Abstract
Cerebral dopamine neurotrophic factor (CDNF) shows beneficial effects in rodent models of Parkinson’s and Alzheimer’s disease. The brain is a challenging target for protein therapy due to its exclusive blood–brain barrier. Hence, the therapeutic protein should be delivered directly to the brain parenchyma. Implantation of encapsulated mammalian cells that constantly secrete CDNF is a potential approach for targeted and long-term protein delivery to the brain. In this study, we generated several CDNF-secreting cell clones derived from human retinal pigment epithelial cell line ARPE-19, and studied CDNF secretion from the clones maintained as monolayers and in polymeric microcapsules. The secretion of wild type (wt) CDNF transgene was low and the majority of the produced protein remained intracellular, locating mainly to the endoplasmic reticulum (ER). The secretion of wtCDNF decreased to even lower levels when the clones were in a non-dividing state, as in the microcapsules. Both codon optimization and deletion of the putative ER-retrieval signal (four last amino acids: KTEL) improved CDNF secretion. More importantly, the secretion of KTEL-deleted CDNF remained constant in the non-dividing clones. Thus, cells expressing KTEL-deleted CDNF, in contrast to wtCDNF, can be considered for cell encapsulation applications if the KTEL-deleted CDNF is proven to be biologically active in vivo.
Keywords: cell encapsulation, CDNF, secretion, ER localization, codon optimization
Introduction
Cerebral dopamine neurotrophic factor (CDNF)1 and its homolog mesencephalic astrocyte-derived neurotrophic factor (MANF)2 form a neurotrophic factor family, which differs structurally, and most probably functionally, from the classical neurotrophic factor families3. CDNF was first described as being able to protect and rescue midbrain dopamine neurons in vivo in a rat 6-hydroxydopamine (6-OHDA) model of Parkinson’s disease when delivered into brain parenchyma as a single injection1. Later, the potency of CDNF to promote the survival and recovery of dopamine neurons has been verified in the rat 6-OHDA model after continuous protein infusion4, and viral gene delivery5 –7. CDNF has also been demonstrated to be neuroprotective and neurorestorative after single injection in a mouse 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) model of PD8. The potential role of CDNF in long-term memory was shown in a mouse model for Alzheimer’s disease after intrahippocampal injections of purified protein or adeno-associated virus serotype 2 (AAV2)-CDNF9. Both Parkinson’s and Alzheimer’s diseases lack restorative therapy.
The central nervous system is surrounded by the blood–brain barrier, which restricts the diffusion of large hydrophilic molecules from the blood circulation. Thus, the therapeutic protein in many cases needs to be delivered directly to the target site in the brain. This can be achieved by administration of the protein by injection or infusion, by gene therapy, or by the aid of implanted cells secreting the therapeutic protein10. Therapeutic protein-secreting cells can be enclosed within a semipermeable membrane, which allows the diffusion of the protein and nutrients, but restricts host immune rejection. A device carrying the encapsulated cells is then delivered to the treatment site. This approach provides constant release of therapeutically active protein from mammalian cells to the site of action, but the encapsulated cells can be retrieved if needed11.
In this study, we sought to generate CDNF-secreting ARPE-19 cell clones for therapeutic cell encapsulation purposes. The ARPE-19 cell line was chosen as the parental cell line because of multiple beneficial characteristics in terms of encapsulated cell technology. The cell line has been shown to endure in alginate microcapsules for several months in vitro12,13, and it demonstrates contact-inhibited growth14. This contact-inhibited growth is important, as continuous cell proliferation could accumulate pressure in the capsule and possibly rupture the capsule membrane; similarly, it could lead to the necrosis of the cells, as seen in case of encapsulated fibroblasts11. The ARPE-19 cell line is of human origin, and it has been used in clinical cell therapy applications without major safety concerns15–18.
In contrast to other proteins expressed in the same cell line for cell encapsulation purposes19,20, we observed poor secretion of wild type (wt) CDNF transgene from the ARPE-19 cell clones. We here report the effects of two different methods employed aiming for improved CDNF expression and secretion. Firstly, we used codon optimization, where the codons are changed to represent a more ubiquitous transfer RNA in the host while keeping the resulting amino acid sequence unchanged21. Secondly, since majority of the produced wtCDNF remained intracellular, locating mainly to the endoplasmic reticulum (ER), we studied the effect of removing the putative C-terminal ER-retrieval signal on CDNF secretion. The secreted and intracellular CDNF was quantified by a novel in-house-built enzyme-linked immunosorbent assay (ELISA).
Materials and Methods
Cell Lines
Human retinal pigment epithelial cell line, ARPE-19 (ATCC number CRL-2302), was cultured in DMEM-F12 (GibcoTM 31330), 10% fetal bovine serum (FBS), 100 U/ml penicillin and 100 µg/ml streptomycin (all from Thermo Fisher Scientific, Waltham, MA, USA). HeLa (CRM-CCL-2), EA.hy926 (CRL-2922), SH-SY5Y (CRL-2266) and U-87 MG (HTB-14) cell lines were cultured in DMEM (D7777, Sigma-Aldrich, Saint Louis, MO, USA), 10% FBS, 100 U/ml penicillin and 100 µg/ml streptomycin. All the cells were cultured in a cell incubator at 37°C and 5% CO2.
Plasmids
Mammalian expression plasmids pCI-neo (Promega, Madison, WI, USA) and pCR3.1 (Invitrogen, Carlsbad, CA, USA) were used in this study. Both vectors contain CMV-promoter for the transgene expression and neomycin resistance gene for clone selection. Human CDNF cDNA in pCR3.1 vector1 was subcloned into pCI-neo vector at restriction sites NheI and XbaI. Codon-optimized (opti) human CDNF cDNA (in BV-vector, obtained from Icosagen, Tartu, Estonia) was amplified by PCR using primers 5’GCTAGCGCCACCATGTGGTGCGCCTC-3’ and 5’-CCAAGACCGAGCTGTGATAACTCGAG-3’ and inserted into pCR3.1 vector.
To produce plasmids expressing CDNF lacking the putative ER-retrieval sequence (KTELdel), we performed inverse PCR mutagenesis on plasmids wtCDNF-pCR3.1 and optiCDNF-pCR3.1 using primers 5’-GGGGTGTGTCGCTGCATACT-3’ and 5’-TGATCAAGCCGAATTCTGCA-3’; and 5’-GGGGTGGGTAGCGGCGTACTTA-3’ and 5’-TGATAACTCGAGAAGCCGAATTCTGCAG-3’, respectively, with Phusion™ high-fidelity DNA polymerase (Thermo Fisher Scientific). After the PCR reaction, the plasmid template was digested with DpnI treatment (Thermo Fisher Scientific). PCR products were purified using QIAquick PCR Purification Kit (Qiagen, Hilden, Germany), and phosphorylated with T4 polynucleotide kinase (Thermo Fisher Scientific) before ligation reaction. After amplification in E. coli DH5α cells, plasmids were purified using QIAGEN Plasmid purification kit (Qiagen). All constructs were confirmed by sequencing.
