Abstract
Enzymes possess unique qualities that make them ideal regulators of supramolecular assembly. They are uniquely sensitive to biomolecules and biological compartments, catalytic in effecting chemical reactions, and present a biocompatible and degradable platform for assembly regulation. Here, we demonstrate the novel utility of Histidine Triad Nucleotide Binding Protein 1 (HINT1) in regulating supramolecular hydrogel formation. We synthesized nucleoside phosphoramidate functionalized self-assembling peptides which we observed to form nanofibers. We found HINT1’s catalytic hydrolysis of the nucleoside phosphoramidate moieties within the nanofiber structures to induce nanofiber organization and higher ordered assembly. The role of HINT1 in effecting this structural change was confirmed with experiments utilizing a high affinity HINT1 inhibitor and catalytically dead HINT1 mutant. In addition, the kinetics and morphology of hydrogel formation were found to be dependent on the structure of the released nucleoside monophosphate. This work highlights the self-assembly of phosphoramidate nanofibers and their higher organization triggered by HINT1 enzymatic activity.
Keywords: Self-Assembly, Hydrogel, Phosphoramidate, HINT1
Graphical Abstract
INTRODUCTION.
The supramolecular assembly of biomolecules is a ubiquitous feature of life. Protein and enzyme complexes, components of the extracellular matrix, phospholipids of cellular membranes, and duplexed or multi-plexed nucleic acid polymers are all supramolecular structures. Each of these biomolecular assemblies consists of monomeric building blocks assembled through physical interactions.1,2 Electrostatic, inductive, and dispersive forces may all contribute to the assembly of monomer building blocks into higher ordered architectures. Although individually weak, these forces act cooperatively to confer significant stability to the nucleic acid structures containing genetic information and the multiprotein complexes required for cellular function4. Nevertheless, the weak and transient nature of individual non-covalent interactions confers a responsiveness of cellular architectures to stimuli.1,4 Weak interactions allow biomolecules to rapidly change conformation, assemble, and disassemble to meet the needs of living systems in changing environments.1,4,14
Inspired by biological supramolecular assemblies, a number of approaches for the preparation of synthetic molecular assemblies whose structure and function are regulated by diverse external stimuli have been explored. 3,5,6,7 Enzymes are particularly well suited for regulating assembly as they are catalytic in nature, biologically compatible, and are dynamically responsive to endogenous regulatory factors.8 Enzyme activators present an ideal platform for the potential regulation of supramolecular biomaterials due to the potential for engineering multi-enzyme responsiveness and for incorporation into wide variety of biocompatible formats. Additionally, a number of enzymes have been utilized to regulate the structures of supramolecular materials.8
HINT1 is a human nucleoside phosphoramidase and acyl-nucleotide hydrolase.18 The enzyme’s catalytic mechanism proceeds through nucleophilic attack by His-112 onto the phosphorus center of a nucleoside phosphoramidate or acyl adenylate.18 Consequently, we hypothesized that conjugation of a polar nucleoside phosphoramidate group to a hydrophobic self-assembling peptide would disrupt non-covalent interactions necessary for assembly of substrate monomers. Upon hydrolysis of the nucleoside phosphoramidate by HINT1, we anticipated supramolecular association of free peptide monomers would result in nanofiber formation. With this goal in mind, we designed peptide-nucleotide conjugates consisting of an N-terminally capped diphenylalanine peptide linked to each of the four RNA nucleoside phosphoramidates and assessed their gelation properties in the presence of HINT1 enzyme catalyst.
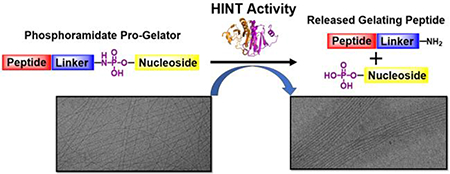
In this work, we provide the first disclosure of HINT1 as an enzymatic tool for the regulation of supramolecular assembly. Previous investigations by our lab established nucleoside phosphoramidates as potent delivery vehicles for the intracellular delivery of therapeutic nucleosides.25,26 In this approach, amine containing side chains act as charge masking groups for the negatively charged nucleoside monophosphate enabling penetration of the cellular plasma membrane.27 In the present work, we extend the use of the nucleoside phosphoramidate for the modification and structural regulation of self-assembled materials. Self-assembling peptides bearing the nucleoside phosphoramidate moiety readily formed highly regular nanofibers in aqueous buffer. Higher ordered association of the nanofibers was achieved with HINT1 enzymatic activity, demonstrating the potential utility of this system for regulation of noncovalent interactions in nanofibers and polymers (Figure 1).
Figure 1.
