; Gänzle MG Influence of Cyclopropane Fatty Acids on Heat, High Pressure, Acid and Oxidative Resistance in Escherichia Coli. Int. J. Food Microbiol. 2016, 222, 16–22. [DOI] [PubMed] [Google Scholar]
- (26).Royce LA; Boggess E; Fu Y; Liu P; Shanks JV; Dickerson J; Jarboe LR Transcriptomic Analysis of Carboxylic Acid Challenge in Escherichia Coli: Beyond Membrane Damage. PLoS One 2014, 9, e89580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Barkan D; Liu Z; Sacchettini JC; Glickman MS Mycolic Acid Cyclopropanation Is Essential for Viability, Drug Resistance, and Cell Wall Integrity of Mycobacterium Tuberculosis. Chem. Biol. 2009, 16, 499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Loffhagen N; Härtig C; Geyer W; Voyevoda M; Harms H Competition between Cis, Trans and Cyclopropane Fatty Acid Formation and Its Impact on Membrane Fluidity. Eng. Life Sci. 2007, 7, 67–74. [Google Scholar]
- (29).Watanabe M; Aoyagi Y; Ridell M; Minnikin DE Separation and Characterization of Individual Mycolic Acids in Representative Mycobacteria. Microbiology (Reading, Engl.) 2001, 147, 1825–1837. [DOI] [PubMed] [Google Scholar]
- (30).Lahiri N; Shah RR; Layre E; Young D; Ford C; Murray MB; Fortune SM; Moody DB Rifampin Resistance Mutations Are Associated with Broad Chemical Remodeling of Mycobacterium Tuberculosis. J. Biol. Chem. 2016, 291, 14248–14256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Glickman MS; Cox JS; Jacobs WR A Novel Mycolic Acid Cyclopropane Synthetase Is Required for Cording, Persistence, and Virulence of Mycobacterium Tuberculosis. Mol. Cell 2000, 5, 717–727. [DOI] [PubMed] [Google Scholar]
- (32).Rao V; Fujiwara N; Porcelli SA; Glickman MS Mycobacterium Tuberculosis Controls Host Innate Immune Activation through Cyclopropane Modification of a Glycolipid Effector Molecule. J. Exp. Med. 2005, 201, 535–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Ghazaei C Mycobacterium Tuberculosis and Lipids: Insights into Molecular Mechanisms from Persistence to Virulence. J. Res. Med. Sci. 2018, 23, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Ndlandla FL; Ejoh V; Stoltz AC; Naicker B; Cromarty AD; van Wyngaardt S; Khati M; Rotherham LS; Lemmer Y; Niebuhr J; et al. Standardization of Natural Mycolic Acid Antigen Composition and Production for Use in Biomarker Antibody Detection to Diagnose Active Tuberculosis. J. Immunol. Methods 2016, 435, 50–59. [DOI] [PubMed] [Google Scholar]
- (35).Lemmer Y; Thanyani ST; Vrey PJ; Driver CHS; Venter L; van Wyngaardt S; ten Bokum AMC; Ozoemena KI; Pilcher LA; Fernig DG; et al. Chapter Five - Detection of Antimycolic Acid Antibodies by Liposomal Biosensors In Methods in Enzymology; Düzgünes N, Ed.; Liposomes, Part F; Academic Press, 2009; Vol. 464, pp 79–104. [DOI] [PubMed] [Google Scholar]
- (36).Layre E; Al-Mubarak R; Belisle JT; Branch Moody D Mycobacterial Lipidomics. Microbiol. Spectr. 2014, 2, MGM2-0033-2013. [DOI] [PubMed] [Google Scholar]
- (37).Harvey DJ Picolinyl Derivatives for the Characterization of Cyclopropane Fatty Acids by Mass Spectrometry. Bio. Mass Spectrom. 1984, 11, 187–192. [Google Scholar]