Transient and Stable Transfections
For the analysis of transgene expression and secretion under transient transfection, ARPE-19 cells were transfected with pCR3.1 vectors encoding either wtCDNF, wtCDNF-KTELdel, optiCDNF, or optiCDNF-KTELdel with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Samples were collected 48 h post-transfection and analyzed on CDNF ELISA, by western blotting, and by immunocytochemistry.
For the production of stable clones, cells were transfected as stated above, except wtCDNF was in pCI-neo expression vector. Then, 24 h after transfection, the cells were seeded on new culture plates at different densities and selection antibiotic G418 (at 0.8 mg/ml; Geneticin, Thermo Fisher Scientific) was added 48 h after transfection. The concentration of G418 was gradually decreased to the maintenance level of 0.4 mg/ml within 4 weeks. During the selection period, cell colonies originating from single cell were isolated and expanded.
Protein Samples
Conditioned media from cultured cells were collected, centrifuged 2000 rpm for 5 min at +4°C, and the supernatants were collected. Cells were washed twice with phosphate buffered saline (PBS), and incubated for 30 min in ice-cold lysis buffer (137 mM NaCl, 20 mM Tris-HCl (pH 8.0), 1% Igepal, 10% glycerol, 2.5 mM EDTA, 0.5 mM Na3VO4, and protease inhibitors (Complete mini protease inhibitor cocktail, Roche, Basel, Switzerland)). The lysates were centrifuged at 12,000 rpm for 20 min at +4°C and the supernatants were collected. For the measurement of endogenous CDNF and MANF, cells were divided on 6-well plates at concentration of 0.3×106 cells/well. The next day, 0.6 ml of growth medium was applied to the cells for 72 h. After incubation, 0.45 ml of conditioned medium was collected and the cells were subsequently lysed in 0.45 ml of lysis buffer. For the analysis of protein expression after transient transfection, cells were incubated in Opti-MEM for 24 h after which the conditioned medium was collected. Cells were lysed in lysis buffer, using volume equal to that of the conditioned medium. In case of stable clones, cells were seeded on 12-well plates at density of 0.4×106 cells/well and 24 h after incubated with 0.5 ml of Opti-MEM or growth medium for 30 to 60 min. Cells were collected in 0.5 ml of lysis buffer. Total protein concentration in the cell lysates was measured using DCTM Protein Assay (Bio-Rad, Hercules, CA, USA).
CDNF ELISA
Human CDNF levels in cell lysates and conditioned medium samples were analyzed by in-house-built CDNF ELISA. In brief, MaxiSorp (Nunc, Thermo Fisher Scientific) 96-well plates were coated overnight at +4°C with mouse anti-human CDNF mAb, clone 7D6 (301-100, Icosagen), at 1 μg/ml in 50 mmol/l carbonate coating buffer (35 mmol/l sodium bicarbonate, 15 mmol/l sodium carbonate; pH 9.6). The plate was washed once with PBS/0.05% Tween 20 (PBST) and incubated with blocking buffer (3% BSA (Probumin® Merck Millipore, Burlington, MA, USA) in PBS) at RT for 2 h. After washing with PBST, standard samples of recombinant human CDNF (P-100-100, Icosagen), ranging from 31.25 to 2000 pg/ml, and study samples diluted in blocking buffer, were added to the plate in duplicate and incubated overnight at +4°C with agitation (100 rpm). The detection antibody, horseradish peroxidase (HRP)-conjugated mouse anti-human CDNF mAb, clone 6G5 (302-100, Icosagen), was incubated on the plate at a concentration of 1 μg/ml for 5 h in agitation at RT. Washing with PBST was repeated four times before and after the antibody incubation. For detection, 3,3’,5,5’-tetramethylbenzidine was used according to the manufacturer’s instructions (DuoSet ELISA Development System, R&D Systems, Minneapolis, MN, USA). The absorbance was read using a plate reader (VICTOR3, Perkin Elmer, Waltham, MA) at 450 nm and 540 nm (for wavelength correction).
Sensitivity of the CDNF ELISA was determined by the mean response of 10 blank samples added with three standard deviations (SD). Within the assay dynamic range, the individual back-calculated accuracy values were within 20% relative error (RE = derived concentration / expected concentration × 100%) and precision within 15% coefficient of variation (CV = SD / mean × 100%). Intra-assay variation was determined by measuring three samples with varying CDNF concentration in replicates of 10 in different parts of a plate. Interassay precision was determined by running same three samples in duplicate in multiple independent assays. Specificity of the ELISA was tested by measuring cross reactivity to recombinant mouse CDNF (R&D Systems) and recombinant human MANF (Icosagen).
MANF ELISA
Endogenous human MANF concentration in the cell samples was analyzed by an in-lab-built MANF ELISA22 with a sensitivity of 45 pg/ml and 8.1 ± 2.1% CV intra-assay and 5.5 ± 1.2% CV interassay reproducibility. The MANF ELISA does not detect human CDNF.
Western Blotting
Conditioned medium samples were concentrated 20-fold using Amicon Ultra centrifugal filters with 10 kDa cut off (UFC501024, Merck Millipore). The concentrated media and cell lysates were mixed with reducing loading buffer (containing 5% β-mercaptoethanol as final concentration), and proteins were denatured by heating at +95°C for 5 min. Proteins were separated on 15% SDS-PAGE and transferred on nitrocellulose membrane (0.45 μm, Amersham HybondTM ECLTM, GE Healthcare, Chicago, IL, USA). The proteins were detected with rabbit anti-CDNF (0.4 μg/ml1) and by mouse anti-actin as a loading control (1:1 000, A-4700, Sigma-Aldrich), followed by HRP-linked secondary antibody. Signal was visualized using ECL Western Blotting Substrate kit (Thermo Fisher Scientific).
Immunocytochemistry
The parental cells or stable cell clones were seeded on coverslips, transfected in case of parental cell line, fixed with 4% PFA (48 h after transfection), permeabilized with 0.1% Triton-X and stained with following antibodies: rabbit anti-CDNF (0.4 μg/ml1), mouse anti-PDI, 1:150 (Enzo Life Sciences, Farmingdale, NY, USA), or mouse anti-GM130, 1:100 (BD Transduction LaboratoriesTM, Franklin Lakes, NJ, USA). Secondary antibodies goat anti-rabbit Alexa 568, 1:400), and donkey anti-mouse Alexa 488, 1:400 (both from Life Technologies, Thermo Scientific) were used for detection of the corresponding primary antibodies. Nuclei were stained with Hoechst 3345 (Sigma-Aldrich). Fluorescent microscopy images were captured using confocal microscope (Leica TCS SP5, Wetzlar, Germany), or epifluorescent microscope (Olympus AX 70 Provis, Tokyo, Japan).
For immunocytochemistry, following controls were included: (1) omission of the primary antibodies, for analyzing possible unspecificity of the secondary antibodies used, and (2) separate single antibody stainings, for analyzing possible channel leakage. Non-transfected wtARPE-19 cells stained with the in-house-produced CDNF antibody did not give background with the light exposure time used (see Fig. 3b, Results), reflecting low endogenous CDNF levels and high specificity of the antibody used.