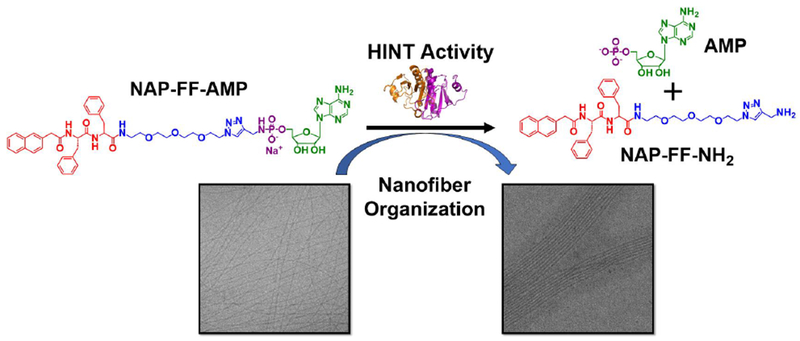
Schematic of HINT1 activated nanofiber organization
EXPERIMENTAL SECTION.
Materials and General Methods
Commercially available chemicals were utilized without additional purification. Fmoc-Phe-OH, nucleoside-5’-monophosphates, propargyl amine, 2-naphthylacetic acid, CuSO4·5xH2O, Na Ascorbate were purchased from Sigma-Aldrich. The amide coupling reagents 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC), (1-[Bis(dimethylamino)methylene]-1H-1,2,3-triazolo[4,5-b]pyridinium 3-oxid hexafluorophosphate (HATU), and (2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU) were purchased from Oakwood Chemical. Fmoc-Phe pre-loaded Wang resin was purchased from Bachem. Amino-PEG3-Azide was purchased from Quanta Biodesign. Diisopropylethylamine, tert-butanol, and piperidine were purchased from Sigma-Aldrich. All bulk solvents were sourced from Fisher Scientific and were of high pressure liquid chromatography (HPLC) grade. HINT1 wild type (WT) and H112N mutant were expressed and purified as previously described.28 Normal and reverse phase chromatographic separations were performed on a Teledyne Isco CombiFlash system. Analytical HPLC was performed on an Ultimate 3000 System (Agilent) and Higgins Analytical Targa C18 5 μm column with 50 mM triethylammonium bicarbonate (TEAB) buffer and acetonitrile (30%−100% Acetonitrile). HINT1 enzymatic activity studies were monitored by HPLC at wavelength 280 nm. Quantitative experiments were performed by diluting hydrogel samples (0.9% phosphoramidate pro-gelator Substrate, 6 μM in HINT1, in Dulbecco’s phosphate buffered saline (DPBS)) 10x by volume in dimethylsulfoxide (DMSO) after 24 h. Aliquots were taken from the DMSO solutions and diluted 10x with 50 mM TEAB Buffer for HPLC injection. Electrospray Ionization Mass Spectrometry (ESI-MS) was performed on an Agilent MSD SL system and high resolution MS was performed on an LTQ Orbitrap Velos (Thermo Scientific). Dowex 50wx8 cation exchange resin was sourced from Sigma-Aldrich. Nuclear magnetic resonance imaging (NMR) of all compounds was performed at 25°C utilizing an Ascend 500 MHz Bruker spectrometer (d6-DMSO, Cambridge Isotope Laboratories). 31PNMR to observe HINT1 catalysis was performed using 5 mM phosphoramidate substrate in HINT1 activity buffer (20 mM HEPES, 1 mM MgCl2, pH 7.3) with 6 uM HINT1 and 5% D2O.
Synthetic Procedures and Preparation of Gels
Detailed synthetic procedures are provided in the Supplemental Information. In brief, the NAP-FF peptide was synthesized using standard Fmoc-based solid phase procedures. Subsequent amide coupling between the carboxyl of NAP-FF with the primary amine of Amino-PEG3-Azide yielded compound 1. Nucleoside propargyl phosphoramidates bearing either A,U,G, or C bases were conjugated to compound 1 through the copper (I) catalyzed Huisgen 1,3-dipolar cycloaddition to yield phosphoramidate pro-gelators 2 (NAP-FF-AMP), 3 (NAP-FF-UMP), 4 (NAP-FF-GMP), and 5 (NAP-FF-CMP) (Figure 2).
Figure 2.
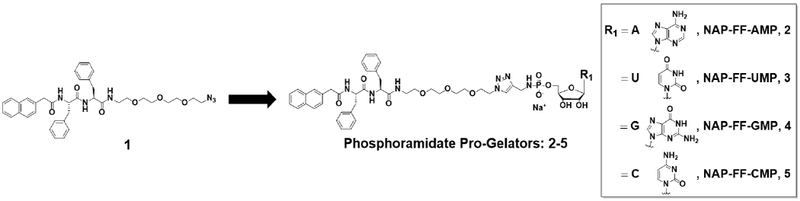
Chemical structures of Phosphoramidate Pro-Gelators 2-5
Precursor and gel samples were prepared as follows: Lyophilized foams of each phosphoramidate pro-gelator were weighed into scintillation vials and dissolved in HINT1 activity buffer resulting in 1% wt/vol solutions. Samples were mixed with vortexing to ensure complete dissolution of the material. Final working solutions of 0.9% wt/vol concentration were obtained by mixing 450 μL of each 1 % wt/vol pro-gelator solution with 50 additional μL of HINT1 activity buffer. Hydrogel samples were generated in a similar manner by adding 50 μL of HINT1 in activity buffer to 450 μL of 1% wt/vol pro-gelator solution to generate samples containing 6 μM concentrations of enzyme and 0.9% wt/vol.