- (38).Oursel D; Loutelier-Bourhis C; Orange N; Chevalier S; Norris V; Lange CM Identification and Relative Quantification of Fatty Acids in Escherichia Coli Membranes by Gas Chromatography/Mass Spectrometry. Rapid Commun. Mass Spectrom. 2007, 21, 3229–3233. [DOI] [PubMed] [Google Scholar]
- (39).Hsu F-F; Turk J Toward Total Structural Analysis of Cardiolipins: Multiple-Stage Linear Ion-Trap Mass Spectrometry on the [M − 2H + 3Li]+ Ions. J. Am. Soc. Mass Spectrom. 2010, 21, 1863–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Hsu F-F; Kuhlmann FM; Turk J; Beverley SM Multiple-Stage Linear Ion-Trap with High Resolution Mass Spectrometry towards Complete Structural Characterization of Phosphatidylethanolamines Containing Cyclopropane Fatty Acyl Chain in Leishmania Infantum. J. Mass Spectrom. 2014, 49, 201–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Crick PJ; Guan XL Lipid Metabolism in Mycobacteria—Insights Using Mass Spectrometry-Based Lipidomics. BBA - Mol. Cell Biol. L. 2016, 1861, 60–67. [DOI] [PubMed] [Google Scholar]
- (42).Watanabe M; Aoyagi Y; Mitome H; Fujita T; Naoki H; Ridell M; Minnikin DE Location of Functional Groups in Mycobacterial Meromycolate Chains; the Recognition of New Structural Principles in Mycolic Acids. Microbiology 2002, 148, 1881–1902. [DOI] [PubMed] [Google Scholar]
- (43).Butler WR; Guthertz LS Mycolic Acid Analysis by High-Performance Liquid Chromatography for Identification of Mycobacterium Species. Clin. Microbiol. Rev 2001, 14, 704–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Shui G; Bendt AK; Jappar IA; Lim HM; Laneelle M; Hervé M; Via LE; Chua GH; Bratschi MW; Rahim SZZ; et al. Mycolic Acids as Diagnostic Markers for Tuberculosis Case Detection in Humans and Drug Efficacy in Mice. EMBO Mol. Med. 2012, 4, 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Song SH; Park KU; Lee JH; Kim EC; Kim JQ; Song J Electrospray Ionization-Tandem Mass Spectrometry Analysis of the Mycolic Acid Profiles for the Identification of Common Clinical Isolates of Mycobacterial Species. J. Microbiol. Methods 2009, 77, 165–177. [DOI] [PubMed] [Google Scholar]
- (46).Szewczyk R; Kowalski K; Janiszewska-Drobinska B; Druszczyńska M Rapid Method for Mycobacterium Tuberculosis Identification Using Electrospray Ionization Tandem Mass Spectrometry Analysis of Mycolic Acids. Diagn. Microbiol. Infect. Dis. 2013, 76, 298–305. [DOI] [PubMed] [Google Scholar]
- (47).Borowska-Strugiń B; Druszczyńska M; Lorkiewicz W; Szewczyk R; Ządzińska E Mycolic Acids as Markers of Osseous Tuberculosis in the Neolithic Skeleton from Kujawy Region (Central Poland). Anthropological Review 2014, 77, 137–149. [Google Scholar]
- (48).Laval F; Lanéelle M-A; Déon C; Monsarrat B; Daffé M Accurate Molecular Mass Determination of Mycolic Acids by MALDI-TOF Mass Spectrometry. Anal. Chem. 2001, 73, 4537–4544. [DOI] [PubMed] [Google Scholar]