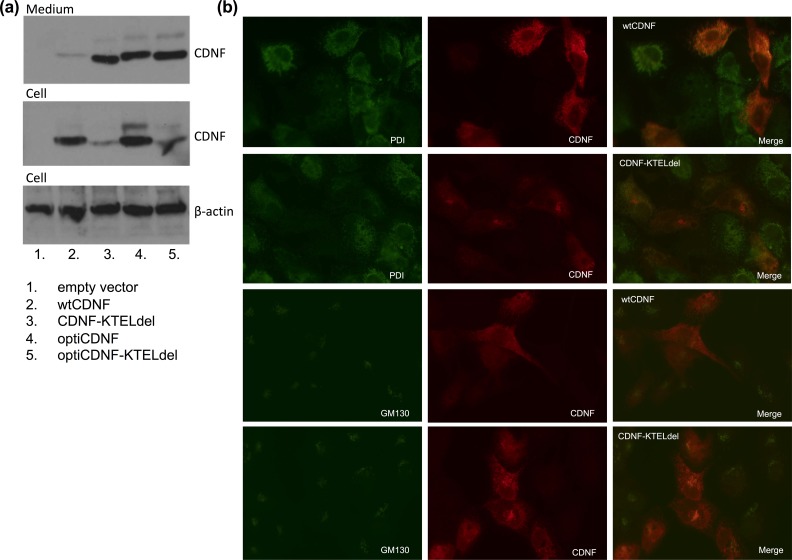
Figure 3.
Codon optimization and deletion of the putative ER-retrieval signal (KTEL) increases the CDNF secretion. (a) Immunoblot showing increased CDNF secretion with the modified CDNF constructs, and lower intracellular protein amount in case of KTEL deletion. Samples were collected 48 h after transient transfection of ARPE-19 cells with empty expression vector, vector with wild type CDNF (wtCDNF), KTEL-deleted CDNF (CDNF-KTELdel), codon-optimized CDNF (optiCDNF), or KTEL-deleted codon-optimized CDNF (optiCDNF-KTELdel). (b) Epifluorescent microscope images on ARPE-19 cells double stained with anti-CDNF and anti-PDI or GM130, for the detection of ER and Golgi apparatus, respectively. Images were captured using same exposure times. The cells were transfected either with wild type CDNF (wtCDNF) or KTEL-deleted CDNF (CDNF-KTELdel).
Cell Encapsulation
Encapsulation of cell clones was performed by an in-house-built microencapsulation device12 using alginate-poly-L-lysine-alginate (APA) protocol12,13. In brief, cells growing on culture dishes were detached and suspended in 1.2% alginate (UP LVG, FP-303-02, NovaMatrix, Sandvika, Norway) at three different cell densities: 2, 4, and 6 million cells/ml of alginate. The microcapsules were produced by dispersing syringe-extruded alginate-cell suspension with nitrogen gas using a nozzle that allows co-laminar flow. The alginate was cross-linked with divalent cations by incubating the capsules first in 68 mM CaCl2 solution (3 min), followed by incubation in 20 mM BaCl2 solution (5 min). Cross-linked alginate capsules were incubated in 0.1% poly-L-lysine solution (15-30 kDa, Sigma-Aldrich) (5 min) for increased durability. A final coating in 0.125% alginate solution (5 min) was added to ensure biocompatibility of the capsules. The capsules produced this way have a molecular weight cut off at 50 kDa13. Thus, 18 kDa CDNF should be able to diffuse from the capsules freely.
The average diameter of the produced capsules was determined from images taken with a digital camera connected to a phase contrast microscope (Leica). The images were processed with LAS EZ program and a minimum of 100 capsules/cell density was analyzed.
Viability of the encapsulated cells was assessed by fluorescent LIVE/DEAD staining (Molecular Probes, Thermo Fisher Scientific) and by Alamar Blue reagent reflecting the activity of mitochondria (Invitrogen). LIVE/DEAD staining with calcein/ethidium homodimer-1 was performed 24 h after cell encapsulation as described earlier for APA-encapsulated ARPE-19 cells12,13. In the Alamar Blue test, capsules were incubated with 400 μl of 10% Alamar Blue reagent for 5 h, after which an aliquot of the incubation medium was transferred into a black 96-well plate and analyzed on plate reader (VarioskanTM Flash, Thermo Fisher Scientific) for fluorescence emission using excitation/emission wavelengths of 530/590 nm.
CDNF secretion from the encapsulated cells in Opti-MEM after 6 h incubation was studied by CDNF ELISA. For Alamar Blue and CDNF secretion studies, capsules were transferred into 24-well plates and the capsule number in each well was calculated from pictures taken with Canon system camera connected to a microscope (Meiji, San Jose, CA, USA). The results were normalized to the capsule number in each sample and expressed as mean ± SEM of three parallel samples with >100 microcapsules. Alamar Blue results are expressed in fluorescence intensity/capsule and CDNF secretion in pg/100 capsules/24 hours. The assays were performed at days 1, 4, 7, 14, and 28 after encapsulation.
Statistics
The data are presented as mean ± SEM. Differences between two related groups were analyzed by paired samples t-test. Differences between three or more groups were analyzed by one-way analysis of variance (ANOVA), followed by Dunnett’s post hoc test. Multiple timepoints were compared by repeated measures ANOVA with Bonferroni's correction. A p-value of less than 0.05 was considered statistically significant. Statistical analysis was performed using IBM SPSS software.
Results
Characterization of Human CDNF ELISA
Sensitivity of the in-lab-built mAb human CDNF ELISA was 30.5 pg/ml, and dynamic range was 31.25–2 000 pg/ml. Average intra- and interassay variations were 7.1% coefficient of variation, CV (range 6.4–8.6), and 8.8% CV (5.5–11.2), respectively. The assay did not detect mouse CDNF nor human MANF (highest tested concentration was 500 ng/ml). The ELISA recognized KTEL-deleted human CDNF as well as wtCDNF. As the antibodies used in the ELISA are monoclonal, the detection of both wtCDNF and KTEL-deleted CDNF should be equal.
Endogenous CDNF is not Found in the Conditioned Medium
Five human cell lines were studied for secreted endogenous CDNF in the culture media: epithelial-like ARPE-19 and HeLa, endothelial-like EA.hy926, neuronal-like SH-SY5Y, and glial-like U-78 MG. In all cases, CDNF remained under the detection limit (<30.5 pg/ml) in the conditioned media of dividing cells at 72 h (Table 1). Instead, MANF was found in the media of all the studied cell lines at concentrations ranging from 0.41 to 4.20 ng/ml. In the cell lysates, MANF was found at 160 to 230 times higher concentrations compared with CDNF. CDNF concentration in cell lysate was 0.36 to 0.73 ng/ml, whereas MANF was found at levels from 57.2 to 128.3 ng/ml.
Table 1.