Conventional and Cryo-Transmission Electron Microscopy
Imaging was performed using an FEI Technai Spirit Bio-Twin instrument. Conventional TEM hydrogelation samples contained 0.9% by wt/vol phopshoramidate pro-gelator and 19 μM HINT1 in activity buffer except NAP-FF-AMP which contained 36 uM HINT1. Gelation samples cured on grid for 10 minutes before blotting and staining. Substrate only samples contained 0.9% by wt/vol phosphoramidate pro-gelator in activity buffer. Conventional TEM samples were stained with 2% uranyl acetate for negative imaging.
Hydrogel samples analyzed with Cryo-TEM contained 0.9% by wt/vol phosphoramidate pro-gelator and 6 μM HINT1 in activity buffer. The gel forming reaction was allowed to proceed for at least 10 minutes before vitrification. Substrate only samples contained 0.9% by wt/vol phosphoramidate pro-gelator in activity buffer. Cyro-TEM of 1% wt/vol NAP-FF-NH2 in DPBS was performed with or without 1% wt/vol adenosine monophosphate.
Oscillatory Rheometry
Rheological experiments were performed on a TA Instruments AR-G2 Rheometer. The 25 mm geometry was utilized with upper and lower geometries modified with 600 grit sandpaper to aid in sample adhesion. All samples were 500 μL in volume and were prepared by combining 450 μL of 1% wt/vol substrate in HINT1 activity buffer with up to 10 μL of Enzyme (Final Concentration in Sample 6 μM), and 40 uL of HINT1 activity buffer to maintain constant volumes between samples. The Peltier temperature was set to a constant temperature of 23oC. Time course experiments were performed at 0.1% strain and 1 rad/s for all experiments with both values within the linear viscoelastic range. Experiment time lengths were kept to a minimum to observe sample curing while avoiding evaporation, the effects of which were mitigated by use of humidity chamber lined with moistened paper towels. In samples demonstrating a clear sol-gel transition with G’ becoming greater than G’’, gelation times are defined as the time point at the crossover. For samples where G’ was greater than G’’ for the entire time course, a time point was estimated by constructing linear curves before and after the G’ increase and determining the time point at which both linear functions possessed equal values to estimate the inflection point. All reported gelation time points are averages of at least 3 individual experiments.
RESULTS AND DISCUSSION
Synthesis and Preparation of Hydrogels
A naphthyl N-terminal cap was chosen over the fluroenylmethoxycarbonyl (Fmoc) group common in self-assembling peptides due to enhanced chemical stability and retention of self-assembly through aromatic interactions.12,20 Previously, the NAP-FF peptide was characterized as a self-assembling motif and its derivates have demonstrated great versatility in the study of enzyme regulated self-assembly by the Xu group. 9,12,13,,19 The pure NAP-FF peptide obtained through standard Fmoc-solid phase synthesis, was coupled to an amino-PEG3-azide linker yielding the azido-labeled self-assembling peptide in good yield.21 Nucleoside phosphoramidates were synthesized through the condensation of propargyl amine and the desired nucleoside monophosphate mediated by EDCI.17 Azido-labeled peptide and corresponding propargyl phosphoramidates were coupled efficiently utilizing the copper catalyzed Huisgen cycloaddition reaction in the presence of Na Ascorbate and Copper (II) sulfate in 2:1 tBuOH/H2O as the solvent system.10
Each substrate was subsequently assessed for its ability to self-assemble and form hydrogels in the presence of HINT1 enzyme using visual inspection. Solutions containing 0.9% NAP-FF-AMP, NAP-FF-UMP, NAP-FF-GMP, and NAP-FF-CMP were prepared with HINT1 activity buffer . HINT1 enzyme in activity buffer was added to generate solutions of 6 μM HINT1 and 0.9% wt/vol substrate. Samples were briefly vortexed to ensure proper mixing and allowed to cure at room temperature. Samples were checked after 15 min., at which time vials were inverted. All four substrates samples were observed to remain viscous solutions in the absence of HINT (Figure 3a–d insets). During the same period, all four substrate solutions containing HINT1 underwent hydrogel formation (Figure 3e–h insets). The four hydrogels remained stable for 24 hours after repeating the inversion test. Hydrogels originating from NAP-FF-AMP, NAP-FF-GMP, and NAP-FF-UMP were all opaque in nature, with NAP-FF-CMP forming translucent hydrogels.