- (49).Williams PE; Klein DR; Greer SM; Brodbelt JS Pinpointing Double Bond and Sn-Positions in Glycerophospholipids via Hybrid 193 Nm Ultraviolet Photodissociation (UVPD) Mass Spectrometry. J. Am. Chem. Soc. 2017, 139, 15681–15690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Klein DR; Brodbelt JS Structural Characterization of Phosphatidylcholines Using 193 Nm Ultraviolet Photodissociation Mass Spectrometry. Anal. Chem. 2017, 89, 1516–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Liebisch G; Vizcaíno JA; Köfeler H; Trötzmüller M; Griffiths WJ; Schmitz G; Spener F; Wakelam MJO Shorthand Notation for Lipid Structures Derived from Mass Spectrometry. J. Lipid Res. 2013, 54, 1523–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Damen CWN; Isaac G; Langridge J; Hankemeier T; Vreeken RJ Enhanced Lipid Isomer Separation in Human Plasma Using Reversed-Phase UPLC with Ion-Mobility/High-Resolution MS Detection. J. Lipid Res. 2014, 55, 1772–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Sartain MJ; Dick DL; Rithner CD; Crick DC; Belisle JT Lipidomic Analyses of Mycobacterium Tuberculosis Based on Accurate Mass Measurements and the Novel “Mtb LipidDB.” J. Lipid Res. 2011, 52, 861–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Layre E; Sweet L; Hong S; Madigan CA; Desjardins D; Young DC; Cheng T-Y; Annand JW; Kim K; Shamputa IC; et al. A Comparative Lipidomics Platform for Chemotaxonomic Analysis of Mycobacterium Tuberculosis. Chem. Biol. 2011, 18, 1537–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Marrakchi H; Lanéelle M-A; Daffé M Mycolic Acids: Structures, Biosynthesis, and Beyond. Chem. Biol. 2014, 21, 67–85. [DOI] [PubMed] [Google Scholar]
- (56).Brodbelt JS Photodissociation Mass Spectrometry: New Tools for Characterization of Biological Molecules. Chem. Soc. Rev. 2014, 43, 2757–2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Feng Y; Chen B; Yu Q; Li L Identification of Double Bond Position Isomers in Unsaturated Lipids by M-CPBA Epoxidation and Mass Spectrometry Fragmentation. Anal. Chem. 2019, 91, 1791–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Ambasta DBK. Chemistry for Engineers; Laxmi Publications, 2008. [Google Scholar]
- (59).Bach RD; Dmitrenko O Strain Energy of Small Ring Hydrocarbons. Influence of C–H Bond Dissociation Energies. J. Am. Chem. Soc. 2004, 126, 4444–4452. [DOI] [PubMed] [Google Scholar]
- (60).Bifani P; Moghazeh S; Shopsin B; Driscoll J; Ravikovitch A; Kreiswirth BN Molecular Characterization of Mycobacterium Tuberculosis H37Rv/Ra Variants: Distinguishing the Mycobacterial Laboratory Strain. J. Clin. Microbiol. 2000, 38, 3200–3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Ordway D; Henao-Tamayo M; Harton M; Palanisamy G; Troudt J; Shanley C; Basaraba RJ; Orme IM The Hypervirulent Mycobacterium Tuberculosis Strain HN878 Induces a Potent TH1 Response Followed by Rapid Down-Regulation. J. Immunol. 2007, 179, 522–531. [DOI] [PubMed] [Google Scholar]
- (62).Manca C; Tsenova L; Barry CE; Bergtold A; Freeman S; Haslett PAJ; Musser JM; Freedman VH; Kaplan G Mycobacterium Tuberculosis CDC1551 Induces a More Vigorous Host Response In Vivo and In Vitro, But Is Not More Virulent Than Other Clinical Isolates. J. Immunol. 1999, 162, 6740–6746. [PubMed] [Google Scholar]
- (63).Portevin D; Sukumar S; Coscolla M; Shui G; Li B; Guan XL; Bendt AK; Young D; Gagneux S; Wenk MR Lipidomics and Genomics of Mycobacterium Tuberculosis Reveal Lineage-Specific Trends in Mycolic Acid Biosynthesis. Microbiologyopen 2014, 3, 823–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