Endogenous CDNF and MANF Protein Concentrations in Cell Lysates and Conditioned Media of Selected Cell Lines of Human Origin. Results are Shown as Average ± SEM (n=3).
| Human cell line | ARPE-19 | HeLa | EA.hy926 | SH-SY5Y | U-87 MG |
|---|---|---|---|---|---|
| MANF | |||||
| Conditioned medium (ng/ml) | 0.41 ± 0.03 | 3.99 ± 0.07 | 2.15 ± 0.16 | 1.27 ± 0.08 | 4.20 ± 0.46 |
| Cell lysate (ng/ml) | 91.7 ± 2.1 | 128.3 ± 2.4 | 57.2 ± 2.7 | 85.3 ± 1.1 | 70.5 ± 2.2 |
| % secreted | 0.4 | 3.0 | 3.6 | 1.5 | 5.6 |
| CDNF (ng/ml) | |||||
| Conditioned medium (ng/ml) | n.d. | n.d. | n.d. | n.d. | n.d. |
| Cell lysate (ng/ml) | 0.49 ± 0.02 | 0.73 ± 0.03 | 0.36 ± 0.01 | 0.37 ± 0.01 | 0.43 ± 0.02 |
| % secreted | n.a. | n.a. | n.a. | n.a. | n.a. |
| MANF vs. CDNF in cell lysate (times) | 187 | 176 | 159 | 231 | 164 |
n.d. = not detected, n.a. = not applicable.
High Concentration of Intracellular wtCDNF in ARPE-19 Clones
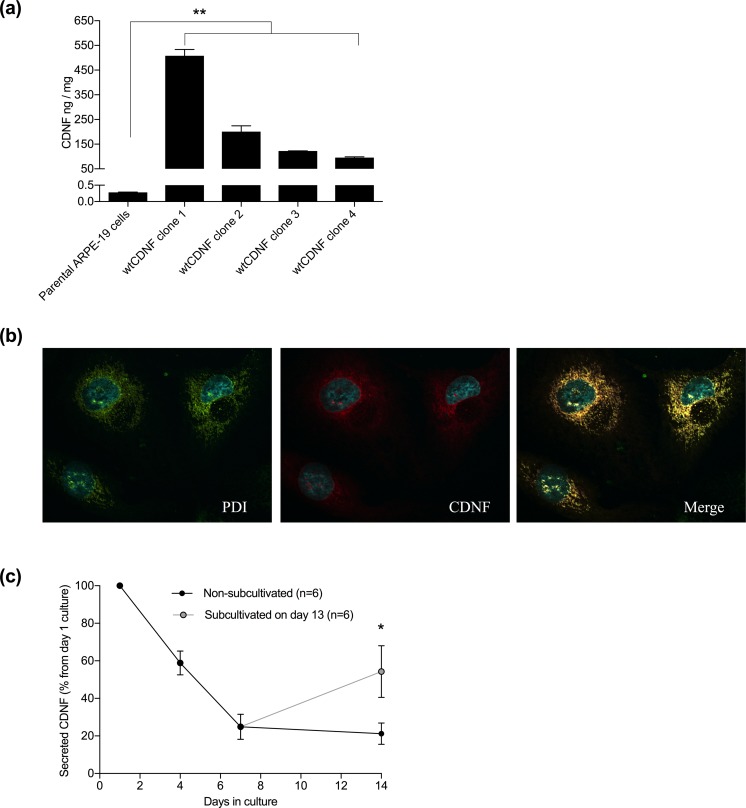
Several wtCDNF-expressing and secreting ARPE-19 cell clones were isolated after transfection of the parental cell line with pCI-neo vector carrying CDNF cDNA sequence under a CMV-promoter, and neomycin phosphotransferase gene for selection. Endogenous CDNF concentration in the parental ARPE-19 cell lysate, representing the intracellular and possibly membrane-bound CDNF, was 0.28 ± 0.01 ng/mg total protein (mean ± SEM, n=4). Differently from the parental cell line, intracellular CDNF concentration in four tested ARPE-19 cell clones expressing and secreting wtCDNF ranged from 95.9 ± 2.9 to 508.3 ± 25.1 ng/mg (n = 2–3/clone). This means that the intracellular CDNF concentration in the clones was increased up to 1800-fold compared with the parental cell line (p=0.001–0.007, one-way ANOVA + Dunnett’s test, Fig. 1a). Strong co-localization of CDNF and the ER-resident enzyme PDI was found by confocal microscopy of immunofluorescent double-labeled cells, indicating that intracellular CDNF mainly resided in the ER in the wtCDNF ARPE-19 clones (Fig. 1b).
Figure 1.
High levels of intracellular CDNF, which localize to the ER in the stable wtCDNF ARPE-19 cell clones. (a) Intracellular CDNF concentration in the clones ranged from 95.9 ± 2.9 to 508.3 ± 25.1 ng/mg (mean ± SEM, n=2–3/clone), whereas, in the parenteral ARPE-19 cell line it was 0.28 ± 0.01 ng/mg total protein (n=4). The increase in the clone intracellular CDNF concentration was up to 1800-fold compared with the parenteral cell line (p=0.001–0.007, one-way ANOVA + Dunnett’s test). (b) Immunofluorescent image of a wtCDNF ARPE-19 cell clone double stained to show the ER (PDI, green) and CDNF (red) and imaged by confocal microscopy. Merged image shows co-localization of CDNF and ER-marker (yellow). (c) CDNF secretion decreased down to 21.2 ± 5.7% from the initial secretion when the wtCDNF ARPE-19 clones were held without sub-cultivation for 14 days. Parallel to non-subcultivated, the same clones were subcultivated 1:2 on day 13 and CDNF secretion was analyzed at day 14. In the subcultivated cells CDNF secretion was 54.3 ± 13.8% from the initial secretion (p=0.036, paired samples t-test, n=6). ** p<0.01, * p<0.05.
Secretion of wtCDNF is Inhibited When the Cells are Confluent
Average CDNF secretion from the wtCDNF ARPE-19 clones was 0.48 ± 0.08 ng/0.4×106 cells/30 min (min–max: 0.02–0.85 ng/0.4×106 cells/30 min, depending on the clone, n=12). CDNF secretion decreased when cell clones reached confluence. The average CDNF medium concentration in cultures held for 14 days without sub-cultivation was only 18.5 ± 4.4% (min–max: 3.3–46.3%) from the initial (p<0.001, paired samples t-test, n=11).
Differently from secreted CDNF, intracellular CDNF concentration did not decrease in the confluent wtCDNF ARPE-19 cultures. Cells growing for 14 days without sub-cultivation had intracellular CDNF concentration of 114.7 ± 17.1% from that of parallel cultures subcultivated 1:2 1 day before sample collection (p=0.79, paired samples t-test, n=5 clones). Thus, the observed drop in CDNF secretion from the clones in confluent state seems not to be caused by decreased CDNF synthesis, but rather decreased secretion in the non-dividing cells. One day after sub-cultivation of confluent cultures, CDNF concentration in the medium was 54.3 ± 13.8% from the initial. In the parallel samples held without sub-cultivation, CDNF medium concentration was 21.2 ± 5.7% from the initial (p=0.036, paired samples t-test, n=6) (Fig. 1c), indicating that the secretion of CDNF can be re-induced in the cells after sub-cultivation.