Figure 3.
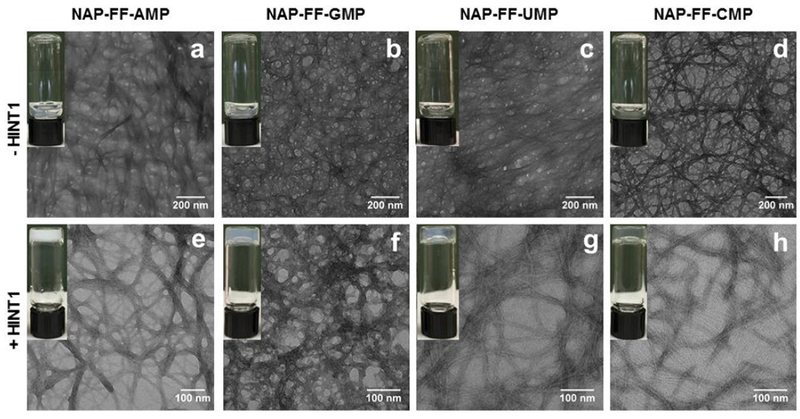
Top Panel: Transmission electron microscopy (TEM) of substrate nanofiber solutions in the absence of HINT1 (scale bars represent 200 nm) (a). NAP-FF-AMP, (b). NAP-FF-GMP, (c). NAP-FF-UMP, and (d) NAP-FF-CMP. Bottom Panel: TEM of HINT1 formed hydrogels (scale bars represent 100 nm) (e). NAP-FF-AMP, (f). NAP-FF-GMP, (g) NAP-FF-UMP, and (h) NAP-FF-CMP.
Structural Characterization of Substrate Nanofibers and Hydrogels
Transmission electron microscopy (TEM) was utilized to investigate the nanostructures of both substrate and hydrogel samples. Interestingly, structures ranging from amorphous aggregates to fibrous networks were observed in the absence of HINT1 (Figure 3a–d). Extensive fiber formation was observed in the NAP-FF-CMP substrate sample with fibrous structures also evident in the NAP-FF-UMP and NAP-FF-GMP substrate samples. Within both the amorphous and fibrillar structures, portions of individual nanofibers could be resolved in samples of all for substrates and were determined to be 6-8 nm in diameter. Hydrogel samples presented clear and defined nanofiber formation for all four substrates (Figure 3e–h). In comparison to the substrate only samples, the fibrous networks formed from HINT1 activity were clearly defined with individual nanofibers possessing diameters between 7-8 nm. Not only were all hydrogel samples observed to exhibit nanofiber formation upon HINT1 activation, but much of the nanofiber content was contained within tightly packed nanofiber bundles. NAP-FF-AMP and NAP-FF-GMP exhibited extensive nanofiber association with NAP-FF-UMP and NAP-FF-CMP samples exhibiting fewer associated nanofibers and a prevalence of single nanofibers. Cryo-TEM was essential for investigating the hydrated structures of the nanoarchitectures formed from the phosphoramidate precursors and their respective hydrogels. This contrasts with conventional TEM which can affect nanostructure observation and characterization due to the necessity for extensive drying of the sample.11 Additionally, conventional TEM imaging necessitates sample staining which can introduce interfering artifacts and background.11 Consequently, all four phosphoramidate pro-gelator substrates were analyzed by cryo-TEM, which elaborated self-assembled fibrillar structures. Consistent with the results of the TEM, solutions of all four substrates in HINT1 activity buffer were found to contain highly regular and continuous nanofibers approximately 7-8 nm in width (Figure 4e–h). The nanofibers formed by all four pro-gelators are largely unassociated and likely only align with lacey carbon of the cryo-TEM sample grid due to sample ice thickness variation. These results indicate the phosphoramidate pro-gelators form organized supramolecular structures in solution prior to HINT1 enzyme activation.
Figure 4.
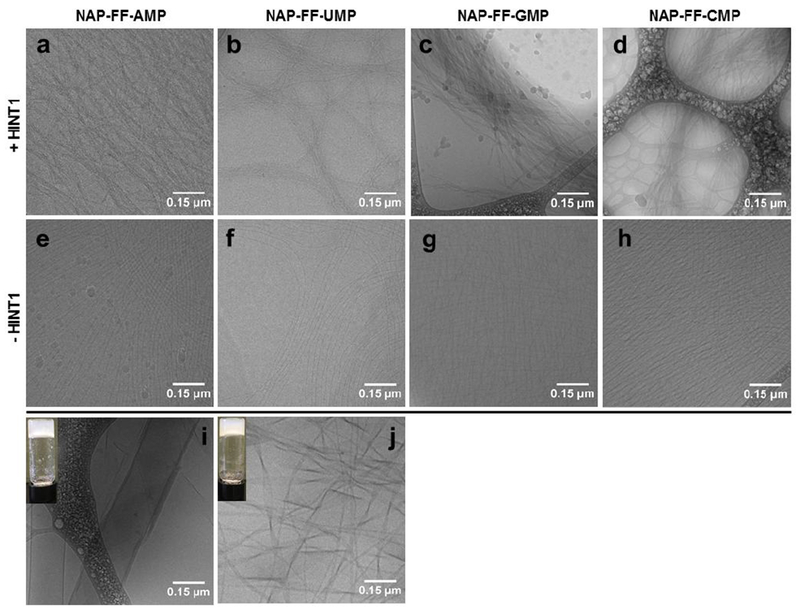
Cryo-TEM of Hydrogels and Substrates. Top Panel: Hydrogels formed in the presence of HINT1 (a) NAP-FF-AMP, (b) NAP-FF-UMP, (c) NAP-FF-GMP, (d) NAP-FF-CMP. Middle Panel: Phosphoramidate Pro-Gelator Nanofibers in Buffer (e) NAP-FF-AMP, (f) NAP-FF-UMP, (g) NAP-FF-GMP, (h) NAP-FF-CMP. Bottom Panel: (i) NAP-FF-NH2 structure in the presence of PBS only, (j) and in the presence of AMP. (All scale bars represent 0.15 μM)
From conventional TEM, the predominating hydrogel structure observed for all for phosphoramidate pro-gelators were bundled nanofiber networks. These observations, combined with the pre-assembly of nanofibers observed with cryo-TEM led to the hypothesis that the phosphoramidate moiety present in the pro-gelator substrate may prevent nanofibers from participating in higher ordered assemblies. To observe what change HINT1 was imparting onto the substrate nanofibers upon initiation of hydrogelation, the hydrogels formed from all four substrates were examined with cryo-TEM. Hydrogel samples were prepared in HINT1 activity buffer at a concentration of 0.9% by wt/vol. with 6 uM HINT1 (Figure 4a–d). Clear nanofiber association was observed in all four hydrogel samples. Consistent with the results from TEM studies, this demonstrates cleavage of the nucleoside phosphoramidate moiety results in self-association of the nanofibers leading to hydrogel formation. Measurements of individual nanofibers present in both the substrate and hydrogel samples of NAP-FF-AMP, NAP-FF-UMP, NAP-FF-GMP, and NAP-FF-CMP were approximately identical and between 7-8 nm in diameter. Additionally, the nanofibers present in bundles were determined to be of equivalent diameter despite their induced assembly into dense association networks. Bundle diameters ranged from 14 nm to over 200 nm. The released peptide generated from HINT1 activity on the substrate nanofibers was also assessed for its gelation ability and nanostructure. NAP-FF-NH2 (Structure in Figure 1) was observed to form hydrogels at a concentration of 1% wt/vol in both water and PBS buffer. Addition of AMP at a concentration of 1% wt/vol also resulted in hydrogel formation. NAP-FF-NH2 exhibited extensive ribbon-like structures exceeding 200 nm in width alone (Figure 4i). Gels in the presence of AMP were observed to form a range of structures including individual nanofibers, twisted nanofiber ribbons approximately 35-55 nm in width, and larger ribbons (Figure 4j). Both gel structures contrast with the highly ordered substrate nanofibers and resulting structures formed from HINT1 activity.