Good Cell Viability, but Poor wtCDNF Secretion from the Encapsulated Cells
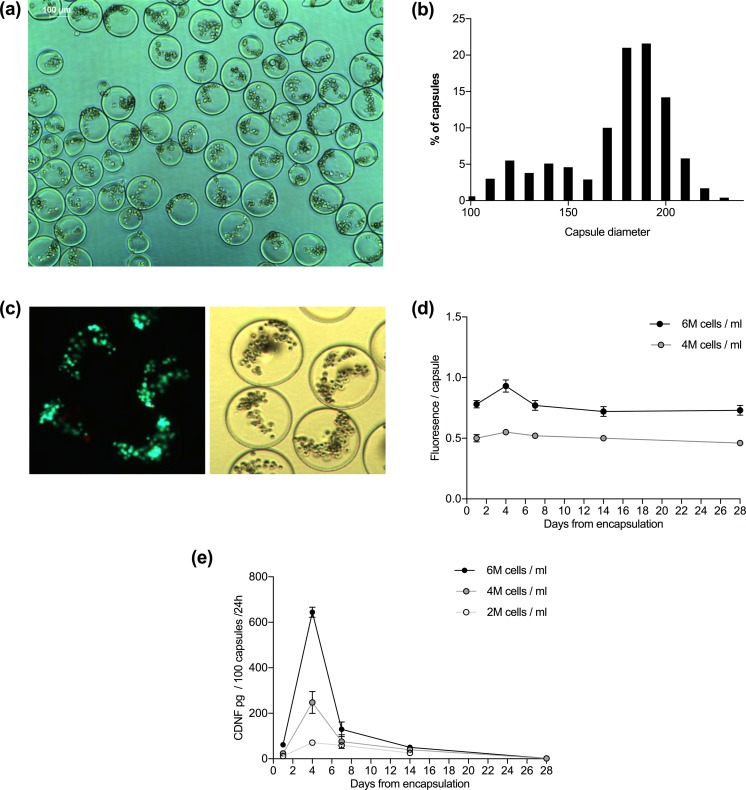
Cell clones were encapsulated in alginate at three different cell densities: 2, 4, and 6 million cells/ml of alginate. An image taken on an inverted phase microscope of the capsules is shown in Fig. 2a. The mean diameter of the produced microcapsules was 169 ± 32, 161 ± 22, and 179 ± 23 μm for 2, 4, and 6 million cells/ml alginate, respectively. Mean diameter of all microcapsules was 169 ± 26 μm (range 99–222 μm, Fig. 2b). Staining with LIVE/DEAD kit showed that the cells endured the encapsulation well, as only a few dead cells were detected (Fig. 2c). Analysis with Alamar Blue reagent showed steady metabolic function of the encapsulated cells up to 28 days, which was the duration of analysis (Fig. 2d).
Figure 2.
Microencapsulated wtCDNF ARPE-19 cell clones are viable but the secretion of CDNF decreases with time. (a) APA-microcapsules with cell density of 2 million cells/ml of alginate. Scale bar 100 μm. (b) A density histogram showing the size distribution of the prepared microcapsules. Mean capsule diameter for all the produced capsules was 169 ± 26 μm (range 99–222 μm). (c) Cells stained with LIVE/DEAD kit. Most of the encapsulated cells stained with calcein (live, green), and only a few with ethidium homodimer-1 (dead, red), implying that the cells survived the encapsulation procedure well. (d) Viability test using resazurin solution (Alamar Blue reagent) shows steady metabolic function of the encapsulated cells. (e) wtCDNF release measured in the conditioned media of encapsulated cells analyzed on CDNF ELISA.
CDNF was found in the conditioned media of encapsulated cells. Secreted CDNF amount was dependent on the cell density in the capsules (Fig. 2e). Four days after encapsulation, CDNF secretion was at its peak. At this time point, CDNF was secreted into the media at levels of 71 ± 14, 248 ± 68, or 644 ± 43 pg/100 capsules/24 h (n=3 replicates/group), in the case of cell encapsulation densities of 2, 4, and 6 million cells/ml, respectively. CDNF secretion decreased close to zero within 28 days from capsulation.
Improved CDNF Secretion with Codon-Optimized CDNF and KTEL Deletion
Since wtCDNF secretion was low, we studied the effects of codon optimization, deletion of the putative ER-retrieval signal, and their combination to improve CDNF secretion from the cells. After transient transfection of wtCDNF-pCR3.1 to ARPE-19 cells, only 8% of the total cellular wtCDNF was secreted within 24 h. Codon optimization resulted in a 3-fold increase in the secreted CDNF concentration, as shown by transient transfection of optiCDNF-pCR3.1 (Fig. 3a, Table 2). The intracellular CDNF concentration was almost doubled in the cells transfected with optiCDNF compared with wtCDNF. Thus, the increase in CDNF concentration in the medium is rather due to the increased CDNF expression by codon optimization than improved secretion.
Table 2.
CDNF Concentrations Analyzed by ELISA in the Conditioned Media and Cell Lysates After Transient Transfection of ARPE-19 Cells. Cells Were Transfected with Empty Expression Vector, Vector Carrying Wild Type CDNF (wtCDNF), Codon-Optimized CDNF (optiCDNF), KTEL-Deleted CDNF (CDNF-KTELdel), or KTEL-Deleted Codon-Optimized CDNF (optiCDNF-KTELdel). All Sequences were Cloned Into pCR3.1 Transfection Vector.
| Empty vector | wtCDNF | optiCDNF | CDNF- KTELdel | optiCDNF-KTELdel | |
|---|---|---|---|---|---|
| CDNF in medium (ng/ml) | n.d. | 142.1 | 474.6 | 409.3 | 687.2 |
| CDNF in cells (ng/ml) | 0.5 | 1566.1 | 2956.5 | 442.4 | 574.9 |
| Times secreted vs. wtCDNF | 1 | 3.3 | 2.9 | 4.8 | |
| Times intracellular vs. wtCDNF | 1 | 1.9 | 0.3 | 0.4 | |
| % secreted | 0 | 8.3 | 13.8 | 48.1 | 54.4 |
n.d. = not detected.
CDNF secretion from cells transfected with wtCDNF-KTELdel was approximately three times higher compared with cells transfected with the wtCDNF, but the intracellular CDNF concentration of wtCDNF-KTELdel cells was only 30% from that of wtCDNF transfected cells (Fig. 3a, Table 2). This implies that the change seen in CDNF medium concentration after KTEL deletion was a consequence of improved secretion and not due to differential CDNF expression. The immunofluorescence images of transiently transfected cells expressing KTEL-deleted CDNF showed different staining patterns with CDNF antibody compared with cells expressing wtCDNF (Fig. 3b). Epifluorescence microscope images showed some co-localization of the KTEL-deleted CDNF with a Golgi marker GM130. This contrasts wtCDNF that colocalized only with the ER marker PDI.
Combination of KTEL deletion and codon optimization resulted in 5-fold increased CDNF secretion compared with that of wtCDNF. Intracellular concentration of optiCDNF-KTELdel was only 20% of that seen for codon-optimized CDNF, implying that KTEL deletion again increased secretion of CDNF.
Secretion of KTEL-Deleted Codon-Optimized CDNF is Stable in 2D Cultures
Stable ARPE-19 cell clones were isolated from cultures transfected with pCR3.1 plasmids carrying either codon-optimized CDNF or KTEL-deleted codon-optimized CDNF. Altogether 5 or 9 out of total 10 isolated clones transfected with optiCDNF or optiCDNF-KTELdel, respectively, secreted sufficient CDNF. CDNF secretion was on average 0.81 ± 0.33 ng/0.4×106 cells/30 min (min–max: 0.07–1.81 ng/0.4×106 cells/30 min) in the case of optiCDNF, and 1.75 ± 0.44 ng/0.4×106 cells/30 min (min–max: 0.30–4.88 ng/0.4×106/30 min) in the case of optiCDNF-KTELdel.