From combined conventional and cryo-TEM results, it was hypothesized that HINT1 catalyzes the transition between the phosphoramidate pro-gelator nanofibers and supramolecular assembly of the nanofiber bundle networks. Presumably, this mechanism of hydrogelation results from hydrolysis of the phosphoramidate moieties present along assembled substrate nanofibers, allowing the “deprotected” nanofibers to self-assemble. Inorganic phosphate has been previously observed to initiate association of primary amine containing supramolecular structures through charge coordination of self-assembled nanofibers.22 It is possible the nucleoside monophosphate generated from HINT1 activity cross-links the poly-cationic self-assembled nanofibers and may contribute additional non-covalent interactions through nucleobase hydrogen bonding and aromatic interactions. Nucleobases have long been investigated as self-assembling motifs and have been incorporated into a wide variety of synthetic and biologically sourced polymers to impart their hydrogen bonding propensity into higher ordered structures.23,24
Role of HINT1 in Hydrogelation
Oscillatory rheometry time sweep experiments were performed to characterize hydrogel network assembly in response to enzymatic activity and nanofiber association.16 In the presence of HINT1, NAP-FF-AMP underwent rapid gelation by 3 μM HINT1 resulting in increases of both G’ and G’’ at an average time of 238 ± 30 s (Supplemental Figure S1). However, hydrogels formed from this substrate exhibited marked syneresis and loss of geometry adhesion over extended time sweep experiments. Two concentrations of HINT1 were utilized to investigate NAP-FF-UMP gelation kinetics (Figure 5a). At a concentration of 6 μM, HINT1 was able to induce hydrogel formation from 0.9% wt/vol of NAP-FF-UMP at an average time of 157 ± 67 s. Reducing the concentration of HINT1 to 3 μM resulted in a modest delay of observed gelation to an average time of 458 ± 130 s. Additionally, decreasing the concentration of NAP-FF-UMP substrate with constant HINT1 (6 μM) resulted in hydrogels of lower moduli indicating a dependence of material rigidity on substrate concentration (Figure 5b). To elucidate the role of HINT1’s active site in initiation of hydrogelation, additional time course experiments were performed in the presence of a competitive HINT1 small molecule inhibitor, HNTI-3a (Kd = 230 nM).15 Samples of 0.9% NAP-FF-AMP and NAP-FF-UMP were prepared in HINT1 activity buffer containing 2% DMSO alone or with 6 mM HNTI-3a. HINT1 was added to the solutions at a concentration of 6 μM and moduli were monitored over time courses to observe the occurrence of hydrogel formation. HNTI-3a was found to completely ablate hydrogelation of NAP-FF-UMP for over 26 min. In contrast, clear hydrogel formation was observed in the absence of inhibitor and in the presence of DMSO only (Figure 5c). Activation of NAP-FF-AMP by HINT1 was not prevented by HNTI-3a. In comparison to DMSO only control which exhibited instantaneous gelation, HNTI-3a resulted in a delay of substrate activation with sharp increases of moduli occurring at an average time of 346 ± 39 s (n = 3) in the presence of the inhibitor (Supplemental Figure S2). It is likely that the relative affinity of HINT1 for purine nucleobase substrates and inhibitors over pyrimidines contributes to the enhanced inhibition of NAP-FF-UMP gelation in comparison to NAP-FF-AMP.17 Additionally, the differences we observe in hydrogel formation kinetics could result from different nucleobases contributing varying levels of hydrogen bonding, base stacking, or a combination of both as mediators of nanofiber interaction.
Figure 5.
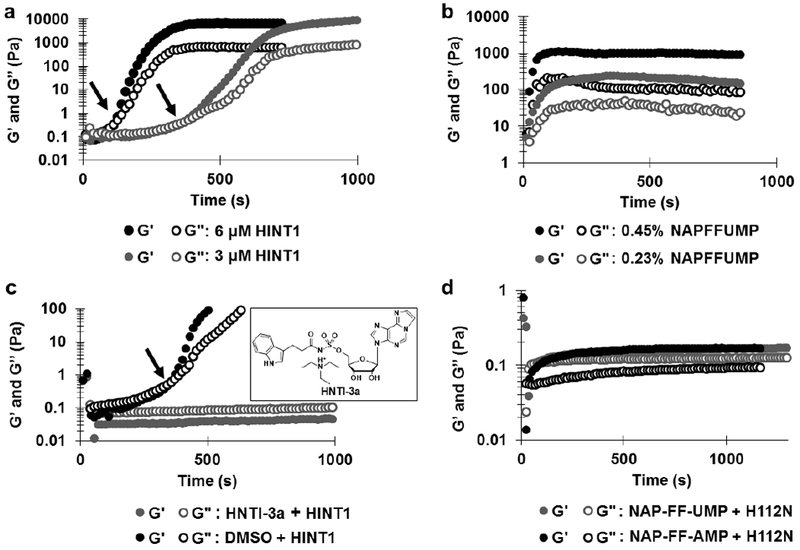
Oscillatory Rheometry on NAP-FF-UMP Hydrogelation (a) Time sweeps in the presence of 6 μM and 3 μM HINT1. Gelation times determined from at least 3 experiments, (b) Time sweeps at 0.45% and 0.23% wt/vol concentrations of NAP-FF-UMP with 6 μM HINT1 (c) Time sweeps in the presence of HINT1 inhibitor or DMSO control. (d) Time sweeps in the presence of H112N HINT1 mutant with NAPFFUMP and NAPFFAMP.
Additional time sweep experiments were conducted in the presence of a HINT1 H112N active site mutant to confirm catalytic activity as the initiator of hydrogelation (Figure 5d). This mutant retains high binding affinity for nucleoside phosphoramidates but lacks the histidine residue necessary for nucleophilic catalysis15. In the presence of 11 μM H112N mutant, NAP-FF-AMP or NAP-FF-UMP demonstrated only weak hydrogel formation with moduli never rising above 0.2 Pa. The H112N mutant was found to be unable to initiate the highly ordered and rigid structures observed with wild-type HINT1 catalysis on either the NAP-FF-AMP or NAP-FF-UMP nanofibers. Consequently, it appears that catalytically active HINT1 is necessary for hydrolyzing the phosphoramidate moieties along substrate nanofibers resulting in hydrogel formation through non-covalent crosslinking of supramolecular nanofibers. Further confirmation of HINT1 remaining catalytically active during the hydrogelation process is evident in 31P NMR experiments that demonstrate the rapid hydrolysis of NAP-FF-AMP to AMP within minutes of HINT1 addition (Figure S3). Quantification of the percent NAP-FF-AMP remaining within HINT1 formed hydrogels after 24 hours was determined to be less than 1% (Figure S4).
CONCLUSIONS
In summary, we have shown the assembly of phosphoramidate pro-gelators into supramolecular nanofibers by Cryo-TEM. In the presence of the phosphoramidase HINT1, these solutions yielded hydrogels consisting of densely associated nanofiber bundles characterized by conventional and Cryo-TEM. The kinetics, sensitivity to inhibitor, and morphology of material formation were found to be dependent on the structure of the released nucleoside monophosphate. A dependence of HINT1 catalytic activity on hydrogelation was established with oscillatory rheometry time course experiments in combination with chemical biological tools and mutant enzyme studies. It is hypothesized that the nucleoside monophosphate produced through HINT1 catalytic activity is responsible for charge coordination of the polycationic product nanofibers, with the nucleobase contributing to yet unknown secondary interactions further affecting the final gel morphology. While the dependence of the initial nanofiber assembly on subsequent hydrogel formation remains to be determined, the potential tunability of the material characteristics and control of gelation by enzyme activation and substrate structure could prove useful for applications requiring greater chemical functionality and control over supramolecular assembly.
Supplementary Material
ACKNOWLEDGMENT
Parts of this work were carried out in the Characterization Facility, University of Minnesota, which receives partial support from NSF through the MRSEC program. The authors gratefully acknowledge Dr. Bob Hafner (Characterization Facility, University of Minnesota) for his assistance with the transmission electron microscopy experiments. Oscillatory rheometry was performed in the University of Minnesota Polymer Characterization Facility. The authors gratefully acknowledge David Giles for assistance with these experiments.
Funding Sources
This work was supported by the National Institutes of Health R21 CA185627 (CRW), F30 CA210345 (CMC), NIH T32 GM008244 (CMC), and the University of Minnesota.
Footnotes
Supporting Information.
The following files are available free of charge. Synthetic Procedures for Phosphoramidate Pro-Gelators 2-5, Characterizations of Phosphoramidate Pro-Gelators 2-5, Oscillatory Rheometry on NAP-FF-AMP, and HINT1 Catalytic Hydrolysis of NAP-FF-AMP (PDF)
REFERENCES
- 1.Tu Y; Peng F; Adawy A; Men Y; Abdelmohsen LK; Wilson DA Mimicking the cell: bio-inspired functions of supramolecular assembly. Chem. Rev. 2016, 116, 2023. [DOI] [PubMed] [Google Scholar]
- 2.Pieters BJGE; van Eldijk MB; Nolte RJ M; Mecinovic, J. Natural supramolecular protein assemblies Chem. Soc. Rev. 2016, 45, 24. [DOI] [PubMed] [Google Scholar]
- 3.Shigemitsu H; Hamachi I Design strategies of stimuli-responsive supramolecular hydrogels relying on structural analyses and cell-mimicking approaches. Acc. Chem. Res. 2017, 50, 740. [DOI] [PubMed] [Google Scholar]
- 4.Mahadevi AS; Sastry GN Cooperativity in noncovalent interactions Chem. Rev. 2016, 116, 2775. [DOI] [PubMed] [Google Scholar]
- 5.Webber MJ Engineering responsive supramolecular biomaterials: toward smart therapeutics. Bioeng. Transl. Med. 2016, 1, 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Webber MJ; Appel EA; Meijer EW; Langer R Supramolecular biomaterials. Nat. Mater. 2016, 13. [DOI] [PubMed] [Google Scholar]
- 7.Busseron E; Ruff Y; Moulin E; Giuseppone N Supramolecular self-assemblies as functional nanomaterials. Nanoscale. 2013, 5, 7098. [DOI] [PubMed] [Google Scholar]
- 8.Zelzer M; Todd SJ; Hirst AR; McDonald TO.; Ulijn RV. Enzyme responsive materials: design strategies and future developments. Biomater. Sci. 2013, 1, 11. [DOI] [PubMed] [Google Scholar]
- 9.Yang Z; Gu H; Fu D; Gao P; Lam JK; Xu B Enzymatic formation of supramolecular hydrogels. Adv. Mater. 2004, 16, 1440 [Google Scholar]
- 10.Himo F; Lovell T; Hilgraf R; Rostovtsev VV; Noodleman L; Sharpless KB; Fokin VV Copper(I)-catalyzed synthesis of azoles. dft study predicts unprecedented reactivity and intermediates. J. Am. Chem. Soc. 2005, 127, 210. [DOI] [PubMed] [Google Scholar]