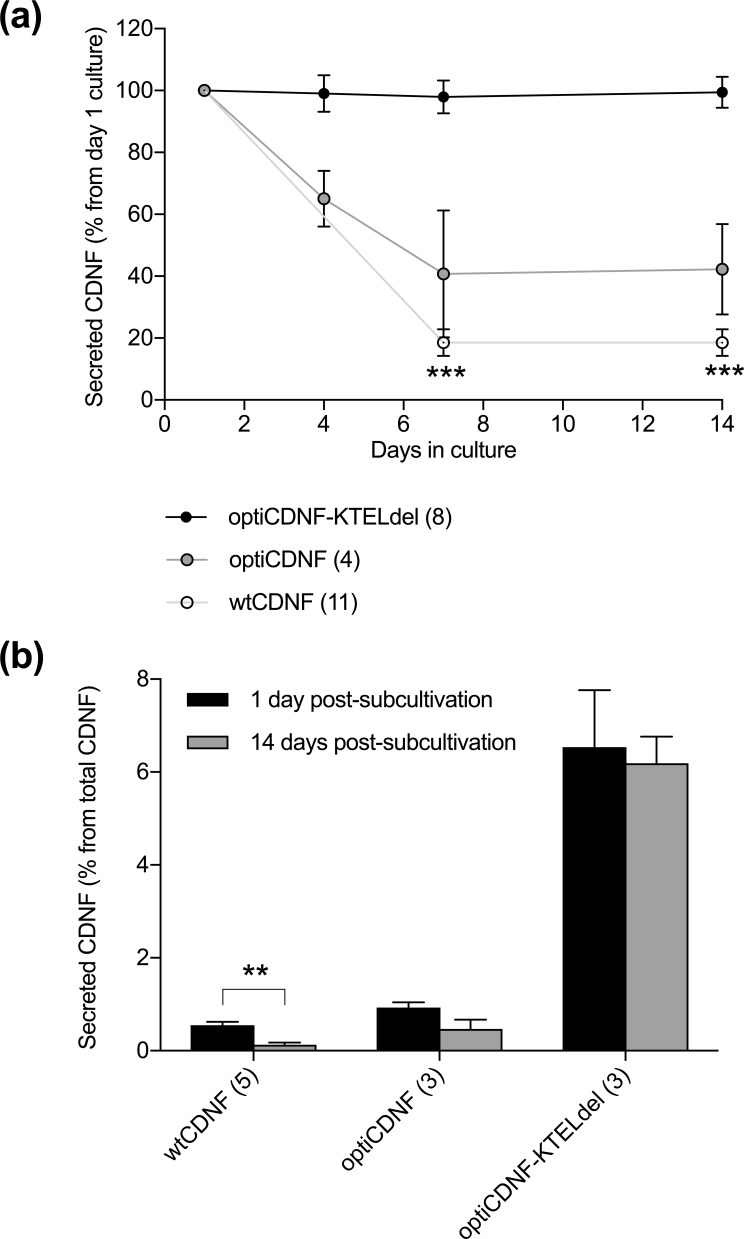
The secretion of CDNF from the KTEL-deleted codon-optimized CDNF clones remained stable in cultures held without sub-cultivation for up to 14 days. The observed secretion of optiCDNF-KTELdel was 99.1 ± 5.9%, 97.9 ± 5.3%, and 99.4 ± 5.0% (p=0.95, repeated measures ANOVA, n=8) from the initial secreted amounts at days 4, 7, and 14 from sub-cultivation, respectively (Fig. 4a). The secretion of optiCDNF was 65.0 ± 9.0%, 40.7 ± 20.5% and 42.2 ± 14.6% (p=0.46, n=4) at the same time points. For reference, CDNF secretion from wtCDNF-expressing clones was 18.5 ± 4.4% (p<0.001, n=11) at 7 and 14 days from the initial secretion.
Figure 4.
Secretion of KTEL-deleted codon-optimized CDNF is stable in clones cultured on Petri dish. (a) The secretion of KTEL-deleted codon-optimized CDNF was stable for 14 days in non-subcultivated cell cultures. In contrast, the secretion of wtCDNF decreased during the time course in cell cultures, which were not subcultivated during the experiment (p<0.001, repeated measures ANOVA with Bonferroni correction post hoc). (b) Secreted portion of CDNF during 1 h incubation from the total expressed CDNF (CDNF concentration in the medium + cell lysate) was similar in case of subcultivated and non-subcultivated clones expressing KTEL-deleted codon-optimized CDNF. Instead, the secreted portion of wtCDNF increased in the subcultivated cells (p=0.005, paired samples t-test). Data are presented as mean ± SEM. *** p<0.001, ** p<0.01.
When the confluent ARPE-19 clones were divided after 14 days without sub-cultivation, the secreted CDNF amount increased 4-fold from 0.13 ± 0.05% to 0.55 ± 0.07% of total CDNF (p=0.005, paired samples t-test, n=5, Fig. 4b) in clones overexpressing wtCDNF. The observed CDNF secretion from optiCDNF cell clones increased 2-fold, but the difference was statistically insignificant (from 0.47 ± 0.20% to 0.93 ± 0.11%, p=0.094, n=3). No increase of CDNF secretion was observed in the case of optiCDNF-KTELdel cell clones (6.19 ± 0.57% vs. 6.54 ± 1.22%, p=0.77, n=3).
Discussion
In this study, we have produced several CDNF-overexpressing ARPE-19 cell clones for the purpose of encapsulated cell therapy. Intracellular endogenous CDNF was detected in the parental ARPE-19 cell line, but no CDNF was found in the conditioned medium. Likewise, secreted endogenous CDNF was not found in any of the other four tested cell lines, although all cell lines expressed CDNF. In contrast, endogenous MANF was found in the conditioned media of all the cell lines. The level of endogenous human CDNF and MANF secretion has not previously been compared. However, in the overexpression situation Flag-tagged mouse CDNF and MANF were secreted at a comparative level23. Because the intracellular concentration of endogenous MANF was approximately 200 times higher than that of CDNF in the cell lines analyzed, the absence of CDNF in the media could be explained by limited ELISA sensitivity. As the measured concentration of MANF in the medium samples was up to 4000 pg/ml, CDNF secreted at the same proportion (appr. 20 pg/ml) would not be detected with the used ELISA (sensitivity 30.5 pg/ml).
The poor secretion can also be a biological characteristic of CDNF, as we encountered challenges in the secretion of wtCDNF in overexpressing ARPE-19 clones as well. Only a minor portion of the synthetized CDNF was secreted, while most of the CDNF remained within the cells. Intracellular CDNF concentration in the clones was increased up to 1800-fold when compared with the endogenous CDNF concentration in the parental ARPE-19 cell line. The secretion of wtCDNF decreased further when the clones were in a non-dividing state. The same was observed in all wtCDNF-expressing cell clones growing as a monolayer, as well as in alginate capsules, in which the cells are unable to divide due to contact-inhibited growth14. This is in sharp contrast to overexpression of ciliary neurotrophic factor (CNTF)19 or nerve growth factor (NGF)20 in the same cell line, where a stable level of secreted transgene was observed for months in non-dividing cells after encapsulation.