- 11.Newcomb CJ; Moyer TJ; Lee SS; Stupp SI Advances in cryogenic transmission electron microscopy for the characterization of dynamic self-assembling nanostructures. Curr. Opin. Colloid Interface Sci. 2012, 17, 350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y; Kuang Y; Gao Y; Xu B Versatile small-molecule motifs for self-assembly in water and the formation of biofunctional supramolecular hydrogels. Langmuir. 2011, 27, 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Z; Ho P-L; Liang G; Chow KH; Wang Q; Cao Y; Guo Z; Xu B Using β-lactamase to trigger supramolecular hydrogelation. J. Am.Chem. Soc. 2007, 129, 266. [DOI] [PubMed] [Google Scholar]
- 14.Nussinov R; Jang H Dynamic multiprotein assemblies shape the spatial structure of cell signaling Prog. Biophys. Mol. Biol. 2014, 116, 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shah R; Strom A; Zhou A; Maize KM; Finzel BC; Wagner CR Design, synthesis, and characterization of sulfamide and sulfamate nucleotidomimetic inhibitors of hHint1. ACS Med. Chem. Lett. 2016, 7, 780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan C; Pochan DJ Rheological properties of peptide-based hydrogels for biomedical and other applications. Chem. Soc. Rev. 2010, 39, 3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chou T-F; Baraniak J; Kaczmarek R; Zhou X; Cheng J; Ghosh B; Wagner CR Phosphoramidate pronucleotides: a comparison of the phosphoramidase substrate specificity of human and escherichia coli histidine triad nucleotide binding proteins. Mol. Pharm. 2007, 4, 208. [DOI] [PubMed] [Google Scholar]
- 18.Zhou X; Chou T-F; Aubol BE; Park CJ; Wolfenden R; Adams J; Wagner CR Kinetic mechanism of human histidine triad nucleotide binding protein 1. Biochemistry. 2013, 52, 3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao Y; Long M; Shi J; Hedstrom L; Xu B Using supramolecular hydrogels to discovery the interactions between proteins and molecular nanofibers of small molecules. Chem. Comm. 2012, 48, 8404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahler A; Reches M; Rechter M; Cohen S; Gazit E Rigid, self-assembled hydrogel composed of a modified aromatic dipeptide. Adv. Mater. 2006, 18, 1365 [Google Scholar]
- 21.Wang S-S p-Alkoxybenzyl alcohol resin and p-alkoxybenzyloxycarbonylhydrazide resin for solid phase synthesis of protected peptide fragments. J. Am. Chem. Soc. 1973, 95, 1328. [DOI] [PubMed] [Google Scholar]
- 22.Aulisa L; Dong H; Hartgerink JD Self-assembly of multidomain peptides: sequence variation allows control over cross-linking and viscoelasticity. Biomacromolecules. 2009, 10, 2694. [DOI] [PubMed] [Google Scholar]
- 23.Karikari AS; Mather BD; Long TE Association of star-shaped poly(d,l-lactide)s containing nucleobase multiple hydrogen bonding. Biomacromolecules. 2007, 8, 302. [DOI] [PubMed] [Google Scholar]
- 24.Kang Y; Pitto-Barry A; Willcock H; Quan WD; Kirby N; Sanchez AM; O’Reilly RK Exploiting nucleobase-containing materials-from monomers to complex morphologies using RAFT dispersion polymerization. Polym. Chem. 2015, 6, 106 [Google Scholar]
- 25.Kim J; Chou T-F; Griesgraber GW; Wagner CR Direct measurement of nucleoside monophosphate delivery from a phosphoramidate pronucleotide by stable isotope labeling and LC-ESI—MS/MS. Mol. Pharm. 2004, 1, 102. [DOI] [PubMed] [Google Scholar]
- 26.Kim J; Park S; Tretyakova NY; Wagner CR A method for quantitating the intracellular metabolism of AZT amino acid phosphoramidate pronucleotides by capillary high-performance liquid chromatography-electrospray ionization mass spectrometry. Mol. Pharm. 2005, 2, 233. [DOI] [PubMed] [Google Scholar]
- 27.Wagner CR; Iyer VV; McIntee EJ Pronucleotides: toward the in vivo delivery of antiviral and anticancer nucleotides. Med. Res. Rev. 2000, 20, 417. [DOI] [PubMed] [Google Scholar]
- 28.Shah R; Maize KM; Zhou X; Finzel BC; Wagner CR Caught before released: structural mapping of the reaction trajectory for the sofosbuvir activating enzyme, human histidine triad nucleotide binding protein 1 (hHint1). Biochem. 2017, 56, 3559. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