CDNF secretion from the cell clones was found to increase after sub-cultivation. As the expression cassette is integrated into the cellular genome in the stable cell clones, this could be a consequence of quiescent chromatin status in the non-dividing cells. When the cell division is triggered, expression of the genome-integrated transgene can be reactivated. However, two aspects argue against this. First, the expression cassette also contains a neomycin resistance gene allowing for positive selection with the presence of selection antibiotic, which was present in the culture medium all the time. This means that the neomycin resistance gene was expressed all the time from the genomic location where CDNF cDNA was inserted. Second, the intracellular CDNF amount in the non-dividing cells was not decreased in the non-dividing wtCDNF ARPE-19 clones. Thus, the clones most likely continued to express CDNF, but its secretion was inhibited.
The observations that intracellular CDNF resided within the ER of the ARPE-19 clones and that the clones secreted only minor amounts of the expressed transgene could indicate that the ER of the cell clones was not functioning properly. However, in the case of CDNF, cell retention may be caused by an ER-retrieval sequence, the C-terminal amino acids: KTEL. The KTEL sequence is recognized by KDEL receptors locating at the Golgi apparatus and retrieved back to the ER24. Also in the case of MANF, the four last amino acids (RTDL, in humans) bind to KDEL receptors25, and the deletion of C-terminal RTDL increases MANF secretion, as shown in vitro in multiple cell lines25–27. The same has been shown for mouse and rat CDNF with the deletion of the C-terminal QTEL23,28.
Under transient transfections, human CDNF secretion was enhanced up to 3-fold when the putative ER-retrieval sequence was deleted. The KTEL-deleted CDNF gave a different staining pattern in immunocytochemistry compared with the wtCDNF, indicating its localization to Golgi apparatus as well as to the ER. The results imply that the ER-retention of wtCDNF was caused, at least in part, by the biological character of CDNF and not due to improper ER function of the produced cell clones.
In addition to deleting the putative ER-retrieval signal, we studied codon-optimized CDNF in relation to improved CDNF secretion. In codon optimization, rare codons are changed into more ubiquitously used ones without changing the resultant amino acid that the codon stands for21. Thus, the translated protein amount increases without any changes in the resultant protein amino acid sequence. With codon optimization, the CDNF secretion was enhanced up to 3-fold, but the intracellular CDNF load increased 2-fold compared with transfection with wtCDNF, which itself was largely retained intracellularly. When combining both of the methods, CDNF secretion increased up to 5-fold, and the intracellular amount of KTEL-deleted codon-optimized CDNF was only 40% from that of wtCDNF.
Additional cell clones were produced with plasmids carrying codon-optimized CDNF or KTEL-deleted codon-optimized CDNF cDNA sequence. On average, the new clones secreted 1.7 or 3.6 times more CDNF, respectively, compared with the wtCDNF clones. More importantly, the secretion of KTEL-deleted codon-optimized CDNF did not decrease in confluent cultures, contrary to what was observed in the case of wtCDNF and codon-optimized CDNF. However, the biological activity of KTEL-deleted CDNF is currently unknown. In the case of MANF, the corresponding RTDL deletion did not abolish the neuroprotective effect of extracellularly administered MANF in a stroke model in vivo27.
According to multiple studies performed on rodents1,4–9, CDNF has potential in the treatment of Parkinson’s and Alzheimer’s diseases. Continuous and de novo synthetized, but retrievable, delivery of CDNF to the target site in the brain can be achieved with polymer-encapsulated cells. According to our results on ARPE-19 cells clones, the secretion of wtCDNF is sub-optimal because it is highly retained intracellularly, and its secretion is further decreased in non-dividing cells. KTEL deletion improved both aspects, and thus could be considered for the delivery of CDNF with encapsulated cells. However, the in vivo biological activity of the mutant protein has to be determined.
Acknowledgements
This study was supported by the Graduate School in Pharmaceutical Research and by The Finnish Funding Agency for Technology and Innovation, Tekes (PrinCell project).
Footnotes
Author Contributions: E.G. designed and performed research, analyzed data and wrote manuscript, P.L. and L-S.K. designed and performed research and commented on manuscript, M.S., A.U., and M.Y. designed research and commented on manuscript.
Ethical Approval: Ethical Approval is not applicable for this article.
Statement of Human and Animal Rights: This article does not contain any studies with human or animal subjects.
Statement of Informed Consent: There are no human subjects in this article and informed consent is not applicable.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Emilia Galli  https://orcid.org/0000-0002-7419-611X
https://orcid.org/0000-0002-7419-611X
References
- 1. Lindholm P, Voutilainen MH, Laurén J, Peränen J, Leppänen VM, Andressoo JO, Lindahl M, Janhunen S, Kalkkinen N, Timmusk T, Tuominen RK, Saarma M. Novel neurotrophic factor CDNF protects and rescues midbrain dopamine neurons in vivo. Nature. 2007;448(7149):73–77. [DOI] [PubMed] [Google Scholar]
- 2. Petrova P, Raibekas A, Pevsner J, Vigo N, Anafi M, Moore MK, Peaire AE, Shridhar V, Smith DI, Kelly J, Durocher Y, Commissiong JW. MANF: A new mesencephalic, astrocyte-derived neurotrophic factor with selectivity for dopaminergic neurons. J Mol Neurosci. 2003;20(2):173–188. [DOI] [PubMed] [Google Scholar]
- 3. Lindahl M, Saarma M, Lindholm P. Unconventional neurotrophic factors CDNF and MANF: Structure, physiological functions and therapeutic potential. Neurobiol Dis. 2017;97(Pt B):90–102. [DOI] [PubMed] [Google Scholar]
- 4. Voutilainen MH, Bäck S, Peränen J, Lindholm P, Raasmaja A, Männistö PT, Saarma M, Tuominen RK. Chronic infusion of CDNF prevents 6-OHDA-induced deficits in a rat model of Parkinson’s disease. Exp Neurol. 2011;228(2):99–108. [DOI] [PubMed] [Google Scholar]
- 5. Bäck S, Peränen J, Galli E, Pulkkila P, Lonka-Nevalaita L, Tamminen T, Voutilainen MH, Raasmaja A, Saarma M, Männisto PT, Tuominen RK. Gene therapy with AAV2-CDNF provides functional benefits in a rat model of Parkinson’s disease. Brain Behav. 2013;3(2):75–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ren X, Zhang T, Gong X, Hu G, Ding W, Wang X. AAV2-mediated striatum delivery of human CDNF prevents the deterioration of midbrain dopamine neurons in a 6-hydroxydopamine induced parkinsonian rat model. Exp Neurol. 2013;248:148–156. [DOI] [PubMed] [Google Scholar]
- 7. Cordero-Llana Ó, Houghton BC, Rinaldi F, Taylor H, Yáñez-Muñoz RJ, Uney JB, Wong LF, Caldwell MA. Enhanced efficacy of the CDNF/MANF family by combined intranigral overexpression in the 6-OHDA rat model of Parkinson’s disease. Mol Ther. 2015;23(2):244–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Airavaara M, Harvey BK, Voutilainen MH, Shen H, Chou J, Lindholm P, Lindahl M, Tuominen RK, Saarma M, Wang Y, Hoffer B. CDNF protects the nigrostriatal dopamine system and promotes recovery after MPTP treatment in mice. Cell Transplant. 2012;21(6):1213–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kemppainen S, Lindholm P, Galli E, Lahtinen HM, Koivisto H, Hämäläinen E, Saarma M, Tanila H. Cerebral dopamine neurotrophic factor improves long-term memory in APP/PS1 transgenic mice modeling Alzheimer’s disease as well as in wild-type mice. Behav Brain Res. 2015;291:1–11. [DOI] [PubMed] [Google Scholar]
- 10. Lindvall O, Wahlberg LU. Encapsulated cell biodelivery of GDNF: a novel clinical strategy for neuroprotection and neuroregeneration in Parkinson’s disease? Exp Neurol. 2008;209(1):82–88. [DOI] [PubMed] [Google Scholar]
- 11. Emerich DF, Orive G, Thanos C, Tornøe J, Wahlberg LU. Encapsulated cell therapy for neurodegenerative diseases: from promise to product. Adv Drug Deliv Rev. 2014;67-68:131–141. [DOI] [PubMed] [Google Scholar]
- 12. Kontturi LS, Yliperttula M, Toivanen P, Määttä A, Määttä AM, Urtti A. A laboratory-scale device for the straightforward production of uniform, small sized cell microcapsules with long-term cell viability. J Control Release. 2011;152(3):376–381. [DOI] [PubMed] [Google Scholar]
- 13. Wikström J, Elomaa M, Syväjärvi H, Kuokkanen J, Yliperttula M, Honkakoski P, Urtti A. Alginate-based microencapsulation of retinal pigment epithelial cell line for cell therapy. Biomaterials. 2008;29(7):869–876. [DOI] [PubMed] [Google Scholar]
- 14. Dunn KC, Aotaki-Keen AE, Putkey FR, Hjelmeland LM. ARPE-19, a human retinal pigment epithelial cell line with differentiated properties. Exp Eye Res. 1996;62(2):155–170. [DOI] [PubMed] [Google Scholar]
- 15. Sieving PA, Caruso RC, Tao W, Coleman HR, Thompson DJ, Fullmer KR, Bush RA. Ciliary neurotrophic factor (CNTF) for human retinal degeneration: phase I trial of CNTF delivered by encapsulated cell intraocular implants. Proc Natl Acad Sci USA. 2006;103(10):3896–3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Eriksdotter-Jönhagen M, Linderoth B, Lind G, Aladellie L, Almkvist O, Andreasen N, Blennow K, Bogdanovic N, Jelic V, Kadir A, Nordberg A, Sundström E, Wahlund LO, Wall A, Wiberg M, Winblad B, Seiger A, Almqvist P, Wahlberg L. Encapsulated cell biodelivery of nerve growth factor to the basal forebrain in patients with Alzheimer’s disease. Dement Geriatr Cogn Disord. 2012;33(1):18–28. [DOI] [PubMed] [Google Scholar]
- 17. Kauper K, McGovern C, Sherman S, Heatherton P, Rapoza R, Stabila P, Dean B, Lee A, Borges S, Bouchard B, Tao W. Two-year intraocular delivery of ciliary neurotrophic factor by encapsulated cell technology implants in patients with chronic retinal degenerative diseases. Invest Ophthalmol Vis Sci. 2012;53(12):7484–7491. [DOI] [PubMed] [Google Scholar]
- 18. Eyjolfsdottir H, Eriksdotter M, Linderoth B, Lind G, Juliusson B, Kusk P, Almkvist O, Andreasen N, Blennow K, Ferreira D, Westman E, Nennesmo I, Karami A, Darreh-Shori T, Kadir A, Nordberg A, Sundström E, Wahlund LO, Wall A, Wiberg M, Winblad B, Seiger Å, et al. Targeted delivery of nerve growth factor to the cholinergic basal forebrain of Alzheimer’s disease patients: application of a second-generation encapsulated cell biodelivery device. Alzheimer’s Res Ther. 2016;8(1):30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tao W, Wen R, Goddard MB, Sherman SD, O’Rourke PJ, Stabila PF, Bell WJ, Dean BJ, Kauper KA, Budz VA, Tsiaras WG, Acland GM, Pearce-Kelling S, Laties AM, Aguirre GD. Encapsulated cell-based delivery of CNTF reduces photoreceptor degeneration in animal models of retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2002;43(10):3292–3298. [PubMed] [Google Scholar]
- 20. Fjord-Larsen L, Kusk P, Tornøe J, Juliusson B, Torp M, Bjarkam CR, Nielsen MS, Handberg A, Sørensen JC, Wahlberg LU. Long-term delivery of nerve growth factor by encapsulated cell biodelivery in the Göttingen minipig basal forebrain. Mol Ther. 2010;18(12):2164–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Quax TE, Claassens NJ, Söll D, van der Oost J. Codon bias as a means to fine-tune gene expression. Mol Cell. 2015;59(2):149–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Galli E, Härkonen T, Sainio MT, Ustav M, Toots U, Urtti A, Yliperttula M, Lindahl M, Knip M, Saarma M, Lindholm P. Increased circulating concentrations of mesencephalic astrocyte-derived neurotrophic factor in children with type 1 diabetes. Sci Rep. 2016;6:29058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Norisada J, Hirata Y, Amaya F, Kiuchi K, Oh-hashi K. A comparative analysis of the molecular features of MANF and CDNF. PLoS One. 2016;11(1):e0146923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Raykhel I, Alanen H, Salo K, Jurvansuu J, Nguyen VD, Latva-Ranta M, Ruddock L. A molecular specificity code for the three mammalian KDEL receptors. J Cell Biol. 2007;179(6):1193–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Henderson MJ, Richie CT, Airavaara M, Wang Y, Harvey BK. Mesencephalic astrocyte-derived neurotrophic factor (MANF) secretion and cell surface binding are modulated by KDEL receptors. J Biol Chem. 2013;288(6):4209–4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Glembotski CC, Thuerauf DJ, Huang C, Vekich JA, Gottlieb RA, Doroudgar S. Mesencephalic astrocyte-derived neurotrophic factor protects the heart from ischemic damage and is selectively secreted upon sarco/endoplasmic reticulum calcium depletion. J Biol Chem. 2012;287(31):25893–25904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mätlik K, Yu LY, Eesmaa A, Hellman M, Lindholm P, Peränen J, Galli E, Anttila J, Saarma M, Permi P, Airavaara M, Arumäe U. Role of two sequence motifs of mesencephalic astrocyte-derived neurotrophic factor in its survival-promoting activity. Cell Death Dis. 2015;6:e2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Arancibia D, Zamorano P, Andrés ME. CDNF induces the adaptive unfolded protein response and attenuates endoplasmic reticulum stress-induced cell death. Biochim Biophys Acta Mol Cell Res. 2018;1865(11 Pt A):1579–1589. [DOI] [PubMed] [Google Scholar]